Abstract
Naive CD4+ T cells become memory cells after proliferating in response to their cognate major histocompatibility complex class II (MHCII)-bound peptide and passing through an effector cell stage. The process by which CD4+ memory T cells emerge from the effector cell pool, however, is less well understood than in the case of CD8+ T cells. During certain acute infections, naive CD4+ T cells proliferate and differentiate into various forms of type 1 (Th1) and follicular helper (Tfh) effector cells. We review the evidence that about 10% of the cells in each of these subsets survive to become memory cells that resemble their effector cell precursors. The roles that asymmetric cell division, the TCF-1 transcription factor, metabolic activity, reactive oxygen species, and the IL-7 receptor play in the effector to memory cell transition are discussed. We propose a speculative model in which the metabolic activity needed for rapid clonal expansion also generates toxic products that induce apoptosis in most effector cells. Memory cells then arise from the effector cells in each subset that are at the low end of the metabolic activity spectrum.
Memory T cells are derived from naive T cells that proliferate in response to foreign antigens. Each nascent CD4+ T cell produces a unique α-β T-cell receptor (TCR) in the thymus via somatic recombination of TCR gene segments (Rothenberg 2019). The ligands for TCRs on CD4+ T cells are peptides bound to host major histocompatibility complex class II (MHCII) molecules (Rudolph et al. 2006). These ligands are generated by MHCII-expressing cells that take up extracellular proteins into endosomes or phagosomes where cathepsins cleave the internalized proteins into peptides (Neefjes et al. 2011). These vesicles also contain newly synthesized MHCII molecules that bind peptides via nine amino acid core sequences with certain key anchor residues. The peptide:MHCII complexes are then trafficked to the cell surface.
CD4+ T-cell development depends on TCR recognition of MHCII-bound host peptides displayed in the thymus (Klein et al. 2014). TCR V β chains have a germ line–encoded affinity for MHC molecules (Marrack et al. 2008). Most thymocytes that have TCRs with high affinity for MHCII-bound host peptides that are abundantly displayed on thymic epithelial cells and dendritic cells are deleted. The remaining cells must then pass through an MHC-dependent positive selection step. Cells with TCRs that bind weakly to MHCII-bound host peptides on thymic epithelial cells receive positive selection signals and leave the thymus as naive cells to circulate through the secondary lymphoid organs. This process creates a large and diverse set of self-tolerant naive CD4+ T cells that is likely to contain a few cells that by chance have TCRs with high affinity for any MHCII-bound “foreign” peptides that are not present in the host's peptidome, for example, peptides from microbes, plants, parasites, food, or tumor neoantigens (Jenkins et al. 2010). On average, about one in 105–106 CD4+ T cells expresses a TCR capable of strong binding to any given MHCII-bound foreign peptide (Jenkins and Moon 2012). This amounts to about 100 cells in a mouse or 100,000 in a human, each cell expressing a different TCR (Moon et al. 2011).
Memory cells form from the effector cell progeny of naive T cells (Jameson and Masopust 2018). Naive CD4+ T cells that express TCRs that allow strong binding to an MHCII-bound peptide in the context of infection or immune adjuvants proliferate rapidly in the secondary lymphoid organs for about a week, increasing about 1000-fold to produce a large population of lymphoblasts known as effector cells (Pepper and Jenkins 2011). The effector cells stop dividing as their numbers peak and about 90% die over the next several weeks (Homann et al. 2001; Pepper et al. 2010; Marshall et al. 2011) during what is known as the contraction phase (Badovinac et al. 2004). The cells that survive the contraction phase are considered to be memory cells, which are quiescent cells that retain the CD44hi phenotype of antigen-experienced cells and persist for months but decline slowly with a half-life of 0.05 lifetimes (Homann et al. 2001; Pepper et al. 2010, 2011). The memory CD4+ T-cell population is maintained by slow IL-15-driven homeostatic proliferation (Purton et al. 2007; Pepper et al. 2010), but evidently not at a high enough rate to achieve the numerical stability typical of CD8+ memory T-cell populations (Murali-Krishna et al. 1999).
CD4+ memory T cells that form during acute viral and bacterial infections will be the focus of this review because these infections are cleared from the body and thus have to be “remembered” by the CD4+ T-cell repertoire if the host is to mount an enhanced response to another occurrence of the infection. Although CD4+ T cells are critical for controlling certain intracellular chronic infections of macrophages such as those caused by Mycobacterium tuberculosis (Tubo and Jenkins 2014), the concept of immune memory is more difficult to apply to these situations because the TCR stimulus persists. We will also focus on in vivo studies of polyclonal memory cells generated in diverse T-cell repertoires since this is the natural situation in normal individuals.
EFFECTOR CELL HETEROGENEITY IN THE PRIMARY RESPONSE
There has been great interest in the immune memory field in assessing phenotypic heterogeneity in CD4+ effector T-cell populations since it is believed that effector cells are the precursors of memory cells. Much of the current understanding of CD4+ effector T-cell heterogeneity is based on studies of mice infected with lymphocytic choriomeningitis virus (LCMV) or a vaccine strain of Listeria monocytogenes (Lm) bacteria, both of which are cleared from the host in about a week (Ahmed et al. 1984; Portnoy et al. 2002). Many of these studies have assessed heterogeneity using the T follicular helper (Tfh) cell markers CXCR5 and PD-1 or BCL6 (Johnston et al. 2009; Marshall et al. 2011; Pepper et al. 2011; Hale et al. 2013; Tubo et al. 2013; Marriott et al. 2014; Künzli et al. 2020). The peptide:MHCII-specific effector cell populations induced by LCMV or Lm infection consist of roughly equal fractions of three subsets with CXCR5– PD-1– BCL6–, CXCR5int PD-1– BCL6lo, and CXCR5hi PD-1+ BCL6hi phenotypes. The CXCR5– PD-1– BCL6– cells are Th1 cells based on uniform expression of large amounts of the Th1-defining transcriptional activator TBET (Szabo et al. 2000) and the capacity to produce interferon γ (IFN-γ) (Pepper et al. 2011). The main function of these cells is to produce IFN-γ and other Th1 cytokines to enhance the microbicidal activities of phagocytes (Tubo and Jenkins 2014). The CXCR5hi PD-1+ BCL6hi cells are germinal center Tfh (GC-Tfh) cells (Johnston et al. 2009; Nurieva et al. 2009; Yu et al. 2009; Eto et al. 2011; Pepper et al. 2011; Liu et al. 2012) based on uniform expression of the Tfh-defining transcriptional repressor BCL6 and residence in GCs (Crotty 2011; Shulman et al. 2013). GC-Tfh cells help GC B cells undergo affinity maturation thereby improving the quality of the humoral immune response. The CXCR5int PD-1–BCL6lo subset expresses CCR7 (at least in Lm infection) and resides in the T-cell zones (Pepper et al. 2011) or mantle regions of the follicles (Shulman et al. 2013). This population contains precursors of GC-Tfh cells (Kitano et al. 2011; Liu et al. 2012; Choi et al. 2013; Takebe et al. 2018).
