Abstract
Background
In the phase III IMpassion130 study, atezolizumab plus nab-paclitaxel (A+nP) showed clinical benefit in advanced or metastatic triple-negative breast cancer patients who were programmed death-ligand 1 (PD-L1)+ (tumor-infiltrating immune cells [IC] ≥1%) using the SP142 immunohistochemistry assay. Here we evaluate 2 other PD-L1 assays for analytical concordance with SP142 and patient-associated clinical outcomes.
Methods
Samples from 614 patients (68.1% of intention-to-treat population) were centrally evaluated by immunohistochemistry for PD-L1 status on IC (VENTANA SP142, SP263, Dako 22C3) or as a combined positive score (CPS; 22C3).
Results
Using SP142, SP263, and 22C3 assays, PD-L1 IC ≥1% prevalence was 46.4% (95% confidence interval [CI] = 42.5% to 50.4%), 74.9% (95% CI = 71.5% to 78.3%), and 73.1% (95% CI = 69.6% to 76.6%), respectively; 80.9% were 22C3 CPS ≥1. At IC ≥1% (+), the analytical concordance between SP142 and SP263 and 22C3 was 69.2% and 68.7%, respectively. Almost all SP142+ cases were captured by other assays (double positive), but several SP263+ (29.6%) or 22C3+ (29.0%) cases were SP142– (single positive). A+nP clinical activity vs placebo+nP in SP263+ and 22C3+ patients (progression-free survival [PFS] hazard ratios [HRs] = 0.64 to 0.68; overall survival [OS] HRs = 0.75 to 0.79) was driven by double-positive cases (PFS HRs = 0.60 to 0.61; OS HRs = 0.71 to 0.75) rather than single-positive cases (PFS HRs = 0.68 to 0.81; OS HRs = 0.87 to 0.95). Concordance for harmonized cutoffs for SP263 (IC ≥4%) and 22C3 (CPS ≥10) to SP142 (IC ≥1%) was subpar (approximately 75%).
Conclusions
22C3 and SP263 assays identified more patients as PD-L1+ (IC ≥1%) than SP142. No inter-assay analytical equivalency was observed. Consistent improved A+nP efficacy was captured by the SP142 PD-L1 IC ≥1% subgroup nested within 22C3 and SP263 PD-L1+ (IC ≥1%) populations.
Immune checkpoint inhibitors targeting the programmed death-ligand 1 (PD-L1) or programmed death-1 pathway have shown clinical benefit for the treatment of triple-negative breast cancer (TNBC), a disease accounting for 15% to 20% of all breast cancer cases and characterized by an aggressive disease course and poor prognosis (1-3). In metastatic TNBC (mTNBC), studies have shown that clinical activity of these agents may be enhanced in patients whose tumors express PD-L1 on either tumor cells (TC) or tumor-infiltrating immune cells (IC) (1,4-7).
Based on evidence from the IMpassion130 trial, the anti-PD-L1 antibody atezolizumab combined with nab-paclitaxel (A+nP) was approved by regulatory authorities and endorsed by experts as a standard-of-care treatment for patients with mTNBC and PD-L1–expressing IC covering at least 1% of the tumor area (PD-L1 IC+) (8-12). In IMpassion130, patients with unresectable locally advanced or mTNBC whose tumors were PD-L1 IC+, identified using the VENTANA PD-L1 SP142 immunohistochemistry (IHC) assay (Ventana Medical Systems; Oro Valley, AZ), demonstrated statistically significantly improved progression-free survival (PFS; hazard ratio [HR] = 0.62, 95% confidence interval [CI] = 0.49 to 0.78, P < .001) and clinically meaningful overall survival (OS; HR = 0.71, 95% CI = 0.54 to 0.94) with A+nP compared with placebo plus nab-paclitaxel (P+nP) (2,13).
The SP142 IHC assay is approved by the US Food and Drug Administration (FDA) to identify patients with mTNBC for treatment with A+nP using a ≥1% expression cutoff for PD-L1 on IC (14). Although SP142 is clinically validated, other commercially available PD-L1 IHC assays, including Dako 22C3 (Dako; Carpinteria, CA, USA) and VENTANA SP263 (15,16), are widely used in laboratories worldwide for TNBC and non-TNBC indications. However, these assays use scoring algorithms different from SP142 and may differ in analytical concordance and/or clinical utility for patients with TNBC treated with A+nP. Comparative assay data provide value in guiding clinical decision making and treatment guidelines globally.
In this post hoc, exploratory substudy of IMpassion130, we investigated analytical concordance and clinical utility among the VENTANA SP142, VENTANA SP263, and Dako 22C3 PD-L1 IHC assays.
Methods
Patients and Treatment
IMpassion130 (NCT02425891) is an international, randomized, double-blind, placebo-controlled, phase III study evaluating first-line A+nP vs P+nP in patients with unresectable locally advanced or mTNBC (2). Eligibility criteria and methodology are described elsewhere (2). Patients were randomly assigned 1:1 to atezolizumab 840 mg or placebo every 2 weeks plus nab-paclitaxel 100 mg/m2 on days 1, 8, and 15 of every 28-day cycle intravenously until disease progression (per Response Evaluation Criteria in Solid Tumors version 1.1) or intolerable toxicity. IMpassion130 was conducted in accordance with Good Clinical Practice guidelines and the Declaration of Helsinki. Protocol approval was obtained from independent review boards or ethics committees for each site; all patients provided written informed consent. The clinical data cutoff date was January 2, 2019.
Prespecified coprimary efficacy endpoints were investigator-assessed PFS per Response Evaluation Criteria in Solid Tumors 1.1 and OS in the intention-to-treat (ITT) population and in PD-L1 IC+ patients (tumors with PD-L1–expressing IC covering ≥1% of tumor area), assessed using the SP142 IHC assay (2). Exploratory analyses reported here were performed in the biomarker-evaluable population (BEP), with available tumor tissue for biomarker analysis (Supplementary Figure 1, available online).
IHC Assays
Histologic sections from formalin-fixed paraffin-embedded tumor samples were centrally evaluated for PD-L1 expression at HistoGeneX (now Cellcarta NV) laboratory locations (Antwerpen, Belgium, and Naperville, IL, USA) using VENTANA SP142 (14), VENTANA SP263 (15), and Dako 22C3 (16) IHC assays on their respective platforms (BenchMark ULTRA instrument [VENTANA] and Autostainer Link 48 platform [Dako]).
Eight HistoGeneX pathologists scored the samples, and each had been trained on the VENTANA or Dako IHC assay according to prespecified algorithms and cutoff values (Table 1). Supplementary Figure 2 (available online) depicts a routine training program, including testing requirements. Each sample was read once by a single pathologist, and several pathologists may have scored more than 1 type of assay or algorithm. Supplementary Table 1 (available online) includes information about the pathologists’ specific training as well as sample types scored by each pathologist. Briefly, 5 pathologists evaluated SP142-stained specimens at the IC 1% cutoff and had received SP142 TNBC training by VENTANA. Three pathologists evaluated SP263-stained samples and had received SP263 non-TNBC training by VENTANA. Two pathologists analyzed 22C3-stained sections at the combined positive score (CPS) 1 cutoff; 1 had been trained in non-TNBC (cervical cancer) by Dako, and the other was trained by the HistoGeneX internal reader-reader training program. Evaluation of 22C3-stained specimens at the nonstandard IC 1% cutoff (an assay or algorithm combination for which no formal training program is currently available) was undertaken by 6 of the 8 pathologists who were trained in SP142 IC 1% scoring (TNBC and other solid tumors). Further details may be found in Supplementary Figure 2 and Supplementary Table 1 (available online).
