Abstract
Purpose:
Enfortumab vedotin (EV) is an antibody-drug conjugate (ADC) targeting NECTIN4 (encoded by the PVRL4/NECTIN4 gene) approved for treatment-refractory metastatic urothelial cancer. Factors that mediate sensitivity or resistance to EV are unknown. In the present study, we sought to 1) examine heterogeneity of NECTIN4 gene expression across molecular subtypes of bladder cancer and 2) determine if NECTIN4 expression mediates EV sensitivity or resistance.
Experimental Design:
Molecular subtyping and NECTIN4 expression data from seven muscle-invasive bladder cancer clinical cohorts (n=1,915 total specimens) were used to assess NECTIN4 expression across molecular subtypes. The outcome of the transcriptomic analysis was relative NECTIN4 expression in the consensus molecular subtypes of bladder cancer. Expression of NECTIN4 was validated in bladder cancer cell lines. NECTIN4 was stably over-expressed or knocked down in basal and luminal bladder cancer cell lines and EV drug sensitivity assays were performed, as measured by cell proliferation and clonogenic assays.
Results:
NECTIN4 expression is heterogenous across molecular subtypes of bladder cancer and significantly enriched in luminal subtypes. NECTIN4 expression is positively correlated with luminal markers GATA3, FOXA1, and PPARG across all cohorts. NECTIN4 expression is both necessary and sufficient for EV sensitivity in luminal and basal subtypes of urothelial bladder cancer cells. Downregulation of NECTIN4 leads to EV resistance.
Conclusions:
Sensitivity to EV is mediated by expression of NECTIN4, which is enriched in luminal subtypes of bladder cancer. These findings may have implications for biomarker development, patient selection and the inclusion of molecular subtyping in ongoing and future EV clinical trials.
Keywords: Urothelial cancer, bladder cancer, enfortumab vedotin, luminal subtype, basal subtype, antibody-drug conjugate
INTRODUCTION:
The surface protein NECTIN4, encoded by the gene NECTIN4 (previously known as PVRL4) is an immunoglobulin-like, transmembrane protein which is highly expressed in urothelial carcinoma and has recently emerged as a new drug target. Enfortumab vedotin (EV; previously ASG-22CE) is an antibody-drug conjugate (ADC) that delivers monomethyl auristatin E (MMAE), a microtubule destabilizing agent, to tumor cells expressing NECTIN4.[1] A recent phase II clinical trial of EV monotherapy (EV-201) demonstrated a 44% overall response rate in patients with heavily pre-treated, metastatic or locally advanced urothelial carcinoma, which led to expedited FDA approval.[2] Multiple additional trials are underway to evaluate EV with or without immune checkpoint inhibitors and/or platinum-based chemotherapy for locally advanced and metastatic urothelial carcinoma.[3] As EV moves into earlier disease states and incorporated into the frontline setting, understanding the molecular characteristics that predict response and delineating mechanisms of resistance to EV are critical to maximizing clinical efficacy.
NECTIN4 expression has not been well defined in bladder cancer, and the limited studies to date demonstrate conflicting results. While nearly all (97%) patients enrolled in EV-201 had detectable NECTIN4 H-scores (median 290; range, 14-300),[2] variation in expression appears to exist. In a study of 524 primary bladder tumors, moderate to strong H-scores (H≥100) were found in only 63% of samples.[1] Moreover, a recent case series found NECTIN4 staining to be highly variable—only 24% of pure urothelial cancers had moderate to strong H-scores (H≥100), and the urothelial component of tumors with mixed histology featured weak and heterogeneous staining.[4]
Molecular subtyping of urothelial cancer has highlighted fundamental differences in the oncogenic mechanisms, immune infiltration, and clinicopathologic features.[5,6] Most recently, six key consensus subtypes have been described: luminal papillary (LumP), luminal nonspecified (LumNS), luminal unstable (LumU), basal/squamous (Bas/Sq), stroma-rich, and neuroendocrine-like (NE-like).[7] Whether NECTIN4 is ubiquitously and uniformly expressed across the six molecular subtypes of urothelial cancer remains unknown. Herein, we interrogated NECTIN4 gene expression in seven clinical datasets, including a post-cisplatin neoadjuvant chemotherapy cohort.[8-12] Our objectives were to examine variability in expression across bladder cancer subtypes and determine whether modulating NECTIN4 expression levels alone alters sensitivity and resistance to EV.