Ly6C and p-selectin glycoprotein ligand 1 (PSGL1), and in some cases folate receptor 4 (FR4), have also been used to study the heterogeneity of CD4+ effector T-cell populations induced by LCMV infection (Marshall et al. 2011; Iyer et al. 2013; Künzli et al. 2020). This gating scheme identifies three effector cell populations with Ly6C+ PSGL1+, Ly6C– PSGL1+, and Ly6C– PSGL1– phenotypes. The Ly6C– PSGL1– population consists mainly of CXCR5hi PD-1+ BCL6hi GC-Tfh cells, which are the only cells that express large amounts of FR4 (Künzli et al. 2020). Th1 cells comprise the Ly6C+ PSGL1+ population (Marshall et al. 2011; Hale et al. 2013). The Ly6C– PSGL1+ effector cell population contains a mixture of CXCR5int and CXCR5– cells (Künzli et al. 2020), which are probably Ly6C– Th1 cells since all peptide:MHCII-specific CXCR5– cells in LCMV-infected mice have the TBEThi phenotype (Hale et al. 2013). Viral peptide:MHCII-specific Ly6C+ Th1 cells produce more IFN-γ than Ly6C– Th1 and may be more terminally differentiated (Hu et al. 2015). In summary, the two gating schemes identify four prominent effector cell subsets common to LCMV and Lm infections: CXCR5hi GC-Tfh cells, CXCR5int GC-Tfh precursors, Ly6C+ Th1 cells, and Ly6C– Th1 cells (Fig. 1).
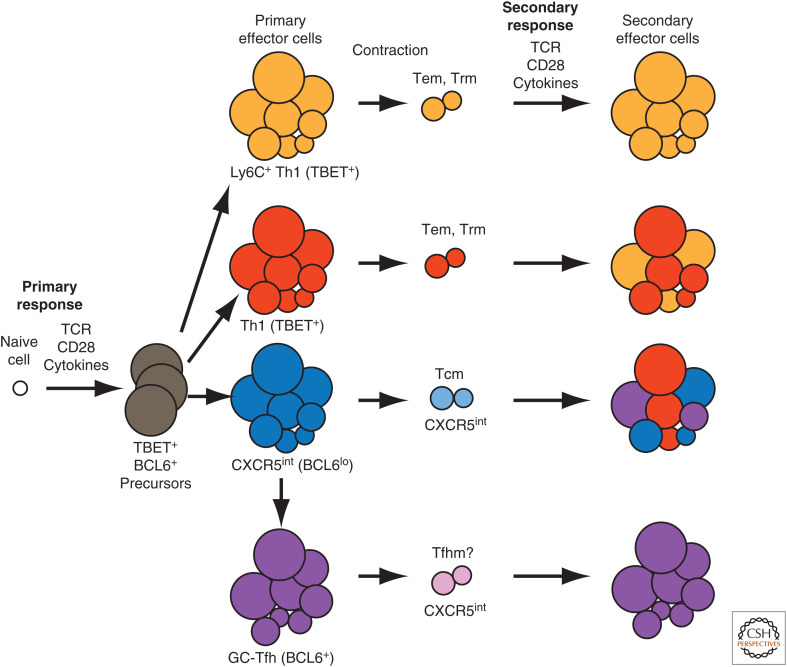
Figure 1.
Model of CD4+ memory T-cell formation. The diagram shows the primary response of a single naive CD4+ T cell exposed to the relevant peptide:MHCII ligand on a CD28 ligand-expressing antigen-presenting cell in an inflammatory cytokine-rich environment produced by an acute infection that is cleared. The cell proliferates to produce TBET+ BCL6+ precursors that differentiate into Ly6C+ Th1, Ly6C– Th1, germinal center (GC)-Tfh, or CXCR5int early effector cells. The cells in each subpopulation have a range of cell sizes and metabolic activities driven by differences in mTOR and c-Myc production. The largest cells, which suffer the most mitochondrial damage during the expansion phase, then preferentially die in each population during the contraction phase. Ly6C+ and Ly6C– Th1 effector cells generate Ly6C+ and Ly6C– Th1 effector (Tem) and resident memory (Trm) cells, CXCR5int effector cells generate CXCR5int memory cells, which may express CCR7 and thus resemble central memory (Tcm) cells, and GC-Tfh effector cells generate Tfh memory (Tfhm) cells. During secondary responses, Ly6C+ Th1 memory cells produce only Ly6C+ effector cells, less differentiated Ly6C– Th1 memory cells generate a mixture of Ly6C+ and Ly6C– Th1 effector cells, CXCR5int Tcm-like memory cells produce Th1, GC-Tfh, and CXCR5int effector cells, and Tfhm cells produce GC-Tfh effector cells. (TCR) T-cell receptor.
MECHANISMS THAT PRODUCE CD4+ EFFECTOR T-CELL HETEROGENEITY
Th1 cell formation has been studied extensively by culturing naive T cells with mitogenic CD3 and CD28 antibodies, IL-2, IL-12, and IL-4 antibodies. TCR, IFN-γ receptor, and STAT1 signaling in the absence of IL-4 causes the T cells to proliferate and undergo STAT4-independent expression of TBET (Mullen et al. 2001; Afkarian et al. 2002; Schulz et al. 2009). As TCR signaling wanes (Schulz et al. 2009), IL-12 receptor (IL-12R) is induced and IL-12R signaling causes STAT4-dependent proliferation of the proto-Th1 cells (Mullen et al. 2001) and enhances their TBET expression and IFN-γ production capacity and induces epigenetic modifications that fully commit the cells to the Th1 fate (Mullen et al. 2001; Afkarian et al. 2002; Schulz et al. 2009; O'Shea and Paul 2010). STAT5 activation downstream of IL-2R signaling (Johnston et al. 2009, 2012) or the KLF2 transcription factor (Lee et al. 2015) also favor Th1 formation by inducing BLIMP1, an inhibitor of BCL6 expression and Tfh formation.
Formation of Tfh cells is promoted by signaling through IL-6R and IL-21R, inducible costimulatory receptor (ICOS) (Choi et al. 2011; Eto et al. 2011; Pepper et al. 2011; Xu et al. 2013), and the CD4+ T-cell-specific transcription factor Thpok (Vacchio et al. 2019), all of which turn on the transcriptional repressor BCL6. The LEF1 and TCF-1 transcription factors coordinate this process by sustaining expression of IL-6R and enhancing expression of ICOS (Choi et al. 2015; Wu et al. 2015; Gullicksrud et al. 2017). BCL6 induces Tfh formation by acting as a transcriptional repressor in two ways. First, it shuts off paths to other lineages. For example, BCL6 represses TBET and RORγt that promote the Th1 and Th17 options (Crotty 2014), thereby opening the door to the Tfh fate. Importantly, however, BCL6 also represses repressors of the genes that positively regulate Tfh formation (Choi et al. 2020). For example, BCL6 represses Id2, a repressor of Cxcr5, and other transcription factors that repress other important Tfh genes, including those encoding PD-1, IL-6R, and ICOS. CXCR5int cells, which express less BCL6 than GC-Tfh cells (Pepper et al. 2011; Hale et al. 2013), are probably cells in which BCL6 has repressed some but not all of the Tfh repressors, thereby allowing intermediate expression of CXCR5 but not the full set of positive regulators of molecules needed for GC-Tfh cell development. CXCR5int cells are thus poised to become GC-Tfh cells if signaled to express the larger amounts of BCL6 needed to repress all the key Tfh repressors.
