ABSTRACT
The emergence of high-level tigecycline resistance mediated by plasmid-borne tet(X) genes greatly threatens the clinical effectiveness of tigecycline. However, the dissemination pattern of plasmid-borne tet(X) genes remains unclear. We here recovered tet(X)-positive Acinetobacter isolates from 684 fecal and environmental samples collected at six livestock farms. Fifteen tet(X)-positive Acinetobacter isolates were identified, mainly including 9 tet(X3)- and 5 tet(X6)-positive Acinetobacter towneri isolates. A clonal dissemination of tet(X3)-positive A. towneri was detected in a swine farm, while the tet(X6)-positive A. towneri isolates mainly disseminated sporadically in the same farm. A tet(X3)-carrying plasmid (pAT181) was self-transmissible from a tigecycline-susceptible A. towneri strain to Acinetobacter baumannii strain ATCC 17978, causing 64- to 512-fold increases in the MIC values of tetracyclines (including tigecycline). Worrisomely, pAT181 was stably maintained and increased the growth rate of strain ATCC 17978. Further identification of tet(X) genes in 10,680 Acinetobacter genomes retrieved from GenBank revealed that tet(X3) (n = 249), tet(X5)-like (n = 61), and tet(X6) (n = 53) were the prevalent alleles mainly carried by four species, and most of them were livestock associated. Phylogenetic analysis showed that most of the tet(X3)- and tet(X6)-positive isolates disseminated sporadically. The structures of the tet(X3), and tet(X6) plasmidomes were highly diverse, and no epidemic plasmids were detected. However, cross-species and cross-region transmissions of tet(X3) might have been mediated by several plasmids in a small proportion of strains. Our study implies that horizontal plasmid transfer may be insignificant for the current dissemination of tet(X3) and tet(X6) in Acinetobacter strains. Continuous surveillance for tet(X) genes in the context of One Health is necessary to prevent them from transmitting to humans.
IMPORTANCE Recently identified plasmid-borne tet(X) genes have greatly challenged the efficiency of tigecycline, a last-resort antibiotic for severe infection, while the dissemination pattern of the plasmid-borne tet(X) genes remains unclear. In this study, we identified a clonal dissemination of tet(X3)-positive A. towneri isolates on a swine farm, while the tet(X6)-positive A. towneri strains mainly disseminated sporadically on the same farm. Of more concern, a tet(X3)-carrying plasmid was found to be self-transmissible, resulting in enhanced tigecycline resistance and growth rate of the recipient. Further exploration of a global data set of tet(X)-positive Acinetobacter genomes retrieved from GenBank revealed that most of the tet(X3)- and tet(X6)-positive isolates shared a highly distant relationship, and the structures of tet(X3) and tet(X6) plasmidomes exhibited high mosaicism. Notably, some of the isolates belong to Acinetobacter species that are opportunistic pathogens and have been identified as sources of nosocomial infections, raising concerns about transmission to humans in the future. Our study evidenced the sporadic dissemination of tet(X3) and tet(X6) in Acinetobacter strains and the necessity of continuous surveillance for tet(X) genes in the context of One Health.
KEYWORDS: plasmid-borne tigecycline resistance, tet(X3), tet(X6), Acinetobacter, self-transmissible plasmid
INTRODUCTION
Tigecycline is used to treat a wide range of clinical infections caused by Gram-positive and Gram-negative bacteria with multidrug resistance (MDR). With the global dissemination of carbapenemases and mobilized colistin resistance (mcr) genes in recent years, this broad-spectrum tetracycline family antibiotic has been raised to be a last-line treatment regimen in clinical settings (1–6). However, the increasing occurrence of transferable tigecycline inactivation genes [tet(X) genes] is threatening the clinical efficacy of tigecycline (7, 8).
The first flavin-dependent monooxygenase gene, named tet(X), was identified in Tn4351 and Tn4400 on the chromosome of Bacteroides fragilis in 1990 (9). Subsequently, 14 chromosome-carried and plasmid-mediated tet(X) genes, tet(X1) to tet(X14), have been reported in various species originating from animals, humans, and the environment (10–12). These Tet(X) enzymes, except for Tet(X1), exhibited different levels of activity against almost all tetracyclines, including a new tetracycline, eravacycline, that was approved by the U.S. Food and Drug Administration (FDA) in 2018 (4, 12, 13). The first plasmid-borne tet(X3) and tet(X4) genes were found in livestock-associated Acinetobacter baumannii and Escherichia coli strains, respectively, in 2019 (7), raising the concern of horizontal transfer of tigecycline resistance. Since then, additional tet(X) alleles have been reported to be plasmid borne, including tet(X5) and tet(X6) and their variants. Epidemiological studies reveal that these novel tet(X) orthologs have mainly circulated in animals in China due to the heavy use of tetracyclines in husbandry (8). In some pioneering studies, ISCR2 was highlighted as the key element facilitating the horizontal transfer of tet(X) genes, through circular intermediates (14–17). However, the role of mobile elements in the dissemination of tet(X) genes remains obscure.
The tet(X) genes have been detected in over 16 bacterial species, and Acinetobacter spp. were among the major hosts (7, 11, 17–20). Currently, most of the tet(X)-positive Acinetobacter species isolates have been associated with livestock, and very few have been found in humans (16, 21). A surveillance study at avian farms in China showed that 1.6% to 18.3% of Acinetobacter species strains were tet(X) positive (22). Another surveillance study for tigecycline-resistant Acinetobacter spp. from 2015 to 2018 in 14 provinces and municipalities in China identified 2.3% to 25.3% tet(X)-positive isolates from pig farms, migratory birds, and samples from human (20). Plasmid-borne and/or chromosome-carried tet(X3) and tet(X6) were prevalent in livestock-associated Acinetobacter species isolates, and tet(X5) has so far only been detected in an A. baumannii strain from humans (7, 16, 20, 22, 23).
In this study, surveillance of tet(X)-positive Acinetobacter species isolates recovered from livestock and their surrounding environmental sources was performed at six livestock farms located in Zhejiang Province in 2019. The epidemiological and genetic characterizations of tet(X)-positive isolates and tet(X)-harboring plasmids were dissected. We further investigated the population structure and distribution of tet(X)-positive Acinetobacter strains identified in a public database, as well as the plasmidomes of tet(X3) and tet(X6).
RESULTS
A. towneri was the prevalent species carrying tet(X) genes among Acinetobacter strains collected in this study.
Two hundred ninety-two isolates were recovered from 534 stool samples and 150 environmental samples collected from 2 swine farms, 2 dairy farms, and 2 sheep farms, including 215 isolates of Acinetobacter spp. and 77 isolates belonging to other species. Twenty-three tet(X)-positive isolates were identified (7.88%; 23/292), including 15 Acinetobacter species isolates (6.97%; 15/215), 5 Empedobacter stercoris isolates, and 3 Myroides odoratimimus isolates (Table 1). The 23 tet(X)-positive isolates were exclusively isolated from swine farms. The Acinetobacter spp. and E. stercoris isolates were all recovered from the fecal samples of swine farm 1, and the 3 M. odoratimimus isolates were from the soil samples of swine farm 2.
TABLE 1.
tet(X)-positive strains isolated in this study
| Strain | Species | Gene | Location | Source | Sequencing platform | Genome accession no. |
|---|---|---|---|---|---|---|
| ZJ202 | Empedobacter stercoris | tet(X2) | Chromosome | Fecal, swine farm 1 | Illumina | JABFOQ000000000 |
| ZJ180 | E. stercoris | tet(X2) | Chromosome | Fecal, swine farm 1 | Illumina | JACXZB000000000 |
| ZJ215 | E. stercoris | tet(X2) | Chromosome | Fecal, swine farm 1 | Illumina | JACXZC000000000 |
| ZJ286 | Myroides odoratimimus | tet(X2) | NAa | Soil, swine farm 2 | Illumina | JACXZD000000000 |
| ZJ291 | M. odoratimimus | tet(X2) | NA | Soil, swine farm 2 | Illumina | JACXZE000000000 |
| ZJ295 | M. odoratimimus | tet(X2) | NA | Soil, swine farm 2 | Illumina | JACXZF000000000 |
| AT184 | Acinetobacter towneri | tet(X3) | Plasmid | Fecal, swine farm 1 | Nanopore | JACXZG000000000 |
| ZJ199 | Acinetobacter sp. | tet(X3) | Chromosome | Fecal, swine farm 1 | Nanopore | CP062182 |
| AT200 | A. towneri | tet(X3) | Plasmid | Fecal, swine farm 1 | Illumina | JACXZH000000000 |
| AT216 | A. towneri | tet(X3) | Plasmid | Fecal, swine farm 1 | Illumina | JACXZI000000000 |
| AT217 | A. towneri | tet(X3) | Plasmid | Fecal, swine farm 1 | Illumina | JACXZJ000000000 |
| AT181 | A. towneri | tet(X3) | Plasmid | Fecal, swine farm 1 | Nanopore | JACXZK000000000 |
| AT209 | A. towneri | tet(X3) | Plasmid | Fecal, swine farm 1 | Illumina | JACXZL000000000 |
| AT211 | A. towneri | tet(X3) | Plasmid | Fecal, swine farm 1 | Illumina | JACXZM000000000 |
| AT213 | A. towneri | tet(X3) | Plasmid | Fecal, swine farm 1 | Illumina | JACXZN000000000 |
| AT214 | A. towneri | tet(X3) | Plasmid | Fecal, swine farm 1 | Illumina | JACXZO000000000 |
| AT185 | A. towneri | tet(X6), tet(X6) | Plasmid | Fecal, swine farm 1 | Illumina | JACXZP000000000 |
| AT208 | A. towneri | tet(X6) | Plasmid | Fecal, swine farm 1 | Illumina | JACXZQ000000000 |
| AT232 | A. towneri | tet(X6) | Plasmid | Fecal, swine farm 1 | Nanopore | CP062183-CP062184 |
| AT235 | A. towneri | tet(X6) | Plasmid | Fecal, swine farm 1 | Nanopore | CP062185-CP062186 |
| AT205 | A. towneri | tet(X6) | Plasmid | Fecal, swine farm 1 | Nanopore | CP048014-CP048018 |
| ZJ183 | E. stercoris | tet(X14), tet(X2), tet(X2) | Chromosome | Fecal, swine farm 1 | Nanopore | CP053698-CP053701 |
| ZJ182 | E. stercoris | tet(X14)-tet(X2) | Chromosome | Fecal, swine farm 1 | Illumina | JACXZR000000000 |
NA, not available: the location of tet(X) gene cannot be resolved in this genome.
