Abstract
Objective:
To evaluate the function of Piezo1, an evolutionarily conserved mechanically activated channel, in periodontal ligament (PDL) tissue homeostasis under compressive loading.
Materials and Methods:
Primary human PDL cells (hPDLCs) were isolated, cultured, and then subjected to 2.0 g/cm2 static compressive loading for 0.5, 3, 6, and 12 hours, respectively. The expressions of Piezo1 and osteoclastogenesis marker gene were assessed by semiquantitative reverse transcription–polymerase chain reaction. In addition, Piezo1 inhibitor, GsMTx4, was used to block the function of Piezo1, and tumor necrosis factor–α was also used as a positive control. After 12 hours of compressive loading the PDLCs were co-cultured with murine monocytic cell line RAW264.7. Immunofluorescence, western blot, enzyme-linked immunosorbent assay, and tartrate-resistant acid phosphatase staining were also used to test the potency of PDLCs to induce osteoclastogenesis and the activation of nuclear factor (NF)-κB.
Results:
Piezo1, cyclooxygenase-2, receptor activator of NF-κB ligand, and prostaglandin E2 were significantly upregulated under static compressive stimuli. GsMTx4 repressed osteoclastogenesis in the mechanical stress–pretreated PDLCs-RAW264.7 co-culture system. Furthermore, NF-κB signaling pathway was involved in the mechanical stress–induced osteoclastogenesis.
Conclusions:
Piezo1 exerts a transduction role in mechanical stress–induced osteoclastogenesis in hPDLCs.
Keywords: Piezo channel, Periodontal ligament, Mechanical loading, Osteoclastogenesis, NF-κB
INTRODUCTION
Periodontal ligament (PDL) tissue, which lies between tooth cementum and alveolar bone, is of fundamental importance in bone homeostasis. Proper orthodontic forces and occlusal forces lead to a balanced bone remodeling. Previous studies showed that PDL cells (PDLCs) can perceive the mechanical signals and transfer them to the progenitor cells, which can differentiate into osteoclasts or osteoblasts.1–4 Recently, we found that the expression of potential osteoclastogenesis inducers was upregulated in a compressed PDLCs model, including receptor activator of nuclear factor κB Ligand (RANKL), cyclooxygenase-2 (Cox2), while the expression of osteoprotegerin (OPG) was downregulated.5,6 Furthermore, prostaglandin E2 (PGE2) may increase RANKL mRNA expression in compressed PDLCs.3 However, the mechanism by which PDLCs sense and transduce mechanical signals is still unclear.
Recently, Coste7 and Coste et al.8 found that Piezo channel is an evolutionarily conserved ion channel family involved in mechanotransduction. It contains 24–32 transmembrane regions. Many other researchers9–12 also reported the Piezo channel to be a key player in cellular response to mechanical stimuli in different organs. PDLCs are mechanosensitive cells, so it is hypothesized that Piezo might also play a role in the response of PDLCs to mechanical stimuli.
Nuclear factor (NF)-κB is important in regulating cellular responses to mechanical stress.13–15 The signals induced by proinflammatory cytokines such as tumor necrosis factor (TNF)-α and other stimuli, are transmitted to the nucleus through nuclear translocation of NF-κB.16 Then nuclear NF-κB binds to consensus sequences of proinflammatory genes to initiate mRNA transcription.17 Mechanical strain augments inflammation and osteoclastogenesis; we speculated that mechanical stimulus–induced osteoclastogenesis might be associated with NF-κB activation, which might in turn be due to altered Piezo1 expression. Therefore, this study aimed to examine the function of Piezo1 in human PDLCs (hPDLCs) under mechanical stress and to explore its putative downstream signaling pathway.
