Abstract
Despite the scientific advances observed in the recent decades and the emergence of new methodologies, the diagnosis of systemic fungal infections persists as a problematic issue. Fungal cultivation, the standard method that allows a proven diagnosis, has numerous disadvantages, as low sensitivity (only 50% of the patients present positive fungal cultures), and long growth time. These are factors that delay the patient's treatment and, consequently, lead to higher hospital costs. To improve the accuracy and quickness of fungal infections diagnosis, several new methodologies attempt to be implemented in clinical microbiology laboratories. Most of these innovative methods are independent of pathogen isolation, which means that the diagnosis goes from being considered proven to probable. In spite of the advantage of being culture-independent, the majority of the methods lack standardization. PCR-based methods are becoming more and more commonly used, which has earned them an important place in hospital laboratories. This can be perceived now, as PCR-based methodologies have proved to be an essential tool fighting against the COVID-19 pandemic. This review aims to go through the main steps of the diagnosis for systemic fungal infection, from diagnostic classifications, through methodologies considered as “gold standard”, to the molecular methods currently used, and finally mentioning some of the more futuristic approaches.
Keywords: Fungal infections diagnosis, Proven diagnosis, Probable diagnosis, Gold standard methodologies, PCR-based methodologies
1. Introduction
Throughout the years the estimated number of fungal species around the world had an explosive growth. In 2015, based on morphological, physiological, and molecular characteristics, this estimation reached 100,000 fungal species [1]. In the same year, the International Code of Nomenclature (ICN) reported that about 1000 to 1500 fungal species were being described and identified every year. By 2020, the number of identified fungal species was 140,000, representing only 12 to 1% of the estimated fungal species present on Earth [2]. Despite the high number of described fungal species, it is estimated that over 700 species are associated with humans, either as commensal or as pathogenic [[3], [4], [5]].
During the past decade, advances in molecular phylogenetic approaches for fungal identification led to significant changes in fungal taxonomy/nomenclature. Borman et al. [6] addressed the recent taxonomy updates for clinically relevant fungi, that comprise new and often cryptic species in fungal genera such as Aspergillus and Fusarium, and also revised names for existing species, like Pichia kudriavzevii (previously Candida krusei). Although C. krusei has been renamed, since clinicians remain reluctant to use it and the majority of the commercially available kits still mentions C. krusei, we will indicate the commercially described name.
Nowadays, people benefit from the progression of medicine, providing an increase in the average life expectancy, as well as the improvement of treatments for various diseases. However, the development of medicine also increased the susceptibility of humans to infections, including fungal infections, especially due to the use of immunosuppressive therapies. These infections, whether caused by opportunistic fungi or by primary pathogens, are divided into superficial mycoses, allergic diseases and mycoses with an invasive character [7,8]. Fungal infections continue to be undervalued and underestimated both by the population and by public health organizations. Diseases caused by protozoa, bacteria and viruses have been recognized as a public health issue over the centuries, but systemic fungal infections were only considered as a relevant issue since the 1980′s [9].
The Global Action Fund for Fungal Infections reported that, annually, more than 300 million people suffer from systemic fungal infections and, from these, about 1.5 million succumb to a fungal infection [10]. The most prevalent fungal pathogens in underdeveloped countries are Cryptococcus spp. and Pneumocystis spp., generally associated with AIDS, but in developed countries, the most frequently diagnosed invasive infections are those caused by Candida spp., Aspergillus spp. and also Cryptococcus spp [11]. Blastomyces, Histoplasma, Paracoccidioides, and Coccidioides are endemic fungi that can cause localized infections, yet they can progress into systemic and have much more severe clinical implications in high-risk patients. Most fungi in this endemic group are dimorphic, growing in the infectious mould form in the environment but switching to a yeast or other form in tissues to produce infection. They differ from the opportunistic fungi in their ability to cause disease in previously healthy persons, but the most serious disseminated disease still occurs in immunocompromised individuals [12].
In 2021 there has been an increased concern related to the COVID-19 pandemic caused by the SARS-CoV-2 virus. According to Sharma and co-workers [13], infection by SARS-CoV-2 leads to a decrease in T cells, namely CD4+T and CD8+T, resulting in a debilitated immune system that makes the patients more susceptible to contracting fungal infections. One of the fungal orders commonly linked to post-COVID-19 infections is Mucorales, being Rhizopus arrhizus responsible for most cases of mucormycosis worldwide [14,15], followed by Rhizomucor pusillus, Apophysomyces, variabilis and Lichtheimia corymbifera [16]. Between 5th May and 3rd August of 2021, approximately 47,508 cases and 4425 deaths of mucormycosis as COVID-19 associated infection, were reported in India [17]. Also during that period, the admission of patients to intensive care units with COVID-19 associated mucormycosis, increased from 2 per month to 600 per month [18]. These high rates of incidence and mortality, led the Central Government of India to declare it as an epidemic [18]. Mucormycosis combined with COVID-19 infection results in a more aggressive fungal infection, thus linked to higher mortality rates. Those can be due to the overload of the health system, poor hygiene in healthcare environments, late diagnosis, and the weakened patient's immune system that results in more critical fungal infections [19]. Specific glucocorticoids steroids, as methylprednisolone and dexamethasone, that are being administrated to COVID-19 patients are linked to immunosuppressive effects [20], as well as rising of blood sugar levels [21], predisposing patients to fungal infections.
The COVID-19 pandemic might have increased the transmission of other nosocomial fungal infections, like those caused by Candida auris that is considered a serious global health threat. This emerging pathogen can cause serious invasive infections and even death, particularly in patients with other comorbidities. Since its first identification in 2009, C. auris outbreaks have been reported in several healthcare facilities [22,23], raising major concerns due to its frequent high antifungal resistance and rapid transmission in hospital environments. There are common risk factors for infections caused by SARS-CoV-2 and C. auris, such as diabetes, contact with intubation systems, mechanical ventilation, and exposure to broad-spectrum antibiotics. Therefore, C. auris outbreaks have been reported in COVID-19 intensive care units [[24], [25], [26]]. Bayona and co-workers reported an increase of C. auris candidaemia cases during the pandemic, in a Spanish hospital. From 2019 to march 2021, the 28-day mortality rate for C. auris candidaemia increased from 33.3% to 57.1% [25].
To prevent epidemics, it seems clear that public health organizations need to consider systemic fungal infections as contemporary and a real problem, as has been observed previously in other models of infectious diseases. In addition, since these infections are less known and caused by less-studied pathogens, they represent a greater risk to public health, and should concentrate higher attention [27].
2. Fungal infections diagnosis
The European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group (EORTC) and the Mycoses Study Group – Education & Research Consortium (MSG-ERC) established definitions incorporating the parameters of the diagnosis of fungal infections at a clinical level. Those have been extremely useful for researchers conducting epidemiologic studies, diagnostic assays, and antifungals clinical trials. The 3 levels of classification of Invasive Fungal Infection (IFI) diagnosis are proven, probable, and possible [[28], [29], [30]]. These definitions, established in 2002, only covered the diagnosis of fungal infections related to immunocompromised, oncological, and hematopoietic stem cell transplant patients [28]. But in 2019, a new revision and updating of the consensus definitions established that the proven IFI classification could be applied to any patient (immunocompromised or not) and that the probable and possible classifications were only projected for immunocompromised patients [30].
The proven diagnosis requires the detection of the pathogenic fungi through histopathological or culture methods from sterile sites [29,30]. For the probable and possible diagnosis to be attributed, 3 variables have to be analysed: (i) the host factors, that are related to the patient's risk of contracting a fungal infection, thus several parameters are evaluated, such a recent history of neutropenia, receipt of an allogeneic stem cell transplant, prolonged use of corticosteroids, immunosuppressants therapy, and inherent immunodeficiency; (ii) the clinical signs and symptoms related to the fungal infection, so some clinical manifestations are taken into consideration as tracheobronchitis, sinonasal infection and central nervous system infection; (iii) mycological evidences, accompanied by the positive result of a diagnostic test, either conventional, molecular or imaging [29,30].
A probable diagnosis is attributed to a patient when the parameters of host factors, clinical signs and mycological evidences are present. In this case, when the positive mycological evidence is achieved through molecular methodologies, such as Polymerase Chain Reaction (PCR)-based methods or serological tests, a probable diagnosis is immediately attributed. On the other hand, a possible diagnosis is attributed to patients who meet 2 of the 3 variables; the host factors and clinical signs strongly indicate an invasive fungal infection, but the mycological evidence parameter remains negative [29].
Pathogenic fungi identification can be obtained through several approaches, from traditional fungal cultures to PCR-based methods [29,30]. A variety of tests are available and, preferably, more than one type of test should be applied to the patient if an invasive fungal infection is suspected so that a more effective and robust diagnosis is achieved. Molecular methodologies allowed to significantly reduce the turn-around time, by introducing methodologies that permit to obtain more specific, efficient, fast, and accurate results. This means that the overall diagnosis process is faster, which allows an adequate and timely delineation of the therapeutic plan, increasing the survival rate. This also leads to a reduction of people admitted to intensive care units, which can yield the hospital approximately $30,000 per patient, according to a study developed in the United States [31].
With this review we plan to provide a framework to optimally choose among the existing molecular methods. So, the advantages and disadvantages, time consumption, sensitivity, specificity, and automation of the most frequently used methods will be provided. Since host factors, clinical signs and symptoms are not under the scope of this review, we will focus on methods for mycological evidences. For further analysis on the previous parameters some reviews are available. Zhang and co-workers [32] analysed the clinical characteristics of 145 cases of invasive fungal infections. Webb and colleagues [33], analysed the incidence, clinical features and outcomes of invasive fungal infections in the US health care network, according to 3374 episodes in 3154 patients.
3. Framework of clinical diagnosis
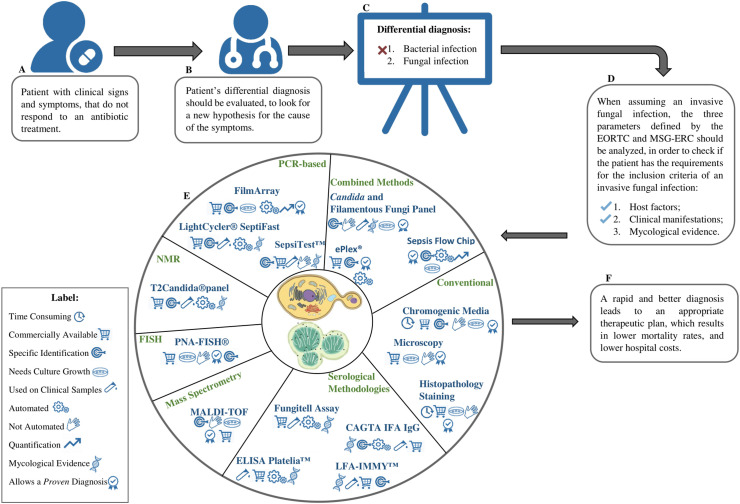
The diagnosis of a fungal infection is a lengthy process, especially due to the symptomatic similarities between a bacterial and fungal infection. Fig. 1 provides a simplified framework reviewing the steps for the diagnosis of systemic fungal infections. When a patient that presents clinical signs of infections does not respond to an antibiotic treatment (Fig. 1A), an invasive fungal infection should be included in the patient's differential diagnosis (Fig. 1B and C). The host factors and clinical signs defined by the EORTC and MSG-ERC are analysed (Fig. 1D) and methodologies available for mycological evidences should be selected accordingly. There are several methodologies available for mycological evidences but it is important to notice that the type of methodology used will dictate the diagnosis classification (Fig. 1E). Methodologies that are capable of detecting the fungal pathogen through histopathological or culture methods from sterile sites provide a proven diagnosis. However, if positive mycological evidence is achieved through molecular methodologies, such as PCR-based methods or serological tests, a probable diagnosis is attributed. Still, it is of utmost importance to achieve specific identifications to establish the most appropriate therapeutic plan, which can lead to higher survival rates and decrease hospital costs (Fig. 1F).
Fig. 1.
Systemic fungal infection diagnosis framework. When a patient does not respond to the antibiotic treatment (A), a systemic fungal infection should be included in the differential diagnosis (B–C). After evaluating the 3 parameters defined by the EORTC and MSG-ERC (D) (host factors, clinical manifestations and mycological evidence), and if there is a strong evidence for a systemic fungal infection, mycological evidences should be assessed (E). The main goal is that the method used to identify the pathogenic fungi delivers fast and accurate results, which leads to a better therapeutic plan and lower hospital costs (F). Abbreviations: EORTC, European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group; MSG-ERC, Mycoses Study Group – Education & Research Consortium; NMR, Nuclear Magnetic Resonance; FISH, Fluorescence in situ hybridization; CAGT, Candida albicans Germ Tube Antibody Assay; MALDI-TOF, Matrix-Assisted Laser Desorption/Ionization – Time of Flight.
4. Methods used for proven diagnosis
For gathering mycological evidences to obtain a definitive proven diagnosis, the fungal culture, microscopy, and histopathology remain the gold standard methods for identification of the pathogenic fungi (Table 1 ). The major advantages of these techniques are the possibility of obtaining the pathogenic fungi, which will enable not only the further identification of the species but also the evaluation of the antifungal resistance (Table 1). The major disadvantages include long turn-around-time and the low fungal culture sensitivity (Table 1). Even when other more modern techniques are available, these conventional methodologies continue to be employed for comparison and confirmation [34].
Table 1.
Overview of advantages and disadvantages of the most commonly used methodologies for mycological evidences.
| Methodologies | Advantages | Disadvantages | References | |
|---|---|---|---|---|
| Proven diagnosis | Fungal culture |
|
|
[35,50,173] |
| Microscopy |
|
|
[34,35,174] | |
| Histopathology |
|
|
[34,35] | |
| Chromogenic mediaa |
|
|
[59,62,63] | |
| Fluorescence in situ hybridizationa(FISH) |
|
|
[34,175] | |
| Matrix-Assisted Laser Desorption Ionization-Time of Flight - Mass spectrometrya(MALDI-TOF) |
|
|
[34,35,77,[79], [80], [81],176] | |
| Biochemical Phenotypic Identification Systemsa |
|
|
[64,[66], [67], [68], [69],72,73,177] | |
| Probable diagnosis (Serological methods) | 1,3 β-D-glucan |
|
|
[35,106,107,166,178] |
| Mannan antigen and antimannan antibody |
|
|
[35,166,178,179] | |
| Galactomannan |
|
|
[35,[124], [125], [126],178,179] | |
| Antibody-based (Immunofluorescence, ELISA, Lateral flow assay, Latex agglutination assay) |
|
|
[35,127,130,131,133,174] | |
| Probable diagnosis | Invasive Fungal Infections Imaging |
|
|
[93,95,96] |
| PCR-based methods |
|
|
[34,35,82,136,143,149] |
Methodologies to achieve specific identification after the isolation of pathogenic fungi.
4.1. Fungal cultures
If the host factors and the clinical signs and symptoms point to an invasive fungal infection, the start-off is to try to isolate the pathogenic fungi. For this, sterile liquids, such as blood, urine and/or cerebrospinal fluid are collected. When the growth of the microorganism in cultures in these fluids is possible, the diagnosis is proven. On the other hand, when using non-sterile fluids, like bronchoalveolar fluid, commensalism and/or environmental contamination needs to be considered. Despite cultivation being a gold standard methodology, this method is associated with low sensitivity. The overall sensitivity of blood cultures for yeasts is about 50–95%, and for moulds the situation is even more complicated, presenting sensitivity values of 1–5% [35,36].
For invasive candidiasis, the golden standard approach for yeast detection is the blood culture. The sensitivity of this technique varies from 50 to 70%, when sampling recommendations are in agreement with the Clinical and Laboratory Standards Institute guidelines (40–60 mL of blood) [37]. Ericson and colleagues [38] evaluated the effectiveness of several commercially available blood culture vials at detecting Candida species. BacT/Alert FA vials detected 144 of 179 samples (80.45%), proving to be the most efficient when compared to Bactec Mycosis IC/F and BacT/Alert FN. This study also reported that anaerobic vials (BacT/Alert FN) were not successful for Candida spp. growth, but Candida glabrata growth was detected (8 out of 179 samples) [38]. Additionally, that the vast majority of the blood culture vials took about 14–72 h to become positive [39]. A comparison between Candida albicans, Candida dubliniensis, C. krusei, C. lusitaniae, Candida parapsilosis and Candida tropicalis growth times, showed no significant differences regardless of the culture used (40 h–57 h). However, C. glabrata had an overall lower growth time (12 h–37 h) and the culture vial was an important factor since significant differences in growth time were observed (Bactec Mycosis IC/F 12 h, and BacT/Alert FN 37 h) [38].
Candida spp. in the bloodstream (candidemia) is associated with around 40–68% of cases of candiduria (presence of Candida species in the urinary tract) [40,41]. Therefore, in suspected candidemia, the use of urine cultures can also be considered [42]. Once more the culture medium has a significant effect on the performance of detection. The standard urine culture (blood and MacConkey agars) showed only 37% of detection while the fungal culture medium (Sabouraud dextrose agar) showed detected 98% of Candida spp. in urine [43].
Cerebrospinal fluid samples (CSF) cultivation is often used in suspected fungal infections on the central nervous system but it detects mainly Cryptococcus and Candida [44,45]. The most appropriate media for pathogens growth from CSF are Sabouraud 4% dextrose and sheep blood agar plates [44]. These pathogens usually take about 3–7 days to grow, with very good sensitivity for Cryptococcus, >95%, while for Candida only 37% [35,46]. However, due to the risk associated with CSF collection, that includes nerve damage, the possibility of an infection, discomfort and/or pain, and bleeding into the spinal cord, CSF culture is not the first option in sample collection [47].
