Abstract
Over the past several decades, RNA modifications have rapidly emerged as an indispensable topic in epitranscriptomics. N6-methyladenosine (m6A), namely, methylation at the sixth position of an adenine base in an RNA molecule, is the most prevalent RNA modification in both coding and noncoding RNAs. m6A has emerged as a crucial posttranscriptional regulator involved in both physiological and pathological processes. Based on accumulating evidence, m6A participates in the pathogenesis of immune-related diseases by regulating both innate and adaptive immune cells through various mechanisms. Autoimmune diseases are caused by a self-destructive immune response in the setting of genetic and environmental factors, and recent studies have discovered that m6A may play an essential role in the development of autoimmune diseases. In this review, we focus on the important role of m6A modification in biological functions and highlight its contributions to immune cells and the development of autoimmune diseases, thereby providing promising epitranscriptomic targets for preventing and treating autoimmune disorders.
Keywords: RNA modifications, N6-methyladenosine, autoimmune disorders, innate immunity, adaptive immunity
Introduction
Epigenetics, a link between genetic factors and environmental factors, refers to heritable modifications that regulate gene expression in the absence of nucleotide sequence alterations. Classical epigenetic mechanisms comprise DNA modifications, histone modifications and noncoding RNAs (ncRNAs). Over the past several decades, RNA modifications have emerged as new epitranscriptomic modifications, enriching the regulatory mechanisms of gene expression and providing novel insights into and strategies for exploring the underlying pathogenesis of diseases. N6-methyladenosine (m6A), the most abundant and widespread RNA modification, has been identified in coding RNAs (messenger RNAs, mRNAs) and ncRNAs, including transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), microRNAs (miRNAs), small nuclear RNAs (snRNAs), long noncoding RNAs (lncRNAs) and circular RNAs (circRNAs) (Niu et al., 2013; Chen et al., 2019; Lence et al., 2019; Li et al., 2020a). m6A is highly conserved and is installed predominantly in specific regions near stop codons, in internal long exons and in 3′ untranslated regions (3′UTRs) (Dominissini et al., 2012; Meyer et al., 2012; Wang et al., 2020a). More specifically, m6A is preferentially installed at the consensus motif RR-m6A-CH (R = G/A; H = A/C/U)(Batista, 2017).
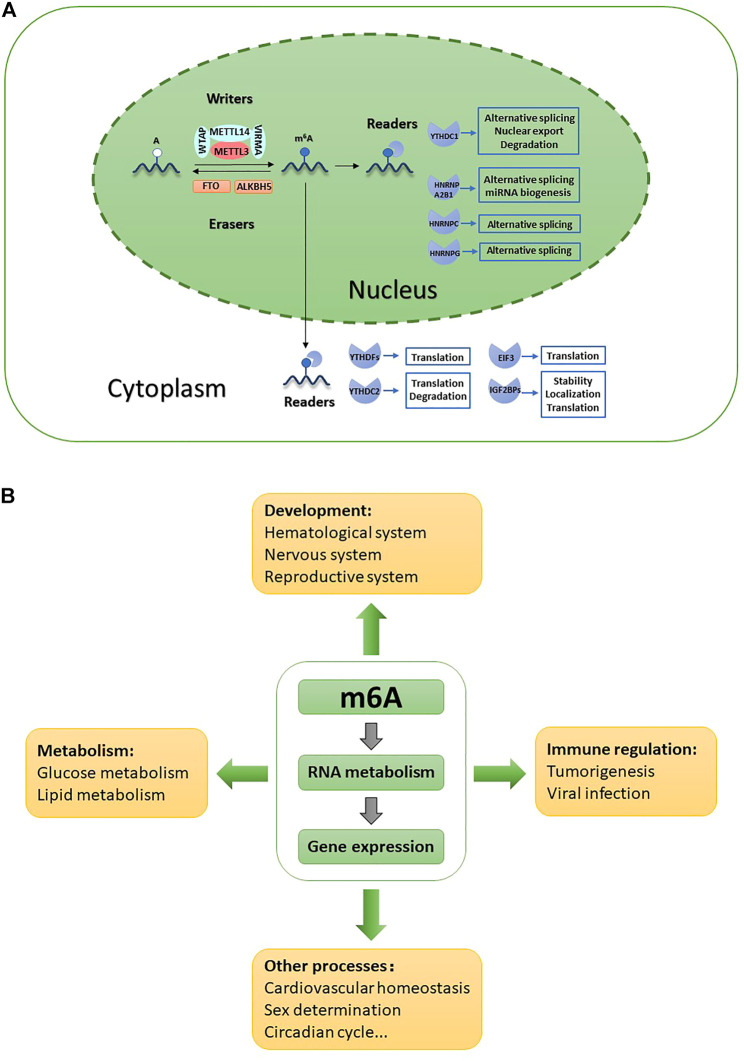
The regulatory proteins involved in m6A modification fall into three categories: “writers,” “erasers” and “readers” (Table 1; Figure 1A). m6A writers are methyltransferase complexes containing multiple subunits that install m6A cotranscriptionally at specific sites in target mRNAs. Methyltransferase-like 3 (METTL3), the only active catalytic component of the writer complex, exhibits catalytic activity independently and functions synergistically with METTL14 by forming a stable heterodimer. Many auxiliary subunits ensure efficient installation of m6A modification and determine the specificity of writers, including Wilms tumor 1-associated protein (WTAP), Vir-like m6A methyltransferase associated (VIRMA), and RNA binding motif protein 15/15B (RBM15/15B) (Shi et al., 2019). Erasers are RNA demethylases that remove the methyl group from m6A. The discovery of RNA demethylases suggested that m6A modification may be a reversible and dynamic process. Only two natural RNA demethylases have been identified to date, namely, fat mass and obesity-associated protein (FTO) and ALKB homolog 5 (ALKBH5), both of which belong to the ALKB family of proteins (Jia et al., 2011; Zheng et al., 2013). Recently, flavin mononucleotide (FMN) was identified as a novel artificial molecular demethylase (Xie et al., 2019). Readers are RNA binding proteins that mediate the fate of target transcripts and regulate downstream biological functions by preferentially recognizing and binding to modified sites (Batista, 2017; He et al., 2019; Zhang et al., 2019). Readers are classified as direct readers or indirect readers according to their interaction patterns with RNAs. Direct readers, such as YTH family members, selectively and directly bind to m6A sites, while indirect readers, such as heterogeneous nuclear ribonucleoprotein G (HNRNPG), indirectly bind to m6A sites based on the ‘‘m6A switch” mechanism (Dai et al., 2018; Zaccara et al., 2019).
TABLE 1.