MATERIALS AND METHODS
Cell Culture
hPDLCs were isolated from the root surface of healthy extracted premolars for orthodontic reasons. This study was approved by the Research Ethics Committee of West China Hospital, Sichuan University, PR China. Written informed consent was obtained from all of the patients. The periodontal tissue was digested with 2 mg/mL collagenase type I (Sigma, St. Louis, MO, USA) in phosphate-buffered saline (PBS) for 30 minutes at 37°C and then centrifuged at 400 g for 8 minutes at room temperature. Cells were then resuspended in α–minimum essential medium (α-MEM) (Hyclone, Shanghai, China) supplemented with 20% fetal bovine serum (FBS) (Gibco, Logan, UT, USA), 100 U/mL penicillin, and 100 µg/mL streptomycin (Hyclone, Shanghai, China) and incubated at 37°C in a 5% CO2 atmosphere. After reaching confluence, culture medium was changed with α-MEM containing 10% FBS. The cells obtained between the third and fifth passages were used for all experiments.
Murine monocytic cell line RAW264.7 (Sciencell, Carlsbad, CA, USA) was maintained in α-MEM containing 10% FBS with addition of 100 U/mL penicillin and 100 µg/mL streptomycin at 37°C in a 5% CO2 atmosphere.
Mechanical Stimulation: Application of Static Compressive Force
Prior to mechanical strain the suspension of hPDLCs (1 × 105 cells in 2 mL media/well) was dripped into a 6-well plate, and 72 hours were given to fully stabilize the PDLCs. The PDLCs were compressed continuously using the same compression method described previously. This method comprises a modification of the method described by Hiroyuki and Li.3,5,18 Briefly, a cover glass and a glass bottle are placed on the monolayer hPDLCs. The compressive force is adjusted by adding lead granules to the bottle to allow PDLCs to be subjected to 2.0 g/cm2 of compressive force, and the force is maintained for 0.5, 3, 6, or 12 hours, respectively.3,18 These time points were selected according to the method described in our previously published article.19 Two and a half microliters of GsMTx4 (Abcam, San Francisco, CA, USA), a specific inhibitor of cationic mechanosensitive ion channels, was added, and 0.8 ng/mL of TNF-α (R&D, Shanghai, China) was added to the indicated groups.20
Immunofluorescence
After mechanical stimulation for 0.5 hours, the hPDLCs were rinsed with PBS twice and fixed with 4% paraformaldehyde (Hyclone, Beijing, China) and then treated with 0.2% Triton X-100 (MP Biomedicals, Santa Ana, CA, USA) for 10 minutes at room temperature. Slices were treated with 5% normal goat serum in PBS for 1 hour at room temperature to block nonspecific immunoreaction. After thorough rinsing, the slices were incubated with primary antibodies of Piezo1 (Novus Biologicals, Littleton, CO, USA) and subsequently with anti-rabbit secondary antibody conjugated with Alexa 488 (diluted 1∶2000; Invitrogen, Carlsbad, CA, USA) to accomplish immunofluorescence. Then the slices were counterstained with Hoechst 33342 to visualize nuclei (Thermo Scientific, Shanghai, China).
Reverse Transcription–Polymerase Chain Reaction
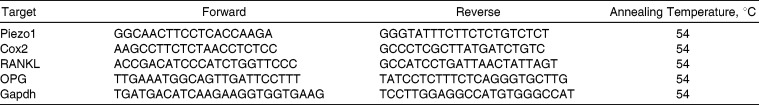
After hours of compressive stress, total RNA was extracted from three replicas using the reagent TRIzol. Semiquantitative reverse transcription–polymerase chain reaction (RT-PCR) was performed and analyzed to assess the expression of Piezo1, RANKL, OPG, and Cox2. The oligonucleotide sequences of the primers were shown in Table 1. RT-PCR was carried out using the RNA PCR kit on a FTC-2000 Real Time PCR system (Funglyn Biotech Inc, Shanghai, China). The 2−ΔΔCt method was used for analysis of gene expression, as described in other studies.21,22 Briefly, equation ΔCt was Ct of each target gene transcript − Ct of Gapdh, and ΔΔCt was calculated using the following formula: ΔΔCt = ΔCt [target gene in experiment group] − ΔCt [target gene in control group], then the relative fold difference was calculated using the formula 2−ΔΔCt.
Table 1.
