Abstract
Background
The causative factors for the recent increase in early-onset colorectal cancer (EO-CRC) incidence are unknown. We sought to determine if early-onset disease is clinically or genomically distinct from average-onset colorectal cancer (AO-CRC).
Methods
Clinical, histopathologic, and genomic characteristics of EO-CRC patients (2014-2019), divided into age 35 years and younger and 36-49 years at diagnosis, were compared with AO-CRC (50 years and older). Patients with mismatch repair deficient tumors, CRC-related hereditary syndromes, and inflammatory bowel disease were excluded from all but the germline analysis. All statistical tests were 2-sided.
Results
In total, 759 patients with EO-CRC (35 years, n = 151; 36-49 years, n = 608) and AO-CRC (n = 687) were included. Left-sided tumors (35 years and younger = 80.8%; 36-49 years = 83.7%; AO = 63.9%; P < .001 for both comparisons), rectal bleeding (35 years and younger = 41.1%; 36-49 years = 41.0%; AO = 25.9%; P = .001 and P < .001, respectively), and abdominal pain (35 years and younger = 37.1%; 36-49 years = 34.0%; AO = 26.8%; P = .01 and P = .005, respectively) were more common in EO-CRC. Among microsatellite stable tumors, we found no differences in histopathologic tumor characteristics. Initially, differences in TP53 and Receptor Tyrosine Kinase signaling pathway (RTK-RAS)alterations were noted by age. However, on multivariate analysis including somatic gene analysis and tumor sidedness, no statistically significant differences at the gene or pathway level were demonstrated. Among advanced microsatellite stable CRCs, chemotherapy response and survival were equivalent by age cohorts. Pathogenic germline variants were identified in 23.3% of patients 35 years and younger vs 14.1% of AO-CRC (P = .01).
Conclusions
EO-CRCs are more commonly left-sided and present with rectal bleeding and abdominal pain but are otherwise clinically and genomically indistinguishable from AO-CRCs. Aggressive treatment regimens based solely on the age at CRC diagnosis are not warranted.
In the United States, colorectal cancer (CRC) incidence and mortality have declined following the implementation of CRC screening based on standardized guidelines adopted in the late 1990s (1-4). However, in adults aged younger than 50 years, for whom routine CRC screening is not recommended, the incidence of CRC has been increasing steadily by 1%-2% annually since the 1990s (2). The greatest increase appears to be in patients aged 20-29 years; incidence in this group has increased by 3.8% annually since 1987, especially for distal colon and rectal cancers (4‐6). It is estimated that, by 2030, 10.9% of all colon cancers and 22.9% of all rectal cancers will be diagnosed in patients younger than 50 years, compared with 4.8% and 9.5%, respectively, in 2010 (7).
The etiology of this increase in CRC among younger patients (early-onset CRC [EO-CRC]) is unknown, and it is unclear whether EO-CRC has a unique biology, compared with average-onset CRC (AO-CRC; aged 50 years and older). EO-CRC may be associated with more aggressive disease biology, resulting in more advanced stages at diagnosis (8‐11). However, because patients aged 50 years and younger do not routinely undergo CRC screening, advanced stage at diagnosis may result from selection bias, wherein EO-CRCs are diagnosed only upon development of symptoms (12). Previous results on EO-CRC tumor genomics have been mixed and partially confounded by the inclusion of patients with EO-CRC with well-established genetic and/or medical predispositions for CRC (7,9,13). A recent genomic report suggested there are few molecular differences between EO-CRC and AO-CRC, besides an enrichment for high-frequency microsatellite instability (MSI) in EO-CRC, consistent with the expected higher prevalence of Lynch syndrome (LS) (14). The majority of cases of EO-CRC are seemingly sporadic, occurring in patients without an identifiable genetic predisposition (15). Established AO-CRC risk factors, such as obesity, diet high in red meat and low in fiber, excess alcohol consumption, physical inactivity, and smoking, do not adequately explain the increase in EO-CRC (16). Dietary and lifestyle factors have been associated with an increase in predominantly right-sided CRCs (17); however, the greatest increase in young patients has been in left-sided CRCs (3‐5).
Whether EO-CRC represents a disease distinct from AO-CRC is an important clinical question with critical implications for the oncological management of these young adults. To elucidate the clinical profile and molecular underpinnings of EO-CRC, we compared the clinical, somatic, and germline characteristics of EO-CRC and AO-CRC. As the prevalence of CRC is most drastically increasing among young individuals (age 35 years and younger), possibly representing a different subgroup of EO-CRC (18‐21), we further divided EO-CRC patients by age at diagnosis (35 years and younger and 36-49 years).
Methods
Patients
We identified all patients aged younger than 50 years with a pathologic diagnosis of CRC (EO-CRC) at Memorial Sloan Kettering Cancer Center (MSK) from January 2014 to June 2019. Patients with a known predisposition syndrome for CRC and those with inflammatory bowel disease were excluded from the clinical and tumor genomic comparisons but were included in the germline analyses, to provide a comprehensive genetic landscape. Data including demographic information, family and medical history, pathology, and presenting symptoms were abstracted from the electronic medical record. The study was approved by the MSK Institutional Review Board. Written informed consent was obtained from patients for all genomic analysis.
Clinical Comparison
Patients with EO-CRC were compared with a previously reported cohort of patients with AO-CRC who were treated at MSK during a similar time frame with identically annotated information (Supplementary Methods, available online) (22). For all comparisons, patients with EO-CRC were further stratified by age at diagnosis: 35 years and younger vs 36-49 years. This was in accordance with Surveillance, Epidemiology, and End Results data, which demonstrate that ages 52 and 36 years are 1 and 2 standard deviations below the mean age at diagnosis of CRC, respectively. This enabled a separate analysis in the 35 years and younger age group where the incidence of CRC is increasing the most and where more aggressive tumor biology has been suggested (4‐6,18‐21).
Tumor Genomic Analyses
Formalin-fixed, paraffin-embedded tumor samples and matched normal blood samples were analyzed prospectively in a Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory using an onsite 341- to 468-gene next-generation sequencing assay (MSK-IMPACT) (23‐25). MSK-IMPACT detects mutations, small insertions and deletions, copy number alterations, and select structural rearrangements and is a validated method to detect MSI (23‐26). Genomic alterations were filtered for oncogenic variants using OncoKB (27). Genes were grouped into signaling pathways using curated pathway templates (28). Genomic data are available in Supplementary Table 1 (available online) and www.cbioportal.org/study/summary?id=crc_eo_2020.
Germline Analyses
Prospective secondary germline analysis was offered to patients who consented to tumor genetic analysis using an institutional review board–approved protocol (23,24). Germline analysis using blood-derived DNA included a 76- to 88-gene MSK-IMPACT panel (Supplementary Table 2, available online), including all cancer-predisposing genes identified by the American College of Medical Genetics and Genomics guidelines (29). Variants were reported and interpreted as described previously (Supplementary Methods, available online) (30‐32).
