ABSTRACT
Background
The human milk microbiome may contribute to the benefits of breastfeeding by providing bacteria to the infant gastrointestinal tract. Many women pump their milk, but the effect of pumping on the milk microbiome is unknown.
Objectives
Our objective was to determine the effects of pumping supplies on the pumped human milk microbiome.
Methods
This was an in-home, randomized, crossover trial of 2 collection methods. Women (n = 52) pumped twice within 3.5 h, once with their own breast pumps and milk collection supplies (OWN SUPP) and once with a hospital-grade pump and sterile collection supplies (STER SUPP). Pumping order was randomized. The milk microbiome was characterized by aerobic culturing and 16S ribosomal RNA gene sequencing.
Results
Milk collected with OWN SUPP yielded more total aerobic and gram-negative bacteria than milk collected with STER SUPP, reflecting a 6.6 (adjusted OR; 95% CI: 1.7, 25; P = 0.006) higher odds of containing >104 total aerobic CFU/mL and 19 (adjusted OR; 95% CI: 4.1, 88; P < 0.0001) higher odds of yielding culturable gram-negative bacteria. Milk collected with OWN SUPP yielded more Proteobacterias , including higher relative abundances of Acinetobacter and Stenotrophomonas, compared to milk collected with STER SUPP. Results were consistent across pumping-order groups.
Conclusions
We demonstrated that pumping supplies altered the milk microbiome. On average, milk collected with OWN SUPP resulted in elevated levels of culturable total and gram-negative bacteria and proteobacterial DNA compared to milk collected with STER SUPP. More research is needed to assess implications for infant health.
Keywords: breastmilk expression/methods, breastmilk collection, gram-negative bacteria, microbiota, bacterial counts, crossover study, Proteobacteria, aerobic bacteria
Introduction
Given the many health benefits associated with breastfeeding (1, 2), human milk is recommended as the only source of nutrition for the first 6 mo and is important in infants’ diets for at least the first year (3). Feeding pumped milk is commonly used as a sole or ancillary strategy to meet breastfeeding recommendations and goals (4–7). In the United States, ∼85% of breastfeeding women pump their milk; 25% do so regularly (4). Evidence-based guidelines for pumping are essential to ensure pumped milk is safe to consume (8). Even so, existing evidence about how pumping and storage practices influence the bacterial composition of pumped milk is largely restricted to nonrandomized and observational studies, many of which were conducted in clinical settings (9–13). Thus, adequate evidence about the effects of at-home pumping practices on the bacterial composition of pumped milk is unavailable for such guidelines.
Although long considered sterile, human milk is now known to harbor a relatively unique microbiome (14–19). The human milk microbiome may contribute to the benefits of breastfeeding by providing bacteria to the infant gastrointestinal (GI) tract (14, 20–22). Human milk–derived bacteria along with other milk components, such as human milk oligosaccharides, support the growth of beneficial bacteria such as Bifidobacterium spp., which support the development of the infant immune system (22). Much research has focused on identifying biological, behavioral, and environmental factors associated with variation in the human milk microbiome. Biological correlates of variation include milk composition (e.g., the profiles of fatty acids and human milk oligosaccharides), gestational age, and maternal race/ethnicity (19, 23). Behavioral and environmental correlates of variation include breastfeeding exclusivity, geography, and maternal diet (15, 19, 24). Little is known, however, about how pumping practices influence the microbiome of pumped human milk.
Previous studies have demonstrated that women's pumping and milk storage practices vary greatly (5–7, 25). Most women in developed countries who express their milk use an electric breast pump (4). During pumping, milk passes from the nipple down a plastic flange, through a valve, and is collected in a bottle. If not cleaned thoroughly, these surfaces may harbor bacteria that may be transferred to milk during pumping (26, 27). Yet how these surfaces are cleaned at home varies widely (7, 25). For example, some women only wipe off milk collection containers and flanges after pumping, whereas others sterilize them (7, 25). To our knowledge, the effects of pumping supplies on the microbiome of pumped human milk have not been rigorously evaluated. To address this research gap, we conducted an in-home, randomized, crossover trial to determine the effect of pumping with a woman's own pump and collection supplies on the microbiome of pumped human milk compared with pumping with a hospital-grade pump and sterile supplies.
Methods
Study design
The Milk in Life Conditions (MiLC) trial was an in-home randomized crossover trial of 2 milk collection methods among women who pump. Participants provided written informed consent according to the study protocol approved by the Cornell University Institutional Review Board (1608006566). The study was registered at clinicaltrials.gov as NCT03123874.
Population
Women were eligible to participate if they were self-described as healthy, lactating, and ≥18 y old; lived within a 45-min drive of Cornell University in Ithaca, New York; used an electric breast pump; and were confident they could donate 30 mL of milk from 1 breast twice within 3.5 h. Women were excluded if their infants consumed formula in the past 2 wk or they reported any signs or symptoms of current breast infection (e.g., breast pain, discomfort, lumps, mastitis with fever, red streaks or hard red portions on their breast) or acute illness (mother or her infant) in the previous 7 d (e.g., rectal or temporal temperature ≥37.5°C, dark green nasal discharge, diarrhea defined as abrupt onset of ≥3 excessively loose stools in 1 d, infant vomiting not associated with feeding, or severe cough). Eligibility was assessed with an online survey when women initially inquired about study participation.
Intervention
All participants expressed their milk once with their own electric breast pumps and milk collection supplies (OWN SUPP) and once with a hospital-grade pump (Medela Symphony; Medela, Inc.) and previously unopened (new), sterile collection kits (STER SUPP). All pumping sessions occurred in participants’ homes within 3.5 h between 07:00 and 11:00 h.
Randomization
Following eligibility screening, participants were randomly assigned to pump with OWN SUPP or STER SUPP first. To account for the possibility that the infant oral microbiome, via retrograde inoculation, influences the milk microbiome (28, 29), the randomization was stratified according to infant diet [human milk only (HM only) or HM and complementary foods (HM + CF)] with an approximately equal allocation ratio. The randomization schedule was produced with the sample function in R (v. 3.4.4) (30). Only women who participated in the study received assignment; allocation (done by the study director, SMR) occurred immediately before participation began.
One woman did not pump in the order of her allocated randomized assignment. She did, however, provide a milk sample from both collection methods and was included in our intention-to-treat analysis (31).
Outcomes
The a priori primary outcomes were total aerobic bacterial counts (CFU/mL, detected via aerobic culturing), 16S-based bacterial richness [the number of different bacterial taxa estimated via amplicon sequence variants (ASVs)] (32), and α diversity (Shannon diversity index) (33).
We also report counts for gram-negative bacteria (CFU/mL) and, for 16S data, the relative abundances of bacterial taxa, Faith's phylogenetic diversity index (34), and β diversity using weighted UniFrac distances (35).
Milk collection
Milk collection occurred in participants’ homes. Milk from the first pumping session was transported back to the laboratory by the study director. Then, the study director returned ∼3 h later for the second pumping session.