Main functions of the three major groups of m6A-related proteins.
| Category | Proteins | Main functions | References |
|---|---|---|---|
| Writers | METTL3 | Catalyzes m6A formation | Tong et al. (2018a); Zhang et al. (2019) |
| METTL14 | Promotes the catalytic activity of METTL3 | Wang et al. (2016); Batista (2017); Tong et al. (2018a); Zaccara et al. (2019); Zhang et al. (2019) | |
| WTAP | Ensures proper localization of the METTL3-METTL14 complex | Ping et al. (2014); Zhang et al. (2019) | |
| Recruits target RNAs and other functional factors | Zhang et al. (2019) | ||
| VIRMA | Mediates m6A deposition at specific regions of target mRNAs | Yue et al. (2018) | |
| RBM15/15B | Recruits the writer complex to specific regions | Patil et al. (2016) | |
| Modulates m6A deposition on XIST | Patil et al. (2016) | ||
| METTL16 | Installs m6A on snRNAs and mRNAs to regulate SAM homeostasis | Pendleton et al. (2017) | |
| CBLL1 | Interacts with WTAP | Růžička et al. (2017) | |
| ZC3H13 | Facilitates the nuclear localization of the writer complex | Knuckles et al. (2018); Wen et al. (2018) | |
| ZCCHC4 | Installs a single m6A modification on rRNA | Pinto et al. (2020) | |
| Modulates mRNA translation | Ma et al. (2019) | ||
| Erasers | FTO | Removes m6A | Jia et al. (2011) |
| Removes m6Am | Mauer et al. (2019) | ||
| Demethylates other sites in certain snRNAs and specific tRNAs | Wang et al. (2014); Mauer et al. (2019) | ||
| ALKBH5 | Removes m6A | Zheng et al. (2013) | |
| Targets ncRNAs separately from mRNAs | Wang et al. (2014) | ||
| Participates in germ cell development | Zheng et al. (2013) | ||
| Modulates RNA metabolism and nuclear RNA export | Zheng et al. (2013); Zhang et al. (2017a) | ||
| FMN | A newly identified artificial molecular demethylase | Xie et al. (2019) | |
| Readers | YTHDF1 | Promotes the efficient translation of target mRNAs | Meyer et al. (2015); Wang et al. (2015) |
| YTHDF2 | Accelerates RNA degradation and inhibits the translation of mRNAs | Meyer et al. (2015); Wang et al. (2015) | |
| YTHDF3 | Promotes RNA translation with YTHDF1 | Shi et al. (2017) | |
| Inhibits the translation of mRNAs with YTHDF2 | Shi et al. (2017) | ||
| YTHDC1 | Modulates alternative RNA splicing | Xiao et al. (2016) | |
| Regulates nuclear export | Xiao et al. (2016) | ||
| Accelerates the decay of transcripts to maintain SAM levels | Shima et al. (2017) | ||
| Suppresses gene expression involving X chromosome inactivation | Patil et al. (2016) | ||
| YTHDC2 | Promotes the efficient translation of its target transcripts | Kretschmer et al. (2018) | |
| Mediates the subsequent degradation of its target transcripts | Kretschmer et al. (2018) | ||
| IGF2BPs | Regulates the stability, localization and translation of target RNAs | Degrauwe et al. (2016); Huang et al. (2018) | |
| EIF3 | Initiates and promotes cap-independent translation | Meyer et al. (2015) | |
| HNRNPA2B1 | Modulates alternative splicing | Alarcón et al. (2015a) | |
| Promotes mature miRNA biogenesis | Alarcón et al. (2015a); Coker et al. (2019) | ||
| HNRNPC | Involved in premRNA processing, including splicing | Liu et al. (2015) | |
| HNRNPG | Involved in premRNA processing, including splicing | Liu et al. (2017) |
ALKBH5, ALKB homolog 5; CBLL1, Casitas B-lineage lymphoma-transforming sequence-like protein 1; FMN, flavin mononucleotide; FTO, fat mass and obesity-associated protein; HNRNPA2B1/C/G, heterogeneous nuclear ribonucleoprotein A2B1/C/G; METTL3/14/16, methyltransferase-like 3/14/16; IGF2BPs, insulin-like growth factor 2 binding proteins; RBM15/15B, RNA binding motif protein 15/15B; VIRMA, Vir-like m6A methyltransferase associated; WTAP, Wilms tumor 1-associated protein; YTHDF1/2/3, YTH domain-containing family 1/2/3, YTHDC1/2, YTH domain-containing 1/2; ZC3H13, zinc finger CCCH domain-containing protein 13; ZCCHC4, zinc finger CCHC-type containing 4.
FIGURE 1.
Role of m6A in various biological functions. (A) m6A is installed by writers, removed by erasers and recognized by nuclear readers and cystoplasmic readers. m6A is involved in all aspects of RNAs metabolism and activity. (B) m6A is involved in both physiological and pathological processes through modulating gene expression.
Autoimmune diseases result from a self-destructive immune response initiated by an impaired immune tolerance mechanism. This group of diseases imposes a substantial burden on health services, economic development and quality of life due to their slow progression, the difficulty in diagnosis because of their heterogeneous clinical manifestations and the numerous side effects occurring during immunosuppressive therapy (Anaya, 2012; Wardowska, 2021). However, the precise cellular and molecular mechanisms underlying autoimmune diseases have remained poorly understood until recently. Epitranscriptomic mechanisms have been widely recognized to play fundamental roles in the pathogenesis of immune-related diseases (Wu et al., 2017; Li et al., 2021a). Both innate and adaptive immunity clearly participate in the occurrence and progression of autoimmune diseases (Bluestone and Bour-Jordan, 2012; Wahren-Herlenius and Dörner, 2013). Numerous studies have recently characterized an essential role for m6A in many aspects of the immune system, including cell development, differentiation, activation, migration and function, indicating that m6A may contribute to the pathogenesis of autoimmune disorders.
In this review, we summarize the crucial role of the m6A modification in regulating cellular biological functions and highlight its contributions to the immune system and the development of autoimmune diseases, thereby providing novel insights into the pathogenesis of autoimmune disorders and potential targets for epitranscriptomic therapy.
Roles of m6A in Various Biological Functions
m6A modification is precisely regulated by writers, erasers and readers and is involved in all aspects of RNA metabolism; moreover, its effects are not limited to mRNAs (Figure 1A). Through regulating gene expression, m6A is involved in diverse biological process including development, metabolism, immunity regulation, sex determination, circadian rhythms, and cardiovascular system homeostasis (Hu et al., 2020; Gu et al., 2021) (Figure 1B).
Roles of m6A in RNA Metabolism and Activity
m6A and related proteins regulate almost all aspects of RNA metabolism and activity, thus modulating gene expression under physiological and pathological conditions. On the one hand, m6A is involved in the processing, alternative splicing, nuclear export, translation and degradation of mRNAs. FTO cooperates with METTL3 to regulate poly(A) sites and change the length of the 3′UTR (Bartosovic et al., 2017). In addition, FTO regulates alternative mRNA splicing not only by inhibiting the binding ability of serine and arginine-rich splicing factor (SRSF) 2 protein in an m6A-dependent manner but also by targeting m6Am during the biogenesis of snRNAs, which are integral spliceosome components and are involved in regulating premRNA splicing (Zhao et al., 2014; Mauer et al., 2019). YTH domain-containing 1 (YTHDC1) regulates alternative splicing by recruiting SRSF3 to promote exon inclusion and facilitates the binding of methylated mRNAs to nuclear export factor 1 (NXF1) to modulate their nuclear export (Xiao et al., 2016). YTH domain-containing family 1 (YTHDF1) promotes the efficient translation of target mRNAs in a cap-independent manner, particularly through its interaction with eukaryotic initiation factor 3 (eIF3), while YTHDF2 accelerates RNA degradation and inhibits protein translation by preferentially binding m6A in the 3’ UTR and then recruiting the CCR4-NOT complex (Meyer et al., 2015; Wang et al., 2015; Du et al., 2016).