Despite advances in fungal cultures, the majority of moulds are rarely isolated from CSF or blood cultures, however in cases of disseminated infection, the Fusarium spp. blood cultures are often positive [36,45,48]. Therefore, particularly for invasive aspergillosis (IA), detection of Aspergillus spp. from bronchoalveolar fluid (BAL) is often used. Guegan and colleagues [49] processed 1555 BAL samples from 1336 patients, including 61 diagnosed with invasive aspergillosis (1 proven, 37 probable, and 23 possible). For invasive aspergillosis patients, the detection of Aspergillus spp. from BAL culture increase the chance of detecting the fungal pathogen from 1.6% to 47.4% (18/38 proven or probable) [49]. It is important to consider environmental contamination at the local of isolation when using BAL samples.
To improve mould detection by culture, Bao et al. [50] analysed the effect of different culture media and time of incubation of 1821 moulds (hyaline, dematiaceous, Onygenales and Mucorales) isolated from 1687 patients. Regarding medium, the inhibitory mould agar (IMA), Sabouraud dextrose agar Emmons (SDAE), and malt-yeast agar (MYA) were tested and results showed that 55.6% of moulds were isolated using IMA, 18.5% using SDAE, and only 3.2% with MYA [50]. This study showed that SDAE, the often-used culture medium to isolate moulds, did not meet the expectations. IMA had the best performance, which can be linked to the fact that this medium inhibits bacterial growth [50]. Concerning incubation time, Mucorales had the fastest growth time, with a growth peak at the 3rd day. Hyaline moulds had the growth peak at 4th day, and 85.4% of Aspergillus fumigatus were mostly isolated after the first week, and 97.3% after two weeks. Contrarily, Histoplasma capsulatum and Trichophyton isolates were isolated during the 4th week of growth [50]. Concerning temperature, Tarrand and co-workers [51] demonstrated that incubation of the 10,062 clinical specimens cultured (sputum, bronchial wash or respiratory lavages) from patients with different pathologies, only 344 (3.4%) showed fungal growth, of Aspergillus species. However, cultures incubated at 35 °C provided higher Aspergillus-positive rate when compared to incubation at 25 °C (31% increase). This is explained by the fact that there is a greater similarity between the incubation environment and the environment within the host, from which they were just isolated at 35 °C [51].
Despite being a gold standard method, fungal cultures are associated with several drawbacks, where the most evident are the long turn-around time which delays patient's treatment, susceptibility to contamination and low sensitivities values. However, this method provides a proven diagnosis, and it can lead to antifungal susceptible tests and identification at the species level (Table 1).
4.2. Direct microscopy and histopathology
Direct microscopy is used to look for fungi morphological structures in a portion of infected biopsy tissue or fluid. This allows to evaluate whether the infection is triggered by a septate mould (such as Aspergillus spp.), a non-septate mould (for example Mucorales), or a yeast (for example a Candida spp.) [52]. Throughout the visualization of the fungi's appearance in the tissue section and identification of specific morphological patterns, it is possible to differentiate between different histopathological diagnoses associated with invasive fungal infections. However, the visualization of those structures alone does not provide a specific identification since the analysed structures are similar in various fungal species [53]. Additionally, it is very important to assess tissue invasion to understand the significance of the isolate (pathogenic fungus/normal microbiota/environmental contamination). Visualization of fungal structures by histopathology and direct microscopy techniques can be improved, through the use of stains, such as Gomori's methenamine silver, the periodic acid–Schiff reaction [53], or India ink [54], and fluorescent brighteners, such as Calcofluor White [55].
Biological samples from patients with clinical diagnosis of cryptococcosis (CSF samples) can be examined directly for the presence of encapsulated budding yeast by India ink preparation. However, the agreement between direct detection of encapsulated yeast cells with India ink and fungal culture (“gold standard”) varies, Martins et al. [56] showed an 80% (8 with India ink test out of 10 positive fungal culture), Sato and colleagues [57] showed only 44% (7 out of 16), while Silva et al. [54] presented 95% (46 out of 48). Fluorescence in situ hybridisation (FISH) can also be used directly in CSF samples. In this technique, fluorescent probes that bind to specific targets on ribosomal RNA allow the direct microscopic visualisation of individual fungal cells. Silva et al. [54] study indicates that FISH directly used on CSF samples presents a 100% concordance with PCR results, at detecting Cryptococcus.
A 10-year retrospective study, analysing surgical pathology evaluation from patients with concurrent positive mould and yeast cultures, without known history of a fungal infection, showed a 79% accuracy of the histologic diagnosis of fungal infections vs culture [58]. However, a specific identification based only on the visualization of those structures is difficult. Discrepant diagnoses included misidentification of septate and nonseptate hyphal organisms and yeast forms, and also from morphologic mimics, use of inappropriate terminology, and incomplete knowledge in mycology [53,58]. Nevertheless, these techniques are very useful to avoid false negative results from the fungal culture or cases of uncultivable fungi and, implementation of a standardized reporting format should improve diagnostic accuracy and prevent adverse outcomes [58]. Still, microscopy requires a trained mycologist to differentiate different species, or even genus, especially due to the similar microscopic appearance of several fungus (Table 1).
5. Methods for identification after pathogen isolation
The isolation of the fungal pathogen from the cultures allows the evaluation of several relevant parameters as their antifungals resistance and species identification [55]. Despite the growth of the pathogenic fungi, the culture media provides only information about the presence/absence of fungus. Therefore, in order to be able to identify the fungal species behind the infection, there are complementary methodologies to achieve a specific identification, leading to a better therapeutic plan.
5.1. Chromogenic media
Considering the unspecific clinical scenarios of fungal infections, the detection of the presence or absence of a fungal pathogen is frequently insufficient, thus chromogenic media can be used to overcome this limitation [59]. Chromogenic media has been widely used in clinical microbiology to detect and identify either bacterial or fungal pathogens [60] being used for Candida identification since 1994. They allow the growth of a specific microorganism, and with the inclusion of chromogenic or fluorescent enzyme subtracts targeting microbial enzymes, such media are able to target pathogens with high specificity [59]. These culture media are suitable for non-sterile samples as they stimulate the growth of a specific genus, inhibiting the growth of other microorganisms [34]. There are several chromogenic media available for the detection of yeasts, and these media often include a chromogenic substrate for β-hexosaminidase, which allows the differentiation of the most frequent and clinically important species, C. albicans. Combining chromogenic substrates, colonies from different species present different colours, enhancing the differentiation of yeast species. CHROMagar® Candida Plus (CHROMagar, Paris, France), chromID™ Candida Agar (CCA, bioMérieux, Marcy-l’Étoile, France), HiCrome® Candida (HiMedia, Mumbai, India), CandiSelect™ 4 (CS4, Bio-Rad, Marnes-la-Coquette, France) and Brilliance™ Candida Agar (BCA, Oxoid, Basingstoke, UK) are examples of commercially available selective and differential media which facilitates differentiation of Candida species, namely C. albicans, C. tropicalis, C. glabrata, C. parapsilosis and C. krusei, on the basis of colouration and colony morphology [59,61].
Even though chromogenic media can provide rapid and direct identification by colony colour and morphology observation, they were not able to differentiate emerging Candida species, such as C. auris, until very recently. In 2020, Borman et al. [62] conducted a study that aimed to test C. auris detection and specificity of a novel CHROMagar™ Candida Plus, using 52 yeast species recovered from clinical samples. The authors reported that CHROMagar™ Candida Plus was able to successfully distinguish C. auris from all other species tested due to the distinct appearance of its pale cream colony with a blue halo, except for Candida diddensiae, which showed a similar appearance to C. auris [62]. De Jong and colleagues [63] compared five commercially available chromogenic media, including the two novel chromogenic formulations designed to differentiate C. auris, the CHROMagar™ Candida Plus and the HiCrome C. auris MDR selective agar (HAMA) that only allows the growth of multidrug-resistant C. auris [63]. CHROMagar™ Candida Plus medium had a better performance in discriminating C. auris from other Candida species tested, with an accuracy of (91.8%) compared to 32.7–87.8% for the other media studied. However, Candida vulturna and Candida pseudohaemulonii showed phenotypic similarities to C. auris colonies [63]. Therefore, the novel chromogenic medium CHROMagar™ Candida Plus is considered a valuable tool for detection and identification of C. auris and other yeast species in clinical contexts. Due to the occasional appearance of similar colony phenotypes, additional confirmation methods may be necessary.
Chromogenic media are capable of delivering fast results in a cost-efficient way. There are multiple assays commercially available, some of them capable of detecting polymicrobial samples, providing identification at species level. Yet, there are some difficulties in distinguishing Candida non-albicans species, and emergent species, such as C. auris (Table 1).
5.2. Biochemical phenotypic identification systems
Several biochemical phenotypic systems have been developed and are commercially available for the identification of microorganisms. These systems evaluate the ability of the microorganism to assimilate different types of nutrients in microwells, including sugars and organic acids, substrates for specific enzymatic activities, and antimicrobial agents. These are turbidimetric methods organized in cards with different microwells and the results are barcoded and compared with a database. However, before performing these methods, it is necessary to obtain a pure culture of the pathogen [64]. Considering fungal infections, these systems are most suitable for yeast species as for instance the manual API 20C AUX and the automated VITEK® 2 YST ID card (bioMérieux, Marcy-l’Étoile, France). API 20C AUX system properly identifies around 97% of the most commonly detected species [65,66].
The accuracy of these two systems (VITEK® 2 and API 20C AUX) for yeast detection is better for commonly detected species (76–95% for VITEK® and 96% for API) than for uncommon yeast species (58–78% for VITEK® 2 and 72% for API) [[66], [67], [68], [69]]. Misidentifications or identifications were mostly detected for C. parapsilosis, C. glabrata, C. dubliniensis, C. norvegensis and C. pelliculosa [66,67]. VITEK® 2 system database has been recently updated (version 8.01) to include C. auris, increasing the accuracy of identification of this emergent pathogen. However, the updated version showed limited ability to distinguish between C. auris and closely related species, only identifying correctly about 52% of the C. auris isolates [70,71]. These biochemical assays provide inaccurate identification of C. auris, mistaken this species with for instance C. haemulonii, Rhodotorola glutinis, C. famata, or C. sake [72,73].
5.3. Matrix-Assisted Laser Desorption/Ionization – Time of Flight (MALDI-TOF MS)
In recent decades, mass spectrometry-based methodologies gained popularity in microbiology laboratories because they provide fast identification at low costs, with easy accessibility and great applicability to several microorganisms. For fungal species identification, the variation of mass spectrometry most widely used is Matrix-assisted laser desorption/ionization (MALDI-TOF MS), which is based on the identification of fingerprints of extracted proteins, mainly ribosomal and membrane proteins. The protein profile obtained for each isolate is compared with universal profile databases, enabling identification at the species and genus level [34,35].
Becker and colleagues [74] identified 290 fungal isolates, at species level, including filamentous fungi and yeasts, belonging to 69 different species, through conventional culture methodologies and by MALDI-TOF MS and confirmed by DNA sequencing. MALDI-TOF MS was able to correctly identify 89% of the species, while conventional cultures only achieved 69% of correct identifications [74]. Lau and co-workers [75] developed a spectra database according to 249 fungal isolates, which was used to identify 421 clinical isolates, through MALDI-TOF. This database was able to correctly identify about 90% of the isolates when compared with the results obtained from DNA sequencing.
The performance of some commercially available MALDI-TOF MS systems, such as VITEK MS (bioMérieux, Marcy l’Etoile, France), MALDI Biotyper (Bruker Daltonics, Bremen, Germany) and Autof MS 1000 (Autobio Diagnostics, Zhengzhou, China) has been evaluated. For instance, Teke et al. [76] conducted a comparative study of the VITEK MS and MALDI Biotyper performance for the identification of 157 isolates, representing 23 yeast species (non-albicans Candida and rare yeast species), and both systems showed high sensitivity for yeast identification (96.8% and 98.7%, respectively). Yi et al. [77] also compared the performance of the Autof MS 1000 and Vitek MS systems for yeasts identification within closely related species complexes that cause invasive fungal infections. This study included the evaluation of 1228 yeast isolates representing 14 different species. The identification accuracy of all species complexes ranged from 98.9% to 100% and from 79.1% to 96.3%, for Autof MS 1000 and VITEK MS, respectively. Both systems showed good performance for C. auris identification [77].
MALDI-TOF MS is now commonly used as a first-line identification method for common yeasts. However, the identification of pathogenic filamentous fungi remains difficult. Mould often grow inside the solid media, which makes harvesting very difficult, leading to agar contamination of the spectra and hampers MALDI-TOF MS identification. In this way, a novel culture medium, ID Fungi Plates (Conidia, Quincieux, France) was developed to allow easier and faster harvesting of the isolates [78]. Overall, common yeast and moulds are mostly well covered in commercially available databases, but the same does not happen for new, rarer, or cryptic species. It seems that the best approach to improve the identification of these species is to continuously update the commercial databases or to complement them with homemade reference libraries and other online databases, such as MicrobeNet (developed by CDC), and the Mass Spectrometry Identification platform [79].
In addition to species-level identification, assessing the antifungal resistance is essential to select the most suitable antifungal therapeutic approach. Several studies have demonstrated that MALDI-TOF MS has the potential to be adapted for antimicrobial susceptibility testing. Vatanshenassan and colleagues recently introduced the MALDI Biotyper antibiotic susceptibility test-rapid assay (MBT ASTRA) to detect echinocandin resistant C. albicans, C. glabrata and C. auris isolates [80,81].
MALDI-TOF MS is associated with several advantages such as being capable of identifying the pathogen at the genus and species level with accurate and fast results, with easy handling and reduced costs. It is applicable to a vast variety of microorganisms and allows antifungal susceptibility detection. Still, it is incapable of performing quantification, and the coverage of databases available is limited, so there must be a continuous update to cover the rarest and emergent species (Table 1).
5.4. Fluorescence in situ hybridization (FISH)
FISH technique had also been widely used in microbiology laboratories to detect pathogenic microorganisms from positive blood cultures. Currently, this technique is not routinely used, but it continues to be useful for certain applications [54]. Although more expensive PNA-based FISH probes are appearing more frequently on the market compared with DNA-based FISH probes since their complementarity has higher affinity and specificity, and are more resistant to degradation [82]. Silva and co-workers [54] evaluated the potential of the FISH methodology in identifying fungal species from blood cultures. They showed that results obtained with FISH analysis were in complete agreement with fungal culture. However, microscopy identification after culture needs specialized clinics to carry and are time-consuming (3–10 days). In contrast, the FISH methodology presented the same results within 5 h [54]. Thus, PNA-FISH® platform that detects several sequences of pathogenic microorganisms, such as Staphylococcus aureus, various Enterococcus spp., gram-negative bacteria, and several Candida spp. was used in the hospital routine [82]. More recently, a new PNA FISH platform, Yeast Traffic Light PNA FISH (AdvanDx, Inc., Woburn, MA; YTL PNA FISH), was developed combining the identification of 5 Candida spp. with their susceptibility to fluconazole, according to different fluorescence dyes. PNA FISH probes that show green fluorescence stands for susceptible to fluconazole treatment (C. albicans and C. parapsilosis), yellow means that a higher dose of fluconazole must be administrated (C. tropicalis), and red means that there is a natural resistance to fluconazole (C. krusei and C. glabrata) [83]. A study using this new platform showed that of 137 patients with positive blood cultures without antifungal treatment, the YTL PNA FISH was able to correctly target the treatment of 132 patients (96.4%), and distinguish between bacteria and yeasts in a concomitant growth (95.8%) [83]. These platforms are capable of displaying results within 2 h, with high sensitivity and specificity values (above 95%) [82]. In microbiology laboratories this assay was replaced by more efficient methodologies especially because FISH platforms presented a high limit of detection, and there was a reduced number of PNA probes commercially available (Table 1).
5.5. PCR-based methodologies
A large variety of PCR-based methodologies are available to identify the pathogenic fungi, after obtaining the fungal isolate. FilmArray® (bioMérieux, Marcy l’Etoile, France) are fully automated platforms that incorporates steps from sample preparation to multiplex PCR amplification and detection/identification of pathogens, in about 1 h [82,84,85]. FilmArray® was the first to be developed, allowing the detection of pathogenic microorganisms, according to different panels, which correlate to sample types. The first panel for blood cultures, Blood Culture Identification Panel, allowed the detection of 19 species of bacteria, 5 Candida species (C. albicans, C. glabrata, C. krusei, C. parapsilosis, and C. tropicalis), and 3 antimicrobial resistance genes, with sensitivity >96% and specificity of 99% [84]. It is also effective in identifying mixed infections, being able to detect all microorganisms in 71% of the samples. The BioFire® FilmArray® Torch System, is a new version of this platform that allows the inclusion of 2–12 modules, using the same developed Panels, offering a maximum of 264 samples per day with 12 modules [82]. Additionally, an updated version of the blood culture Panel, the BioFire® FilmArray® Blood Culture Identification 2 Panel provides results for 26 bacterial, 7 yeast species (with the addition of C. auris and Cryptococcus gattii/C. neoformans species complexes) and 10 antimicrobial resistance genes [86,87]. The identification of microorganisms through this panel yielded higher values of sensitivity and specificity (>98% and >99%, respectively) when compared to cultures [87].