Participants fully expressed milk from the same breast during both pumping sessions. Women were instructed to pump until the flow of milk had stopped. Women's breasts were not cleaned prior to pumping. Women were instructed not to nurse from the study breast for the duration of the study period (2 h before the first pumping session until after the second session) but were able to feed their infants on the other breast during this time.
The study director was present during both pumping sessions for all participants to ensure that the study protocol was followed. For the collection with OWN SUPP, and consistent that milk collection was conducted under “real-life” conditions, women were encouraged to follow their usual practices. For the collection with STER SUPP, the study director wore nitrile gloves treated with hand sanitizer after being donned. Using aseptic technique, the sterile collection kit was passed to the participant, who was instructed not to touch the inner surface of the flange with her hands and immediately proceeded to express her milk. From each full expression, ∼30 mL milk was reserved for analyses; mothers kept any additional pumped milk. Milk samples were aliquoted into 5 sterile containers by the study director using a single-use, sterile transfer pipette. One container was taken to the laboratory on ice immediately after expression. The other 4 containers were stored at the participants’ homes for various durations and storage temperatures. Here, we report only the results obtained from the first container. For practical reasons, the trial was not blinded.
Bacterial analyses
Culture-dependent analyses and storage of the remaining milk at −20°C for later 16S sequencing were completed within 2 h of sampling. After all samples were collected and between 2 wk and 3 mo after expression, milk samples were shipped on dry ice to the University of Idaho for 16S ribosomal RNA (rRNA) gene sequencing.
Aerobic culturing
Milk was cultured on plate count agar and MacConkey agar (both from Becton Dickinson). Serial 10-fold dilutions were prepared using 0.1% sterile peptone water (Hardy Diagnostics) and inoculated in triplicate with 10-μL drops using the drop plate method (36). For each media type, 100 μL undiluted milk was also spread-plated on 100-mm plates for each of the above media. Plates were aerobically incubated for 48 h at 37°C. CFU/mL were calculated from plates with ≥1 colony, as typical for the drop-plate method (36). The sample was considered culture negative if no colony was detected on both drop and spread plates.
16S rRNA gene sequencing
DNA extraction, amplification, and sequencing of milk samples were conducted as previously described (15, 24). Briefly, 1.25 mL milk was thawed for 5 min at 37°C, then centrifuged at 13,000 × g for 10 min at 4°C. The lipid layer and supernatant were discarded, and then the remaining cell pellet was resuspended in 500 μL TE50 buffer, followed by enzymatic lysis for 1 h at 37°C. Total DNA was then extracted from the resuspended pellet using QIAamp DNA Mini Kits (Qiagen) following the manufacturer's instructions, with minor modifications as described previously (15). The V1–V3 hypervariable region of the bacterial 16S rRNA gene was amplified using a dual-barcoded, 2-step, 30-cycle PCR protocol with subsequent quality control as described previously (15). For the first step, we used a 7-fold degenerate forward primer targeting position 27 and a reverse primer targeting position 534 (positions numbered according to the Escherichia coli rRNA) (24). Nuclease-free, sterile water (2 µL), and E. coli DNA (2 µL; 221 ng/mL) served as controls.
Amplicon quality was assessed using the QIAxcel DNA screening cartridge (Qiagen), with quantity assessed using a Qubit 2.0 fluorometer and the DS High Sensitivity Assay (Thermo Fisher Scientific) as previously described (15). Amplicons (50 ng) from each sample were pooled and sent to the Institute for Bioinformatics and Evolutionary Studies Genomics Resources Core at the University of Idaho for additional processing and sequencing using previously described methods (15). Also sequenced was a mock community consisting of DNA extracted from 10 known species and their theoretical relative abundances (Zymo Research). Amplicons were sequenced using an Illumina MiSeq v3 paired-end 300-bp protocol for 600 cycles.
Processing of bacterial sequences
Sequences were demultiplexed using dbcAmplicons (37), splitting them into separate files for forward and reverse sequences using splitReadsBySample.py (38) as previously described (15). Overlapping paired-end sequences underwent sorting, filtering, trimming, and merging using the DADA2 (39) plug-in in Quantitative Insights Into Microbial Ecology 2 (v. 2018.2) (40). The resulting ASVs were taxonomically classified using the SILVA 16S rRNA reference database (v. 128). The decontam package (41) allowed for the identification of universal contaminants, with the corresponding ASV subsequently removed. Additional filtering removed nonbacterial (mitochondria and chloroplasts) and ambiguous sequences (“unassigned” taxonomy at the domain level). Finally, because features present in only 1 sample are not statistically informative, removing them increased our power to declare differences significant (42). After filtering, a total of 2,229,832 sequences were retained (median = 20,525 per sample; range: 1041–60,479), representing 997 ASVs (Supplemental Figure 1). Data were rarefied to 1041 sequences per sample for diversity metrics only.
Sample size justification
Insufficient information was available to estimate a clinically meaningful difference in the milk microbiome. Rather, we aimed to enroll at least 50 women with equal numbers in each infant diet group based on sample sizes of earlier studies (15, 16, 24).
Statistical analyses
We conducted an intention-to-treat analysis (31) between April 1, 2018, and September 22, 2020, using R (v. 3.4.4 and 3.5.3) (30). The effect of pump supplies on aerobic bacterial counts was tested by linear mixed-effects regression. A log(x + 1) transformation was applied to bacterial counts to account for data skew and zeros in the data (43). The sample variance was estimated using the Satterthwaite approximation to account for unequal variance (44). Models included infant diet and randomly assigned pumping order as fixed effects, participant as a random effect, and all possible 2- and 3-way interaction terms among the type of pumping supplies, infant diet group, and randomly assigned pumping order. The effect of pump supplies on dichotomous outcomes (>104 CFU/mL total aerobic bacteria or presence of viable gram-negative bacteria) was tested by logistic mixed-effects regression with the same adjustments, excluding interactions. Linear mixed-effects regression with the same adjustments was also used to test the effect of pump supplies on α diversity metrics. A log transformation was applied to Faith's phylogenetic diversity index and the observed ASV to account for data skew. The effect of pump supplies on β diversity was tested by permutational ANOVA (PERMANOVA) with 999 permutations on weighted UniFrac distances adjusted for infant diet and randomly assigned pumping order. Differentially abundant phyla and genera were identified using DESeq2 (v. 1.22.2), which uses a negative binomial distribution (45). Spearman correlations were calculated between bacterial counts and relative abundances of bacterial phyla. Results are reported as least squares (LS) means and standard errors, adjusted odds ratios (aORs) with 95% CI, or observed means and standard deviations as indicated. Results were considered significant at P < 0.05, except for DESeq2 results, which were considered significant at P < 0.05 after being adjusted with the false discovery method for multiple comparisons (46).