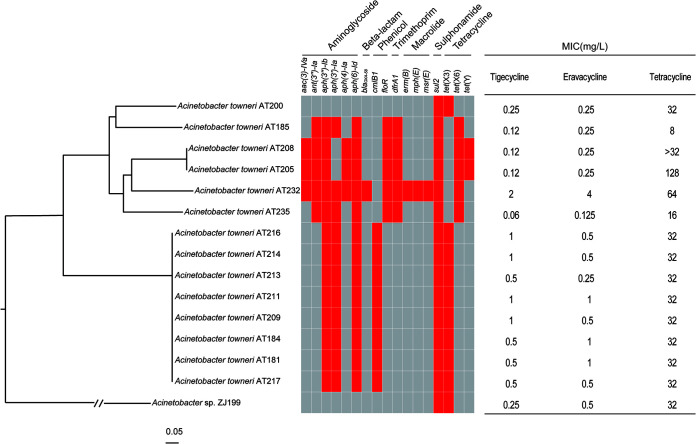
The 15 tet(X)-positive Acinetobacter species isolates were assigned by average nucleotide identity (ANI) analysis to Acinetobacter towneri (n = 14) and an unclassified species (n = 1), and the other 8 tet(X)-positive isolates were E. stercoris (n = 5) and M. odoratimimus (n = 3) (Table 1). Four different tet(X) genes [tet(X2), tet(X3), tet(X6), and tet(X14)] were identified in the 23 tet(X)-positive isolates (Table 1). tet(X2) was exclusively detected in the 8 non-Acinetobacter isolates, and tet(X3) was in 9 A. towneri isolates and 1 unclassified species isolate (ZJ199). tet(X6) and tet(X14) were found in 5 A. towneri and 2 E. stercoris isolates, respectively. Notably, two copies of tet(X6) were carried by an A. towneri isolate (AT185). Eight of the tet(X3)-positive Acinetobacter species isolates clustered together, with 3 to 36 single-nucleotide polymorphisms (SNPs) (Fig. 1), suggesting the clonal dissemination of one strain. Two of the tet(X6)-positive isolates were also clonally related (1 SNP). The remaining five isolates showed distant relationships (26,876 to 31,071 SNPs), indicating sporadic dissemination of these strains.
FIG 1.
Phylogenetic analysis of tet(X)-positive Acinetobacter isolates collected in this study. The core-genome SNPs of tet(X)-encoding strains were used to generate the phylogenetic tree. The tree is rooted at strain ZJ199. The ARGs of each strain are exhibited by the heatmap, and the existence of ARGs is in red. MIC values of each strain against tetracyclines are listed. AT205 has been reported previously (26).
Antimicrobial resistance profiles of tet(X)-carrying isolates.
Eight of the 23 tet(X)-positive isolates (34.78%) were resistant to tigecycline, with MIC values at 1 to 2 mg/liter, encompassing 4 tet(X3)-positive A. towneri isolates, 1 tet(X6)-positive A. towneri isolate, 2 tet(X2)- and tet(X14)-positive E. stercoris isolates, and 1 tet(X2)-positive M. odoratimimus isolate (Table 2). Five tigecycline-resistant isolates (3 A. towneri isolates and 2 E. stercoris isolates) additionally exhibited resistance to the newly FDA-approved eravacycline, with MIC values at 1 to 4 mg/liter. Except for the isolate carrying 2 copies of tet(X6), the other 14 Acinetobacter species isolates were resistant to tetracycline with MIC values of ≥16 mg/liter (Table 2). Strain AT232 showed significantly higher resistance to tetracyclines than the other 13 isolates, which might be caused by the presence of a two-component system, AdeSR, involved in the expression of the AdeABC efflux pump (24). Four and two Acinetobacter species isolates additionally showed resistance to ciprofloxacin and doxycycline, respectively (Table 2). All of the tet(X)-positive Acinetobacter species isolates were susceptible to colistin and carbapenems. The M. odoratimimus isolates were resistant to colistin and carbapenems due to intrinsic resistance (25).
TABLE 2.
MIC values of antibiotics tested in this study
| Strain | MIC (mg/liter) ofa: |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAZ | CRO | FEP | IPM | MEM | CIP | LVX | AMK | GEN | SXT | CSL | COL | TGC | OTC | CTC | DMC | DOX | MIN | ERV | TET | |
| ZJ202 | 4 | 2 | 0.125 | 0.25 | 0.125 | 1 | 0.5 | 16 | 8 | 0.25 | 2 | 16 | 0.5 | 32 | 4 | 4 | 1 | 0.5 | 0.5 | 16 |
| ZJ180 | 2 | 2 | 0.125 | 0.5 | 0.25 | 1 | 0.5 | 16 | 4 | 0.06 | 4 | 32 | 0.5 | 16 | 4 | 2 | 0.5 | 0.25 | 1 | 8 |
| ZJ215 | 2 | 2 | 0.25 | 0.5 | 0.125 | 0.125 | 0.5 | 2 | 4 | >8 | 0.25 | 16 | 0.5 | 32 | 4 | 4 | 1 | 0.5 | 1 | 16 |
| ZJ286 | 64 | >64 | 8 | >32 | 2 | >32 | 8 | >128 | >128 | 1 | >128 | >32 | 0.5 | >128 | >128 | >128 | >128 | 128 | 1 | >128 |
| ZJ291 | 64 | >64 | 8 | >32 | 2 | >32 | 16 | >128 | >128 | >8 | >128 | >32 | 2 | >128 | >128 | >128 | 64 | 32 | 1 | >128 |
| ZJ295 | 64 | >64 | 8 | >32 | 2 | >32 | 8 | >128 | >128 | 0.5 | >128 | >32 | 0.5 | >128 | >128 | >128 | >128 | 16 | 0.5 | >128 |
| AT184 | 2 | 4 | 0.5 | 0.125 | 0.03 | 1 | 1 | 1 | 1 | >8 | 1 | 0.5 | 0.5 | 128 | 16 | 16 | 1 | 0.5 | 1 | 32 |
| ZJ199 | 0.25 | 0.25 | 0.06 | 0.03 | 0.03 | 4 | 2 | 0.06 | 0.125 | >8 | 0.06 | 1 | 0.25 | 128 | 16 | 8 | 2 | 0.25 | 0.5 | 32 |
| AT200 | 2 | 4 | 0.25 | 0.125 | 0.03 | 0.03 | 0.06 | 0.25 | 0.125 | >8 | 0.5 | 2 | 0.25 | 64 | 8 | 4 | 0.5 | 0.5 | 0.25 | 32 |
| AT216 | 2 | 4 | 0.5 | 0.125 | 0.06 | 2 | 0.5 | 1 | 0.25 | >8 | 0.25 | 1 | 1 | 64 | 16 | 8 | 0.5 | 0.25 | 0.5 | 32 |
| AT217 | 2 | 4 | 0.5 | 0.125 | 0.06 | 2 | 0.5 | 1 | 0.25 | 8 | 0.25 | 1 | 0.5 | 128 | 16 | 16 | 0.5 | 0.25 | 0.5 | 32 |
| AT181 | 2 | 4 | 0.25 | 0.125 | 0.06 | 1 | 0.5 | 1 | 0.5 | >8 | 1 | 1 | 0.5 | 128 | 16 | 16 | 1 | 0.5 | 1 | 32 |
| AT209 | 2 | 4 | 0.25 | 0.125 | 0.03 | 0.03 | 0.5 | 1 | 0.5 | >8 | 1 | 0.5 | 1 | 128 | 16 | 8 | 0.5 | 0.5 | 0.5 | 32 |
| AT211 | 2 | 4 | 0.25 | 0.125 | 0.03 | 0.03 | 0.5 | 1 | 0.5 | >8 | 1 | 1 | 1 | 128 | 16 | 8 | 1 | 0.25 | 1 | 32 |
| AT213 | 2 | 4 | 0.25 | 0.125 | 0.03 | 0.03 | 0.5 | 2 | 0.5 | >8 | 1 | 1 | 0.5 | 128 | 16 | 8 | 0.5 | 0.5 | 0.25 | 32 |
| AT214 | 2 | 4 | 0.25 | 0.125 | 0.03 | 0.03 | 0.5 | 2 | 0.5 | >8 | 1 | 1 | 1 | 64 | 8 | 8 | 0.25 | 0.5 | 0.5 | 32 |
| AT185 | 2 | 4 | 0.5 | 0.25 | 0.03 | 1 | 0.5 | 0.5 | 0.25 | >8 | 1 | 2 | 0.12 | 32 | 8 | 4 | 0.25 | 0.25 | 0.25 | 8 |
| AT208 | 2 | 4 | 0.25 | 0.25 | 0.03 | 0.03 | 1 | 1 | 8 | >8 | 1 | 2 | 0.12 | >128 | 128 | 128 | 16 | 2 | 0.25 | >32 |
| AT232 | 2 | 4 | 0.5 | 0.25 | 0.06 | 4 | 1 | 0.5 | 4 | 8 | 0.5 | 2 | 2 | 128 | 64 | 32 | 4 | 2 | 4 | 64 |
| AT235 | 2 | 4 | 0.5 | 0.125 | 0.03 | 4 | 1 | 0.5 | 0.125 | 8 | 0.25 | 2 | 0.06 | 32 | 4 | 2 | 0.25 | 0.25 | 0.125 | 16 |
| AT205 | 4 | 8 | 0.5 | 0.5 | 0.06 | 4 | 1 | 1 | 8 | >8 | 1 | 2 | 0.12 | 128 | 128 | 128 | 32 | 0.5 | 0.25 | 128 |