This model is supported by the observation that CXCR5int and GC-Tfh cells disappear in mice lacking both copies of the Bcl6 gene (Pepper et al. 2011; Liu et al. 2012), indicating that both populations are in the Tfh lineage. CXCR5int effector cells, however, appear in normal numbers in mice lacking one copy of the Bcl6 gene (Pepper et al. 2011) while GC-Tfh cells are greatly reduced in these instances. The fact that CXCR5int cells can form under conditions of suboptimal BCL6 induction is consistent with partial repression of Tfh repressors and initiation but not completion of the full GC-Tfh program.
MECHANISMS OF EFFECTOR CELL SPECIFICATION
It is perplexing that Th1 and Tfh effector cells can form simultaneously in the same primary immune response given that they depend on different signals. Experiments based on single-cell methods (Tubo et al. 2013; Becattini et al. 2015) indicate that most single naive T cells generate mixtures of Th subsets, ruling out the possibility that certain subsets are derived from different specialized naive cells. The four effector cell types that are present at the peak of clonal expansion are the progeny of an even earlier homogenous effector cell population that expresses TBET and BCL6 (Nakayamada et al. 2011; Oestreich et al. 2011). Further differentiation of cells from this population could relate to proximity to cytokines. Early TBET+ BCL6+ effector cells that happen to reside near sources of IL-12 and/or IL-2 might become Th1 cells, while cells in places containing abundant IL-6, IL-21, and B cells might become Tfh cells. TCR signaling is another possible heterogeneity generator. The IRF4 transcription factor and the IL-2R are induced in proportion to TCR signal strength (Nayar et al. 2014; Krishnamoorthy et al. 2017; Kotov et al. 2019). High levels of IRF4 and IL-2R signaling favor induction of BLIMP1 over BCL6 (Krishnamoorthy et al. 2017), and evidence in the literature supports the idea that strong TCR, IRF4, and IL-2R signaling favors Th1 formation (Kim et al. 2013; Tubo et al. 2013; Krishnamoorthy et al. 2017; Snook et al. 2018), although it has been reported that IRF4 favors CXCR5+ cells in humans (Schmitt et al. 2016). Thus, naive murine T cells that have TCRs with high affinity for MHCII-bound peptides or happen to engage with antigen-presenting cells displaying large amounts of these ligands may induce large amounts of IRF4 and BLIMP1, sustain IL-2R expression, and become Th1 effector cells. Other naive T cells that have TCRs with lower affinity for the MHCII-bound peptide or happen to engage with dendritic cells displaying lower amounts of this ligand may not sustain the IL-2R or BLIMP1, express BCL6, and generate GC-Tfh effector cells.
It should be noted, however, that not all of the evidence supports the idea that strong TCR signaling favors Th1 formation. McHeyzer-Williams and colleagues found that the effector cells induced by immunization with a protein and adjuvant bound more peptide:MHCII tetramer, a correlate of high TCR affinity, than other effector cells (Fazilleau et al. 2009). Although we found that increasing the antigen dose, and presumably TCR signaling, increased Th1 formation up to a point, the largest antigen doses actually favored GC-Tfh formation (Tubo et al. 2013). Similarly, Weaver and colleagues found that cells receiving the strongest TCR signals produced IL-2 but did not respond to it and became CXCR5+ cells (DiToro et al. 2018). King and colleagues provided some clarity to this confusing issue by showing that TCR affinity and antigen dose, which one might think would influence TCR signaling in the same direction, actually have separate and somewhat opposite effects. They found that although increasing TCR affinity increases Th1 formation, increasing antigen dose increases GC-Tfh formation (Keck et al. 2014). Another complexity is that the amount of tonic signaling that a naive T cell receives in the absence of its cognate MHCII-bound foreign peptide can vary depending on the affinity of its TCR for the relevant MHCII-bound host peptide. This variation in tonic signaling can influence the probability that the naive CD4+ T cell will produce Tfh or non-Tfh progeny (Bartleson et al. 2020). In summary, although the literature suggests that TCR signal strength influences the bifurcation of GC-Tfh and non-Tfh cells, the complex interaction of TCR affinity of MHCII-bound self and foreign peptides and antigen dose have made it difficult to resolve the direction of this effect.
MEMORY CELL FORMATION AND HETEROGENEITY
The CD4+ memory T-cell populations that emerge from LCMV- or Lm-induced effector cell populations contract have also been studied extensively. These memory populations contain CXCR5– TBEThi and CXCR5int TBETlo CCR7+ cells as in the effector phase but lack CXCR5hi PD-1+ BCL6hi cells (Marshall et al. 2011; Pepper et al. 2011; Hale et al. 2013; Ciucci et al. 2019; Künzli et al. 2020). In addition, the Ly6C+ PSGL1+, Ly6C– PSGL1+, and Ly6C– PSGL1– subsets that are found in the effector phase are also found in the memory phase (Marshall et al. 2011; Hale et al. 2013; Künzli et al. 2020). CXCR5– TBEThi memory cells are located in the secondary lymphoid organs (Pepper et al. 2011) where they are generated but also migrate into nonlymphoid organs (Tubo et al. 2013) where a CD69+ fraction permanently resides (Romagnoli et al. 2017; Beura et al. 2019; Malhotra et al. 2020). Thus, the CXCR5– TBEThi population that survives the contraction phase consists of Th1 memory cells, some expressing Ly6C (Marshall et al. 2011; Hale et al. 2013; Künzli et al. 2020), some of the effector memory type (Sallusto et al. 1999) recirculating between lymphoid and nonlymphoid organs via the blood, and some of the resident memory type (Schenkel and Masopust 2014) locked mainly in nonlymphoid organs. CXCR5+ memory cells are located primarily in the secondary lymphoid organs (Fazilleau et al. 2009; Takebe et al. 2018).
The evidence suggests that each of the effector cell populations contributes to the CD4+ memory T-cell pool. Fate-mapping experiments with TCR transgenic T cells showed that IFN-γ-producing effector cells produce IFN-γ-producing memory cells (Harrington et al. 2008; Lohning et al. 2008). Therefore, the polyclonal CXCR5– TBEThi effector T cells induced by LCMV and Lm infection are likely the precursors of the CXCR5– TBEThi memory T cells in the postcontraction population.
We have also shown that CXCR5int PD-1– effector cells and CXCR5hi PD-1+ GC-Tfh cells both yield CXCR5int PD-1– memory cells (Tubo et al. 2016) after transfer into naive mice indicating that these two effector cell populations both contribute to the CXCR5int memory cell population, although GC-Tfh-derived memory cells reduce expression of CXCR5 and PD-1. This contention is consistent with the finding that the Ly6C– PSGL1– effector cell population consists of CXCR5hi PD-1+ BCL6hi GC-Tfh cells, while Ly6C– PSGL1– memory cells are CXCR5int PD-1– BCL6lo (Marshall et al. 2011). It should be noted, however, that most studies may have underestimated CXCR5hi PD-1+ BCL6hi memory cells because this population is preferentially susceptible to P2RX7-dependent death during tissue processing before flow cytometry analysis (Künzli et al. 2020). Thus, some murine CXCR5hi PD-1+ BCL6hi GC-Tfh memory cells can be found if P2RX7 is blocked (Künzli et al. 2020). This observation fits with the finding that human memory cell populations contain a BCL6hi CCR7lo PD-1hi subset resembling murine GC-Tfh cells and BCL6lo CCR7hi PD-1lo subset resembling the CXCR5int memory cells of mice (He et al. 2013).