In post hoc analyses, the association between whether milk yielded viable gram-negative bacteria and relative abundances of Proteobacteria a16S sequences was tested by PERMANOVA with 999 permutations on Euclidean distances. A volcano plot was used to visualize the fold change of all genera of Proteobacteria that differed significantly between samples that yielded viable gram-negative bacteria and those that did not.
Results
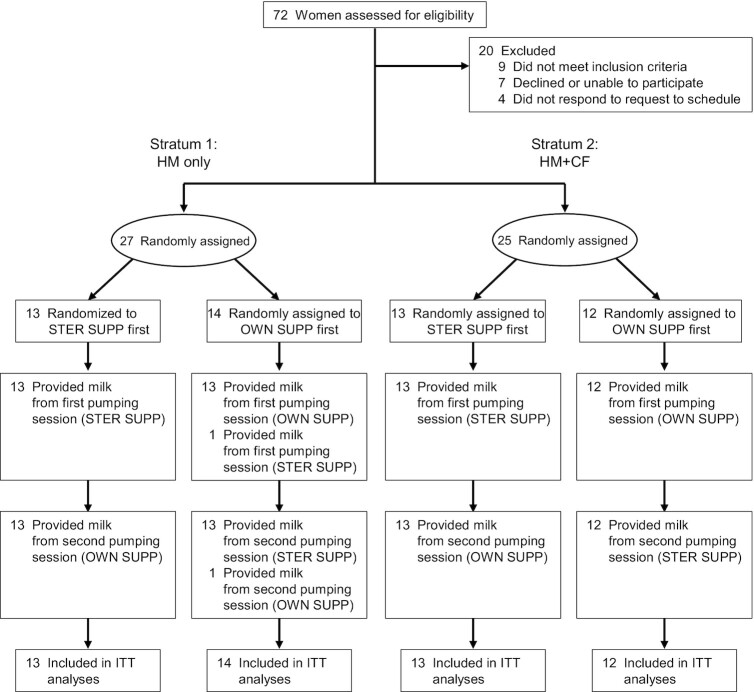
The MiLC trial was conducted from June to October 2017. Fifty-two women were randomly assigned to treatment order, with equal numbers allocated to pumping with STER SUPP and OWN SUPP first (Figure 1). Each woman served as her control, resulting in 104 paired milk samples. Analyses were conducted as intention to treat. Participants were on average 34 ± 4.0 y old and 6.5 ± 4.2 mo postpartum (Table 1). Maternal age and race were similar between groups. As anticipated, mothers in the HM-only group were fewer months postpartum than those in the HM + CF group.
FIGURE 1.
Participant flow diagram. Each woman contributed 2 milk samples, one from each collection method. One participant pumped in the opposite order of her randomly assigned assignment. All samples were analyzed by intention-to-treat analysis. HM, human milk; HM + CF, human milk and complementary foods; ITT, intention to-treat; OWN SUPP, personal electric breast pump and milk collection kits; STER SUPP, hospital-grade pump and new, sterile collection kits.
TABLE 1.
Participant characteristics according to infant diet and randomly assigned pumping order for the 52 participants of the MiLC trial1
| HM only | HM + CF | ||||
|---|---|---|---|---|---|
| Participant characteristic | All participants (N = 52) | STER SUPP First (n = 13) | OWN SUPP First (n = 14) | STER SUPP First (n = 13) | OWN SUPP First (n = 12) |
| Maternal age, y | 34 ± 4.0 | 32 ± 4.0 | 32 ± 4.5 | 35 ± 4.1 | 34 ± 3.3 |
| Caucasian | 45 (86) | 11 (85) | 13 (93) | 11 (85) | 10 (83) |
| Time postpartum, mo | 6.5 ± 4.2 | 3.6 ± 1.3 | 3.0 ± 1.7 | 11 ± 3.8 | 8.7 ± 2.3 |
Values are means ± SDs for continuous variables or n (%) for categorical variables. CF, complementary foods; HM, human milk; OWN SUPP First, randomly assigned to use own pumping supplies first; STER SUPP First, randomly assigned to use sterile pumping supplies first.
Effects of OWN SUPP on viable total aerobic and gram-negative bacteria in milk
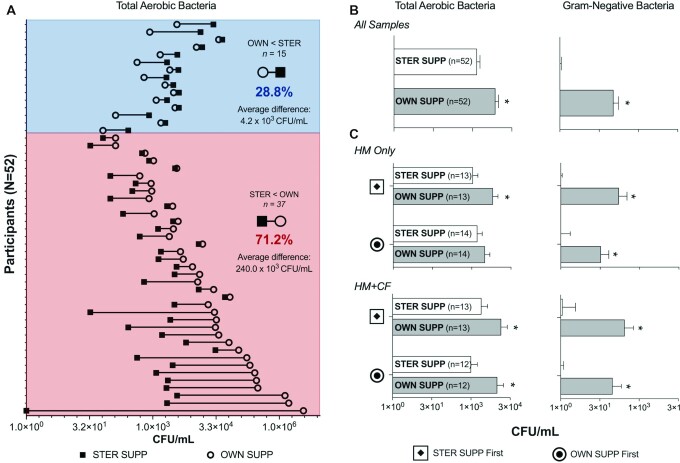
Although STER SUPP yielded more culturable bacteria in milk than OWN SUPP for 28.8% of women, pumping with OWN SUPP significantly increased the average levels of culturable bacteria in milk samples (Figure 2, Supplemental Tables 1 and 2). On average, milk collected with STER SUPP yielded 1.5 × 103 ± 4.8 × 102 CFU/mL total aerobic bacteria and rarely grew gram-negative bacteria. In contrast, milk collected with OWN SUPP yielded 5.11 ± 2.15 times more total CFU/mL and 82.5 ± 61.8 times more gram-negative CFU/mL than milk collected with STER SUPP (P ≤ 0.0003, ratios of LS means from linear mixed regression) (Figure 2B).
FIGURE 2.
Effect of OWN SUPP on cultured bacteria in pumped human milk (n = 104), among all samples (A, B) and according to infant diet and randomly assigned pumping order (C). LS means and SEM from linear mixed-effects regression are shown. *Difference compared with STER SUPP (P ≤ 0.04). All other comparisons were not significant (P ≥ 0.20). HM, human milk; HM + CF, human milk and complementary foods; OWN SUPP, personal electric breast pump and milk collection kits; OWN SUPP First, randomly assigned to use own pumping supplies first; STER SUPP, hospital-grade pump and new, sterile collection kits; STER SUPP First, randomly assigned to use sterile pumping supplies first.
The effect of pumping supplies on average total aerobic and gram-negative bacteria was unaffected by infant diet group or randomly assigned pumping order, as indicated by the lack of significant 2- and 3-way interactions as well as lack of significant main effects of infant diet group and randomly assigned pumping order in linear mixed-effects regression models. The only exception was among women in the HM-only stratum and randomly assigned to pumping with OWN SUPP first, for which no effect of the pumping supplies was observed for total aerobic bacterial counts (Figure 2C).
