Abstract
The epidemic coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has now spread worldwide and efficacious therapeutics are urgently needed. 3-Chymotrypsin-like cysteine protease (3CLpro) is an indispensable protein in viral replication and represents an attractive drug target for fighting COVID-19. Herein, we report the discovery of 9,10-dihydrophenanthrene derivatives as non-peptidomimetic and non-covalent inhibitors of the SARS-CoV-2 3CLpro. The structure-activity relationships of 9,10-dihydrophenanthrenes as SARS-CoV-2 3CLpro inhibitors have carefully been investigated and discussed in this study. Among all tested 9,10-dihydrophenanthrene derivatives, C1 and C2 display the most potent SARS-CoV-2 3CLpro inhibition activity, with IC50 values of 1.55 ± 0.21 μM and 1.81 ± 0.17 μM, respectively. Further enzyme kinetics assays show that these two compounds dose-dependently inhibit SARS-CoV-2 3CLprovia a mixed-inhibition manner. Molecular docking simulations reveal the binding modes of C1 in the dimer interface and substrate-binding pocket of the target. In addition, C1 shows outstanding metabolic stability in the gastrointestinal tract, human plasma, and human liver microsome, suggesting that this agent has the potential to be developed as an orally administrated SARS-CoV-2 3CLpro inhibitor.
Keywords: COVID-19; SARS-CoV-2 3CLpro; 9,10-Dihydrophenanthrenes; Structure-activity relationships
Graphical abstract
1. Introduction
Since the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the public around the world is plagued with Coronavirus Disease 2019 (COVID-19) [1,2]. Accordingly, many worldwide endeavors were made to effectively prevent or treat COVID-19, boosting several vaccines that were successfully administered applied to the public [[3], [4], [5]]. However, SARS-CoV-2 is constantly mutating for better survival in hosts [6]. For example, the B.1.617.2 (Delta) variant appears with a faster transmission capacity and stronger toxicity, and has become dominant in the current pandemic [7,8]. Nevertheless, the latest reports manifest that the effectiveness of COVID-19 vaccines is reduced against the Delta variant, and people who have been vaccinated are still facing the risk of being infected [9,10]. Until now, there are few effective antiviral medications to cure CoV-related diseases, such as COVID-19. We urgently need novel antiviral agents to fight against the ongoing SARS-CoV-2 and other unknown coronaviruses in the long term. At the same time, scientists are working against time to discover potential anti-SARS-CoV-2 drugs by targeting the key proteases to obstruct the viral life cycle [11,12]. 3-Chymotrypsin-like cysteine protease (3CLpro), an indispensable tool for an intact coronaviral replication cycle, can convert the viral polyproteins into functional proteins [13]. Besides this important function, 3CLpros share a highly conserved gene sequence among several pathogenic human coronaviruses (such as SARS-CoV-2, MERS-CoV, and SARS-CoV) [14]; accordingly, 3CLpro is deemed to be an attractive target for developing broad-spectrum antiviral drugs. In addition, potent 3CLpro inhibition can reduce the risk of mutation-mediated drug resistance [15].
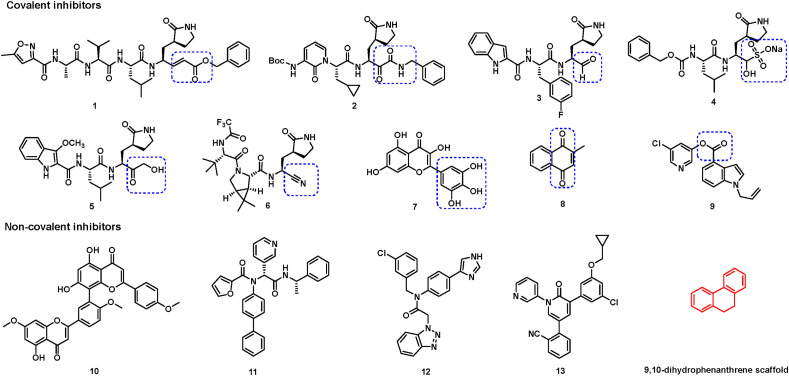
Up to now, a variety of SARS-CoV-2 3CLpro inhibitors have been developed using multiple drug discovery strategies, which can be divided into covalent inhibitors and non-covalent inhibitors according to their inhibitory mechanisms [[16], [17], [18]]. Covalent SARS-CoV-2 3CLpro inhibitors, including peptidomimetics and small molecules, generally bear different covalent warheads and act through a two-step mechanism [19]. First, these agents bind to the active site and form a non-covalent complex with the target protease. Then, the warhead forms a covalent bond with the nucleophilic residues, especially catalytic Cys145 or other key cysteines (such as Cys300 [20] and Cys44 [20,21]), whose covalent modifications can further elicit the inactivation of SARS-CoV-2 3CLpro. Several chemical warheads, such as Michael acceptor (compound 1) [22], α-ketoamide (compound 2) [23], aldehyde (compounds 3) [24], α-hydroxy bisulfite (compound 4, GC-376) [25], hydroxymethyl ketone (compound 5, PF-00835231) [26], cyano (compound 6, PF-07321332) [27], pyrogallol (compound 7, myricetin) [20,28], quinone (compound 8, Vitamin K3) [29], and ester carbonyl group (compound 9) have been explored by pharmaceutical companies or research groups (Fig. 1 ) [30]. Among these inhibitors, only compounds 5 and 6 from Pfizer Inc. entered the clinical trial stage. Additionally, some natural products or small molecules, such as sciadopitysin (compound 10) [31], 23R (compound 11) [32], CCF981 (compound 12) [33], and pyridine 13 [34], were found to have strong inhibitory activities against SARS-CoV-2 3CLpro via non-covalent modes. Candidates for use as anti-COVID-19 agents are expected to possess several positive traits, such as excellent pharmacokinetic property and good cell permeability, besides the superior biological activities. However, most of these peptidomimetics or natural products exhibit poor membrane permeability and poor oral bioavailability. Further, the potential off-target issue of covalent inhibitors may lead to unpredictable toxic side effects clinically [19,35]. Thus, there is a strong desire for novel potent non-covalent SARS-CoV-2 3CLpro inhibitors with drug-like properties.
Fig. 1.
Representative SARS-CoV-2 3CLpro inhibitors. For covalent inhibitors, different covalent warheads are emphasized by dashed rectangles.
Herein, we report the discovery of non-covalent SARS-CoV-2 3CLpro inhibitors with 9,10-dihydrophenanthrene scaffold using an established fluorescence resonance energy transfer (FRET) biochemical assay. We also carefully carried out the structure-activity relationship (SAR) and inhibitory mechanism studies. Furthermore, the in vitro metabolic stability in the gastrointestinal tract, human plasma, and liver microsome validated that the newly identified SARS-CoV-2 3CLpro inhibitor C1 has the potential to be developed into an orally administered agent.
2. Results and discussion
2.1. Screening and SARs analysis
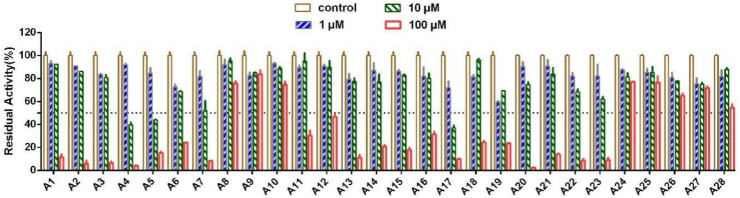
To obtain potent SARS-CoV-2 3CLpro inhibitors, we first screened our in-house compound library (compounds containing 9,10-dihydrophenanthrene scaffold) using established fluorescence resonance energy transfer (FRET) assay at final concentrations of 1 μM, 10 μM, and 100 μM (Fig. 2 ) [31,36]. Briefly, the fluorescent substrate can be recognized and cleaved by 3CLpro, and the products emit a strong fluorescence at 490 nm after being excited at 340 nm. Disulfiram (a reported SARS-CoV-2 3CLpro inhibitor) was used as the positive control compound [22]. Of them, some compounds showed good inhibitory activities and were selected for further IC50 values investigation. It was worth noting that the functional groups of our screened compounds varied at the positions R1, R2, and R3, which was because of the limitations of our invented synthetic methodology [37]. As shown in Table 1 , we summarized the SARs of the tested 9,10-dihydrophenanthrene derivatives. For R1 substituent, the activities against SARS-CoV-2 3CLpro were improved along with an increasing volume of the substituted groups, such as methyl (A1, IC50 = 61.15 μM) to ethyl (A2, IC50 = 33.06 μM), isopropyl (A3, IC50 = 29.46 μM), cyclohexyl (A4, IC50 = 9.06 μM), and 4-bromine phenyl (A5, IC50 = 6.44 μM). When we kept increasing the size of the R 1 group, such as 4- cyanophenyl (A6, IC50 = 19.32 μM) and benzyl (A7, IC50 = 11.39 μM), the inhibitory activities were decreased. For the R2 substituent, introducing methyl at C-2 or C-3 position generated compounds A13 and A14, which displayed inhibitory activities similar to A1 (A13, IC50 = 57.67 μM, A14, IC50 = 53.81 μM). The presence of substituent groups such as methyl (A8), methoxyl (A9), fluorine (A10), or bromine (A12) at the C-4 position led to a significant loss of potency. For the R3 substituent, the incorporation of 6-methyl (A15), 5-methyl (A16), or 4-fluorine (A18) at pyridyl group exhibited comparable inhibitory activities with A1. The incorporation of 4-chlorine (A19) or 4-bromine (A20) at pyridyl group or replacing pyridine with quinoline (A21, A22, A23) showed increased inhibitory activities. Compound A17 with 5-phenyl at the pyridyl group displayed the best inhibitory activity with an IC50 value of 5.65 μM. In contrast, the substituents of pyrimidine (A24, A25), pyrazole (A26), and purine derivative (A27, A28) demonstrated no inhibitory activity against SARS-CoV-2 3CLpro.
Fig. 2.
Inhibitory effects of A1-A28 against SARS-CoV-2 3CLpro.
Table 1.
Inhibitory activities of 9,10-dihydrophenanthrene derivatives against SARS-CoV-2 3CLpro.
 | ||||
|---|---|---|---|---|
| Compds | R1 | R2 | R3 | IC50 ± SD (μM)a |
| A1 | CH3 | H |  |
61.15 ± 3.60 |
| A2 | C2H5 | H |  |
33.06 ± 8.58 |
| A3 | isopropyl | H | 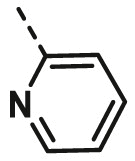 |
29.46 ± 1.64 |
| A4 | cyclohexyl | H | 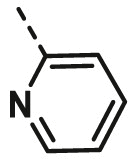 |
9.06 ± 0.34 |
| A5 | 4-Br Phenyl | H | 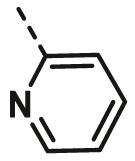 |
6.44 ± 0.64 |
| A6 | 4-CN Phenyl | H | 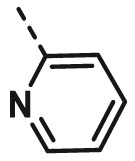 |
19.32 ± 1.92 |
| A7 | benzyl | H | 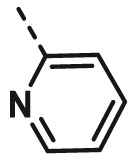 |
11.39 ± 0.64 |
| A8 | CH3 | 4-CH3 | 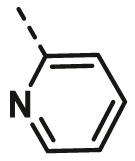 |
>100 |
| A9 | CH3 | 4-OCH3 | 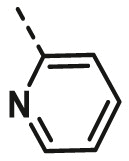 |
>100 |
| A10 | CH3 | 4-F | 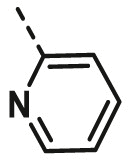 |
>100 |
| A11 | CH3 | 4-Cl | 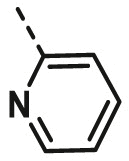 |
85.58 ± 2.36 |
| A12 | CH3 | 4-Br | 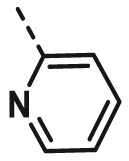 |
>100 |
| A13 | CH3 | 3-CH3 | 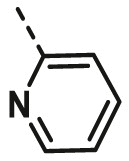 |
57.67 ± 2.78 |
| A14 | CH3 | 2-CH3 | 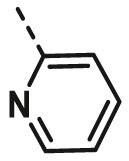 |
53.81 ± 8.49 |
| A15 | CH3 | H | 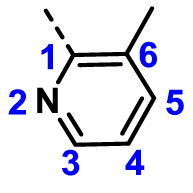 |
58.91 ± 4.58 |
| A16 | CH3 | H | 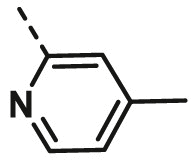 |
68.16 ± 2.64 |
| A17 | CH3 | H | 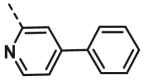 |
5.65 ± 1.40 |
| A18 | CH3 | H | 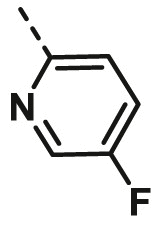 |
66.80 ± 2.74 |
| A19 | CH3 | H | 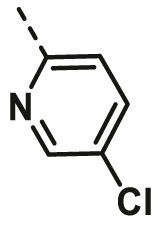 |
35.77 ± 2.96 |
| A20 | CH3 | H | 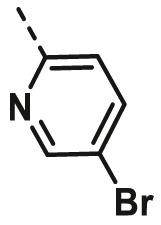 |
18.50 ± 0.76 |
| A21 | CH3 | H | 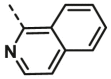 |
14.17 ± 1.30 |
| A22 | CH3 | H | 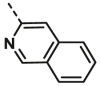 |
11.97 ± 0.68 |
| A23 | CH3 | H | 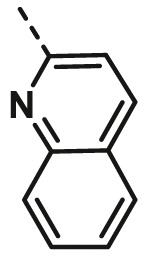 |
13.82 ± 0.90 |
| A24 | CH3 | H | 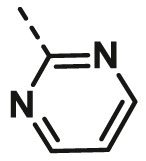 |
>100 |
| A25 | CH3 | H | 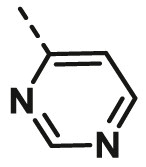 |
>100 |
| A26 | CH3 | H | 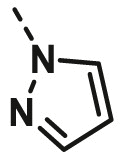 |
>100 |
| A27 | CH3 | H | 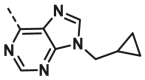 |
>100 |
| A28 | CH3 | H | 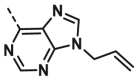 |
>100 |
| disulfiram | – | – | – | 1.04 ± 0.03 |
The inhibitory effects of these compounds against SARS-CoV-2 3CLpro were determined by the FRET assay. The data are the mean ± SD from at least three independent experiments.
The abovementioned analyses suggested the preliminary SARs for the 9,10-dihydrophenanthrene derivatives that can be summarized as follows: (1) the installation of suitable bulkier groups at the R1 position was preferred and, cyclohexyl and 4-Br phenyl displayed better inhibitory activities than others. (2) The conversion of the pyridine group into quinoline at the R3 position was favorable, whereas pyrimidine, pyrazole, and purine derivatives at this position were unfavorable. (3) The incorporation of 5-phenyl at the pyridyl group was of great importance for the inhibition activity.