Table 1.
Immunohistochemistry assays, scoring algorithms and cutoffsa
| PD-L1 assay | Scoring algorithm used | Cutoff used |
|---|---|---|
| SP142 | SP142 IC scoring algorithm (PD-L1 staining of any intensity on IC covering ≥1% of tumor area) | IC 1% |
| SP263 | SP142 IC scoring algorithm | IC 1% |
| 22C3 | SP142 IC scoring algorithm | IC 1% |
| 22C3 | CPS scoring algorithm: enumerating PD-L1–stained cells (TC, lymphocytes, macrophages) divided by total number of viable TC and multiplied by 100 | CPS 1 |
CPS = combined positive score; IC = tumor-infiltrating immune cells; PD-L1 = programmed death-ligand; TC = tumor cells.
For SP263 and SP142, the IC value was recorded as a percentage of tumor area (consisting of TC and associated intra-tumoral and continuous peri-tumoral stroma) occupied by IC with discernible PD-L1 staining of any intensity (14,15). For 22C3, CPS was defined as the number of PD-L1–stained cells (including TC, lymphocytes, and macrophages) divided by the total number of viable TC and multiplied by 100, with a score cutoff of ≥1 (16). In addition to validated scoring algorithms and cutoffs, an IC scoring algorithm was used to score 22C3-stained samples (Table 1). Although reader precision was established at the IC 1% or CPS 1 cutoff point, pathologists also recorded the raw PD-L1 scoring values as a continuous variable.
Stromal tumor-infiltrating lymphocyte (sTIL) evaluations were performed with hematoxylin and eosin by trained pathologists at HistoGeneX (Antwerp, Belgium) in accordance with TIL International Working Group guidelines (17).
Statistical Analysis
Analytical concordance between SP142 (used as reference standard at IC 1%) and the comparator assays was assessed with positive percentage agreement (PPA), negative percentage agreement (NPA), and overall percentage agreement (OPA) at the preselected cutoff values for the alternative assay (SP263 or 22C3). For each metric, 95% confidence intervals were calculated.
Hazard ratio estimates with associated 95% CIs were derived to compare investigator-assessed PFS and OS among biomarker-defined patient subgroups using Cox regression adjusted for key baseline prognostic factors (age, Eastern Cooperative Oncology Group performance status, presence of liver metastases, and prior taxane treatment). Kaplan-Meier estimates and corresponding median survival durations were also evaluated for subgroups identified by each assay at the preselected cutoff values. Comparisons between sTIL counts were performed using the Kruskal-Wallis test with the Dunn’s test for multiple comparisons. Statistical significance tests were 2-sided, and a P value of less than .05 was considered statistically significant.
Receiver operating characteristic (ROC) assessments (R package pROC) were used to compute maximized OPAs, with the clinically validated SP142 IC 1% cutoff as reference standard used to determine SP263 and 22C3 IC and CPS harmonized cutoffs.
Results
Characteristics of the IMpassion130 Biomarker Population
In this study, the SP142 (cutoff: IC ≥1%), SP263 (cutoff: IC ≥1%), and 22C3 (cutoffs: IC ≥1% and CPS ≥1) PD-L1 IHC assays were evaluated in a BEP of 614 patients (68.1% of the 902 in the IMpassion130 ITT population). Baseline demographic and clinical characteristics of the BEP were comparable with the ITT population except for a higher prevalence of PD-L1 IC ≥1% patients using the SP142 IHC assay (46.4% and 40.9% for BEP and ITT, respectively; Supplementary Table 2, available online).
Inter-Assay PD-L1 Prevalence and Analytical Concordance
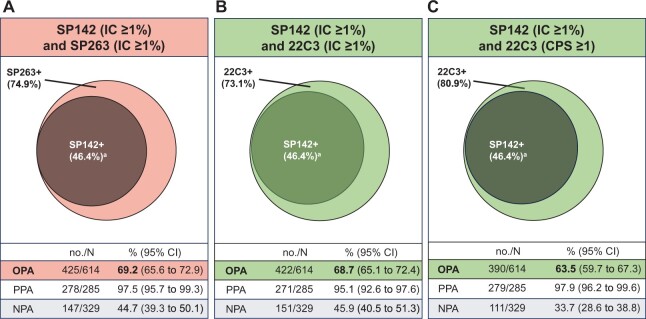
Prevalence rates using the PD-L1 IC ≥1% cutoff for SP142, SP263, and 22C3 were 46.4% (285 of 614, 95% CI = 42.5% to 50.4%), 74.9% (460 of 614, 95% CI = 71.5% to 78.3%), and 73.1% (449 of 614, 95% CI = 69.6% to 76.6%), respectively. The prevalence of PD-L1 22C3 CPS ≥1 was 80.9% (497 of 614, 95% CI = 77.8% to 84.1%).
The correlation between SP142 IC continuous PD-L1 raw scoring values and those for SP263 IC, 22C3 IC, and 22C3 CPS, assessed with the r Spearman correlation index, was 0.69, 0.69, and 0.57, respectively (Supplementary Figure 3, available online). The OPA between SP142 IC ≥1% and SP263 IC ≥1%, 22C3 IC ≥1% and 22C3 CPS ≥1 were 69.2%, 68.7%, and 63.5%, respectively. Although the PPA between SP142 IC ≥1% and the other assays approached complete agreement (97.5%, 95.1%, and 97.9%, respectively), the NPAs were poor (44.7%, 45.9%, and 33.7%, respectively). These data indicate that almost all cases identified as SP142 PD-L1 IC ≥1% were included within the SP263 IC ≥1% and 22C3 CPS ≥1 populations, whereas many of the SP142 PD-L1 IC-negative (IC <1%) cases tested positive with the SP263 and 22C3 assays; 29.6% and 29.0% of cases deemed SP142 IC-negative were designated as positive with SP263 (IC ≥1%) and 22C3 (CPS ≥1) (Figure 1; Table 2). Collectively, the SP142 IC ≥1% subgroup was almost completely captured within the 22C3+ or SP263+ populations. Overall, these data suggest that the assays were not analytically equivalent at the assessed cutoffs.
Figure 1.
Analytical concordance between SP142, SP263, and 22C3 assays. Venn diagrams of the overlap between SP142 ≥1% and SP263 IC ≥1% (A), 22C3 ≥IC 1% (B), and 22C3 CPS ≥1 (C); all programmed death-ligand 1 cutoffs were defined as positive [+]. aGreater than 97% of SP142+ samples were included in 22C3+ and SP263+ samples. CI = confidence interval; NPA = negative percentage agreement; OPA = overall positive agreement; PPA = positive percentage agreement.
Table 2.