Pumping supplies significantly affected the proportion of milk samples with high bacterial loads. Fifteen percent of milk samples collected with STER SUPP yielded >104 total aerobic CFU/mL compared with 42% of milk samples collected using OWN SUPP (aOR: 6.6; 95% CI, 1.7, 25; P = 0.006).
In addition, pumping supplies significantly affected the proportion of milk samples that were culture positive for gram-negative bacteria. Only 5.8% of milk samples collected with STER SUPP yielded gram-negative bacteria compared with 50% of milk samples collected with OWN SUPP (aOR: 19; 95% CI, 4.1, 88; P < 0.001).
Effects of OWN SUPP on 16S rRNA bacterial sequences
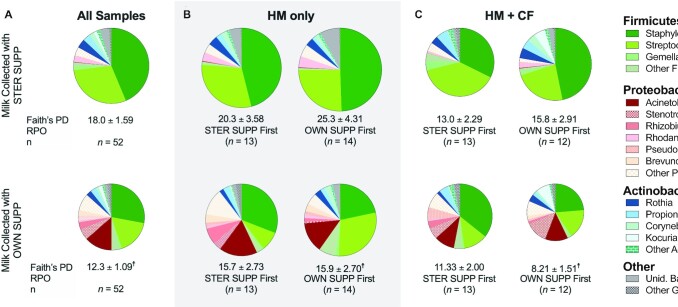
On average, pumping with OWN SUPP decreased the phylogenetic diversity of 16S sequences, although the effect was stronger for some subsets of women. Overall, compared with milk collected using STER SUPP, that collected with OWN SUPP had 32 ± 8.7% lower phylogenetic diversity (Faith's phylogenetic diversity index, P = 0.0005, Figure 3A, Supplemental Table 3). This trend ranged from 13% to 48% and was strongest among women randomly assigned to pumping with OWN SUPP first (37–48%; P ≤ 0.05, Supplemental Table 3). The bacterial richness and Shannon diversity index did not differ between pump and infant diet groups or randomly assigned pumping order (Supplemental Tables 4–5).
FIGURE 3.
Effect of OWN SUPP on the genus-level composition of 16S ribosomal RNA sequences (n = 104), according to infant diet and randomly assigned pumping order (A–C). Asterisks denote differentially abundant genera. Circles scaled by Faith's phylogenetic diversity index. †P < 0.05 compared with STER SUPP using mixed-effects regression. OWN SUPP, personal electric breast pump and milk collection kits; PD, phylogenetic diversity; RPO, randomly assigned pumping order; STER SUPP, hospital-grade pump and new, sterile collection kits; Unid., unidentified.
Pumping with OWN SUPP also altered the community structure of milk (R2 = 0.024, P = 0.03, adjusted for infant diet and randomly assigned pumping order determined by PERMANOVA). Principal components analysis showed clear clustering patterns by the pump group but not by randomly assigned pumping order (Supplemental Figure 2). These clustering patterns coincided with large differences in the milk microbiota composition. Milk collected with STER SUPP was dominated by Staphylococcus and Streptococcus (Figure 3A, Supplemental Table 6) and contained 6.7 ± 10.0% Proteobacteria (median: 3.0%; range: 0–52.9%). In contrast, Acinetobacter was the third most abundant genera in milk collected with OWN SUPP, which contained 13.5 ± 21.0% Proteobacteria (median: 21.3%; range: 0–99.5%). Differential abundance testing revealed milk collected with OWN SUPP contained significantly more Acinetobacter, Stenotrophomonas, Rhizobium, Pseudomonas, and Brevundimonas (all genera of Proteobacteria). Although the specific taxa identified as differentially abundant in milk differed somewhat according to the infant diet group and randomly assigned pumping order, Proteobacteria were overrepresented in milk collected using OWN SUPP across these groups (Supplemental Figure 3).
Correlations between 16S sequences and cultured bacteria in milk
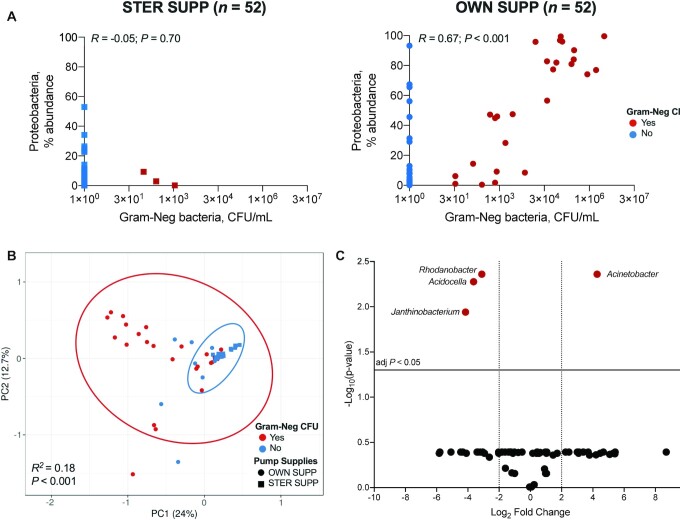
Given these findings, we examined the correlations between 16S sequences and cultured bacteria in milk. In post hoc analyses, results for these correlations were discordant between milk samples collected with STER SUPP and those collected with OWN SUPP. Specifically, there was a positive correlation between total aerobic bacterial counts and relative abundance of Firmicutes for milk collected with STER SUPP (Spearman r = 0.49, P < 0.001; Supplemental Figure 4). This correlation, however, was negative for milk collected with OWN SUPP (Spearman r = –0.40, P = 0.003). In addition, no correlation was observed between gram-negative bacterial counts and relative abundance of proteobacterial 16S sequences for milk collected with STER SUPP (Spearman r = –0.05, P = 0.70), but a strong positive correlation was observed for milk collected with OWN SUPP (Spearman r = 0.67, P < 0.001) (Figure 4A). These findings were strongest among women in the HM-only feeding group and were unaffected by randomly assigned pumping order (Supplemental Table 4).
FIGURE 4.
Correlations between culturable gram-negative bacteria and 16S microbiome. (A) There was no Spearman correlation between Gram-Neg bacterial counts and percent relative abundances of Proteobacteria 16S in STER SUPP (P = 0.7) but a strong correlation for OWN SUPP (P < 0.001). (B) PCA ordination of Proteobacteria 16S sequences; samples with culturable Gram-Neg bacteria had a different 16S community than samples without culturable Gram-Neg bacteria (permutational ANOVA P < 0.001). (C) The volcano plot shows the log2 fold change (x-axis) of Proteobacteria asegenera that differed significantly (FDR-adjusted P value y-axis) between samples yielding culturable gram-negative bacteria and those that did not, regardless of pumping supplies used for collection. FDR, false discovery rate; Gram-Neg, gram-negative bacteria; OWN SUPP, personal electric breast pump and milk collection kits; PCA, Principal component analysis; PC1, Principal component 1; PC2, Principal component 2; STER SUPP, hospital-grade pump and new, sterile collection kits.