Sepsis Flow Chip (Master Diagnóstica, Spain) is a platform that combines multiplex PCR with a reverse dot blot hybridization for the detection of the most common pathogens in systemic infections, from positive blood cultures [82,88]. This methodology is able to identify 36 species of bacteria, 20 antimicrobial resistance genes, C. albicans and Candida spp. (non-albicans), in 3 h [89]. In its validation and verification trial, this platform obtained high values of sensitivity and specificity regarding Candida species: 93 and 100%, respectively [88]. It also showed excellent results when identifying cultures with more than one pathogen, with a sensitivity of 89%. This diagnostic assay was optimized to use samples directly from the blood culture without previous DNA extraction [88].
ePlex® system (GenMark Diagnosis, USA) is a fully automated platform, incorporating all the necessary steps for the analysis of positive blood cultures. It has a sample preparation system, followed by a multiplex PCR amplification system, and finally the amplicon analysis through electrochemical examination [82,90,91]. It has several panels that allow the detection of various pathogens such as gram-negative and gram-positive bacteria, and fungal species, from blood cultures. The fungal pathogens that this panel identifies are Candida species (C. albicans, C. auris, C. dubliniensis, C. famata, C. glabrata, C. guilliermondii, C. kefyr, C. lusitaniae, C. parapsilosis, and C. tropicalis C. krusei) Cryptococcus neoformans sensu lato, C. gattii sensu lato, Fusarium spp, and Rhodotorula spp [90,91]. The sensitivity of ePlex® system from blood cultures ranged from 99.8 to 100% for fungal pathogens, and specificity of 100% [91,92].
PCR-based methods have a short turn-around time, capable of providing fast and accurate results. However, some PCR-based methods do not allow quantification of the amplified DNA, which can be crucial to interpreting the infection severity, moreover contaminations are prone to happen if the PCR-platform does not occur in a closed system, and also the design of primers should be carefully made (Table 1).
6. Probable diagnosis
When no detection of the pathogenic fungi through histopathological or culture methods from sterile sites is possible, but only detection of traces of the pathogen, a probable diagnosis is attributed. Serological, molecular, and other more recent techniques are available to collect evidence of the presence of the pathogenic fungi. However, for a probable diagnosis to be attributed, several aspects must be considered, such as the patients’ clinical signs and symptoms and the host factors (immunocompromised or not). Regarding mycological evidences, there should be a final result that is concordant between at least two different methodologies. Some of these methodologies can also be used after a positive blood culture for species identification.
6.1. Invasive fungal infections imaging
There are numerous benefits linked to imaging towards invasive fungal infections diagnosis, since through this method it is possible to monitor the progression of the infections, as well as patients’ response to the treatment [93,94]. However, according to the EORTC and MSG-ERC guidelines, imaging methods regarding invasive fungal infections, fit only on probable and possible diagnosis [30]. Several imaging techniques are available to support invasive fungal infections diagnosis, being chest X ray (CXR), computed tomography (CT), magnetic resonance (MR), and positron emission tomography (PET) the most commonly used [95].
Infections with these organisms often start in the lungs and become systemic in susceptible individuals, disseminating through the bloodstream to the brain, and susceptible organs.
Infections of the central nervous system (CNS) are rare, often associated with immunocompromised patients and commonly caused by Cryptococcus, Candida spp, Mucorales fungi and Aspergillus spp. The most common CNS manifestations caused by those pathogens are meningitis and focal masses such as cerebral abscesses and granulomas. When meningitis is suspected the CT can be initially used to observe the impaired cerebrospinal fluid absorption caused by the pathogen. MR is the preferred modality, which makes evident the thick meningeal enhancement, at the skull base [93]. Sinus infections are most commonly associated with Mucorales fungi, especially Rhizopus and Rhizomucor species, and Aspergillus spp., which may cause bone destruction, and spread to other areas, as soft tissues and even to intracranial cavity. Initially, CT is often used to analyse bone destruction whereas MR is applied to evaluate the extension of intracranial cavity sinus infections [93]. Concerning pulmonary aspergillosis, the commonly associated organisms are Aspergillus, Candida, Nocardia and Actinomyces species, as well as Mucorales fungi, Pneumocystis jirovecii, Histoplasma species and Cryptococcus species. As for pulmonary infections, a variety of characteristics can be observed, such as nodules, chest wall invasion and lobar consolidations. CT is often applied to visualise parenchymal nodules or consolidation with a surrounding area, called CT “halo”, linked to angioinvasive pulmonary aspergillosis, Candida spp., Coccidioides and Cryptococcus infections. In contrast, pulmonary mucormycosis is associated to a CT “reverse-halo” (consolidation surrounding a central opacity) [93]. Regarding gastrointestinal and genitourinary infections, the most associated fungal pathogen is C. albicans. When using CT and MR imaging these infections usually manifest similar parenchymal microabscesses [93].
The imaging methodologies commonly used to assess and diagnose invasive fungal infections are CT or MRI, since they provide overall information in less time [95]. However, there is a need for multimodal imaging to overcome the limitations that exist for the individual use of each imaging method (Table 1) [96].
6.2. Serological methodologies
The development of laboratory biological markers and the launching of antigen testing have improved the diagnosis of invasive fungal infections regarding quickness and efficiency. Fungal antigens, metabolites, or antibodies produced by the host's immune system can be detected in several serum samples, but also urine and bronchoalveolar fluid [53]. In this review, the most frequently used techniques will be presented.
6.2.1. β-(1,3)-D-glucan assay
β-(1,3)-D-glucan is a polysaccharide present in the cell wall of several fungi, and the detection of this antigenic marker can indicate a variety of infections, from invasive candidiasis, to invasive aspergillosis and also P. jirovecii pneumonia [35]. The assays developed specifically to measure fungal β-(1,3)-D-glucan typically depends on the activation, by the serine protease zymogen Factor G, of a clotting cascade present in the limulus amebocyte lysate (LAL). The developed assays use serum samples and rely on the activation of the LAL clotting cascade that ultimately cleaves a chromogenic substance, p-nitroanilide, from a synthetic peptide in the beta-glucan LAL, changing to a yellow colour. However, it has been observed that serum samples have an inherent yellow colour that may overestimate quantification therefore, a variation of the β-(1,3)-D-glucan assay uses the diazo derivative of p-nitroanilide which by cleavage is purple [97]. There several β-(1,3)-D-glucan assays are available for in vitro diagnosis of invasive fungal infection in clinical samples: Fungitell (Associates of Cape Cod, East Falmouth, USA), Fungitec-G (Seikagaku Biobusiness, Japan), beta-Glucan Test (Waco Pure Chemical Industries, Japan), and BGSTAR β-Glucan Test (Maruha, Japan), which have different measurement methods and cut-off values. The overall sensitivity and specificity values for these assays range from 58 to 100% and 57–100%, respectively [[98], [99], [100]]. Still, only Fungitell is FDA approved and it is the commonly used assay in hospital settings, however beta-Glucan Test became commercially available in Europe. Fungitell assay presented sensitivity and specificity values of 69.9–100% and 73–97.3%, respectively, for invasive candidiasis [[99], [100], [101], [102]], and for invasive aspergillosis showed sensitivity and specificity values of 81–93.3% and 77.2–99.5% [99,103,104]. Beta–Glucan Test, on the other hand, showed sensitivity and specificity values of 42.5–98.7% and 98–98% respectively, for invasive candidiasis, and for invasive aspergillosis values of 80–80.3% and specificity of 97.3%, respectively [99,105,106].
Although serum (1–3)-β-D- glucan testing is an effective panfungal marker to aid in the screening and diagnosis of invasive fungal infections, a major problem is the high number of false-positives. Racil et al. [107] reported a value of 75% of false positives when analysing 1143 samples, and most of the false-positive results were attributed to concurrent bacteremia, use of hemodialysis, or treatment involving the use of human immunoglobulin. Mennink-Kersten et al. [108] reported that bacteria such as Alcaligenes faecalis, Streptococcus pneumoniae and Pseudomonas aeruginosa showed β-(1,3)-D-glucan reactivity with the Fungitell assay, which can provide false-positive results. According to Hammarström and colleagues [109], patients receiving treatment with pegylated asparaginase and ICU patients treated with plasma, albumin, or coagulation factors, showed elevated levels of β-(1,3)-D-glucan, being more likely to test positive for the β-glucan assays. Additionally, some drugs (lentinan, crestin, scleroglucan, and schizophyllan) used in intensive care units contain glucans that may also cause false positive test results. Thus, due to the excellent negative predictive values of the assay (90.57–99.8%) [106,107] and high false-positives, Fungitell assay serve best to identify those patients without invasive fungal infection rather than identifying those for whom infection has actually been detected.
Despite β-(1,3)-D-glucan assays provides fast results regarding detection of a broad range of fungal pathogens, in a cost-effective and non-invasive way. It is more effective in excluding an invasive fungal infection than diagnosing it. Even so, it is a nonspecific panfungal test, associated with low sensitivity values, and with several issues regarding false-positive results. Moreover, some fungal species produce less β-D-glucan, like Cryptococcus spp, or even any such Blastomyces and Mucorales moulds (Table 1).
6.2.2. Mannan antigen and antimannan antibody detection
Mannan is one of the major components of the Candida cell wall that has been used as a diagnostic biomarker of invasive candidiasis. There are some commercially available serologic tests for mannan antigen and anti-mannan antibody detection, such as Platelia™ Candida Ag/Ab Plus (Bio-Rad, France), which is the most well-studied, and Serion ELISA Antigen Candida Assay (Serion GmbH, Germany) [110,111]. In several studies with different designs and populations, the clinical value of mannan detection assays showed variable sensitivity (52%–85%) and specificity (86.8%–98%) [[111], [112], [113]]. The anti-mannan antibodies detection specificity and sensitivity ranged from 57.7 to 80.4% and the specificity from 60 to 87% [112,113].
These serologic tests present some disadvantages (Table 1), such as limited specificity due to the presence of Candida species in the host normal flora, limited sensitivity of antibody assays for high-risk immunocompromised patients, and also different sensitivity for different species (lower for C. krusei and C. parapsilosis).
6.2.3. Candida albicans germ tube antibody (CAGTA) assay
Regarding Candida species, a broad range of serologic tests is also available, as for instance the CAGTA [35]. Invasive candidiasis CAGTA IFA IgG (Vircell Microbiologist S.L., Spain) is a commercially available indirect immunofluorescence kit that detects antibodies against antigens in the cell wall surface of the mycelium of C. albicans, in human serum or plasma. This test was then adapted to an automatic chemiluminescence assay, the Invasive Candidiasis (CAGTA) VirCLia® IgG Monotest (Vircell Microbiologist S.L., Spain), which provides faster results [114]. For invasive candidiasis, the CAGTA IFA IgG assay showed sensitivity values ranging from 51.7 to 69.2%, and specificity values from 75 to 80.3%, while for VirCLia®, these values were 76.9% and 75.8%, respectively [115,116]. Wei et al. performed a meta-analysis of data from 7 different studies, using CAGTA IFA IgG assay, and results showed a pooled sensitivity of 66%, and specificity of 76%, concluding that this assay has moderate accuracy for invasive candidiasis diagnosis [117].
CAGTA assay despite being easy handling, with a fast performance at low cost, this assay is one of the few antibody-based that falls short in terms of sensitivity and specificity for invasive candidiasis diagnosis, which ends up being a huge drawback (Table 1).
6.2.4. Galactomannan (GM) assay
For invasive aspergillosis, GM is the main cell wall antigen detected in serum, in bronchoalveolar fluid or in cerebrospinal fluid. The overall sensitivity and specificity values of the GM assay ranged from 67 to 100% and 86–100%, respectively [[118], [119], [120], [121]]. The commercially available assay to detect GM, the ELISA Platelia Aspergillus assay™ (Bio-Rad, USA), is the most frequently used in the clinical context to diagnose invasive aspergillosis and optimized for serum. ELISA Platelia Aspergillus assay™ showed sensitivity values ranging from 44% to 100%, and specificity from 78.6% to 100% [122,123]. This assay has a higher sensitivity to Aspergillus non-fumigatus species, which turns out to be a drawback, because A. fumigatus is the prevalent pathogen in invasive aspergillosis [35]. Despite GM being present in the cell wall of Histoplasma spp. and Fusarium spp., this antigen detection assay is mentioned as an Aspergillus-specific methodology [[124], [125], [126]]. However, this kit can be a useful tool for the diagnosis of infections caused by Fusarium spp., since there is no antigen test for this pathogenic species [125], and also for histoplasmosis, since Histoplasma spp. can take up to 4 weeks to grow in culture [50,124,126]. GM assay can be a useful biomarker for the diagnosis of invasive aspergillosis. However, although being the most frequently used assay in hospital settings for invasive aspergillosis, this assay has low sensitivity for early diagnosis and falls short regarding specificity (Table 1).
6.2.5. Lateral-flow devices
Lateral-flow devices detect antibodies, antigens, or metabolites in serum, respiratory samples, or even urine, being applied for the diagnosis of probable infections. Due to its easy performance, these devices can be applied to point-of-care (POC) testing, presenting results in 15 min [127]. A POC test for aspergillosis was recently developed, the LFA-IMMY™ (IMMY, Norman, Oklahoma) assay. This methodology is Aspergillus-specific and detects galactomannan in any respiratory samples, however, it does not discriminate different types of aspergillosis, but only if there is Aspergillus present in respiratory samples [128]. The accuracy of this methodology was compared with conventional ones, such as culture and microscopy, using 398 respiratory samples. It was shown that the LFA-IMMY™ assay achieved an accuracy of 92% in identifying Aspergillus. In other words, LFA-IMMT™ assay led to 48 true-positive results, 44 true-negative results, 4 false-negative results, and 4 false-positive results, presenting sensitivity values ranging from 86 to 96%, and specificity of 84% [128]. Another study evaluated the accuracy of the LFA-IMMY™ in 179 serum samples, and the test performed slightly better, achieving a sensitivity of 96.6% and specificity of 98% [128,129].
Lateral-flow assays are available to detect and diagnose other fungal infections, for example, histoplasmosis. MiraVista Diagnostics recently presented a new, rapid and non-invasive way to diagnose histoplasmosis, the MVista LFA. This is a qualitative assay that uses polyclonal antibodies to detect Histoplasma antigen in urine samples, within 30 min, and when compared to proven cultures achieved sensitivity and specificity of 93.1% and 96.1%, respectively [130,131] Comparing MVista LFA assay, with the traditional enzyme immunoassay in diagnosing histoplasmosis using 352 cases (44 proven histoplasmosis infections, 22 probable infections, and 286 controls) results showed an overall concordant diagnosis between the two methodologies [130]. The diagnostic accuracy using MVista LFA was higher in patients with proven disease (93.2%), when compared with patients with probable disease (78.6%). However, MVista LFA demonstrated 85%–30% cross-reactivity with other endemic fungal infections, such as blastomycosis, paracoccidioidomycosis and coccidioidomycosis. Even though this new methodology provides developing countries with an easy and effective way to diagnose histoplasmosis [130,131].
CrAg® LFA, developed by IMMY is a new assay, also based on lateral flow analysis that aims to detect Cryptococcus antigen (CrAg) in aiding to diagnose cryptococcal infection. This assay provides faster results (10 min), with higher sensitivity and specificity values 100% and 93%, respectively, when compared to the Meridian's enzyme immunoassay [132]. A study testing 4627 samples from 3969 patients with the CrAg® LFA resulted in 55 positive cases. Of this, 38 patients lacked a prior cryptococcal disease history where 20 were confirmed with a positive culture (proven diagnosis), 5 presented cut-of values way above the cut-of value (probable diagnosis) and 13 showed cut-off values in the limit, leading them to consider these patients as false-positive (a rate of 34% of false positives) [133]. Thus, to avoid misdiagnosis, the results from patients with no history of cryptococcal disease should be carefully analysed. There are other commercially available LFA to aid the diagnosis of cryptococcal infection, however they are not as much evaluated as the CrAg® LFA. Shi and colleagues [134] evaluated the effectiveness of other commercially available LFA in detecting Cryptococcus spp., such as CryptoPS (Biosynex, Illkirch-Graffenstaden, France), cryptococcal antigen LFA (Dynamiker Biotechnology, Tianjin, China), and cryptococcal capsular polysaccharide detection K-set (FungiXpert, Genobio Pharmaceutical, Tianjin, China), and results were compared to those presented by CrAg® LFA. For this, 40 cryptococcal strains were used, representing all the 7 recognized species, and interspecies hybrids were also tested. Results showed that CrAg® LFA and cryptococcal capsular polysaccharide detection K-set were able to successfully detect all 7 species. Cryptococcal antigen LFA was not able to detect one strain of C. bacillisporus and 2 strains of C. tetragattii. CyptoPS could not detect one strain of C. bacillisporus, one strain of C. deuterogattii and none of the strains of C. tretagattii. This means that CrAg® LFA and cryptococcal capsular polysaccharide detection K-set should be employed over the other two LFA available, especially CyptoPS since it has a blind spot at detecting C. tretagattii. The major advantage of LFD is that it can be used in POC, capable of providing results in 15 min, at low costs. Still, the majority of LFD commercially available lack specificity, since there is cross-reactivity with other fungal species and, some results should be carefully analysed in patients that lack clinical signs (Table 1).
7. Nucleic acid molecular methodologies
Several studies showed that rapid identification of the infectious agent leads to an appropriate therapeutic plan, which results in lower mortality rates [31,82,135]. Since 1990, thousands of studies referring to the diagnosis of fungal infections through molecular methodologies have been published. However, the use of these techniques in hospital settings, for some years was hampered by the lack of standardization and accreditation [136]. Molecular methodologies have also evolved to be totally independent of the growth of the microorganism in blood culture.
