Abstract
A novel genus-specific PCR for mycobacteria with simple identification to the species level by restriction fragment length polymorphism (RFLP) was established using the 16S-23S ribosomal RNA gene (rDNA) spacer as a target. Panspecificity of primers was demonstrated on the genus level by testing 811 bacterial strains (122 species in 37 genera from 286 reference strains and 525 clinical isolates). All mycobacterial isolates (678 strains among 48 defined species and 5 indeterminate taxons) were amplified by the new primers. Among nonmycobacterial isolates, only Gordonia terrae was amplified. The RFLP scheme devised involves estimation of variable PCR product sizes together with HaeIII and CfoI restriction analysis. It yielded 58 HaeIII patterns, of which 49 (84%) were unique on the species level. Hence, HaeIII digestion together with CfoI results was sufficient for correct identification of 39 of 54 mycobacterial taxons and one of three or four of seven RFLP genotypes found in Mycobacterium intracellulare and Mycobacterium kansasii, respectively. Following a clearly laid out diagnostic algorithm, the remaining unidentified organisms fell into five clusters of closely related species (i.e., the Mycobacterium avium complex or Mycobacterium chelonae-Mycobacterium abscessus) that were successfully separated using additional enzymes (TaqI, MspI, DdeI, or AvaII). Thus, next to slowly growing mycobacteria, all rapidly growing species studied, including M. abscessus, M. chelonae, Mycobacterium farcinogenes, Mycobacterium fortuitum, Mycobacterium peregrinum, and Mycobacterium senegalense (with a very high 16S rDNA sequence similarity) were correctly identified. A high intraspecies sequence stability and the good discriminative power of patterns indicate that this method is very suitable for rapid and cost-effective identification of a wide variety of mycobacterial species without the need for sequencing. Phylogenetically, spacer sequence data stand in good agreement with 16S rDNA sequencing results, as was shown by including strains with unsettled taxonomy. Since this approach recognized significant subspecific genotypes while identification of a broad spectrum of mycobacteria rested on identification of one specific RFLP pattern within a species, this method can be used by both reference (or research) and routine laboratories.
The genus Mycobacterium is represented by a wide range of species. They form a heterogenous group in terms of occurrence in clinical or environmental material, complex phenotypic and genetic data, and disease association (25, 33). Currently, identification of mycobacteria grown in culture is achieved by standard culture and biochemical methods, and for a few species, probes are commercially available (Mycobacterium tuberculosis, Mycobacterium avium, Mycobacterium intracellulare, Mycobacterium gordonae, Mycobacterium kansasii, and Mycobacterium fortuitum) (10, 12). Determination of phenotypic features is time-consuming, difficult to assimilate into a precise diagnosis concerning closely related taxa, and not always highly reproducible (26). The majority of clinically isolated nontuberculous bacteria, such as M. gordonae or rapidly growing species, are not pathogenic or are of doubtful clinical relevance (3, 7). On the other hand, a rise in incidence of nontuberculous bacteria, including newly described species or subspecific phylogenetic lineages of potential clinical significance, and the crucial role of the laboratory in establishing the diagnosis demand methods that provide accurate results in a more timely fashion (7, 26). Therefore, efforts for rapid and accurate molecular identification have been undertaken in recent years (13–17, 21–23, 26, 29, 30; J. L. Miller, training manual, MIDI Inc., Newark, N.J., 1997). Today, sequencing of the 16S RNA gene (rDNA) is regarded as the most suitable method for identification of mycobacteria (14, 32). Even so, the high expense, together with a lack of clinical relevance for most species identified in routine laboratory practice, renders sequencing unacceptable for general use. Limitations of the 16S RNA gene are evident because the number of polymorphic sites within the genus Mycobacterium is rather low (13, 22). Some species have the same sequence or a very high degree of similarity (22). This leads to problems in development of simpler sequence analysis methods, such as restriction fragment length polymorphism (RFLP) analysis or hybridization with probes (3, 6, 15). To meet this need, alternative genetic targets have been studied (13, 16, 22, 27, 28). Of these, the hsp65 gene has so far been best investigated, and the data were recently improved by inclusion of more species, especially some rapidly growing mycobacteria (e.g., Mycobacterium chelonae and Mycobacterium abscessus) (5, 21). However, hsp65 gene-based PCR-RFLP analysis has been impeded by difficulties, such as minor differences of band sizes between some species and the occurrence of new patterns not previously reported (20, 34; B. A. Forbes, K. S. Hicks, and D. L. Kiska, Abstr. 9th Eur. Cong. Clin. Microbiol. Infect. Dis., Clin. Microbiol. Infect. 5:(Suppl. 3), abstr. O37, p. 62, 1999). Moreover, all current molecular approaches to detect mycobacteria have the common disadvantage that the primers used for amplification are not specific for mycobacteria. Thus, undesirable amplification of other gram-positive bacterial contaminants, such as corynebacteria, represents a potential issue of concern in most clinical settings.
In view of this, we aimed to investigate the 16S-23S ribosomal DNA (rDNA) internal transcribed spacers of a larger number of mycobacterial species for their suitability to establish a PCR-RFLP-based identification scheme (16, 23). The first goal was to develop and evaluate novel primers for genus-specific amplification of mycobacteria and, secondly, to establish a reliable diagnostic algorithm for identification of a broad spectrum of mycobacterial species with one to three endonucleases. The interspecies discriminatory power and the degree of intraspecies divergence of patterns of such a new RFLP-based approach were investigated by using 678 mycobacterial strains within 48 species.
MATERIALS AND METHODS
Bacterial strains, identification, and sequencing.
The bacteria used in this study comprised 811 strains listed in Table 1. They constituted 122 species within 37 genera of 286 reference strains and 525 clinical isolates. Six Nocardia clinical isolates were identified to the genus level only. Species other than mycobacteria were chosen mostly from taxa of actinomycetes closely related to mycobacteria. Mycobacteria were represented by a total of 678 strains (179 reference strains and 499 clinical isolates) of 48 defined species and 5 Mycobacterium spp. that failed to match either biochemical species patterns or known 16S rDNA signature sequences (26).
TABLE 1.