The differences in correlation coefficients appeared to be driven by whether milk samples were culture positive for gram-negative bacteria, regardless of pump supplies used for collection. Principal components analysis (PCA) of proteobacterial 16S sequences showed clear clustering patterns by viable gram-negative bacteria status (R2 = 0.18, P < 0.001, determined by PERMANOVA; Figure 4B). These clustering patterns coincided with large differences in the relative abundances of several genera of Proteobacteria (Figure 4C). Notably, the average relative abundance of Acinetobacter was 4.28-log2 fold higher in milk samples that yielded viable gram-negative bacteria than those that did not [false discovery rate (FDR)-adjusted P = 0.004].
Discussion
Our in-home, randomized, crossover trial demonstrated that typical pumping supplies altered the microbiome of pumped human milk, but infant diet and pumping order did not. We provide experimental evidence that viable bacteria, bacterial 16S DNA sequences, and the phylogenetic diversity of milk differed by pumping supplies for most women. We also showed that counts of gram-negative bacteria were correlated with relative abundances of Acinetobacter. This evidence may have important implications for the GI microbiome of infants consuming pumped human milk. It is broadly relevant because most women pump to meet human milk–feeding recommendations and goals (4), and milk banks rely on donations from women who pump at home (47, 48). Our results may inform the development of evidence-based guidelines for pump hygiene practices at home.
MiLC results are in line with previous studies showing that 61–74% of milk collected at home yielded >104 CFU/mL total aerobic bacteria (9, 49) and would therefore exceed recommendations for feeding to preterm infants without pasteurization (47). The source of these bacteria in previous studies was uncertain because collection and handling practices vary greatly among women (5–7, 25). Thus, it remained unclear whether elevated levels of bacteria were a function of collection method or handling practices. Using paired analyses with each woman serving as her own control, we demonstrated that pumping supplies were a significant source of exogenous bacteria. These findings are further supported by our observed differences in correlations between cultured bacteria and 16S sequences, which are likely driven by differences in concentrations of viable bacteria on the pumping supply surfaces.
Our findings improve the scientific premise for the conclusion that women's own pumping supplies were a significant source of exogenous bacteria as suggested by others (26, 29). However, more work is needed to understand the effects of specific hygiene practices on the bacterial composition of pumped human milk. Fifteen women in our study had more culturable total aerobic bacteria in their milk collected with STER SUPP compared with OWN SUPP. Thus, it is plausible that differences in the personal hygiene practices and/or practices used to clean pump parts could have contributed to bacterial composition of pumped milk. These possibilities warrant further research.
The health impact of elevated levels of culturable bacteria in pumped human milk remains unclear for healthy infants. The amount of culturable bacteria in pumped milk has long been of concern for preterm infants, as they often rely on pumped milk and are especially vulnerable to bacterial infection (50, 51). In contrast, the practice of feeding pumped milk to healthy, term infants has increased dramatically since the early 2000s (4, 52, 53). Thus, only limited research exists on the effects of the bacterial load of milk on these infants. Although others have also found milk expressed at home yielded relatively high amounts of viable gram-negative bacteria (9–11, 54), prior evidence was observational. Conversely, our randomized crossover design allowed for causal inference that pumping with OWN SUPP enriched human milk with total culturable and gram-negative bacteria.
The findings of our clinical trial add to previous observational evidence that gram-negative bacteria may be added to human milk during pumping (9–11). Others have shown that this may pose a risk for vulnerable infants (8, 55–60). Specifically, in our study, pumped milk that contained viable gram-negative bacteria also had significantly higher abundances of Acinetobacter 16S sequences than pumped milk that yielded no viable gram-negative bacteria. This may have important clinical implications because Acinetobacter species have been linked to illness in preterm infants (61–63). We recently reported on the genomes of several Acinetobacter bereziniaeisolated from milk collected during this study (64). Importantly, these isolates encoded known pathogenicity determinants, including hemolysins and lipopolysaccharide. They also encoded a ceramidase, which may have an impact on natural antimicrobial lipids in milk. Although more work is required to determine the impact of these bacteria on milk and in the GI tract, A. bereziniae are considered emerging pathogens that have been implicated in nosocomial infections (65, 66).
The presence of exogenous gram-negative bacteria, including Acinetobacter, does not necessarily warrant concern. The mothers in our trial reported that they and their infants were healthy. Acinetobacter were one of the most abundant microbes in the feces of healthy 7-d-old infants (67). The presence of Acinetobacter has also been associated with a reduction in allergy in 6-mo-old infants, suggesting these bacteria may play an important immunomodulatory role (68). Thus, the function and importance of exogenous gram-negative bacteria in human milk, including Acinetobacter, remain an open question that could have different answers depending on the health of the infant consuming it. More genome-derived information on the metabolic potential of exogenous gram-negative bacteria added to human milk, including Acinetobacter, will be essential to future efforts aimed at understanding the potential duality of their effects on infant health.
Strengths and limitations
The main strength of the MiLC trial was our randomized, crossover design in which women served as their own controls. This allowed for causal inference of the effects of women's pumping supplies on the microbiome of their pumped milk that were not obvious in a larger recent study that did not control for milk collection practice (19).
More specifically, our experimental approach and milk collection protocol controlled for interindividual and diurnal variability, which increased statistical power. Our analyses of milk collected with STER SUPP were consistent with previous studies that had stringent milk collection protocols and showed milk to be dominated by Staphylococcus and Streptococcus (15, 16, 24, 69). Moreover, randomization was stratified by infant diet, which increased the generalizability of our findings to women with a range of infant feeding practices. The study design we employed allowed us to demonstrate the importance of milk collection practices for accurately characterizing the human milk microbiome and thus has implications for others seeking to study the impact of pumped human milk on infant health.
The fact that the MiLC trial employed both culture-dependent and culture-independent, next-generation sequencing is also important. Although there was a correlation between culturable bacteria and DNA sequences that demonstrated the impact of collection practice on the microbiome of pumped human milk, we found that culture conditions were insufficient to capture the diversity of bacteria in milk. This is important because the influence of the human milk microbiome on infant health is an active area of research (70). The role of bacterial viability warrants additional investigation.
The main limitation of our study was our inability to determine the viability of some organisms detected via 16S but not via culturing. In addition, the use of short 16S amplicons (∼300 bp) to analyze the composition of milk restricted bacterial identification to the genus level; strain-level information is needed to unambiguously identify pathogens (71). To this end, we recently employed additional culturing coupled with genome sequencing to further characterize 4 isolates of Acinetobacter bereziniae from a single sample of milk collected with OWN SUPP (64). More work is clearly needed to characterize opportunistic pathogens in pumped human milk. Finally, we chose not to clean the participants’ breasts before collection. Although this decision precluded direct comparison with some other studies of the human milk microbiome (15, 16, 72), our design provides an accurate and appropriate characterization of microbiota ingested by infants.