On the other hand, m6A is involved in regulating ncRNA metabolism and activity, including miRNA biogenesis, circRNA translation, and lncRNA stability and localization. With the assistance of METTL3 methylation activity, HNRNPA2B1 promotes mature miRNA biogenesis by recruiting DiGeorge syndrome chromosomal region (DGCR) 8 to primary miRNAs(Alarcón et al., 2015a; Alarcón et al., 2015b). METTL14 promotes the processing of pri-miR126 by directly recruiting DCGR8 (Ma et al., 2017). METTL3 has been reported to indirectly regulate miRNA expression and facilitate the translation initiation of circRNAs through an m6A-dependent mechanism (Karthiya and Khandelia, 2020). RBM15/15B alters the deposition of m6A on X-inactive specific transcript (XIST) by promoting the methylation of XIST, resulting in X-chromosome inactivation and gene silencing (Patil et al., 2016). METTL3 overexpression significantly increases the localization of the lncRNA RP11 in the nucleus, indicating that the localization of lncRNAs may also be regulated by m6A (Wu et al., 2019). ALKBH5 maintains the stability of the lncRNA GAS5-AS1, while YTHDF2/3 reduces its stability and accelerates lncRNA decay (Wang et al., 2019a; Ni et al., 2019). METTL16 introduces m6A into the U6 snRNA and regulates subsequent processing, thus regulating SAM homeostasis (Pendleton et al., 2017).
Role of m6A in the Development of Multiple Organs
The dynamic m6A modification precisely regulates mRNA translation and degradation during early development. METTL3 mutations lead to early developmental stagnation, defects in the transition from mother to zygote and even embryonic lethality (Gu et al., 2021). Recent studies have focused on the role of m6A in the development of three main systems: the hematological system, nervous system and reproductive system. Further investigations are needed to determine whether m6A affects other systems. METTL3 deficiency affects hematopoietic development by significantly inhibiting the transition from endothelial cells to hematopoietic stem cells (HSCs) (Zhang et al., 2017b). In vascular endothelial cells, METTL3 knockout inhibits the function of hematopoietic stem/progenitor cells (HSPCs), while METTL3 knockout in HSPCs promotes differentiation (Vu et al., 2017; Lv et al., 2018).
The development of the nervous system depends on the specific expression of m6A modulators in different regions, cell subtypes and developmental stages of the brain. YTHDF2 deficiency in the embryonic neocortex impairs the self-renewal of neural stem/progenitor cells and special patterns of brain cell generation, leading to a failure of neural development (Yoon et al., 2017). METTL3 overexpression leads to structural disorders in both Purkinje and glial cells, and low METTL3 expression results in severe developmental defects in the cerebellum, indicating that a delicate m6A balance is essential for normal development (Ma et al., 2018; Wang et al., 2018). FTO knockout inhibits the proliferation and neuronal differentiation of adult neural stem cells and suppresses the expression of several crucial proteins involved in the brain-derived neurotrophic factor pathway, indicating its essential role in regulating adult neurogenesis (Li et al., 2017a).
Gametogenesis, a key step in reproductive system development, is also regulated by m6A modifications at the posttranscriptional level. YTHDC1/2 is required for spermatogenesis and oogenesis. YTHDC1 deficiency alters the length of the 3′ UTR by causing extensive alternative polyadenylation and impairs alternative splicing by inhibiting factors associated with premRNA 3′ end processing (Kasowitz et al., 2018). These events block oocytes at the primary follicle stage and eventually result in defective oogenesis. YTHDC2 knockout in germ cells leads to a failure to develop past the zygotene stage, thus resulting in male and female infertility (Hsu et al., 2017).
Roles of m6A in Metabolism and Energy Homeostasis
m6A plays important roles in nutritional metabolism and energy balance, which are related to the pathogenesis of metabolism-related diseases, including type 2 diabetes and obesity (Gu et al., 2021). m6A activates glucose oxidation in rat adipocytes, suggesting that appropriate m6A levels may be essential for maintaining certain blood glucose concentrations (Souness et al., 1982). METTL3 knockout suppresses the expression of genes related to insulin secretion, thus inducing islet β-cell failure (Li et al., 2021b). METTL3 deficiency in mouse hepatocytes improves glucose tolerance and insulin sensitivity and decreases lipid accumulation (Li et al., 2020b). Acute deletion of METTL14 in β-cells reduces insulin secretion by increasing the activity of the IRE1a/sXBP-1 signaling pathway, finally leading to glucose intolerance (Men et al., 2019). Based on these results, METTL3/4 is essential for islet β-cell biology and maintaining the glucose balance. FTO is associated with carbohydrate and lipid metabolism and is involved in energy homeostasis. According to recent evidence, FTO participates in glucose metabolism through both m6A-dependent and nonm6A-dependent pathways (Wu et al., 2020). FTO also regulates adipogenesis by mediating the spicing of adipogenic regulatory factor RUNX1 translocation partner 1 (RUNX1T1), and AMPK positively regulates m6A levels in mRNAs to negatively regulate lipid accumulation in skeletal muscles (Zhao et al., 2014; Wu et al., 2017).
Role of m6A in Immune Regulation
m6A has been reported to exert dual effects on regulating the immune response during both tumorigenesis and viral infection (Li et al., 2021a; Gu et al., 2021). m6A inhibits or promotes cancer progression by exerting dual-directional regulatory effects on apoptosis, autophagy, angiogenesis and the epithelial-mesenchymal transition (EMT). m6A modulators inhibit apoptosis by increasing oncogene expression levels and inhibiting tumor suppressor expression. In contrast, m6A modulators promote apoptosis by inhibiting the expression of oncogenes and promoting the expression of tumor suppressors (Li et al., 2021a). FTO silencing decreases the expression of light chain 3B (LC3B), a membrane marker of autophagy, but increases the expression levels of autophagy substrates by demethylating the UNC-51-like kinase 1 (ULK1) mRNA (Jin et al., 2018). Conversely, FTO promotes autophagy by directly demethylating autophagy-related (ATG) 5 and ATG7 (Wang et al., 2020b). In hepatocellular carcinoma, METTL3 knockdown induces the expression of some angiogenic biomarkers and increases the formation of tubes, indicating that METTL3 inhibits angiogenesis (Lin et al., 2020). IGF2BP3 recognizes m6A sites catalyzed by METTL3 and promotes angiogenesis by increasing the stability of HDGF transcripts (Wang et al., 2020c). Moreover, METTL3 overexpression decreases the expression of vimentin, β-catenin and N-cadherin and increases E-cadherin accumulation in renal cell carcinoma cells, thus promoting the EMT. Accordingly, METTL3 loss resulted in opposite alterations (Li et al., 2017b). Moreover, modulators alter the tumor microenvironment by regulating the expression of regulatory factors, such as lysosomal cathepsins and the TLR4 adaptor protein TIRAP, thus affecting immune escape and cancer immunotherapy (Li et al., 2021a). Based on the mechanism described above, m6A plays dual regulatory roles in tumorigenesis. However, the precise mechanism of the m6A modification in tumorigenesis remains to be fully elucidated.