Analytical evaluation of concordance between SP142, SP263, and 22C3 assays
| Agreement | SP263 |
22C3 |
22C3 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| IC <1% | IC ≥1% | Total | IC <1% | IC ≥1% | Total | CPS <1 | CPS ≥1 | Total | |
| SP142 | |||||||||
| IC <1%, no. (%a) | 147 (23.9) | 182 (29.6) | 329 | 151 (24.6) | 178 (29.0) | 329 | 111 (18.1) | 218 (35.5) | 329 |
| IC ≥1%, no. (%a) | 7 (1.1) | 278 (45.3) | 285 | 14 (2.3) | 271 (44.1) | 285 | 6 (1.0) | 279 (45.4) | 285 |
| Total, no. | 154 | 460 | 614 | 165 | 449 | 614 | 117 | 497 | 614 |
| Positive percentage agreement | |||||||||
| no./N | 278/285 | 271/285 | 279/285 | ||||||
| % (95% CI) | 97.5 (95.7 to 99.3) | 95.1 (92.6 to 97.6) | 97.9 (96.2 to 99.6) | ||||||
| Negative percentage agreement | |||||||||
| no./N | 147/329 | 151/329 | 111/329 | ||||||
| % (95% CI) | 44.7 (39.3 to 50.1) | 45.9 (40.5 to 51.3) | 33.7 (28.6 to 38.8) | ||||||
| Overall positive agreement | |||||||||
| no./N | 425/614 | 422/614 | 390/614 | ||||||
| % (95% CI) | 69.2 (65.6 to 72.9) | 68.7 (65.1 to 72.4) | 63.5 (59.7 to 67.3) | ||||||
Calculated based on the total number of biomarker-evaluable patients. CI = confidence interval; CPS = combined positive score; IC = tumor-infiltrating immune cells.
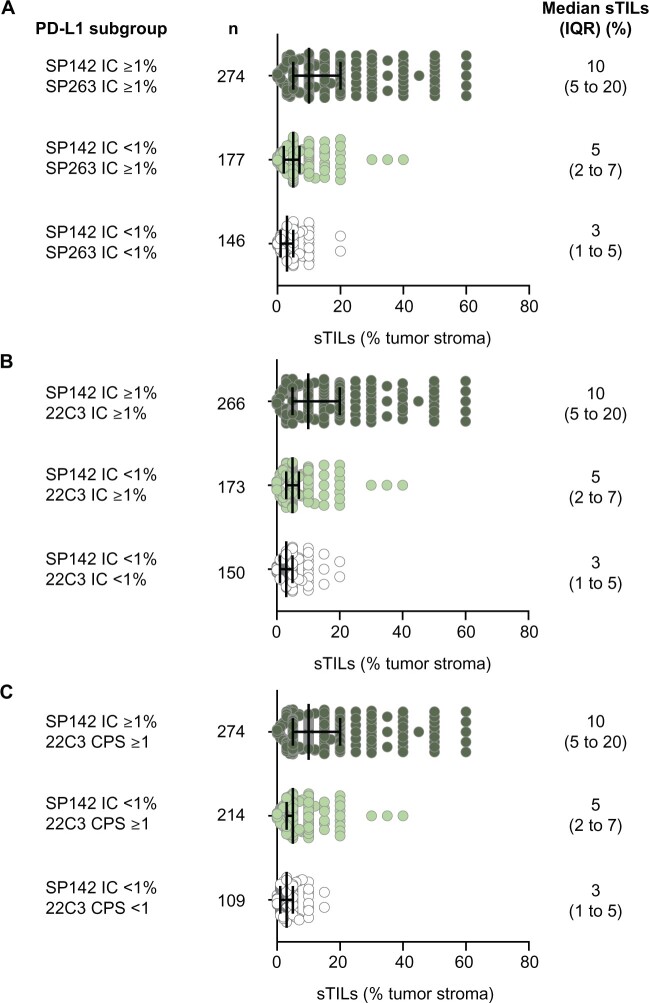
We also observed that the median sTIL count was higher in double-positive subgroups (10% each for SP142 IC ≥1% samples that were also SP263 IC ≥1%, 22C3 IC ≥1, or CPS ≥1) vs single-positive subgroups (5% each for SP142 IC <1% samples that were SP263 IC ≥1%, 22C3 IC ≥1, or CPS ≥1; P < .001 for all) or double-negative subgroups (3% each for samples that were SP142 IC <1% and SP263 IC <1%, 22C3 IC <1%, or CPS <1; P < .01 for all) (Figure 2).
Figure 2.
Stromal tumor-infiltrating lymphocytes (sTIL) in programmed death-ligand 1 (PD-L1) subgroups. Individual and median sTIL counts (as percentage of tumor stroma) by double or single selection of cutoffs for PD-L1 expression for each assay combination: SP142 tumor-infiltrating immune cells (IC) 1% and SP263 IC 1% (A), 22C3 IC 1% (B), and 22C3 combined positive score (CPS) 1 (C). For all panels, 2-sided P values per Kruskal-Wallis test with the Dunn’s test for multiple comparisons were P less than .001. IQR = interquartile range.
Clinical Activity Based on SP142, SP263, and 22C3 PD-L1 Assays
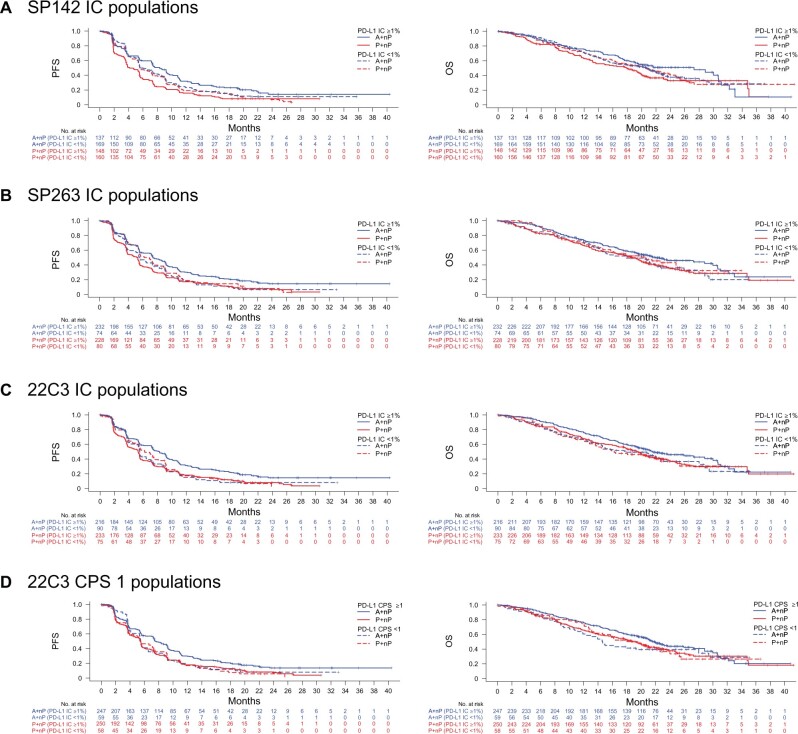
The clinical activity of A+nP and P+nP in the BEP and by PD-L1 SP142 IC subgroup was similar to results in the ITT population (Figure 3, A;Supplementary Figure 4, available online); however, the atezolizumab arm of the BEP slightly overperformed for PFS outcomes vs the ITT population.
Figure 3.
Clinical activity in assay-defined biomarker-evaluable populations (BEPs). Kaplan-Meier plots of progression-free survival (PFS) and overall survival (OS) in BEPs by programmed death-ligand 1 (PD-L1)–positive or –negative status for SP142 tumor-infiltrating immune cells (IC) 1% (A), SP263 IC 1% (B), 22C3 IC 1% (C), and 22C3 combined positive score (CPS) 1 (D). A = atezolizumab; nP = nab-paclitaxel; P = placebo.