Primer Sequences for Reverse Transcription–Polymerase Chain Reaction (RT-PCR) Analyses
Western Blot Analysis
For extraction of whole cellular proteins, PDLCs were rinsed with cold PBS twice and then lysed with cell RIPA buffer (added to 1 mM PMSF, phenylmethylsulfonyl fluoride), on ice for 30 minutes. For fractions of cytosolic and nuclear proteins, an extraction reagent kit (NE-PER Nuclear and Cytoplasmic Extraction Reagents, Thermo, Shanghai, China) was used according to the manufacturer's recommendations. Proteins were boiled in Laemmli sample buffer (Invitrogen, Carlsbad, CA, USA) and separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and then transferred to polyvinylidene fluoride membrane. The membranes were incubated with primary antibodies of Piezo1, NF-κB, β-actin, and histone 3 (Santa Cruz, Dallas, TX, USA) overnight; this step was followed by incubation for 1 hour with secondary antibody (diluted 1∶2000; Bioss, Boston, MA, USA). The membrane was detected with the aid of enhanced chemiluminescent detection reagents using a fluorescence imaging system (Clinx Science Instruments, Beijing, China).
PGE2 Measurement by Enzyme-Linked Immunosorbent Assay
The amount of secreted PGE2 in the cell culture supernatant was determined using a PGE2 enzyme-linked immunosorbent assay kit (Mybiosource, Saint Louis, MO, USA), per the manufacturer's protocol.
Co-Culture of RAW264.7 and PDLCs
In order to test the ability of stressed-induced PDLCs to induce osteoclastogenesis, we co-cultured the RAW264.7 cells with PDLCs after compressive stimulation or TNF-α treatment.23 After 12 hours of compressive stimulus, the glass bottle was removed carefully. We subsequently co-cultured 1 × 105 RAW264.7 cells with PDLCs for 4 days in 10% FBS α-MEM.
Tartrate-Resistant Acid Phosphatase Staining
After co-culturing of PDLCs and RAW264.7 for 4 days, cells were rinsed twice with PBS and fixed with 4% paraformaldehyde, then were stained for tartrate-resistant acid phosphatase (TRAP) activity by using an acid phosphatase leukocyte kit (Sigma, Saint Louis, MO, USA) per the manufacturer's protocol. Multinucleated cells having three or more nuclei were counted as osteoclasts, and three researchers counted independently under low-power microscopy (4×). After training in calibration, they randomly picked three view fields in each slice and calculated the average number of the osteoclasts under one view field.
Statistical Evaluation
Quantitative data were analyzed for statistical difference using a Mann-Whitney test at a significance level of P < .05 (n = 3 for each group). The values are expressed as the means ± the standard error of the mean (SEM).
RESULTS
Mechanical Loading Induces Both Osteoclastogenesis Marker Gene and Piezo1 Upregulation
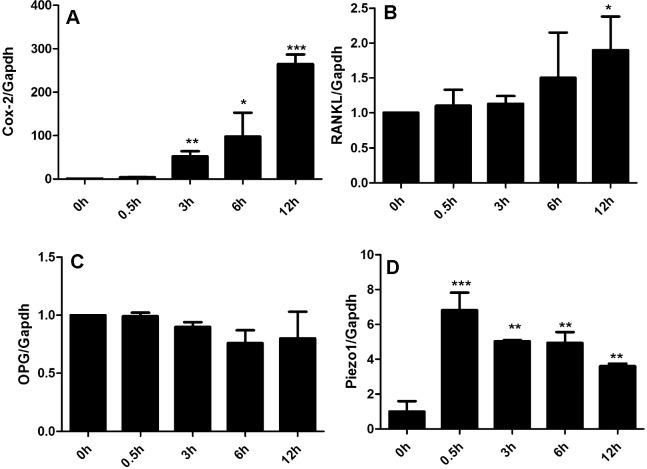
As expected, exposure to mechanical stimuli significantly increased Cox2 expression over 12 hours in a time-dependent manner (Figure 1A). Furthermore, RANKL mRNA in tested hPDLCs at 12 hours was significantly higher than that measured at the earlier time point (Figure 1B). In contrast, OPG mRNA in loaded or controlled hPDLCs showed no significant difference, at least in our system (Figure 1C), resulting in an increased ratio of RANKL to OPG.
Figure 1.