Conclusions
Through paired analyses and a randomized, crossover design clinical trial, we provide the strongest level of evidence to date that pumping supplies can drastically change the microbiome of pumped human milk. On average, milk collected with women's own pumping supplies resulted in elevated levels of viable total aerobic and gram-negative bacteria and proteobacterial DNA compared with sterile control. More research is needed to understand the implications, if any, of our findings on infant health. These results, if replicated by others in a broader population, may have implications for several stakeholders, including policymakers, women who rely on pumping their milk to meet their breastfeeding goals, and milk banks that rely on donations from women who pump at home. Questions could include the frequency and degree of pumping, as well as what interventions could reduce the impact of pumping on the human milk microbiome.
Supplementary Material
Acknowledgments
We thank all the women who participated in this study and our helpful staff in the Hay and McGuire laboratories, especially Morgan Potton, Romana Hyde, and Claire Concepcion. We also thank Dr. Ariel Ganz and the Cornell University Statistical Consulting Unit for their constructive insights for data analysis. We also acknowledge the generosity of Medela in supplying in-kind contributions, namely, loaning us Medela Symphony breast pumps and donating the sterile milk collection kits. The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication. The authors' responsibilities were as follows—SMR, AGH, and KMR: conceptualized, designed, obtained funding for the study; SMR: conducted the study and had full access to the data for the study and takes responsibility for the integrity of the data and the accuracy of the data analysis; AGH and KMR: supervised the study; JEW and MAM: supervised 16S rRNA analyses; SMR and DLA: conducted culture experiments; DLA and JEW: provided administrative and technical support; SMR and AGH: analyzed the data and drafted the initial manuscript; SMR, AGH, JEW, MAM, MKM, and KMR: assisted in the interpretation of the data; and all authors: provided feedback and read and approved the final manuscript. The authors report no conflicts of interest.
Notes
Supported, in part, by NIH T32-DK007158, USDA National Institute of Food and Agriculture (Hatch NYC-399346 and IDA01643), a travel grant from International Society for Research in Human Milk and Lactation and the Family Larsson-Rosenquist Foundation, McNair Scholars Program at Cornell University, NIH P30 GM103324, and in-kind contributions from Medela.
Supplemental Figures 1–4 and Supplemental Tables 1–6 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/ajcn/.
Abbreviations used: aOR, adjusted odds ratio; ASV, amplicon sequence variant; FDR, false discovery rate; GI, gastrointestinal; HM, human milk; HM + CF, human milk and complementary foods; ITT, intention-to-treat analysis; LPS, lipopolysaccharide; LS, least squares; MiLC, Milk in Life Conditions trial; OWN SUPP, personal electric breast pump and own milk collection kit; PCA, principal components analysis; PERMANOVA, permutational ANOVA; rRNA, ribosomal RNA; STER SUPP, hospital-grade breast pump and new, sterile collection kit.
Contributor Information
Sarah M Reyes, Division of Nutritional Sciences, Cornell University, Ithaca, NY, USA.
Dainelle L Allen, Division of Nutritional Sciences, Cornell University, Ithaca, NY, USA.
Janet E Williams, Department of Animal, Veterinary, and Food Sciences, University of Idaho, Moscow, ID, USA.
Mark A McGuire, Department of Animal, Veterinary, and Food Sciences, University of Idaho, Moscow, ID, USA.
Michelle K McGuire, Margaret Ritchie School of Family and Consumer Sciences, University of Idaho, Moscow, ID, USA.
Anthony G Hay, Department of Microbiology, Cornell University, Ithaca, NY, USA.
Kathleen M Rasmussen, Division of Nutritional Sciences, Cornell University, Ithaca, NY, USA.
Data Availability
Data described in the manuscript, code book, and analytic code will be made available upon request pending application and approval. Sequence data will be made available on the National Center for Biotechnology Information's Sequence Read Archive pending publication.
References
- 1. Ip S, Chung M-Y, Raman G, Chew P, Magula N, DeVine D, Trikalinos T, Lau J. Breastfeeding and maternal and infant health outcomes in developed countries. Evid Rep Technol Assess (Full Rep). 2007;153:1–186. [PMC free article] [PubMed] [Google Scholar]
- 2. Horta B, Bahl R, Martinés J, Victora CG. Evidence on the long-term effects of breastfeeding: systematic reviews and meta-analysis. Geneva (Switzerland): World Health Organization, 2007. [Google Scholar]
- 3. Eidelman AL, Johnston M, Landers S, Noble L, Szucs K, Viehmann L. Breastfeeding and the use of human milk. Pediatrics. 2012;129(3):e827–41. [DOI] [PubMed] [Google Scholar]
- 4. Labiner-Wolfe J, Fein SB, Shealy KR, Wang C. Prevalence of breast milk expression and associated factors. Pediatrics. 2008;122(Suppl 2):S63–8. [DOI] [PubMed] [Google Scholar]
- 5. O'Sullivan EJ, Geraghty SR, Rasmussen KM. Human milk expression as a sole or ancillary strategy for infant feeding: a qualitative study. Matern Child Nutr. 2017;13(3):e12332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Felice JP, Geraghty SR, Quaglieri CW, Yamada R, Wong AJ, Rasmussen KM. “Breastfeeding” without baby: a longitudinal, qualitative investigation of how mothers perceive, feel about, and practice human milk expression. Matern Child Nutr. 2017;13(3):e12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Felice JP, Geraghty SR, Quaglieri CW, Yamada R, Wong AJ, Rasmussen KM. “Breastfeeding” but not at the breast: mothers' descriptions of providing pumped human milk to their infants via other containers and caregivers. Matern Child Nutr. 2017;13(3):e12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bowen A, Wiesenfeld HC, Kloesz JL, Pasculle AW, Nowalk AJ, Brink L, Elliot E, Martin H, Tarr CL. Notes from the field: Cronobacter sakazakii infection associated with feeding extrinsically contaminated expressed human milk to a premature infant—Pennsylvania, 2017. MMWR Morbid Mortal Wkly Rep. 2017;66(28):761–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Keim SA, Hogan JS, McNamara KA, Gudimetla V, Dillon CE, Kwiek JJ, Geraghty SR. Microbial contamination of human milk purchased via the Internet. Pediatrics. 2013;132(5):e1227–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Serra VV, Teves S, Lopez de Volder A, Ossorio F, Aguilar N, Armadans M. Comparison of the risk of microbiological contamination between samples of breast milk obtained at home and at a healthcare facility. Arch Argent Pediatr. 2013;111(2):115–9. [DOI] [PubMed] [Google Scholar]