MATERIALS AND METHODS:
Patient populations and transcriptome profiling:
Seven retrospective cohorts were analyzed, including five publicly available cohorts. The Sjödahl 2012 (n=93) dataset [8] (obtained from NCBI Gene Expression Omnibus (GEO), accession number GSE32894) had undergone batch effect correction, quantile normalization, log2-transformation and gene centering by the authors. The Sjödahl 2017 (n=243) dataset [9] (obtained from GEO, accession number GSE83586) had been pre-processed including RMA-normalization and gene centering by the authors. TCGA (n=406) dataset [10] was obtained from cBioPortal and had been normalized by RSEM and was further transformed by log2(RSEM+1). [13,14] The Seiler 2017 (n=305) dataset [11] (obtained from GEO, accession number GSE87304) had been SCAN-normalized by the authors. Three additional cohorts were also analyzed: a post-NAC radical cystectomy cohort (n=133) (obtained from GEO, accession number GSE124305), [15] a cohort of localized TURBT specimens (MOL) (n=206), [12] and a commercial cohort (n=529) collected as part of the Decipher GRID registry (NCT02609269) (Supplementary Table 1). For the retrospective post-NAC and MOL cohorts, the Institutional Review Board (IRB) for each institution approved the respective studies and all patients provided written informed consent to analysis of their tumor tissues when required by each institution. The commercial cohort was collected from routine clinical use of the Decipher Bladder test ordered by US-based physicians between July 2017 and September 2020. These data consisted of de-identified and anonymized gene expression profiles that were available as part of the Decipher Genomic Resource Information Database (GRID) prospective registry study (NCT02609269). No follow up data was collected. The studies were conducted in accordance with the ethical guidelines of the Declaration of Helsinki. All cohorts underwent consensus molecular cluster subtyping as previously described.[7] All further analyses were made in the R statistical environment v3.6.2 [16] using RStudio v1.2.5033.[17]
Cell culture:
HT-1376, HT-1197, 647V and TCCSUP cells were obtained from the UCSF Cell Culture Facility. UMUC-3 cells were from the American Type Culture Collection (ATCC). UMUC-9, UMUC-14 and 253JBV cells were gifts from David McConkey (Pathology Core, Bladder Cancer SPORE, MD Anderson Cancer Center). Cells were grown in standard MEM media (Life Technologies) supplemented with 10% FBS (Seradigm). Cells were routinely tested for mycoplasma (last tested December 2020, MycoAlert Lonza) and validated by STR DNA profiling (Genetica). All experiments were conducted within 20 passages from the parental stock.
Plasmids and Cloning:
The pHR-SFFV-dCas9-BFP-KRAB and pCRISPRia_v2 plasmids were gifts from Jonathan Weissman and Luke Gilbert (Addgene #46911 and #84832). Five sgRNA guide sequences targeting NECTIN4 were cloned into pCRISPRi using BstX1 and Blp1 and Quick Ligation Kit (New England BioLabs). sgRNA guide sequences for NECTIN4 are listed in Supplementary Table 2. The NECTIN4 open reading frame (Horizon) was cloned into lentiviral vectors pCMV-V5-Blast (gift from Minkyu Kim) using Gateway cloning protocols (ThermoFisher).
CRISPRi/dCAS9 cell lines and Fluorescence-Activated Cell Sorting (FACS):
To generate the CRISPRi cell lines, cells were transduced with pHR-SFFV-dCas9-BFP-KRAB lentivirus and selected by FACS (BD Aria). Multiple sorts were performed until there was a uniform BFP peak. To generate knockdown cells, cells were transduced with lentivirus containing the sgRNA. Stably transduced cells were selected with puromycin (2μg/ml) or blasticidin (6 μg/ml) for at least 5 days.
Lentiviral production:
Lentiviral production was carried out using calcium-phosphate-mediated transfection of HEK293T-Lenti-X cells and collected at 48-96 hours. Lentivirus was concentrated using the Lenti-X Concentrator (Takara).
NECTIN4 surface staining and western blotting:
Cells were trypsinized, washed, incubated with Fc block (Fisher Scientific, Catalog #BDB564220, 1.25 ug/1x106 cells) for 10 minutes then incubated with anti-NECTIN4 PE antibody (Miltenyi Biotec #130-116-027, 1:100) for 15-20 minutes on ice. Cells were analyzed using an Attune NxT Flow and data were analyzed using FlowJo software. For western blotting, cells were lysed in RIPA buffer containing Halt protease and phosphatase inhibitor cocktail (Thermo Scientific). Lysates were subjected to SDS-PAGE, transferred to PVDF membranes, blocked in 5% w/v BSA, and incubated with primary antibody overnight. The blot was visualized using ECL Detection Reagents after 24h (Genesee Scientific). Antibodies against GAPDH (Cell Signaling Technology), NECTIN4 (Abcam) HRP-anti-rabbit (Cell Signaling Technology) were used according to manufacturer’s recommended dilutions.
Drug dose-response assays:
Cells were seeded in a 96-well plate in sextuplicate. After 24h, EV (Seattle Genetics) was added at indicated concentrations for 7-12 days and then incubated with Cell Proliferation Reagent WST-1 (Roche). Absorbance values were obtained using an Epoch Microplate Spectrophotometer and analyzed using Prism8 (GraphPad). For clonogenic assays, 2,000-5,000 cells were plated and EV was added in serial dilution. After 9-12 days, colonies were stained with 0.05% crystal violet and quantified on Oxford Optronix GelCount machine (Abingdon, UK). Assays were performed in triplicate.