Statistical Analysis
Baseline clinical characteristics and genomic frequencies were compared using a 2-sided Fisher exact test. Given the expected higher prevalence of germline alterations in EO-CRCs, for the germline comparisons, a 1-sided Fisher exact test was applied. Continuous variables were compared using Wilcoxon test. Kaplan-Meier curves were generated and compared using log-rank test. Cox proportional hazards models were used to generate hazard ratios (HRs) and confidence intervals (CIs) (Supplementary Methods, available online). Clinical and genomic features statistically significant on univariate analysis were included in a multivariate model. Multiple testing correction was performed using the Benjamini-Hochberg method (q-value cutoff of 0.1). R version 3.6.1 statistical software was used for analysis. Clinical and genomic characteristics were statistically compared independently between the 35 years and younger group and AO and the 36-49 years group and AO CRCs (P value cutoff of .05). Unless otherwise indicated, all statistical tests were 2-sided.
Results
Cancer-Specific Features of EO-CRC
We compared the demographic and clinical features of patients with EO-CRC (n = 759) and patients with AO-CRC (n = 687) (Table 1 and Figure 1; Supplementary Figure 1 and Supplementary Tables 3 and 4, available online). To elucidate differences by age at diagnosis of CRC, clinical characteristics were assessed across 3 groups: 35 years and younger (n = 151), 36-49 years (n = 608), and 50 years and older (AO). Sex, race, tumor grade, and stage at presentation were not statistically different between the cohorts (Table 1 and Figure 2, A). The majority of patients in all groups had stage IV disease, reflective of the CRC population that undergoes next-generation sequencing tumor genomic testing (Table 1).
Table 1.
Clinical and tumor characteristics of patients with early-onset and average-onset colorectal cancer
| Characteristic | EO-CRC, ≤35 years (n = 151) | EO-CRC, 36-49 years (n = 608) | AO-CRC (n = 687) | ≤35 years vs AO |
36-49 years vs AO |
||
|---|---|---|---|---|---|---|---|
| P a | q | P a | q | ||||
| Age at diagnosis, y | |||||||
| Mean (SD) | 30.8 (3.8) | 43.8 (3.8) | 62.1 (8.5) | — | — | — | — |
| Median (range) | 31 (14-35) | 44 (36-49) | 61 (50-93) | — | — | — | — |
| Sex, No. (%) | |||||||
| Male | 74 (49) | 347 (57.1) | 370 (53.9) | .28 | — | .26 | — |
| Female | 77 (51) | 261 (42.9) | 317 (46.1) | — | — | — | — |
| Race, No. (%) | |||||||
| White | 113 (74.8) | 475 (78.1) | 561 (81.7) | .07 | 0.266 | .13 | 0.204 |
| Black or African American | 11 (7.3) | 29 (4.8) | 46 (6.7) | .86 | 0.858 | .15 | 0.204 |
| Asian or Indian subcontinent | 14 (9.3) | 59 (9.7) | 40 (5.8) | .14 | 0.266 | .01 | 0.046 |
| Other | 13 (8.6) | 45 (7.4) | 40 (5.8) | .20 | 0.266 | .26 | 0.263 |
| BMI, No. (%) | |||||||
| Normal | 81 (53.6) | 233 (38.3) | 211 (30.7) | <.001 | <0.001 | .006 | 0.023 |
| Underweight | 4 (2.6) | 13 (2.1) | 11 (1.6) | .33 | 0.325 | .54 | 0.539 |
| Overweight | 39 (25.8) | 192 (31.6) | 248 (36.1) | .02 | 0.023 | .08 | 0.155 |
| Obese | 25 (16.6) | 168 (27.6) | 211 (30.7) | <.001 | <0.001 | .22 | 0.294 |
| Unknown | 2 (1.4) | 2 (0.4) | 6 (0.9) | — | — | — | — |
| Median BMI | 24.3 | 26.5 | 27.0 | <.001 | — | .07 | — |
| Smoking history, No. (%) | |||||||
| Ever | 25 (16.6) | 194 (31.9) | 310 (45.1) | <.001 | — | <.001 | — |
| Never | 124 (82.1) | 414 (68.1) | 367 (53.4) | — | — | — | — |
| Unknown | 2 (1.3) | 0 (0) | 10 (1.5) | — | — | — | — |
| Hypertension, No. (%) | |||||||
| Yes | 2 (1.3) | 69 (11.3) | 300 (43.7) | <.001 | — | <.001 | — |
| No | 148 (98) | 531 (87.3) | 385 (56) | — | — | — | — |
| Unknown | 1 (0.7) | 8 (1.4) | 2 (0.3) | — | — | — | — |
| Diabetes, No. (%) | |||||||
| Yes | 4 (2.6) | 33 (5.4) | 80 (11.6) | <.001 | — | <.001 | — |
| No | 146 (96.7) | 574 (94.4) | 605 (88.1) | — | — | — | — |
| Unknown | 1 (0.7) | 1 (0.2) | 2 (0.3) | — | — | — | — |
| Family history of CRC, No. (%) | |||||||
| Yes | 74 (49) | 306 (50.3) | NA | — | — | — | — |
| No | 76 (50.3) | 295 (48.5) | NA | — | — | — | — |
| Unknown | 1 (0.7) | 7 (1.2) | NA | — | — | — | — |
| Stage, No. (%) | |||||||
| I | 2 (1.3) | 22 (3.6) | 29 (4.2) | .10 | 0.131 | .67 | 0.668 |
| II | 12 (7.9) | 53 (8.7) | 93 (13.5) | .08 | 0.131 | .006 | 0.025 |
| III | 47 (31.1) | 165 (27.1) | 161 (23.4) | .06 | 0.131 | .14 | 0.28 |