The majority of molecular methodologies used in clinical context were first developed in research laboratories and entitled “research use only” (RUOs) [136]. In order to reach bioindustry and clinical laboratories, those methodologies must undergo a rigorous process of verification and validation controlled by several entities [[137], [138], [139]]. Throughout the verification process, the new method is defined, characterized, and compared with the gold standard methodology, considering the disease or condition it aims to diagnose. This process allows the research centre to evaluate the limitations, risks of error, and the likelihood of causing changes in the interpretation of the test results or treatment decisions. The validation process incorporates the methodology quality control, that is assessed during the time it is commercially available, to guarantee that it works the way it was intended [[137], [138], [139]]. There is a special concern regarding the validation and verification of molecular methodologies for invasive fungal infections since gold standard techniques show inconsistent results, associated with lower rates of specificity and sensitivity. So, comparing a new molecular methodology with the gold standard, as for example cultivation in appropriate media, may result in the conclusion that the new methodology is not suitable [35,136,137].
7.1. PCR-based methods
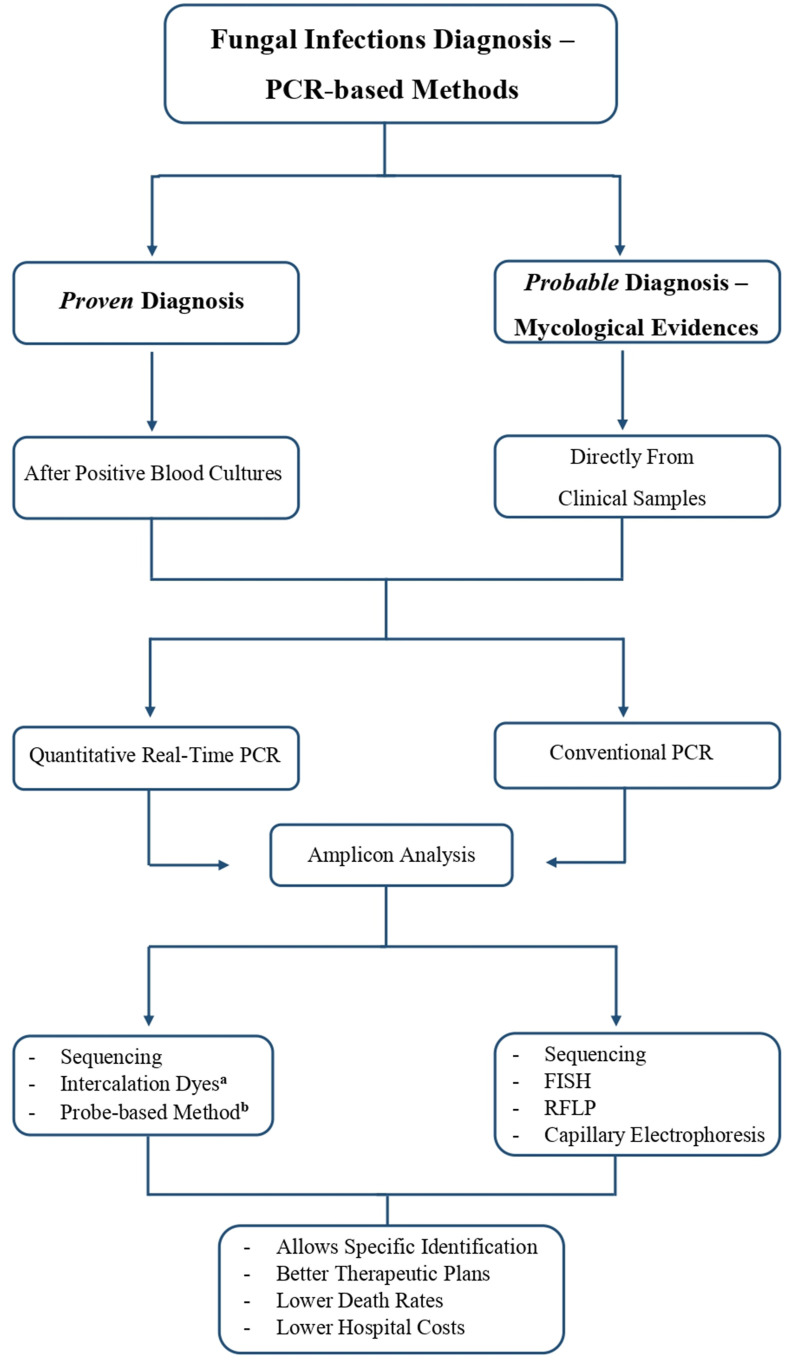
In clinical terms, PCR-based methodologies are commonly associated with the direct use of samples from sterile sites such as whole blood and cerebrospinal fluid, or from nonsterile sites like bronchoalveolar lavage, to detect fungal DNA (Fig. 2 ).
Fig. 2.
PCR-based methods framework for fungal infections diagnosis and the possible outcomes. aQuantitative real-time PCR using intercalation dyes may require melting curve analysis of amplicons. bProbe-based qPCR uses probes with a specific fluorescence to distinguish different amplicons, being ideal for multiplex situations. Abbreviations: PCR, Polymerase Chain Reaction; FISH, Fluorescence in situ Hybridization; RFLP, restriction fragment length polymorphism.
Nucleic acid amplification-based methodologies consist of enzymatic processes in which one or more enzymes can synthesize copies of target sequences. That is achieved through a pair of primers, which specifically bind to the target sequence, resulting in the amplification of that sequence. The biggest drawback of these methods is contamination, which may lead to the amplification of unwanted sequences [136]. PCR was the first nucleic acid amplification methodology to be developed. Throughout the years, novel and more sophisticated variants of conventional PCR have been developed, namely nested PCR, reverse transcriptase-PCR and real-time PCR. Regarding fungal pathogens detection, conventional PCR and real-time PCR are the most widely used, presenting high sensitivity, easy handling, and allowing identification of the pathogen in a short time [35,136,140].
For a few years, the scientific community faced some obstacles related to the manipulation of PCR methodologies in hospital microbiology laboratories. For instance, the fungal burden associated with invasive fungal infections was considered very close to the limit of detection of PCR methodologies, so DNA extraction was a crucial step in the diagnosis [35,136]. Fungi, especially moulds, have a rigid cell wall, which poses an obstacle for fungal DNA isolation and detection. Another complication is the omnipresence of fungi which increases the risk of contamination and false-positive results. Also, human DNA and other components in clinical samples can inhibit or interfere with the PCR reaction [35]. Despite the challenges faced, the obstacles were eventually overcome.
In clinical contexts, the use of conventional PCR to detect and identify pathogenic microorganisms is linked to an extra step for PCR product analysis, which increases the risk of contamination by external factors. Another disadvantage is the lack of quantification of the PCR products, precluding the differentiation between commensal colonization and active infection [35].
Amplicon analysis after conventional PCR can be done through (i) sequencing - amplified products are sequenced to identify pathogenic fungi at species or genus level [136,141]; (ii) FISH - this methodology is used for amplicon analysis by adding specific fluorescent DNA probes to the PCR products, visualized by fluorescent microscopy [54]; (iii) restriction fragment length polymorphism (RFLP) - PCR-RFLP allows the rapid differentiation of fungal species, through the analysis of the patterns obtained and the size of the PCR product after digestion of the PCR product with restriction enzymes, such as MspI [34,142]; and (iv) capillary electrophoresis - the PCR fragments are analysed according to their size. Products with close sizes can be distinguished by introducing different fluorescent labels in one of the primers [65].
Quantitative real-time PCR (qPCR) enables the monitorization and quantification of the DNA during the amplification process, implying that the data is collected and visualized as the PCR reaction proceeds. This methodology occurs entirely in a closed system, with no transfer of samples, no addition of reagents, or electrophoresis [35,136,143]. Several fluorescent reporters are used to monitor qPCR reactions, being divided into intercalation dyes and probe-based qPCR [136,144].
Intercalation dyes become fluorescent in the presence of dsDNA. The amount of DNA present in the sample is proportional to the fluorescence emitted and observed on the monitor. However, intercalation dyes, like SYBR Green and EvaGreen, bind to any dsDNA, which is also the case of primer-dimers. Nevertheless, these dyes are low-cost and prevent the need to resort to probe design [136,[144], [145], [146]]. The major downside associated with intercalation dyes qPCR, is that an extra step is needed to perform amplicon analysis that is a melting curve analysis, which takes extra time to perform. Melting curve analysis (MCA) is a methodology that can be automatically performed after qPCR reaction, with high sensitivity values, based on the association of different amplicons to different melting temperatures. Melting temperatures are mainly determined by the guanine and cytosine content, but also by the size of the amplicon [136,147]. MCA usually accompanies the use of intercalation dyes like SYBR Green, since it binds to all dsDNA present in the sample, all amplicons will be analysed through MCA, and for this reason, it requires a more careful analysis [136,141,147]. This approach is not the most indicated to medical diagnostics, and so currently for a rigorous monitoring of the amplification in real-time, the probe-based qPCR is used instead.
Probe-based qPCR techniques are highly specific since they combine the specificities of the primers and probes, and due to different dyes available, they can also be used in a multiplex system [[144], [145], [146]]. Several types of probes are available: (i) Hydrolysis (TaqMan) probes are related to the phenomenon of fluorescence resonance energy transfer (FRET) between a reporter and a quencher present in the probe. They are able to bind to the target sequence and since the reporter and the quencher are close no fluorescence is emitted. When DNA polymerase begins to synthesize a new sequence, the probe is cleaved and the reporter and quencher are separated, and so the fluorescence emitted by the reporter is detected by the device [136,141]; (ii) Molecular beacons (harpin probes) are also incorporated with a reporter and a quencher for monitoring fluorescence but contrary to the hydrolysis probes they are not degraded. These probes form hairpin systems that when in secondary structure the reporter and quencher are close together and do not emit fluorescence, but since the sequence of the probe is complementary to the target sequence, when the probe denatures and hybridizes emits fluorescence [136,145]; (iii) Scorpion primers are also incorporated with a reporter and a quencher for fluorescence monitorization. These probes also form a loop, however the hairpin loop is directly linked to the 5′-end of a primer. The extension of primers allows the hairpin loop to open and bind to the complementary sequence, in this way the fluorescence is no longer quenched and can be measured [136,145,148].
Probe-based qPCR techniques can be applied in multiplex situations, although they depend on the capacity of the equipment to read fluorescence at different wavelengths [136]. In this case, each probe would be associated with the detection of a specific target, with a specific fluorescence, and the equipment would have to be able to detect different fluorescence simultaneously [136,147].
PCR methodologies can be utilized to detect all fungi (Panfungal PCR) by using universal primers for highly conserved regions present in all fungal genomes, thus being possible to detect any fungal DNA in a sample, even the rarest species. The specific identification of the fungal pathogen can be achieved by sequencing, which increases the risks of contamination, or by a specific PCR [141,149]. Usually, the medical procedure in suspicion of a systemic fungal infection, is to perform a specific Candida or Aspergillus PCR test. However, in case of a negative result, only a panfungal assay would eliminate the hypothesis of fungal infection, and then direct the diagnosis to bacterial infection [136,149]. In a study conducted by Camp and co-workers [149], the overall accuracy of Fungi Assay (real-time panfungal PCR) was compared with fungal cultures. For this study, 265 clinical samples (blood, CSF, BAL, and tissue) were used, and a 55.1% agreement between Fungi Assay and fungal cultures was observed (146 out of 265). Blood samples presented the best results regarding concordance between Fungi Assay and fungal cultures, 43 out of 59 (72.9%), while CSF, Tissue and BAL samples presented lower concordance rates, 60% (36 out of 60), 60% (30 out of 50) and 38.5% (37 out of 96), respectively. Fungi Assay was able to detect 3 true-positive blood samples, while fungal cultures remained negative [149]. This study indicated that Fungi Assay has a great potential in diagnosing cases where there are strong evidences of fungal infection, providing better results with blood samples.
Since quantitative real-time PCR methodologies are the ones accepted for clinical use, Table 2 summarizes the methodologies commercially available, reviewing their most important features (PCR targets, detected species and resistances, specimens used, assay time, sensitivity and specificity).
Table 2.
List of commercially available real-time PCR-based assays for detection of fungi.
| Product (Manufacturer) | Assay method | PCR targets | Detected species | Detected resistance mutations | Specimens | Assay Time | Sensitivity/Specificitya | References |
|---|---|---|---|---|---|---|---|---|
| SeptiFast LightCycler (Roche) | Multiplex Real-time PCR (DNA melt curve analysis) | ITS region |
|
– | WB | 6–7 h | 60–86%/96.1–100% | [[180], [181], [182], [183]] |
| Magicplex Sepsis Real–Time Test (Seegne) | Multiplex real-time PCR | Unknown |
|
– | WB | 6 h (including DNA extraction) |
29%/95% | [184,185] |
| A. fumigatus Bio-Evolution (Bio-Evolution) | Real-time PCR | ITS1 region |
|
– | BAL | <80 min (excluding DNA extraction) |
81%/100% | [186,187] |
| MycAssay Aspergillus (Myconostica) | Real-time PCR with molecular beacons | 18 S rDNA | Eighteen Aspergillus species including:
|
– | Serum BAL |
4 h (after sample collection) |
80–100%/82.4–98.6% | [180,[187], [188], [189]] |
| AsperGenius® (PathoNostics) | Multiplex real-time PCR | 28 S rRNA |
Aspergillus spp. including:
|
Cyp51 A gene:
|
BAL Serum Plasma Biopsy CSF |
<3 h (after sample collection) |
65.5–88.9%/77.8–93.3% | [180,187,[190], [191], [192], [193]] |
| Fungiplex® Aspergillus and Fungiplex® Aspergillus Azole-R (Bruker Daltonics) | Multiplex real-time PCR | Unknown |
|
Cyp51 gene:
|
WB Serum Plasma BAL |
2 h (excluding DNA extraction) |
60%/91.2% | [194,195] |
| Aspergillus spp. ELITe MGB® Kit (ELITechGroup) | Quantitative real-time PCR | 18 S rDNA |
Aspergillus spp. including:
|
– | BAL BA |
NA | 90–100%/97–97.8% | [196,197] |
| MycoReal Aspergillus (Ingenetix) | Real-time PCR (melt curve Analysis) |
ITS2 region |
|
– | BAL Blood CSF Tissues |
NA | NA | [189,198,199] |
| MycoGENIE® Aspergillus Species and MycoGENIE® Aspergillus fumigatus and resistance TR34/L98H (Ademtech) | Quadruplex real-time PCR | 28 S rRNA |
Aspergillus spp. including:
|
TR34/L98H mutations | Serum BAL Biopsy |
NA | 71–100%/84.6–100% | [195,200,201] |
| AspID (OlmDiagnostics) | Multiplex real-time PCR | Unknown |
Aspergillus spp. including:
|
– | BAL | 90 min (excluding DNA extraction) |
94.1%/76.5% | [[200], [201], [202]] |
| CandID® and AurisID® (OlmDiagnostics) | Multiplex real-time PCR | Unknown |
|
– |
CandID: Plasma Synthetic BAL AurisID: Blood |
45 min (excluding DNA extraction) |
CandID: NA AurisID: 96.6%/100% |
[185,203] |
| FungiPlex® Candida (Bruker Daltonics) |
Multiplex real-time PCR | Unknown |
|
– | WB Serum Plasma |
<2 h (excluding DNA extraction) |
98.4–100%/94.1–99.8% | [183,185] |
| PneumoGenius (PathoNostics) | Real-time PCR | Mitochondrial ribosomal large subunit (rLSU) and two dihydropteroate synthase (DHPS) gene mutations |
|
DHPS mutations:
|
BAL | <3 h (after sample collection) |
70%/82% | [202,203] |
| AmpliSens Pneumocystis jirovecii (carinii)-FRT (AmpliSens) |
Real-time PCR | Mitochondrial large subunit ribosomal(rLSU) RNA gene |
|
– | BAL BA Biopsy |
130 min (excluding DNA extraction) |
100%/83% | [204] |
|
Pneumocysist jiorovecii Bio-Evolution (Bio-Evolution) |
Real-time PCR | Unknown |
|
– | BAL BA |
80 min (excluding DNA extraction) |
72–95%/82–100% | [204,205] |
| PneumID® (OlmDiagnostics) | Multiplex real-time PCR | Unknown |
|
– | BAL BA |
45 min (excluding DNA extraction) |
-/90% | [206] |
| MucorGenius® (PathoNostics) | Real-time PCR | Unknown |
|
– | BAL Biopsy Serum |
<3 h (after sample collection) |
75–90%/97.9% | [[207], [208], [209]] |
Abbreviations: BA, bronchial aspirate; BAL, bronchoalveolar lavage; CSF, cerebrospinal fluid; WB, whole blood; NA, not available.
Sensitivity and specificity vary according to the specimen, as well as the clinical context of the patients.
PCR-based methods applied directly to clinical samples have several advantages, to provide a probable diagnosis, such as short turn-around time, and accurate identification with high sensitivity and specificity values, especially in blood samples. qPCR allows quantification of the amplified DNA, which can be critical in the analysis of infection severity. The new platforms are capable of including all steps in this analysis, from clinical sample preparation to visualization of the result, by adding just the sample, which reduces contamination since no further input of the technician is needed, and the circuit is closed (Table 1).