| ZJ183 | 2 | 4 | 0.5 | 0.25 | 0.125 | 1 | 1 | 32 | 16 | 0.06 | 4 | 32 | 1 | 128 | 8 | 8 | 4 | 0.125 | 1 | 16 |
| ZJ182 | 1 | 1 | 0.06 | 0.125 | 0.125 | 2 | 1 | 16 | 8 | 0.06 | 2 | 32 | 1 | 64 | 8 | 8 | 2 | 1 | 2 | 16 |
CAZ, ceftazidime; CRO, ceftriaxone; FEP, cefepime; IPM, imipenem; MEM, meropenem; CIP, ciprofloxacin; LVX, levofloxacin; AMK, amikacin; GEN, gentamycin; SXT, sulfamethoxazole-trimethoprim; CSL, cefoperazone-sulbactam; COL, colistin; TGC, tigecycline; OTC, oxytetracycline; CTC, chlortetracycline; DMC, demeclocycline; DOX, doxycycline; MIN, minocycline; ERV, eravacycline; TET, tetracycline.
The 23 tet(X)-positive isolates were subjected to whole-genome sequencing (WGS) (Table S1 in the supplemental material). All of the A. towneri strains were multidrug resistant (MDR), and more antibiotic resistance genes (ARGs) were detected in the tet(X6)-carrying clone (mean = 8.67; median = 9) than in the tet(X3)-carrying clone (mean = 6; median = 6), albeit the difference was not significant (P > 0.05) (Fig. 1). The 8 strains of the tet(X3)-carrying clone shared an identical resistome [aph(3″)-Ib, aph(3′)-Ia, aph(6)-Id, cmlB1, sul2, and tet(X3)], further supporting the aforementioned clonal dissemination (Fig. 1), while the resistomes of the tet(X6)-carrying strains were highly diverse, with genes that included the following: aacC4, ant(3″)-Ia, and aph(4)-Ia, encoding resistance to aminoglycosides; blaOXA-58, encoding resistance to beta-lactams; floR, encoding resistance to phenicols; dfrA1, encoding resistance to trimethoprim; erm(B), mph(E), and msr(E), encoding resistance to macrolides; and tet(X6) and tet(Y), encoding resistance to tetracyclines (Fig. 1). Only two ARGs were detected in strain ZJ199 [sul2 and tet(X3)]. The resistomes of E. stercoris and M. odoratimimus were different from those of Acinetobacter spp. (Table S2).
tet(X3) and tet(X6) were harbored by various plasmids.
To understand the vectors of the two prevalent tet(X) genes, i.e., tet(X3) and tet(X6), five representative strains [AT181 and AT184 for tet(X3) and ZJ199, AT232, and AT235 for tet(X6)] were additionally chosen for long-read sequencing based on their antimicrobial resistance profiles and the genetic environments of their tet(X) genes (Table S1). The hybrid assembly confirmed that tet(X3) and tet(X6) were plasmid borne in the four A. towneri isolates, and a chromosome-carried tet(X3) was detected in ZJ199.
The tet(X3)-carrying plasmids detected in AT181 (pAT181) and AT184 (pAT184) were circularized (confirmed by PCR) and identical, with a size of 75,969 bp. These two plasmids were untypeable, with an average GC content of 42.5%. Multiple ARGs were carried by the two plasmids, including aph(3′)-Ia, aph(3″)-Ib, aph(6)-Id, sul2, and tet(X3). BLAST analysis of the nucleotide sequence of pAT181 in GenBank showed that the best match was a transferable tet(X3)-harboring plasmid, p10FS3-1-3 (accession number CP039146) (100% identity and 97% coverage) carried by a novel species of Acinetobacter (20). Others sharing a high similarity with pAT181 included a tet(X5)-harboring plasmid, pAB17H194-1 (accession number CP040912; 99.95% identity and 86% coverage), carried by an A. pittii strain and a tet(X3)-harboring plasmid, p18TQ-X3 (accession number CP045132; 99.99% identity and 80% coverage), carried by an A. indicus strain. These data suggested that pAT181-like plasmids have disseminated among various species of Acinetobacter.
In accordance with the phylogeny, the tet(X3)-carrying plasmids carried by the 8 clonal isolates were all homologous to pAT181, with >90% coverage and nucleotide identity (Fig. S1A), and the tet(X3)-carrying plasmid carried by AT200 was different from pAT181, with <50% coverage and >90% identity (Fig. S1A). The best match for pAT200 in GenBank was p10FS3-1-3, with 58.77% coverage and 70% identity.
The two tet(X6)-harboring circularized plasmids pAT232 and pAT235 shared as little as 38% coverage and 99.95% identity; however, the sequences of their rep genes were identical, indicating that they might originate from a common ancestor. pAT232 was 186,508-bp in length, with a GC content of 41.03%. A BLAST search against GenBank showed that the best matches for pAT232 were a tet(X6)-carrying plasmid, pAT205 (accession number CP048015) (76% coverage and 99.99% identity), carried by A. towneri strain AT205 isolated on the same swine farm (26), and a tet(X)-negative plasmid, p19110F47-2 (accession number CP046044) (70% coverage and 99.99% identity), carried by an A. towneri strain isolated from pigs. pAT235 was 124,466 bp in length, with a GC content of 41.16%. The best matches for pAT235 were pAT205 (49% coverage and 100% identity) and a tet(X3)-harboring plasmid, pGX7 (accession number CP071772) (44% coverage and 99.95% identity), detected in an A. towneri strain isolated from pigs in China. These data suggest that pAT232 and pAT235 might originate from A. towneri strains associated with pigs.
When pAT232 was used as a reference to identify the plasmids bearing tet(X6) in the other tet(X6)-positive isolates collected here, AT208 showed the highest similarity to pAT232 (77.84% coverage and 99.16% identity) (Fig. S1B). When pAT235 was used as a reference, AT185 shared 100% coverage and 94.51% identity (Fig. S1C), suggesting that a pAT235-like tet(X6)-encoding plasmid was harbored in AT185. Of note, AT185 was genetically distant from AT235, with 30,097 SNPs (Fig. 1). A pAT205-like tet(X6)-harboring plasmid was detected in AT208 when pAT205 was used as a reference (100% coverage and 96.48% identity) (Fig. S1D). These results reveal that horizontal transfers of tet(X6)-carrying plasmids might have occurred sporadically.
Genetic environments of tet(X3) and tet(X6).