Similarly, m6A promotes and inhibits immunity against viruses by modulating the lifecycles of viruses and immune responses of hosts (Gu et al., 2021). METTL3 increases replication efficiency by increasing the SUMOylation and ubiquitination of RNA polymerase 3D and is recruited to replication sites of viral RNAs(Hao et al., 2019). Moreover, writer and eraser knockdown promotes and inhibits the infection rate of the hepatitis C virus by increasing or decreasing the production of infectious viral particles, respectively (Gokhale et al., 2016). High METTL14 expression maintains the stability of latent Epstein-Barr virus transcripts (Lang et al., 2019). In hosts, METTL3 regulates innate immunity and adaptive immunity, including macrophages, dendritic cells (DCs), to exert a regulatory effect on viral infection (Gu et al., 2021).
Role of m6A in Immune Cells
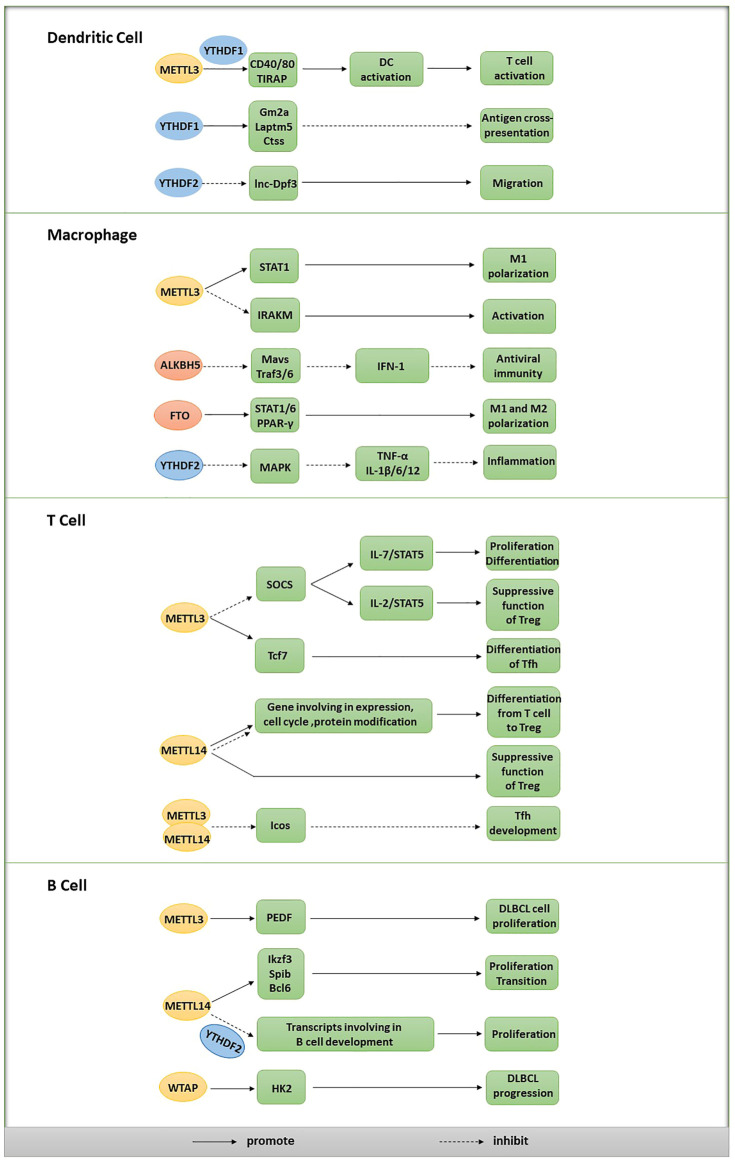
The precursor hematopoietic stem cells located in the bone marrow are the original source of all immune cells in the blood, lymph and immune organs. Immune cells are classified as innate immune cells and adaptive immune cells according to their functions and patterns in the immune response. Innate immune cells primarily include DCs, macrophages, granulocytes and mast cells that respond rapidly to antigenic stimuli. Adaptive immune cells primarily include T lymphocytes (CD4+ T cells and CD8+ T cells) and B lymphocytes that initially exhibit a delayed response but are involved in the formation of immunological memory to respond strongly and rapidly to repeated stimulation with the same antigen. In addition, other types of immune cells, such as natural killer (NK) cells and NK-T cells, are components of both immune systems (McComb et al., 2019). Based on accumulating evidence, m6A is required for many processes in immune cells, including development, differentiation, activation, migration and polarization, thereby modulating the immune response (Figure 2).
FIGURE 2.
Effects of m6A on different types of immune cells.
Role of m6A in DCs
Antigens are efficiently phagocytosed, processed and presented by DCs, leading to the activation of T cells and initiation of the immune response. METTL3, the catalytic subunit of the writer complex, is essential for the maturation and functional activation of DCs. METTL3 in DCs promotes T cell activation by catalyzing the formation of m6A in signaling molecule transcripts, including CD40, CD80 and TLR4 signaling adaptor (TIRAP). Then, these m6A-modified transcripts are recognized by YTHDF1 to increase their downstream translation, thus promoting DC activation and subsequent T cell responses (Wang et al., 2019b). On the other hand, YTHDF1 recognizes m6A-modified transcripts encoding lysosomal proteases and promotes their translation, thus limiting antigen cross-presentation by degrading protein antigens (Han et al., 2019). Therefore, YTHDF1 deficiency in DCs enhances antitumor immunity by promoting the cross-presentation of tumor antigens and cross-priming of CD8+ T cells, indicating that YTHDF1 is likely to become a promising antitumor target (Karthiya and Khandelia, 2020). CCR7 induces lnc-Dpf3, a lncRNA, to bind HIF-1α and suppress HIF-1-dependent transcription of glycolytic genes, thus suppressing CCR7-dependent DC migration. YTHDF2 recognizes m6A-modified lnc-Dpf3 and accelerates its degradation, which may exacerbate the inflammatory response and disrupt immune homeostasis by promoting DC migration (Liu et al., 2019a).
Role of m6A in Macrophages
Macrophages perform various functions, including removing damaged, dead or dying cells and other debris, presenting antigens to cells and producing cytokines and other regulatory factors, similar to secretory cells, to modulate the immune response. Type I interferon (IFN-1) production is inhibited by DDX46, a DDX helicase, through its interaction with Mavs, Traf3 and Traf6 transcripts, which encode signaling molecules essential for IFN-1 production under viral stimulation. In infected macrophages, DDX46 recruits ALKBH5 through its DEAD helicase domain to catalyze the demethylation of these methylated transcripts, which leads to the retention of the unmodified transcripts in the nucleus, prevents their efficient translation, inhibits IFN-1 production and finally suppresses antiviral immunity (Zheng et al., 2017).