| IV | 90 (59.7) | 368 (60.6) | 404 (58.9) | .93 | 0.927 | .53 | 0.668 |
| Primary tumor location, No. (%) | |||||||
| Cecum | 15 (9.9) | 39 (6.4) | 120 (17.5) | .02 | 0.091 | <.001 | <0.001 |
| Ascending colon | 9 (6) | 37 (6.1) | 80 (11.6) | .04 | 0.123 | <.001 | <0.001 |
| Hepatic flexure | 1 (0.7) | 11 (1.8) | 23 (3.3) | .10 | 0.184 | .08 | 0.125 |
| Transverse colon | 6 (4) | 18 (3) | 30 (4.4) | 1.00 | 1.000 | .19 | 0.209 |
| Splenic flexure | 5 (3.3) | 18 (3) | 9 (1.3) | .15 | 0.227 | .05 | 0.092 |
| Descending colon | 9 (6) | 41 (6.7) | 39 (5.7) | .85 | 0.956 | .49 | 0.489 |
| Sigmoid | 47 (31.1) | 197 (32.4) | 160 (23.3) | .06 | 0.139 | <.001 | <0.001 |
| Rectosigmoid | 8 (5.3) | 41 (6.7) | 60 (8.7) | .19 | 0.243 | .18 | 0.209 |
| Rectum | 51 (33.7) | 205 (33.7) | 155 (22.6) | .007 | 0.061 | <.001 | <0.001 |
| Colon, NOS | 0 (0) | 1 (0.2) | 11 (1.6) | — | — | — | — |
| Metastasectomy, No. (%) | |||||||
| Yes | 78 (51.7) | 280 (46.1) | 338 (49.2) | .41 | — | .69 | — |
| No | 68 (45) | 303 (49.8) | 348 (50.7) | — | — | — | — |
| Unknown | 5 (3.3) | 25 (4.1) | 1 (0.1) | — | — | — | — |
| Molecular subtype, No. (%) | |||||||
| MSS | 139 (92.1) | 583 (95.8) | 626 (91.1) | .87 | 0.873 | <.001 | <0.001 |
| MSI | 8 (5.3) | 21 (3.5) | 56 (8.2) | .31 | 0.464 | <.001 | <0.001 |
| POLE | 4 (2.6) | 4 (0.7) | 5 (0.7) | .06 | 0.182 | 1.00 | 1.000 |
| Tumor grade (MSS only), No. (%)b | |||||||
| Well differentiated | 1 (0.7) | 10 (1.7) | 1 (0.2) | .34 | 0.344 | .005 | 0.016 |
| Moderately differentiated | 116 (83.5) | 454 (77.9) | 477 (76.2) | .34 | 0.344 | .72 | 0.827 |
| Poorly differentiated | 22 (15.8) | 114 (19.6) | 122 (19.5) | .28 | 0.344 | .83 | 0.827 |
| Unknown | 0 (0) | 5 (0.8) | 26 (4.1) | — | — | — | — |
| Symptoms | |||||||
| BRBPR | 62 (41.1) | 249 (41.0) | 178 (25.9) | <.001 | 0.002 | <.001 | <0.001 |
| Abdominal pain, bloating, pelvic pain | 56 (37.1) | 207 (34.0) | 184 (26.8) | .01 | 0.049 | .005 | 0.011 |
| Change in bowel habits | 36 (23.8) | 114 (18.8) | 188 (27.4) | .42 | 0.626 | <.001 | <0.001 |
| Weight loss | 12 (7.9) | 40 (6.6) | 66 (9.5) | .64 | 0.803 | .07 | 0.099 |
| Anemia | 3 (2.0) | 22 (3.6) | 100 (14.6) | <.001 | <0.001 | <.001 | <0.001 |
| Fatigue | 3 (2.0) | 25 (4.1) | 47 (6.9) | .02 | 0.064 | .04 | 0.072 |
| Screening colonoscopy | 3 (2.0) | 17 (2.8) | 33 (4.8) | .18 | 0.338 | .08 | 0.105 |
| Upper GI symptoms | 3 (2.0) | 8 (1.3) | 26 (3.8) | .34 | 0.558 | .005 | 0.011 |
| Rectal pain | 22 (15.8) | 114 (19.6) | 122 (19.5) | .28 | 0.344 | .83 | 0.827 |
| Back, shoulder, bone pain | 1 (0.7) | 12 (2.0) | 6 (0.9) | 1.00 | 1.00 | .10 | 0.117 |
| Obstruction | 0 (0.0) | 4 (0.7) | 13 (1.9) | .14 | 0.302 | .08 | 0.105 |
A 2-sided Fisher exact test was used. AO = average-onset; BMI = body mass index; CRC = colorectal cancer; EO = early-onset; GI = gastrointestinal; MSI = microsatellite instability; MSS = microsatellite stable; NA = not available; NOS = not otherwise specified; POLE = polymerase epsilon; BRBPR = bright red blood per rectum.
For tumor grade, only MSS tumors were compared, given that MSI tumors are known to be associated with a higher prevalence of poorly differentiated tumors (35 years and younger, n = 139; 36-49 years, n = 583; AO, n = 626). For classification of tumor grade, tumors were categorized according to the most aggressive differentiation exhibited within any given tumor.
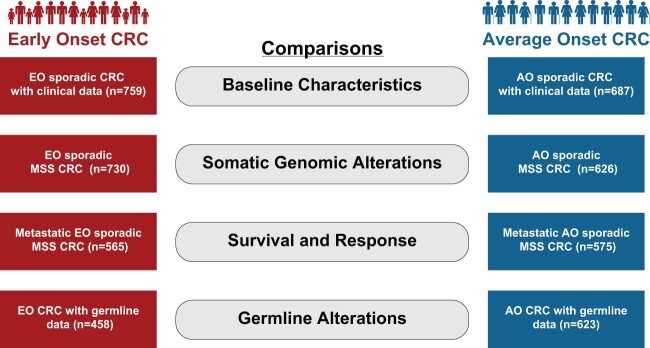
Figure 1.
Overview of comparison groups. The figure shows the comparisons made between patients with EO-CRC and AO-CRC. AO = average onset; CRC= colorectal cancer; EO = early-onset; MSS = microsatellite stable.
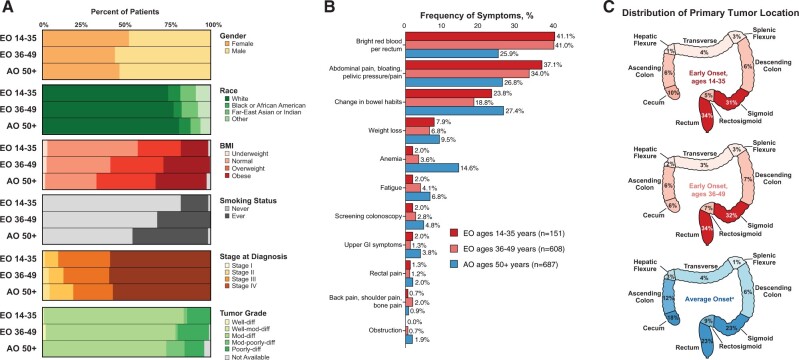
Figure 2.
Clinical and tumor characteristics. Cancer-specific features of early-onset colorectal EO-CRC and AO-CRC by age at diagnosis: 35 years and younger, 36-49 (EO-CRC) and 50 years and older AO-CRC. A) Comparison of demographic, clinical, and tumor characteristics demonstrates that there is no significant difference in several characteristics, including sex and tumor grade distribution. Median body mass index was lower in the 35 years and younger cohort than in the AO-CRC cohort. B) Frequency of cancer-related presenting symptoms. C) Colorectal primary tumor location. AO = average onset; CRC = colorectal cancer; EO = early-onset; NOS = not otherwise specified.