Mechanical stress is associated with increased osteoclastogenesis and Piezo1 activation in hPDLCs. (A) Cox-2 mRNA expression significantly increased after 3 hours of loading. (B) RANKL mRNA significantly increased after 12 hours of loading. (C) OPG mRNA expression did not change after mechanical stress. (D) Piezo1 mRNA expression significantly increased after 0.5 hours of loading and over the tested 12 hours. Values show as mean ± SEM. n = 3. * P = .05; ** P = .01; *** P = .001; **** P = .0001.
Consistent with increased osteoclastogenesis-related genes, the Piezo1 mRNA profile in loaded hPDLCs increased at 0.5 hours and lasted 12 hours. Immunostaining and Western blot further confirmed the enhancement of Piezo1 production in tested hPDLCs (Figures 2A–F and 3A).
Figure 2.
Piezo1 expression in isolated hPDLCs. Piezo1 expression is much stronger in hPDLCs loaded for 3 hours (D–F) than in normal hPDLCs (A–C). Scale bar: 100 µm.
Figure 3.
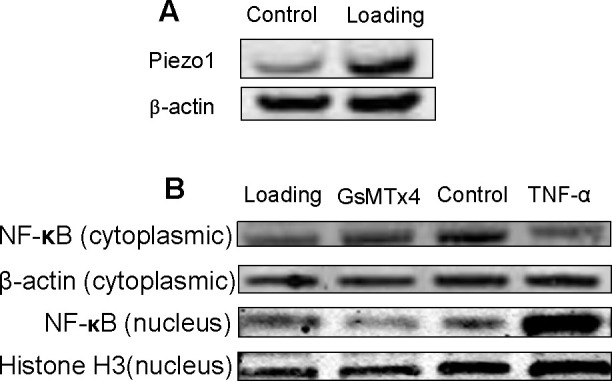
Western blot analysis of Piezo1 and NF-κB in hPDLCs. (A) Western blot analysis showing the effect of mechanical stress on Piezo1. (B) Western blot analysis showing the effect of GsMTx4 and TNF-α on nuclear NF-κB; Western immunoblots probed with antibodies against NF-κB from nuclear and cytosolic proteins. Histone 3 and β-actin were used as internal controls.
GsMTx4 Suppresses NF-κB Activation
To investigate the intracellular mechanisms by which Piezo1 may facilitate mechanical stress-induced osteoclastogenesis responses, we evaluated the nuclear translocation of NF-κB, the major transcription factor involved in the signal transduction of proinflammatory cytokines. Both cytoplasmic and nuclear NF-κB were investigated using Western immunoblots. Nuclear NF-κB was significantly increased 3 hours after mechanical loading. Addition of TNF-α served as a positive control. GsMTx4 suppressed the increase of nuclear NF-κB induced by mechanical loading. In contrast, cytoplasmic NF-κB decreased after mechanical loading, which was recovered by GsMTx4 (Figure 3B).
GsMTx4 Inhibits Osteoclastogenesis
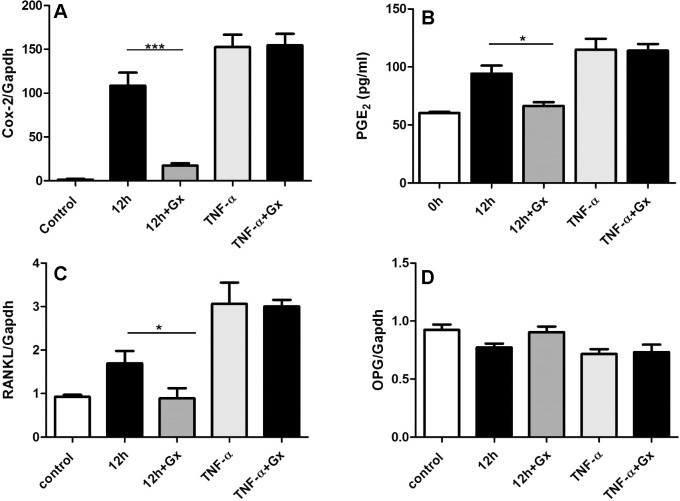
The effects of GsMTx4 on osteoclastogenesis were then explored by measuring Cox2, RANKL, and OPG mRNA amounts as well as synthesized PGE2 amount. GsMTx4 significantly reduced the mechanical stress induced, but it did not influence TNF-α–induced upregulation of Cox2 and RANKL mRNA (Figure 4A,C). Consistently, GsMTx4 also inhibited mechanical stress–induced PGE2 synthesis but not TNF-α–induced PGE2 synthesis (Figure 4B). However, OPG mRNA shows no significant difference among all groups (Figure 4D). Furthermore, we also tested the formation of osteoclast-like cells after 4 days of co-culture. As shown in Figure 5A, mechanically loaded PDLCs induced the differentiation of RAW264.7 into osteoclast-like cells (shown as multinucleated TRAP-positive cells in image), and GsMTx4 inhibited the effect of mechanical stimulus. However, GsMTx4 could not affect the function of TNF-α in inducing osteoclastogenesis. The quantification of osteoclasts numbers shows GsMTx4 inhibits the effect of mechanical stress by 65% (Figure 5B). Thus, those observations indicate that GsMTx4 exerts a restraining effect on mechanical stress–induced osteoclastogenesis.