Macrophages can be polarized into the M1 and M2 phenotypes; M1 macrophages produce interferon γ (IFN-γ) to mediate proinflammatory activities, while M2 macrophages produce the cytokine interleukin-4 (IL-4) to mediate anti-inflammatory activities. The functional status of macrophages changes substantially with alterations between M1 and M2 polarization. The level of the METTL3 protein was reported to be specifically upregulated after M1 polarization of mouse macrophages. Furthermore, METTL3 directly methylates the mRNA encoding signal transducer and activator of transcription 1 (STAT1), an essential modulator of M1 polarization. Then, the stability of the methylated STAT1 mRNA is increased, and the STAT1 protein level is accordingly upregulated, thus driving M1 macrophage polarization. However, METTL3 deletion exerts opposing effects on macrophage polarization, decreasing M1 polarization but increasing M2 polarization and thus promoting an anti-inflammatory response (Liu et al., 2019b). In another study, FTO silencing induced the downregulation of STAT1 expression in M1-polarized macrophages and decreased the expression of STAT6 and PPAR-γ in M2-polarized macrophages. Specifically, FTO knockdown suppressed NF-κB signaling by downregulating the phosphorylation of related proteins and decreasing the mRNA stability of STAT1 and PPAR-γ through a mechanism dependent on the effect of YTHDF2, thereby preventing both M1 and M2 polarization of macrophages (Gu et al., 2020).
IRAKM is postulated to negatively regulate TLR4 signaling, which promotes macrophage activation. In another study, METTL3 deficiency led to impaired m6A formation in the IRAKM mRNA and slowed IRAKM degradation, ultimately inhibiting macrophage activation mediated by the suppression of TLR signaling (Tong et al., 2021). Upon lipopolysaccharide (LPS) stimulation, YTHDF2-deficient macrophages exhibited increased stability of the MAPK mRNA and its upregulated expression; the MAPK and NF-κB signaling pathways were subsequently activated to increase the expression levels of signaling molecules, including TNF-α, IL-1β, IL-6 and IL-12 (Liu et al., 2021).
Role of m6A in T Cells
CD4+ T cells and CD8+ T cells are the primary classes of T cells. CD8+ T cells (cytotoxic T lymphocytes, CTLs) secrete cytotoxic granules and perforin into the immune synapse to induce apoptosis in target cells, including infected cells and tumor cells. CD4+ T cells (helper T cells, Th) differentiate into different phenotypes upon stimulation with different cytokines, and the differentiated cells contribute to cellular immunity or humoral immunity by secreting different cytokines. CD4+ T cells are activated by IL-12 and IFN-γ stimulation to differentiate into Th1 cells and secrete IFN-γ and lymphotoxin-alpha (LT-α) to induce inflammation and support cellular immunity, while CD4+ T cells are activated by IL-4 stimulation to differentiate into Th2 cells and secrete IL-4 to support humoral immunity and antibody production (McComb et al., 2019). In summary, Th1 cells induce a proinflammatory response, while Th2 cells induce an anti-inflammatory response. As another Th cell subtype, Th-17 cells produce IL-17 and promote inflammation and autoimmunity, while regulatory T cells (Tregs) promote immune tolerance, maintain immune homeostasis and suppress autoimmunity (Bettelli et al., 2007; Sakaguchi et al., 2008; Wan, 2010).
SOCS family proteins compete with IL-7 for binding to the IL-7 receptor, resulting in a failure to activate STAT5 and downstream signals that are important for the differentiation and proliferation of naïve T cells. METTL3 deletion reduces the m6A level in the SOCS mRNA and decreases SOCS mRNA degradation, leading to increased SOCS mRNA and protein levels. Accordingly, upregulated SOCS family activity inhibits STAT5 activation mediated by IL-7, eventually preventing the normal proliferation and differentiation of T cells. Moreover, m6A may contribute to the induction of SOCS mRNA degradation upon IL-7 stimulation to promote proliferation and differentiation by reprogramming naïve T cells (Li et al., 2017c). Another study showed that the depletion of METTL3 in Tregs increased the mRNA levels of SOCS family genes, suppressed the IL-2/STAT5 signaling pathway and impaired the suppressive function of Tregs (Tong et al., 2018b). Based on these results, the m6A RNA modification regulates the differentiation of naïve T cells and sustains the suppressive functions of Tregs by specifically targeting the same family of genes in different T cell subtypes. Additionally, in a mouse model of colitis, METTL14 deficiency in T cells increases inflammatory cell infiltration, increases cytokine release from Th1 and Th17 cells and prevents the differentiation of naïve T cells into Tregs (Lu et al., 2020).
T follicular helper (Tfh) cells are critical for the formation of germinal centers (GCs) and effective humoral immunity. METTL3 has been suggested to play a key role in modulating the expression of important Tfh signature genes, including Tcf7 and Icos, which are related to the development and differentiation of Tfh cells. The m6A modification was reported to increase the stability of Tcf7 transcripts to promote Tfh cell differentiation programs in a METTL3-dependent manner (Yao et al., 2021), and the METTL3/METTL14 complex was shown to catalyze m6A installation on the Icos mRNA and subsequently cause GAPDH protein-induced suppression of Icos expression, thereby inhibiting Tfh cell development (Zhu et al., 2019).
Role of m6A in B Cells
B cells are the main cells mediating humoral immunity. B cells depend on their B cell receptors (BCRs) to recognize specific antigens and differentiate into plasma cells, which produce and secrete specific antibodies that bind to the target antigen (McComb et al., 2019).
According to recent studies, the m6A modification and its regulators may be involved in the early development and proliferation of B cells. For example, METTL14 deficiency dramatically decreases the m6A level in mRNAs and causes the aberrant expression of genes essential for B cell development, eventually inhibiting IL-7-induced pro-B cell proliferation and the transition from large pre-B cells to small pre-B cells (Zheng et al., 2020). Moreover, IL-7-induced pro-B cell proliferation depends on transcriptional suppression mediated by YTHDF2, while the failure to transition from large pre-B cells to small pre-B cells is independent of both YTHDF1 and YTHDF2 (Zheng et al., 2020). METTL3 is expressed at high levels in diffuse large B cell lymphoma (DLBCL) cell lines and patient tissues and increases the m6A levels in the pigment epithelium-derived factor (PEDF) transcript to promote DLBCL cell proliferation (Cheng et al., 2020). Similarly, WTAP upregulation induced by piRNA-30473 promotes DLBCL progression by increasing the m6A level in HK2 transcripts and the subsequent expression of HK2(Han et al., 2021).
Role of m6A in Autoimmune Diseases
The role of immune cells in the pathogenesis of autoimmune diseases has been extensively studied, and these cells have proven to be involved in the development of autoimmune disorders. Interestingly, some immune cells, including macrophages, T cells and NK cells, exert dual effects: disease promotion and disease prevention (Table 2). This dual function may be attributed to differences in subsets of immune cells, the tissue microenvironment, and stages of autoimmune disease and to interactions between immune cells. Recent evidence shows that m6A may be involved in the development of autoimmune diseases. Moreover, some studies strongly indicate that m6A regulates the functions of immune cells, thereby affecting autoimmune diseases.
TABLE 2.