2.2. Rational design of SARS-CoV-2 3CLpro inhibitors
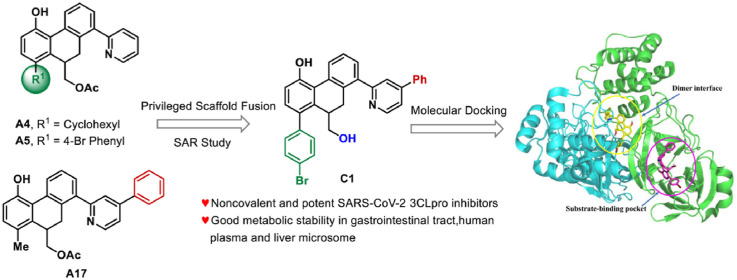
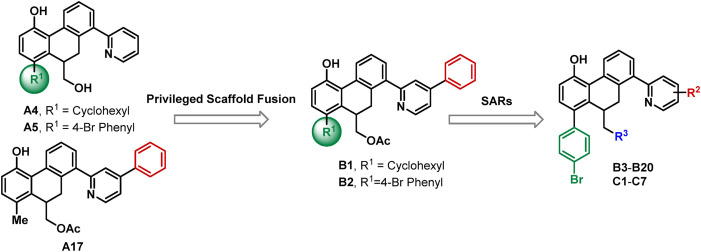
The abovementioned structure–activity relationships showed that cyclohexyl and 4-Br phenyl groups at R1 position (A4 and A5) and 5-phenyl at the pyridyl group (A17) were optimal options for maintaining the SARS-CoV-2 3CLpro inhibition activity. To improve the biological activity of the identified hit compounds, we maintained those privileged fragments and designed compounds B1 and B2 (as shown in Fig. 3 ). On the basis of the optimized 4-Br phenyl group, SARs of various substitutions at the pyridine ring were explored again (B3–B20). Furthermore, the ethyl ester group was hydrolyzed to investigate its effect on inhibitory activity (C1–C7).
Fig. 3.
Rational design of SARS-CoV-2 3CLpro inhibitors by privileged scaffold fusion and structure-activity relationships study.
2.3. Chemistry
The designed compounds were prepared according to our previously reported synthetic method, which is shown in Scheme 1 [37]. First, the substrates cyclohexadienone-containing 1,6-enynes 16 and 17 were prepared from commercially available 4-bromo-4′-hydroxybiphenyl and 4-cyclohexylphenol, respectively, using propargyl alcohol and phenyliodine (III) diacetate (PIDA). In addition, 2-arylazaarenes 18a-18v were purchased directly from commercial companies or generated via a Suzuki cross-coupling reaction with the use of phenylboronic acid and substituted 2-bromopyridine. Subsequently, an efficient and one-pot synthesis of 9,10-dihydrophenanthrenes from 16, 17, and 18a-18v was conducted using rhodium (III)-catalyzed C–H activation and relay Diels–Alder reaction to obtain target compounds B1–B20. Subsequently, compounds B2, B6, B10, B11, B12, B14, and B18 were hydrolyzed under 1 M NaOH and CH3OH solution to afford C1–C7.
Scheme 1.
Reagents and conditions: (a) [Cp∗RhCl2]2, NaBArF4, CH3COOH, 100 °C, 8 h, 20.0%–87.0%; (b) 1 M NaOH (aq.), CH3OH, 50 °C, 2 h, 37.0%–99.0%.
2.4. Biological assays
2.4.1. SARS-CoV-2 3CLpro inhibition assays
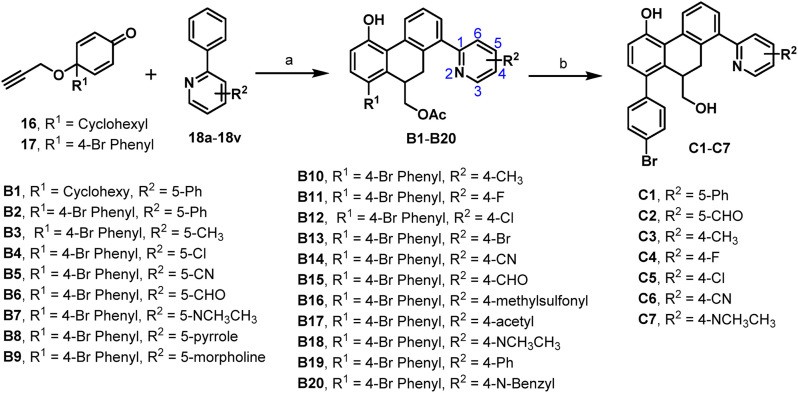
To measure the inhibitory activity of our designed compounds against SARS-CoV-2 3CLpro, a fluorescence resonance energy transfer (FRET) protease assay was performed as described previously [31,36]. As seen in Table 2 , compound B2 with the privileged fragment 4-Br phenyl group at the R1 position and 5-phenyl at the R2 position could improve the activity as expected with an IC50 value of 2.46 μM. However, compound B1 with a cyclohexyl at the R1 position and 5-phenyl at the R2 position failed to enhance the activity with an IC50 value of 8.54 μM. Therefore, we fixed 4-Br phenyl group at the R1 position and changed the group at pyridine region to examine the influence on the inhibitory activity. When the substituent groups, such as methyl (B3), chlorine (B4), cyano (B5), aldehyde (B6), N, N-dimethyl (B7), pyrrole (B8), and morpholine (B9), were located at the C-5 position of the pyridine ring, compounds B4, B6, and B9 displayed better inhibitory activities with IC50 values of 4.79 μM, 3.32 μM, and 7.39 μM, respectively. Other compounds exhibited moderate activities with IC50 values more than 10 μM. Incorporation of the substituent groups, such as methyl (B10), fluorine (B11) chlorine (B12), bromine (B13), cyano (B14), aldehyde (B15), methylsulfonyl (B16), acetyl (B17), N, N-dimethyl (B18), phenyl (B19), and N-benzyl (B20) at the C-4 position, all the compounds demonstrated potent activities with IC50 values ranging from 2.72 μM to 8.89 μM. To further explore the antiviral activity of the ethyl ester group, compounds B2, B6, B10, B11, B12, B14, and B18 with better activity described above were hydrolyzed to the provided compounds C1–C7, containing a typical hydroxyl pharmacophore. Interestingly, only compounds C1 and C2 showed a slight increase in the inhibitory activity with IC50 values of 1.55 μM and 1.81 μM, respectively (Table 3 ). However, compounds C4, C5, and C7 led to slight decreases in the inhibitory activity, whereas C6 with a 4-cyano group resulted in a substantial decrease of the activity (IC50 = 10.68 μM). Taken together, compounds C1 and C2 showed better in vitro inhibitory activities against SARS-CoV-2 3CLpro and were selected for further evaluation.
Table 2.
Inhibitory activities of B1–B20 against SARS-CoV-2 3CLpro.
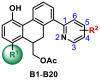 | |||
|---|---|---|---|
| Compds | R1 | R2 | IC50 ± SD (μM)a |
| B1 | Cyclohexyl | 5-Ph | 8.54 ± 0.94 |
| B2 | 4-Br Phenyl | 5-Ph | 2.46 ± 0.14 |
| B3 | 4-Br Phenyl | 5-CH3 | 9.82 ± 1.16 |
| B4 | 4-Br Phenyl | 5-Cl | 4.79 ± 0.22 |
| B5 | 4-Br Phenyl | 5-CN | 10.26 ± 0.66 |
| B6 | 4-Br Phenyl | 5-CHO | 3.32 ± 0.25 |
| B7 | 4-Br Phenyl | 5-NCH3CH3 | 10.00 ± 0.47 |
| B8 | 4-Br Phenyl | 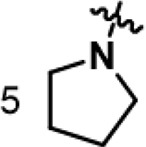 |
11.23 ± 0.70 |
| B9 | 4-Br Phenyl | 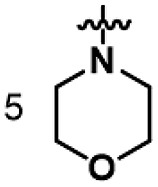 |
7.39 ± 0.11 |
| B10 | 4-Br Phenyl | 4-CH3 | 5.25 ± 0.24 |
| B11 | 4-Br Phenyl | 4-F | 4.29 ± 0.62 |
| B12 | 4-Br Phenyl | 4-Cl | 2.72 ± 0.23 |
| B13 | 4-Br Phenyl | 4-Br | 6.73 ± 1.29 |
| B14 | 4-Br Phenyl | 4-CN | 2.78 ± 0.18 |
| B15 | 4-Br Phenyl | 4-CHO | 8.89 ± 3.34 |
| B16 | 4-Br Phenyl | 4-methylsulfonyl | 6.65 ± 0.26 |
| B17 | 4-Br Phenyl | 4-acetyl | 5.83 ± 0.34 |
| B18 | 4-Br Phenyl | 4-NCH3CH3 | 3.66 ± 0.17 |
| B19 | 4-Br Phenyl | 4-Ph | 8.28 ± 1.88 |
| B20 | 4-Br Phenyl | 4-N-Benzyl | 3.31 ± 0.36 |
| disulfiram | – | – | 1.04 ± 0.03 |
The inhibitory effects of these compounds against SARS-CoV-2 3CLpro were determined by the FRET assay. The data are the mean ± SD from at least three independent experiments.
Table 3.
Inhibitory activities of C1–C7 against SARS-CoV-2 3CLpro.
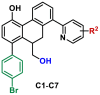 | ||||
|---|---|---|---|---|
| Compds | R2 | IC50 ± SD (μM)a | Ki (μM) | Inhibition mode |
| C1 | 5-Ph | 1.55 ± 0.21 | 6.09 | Mixed |
| C2 | 5-CHO | 1.81 ± 0.17 | 7.64 | Mixed |
| C3 | 4-CH3 | 10.23 ± 3.50 | – | – |
| C4 | 4-F | 5.39 ± 0.65 | – | – |
| C5 | 4-Cl | 4.71 ± 0.20 | – | – |
| C6 | 4-CN | 10.68 ± 0.46 | – | – |
| C7 | 4-NCH3CH3 | 5.50 ± 0.35 | – | – |
| disulfiram | – | 1.04 ± 0.03 | – | – |
The inhibitory effects of these compounds against SARS-CoV-2 3CLpro were determined by the FRET assay. The data are the mean ± SD from at least three independent experiments.
2.4.2. Inhibition kinetics assays of C1 and C2 against SARS-CoV-2 3CLpro
To uncover the inhibitory mechanisms of two newly identified SARS-CoV-2 3CLpro inhibitors (C1 and C2), the inhibition kinetics assays of these two agents were carefully evaluated in vitro. As shown in Fig. 4 , the Lineweaver–Burk plots showed that both C1 and C2 could dose-dependently inhibit SARS-CoV-2 3CLpro via a mixed-inhibition manner, with K i values of 6.09 μM and 7.64 μM, respectively. In addition, the corresponding second plot of the Lineweaver–Burk plots were presented in Fig. S4, showing good linearity (R2 > 0.95).
Fig. 4.
The dose-dependent inhibition curves (left) and the second plot of Lineweaver–Burk plots (right) for C1 (A, B) and C2 (C, D) against SARS-CoV-2 3CLpro.
2.4.3. SARS-CoV 3CLpro inhibition assays
To verify whether these series compounds have a broad-spectrum inhibitory effect on 3CLpro, we obtained highly purified SARS-CoV 3CLpro according to the previously reported expression and purification procedures of SARS-CoV-2 3CLpro [31]. Subsequently, 11 compounds that exhibited potent inhibitory effects (IC50 < 5 μM) against SARS-CoV-2 3CLpro were chosen for SARS-CoV 3CLpro inhibition assay. It is clearly shown in Table 4 and Fig. S8 that most of the tested compounds show relatively strong inhibitory activities against SARS-CoV 3CLpro, with IC50 values less than 10 μM. These results suggest that the 9,10-dihydrophenanthrene derivatives can easily be developed as broad-spectrum 3CLpro inhibitors for the treatment of coronavirus-related diseases.
Table 4.
Inhibitory activities of 11 compounds against SARS-CoV 3CLpro.
| Compds | R1 | R2 | IC50 ± SD (μM)a |
|---|---|---|---|
| B2 | 4-Br Phenyl | 5-Ph | 17.66 ± 1.42 |
| B4 | 4-Br Phenyl | 5-Cl | 5.44 ± 0.36 |
| B6 | 4-Br Phenyl | 5-CHO | 3.20 ± 0.25 |
| B11 | 4-Br Phenyl | 4-F | 7.81 ± 0.51 |
| B12 | 4-Br Phenyl | 4-Cl | 4.06 ± 0.17 |
| B14 | 4-Br Phenyl | 4-CN | 7.12 ± 0.32 |
| B18 | 4-Br Phenyl | 4-NCH3CH3 | 5.92 ± 0.22 |
| B20 | 4-Br Phenyl | 4-Benzyl | 5.79 ± 0.35 |
| C1 | 4-Br Phenyl | 5-Ph | 5.35 ± 0.30 |
| C2 | 4-Br Phenyl | 5-CHO | 5.32 ± 0.22 |
| C5 | 4-Br Phenyl | 4-Cl | 10.49 ± 0.23 |
| Disulfiram | – | – | 6.72 ± 1.02 |
The inhibitory effects of these compounds against SARS-CoV 3CLpro were determined by the FRET assay. The data are the mean ± SD from at least three independent experiments.
2.5. Molecular docking simulations
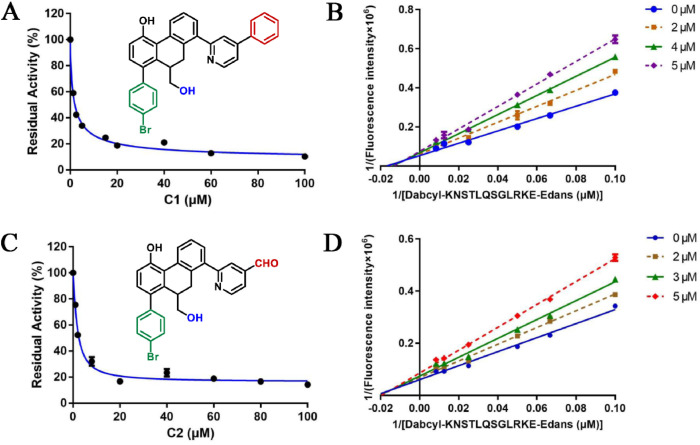
As C1 inhibits SARS-CoV-2 3CLpro via mixed inhibition, which means this small molecule could bind at least two sites in the target protein. Recent studies indicated that the dimerization of SARS-CoV-2 3CLpro was crucial to enzymatic activity, the monomers were catalytically inactive [38]. Therefore, there are at least two potential sites that can be employed to develop inhibitors against this enzyme: the substrate-binding pocket and the dimer interface of SARS-CoV-2 3CLpro, both of whose key interactions with the inhibitors may affect the catalytic activity. After failing to obtain the crystal structure of C1-SARS-CoV-2-3CLpro complex, the molecular docking study was conducted to predict the binding modes at the substrate binding pocket and the dimer interface (Fig. 5 A).