Quantitative PCR (QPCR):
Total RNA was isolated from cells using the Quick RNA Kit (Zymo Research). cDNA was synthesized using the Superscript III RT First Strand Kit (Invitrogen). QPCR was performed using SYBR Green master mix (Roche) in Applied Biosystems QuantStudio7. Ct values were normalized to HPRT and GAPDH, and relative expression was calculated using the 2ddCt method. Primer sequences were found using the Harvard Primer Bank (Supplementary Table 2).
Statistical analysis:
For the transcriptomic analysis, correlation was calculated by Spearman’s rank correlation. The Kruskal-Wallis test was used to test for differences when there were more than two groups, and the Wilcoxon rank-sum test was used to test for differences between two groups unless otherwise stated. p<0.05 was considered significant. Violin plots were created using the ggplot2 package. [18] Kaplan-Meier survival analyses were performed with overall survival as endpoint using the survival package [19,20] and rms package. [21] Hazard ratios (HR) were calculated using the Cox proportional hazards regression model. Centroid values for selected genes were retrieved from the consensus MIBC R package. [7] For the in vitro assays, statistical analysis was performed using Prism8. All data are presented as mean +/− SEM, unless otherwise stated. When only two groups were compared, the two-tailed Student t-test was used.
Tumor xenografts, immunostaining and histology.
Mouse experiments were reviewed and approved by the UCSF IACUC. NSG (NOD/SCID/gamma) mice were housed in the UCSF barrier facility under pathogen-free conditions. 1x106 UMUC3-control and UMUC3-NECTIN4 overexpressing cells were injected subcutaneously in a suspension of 1:1 matrigel and serum free media. Tumor samples were collected and fixed in 10% neutral buffered formalin overnight and paraffin embedded. A bladder cancer tissue microarray (TMA), which contains 80 formalin-fixed, paraffin-embedded (FFPE) biopsy specimens in duplicate, was obtained from the University of British Columbia and previously described [15]. Antigen retrieval for immunohistochemistry was performed using EDTA buffer. Primary antibodies were incubated overnight at 4°C and secondary antibodies were incubated for 2 hours at room temperature. H-scores for NECTIN4 were assigned in a blinded manner and an average score was taken for each sample. The following antibodies were used: rabbit anti-NECTIN4 (abcam), normal rabbit IgG (Santa Cruz), biotinylated anti-rabbit (Jackson Lab), and HRP-Streptavidin (ThermoFisher). The DAB developing kit (Vector Laboratories) was used according to manufacturer’s instructions, and slides were counterstained with hematoxylin (Sigma-Aldrich).
RESULTS:
NECTIN4 is enriched in luminal subtypes of muscle-invasive bladder cancer
To assess NECTIN4 mRNA expression across molecular subtypes, we analyzed NECTIN4 mRNA expression in seven cohorts of patients with localized bladder tumors (Supplementary Table 1). Using the consensus classifier subtypes, we found luminal tumors (LumP, LumNS, LumU) had higher NECTIN4 mRNA gene expression compared to basal, NE-like, or stroma-rich subtypes (Figure 1A-1B, Supplementary Table 3). Molecular subtyping using the TCGA, Lund and Decipher classifiers also demonstrated high NECTIN4 mRNA expression in luminal subtypes (Supplementary Figure 1). In patients previously treated with cisplatin-based neoadjuvant chemotherapy (post-NAC), NECTIN4 mRNA expression was also highest in the luminal-like cluster (CC2) on radical cystectomy specimens (Figure 1C, Supplementary Table 3). [15] Interestingly, the expression levels of transcription factors which drive luminal cell fate in bladder cancer (GATA3, PPARG, FOXA1) positively correlated with NECTIN4 expression across multiple cohorts (Figure 1D, Supplementary Figure 2A-C). [22] Centroid values for NECTIN4, GATA3, PPARG, and FOXA1 in the consensus molecular classifier are shown in Supplementary Figure 2D. No significant associations between quartiles of NECTIN4 expression and overall survival were seen in cohorts for which retrospective clinical data were available (Supplementary Figure 2E). Immunohistochemistry staining for NECTIN4 protein in a bladder cancer tissue microarray (TMA) also demonstrated significant heterogeneity in expression. NECTIN4 protein was primarily localized to the cell surface in tumors that were positive, and the H-score was associated with the molecular subtype, with highest NECTIN4 protein expression found in the luminal subtypes (Supplementary Figure 3).
FIGURE 1:
NECTIN4 mRNA expression across the consensus molecular subtypes of muscle invasive bladder cancers (MIBC) and association with luminal markers GATA3, FOXA1, PPARG. (A) Violin plots showing NECTIN4 mRNA expression levels by consensus molecular subtypes in four public clinical cohorts of MIBC. (B) NECTIN4 mRNA expression in the commercial Decipher cohort (n=529) by consensus molecular subtype. (C) NECTIN4 mRNA expression across consensus clustering (CC) subtypes in the post-neoadjuvant chemotherapy (NAC) cohort (n=133). The p values from Kruskal-Wallis testing for each cohort are shown in A-C, and pairwise comparisons using the Wilcoxon rank-sum test are shown in Supplementary Table 2. (D) Scatter plot showing the mRNA expression of the luminal markers GATA3 (top), FOXA1 (middle), and PPARG (bottom) versus NECTIN4 in the Decipher cohort, color coded by molecular subtype. Spearman’s rank correlation is shown for NECTIN4 versus GATA3 (r = 0.39), FOXA1 (r = 0.36), and PPARG (r = 0.43).