Contribution of immune cells to the development of autoimmune diseases.
| Cell type | Contribution | Autoimmune disease | References |
|---|---|---|---|
| T cell (except Treg) | Promotion | Rheumatoid arthritis | Cope et al. (2007); Han et al. (2015) |
| Systemic lupus erythematosus | Han et al. (2015); Shao et al. (2020) | ||
| Sjogren’s syndrome | Han et al. (2015) | ||
| Inflammatory bowel disease | Han et al. (2015) | ||
| Multiple sclerosis | Han et al. (2015) | ||
| Type 1 diabetes | Shao et al. (2020) | ||
| Treg | Protection | Rheumatoid arthritis | Cope et al. (2007) |
| Systemic lupus erythematosus | Ohl and Tenbrock, (2015) | ||
| Sjogren’s syndrome | Alunno et al. (2015) | ||
| Multiple sclerosis | Danikowski et al. (2017) | ||
| Inflammatory bowel disease | Műzes et al. (2012) | ||
| Type 1 diabetes | ElEssawy and Li, (2015) | ||
| B cell | Promotion | Rheumatoid arthritis | Rubin et al. (2019) |
| Systemic lupus erythematosus | Rubin et al. (2019) | ||
| Sjogren’s syndrome | Rubin et al. (2019) | ||
| Type 1 diabetes | Rubin et al. (2019) | ||
| Multiple sclerosis | Sabatino et al. (2019) | ||
| Dendritic cell | Promotion | Rheumatoid arthritis | Worbs et al. (2017) |
| Systemic lupus erythematosus | Worbs et al. (2017) | ||
| Psoriasis | Worbs et al. (2017) | ||
| Type 1 diabetes | Diana et al. (2013) | ||
| Multiple sclerosis | Mohammad et al. (2012) | ||
| Macrophage | Promotion | Rheumatoid arthritis | Funes et al. (2018); Shapouri-Moghaddam et al. (2018) |
| Systemic lupus erythematosus | Funes et al. (2018) | ||
| Autoimmune neuritis | Funes et al. (2018) | ||
| Inflammatory bowel disease | Funes et al. (2018) | ||
| Systemic sclerosis | Funes et al. (2018) | ||
| Autoimmune hepatitis | Shapouri-Moghaddam et al. (2018) | ||
| Crohn’s disease | Shapouri-Moghaddam et al. (2018) | ||
| Multiple sclerosis | Shapouri-Moghaddam et al. (2018) | ||
| Protection | Systemic lupus erythematosus | Funes et al. (2018) | |
| Autoimmune neuritis | Funes et al. (2018) | ||
| Inflammatory bowel disease | Funes et al. (2018) | ||
| NK cell | Promotion | Multiple sclerosis | Fogel et al. (2013) |
| Rheumatoid arthritis | Palm et al. (2015) | ||
| Psoriasis | Ottaviani et al. (2006) | ||
| Primary biliary cirrhosis | Shimoda et al. (2011) | ||
| Type 1 diabetes | Dotta et al. (2008) | ||
| Protection | Multiple sclerosis | Shi et al. (2011) | |
| Type 1 diabetes | Dotta et al. (2008) |
Rheumatoid Arthritis
High-throughput m6A sequencing revealed a potential relationship between RNA methylation and rheumatoid arthritis (RA)-related genes, suggesting that m6A may contribute to the initiation and development of RA. Indeed, the global m6A content in peripheral blood is significantly increased in patients with RA compared to healthy people. Quantitative real-time polymerase chain reaction showed that the mRNA expression levels of ALKBH5, FTO and YTHDF2 were decreased in peripheral blood mononuclear cells (PBMCs) isolated from patients with RA. However, ALKBH5 expression is upregulated in patients with RA after treatment with the appropriate drug therapy. In addition, associations were identified between FTO mRNA expression and some indicative markers of RA activity, including the IgG level, C3 level, disease activity score 28 (DAS28) score and lymphocyte-to-monocyte ratio (LMR). Moreover, associations were observed between YTHDF2 mRNA expression and the red blood cell (RBC) count, lymphocyte percentage (L%), neutrophil percentage (N%), neutrophil-to-lymphocyte ratio (NLR), and LMR. In summary, m6A and regulators such as ALKBH5, FTO and YTHDF2 may be promising candidates for assessing the risk and progression of RA (Luo et al., 2020a).
METTL3 expression is significantly upregulated in patients with RA. In addition, positive correlations were found between METTL3 expression and biochemical indexes, including the C-reactive protein (CRP) level and erythrocyte sedimentation rate (ESR), which suggested alterations in RA disease activity. LPS stimulation of macrophages increases the expression and biological effects of METTL3. Moreover, METTL3 overexpression significantly inhibits the LPS-induced inflammatory response in macrophages through the NF-κB pathway (Wang et al., 2019). However, METTL3 may promote the activation of fibroblast-like synoviocytes (FLSs) and the inflammatory response through the NF-κB pathway, thus accelerating the initiation and progression of RA (Shi et al., 2021). Therefore, the precise role of METTL3 in the pathogenesis of RA remains to be further investigated.
Systemic Lupus Erythematosus
A comprehensive review first proposed that a link between the m6A modification and systemic lupus erythematosus (SLE) is reasonable based on the observation that m6A effectively regulates gene expression and the immune system (Li et al., 2018). Other scientists then observed downregulated mRNA expression of m6A regulators, including METTL3, METTL14, WTAP, FTO, ALKBH5 and YTHDF2, in patients with SLE (Luo et al., 2020b; Luo et al., 2020c). These decreases correlated with the index used to predict SLE disease activity. Specifically, levels of the METTL14 and YTHDF2 mRNAs in patients with SLE were associated with CRP and C3 levels, while the ALKBH5 mRNA levels of in patients with SLE were associated with C3, CRP and autoantibody levels and skin manifestations. In addition, positive correlations were observed among mRNA levels of three different regulators in PBMCs from patients with SLE (Luo et al., 2020b; Luo et al., 2020c). In addition, logistic regression and multivariate logistic regression analyses revealed that downregulated expression of the YTHDF2 or ALKBH5 mRNA may be associated with an increased risk of developing SLE (Luo et al., 2020b; Luo et al., 2020c). These findings indicate that the m6A regulators ALKBH5 and YTHDF2 are likely to be involved in the pathogenesis of SLE and are expected to be effective biomarkers to assess the SLE risk and disease activity (Luo et al., 2020c).
Multiple Sclerosis
In a comprehensive analysis of DNA methylation and gene expression data, Mo et al. identified rs923829 in METTL21B and rs2288481 in the DKKL1 gene as strongly correlated with multiple sclerosis (MS). An analysis of the HaploReg database showed that these two m6A-related SNPs regulate the expression of the METTL21B and DKKL1 genes. Then, the researchers selected PBMCs from a small group of Chinese participants to validate the association between rs923829 and METTL21B expression and between rs2288481 and DKKL1 expression. Importantly, rs923829 is strongly associated with METTL21B expression, while a significant statistical association is not observed between rs2288481 and DKKL1 expression. This group proved that these m6A-related SNPs may be related to the pathogenesis of MS(Mo et al., 2019).
Experimental autoimmune encephalomyelitis (EAE) is internationally recognized as an animal model for studying MS. More recently, specific ablation of ALKBH5 in T cells conferred protection against EAE. Mechanistically, the m6A eraser ALKBH5 decreased the m6A levels in the CXCL2 and IFN-γ mRNAs and subsequently increased their transcript stability and protein expression, thereby enhancing CD4+ T cell-mediated responses and inflammatory cell infiltration in the central nervous system to induce neuroinflammation (Zhou et al., 2021). This study was the first to prove a direct link between autoimmunity and m6A-mediated functions of immune cells.