Figure 4.
Piezo1 inhibitor, GsMTx4 (Gx), treatment is associated with reduced mechanical stress–induced osteoclastogenesis but not with TNF-α–induced osteoclastogenesis. Changes of mRNA profile of Cox2 (A), RANKL (C), OPG (D), and protein production of PGE2 (B) in hPDLCs after mechanical stress (12 hours) or TNF-α induction with or without addition of Gx. Values shown as mean ± SEM. n = 3, * P = .05; ** P = .01; *** P = .001; **** P = .0001.
Figure 5.
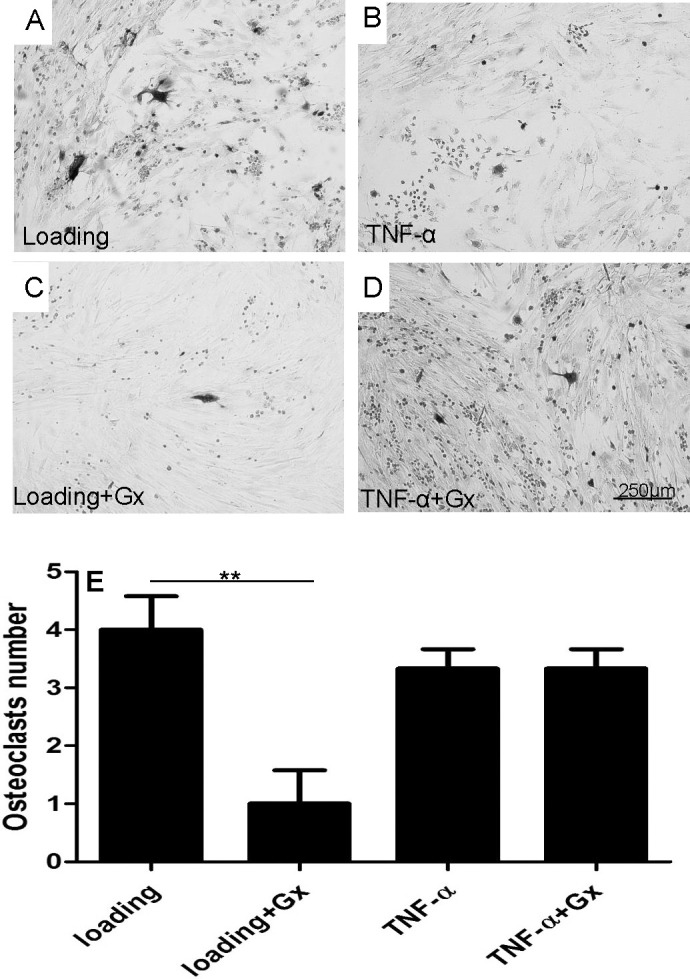
TRAP staining analyzes the effects of Gx on mechanical stress–induced and TNF-α–induced osteoclastogenesis in a PDL-RAW264.7 co-culture system. (A,C) Gx reduced numbers of osteoclast-like (TRAP+) cells in mechanical stress–pretreated PDL-RAW264.7 co-culture system. (B,D) Gx did not alter numbers of osteoclast-like cells in a TNF-α–pretreated PDL-RAW264.7 co-culture system. (E) Quantification of TRAP staining results. Values show as mean ± SEM. Scale bar = 250 µm. * P = .05; ** P = .01; *** P = .001; **** P = .0001.