Reference strains and clinical isolates of bacteria used in the present study
| Genus | Species | No. of strains | Reference strain(s)a | No. of clinical isolates |
|---|---|---|---|---|
| Actinomadura | madurae | 1 | DSM 43067T | |
| Actinomadura | pelletieri | 1 | DSM 43383T | |
| Actinomyces | israelii | 3 | DSM 43320T, S293 | 1 |
| Actinomyces | meyeri | 1 | 1 | |
| Actinomyces | naeslundii | 2 | DSM 43013T | 1 |
| Actinomyces | neuii | 1 | DSM 8577T | |
| Actinomyces | viscosus | 2 | DSM 43327T | 1 |
| Amycolatopsis | orientalis | 2 | DSM 40040T, 43387 | |
| Arcanobacterium | haemolyticum | 3 | DSM 20595T, S333 | 1 |
| Arcanobacterium | pyogenes | 1 | DSM 20630T | |
| Prevotella | buccae | 1 | 1 | |
| Prevotella | disiens | 1 | 1 | |
| Bacteroides | fragilis | 1 | DSM 2151T | |
| Cellulomonas | cellulans | 1 | DSM 43879T | |
| Cellulomonas | turbata | 2 | DSM 20577T, S330 | |
| Clostridium | glycolicum | 1 | 1 | |
| Clostridium | perfringens | 1 | 1 | |
| Corynebacterium | diphtheriae | 2 | DSM 44123T | 1 |
| Corynebacterium | jeikeium | 2 | DSM 7171T | 1 |
| Corynebacterium | pseudodiphthericum | 1 | 1 | |
| Corynebacterium | renale | 1 | DSM 20688T | |
| Corynebacterium | striatum | 2 | DSM 20668T | 1 |
| Corynebacterium | urealyticum | 2 | DSM 7109T | 1 |
| Corynebacterium | xerosis | 5 | DSM 20743T, 20170 | 3 |
| Dermatophilus | congolensis | 1 | DSM 44180T | |
| Dietzia | maris | 1 | DSM 43672T | |
| Escherichia | coli | 3 | DSM 1103, 5923, ATCC 9637 | |
| Enterococcus | faecalis | 1 | DSM 2570 | |
| Fusobacterium | nucleatum | 1 | DSM 20482T | |
| Gordonia | aichensis | 1 | DSM 43978T | |
| Gordonia | amarae | 2 | DSM 43392T, 43391 | |
| Gordonia | bronchialis | 1 | DSM 43247T | |
| Gordonia | hirsuta | 1 | DSM 44140T | |
| Gordonia | hydrophobica | 1 | DSM 44015T | |
| Gordonia | rubropertinctus | 2 | DSM 43197T, 10347 | |
| Gordonia | sputi | 2 | DSM 43896T, 43979 | |
| Gordonia | terrae | 5 | DSM 43249T, 46040, 43342, 43568, 43569 | |
| Haemophilus | influenzae | 1 | ATCC 49247 | |
| Lactobacillus | rhamnosus | 1 | S331 | |
| Listeria | ivanovii | 1 | S302 | |
| Listeria | monocytogenes | 3 | S303, 305, 306 | |
| Mycobacterium | abscessus | 9 | DSM 44196T, 43492, 43493, S322–324 | 3 |
| Mycobacterium | asiaticum | 3 | DSM 44056, 44292, 44297 | |
| Mycobacterium | aurum | 2 | DSM 43999T, S283 | |
| Mycobacterium | avium | 111 | DSM 44156T, DSM 44133T | 109 |
| Mycobacterium | bohemicum | 2 | DSM 44277T | 1 |
| Mycobacterium | celatum | 15 | DSM 44243T, S274, 275 | 12 |
| Mycobacterium | chelonae | 14 | DSM 43804T, 43217, 43276, 43483, 43484, 43487, 43488–43490, 46626, S268 | 3 |
| Mycobacterium | chlorophenolicum | 1 | DSM 43826T | |
| Mycobacterium | conspicuum | 1 | DSM 44136T | |
| Mycobacterium | duvalii | 1 | DSM 44244T | |
| Mycobacterium | farcinogenes | 20 | DSM 43637T, M9, 15, 16, 39, 52, 57, 191, 217, 269, 274, 275, 281, 285, 612, 687, 785, N710, 715, 725 | |
| Mycobacterium | flavescens | 7 | DSM 43991T, S318, 523–527 | |
| Mycobacterium | fortuitum | 26 | DSM 46621T, 44220T, M205, 368, 390, N723, S113, S485, 487–493 | 11 |
| Mycobacterium | gastri | 5 | DSM 43505T, 43506, S227–229 | |
| Mycobacterium | genavense | 11 | 11 | |
| Mycobacterium | gordonae | 48 | DSM 44160T | 47 |
| Mycobacterium | haemophilum | 3 | 3 | |
| Mycobacterium | hassiacum | 1 | DSM 44199T | |
| Mycobacterium | hodleri | 1 | DSM 44183T | |
| Mycobacterium | interjectum | 1 | DSM 44064T | |
| Mycobacterium | intermedium | 4 | DSM 44049T | 3 |
| Mycobacterium | intracellulare | 34 | DSM 43223T, S138, 347 | 31 |
| Mycobacterium | kansasii | 52 | DSM 44162T | 51 |
| Mycobacterium | lentiflavum | 5 | DSM 44195T, 44194, S136, 360 | 1 |
| Mycobacterium | malmoense | 11 | DSM 44163T, S217 | 9 |
| Mycobacterium | marinum | 16 | DSM 44344T, 43518, 43519, 43824, 44345, S287 | 10 |
| Mycobacterium | mucogenicum | 1 | DSM 44124 | |
| Mycobacterium | neoaurum | 1 | DSM 44074T | |
| Mycobacterium | nonchromogenicum | 4 | DSM 44164T, S264–266 | |
| Mycobacterium | obuense | 1 | DSM 44075T | |
| Mycobacterium | parafortuitum | 4 | DSM 43528T, 43526, 43527, S528 | |
| Mycobacterium | peregrinum | 17 | DSM 43271T, M418–420, S254, 486, 494–496 | 8 |
| Mycobacterium | phlei | 5 | DSM 43239T, 43214, 44018 | 2 |
| Mycobacterium | porcinum | 1 | DSM 44242T | |
| Mycobacterium | rhodesiae | 1 | DSM 44223T | |
| Mycobacterium | scrofulaceum | 11 | DSM 43992T, 43226, 43512, 43513, S343, 244 | 5 |
| Mycobacterium | senegalense | 10 | DSM 43656T, 43658, M266, 555, N714, 717–718, 721, 728, S114 | |
| Mycobacterium | shimoidei | 3 | DSM 44152T, S234 | 1 |
| Mycobacterium | simiae | 15 | DSM 44165T, S137, 140, 141, 146, 148, 149 | 8 |
| Mycobacterium | smegmatis | 2 | DSM 43756T, 43299 | |
| Mycobacterium | sp. | 2 | M511, 516 | |
| Mycobacterium | sp. (gastri)b | 10 | DSM 43221, 43507, S230–233, R230–233 | |
| Mycobacterium | sp. (malmoense) | 2 | S222, 279 | 2 |
| Mycobacterium | sp. (scrofulaceum) | 5 | S245, 313–314, 316, R39 | 5 |
| Mycobacterium | sp. (xenopi) | 2 | S369, 504 | 2 |
| Mycobacterium | szulgai | 8 | DSM 44166T, S97 | 6 |
| Mycobacterium | terrae | 9 | DSM 43227T, S280–281, 353 | 5 |
| Mycobacterium | triplex | 2 | S139 | 1 |
| Mycobacterium | triviale | 2 | DSM 44153T | 1 |
| Mycobacterium | tuberculosis | 90 | DSM 44156 | 89 |
| Mycobacterium | ulcerans | 3 | ATCC 19423T, S219 | 1 |
| Mycobacterium | vaccae | 4 | DSM 43292T, 43229, 43514, S345 | |
| Mycobacterium | xenopi | 59 | DSM 43995T | 58 |
| Nocardia | asteroides | 12 | DSM 43757T, 43003–43005, 43208, 43244, 43289, S308, 328, ATCC 23824 | 2 |
| Nocardia | brasiliensis | 2 | DSM 43758T, 43009 | |
| Nocardia | brevicatena | 1 | DSM 43024T | |
| Nocardia | carnea | 2 | DSM 43397T, 40840 | |
| Nocardia | corynebacterioides | 1 | DSM 20151T | |
| Nocardia | farcinica | 4 | DSM 43665T, 43666, S304, 327 | |
| Nocardia | nova | 1 | ATCC 33726 | |
| Nocardia | otitidiscaviarum | 3 | DSM 43242T, 43010, 43398 | |
| Nocardia | pseudobrasiliensis | 1 | DSM 44290T | |
| Nocardia | seriolae | 1 | DSM 44129T | |
| Nocardia | sp. | 6 | 6 | |
| Nocardioides | albus | 1 | DSM 43109T | |
| Nocardiopsis | dassonvillei | 1 | DSM 43111T | |
| Peptostreptococcus | anaerobius | 1 | DSM 2949 | |
| Pseudomonas | aeruginosa | 1 | DSM 1117 | |
| Pseudonocardia | autotrophica | 2 | DSM 535T, 43082 | |
| Rhodococcus | equi | 2 | DSM 20307T, S307 | |
| Rhodococcus | rhodochrous | 1 | DSM 43241T | |
| Rhodococcus | ruber | 1 | DSM 43338T | |
| Rothia | dentocariosa | 3 | DSM 43762T, S325, 326 | |
| Saccharomonospora | glauca | 1 | DSM 43769T | |
| Saccharomonospora | viridis | 1 | DSM 43017T | |
| Saccharopolyspora | rectivirgula | 1 | DSM 43747T | |
| Saccharothrix | aerocolonigenes | 1 | DSM 40034T | |
| Skermania | piniformis | 1 | DSM 43998T | |
| Staphylococcus | aureus | 2 | DSM 2569, 1104 | |
| Streptococcus | pneumoniae | 2 | ATCC 6303 | 1 |
| Streptomyces | albus | 1 | DSM 40313T | |
| Streptomyces | somaliensis | 1 | DSM 40738T | |
| Thermoactinomyces | vulgaris | 1 | DSM 43016T | |
| Tsukamurella | paurometabolum | 3 | DSM 20162T, 44119, S329 | |
| Tsukamurella | pulmonis | 1 | DSM 44142T | |
| Tsukamurella | tyrosinosolvens | 1 | DSM 44234T | |
| Turicella | otitidis | 1 | DSM 8821T | |
| Total | 811 | 525 |
ATCC, American Type Culture Collection, Manassas, Va.; DSM, Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany; M and N, strain collection, Department of Microbiology, The Medical School, University of Newcastle, Newcastle upon Tyne, United Kingdom; R, strain collection, Institut für Mikrobiologie und Hygiene, Universitätsklinikum Regensburg, Regensburg, Germany; S, strain collection, Institut für Mikrobiologie und Immunologie, Krankenhaus Zehlendorf, Berlin, Germany.