Fig. 5.
Predicted SARS-CoV-2 3CLpro inhibitor binding modes for C1. (A) Cartoon representation of the docking results. Compound C1 at the dimer interface position is shown as a yellow stick; Compound C1 at substrate-binding pocket is shown as a magenta stick; (B) Predicted interactions of C1 at the dimer interface position; (C) Surface representation of C1 in the substrate-binding pocket. Four subsites, S1’, S1, S2, and S4, are labeled; (D) Predicted interactions of C1 at the substrate-binding pocket. Hydrogen bonds are indicated as dashed lines.
For the dimer interface position (Fig. 5B), the phenolic hydroxyl group of C1 formed a hydrogen bond with the crucial residue Glu290, which may disrupt the salt bridge interaction with Arg4 in another monomer. Furthermore, a benzene ring generated a cation-π interaction with Lys5. Meanwhile, the binding affinity of C1 was enhanced through some hydrophobic interactions with Phe3 and Tyr126 residues. For the substrate-binding pocket (Fig. 5C and D), the 4-bromine phenyl moiety deeply inserted into the big S2 site, which was usually large enough to accommodate the larger hydrophobic fragment. The phenol group occupied the S1′ site and was stacked with the imidazole ring of His41. Notably, this hydroxyl group formed two hydrogen bonds with Gly143 and Cys145. The 5-phenyl group at the pyridine ring fitted well into the S4 pocket and formed multiple hydrophobic interactions with the target. In addition, the hydroxymethyl group could form a 2.3 Å hydrogen bond interaction with Gln189, which was consistent with the observation that the hydroxyl group worked better than ethyl ester group (B2 vs C1). In short, these interactions between C1 and the target are responsible for its potent SARS-CoV-2 3CLpro inhibitory activity.
2.6. In vitro gastrointestinal, plasma, and microsome stability assays
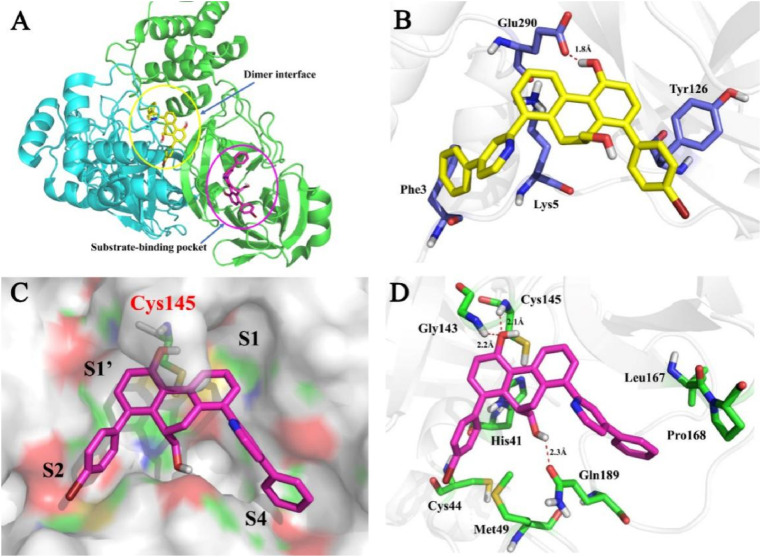
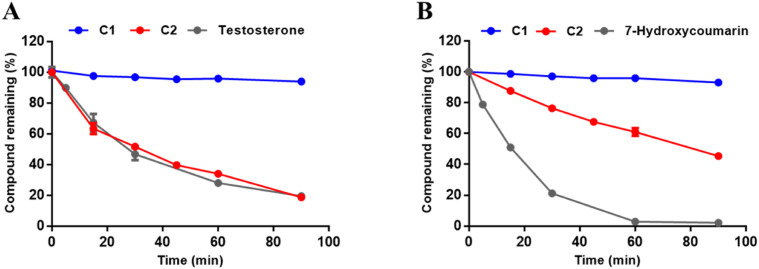
Next, the potential of compounds C1 and C2 as orally administered agents was investigated by performing gastrointestinal stability and metabolic stability assays. Firstly, the gastrointestinal stabilities of C1 and C2 in simulated gastric fluid (SGF) and simulated intestinal fluid (SIF) systems were assayed. As presented in Fig. 6 , both C1 and C2 showed good gastrointestinal stabilities in 90 min. Moreover, the plasma stability of C1 was also prominent, whereas the half-life of C2 in human plasma was 8.97 h.
Fig. 6.
(A–B) The gastrointestinal stabilities of C1 and C2 in SGF (A) and SIF (B); (C) The plasma stabilities of C1 and C2 in human blood plasma.
Meanwhile, the in vitro metabolic stability assays for compounds C1 and C2 in human liver microsome were also investigated and the results are shown in Fig. 7 and Table 5 . Notably, C1 was hardly metabolized by the human cytochrome P450 enzymes (hCYPs) or the human hepatic UDP-Glucuronosyltransferases (hUGTs) in the presence of sugar donor UDPGA (Uridine-Diphosphate-Glucuronic Acid trisodium salt). In contrast, C2 could be readily metabolized by hCYPs and moderately metabolized by hUGTs, with the in vitro half-life of 32.09 min (in the phase I metabolism system) and 81.88 min (in the phase II metabolism system). Moreover, as shown in Fig. S11, C1 showed moderate inhibitory effects against most CYPs except for CYP2C8 at 100 μM. These results indicate that C1 possessed excellent plasma stability and good gastrointestinal and metabolic stability, suggesting that this agent has a great potential as a novel orally administered agent to fight against COVID-19 via targeting the key SARS-CoV-2-3CLpro.
Fig. 7.
Metabolic stabilities of C1 and C2 in phase I (A) and phase II (B) metabolic system in human liver microsomes.
Table 5.
The half-life of C1 and C2 in phase I or phase II metabolic system in human liver microsomes.
| No. | Compound |
T1/2 (min) |
|
|---|---|---|---|
| phase I | phase II | ||
| 1 | C1 | >90 | >90 |
| 2 | C2 | 32.09 | 81.88 |
| 3 | Testosterone | 27.62 | ND |
| 4 | 7-Hydroxycoumarin | ND | 15.53 |
a ND = Not detected.
3. Conclusions
In summary, this study reported the discovery of 9,10-dihydrophenanthrene derivatives as potent noncovalent SARS-CoV-2 3CLpro inhibitors for the treatment of COVID-19. Systematic SAR explorations resulted in the discovery of two efficacious lead compounds (C1 and C2) for further development. Enzyme kinetic analyses revealed that both C1 and C2 dose-dependently inhibited SARS-CoV-2 3CLpro through a mixed-inhibition manner. Molecular docking simulations elucidated the possible binding mode of C1 in the substrate-binding pocket and the dimer interface of the target. Furthermore, compound C1 showed good metabolic stability in the human gastrointestinal tract, plasma, and liver microsomes, which was a great advantage compared with covalent peptides or peptidomimetics. We hope that this work can provide more structural reference for the development of SARS-CoV-2 3CLpro inhibitors for combating this rapidly evolving virus and further in-depth research will be reported in due course.
4. Experimental section
4.1. Chemistry
The reagents and solvents were purchased from commercial companies, such as Adamas-beta®, Bide pharmatech etc., and used without further purification. The nonaqueous reactions were performed under nitrogen atmosphere. The reactions were monitored by Shimadzu 8040 quadrupole LC/MS system or TLC and terminated as judged by the consumption of the starting material. Flash chromatography was performed on silica gel (200–300 mesh) using SepaBean® machine and visualized under UV light monitor. 1HNMR (600 MHz) and 13CNMR (150 MHz) spectra were measured on Bruker 600 MHz spectrometer using TMS as internal standard at ambient temperature. The chemical shifts (δ) were described as parts per million (ppm) downfield and coupling constants (J) values were expressed as hertz (Hz). High-resolution mass spectra (HRMS) data were reported by Agilent 6545 Accurate-Mass QTOF LC/MS system.
4.1.1. General procedures for the synthesis of compounds18a-18e, 18j, 18k, 18m-18p, 18r, 18s [39]
Reactions were carried out with Phenylboronic acid (146 mg, 1.2 mmol), 2-bromo-pyridine derivatives (234 mg, 1.0 mmol, 2-bromo-4-phenylpyridine for 18a; 172 mg, 1.0 mmol, 2-bromo-4-methylpyridine for 18b; 192 mg, 1.0 mmol, 2-bromo-4-chloropyridine for 18c; 183 mg, 1.0 mmol, 2-bromoisonicotinonitrile for 18d; 186 mg, 1.0 mmol, 2-bromoisonicotinaldehyde for 18e; 176 mg, 1.0 mmol, 2-bromo-5-fluoropyridine for 18j; 192 mg, 1.0 mmol, 2-bromo-5-chloropyridine for 18k; 183 mg, 1.0 mmol, 6-bromonicotinonitrile for 18m; 186 mg, 1.0 mmol, 6-bromonicotinaldehyde for 18n; 236 mg, 1.0 mmol, 2-bromo-5-(methylsulfonyl)pyridine for 18o, 200 mg, 1.0 mmol, 1-(6-bromopyridin-3-yl)ethan-1-one for 18p, 234 mg, 1.0 mmol, 2-bromo-5-phenylpyridine for 18r; 263 mg, 1.0 mmol, N-benzyl-6-bromopyridin-3-amine for 18s), Pd(PPh3)2Cl2 (3.5 mg, 0.5 mol %), K2CO3 (387 mg, 2.8 mmol) in DME/H2O (8 mL, 3:1) under nitrogen atmosphere. The mixture was stirred at 80 °C for about 14 h and the completion of the reaction was monitored by TLC. After cooling to room temperature, the mixture was extracted with ethyl acetate. The combined organic layers were washed with brine and the organic phase was dried with anhydrous sodium sulfate and concentrated in vacuo. The crude compound was purified by silica gel column chromatography to give corresponding intermediates.
4.1.1.1. 2,4-diphenylpyridine (18a)
Colorless oil, 57.0% yield. 1H NMR (600 MHz, CDCl3) δ 8.71 (d, J = 5.1 Hz, 1H), 8.04 (d, J = 7.5 Hz, 2H), 7.90 (s, 1H), 7.65 (d, J = 7.5 Hz, 2H), 7.49–7.45 (m, 4H), 7.43 (d, J = 7.4 Hz, 1H), 7.40 (d, J = 6.8 Hz, 2H). ESI-HRMS [M + H]+ calcd for C17H14N: 232.1121, found: 232.1131.
4.1.1.2. 4-methyl-2-phenylpyridine (18b)
White solid, 42.0% yield. 1H NMR (600 MHz, CDCl3) δ 8.55 (d, J = 5.0 Hz, 1H), 7.97 (d, J = 7.4 Hz, 2H), 7.55 (s, 1H), 7.47 (t, J = 7.6 Hz, 2H), 7.40 (t, J = 7.3 Hz, 1H), 7.06 (d, J = 4.9 Hz, 1H), 2.42 (s, 3H). ESI-HRMS [M + H]+ calcd for C12H12N: 170.0964, found: 170.0965.
4.1.1.3. 4-chloro-2-phenylpyridine (18c)
Colorless oil, 54.0% yield. 1H NMR (600 MHz, CDCl3) δ 8.57 (d, J = 5.1 Hz, 1H), 7.96 (d, J = 7.3 Hz, 2H), 7.72 (s, 1H), 7.49–7.42 (m, 3H), 7.23 (d, J = 3.6 Hz, 1H). ESI-HRMS [M + H]+ calcd for C11H9ClN: 190.0418, found: 190.0421.
4.1.1.4. 2-Phenylisonicotinonitrile (18d)
White solid, 77.8% yield. 1H NMR (600 MHz, CDCl3) δ 8.78 (d, J = 4.3 Hz, 1H), 7.92 (dd, J = 8.1, 1.4 Hz, 2H), 7.86 (s, 1H), 7.45–7.39 (m, 3H), 7.37 (dd, J = 4.9, 1.3 Hz, 1H). ESI-HRMS [M + H]+ calcd for C12H9N2: 181.0760, found: 181.0772.
4.1.1.5. 2-Phenylisonicotinaldehyde (18e)
Colorless oil, 95.0% yield. 1H NMR (600 MHz, CDCl3) δ 10.02 (s, 1H), 8.83 (d, J = 4.8 Hz, 1H), 8.02 (s, 1H), 7.96 (d, J = 7.3 Hz, 2H), 7.52 (dd, J = 4.8, 1.1 Hz, 1H), 7.41 (t, J = 7.4 Hz, 2H), 7.37 (d, J = 7.2 Hz, 1H). ESI-HRMS [M + H]+ calcd for C12H10NO: 184.0757, found: 184.0758.
4.1.1.6. 4-fluoro-2-phenylpyridine (18j)
White solid, 82.9% yield. 1H NMR (600 MHz, CDCl3) δ 8.46 (d, J = 2.9 Hz, 1H), 7.85 (d, J = 7.4 Hz, 2H), 7.63 (dd, J = 8.7, 4.3 Hz, 1H), 7.42–7.36 (m, 3H), 7.33 (t, J = 7.3 Hz, 1H). ESI-HRMS [M + H]+ calcd for C11H9FN: 174.0714, found: 174.0716.
4.1.1.7. 5-chloro-2-phenylpyridine (18k)
Colorless oil, 82.0% yield. 1H NMR (600 MHz, CDCl3) δ 8.56 (d, J = 2.4 Hz, 1H), 7.89–7.86 (m, 2H), 7.64 (dd, J = 8.5, 2.5 Hz, 1H), 7.59 (d, J = 8.5 Hz, 1H), 7.40 (t, J = 7.5 Hz, 2H), 7.36–7.33 (m, 1H). ESI-HRMS [M + H]+ calcd for C11H9NCl: 190.0418, found: 190.0420.
4.1.1.8. 5-Phenylnicotinonitrile (18m)
White solid, 93.4% yield. 1H NMR (600 MHz, CDCl3) δ 8.94 (dd, J = 2.1, 0.7 Hz, 1H), 8.04 (dd, J = 7.9, 1.7 Hz, 2H), 8.01 (dd, J = 8.3, 2.2 Hz, 1H), 7.85 (dd, J = 8.3, 0.8 Hz, 1H), 7.53–7.50 (m, 3H). ESI-HRMS [M + H]+ calcd for C12H9N2: 181.0760, found: 181.0763.