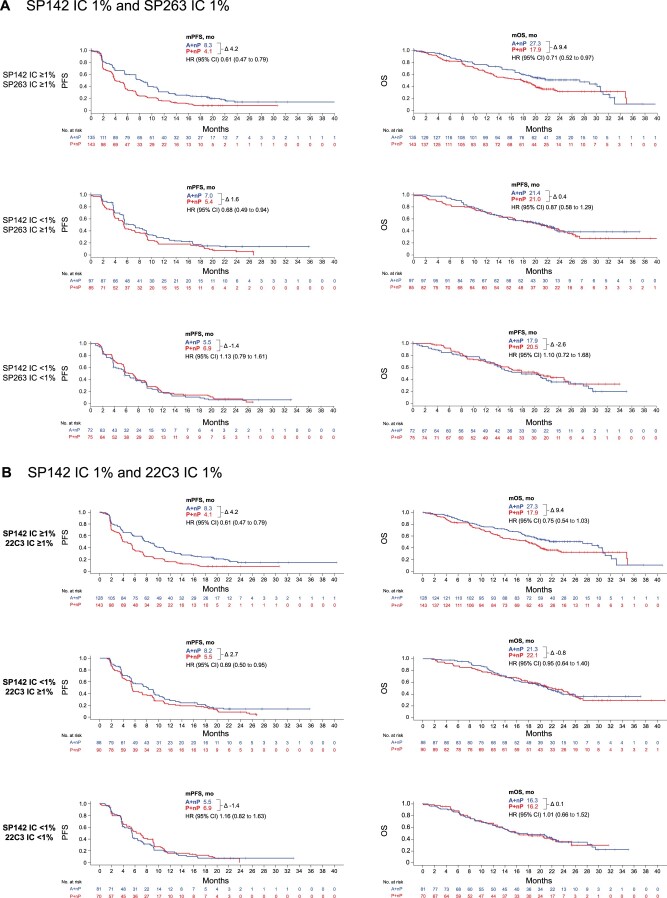
Despite prevalence differences between the assays, similar clinical activity was observed with A+nP vs P+nP in SP142 IC ≥1%, SP263 IC ≥1%, 22C3 IC ≥1%, and 22C3 CPS ≥1 patients (PFS HR = 0.60 to 0.68; OS HR = 0.74 to 0.79; Table 3), although median PFS and OS improvements were higher for SP142 IC ≥1% patients compared with patients selected by the other assays (difference in median values between treatment arms PFS = 4.2 vs 2.1 to 3.0 months; difference in median values of OS between treatment arms = 9.4 vs 2.2 to 3.3 months; Table 3). When analyzing clinical activity based on combinations of SP142 IC ≥1% and either SP263 IC ≥1%, 22C3 IC ≥1%, or 22C3 CPS ≥1, PFS and OS clinical activity with A+nP were highest in the double-positive subgroups (HR = 0.60 to 0.61 and 0.71 to 0.75, respectively), and PFS and OS improvements were modest in single-positive subgroups (HRs = 0.68 to 0.81 and 0.87 to 0.95, respectively; Figure 4; Supplementary Figure 5, available online). Little to no benefit (HRs ≥ 1.0) was observed in subgroups identified as PD-L1 negative by both SP142 and 22C3 or SP263. Of note, clinical activity favoring A+nP in the double-positive subgroups recapitulated activity of the SP142+ cases.
Table 3.
Clinical activity of A+nP with standard and alternative PD-L1 immunohistochemical assays, including model-derived cutoffs
| Population | No. (%) | PFS |
OS |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Median, mo |
HRa (95% CI) | Median, mo |
HRa (95% CI) | ||||||
| A+nP | P+nP | Δ | A+nP | P+nP | Δ | ||||
| BEP | 614 (100) | 7.4 | 5.4 | 2.0 | 0.72 (0.61 to 0.86) | 21.1 | 19.2 | 1.9 | 0.84 (0.68 to 1.03) |
| SP142 | |||||||||
| IC ≥1% | 285 (46.4) | 8.3 | 4.1 | 4.2 | 0.60 (0.47 to 0.78) | 27.3 | 17.9 | 9.4 | 0.74 (0.54 to 1.01) |
| IC <1% | 329 (53.6) | 5.7 | 5.6 | 0.1 | 0.86 (0.68 to 1.09) | 20.8 | 20.7 | 0.1 | 0.95 (0.72 to 1.27) |
| SP263 | |||||||||
| IC ≥1% | 460 (74.9) | 7.5 | 5.3 | 2.2 | 0.64 (0.53 to 0.79) | 22.0 | 18.7 | 3.3 | 0.75 (0.59 to 0.96) |
| IC <1% | 154 (25.1) | 5.5 | 6.9 | −1.4 | 1.08 (0.77 to 1.51) | 17.9 | 20.5 | −2.6 | 1.15 (0.76 to 1.74) |
| 22C3 | |||||||||
| IC ≥1% | 449 (73.1) | 8.3 | 5.3 | 3.0 | 0.64 (0.52 to 0.78) | 21.6 | 19.4 | 2.2 | 0.79 (0.62 to 1.01) |
| IC <1% | 165 (26.9) | 5.5 | 6.2 | −0.7 | 1.08 (0.78 to 1.50) | 17.8 | 16.2 | 1.6 | 0.97 (0.65 to 1.45) |
| 22C3 | |||||||||
| CPS ≥1 | 497 (80.9) | 7.5 | 5.4 | 2.1 | 0.68 (0.56 to 0.82) | 21.6 | 19.2 | 2.4 | 0.78 (0.62 to 0.99) |
| CPS <1 | 117 (19.1) | 5.5 | 5.5 | 0 | 1.00 (0.68 to 1.49) | 14.7 | 19.6 | −4.9 | 1.12 (0.70 to 1.77) |
| SP263 | |||||||||
| IC ≥4% | 286 (46.6) | 8.7 | 5.5 | 3.2 | 0.64 (0.49 to 0.83) | 28.9 | 19.6 | 9.3 | 0.71 (0.51 to 0.98) |
| IC <4% | 328 (53.4) | 5.6 | 5.4 | 0.2 | 0.82 (0.65 to 1.03) | 17.9 | 18.0 | −0.1 | 0.97 (0.73 to 1.28) |
| 22C3 | |||||||||
| CPS ≥10 | 325 (52.9) | 7.5 | 5.5 | 2.0 | 0.71 (0.56 to 0.91) | 22.0 | 18.7 | 3.3 | 0.77 (0.57 to 1.03) |
| CPS <10 | 289 (47.1) | 5.8 | 5.4 | 0.4 | 0.73 (0.57 to 0.93) | 20.2 | 19.4 | 0.8 | 0.94 (0.69 to 1.26) |
| 22C3 | |||||||||
| IC ≥4%b | 278 (45.3) | 8.2 | 5.4 | 2.8 | 0.64 (0.49 to 0.83) | 27.3 | 19.2 | 8.1 | 0.75 (0.55 to 1.04) |
| IC <4% | 336 (54.7) | 5.7 | 5.5 | 0.2 | 0.80 (0.64 to 1.01) | 19.6 | 18.0 | 1.5 | 0.92 (0.70 to 1.21) |
Hazard ratios (HRs) were adjusted for prior taxanes, presence of liver metastases, and ECOG PS. A = atezolizumab; BEP = biomarker-evaluable population; CI = confidence interval; CPS = combined positive score; ECOG PS = Eastern Cooperative Oncology Group performance status; Δ = difference; nP = nab-paclitaxel; OS = overall survival; P = placebo; PD-L1 = programmed death-ligand 1; PFS = progression-free survival.