Provisionally termed Mycobacterium sp. on the basis of phenotypic and genotypic data (for details, see the text).
All clinical isolates were identified to the species level by standard biochemical methods and/or AccuProbes (12, 25). Most of the reference strains and all clinical isolates—with the exception of M. avium, M. tuberculosis, 23 M. gordonae isolates, and 14 Mycobacterium xenopi isolates—were sequenced in the variable regions A and B within the 16S RNA gene (14, 22, 25). To allow for a better understanding, we sequenced the nearly 1.5-kbp 16S rDNA in a few strains with unique or discordant RFLP patterns (one clinical isolate each of M. kansasii, Mycobacterium phlei, and Mycobacterium triviale, and the reference strains Mycobacterium flavescens S526 and S318 and Mycobacterium parafortuitum DSM 43526) and some of those with unsettled taxonomic status (strains M511, S245, S279, S369, and S504) using a method described elsewhere (24). To obtain more 16S-23S spacer sequence data, a selection of strains were also sequenced within the 16S-23S spacer (22). A few DSM reference strains with apparently wrong designation according to the RFLP results were subsequently reclassified after partial 16S rDNA sequencing and analyses of fatty acids by gas chromatography and of mycolic acids by high-performance liquid chromatography (17; Miller, 1997).
PCR amplification.
Chromosomal DNA was released from bacterial suspensions by sonication with glass beads according to methods described elsewhere (22). Amplification of a part of the 16S-23S spacer was performed with primers Sp1 (5′-ACC TCC TTT CTA AGG AGC ACC-3′) (AAGGA corresponds to the beginning of the spacer sequence) and Sp2 (5′-GAT GCT CGC AAC CAC TAT CCA-3′) (positions 210 to 190 of the M. tuberculosis spacer sequence; EMBL accession number L15623). The amplification was done with a 50-μl reaction mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, 200 μM (each) deoxynucleoside triphosphate (dATP, dGTP, dCTP, and dUTP), 75 ng of each primer, 1 U of Thermus aquaticus DNA polymerase (all reagents were from Pharmacia Biotech, Freiburg, Germany), and 5 μl of DNA. The thermal profile involved initial denaturation for 5 min at 96°C and 38 cycles with the following steps: 1-min denaturation at 94°C, annealing at 59°C, and extension at 72°C.
RFLP analysis.
The amplified products were digested separately with 2 U of restriction enzyme HaeIII, CfoI, TaqI, MspI (Sigma, Diesenhofen, Germany), DdeI (Promega, Madison, Wis.), or AvaII and HinfI (Amersham, Braunschweig, Germany) according to the recommendations of the manufacturers and electrophoresed in 4% Small agarose (Biozym, Oldendorf, Germany) in the presence of ethidium bromide at 65 V for 2.0 to 3.0 h. For restriction with HinfI, dUTP in the PCR mixture was replaced by dTTP. Fragment band sizes were estimated visually by comparison with appropriate controls run in parallel (type strains of M. avium, M. intracellulare, and M. kansasii) and a 100-bp ladder. All restriction fragment sizes of patterns shown for slowly growing mycobacteria rely on sequence data (fragments smaller than 30 bp are not shown). Due to unavailable sequence data for most rapidly growing representatives, their RFLP fragment sizes were estimated visually without computerized help and rounded to the nearest 5 bp.
Nucleotide sequence accession numbers.
The 16S RNA gene sequences of Mycobacterium spp. strains S245 (MCRO 33; scrofulaceum), S318 (M. flavescens), and S369 (M. xenopi) were deposited in the GenBank database under the accession numbers AF152559, AF174289, and AF174290, respectively. Cultures of S245, S279, S369, S318, and S522 were deposited in the strain collection of the Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany under the numbers 44427, 44429, 44428, 44430, and 44431.
RESULTS
Specificity of primers and RFLP patterns.
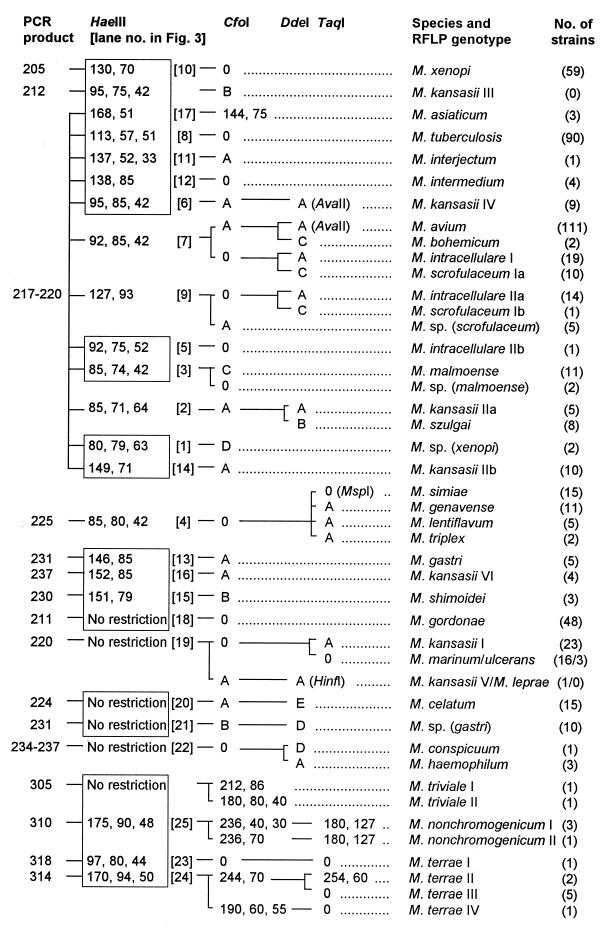
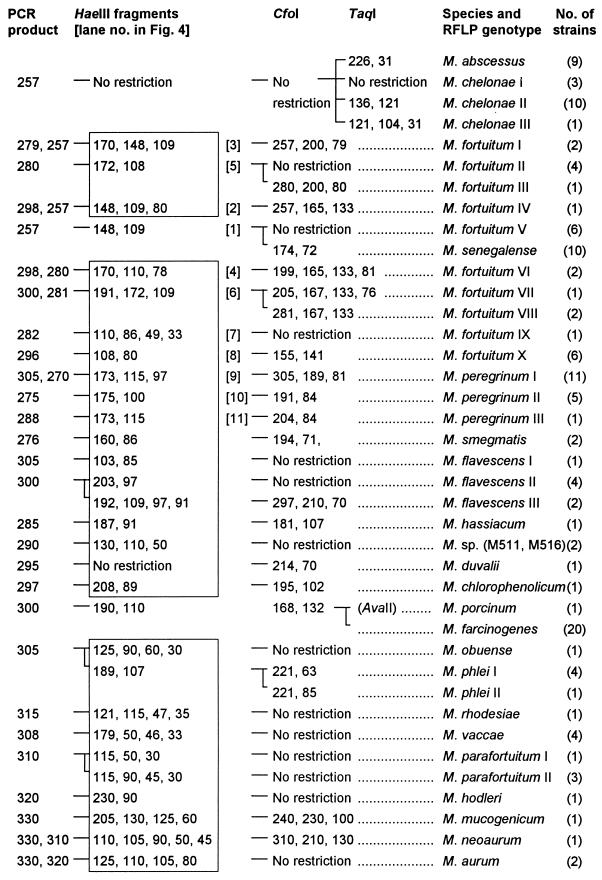
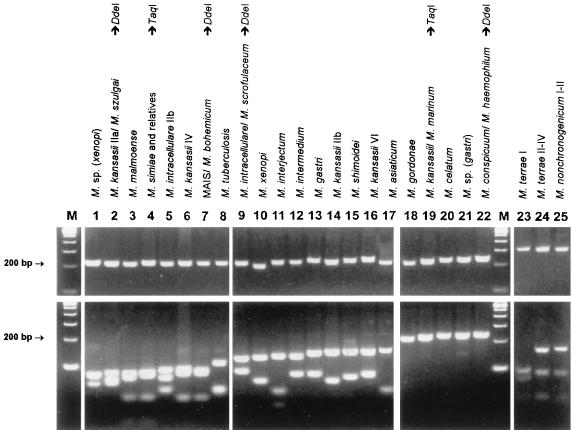
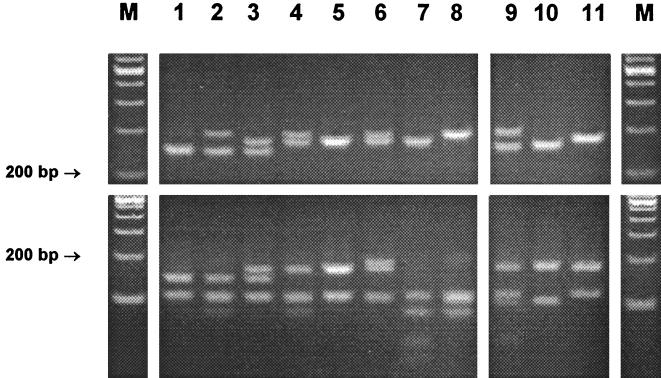
With the exception of Gordonia terrae, none of the bacteria other than mycobacteria were amplifiable. Amplification products of varying sizes were obtained from all mycobacteria tested. Amplicon sizes varied from 200 bp (M. xenopi) to 330 bp (Mycobacterium neoaurum). Mycobacterium nonchromogenicum, Mycobacterium terrae, M. triviale, and rapidly growing species showed fragments larger than 250 bp. HaeIII was selected as the first-line enzyme that, together with the knowledge about amplicon sizes, would produce the most discriminative RFLP patterns. Of 58 discernible HaeIII patterns, 49 (84%) were unique and thus indicative and sufficient for identification to the species level. HaeIII species-specific patterns are highlighted in Fig. 1 and 2. The HaeIII patterns of slowly growing mycobacteria, M. fortuitum, and Mycobacterium peregrinum are displayed in Fig. 3 and 4. Except for the patterns shown in Fig. 3, lanes 3 and 4, 6 and 7, 13 and 16, 18 and 19, or 24 and 25 exhibiting minor differences of 3 to 9 bp, HaeIII restriction produced mostly two to three DNA fragments whose sizes could be easily estimated by visual inspection of the gels. Since primer-dimer formation was never noticed, fragments as small as 30 bp were also used for classification of RFLP patterns.