4.1.1.9. 5-Phenylnicotinaldehyde (18n)
White solid, 79.1% yield. 1H NMR (600 MHz, CDCl3) δ 10.16 (s, 1H), 9.15 (d, J = 1.9 Hz, 1H), 8.26 (dd, J = 8.2, 2.2 Hz, 1H), 8.11 (dd, J = 8.0, 1.5 Hz, 2H), 7.93 (d, J = 8.2 Hz, 1H), 7.56–7.51 (m, 3H). ESI-HRMS [M + H]+ calcd for C12H10NO: 184.0757, found: 184.0765.
4.1.1.10. 4-(methylsulfonyl)-2-phenylpyridine (18o)
White solid, 99.0% yield. 1H NMR (600 MHz, CDCl3) δ 9.20 (d, J = 2.3 Hz, 1H), 8.27 (dd, J = 8.4, 2.4 Hz, 1H), 8.07 (dd, J = 7.7, 1.9 Hz, 2H), 7.92 (d, J = 8.4 Hz, 1H), 7.55–7.50 (m, 3H), 3.15 (s, 3H). ESI-HRMS [M + H]+ calcd for C12H12NO2S: 234.0583, found: 234.0591.
4.1.1.11. 1-(6-phenylpyridin-3-yl)ethan-1-one (18p)
White solid, 75.0% yield. 1H NMR (600 MHz, CDCl3) δ 9.26 (d, J = 1.9 Hz, 1H), 8.32 (dd, J = 8.3, 2.3 Hz, 1H), 8.09 (d, J = 7.0 Hz, 2H), 7.87 (d, J = 8.3 Hz, 1H), 7.55–7.49 (m, 3H), 2.69 (s, 3H). ESI-HRMS [M + H]+ calcd for C13H12NO: 198.0913, found: 198.0915.
4.1.1.12. 2,5-diphenylpyridine (18r)
Colorless oil, 44.2% yield. 1H NMR (600 MHz, CDCl3) δ 8.86 (d, J = 2.3 Hz, 1H), 7.97 (d, J = 7.1 Hz, 2H), 7.88 (dd, J = 8.2, 2.4 Hz, 1H), 7.73 (dd, J = 8.2, 0.6 Hz, 1H), 7.56 (d, J = 7.1 Hz, 2H), 7.42 (t, J = 7.6 Hz, 4H), 7.37–7.32 (m, 2H). ESI-HRMS [M + H]+ calcd for C17H14N: 232.1121, found: 232.1172.
4.1.1.13. N-benzyl-6-phenylpyridin-3-amine (18s)
Brown oil, 48.0% yield. 1H NMR (600 MHz, CDCl3) δ 8.16 (d, J = 2.8 Hz, 1H), 7.89 (d, J = 7.2 Hz, 2H), 7.54 (d, J = 8.6 Hz, 1H), 7.42 (t, J = 7.7 Hz, 2H), 7.40–7.35 (m, 4H), 7.33–7.29 (m, 2H), 6.95 (dd, J = 8.6, 2.9 Hz, 1H), 4.39 (s, 2H). ESI-HRMS [M + Na]+ calcd for C18H16N2Na: 283.1206, found: 283.1209.
4.1.2. General procedures for the synthesis of compounds18f-18h, 18q [40]
Reactions were carried out with Phenylboronic acid (146 mg, 1.2 mmol), 2-Bromo-pyridine derivatives (201 mg, 1.0 mmol, 2-bromo-N,N-dimethylpyridin-4-amine for 18f; 227 mg, 1.0 mmol, 2-bromo-4-(pyrrolidin-1-yl)pyridine for 18g, 243 mg, 1.0 mmol, 4-(2-bromopyridin-4-yl)morpholine for 18h, 201 mg, 1.0 mmol, 201 mg, 1.0 mmol, 6-bromo-N,N-dimethylpyridin-3-amine for 18q), Pd(PPh3)4 (12 mg, 1.0 mol %), K2CO3 (276 mg, 2.0 mmol) in C2H5OH (5 mL) under nitrogen atmosphere. The mixture was stirred at 75 °C for about 15 h and the completion of the reaction was monitored by TLC. After cooling to room temperature, the mixture was extracted with ethyl acetate. The combined organic layers were washed with brine and the organic phase was dried with anhydrous sodium sulfate and concentrated in vacuo. The crude compound was purified by silica gel column chromatography to give corresponding intermediates.
4.1.2.1. N, N-dimethyl-2-phenylpyridin-4-amine (18f)
White solid, 83.0% yield. 1H NMR (600 MHz, CDCl3) δ 8.25 (d, J = 6.0 Hz, 1H), 7.89 (d, J = 8.1 Hz, 2H), 7.40 (t, J = 7.5 Hz, 2H), 7.35 (d, J = 7.2 Hz, 1H), 6.81 (s, 1H), 6.38 (d, J = 5.9 Hz, 1H), 2.93 (s, 6H). ESI-HRMS [M + H]+ calcd for C13H15N2: 199.1230, found: 199.1231.
4.1.2.2. 2-phenyl-4-(pyrrolidin-1-yl)pyridine (18g)
White solid, 75% yield. 1H NMR (600 MHz, CDCl3) δ 8.31 (d, J = 5.9 Hz, 1H), 7.93 (d, J = 7.6 Hz, 2H), 7.45 (t, J = 7.5 Hz, 2H), 7.40 (t, J = 7.3 Hz, 1H), 6.76 (d, J = 2.0 Hz, 1H), 6.38 (dd, J = 5.9, 2.1 Hz, 1H), 3.39 (t, J = 6.5 Hz, 4H), 2.06 (t, J = 6.6 Hz, 4H). ESI-HRMS [M + H]+ calcd for C15H18N2: 225.1386, found: 225.1388.
4.1.2.3. 4-(2-phenylpyridin-4-yl)morpholine (18h)
Colorless oil, 61.0% yield.1H NMR (600 MHz, CDCl3) δ 8.40 (d, J = 5.9 Hz, 1H), 7.91 (d, J = 7.6 Hz, 2H), 7.45 (t, J = 7.5 Hz, 2H), 7.39 (t, J = 7.3 Hz, 1H), 7.08 (d, J = 2.4 Hz, 1H), 6.65 (dd, J = 5.9, 2.5 Hz, 1H), 3.90–3.85 (m, 4H), 3.39–3.33 (m, 4H). ESI-HRMS [M + H]+ calcd for C15H17N2O: 241.1335, found: 241.1338.
4.1.2.4. N, N-dimethyl-6-phenylpyridin-3-amine (18q)
White solid, 83.0% yield. 1H NMR (600 MHz, Acetone-d6) δ 8.08 (s, 1H), 7.91–7.84 (m, 2H), 7.60 (d, J = 8.8 Hz, 1H), 7.28–7.25 (m, 2H), 7.15 (t, J = 6.6 Hz, 1H), 7.04 (dd, J = 9.6, 2.3 Hz, 1H), 2.90 (s, 6H). ESI-HRMS [M + H]+ calcd for C13H15N2: 199.1230, found: 199.1231.
4.1.3. General procedures for the synthesis of B1–B20
The synthesis of the compounds B1–B20 was previously reported in ref. 37.
Reactions were carried out with Phenylpyridine analogs (0.4 mmol), 1,6-enynes (60 mg, 0.2 mmol), [Cp∗RhCl2]2 (8 mg, 5 mol %), NaBArF4 (55 mg, 24 mol %) in CH3COOH (1.0 mL) under nitrogen atmosphere, The mixture was stirred at 100 °C for about 8 h and the completion of the reaction was monitored by TLC. After cooling to room temperature, the NaHCO3 aqueous solution was added to adjust the pH to weak alkaline. The mixture was extracted with ethyl acetate. The combined organic layers were washed with brine and the organic phase was dried with anhydrous sodium sulfate and concentrated in vacuo. The crude compound was purified by silica gel. column chromatography to give corresponding products.
4.1.3.1. (8-cyclohexyl-5-hydroxy-1-(4-phenylpyridin-2-yl)-9,10-dihydrophenanthren-9-yl)methyl acetate (B1)
White solid, yield: 45.0%. 1H NMR (600 MHz, CDCl3) δ 8.79 (d, J = 5.2 Hz, 1H), 8.16 (d, J = 7.7 Hz, 1H), 7.71 (d, J = 7.2 Hz, 2H), 7.67 (d, J = 1.0 Hz, 1H), 7.56 (dd, J = 5.2, 1.8 Hz, 1H), 7.56 (dd, J = 5.2, 1.8 Hz, 1H), 7.51 (t, J = 7.4 Hz, 2H),7.47 (d, J = 7.2 Hz, 1H), 7.32 (dd, J = 7.5, 0.8 Hz, 1H), 7.24 (d, J = 7.7 Hz, 1H), 7.00 (d, J = 8.6 Hz, 1H), 6.74 (d, J = 8.5 Hz, 1H), 3.87 (dd, J = 10.8, 5.7 Hz, 1H), 3.48 (t, J = 10.5 Hz, 1H), 3.43–3.37 (m, 1H), 2.92 (dd, J = 15.5, 2.0 Hz, 1H), 2.78–2.69 (m, 2H), 1.86–1.67 (m, 8H), 1.57–1.51 (m, 1H), 1.44–1.33 (m, 2H), 1.29–1.22 (m, 2H). 13C NMR (150 MHz, CDCl3) δ 170.78, 160.42, 151.69, 149.05, 148.99, 139.79, 137.99, 137.05, 135.64, 133.24, 132.79, 129.31, 129.23, 128.08, 128.01, 127.10, 126.07, 125.92, 122.61, 121.71, 119.87, 116.46, 63.65, 38.97, 35.90, 33.62, 33.08, 28.15, 27.10, 26.25, 20.51. ESI-HRMS [M + H]+ calcd for C34H34NO3: 504.2533, found: 504.2540.
4.1.3.2. (8-(4-bromophenyl)-5-hydroxy-1-(4-phenylpyridin-2-yl)-9,10-dihydrophenanthren-9-yl)methyl acetate (B2)
White solid, yield: 68.5%. 1H NMR (600 MHz, CDCl3) δ 8.78 (d, J = 5.3 Hz, 1H), 8.26 (d, J = 7.7 Hz, 1H), 7.70 (d, J = 7.2 Hz, 2H), 7.63 (s, 1H), 7.58 (dd, J = 5.3, 1.6 Hz, 1H), 7.52–7.45 (m, 6H), 7.24 (d, J = 7.7 Hz, 1H), 7.11 (d, J = 8.3 Hz, 2H), 6.86 (d, J = 8.3 Hz, 1H), 6.71 (d, J = 8.3 Hz, 1H), 3.65 (dd, J = 10.8, 5.6 Hz, 1H), 3.49 (t, J = 10.5 Hz, 1H), 3.13–3.09 (m, 1H), 2.88 (dd, J = 15.6, 4.5 Hz, 1H), 2.77 (dd, J = 15.6, 2.2 Hz, 1H), 1.50 (s, 3H)·13C NMR (150 MHz, CDCl3) δ 170.42, 153.43, 148.86, 140.48, 137.81, 136.27, 133.13, 132.78, 132.44, 131.23, 131.19, 129.71, 129.42, 129.26, 128.48, 128.09, 127.07, 125.91, 122.56, 121.89, 120.97, 120.01, 118.12, 116.25, 63.46, 34.14, 28.30, 20.42. ESI-HRMS [M + H]+ calcd for C34H27BrNO3: 576.1169, found: 576.1186.
4.1.3.3. (8-(4-bromophenyl)-5-hydroxy-1-(4-methylpyridin-2-yl)-9,10-dihydrophenanthren-9-yl)methyl acetate (B3)
White solid, yield: 40.9%, 1H NMR (600 MHz, DMSO‑d 6) δ 10.12 (s, 1H), 8.56 (d, J = 5.0 Hz, 1H), 8.49 (d, J = 7.8 Hz, 1H), 7.64 (d, J = 8.4 Hz, 2H), 7.42 (t, J = 7.7 Hz, 1H), 7.37 (dd, J = 7.6, 1.2 Hz, 1H), 7.35 (s, 1H), 7.33 (d, J = 8.4 Hz, 2H), 7.26 (d, J = 5.0 Hz, 1H), 7.04 (q, J = 8.4 Hz, 2H), 3.54 (dd, J = 10.8, 6.2 Hz, 1H), 3.44 (t, J = 10.2 Hz, 1H), 3.24–3.19 (m, 1H), 2.92–2.84 (m, 2H), 2.43 (s, 3H), 1.64 (s, 3H)·13C NMR (150 MHz, DMSO‑d 6) δ 170.00, 159.24, 155.01, 149.13, 147.43, 140.93, 140.65, 136.57, 133.26, 132.51, 132.13, 131.62, 131.49, 130.28, 128.80, 128.60, 125.96, 125.36, 123.26, 121.12, 120.61, 115.99, 63.25, 33.82, 28.25, 21.00, 20.60. ESI-HRMS [M + H]+ calcd for C29H25BrNO3: 514.1012, found; 514.1020.
4.1.3.4. (8-(4-bromophenyl)-1-(4-chloropyridin-2-yl)-5-hydroxy-9,10-dihydrophenanthren-9-yl)methyl acetate (B4)
White solid, yield: 27.3%, 1H NMR (600 MHz, Acetone-d 6) δ 9.03 (s, 1H), 8.66 (d, J = 5.4 Hz, 1H), 8.59 (dd, J = 5.4, 3.8 Hz, 1H), 7.62 (d, J = 8.3 Hz, 2H), 7.60 (d, J = 1.8 Hz, 1H), 7.48 (dd, J = 5.3, 1.9 Hz, 1H), 7.42 (d, J = 1.8 Hz, 1H), 7.41 (s, 1H), 7.33 (d, J = 8.3 Hz, 2H), 7.06 (q, J = 8.3 Hz, 2H), 3.65 (dd, J = 10.9, 5.8 Hz, 1H), 3.52 (t, J = 10.5 Hz, 1H), 3.31–3.26 (m, 1H), 2.98 (dd, J = 15.7, 2.3 Hz, 1H), 2.91–2.88 (m, 1H), 1.66 (s, 3H)·13C NMR (150 MHz, CDCl3) δ 170.42, 153.43, 140.48, 137.81, 137.73, 136.27, 136.23, 133.13, 132.78, 132.44, 132.33, 131.19, 129.71, 129.26, 128.48, 127.07, 125.91, 122.56, 121.89, 120.97, 120.01, 116.25, 63.46, 34.14, 28.30, 20.42. ESI-HRMS [M + H]+ calcd for C28H22BrClNO3: 534.0466, found: 534.0475.