We also found that NECTIN4 mRNA expression was higher in tumors from male patients compared to females in the Seiler cohort (Supplementary Figure 4) and in patients with a pathologic T1 stage tumors compared to pT2 tumors in the MOL cohort (Supplementary Figure 5). Differences in NECTIN4 mRNA expression across other clinical variables, including age and clinical stage were not significant in these cohorts (Supplementary Figures 4B-D, 5A-C).
NECTIN4 expression is enriched in human luminal bladder cancer cells
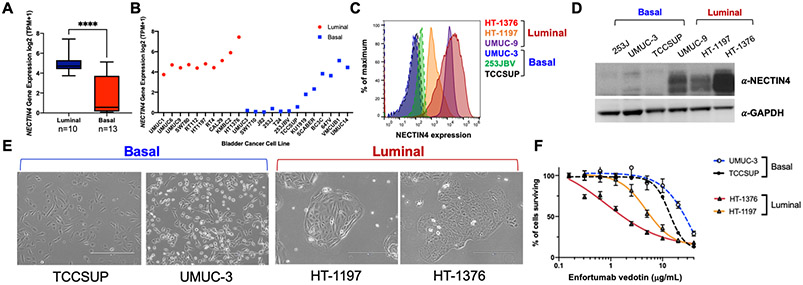
Human bladder cancer cell lines have been classified into luminal and basal subtypes.[23] We found that NECTIN4 mRNA expression was also enriched in luminal bladder cancer cell lines (p<0.0001, Figure 2A-2B), correlated strongly with expression of the luminal transcription factor GATA3 (n=36 urothelial cancer cell lines, p=0.00007) and not essential for cell survival (based on RNAi knockdown and CRISPR knockouts across hundreds of cell lines) (Supplementary Figure 6A-6B). Endogenous NECTIN4 protein levels on the cell surface were also significantly higher in luminal cell lines (UMUC-9, HT-1197 and HT-1376) compared to basal cell lines (253JBV, UMUC-3 and TCCSUP), which correlated with total protein expression levels (Figure 2B-2D). Luminal cells grew in cohesive clusters with a more epithelial morphology, whereas basal cells had a spindle-shaped, mesenchymal morphology (Figure 2E). Interestingly, some bladder cancer cell lines classified as basal (i.e. UMUC-14, VMCUB1 and 647V) also expressed NECTIN4, based on mRNA expression (Figure 2B). We validated cell surface and total protein expression of NECTIN4 in two of these lines (UMUC-14 and 647V), which expressed detectable but lower NECTIN4 protein on the cell surface compared to HT-1376 cells (Supplementary Figure 7A-7C). 647V cells appeared more luminal-like whereas the UMUC-14 cells exhibited an intermediate morphology compared to the UMUC-3 and TCCSUP cells (Supplementary Figure 7D), consistent with NECTIN4 playing a role in cell adhesion [3].
FIGURE 2:
NECTIN4 expression varies across bladder cancer cell lines and is associated with phenotypic differences and variable EV sensitivity. (A) NECTIN4 mRNA expression between luminal (n=10) and basal (n=13) bladder cancer subtypes (Source: DepMap database) (****p<0.0001). (B) NECTIN4 mRNA expression for individual bladder cancer cell lines are shown. Red denotes luminal cell lines and blue denotes basal cell lines (Source: DepMap database). (C) Surface staining for NECTIN4 protein in representative luminal (HT-1376, HT-1197, and UMUC-9) and basal (UMUC-3, 253JBV, and TCCSUP) bladder cancer cell lines. (D) Western blot demonstrating NECTIN4 protein expression in whole cell lysates across representative luminal (HT-1376, HT-1197, and UMUC-9) and basal (UMUC-3, 253JBV, and TCCSUP) bladder cancer cell lines. GAPDH is shown as the loading control. (E) Brightfield microscopy of luminal (HT-1197 and HT-1376) and basal (TCCSUP and UMUC-3) cells depicting the morphologic differences between basal cells (left) and luminal cells (right). Scale bar = 400 mm. (F) EV dose-response curves for basal cells (UMUC-3 and TCCSUP) and luminal cells (HT-1376 and HT-1197).