FIG. 1.
Algorithm of RFLP patterns of 28 slowly growing mycobacterial species and 4 Mycobacterium spp. of uncertain taxonomic status from PCR-amplified 16S-23S rDNA spacer sequences (547 strains). PCR products and restriction fragments are designated by molecular sizes in base pairs. HaeIII species-specific patterns are highlighted by boxes. CfoI patterns A to D are as follows: A, 126 to 144 and 91 to 96 (digest size varies depending on the PCR product size); B, 129 to 146 and 83; C, 126, 63, and 30; D, 160 and 62. DdeI patterns A-E are as follows: A, 120 and 90; B, 120 and 80; C, 120 and 70; D, 120 and 100; E, 214. TaqI pattern A is 155 and 70; 0, no restriction. Type strains were assigned to genotype I if more than one pattern occurred in a species. Genotypes Ia and Ib or IIa and IIb indicate that the strains are genetically very similar but new RFLP genotypes have occurred after loss or acquisition of one HaeIII restriction site due to allelic microheterogeneity. M. leprae and M. kansasii III RFLP patterns were deduced from nucleotide sequence accession no. X56657 (EMBL) and the M. kansasii genotype III sequence published by Alcaide et al. (1). For details concerning AvaII, HinfI, and MspI patterns and descriptions of Mycobacterium spp., see the text.
FIG. 2.
Algorithm of RFLP patterns of 21 rapidly growing mycobacterial species and one rapidly growing Mycobacterium sp. of unknown taxonomic status from PCR-amplified 16S-23S rDNA spacer sequences. Details are given in the legend to Fig. 1.
FIG. 3.
Gel electrophoresis and HaeIII RFLP patterns of slowly growing mycobacteria from PCR-amplified 16S-23S rDNA spacer sequences (the upper panel shows PCR products without restriction). The molecular sizes of the fragments are given in Fig. 1. The patterns are displayed in order of increasing size of the biggest fragment. M, molecular size marker (100-bp ladder). MAIS, M. avium-M. intracellulare-M. scrofulaceum.
FIG. 4.
Gel electrophoresis and HaeIII RFLP patterns of M. fortuitum (lanes 1 to 8) and M. peregrinum (lanes 9 to 11) from PCR-amplified 16S-23S rDNA spacer sequences (the upper panel shows PCR products without restriction). The patterns are described in the legend to Fig. 2. M, molecular size marker (100-bp ladder).
The remaining nine HaeIII patterns that did not give a final species assignment needed further analysis with additional endonucleases. Therefore, all test organisms were subjected to CfoI digestion. Although CfoI patterns were not necessary for final identification of most species (in particular, rapidly growing ones), as a practical routine, immediate restriction with both HaeIII and CfoI may be advisable (and thus it is shown for all species) because the reliability of results for one enzyme is augmented if confirmed by a second endonuclease analysis. Following the established algorithm for slowly growing mycobacteria (Fig. 1), precise estimates of fragments after CfoI restriction were not generally required. Rather, the distinguishing feature of this enzyme was mostly confined to the question of whether the amplicon was cut or not. Owing to the high sequence similarity of some species such as M. avium, M. chelonae, M. kansasii, and Mycobacterium simiae to their nearest relatives, these species formed groups that needed further analysis by DdeI, TaqI, or AvaII for accurate identification as shown in Fig. 1, 2, and 3. Four of these clusters (Fig. 3, lanes 2, 7, 9, and 22) were readily resolved using DdeI, and three were resolved using TaqI, but the latter enzyme could not distinguish Mycobacterium genavense, Mycobacterium lentiflavum, and Mycobacterium triplex. If necessary, these species, which have a high spacer sequence similarity of 95%, can be separated by restriction with MspI: M. simiae and M. lentiflavum strains were cut once (139 and 86 bp), and M. genavense and M. triplex were cut twice (114, 86, and 25 and 86, 79, and 60 bp). The endonucleases AvaII and HinfI may be of use in exceptional cases, notably, in research or reference facilities, for separation of M. kansasii V from Mycobacterium leprae (HinfI), or Mycobacterium porcinum from Mycobacterium farcinogenes (AvaII). One isolate of this specific M. kansasii genotype and the M. porcinum type strain (Table 1) tested were cut, with resulting fragments of 116 and 105 bp and 215 and 85 bp, respectively. M. farcinogenes isolates were not cut, nor was M. leprae according to the database sequence. Finally, AvaII is shown in Fig. 1 for optional application in connection with M. avium, M. kansasii IV, and Mycobacterium bohemicum because these showed HaeIII patterns with the highest degree of similarity. The first was digested by AvaII (144 and 75 bp); the latter two taxons were not. Mycobacterium marinum and Mycobacterium ulcerans possess identical spacer sequences (1, 22). Therefore, these organisms could not be separated by this method.