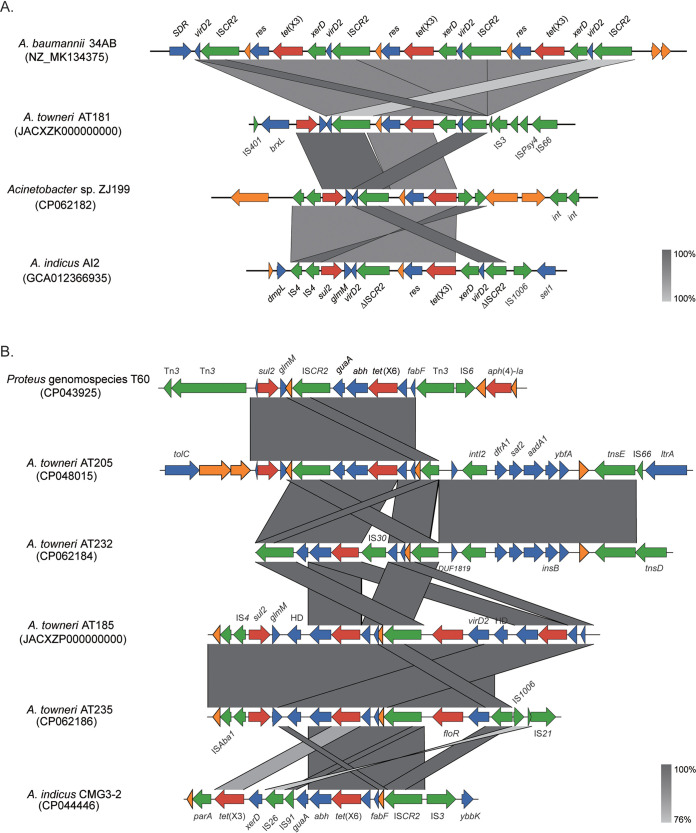
The genetic environments of plasmid-borne tet(X3) [ΔISCR2-xerD-tet(X3)-res-ISCR2] detected in 8 of 9 A. towneri strains were identical and highly similar to that of the prototype detected in A. baumannii strain 34AB (Fig. 2A) (7). To fully understand the distribution of this genetic environment among tet(X3)-carrying Acinetobacter strains, we used BLAST to compare it to 249 tet(X3)-carrying Acinetobacter genomes retrieved from GenBank (see below). The fragment ΔISCR2-xerD-tet(X3)-res-ISCR2 was detected on a single contig of 21.3% (53/249) of genomes with >90% coverage and identity. The proportion increased to 86.35% (215/249) when matches on different contigs were counted together, implying a major structure encoding tet(X3) in Acinetobacter spp. A different tet(X3) genetic environment [IS4-IS4-tet(X3)-res-ΔISCR2] was detected on the chromosome of strain ZJ199 (Fig. 2A).
FIG 2.
Genetic context of tet(X3) and tet(X6) genes identified in Acinetobacter spp. (A) Comparison of the genetic contexts of tet(X3). The genomic contexts of tet(X3) identified in A. baumannii strain 34AB (accession number MK134375) and A. indicus strain AI2 (accession number GCA_012366935) are used as the reference sequences. (B) Comparison of the genetic contexts of tet(X6). The genomic contexts of tet(X6) identified in Proteus genomospecies 6 T60 (accession number CP043925) and A. indicus strain CMG3-2 (accession number CP044446) are used as the reference sequences. Genes are indicated by color-coded arrows dependent on the functional annotations and direction of transcription. ARGs are in red; mobile genetic element genes are in green; genes with other functions are in blue; hypothetical genes are in orange.
The genetic environments of tet(X6) were much more diverse than those of tet(X3) detected in our collection (Fig. 2B). A 7,270-bp composite structure [ΔISCR2-IS30-tet(X6)-abh-guaA-ISCR2] was detected in pAT232, which is similar to the prototype [ΔISCR2-tet(X6)-abh-guaA-ISCR2] identified in pAT205 and a Proteus genomospecies 6 strain (26, 27), except for an insertion of an IS30. The tet(X6) located within a 6,885-bp region [ISCR2-fabF-tet(X6)-abh-glmM-sul2] in pAT235 (Fig. 2B) was almost identical to that detected on the chromosome of A. indicus strain Q186-3_T (100% coverage and 99.58% identity) and on pABF9692, carried by an A. baumannii strain (accession number CP048828) (100% coverage and 98.70% identity). In strain AT185, the genetic context of one copy of tet(X6) was identical to that detected in pAT235, and a truncated structure was found for the other copy (Fig. 2B). The ISCR2-fabF-tet(X6)-abh fragment was also found on the chromosomes of A. indicus strain LYS68A (CP070997) and A. baumannii strain 31FS3-2 (CP0445177), indicating that this structure might mediate the mobilization of tet(X6) between plasmids and chromosomes in Acinetobacter spp.
A tet(X3)-carrying plasmid was self-transmissible from A. towneri to A. baumannii and increased its resistance to tetracyclines and growth rate.
A conjugation assay was performed to test the transferability of tet(X)-encoding plasmids. We only obtained tigecycline-resistant A. baumannii transconjugants from A. towneri strain AT181, with frequencies of 1.85 × 10−6 per recipient cell. Multiple attempts at plasmid transfers failed when E. coli strain EC600 was used as a recipient. Compared with those of the recipient strain ATCC 17978, the MIC values of tigecycline and the other tetracyclines against the transconjugant ATCC 17978-pAT181 increased by 128-fold and ∼64- to 512-fold, respectively (Table S3). WGS was performed for ATCC 17978-pAT181 and ATCC 17978 to detect the transferable structure of tet(X3). A unique plasmid, pAT181, was detected in the transconjugant ATCC 17978-pAT181, demonstrating that the transmission of tigecycline resistance was mediated by pAT181 (Fig. S2). This is different from another self-transmissible tet(X3)-harboring plasmid p10FS3-1-3 in that the transfer of p10FS3-1-3 into Acinetobacter baylyi strain ADP1 did not bring a significant additive effect to the resistance to tetracyclines (20). To the best of our knowledge, this is the first report showing the horizontal transfer of a tet(X3)-carrying plasmid conferring tetracycline resistance to the recipient.
tet(X3) was stable in the recipient strain ATCC 17978 without antibiotic stress during 10 days of passage, with a 100% retention rate. Compared with that of ATCC 17978, the doubling time of ATCC 17978-pAT181 was shortened from 4.59 h to 2.91 h (Fig. 3). These results suggest that pAT181 could facilitate the dissemination of tet(X3) among Acinetobacter spp. strains.
FIG 3.
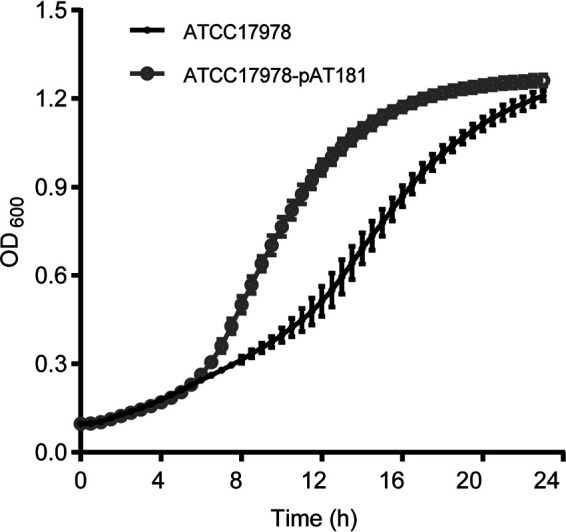
Growth curves of the recipient strain A. baumannii ATCC 17978 and the transconjugant strain ATCC 17978-pAT181 at 37°C. The optical density at 600 nm was recorded every 30 min. The assay was in triplicate.
tet(X3), tet(X5)-like, and tet(X6) are the prevalent alleles among the tet(X) family and disseminate sporadically in four species of Acinetobacter spp.
To fully understand the distribution of tet(X) genes among Acinetobacter spp., a BLAST comparison of the nucleotide sequences of 15 known tet(X) alleles and their variants to 10,680 Acinetobacter genomes retrieved from GenBank was performed. tet(X3) was found in 249 genomes, tet(X4) in 9 genomes, tet(X5) (n = 2), tet(X5.2) (n = 53), and tet(X5.3) (n = 6) in 61 genomes, tet(X6) in 53 genomes, and tet(X13), a 1-residue variant of tet(X6), in 4 genomes. These data reveal that tet(X3), tet(X5.2), and tet(X6) are the prevalent tet(X) genes among Acinetobacter spp.
Species identification by ANI analysis showed three predominant Acinetobacter species carrying tet(X3), i.e., A. indicus (27.71%; 69/249), Acinetobacter sp002018365 (26.51%; 66/249) (an unclassified species with Acinetobacter sp. ANC 4845 as the reference), and A. towneri (12.85%; 32/249) (Table S4). Except for A. variabilis (11.32%; 6/53), A. indicus (22.64%; 12/53), Acinetobacter sp002018365 (20.75%; 11/53), and A. towneri (11.32%; 6/53) are also the predominant species carrying tet(X6). The distribution of tet(X5.2)-harboring species was similar to that of species carrying tet(X6), including A. indicus (22.64%; 12/53), Acinetobacter sp002018365 (20.75%; 11/53), A. towneri (11.32%; 6/53), A. variabilis (11.32%; 6/53), and A. lwoffii (11.32%; 6/53). These results indicate that A. indicus and Acinetobacter sp002018365 are the most prevalent species carrying tigecycline-resistant tet(X) genes.