Another indication that CD4+ memory T-cell subsets are related to similar effector cell subsets comes from in vivo limiting dilution experiments. As mentioned above, different single naive T cells generate different proportions of Th1, CXCR5int, and GC-Tfh effector cells. We (Tubo et al. 2016) used limiting dilution of naive T cells and a hemi-splenectomy approach to study clonally derived Lm peptide:MHCII-specific T cells at effector and memory time points after infection. We found that the effector cell populations generated from about 150 different naive clones all underwent contraction but yielded memory cell populations, although as observed for CD8+ T cells (Buchholz et al. 2013), large effector cells populations tended to contract more than small ones. Remarkably, although the proportions of Th1, CXCR5int, and GC-Tfh (identified by PD-1 expression) types varied between clonally derived effector cell populations, their memory cell populations retained their subset distribution when loss of PD-1 by the memory cell progeny of GC-Tfh cells was accounted for. For example, one clonally derived effector cell population of 40% Th1 cells, 30% CXCR5int cells, and 30% GC-Tfh cells might yield a memory cell population of 40% CXCR5– Th1 cells and 60% CXCR5+ cells. The fact that the idiosyncrasies of each effector population were reflected in the descended memory population favors the idea that memory cells are simply effector cells that survive contraction while retaining many of their phenotypic characteristics.
CD4+ MEMORY T-CELL POTENTIAL DURING SECONDARY RESPONSES
There has also been great interest in the types of effector cell progeny that CD4+ memory T-cell subsets produce during subsequent antigen exposures as this issue has relevance to vaccine design. For example, prime-boost strategies aimed at generating antibodies with dozens of somatic mutations will depend on generation of GC-Tfh cells from memory cells. The issue of memory cell response potential has been addressed experimentally by transferring purified CD4+ memory T cell types from infected mice into naive recipients, immediately infecting the recipients with the initial microbe, and then assessing the phenotypes of the donor memory cell–derived effector cells.
In the Lm model, we found that purified CXCR5– TBEThi memory cells generated a secondary effector population consisting of >95% CXCR5– TBEThi cells. Ly6C expression by the transferred memory cells or their effector cell progeny was not measured in these experiments. In the LCMV model, several groups (Hale et al. 2013; Künzli et al. 2020) showed that purified Ly6C+ PSGL1+ memory populations, which contain mainly CXCR5– cells and some CXCR5int cells, generated secondary effector cell populations consisting of 80% CXCR5– Ly6C+ PSGL1+ Th1 cells and 20% CXCR5int Ly6C+ PSGL1+ cells. Similar results were obtained in the herpes virus model system (Hu et al. 2015). The results from the Lm model suggest that the CXCR5– Ly6C+ PSGL1+ Th1 memory cells in the transferred population were the major source of the CXCR5– Ly6C+ PSGL1+ Th1 effector cells. If so, then the CXCR5int effector cells must have come from the transferred CXCR5int Ly6C+ PSGL1+ memory cells, although this experiment does not rule out the possibility that these memory cells also generated some of the CXCR5– Th1 effector cells. Together, the evidence indicates that Th1 memory cells are committed to producing only Th1 effector cells during secondary responses.
We explored the effector cell generation potential of the CXCR5+ memory cells in the Lm model. These cells produced IL-2 but very little IFN-γ (Pepper et al. 2011) and generated CXCR5– Th1, CXCR5int, and GC-Tfh effector cells after transfer into naive recipients then infected with Lm (Pepper et al. 2011). This result showed that the CXCR5+ memory T-cell population is more multipotent than the CXCR5– Th1 memory population, which could only produce Th1 effector cells, and, for this reason, we referred to the CXCR5+ memory cells as central memory cells. Because the CXCR5+ memory T-cell population was later recognized to contain descendants of CXCR5int pre-GC-Tfh and CXCR5hi GC-Tfh effector cells (Hale et al. 2013; Tubo et al. 2016), it was necessary to determine the secondary effector cell-generation potential of the individual memory cell types. Künzli et al. (2020) addressed this issue by focusing on Ly6C– PSGL-1– memory cells, which in the LCMV system consist mainly of GC-Tfh-derived memory cells. They found that these memory cells generated CXCR5– Th1, CXCR5int, and GC-Tfh effector cells after transfer into naive recipients then challenged with LCMV, while Ly6C– PSGL1+ memory cells, a mixture of CXCR5int and Ly6C– Th1 cells generated CXCR5– Th1, CXCR5int, but not GC-Tfh effector cells. These results indicate that GC-Tfh-derived memory cells can produce all the effector cell types and are the only memory cells that can produce GC-Tfh effector cells. The true potential of memory cells derived from CXCR5int effector cells awaits an experiment in which these cells are studied in the absence of contributions from other memory cells.
WHY DO EFFECTOR CELLS DISAPPEAR DURING THE CONTRACTION PHASE?
The appearance of CD4+ memory T cells is always preceded by the loss of most of the effector cells during the contraction phase. This loss is not simply due to migration from the secondary lymphoid organs to nonlymphoid organs since effector cells disappear from both sites (Reinhardt et al. 2001). Rather, effector cells disappear during the contraction phase because they undergo apoptosis as evidenced by the fact that the process is blunted in mice lacking the proapoptotic factors BIM or p27(Kip1) (Wojciechowski et al. 2006; Prlic and Bevan 2008; Jatzek et al. 2012). In addition, about 10% of TCR transgenic T cells undergoing contraction express the apoptosis-inducing enzyme CASPASE3 and die at a high rate after transfer into adoptive hosts (Garrod et al. 2012). CD8+ T cells in mice with a defect in autophagy also undergo an exaggerated contraction phase, in this case associated with impaired mitochondrial function (Xu et al. 2014). This result together with the fact that BIM and CASPASE3 act on mitochondria (Harada and Grant 2003) suggest that mitochondrial damage is a mechanism of cell death during the contraction phase.
Reactive oxygen species (ROS) may produce the mitochondrial damage that triggers apoptosis during the contraction phase. Effector T cells rely on anaerobic glycolysis to produce the substrates for the macromolecular synthesis needed for cell growth and division (Buck et al. 2015). But these cells also ramp up the respiratory chain in mitochondria to generate ATP via oxidative phosphorylation and in the process produce ROS that could damage macromolecules and induce apoptosis (Hildeman et al. 1999; Belikov et al. 2015). The observations that effector cells survive poorly in mice lacking mitochondrial superoxide dismutase 2 (Case et al. 2011), an enzyme that reduces mitochondrial ROS, or have reduced intracellular glutathione (Mak et al. 2017), a molecule that buffers intracellular ROS, are consistent with this possibility. The finding that a superoxide dismutase-inhibiting chemical blocked in vitro death of T cells induced to die in vivo (Hildeman et al. 1999) also suggests that ROS could trigger the contraction phase, perhaps by damaging mitochondria.
WHY DO SOME CELLS SURVIVE THE CONTRACTION PHASE?