The presence of hypertension (35 years and younger = 1.3%; 36-49 years = 11.3%; AO = 43.7%; P < .001 for both comparisons), diabetes (35 years and younger = 2.6%; 36-49 years = 5.4%; AO = 11.6%; P < .001 for both comparisons), and smoking history (35 years and younger = 16.6%; 36-49 years = 31.9%; AO = 45.1%; P < .001 for both comparisons) progressively increased with age at diagnosis (Table 1). Median body mass index was lower in the 35 years and younger cohort than in the AO cohort (24.3 vs 27.0 kg/m2; P < .001); the difference in body mass index between the 36-49 years and AO cohorts was not statistically significant (26.5 vs 27.0 kg/m2; P = .07) (Table 1).
Rectal bleeding, a presenting symptom, was more common in the EO (35 years and younger = 41.1%; 36-49 years = 41.0%) than in the AO cohort (25.9%, P = .001 and P < .001, respectively). Abdominal pain or bloating, another presenting symptom, was also more common in the EO cohorts (≤35 years and younger = 37.1%; 36-49 years = 34.0%; AO = 26.8%; P = .01 and P = .005, respectively). Accounting for tumor-sidedness, rectal bleeding was still more common in EO than AO left-sided CRCs; abdominal pain was more common in EO than AO right-sided CRCs. Anemia was more common in the AO cohort (AO = 14.6%; 35 years and younger = 2.0%; 36-49 years = 3.6%; P < .001 for both comparisons) (Figure 2, B, and Table 1; Supplementary Table 5, available online) irrespective of tumor sidedness. Left-sided tumors were more common in the EO cohorts (35 years and younger = 80.8%; 36-49 years = 83.7%; AO = 63.9%; P < .001 for both comparisons), as was rectal cancer (both EO cohorts = 33.7%; AO = 22.6%; P = .007 and P < .001, respectively) (Figure 2, C).
Somatic Mutation Analyses of EO-CRC of Microsatellite Stable (MSS) CRC
Using MSK-IMPACT, we assessed somatic alterations in EO-CRC and AO-CRC. To eliminate bias in the genomic analysis introduced by hypermutated tumors with MSI, the comparison was limited to MSS tumors (35 years and older, n = 142; 36-49 years, n = 588; AO, n = 626) (Figure 1; Supplementary Figure 1, available online).
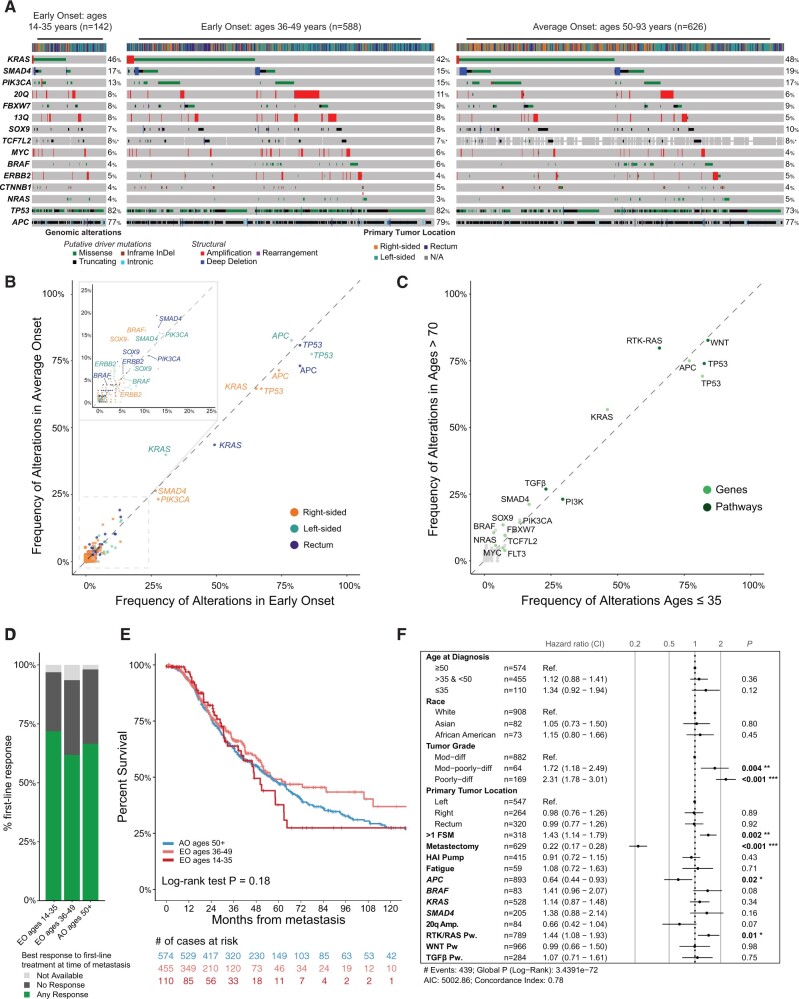
The most common alterations in EO-CRCs were APC (78.7%), TP53 (82.1%), KRAS (42.5%), SMAD4 (15.5%), PIK3CA (14.9%), FBXW7 (8.9%), SOX9 (7.7%), TCF7L2 (7.2%), and BRAF (5.5%). The frequency of oncogenic driver mutations did not differ between the 3 cohorts, except that TP53 alterations were enriched in the 36-49 years cohort, compared with the AO cohort (82.1% vs 73.5%; q = 0.1). Receptor Tyrosine Kinase signaling pathway (RTK‐RAS) alterations were less frequent in the 36-49 cohort than in the AO cohort (64.6% vs 71.7%; q = 0.04) (Figure 3, A; Supplementary Figure 2, A, available online). However, after adjustment for tumor location, the differences in the prevalence of TP53 and RTK-RAS pathway alterations, among all cohorts, were no longer statistically significant at the gene or pathway level (Figure 3, B and C; Supplementary Figure 2, B, available online). To assess for genomic differences possibly masked by the inclusion of patients close to age 50 years at diagnosis, we compared extreme age cohorts (35 years and younger vs older than 70 years); no statistically significant differences were observed (Figure 3, C). No statistically significant differences in tumor mutational burden, fraction of genome altered, whole-genome duplication, or loss of heterozygosity (LOH) were observed (Supplementary Figure 2, C, available online).
Figure 3.
Somatic mutation analyses. Comparison of somatic tumor mutation analyses between EO-CRC and AO-CRC. A) Frequency of oncogenic alterations by age group (35 years and younger, 36-49, 50 years and older [AO-CRC]). B) Frequency of oncogenic alterations adjusted for primary tumor location (left vs right vs rectum) between the 35 years and younger and AO cohorts. C) Frequency of oncogenic alterations adjusted for primary tumor location (left vs right vs rectum) between the 36-49 and AO cohorts. D) Response to first-line chemotherapy. E) Survival outcomes by age groups. F) Multivariate survival analysis incorporating clinical and genomic characteristics that were statistically significant on univariate analysis, as well as age. All statistical tests were 2-sided. AIC = Akaike information criterion; AO = average onset; CI = confidence interval; CRC = colorectal cancer; EO = early-onset; FSM = first sites of metastasis; HAI = hepatic arterial infusion; Dif = differentiated; Mod = moderately; Pw = Pathway.