- 11. Haiden N, Pimpel B, Assadian O, Binder C, Kreissl A, Repa A, Thanhauser M, Roberts CD, Berger A. Comparison of bacterial counts in expressed breast milk following standard or strict infection control regimens in neonatal intensive care units: compliance of mothers does matter. J Hosp Infect. 2016;92(3):226–8. [DOI] [PubMed] [Google Scholar]
- 12. Pittard WB, Geddes KM, Brown S, Mintz S, Hulsey TC. Bacterial contamination of human milk: container type and method of expression. Am J Perinatol. 1991;8(1):25–7. [DOI] [PubMed] [Google Scholar]
- 13. Karimi M, Eslami Z, Shamsi F, Moradi J, Ahmadi AY, Baghianimoghadam B. The effect of educational intervention on decreasing mothers' expressed breast milk bacterial contamination whose infants are admitted to neonatal intensive care unit. J Res Health Sci. 2012;13(1):43–7. [PubMed] [Google Scholar]
- 14. Williams JE, Carrothers JM, Lackey KA, Beatty NF, Brooker SL, Peterson HK, Steinkamp KM, York MA, Shafii B, Price WJet al. . Strong multivariate relations exist among milk, oral, and fecal microbiomes in mother-infant dyads during the first six months postpartum. J Nutr. 2019;149(6):902–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lackey KA, Williams JE, Meehan CL, Zachek JA, Benda ED, Price WJ, Foster JA, Sellen DW, Kamau-Mbuthia EW, Kamundia EWet al. . What's normal? Microbiomes in human milk and infant feces are related to each other but vary geographically: the INSPIRE study. Front Nutr. 2019;6:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hunt KM, Foster JA, Forney LJ, Schutte UME, Beck DL, Abdo Z, Fox LK, Williams JE, McGuire MK, McGuire MA. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS One. 2011;6(6):e21313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marin ML, Arroyo R, Jimenez E, Gomez A, Fernandez L, Rodriguez JM. Cold storage of human milk: effect on its bacterial composition. J Pediatr Gastroenterol Nutr. 2009;49(3):343–8. [DOI] [PubMed] [Google Scholar]
- 18. Fernandez L, Langa S, Martin V, Maldonado A, Jimenez E, Martin R, Rodriguez JM. The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res. 2013;69(1):1–10. [DOI] [PubMed] [Google Scholar]
- 19. Moossavi S, Sepehri S, Robertson B, Bode L, Goruk S, Field CJ, Lix LM, de Souza RJ, Becker AB, Mandhane PJet al. . Composition and variation of the human milk microbiota are influenced by maternal and early-life factors. Cell Host Microbe. 2019;25(2):324–335.e4. [DOI] [PubMed] [Google Scholar]
- 20. Pannaraj PS, Li F, Cerini C, Bender JM, Yang S, Rollie A, Adisetiyo H, Zabih S, Lincez PJ, Bittinger Ket al. . Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 2017;171(7):647–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fehr K, Moossavi S, Sbihi H, Boutin RCT, Bode L, Robertson B, Yonemitsu C, Field CJ, Becker AB, Mandhane PJet al. . Breastmilk feeding practices are associated with the co-occurrence of bacteria in mothers' milk and the infant gut: the CHILD Cohort Study. Cell Host Microbe. 2020;28(2):285–297.e4. [DOI] [PubMed] [Google Scholar]
- 22. Moossavi S, Miliku K, Sepehri S, Khafipour E, Azad MB. The prebiotic and probiotic properties of human milk: implications for infant immune development and pediatric asthma. Front Pediatr. 2018;6:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bode L, McGuire M, Rodriguez JM, Geddes DT, Hassiotou F, Hartmann PE, McGuire MK. It's alive: microbes and cells in human milk and their potential benefits to mother and infant. Adv Nutr. 2014;5(5):571–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Williams JE, Carrothers JM, Lackey KA, Beatty NF, York MA, Brooker SL, Shafii B, Price WJ, Settles ML, McGuire MAet al. . Human milk microbial community structure is relatively stable and related to variations in macronutrient and micronutrient intakes in healthy lactating women. J Nutr. 2017;147(9):1739–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Labiner-Wolfe J, Fein SB. How US mothers store and handle their expressed breast milk. J Hum Lact. 2013;29(1):54–8. [DOI] [PubMed] [Google Scholar]
- 26. Price E, Weaver G, Hoffman P, Jones M, Gilks J, O'Brien V, Ridgway G. Decontamination of breast pump milk collection kits and related items at home and in hospital: guidance from a Joint Working Group of the Healthcare Infection Society and Infection Prevention Society. J Hosp Infect. 2016;92(3):213–21. [DOI] [PubMed] [Google Scholar]
- 27. Froh EB, Vanderpool J, Spatz DL. Best practices to limit contamination of donor milk in a milk bank. J Obstet Gynecol Neonatal Nurs. 2018;47(4):547–55. [DOI] [PubMed] [Google Scholar]
- 28. Oba PM, Holscher HD, Mathai RA, Kim J, Swanson KS. Diet influences the oral microbiota of infants during the first six months of life. Nutrients. 2020;5(12):3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moossavi S, Azad MB. Origins of human milk microbiota: new evidence and arising questions. Gut Microbes. 2020;12(1):1667722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. The R Project for Statistical Computing. [Internet]. Available from: https://www.R-project.org/ (accessed 25 March 2017).
- 31. Gupta SK. Intention-to-treat concept: a review. Perspect Clin Res. 2011;2(3):109–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Whittaker RH. Evolution and measurement of species diversity. Taxon. 1972;21(2–3):213–51. [Google Scholar]
- 33. Morris EK, Caruso T, Buscot F, Fischer M, Hancock C, Maier TS, Meiners T, Muller C, Obermaier E, Prati Det al. . Choosing and using diversity indices: insights for ecological applications from the German Biodiversity Exploratories. Ecol Evol. 2014;4(18):3514–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61(1):1–10. [Google Scholar]
- 35. Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Naghili H, Tajik H, Mardani K, Razavi Rouhani SM, Ehsani A, Zare P. Validation of drop plate technique for bacterial enumeration by parametric and nonparametric tests. Vet Res Forum. 2013;4(3):179–83. [PMC free article] [PubMed] [Google Scholar]
- 37. Settles M. dbcAmplicons. [Internet]. Available from: https://github.com/msettles/dbcAmplicons (accessed 14 December 2017).
- 38. Settles M. Split Reads By Sample. [Internet]. Available from:https://github.com/msettles/dbcAmplicons/blob/master/scripts/python/splitReadsBySample.py(accessed 14 December 2017).