DISCUSSION
Orthodontic force is believed to be sensed and transduced by PDLCs. Progenitor cells in the periodontium then differentiate into osteoclasts and osteoblasts, thereby causing bone remodeling and tooth movement. In the present study, we first investigated whether osteoclastogenesis-associated genes are activated following mechanical force administration in hPDLCs. We tested time-course mRNA profiles of Cox2, RANKL, and OPG. As many other studies and our previous microarray data show, osteoclastogenesis inducer gene expression increased and osteoclastogenesis inhibitors dropped down under static mechanical compression.5,6 In the present study, Cox2 and RANKL mRNA also increased significantly after hours of mechanical stress. However, in the present study OPG mRNA was not changed in loaded hPDLCs.
Next, we tested the role of Piezo channels in PDL tissues. To the best of our knowledge, few reports have involved study of the role of Piezo channels, although there are several studies that mention other ion channels in PDL tissues, K+ channels, like TREK-1.24,25 Thus, in the present study, we tested time-course Piezo1 gene expression in primary hPDLCs under compressive loading. We found that Piezo1 mRNA increased after 0.5 hours of mechanical loading and lasted for 12 hours. Immunofluorescence and Western blot analysis reconfirm the increases of Piezo1 expression in hPDLCs after stimulation.
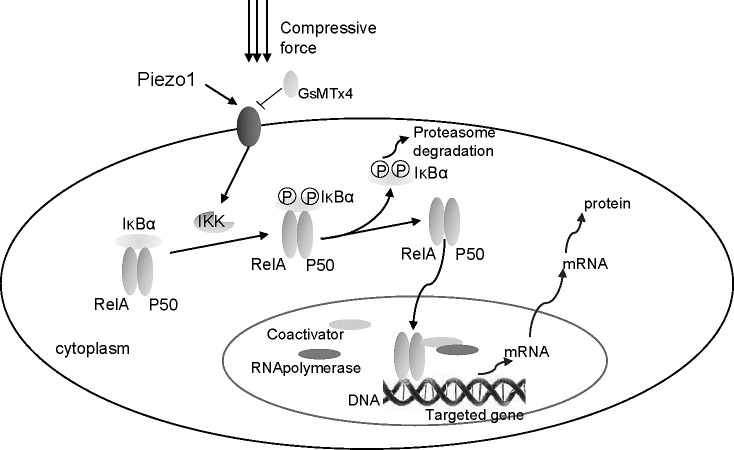
NF-κB transcription factors are involved in highly conserved signaling pathways that respond to proinflammatory cytokines and mechanical signals through their nuclear translocation and subsequent activation of a plethora of genes.13–15 NF-κB consists of the transactivation subunit RelA/p65 and the DNA-binding subunits.16 We demonstrate that mechanical stress induces nuclear translocation of NF-κB, which affects the expression of target genes such as Cox2 and RANKL (Figure 6), while the underlying mechanism remains unknown. We also used Piezo1 channel inhibitor (GsMTx4) to block the function of Piezo1. After utilizing GsMTx4, mechanical stress–induced NF-κB nuclear translocation and osteoclastogenesis-associated genes such as Cox2, RANKL were restrained. In addition to the inhibition of Cox2 and RANKL gene expression, we also found that static mechanical compression promotes the secretion of PGE2, which is consistent with the findings of previous studies26,27 that claim that PGE2 has a role in osteoclastogensis.26,27 Furthermore, GsMTx4 can also restrain the upregulation of PGE2.
Figure 6.
Mechanical stress activates NF-κB signaling via Piezo1-dependent pathway.
In the present study, administration of GsMTx4 also diminished the potency of mechanically loaded but not TNF-α–treated hPDLCs differentiating RAW264.7 into osteoclast-like cells.