According to the spacer RFLP method, four reference strains deposited as M. fortuitum (DSM 43276 and 46626), Nocardia farcinica (DSM 43231), and Mycobacterium thermoresistibile (DSM 43644) were diagnosed as M. chelonae (the first two isolates), Mycobacterium senegalense, and M. phlei, respectively. Partial 16S rDNA sequencing and analysis of fatty and mycolic acids were in full agreement with these findings, and these strains were thereafter reclassified (the names used in Table 1 are those after reclassification). Of five G. terrae strains, three showed a 315-bp and two showed a 330-bp PCR product, and the respective HaeIII RFLP patterns were 200, 170, and 130 and 185 and 160 bp, respectively.
Taxonomically uncertain strains.
Two rapidly photochromogenic strains deposited as Mycobacterium sp. (reference strains M511 and M516) had a unique RFLP pattern compared to other rapidly growing mycobacteria. Data on the exact phenotype were not available, but complete 16S rDNA sequencing revealed three substitutions compared to Mycobacterium smegmatis: ACA → ATA, TAG → TGG, and TTA → TGA at positions 137, 162, and 1075 of the reference sequence (EMBL X52922). Four groups comprising slowly growing strains with uncertain taxonomic status were formed and tentatively named Mycobacterium sp. (Table 1). Although this nomenclature has no taxonomic standing (the names of the most closely related species are provisionally added in parentheses), phenotypic and genotypic data together with the finding that all showed unique spacer RFLP patterns obviated the need to separate these groups from established species. The following is a detailed description of the results.
(i) Mycobacterium sp. (gastri).
Ten reference strains originally deposited as either M. kansasii or Mycobacterium gastri clustered in one RFLP genotype (Fig. 3, lane 21). All these strains were sequenced in the spacer except for strains S230 to S233, which had previously been characterized as M. gastri spacer genotype Mga B (22). This revealed that the four M. kansasii strains and two M. gastri strains (DSM 43221 and 43507) were attributable to spacer M. kansasii genotype IV described by Alcaide et al. (1) (Table 2 gathers available data together with the findings of this study). The spacer genotype sequences IV and Mga B have a similarity value of 99.9% due to a 1-nucleotide substitution at position 223 (ACT → AAT), and sequences Mga B and Mga A (M. gastri type strain sequence) display a similarity value of 98% (4-nucleotide difference). Careful reassessment of biochemical tests showed that these strains were incapable of hydrolyzing Tween 80 (both M. gastri and M. kansasii hydrolyze Tween), while tests clearly positive for M. kansasii (nitrate reduction, catalase, and photochromogenicity) were weakly positive (delayed colony pigment formation). Thus, these strains were assumed to be an indeterminate (or new) taxon near to the species M. gastri, as evidenced by their 16S-23S rDNA sequences. In contrast, all M. kansasii strains hydrolyzed Tween, and their spacer sequences showed sufficient diversity to emerge as three distinct subgroups (dendrogram not shown): sequence genotypes (sequevars) I together with II, and III together with Mka B and Mka C as separate clusters, while type V showed the highest degree of similarity to M. kansasii type IV (90%). Resolution of 16S rDNA sequences within these entities was poor (Table 2). We found and thus confirmed minor variants in the variable region B recently described by Richter et al. (20). Full-length 16S rDNA sequencing of one strain of concern because of its negative AccuProbe result (strain S522 with spacer genotype Mka C) flawed the probability that this strain could be an unrecognized new species, since it showed complete identity with the M. kansasii type strain sequence downstream from variable region B.
TABLE 2.
Spacer genotyping results for M. kansasii and M. gastrii compared to 16S RNA and hsp65 gene
| Species | Results
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 16S-23S spacer
|
16S rDNA sequence variants (nt 461–469)b | AccuProbe (new version) |
hsp65 genec
|
|||||||
| RFLP genotype | No. of strains analyzed | Sequence sequevara | No. of strains analyzed | EMBL no. | Genotype | Restriction fragments (bp)
|
||||
| BstEII | HaeIII | |||||||||
| M. kansasii | I | 23 | I/Mka A | 6 | X97632/L42262 | M. kansasii/gastri CGG GTT CTC | Positive | I | 231, 212 | 127, 103, 78 |
| II | 15 | II | 7 | CGG GTT GTC | Positive | II | 231, 133, 79 | 127, 103 (70) | ||
| IV | 9 | Mka B | 2 | L42263 | CGG GTT TTC | Positive | III | 231, 133, 79 | 127, 94, 69 | |
| V | 1 | Mka C | 1 | L42264 | CGG GTT TCC | Negative | VI | 231, 133, 79 | 127, 103, 69 | |
| VI | 4 | V | 1 | M. kansasii/gastri | Positive | V | 325, 125 | 140, 100, 80 | ||
| Mycobacterium sp. (gastri) | 10 | IV | 6 | M. kansasii/gastri | Positive | IV | 231, 118, 79 | 127, 112, 69 | ||
| Mga B | 4 | Y14182 | M. kansasii/gastri | Positive | Unknown | Unknown | ||||
| M. gastri | 5 | Mga A | 4 | X97633 | M. kansasii/gastri | Negative | 231, 133, 79 | 127, 103, 69 | ||
Genotypes I to IV and Mga A and B in accordance with references 1 and 22, respectively. Sequevar Mka B and C sequences were deposited in the EMBL database in 1995 (unpublished data) and were later confirmed by Richter et al. (20). Sequence sequevar III (1) is not shown because the genotype was not found in this study.
Refers to partial 16S rDNA sequencing within the variable region B only. Underlined bases indicate positions of substitutions.
(ii) Mycobacterium sp. (malmoense).
Two clinical scotochromogenic isolates phenotypically resembling Mycobacterium malmoense exhibited a 16S rDNA sequence with nine substitutions compared to that of M. malmoense: CCC CGA → CCA CTT, GGG → GTG, ACG → ATG, TGG → TAG, CCT TGT → CCC CGT, and TCG → TTG at positions 141, 159, 220, 601, 1062, and 1403 of the reference sequence (EMBL X52930). These strains could represent subspecies of M. malmoense and, interestingly, they emerged as a distinct RFLP genotype in the vicinity of M. malmoense.
(iii) Mycobacterium sp. (scrofulaceum).
Similarly to the former case, five clinical isolates phenotypically very closely related to Mycobacterium scrofulaceum (the only physiological difference was a lack of growth at 25°C) showed an RFLP genotype near to but distinguishable from that of M. scrofulaceum, and in good correlation with this, possessed a distinctive 16S rDNA sequence. The latter was identical to the MCRO 33 sequence published previously (26), which typically shows identity with M. scrofulaceum in variable region A and identity with M. simiae in region B (deletion of 12 nucleotides).