NECTIN4 expression is necessary and sufficient for EV-induced cell death
We then investigated whether luminal and basal cells had differential baseline sensitivity to EV and found that HT-1376 and HT-1197 luminal cells (high endogenous NECTIN4) were more sensitive to EV than UMUC-3 and TCCSUP basal cells (little to no endogenous NECTIN4), which were highly resistant (Figure 2F). In addition, despite detectable levels of NECTIN4 expression, the basal UMUC-14 cells were resistant to EV-induced cell death, whereas 647V cells were moderately sensitive (IC50 ~3 μg/mL) compared to HT-1376 cells (Supplementary Figure 7E). Taken together, these results demonstrate that NECTIN4 mRNA and protein expression are higher in luminal bladder cancer cell lines, which accordingly, are also more sensitive to EV-induced cell death.
Next, we asked whether knockdown of NECTIN4 in luminal HT-1376 cells could abrogate the sensitivity to EV. To do this, we generated HT-1376 cells expressing the catalytically-inactive dCas9 (known as CRISPR interference or CRISPRi)[24] and used 5 unique sgRNAs to silence NECTIN4 (Supplementary Table 2). We found that all 5 sgRNAs efficiently silenced NECTIN4 mRNA and NECTIN4 cell surface protein expression by >90%, as detected by quantitative polymerase chain reaction (qPCR) and surface protein staining (Figure 3A, Supplementary Figure 8A-8B). NECTIN4 knockdown cells retained a generally similar luminal morphology with some spindle-shaped features (Supplementary Figure 8C).
FIGURE 3:
NECTIN4 knockdown in HT-1376 cells decreases sensitivity to EV. (A) NECTIN4 surface protein expression in HT-1376 control and CRISPRi NECTIN4 knockdown (KD) HT-1376 cells. (B) EV dose-response WST proliferation assay in HT-1376 control and NECTIN4 KD cells, normalized to the mean absorbance of untreated cells. (C) Clonogenic assay quantification of EV sensitivity in HT-1376 control and HT-1376 NECTIN4 KD cells. **p<0.01 (D) Representative images of the clonogenic assay in HT-1376 control and HT-1376 NECTIN4 KD cells treated with EV.
Importantly, loss of NECTIN4 led to EV resistance. Whereas HT-1376 control cells with robust NECTIN4 expression were completely inhibited at 0.02 μg/ml of EV, HT-1376 NECTIN4 knockdown cells required 1 μg/ml of EV to be inhibited, representing a 25-fold difference in sensitivity (Figure 3B). Clonogenic assays confirmed the observation that loss of NECTIN4 conferred resistance to EV (Figure 3C-D). Loss of NECTIN4 also led to EV resistance in a second luminal bladder cancer cell line, HT-1197 (Supplementary Figure 8D-8E).
To investigate whether NECTIN4 expression alone mediated EV-induced cell death, we constitutively expressed NECTIN4 in the basal UMUC-3 cell line. Surface protein staining and qPCR demonstrated that NECTIN4 was robustly expressed (Figure 4A-4B). In addition, NECTIN4 protein was confirmed to be primarily localized to the cell surface of UMUC3-NECTIN4 overexpressing (OE) tumor xenografts by immunohistochemistry; no detectable endogenous NECTIN4 protein was found in the basal UMUC3 control tumor xenografts (Supplementary Figure 9). Importantly, upregulating NECTIN4 expression in UMUC-3 cells resulted in markedly increased sensitivity to EV as evidenced by proliferation (Figure 4C) and clonogenic assays (Figure 4D-4E). We found similar results in a second basal bladder cancer line, TCCSUP (Supplementary Figure 10). Taken together, our results demonstrate that NECTIN4 expression is required for EV-induced cell death and underscore the findings that i) downregulation or loss of NECTIN4 may be an important mechanism of resistance and that ii) inducing NECTIN4 expression increases EV sensitivity.
FIGURE 4:
NECTIN4 overexpression in UMUC-3 cells increases sensitivity to EV. (A) NECTIN4 surface protein expression in UMUC-3 control and NECTIN4 overexpressing (OE) cells. (B) NECTIN4 RNA expression in UMUC-3 Control and UMUC-3 NECTIN4 OE cells by qPCR. **p<0.001 (C) EV dose-response WST assay in UMUC-3 control and NECTIN4 OE cells, normalized to mean survival of untreated cells. (D) Clonogenic assay quantification of EV sensitivity in UMUC-3 Control and UMUC-3 NECTIN4 OE cells. **p<0.01 (E) Representative images of the clonogenic assay in UMUC-3 Control and NECTIN4 OE cells treated with EV.
DISCUSSION:
Expanding treatment options for advanced urothelial carcinoma remains a critical unmet need, as shown by the rapid incorporation of EV into the third-line metastatic setting. The neoadjuvant space is primed for additional therapies as well—60% of patients treated with existing cisplatin-based regimens are found to have residual invasive disease at radical cystectomy, [25] with less apparent benefit in luminal-type tumors. [26] In this study, we found significant heterogeneity in NECTIN4 expression across the molecular subtypes, with highest expression in luminal cancers. NECTIN4 was also strong correlated with GATA3, FOXA1, and PPARG, which are transcription factors known to determine luminal cell fate. Notably, NECTIN4 was lowest in the NE-like subtype. We also found that NECTIN4 expression remains highest in a post-NAC CC2 luminal molecular subtype. Taken together, this variability may reflect differences in underlying tumor biology and have important implications for the susceptibility of luminal tumors to EV-based neoadjuvant or adjuvant regimens. Interestingly, >90% of upper tract urothelial tumors are also luminal, [27] which may express higher levels of NECTIN4.