One limitation to this study worth noting is that GsMTx4 is not specific for Piezo1, although this inhibitor indeed can only block mechanosensitive ion channels, but not all ion channels. Thus, further study with RNA interference is needed to reconfirm it.
In conclusion, we found that Piezo1 exerted a transduction role in mechanical stimuli–induced osteoclastogenesis with the involvement of NF-κB activation. A better understanding of the biological behavior of PDLCs responding to mechanical stress might facilitate the development of pharmacological therapy for accelerating orthodontic tooth movement or preventing postorthodontic treatment relapse.
CONCLUSIONS
The present data demonstrate for the first time that PDLC exposure to mechanical loading is associated with the activation of mechanically sensitive ion channel Piezo1. The pharmacological blockade of Piezo1 by GsMTx4 might inhibit the potency of PDLCs inducing osteoclastogenesis.
Our results indicate that osteoclastogenesis induced by mechanical stress is transduced by Piezo1 ion channel and mediated by NF-κB signaling pathway.
ACKNOWLEDGMENT
This work is supported by the National Nature Science Foundation of China (10772128 to Dr Zhihe Zhao).
REFERENCES
- 1.Idris AI, Landao-Bassonga E, Ralston SH. The TRPV1 ion channel antagonist capsazepine inhibits osteoclast and osteoblast differentiation in vitro and ovariectomy induced bone loss in vivo. Bone. 2010;46:1089–1099. doi: 10.1016/j.bone.2010.01.368. [DOI] [PubMed] [Google Scholar]
- 2.Kanjanamekanant K, Luckprom P, Pavasant P. Mechanical stress-induced interleukin-1beta expression through adenosine triphosphate/P2X7 receptor activation in human periodontal ligament cells. J Periodont Res. 2013;48:169–176. doi: 10.1111/j.1600-0765.2012.01517.x. [DOI] [PubMed] [Google Scholar]
- 3.Kanzaki H, Chiba M, Shimizu Y, Mitani H. Periodontal ligament cells under mechanical stress induce osteoclastogenesis by receptor activator of nuclear factor kappaB ligand up-regulation via prostaglandin E2 synthesis. J Bone Miner Res Off J Am Soc Bone Miner Res. 2002;17:210–220. doi: 10.1359/jbmr.2002.17.2.210. [DOI] [PubMed] [Google Scholar]
- 4.Kim SJ, Park KH, Park YG, Lee SW, Kang YG. Compressive stress induced the up-regulation of M-CSF, RANKL, TNF-alpha expression and the down-regulation of OPG expression in PDL cells via the integrin-FAK pathway. Arch Oral Biol. 2013;58:707–716. doi: 10.1016/j.archoralbio.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Zheng W, Liu JS, et al. Expression of osteoclastogenesis inducers in a tissue model of periodontal ligament under compression. J Dent Res. 2011;90:115–120. doi: 10.1177/0022034510385237. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Li M, Tan L, et al. Analysis of time-course gene expression profiles of a periodontal ligament tissue model under compression. Arch Oral Biol. 2013;58:511–522. doi: 10.1016/j.archoralbio.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Coste B. Piezo proteins form a new class of mechanically activated ion channels. Med Sci. 2012;28:1056–1057. doi: 10.1051/medsci/20122812012. [DOI] [PubMed] [Google Scholar]
- 8.Coste B, Xiao B, Santos JS, et al. Piezo proteins are pore-forming subunits of mechanically activated channels. Nature. 2012;483:176–181. doi: 10.1038/nature10812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eijkelkamp N, Linley JE, Torres JM, et al. A role for Piezo2 in EPAC1-dependent mechanical allodynia. Nat Commun. 2013;4:1682. doi: 10.1038/ncomms2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisenhoffer GT, Loftus PD, Yoshigi M, et al. Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature. 2012;484:546–549. doi: 10.1038/nature10999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottlieb PA, Sachs F. Piezo1: properties of a cation selective mechanical channel. Channels. 