4.1.3.5. (8-(4-bromophenyl)-1-(4-cyanopyridin-2-yl)-5-hydroxy-9,10-dihydrophenanthren-9-yl)methyl acetate (B5)
White solid, yield: 69.9%, 1H NMR (600 MHz, Acetone-d 6) δ 9.05 (d, J = 16.6 Hz, 1H), 8.61 (dd, J = 7.4, 1.4 Hz, 1H), 8.33 (dd, J = 8.2, 2.1 Hz, 1H), 7.78 (d, J = 8.2 Hz, 1H), 7.61 (d, J = 8.3 Hz, 2H), 7.46–7.42 (m, 2H), 7.32 (d, J = 8.3 Hz, 2H), 7.08–7.02 (m, 2H), 3.64 (dd, J = 10.9, 5.9 Hz, 1H), 3.49 (t, J = 10.4 Hz, 1H), 3.30–3.25 (m, 1H), 3.00 (dd, J = 15.7, 2.2 Hz, 1H), 2.94 (dd, J = 13.5,4.5 Hz, 1H), 1.62 (s, 3H). 13C NMR (150 MHz, Acetone-d 6) δ 171.11, 164.81, 156.25, 153.61, 142.61, 141.50, 140.93, 138.50, 135.36, 134.69, 134.00, 133.43, 132.98, 131.92, 131.61, 130.30, 127.59, 126.25, 122.92, 122.32, 118.60, 117.44, 109.53, 64.54, 35.90, 29.80, 21.33. ESI-HRMS [M + Na]+ calcd for C29H21BrN2O3Na: 547.0628, found: 547.0635.
4.1.3.6. (8-(4-bromophenyl)-1-(4-formylpyridin-2-yl)-5-hydroxy-9,10-dihydrophenanthren-9-yl)methyl acetate (B6)
Yellow solid, yield: 35.8%, 1H NMR (600 MHz, Acetone-d 6) δ 10.23 (s, 1H), 9.03 (s, 1H), 8.97 (d, J = 4.9 Hz, 1H), 8.60 (d, J = 7.7 Hz, 1H), 7.96 (s, 1H), 7.80 (d, J = 4.8 Hz, 1H), 7.60 (d, J = 8.1 Hz, 2H), 7.48–7.40 (m, 2H), 7.31 (d, J = 8.1 Hz, 2H), 7.04 (q, J = 8.3 Hz, 2H), 3.64 (dd, J = 10.8, 5.9 Hz, 1H), 3.51 (t, J = 10.4 Hz, 1H), 3.31–3.23 (m, 1H), 3.00 (d, J = 15.6 Hz, 1H), 2.94–2.90 (m, 1H), 1.57 (s, 3H). 13C NMR (150 MHz, Acetone-d 6) δ 192.34, 169.32, 161.24, 154.49, 150.52, 142.34, 140.88, 139.77, 136.73, 133.43, 132.75, 132.16, 131.65, 131.17, 130.01, 129.30, 128.48, 125.69, 122.84, 121.30, 120.48, 119.83, 115.63, 62.80, 48.89, 34.15, 19.49. ESI-HRMS [M + Na]+ calcd for C29H22BrNO4Na: 550.0624, found: 550.0631.
4.1.3.7. (8-(4-bromophenyl)-1-(4-(dimethylamino)pyridin-2-yl)-5-hydroxy-9,10-dihydrophenanthren-9-yl)methyl acetate (B7)
White solid, yield: 36.0%, 1H NMR (600 MHz, CDCl3) δ 8.30 (s, 1H), 8.26 (d, J = 5.8 Hz, 1H), 7.46 (d, J = 7.7 Hz, 2H), 7.09 (d, J = 7.7 Hz, 2H), 7.00 (d, J = 3.7 Hz, 2H), 6.77 (s, 2H), 6.60 (d, J = 4.4 Hz, 1H), 6.54 (s, 1H), 3.60 (dd, J = 10.6, 5.7 Hz, 1H), 3.46 (t, J = 9.8 Hz, 1H), 3.10 (s, 7H), 2.88 (d, J = 14.9 Hz, 1H), 2.56 (d, J = 15.1 Hz, 1H), 1.63 (s, 3H). 13C NMR (150 MHz, CDCl3) δ 170.37, 155.76, 155.00, 141.05, 135.58, 133.32, 132.70, 131.39, 131.23, 131.04, 129.46, 127.48, 125.32, 121.54, 120.62, 116.97, 107.22, 105.24, 64.07, 39.55, 33.98, 29.71, 28.44, 20.64. ESI-HRMS [M + H]+ calcd for C30H28BrN2O3: 543.1278, found: 543.1288.
4.1.3.8. (8-(4-bromophenyl)-5-hydroxy-1-(4-(pyrrolidin-1-yl)pyridin-2-yl)-9,10-dihydrophenanthren-9-yl)methyl acetate (B8)
White solid, yield: 68.5%, 1H NMR (600 MHz, CDCl3) δ 8.41–8.36 (m, 1H), 8.11 (d, J = 6.0 Hz, 1H), 7.46 (d, J = 8.3 Hz, 2H), 7.09 (d, J = 8.3 Hz, 2H), 7.07 (d, J = 4.1 Hz, 2H), 7.03 (d, J = 8.0 Hz, 1H), 6.82 (d, J = 8.3 Hz, 1H), 6.55 (dd, J = 6.8, 2.2 Hz, 1H), 6.43 (d, J = 2.2 Hz, 1H), 3.60 (dd, J = 10.9, 5.9 Hz, 1H), 3.46 (d, J = 10.5 Hz, 5H), 3.12–3.16 (m, 1H), 3.02 (d, J = 13.6 Hz, 1H), 2.55 (d, J = 14.3 Hz, 1H), 2.09 (s, 4H), 1.62 (s, 3H)·13C NMR (150 MHz, CDCl3) δ 170.36, 154.85, 153.92, 140.77, 135.56, 133.76, 132.74, 131.47, 131.38, 131.12, 130.62, 130.04, 127.64, 126.05, 120.96, 120.74, 116.92, 107.77, 105.99, 64.05, 48.14, 33.98, 28.62, 25.24, 20.68. ESI-HRMS [M + Na]+ calcd for C32H29BrN2O3Na: 591.1254, found: 591.1259.
4.1.3.9. (8-(4-bromophenyl)-5-hydroxy-1-(4-morpholinopyridin-2-yl)-9,10-dihydrophenanthren-9-yl)methyl acetate (B9)
White solid, yield: 23.0%, 1H NMR (600 MHz, CDCl3) δ 8.41 (d, J = 6.0 Hz, 1H), 8.26 (d, J = 7.8 Hz, 1H), 7.47 (d, J = 8.3 Hz, 2H), 7.09 (d, J = 8.3 Hz, 2H), 7.05–6.98 (m, 2H), 6.76–6.71 (m, 3H), 6.63 (d, J = 8.2 Hz, 1H), 3.87–3.82 (m, 4H), 3.56 (dd, J = 10.8, 6.0 Hz, 1H), 3.49 (t, J = 10.1 Hz, 1H), 3.41–3.35 (m, 4H), 3.06 (s, 1H), 2.86 (dd, J = 15.5, 4.0 Hz, 1H), 2.59 (d, J = 14.1 Hz, 1H), 1.65 (s, 3H)·13C NMR (150 MHz, CDCl3) δ 170.35, 156.05, 154.52, 141.03, 135.76, 132.89, 131.52, 131.35, 131.06, 129.25, 127.62, 125.09, 121.87, 120.67, 116.75, 108.66, 106.49, 66.33, 64.09, 46.10, 34.01, 28.39, 20.71. ESI-HRMS [M + H]+ calcd for C32H30BrN2O4: 585.1383, found: 585.1396.
4.1.3.10. (8-(4-bromophenyl)-5-hydroxy-1-(5-methylpyridin-2-yl)-9,10-dihydrophenanthren-9-yl)methyl acetate (B10)
White solid, yield: 44.6%, 1H NMR (600 MHz, CDCl3) δ 8.57 (s, 1H), 8.20 (t, J = 4.6 Hz, 1H), 7.62 (d, J = 7.8 Hz, 1H), 7.48 (d, J = 8.3 Hz, 2H), 7.31 (d, J = 7.9 Hz, 1H), 7.16 (d, J = 4.4 Hz, 2H), 7.09 (d, J = 8.3 Hz, 2H), 6.82 (d, J = 8.3 Hz, 1H), 6.67 (d, J = 8.3 Hz, 1H), 3.64 (dd, J = 10.9, 5.9 Hz, 1H), 3.44 (t, J = 10.4 Hz, 1H), 3.12–3.07 (m, 1H), 2.82 (dd, J = 15.5, 4.5 Hz, 1H), 2.70 (dd, J = 15.5, 2.0 Hz, 1H), 2.44 (s, 3H), 1.66 (s, 3H). 13C NMR (150 MHz, CDCl3) δ 170.39, 153.44, 148.78, 140.54, 137.28, 136.31, 133.06, 132.69, 132.33, 131.66, 131.26, 131.17, 129.63, 128.51, 127.78, 125.80, 123.98, 121.91, 120.92, 116.18, 63.66, 34.08, 28.35, 20.63, 18.25. ESI-HRMS [M + H]+ calcd for C29H25BrNO3: 514.1012, found: 514.1020.
4.1.3.11. (8-(4-bromophenyl)-1-(5-fluoropyridin-2-yl)-5-hydroxy-9,10-dihydrophenanthren-9-yl)methyl acetate (B11)
White solid, yield: 36.0%, 1H NMR (600 MHz, CDCl3) δ 8.57 (d, J = 2.6 Hz, 1H), 8.21 (dd, J = 6.8, 2.0 Hz, 1H), 7.50 (d, J = 8.1 Hz, 3H), 7.42 (dd, J = 8.6, 4.4 Hz, 1H), 7.33–7.29 (m, 2H), 7.13 (d, J = 8.2 Hz, 2H), 6.94 (d, J = 8.3 Hz, 1H), 6.77 (d, J = 8.3 Hz, 1H), 3.69 (dd, J = 10.9, 5.8 Hz, 1H), 3.48 (t, J = 10.4 Hz, 1H), 3.17 (s, 1H), 2.88–2.78 (m, 2H), 1.68 (s, 3H)·13C NMR (150 MHz, CDCl3) δ 170.47, 155.90, 153.06, 140.27, 139.16, 137.11, 136.96, 136.42, 133.05, 132.81, 131.28, 131.22, 129.96, 128.71, 128.02, 126.19, 125.32, 123.39, 123.26, 121.69, 121.11, 115.99, 63.50, 34.13, 28.34, 20.64. 19F NMR (565 MHz, CDCl3) δ −129.13. ESI-HRMS [M + Na]+ calcd for C28H21BrFNO3Na: 540.0581, found: 540.0590.
4.1.3.12. (8-(4-bromophenyl)-1-(5-chloropyridin-2-yl)-5-hydroxy-9,10-dihydrophenanthren-9-yl)methyl acetate (B12)
White solid, yield: 25.5%, 1H NMR (600 MHz, CDCl3) δ 8.67 (d, J = 2.1 Hz, 1H), 8.22 (dd, J = 6.9, 1.7 Hz, 1H), 7.76 (dd, J = 8.3, 2.4 Hz, 1H), 7.51 (d, J = 8.2 Hz, 2H), 7.38 (d, J = 8.3 Hz, 1H), 7.34–7.29 (m, 2H), 7.13 (d, J = 8.2 Hz, 2H), 6.94 (d, J = 8.3 Hz, 1H), 6.77 (d, J = 8.3 Hz, 1H), 6.71 (s, 1H), 3.69 (dd, J = 11.0, 5.8 Hz, 1H), 3.47 (t, J = 10.4 Hz, 1H), 3.17 (s, 1H), 2.88–2.80 (m, 2H), 1.68 (s, 3H)·13C NMR (150 MHz, CDCl3) δ 170.44, 157.72, 152.96, 147.78, 140.21, 139.07, 136.45, 136.17, 133.04, 132.84, 132.79, 131.27, 131.19, 130.64, 130.01, 128.67, 128.11, 126.28, 125.12, 121.61, 121.11, 115.96, 63.47, 34.11, 28.36, 20.64. ESI-HRMS [M + H]+ calcd for C28H22BrClNO3: 534.0466, found: 534.0475.
4.1.3.13. (8-(4-bromophenyl)-1-(5-bromopyridin-2-yl)-5-hydroxy-9,10-dihydrophenanthren-9-yl)methyl acetate (B13)
Yellow solid, yield: 20.0%, 1H NMR (600 MHz, CDCl3) δ 8.77 (d, J = 2.0 Hz, 1H), 8.22 (dd, J = 7.2, 1.6 Hz, 1H), 7.90 (d, J = 8.3 Hz, 1H), 7.51 (d, J = 8.3 Hz, 2H), 7.33 (t, J = 8.7 Hz, 3H), 7.14 (d, J = 8.2 Hz, 2H), 6.96 (dd, J = 8.3, 1.8 Hz, 1H), 6.80 (dd, J = 8.3, 2.5 Hz, 1H), 6.40 (s, 1H), 3.69 (dd, J = 10.9, 5.7 Hz, 1H), 3.48 (t, J = 10.4 Hz, 1H), 3.19 (s, 1H), 2.86 (d, J = 3.3 Hz, 2H), 1.69 (s, 3H)·13C NMR (150 MHz, CDCl3) δ 170.41, 157.98, 152.86, 150.05, 140.16, 138.96, 136.53, 133.04, 132.90, 132.84, 131.29, 131.19, 130.09, 128.69, 128.00, 127.13, 126.40, 125.59, 121.59, 121.14, 119.26, 115.93, 63.44, 34.13, 28.38, 20.65. ESI-HRMS [M + Na]+ calcd for C28H21Br2NO3Na: 599.9780, found: 599.9788.
4.1.3.14. (8-(4-bromophenyl)-1-(5-cyanopyridin-2-yl)-5-hydroxy-9,10-dihydrophenanthren-9-yl)methyl acetate (B14)
White solid, yield: 26.9%, 1H NMR (600 MHz, Acetone-d 6) δ 9.05 (s, 1H), 8.94 (d, J = 5.0 Hz, 1H), 8.61 (d, J = 7.6 Hz, 1H), 7.92 (s, 1H), 7.75 (dd, J = 5.0, 1.3 Hz, 1H), 7.61 (d, J = 8.3 Hz, 2H), 7.46–7.41 (m, 2H), 7.32 (d, J = 8.3 Hz, 2H), 7.07–7.03 (m, 2H), 3.66 (dd, J = 10.9, 5.8 Hz, 1H), 3.50 (t, J = 10.4 Hz, 1H), 3.30–3.25 (m, 1H), 2.99–2.90 (m, 2H), 1.63 (s, 3H)·13C NMR (150 MHz, Acetone-d 6) δ 169.34, 160.87, 154.52, 150.32, 140.84, 138.93, 136.66, 133.50, 132.80, 132.15, 131.65, 131.18, 130.09, 129.62, 128.50, 125.88, 125.76, 123.30, 121.16, 120.50, 120.35, 116.61, 115.65, 62.78, 34.13, 27.89, 19.55. ESI-HRMS [M + H]+ calcd for C29H22BrN2O3: 525.0808, found: 525.0822.