- 39. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet C, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar Fet al. . Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Davis NM, Proctor D, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6(1):226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bourgon R, Gentleman R, Huber W. Independent filtering increases detection power for high-throughput experiments. Proc Natl Acad Sci U S A. 2010;107(21):9546–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fletcher D, Mackenzie D, Villouta E. Modelling skewed data with many zeros: a simple approach combining ordinary and logistic regression. Environ Ecol Stat. 2005;12(1):45–54. [Google Scholar]
- 44. Satterthwaite FE. An approximate distribution of estimates of variance components. Biometrics Bull. 1946;2(6):110–4. [PubMed] [Google Scholar]
- 45. Love MI, Wolfgang H, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 47. Jones F. Best practices for expressing, storing, and handling human milk in hospitals, homes, and child care settings. 4th ed. Fort Worth, TX: Human Milk Bank Association of North America; 2019. [Google Scholar]
- 48. Human Milk Banking Association of North America. Guidelines for the establishment and operation of a donor human milk bank. 16th ed. Fort Worth, TX: Human Milk Banking Association of North America; 2011. [Google Scholar]
- 49. Landers S, Updegrove K. Bacteriological screening of donor human milk before and after Holder pasteurization. Breastfeed Med. 2010;5(3):117–21. [DOI] [PubMed] [Google Scholar]
- 50. Platt MJ. Outcomes in preterm infants. Public Health. 2014;128(5):399–403. [DOI] [PubMed] [Google Scholar]
- 51. Narayanan I, Prakash K, Bala S, Verma RK, Gujral VV. Partial supplementation with expressed breast-milk for prevention of infection in low-birth-weight infants. Lancet North Am Ed. 1980;316(8194):561–3. [DOI] [PubMed] [Google Scholar]
- 52. Binns CW, Win NN, Zhoa Y, Scott JA. Trends in the expression of breastmilk 1993–2003. Breastfeed Rev. 2006;14(3):5–9. [PubMed] [Google Scholar]
- 53. Bai DL, Fong DY, Lok KY, Wong JY, Tarrant M. Practices, predictors and consequences of expressed breast-milk feeding in healthy full-term infants. Public Health Nutr. 2017;20(3):492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ng DK, Lee SYR, Leung LCK, Wong SF, Ho JCS. Bacteriological screening of expressed breast milk revealed a high rate of bacterial contamination in Chinese women. J Hosp Infect. 2004;58(2):146–50. [DOI] [PubMed] [Google Scholar]
- 55. Ibrahim J, Hamwi N, Rabei H, Abdelghafar M, Al-Dulaimi Z, Al Tatari H. Stenotrophomonas maltophilia meningitis in a term healthy neonate: a case report and literature review. Case Rep Pediatr. 2018;2018:1543934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bonifacio SL, Kitterman JA, Ursell PC. Pseudomonas pneumonia in infants: an autopsy study. Hum Pathol. 2003;34(9):929–38. [DOI] [PubMed] [Google Scholar]
- 57. Rizzatti G, Lopetuso LR, Gibiino G, Binda C, Gasbarrini A. Proteobacteria: a common factor in human diseases. Biomed Res Int. 2017;2017:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dorota P, Chmielarczyk A, Katarzyna L, Piotr M, Jan L, Renata R, Mach Jadwiga W. Klebsiella pneumoniae in breast milk: a cause of sepsis in neonate. Arch Med. 2017;9:1. [Google Scholar]
- 59. Ramachandran G. Gram-positive and gram-negative bacterial toxins in sepsis: a brief review. Virulence. 2014;5(1):213–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Patel SJ, Green N, Clock SA, Paul DA, Perlman JM, Zaoutis T, Ferng YH, Alba L, Jia H, Larson ELet al. . Gram-negative bacilli in infants hospitalized in the neonatal intensive care unit. J Pediatric Infect Dis Soc. 2017;6(3):227–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Touati A, Achour W, Cherif A, Hmida HB, Afif FB, Jabnoun S, Khrouf N, Hassen AB. Outbreak of Acinetobacter baumannii in a neonatal intensive care unit: antimicrobial susceptibility and genotyping analysis. Ann Epidemiol. 2009;19(6):372–8. [DOI] [PubMed] [Google Scholar]
- 62. Engur D, Cakmak BC, Turkmen MK, Telli M, Eyigor M, Guzunler M. A milk pump as a source for spreading Acinetobacter baumannii in a neonatal intensive care unit. Breastfeed Med. 2014;9(10):551–4. [DOI] [PubMed] [Google Scholar]
- 63. Kilic A, Li H, Mellmann A, Basustaoglu AC, Kul M, Senses Z, Aydogan H, Stratton CW, Harmsen D, Tang Y-W. Acinetobacter septicus sp. nov. association with a nosocomial outbreak of bacteremia in a neonatal intensive care unit. J Clin Microbiol. 2008;46(3):902–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Reyes SM, Bolettieri E, Allen D, Hay AG. Genome sequences for four strains of Acinetobacter bereziniae isolated from human milk pumped with a personal breast pump and hand-washed milk collection supplies. Microbiol Resource Announcements. 2020;9(44):e00770–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Grosso F, Silva L, Sousa C, Ramos H, Quinteira S, Peixe L. Extending the reservoir of bla IMP-5: the emerging pathogen Acinetobacter bereziniae. Future Microbiol. 2015;10(10):1609–13. [DOI] [PubMed] [Google Scholar]
- 66. Kuo S-C, Fung C-P, Lee Y-T, Chen C-P, Chen T-L. Bacteremia due to Acinetobacter genomic species 10. J Clin Microbiol. 2010;48(2):586–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pandey PK, Verma P, Kumar H, Bavdekar A, Patole MS, Shouche YS. Comparative analysis of fecal microflora of healthy full-term Indian infants born with different methods of delivery (vaginal vs cesarean): A cinetobacter sp. prevalence in vaginally born infants. J Biosci. 2012;37(Suppl 1):989–98. [DOI] [PubMed] [Google Scholar]
- 68. Ruokolainen L, Parkkola A, Karkman A, Sinkko H, Peet A, Hämäläinen AM, von Hertzen L, Tillmann V, Koski K, Virtanen SMet al. . Contrasting microbiotas between Finnish and Estonian infants: exposure to Acinetobacter may contribute to the allergy gap. Allergy. 2020;75(9):2342–51. [DOI] [PubMed] [Google Scholar]
- 69. McGuire MK, McGuire MA. Got bacteria? The astounding, yet not-so-surprising, microbiome of human milk. Curr Opin Biotechnol. 2017;44:63–8. [DOI] [PubMed] [Google Scholar]
- 70. McGuire MK, McGuire MA. Mapping the human milk microbiome: impetus for a long-awaited renaissance in maternal and infant nutrition research?. J Nutr. 2021;151(2):278–80. [DOI] [PubMed] [Google Scholar]
- 71. Couto N, Schuele L, Raangs EC, Machado MP, Mendes CI, Jesus TF, Chlebowicz M, Rosema S, Ramirez M, Carrico JAet al. . Critical steps in clinical shotgun metagenomics for the concomitant detection and typing of microbial pathogens. Sci Rep. 2018;8(1):13767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr. 2012;96(3):544–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data described in the manuscript, code book, and analytic code will be made available upon request pending application and approval. Sequence data will be made available on the National Center for Biotechnology Information's Sequence Read Archive pending publication.