Here, we found that NECTIN4 expression is critical for EV-induced cell death. Using representative luminal (HT-1376, HT-1197) and basal (UMUC-3, TCCSUP) cell lines, we demonstrate that knockdown of NECTIN4 in luminal cells leads to EV resistance, while expression of NECTIN4 in basal cells leads to EV sensitivity. To the best of our knowledge, these data are the first to formally demonstrate that expression of NECTIN4 is a key determinant of EV treatment efficacy in pre-clinical cell line models of urothelial cancer. Our data confirm and build upon existing pre-clinical in vitro and in vivo data [1]. Interestingly, we found that some basal cell lines expressed NECTIN4 mRNA and protein, including the 647V and UMUC-14 cells. Whereas 647V cells were relatively sensitive to EV, UMUC-14 cells were highly resistant, suggesting that there are also likely additional factors that mediate EV sensitivity (i.e., sensitivity to the MMAE payload).
Our study has several important limitations. Firstly, our gene expression data in >1,900 clinical samples may not necessarily reflect NECTIN4 surface protein expression. Nonetheless, our surface protein expression data on several available cell lines (Figure 2C and Supplementary Figures 7 and 9) confirm that mRNA and protein expression levels correlate strongly, which is in agreement with prior studies demonstrating correlation between mRNA and protein levels [28-30]. In addition, immunohistochemistry staining for NECTIN4 protein using a bladder cancer TMA demonstrated higher NECTIN4 H-scores in luminal subtypes, in accordance with the transcriptomic expression data (Supplementary Figure 3). Secondly, the samples analyzed in these cohorts were obtained from primary tumor tissue (due to data availability), and may not necessarily represent expression levels in metastatic tumors. Additional study of biopsies from metastatic sites is needed to better characterize the similarities and differences in NECTIN4 expression between primary tumors and metastases. In addition, there may be intrapatient heterogeneity of expression amongst different metastatic lesions that is underappreciated when profiling single biopsies. While our data demonstrate a correlation between NECTIN4 expression and response to EV in experimental systems, future studies should aim to correlate our findings with clinical patient responses to EV in both the localized and metastatic settings and to identify the threshold of expression required for response. Lastly, the inherent limitations of two-dimensional cell culture must be acknowledged as these conditions do not recapitulate in vivo tumor microenvironments and cell-cell interactions. Future study in three-dimensional cell culture and patient-derived organoid models should be pursued.
Multiple resistance mechanisms to ADC treatment likely exist, including downregulation of the target surface protein expression (i.e., “antigen escape”). For example, decreased HER2 expression has been shown to be a potential resistance mechanism to HER2-targeted ADCs such as trastuzumab emtansine for metastatic breast cancer.[31] Loss of CD19 in patients with B cell malignancies treated with CD19-targeting ADCs and chimeric antigen receptor (CAR)-T cells has been described.[32] Given the apparent heterogeneity of NECTIN4 expression, the threshold of expression required to elicit cell death will also be important to elucidate. Other resistance mechanisms likely co-exist, including decreased receptor internalization, increased drug efflux pumps, changes in endosomal trafficking, or decreased sensitivity to the payload.[33] A recent CRISPR screen identified a number of endosomal proteins, including C18ORF8/RMC1, required for ADC efficacy.[34] Further investigation of pre- and post-EV treated tumors is warranted to identify additional resistance mechanisms. Whether patients with basal and other non-luminal subtypes of urothelial carcinoma are more resistant to EV remains to be determined. Furthermore, in the neoadjuvant setting, strategies that target luminal subtypes are urgently needed, as these tumors derive less benefit from platinum-based chemotherapy.[11] Future EV clinical trials should consider incorporating molecular subtypes to predict treatment response, as well as explore the efficacy of EV in earlier disease states in the neoadjuvant, adjuvant, and intravesical treatment settings.
CONCLUSION:
Our study demonstrates that NECTIN4 mRNA expression is highest in the luminal subtypes of bladder cancer (LumNS, LumP, and LumU) and remain persistently elevated in post-neoadjuvant cisplatin-treated luminal tumors. NECTIN4 expression correlates with expression of several luminal transcription factors, including GATA3, FOXA1 and PPARG. In cell line models of bladder cancer, NECTIN4 expression levels correlate with EV-sensitivity. Moreover, NECTIN4 is required for EV-induced cell death in human luminal bladder cancer cell lines, and EV-sensitivity can be induced by expressing NECTIN4 in basal bladder cancer cell lines. Clinical trials evaluating the efficacy of EV should strongly consider molecular subtyping for stratifying patients prior to treatment.