4.1.3.15. (8-(4-bromophenyl)-1-(5-formylpyridin-2-yl)-5-hydroxy-9,10-dihydrophenanthren-9-yl)methyl acetate (B15)
White solid, yield: 28.3%, 1H NMR (600 MHz, CDCl3) δ 10.18 (s, 1H), 9.17 (s, 1H), 8.29–8.26 (m, 2H), 7.62 (d, J = 8.0 Hz, 1H), 7.51 (d, J = 8.0 Hz, 2H), 7.43–7.36 (m, 2H), 7.14 (d, J = 7.9 Hz, 2H), 6.97 (d, J = 8.2 Hz, 1H), 6.81 (d, J = 8.2 Hz, 1H), 6.40 (s, 1H), 3.71 (dd, J = 10.8, 5.7 Hz, 1H), 3.49 (t, J = 10.4 Hz, 1H), 3.19 (s, 1H), 2.94–2.86 (m, 2H), 1.66 (s, 3H)·13C NMR (150 MHz, CDCl3) δ 190.36, 170.36, 164.75, 152.84, 151.49, 140.07, 139.26, 136.54, 136.09, 133.25, 133.04, 132.98, 131.32, 131.17, 130.20, 129.67, 128.87, 128.75, 126.51, 124.86, 121.46, 121.20, 115.96, 63.40, 34.11, 28.51, 20.62. ESI-HRMS [M + Na]+ calcd for C29H22BrNO4Na: 550.0624, found: 500.0632.
4.1.3.16. (8-(4-bromophenyl)-5-hydroxy-1-(5-(methylsulfonyl)pyridin-2-yl)-9,10-dihydrophenanthren-9-yl)methyl acetate (B16)
White solid, yield: 39.0%, 1H NMR (600 MHz, Acetone-d 6) δ 9.17 (s, 1H), 9.04 (s, 1H), 8.62 (d, J = 7.6 Hz, 1H), 8.40 (dd, J = 8.2, 2.2 Hz, 1H), 7.81 (d, J = 8.2 Hz, 1H), 7.60 (d, J = 8.2 Hz, 2H), 7.47–7.42 (m, 2H), 7.31 (d, J = 8.3 Hz, 2H), 7.07–7.03 (m, 2H), 3.64 (dd, J = 10.9, 5.9 Hz, 1H), 3.50 (t, J = 10.4 Hz, 1H), 3.29 (s, 4H), 3.00 (d, J = 15.7 Hz, 1H), 2.93 (dd, J = 15.7, 4.5 Hz, 1H), 1.61 (s, 3H). 13C NMR (150 MHz, Acetone-d 6) δ 170.29, 165.03, 155.44, 148.65, 141.80, 140.11, 137.69, 136.53, 136.50, 134.52, 133.92, 133.17, 132.61, 132.15, 131.08, 130.72, 129.51, 126.75, 125.50, 122.11, 121.48, 116.63, 63.79, 44.73, 35.06, 29.01, 20.54. ESI-HRMS [M + H]+ calcd for C29H25BrNO5S: 578.0631, found: 578.0636.
4.1.3.17. (1-(5-acetylpyridin-2-yl)-8-(4-bromophenyl)-5-hydroxy-9,10-dihydrophenanthren-9-yl)methyl acetate (B17)
Yellow solid, yield: 33.8%, 1H NMR (600 MHz, CDCl3) δ 9.27 (s, 1H), 8.33 (d, J = 5.9 Hz, 1H), 8.28 (d, J = 6.5 Hz, 1H), 7.56 (d, J = 8.1 Hz, 1H), 7.50 (d, J = 8.3 Hz, 2H), 7.31 (q, J = 4.7 Hz, 2H), 7.11 (d, J = 8.3 Hz, 2H), 6.91 (d, J = 8.3 Hz, 1H), 6.73 (d, J = 8.3 Hz, 1H), 3.68 (dd, J = 11.0, 5.8 Hz, 1H), 3.47 (t, J = 10.4 Hz, 1H), 3.19–3.14 (m, 1H), 2.91 (dd, J = 15.6, 4.5 Hz, 1H), 2.81 (d, J = 15.6 Hz, 1H), 2.70 (s, 3H), 1.66 (s, 3H). 13C NMR (150 MHz, CDCl3) δ 196.40, 170.45, 163.70, 153.22, 149.11, 140.25, 138.99, 136.36, 136.08, 133.06, 133.04, 132.62, 131.27, 131.19, 130.55, 129.95, 128.88, 128.57, 126.18, 124.46, 121.53, 121.09, 116.04, 63.53, 34.05, 28.46, 26.82, 20.65. ESI-HRMS [M + H]+ calcd for C30H25BrNO4: 542.0961, found: 542.0979.
4.1.3.18. (8-(4-bromophenyl)-1-(5-(dimethylamino)pyridin-2-yl)-5-hydroxy-9,10-dihydrophenanthren-9-yl)methyl acetate (B18)
White solid, yield: 48.1%, 1H NMR (600 MHz, CDCl3) δ 8.24 (s, 1H), 8.18 (d, J = 6.5 Hz, 1H), 7.46 (d, J = 6.5 Hz, 2H), 7.24 (s, 1H), 7.14–7.04 (m, 5H), 6.75 (d, J = 8.2 Hz, 1H), 6.59 (d, J = 8.2 Hz, 1H), 3.61–3.59 (m, 1H), 3.44 (t, J = 10.2 Hz, 1H), 3.06 (s, 7H), 2.85 (d, J = 13.0 Hz, 1H), 2.70 (d, J = 15.5 Hz, 1H), 1.66 (s, 3H). 13C NMR (150 MHz, CDCl3) δ 170.48, 154.12, 147.61, 145.06, 140.97, 138.80, 136.05, 133.11, 132.78, 132.51, 131.70, 131.34, 131.06, 129.20, 128.22, 127.88, 125.21, 124.50, 122.18, 120.69, 119.67, 116.45, 64.00, 40.19, 34.10, 28.38, 20.69. ESI-HRMS [M + H]+ calcd for C30H28BrN2O3: 543.1278, found: 543.1290.
4.1.3.19. (8-(4-bromophenyl)-5-hydroxy-1-(5-phenylpyridin-2-yl)-9,10-dihydrophenanthren-9-yl)methyl acetate (B19)
White solid, yield:43.8%, 1H NMR (600 MHz, CDCl3) δ 8.99 (s, 1H), 8.25 (d, J = 7.7 Hz, 1H), 8.02 (d, J = 7.6 Hz, 1H), 7.79 (s, 1H), 7.67 (d, J = 7.3 Hz, 2H), 7.56–7.44 (m, 6H), 7.34–7.27 (m, 2H), 7.12 (d, J = 7.5 Hz, 2H), 6.89 (d, J = 8.2 Hz, 1H), 6.75 (d, J = 8.2 Hz, 1H), 3.71–3.65 (m, 1H), 3.49 (t, J = 10.3 Hz, 1H), 3.16 (s, 1H), 2.88 (dd, J = 44.9, 15.4 Hz, 2H), 1.67 (s, 3H). 13C NMR (150 MHz, CDCl3) δ 170.42, 153.45, 140.45, 136.31, 135.19, 133.10, 132.90, 132.42, 131.25, 131.20, 129.77, 129.28, 128.56, 128.45, 128.34, 127.10, 126.02, 124.62, 121.78, 120.97, 116.22, 63.63, 34.10, 28.45, 20.66. ESI-HRMS [M + H]+ calcd for C34H27BrNO3: 576.1169, found: 576.1186.
4.1.3.20. (1-(5-(benzylamino)pyridin-2-yl)-8-(4-bromophenyl)-5-hydroxy-9,10-dihydrophenanthren-9-yl)methyl acetate (B20)
Yellow solid, yield:87.0%, 1H NMR (600 MHz, MeOD) δ 8.45 (d, J = 7.9 Hz, 1H), 7.96 (d, J = 2.7 Hz, 1H), 7.53 (d, J = 8.2 Hz, 2H), 7.38 (d, J = 7.5 Hz, 2H), 7.31 (t, J = 7.7 Hz, 3H), 7.25–7.21 (m, 2H), 7.20–7.16 (m, 3H), 7.07 (dd, J = 8.5, 2.8 Hz, 1H), 6.96 (d, J = 8.3 Hz, 1H), 6.90 (d, J = 8.3 Hz, 1H), 4.40 (d, J = 2.0 Hz, 2H), 3.63 (dd, J = 10.8, 5.4 Hz, 1H), 3.40 (t, J = 10.6 Hz, 1H), 3.17–3.12 (m, 1H), 2.79 (s, 2H), 1.56 (s, 3H). 13C NMR (150 MHz, MeOD) δ 170.80, 154.53, 146.72, 143.93, 140.78, 139.63, 139.11, 136.15, 133.22, 132.28, 131.86, 131.19, 130.91, 129.54, 128.31, 128.23, 127.87, 126.87, 126.73, 125.28, 124.88, 121.46, 120.50, 119.49, 115.06, 62.88, 46.55, 34.31, 27.66, 19.16. ESI-HRMS [M + H]+ calcd for C35H30BrN2O3: 605.1434, found: 605.1443.
4.1.4. General procedures for the synthesis ofC1–C7
NaOH (1 M) water solution (3 mL, 3.00 mmol) and CH3OH (3 mL) were added to the 0.2 mmol corresponding intermediate B2, B6, B10, B11, B12, B14 or B18 and stirred for 2 h. Then the methanol was removed under vacuum, and the residue was acidified to pH = 2 or below with HCl (1 M). Then the solution was extracted with ethyl acetate (20 mL × 3) and the combined organic solvents were dried with sodium sulfate and concentrated in vacuo. The crude compound was purified by silica gel column chromatography to give the desired compounds C1–C7.
4.1.4.1. 1-(4-bromophenyl)-10-(hydroxymethyl)-8-(4-phenylpyridin-2-yl)-9,10-dihydrophenanthren-4-ol (C1)
White solid, yield:78.8%, 1H NMR (600 MHz, MeOD) δ 8.59 (d, J = 5.3 Hz, 1H), 8.56 (d, J = 7.8 Hz, 1H), 7.89 (s, 1H), 7.80 (d, J = 7.3 Hz, 2H), 7.68 (dd, J = 5.3, 1.6 Hz, 1H), 7.54 (d, J = 8.3 Hz, 2H), 7.51 (t, J = 7.4 Hz, 2H), 7.46 (d, J = 7.1 Hz, 1H), 7.38 (t, J = 7.7 Hz, 1H), 7.35 (d, J = 7.5 Hz, 1H), 7.21 (d, J = 8.3 Hz, 2H), 6.95 (d, J = 8.3 Hz, 1H), 6.88 (d, J = 8.3 Hz, 1H), 4.60 (s, 1H), 3.29–3.27 (m, 1H), 3.21 (d, J = 14.3 Hz, 1H), 3.11–3.03 (m, 2H), 2.72 (dd, J = 15.1, 3.4 Hz, 1H). 13C NMR (150 MHz, MeOD) δ 160.19, 154.61, 149.82, 148.39, 141.03, 139.66, 137.59, 137.47, 133.96, 132.52, 131.79, 131.25, 130.88, 129.60, 129.11, 128.91, 128.01, 126.85, 125.37, 122.50, 121.16, 120.44, 119.88, 114.69, 60.29, 37.69, 26.77. ESI-HRMS [M + H]+ calcd for C32H25BrNO2, 534.1063, found, 534.1078.
4.1.4.2. 2-(8-(4-bromophenyl)-5-hydroxy-9-(hydroxymethyl)-9,10-dihydrophenanthren-1-yl)isonicotinaldehyde (C2)
Colorless oil, yield:37.0%, 1H NMR (600 MHz, CDCl3) δ 10.16 (s, 1H), 8.82 (d, J = 5.0 Hz, 1H), 8.38 (d, J = 7.9 Hz, 1H), 7.96 (s, 1H), 7.72 (d, J = 5.0 Hz, 1H), 7.51 (d, J = 8.2 Hz, 2H), 7.44 (t, J = 7.8 Hz, 1H), 7.32 (d, J = 7.6 Hz, 1H), 7.17 (d, J = 8.2 Hz, 2H), 6.99 (d, J = 8.2 Hz, 1H), 6.83 (d, J = 8.2 Hz, 1H), 6.17 (s, 1H), 3.47–3.40 (m, 2H), 3.27 (t, J = 11.4 Hz, 1H), 3.13–3.11 (m, 1H), 2.55 (dd, J = 14.7, 3.4 Hz, 1H)·13C NMR (150 MHz, CDCl3) δ 191.09, 161.55, 152.96, 149.62, 143.08, 140.11, 138.63, 134.09, 133.22, 133.07, 131.33, 131.12, 130.25, 129.15, 128.31, 126.69, 123.09, 121.55, 121.30, 120.78, 115.30, 60.91, 37.91, 26.88. ESI-HRMS [M + Na]+ calcd for C27H20BrNO3Na: 508.0519, found: 508.0534.
4.1.4.3. 1-(4-Bromophenyl)-10-(hydroxymethyl)-8-(5-methylpyridin-2-yl)phenanthren-4-ol (C3)
White solid, yield:99.0%,1H NMR (600 MHz, CDCl3) δ 8.37 (s, 1H), 8.28 (d, J = 7.9 Hz, 1H), 7.66 (dd, J = 7.9, 1.5 Hz, 1H), 7.50 (d, J = 8.2 Hz, 2H), 7.44 (d, J = 7.9 Hz, 1H), 7.39 (t, J = 7.8 Hz, 1H), 7.25 (s, 1H), 7.17 (d, J = 8.2 Hz, 2H), 6.97 (d, J = 8.2 Hz, 1H), 6.82 (d, J = 8.2 Hz, 1H), 6.26 (s, 1H), 3.52 (dd, J = 14.6, 2.6 Hz, 1H), 3.41 (dd, J = 11.6, 5.1 Hz, 1H), 3.27 (t, J = 11.4 Hz, 1H), 3.12–3.07 (m, 1H), 2.50 (dd, J = 14.6, 3.4 Hz, 1H), 2.37 (s, 3H). 13C NMR (150 MHz, CDCl3) δ 156.68, 152.91, 148.35, 140.22, 139.70, 138.89, 138.34, 133.82, 133.18, 133.15, 131.82, 131.26, 131.14, 130.06, 129.11, 127.19, 126.43, 123.76, 121.78, 121.20, 115.17, 60.75, 38.11, 26.96, 18.19. ESI-HRMS [M + H]+ calcd for C27H23BrNO2 + 472.0907, Found 472.0914.