Clinical Outcomes of Metastatic EO-CRC
We then compared clinical outcomes between EO-CRC and AO-CRC by evaluating response to therapy and survival. Our analysis focused on patients with MSS tumors who had metastatic disease (35 years and younger, n = 110; 36-49 years, n = 455; AO, n = 574) (Figure 1; Supplementary Figure 1, available online). We found that the use and type of first-line chemotherapy, the site of first metastases, and metastastectomy frequency were similar between cohorts and therefore did not confound survival data (Table 1). The majority of patients (69.6%) EO 35 years and younger, (66.3%), EO 36-49 years, and (72.4%) AO received fluoropyrimidine plus oxaliplatin with or without bevacizumab as first-line chemotherapy. Radiographic response to first-line chemotherapy (35 years and younger = 71.9%; 36-49 years = 61.8%; AO = 66.5%; P = .36 and P = .70, respectively) and median overall survival (35 years and younger = 46.9 months; 36-49 years = 56.4 months; AO = 54.5 months; 35 years and younger vs AO, P = .90; and 36-49 years vs AO P = .17, respectively) were not statistically different among the 3 cohorts (Figure 3, D and E).
On univariate analysis, metastasectomy, tumor grade, and presence of APC alterations or 20q amplification in the tumor, BRAF alterations, and SMAD4 alterations were statistically significantly associated with survival. Notably, age was not statistically significantly associated with survival. Variables statistically significant on univariate analysis were incorporated into a multivariate model with the additional inclusion of age. Again, after adjustment for the statistically significant variables, age was not statistically significantly associated with outcome (Figure 3, F). An association between age 35 years and younger and worse outcomes was noted, although it was not statistically significant (HR = 1.43, 95% CI = 0.99 to 2.07; P = .06).
Germline Analysis of EO-CRC
Germline genomic analysis was performed using a 76-gene (n = 351) or 88-gene (n = 730) panel (35 years and younger, n = 116; 36-49 years, n = 342; AO, n = 623; Supplementary Table 2, available online). This analysis included patients regardless of microsatellite status or known risk factors for CRC (Figure 1; Supplementary Figure 1, available online).
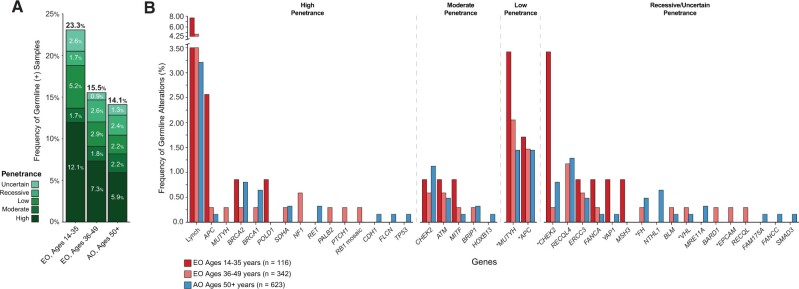
Germline pathogenic (P) and likely pathogenic (LP) variant prevalence in patients with EO-CRC was 17.5%. LS was the most common cancer predisposition syndrome, accounting for 5.5% of all EO-CRCs (Figure 4). By age group, the highest mutation prevalence was identified in the 35 years and younger cohort with 23.3% harboring an LP or P variant, compared with 14.1% of the AO cohort (P = .01), and was driven by an enrichment of high-penetrance gene variants (35 years and younger = 12.1%; AO = 5.9%; P = .02) (Figure 4, A). Notably, 92.8% (13 of 14) of these high-penetrance germline mutation carriers harbored LP or P variants in known CRC-associated cancer predisposition genes, including DNA mismatch repair genes (n = 9), APC (n = 3), and POLD1 (n = 1). The distribution of germline variants by gene and penetrance is shown in Figure 4, B. The prevalence of germline variants was equivalent between rectal and colon cancer patients.
Figure 4.
Germline mutation analyses. A) Germline mutations by age group. B) Distribution of germline mutations by gene and penetrance by age group. An asterisk (*) next to a gene name designates the following: MUTYH, under low penetrance, represents the presence of monoallelic MUTYH variant; APC, under low penetrance, represents the APC p. Ile130Lys germline variant, not associated with classical or attenuated familial adenomatous polyposis; CHEK2, under uncertain penetrance, represents the CHEK2 p. Ile15Thr germline variant of uncertain clinical actionability; FH, under uncertain or recessive penetrance, represents the FH p. Lys477dup variant, not associated with the high-penetrance hereditary leiomyomatosis and renal cell carcinoma syndrome; VHL, under recessive penetrance, represents VHL p. Arg200Trp, associated with a recessive form of VHL-dependent polycythemia and not Von Hippel-Lindau syndrome; EPCAM, deletion exons 2-7, under recessive penetrance, represents variant possibly associated with autosomal recessive congenital tufting enteropathy, but not Lynch syndrome.
Using matched tumor samples, we interrogated somatic genomic data to assess biallelic inactivation—somatic mutations or LOH—at the implicated germline region. Overall, 36% of all LP and P germline mutations exhibited biallelic inactivation. The 35 years and younger cohort had the highest rate of biallelic inactivation (35 years and younger = 51.7%; AO = 32.6%; P = .04), suggesting that these germline events were driving CRC carcinogenesis (Supplementary Figure 3, available online).
Discussion
Our study represents the largest and most comprehensive evaluation of the clinical characteristics and genomic features of patients with EO-CRC to date and includes an independent evaluation of patients with very early-onset disease (35 years and younger at the time of diagnosis). In contrast to prior reports, after adjustment for known confounders, neither genomic tumor profiles nor clinical outcome data support the hypothesis that sporadic EO-CRC is biologically distinct from sporadic AO-CRC. Despite no genomic or biologic tumor differences, patients with EO-CRC do have different clinical findings. Exclusive of patients with a hereditary predisposition for CRC, we found that more than 80% of EO-CRC patients presented with left-sided CRCs—and nearly one-third presented with rectal cancer—which is consistent with prior reports (5,33). Patients with EO-CRC were more likely to present with rectal bleeding and abdominal pain but were less likely to present with anemia, which may reflect tumor location and possibly more routine blood count evaluations in older individuals.