Rounded from IC ≥3.5%.
Figure 4.
Clinical outcomes in biomarker-evaluable population (BEP) double-selected populations defined by different assay combinations. Kaplan-Meier plots of progression-free survival (PFS) and overall survival (OS) in BEP double-selected populations defined by SP142 tumor-infiltrating immune cells (IC) 1% and SP263 IC 1% (A) and 22C3 IC 1% (B) cutoffs. A = atezolizumab; CI = confidence interval; HR = hazard ratio; nP = nab-paclitaxel; P = placebo.
Analytical Harmonization to SP142 IC 1%
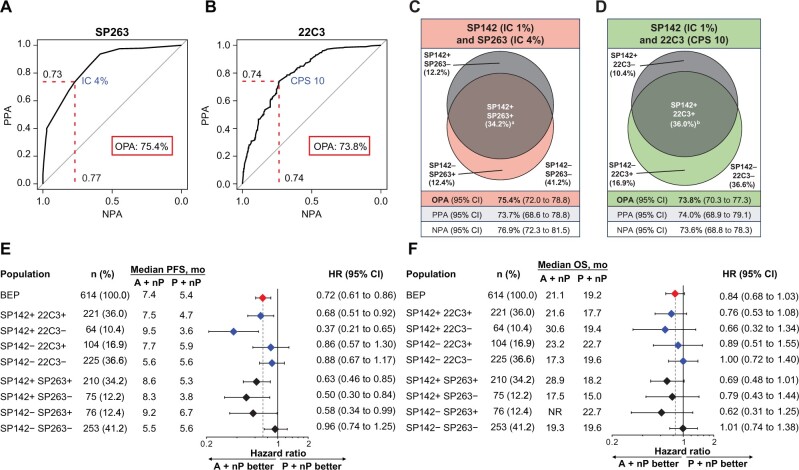
The greatest clinical activity favoring A+nP with the PD-L1 assays was derived from the nested SP142+ cases. To determine whether a cutoff could be identified for SP263 and 22C3 that replicated the patient populations with the SP142 IC ≥1% cutoff in IMpassion130 patients (“analytical harmonization” between SP142 and SP263 or 22C3), an ROC mathematical approach was applied to maximize OPA among assays using SP142 IC ≥1% as reference standard. The cutoffs with the highest combined OPA, NPA, and PPA between SP142 IC ≥1% and SP263 and 22C3 were IC ≥4% and CPS ≥10, respectively (Figure 5, A and B). Although the prevalence of PD-L1 SP263 IC ≥4% and 22C3 CPS ≥10 cases was 46.6% and 52.9%, respectively, concordance with SP142 IC ≥1% only slightly improved, with OPAs of 75.4% and 73.8%, respectively (Table 3; Figure 5, C and D). Moreover, SP263 IC ≥4% and 22C3 CPS ≥10 did not identify the same population as the SP142 assay at IC ≥1%, missing a proportion of SP142 IC ≥1% patients (75 of 285 [26.3%] and 64 of 285 [22.4%], respectively). Using the IC algorithm, the identified cutoff for 22C3 that replicated the SP142 IC ≥1% cutoff was IC ≥3.5%, rounded to IC ≥4% (Supplementary Figure 6, A, available online).
Figure 5.
Identification and clinical activity of model-derived optimized cutoffs for 22C3 combined positive score (CPS) and SP263 tumor-infiltrating immune cells (IC) that maximize analytical concordance with SP142 IC 1%. Receiver operating characteristic curve analysis (area under the curve) of optimal overall percentage agreement (OPA) based on negative percentage agreement (NPA) and positive percentage agreement (PPA) values for (A) SP263 IC and (B) 22C3 CPS using SP142 IC 1% as the reference standard. Venn diagrams of the overlap between SP142 IC ≥1% and exploratory model-derived optimized cutoffs of SP263 IC ≥4% (C) and 22C3 CPS ≥10 (D). Forest plots of (E) progression-free survival (PFS) and (F) overall survival (OS) in biomarker-evaluable population (BEP) subpopulations defined by SP142 IC 1% and exploratory nonstandard model-derived cutoffs of SP263 IC 4% or 22C3 CPS 10. a73.7% of SP142+ samples included in SP263+ samples. b77.5% of SP142+ samples included in 22C3+ samples. CI = confidence interval; HR = hazard ratio; NR = not reached.
Clinical activity of A+nP using the cutoffs for 22C3 CPS ≥10, SP263 IC ≥4%, and 22C3 IC ≥4% was inconsistent among assays defined by PD-L1 status (Figure 5, E and F; Table 3; Supplementary Figure 6, B and C, available online). Despite analytical discordance, A+nP PFS and OS clinical activity in SP263 IC ≥4% cases had similar hazard ratios as the SP142 IC ≥1% subgroup. Improvements in PFS and OS were lower for PD-L1 22C3 CPS ≥10 cases vs those identified with the SP142 IC ≥1% cutoff in this study.
Discussion
To our knowledge, IMpassion130 was the first phase III study of a checkpoint inhibitor to demonstrate clinical benefit for mTNBC, specifically in PD-L1 IC ≥1% patients identified using the FDA-approved VENTANA SP142 IHC assay (2). This retrospective exploratory analysis of the IMpassion130 BEP (n = 614) demonstrated that, at the IC ≥1% cutoff point (the only clinically validated cutoff for SP142), the SP263 and 22C3 PD-L1 IHC assays showed subpar analytical concordance (<90% OPA, NPA, and PPA) and were not considered analytically equivalent. Notably, analyses of clinical activity according to biomarker-defined subpopulations revealed that OS benefit favoring A+nP vs P+nP in the SP263 IC ≥1% and 22C3 IC ≥1% or CPS ≥1 subgroup generally appeared to be driven by the SP142 IC ≥1% population. It was encouraging that PFS benefit with A+nP vs P+nP was seen across assay-defined subgroups.
In this study, in attempts to harmonize the 3 assays, we aimed to determine the analytical cutoffs for 22C3 and SP263 that best captured patients at the SP142 IC 1% subgroup using ROC. However, concordance remained subpar between SP142 IC 1% and SP263 and 22C3 at the new cutoffs (SP263 IC ≥4% and 22C3 CPS ≥10 or IC ≥4%). Consistent patterns of PFS or OS benefit using the ROC-derived cutoffs were not seen in this data set. It should be noted that unlike the standard IC 1% and CPS 1 cutoffs, pathologist evaluation at the ROC-derived IC 4% or CPS 10 cutoff points was performed in an exploratory manner. Interestingly, SP263 IC ≥4% identified an additional population that might achieve PFS benefit; however, although this observation could potentially help expand the group of patients who could benefit, it should be noted that IC ≥4% excluded one-quarter (26.3%) of SP142 IC ≥1% patients who may have benefited from A+nP.