4.1.4.4. 1-(4-bromophenyl)-8-(5-fluoropyridin-2-yl)-10-(hydroxymethyl)-9,10-dihydrophenanthren-4-ol (C4)
White solid, yield:83.0%, 1H NMR (600 MHz, CDCl3) δ 8.42 (d, J = 2.3 Hz, 1H), 8.30 (d, J = 7.9 Hz, 1H), 7.60–7.53 (m, 2H), 7.52 (d, J = 8.2 Hz, 2H), 7.41 (t, J = 7.8 Hz, 1H), 7.24 (s, 1H), 7.16 (d, J = 8.2 Hz, 2H), 7.00 (d, J = 8.2 Hz, 1H), 6.84 (d, J = 8.2 Hz, 1H), 5.73 (s, 1H), 3.46–3.37 (m, 2H), 3.23 (t, J = 11.4 Hz, 1H), 3.10 (dd, J = 7.0, 3.8 Hz, 1H), 2.52 (dd, J = 14.7, 3.4 Hz, 1H). 13C NMR (150 MHz, CDCl3) δ 152.65, 140.09, 138.81, 136.62, 136.46, 133.83, 133.39, 133.23, 131.33, 131.11, 130.20, 129.14, 127.53, 126.55, 125.38, 124.67, 121.65, 121.31, 115.26, 60.86, 37.99, 26.80. 19F NMR (565 MHz, CDCl3) δ −128.27. ESI-HRMS [M + H]+ calcd for C26H20BrFNO2: 476.0656, found: 476.0667.
4.1.4.5. 1-(4-bromophenyl)-8-(5-chloropyridin-2-yl)-10-(hydroxymethyl)-9,10-dihydrophenanthren-4-ol (C5)
White solid, yield:84.4%, 1H NMR (600 MHz, CDCl3) δ 8.52 (d, J = 2.3 Hz, 1H), 8.32 (d, J = 7.9 Hz, 1H), 7.83 (dd, J = 8.3, 2.5 Hz, 1H), 7.50 (t, J = 8.2 Hz, 3H), 7.41 (t, J = 7.8 Hz, 1H), 7.25 (d, J = 8.1 Hz, 1H), 7.16 (d, J = 8.3 Hz, 2H), 6.99 (d, J = 8.2 Hz, 1H), 6.82 (d, J = 8.3 Hz, 1H), 6.02 (s, 1H), 5.27 (s, 1H), 3.45 (dd, J = 14.8, 2.7 Hz, 1H), 3.39 (s, 1H), 3.24 (t, J = 11.4 Hz, 1H), 3.13–3.09 (m, 1H), 2.53 (dd, J = 14.8, 2.5 Hz, 1H). 13C NMR (150 MHz, CDCl3) δ 157.82, 152.83, 147.15, 140.10, 138.71, 138.58, 137.54, 133.93, 133.27, 133.11, 131.32, 131.11, 130.81, 130.23, 129.03, 127.89, 126.59, 125.20, 121.59, 121.29, 115.26, 60.88, 37.93, 26.84. ESI-HRMS [M + H]+ calcd for C26H20BrClNO2: 492.0360, found: 492.0373.
4.1.4.6. 6-(8-(4-bromophenyl)-5-hydroxy-9-(hydroxymethyl)-9,10-dihydrophenanthren-1-yl)nicotinonitrile (C6)
White solid, yield:80.0%, 1H NMR (600 MHz, MeOD) δ 8.93 (s, 1H), 8.58 (d, J = 7.7 Hz, 1H), 8.23 (d, J = 8.1 Hz, 1H), 7.79 (d, J = 8.2 Hz, 1H), 7.54 (d, J = 8.0 Hz, 2H), 7.42–7.34 (m, 2H), 7.21 (d, J = 8.0 Hz, 2H), 6.96 (d, J = 8.3 Hz, 1H), 6.88 (d, J = 8.3 Hz, 1H), 3.25 (dd, J = 10.3, 4.1 Hz, 1H), 3.09 (d, J = 15.3 Hz, 1H), 3.02 (s, 1H), 2.97 (t, J = 10.6 Hz, 1H), 2.76 (dd, J = 15.3, 3.7 Hz, 1H) ppm. 13C NMR (150 MHz, MeOD) δ 163.20, 154.58, 151.25, 140.95, 140.17, 138.47, 137.36, 134.16, 132.81, 131.76, 131.23, 130.90, 129.75, 127.97, 125.48, 124.84, 120.84, 120.47, 116.28, 114.71, 107.97, 60.28, 37.61, 26.88.ESI-HRMS [M + H]+ calcd for C27H20BrN2O2: 483.0703, found: 483.0724.
4.1.4.7. 1-(4-bromophenyl)-8-(5-(dimethylamino)pyridin-2-yl)-10-(hydroxymethyl)-9,10-dihydrophenanthren-4-ol (C7)
White solid, yield:81.3%, 1H NMR (600 MHz, CDCl3) δ 8.21 (d, J = 7.8 Hz, 1H), 8.04 (d, J = 2.9 Hz, 1H), 7.50 (d, J = 8.2 Hz, 2H), 7.37 (dd, J = 18.0, 8.3 Hz, 2H), 7.25 (d, J = 7.7 Hz, 1H), 7.17 (d, J = 8.2 Hz, 2H), 7.13 (dd, J = 8.7, 2.9 Hz, 1H), 6.97 (d, J = 8.2 Hz, 1H), 6.84 (d, J = 8.2 Hz, 1H), 3.57 (d, J = 14.2 Hz, 1H), 3.40 (dd, J = 11.6, 5.1 Hz, 1H), 3.26 (t, J = 11.4 Hz, 1H), 3.13–3.07 (m, 1H), 2.99 (s, 6H), 2.49 (dd, J = 14.5, 3.3 Hz, 1H). 13C NMR (150 MHz, CDCl3) δ 152.85, 144.97, 140.30, 139.02, 133.71, 133.30, 133.19, 131.24, 131.16, 129.94, 129.09, 126.37, 124.07, 121.95, 121.17, 115.18, 60.73, 40.06, 38.23, 27.07 ESI-HRMS [M + H]+ calcd for C28H26BrN2O2: 501.1172, found: 501.1180.
4.2. Biological evaluations
4.2.1. Chemicals and reagents
SARS-CoV-2 3CLpro and SARS-CoV 3CLpro were expressed and purified by ourselves. All tested compounds were dissolved in DMSO and stored at 4 °C. Dabcyl-KNSTLQSGLRKE-Edans is the fluorescent probe for SARS-CoV-2 3CLpro inhibitory assessments [31,36], which was ordered from Shanghai Sangon Biological Engineering & Technology and Service Co. Ltd. (Shanghai, China). Ethylene Diamine Tetraacetic Acid (EDTA), disulfiram (the positive inhibitor), β-NADP+, d-glucose-6-phosphate (G-6-P), glucose-6-phosphate dehydrogenase (G-6-PDH), testosterone, phenacetin, coumarin, paclitaxel, omeprazole, dextromethorphan, chlorzoxazone, pancreatin, as well as pepsin were provided by Dalian Meilun Biotechnology Co. LTD. (Dalian, China). Diclofenac was obtained from Ark Pharm (Wuhan, China). Lansoprazole was purchased from Hairong (Sichuan, China). KH2PO4, MgCl2, NaOH and HCl were bought from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). Uridine-50-diphosphoglucuronic acid (UDPGA), tris mefenamic acid and dodecyl polyglycol ether were purchased from Sigma-Aldrich (St. Louis, MO, USA). 7-hydroxycoumarin was gained from AlfaAesar (Shanghai, China). Warfarin sodium was purchased from J&K Scientific (Beijing, China). The pooled human liver microsomes (Lot No. H0610) from 50 individual donors were supplied by XenoTech (USA). The pooled human plasma samples were provided by Putuo Hospital, Shanghai University of Traditional Chinese Medicine (Shanghai, China), with the reference number of RTEC-A-2020-59-1. The stock solution of the fluorescent substrate, PBS buffer, Tris-HCl buffer were prepared by Millipore water (Millipore, Bedford, USA) and stored at 4 °C for further use. HPLC grade DMSO (Tedia, USA) was used throughout.
4.2.2. 3CLpro inhibitory assays
The SARS-CoV-2 3CLpro and SARS-CoV 3CLpro inhibitory assays were conducted according to the reported procedure [31,36], which were established by the 3CLpro-mediated Dabcyl-KNSTLQSGLRKE-Edans hydrolytic reaction. In brief, the tested compound, SARS-CoV-2 3CLpro (4 μg/mL, final concentration) or SARS-CoV 3CLpro (30 μg/mL, final concentration) were added into a black 96-well plate that filled with PBS buffer (0.1 M, pH 7.4, 1 mM EDTA) to co-incubate at 37 °C for 30 min. After the peptide-like substrate (20 μM or 40 μM, final concentration) was added to initiate the reaction, the produced fluorescent values were continuously detected by a microplate reader (SpectraMax® iD3, Molecular Devices, Austria) for 20 min at the excitation wavelength of 340 nm and emission wavelength of 490 nm.
4.2.3. Determination of inhibition constants of C1and C2
The inhibition constants (K i) and inhibition modes for C1 and C2 against SARS-CoV-2-3CLpro-mediated-Dabcyl-KNSTLQSGLRKE-Edans cleavage were determined by inhibition kinetics. According to the previously reported methodology [41,42], increasing concentrations of C1 and C2 were separately co-incubated with various concentrations of Dabcyl-KNSTLQSGLRKE-Edans, and the generated fluorescence signals were analyzed to determine the inhibition modes and the K i values.
4.2.4. Gastrointestinal stability assays
The simulated intestinal fluid (SIF) was confected by pancreatin (400 mg), KH2PO4 (272 mg), as well as NaOH (0.4%) in 40 mL water, with the pH value of 6.8. The simulated gastric fluid (SGF) was prepared by water (40 mL) that contained pepsin (400 mg) and HCl (656 μL, 1 M). The gastrointestinal metabolic system (100 μL) included SIF or SGF, and the analyte (20 μM). After the reaction was proceed for 0 min, 5 min, 10 min, 15 min, 20 min, 30 min, 45 min, 60 min, 90 min at 37 °C, equal volume of pre-cooling acetonitrile (100 μL) was added to quench the reaction. Then the samples were centrifuged at 20,000×g, 4 °C for 30 min, 100 μL of which were transferred for LC-UV analysis.
4.2.5. Plasma stability assays
The stabilities of C1 and C2 were proceed in PBS buffer (100 mM, pH 7.4) at 37 °C. After the co-incubation was proceed for 0 min, 15 min, 30 min, 1 h, 2 h, 4 h, 6 h, 8 h, 10 h, 12 h, the pre-cooling acetonitrile (300 μL) was added to precipitate the proteins. Then the samples were centrifuged at 20,000×g, 4 °C for 30 min, 100 μL of which were transferred for LC-UV analysis [43].
4.2.6. Metabolic stability assays
The phase I metabolic assessments were proceed in PBS buffer (100 mM, pH 7.4) that comprised with C1 or C2 or testosterone (20 μM), HLMs (0.2 μg/mL), as well as specific cofactors (4 mM MgCl2, 10 mM G-6-P, 1 unit/mL G-6-PDH). After equilibrated at 37 °C for 3 min, the samples were initiated by β-NADP+ (1 mM). While the phase Ⅱ metabolic assessments were conducted in Tris-HCl buffer (50 mM, pH 7.4), which contained C1 or C2 or 7-Hydroxycoumarin (20 μM), HLMs (0.2 mg/mL), as well as MgCl2 (5 mM). The reaction mixture was incubated at 37 °C for 3 min, then added with UDPGA (2 mM) to initiate the reaction. The reactions of phase I and phase Ⅱ metabolic reactions were quenched by pre-cooling acetonitrile (100 μL) after the reactions proceed for different times (0 min, 5 min, 15 min, 30 min, 45 min, 60 min, 90 min). All the quenched mixtures were centrifuged at 20,000×g, 4 °C for 30 min, the supernatant was transferred for LC-UV analysis [42].