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE:
Enfortumab vedotin (EV), an antibody-drug conjugate targeting NECTIN4, was recently approved for treatment-refractory metastatic urothelial cancer. NECTIN4 is a cell surface protein implicated in cell-cell adhesion and thought to be ubiquitously expressed in primary bladder tumors. In early clinical trials, however, EV response rates have been variable. We sought to determine whether NECTIN4 expression could be heterogeneous across molecular subtypes of bladder cancer. Using gene expression data from seven clinical cohorts with >1,900 patient samples, we found that NECTIN4 expression is highest in the luminal molecular subtypes of bladder cancer when compared to basal, neuroendocrine, or stroma-rich subtypes. NECTIN4 expression was both necessary and sufficient for EV sensitivity in luminal and basal cell line models of urothelial cancer, and downregulation of NECTIN4 induced resistance to EV. Our findings support the use of molecular subtyping to predict EV response and may have implications for patient selection and biomarker development in clinical trials.
ACKNOWLEDGEMENTS:
CC was supported by a CSTI (Clinical & Translational Science Institute) TL-1 grant.
MS was supported by the Swedish Research Council (Vetenskapsrådet) with grant number 2018-00382 and the Swedish Society of Medicine (Svenska Läkaresällskapet).
JC was supported by the A.P. Giannini Foundation Postdoctoral Fellowship.
FYF was supported by the Benioff-Goldberg Fund for Translational Research and NIH R01 CA235741.
Footnotes
DISCLOSURES:
FYF has consulted for Astellas, Bayer, BlueEarth Diagnostics, Celgene, Clovis, EMD Serono, Genentech, Janssen, Myovant, Ryovant and Sanofi, is a co-founder of PFS Genomics, and serves on the scientific advisory board of SerImmune.
EAG is an employee of Decipher Biosciences, Inc.
SPP has consulted for ProTara, Merck and Photocure.
PCB has consulted for AbbVie, Astellas Pharma, Janssen Oncology, Amgen, Bayer, Merck, Sanofi Canada, Biosyent, Ferring, Roche Canada, MDxHealth, AstraZeneca, Urogen Pharma, Asieris, Bristol-Myers Squibb and has received research funding from Genome Dx, and Sitka.
YL has consulted for Cepheid, Pacific Edge, Photocure, AstraZeneca, Merck, Fergene, Ferring Research and has received research funding from Abbott, MDxHealth, Cepheid, Pacific Edge Bladder cancer, Genome Dx, Storz, FKD.
TWF has consulted for Seattle Genetics, EMD Serono, Astra Zeneca, and Merck and has received research funding from Seattle Genetics and Roche/Genentech.
REFERENCES:
- [1].Challita-Eid PM, Satpayev D, Yang P, An Z, Morrison K, Shostak Y, et al. Enfortumab Vedotin Antibody-Drug Conjugate Targeting Nectin-4 Is a Highly Potent Therapeutic Agent in Multiple Preclinical Cancer Models. Cancer Res 2016;76:3003–13. 10.1158/0008-5472.CAN-15-1313. [DOI] [PubMed] [Google Scholar]
- [2].Rosenberg JE, O’Donnell PH, Baalr A V, McGregor BA, Heath EI, Yu EY, et al. Pivotal Trial of Enfortumab Vedotin in Urothelial Carcinoma After Platinum and Anti-Programmed Death 1/Programmed Death Ligand 1 Therapy. J Clin Oncol 2019;37:2592–600. 10.1200/JCO.19.01140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Heath EI, Rosenberg JE. The biology and rationale of targeting nectin-4 in urothelial carcinoma. Nat Rev Urol 2020:1–11. 10.1038/s41585-020-00394-5. [DOI] [PubMed] [Google Scholar]
- [4].Hoffman-Censits JH, Choi W, Lombardo K, Hahn NM, McConkey DJ, McGuire B, et al. Expression of nectin-4 in bladder cancer with variant histology. J Clin Oncol 2020;38:546–546. 10.1200/JCO.2020.38.6_suppl.546. [DOI] [Google Scholar]
- [5].Choi W, Porten S, Kim S, Willis D, Plimack ER, Hoffman-Censits J, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 2014;25:152–65. 10.1016/j.ccr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2017;171:540–556.e25. 10.1016/j.cell.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kamoun A, de Reyniès A, Allory Y, Sjödahl G, Robertson AG, Seiler R, et al. A Consensus Molecular Classification of Muscle-invasive Bladder Cancer. Eur Urol 2020;77:420–33. 10.1016/j.eururo.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sjödahl G, Lauss M, Lövgren K, Chebil G, Gudjonsson S, Veerla S, et al. A molecular taxonomy for urothelial carcinoma. Clin Cancer Res 2012;18:3377–86. 10.1158/1078-0432.CCR-12-0077-T. [DOI] [PubMed] [Google Scholar]
- [9].Sjödahl G, Eriksson P, Liedberg F, Höglund M. Molecular classification of urothelial carcinoma: global mRNA classification versus tumour-cell phenotype classification. J Pathol 2017;242:113–25. 10.1002/path.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2017;171:540–556.e25. 10.1016/j.cell.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Seiler R, Ashab HAD, Erho N, van Rhijn BWG, Winters B, Douglas J, et al. Impact of Molecular Subtypes in Muscle-invasive Bladder Cancer on Predicting Response and Survival after Neoadjuvant Chemotherapy. Eur Urol 2017;72:544–54. 10.1016/j.eururo.2017.03.030. [DOI] [PubMed] [Google Scholar]
- [12].Lotan Y, Boorjian SA, Zhang J, Bivalacqua TJ, Porten SP, Wheeler T, et al. Molecular Subtyping of Clinically Localized Urothelial Carcinoma Reveals Lower Rates of Pathological Upstaging at Radical Cystectomy Among Luminal Tumors. Eur Urol 2019;76:200–6. 10.1016/J.EURURO.2019.04.036. [DOI] [PubMed] [Google Scholar]
- [13].Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Seiler R, Gibb EA, Wang NQ, Oo HZ, Lam H-M, van Kessel KE, et al. Divergent Biological Response to Neoadjuvant Chemotherapy in Muscle-invasive Bladder Cancer. Clin Cancer Res 2019;25:5082–93. 10.1158/1078-0432.CCR-18-1106. [DOI] [PubMed] [Google Scholar]
- [16].R Core Team. R: A language and environment for statistical computing. 2019.