Multiple studies have suggested that patients with EO-CRC present with more advanced disease (8‐11,34), owing to a tumor biology that is inherently more aggressive (8,35). This, in turn, has led to more aggressive treatment regimens in young patients, with chemotherapy overuse but without matched survival improvement (36,37). Our study found no statistically significant difference in tumor grade—a marker of tumor biology—between patients with EO-CRC and patients with AO-CRC. MSI CRCs, which are often enriched in poorly differentiated adenocarcinomas (38), were excluded from this assessment; prior studies may have been confounded by the expected higher incidence of LS-associated MSI tumors among patients with EO-CRC (8,15). Similarly, our tumor genomic analysis was limited to MSS CRCs; no statistically significant differences in CRC-driver pathway gene alterations were observed between cohorts. In contrast to the lower rates of BRAF V600E and APC variants observed in EO-CRCs (14), no statistically significant differences in the prevalence of these mutations were observed after adjustment for MSI status and tumor sidedness, a factor associated with distinct molecular characteristics (22). The higher prevalence of TP53 mutations among MSS EO-CRCs, compared with AO-CRCs, was not statistically significant after adjustment for tumor side; this may account for contrasting results with prior studies that noted increased rates of APC and TP53 mutations among EO-CRCs when sidedness was not considered (39).
A key strength of our study is its focus on patients with CRC without known clinical predispositions to CRC; thus, we evaluated the exact EO-CRC population with the most dramatic increases in the incidence of CRC. Contrary to previous results (40,41), we observed no statistically significant differences in clinical outcomes between metastatic MSS EO-CRC and AO-CRC. Importantly, this lack of difference could not be attributed to more aggressive therapy in patients with EO-CRC, as we assessed the type of chemotherapy and response to first-line chemotherapy and found no statistically significant differences between cohorts. Moreover, the use of locoregional interventions, such as surgery, were not statistically different between the cohorts. Prior studies have also demonstrated comparable outcomes between patients with EO-CRC and those with AO-CRC (42,43). Late stage of presentation noted in prior studies of EO-CRC may reflect a delay in diagnosis, as opposed to a different disease biology. Indeed, more than half of EO-CRC patients wait up to 1 year from the onset of presenting symptoms to seek medical care and are evaluated by an average of 3 medical providers before diagnosis (34,44).
Our germline analysis revealed that, among patients with very early-onset CRC, the prevalence of germline mutations was especially high at 23% because of a near doubling of mutations in high-penetrance cancer susceptibility genes. The rate of mutations in high- and moderate-penetrance genes was similar to that in prior publications, after adjustment for number of genes tested (45). Our integrated germline and somatic analysis helped further elucidate the role of these germline variants in CRC carcinogenesis. Biallelic inactivation, resulting from somatic mutation or LOH in the tumor, was present in nearly all patients with mutations in a high-penetrance CRC susceptibility gene and in a higher proportion of patients aged 35 years and younger, compared with older patients. Universal tumor screening of all CRCs for LS has been incorporated into national guidelines (46). However, given the high prevalence of germline variants in patients aged 35 years and younger, consideration of other markers of genetic risk (ie, colonic polyposis, strong CRC family history) and a low clinical threshold for pursuing multigene panel genetic testing is especially important in this high-risk population.
Established risk factors for CRC in older individuals, such as obesity (47), a Western diet (17,48,49), and diabetes (50), have not been definitively established as contributors to the increasing incidence of EO-CRC. Assessment of such risk factors is beyond the scope of our study, as we did not compare EO-CRC patients with age- and population-matched controls. As is expected in an older population, our AO-CRC patients had higher rates of chronic diseases, such as diabetes, hypertension, and obesity. Importantly, the selective pressure or insult that has shifted the development of CRC to younger populations over the past few decades appears to be steady, persistent, and globally occurring (16). It therefore seems to reason that external or environmental factors are likely to be driving earlier CRC development, and further investigations, using appropriately matched control populations, are necessary.
Although our study represents the largest comprehensive analysis of EO-CRCs to date, it is a retrospective, single-institution analysis. To attempt to overcome potential referral and other demographic biases pertinent to our tertiary-care cancer center, EO-CRC patients were compared with AO-CRC patients who were treated at our institution, with identical clinical annotations and genomic profiling, during a similar time frame. Furthermore, although the age groups are balanced ethnically, representation of Black and Hispanic patients is low. To date, no statistically significant differences have been noted; however, we and other researchers are actively pursuing national and international collaborations evaluating EO-CRC genomics in more diverse populations (51).
Unlike prior reports, our study demonstrates that, after adjustment for known confounders, sporadic EO-CRCs are genomically equivalent to AO-CRCs. Inherited genetic susceptibility, although higher in EO-CRCs, explains only a fraction of all EO-CRCs. We highlight the importance of the lack of genomic and biological differences in this disease, as initial reports described EO-CRC as a potentially different and more aggressive disease entity leading many clinicians to select more intense treatments (36,37). Our results demonstrate that clinical outcomes and response to chemotherapy are the same and that aggressive treatment regimens based solely on the age at CRC diagnosis are not warranted. Further research should be focused on evaluating diverse populations and identifying potential environmental risk factors that are contributing to this shift in incidence to better distinguish the young population at risk and improve outcomes in this disease.
Funding
This work was supported by the National Cancer Institute (P30 CA008748); the National Institutes of Health (T32 GM132083; R25 CA233208); Stand Up to Cancer Colorectal Cancer Dream Team Translational Research Grant (SU2C-AACR-DT22-17). Stand Up to Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research, the Marie-Josée and Henry R. Kravis Center for Molecular Oncology; the Precision, Interception and Prevention Program at MSK; and the Romeo Milio Lynch Syndrome Foundation.
Notes
Role of the funders: The funders had no role in the design of the study, the the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
Disclosures: AC is on advisory boards for Bayer and Array BioPharma, and receives research funding from Seattle Genetics, Tesaro/GSK and RGenix. RY receives research funding from Array BioPharma, Novartis, and Boehringer Ingelheim, and participates in consulting for Array BioPharma. LAD is a member of the board of directors of Personal Genome Diagnostics (PGDx) and Jounce Therapeutics. He is a paid consultant to PGDx and Neophore. He is an uncompensated consultant for Merck but has received research support for clinical trials from Merck. LAD is an inventor of multiple licensed patents related to technology for circulating tumor DNA analyses and mismatch repair deficiency for diagnosis and therapy from Johns Hopkins University. Some of these licenses and relationships are associated with equity or royalty payments directly to Johns Hopkins and LAD. He holds equity in PGDx, Jounce Therapeutics, Thrive Earlier Detection, and Neophore. His wife holds equity in Amgen. The terms of all these arrangements are being managed by Johns Hopkins and Memorial Sloan Kettering in accordance with their conflict-of-interest policies. ZKS has immediate family members who hold consulting/advisory roles in Ophthalmology with Allergan, Adverum Biotechnologies, Alimera Sciences, Biomarin, Fortress Biotech, Genentech/Roche, Novartis, Optos, Regeneron, Regenxbio, and Spark Therapeutics. All other authors have nothing to disclose.