4.3. Molecular docking
Molecular docking study was conducted using Schrodinger software package. Firstly, the SARS-CoV-2 3CLpro was downloaded from Protein Data Bank (PDB code: 7BE7) [44] and prepared with the Protein Preparation Wizard model including the removal of waters, the addition of missing hydrogen atoms, the generation of heteroatom states using Epik: pH 7.0 ± 2.0 and the assignment of bond order. Then, a restrained minimization to relieve static clashes was conducted using the OPLS-2005 force field (converge heavy atoms to RMSD 0.30 Å). For the docking study at the dimer interface position, the chain B should be deleted except for residues 1–8. Then, the receptor grid was generated at the centroid of residues 1–8 and the grid box size was set to 20 Å × 20 Å × 20 Å. For the docking study at the substrate-binding pocket, the receptor grid was generated at the centroid of ligand and the grid box size was set to 20 Å × 20 Å × 20 Å. After preparing ligands using LigPrep module, ligand docking was carried out using the standard precision (SP) with the default settings. Finally, the pictures were generated using pymol software.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
Financial support was generously provided by the National Natural Science Foundation of China (Nos.: 81903423, 21871184, 22071155, 81922070 and 81973286), the Shanghai Sailing Program (No: 19YF1449300), the Shanghai Municipal Education Commission (No: 2019-01-07-00-10-E00072), the Science and Technology Commission of Shanghai Municipality (Nos.: 18401933500 and 20400750300), Shanghai Science and Technology Innovation Action Plan biomedical technology support special project (No: 21S21900600), the Innovation Team and Talents Cultivation Program of National Administration of Traditional Chinese Medicine (No: ZYYCXTD-D-202004).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejmech.2021.114030.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Otto S.P., Day T., Arino J., Colijn C., Dushoff J., Li M., Mechai S., Domselaar G.V., Wu J.H., Earn D.J.D., Ogden N.H. The origins and potential future of SARS-CoV-2 variants of concern in the evolving COVID-19 pandemic. Curr. Biol. 2021;31:R918–R929. doi: 10.1016/j.cub.2021.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma A., Ahmad Farouk I., Lal S.K. COVID-19: a review on the novel coronavirus disease evolution, transmission, detection, control and prevention. Viruses. 2021;13:202. doi: 10.3390/v13020202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu T., Kang S.T., Peng W.Y., Zuo C.Z., Zhu Y.H., Pan L.Y., Fu K.Y., You Y.X., Yang X.Y., Luo X., Jiang L.P., Deng M.C. Original hosts, clinical features, transmission routes, and vaccine development for coronavirus disease (COVID-19) Front. Med. 2021;8:702066. doi: 10.3389/fmed.2021.702066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Awadasseid A., Wu Y., Tanaka Y., Zhang W. Current advances in the development of SARS-CoV-2 vaccines. Int. J. Biol. Sci. 2021;17:8–19. doi: 10.7150/ijbs.52569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cihan P. Forecasting fully vaccinated people against COVID-19 and examining future vaccination rate for herd immunity in the US, Asia, Europe, Africa, South America, and the World. Appl. Soft Comput. 2021;111:107708. doi: 10.1016/j.asoc.2021.107708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Badua C.L.D.C., Baldo K.A.T., B Medina P.M. Genomic and proteomic mutation landscapes of SARS-CoV-2. J. Med. Virol. 2021;93:1702–1721. doi: 10.1002/jmv.26548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scudellari M. How the coronavirus infects cells - and why Delta is so dangerous. Nature. 2021;595:640–644. doi: 10.1038/d41586-021-02039-y. [DOI] [PubMed] [Google Scholar]
- 8.Singh J., Rahman S.A., Ehtesham N.Z., Hira S., Hasnain S.E. SARS-CoV-2 variants of concern are emerging in India. Nat. Med. 2021;7:1131–1133. doi: 10.1038/s41591-021-01397-4. [DOI] [PubMed] [Google Scholar]
- 9.Liu C., Ginn H.M., Dejnirattisai W., Supasa P., Wang B., Tuekprakhon A., Nutalai R., Zhou D., Mentzer A.J., Zhao Y.G., Duyvesteyn H.M.E., López-Camacho C., Slon-Campos J., Walter T.S., Skelly D., Johnson S.A., Ritter T.G., Mason C., Clemens S.A.C., Naveca F.G., Nascimento V., Nascimento F., Costa C.F.D., Resende P.C., Pauvolid-Correa A., Siqueira M.M., Dold C., Temperton N., Dong T., Pollard A.J., Knight J.C., Crook D., Lambe T., Clutterbuck E., Bibi S., Flaxman A., Bittaye M., Belij-Rammerstorfer S., Gilbert S.C., Malik T., Carroll M.W., Klenerman P., Barnes E., Dunachie S.J., Baillie V., Serafin N., Ditse Z., Silva K.D., Paterson N.G., Williams M.A., Hall D.R., Madhi S., Nunes M.C., Goulder P., Fry E.E., Mongkolsapaya J., Ren J., Stuart D.I., Screaton G.R. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. 2021;184:4220–4236. doi: 10.1016/j.cell.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez Bernal J., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., Stowe J., Tessier E., Groves N., Dabrera G., Myers R., Campbell C.N.J., Amirthalingam G., Edmunds M., Zambon M., Brown K.E., Hopkins S., Chand M., Ramsay M. Effectiveness of Covid-19 vaccines against the B.1.617.2 (Delta) varian. N. Engl. J. Med. 2021;358:585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shamsi A., Mohammad T., Anwar S., Amani S., Khan M.S., Husain F.M., Rehman M.T., Islam A., Hassan M.I. Potential drug targets of SARS-CoV-2: from genomics to therapeutics. Int. J. Biol. Macromol. 2021;177:1–9. doi: 10.1016/j.ijbiomac.2021.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pillaiyar T., Meenakshisundaram S., Manickam M. Recent discovery and development of inhibitors targeting coronaviruses, Drug Discov. Today Off. 2020;25:668–688. doi: 10.1016/j.drudis.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q., Kang C. Progress in developing inhibitors of SARS-CoV-2 3C-Like protease. Microorganisms. 2020;8:1250. doi: 10.3390/microorganisms8081250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu R.J., Zhao X., Li J., Niu P.H., Yang B., Wu H.L., Wang W.L., Song H., Huang B.Y., Zhu N., Bi Y.H., Ma X.J., Zhan F.X., Wang L., Hu T., Zhou H., Hu Z.H., Zhou W.M., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J.Y., Xie Z.H., Ma J.M., Liu W.J., Wang D.Y., Xu W.B., Holmes E.C., Gao G.F., Wu G., Chen W.J., Shi W.F., Tan W.J. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deshmukh M.G., Ippolito J.A., Zhang C.H., Stone E.A., Reilly R.A., Miller S.J., Jorgensen W.L., Anderson K.S. Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease. Structure. 2021;8:823–833. doi: 10.1016/j.str.2021.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.R. Cannalire, C. Cerchia, A.R. Beccari, F.S.D. Leva, V. Summa, Targeting SARS-CoV-2 proteases and polymerase for COVID-19 treatment: state of the art and future opportunities, J. Med. Chem. doi: 10.1021/acs.jmedchem.0c01140. [DOI] [PMC free article] [PubMed]
- 17.Shagufta I. Ahmad. The race to treat COVID-19: potential therapeutic agents for the prevention and treatment of SARS-CoV-2. Eur. J. Med. Chem. 2021;213:113157. doi: 10.1016/j.ejmech.2021.113157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y.Z., Liang C.Y., Xin L., Ren X.D., Tian L., Ju X.K., Li H., Wang Y.B., Zhao Q.Q., Liu H., Cao W.Q., Xie X.L., Zhang D.Z., Wang Y., Jian Y.L. The development of Coronavirus 3C-Like protease (3CLpro) inhibitors from 2010 to 2020. Eur. J. Med. Chem. 2020;206:112711. doi: 10.1016/j.ejmech.2020.112711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vita E.D. 10 years into the resurgence of covalent drugs. Future Med. Chem. 2021;13:193–210. doi: 10.4155/fmc-2020-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiong Y., Zhu G.H., Zhang Y.N., Hu Q., Wang H.N., Yu H.N., Qin X.Y., Guan X.Q., Xiang Y.W., Tang H., Ge G.B. Flavonoids in Ampelopsis grossedentata as covalent inhibitors of SARS-CoV-2 3CLpro: inhibition potentials, covalent binding sites and inhibitory mechanisms. Int. J. Biol. Macromol. 2021;187:976–987. doi: 10.1016/j.ijbiomac.2021.07.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verma N., Henderson J.A., Shen J. Proton-coupled conformational activation of SARS coronavirus main proteases and opportunity for designing small-molecule broad-spectrum targeted covalent inhibitors. J. Am. Chem. Soc. 2020;142:21883–21890. doi: 10.1021/jacs.0c10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 23.Zhang L.L., Lin D.Z., Sun X.Y.Y., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.(a) Dai W.H., Zhang B., Jiang X.M., Su H.X., Li J., Zhao Y., Xie X., Jin Z.M., Peng J.J., Liu F.J., Li C.P., Li Y., Bai F., Wang H.F., Cheng X., Cen X.B., Hu S.L., Yang X.N., Wang J., Liu X., Xiao G.F., Jiang H.L., Rao Z.H., Zhang L.K., Xu Y.C., Yang H.T., Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Qiao J.X., Li Y.S., Zeng R., Liu F.L., Luo R.H., Huang C., Wang Y.F., Zhang J., Quan B.X., Shen C.J., Mao X., Liu X.L., Sun W.N., Yang W., Ni X.C., Wang K., Xu L., Duan Z.L., Zou Q.C., Zhang H.L., Qu W., Long Y.H.P., Li M.H., Yang R.C., Liu X.L., You J., Zhou Y.L., Yao R., Li W.P., Liu J.M., Chen P., Liu Y., Lin G.F., Yang X., Zou J., Li L.L., Hu Y.G., Lu G.W., Li W.M., Wei Y.Q., Zheng Y.T., Lei J., Yang S.Y. SARS-CoV-2 Mpro inhibitors with antiviral activity in a transgenic mouse model. Science. 2021;371:1374–1378. doi: 10.1126/science.abf1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y.M., Szeto T., Zhang X.J., Tarbet B., Marty M.T., Chen Y., Wang J. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boras B., Jones R.M., Anson B.J., Arenson D., Aschenbrenner L., Bakowski M.A., Beutler N., Binder J., Chen E., Eng H., Hammond H., Hammond J., Haupt R.E., Hoffman R., Kadar E.P., Kania R., Kimoto E., Kirkpatrick M.G., Lanyon L., Lendy E.K., Lillis J.R., Logue J., Luthra S.A., Ma C., Mason S.W., McGrath M.E., Noell S., Obach R.S., O'Brien M.N., O'Connor R., Ogilvie K., Owen D., Pettersson M., Reese M.R., Rogers T.F., Rossulek M.I., Sathish J.G., Shirai N., Steppan C., Ticehurst M., Updyke L.W., Weston S., Zhu Y., Wang J., Chatterjee A.K., Mesecar A.D., Frieman M.B., Anderson A.S., Allerton C. Discovery of a novel inhibitor of coronavirus 3CL protease as a clinical candidate for the potential treatment of COVID-19. BioRxiv. 2020 doi: 10.1101/2020.09.12.293498. [DOI] [Google Scholar]
- 27.Pfizer Unveils its Oral SARS-CoV-2 Inhibitor. 2021. https://cen.acs.org/acs-news/acs-meeting-news/Pfizerunveils-oral-SARS-CoV/99/i13 Available from: [Google Scholar]
- 28.Su H.X., Yao S., Zhao W.F., Zhang Y.M., Liu J., Shao Q., Wang Q.X., Li M.J., Xie H., Shang W.J., Ke C.Q., Feng L., Jiang X.R., Shen J.S., Xiao G.F., Jiang H.L., Zhang L.K., Ye Y., Xu Y.C. Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease. Nat. Commun. 2021;12:3623. doi: 10.1038/s41467-021-23751-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang R.Y., Hu Q., Wang H.N., Zhu G.H., Wang M.G., Zhang Q., Zhao Y.S., Li C.Y., Zhang Y.N., Ge G.B., Chen H.Z., Chen L.L. Identification of Vitamin K3 and its analogues as covalent inhibitors of SARS-CoV-2 3CL. 2021;183:182–192. doi: 10.1016/j.ijbiomac.2021.04.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.A.K. Ghosh, J. Raghavaiah, D. Shahabi, M. Yadav, B.J. Anson, E.K. Lendy, S. Hattori, N. Higashi-Kuwata, H. Mitsuya, A.D. Mesecar, Indole chloropyridinyl ester-derived SARS-CoV-2 3CLpro inhibitors: enzyme inhibition, antiviral efficacy, structure-activity relationship, and X-ray structural studies, J. Med. Chem. doi:10.1021/acs.jmedchem.1c01214. [DOI] [PMC free article] [PubMed]
- 31.Xiong Y., Zhu G.H., Wang H.N., Hu Q., Chen L.L., Guan X.Q., Li H.L., Chen H.Z., Tang H., Ge G.B. Discovery of naturally occurring inhibitors against SARS-CoV-2 3CL from Ginkgo biloba leaves via large-scale screening. Fitoterapia. 2021;152:104909. doi: 10.1016/j.fitote.2021.104909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.N. Kitamura, M.D. Sacco, C. Ma, Y.M. Hu, J.A. Townsend, X.Z. Meng, F.S. Zhang, X.J. Zhang, M. Ba, T. Szeto, A. Kukuljac, M.T. Marty, D. Schultz, S. Cherry, Y. Xiang, Y. Chen, J. Wang, Expedited approach toward the rational design of noncovalent SARS-CoV-2 main protease inhibitors, J. Med. Chem.. doi: 10.1021/acs.jmedchem. 1C00509. [DOI] [PMC free article] [PubMed]
- 33.Han S.H., M Goins C., Arya T., Shin W.J., Maw J., Hooper A., Sonawane D.P., Porter M.R., Bannister B.E., Crouch R.D., Lindsey A.A., Lakatos G., Martinez S.R., Alvarado J., Akers W.S., Wang N.S., Jung J.U., Macdonald J.D., Stauffer S.R. Structure-based optimization of ML300-derived, noncovalent inhibitors targeting the severe acute respiratory syndrome coronavirus 3CL protease (SARS-CoV-2 3CLpro) J. Med. Chem. 2021 doi: 10.1021/acs.jmedchem.1c00598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang C.H., Stone E.A., Deshmukh M., Ippolito J.A., Ghahremanpour M.M., Tirado-Rives J., Spasov K.A., Zhang S., Takeo Y., Kudalkar S.N., Liang Z., Isaacs F., Lindenbach B., Miller S.J., Anderson K.S., Jorgensen W.L. Potent noncovalent inhibitors of the main protease of SARS-CoV-2 from molecular sculpting of the drug perampanel guided by free energy perturbation calculations. ACS Cent. Sci. 2021;7:467–475. doi: 10.1021/acscentsci.1c00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erak M., Bellmann-Sickert K., Els-Heindl S., Beck-Sickinger A.G. Peptide chemistry toolbox – transforming natural peptides into peptide therapeutics. Bioorg. Med. Chem. 2018;26:2759–2765. doi: 10.1016/j.bmc.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 36.Chen L.L., Chen S., Gui C., Shen J., Shen X., Jiang H. Discovering severe acute respiratory syndrome coronavirus 3CL protease inhibitors: virtual screening, surface plasmon resonance, and fluorescence resonance energy transfer assays. J. Biomol. Screen. 2006;118:915–921. doi: 10.1177/1087057106293295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao D.D., Liu X.Y., Xu H., Tan Y.X., Liao Q., Li Q.H., Yang X.D., Lin G.Q., Tian P. One-pot preparation of 9,10-dihydrophenanthrenes initiated by rhodium (III)-catalyzed C−H activation and relay Diels-Alder reaction. Org. Lett. 2020;22:4300–4305. doi: 10.1021/acs.orglett.0c01339. [DOI] [PubMed] [Google Scholar]
- 38.Goyal B., Goyal D. Targeting the dimerization of the main protease of coronaviruses: a potential broad-spectrum therapeutic strategy. ACS Comb. Sci. 2020;22:297–305. doi: 10.1021/acscombsci.0c00058. [DOI] [PubMed] [Google Scholar]
- 39.Paterson A.J., Heron C.J., McMullin C.L., Mahon M.F., Pressc N.J., Frost C.G. α-Halo carbonyls enable meta selective primary, secondary and tertiary C-H alkylations by ruthenium catalysis. Org. Biomol. Chem. 2017;15:5993–6000. doi: 10.1039/c7ob01192j. [DOI] [PubMed] [Google Scholar]
- 40.Brunner F., Marinakis N., Wobill C. Modular synthesis of simple cycloruthenated complexes with state-of-the-art performance in p-type DSCs. J. Mater. Chem. C. 2016;4:9823–9833. [Google Scholar]
- 41.Song Y.Q., Weng Z.M., Finel M., Wang Y.Q., Ding L.L., Jin Q., Wang D.D., Fang S.Q., Cao Y.F., Hou J., Ge G.B. Inhibition of human carboxylesterases by magnolol: kinetic analyses and mechanism. Chem. Biol. Interact. 2019;308:339–349. doi: 10.1016/j.cbi.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 42.Huo P.C., Guan X.Q., Liu P., Song Y.Q., Sun M.R., He R.J., Zou L.W., Xue L.J., Shi J.H., Zhang N., Liu Z.G., Ge G.B. Design, synthesis and biological evaluation of indanone-chalcone hybrids as potent and selective hCES2A inhibitors. Eur. J. Med. Chem. 2021;209:112856. doi: 10.1016/j.ejmech.2020.112856. [DOI] [PubMed] [Google Scholar]
- 43.Chung T.D.Y., Terry D.B., Smith L.H. 2004. In Vitro and in Vivo Assessment of ADME and PK Properties during Lead Selection and Lead Optimization – Guidelines, Benchmarks and Rules of Thumb; pp. 1307–1320.https://www.ncbi.nlm.nih.gov/books/NBK326710/ Bethesda (MD) [PubMed] [Google Scholar]
- 44.Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. https://www.rcsb.org/structure/7BE7 Available from.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.