Collectively, results from the analytical concordance, clinical activity, and harmonization analyses suggest that the 3 PD-L1 IHC assays are not analytically interchangeable, consistent with previous findings in TNBC (18). Although a small study of 95 TNBC samples reported interchangeable performance between SP142 and SP263 at IC ≥1% (OPA of 91.2) (19), that study is not in line with ours or other analyses. The observed analytical and clinical divergence in this study may be attributed to different assay sensitivities as well as different immunostaining patterns among assays; these have not been explored here but warrant further study. Indeed, differences in expression patterns between SP263 and 22C3 were seen in a study of 136 invasive ductal carcinoma samples (20). The SP263 and 22C3 PD-L1 assays identified larger patient populations, within which almost all SP142+ cases were captured. Evidence has shown that sTILs mainly comprise CD4 and CD8 T cells (21), and in this study, sTIL counts were higher among SP142 IC ≥1% subgroups (relative to SP142 IC <1% subgroups), suggesting that the PD-L1 IC ≥1% population selected by SP142 staining may be more T-cell rich compared with SP142 IC <1% populations. We cannot rule out the possibility that SP142 is less sensitive than the other assays, which could explain the higher association of sTILs with SP142-positive cases compared with SP263 or 22C3. Notably, a separate IMpassion130 substudy showed clinical benefit with A+nP in patients who were both sTIL+ and PD-L1 IC ≥1% (6).
PD-L1 expression and immune biology vary by tumor type. For instance, in non-small cell lung cancer (NSCLC), both IC and TC biology seem to influence predictiveness of clinical benefit with immunotherapy (22), whereas in TNBC and other cancers such as urothelial carcinoma and small-cell lung cancer, IC biology may be more relevant (23,24). Despite demonstrating lower sensitivity for TC than IC (25,26), the SP142 assay is able to stain for PD-L1–expressing TC and/or IC across expression levels (27), including those in TNBC (6), and is currently used as a complementary diagnostic in NSCLC and a companion diagnostic in urothelial carcinoma and TNBC (28). Concordance and noncomparative PD-L1 expression studies with SP142, SP263, and 22C3 in these other disease areas, including NSCLC, have been conducted (26,29-35) and showed high similarity between antibodies in their ability to bind PD-L1 (36). However, similar to TNBC, in NSCLC the different PD-L1 assays did not appear analytically interchangeable, although measurements of concordance based on TC and/or IC were variable (25,30,31). Although not all NSCLC studies have evaluated how analytical differences manifest into clinical outcomes, there has been some indication that interassay biomarker predictiveness for survival outcomes with immunotherapy is more similar in NSCLC, in contrast to our findings in TNBC (30,31). Together, these observations indicate that PD-L1 assay findings across tumor types cannot be fully translated to TNBC.
This study has several limitations. Consistent with the exploratory nature of this substudy, these results should be considered hypothesis generating, with clinical activity in the BEP evaluated in small subgroups that were not predefined in the statistical analysis plan. Clinical outcomes in the BEP were not unequivocally comparable with those in the overall population. Further, it was not possible to validate the 22C3 IC 1% cutoff compared with the other assays and cutoffs because there are no formal training programs for this nonstandard evaluation, reducing the precision of this analysis. In 2020, the KEYNOTE-355 study showed that first-line pembrolizumab plus chemotherapy was beneficial in a mTNBC PD-L1+ population per 22C3 CPS ≥10, leading to FDA approval of the combination in the United States (1,4-7). CPS ≥10 represents the cutoff identified in our harmonization analysis, wherein concordance remained subpar between SP142 IC ≥1% and 22C3 CPS ≥10; thus, these data are of interest given the changes in TNBC testing landscape. Tumor tissue source differences between KEYNOTE-355 (all samples were metastatic) and IMpassion130 (both primary and metastatic samples), as well as lack of reader-validated precision on the CPS 10 cutoff (pathologists in this study were not trained by the vendor for the TNBC indication based on 22C3 using CPS), may account for interstudy variations in CPS ≥10 prevalence and outcomes. Regarding study validity, each immunostained slide was read by a single trained pathologist per scoring algorithm, as is commonplace in clinical practice; the VENTANA and Dako IHC assays have demonstrated high reader-reader precision and interlaboratory reproducibility (14-16). It should be noted that scoring inconsistency has been observed in the real-world setting (37); however, 7 of the 8 pathologists in this study received formal vendor training, and all had high performance standards.
Overall, these findings represent a robust data set derived from a large, randomized, phase III study as opposed to studies that have relied on arbitrarily selected specimens from tumor banks (25). To our knowledge, this is the first PD-L1 assay comparison of both concordance and associated clinical survival outcomes in patients with mTNBC. Further prospective studies to understand the biological explanation and clinical relevance of inter-assay differences are warranted.
Funding
This work was supported by F. Hoffmann-La Roche, Ltd/Genentech, Inc, a member of the Roche Group, which also funded medical writing support.
Notes
Role of the funder: F. Hoffmann-La Roche/Genentech provided atezolizumab and the matched placebo and collaborated with academic authors regarding the trial design, data collection, analysis and interpretation. Celgene provided nab-paclitaxel but had no role in trial design, data collection or analysis, although the company did review the manuscript.
Disclosures: All authors received grants and non-financial support from F. Hoffmann-La Roche during the conduct of the study. Editorial support, funded by the sponsor, was provided by an independent medical writer under the guidance of the authors. Hope S. Rugo reports research support for clinical trials through the University of California from AstraZeneca, Boehringer Ingelheim, Daiichi Sankyo, Immunomedics, Lilly, Macrogenics, Merck, Novartis, Odonate, Pfizer, Polyphor, Roche, Seattle Genetics and Sermonix; and honoraria from Mylan, Puma and Samsung. Sherene Loi reports research funding to her institution from Novartis, Bristol Myers Squibb, Merck, Roche, Genentech, Puma, Pfizer and Eli Lilly; has acted as an unpaid consultant to Seattle Genetics, Pfizer, Novartis, Bristol Myers Squibb, Merck, AstraZeneca and Roche/Genentech; and acted as consultant to Aduro Biotech (fees paid to her institution). Sylvia Adams reports uncompensated consulting or advisory roles with Bristol-Meyers Squibb, Genentech and Merck and reports research funding to her institution from Amgen, Bristol-Meyers Squibb, Celgene, Genentech, Merck and Novartis. Peter Schmid reports research support (grants) to his institution from Genentech, F. Hoffmann-La Roche, OncoGenex and Novartis; reports honoraria from AstraZeneca, F. Hoffmann-La Roche, Medscape and G1 Therapeutics; reports a consulting or advisory role with AstraZeneca, Novartis, F. Hoffmann-La Roche, Merck, Boehringer Ingelheim, Bayer, Eisai, Celgene, Pfizer and Puma; is an uncompensated steering committee member for the IMpassion130 trial; and reports that his spouse has a consulting role for Genentech and F. Hoffmann-La Roche. Andreas Schneeweiss reports research grants from Celgene, F. Hoffmann-La Roche, AbbVie and Molecular Partners; consulting fees and travel expenses from F. Hoffmann-La Roche and AstraZeneca; honoraria from F. Hoffmann-La Roche, Celgene, AstraZeneca, Novartis, Merck Sharp and Dohme, Tesaro and Eli Lilly; and