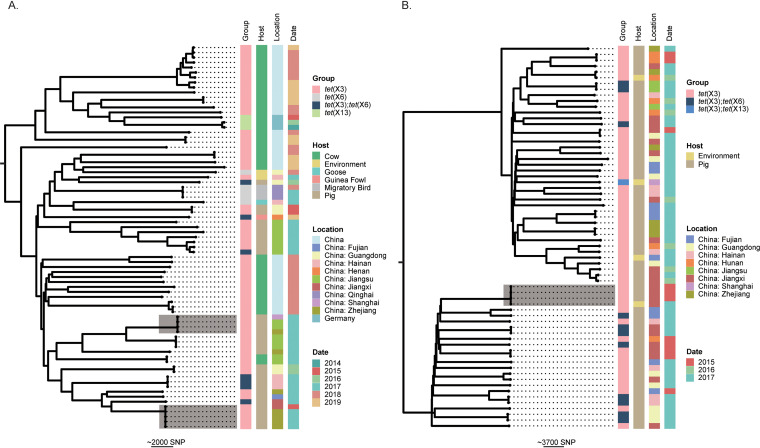
To further evaluate the patterns of dissemination of tet(X3) and tet(X6) among Acinetobacter populations, we performed phylogenomic analysis for tet(X3)-/tet(X6)-positive isolates carried by four major species as representatives, i.e., A. indicus, Acinetobacter sp002018365, A. towneri, and A. variabilis (Fig. 4; Fig. S3). Most isolates of each species shared a distant relationship, and no epidemic clones were detected. Two interregional transmission events were detected for 4 (no SNPs) and 5 (0 or 1 SNP) isolates of A. indicus, and one cross-host event (pig and environment) was detected for 4 isolates (1 to 44 SNPs) of Acinetobacter sp002018365 (Fig. 4). The data suggested that tet(X3) and tet(X6) mainly disseminated sporadically among Acinetobacter populations.
FIG 4.
Phylogenetic analysis of genomes carrying tet(X3)/tet(X6)/tet(X13) retrieved from GenBank. (A) The phylogenetic tree of A. indicus. (B) The phylogenetic tree of Acinetobacter sp002018365. The core-genome SNPs were used to calculate the phylogenetic trees. The trees are midpoint rooted. The tet(X) genes (Group), isolate source (Host), sampling location (Location), and years of isolation (Date) of strains are shown at the right side of each tree as indicated in the color keys. Two interregional transmission events for 4 and 5 strains of A. indicus and one cross-host event for 4 strains of Acinetobacter sp002018365 are highlighted by shading. The scale bar represents the number of SNPs.
The structures of tet(X3)/tet(X6) plasmidomes are highly diverse, and no epidemic plasmids have yet been detected among Acinetobacter.
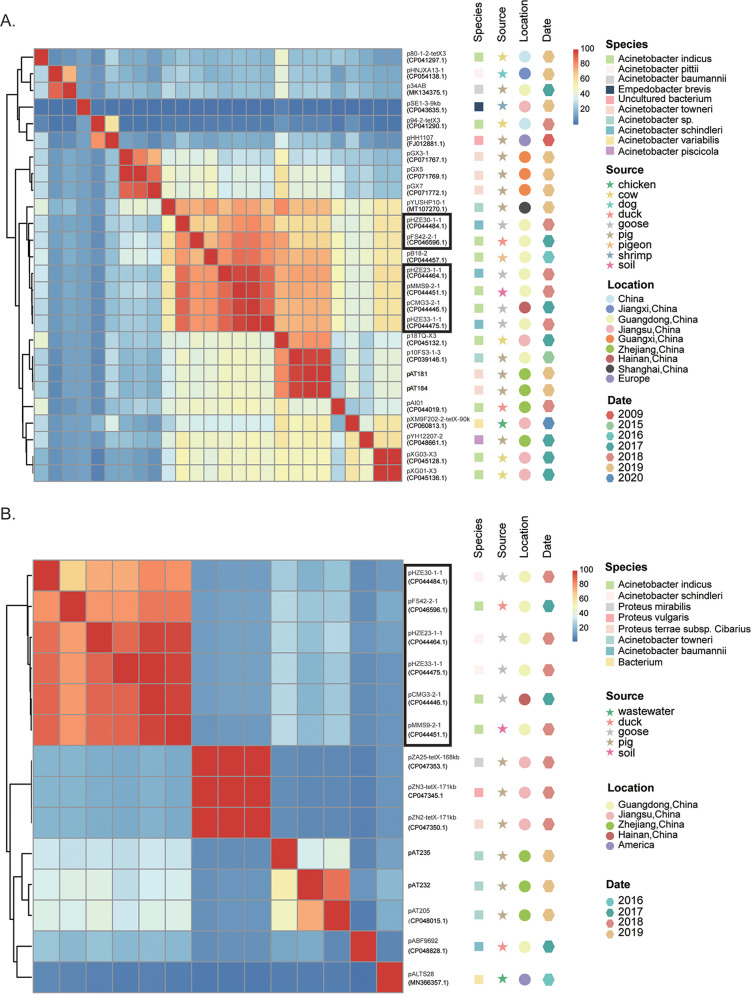
To explore the role of plasmids in the dissemination of tet(X3) and tet(X6) in Acinetobacter spp., we here intended to dissect the genetic relatedness of tet(X3)- and tet(X6)-harboring plasmids. Four circularized tet(X3)-/tet(X6)-harboring plasmids were obtained in this study, and all finished tet(X3)-/tet(X6)-harboring plasmids deposited in GenBank [n = 30; 18 for tet(X3), 6 for tet(X6), and 6 for tet(X3) and tet(X6)] were analyzed first. Twenty-five of the 30 publicly available plasmids were carried by Acinetobacter spp. Most of the 26 tet(X3)-harboring plasmids [including the 6 tet(X3)-tet(X6)-harboring plasmids] shared a coverage lower than 65%, indicating a highly diverse structure for the plasmidome of tet(X3) (Fig. 5A). Four of the 6 tet(X3)-tet(X6)-positive plasmids shared high similarity (>89.8% coverage and >85% identity), suggesting that they were derived from an ancestor. The four plasmids were hosted in A. schindleri and A. indicus strains isolated from goose and soil samples collected in different provinces of China (Fig. 5A), indicating that cross-species, cross-sector (poultry and environment), and/or cross-region transmission has occurred for these plasmids. A similar transmission event was observed for another three tet(X3)-encoding plasmids (pAT181, pAT184, and p10FS3-1-3) carried by A. towneri and a novel species of Acinetobacter as mentioned above (Fig. 5A).
FIG 5.
Pairwise sequence comparisons between circularized tet(X3)-/tet(X6)-carrying plasmids. (A) The percentages of aligned bases between pairs of tet(X3)-carrying plasmids. (B) The percentages of aligned bases between pairs of tet(X6)-carrying plasmids. The row and column orders are the same. The host species, sampling source, sampling location, and year of isolation are shown by colored symbols to the right of the phylogenetic trees as indicated in the color keys. The six plasmids that coharbored tet(X3) and tet(X6) genes are boxed.
The 5 tet(X6)-harboring plasmids carried by Acinetobacter and an unknown species share low similarities, except for pAT232 and pAT205, as mentioned above (Fig. 5B). They are different from the 3 tet(X6)-harboring plasmids (pAZ25, pZN3, and pZN2) carried by Proteus species and from the 6 tet(X3)-tet(X6)-harboring plasmids (Fig. 5B). Hence, the tet(X3)-tet(X6)-harboring plasmids might have resulted from the capture of tet(X6) by tet(X3)-harboring plasmids.
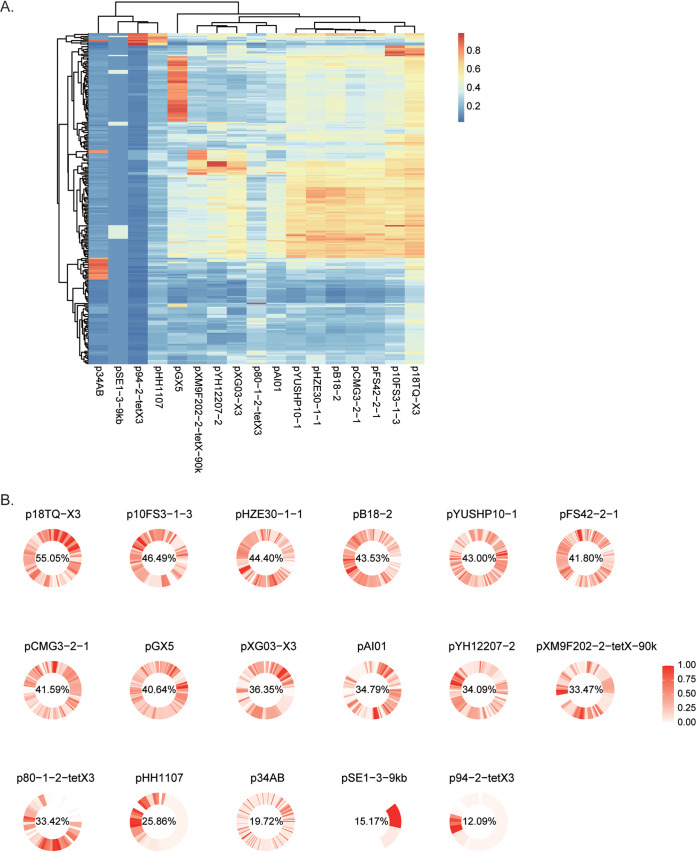
To further estimate the distribution of tet(X3)-harboring plasmids among Acinetobacter spp., we selected 17 plasmids out of 26 tet(X3)-harboring plasmids as references according to their similarities (<80% coverage and identity). The 17 plasmids were compared to the 243 tet(X3)-positive genomes [6 genomes with chromosome-carried tet(X3) were excluded] by using BLAST, and no epidemic plasmids were found (Fig. 6A). We further mapped the 243 genomic sequences against the 17 representative plasmids (Fig. 6B), and this revealed that tet(X3) plasmid structures were highly diverse among isolates (mean plasmid coverage range of 12.09% to 55.05%). Using a cutoff range of >80% coverage and >90% identity, we found that pGX5-like plasmids were hosted in 36 strains belonging to different species (20 A. towneri strains, 10 A. variabilis strains, 4 Acinetobacter sp002018365 strains, and 2 A. indicus strains), and p34AB-like, p94-2-tetX3-like, pXM9F202-2-tetX-90k-like, and p10FS3-1-3-like plasmids were found in 17, 9, 8, and 7 strains belonging to different species, respectively (Fig. 6A). These data suggest that the current dissemination of tet(X3) in Acinetobacter is mainly mediated by various plasmids and that cross-species transmissions mediated by a few of them might have occurred in a small proportion of cases.