Author contributions: AC: Conceptualization, Methodology, Investigation, Resources, Data Curation, Writing—Original Draft, Writing—Review & Editing, Supervision, Project Administration, Funding acquisition. WKC: Methodology, Software, Formal analysis, Writing—Original Draft, Writing—Review & Editing, Visualization. RY: Conceptualization, Methodology, Writing—Review & Editing. HW: Methodology, Software, Formal analysis, Writing—Review & Editing, Visualization. GDSF: Methodology, Data Curation. AK: Data Curation. LP: Investigation, Data Curation, Project Administration. AM: Writing—Review & Editing. YK: Investigation, Data Curation. PS: Investigation. CB: Investigation. ES-M: Investigation. PRT: Investigation. KB: Investigation. JG: Investigation. VJ: Investigation. NS: Writing—Review & Editing. AV: Writing—Review & Editing. DR-L: Writing—Review & Editing. JS: Writing—Review & Editing. EV: Writing—Review & Editing. SM: Investigation, Data Curation, Project Administration. RM: Writing—Review & Editing. MAL: Writing—Review & Editing. FS: Formal analysis. NK: Writing—Review & Editing. LC: Writing—Review & Editing. KG: Writing—Review & Editing. AM: Writing—Review & Editing. GN: Writing—Review & Editing. JG: Writing—Review & Editing.JJS: Writing—Review & Editing. PBP: Writing—Review & Editing. LZ: Writing—Review & Editing. DM: Writing—Review & Editing. OB: Writing—Review & Editing. MR: Writing—Review & Editing. KO: Writing—Review & Editing. BT: Investigation. MB: Investigation. DS: Investigation. MW: Writing—Review & Editing. LBS: Writing—Review & Editing. JGA: Writing—Review & Editing. NS: Methodology, Formal analysis, Visualization. LAD : Conceptualization, Methodology, Formal analysis, Writing—Review & Editing. ZKS: Conceptualization, Methodology, Formal analysis, Investigation, Writing—Original Draft, Writing—Review & Editing.
Disclaimers: None.
Prior presentations: Partial results of the somatic genomic profiling were presented at the American Society of Clinical Oncology and the European Society for Medical Oncology meetings in 2018, and the germline analysis was a platform presentation at the annual meeting of the Collaborative Group of the Americas on Inherited Gastrointestinal Cancer in 2019.
Data Availability
Genomic data are available at www.cbioportal.org/study/summary?id=EO_CRC_2020. The authors confirm that the remainder of the data supporting the findings of this study are available within the article and/or its Supplementary Materials.
Supplementary Material
References
- 1. Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA A Cancer J Clin. 2020;70(3):145–164. [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Jemal A, Ward EM.. Increase in incidence of colorectal cancer among young men and women in the United States. Cancer Epidemiol Biomarkers Prev. 2009;18(6):1695–1698. [DOI] [PubMed] [Google Scholar]
- 3. Winawer SJ, Fletcher RH, Miller L, et al. Colorectal cancer screening: clinical guidelines and rationale. Gastroenterology. 1997;112(2):594–642. [DOI] [PubMed] [Google Scholar]
- 4. Smith RA, Cokkinides V, Eyre HJ; for the American Cancer Society. American Cancer Society guidelines for the early detection of cancer, 2003. CA Cancer J Clin. 2003;53(1):27–43. [DOI] [PubMed] [Google Scholar]
- 5. Meyer JE, Narang T, Schnoll-Sussman FH, Pochapin MB, Christos PJ, Sherr DL.. Increasing incidence of rectal cancer in patients aged younger than 40 years: an analysis of the surveillance, epidemiology, and end results database. Cancer. 2010;116(18):4354–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Howlader N, Noone AM, Krapcho M, et al. , eds. National Cancer Institute Surveillance, Epidemiology, and End Results Program, Nov 2008 Sub (1975-2016). National Cancer Institute, DCCPS, Surveillance Research Program, Surveillance Systems Branch. https://seer.cancer.gov/csr/1975_2016/. Published 2019. Accessed October 01, 2020.
- 7. Bailey CE, Hu C-Y, You YN, et al. Increasing disparities in the age-related incidences of colon and rectal cancers in the United States, 1975-2010. JAMA Surg. 2015;150(1):17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fu J, Yang J, Tan Y, et al. Young patients (≤ 35 years old) with colorectal cancer have worse outcomes due to more advanced disease: a 30-year retrospective review. Medicine (Baltimore). 2014;93(23):e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yantiss RK, Goodarzi M, Zhou XK, et al. Clinical, pathologic, and molecular features of early onset colorectal carcinoma. Am J Surg Pathol. 2009;33(4):572–582. [DOI] [PubMed] [Google Scholar]
- 10. Chang DT, Pai RK, Rybicki LA, et al. Clinicopathologic and molecular features of sporadic early onset colorectal adenocarcinoma: an adenocarcinoma with frequent signet ring cell differentiation, rectal and sigmoid involvement, and adverse morphologic features. Mod Pathol. 2012;25(8):1128–1139. [DOI] [PubMed] [Google Scholar]
- 11. Kirzin S, Marisa L, Guimbaud R, et al. Sporadic early onset colorectal cancer is a specific sub-type of cancer: a morphological, molecular and genetics study. PLoS One. 2014;9(8):e103159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen FW, Sundaram V, Chew TA, Ladabaum U.. Advanced-stage colorectal cancer in persons younger than 50 years not associated with longer duration of symptoms or time to diagnosis. Clin Gastroenterol Hepatol. 2017;15(5):728–737.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yeo H, Betel D, Abelson JS, Zheng XE, Yantiss R, Shah MA.. Early onset colorectal cancer is distinct from traditional colorectal cancer. Clin Colorectal Cancer. 2017;16(4):293–299.e6. [DOI] [PubMed] [Google Scholar]
- 14. Willauer AN, Liu Y, Pereira AAL, et al. Clinical and molecular characterization of early onset colorectal cancer. Cancer. 2019;125(12):2002–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Silla IO, Rueda D, Rodríguez Y, García JL, de la Cruz Vigo F, Perea J.. Early onset colorectal cancer: a separate subset of colorectal cancer. World J Gastroenterol. 2014;20(46):17288–17296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Siegel RL, Torre LA, Soerjomataram I, et al. Global patterns and trends in colorectal cancer incidence in young adults. Gut. 2019;68(12):2179–2185. [DOI] [PubMed] [Google Scholar]
- 17. Mehta RS, Song M, Nishihara R, et al. Dietary patterns and risk of colorectal cancer: analysis by tumor location and molecular subtypes. Gastroenterology. 2017;152(8):1944–1953.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao L, Bao F, Yan J, et al. Poor prognosis of young patients with colorectal cancer: a retrospective study. Int J Colorectal Dis. 2017;32(8):1147–1156. [DOI] [PubMed] [Google Scholar]
- 19. Bleyer A, Barr R, Hayes-Lattin B, et al. ; for the Biology and Clinical Trials Subgroups of the US National Cancer Institute Progress Review Group in Adolescent and Young Adult Oncology. The distinctive biology of cancer in adolescents and young adults. Nat Rev Cancer. 2008;8(4):288–298. [DOI] [PubMed] [Google Scholar]