honoraria and travel expenses from Pfizer. Carlos H. Barrios reports research grants from Pfizer, Novartis, Amgen, AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Roche/Genentech, Eli Lilly, Sanofi, Taiho Pharmaceutical, Mylan, Merrimack, Merck, AbbVie, Astellas, BioMarin, Bristol Myers Squibb, Daiichi Sankyo, Abraxis BioScience, AB Science, Asana BioSciences, Medivation, Exelixis, ImClone Systems, LEO Pharma and Millennium; and consulting fees from Roche/Genentech, Boehringer Ingelheim, GlaxoSmithKline, Novartis, Pfizer, Eisai, Bayer, Merck Sharp and Dohme doi:10.1093/jnci/djz218.and AstraZeneca. Hiroji Iwata reports honoraria, consulting fees and research support for the IMpassion130 study; editorial support from F. Hoffmann-La Roche and Chugai; serves as an uncompensated member of the steering committee for the IMpassion130 trial; and reports honoraria and consulting fees from Novartis, AstraZeneca, Pfizer, Eli Lilly and Daiichi Sankyo. Véronique Diéras reports honoraria for serving on advisory boards for F. Hoffmann-La Roche, Genentech, Pfizer, Eli Lilly, Novartis, Daiichi Sankyo, AstraZeneca, MSD, AbbVie Seattle Genetics and Odonate. Eric P. Winer reports honoraria from Eli Lilly, Leap, Genentech, Infinite MD, Carrick Therapeutics, GlaxoSmithKline, Jounce, Genomic Health, Merck and Seattle Genetics and is a scientific advisory board member for Leap. Mark M. Kockx and Dieter Peeters report employment at HistoGeneX. Stephen Y. Chui reports employment at and stock from Genentech/Roche during the conduct of the study and has a use patent to disclose with Roche/Genentech. Jennifer C. Lin and Anh Nguyen Duc report employment at and stock from Genentech/Roche during the conduct of the study. Giuseppe Viale reports honoraria for speaker engagements and consultancy and advisory roles from Roche/Genentech, Ventana, MSD Oncology, Bayer, Novartis, Menarini, Dako/Agilent, Daiichi Sankyo, AstraZeneca and Eli Lilly; support for contracted research at his institution from Roche/Genentech, Ventana, Cepheid and Dompé. Luciana Molinero is an employee of Roche/Genentech; reports stock ownership in Roche and has a use patent to disclose with Roche/Genentech. Leisha A. Emens is co-chair of the steering committee for the IMpassion130 study and chair of the KATE2 study steering committee; reports honoraria from AbbVie, Amgen, Celgene, Chugai, Gritstone, MedImmune, Peregrine, Shionogi and Syndax; honoraria and travel support from AstraZeneca, Bayer, MacroGenics, Replimune and Vaccinex; travel support from Bristol Myers Squibb, Genentech/Roche and Novartis; has potential future stock from MolecuVax; reports institutional support from Aduro Biotech, AstraZeneca, the Breast Cancer Research Foundation, Bristol Myers Squibb, Bolt Therapeutics, Corvus, the US Department of Defense, EMD Serono, Genentech, MaxCyte, Merck, the National Cancer Institute, the National Surgical Adjuvant Breast and Bowel Project Foundation, Roche, Silverback, the Translational Breast Cancer Research Consortium, Tempest and HeritX; and reports royalties from Aduro.
Author contributions: Hope S. Rugo: Conceptualization, Investigation, Resources, Writing—Original Draft, Writing—Review and Editing. Sherene Loi: Conceptualization, Methodology, Formal analysis, Investigation, Resources, Writing—Original Draft, Writing—Review and Editing, Supervision. Sylvia Adams: Conceptualization, Investigation, Resources, Writing—Review and Editing, Project administration. Peter Schmid: Conceptualization, Methodology, Writing—Review and Editing, Supervision. Andreas Schneeweiss: Conceptualization, Methodology, Investigation, Resources, Writing—Review and Editing, Visualization. Carlos H. Barrios: Conceptualization, Methodology, Formal analysis, Investigation, Writing—Original Draft, Writing—Review and Editing, Visualization, Supervision. Hiroji Iwata: Investigation, Resources, Writing—Review and Editing. Véronique Diéras: Conceptualization, Resources, Writing—Review and Editing. Eric P. Winer: Writing—Review and Editing. Mark M. Kockx: Conceptualization, Methodology, Investigation, Resources, Writing—Review and Editing, Visualization, Supervision. Dieter Peeters: Validation, Investigation, Writing—Review and Editing. Stephen Y. Chui: Conceptualization, Methodology, Investigation, Writing—Original Draft, Writing—Review and Editing, Supervision, Funding acquisition. Jennifer C. Lin: Conceptualization, Methodology, Writing—Review and Editing; Anh Nguyen Duc: Validation, Formal analysis, Writing—Review and Editing. Giuseppe Viale: Conceptualization, Writing—Review and Editing, Visualization. Luciana Molinero: Conceptualization, Validation, Formal analysis, Investigation, Resources, Data Curation, Writing—Review and Editing, Visualization, Supervision, Project administration, Funding acquisition. Leisha A. Emens: Conceptualization, Investigation, Resources, Writing—Review and Editing, Supervision.
Prior presentations: Data from the analysis were presented in part at ESMO 2019 and SABCS 2019.
Acknowledgements: This study was sponsored by F. Hoffmann-La Roche, Ltd/Genentech, Inc, a member of the Roche Group. We thank the patients and their families. Medical writing assistance for this manuscript was provided by Anusha Bolonna, PhD, and Ashley Pratt, PhD, of Health Interactions and funded by F. Hoffmann-La Roche, Ltd.
Data Availability
Qualified researchers may request access to individual patient-level data through the clinical study data request platform (https://vivli.org/). Further details on Roche's criteria for eligible studies are available here (https://vivli.org/members/ourmembers/). For further details on Roche's Global Policy on the Sharing of Clinical Information and how to request access to related clinical study documents, see here (https://www.roche.com/research_and_development/who_we_are_how_we_work/clinical_trials/our_commitment_to_data_sharing.htm).
Supplementary Material
†Formerly Caprion HistoGeneX.
References
- 1. Marra A, Viale G, Curigliano G.. Recent advances in triple negative breast cancer: the immunotherapy era. BMC Med. 2019;17(1):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schmid P, Adams S, Rugo HS, et al. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med. 2018;379(22):2108–2121. [DOI] [PubMed] [Google Scholar]
- 3. Schmid P, Cortes J, Pusztai L, et al. Pembrolizumab for early triple-negative breast cancer. N Engl J Med. 2020;382(9):810–821. [DOI] [PubMed] [Google Scholar]
- 4. Emens LA, Cruz C, Eder JP, et al. Long-term clinical outcomes and biomarker analyses of atezolizumab therapy for patients with metastatic triple-negative breast cancer: a phase 1 study. JAMA Oncol. 2019;5(1):74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cortés J, Lipatov O, Im S, et al. KEYNOTE-119: phase 3 study of pembrolizumab (pembro) versus single-agent chemotherapy (chemo) for metastatic triple-negative breast cancer (mTNBC). Ann Oncol. 2019;30:83. [Google Scholar]
- 6. Emens LA, Molinero L, Loi S, et al. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer: biomarker evaluation of the IMpassion130 study [published online ahead of print]. J Natl Cancer Inst. 2021;113(8):1005–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cortes J, Cescon DW, Rugo HS, et al. KEYNOTE-355: randomized, double-blind, phase III study of pembrolizumab + chemotherapy versus placebo + chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer. J Clin Oncol. 2020;38:1. [DOI] [PubMed] [Google Scholar]
- 8.TECENTRIQ. (Atezolizumab) [summary of product characteristics]. Welwyn Garden City, UK: Roche Registration Limited; 2019. [Google Scholar]
- 9.TECENTRIQ. (Atezolizumab) [package insert]. South San Francisco, CA: Genentech, Inc.; 2019. [Google Scholar]
- 10.National Comprehensive Cancer Network. NCCN clinical practice guidelines in oncology. Breast cancer. V3. 2020. https://www.nccn.org/professionals/physician_gls/pdf/breast.pdf. Accessed July 23, 2020.