If metabolically active effector cells produce enough ROS to damage themselves to the point of undergoing apoptosis, why do all the effector cells not succumb to this process? There must be a brake on the contraction mechanism that allows a minority of effector cells to survive to become memory cells.
The IL-7R could be such a brake. Extensive research on CD8+ T cells has led to a model in which the surface protein KLRG1 marks effector cells that are likely to die while IL-7R marks memory precursor cells (Kaech and Wherry 2007). IL-7R, which is expressed by all naive CD4+ T cells and helps maintain their survival, is lost on early effector cells as a result of TCR signaling (Barata et al. 2019). Cells in the effector cell population then begin to reexpress the IL-7R such that about 20% of the cells are IL-7R+ at the time of peak clonal expansion (Marshall et al. 2011). IL-7R+ cells become more prevalent in the population as the contraction phase proceeds until all the cells are IL-7R+ when the contraction phase ends, and the memory phase begins. IL-7R then becomes critical for the survival of the memory cells (Seddon et al. 2003).
These results are consistent with the possibility that reexpression of the IL-7R improves the chances that an effector cell will survive the contraction phase. As attractive as this model is, it does not square with cell-sorting experiments, indicating that IL-7R+ CD4+ effector cells do not have an advantage over IL-7R– cells at forming memory cells after transfer into adoptive hosts (Marshall et al. 2011), that a TCR transgenic population undergoing contraction contains CASPASE3+ IL-7R+ cells (Garrod et al. 2012), and that forced expression of IL-7R during the expansion phase does not prevent the contraction of CD8+ effector cells (Hand et al. 2007). Given the complexities of adoptive transfer and overexpression experiments and the fact that IL-7R is clearly critical for the maintenance of established memory T cells, however, this issue should probably be revisited using mice in which the Il7ra gene can be ablated in effector cells just as IL-7R normally begins to be reexpressed.
CONCLUDING REMARKS: MODEL OF CD4+ MEMORY T-CELL GENERATION
Any model of CD4+ memory T-cell formation must describe the heterogeneity that causes most effector cells of all types to die and a minority to live on as memory cells. If effector cell contraction and memory cell formation are controlled by ROS, then what mechanism creates heterogenous expression of this factor in the effector cell pool from which memory cells emerge?
Asymmetric division is an attractive, albeit controversial, possibility. Reiner and colleagues championed the idea that a T cell that undergoes cell division while attached to an antigen-presenting cell will asymmetrically partition molecules between the proximal and distal daughter cell and that this partitioning could affect T-cell fate (Reiner and Adams 2014). Green and colleagues showed that proximal daughter cells preferentially acquire the amino acid transporter SLC1A5, which increases the activities of mTORC1 and c-Myc, master regulators of cellular metabolism (Verbist et al. 2016). In this study, proximal daughters had higher metabolic activities than distal daughters and were more likely to produce short-lived effector cells. These results lead to the conclusion that the progeny of naive T cells are destined at the first cell division to die as effector cells or live as memory cells depending on their relationship to the antigen-presenting cell. Another possibility follows, however, if the fates of the proximal and distal daughter cells are not completely sealed at the first division. In this case, unequal partitioning of c-Myc would be a heterogeneity driver, as some distal daughters from the first division could by chance be proximal daughters as they encounter new antigen-presenting cells and divide again as the expansion phase proceeds. Repetition of this process over many rounds of division would be expected to produce considerable variation in the metabolic activities of effector cells in the population. Indeed, the cells in early effector cell populations are very heterogenous in size, a feature that is proportional to c-Myc levels at least in B cells (Finkin et al. 2019). Even without invoking asymmetric cell division, it is easy to imagine that TCR, costimulatory, or cytokine receptor signals and downstream metabolic activities could vary widely between effector cells based simply on variability in the provision of these signals by the antigen-presenting cells the T cells happen to interact with.
We speculate that variability in metabolic activities and ROS levels in the effector cell population could produce the conditions for the contraction phase along the lines suggested by Buchholz et al. (Fig. 1; Buchholz et al. 2013). Effector cells that receive strong TCR (Snook et al. 2018), CD28, or cytokine signals and thus have high metabolic activity and ROS levels would damage themselves to death while cells with lower levels would survive to become memory cells. The observation that large, clonally derived effector cell populations undergo more contraction than small ones (Buchholz et al. 2013; Tubo et al. 2016) is consistent with the model.
We further propose that this process acts on all effector cell types similarly such that 10% of each subset survives to become quiescent memory cells that retain aspects of the phenotypes they had as effector cells (Fig. 1). The Th1 memory cells then recirculate through lymphoid and nonlymphoid organs via the blood or become resident in these locations. During a secondary response, Ly6C+ Th1 memory cells generate primarily Ly6C+ Th1 effector cells, while Ly6C– Th1 memory cells generate primarily Ly6C– Th1 effector cells. The memory cells derived from CXCR5int effector cells have the properties of Tcm cells including recirculation through lymphoid organs and the plasticity to produce Th1, CXCR5int, and GC-Tfh effector cells after reexposure to antigen, while the memory cell progeny of the GC-Tfh cells primarily produce GC-Tfh cells during secondary responses.
Footnotes
Editors: David Masopust and Rafi Ahmed
Additional Perspectives on T-Cell Memory available at www.cshperspectives.org
REFERENCES
- Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. 2002. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat Immunol 3: 549–557. 10.1038/ni794 [DOI] [PubMed] [Google Scholar]
- Ahmed R, Salmi A, Butler LD, Chiller JM, Oldstone MB. 1984. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J Exp Med 160: 521–540. 10.1084/jem.160.2.521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badovinac VP, Porter BB, Harty JT. 2004. CD8+ T cell contraction is controlled by early inflammation. Nat Immunol 5: 809–817. 10.1038/ni1098 [DOI] [PubMed] [Google Scholar]
- Barata JT, Durum SK, Seddon B. 2019. Flip the coin: IL-7 and IL-7R in health and disease. Nat Immunol 20: 1584–1593. 10.1038/s41590-019-0479-x [DOI] [PubMed] [Google Scholar]