FIG 6.
Analysis of tet(X3) plasmidome. (A) Results for BLAST analysis of the 17 representative tet(X3)-carrying plasmids versus 243 tet(X3)-positive genomes. The heat map shows the percentages of aligned bases between pairs of tet(X3)-positive plasmids and genomes. (B) Conservation of reference plasmid genes among 243 genome sequences of tet(X3)-carrying Acinetobacter spp. The frequency of each gene in the reference plasmid is shown in circularized heatmaps. Genes are ordered according to the sequence of the corresponding reference plasmid. The mean coverage (%) of the reference plasmid sequence is indicated for each plasmid.
DISCUSSION
Recently identified plasmid-borne tet(X) genes causing the horizontal transfer of tigecycline resistance have significantly compromised the treatment effectiveness of tigecycline and, thus, have aroused considerable concern. A set of surveillance studies revealed the wide range of ecosystems in which tet(X) genes can be found, including soil, sewage, animals, hospitals, livestock farms, and the human gut (14–16, 19). tet(X)-positive isolates are especially prevalent in livestock and poultry, such as pigs, cows, and chickens, and less so in shrimp, migratory birds, and waterfowl (7, 16, 18, 19, 22, 28–30). Understanding the distribution and transmission of tet(X) genes in the context of One Health is imperative to efficiently control their further dissemination. In this study, we isolated tet(X)-positive Acinetobacter spp. from livestock and their surrounding environmental sources and comprehensively investigated their population structures and genetic characterizations.
According to our surveillance data, 23 tet(X)-positive isolates were recovered from 2 different swine farms but not from dairy farms or sheep farms. A. towneri was the most prevalent species carrying tet(X) genes in Acinetobacter spp., with tet(X3) and tet(X6) being the prevalent alleles (Table 1). A similar finding that tet(X3)-positive Acinetobacter species isolates were exclusively detected in intensive pig farms in China has been reported recently (20). These results suggest that the risk of dissemination of tet(X) genes to humans from pigs could be much higher than the risk of dissemination from other kinds of livestock.
Acinetobacter spp. are ubiquitous in the natural environment, and some of them, e.g., A. baumannii, A. indicus, and A. lwoffii, have become important opportunistic pathogens in clinical settings. Our and other studies showed that Acinetobacter spp. was the major reservoir of tigecycline-resistant tet(X) genes (17, 20, 22). Through searching tet(X) genes in GenBank, we found that A. indicus, Acinetobacter sp002018365, and A. towneri were the prevalent species carrying tet(X3) and tet(X6). Likewise, a national surveillance of tet(X)-positive Acinetobacter isolates from humans, animals, and their surrounding environments conducted between 2015 and 2018 showed that, after a novel species of Acinetobacter, A. towneri and A. indicus were the major hosts of tet(X3), tet(X4), and tet(X5) (20). Notably, most of the tet(X)-positive Acinetobacter isolates were livestock associated, raising concerns that the tigecycline-resistant tet(X) genes could be transmitted to humans from livestock via opportunistic pathogens of Acinetobacter. Our analysis showed that most of the tet(X)-positive Acinetobacter isolates disseminated sporadically; however, few interregional transmission events were detected here, highlighting the need for controlling the dissemination of tet(X3)- and tet(X6)-positive Acinetobacter species isolates.
Although numerous tet(X) genes have been continuously identified either on chromosomes or on plasmids in various bacterial species, the major vectors of tigecycline-resistant tet(X) genes remain unclear. Pioneering studies have shown the importance of the ISCR2-mediated tet(X) transposition structure (7, 17). The rolling-circle transposition has been experimentally confirmed by using the ΔtpnF-tet(X3)-hp-hp-ISCR2 cassette clone, and inverse PCR assays identified ISCR2-xerD-tet(X3)-res-ORF1 and ISCR2-ORF2-abh-tet(X4) minicircles in different studies (7, 20). In our study, ISCR2 was found upstream or downstream from tet(X3) and tet(X6) genes. Albeit we did not test the transferability of the ISCR2-mediated tet(X) transposition structure, the genetic contexts of tet(X3) carried by 249 genomes of Acinetobacter species were comprehensively compared. The proportion of the structure ISCR2-xerD-tet(X3)-res-ISCR2 might be up to 86.35% (215/249), implying a critical role of ISCR2 in the dissemination of tet(X3).
Of note, we found that a tet(X3)-encoding plasmid, pAT181, was self-transmissible from A. towneri to A. baumannii and conferred tetracycline resistance to the recipient. Currently, very few studies have identified self-transmissible plasmids carrying tet(X) genes. Chen et al. reported the conjugability of a tet(X3)- and tet(X5.3)-harboring plasmid, pYH12207-2, from Acinetobacter piscicola to A. baylyi strain ADP1 and the conjugability of a tet(X3)-harboring plasmid, p10FS3-1-3, from a novel Acinetobacter species to A. baylyi ADP1. However, these two plasmids did not enhance the resistance to tetracyclines in the recipient strain (20), which is different from our findings. Concerningly, pAT181, with a relatively high transfer frequency (10−6), did not impose a fitness cost but increased the growth rate of the recipient. It is suggested that successful dissemination of resistance plasmids largely depends on the fitness cost imposed on hosts (31). No fitness cost imposed on hosts by obtaining pAT181-like plasmids would greatly facilitate their spread, and thus, might contribute to the propagation of the tet(X3) gene in the future. Additionally, although no epidemic plasmids carrying tet(X3) have been detected currently, several plasmids were found to be circulating in a small proportion of strains. These plasmids could become epidemic after transmitting to other hosts in the future.
Conclusions.
Our study provides evidence that the predominant tet(X) alleles, tet(X3) and tet(X6), disseminate sporadically in Acinetobacter populations. Currently, the dissemination of tet(X3) and tet(X6) is mainly limited to livestock-associated sites. Continuous surveillance for tet(X) genes in the context of One Health is necessary to prevent them from transmitting to humans.
MATERIALS AND METHODS
Screenings of tet(X)-positive Acinetobacter strains.
Five hundred thirty-four nonrepetitive fecal samples were collected from 6 livestock farms located in Zhejiang Province in 2019, including 2 swine farms, 2 dairy farms, and 2 sheep farms. In addition, environmental samples were collected from soil (n = 72) and water (n = 78) surrounding the farms. All the samples were initially enriched in LB medium (5 g/liter yeast extract, 10 g/liter tryptone, 10 g/liter NaCl) for 6 h and spread on CHROMagar Acinetobacter medium plates (CHROMagar, Paris, France) to recover Acinetobacter species isolates. PCR screens of tet(X) alleles were performed as previously described (26).
Antimicrobial susceptibility testing (AST).
The MICs for all the tet(X)-positive isolates were determined using the broth microdilution method according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (32). The tested drugs included tigecycline, tetracycline, eravacycline, minocycline, doxycycline, demeclocycline, chlortetracycline, oxytetracycline, colistin, cefoperazone-sulbactam, trimethoprim-sulfamethoxazole, gentamicin, amikacin, levofloxacin, ciprofloxacin, meropenem, cefepime, ceftriaxone, and ceftazidime. The resistance breakpoint for tetracycline was defined as ≥16 mg/liter for Acinetobacter spp., Enterobacteriaceae, and non-Enterobacteriaceae according to CLSI (32). The breakpoint for tigecycline and eravacycline was delineated as >0.5 mg/liter for Enterobacteriaceae according to EUCAST V10 (33). E. coli strain ATCC 25922 was used as the quality control strain.
WGS and bioinformatic analysis.