The upcoming In Vitro Diagnostic Medical Device Regulation (EU IVDR 2017/746) will implement significant changes and fully replace the Directive 98/79/EC on in vitro diagnostic medical devices (IVDD) on May 26, 2022. From that date on, IVD manufacturers' devices in Europe have to comply with IVDR requirements, to achieve the “Conformité Européenne” (CE) certification [150]. EU IVDR aims to regulate CE-IVDs (will have to be certified by notified bodies and present post-market performance data) and comprises the (re)classification of both existing and new IVDs using a risk-based system, which ranges from Class A (lowest risk) to Class D (highest risk). Additionally, the laboratory's ability to manufacture and implement in-house IVDs (IH-IVDs) will be more restricted under the EU IVDR, which will only allow the use of IH-IVDs when no equivalent CE-IVD is available. BioMed Alliance (which represents 36 European medical professional societies), in collaboration with the European Haematology Association and the European Federation of Clinical Chemistry and Laboratory Medicine, released a statement indicating some apprehensions regarding the upcoming implementation of the new EU IVDR [151]. This statement details the concerns about the impact of the new EU regulation in the availability of essential medical diagnostic laboratory tests and highlights the crucial role of IH-IVDs in complementing the CE-marked IVDs. Thus appealing to the European Commission and Member States to assure the availability of any essential medical diagnostic laboratory tests and to provide appropriate guidance to the laboratories work towards EU IVDR compliance [151].
This regulation will have a huge impact on molecular diagnostics, and since most medical decisions are based on diagnostic tests, it can bring significant consequences to the European healthcare system and patient care [150,151].
8. Combined new methodologies
Recently, combinations of the most innovative and positive aspects of various methodologies have emerged, to ensure a quick and efficient diagnosis [82,143]. Scientific advances, which have been felt in recent decades, were the main driving force behind the emergence of these combined methodologies, gathering several advantages in a single methodology. Some examples are the Sepsis Flow Chip platform (real-time PCR combined with reverse dot blot hybridization), and ePlex® (PCR combined with electrochemical examination), which were previously described in this review. However, new methodologies for the diagnosis of fungal species continue to appear, some emerging from the positive aspects of previous methodologies, and others with a completely innovative character.
8.1. Candida panel and Filamentous fungi panel
Candida panel and Filamentous fungi panel is a recent technique proposed by Carvalho-Pereira et al. [152] based on a multiplex PCR methodology coupled with capillary electrophoresis, for the separation of PCR products, and product size determination by GeneScan. Candida panel uses specific primers to identify the 5 most common species related to infections by Candida (C. albicans, C. glabrata, C. tropicalis, C. parapsilosis and C. krusei), and the Filamentous Fungi Panel uses specific primers that identify the most prevalent species in infections caused by Aspergillus spp. (A. fumigatus, Aspergillus flavus, Aspergillus terreus and Aspergillus niger) and R. arrhizus. The diagnosis is made through the visualization of the panel, based on the analysis of peaks with a combination of different molecular weights and fluorescence. The innovative character of the work developed is the use of specific primers that result in different and specific amplicon lengths for each species combined with different fluorochromes. The designed panels allow a practical and direct interpretation of the results by the visualization/identification of the specific amplicons in the panel. The analysis also allows identifying any peak that is not in the right position/fluorescence in comparison with the panel, aiding the interpretation of false-positives. Although not yet commercially available, the methodology showed a sensitivity of 89% and specificity of 100%, when using whole blood or serum [152].
8.2. Solid-phase cytometry
Solid-phase cytometry emerged from the combined use of two existing methodologies, fluorescence microscopy, and flow cytometry. This innovative methodology allows the detection and quantification of various microorganisms, such as fungi and bacteria [153]. This methodology delivers fast results, in a fully automated way, directly through clinical samples. However, solid-phase cytometry still faces some obstacles in clinical microbiology laboratories, especially associated with the validation and verification of the methodology [153].
Until the final result of the microorganism identification, the sample goes through a series of steps [154]. The sample is first filtered on a membrane and then retained cells are fluorescently labelled. Fluorescent cells are analysed using a solid-phase cytometer, where background signals are distinguished from specific signals referring to target cells. Finally, the sample is analysed using fluorescence microscopy, in order to validate and examine the target cells [153,154].
In a study conducted by Lies et al. [155], solid-phase cytometry methodology was used to identify A. fumigatus in air samples, since the control of spores in the air is an important epidemiological factor. The results obtained through this methodology presented several advantages when compared to traditional culture methods. Solid-phase cytometry has a low detection limit (4 cells per m3), and provides results within 24 h [155].
The effectiveness of solid-phase cytometry was analysed in 16 clinical samples, with the objective of identifying Candida cells present in the whole blood of patients diagnosed or suspected of candidemia [154]. Despite the low number of clinical samples used in the study, several advantages of this methodology when compared to blood cultures are that solid-phase cytometry was able to provide faster results (4 h), and also an accurate quantification of Candida cells, since all the patients previously diagnosed with candidemia presented positive results. This methodology was also able to identify mixed infections, present in 5 of the 16 clinical samples used, which suggests that it is a more common phenomenon than the one that diagnosis through blood cultures suggests [154].
8.3. Fourier transform infrared (FTIR)
The Fourier transform infrared (FTIR) methodology is based on the principles of spectroscopy. The functionality of this methodology is based on passing infrared radiation through the sample, where some radiation ends up being absorbed. The equipment's detector produces a spectrum that represents the molecular fingerprint of the analysed sample. In clinical terms, different microorganisms will produce different fingerprints, and their distinction is possible through the analysis of the spectra produced [156].
Potocki and co-workers [157] used FTIR methodology with the main objective of distinguishing Candida non-albicans from C. albicans species, since non-albicans species are mostly associated with resistance to antifungal agents used. FTIR was used in 25 clinical isolates of Candida spp. and the distinction of each isolate was possible due to the diversity of spectra produced by each species. The methodology also appears promising regarding the search for antifungal resistance genes, since resistant species will produce a different spectrum than a non-resistant species [157]. According to Erukhimovitch [158], the distinction between a bacterial and fungal infection remains a problem, especially due to the symptomatic similarities. Therefore, FTIR methodology is considered a great screening tool in these situations since bacteria and fungi produce completely different spectra. This study used fungal and bacterial strains to evaluate the applicability of FTIR in distinguishing these pathogens, and results showed that distinction was possible in just 1 h, which turns out to be a huge advantage [158].
8.4. Surface-enhanced Raman scattering (SERS)
Surface-enhanced Raman scattering (SERS) is a combination of Raman spectroscopy and the use of nanoparticles, which has been previously used to detect several pathogenic organisms, including fungi. This technique provides qualitative and quantitative analysis, allows to trace clinically relevant biomolecules, and establishes molecular profiles that can be important to determine the severity of fungal infections [159]. Moreover, a recent study conducted by Hu et al. [160] aimed to directly detect and identify Candida species in serum, by combining nanoparticles, SERS spectrum, and OPLS-DA multivariate statistical analysis. In this experiment, Fe3O4@PEI magnetic nanoparticles showed high capture efficiency of Candida cells in serum, due to electrostatic attraction, producing the Fe3O4@PEICandida complex. Then, positively charged silver nanoparticles (AgNPs+) were used as the substrate for SERS, to enhance the intensity of the signal. This method is described as fast, affordable, and non-destructive, as does not require pure cultures, cell wall lysis, or DNA extraction. This method presented an average accuracy of 99.8%, and the capture efficiency of Fe3O4@PEI in the serum samples was 95.9%, 98.0%, and 79.6% for C. albicans, C. tropicalis, and C. krusei, respectively [160].
8.5. Nanotechnology
Nanotechnology has increasingly contributed to the development and evolution of health-related fields. For instance, the application of gold nanoparticles has been intensively studied, being applied in vaccines as preventive agents, used as drug delivery systems in cancer or other health conditions therapies, and also in diagnostic approaches [161]. Sojinrin and co-workers [162] developed a protocol to detect the presence of spore-forming fungi based on gold nanoparticles. Essentially, when the gold nanoparticles enter in contact, for example, with A. niger, they endure structural and morphological changes, from spherical to star-shaped, and change of colour from red to blue. This technique showed sensitivity of 80% and specificity of 95% for athlete's foot diagnosis. This is a fast, straightforward and low-priced method, yet does not allow specific identification of pathogens [162].
8.6. Nuclear Magnetic Resonance (NMR)
Since 2001, NMR has been useful in the microbiology field for species identification and detection, through the use of nanoparticles, with subsequent analysis by magnetic resonance [82]. In this case, the detection of the target organism is done by beads that have a complementary sequence to the organism's DNA, allowing the binding. This binding allows the aggregation of beads, which can be observed through magnetic resonance. NMR methodologies can be used alone, or following a conventional PCR, for product analysis [34,35]. T2Candida® was the first methodology to be verified and validated by the FDA (Food and Drug Administration) for invasive candidiasis diagnosis. It is an automated platform based on NMR, which allows to identify 5 Candida spp. (C. albicans, C. glabrata, C. tropicalis, C. parapsilosis and C. krusei) directly from clinical samples of whole blood or serum, within 3–5 h [82,143]. Firstly, the clinical sample is inserted into the platform, yielding an automated DNA extraction, which is then analysed by magnetic resonance, detecting pathogenic Candida spp [[163], [164], [165]]. In the clinical trial study, T2Candida® demonstrated a sensitivity of 91.1% and specificity of 99.4% which was a major achievement regarding molecular diagnosis [165].
8.7. Biosensors
Other research area under constant development consists in the use of biosensors. Those are designed as portable devices that convert biological and biochemical information into an output analytical signal [166]. Fungal biosensors produced for clinic diagnosis have to fulfil several requirements, such as the careful selection of a specific biomarker of the target pathogen, which has to be suitable for the biological recognition system and to hold measurable features associated with normal conditions or with infection [166]. Pla et al. [167] described an innovative nanosensor to detect C. auris based on biocompatible nanoporous anodic alumina (NAA) supports, with the pores loaded with fluorophores and oligonucleotides attached. The oligonucleotides are specially selected in order to make the sensor completely specific for C. auris. When this pathogen is present in a sample, the oligonucleotide hybridizes to its genomic DNA exclusively, thus opening the pore and releasing the trapped fluorophore. This system presented high sensitivity (85%) and selectivity (100%) for C. auris detection from blood culture samples, also the results can be obtained within an hour, and previous steps such as DNA extraction are not required [167].
8.8. Volatile organic compounds (VOC) assay
Volatile organic compounds assay is a new type of methodology for the diagnosis of invasive aspergillosis, with sensitivity rates above 90%. In this assay, several metabolites characteristic of A. fumigatus are detected from the patient's exhaled air [35,168]. The innovative character of this assay is that it uses an artificial olfactory system that distinguishes several VOCs produced by the pathogen, called “breathprints” [[168], [169], [170]]. The majority of VOCs produced by A. fumigatus that are identified by this assay are 3-octanone, 2-pentylfuran, isoamyl alcohol, ethanol and others [171,172]. However, the detection of these metabolites is often associated with pulmonary aspergillosis [170].
9. Conclusion and final remarks
The scientific community has played a very important role in improving diagnostic methodologies in order to achieve accurate detection and identification of clinically relevant fungal pathogens. This development was mainly due to technological advancements in the last two decades, but also to the greater knowledge of molecular genetics. Another fundamental factor is the increasing interaction between humans and wildlife, which enhances the appearance of new pathogenic species.
Real-time PCR methodologies are becoming increasingly more valued for the diagnosis of fungal infections. This preference is mainly due to the easy handling of the methodology, and also because the reaction occurs in a closed system, which makes external contamination more difficult. For those reasons, the real-time PCR methodology remains the most widely used in the hospital environment for diagnosing numerous infectious diseases.
Regarding the identification of fungal pathogens, it is of utmost importance to achieve specific identifications, in order to establish an adequate therapeutic plan, increasing the patient's chance of survival. In the treatment of systemic fungal infections, identification at the species level is essential, because different fungal species have distinct antifungal susceptibilities. Therefore, a specific antifungal, with a specific concentration should be used. For example, C. auris is resistant to the majority of antifungals, C. glabrata easily acquire resistance to fluconazole, and C. krusei has intrinsic resistance to azoles.
The development of more sophisticated and automated molecular methodologies that deliver faster results represents a huge improvement in the clinical management of fungal infections. However, there is a long way to go to accomplish the global standardization of such methodologies.
Funding
This work was supported by the UID/BIA/04050/2013 (POCI-01-0145-FEDER-007569) and UID/BIA/04050/2019 strategic programs, which is funded by national funds through the FCT-Fundação para a Ciência e Tecnologia, and by the ERDF- European Regional Development Fund through the COMPETE2020–Programa Operacional Competitividade e Internacionalização (POCI) and Sistema de Apoio à Investigação Científica e Tecnológica (SAICT).
Declaration of competing interest
All authors report no conflicts of interest relevant to this article.