- 20. Shida D, Ahiko Y, Tanabe T, et al. Shorter survival in adolescent and young adult patients, compared to adult patients, with stage IV colorectal cancer in Japan. BMC Cancer. 2018;18(1):334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chiang J-M, Chen M-C, Changchien CR, et al. Favorable influence of age on tumor characteristics of sporadic colorectal adenocarcinoma: patients 30 years of age or younger may be a distinct patient group. Dis Colon Rectum. 2003;46(7):904–910. [DOI] [PubMed] [Google Scholar]
- 22. Yaeger R, Chatila WK, Lipsyc MD, et al. Clinical sequencing defines the genomic landscape of metastatic colorectal cancer. Cancer Cell. 2018;33(1):125–136.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17(3):251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Middha S, Zhang L, Nafa K, et al. Reliable pan-cancer microsatellite instability assessment by using targeted next-generation sequencing data. J Clin Oncol Precis Oncol. 2017;2017(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stadler ZK, Battaglin F, Middha S, et al. Reliable detection of mismatch repair deficiency in colorectal cancers using mutational load in next-generation sequencing panels. J Clin Oncol. 2016;34(18):2141–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chakravarty D, Gao J, Phillips SM, et al. OncoKB: a precision oncology knowledge base. J Clin Oncol Precis Oncol. 2017;2017(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sanchez-Vega F, Mina M, Armenia J, et al. ; for the Cancer Genome Atlas Research Network. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173(2):321–337.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kalia SS, Adelman K, Bale SJ, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19(2):249–255. [DOI] [PubMed] [Google Scholar]
- 30. Schrader KA, Cheng DT, Joseph V, et al. Germline variants in targeted tumor sequencing using matched normal DNA. JAMA Oncol. 2016;2(1):104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mandelker D, Zhang L, Kemel Y, et al. Mutation detection in patients with advanced cancer by universal sequencing of cancer-related genes in tumor and normal DNA vs guideline-based germline testing. JAMA. 2017;318(9):825–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Richards S, Aziz N, Bale S, et al. ; for the ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Glover M, Mansoor E, Panhwar M, Parasa S, Cooper GS.. Epidemiology of colorectal cancer in average risk adults 20-39 years of age: a population-based national study. Dig Dis Sci. 2019;64(12):3602–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yarden RI, Newcomer KL for the Never to Young Advisory Board and Colorectal Cancer Alliance. Young onset Colorectal Cancer Alliance. Young onset colorectal cancer patients are diagnosed with advanced disease after multiple misdiagnoses. In: Science and Health Policy. American Association for Cancer Research; 2019:3347-3347. Abstract 3347. doi:10.1158/1538-7445.AM2019-3347
- 35. You YN, Xing Y, Feig BW, Chang GJ, Cormier JN.. Young-onset colorectal cancer: is it time to pay attention? Arch Intern Med. 2012;172(3):287–289. [DOI] [PubMed] [Google Scholar]
- 36. Kneuertz PJ, Chang GJ, Hu C-Y, et al. Overtreatment of young adults with colon cancer: more intense treatments with unmatched survival gains. JAMA Surg. 2015;150(5):402–409. [DOI] [PubMed] [Google Scholar]
- 37. Manjelievskaia J, Brown D, McGlynn KA, Anderson W, Shriver CD, Zhu K.. Chemotherapy use and survival among young and middle-aged patients with colon cancer. JAMA Surg. 2017;152(5):452–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349(3):247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lieu CH, Golemis EA, Serebriiskii IG, et al. Comprehensive genomic landscapes in early and later onset colorectal cancer. Clin Cancer Res. 2019;25(19):5852–5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sanford SD, Zhao F, Salsman JM, Chang VT, Wagner LI, Fisch MJ.. Symptom burden among young adults with breast or colorectal cancer. Cancer. 2014;120(15):2255–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lieu CH, Renfro LA, de Gramont A, et al. Association of age with survival in patients with metastatic colorectal cancer: analysis from the ARCAD Clinical Trials Program. J Clin Oncol. 2014;32(27):2975–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Steinhagen E, Shia J, Riedel E, et al. Response to neoadjuvant therapy in patients with early age-of-onset rectal cancer. Dis Colon Rectum. 2013;56(1):58–63. [DOI] [PubMed] [Google Scholar]
- 43. Quah HM, Joseph R, Schrag D, et al. Young age influences treatment but not outcome of colon cancer. Ann Surg Oncol. 2007;14(10):2759–2765. [DOI] [PubMed] [Google Scholar]
- 44. Sheneman DW, Finch JL, Messersmith WA, et al. The impact of young adult colorectal cancer: incidence and trends in Colorado. Colorectal Cancer. 2017;6(2):49–56. [Google Scholar]
- 45. Pearlman R, Frankel WL, Swanson B, et al. ; for the Ohio Colorectal Cancer Prevention Initiative Study Group. Prevalence and spectrum of germline cancer susceptibility gene mutations among patients with early onset colorectal cancer. JAMA Oncol. 2017;3(4):464–471. doi:10.1001/jamaoncol.2016.5194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gupta S, Provenzale D, Llor X, et al. ; for the CGC. NCCN guidelines insights: genetic/familial high-risk assessment: colorectal, version 2.2019. J Natl Compr Canc Netw. 2019;17(9):1032–1041. [DOI] [PubMed] [Google Scholar]
- 47. Bardou M, Barkun AN, Martel M.. Obesity and colorectal cancer. Gut. 2013;62(6):933–947. [DOI] [PubMed] [Google Scholar]
- 48. Beresford SAA, Johnson KC, Ritenbaugh C, et al. Low-fat dietary pattern and risk of colorectal cancer: the Women’s Health Initiative Randomized Controlled Dietary Modification trial. JAMA. 2006;295(6):643–654. [DOI] [PubMed] [Google Scholar]
- 49. Park S-Y, Boushey CJ, Wilkens LR, Haiman CA, Le Marchand L.. High-quality diets associate with reduced risk of colorectal cancer: analyses of diet quality indexes in the multiethnic cohort. Gastroenterology. 2017;153(2):386–394.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tsilidis KK, Kasimis JC, Lopez DS, Ntzani EE, Ioannidis JPA.. Type 2 diabetes and cancer: umbrella review of meta-analyses of observational studies. BMJ. 2015;350:g7607. [DOI] [PubMed] [Google Scholar]
- 51. Kingham TP, Alatise OI.. Establishing translational and clinical cancer research collaborations between high- and low-income countries. Ann Surg Oncol. 2015;22(3):741–746. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic data are available at www.cbioportal.org/study/summary?id=EO_CRC_2020. The authors confirm that the remainder of the data supporting the findings of this study are available within the article and/or its Supplementary Materials.