Genomic DNAs of the tet(X)-positive isolates were extracted using the Puregene yeast/bact. kit B (Qiagen, Gaithersburg, MD) according to the instructions of the manufacturer and were sequenced by using the HiSeq 4000 system (Illumina, San Diego, United States). The isolates were taxonomically assigned using GTDB-Tk (version 1.3.0) with the Genome Taxonomy Database (release 95) (34). The sequence similarities of tet(X)-harboring plasmids were analyzed using BRIG version 0.95 (35). Representative strains with various genetic contexts of tet(X) genes were selected to be further sequenced using the PromethION platform (Nanopore, Oxford, UK). Hybrid assembly of short-read and long-read sequencing data was performed using Unicycler version 0.4.8 (36).
Phylogenetic analysis was performed using Parsnp version 1.2 (37), and the numbers of single-nucleotide polymorphisms (SNPs) among the core genomes were determined by using MEGA X (38). Functional annotation was performed using the RAST server (39). Antibiotic resistance genes (ARGs) were identified using ResFinder 4.0 (40) and CARD (https://card.mcmaster.ca/) with a threshold of nucleotide identity of >90% and coverage of >90%. Synteny analysis was performed using Easyfig (41).
Compilation of genomic data set and plasmidome analysis.
All assembled genomes of Acinetobacter spp. (n = 10,680) deposited in GenBank (as of 31 May 2021) were downloaded to search for tet(X) genes. The 15 tet(X) alleles were queried in these genomes by BLAST comparison to their nucleotide sequences, using 99% identity and 100% coverage as the cutoff (42).
Conservation of reference plasmid genes was calculated as previously described (43). Briefly, the RedDog pipeline (https://github.com/katholt/RedDog) was used to simulate 100-bp reads from tet(X3)-carrying genomes. To calculate the coverage of each representative plasmid in each genome, those 100-bp reads were mapped against representative tet(X3)-harboring plasmids by using Bowtie2 version 2.2.9 (44). The proportion of tet(X3)-carrying genomes containing annotated genes of each reference plasmid was calculated according to the gene presence/absence table reported by RedDog (at least five reads covering ≥95% of the length of the gene was defined as presence), and the results were plotted as circular heatmaps using ggplot2 in R (geom_tile for heatmap grid and coord_polar for circularization).
Pairwise sequence comparison of circularized plasmids was performed as previously described (45). Briefly, the lengths of nucleotide sequences that could be aligned between pairs of plasmids and the numbers of SNPs among the aligned regions were determined by using NUCmer version 3.1 (46) from the MUMmer package. The percentages of aligned bases between pairs of complete plasmids were shown in a heatmap generated by the “gplots” package (version 3.1.1) in R version 4.0.5 (https://www.r-project.org/).
Conjugation assay.
The transmissibility of tet(X3) and tet(X6) was evaluated by a conjugation assay. Briefly, a donor tet(X)-carrying Acinetobacter isolate (AT181) was mixed with the rifampicin-resistant A. baumannii strain ATCC 17978 or rifampicin-resistant E. coli strain EC600 as a recipient strain at a ratio of 1:1 by conjugational mating at 37°C without shaking overnight. The transconjugants were selected on LB agar plates containing rifampicin (600 mg/liter) and tigecycline (2 mg/liter). The species of all putative transconjugants were verified by using matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (Hexin, Guangzhou, China). PCR verifications of tet(X) genes were performed for the putative transconjugants for which the species was confirmed as A. baumannii or E. coli. Transfer frequency was calculated as the number of transconjugants obtained per donor. The growth of the donor strain and transconjugants was measured by determining the optical density at 600 nm (OD600) every 30 min. The assay was in triplicate.
Plasmid stability testing.
Plasmid stability was estimated according to the method of a previous study with minor modifications (47). Transconjugants were cultured in antibiotic-free LB broth at 37°C for 24 h. The 24-h cultures were diluted at a ratio of 1:100 in fresh LB medium. These freshly inoculated cultures constituted time point zero, and cultures were grown at 37°C in a shaking bath (200 rpm) and serially passaged for 10 days (approximately 200 generations). Cultures were diluted and plated onto antibiotic-free LB plates every 24 h. The colonies growing on antibiotic-free LB agar plates were randomly selected (∼50 colonies per day) for tet(X)-specific PCRs to determine the proportion of tet(X)-positive bacteria in each population. Plasmids were considered stable when the retention rates were still over 80% at the end of the experiment. The plasmid stability was evaluated in triplicate.
Statistical analysis.
The unpaired t test was performed to compare the number of ARGs in the tet(X6)-carrying clone and the tet(X3)-carrying clone, and statistical significance was taken as a P value of <0.05.
Availability of data.
The genome sequences of tet(X)-positive strains have been submitted to GenBank under BioProject accession number PRJNA631342, and the accession number of each genome is listed in Table 1.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Program of China (grant number 2017YFC1200200); the National Natural Science Foundation of China (grants number 81902029 and 82172330); Shenzhen Basic Research Key projects (grant number JCYJ20200109144220704), and Shenzhen Basic Research projects (grants number JCYJ20190807144409307 and JCYJ20190807150401657).
K.Z., Y.-Y.C., Y.-H.X., and R.-C.C. designed the study. Y.-Y.C., Y.C., and F.-M.H. collected the data. Y.-Y.C. and Y.L. analyzed and interpreted the data. Y.-Y.C. and K.Z. wrote and revised the manuscript. All authors reviewed, revised, and approved the final report.
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Kai Zhou, Email: Kai_Zhou@zju.edu.cn.
Tim Downing, Dublin City University.
Miguel Cevallos, Centro de Ciencias Genómicas, UNAM.
Dayananda siddavattam, University of HYderabad.
Nabil Karah, Umeå University.
REFERENCES
- 1.Wu W, Feng Y, Tang G, Qiao F, McNally A, Zong Z. 2019. NDM metallo-β-lactamases and their bacterial producers in health care settings. Clin Microbiol Rev 32:e00115-18. doi: 10.1128/CMR.00115-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nang SC, Li J, Velkov T. 2019. The rise and spread of mcr plasmid-mediated polymyxin resistance. Crit Rev Microbiol 45:131–161. doi: 10.1080/1040841X.2018.1492902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang C, Feng Y, Liu L, Wei L, Kang M, Zong Z. 2020. Identification of novel mobile colistin resistance gene mcr-10. Emerg Microbes Infect 9:508–516. doi: 10.1080/22221751.2020.1732231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doan TL, Fung HB, Mehta D, Riska PF. 2006. Tigecycline: a glycylcycline antimicrobial agent. Clin Ther 28:1079–1106. doi: 10.1016/j.clinthera.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Wang Y, Zhou Y, Li J, Yin W, Wang S, Zhang S, Shen J, Shen Z, Wang Y. 2018. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect 7:122. doi: 10.1038/s41426-018-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang H, Dong N, Shu L, Lu J, Sun Q, Chan EW, Chen S, Zhang R. 2020. Colistin-resistance gene mcr in clinical carbapenem-resistant Enterobacteriaceae strains in China, 2014–2019. Emerg Microbes Infect 9:237–245. doi: 10.1080/22221751.2020.1717380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He T, Wang R, Liu D, Walsh TR, Zhang R, Lv Y, Ke Y, Ji Q, Wei R, Liu Z, Shen Y, Wang G, Sun L, Lei L, Lv Z, Li Y, Pang M, Wang L, Sun Q, Fu Y, Song H, Hao Y, Shen Z, Wang S, Chen G, Wu C, Shen J, Wang Y. 2019. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat Microbiol 4:1450–1456. doi: 10.1038/s41564-019-0445-2. [DOI] [PubMed] [Google Scholar]
- 8.Fang LX, Chen C, Cui CY, Li XP, Zhang Y, Liao XP, Sun J, Liu YH. 2020. Emerging high-level tigecycline resistance: novel tetracycline destructases spread via the mobile tet(X). Bioessays 42:e2000014. doi: 10.1002/bies.202000014. [DOI] [PubMed] [Google Scholar]