Psoriasis
Transcriptome-wide m6A profiling revealed that transcripts from psoriatic skin had the fewest m6A peaks and lowest m6A peak density compared with transcripts from uninvolved psoriatic skin and healthy skin. Bioinformatics pathway analyses indicated that transcripts that were hypermethylated in psoriatic skin were primarily correlated with inducing various responses, including immune responses, cytokine production and olfactory signal transduction, while transcripts that were hypomethylated in psoriatic skin were strongly related to the Wnt signaling pathway and development-related processes. Transcripts with lower expression levels were preferentially modified with m6A. Moreover, gene expression was upregulated in psoriatic skin, accompanied by increased m6A levels, indicating that alterations in m6A methylation affect the gene expression pattern (Wang and Jin, 2020).
Other Autoimmune Diseases
T cell-specific METTL14 deficiency prevents the differentiation of naïve T cells into Tregs, leading to an imbalance in Th17 cells and Tregs, and thereby inducing spontaneous colitis. Considering that dysregulation of the balance between Th17 cells and Tregs is strongly associated with the initiation of inflammatory bowel disease (IBD), a reasonable assumption is that METTL14 may be involved in IBD development (Lu et al., 2020). Another study showed that CD4+ T cells induce autoimmune colitis, an ability that might be controlled by ALKBH5. Therefore, ALKBH5-deficient naïve CD4+ T cells failed to migrate into colon tissue, and their ability to promote colitis was reduced (Zhou et al., 2021).
Variants in IGF2BP2 were shown to decrease glucose-stimulated insulin secretion in the first phase of diabetes development. Additionally, IGF2BP2 was found to be downregulated and linked to diabetic nephropathy in male patients with type 1 diabetes (T1D), as well as impaired glucose tolerance in patients with type 2 diabetes (T2D) (Wang et al., 2021b). Correlations were observed between polymorphisms in the ALKBH5 gene, including rs9913266 and rs12936694, and the development of autoimmune thyroid disease (AITD). Therefore, ALKBH5 might be a candidate susceptibility gene for AITD (Song et al., 2021).
Conclusions and Future Perspectives
Over several decades, studies of m6A modifications have resulted in substantial progress in epitranscriptomics. Convincing evidence suggests that reversible m6A modification may be involved in regulating many processes in immune cells, including development, differentiation, activation, and migration. Based on these results, m6A may participate in the pathogenesis of immune-related diseases, including cancers, viral infections, and inflammatory and autoimmune diseases. Currently, the relationships between m6A and cancer and viral infection have received extensive research attention. However, studies examining the essential role of the m6A modification in the pathogenesis of autoimmune diseases are lacking, although existing evidence strongly indicates that m6A may be involved in the development of autoimmune diseases. Direct studies assessing the mechanisms by which and to what extent m6A contributes to autoimmune diseases are urgently needed. In addition to the pathogenesis of autoimmune diseases, we should focus more on achieving the transition from mechanistic research to clinical applications, including diagnosis and treatment. Therefore, the following research gaps still remain to be filled to provide new opportunities for the treatment of immune-related diseases, including autoimmune diseases.
1. Innovative and more advanced technology. Various limitations in current technology exist, including low precision, poor calculation methods, high complexity, low repetition. Scientists must develop more convenient and accurate sequencing and imaging technology for the rapid and quantitative detection of the m6A modification, perform functional analysis and understand the dynamic mechanisms of modified RNAs.
2. Precise regulatory mechanisms among m6A modulators. Although numerous findings related to the function of m6A modulators have been reported, many knowledge gaps remain to be filled. The dynamic expression pattern of modulators makes functional identification more complicated. We should understand the mechanisms mediating the spatiotemporal specificity of m6A, how the function of regulatory proteins is regulated in different cell types, how to precisely regulate different target RNAs, how mediators regulate interactions with other regulatory proteins to perform their respective functions or exert their comprehensive effects, and how their unbalanced deposition leads to pathological processes.
3. Complicated network between m6A and other regulatory factors. m6A modification and other epigenetic regulatory mechanisms, including chromatin state interaction and histone modification, are emerging as a new area in the epitranscriptomic field. In addition, m6A exerts a decisive effect on the fate of noncoding RNAs, including microRNAs, lncRNAs and circRNAs. Their interaction will prompt more studies to obtain an in-depth understanding of m6A. Moreover, the network between m6A and other RNA modifications, including 5-methylcytosine (m5C) and pseudouridine (Ψ), should be further explored.
4. Promizing but difficult clinical applications. m6A provides novel insights into the diagnosis, treatment and prognosis of diseases, especially autoimmune diseases. However, the use of modulators as therapeutic agents remains an important challenge. First, in the present review, m6A exerts dual effects on immune-related diseases, indicating a lack of consistent and consolidated evidence. Second, its safety has not been guaranteed. Finally, few small-molecule stimulants or inhibitors targeting m6A are available. Therefore, efforts are urgently needed to screen molecular drugs targeting m6A. An understanding of the mechanisms by which RNA modifications are introduced, removed or read will reveal the underlying pathogenesis and provide therapeutic targets for autoimmune diseases in the future.