- Bartleson JM, Viehmann Milam AA, Donermeyer DL, Horvath S, Xia Y, Egawa T, Allen PM. 2020. Strength of tonic T cell receptor signaling instructs T follicular helper cell-fate decisions. Nat Immunol 21: 1384–1396. 10.1038/s41590-020-0781-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becattini S, Latorre D, Mele F, Foglierini M, De Gregorio C, Cassotta A, Fernandez B, Kelderman S, Schumacher TN, Corti D, et al. 2015. T cell immunity. Functional heterogeneity of human memory CD4+ T cell clones primed by pathogens or vaccines. Science 347: 400–406. 10.1126/science.1260668 [DOI] [PubMed] [Google Scholar]
- Belikov AV, Schraven B, Simeoni L. 2015. T cells and reactive oxygen species. J Biomed Sci 22: 85. 10.1186/s12929-015-0194-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura LK, Fares-Frederickson NJ, Steinert EM, Scott MC, Thompson EA, Fraser KA, Schenkel JM, Vezys V, Masopust D. 2019. CD4+ resident memory T cells dominate immunosurveillance and orchestrate local recall responses. J Exp Med 216: 1214–1229. 10.1084/jem.20181365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz VR, Flossdorf M, Hensel I, Kretschmer L, Weissbrich B, Graf P, Verschoor A, Schiemann M, Hofer T, Busch DH. 2013. Disparate individual fates compose robust CD8+ T cell immunity. Science 340: 630–635. 10.1126/science.1235454 [DOI] [PubMed] [Google Scholar]
- Buck MD, O'Sullivan D, Pearce EL. 2015. T cell metabolism drives immunity. J Exp Med 212: 1345–1360. 10.1084/jem.20151159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case AJ, McGill JL, Tygrett LT, Shirasawa T, Spitz DR, Waldschmidt TJ, Legge KL, Domann FE. 2011. Elevated mitochondrial superoxide disrupts normal T cell development, impairing adaptive immune responses to an influenza challenge. Free Radic Biol Med 50: 448–458. 10.1016/j.freeradbiomed.2010.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Kageyama R, Eto D, Escobar TC, Johnston RJ, Monticelli L, Lao C, Crotty S. 2011. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity 34: 932–946. 10.1016/j.immuni.2011.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Yang JA, Yusuf I, Johnston RJ, Greenbaum J, Peters B, Crotty S. 2013. Bcl6 expressing follicular helper CD4 T cells are fate committed early and have the capacity to form memory. J Immunol 190: 4014–4026. 10.4049/jimmunol.1202963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Gullicksrud JA, Xing S, Zeng Z, Shan Q, Li F, Love PE, Peng W, Xue HH, Crotty S. 2015. LEF-1 and TCF-1 orchestrate TFH differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat Immunol 16: 980–990. 10.1038/ni.3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Diao H, Faliti CE, Truong J, Rossi M, Bélanger S, Yu B, Goldrath AW, Pipkin ME, Crotty S. 2020. Bcl-6 is the nexus transcription factor of T follicular helper cells via repressor-of-repressor circuits. Nat Immunol 21: 777–789. 10.1038/s41590-020-0706-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciucci T, Vacchio MS, Gao Y, Tomassoni Ardori F, Candia J, Mehta M, Zhao Y, Tran B, Pepper M, Tessarollo L, et al. 2019. The emergence and functional fitness of memory CD4+ T cells require the transcription factor Thpok. Immunity 50: 91–105.e4. 10.1016/j.immuni.2018.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. 2011. Follicular helper CD4 T cells (TFH). Annu Rev Immunol 29: 621–663. 10.1146/annurev-immunol-031210-101400 [DOI] [PubMed] [Google Scholar]
- Crotty S. 2014. T follicular helper cell differentiation, function, and roles in disease. Immunity 41: 529–542. 10.1016/j.immuni.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiToro D, Winstead CJ, Pham D, Witte S, Andargachew R, Singer JR, Wilson CG, Zindl CL, Luther RJ, Silberger DJ, et al. 2018. Differential IL-2 expression defines developmental fates of follicular versus nonfollicular helper T cells. Science 361: eaao2933. 10.1126/science.aao2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto D, Lao C, DiToro D, Barnett B, Escobar TC, Kageyama R, Yusuf I, Crotty S. 2011. IL-21 and IL-6 are critical for different aspects of B cell immunity and redundantly induce optimal follicular helper CD4 T cell (Tfh) differentiation. PLoS ONE 6: e17739. 10.1371/journal.pone.0017739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazilleau N, McHeyzer-Williams LJ, Rosen H, McHeyzer-Williams MG. 2009. The function of follicular helper T cells is regulated by the strength of T cell antigen receptor binding. Nat Immunol 10: 375–384. 10.1038/ni.1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkin S, Hartweger H, Oliveira TY, Kara EE, Nussenzweig MC. 2019. Protein amounts of the MYC transcription factor determine germinal center B cell division capacity. Immunity 51: 324–336.e5. 10.1016/j.immuni.2019.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrod KR, Moreau HD, Garcia Z, Lemaître F, Bouvier I, Albert ML, Bousso P. 2012. Dissecting T cell contraction in vivo using a genetically encoded reporter of apoptosis. Cell Rep 2: 1438–1447. 10.1016/j.celrep.2012.10.015 [DOI] [PubMed] [Google Scholar]
- Gullicksrud JA, Li F, Xing S, Zeng Z, Peng W, Badovinac VP, Harty JT, Xue HH. 2017. Differential requirements for Tcf1 long isoforms in CD8+ and CD4+ T cell responses to acute viral infection. J Immunol 199: 911–919. 10.4049/jimmunol.1700595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale JS, Youngblood B, Latner DR, Mohammed AU, Ye L, Akondy RS, Wu T, Iyer SS, Ahmed R. 2013. Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity 38: 805–817. 10.1016/j.immuni.2013.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand TW, Morre M, Kaech SM. 2007. Expression of IL-7 receptor α is necessary but not sufficient for the formation of memory CD8 T cells during viral infection. Proc Natl Acad Sci 104: 11730–11735. 10.1073/pnas.0705007104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Grant S. 2003. Apoptosis regulators. Rev Clin Exp Hematol 7: 117–138. [PubMed] [Google Scholar]
- Harrington LE, Janowski KM, Oliver JR, Zajac AJ, Weaver CT. 2008. Memory CD4 T cells emerge from effector T-cell progenitors. Nature 452: 356–360. 10.1038/nature06672 [DOI] [PubMed] [Google Scholar]
- He J, Tsai LM, Leong YA, Hu X, Ma CS, Chevalier N, Sun X, Vandenberg K, Rockman S, Ding Y, et al. 2013. Circulating precursor CCR7lo PD-1hi CXCR5+ CD4+ T cells indicate Tfh cell activity and promote antibody responses upon antigen reexposure. Immunity 39: 770–781. 10.1016/j.immuni.2013.09.007 [DOI] [PubMed] [Google Scholar]
- Hildeman DA, Mitchell T, Teague TK, Henson P, Day BJ, Kappler J, Marrack PC. 1999. Reactive oxygen species regulate activation-induced T cell apoptosis. Immunity 10: 735–744. 10.1016/S1074-7613(00)80072-2 [DOI] [PubMed] [Google Scholar]
- Homann D, Teyton L, Oldstone MB. 2001. Differential regulation of antiviral T-cell immunity results in stable CD8+ but declining CD4+ T-cell memory. Nat Med 7: 913–919. 10.1038/90950 [DOI] [PubMed] [Google Scholar]