(iv) Mycobacterium sp. (xenopi).
Two strains, S369 and S504, isolated from the sputa of two patients with lung disease, displayed identical complete 16S rDNA sequences, with the highest similarity to that of the M. xenopi type strain (97%). A missing arylsulfatase activity (2 weeks) and negative nicotinamidase and pyrazinamidase were reactions in discordance with M. xenopi. The RFLP results were somewhat different from those of M. xenopi, exhibiting a unique HaeIII pattern.
Intraspecies stability of spacer sequences.
Intraspecies spacer sequence polymorphisms seemed to be more frequent in rapidly growing mycobacteria than in slowly growing species. In fact, many of the rapidly growing representatives for which multiple strains within a species were studied presented more than one RFLP pattern. As expected from the known variability found in the 16S rDNA, M. fortuitum was associated with a considerable variability leading to eight different HaeIII patterns (Fig. 4). They all shared a 108- to 110-bp band, and some of them showed typical PCR products of two different sizes due to interoperon variability. These features, together with the CfoI result, gave the correct species identification in all cases. M. fortuitum subsp. acetamidolyticum (DSM 44220) was assigned to RFLP genotype II, and one strain with a 16S signature of M. fortuitum biovariant 3 was assigned to RFLP genotype VII.
The occurrence of RFLP genotypes different from type strain genotypes in M. flavescens and M. parafortuitum reference strains (Fig. 2) was found to be associated with hitherto unknown sequence polymorphisms in the 16S RNA gene. M. flavescens II (S526) and III (S318) had identical 16S sequences, but they differed by as many as 23 nucleotides from the type strain sequence, which raises the question of the species integrity of these reference strains. M. parafortuitum RFLP genotype II (DSM 43526) displayed six base substitutions compared to the type strain 16S rDNA sequence: five differences in the signature sequence of variable region A (AAT AGG ATC ACT GGC TTC ATG GTC) and one mismatch (GAA → GGA) at position 882 of the reference sequence (EMBL X93183). Full-length 16S rDNA sequences of M. phlei and M. triviale RFLP genotypes II (one strain each) showed 100% identity with the respective type strain sequences.
Of slowly growing mycobacteria, only M. kansasii (as noted), the M. terrae complex, and, to a lesser extent, M. intracellulare and M. scrofulaceum were characterized by a genetic heterogeneity leading to more than one RFLP pattern in a species. Intraspecies sequence variations for many slowly growing mycobacteria, such as the M. avium complex, M. simiae, M. gordonae (the last deposited as EMBL accession numbers L42258 to 42261), and M. xenopi have been described (8, 22). Despite this, allelic heterogeneities had no detrimental impact on the clearly arranged RFLP algorithm. Even so, we sequenced a selection of strains by way of example to obtain a better estimate of the occurrence of sequence diversity within the spacer. Species were chosen in which a diversity might be expected because sequence variations occur both in the 16S rDNA and the hsp65 gene. The results are shown in an alignment with published sequences in Fig. 5. The reproducibility (and thus the degree of stability) of spacer sequences was confirmed because the sequences found were in full agreement with published data (8, 9), albeit some new sequevars were found (Mac J to L and Mgo E and F). Of 81 M. avium strains examined, 44 and 37 fell into the Mav A and B sequevars, respectively. The spacer sequences of six M. celatum isolates were all identical. The positions of restriction sites that generate species-specific or subspecific RFLP genotypes are located in the more conserved stem-loop regions. For example, in the case of M. intracellulare these contain distinct sequence motifs found in subspecific groups related to either Min or Mac sequevars, which in turn have led to the formation of two RFLP genotypes. These clusters consist of a larger number of sequevars, which are characterized by a high rate of substitutions in variable regions, such as the antitermination elements (position 130 to 160) or within helices 2, 5, and 6. The last two lie beyond the part of the spacer amplified by primers Sp1 and Sp2, and a single sporadic mutation with generation of a new restriction site within helix 2 was observed only once (Mac K with RFLP genotype M. intracellulare IIb). Although substitutions in the stem regions can be expected to occur rarely, we found substitutions at the transition from helix 3 to the stem sequence (position 76 [Fig. 5]). Hence, M. kansasii II and M. scrofulaceum were split into two RFLP genotypes attributable to the same mutational event. Ultimately, the high number of strains studied within some species associated with only one RFLP genotype despite sequence microheterogeneities, such as M. gordonae or M. xenopi, provide firm evidence that the degree of stability of the RFLP patterns is very high.
FIG. 5.
Sequence stability and microheterogeneity of 16S-23S rDNA spacer sequences in conserved and more variable regions with relevance for the cleaving action of HaeIII (GGCC) and CfoI (GCGC). Sequences not found in this study but published elsewhere are included (4, 8, 9). The respective sequevar designations are shown in brackets, and the number of strains sequenced for this study are shown in parentheses. Sequevars combined in one line exhibit base substitutions located in other regions of the spacer that are not displayed. Of Mav A to E and Min A to C, only Mav A or B and Min A were found. The sequevar Mgo B was not found among seven M. gordonae isolates examined.
DISCUSSION
We sought to establish a new molecular method for identification of mycobacteria that on the one hand would be capable of identifying all taxons to the species level with high accuracy and reliability and on the other hand would be simple enough for application even in routine laboratories. In view of this, emphasis was laid on inclusion of a broad spectrum of species and, even more important, on examination of a larger number of reference and clinical isolates within a species. First, this was important in order to determine the reliability of new primers chosen within a genetic target with a tendency to show more frequent sequence rearrangements due to a higher evolutionary rate (11, 22). Second, the occurrence of additional RFLP patterns due to sequence polymorphisms not presently recognized due to unavailable clinical isolates may later seriously compromise the devised diagnostic algorithm or even make evaluation of additional enzymes necessary. Besides the genus specifity of the primers shown, the size variations of the PCR amplicons described are particularly useful because they provide a simple means for distinguishing rapidly growing from slowly growing species at first glance, since the latter produce amplicons larger than 250 bp. Furthermore, this valuable new feature of the method can prove helpful in unequivocally recognizing mixed cultures. For example, a culture containing a slowly growing relevant pathogen like M. malmoense can easily be recognized when overgrowth by M. fortuitum or M. terrae occurs because, irrespective of mixed patterns, the latter exhibit PCR products far larger than 220 bp.
Concerning intraspecies stability of RFLP patterns, we can state that the spacer-based method was successfully evaluated with respect to expanded groups of strains within slowly growing species, such as M. tuberculosis, M. avium, M. xenopi, or M. gordonae. These are frequently found in a routine laboratory setting, and we were indeed satisfied to see that, with a few exceptions, all of these strains in a species were associated with only one RFLP pattern. This is in contrast to results obtained using the hsp65 gene-based RFLP method, which exhibits a greater number of RFLP genotypes within one species (e.g., six patterns for M. gordonae) (5, 29). The finding of distinct M. kansasii subgroups accurately defined by unique spacer RFLP genotypes is in perfect correlation with previous reports (1, 18, 20). Of note, the hsp65 gene RFLP method is unable to distinguish the clinically relevant subspecies M. kansasii II and Mka C from the nonpathogenic M. gastri (Table 2). By contrast, the use of the spacer is flawed by the sequence identity of M. marinum and M. ulcerans, but this represents a minor problem from a clinical point of view, since these species appear under completely different epidemiological circumstances (7).