- [17].R Studio Team. RStudio: Integrated Development for R 2019.
- [18].Wickham H. ggplot2: Elegant Graphics for Data Analysis 2016.
- [19].Therneau T. A Package for Survival Analysis in S 2015.
- [20].Therneau Terry M. and Grambsch Patricia M.. Modeling Survival Data: Extending the Cox Model. vol. 20. New York: : Springer-Verlag; 2001. [Google Scholar]
- [21].Harrell FEJ. rms: Regression Modeling Strategies 2019.
- [22].Warrick JI, Walter V, Yamashita H, Chung E, Shuman L, Amponsa VO, et al. FOXA1, GATA3 and PPARɣ Cooperate to Drive Luminal Subtype in Bladder Cancer: A Molecular Analysis of Established Human Cell Lines. Sci Rep 2016;6:38531. 10.1038/srep38531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Earl J, Rico D, Carrillo-de-Santa-Pau E, Rodríguez-Santiago B, Méndez-Pertuz M, Auer H, et al. The UBC-40 Urothelial Bladder Cancer cell line index: a genomic resource for functional studies. BMC Genomics 2015;16:403. 10.1186/s12864-015-1450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, et al. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013;152:1173–83. 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zargar H, Espiritu PN, Fairey AS, Mertens LS, Dinney CP, Mir MC, et al. Multicenter assessment of neoadjuvant chemotherapy for muscle-invasive bladder cancer. Eur Urol 2015;67:241–9. 10.1016/j.eururo.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Seiler R, Ashab HAD, Erho N, van Rhijn BWG, Winters B, Douglas J, et al. Impact of Molecular Subtypes in Muscle-invasive Bladder Cancer on Predicting Response and Survival after Neoadjuvant Chemotherapy. Eur Urol 2017;72:544–54. 10.1016/j.eururo.2017.03.030. [DOI] [PubMed] [Google Scholar]
- [27].Kim K, Hu W, Audenet F, Almassi N, Hanrahan AJ, Murray K, et al. Modeling biological and genetic diversity in upper tract urothelial carcinoma with patient derived xenografts. Nat Commun 2020;11:1975. 10.1038/s41467-020-15885-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Edfors F, Danielsson F, Hallström BM, Käll L, Lundberg E, Pontén F, et al. Gene-specific correlation of RNA and protein levels in human cells and tissues. Mol Syst Biol 2016;12. 10.15252/MSB.20167144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature 2011;473:337–42. 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- [30].Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet n.d.;13:227. 10.1038/NRG3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Li BT, Michelini F, Misale S, Cocco E, Baldino L, Cai Y, et al. HER2-Mediated Internalization of Cytotoxic Agents in ERBB2 Amplified or Mutant Lung Cancers. Cancer Discov 2020. 10.1158/2159-8290.CD-20-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mejstríková E, Hrusak O, Borowitz MJ, Whitlock JA, Brethon B, Trippett TM, et al. CD19-negative relapse of pediatric B-cell precursor acute lymphoblastic leukemia following blinatumomab treatment. Blood Cancer J 2017;7:659. 10.1038/s41408-017-0023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tumey LN. An Overview of the Current ADC Discovery Landscape. Methods Mol Biol 2020;2078:1–22. 10.1007/978-1-4939-9929-3_1. [DOI] [PubMed] [Google Scholar]
- [34].Tsui CK, Barfield RM, Fischer CR, Morgens DW, Li A, Smith BAH, et al. CRISPR-Cas9 screens identify regulators of antibody–drug conjugate toxicity. Nat Chem Biol 2019;15:949–58. 10.1038/s41589-019-0342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.