2012;6:214–219. doi: 10.4161/chan.21050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim SE, Coste B, Chadha A, Cook B, Patapoutian A. The role of Drosophila Piezo in mechanical nociception. Nature. 2012;483:209–212. doi: 10.1038/nature10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 14.Agarwal S, Long P, Seyedain A, Piesco N, Shree A, Gassner R. A central role for the nuclear factor-kappaB pathway in anti-inflammatory and proinflammatory actions of mechanical strain. FASEB J. 2003;17:899–901. doi: 10.1096/fj.02-0901fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ritter N, Mussig E, Steinberg T, Kohl A, Komposch G, Tomakidi P. Elevated expression of genes assigned to NF-kappaB and apoptotic pathways in human periodontal ligament fibroblasts following mechanical stretch. Cell Tissue Res. 2007;328:537–548. doi: 10.1007/s00441-007-0382-x. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109(suppl):S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 17.Karin M, Lin A. NF-kappaB at the crossroads of life and death. Nat Immunol. 2002;3:221–227. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- 18.Nakajima R, Yamaguchi M, Kojima T, Takano M, Kasai K. Effects of compression force on fibroblast growth factor-2 and receptor activator of nuclear factor kappa B ligand production by periodontal ligament cells in vitro. J Periodont Res. 2008;43:168–173. doi: 10.1111/j.1600-0765.2007.01008.x. [DOI] [PubMed] [Google Scholar]
- 19.Tang N, Zhao Z, Zhang L, et al. Up-regulated osteogenic transcription factors during early response of human periodontal ligament stem cells to cyclic tensile strain. Arch Med Sci. 2012;8:422–430. doi: 10.5114/aoms.2012.28810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bae C, Sachs F, Gottlieb PA. The mechanosensitive ion channel Piezo1 is inhibited by the peptide GsMTx4. Biochemistry. 2011;50:6295–6300. doi: 10.1021/bi200770q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes PA, Brierley SM, Young RL, Blackshaw LA. Localization and comparative analysis of acid-sensing ion channel (ASIC1, 2, and 3) mRNA expression in mouse colonic sensory neurons within thoracolumbar dorsal root ganglia. J Comp Neurol. 2007;500:863–875. doi: 10.1002/cne.21204. [DOI] [PubMed] [Google Scholar]
- 22.Swartz MF, Fink GW, Lutz CJ, et al. Left versus right atrial difference in dominant frequency, K(+) channel transcripts, and fibrosis in patients developing atrial fibrillation after cardiac surgery. Heart Rhythm Off J Heart Rhythm Soc. 2009;6:1415–1422. doi: 10.1016/j.hrthm.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lossdorfer S, Gotz W, Jager A. PTH(1-34)-induced changes in RANKL and OPG expression by human PDL cells modify osteoclast biology in a co-culture model with RAW 264.7 cells. Clin Oral Invest. 2011;15:941–952. doi: 10.1007/s00784-010-0456-0. [DOI] [PubMed] [Google Scholar]
- 24.Ohara A, Saeki Y, Nishikawa M, Yamamoto Y, Yamamoto G. Single-channel recordings of TREK-1 K+ channels in periodontal ligament fibroblasts. J Dent Res. 2006;85:664–669. doi: 10.1177/154405910608500716. [DOI] [PubMed] [Google Scholar]
- 25.Saeki Y, Ohara A, Nishikawa M, Yamamoto T, Yamamoto G. The presence of arachidonic acid-activated K+ channel, TREK-1, in human periodontal ligament fibroblasts. Drug Metab Rev. 2007;39:457–465. doi: 10.1080/03602530701498786. [DOI] [PubMed] [Google Scholar]
- 26.Romer P, Kostler J, Koretsi V, Proff P. Endotoxins potentiate COX-2 and RANKL expression in compressed PDL cells. Clin Oral Invest. 2013;17(9):2041–2048. doi: 10.1007/s00784-013-0928-0. [DOI] [PubMed] [Google Scholar]
- 27.Nukaga J, Kobayashi M, Shinki T, et al. Regulatory effects of interleukin-1β and prostaglandin E2 on expression of receptor activator of nuclear factor-κB ligand in human periodontal ligament cells. J Periodontol. 2004;75:249–259. doi: 10.1902/jop.2004.75.2.249. [DOI] [PubMed] [Google Scholar]