The sequence variability of rapid growers was considerable. We can expect that additional spacer RFLP patterns will be found when more strains are analyzed. This may be particularly true for the M. terrae-M. nonchromogenicum complex or rapidly growing species such as M. neoaurum, which exhibited species-specific results, although the small number of strains used probably underestimates their true genetic heterogeneity (32). Hence, we can state that data on most rapid growers are still insufficient and remain to be improved in further studies. Similar observations have been made concerning the hsp65 gene as a genetic target (21). It could be interesting to validate the biological significance of spacer RFLP genotypes in comparison to type strains. Since 16S rDNA sequencing data for rapidly growing mycobacteria are still very incomplete (21, 26), it appears mandatory to look for the possibility that additional RFLP genotypes found may represent unknown infrasubspecific 16S rDNA genotypes. Evidence for this was shown here for M. flavescens and M. parafortuitum, but further investigations of the exact phenotypes are certainly warranted because these reference strains were not reassessed by biochemical tests in this study. Besides phylogenetic or taxonomic considerations, such RFLP subgroups may reflect clinically, physiologically, or epidemiologically significant subdivisions, as has been proposed for M. chelonae (19) or the M. avium complex (4, 8, 9). Some additional RFLP genotypes in a species may not have recognizable phenotypic or genotypic correlates in either the physiological tests usually performed or in their complete 16S rDNAs, respectively, due to the higher phylogenetic resolution of 16S-23S spacer sequences. This was nicely shown by 16S rDNA sequencing of M. phlei and M. triviale RFLP genotypes II.
The high similarity of M. avium to M. bohemicum and M. kansasii IV represents an undesirable shortcoming of the method, since M. avium is the most frequently isolated member of the genus Mycobacterium. If gel electrophoresis was performed carefully, the above-mentioned similar patterns were not confused by technical staff in our laboratory (Fig. 3, compare lanes 6 and 7), and ultimately, application of a third enzyme resulted in a definite correct assignment in all cases. In addition, M. bohemicum and M. kansasii RFLP genotype IV (sequevar Mka B) are very rare in clinical specimens (1, 20). A possible failing of the method in this case can be disregarded in most laboratories that use GenProbes for identification of M. tuberculosis and the M. avium complex. However, if RFLP is used as the sole procedure for identification, investigators should remain vigilant for this pattern by letting gels run longer in comparisons to M. avium as the proposed internal standard. In this context, a conclusion that deserves mention is that judicious inclusion of closely related species is crucial for a complete assessment of the reliability and discriminatory power of these methods. In fact, the necessity to apply additional enzymes in a few groups was only recognized because care was taken to study, if possible, closely related mycobacteria as well (i.e., M. simiae together with M. lentiflavum and M. triplex). Apparently, in contrast to the hsp65 gene, the diversity of spacer sequences is not high enough at a species level in all phylogenetic groups to allow separation by only two digests. Nevertheless, we believe that this disadvantage is compensated for by the overall simplicity of the scheme for the majority of other species and the large amount of information yielded after HaeIII digestion by itself. RFLP results for species such as M. lentiflavum-M. triplex, M. bohemicum (which is closely related to M. avium), Mycobacterium interjectum-Mycobacterium intermedium, M. farcinogenes, or Mycobacterium obuense have not yet been reported for the hsp65 gene (5, 27–30). This issue must keep us alert to the fact that the hsp65 data have yet to be perfected.
A major result that emerges from our study is the fact that the method presented reveals the potential to be used in mycobacterial taxonomy. It is interesting that species closely related to each other clustered in the same or similar HaeIII patterns. Examples are the M. avium complex together with M. scrofulaceum and M. bohemicum, M. simiae and relatives, M. fortuitum and M. senegalense, and similar HaeIII patterns found for M. terrae and M. nonchromogenicum. By contrast, one included taxon that probably represents a new species (termed Mycobacterium sp. xenopi), as indicated by a low 16S rDNA similarity of only 97% with M. xenopi, clearly possessed a distinct HaeIII pattern. Similarly, HaeIII patterns rather different from the type strain pattern found in six M. flavescens reference strains suggested a genetic disintegrity of this group, a finding that was later confirmed by 16S sequencing. Organisms provisionally denoted subspecies, for example, Mycobacterium sp. malmoense, by contrast emerged as unique identifiable entities but shared the same HaeIII pattern with the most closely related species. Irrespective of the taxonomical validity of these observations, they are a reflection of a high degree of spacer sequence conservation and reinforce the previously discussed view that 16S-23S rDNA spacer sequence analysis constitutes an adjunct to mycobacterial phylogeny (8, 9, 22). This study provides additional evidence that spacer sequence analysis results are in good correlation with 16S rDNA data. Valid descriptions of new species or subspecies were not addressed in this study, but the M. kansasii-M. gastri and M. flavescens cases illustrate the usefulness of our method in identifying novel taxons before more accurate but labor-intensive comparative sequencing investigations (or those involving numerical taxonomy) are initiated. The problem of pigmented M. gastri strains was addressed by Anz and Schröder as early as 1970 (2), and this debate was taken up again as the genetic heterogeneity of M. kansasii was recognized later (1, 20). The somewhat confusing nomenclature of different studies is gathered in Table 2, and it can be acknowledged that spacer-based methods are superior to all other genetic approaches, including the 16S rDNA method, which cannot differentiate deeply enough to account for the genotypes found, and the AccuProbe method, which misses one M. kansasii subgroup while the taxonomically indeterminate subgroup with the spacer genotype IV produces positive results. The latter finding has no impact on clinical reports because so far these strains have only been isolated in environmental samples (1).
In conclusion, 16S-23S rDNA PCR-RFLP is a promising new method with consistent advantages over the previously used hsp65 gene-based method for reliable and easy identification of mycobacteria. Compared to new but technically demanding (and thus still cost prohibitive) techniques, such as the development of DNA probe arrays (31), this method has the advantage of being both simple and extensive in its diagnostic spectrum and cost-effective at the same time. Despite allelic diversity, good intraspecies stabilities of recognizable RFLP genotypes were demonstrated. More significant polymorphisms on a subspecies level do not preclude the use of this method; rather, finding correlations of specific genetic subtypes to medically relevant linkages represents an issue worthy of further investigation.