References
- 1.Echavarria M., Robinson C.R.T.A. Manual of clinical microbiology. 11th ed. ASM Press; Washington DC: 2015. Taxonomy and classification of fungi; pp. 1935–1943. [Google Scholar]
- 2.Xu J. Fungal species concepts in the genomics era. Genome. 2020;63 doi: 10.1139/gen-2020-0022. [DOI] [PubMed] [Google Scholar]
- 3.Stajich J.E., Kahmann R., Boone C., Denning D.W., Gow N.A.R., Klein B.S., et al. Threats posed by the fungal kingdom to humans. Wildlife, and Agriculture. 2020;11:1–17. doi: 10.1128/mBio.00449-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Konopka J.B., Casadevall A., Taylor J.W., Heitman J., Cowen L. Colloq. Rep.; 2019. One health: fungal pathogens of humans, animals and plants; pp. 1–3. [Google Scholar]
- 5.Visited on-line: november 17 Clinical fungi. Atlas clin fungi. 2021. www.clinicalfungi.org
- 6.Borman A.M., Johnson E.M. Name changes for fungi of medical importance, 2018 to 2019. J Clin Microbiol. 2021:59. doi: 10.1128/JCM.01811-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hawksworth D.L., Lücking R. Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol Spectr. 2017;15 doi: 10.1128/microbiolspec.FUNK-0052-2016.Correspondence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Köhler J.R., Casadevall A., Perfect J. The spectrum of fungi that infects humans. Cold Spring Harb Perspect Med. 2015;5 doi: 10.1101/cshperspect.a019273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nucci M., Marr K.A. Emerging fungal diseases. Clin Infect Dis. 2005;41 doi: 10.1086/432060. [DOI] [PubMed] [Google Scholar]
- 10.Bongomin F., Gago S., Oladele R.O., Denning D.W. Global and multi-national prevalence of fungal diseases—estimate precision. J Fungi. 2017;3 doi: 10.3390/jof3040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janbon G., Quintin J., Lanternier F., d'Enfert C. Studying fungal pathogens of humans and fungal infections: fungal diversity and diversity of approaches. Genes Immun. 2019;20:403–414. doi: 10.1038/s41435-019-0071-2. [DOI] [PubMed] [Google Scholar]
- 12.Damasceno-Escoura A.H., Mora D.J., Cardeal A.C., Berto-Nascimento J.C., Etchebehere R.M., de Meneses A.C.O., et al. Histoplasmosis in HIV-infected patients: epidemiological, clinical and necropsy data from a Brazilian teaching hospital. Mycopathologia. 2020;185:339–346. doi: 10.1007/s11046-020-00435-y. [DOI] [PubMed] [Google Scholar]
- 13.Sharma S Grover, Bhargava M., S Samdani S., Kataria T. Post coronavirus disease mucormycosis: a deadly addition to the pandemic spectrum. J Laryngol Otol. 2021:135. doi: 10.1017/S0022215121000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gokulshankar S., Bk M. Covid-19 and black fungus. Asian J Med Heal Sci. 2021:4. [Google Scholar]
- 15.Arora V. Mucormycosis – the black fungus. J Cardiol Cardiovasc Res. 2021:25. doi: 10.37191/mapsci-jccr-2(2)-028. [DOI] [Google Scholar]
- 16.Visited on-line: 29 september COVID-19-Associated mucormycosis: triple threat of the pandemic. https://asm.org/Articles/2021/July/COVID-19-Associated-Mucormycosis-Triple-Threat-of
- 17.Visited on-line: 29 september Joint general sessions - emerging pathogens. https://www.worldmicrobeforum.org/index.php/program/joint-general-sessions
- 18.Visited on-line: 29 september Mucormycosis black fungus. https://governmentstats.com/mucormycosis/index.html
- 19.Ahmadikia K., Hashemi S.J., Khodavaisy S., Getso M.I., Alijani N., Badali H., et al. The double-edged sword of systemic corticosteroid therapy in viral pneumonia: a case report and comparative review of influenza-associated mucormycosis versus COVID-19 associated mucormycosis. Mycoses. 2021 doi: 10.1111/myc.13256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chouhan A.S. Overuse of steroid drugs Methylprednisolone and Dexamethasone (Oral) causes a diabetic patient to become infected with the Black Fungus in the corona virus. Sys Rev Pharm. 2021;12(11):630–634. [Google Scholar]
- 21.Visited on-line: 30 september Fighting India's “black fungus” epidemic. https://www.pharmaceutical-technology.com/features/fighting-indias-black-fungus-epidemic/
- 22.Alfouzan W., Ahmad S., Dhar R., Asadzadeh M., Almerdasi N., Abdo N.M., et al. Molecular epidemiology of Candida auris outbreak in a major secondary-care hospital in Kuwait. J Fungi. 2020;6:1–18. doi: 10.3390/jof6040307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Theodoropoulos N.M., Bolstorff B., Bozorgzadeh Adel, Christina Brandeburg M.C., Daly J.S., III Rte, et al. Candida auris outbreak involving liver transplant recipients in a surgical intensive care unit. Am J Transplant. 2020;20:3673–3679. doi: 10.1111/ajt.16144. [DOI] [PubMed] [Google Scholar]
- 24.Prestel C., Anderson E., Forsberg K., Lyman M., de Perio M.A., Kuhar D., et al. Candida auris outbreak in a COVID-19 specialty care unit — Florida, july–august 2020. MMWR Morb Mortal Wkly Rep. 2021;70:56–57. doi: 10.15585/mmwr.mm7002e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mulet Bayona J.V., Tormo Palop N., Salvador García C., Fuster Escrivá B., Chanzá Aviñó M., Ortega García P., et al. Impact of the SARS-CoV-2 pandemic in candidaemia, invasive aspergillosis and antifungal consumption in a tertiary hospital. J Fungi. 2021;7 doi: 10.3390/jof7060440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chowdhary A., Sharma A. The lurking scourge of multidrug resistant Candida auris in times of COVID-19 pandemic. J Glob Antimicrob Resist. 2020;22:175–176. doi: 10.1016/j.jgar.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodrigues M.L., Nosanchuk J.D. Fungal diseases as neglected pathogens: a wake-up call to public health officials. PLoS Negl Trop Dis. 2020;14 doi: 10.1371/journal.pntd.0007964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ascioglu S., Rex J.H., De Pauw B., Bennett J.E., Bille J., Crokaert F., et al. Defining opportunistic invasive fungal infections in immunocompromised patients with cancer and hematopoietic stem cell transplants: an international consensus. Clin Infect Dis. 2002;34 doi: 10.1086/323335. [DOI] [PubMed] [Google Scholar]
- 29.De Pauw B., Walsh T.J., Donnelly J.P., Stevens D.A., Edwards J.E., Calandra T., et al. Revised definitions of invasive fungal disease from the European organization for research and treatment of cancer/invasive fungal infections cooperative group and the national Institute of allergy and infectious diseases mycoses study group (EORTC/MSG) C. Clin Infect Dis. 2008;46 doi: 10.1086/588660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peter Donnelly J., Chen S.C., Kauffman C.A., Steinbach W.J., Baddley J.W., Verweij P.E., et al. Revision and update of the consensus definitions of invasive fungal disease from the european organization for research and treatment of cancer and the mycoses study group education and research consortium. Clin Infect Dis. 2020;71 doi: 10.1093/cid/ciz1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnold H.M., Micek S.T., Shorr A.F., Zilberberg M.D., Labelle A.J., Kothari S., et al. Hospital resource utilization and costs of inappropriate treatment of candidemia. Pharmacotherapy. 2010;30 doi: 10.1592/phco.30.4.361. [DOI] [PubMed] [Google Scholar]
- 32.Zhang H., Zhu A. Emerging invasive fungal infections: clinical features and controversies in diagnosis and treatment processes. Infect Drug Resist. 2020;13 doi: 10.2147/IDR.S237815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Webb B.J., Ferraro J.P., Rea S., Kaufusi S., Goodman B.E., Spalding J. Epidemiology and clinical features of invasive fungal infection in a US health care network. Open Forum Infect Dis. 2018;5 doi: 10.1093/ofid/ofy187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franco-Duarte R., Černáková L., Kadam S., Kaushik K.S., Salehi B., Bevilacqua A., et al. Advances in chemical and biological methods to identify microorganisms—from past to present. Microorganisms. 2019;7 doi: 10.3390/microorganisms7050130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arvanitis M., Anagnostou T., Fuchs B.B., Caliendo A.M., Mylonakis E. Molecular and nonmolecular diagnostic methods for invasive fungal infections. Clin Microbiol Rev. 2014;27 doi: 10.1128/CMR.00091-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Badiee P., Alborzi A., Karimi M., Pourabbas B., Haddadi P., Mardaneh J., et al. Diagnostic potential of nested PCR, galactomannan EIA, and beta-D-glucan for invasive aspergillosis in pediatric patients. J Infect Dev Ctries. 2012;6 doi: 10.3855/jidc.2110. [DOI] [PubMed] [Google Scholar]
- 37.Colombo A.L., de Almeida Júnior J.N., Slavin M.A., Chen S.C.A., Sorrell T.C. Candida and invasive mould diseases in non-neutropenic critically ill patients and patients with haematological cancer. Lancet Infect Dis. 2017;17 doi: 10.1016/S1473-3099(17)30304-3. [DOI] [PubMed] [Google Scholar]
- 38.Ericson E.L., Klingspor L., Ullberg M., Özenci V. Clinical comparison of the Bactec Mycosis IC/F, BacT/Alert FA, and BacT/Alert FN blood culture vials for the detection of candidemia. Diagn Microbiol Infect Dis. 2012;73 doi: 10.1016/j.diagmicrobio.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 39.Park B.R.G., Kim T.H., Kim H.R., Lee M.K. Comparative analysis of simulated candidemia using two different blood culture systems and the rapid identification of Candida albicans. Ann Clin Lab Sci. 2011;41 [PubMed] [Google Scholar]
- 40.Fisher J.F. Candida urinary tract infections - epidemiology, pathogenesis, diagnosis, and treatment: executive summary. Clin Infect Dis. 2011;52 doi: 10.1093/cid/cir108. [DOI] [PubMed] [Google Scholar]
- 41.Gajdács M., Dóczi I., Ábrók M., Lázár A., Burián K. Epidemiology of candiduria and Candida urinary tract infections in inpatients and outpatients: results from a 10-year retrospective survey. Cent Eur J Urol. 2019;72 doi: 10.5173/ceju.2019.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toner L., Papa N., Aliyu S.H., Dev H., Lawrentschuk N., Al-Hayek S. Candida growth in urine cultures: a contemporary analysis of species and antifungal susceptibility profiles. Qjm. 2016;109 doi: 10.1093/qjmed/hcv202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Helbig S., Achkar J.M., Jain N., Wang X., Gialanella P., Levi M., et al. Diagnosis and inflammatory response of patients with candiduria. Bone. 2013;23:1–7. doi: 10.1111/j.1439-0507.2012.02201.x.Diagnosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barenfanger J., Lawhorn J., Drake C. Nonvalue of culturing cerebrospinal fluid for fungi. J Clin Microbiol. 2004;42 doi: 10.1128/JCM.42.1.236-238.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwartz S., Kontoyiannis D.P., Harrison T., Ruhnke M. Advances in the diagnosis and treatment of fungal infections of the CNS. Lancet Neurol. 2018;17:362–372. doi: 10.1016/S1474-4422(18)30030-9. [DOI] [PubMed] [Google Scholar]
- 46.Cohen-Wolkowiez M., Smith P.B., Mangum B., Steinbach W.J., Alexander B.D., Cotten C.M., et al. Neonatal Candida meningitis: significance of cerebrospinal fluid parameters and blood cultures. J Perinatol. 2007;27 doi: 10.1038/sj.jp.7211628. [DOI] [PubMed] [Google Scholar]
- 47.Donohue M. Cerebrospinal Fluid Culture; 2016. Visited On-Line: October 6.https://www.healthline.com/health/cerebrospinal-fluid-culture [Google Scholar]
- 48.Nucci M., Anaissie E. Fusarium infections in immunocompromised patients. Clin Microbiol Rev. 2007;20:695–704. doi: 10.1128/CMR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guegan H., Robert-Gangneux F., Camus C., Belaz S., Marchand T., Baldeyrou M., et al. Improving the diagnosis of invasive aspergillosis by the detection of Aspergillus in broncho-alveolar lavage fluid: comparison of non-culture-based assays. J Infect. 2018;76 doi: 10.1016/j.jinf.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 50.Bao J.R., Master R.N., Jones R.S., Clark R.B., Moore E.C., Shier K.L. Recovery and its dynamics of filamentous fungi from clinical specimen cultures: an extensive study. Microbiol Spectr. 2021;9 doi: 10.1128/spectrum.00080-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tarrand J.J., Han X.Y., Kontoyiannis D.P., May G.S. Aspergillus hyphae in infected tissue: evidence of physiologic adaptation and effect on culture recovery. J Clin Microbiol. 2005;43 doi: 10.1128/JCM.43.1.382-386.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schelenz S., Barnes R.A., Barton R.C., Cleverley J.R., Lucas S.B., Kibbler C.C., et al. British Society for Medical Mycology best practice recommendations for the diagnosis of serious fungal diseases. Lancet Infect Dis. 2015;15:461–474. doi: 10.1016/S1473-3099(15)70006-X. [DOI] [PubMed] [Google Scholar]
- 53.Jensen H.E. Histopathology in the diagnosis of invasive fungal diseases. Curr Fungal Infect Rep. 2021 doi: 10.1007/s12281-021-00412-y. [DOI] [Google Scholar]
- 54.Da Silva R.M., Da Silva Neto J.R., Santos C.S., Frickmann H., Poppert S., Cruz K.S., et al. Evaluation of fluorescence in situ hybridisation (FISH) for the detection of fungi directly from blood cultures and cerebrospinal fluid from patients with suspected invasive mycoses. Ann Clin Microbiol Antimicrob. 2015;14 doi: 10.1186/s12941-015-0065-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jenks J.D., Gangneux J.-P., Schwartz I.S., Alastruey-Izquierdo A., Lagrou K., Thompson G.R., III, et al. Diagnosis of breakthrough fungal infections in the clinical mycology laboratory: an ECMM consensus statement. J Fungi. 2020;6 doi: 10.3390/jof6040216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martins M.L., Ferreira A.S., Sampaio A., Vieira R., Inácio J. Direct and specific identification of Cryptococcus neoformans in biological samples using fluorescently labelled DNA probes. Eur J Clin Microbiol Infect Dis. 2010;29:571–576. doi: 10.1007/s10096-010-0897-z. [DOI] [PubMed] [Google Scholar]
- 57.Sato Y., Osabe S., Kuno H., Kaji M., Oizumi K. Rapid diagnosis of cryptococcal meningitis by microscopic examination of centrifuged cerebrospinal fluid sediment. J Neurol Sci. 1999;164:72–75. doi: 10.1016/S0022-510X(99)00047-7. [DOI] [PubMed] [Google Scholar]
- 58.Sangoi A.R., Rogers W.M., Longacre T.A., Montoya J.G., Baron E.J., Banaei N. Challenges and pitfalls of morphologic identification of fungal infections in histologic and cytologic specimens : a Ten-Year retrospective review at a single institution. Am J Clin Pathol. 2009;131:364–375. doi: 10.1309/AJCP99OOOZSNISCZ. [DOI] [PubMed] [Google Scholar]
- 59.Scharmann U., Kirchhoff L., Chapot V. le S., Dziobaka J., Verhasselt H.L., Stauf R., et al. Comparison of four commercially available chromogenic media to identify Candida albicans and other medically relevant Candida species. Mycoses. 2020;63:823–831. doi: 10.1111/myc.13119. [DOI] [PubMed] [Google Scholar]
- 60.Perry J.D. A decade of development of chromogenic culture media for clinical microbiology in an era of molecular diagnostics impact of laboratory automation on the use of chromogenic media . 468. Clin Microbiol Reviiews. 2017;30:449–479. doi: 10.1128/CMR.00097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pravin Charles M.V., Kali A., Joseph N.M. Performance of chromogenic media for Candida in rapid presumptive identification of Candida species from clinical materials. Pharmacognosy Res. 2015;7:S69–S73. doi: 10.4103/0974-8490.150528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Borman A.M., Fraser M., Johnson E.M. Chromagartmcandida plus: a novel chromogenic agar that permits the rapid identification of Candida auris. Med Mycol. 2021;59:253–258. doi: 10.1093/mmy/myaa049. [DOI] [PubMed] [Google Scholar]
- 63.Jong AW De. 2021. Performance of two novel chromogenic media for the identification of multidrug-resistant Candida Auris compared. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zuluaga A., Arango-Bustamante K., Caceres D.H., Sánchez-Quitian Z.A., Velásquez V., Gómez B.L., et al. Concordance analysis between different methodologies used for identification of oral isolates of Candida species. Colomb Méd. 2018;49:193–200. doi: 10.25100/cm.v49i3.3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aarstehfar A., Daneshnia F., Kord M., Roudbary M., Zarrinfar H., Fang W., et al. Comparison of 21-Plex PCR and API 20C AUX, MALDI-TOF MS, and rDNA sequencing for a wide range of clinically isolated yeast species: improved identification by combining 21-Plex PCR and API 20C AUX as an alternative strategy for developing countries. Front Cell Infect Microbiol. 2019;9 doi: 10.3389/fcimb.2019.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karabiçak N., Uludağ Altun H., Karatuna O., Hazirolan G., Aksu N., Adi̇loğlu A., et al. Evaluation of common commercial systems for the identification of yeast isolates in microbiology laboratories. A Multicenter Study. 2015;49:210–220. doi: 10.5578/mb.9370. [DOI] [PubMed] [Google Scholar]
- 67.Huang Y.S., Wang F Der, Chen Y.C., Huang Y.T., Hsieh M.H., Hii I.M., et al. High rates of misidentification of uncommon Candida species causing bloodstream infections using conventional phenotypic methods. J Formos Med Assoc. 2021;120:1179–1187. doi: 10.1016/j.jfma.2020.11.002. [DOI] [PubMed] [Google Scholar]
- 68.Pulcrano G., Iula D.V., Vollaro A., Tucci A., Cerullo M., Esposito M., et al. Rapid and reliable MALDI-TOF mass spectrometry identification of Candida non-albicans isolates from bloodstream infections. J Microbiol Methods. 2013;94:262–266. doi: 10.1016/j.mimet.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 69.Chao Q.T., Lee T.F., Teng S.H., Peng L.Y., Chen P.H., Teng L.J., et al. Comparison of the accuracy of two conventional phenotypic methods and two MALDI-TOF MS systems with that of DNA sequencing analysis for correctly identifying clinically encountered yeasts. PLoS One. 2014;9 doi: 10.1371/journal.pone.0109376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ambaraghassi G., Dufresne P.J., Dufresne S.F., Vallières É., Muñoz J.F., Cuomo C.A., et al. Identification of Candida auris by use of the updated vitek 2 yeast identification system, version 8.01: a multilaboratory evaluation study. J Clin Microbiol. 2019;57 doi: 10.1128/JCM.00884-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tan Y.E., Teo J.Q.M., Rahman N.B.A., Ng O.T., Kalisvar M., Tan A.L., et al. Candida auris in Singapore: genomic epidemiology, antifungal drug resistance, and identification using the updated 8.01 VITEK2 system. Int J Antimicrob Agents. 2019;54:709–715. doi: 10.1016/j.ijantimicag.2019.09.016. [DOI] [PubMed] [Google Scholar]