- Hu Z, Blackman MA, Kaye KM, Usherwood EJ. 2015. Functional heterogeneity in the CD4+ T cell response to murine γ-herpesvirus 68. J Immunol 194: 2746–2756. 10.4049/jimmunol.1401928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer SS, Latner DR, Zilliox MJ, McCausland M, Akondy RS, Penaloza-Macmaster P, Hale JS, Ye L, Mohammed AU, Yamaguchi T, et al. 2013. Identification of novel markers for mouse CD4+ T follicular helper cells. Eur J Immunol 43: 3219–3232. 10.1002/eji.201343469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson SC, Masopust D. 2018. Understanding subset diversity in T cell memory. Immunity 48: 214–226. 10.1016/j.immuni.2018.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jatzek A, Tejera MM, Singh A, Sullivan JA, Plisch EH, Suresh M. 2012. p27Kip1 negatively regulates the magnitude and persistence of CD4 T cell memory. J Immunol 189: 5119–5128. 10.4049/jimmunol.1201482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins MK, Moon JJ. 2012. The role of naive T cell precursor frequency and recruitment in dictating immune response magnitude. J Immunol 188: 4135–4140. 10.4049/jimmunol.1102661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins MK, Chu HH, McLachlan JB, Moon JJ. 2010. On the composition of the preimmune repertoire of T cells specific for Peptide-major histocompatibility complex ligands. Annu Rev Immunol 28: 275–294. 10.1146/annurev-immunol-030409-101253 [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. 2009. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 325: 1006–1010. 10.1126/science.1175870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. 2012. STAT5 is a potent negative regulator of TFH cell differentiation. J Exp Med 209: 243–250. 10.1084/jem.20111174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech SM, Wherry EJ. 2007. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity 27: 393–405. 10.1016/j.immuni.2007.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck S, Schmaler M, Ganter S, Wyss L, Oberle S, Huseby ES, Zehn D, King CG. 2014. Antigen affinity and antigen dose exert distinct influences on CD4 T-cell differentiation. Proc Natl Acad Sci 111: 14852–14857. 10.1073/pnas.1403271111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Wilson T, Fischer KF, Williams MA. 2013. Sustained interactions between T cell receptors and antigens promote the differentiation of CD4+ memory T cells. Immunity 39: 508–520. 10.1016/j.immuni.2013.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano M, Moriyama S, Ando Y, Hikida M, Mori Y, Kurosaki T, Okada T. 2011. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity 34: 961–972. 10.1016/j.immuni.2011.03.025 [DOI] [PubMed] [Google Scholar]
- Klein L, Kyewski B, Allen PM, Hogquist KA. 2014. Positive and negative selection of the T cell repertoire: what thymocytes see (and don't see). Nat Rev Immunol 14: 377–391. 10.1038/nri3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotov DI, Mitchell JS, Pengo T, Ruedl C, Way SS, Langlois RA, Fife BT, Jenkins MK. 2019. TCR affinity biases Th cell differentiation by regulating CD25, Eef1e1, and Gbp2. J Immunol 202: 2535–2545. 10.4049/jimmunol.1801609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy V, Kannanganat S, Maienschein-Cline M, Cook SL, Chen J, Bahroos N, Sievert E, Corse E, Chong A, Sciammas R. 2017. The IRF4 gene regulatory module functions as a read-write integrator to dynamically coordinate T helper cell fate. Immunity 47: 481–497.e7. 10.1016/j.immuni.2017.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künzli M, Schreiner D, Pereboom TC, Swarnalekha N, Litzler LC, Lötscher J, Ertuna YI, Roux J, Geier F, Jakob RP, et al. 2020. Long-lived T follicular helper cells retain plasticity and help sustain humoral immunity. Sci Immunol 5: eaay5552. 10.1126/sciimmunol.aay5552 [DOI] [PubMed] [Google Scholar]
- Lee JY, Skon CN, Lee YJ, Oh S, Taylor JJ, Malhotra D, Jenkins MK, Rosenfeld MG, Hogquist KA, Jameson SC. 2015. The transcription factor KLF2 restrains CD4+ T follicular helper cell differentiation. Immunity 42: 252–264. 10.1016/j.immuni.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yan X, Zhong B, Nurieva RI, Wang A, Wang X, Martin-Orozco N, Wang Y, Chang SH, Esplugues E, et al. 2012. Bcl6 expression specifies the T follicular helper cell program in vivo. J Exp Med 209: 1841–1852, S1841–1824. 10.1084/jem.20120219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohning M, Hegazy AN, Pinschewer DD, Busse D, Lang KS, Höfer T, Radbruch A, Zinkernagel RM, Hengartner H. 2008. Long-lived virus-reactive memory T cells generated from purified cytokine-secreting T helper type 1 and type 2 effectors. J Exp Med 205: 53–61. 10.1084/jem.20071855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak TW, Grusdat M, Duncan GS, Dostert C, Nonnenmacher Y, Cox M, Binsfeld C, Hao Z, Brüstle A, Itsumi M, et al. 2017. Glutathione primes T cell metabolism for inflammation. Immunity 46: 675–689. 10.1016/j.immuni.2017.03.019 [DOI] [PubMed] [Google Scholar]
- Malhotra D, Burrack KS, Jenkins MK, Frosch AE. 2020. Antigen-specific CD4+ T cells exhibit distinct kinetic and phenotypic patterns during primary and secondary responses to infection. Front Immunol 11: 2125. 10.3389/fimmu.2020.02125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. 2008. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu Rev Immunol 26: 171–203. 10.1146/annurev.immunol.26.021607.090421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marriott CL, Mackley EC, Ferreira C, Veldhoen M, Yagita H, Withers DR. 2014. OX40 controls effector CD4+ T-cell expansion, not follicular T helper cell generation in acute Listeria infection. Eur J Immunol 44: 2437–2447. 10.1002/eji.201344211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall HD, Chandele A, Jung YW, Meng H, Poholek AC, Parish IA, Rutishauser R, Cui W, Kleinstein SH, Craft J, et al. 2011. Differential expression of Ly6C and T-bet distinguish effector and memory Th1 CD4+ cell properties during viral infection. Immunity 35: 633–646. 10.1016/j.immuni.2011.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JJ, Dash P, Oguin TH III, McClaren JL, Chu HH, Thomas PG, Jenkins MK. 2011. Quantitative impact of thymic selection on Foxp3+ and Foxp3– subsets of self-peptide/MHC class II-specific CD4+ T cells. Proc Natl Acad Sci 108: 14602–14607. 10.1073/pnas.1109806108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AC, High FA, Hutchins AS, Lee HW, Villarino AV, Livingston DM, Kung AL, Cereb N, Yao TP, Yang SY, et al. 2001. Role of T-bet in commitment of Th1 cells before IL-12-dependent selection. Science 292: 1907–1910. 10.1126/science.1059835 [DOI] [PubMed] [Google Scholar]
- Murali-Krishna K, Lau LL, Sambhara S, Lemonnier F, Altman J, Ahmed R. 1999. Persistence of memory CD8 T cells in MHC class I-deficient mice. Science 286: 1377–1381. 10.1126/science.286.5443.1377 [DOI] [PubMed] [Google Scholar]
- Nakayamada S, Kanno Y, Takahashi H, Jankovic D, Lu KT, Johnson TA, Sun HW, Vahedi G, Hakim O, Handon R, et al. 2011. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity 35: 919–931. 10.1016/j.immuni.2011.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayar R, Schutten E, Bautista B, Daniels K, Prince AL, Enos M, Brehm MA, Swain SL, Welsh RM, Berg LJ. 2014. Graded levels of IRF4 regulate CD8+ T cell differentiation and expansion, but not attrition, in response to acute virus infection. J Immunol 192: 5881–5893. 10.4049/jimmunol.1303187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neefjes J, Jongsma MLM, Paul P, Bakke O. 2011. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol 11: 823–836. 10.1038/nri3084 [DOI] [PubMed] [Google Scholar]
- Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, Dong C. 2009. Bcl6 mediates the development of T follicular helper cells. Science 325: 1001–1005. 10.1126/science.1176676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich KJ, Huang AC, Weinmann AS. 2011. The lineage-defining factors T-bet and Bcl-6 collaborate to regulate Th1 gene expression patterns. J Exp Med 208: 1001–1013. 10.1084/jem.20102144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea JJ, Paul WE. 2010. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science 327: 1098–1102. 10.1126/science.1178334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper M, Jenkins MK. 2011. Origins of CD4+ effector and central memory T cells. Nat Immunol 12: 467–471. 10.1038/ni.2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper M, Linehan JL, Pagán AJ, Zell T, Dileepan T, Cleary PP, Jenkins MK. 2010. Different routes of bacterial infection induce long-lived TH1 memory cells and short-lived TH17 cells. Nat Immunol 11: 83–89. 10.1038/ni.1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper M, Pagán AJ, Igyártó BZ, Taylor JJ, Jenkins MK. 2011. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity 35: 583–595. 10.1016/j.immuni.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy DA, Auerbuch V, Glomski IJ. 2002. The cell biology of Listeria monocytogenes infection: the intersection of bacterial pathogenesis and cell-mediated immunity. J Cell Biol 158: 409–414. 10.1083/jcb.200205009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prlic M, Bevan MJ. 2008. Exploring regulatory mechanisms of CD8+ T cell contraction. Proc Natl Acad Sci 105: 16689–16694. 10.1073/pnas.0808997105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purton JF, Tan JT, Rubinstein MP, Kim DM, Sprent J, Surh CD. 2007. Antiviral CD4+ memory T cells are IL-15 dependent. J Exp Med 204: 951–961. 10.1084/jem.20061805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner SL, Adams WC. 2014. Lymphocyte fate specification as a deterministic but highly plastic process. Nat Rev Immunol 14: 699–704. 10.1038/nri3734 [DOI] [PubMed] [Google Scholar]
- Reinhardt RL, Khoruts A, Merica R, Zell T, Jenkins MK. 2001. Visualizing the generation of memory CD4 T cells in the whole body. Nature 410: 101–105. 10.1038/35065111 [DOI] [PubMed] [Google Scholar]
- Romagnoli PA, Fu HH, Qiu Z, Khairallah C, Pham QM, Puddington L, Khanna KM, Lefrançois L, Sheridan BS. 2017. Differentiation of distinct long-lived memory CD4 T cells in intestinal tissues after oral Listeria monocytogenes infection. Mucosal Immunol 10: 520–530. 10.1038/mi.2016.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV. 2019. Programming for T-lymphocyte fates: modularity and mechanisms. Genes Dev 33: 1117–1135. 10.1101/gad.327163.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph MG, Stanfield RL, Wilson IA. 2006. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol 24: 419–466. 10.1146/annurev.immunol.23.021704.115658 [DOI] [PubMed] [Google Scholar]
- Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401: 708–712. 10.1038/44385 [DOI] [PubMed] [Google Scholar]
- Schenkel JM, Masopust D. 2014. Tissue-resident memory T cells. Immunity 41: 886–897. 10.1016/j.immuni.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt N, Liu Y, Bentebibel SE, Ueno H. 2016. Molecular mechanisms regulating T helper 1 versus T follicular helper cell differentiation in humans. Cell Rep 16: 1082–1095. 10.1016/j.celrep.2016.06.063 [DOI] [PubMed] [Google Scholar]
- Schulz EG, Mariani L, Radbruch A, Höfer T. 2009. Sequential polarization and imprinting of type 1 T helper lymphocytes by interferon-γ and interleukin-12. Immunity 30: 673–683. 10.1016/j.immuni.2009.03.013 [DOI] [PubMed] [Google Scholar]
- Seddon B, Tomlinson P, Zamoyska R. 2003. Interleukin 7 and T cell receptor signals regulate homeostasis of CD4 memory cells. Nat Immunol 4: 680–686. 10.1038/ni946 [DOI] [PubMed] [Google Scholar]
- Shulman Z, Gitlin AD, Targ S, Jankovic M, Pasqual G, Nussenzweig MC, Victora GD. 2013. T follicular helper cell dynamics in germinal centers. Science 341: 673–677. 10.1126/science.1241680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snook JP, Kim C, Williams MA. 2018. TCR signal strength controls the differentiation of CD4+ effector and memory T cells. Sci Immunol 3: eaas9103. 10.1126/sciimmunol.aas9103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. 2000. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100: 655–669. 10.1016/S0092-8674(00)80702-3 [DOI] [PubMed] [Google Scholar]
- Takebe T, Sakamoto K, Higami Y, Harada Y. 2018. A novel mouse model for tracking the fate of CXCR5-expressing T cells. Biochem Biophys Res Commun 495: 1642–1647. 10.1016/j.bbrc.2017.12.029 [DOI] [PubMed] [Google Scholar]
- Tubo NJ, Jenkins MK. 2014. CD4+ T cells: guardians of the phagosome. Clin Microbiol Rev 27: 200–213. 10.1128/CMR.00097-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubo NJ, Pagán AJ, Taylor JJ, Nelson RW, Linehan JL, Ertelt JM, Huseby ES, Way SS, Jenkins MK. 2013. Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during infection. Cell 153: 785–796. 10.1016/j.cell.2013.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubo NJ, Fife BT, Pagan AJ, Kotov DI, Goldberg MF, Jenkins MK. 2016. Most microbe-specific naive CD4+ T cells produce memory cells during infection. Science 351: 511–514. 10.1126/science.aad0483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacchio MS, Ciucci T, Gao Y, Watanabe M, Balmaceno-Criss M, McGinty MT, Huang A, Xiao Q, McConkey C, Zhao Y, et al. 2019. A Thpok-directed transcriptional circuitry promotes Bcl6 and Maf expression to orchestrate T follicular helper differentiation. Immunity 51: 465–478.e6. 10.1016/j.immuni.2019.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbist KC, Guy CS, Milasta S, Liedmann S, Kamiński MM, Wang R, Green DR. 2016. Metabolic maintenance of cell asymmetry following division in activated T lymphocytes. Nature 532: 389–393. 10.1038/nature17442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowski S, Jordan MB, Zhu Y, White J, Zajac AJ, Hildeman DA. 2006. Bim mediates apoptosis of CD127lo effector T cells and limits T cell memory. Eur J Immunol 36: 1694–1706. 10.1002/eji.200635897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Shin HM, Moseman EA, Ji Y, Huang B, Harly C, Sen JM, Berg LJ, Gattinoni L, McGavern DB, et al. 2015. TCF1 is required for the T follicular helper cell response to viral infection. Cell Rep 12: 2099–2110. 10.1016/j.celrep.2015.08.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Li X, Liu D, Li J, Zhang X, Chen X, Hou S, Peng L, Xu C, Liu W, et al. 2013. Follicular T-helper cell recruitment governed by bystander B cells and ICOS-driven motility. Nature 496: 523–527. 10.1038/nature12058 [DOI] [PubMed] [Google Scholar]
- Xu X, Araki K, Li S, Han JH, Ye L, Tan WG, Konieczny BT, Bruinsma MW, Martinez J, Pearce EL, et al. 2014. Autophagy is essential for effector CD8+ T cell survival and memory formation. Nat Immunol 15: 1152–1161. 10.1038/ni.3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, et al. 2009. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity 31: 457–468. 10.1016/j.immuni.2009.07.002 [DOI] [PubMed] [Google Scholar]