REFERENCES
- 1.Alcaide F, Richter I, Bernasconi C, Springer B, Hagenau C, Schulze-Röbbecke R, Tortoli E, Martin R, Böttger E, Telenti A. Heterogeneity and clonality among isolates of Mycobacterium kansasii: implications for epidemiological and pathogenicity studies. J Clin Microbiol. 1997;35:1959–1964. doi: 10.1128/jcm.35.8.1959-1964.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anz W, Schröder K-H. Photochromogenic strains of Mycobacterium gastri? Zentrabl Bakteriol. 1970;214:553–554. [PubMed] [Google Scholar]
- 3.De Beenhouwer H, Liang Z, de Rijk P, van Eekeren C, Portaels F. Detection and identification of mycobacteria by DNA amplification and oligonucleotide-specific capture plate hybridization. J Clin Microbiol. 1995;33:2994–2998. doi: 10.1128/jcm.33.11.2994-2998.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Smet A L, Brown I N, Yates M, Ivanyi J. Ribosomal internal transcribed spacers are identical among Mycobacterium avium-intracellulare complex isolates from AIDS patients, but vary among isolates from elderly pulmonary disease patients. Microbiology. 1995;141:2739–2747. doi: 10.1099/13500872-141-10-2739. [DOI] [PubMed] [Google Scholar]
- 5.Devallois A, Goh K S, Rastogi N. Rapid identification of mycobacteria to species level by PCR-restriction fragment length polymorphism analysis of the hsp65 gene and proposition of an algorithm to differentiate 34 mycobacterial species. J Clin Microbiol. 1997;35:2969–2973. doi: 10.1128/jcm.35.11.2969-2973.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emler S, Ninet B, Rohner P, Auckenthaler R, Jäger D, Hirschel B. Molecular basis for cross-reactivity between a strain of Mycobacterium terrae and DNA probes for Mycobacterium tuberculosis complex. Eur J Microbiol Infect Dis. 1995;14:627–629. doi: 10.1007/BF01690741. [DOI] [PubMed] [Google Scholar]
- 7.Falkinham J O. Epidemiology of infection by nontuberculous mycobacteria. Clin Microbiol Rev. 1996;9:177–215. doi: 10.1128/cmr.9.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frothingham R, Wilson K H. Sequence-based differentiation of strains in the Mycobacterium avium complex. J Bacteriol. 1993;175:2818–2825. doi: 10.1128/jb.175.10.2818-2825.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frothingham R, Wilson K H. Molecular phylogeny of the Mycobacterium avium complex demonstrates clinically meaningful divisions. J Infect Dis. 1994;169:305–312. doi: 10.1093/infdis/169.2.305. [DOI] [PubMed] [Google Scholar]
- 10.Good R C. Opportunistic pathogens in the genus Mycobacterium. Annu Rev Microbiol. 1985;39:347–369. doi: 10.1146/annurev.mi.39.100185.002023. [DOI] [PubMed] [Google Scholar]
- 11.Gürtler V, Stanisch V A. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology. 1996;142:3–16. doi: 10.1099/13500872-142-1-3. [DOI] [PubMed] [Google Scholar]
- 12.Kent P T, Kubica G P. Public health mycobacteriology—a guide for the level III laboratory. U.S. Department of Health and Human Services publication (CDC) 86-8230. Atlanta, Ga: Centers for Disease Control; 1985. [Google Scholar]
- 13.Kim B-J, Lee S-H, Lyu M-A, Kim S-J, Bai G-H, Kim S-J, Chae G-T, Kim E-C, Cha C-Y, Kook Y-H. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB) J Clin Microbiol. 1999;37:1714–1720. doi: 10.1128/jcm.37.6.1714-1720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirschner P, Springer B, Vogel U, Meier A, Wrede A, Kiekenbeck M, Bange F C, Böttger E C. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J Clin Microbiol. 1993;31:2882–2889. doi: 10.1128/jcm.31.11.2882-2889.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kox L F F, van Leeuwen J, Knijper S, Jansen H M, Kolk A H. PCR assay based on DNA coding for 16S rRNA for detection and identification of mycobacteria in clinical samples. J Clin Microbiol. 1995;33:3225–3233. doi: 10.1128/jcm.33.12.3225-3233.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lappayawichit P, Rienthong S, Rienthong D, Chuchottaworn C, Chaiprasert A, Panbangred W, Saringcarinkul H, Palittapongarnpim P. Differentiation of Mycobacterium species by restriction enzyme analysis of amplified 16S-23S ribosomal DNA spacer sequences. Tubercle Lung Dis. 1996;77:257–263. doi: 10.1016/s0962-8479(96)90010-6. [DOI] [PubMed] [Google Scholar]
- 17.Müller K-D, Schmid E N, Kroppenstedt R M. Improved identification of mycobacteria by using the Microbial Identification System in combination with additional trimethylsulfonium hydroxide pyrolysis. J Clin Microbiol. 1998;36:2477–2480. doi: 10.1128/jcm.36.9.2477-2480.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Picardeau M, Prod'Hom G, Raskine L, LePennec M P, Vincent V. Genotypic characterization of five subspecies of M. kansasii. J Clin Microbiol. 1997;35:25–32. doi: 10.1128/jcm.35.1.25-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Portaeles F, de Rijk P, Jannes G, Lemans R, Mijs W, Riqouts L, Rossau R. The 16S-23S rRNA spacer, a useful tool for taxonomical and epidemiological studies of the M. chelonae complex. Tubercle Lung Dis. 1996;77(Suppl. 2):17–18. [Google Scholar]
- 20.Richter E, Niemann S, Rüsch-Gerdes S, Hoffner S. Identification of M. kansasii by using a DNA probe (AccuProbe) and molecular techniques. J Clin Microbiol. 1999;37:964–970. doi: 10.1128/jcm.37.4.964-970.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ringuet H, Akoua-Koffi C, Honore S, Varnerot A, Vincent V, Berche P, Gaillard J L, Pierre-Audigier C. hsp65 sequencing for identification of rapidly growing mycobacteria. J Clin Microbiol. 1999;37:852–857. doi: 10.1128/jcm.37.3.852-857.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roth A, Fischer M, Hamid H E, Ludwig W, Michalke S, Mauch H. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. J Clin Microbiol. 1998;36:139–147. doi: 10.1128/jcm.36.1.139-147.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanguinetti M, Pasteraro B, Ardito F, Zanetti S, Cingolani A, Sechi L, De Luca A, Ortona L, Fadda G. Routine use of PCR-reverse cross-blot hybridization assay for rapid identification of Mycobacterium species growing in liquid media. J Clin Microbiol. 1998;36:1530–1533. doi: 10.1128/jcm.36.6.1530-1533.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schröder K-H, Naumann L, Kroppenstedt R M, Reischl U. Mycobacterium hassiacum sp. nov., a new rapidly growing thermophilic mycobacterium. Int J Syst Bacteriol. 1997;47:86–91. doi: 10.1099/00207713-47-1-86. [DOI] [PubMed] [Google Scholar]
- 25.Shinnick T M, Good R C. Mycobacterial taxonomy. Eur J Clin Microbiol Infect Dis. 1994;13:884–901. doi: 10.1007/BF02111489. [DOI] [PubMed] [Google Scholar]
- 26.Springer B, Stockman L, Teschner K, Roberts G D, Böttger E C. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J Clin Microbiol. 1996;34:296–303. doi: 10.1128/jcm.34.2.296-303.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steingrube A V, Gibson J L, Brown B A, Zhang Y, Wilson R W, Rajagopalan M, Wallace R J. PCR amplification and restriction endonuclease analysis of a 65-kilodalton heat shock protein gene sequence for taxonomic separation of rapidly growing mycobacteria. J Clin Microbiol. 1995;33:149–153. doi: 10.1128/jcm.33.1.149-153.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steingrube V A, Wilson R W, Brown B A, Jost K C, Blacklock Z, Gibson J L, Wallace R J. Rapid identification of clinically significant species and taxa of aerobic actinomycetes, including Actinomadura, Gordonia, Nocardia, Rhodococcus, Streptomyces, and Tsukamurella isolates, by DNA amplification and restriction endonuclease analysis. J Clin Microbiol. 1997;35:817–822. doi: 10.1128/jcm.35.4.817-822.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor T B, Patterson C, Hlae Y, Safranek W W. Routine use of PCR-restriction fragment length polymorphism analysis for identification of mycobacteria growing in liquid media. J Clin Microbiol. 1997;35:79–85. doi: 10.1128/jcm.35.1.79-85.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Telenti A, Marchesi F, Balz M, Bally F, Böttger E C, Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. 1993;31:175–178. doi: 10.1128/jcm.31.2.175-178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Troesch A, Nguyen H, Miyada C G, Desvarenne S, Gingeras T R, Kaplan P M, Cros P, Mabilat C. Mycobacterium species identification and rifampin resistance testing with high-density DNA probe arrays. J Clin Microbiol. 1999;37:49–55. doi: 10.1128/jcm.37.1.49-55.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torkko P, Suutari M, Suomalainen S, Paulin L, Larsson L, Katila M-L. Separation among species of Mycobacterium terrae complex by lipid analysis: comparison with biochemical tests and 16S rRNA sequencing. J Clin Microbiol. 1998;36:499–505. doi: 10.1128/jcm.36.2.499-505.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wallace R J. Recent changes in taxonomy and disease manifestations of the rapidly growing mycobacteria. Eur J Clin Microbiol Infect Dis. 1994;13:953–960. doi: 10.1007/BF02111497. [DOI] [PubMed] [Google Scholar]
- 34.Wilson R W, Steingrube V A, Brown B A, Wallace R J. Clinical application of PCR-restriction enzyme pattern analysis for rapid identification of aerobic actinomycete isolates. J Clin Microbiol. 1998;36:148–152. doi: 10.1128/jcm.36.1.148-152.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]