- 72.Magobo R.E., Corcoran C., Seetharam S., Govender N.P. Candida auris-associated candidemia, South Africa. Emerg Infect Dis. 2014;20:1250–1251. doi: 10.3201/eid2007.131654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chowdhary A., Sharma C., Duggal S., Agarwal K., Prakash A., Singh P.K., et al. New clonal strain of Candida auris, Delhi, India. Emerg Infect Dis. 2013;19:1670–1673. doi: 10.3201/eid1910.130393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Becker P., Normand A.C., Vanantwerpen G., Vanrobaeys M., Haesendonck R., Vercammen F., et al. Identification of fungal isolates by MALDI-TOF mass spectrometry in veterinary practice: validation of a web application. J Vet Diagn Investig. 2019;31 doi: 10.1177/1040638719835577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lau A.F., Drake S.K., Calhoun L.B., Henderson C.M., Zelazny A.M. Development of a clinically comprehensive database and a simple procedure for identification of molds from solid media by matrix-assisted laser desorption ionization-Time of flight mass spectrometry. J Clin Microbiol. 2013;51 doi: 10.1128/JCM.02852-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Teke L., Barış A., Bayraktar B. Comparative evaluation of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) systems for non-albicans Candida and uncommon yeast isolates. J Microbiol Methods. 2021:185. doi: 10.1016/j.mimet.2021.106232. [DOI] [PubMed] [Google Scholar]
- 77.Yi Q., Xiao M., Fan X., Zhang G., Yang Y., Zhang J.J., et al. Evaluation of Autof MS 1000 and vitek MS MALDI-TOF MS system in identification of closely-related yeasts causing invasive fungal diseases. Front Cell Infect Microbiol. 2021;11:1–7. doi: 10.3389/fcimb.2021.628828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Robert M.G., Romero C., Dard C., Garnaud C., Cognet O., Girard T., et al. Evaluation of ID fungi plates medium for identification of molds by MALDI biotyper. J Clin Microbiol. 2020;58:1–8. doi: 10.1128/JCM.01687-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Robert M.G., Cornet M., Hennebique A., Rasamoelina T., Caspar Y., Pondérand L., et al. MALDI-TOF MS in a medical mycology laboratory: on stage and backstage. Microorganisms. 2021;9:1–16. doi: 10.3390/microorganisms9061283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vatanshenassan M., Boekhout T., Lass-Flörl C., Lackner M., Schubert S., Kostrzewa M., et al. Proof of concept for MBT ASTRA, a rapid matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS)-Based method to detect caspofungin resistance in Candida albicans and Candida glabrata. J Clin Microbiol. 2018;56 doi: 10.1128/jcm.00420-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vatanshenassan M., Boekhout T., Meis J.F., Berman J., Chowdhary A., Ben-Ami R., et al. Candida auris identification and rapid antifungal susceptibility testing against echinocandins by MALDI-TOF MS. Front Cell Infect Microbiol. 2019;9 doi: 10.3389/fcimb.2019.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peker N., Couto N., Sinha B., Rossen J.W. Diagnosis of bloodstream infections from positive blood cultures and directly from blood samples: recent developments in molecular approaches. Clin Microbiol Infect. 2018;24 doi: 10.1016/j.cmi.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 83.Klingspor L., Lindbäck E., Ullberg M., Özenci V. Seven years of clinical experience with the Yeast Traffic Light PNA FISH: assay performance and possible implications on antifungal therapy. Mycoses. 2018;61 doi: 10.1111/myc.12722. [DOI] [PubMed] [Google Scholar]
- 84.Salimnia H., Fairfax M.R., Lephart P.R., Schreckenberger P., DesJarlais S.M., Johnson J.K., et al. Evaluation of the FilmArray blood culture identification panel: results of a multicenter controlled trial. J Clin Microbiol. 2016;54 doi: 10.1128/JCM.01679-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Altun O., Almuhayawi M., Ullberg M., Ozenci V. Clinical evaluation of the Filmarray blood culture identification panel in identification of bacteria and yeasts from positive blood culture bottles. J Clin Microbiol. 2013;51 doi: 10.1128/JCM.01835-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Visited on-line: october 9 BioMérieux diagnostic. https://www.biomerieux-diagnostics.com/biofire-bcid-panel
- 87.Burton T., Anderson A., Seare J., Dillon R.J., Mccann E. Multi-center evaluation of the BioFire® FilmArray® blood culture identification 2 panel for the detection of microorganisms and resistance markers in positive blood cultures. Open Forum Infect Dis. 2019;6:2018–2019. [Google Scholar]
- 88.Galiana A., Coy J., Gimeno A., Guzman N.M., Rosales F., Merino E., et al. Evaluation of the Sepsis Flow Chip assay for the diagnosis of blood infections. PLoS One. 2017;12 doi: 10.1371/journal.pone.0177627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Visited on-line: october 9 MasterDiagnóstica. www.masterdiagnostica.com
- 90.Carroll K.C., Reid J.L., Thornberg A., Whitfield N.N., Trainor D., Lewis S., et al. Clinical performance of the novel Genmark DX eplex blood culture ID Gram-positive panel. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.01730-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bryant S., Almahmoud I., Pierre I., Bardet J., Touati S., Maubon D., et al. Evaluation of microbiological performance and the potential clinical impact of the ePlex® blood culture identification panels for the rapid diagnosis of bacteremia and fungemia. Front Cell Infect Microbiol. 2020;10 doi: 10.3389/fcimb.2020.594951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang S.X., Carroll K.C., Lewis S., Totten M., Mead P., Samuel L., et al. Multicenter evaluation of a PCR-based digital microfluidics and electrochemical detection system for the rapid identification of 15 fungal pathogens directly from positive blood cultures. J Clin Microbiol. 2020;58:1–9. doi: 10.1128/JCM.02096-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Orlowski H.L.P., McWilliams S., Mellnick V.M., Bhalla S., Lubner M.G., Pickhardt P.J., et al. Imaging spectrum of invasive fungal and fungal-like infections. Radiographics. 2017;37:1119–1134. doi: 10.1148/rg.2017160110. [DOI] [PubMed] [Google Scholar]
- 94.Visited on-line: october 10 Inside view. https://blog.radiology.virginia.edu/different-imaging-tests-explained/
- 95.Alexander B.D., Lamoth F., Heussel C.P., Prokop C.S., Desai S.R., Morrissey C.O., et al. Guidance on imaging for invasive pulmonary aspergillosis and mucormycosis: from the imaging working group for the revision and update of the consensus definitions of fungal disease from the EORTC/MSGERC. Clin Infect Dis. 2021;72:S79–S88. doi: 10.1093/cid/ciaa1855. [DOI] [PubMed] [Google Scholar]
- 96.van Dyck K., Rogiers O., Vande Velde G., van Dijck P. Let's shine a light on fungal infections: a noninvasive imaging toolbox. PLoS Pathog. 2020;16:1–6. doi: 10.1371/journal.ppat.1008257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wright W.F., Overman S.B., Ribes J.A. (1-3)-β-D-glucan assay: a review of its laboratory and clinical application. Lab Med. 2011;42:679–685. doi: 10.1309/LM8BW8QNV7NZBROG. [DOI] [Google Scholar]
- 98.Akamatsu N., Sugawara Y., Kaneko J., Tamura S., Makuuchi M. Preemptive treatment of fungal infection based on plasma (1→3)β-D-glucan levels after liver transplantation. Infection. 2007;35:346–351. doi: 10.1007/s15010-007-6240-7. [DOI] [PubMed] [Google Scholar]
- 99.de Carolis E., Marchionni F., Torelli R., Angela M.G., Pagano L., Murri R., et al. Comparative performance evaluation of Wako β-glucan test and Fungitell assay for the diagnosis of invasive fungal diseases. PLoS One. 2020;15 doi: 10.1371/journal.pone.0236095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ostrosky-Zeichner L., Alexander B.D., Kett D.H., Vazquez J., Pappas P.G., Saeki F., et al. Multicenter clinical evaluation of the (1→3) β-D-glucan assay as an aid to diagnosis of fungal infections in humans. Clin Infect Dis. 2005;41 doi: 10.1086/432470. [DOI] [PubMed] [Google Scholar]
- 101.Mohr J.F., Sims C., Paetznick V., Rodriguez J., Finkelman M.A., Rex J.H., et al. Prospective survey of (1→3)-β-D-glucan and its relationship to invasive candidiasis in the surgical intensive care unit setting. J Clin Microbiol. 2011;49:58–61. doi: 10.1128/JCM.01240-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.León C., Ruiz-Santana S., Saavedra P., Castro C., Loza A., Zakariya I., et al. Contribution of Candida biomarkers and DNA detection for the diagnosis of invasive candidiasis in ICU patients with severe abdominal conditions. Crit Care. 2016;20:1–14. doi: 10.1186/S13054-016-1324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sulahian A., Porcher R., Bergeron A., Touratier S., Raffoux E., Menotti J., et al. Use and limits of (1-3)-β-D-glucan assay (fungitell), compared to galactomannan determination (platelia Aspergillus), for diagnosis of invasive aspergillosis. J Clin Microbiol. 2014;52:2328–2333. doi: 10.1128/JCM.03567-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pickering J.W., Sant H.W., Bowles C.A.P., Roberts W.L., Woods G.L. Evaluation of a (1→3)-β-D-glucan assay for diagnosis of invasive fungal infections. J Clin Microbiol. 2005;43:5957–5962. doi: 10.1128/JCM.43.12.5957-5962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Carolis E De, Marchionni F., Rosa M La, Pascale G De, Carelli S., Sanguinetti M., et al. 2019. Evaluation of Wako Beta-Glucan Test Performance in Diagnosing Invasive Fungal Infections. [Google Scholar]
- 106.Friedrich R., Rappold E., Bogdan C. Comparative analysis of the wako -glucan test and the fungitell assay for diagnosis of candidemia and Pneumocystis jirovecii pneumonia. J Clin Microbiol. 2018;56:1–12. doi: 10.1128/JCM.00464-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Racil Z., Kocmanova I., Lengerova M., Weinbergerova B., Buresova L., Toskova M., et al. Difficulties in using 1,3-β-D-glucan as the screening test for the early diagnosis of invasive fungal infections in patients with haematological malignancies - high frequency of false-positive results and their analysis. J Med Microbiol. 2010;59 doi: 10.1099/jmm.0.019299-0. [DOI] [PubMed] [Google Scholar]
- 108.Mennink-Kersten M.A.S.H. Dorien ruegebrink and PEV. Pseudomonas aeruginosa as a cause of (1→3)-β-D-glucan assay reactivity. Clin Infect Dis. 2008;46 doi: 10.1086/588564. [DOI] [PubMed] [Google Scholar]
- 109.Hammarström H., Kondori N., Friman V., Wennerås C. How to interpret serum levels of beta-glucan for the diagnosis of invasive fungal infections in adult high-risk hematology patients: optimal cut-off levels and confounding factors. Eur J Clin Microbiol Infect Dis. 2015;34 doi: 10.1007/s10096-014-2302-9. [DOI] [PubMed] [Google Scholar]
- 110.Hartl B., Zeller I., Manhart A., Selitsch B., Lass-Flörl C., Willinger B. A retrospective assessment of four antigen assays for the detection of invasive candidiasis among high-risk hospitalized patients. Mycopathologia. 2018;183:513–519. doi: 10.1007/s11046-017-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pappas P.G., Kauffman C.A., Andes D.R., Clancy C.J., Marr K.A., Ostrosky-Zeichner L., et al. Clinical practice guideline for the management of candidiasis: 2016 update by the infectious diseases society of America. Clin Infect Dis. 2015;62:e1–e50. doi: 10.1093/cid/civ933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ruan H., Xu W., Xia M., Ma Z., Fu S., Chen X. Analysis of clinical characteristics and diagnostic value of fungal serology in patients with invasive candidiasis. Open J Med Microbiol. 2020;10:222–232. doi: 10.4236/ojmm.2020.104019. [DOI] [Google Scholar]
- 113.Wang K., Luo Y., Zhang W., Xie S., Yan P., Liu Y., et al. Diagnostic value of Candida mannan antigen and anti-mannan IgG and IgM antibodies for Candida infection. Mycoses. 2020;63:181–188. doi: 10.1111/myc.13035. [DOI] [PubMed] [Google Scholar]
- 114.Visited on-line: october 28 Vircell microbiologist. https://en.vircell.com/?fbclid=IwAR2DazI1X4Gafn7bQGoytjD1llhQz5Fz8-ZbXPadwK3O9JNPH4h4tYquBc0
- 115.Parra-Sánchez M., Zakariya-Yousef Breval I., Castro Méndez C., García-Rey S., Loza Vazquez A., Úbeda Iglesias A., et al. Candida albicans germ-tube Antibody: evaluation of a new automatic assay for diagnosing invasive candidiasis in ICU patients. Mycopathologia. 2017;182:645–652. doi: 10.1007/s11046-017-0125-9. [DOI] [PubMed] [Google Scholar]
- 116.Pietro P., Bruna C., Enrico M., Anna C., Claudia V., Mario S., et al. Performance of Candida albicans germ tube antibodies (CAGTA) and its association with (1 → 3)-β-D-glucan (BDG) for diagnosis of invasive candidiasis (IC) Diagn Microbiol Infect Dis. 2019;93:39–43. doi: 10.1016/j.diagmicrobio.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 117.Wei S., Wu T., Wu Y., Ming D., Zhu X. Diagnostic accuracy of Candida albicans germ tube antibody for invasive candidiasis: systematic review and meta-analysis. Diagn Microbiol Infect Dis. 2019;93:339–345. doi: 10.1016/j.diagmicrobio.2018.10.017. [DOI] [PubMed] [Google Scholar]
- 118.Ulusakarya A., Chachaty E., Vantelon J.M., Youssef A., Tancrède C., Pico J.L., et al. Surveillance of Aspergillus galactomannan antigenemia for invasive aspergillosis by enzyme-linked immunosorbent assay in neutropenic patients treated for hematological malignancies. Hematol J. 2000;1(2):111–116. doi: 10.1038/sj.thj.6200009. [DOI] [PubMed] [Google Scholar]
- 119.Maertens J., Verhaegen J., Demuynck H., Brock P., Verhoef G., Vandenberghe P., et al. Autopsy-controlled prospective evaluation of serial screening for circulating galactomannan by a sandwich enzyme-linked immunosorbent assay for hematological patients at risk for invasive aspergillosis. J Clin Microbiol. 1999;37:3223–3228. doi: 10.1128/jcm.37.10.3223-3228.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Maertens J., Verhaegen J., Lagrou K., Van Eldere J., Boogaerts M. Screening for circulating galactomannan as a noninvasive diagnostic tool for invasive aspergillosis in prolonged neutropenic patients and stem cell transplantation recipients: a prospective validation. Blood. 2001;97:1604–1610. doi: 10.1182/blood.V97.6.1604. [DOI] [PubMed] [Google Scholar]
- 121.Bretagne S., Costa J.M., Bart-Delabesse E., Dhédin N., Rieux C., Cordonnier C. Comparison of serum galactomannan antigen detection and competitive polymerase chain reaction for diagnosing invasive aspergiliosis. Clin Infect Dis. 1998;26:1407–1412. doi: 10.1086/516343. [DOI] [PubMed] [Google Scholar]
- 122.Dichtl K., Seybold U., Ormanns S., Horns H. Evaluation of a novel Aspergillus antigen enzyme-linked immunosorbent assay. J Clin Microbiol. 2019;57 doi: 10.1128/JCM.00136-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Visited on-line: october 28 Bio-Rad. https://www.bio-rad.com/SearchResults?search_api_fulltext=elisa+platelia+aspergillus
- 124.Min Z., Baddley J.W., Rodriguez J.M., Moser S.A., Patel M. Cross-reactivity of Aspergillus galactomannan in an HIV-infected patient with histoplasmosis. Med Mycol Case Rep. 2012;1 doi: 10.1016/j.mmcr.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tortorano A.M., Esposto M.C., Prigitano A., Grancini A., Ossi C., Cavanna C., et al. Cross-reactivity of Fusarium spp. in the Aspergillus galactomannan enzyme-linked immunosorbent assay. J Clin Microbiol. 2012;50 doi: 10.1128/JCM.05946-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Vergidis P., Walker R.C., Kaul D.R., Kauffman C.A., Freifeld A.G., Slagle D.C., et al. False-positive Aspergillus galactomannan assay in solid organ transplant recipients with histoplasmosis. Transpl Infect Dis. 2012;14 doi: 10.1111/j.1399-3062.2011.00675.x. [DOI] [PubMed] [Google Scholar]
- 127.White P.L., Parr C., Thornton C., Barnes R.A. Evaluation of real-time PCR, galactomannan enzyme-linked immunosorbent assay (ELISA), and a novel lateral-flow device for diagnosis of invasive aspergillosis. J Clin Microbiol. 2013;51 doi: 10.1128/JCM.03189-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lass-Flörl C., Lo Cascio G., Nucci M., Camargo dos Santos M., Colombo A.L., Vossen M., et al. Respiratory specimens and the diagnostic accuracy of Aspergillus lateral flow assays (LFA-IMMYTM): real-life data from a multicentre study. Clin Microbiol Infect. 2019;25 doi: 10.1016/j.cmi.2019.08.009. [DOI] [PubMed] [Google Scholar]
- 129.White P.L., Price J.S., Posso R., Vale L., Backx M. Evaluation of the performance of the IMMY sona Aspergillus galactomannan lateral flow assay when testing serum to aid in diagnosis of invasive aspergillosis. J Clin Microbiol. 2020;58:1–7. doi: 10.1128/JCM.01006-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Abdallah W., Myint T., LaRue R., Minderman M., Gunn S., Wheat L.J., et al. Diagnosis of histoplasmosis using the MVista Histoplasma galactomannan antigen qualitative lateral flow–based immunoassay: a multicenter study. Open Forum Infect Dis. 2021;8 doi: 10.1093/ofid/ofab454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Visited on-line: october 7 MiraVista diagnostics. https://miravistalabs.com/devices/
- 132.Visited on-line: october 8 IMMY. https://www.immy.com/crag
- 133.Dubbels M., Granger D., Theel E.S. Low cryptococcus antigen titers as determined by lateral flow assay should be interpreted cautiously in patients without prior diagnosis of cryptococcal infection. J Clin Microbiol. 2017;55:2472–2479. doi: 10.1128/JCM.00751-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shi D., Haas P.J., Boekhout T., Hahn R.C., Hagena F. Neglecting genetic diversity hinders timely diagnosis of cryptococcus infections. J Clin Microbiol. 2021;59:1–3. doi: 10.1128/JCM.02837-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Benedict K., Jackson B.R., Chiller T., Beer K.D. Estimation of direct healthcare costs of fungal diseases in the United States. Clin Infect Dis. 2019;68 doi: 10.1093/cid/ciy776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gebhart C. Molecular microbiology: diagnostic principles and practice. 2nd ed. ASM Press; Washington DC: 2014. Molecular detection and identification of fungal pathognes; pp. 489–500. [Google Scholar]