- 9.Speer BS, Bedzyk L, Salyers AA. 1991. Evidence that a novel tetracycline resistance gene found on two Bacteroides transposons encodes an NADP-requiring oxidoreductase. J Bacteriol 173:176–183. doi: 10.1128/jb.173.1.176-183.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng Y, Chen Y, Liu Y, Guo Y, Zhou Y, Xiao T, Zhang S, Xu H, Chen Y, Shan T, Xiao Y, Zhou K. 2020. Identification of novel tetracycline resistance gene tet(X14) and its co-occurrence with tet(X2) in a tigecycline-resistant and colistin-resistant Empedobacter stercoris. Emerg Microbes Infect 9:1843–1852. doi: 10.1080/22221751.2020.1803769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang R, Dong N, Shen Z, Zeng Y, Lu J, Liu C, Zhou H, Hu Y, Sun Q, Cheng Q, Shu L, Cai J, Chan EW, Chen G, Chen S. 2020. Epidemiological and phylogenetic analysis reveals Flavobacteriaceae as potential ancestral source of tigecycline resistance gene tet(X). Nat Commun 11:4648. doi: 10.1038/s41467-020-18475-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasparrini AJ, Markley JL, Kumar H, Wang B, Fang L, Irum S, Symister CT, Wallace M, Burnham CD, Andleeb S, Tolia NH, Wencewicz TA, Dantas G. 2020. Tetracycline-inactivating enzymes from environmental, human commensal, and pathogenic bacteria cause broad-spectrum tetracycline resistance. Commun Biol 3:241. doi: 10.1038/s42003-020-0966-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee YR, Burton CE. 2019. Eravacycline, a newly approved fluorocycline. Eur J Clin Microbiol Infect Dis 38:1787–1794. doi: 10.1007/s10096-019-03590-3. [DOI] [PubMed] [Google Scholar]
- 14.Sun C, Cui M, Zhang S, Wang H, Song L, Zhang C, Zhao Q, Liu D, Wang Y, Shen J, Xu S, Wu C. 2019. Plasmid-mediated tigecycline-resistant gene tet(X4) in Escherichia coli from food-producing animals, China, 2008–2018. Emerg Microbes Infect 8:1524–1527. doi: 10.1080/22221751.2019.1678367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li R, Peng K, Li Y, Liu Y, Wang Z. 2020. Exploring tet(X)-bearing tigecycline-resistant bacteria of swine farming environments. Sci Total Environ 733:139306. doi: 10.1016/j.scitotenv.2020.139306. [DOI] [PubMed] [Google Scholar]
- 16.Zhang R, Dong N, Zeng Y, Shen Z, Lu J, Liu C, Huang ZA, Sun Q, Cheng Q, Shu L, Cai J, Chan EW, Liu D, Chen G, Wang Y, Chen S. 2020. Chromosomal and plasmid-borne tigecycline resistance genes tet(X3) and tet(X4) in dairy cows on a Chinese farm. Antimicrob Agents Chemother 64:e00674-20. doi: 10.1128/AAC.00674-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C, Cui CY, Wu XT, Fang LX, He Q, He B, Long TF, Liao XP, Chen L, Liu YH, Sun J. 2021. Spread of tet(X5) and tet(X6) genes in multidrug-resistant Acinetobacter baumannii strains of animal origin. Vet Microbiol 253:108954. doi: 10.1016/j.vetmic.2020.108954. [DOI] [PubMed] [Google Scholar]
- 18.Chen C, Cui CY, Zhang Y, He Q, Wu XT, Li G, Liao XP, Kreiswirth BN, Liu YH, Chen L, Sun J. 2019. Emergence of mobile tigecycline resistance mechanism in Escherichia coli strains from migratory birds in China. Emerg Microbes Infect 8:1219–1222. doi: 10.1080/22221751.2019.1653795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun J, Chen C, Cui CY, Zhang Y, Liu X, Cui ZH, Ma XY, Feng Y, Fang LX, Lian XL, Zhang RM, Tang YZ, Zhang KX, Liu HM, Zhuang ZH, Zhou SD, Lv JN, Du H, Huang B, Yu FY, Mathema B, Kreiswirth BN, Liao XP, Chen L, Liu YH. 2019. Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat Microbiol 4:1457–1464. doi: 10.1038/s41564-019-0496-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen C, Cui CY, Yu JJ, He Q, Wu XT, He YZ, Cui ZH, Li C, Jia QL, Shen XG, Sun RY, Wang XR, Wang MG, Tang T, Zhang Y, Liao XP, Kreiswirth BN, Zhou SD, Huang B, Du H, Sun J, Chen L, Liu YH. 2020. Genetic diversity and characteristics of high-level tigecycline resistance Tet(X) in Acinetobacter species. Genome Med 12:111. doi: 10.1186/s13073-020-00807-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu D, Zhai W, Song H, Fu Y, Schwarz S, He T, Bai L, Wang Y, Walsh TR, Shen J. 2020. Identification of the novel tigecycline resistance gene tet(X6) and its variants in Myroides, Acinetobacter and Proteus of food animal origin. J Antimicrob Chemother 75:1428–1431. doi: 10.1093/jac/dkaa037. [DOI] [PubMed] [Google Scholar]
- 22.Wang L, Liu D, Lv Y, Cui L, Li Y, Li T, Song H, Hao Y, Shen J, Wang Y, Walsh TR. 2019. Novel plasmid-mediated tet(X5) gene conferring resistance to tigecycline, eravacycline, and omadacycline in a clinical Acinetobacter baumannii isolate. Antimicrob Agents Chemother 64:e01326-19. doi: 10.1128/AAC.01326-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui CY, Chen C, Liu BT, He Q, Wu XT, Sun RY, Zhang Y, Cui ZH, Guo WY, Jia QL, Li C, Kreiswirth BN, Liao XP, Chen L, Liu YH, Sun J. 2020. Co-occurrence of plasmid-mediated tigecycline and carbapenem resistance in Acinetobacter spp. from waterfowls and their neighboring environment. Antimicrob Agents Chemother 64:e02502-19. doi: 10.1128/AAC.02502-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin L, Ling BD, Li XZ. 2009. Distribution of the multidrug efflux pump genes, adeABC, adeDE and adeIJK, and class 1 integron genes in multiple-antimicrobial-resistant clinical isolates of Acinetobacter baumannii-Acinetobacter calcoaceticus complex. Int J Antimicrob Agents 33:27–32. doi: 10.1016/j.ijantimicag.2008.06.027. [DOI] [PubMed] [Google Scholar]
- 25.Hu SH, Yuan SX, Qu H, Jiang T, Zhou YJ, Wang MX, Ming DS. 2016. Antibiotic resistance mechanisms of Myroides sp. J Zhejiang Univ Sci B 17:188–199. doi: 10.1631/jzus.B1500068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng Y, Chen Y, Liu Y, Song J, Chen Y, Shan T, Xiao Y, Zhou K. 2021. Detection of a new tet(X6)-encoding plasmid in Acinetobacter towneri. J Glob Antimicrob Resist 25:132–136. doi: 10.1016/j.jgar.2021.03.004. [DOI] [PubMed] [Google Scholar]
- 27.He D, Wang L, Zhao S, Liu L, Liu J, Hu G, Pan Y. 2020. A novel tigecycline resistance gene, tet(X6), on an SXT/R391 integrative and conjugative element in a Proteus genomospecies 6 isolate of retail meat origin. J Antimicrob Chemother 75:1159–1164. doi: 10.1093/jac/dkaa012. [DOI] [PubMed] [Google Scholar]
- 28.Bai L, Du P, Du Y, Sun H, Zhang P, Wan Y, Lin Q, Fanning S, Cui S, Wu Y. 2019. Detection of plasmid-mediated tigecycline-resistant gene tet(X4) in Escherichia coli from pork, Sichuan and Shandong Provinces, China, February 2019. Euro Surveill 24:1900340. doi: 10.2807/1560-7917.ES.2019.24.25.1900340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen C, Chen L, Zhang Y, Cui CY, Wu XT, He Q, Liao XP, Liu YH, Sun J. 2019. Detection of chromosome-mediated tet(X4)-carrying Aeromonas caviae in a sewage sample from a chicken farm. J Antimicrob Chemother 74:3628–3630. doi: 10.1093/jac/dkz387. [DOI] [PubMed] [Google Scholar]
- 30.Li R, Liu Z, Peng K, Liu Y, Xiao X, Wang Z. 2019. Co-occurrence of two tet(X) variants in an Empedobacter brevis of shrimp origin. Antimicrob Agents Chemother 63:e01636-19. doi: 10.1128/AAC.01636-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andersson DI, Hughes D. 2010. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol 8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 32.CLSI. 2019. Performance standards for antimicrobial susceptibility testing, 29th ed. CLSI supplement M100. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 33.EUCAST. 2020. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters, version 10.0. http://www.eucast.org/clinical_breakpoints/.
- 34.Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH. 2019. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36:1925–1927. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hall RM, Schwarz S. 2016. Resistance gene naming and numbering: is it a new gene or not? J Antimicrob Chemother 71:569–571. doi: 10.1093/jac/dkv351. [DOI] [PubMed] [Google Scholar]
- 43.Lam MMC, Wyres KL, Judd LM, Wick RR, Jenney A, Brisse S, Holt KE. 2018. Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae. Genome Med 10:77. doi: 10.1186/s13073-018-0587-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.David S, Cohen V, Reuter S, Sheppard AE, Giani T, Parkhill J, Rossolini GM, Feil EJ, Grundmann H, Aanensen DM, European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) Working Group, ESCMID Study Group for Epidemiological Markers (ESGEM). 2020. Integrated chromosomal and plasmid sequence analyses reveal diverse modes of carbapenemase gene spread among Klebsiella pneumoniae. Proc Natl Acad Sci USA 117:25043–25054. doi: 10.1073/pnas.2003407117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He T, Wei R, Zhang L, Sun L, Pang M, Wang R, Wang Y. 2017. Characterization of NDM-5-positive extensively resistant Escherichia coli isolates from dairy cows. Vet Microbiol 207:153–158. doi: 10.1016/j.vetmic.2017.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM01141-21_Supp_1_seq12.pdf, PDF file, 6.5 MB (6.6MB, pdf)
Supplemental material. Download SPECTRUM01141-21_Supp_2_seq13.xlsx, XLSX file, 0.3 MB (295.3KB, xlsx)