- 11.Arbeitsgemeinschaft Gynäkologische Onkologie A. Guidelines. Breast. V2020.1. https://www.ago-online.de/en/leitlinien-empfehlungen/leitlinien-empfehlungen/kommission-mamma. Accessed April 1, 2020.
- 12. Heimes AS, Schmidt M.. Atezolizumab for the treatment of triple-negative breast cancer. Expert Opin Investig Drugs. 2019;28(1):1–5. [DOI] [PubMed] [Google Scholar]
- 13. Schmid P, Rugo HS, Adams S, et al. Atezolizumab plus nab-paclitaxel as first-line treatment for unresectable, locally advanced or metastatic triple-negative breast cancer (IMpassion130): updated efficacy results from a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2020;21(1):44–59. [DOI] [PubMed] [Google Scholar]
- 14.VENTANA. PD-L1 (SP142) assay (CE-IVD) [package insert]. Tucson, AZ: Ventana Medical Systems, Inc; 2019. [Google Scholar]
- 15.VENTANA. PD-L1 (SP263) assay (CE-IVD) [package insert]. Tucson, AZ: Ventana Medical Systems, Inc; 2018. [Google Scholar]
- 16.DAKO. PD-L1 IHC 22C3 pharmDx assay [instructions for use]. Carpinteria, CA: Dako North America, Inc; 2018. [Google Scholar]
- 17. Salgado R, Denkert C, Demaria S, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an international TILs working group 2014. Ann Oncol. 2015;26(2):259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scott M, Scorer P, Barker C, et al. Comparison of patient populations identified by different PD-L1 assays in in triple-negative breast cancer (TNBC). Ann Oncol. 2019;30:iii4. [Google Scholar]
- 19. Lee SE, Park HY, Lim SD, et al. Concordance of programmed death-ligand 1 expression between SP142 and 22C3/SP263 assays in triple-negative breast cancer. J Breast Cancer. 2020;23(3):303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Karnik T, Kimler BF, Fan F, Tawfik O.. PD-L1 in breast cancer: comparative analysis of 3 different antibodies. Hum Pathol. 2018;72:28–34. [DOI] [PubMed] [Google Scholar]
- 21. Savas P, Virassamy B, Ye C, et al. ; Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer (kConFab). Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med. 2018;24(7):986–993. [DOI] [PubMed] [Google Scholar]
- 22. Kowanetz M, Zou W, Gettinger SN, et al. Differential regulation of PD-L1 expression by immune and tumor cells in NSCLC and the response to treatment with atezolizumab (anti-PD-L1). Proc Natl Acad Sci USA. 2018;115(43):E10119–E10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Petrylak DP, Powles T, Bellmunt J, et al. A phase Ia study of MPDL3280A (anti-PDL1): updated response and survival data in urothelial bladder cancer (UBC). J Clin Oncol. 2015;33(suppl 15):4501. [Google Scholar]
- 24. Reck M, Liu SV, Mansfield AS, et al. IMpower133: updated overall survival (OS) analysis of first-line (1L) atezolizumab (atezo) + carboplatin + etoposide in extensive-stage SCLC (ES-SCLC). Ann Oncol. 2019;30(suppl 5):v710–v717. [Google Scholar]
- 25. Hirsch FR, McElhinny A, Stanforth D, et al. PD-L1 immunohistochemistry assays for lung cancer: results from phase 1 of the blueprint PD-L1 IHC assay comparison project. J Thorac Oncol. 2017;12(2):208–222. [DOI] [PubMed] [Google Scholar]
- 26. Tsao MS, Kerr KM, Kockx M, et al. PD-L1 immunohistochemistry comparability study in real-life clinical samples: results of blueprint phase 2 project. J Thorac Oncol. 2018;13(9):1302–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vennapusa B, Baker B, Kowanetz M, et al. Development of a PD-L1 complementary diagnostic immunohistochemistry assay (SP142) for atezolizumab. Appl Immunohistochem Mol Morphol. 2019;27(2):92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roche. VENTANA PD-L1 (SP142) Assay (CE IVD). https://diagnostics.roche.com/global/en/products/tests/ventana-pd-l1-_sp142-assay2.html#productSpecs. Accessed April 1, 2020.
- 29. Xu H, Lin G, Huang C, et al. Assessment of concordance between 22C3 and SP142 immunohistochemistry assays regarding PD-L1 expression in non-small cell lung cancer. Sci Rep. 2017;7(1):16956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Herbst RS, Giaccone G, de Marinis F, et al. Atezolizumab for first-line treatment of PD-L1-selected patients with NSCLC. N Engl J Med. 2020;383(14):1328–1339. [DOI] [PubMed] [Google Scholar]
- 31. Gadgeel S, Kowanetz M, Zou W, et al. Clinical efficacy of atezolizumab in PD-L1 selected subgroups defined by SP142 and 22C3 IHC assays in 2L+ NSCLC: results from the randomized OAK trial. Presented at European Society of Medical Oncology Congress; September 8-12, 2017; Madrid, Spain.
- 32. O’Malley DP, Yang Y, Boisot S, et al. Immunohistochemical detection of PD-L1 among diverse human neoplasms in a reference laboratory: observations based upon 62,896 cases. Mod Pathol. 2019;32(7):929–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rimm DL, Han G, Taube JM, et al. Reanalysis of the NCCN PD-L1 companion diagnostic assay study for lung cancer in the context of PD-L1 expression findings in triple-negative breast cancer. Breast Cancer Res. 2019;21(1):72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Torlakovic E, Lim HJ, Adam J, et al. “Interchangeability” of PD-L1 immunohistochemistry assays: a meta-analysis of diagnostic accuracy. Mod Pathol. 2020;33(1):4–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hendry S, Byrne DJ, Wright GM, et al. Comparison of four PD-L1 immunohistochemical assays in lung cancer. J Thorac Oncol. 2018;13(3):367–376. [DOI] [PubMed] [Google Scholar]
- 36. Gaule P, Smithy JW, Toki M, et al. A quantitative comparison of antibodies to programmed cell death 1 ligand 1. JAMA Oncol. 2017;3(2):256–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reisenbichler ES, Han G, Bellizzi A, et al. Prospective multi-institutional evaluation of pathologist assessment of PD-L1 assays for patient selection in triple negative breast cancer. Mod Pathol. 2020;33(9):1746–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Qualified researchers may request access to individual patient-level data through the clinical study data request platform (https://vivli.org/). Further details on Roche's criteria for eligible studies are available here (https://vivli.org/members/ourmembers/). For further details on Roche's Global Policy on the Sharing of Clinical Information and how to request access to related clinical study documents, see here (https://www.roche.com/research_and_development/who_we_are_how_we_work/clinical_trials/our_commitment_to_data_sharing.htm).