- 137.Jones Mark, Marengo S. Acad. Press; 2016. Molecular microbial diagnostic methods; pp. 107–133. [Google Scholar]
- 138.Marilyn M., Lowery-Nordberg L.M., Melissa M., Sue R.C., Rothberg P. 2014. Molecular diagnostic assay validation update to the 2009 AMP molecular diagnostic assay validation white paper. [Google Scholar]
- 139.Miller S.A. 2nd ed. Clin. Lab. Stand. Inst.; 2020. Validation and verification of multiplex nucleic acid assays; pp. 13–62. [Google Scholar]
- 140.Li J., Macdonald J., Von Stetten F. Review: a comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst. 2019;144 doi: 10.1039/c8an01621f. [DOI] [PubMed] [Google Scholar]
- 141.Zeller I., Schabereiter-Gurtner C., Mihalits V., Selitsch B., Barousch W., Hirschl A.M., et al. Detection of fungal pathogens by a new broad range real-time PCR assay targeting the fungal ITS2 region. J Med Microbiol. 2017;66 doi: 10.1099/jmm.0.000575. [DOI] [PubMed] [Google Scholar]
- 142.Jafari Z., Motamedi M., Jalalizand N., Shokoohi G.R., Charsizadeh A., Mirhendi H. A comparison between CHROMagar, PCR-RFLP and PCR-FSP for identification of Candida species. Curr Med Mycol. 2017;3 doi: 10.29252/cmm.3.3.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.White P.L., Alanio A., Cruciani M., Gorton R., Millon L., Rickerts V., et al. Nucleic acid tools for invasive fungal disease diagnosis. Curr Fungal Infect Rep. 2020;14 doi: 10.1007/s12281-020-00374-7. [DOI] [Google Scholar]
- 144.Somogyvari F., Horvath A., Serly J., Majoros H., Vagvolgyi C., Peto Z. vol. 26. In Vivo; Brooklyn: 2012. (Detection of invasive fungal pathogens by real-time PCR and high-resolution melting analysis). [PubMed] [Google Scholar]
- 145.Gudnason H., Dufva M., Bang D.D., Wolff A. Comparison of multiple DNA dyes for real-time PCR: effects of dye concentration and sequence composition on DNA amplification and melting temperature. Nucleic Acids Res. 2007;35 doi: 10.1093/nar/gkm671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bonab M.M., Alimoghaddam K., Talebian F., Ghaffari S.H., Ghavamzadeh A., Nikbin B., et al. Real-time PCR handbook. Lab Chip. 2015;4:10–13. [Google Scholar]
- 147.Wen X., Chen Q., Yin H., Wu S., Wang X. Rapid identification of clinical common invasive fungi via a multi-channel real-time fluorescent polymerase chain reaction melting curve analysis. Med (United States) 2020;99 doi: 10.1097/MD.0000000000019194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Thelwell N., Millington S., Solinas A., Booth J., Brown T. Mode of action and application of Scorpion primers to mutatation detection. Nucleic Acids Res. 2000;28:3752–3761. doi: 10.1093/nar/28.19.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Camp I., Manhart G., Schabereiter-Gurtner C., Spettel K., Selitsch B., Willinger B. Clinical evaluation of an in-house panfungal real-time PCR assay for the detection of fungal pathogens. Infection. 2020;48 doi: 10.1007/s15010-020-01395-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bitter W.M., Dongen Jjm Van . 2021. The new EU regulation on in vitro diagnostic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Visited on-line: october 12 BioMed alliance. https://www.biomedeurope.org/
- 152.Carvalho-Pereira J., Fernandes F., Araújo R., Springer J., Loeffler J., Buitrago M.J., et al. Multiplex PCR based strategy for detection of fungal pathogen dna in patients with suspected invasive fungal infections. J Fungi. 2020;6 doi: 10.3390/jof6040308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Vanhee L.M.E., D'Haese E., Cools I., Nelis H.J., Coenye T. Detection and quantification of bacteria and fungi using solid-phase cytometry. NATO Sci Peace Secur Ser A Chem Biol. 2010 doi: 10.1007/978-90-481-8544-3_2. [DOI] [Google Scholar]
- 154.Vanhe L.M.E., Meersseman W., Lagrou K., Maertens J., Nelis H.J., Coenye T. Rapid and direct quantification of viable Candida species in whole blood by use of immunomagnetic separation and solid-phase cytometry. J Clin Microbiol. 2010;48 doi: 10.1128/JCM.00035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Vanhee L.M.E., Nelis H.J., Coenye T. Rapid detection and quantification of Aspergillus fumigatus in environmental air samples using solid-phase cytometry. Environ Sci Technol. 2009;43 doi: 10.1021/es803435a. [DOI] [PubMed] [Google Scholar]
- 156.Bayu A., Nandiyanto D., Oktiani R., Ragadhita R. How to read and interpret FTIR spectroscope of organic. Material. Indones J Sci Technol. 2019;4 [Google Scholar]
- 157.Potocki L., Depciuch J., Kuna E., Worek M., Lewinska A., Wnuk M. FTIR and Raman spectroscopy-based biochemical profiling reflects genomic diversity of clinical candida isolates that may be useful for diagnosis and targeted therapy of candidiasis. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20040988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Erukhimovitch Vitaly, Pavlov Valentina, Talyshinsky Marina, Yelena Souprun M.H. FTIR microscopy as a method for identification of bacterial and fungal infections. J Pharmaceut Biomed Anal. 2014;3 doi: 10.1016/j.jpba.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 159.Chisanga M., Muhamadali H., Ellis D.I., Goodacre R. Enhancing disease diagnosis: biomedical applications of surface-enhanced Raman scattering. Appl Sci. 2019;9 doi: 10.3390/app9061163. [DOI] [Google Scholar]
- 160.Hu S., Kang H., Gu F., Wang C., Cheng S., Gong W., et al. Rapid detection method for pathogenic Candida captured by magnetic nanoparticles and identified using SERS via AgNPs+ Int J Nanomed. 2021;16:941–950. doi: 10.2147/IJN.S285339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang J., Mou L., Jiang X. Surface chemistry of gold nanoparticles for health-related applications. Chem Sci. 2020;11:923–936. doi: 10.1039/c9sc06497d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sojinrin T., Conde J., Liu K., Curtin J., Byrne H.J., Cui D., et al. Plasmonic gold nanoparticles for detection of fungi and human cutaneous fungal infections. Anal Bioanal Chem. 2017;409:4647–4658. doi: 10.1007/s00216-017-0414-7. [DOI] [PubMed] [Google Scholar]
- 163.Zervou F.N., Zacharioudakis I.M., Kurpewski J., Mylonakis E. T2 magnetic resonance for fungal diagnosis. Methods Mol Biol. 2017;1508 doi: 10.1007/978-1-4939-6515-1_18. [DOI] [PubMed] [Google Scholar]
- 164.Neely L.A., Audeh M., Phung N.A., Min M., Suchocki A., Plourde D., et al. T2 magnetic resonance enables nanoparticle-mediated rapid detection of candidemia in whole blood. Sci Transl Med. 2013;5 doi: 10.1126/scitranslmed.3005377. [DOI] [PubMed] [Google Scholar]
- 165.Mylonakis E., Clancy C.J., Ostrosky-Zeichner L., Garey K.W., Alangaden G.J., Vazquez J.A., et al. T2 magnetic resonance assay for the rapid diagnosis of candidemia in whole blood: a clinical trial. Clin Infect Dis. 2015;60 doi: 10.1093/cid/ciu959. [DOI] [PubMed] [Google Scholar]
- 166.Hussain K.K., Malavia D., Johnson E.M., Littlechild J., Winlove C.P., Vollmer F., et al. Biosensors and diagnostics for fungal detection. J Contr Release. 2020;6 doi: 10.3390/jof6040349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pla L., Santiago-Felipe S., Tormo-Mas M.Á., Ruiz-Gaitán A., Pemán J., Valentín E., et al. Oligonucleotide-capped nanoporous anodic alumina biosensor as diagnostic tool for rapid and accurate detection of Candida auris in clinical samples. Emerg Microb Infect. 2021;10:407–415. doi: 10.1080/22221751.2020.1870411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.De Heer K., Van Der Schee M.P., Zwinderman K., Van Den Berk I.A.H., Visser C.E., Van Oers R., et al. Electronic nose technology for detection of invasive pulmonary aspergillosis in prolonged chemotherapy-induced neutropenia: a proof-of-principle study. J Clin Microbiol. 2013;51 doi: 10.1128/JCM.02838-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lewis N.S. Comparisons between mammalian and artificial olfaction based on arrays of carbon black-polymer composite vapor detectors. Acc Chem Res. 2004;37 doi: 10.1021/ar030120m. [DOI] [PubMed] [Google Scholar]
- 170.Gerritsen M.G., Brinkman P., Escobar N., Bos L.D., De Heer K., Meijer M., et al. Profiling of volatile organic compounds produced by clinical Aspergillus isolates using gas chromatography-mass spectrometry. Med Mycol. 2018;56 doi: 10.1093/mmy/myx035. [DOI] [PubMed] [Google Scholar]
- 171.Chambers S.T., Syhre M., Murdoch D.R., McCartin F., Epton M.J. Detection of 2-Pentylfuran in the breath of patients with Aspergillus fumigatus. Med Mycol. 2009;47 doi: 10.1080/13693780802475212. [DOI] [PubMed] [Google Scholar]
- 172.Perl T., Jünger M., Vautz W., Nolte J., Kuhns M., Borg-von Zepelin M., et al. Detection of characteristic metabolites of Aspergillus fumigatus and Candida species using ion mobility spectrometry - metabolic profiling by volatile organic compounds. Mycoses. 2011;54 doi: 10.1111/j.1439-0507.2011.02037.x. [DOI] [PubMed] [Google Scholar]
- 173.Lion T. vol. 1508. 2017. (Human fungal pathogen identification: methods and protocols). [Google Scholar]
- 174.Jenks J.D., Gangneux J., Schwartz I.S., Alastruey-izquierdo A. Diagnosis of breakthrough fungal infections in the clinical mycology laboratory : an ECMM consensus statement. J Fungi. 2020;6 doi: 10.3390/jof6040216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Florio W., Morici P., Ghelardi E., Barnini S., Lupetti A. Recent advances in the microbiological diagnosis of bloodstream infections. Crit Rev Microbiol. 2018;44 doi: 10.1080/1040841X.2017.1407745. [DOI] [PubMed] [Google Scholar]
- 176.Dingle T.C., Butler-wu S.M. MALDI-TOF mass spectometry for microorganism identification. Clin Lab Med. 2013;33 doi: 10.1016/j.cll.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 177.Visited on-line: october 29 Microbe notes. https://microbenotes.com/category/biochemical-test/
- 178.Lass-fl C., Samardzic E., Knoll M. Serology anno 2021-fungal infections: from invasive to chronic. Clin Microbiol Infect. 2021;27(9):1230–1241. doi: 10.1016/j.cmi.2021.02.005. [DOI] [PubMed] [Google Scholar]
- 179.Mikulska M., Calandra T., Sanguinetti M., Poulain D., Viscoli C. The use of mannan antigen and anti-mannan antibodies in the diagnosis of invasive candidiasis: recommendations from the Third European Conference on Infections in Leukemia. Crit Care. 2010;14 doi: 10.1186/cc9365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wickes B.L., Wiederhold N.P. Molecular diagnostics in medical mycology. Nat Commun. 2018;9 doi: 10.1038/s41467-018-07556-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sancho-Tello S., Bravo D., Borrás R., Costa E., Muñoz-Cobo B., Navarro D. Performance of the lightCycler septiFast test M grade in detecting microbial pathogens in purulent fluids. J Clin Microbiol. 2011;49:2988–2991. doi: 10.1128/JCM.00359-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Steinmann J., Buer J., Rath P.M. Detection of Aspergillus fumigatus in blood samples from critically ill patients in intensive care units by use of the septifast assay. J Clin Microbiol. 2016;54:1918–1921. doi: 10.1128/JCM.00478-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Fuchs S., Lass-Flörl C., Posch W. Diagnostic performance of a novel multiplex PCR assay for candidemia among ICU patients. J Fungi. 2019;5 doi: 10.3390/jof5030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zboromyrska Y., Cillóniz C., Cobos-Trigueros N., Almela M., Hurtado J.C., Vergara A., et al. Evaluation of the MagicplexTM sepsis real-time test for the rapid diagnosis of bloodstream infections in adults. Front Cell Infect Microbiol. 2019;9:1–8. doi: 10.3389/fcimb.2019.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Camp I., Spettel K., Willinger B. Molecular methods for the diagnosis of invasive candidiasis. J Fungi. 2020;6:1–14. doi: 10.3390/jof6030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Visited on-line: march 15. Bio-Evolution. https://www.bio-evolution.net/index.php/en/mycology/17-aspergillus-fumigatus
- 187.Denis J., Forouzanfar F., Herbrecht R., Toussaint E., Kessler R., Sabou M., et al. Evaluation of two commercial real-time PCR kits for Aspergillus DNA detection in bronchoalveolar lavage fluid in patients with invasive pulmonary aspergillosis. J Mol Diagnostics. 2018;20:298–306. doi: 10.1016/j.jmoldx.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Guinea J., Padilla C., Escribano P., Muñoz P., Padilla B., Gijón P., et al. Evaluation of MycAssayTM Aspergillus for diagnosis of invasive pulmonary aspergillosis in patients without hematological cancer. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Rath P.M., Steinmann J. Overview of commercially available PCR assays for the detection of Aspergillus spp. DNA in patient samples. Front Microbiol. 2018;9 doi: 10.3389/fmicb.2018.00740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Chong G.L.M., Van De Sande Wwj, Dingemans G.J.H., Gaajetaan G.R., Vonk A.G., Hayette M.P., et al. Validation of a new Aspergillus real-time PCR assay for direct detection of Aspergillus and azole resistance of Aspergillus fumigatus on bronchoalveolar lavage fluid. J Clin Microbiol. 2015;53:868–874. doi: 10.1128/JCM.03216-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Chong G.M., van der Beek M.T., von dem Borne P.A., Boelens J., Steel E., Kampinga G.A., et al. PCR-based detection of Aspergillus fumigatus Cyp51A mutations on bronchoalveolar lavage: a multicentre validation of the AsperGenius assay® in 201 patients with haematological disease suspected for invasive aspergillosis. J Antimicrob Chemother. 2016;71:3528–3535. doi: 10.1093/JAC/DKW323. [DOI] [PubMed] [Google Scholar]
- 192.Visited On-Line: March 20. Pathonostics. www.pathonostics.com%0A/product/aspergenius%0A.
- 193.White P.L., Posso R.B., Barnes R.A. Analytical and clinical evaluation of the pathonostics aspergenius assay for detection of invasive aspergillosis and resistance to azole antifungal drugs directly from plasma samples. J Clin Microbiol. 2017;55:2356–2366. doi: 10.1128/JCM.00411-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Visited On-Line: March 17. Bruker. www.bruker%0A.com/en/products-and-solutions/microbiology-and-diagnostics/molecular-diagnostics/fungiplex-aspergillus.html%0A.
- 195.Scharmann U., Kirchhoff L., Hain A., Buer J., Koldehoff M., Steinmann J., et al. Evaluation of three commercial PCR assays for the detection of azole-resistant Aspergillus fumigatus from respiratory samples of immunocompromised patients. J Fungi. 2021;7:1–10. doi: 10.3390/jof7020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Visited on-line: march 22. ElitechGroup.elitechgroup.com%0A/product/aspergillus-elite-mgb-kit%0A.
- 197.Grancini A., Orlandi A., Lunghi G., Consonni D., Pozzi C., Rossetti V., et al. Evaluation of Real Time PCR Aspergillus spp. in bronchoalveolar lavage samples. New Microbiol. 2018;41:67–70. [PubMed] [Google Scholar]
- 198.Visited on-line: march 30 Ingenetix. https://www.ingenetix.com/produkt/human/Aspergillus/
- 199.Kidd S.E., Chen S.C.A., Meyer W., Halliday C.L. A new age in molecular diagnostics for invasive fungal disease: are we ready? Front Microbiol. 2020;10:1–20. doi: 10.3389/fmicb.2019.02903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Dannaoui E., Gabriel F., Gaboyard M., Lagardere G., Audebert L., Quesne G., et al. Molecular diagnosis of invasive aspergillosis and detection of azole resistance by a newly commercialized PCR kit. J Clin Microbiol. 2017;55:3210–3218. doi: 10.1128/JCM.01032-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Jenks J.D., Spiess B., Buchheidt D., Hoenigl M. Methods for detection of Aspergillus fumigatus resistance in clinical samples. Curr Fungal Infect Rep. 2019;13:129–136. doi: 10.1007/s12281-019-00342-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Prattes J., Hoenigl M., Zinke S.E.M., Heldt S., Eigl S., Johnson G.L., et al. Evaluation of the new AspID polymerase chain reaction assay for detection of Aspergillus species: a pilot study. Mycoses. 2018;61:355–359. doi: 10.1111/myc.12757. [DOI] [PubMed] [Google Scholar]
- 203.Mulet Bayona J.V., Salvador García C., Tormo Palop N., Gimeno Cardona C. Validation and implementation of a commercial real-time PCR assay for direct detection of Candida auris from surveillance samples. Mycoses. 2021:1–4. doi: 10.1111/myc.13250. [DOI] [PubMed] [Google Scholar]
- 204.Huh H.J., Lim K.R., Ki C.S., Huh K., Shim H.J., Song D.J., et al. Comparative evaluation between the RealStar Pneumocystis jirovecii PCR kit and the AmpliSens Pneumocystis jirovecii (carinii)-FRT PCR Kit for detecting P. Jirovecii in Non-HIV immunocompromised patients. Ann Lab Med. 2019;39:176–182. doi: 10.3343/alm.2019.39.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Montesinos I., Delforge M.L., Ajjaham F., Brancart F., Hites M., Jacobs F., et al. Evaluation of a new commercial real-time PCR assay for diagnosis of Pneumocystis jirovecii pneumonia and identification of dihydropteroate synthase (DHPS) mutations. Diagn Microbiol Infect Dis. 2017;87:32–36. doi: 10.1016/j.diagmicrobio.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 206.Visited on-line: march 28 OlmDiagnostics. http://olmdiagnostics.com/products/pneumid/
- 207.Mercier T., Reynders M., Beuselinck K., Guldentops E., Maertens J., Lagrou K. Serial detection of circulating mucorales DNA in invasive mucormycosis: a retrospective multicenter evaluation. J Fungi. 2019;5:1–10. doi: 10.3390/jof5040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Guegan H., Iriart X., Bougnoux M.E., Berry A., Robert-Gangneux F., Gangneux J.P. Evaluation of MucorGenius® mucorales PCR assay for the diagnosis of pulmonary mucormycosis. J Infect. 2020;81:311–317. doi: 10.1016/j.jinf.2020.05.051. [DOI] [PubMed] [Google Scholar]
- 209.Visited on-line: march 23 Pathonostics. https://www.pathonostics.com/product/mucorgenius