Author Contributions
YW and LL performed the literature search, wrote the first draft of the article, and revised the text. JL, BZ, GH, XL, and ZZ critically revised the text and provided substantial scientific contribution. ZX proposed the project and revised the article. All authors approved the final version of the article.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81873634,82070813 and 81800745), the National Key R&D Program of China (Grant Nos. 2016YFC1305000 and 2016YFC1305001), and the Hunan Province Natural Science Foundation of China (Grant Nos. 2018JJ2573,2020JJ2053 and 2021JJ40826).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Alarcón C. R., Goodarzi H., Lee H., Liu X., Tavazoie S., Tavazoie S. F. (2015). HNRNPA2B1 Is a Mediator of m6A-dependent Nuclear RNA Processing Events. Cell 162, 1299–1308. 10.1016/j.cell.2015.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcón C. R., Lee H., Goodarzi H., Halberg N., Tavazoie S. F. (2015). N6-methyladenosine marks Primary microRNAs for Processing. Nature 519, 482–485. 10.1038/nature14281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alunno A., Carubbi F., Bistoni O., Caterbi S., Bartoloni E., Mirabelli G., et al. (2015). T Regulatory and T Helper 17 Cells in Primary Sjögren's Syndrome: Facts and Perspectives. Mediators Inflamm. 2015, 243723. 10.1155/2015/243723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anaya J.-M. (2012). Common Mechanisms of Autoimmune Diseases (The Autoimmune Tautology). Autoimmun. Rev. 11, 781–784. 10.1016/j.autrev.2012.02.002 [DOI] [PubMed] [Google Scholar]
- Bartosovic M., Molares H. C., Gregorova P., Hrossova D., Kudla G., Vanacova S. (2017). N6-methyladenosine Demethylase FTO Targets Pre-mRNAs and Regulates Alternative Splicing and 3′-end Processing. Nucleic Acids Res. 45, 11356–11370. 10.1093/nar/gkx778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista P. J. (2017). The RNA Modification N 6 -methyladenosine and its Implications in Human Disease. Genomics, Proteomics & Bioinformatics 15, 154–163. 10.1016/j.gpb.2017.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E., Oukka M., Kuchroo V. K. (2007). TH-17 Cells in the circle of Immunity and Autoimmunity. Nat. Immunol. 8, 345–350. 10.1038/ni0407-345 [DOI] [PubMed] [Google Scholar]
- Bluestone J. A., Bour-Jordan H. (2012). Current and Future Immunomodulation Strategies to Restore Tolerance in Autoimmune Diseases. Cold Spring Harb Perspect. Biol. 4. 10.1101/cshperspect.a007542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. G., Chen R., Ahmad S., Verma R., Kasturi S. P., Amaya L., et al. (2019). N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol. Cell 76, 96–109. e9. 10.1016/j.molcel.2019.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Fu Y., Wang Y., Wang J. (2020). The m6A Methyltransferase METTL3 Is Functionally Implicated in DLBCL Development by Regulating m6A Modification in PEDF. Front. Genet. 11, 955. 10.3389/fgene.2020.00955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coker H., Wei G., Brockdorff N. (2019). m6A Modification of Non-coding RNA and the Control of Mammalian Gene Expression. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1862, 310–318. 10.1016/j.bbagrm.2018.12.002 [DOI] [PubMed] [Google Scholar]
- Cope A. P., Schulze-Koops H., Aringer M. (2007). The central Role of T Cells in Rheumatoid Arthritis. Clin. Exp. Rheumatol. 25, S4–S11. [PubMed] [Google Scholar]
- Dai D., Wang H., Zhu L., Jin H., Wang X. (2018). N6-methyladenosine Links RNA Metabolism to Cancer Progression. Cell Death Dis 9, 124. 10.1038/s41419-017-0129-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danikowski K. M., Jayaraman S., Prabhakar B. S. (2017). Regulatory T Cells in Multiple Sclerosis and Myasthenia Gravis. J. Neuroinflammation 14, 117. 10.1186/s12974-017-0892-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degrauwe N., Suvà M.-L., Janiszewska M., Riggi N., Stamenkovic I. (2016). IMPs: an RNA-Binding Protein Family that Provides a Link between Stem Cell Maintenance in normal Development and Cancer. Genes Dev. 30, 2459–2474. 10.1101/gad.287540.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diana J., Simoni Y., Furio L., Beaudoin L., Agerberth B., Barrat F., et al. (2013). Crosstalk between Neutrophils, B-1a Cells and Plasmacytoid Dendritic Cells Initiates Autoimmune Diabetes. Nat. Med. 19, 65–73. 10.1038/nm.3042 [DOI] [PubMed] [Google Scholar]
- Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., et al. (2012). Topology of the Human and Mouse m6A RNA Methylomes Revealed by m6A-Seq. Nature 485, 201–206. 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- Dotta F., Fondelli C., Falorni A. (2008). Can NK Cells Be a Therapeutic Target in Human Type 1 Diabetes? Eur. J. Immunol. 38, 2961–2963. 10.1002/eji.200838851 [DOI] [PubMed] [Google Scholar]
- Du H., Zhao Y., He J., Zhang Y., Xi H., Liu M., et al. (2016). YTHDF2 Destabilizes m6A-Containing RNA through Direct Recruitment of the CCR4-NOT Deadenylase Complex. Nat. Commun. 7, 12626. 10.1038/ncomms12626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ElEssawy B., Li X. C. (2015). Type 1 Diabetes and T Regulatory Cells. Pharmacol. Res. 98, 22–30. 10.1016/j.phrs.2015.04.009 [DOI] [PubMed] [Google Scholar]
- Fogel L. A., Yokoyama W. M., French A. R. (2013). Natural Killer Cells in Human Autoimmune Disorders. Arthritis Res. Ther. 15, 216. 10.1186/ar4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funes S. C., Rios M., Escobar-Vera J., Kalergis A. M. (2018). Implications of Macrophage Polarization in Autoimmunity. Immunology 154, 186–195. 10.1111/imm.12910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale N. S., McIntyre A. B. R., McFadden M. J., Roder A. E., Kennedy E. M., Gandara J. A., et al. (2016). N6 -Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell Host & Microbe 20, 654–665. 10.1016/j.chom.2016.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Zhan Y., Zhuo L., Zhang Q., Li G., Li Q., et al. (2021). Biological Functions of m6A Methyltransferases. Cell Biosci 11, 15. 10.1186/s13578-020-00513-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., Zhang Y., Li D., Cai H., Cai L., Xu Q. (2020). N6-methyladenosine Demethylase FTO Promotes M1 and M2 Macrophage Activation. Cell Signal. 69, 109553. 10.1016/j.cellsig.2020.109553 [DOI] [PubMed] [Google Scholar]
- Han D., Liu J., Chen C., Dong L., Liu Y., Chang R., et al. (2019). Anti-tumour Immunity Controlled through mRNA m6A Methylation and YTHDF1 in Dendritic Cells. Nature 566, 270–274. 10.1038/s41586-019-0916-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H., Fan G., Song S., Jiang Y., Qian C. a., Zhang W., et al. (2021). piRNA-30473 Contributes to Tumorigenesis and Poor Prognosis by Regulating m6A RNA Methylation in DLBCL. Blood 137, 1603–1614. 10.1182/blood.2019003764 [DOI] [PubMed] [Google Scholar]
- Han L., Yang J., Wang X., Li D., Lv L., Li B. (2015). Th17 Cells in Autoimmune Diseases. Front. Med. 9, 10–19. 10.1007/s11684-015-0388-9 [DOI] [PubMed] [Google Scholar]
- Hao H., Hao S., Chen H., Chen Z., Zhang Y., Wang J., et al. (2019). N6-methyladenosine Modification and METTL3 Modulate Enterovirus 71 Replication. Nucleic Acids Res. 47, 362–374. 10.1093/nar/gky1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Li H., Wu A., Peng Y., Shu G., Yin G. (2019). Functions of N6-Methyladenosine and its Role in Cancer. Mol. Cancer 18, 176. 10.1186/s12943-019-1109-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. J., Zhu Y., Ma H., Guo Y., Shi X., Liu Y., et al. (2017). Ythdc2 Is an N6-Methyladenosine Binding Protein that Regulates Mammalian Spermatogenesis. Cell Res 27, 1115–1127. 10.1038/cr.2017.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Wang S., Liu J., Huang Y., Gong C., Liu J., et al. (2020). New Sights in Cancer: Component and Function of N6-Methyladenosine Modification. Biomed. Pharmacother. 122, 109694. 10.1016/j.biopha.2019.109694 [DOI] [PubMed] [Google Scholar]
- Huang H., Weng H., Sun W., Qin X., Shi H., Wu H., et al. (2018). Recognition of RNA N6-Methyladenosine by IGF2BP Proteins Enhances mRNA Stability and Translation. Nat. Cell Biol 20, 285–295. 10.1038/s41556-018-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., et al. (2011). N6-methyladenosine in Nuclear RNA Is a Major Substrate of the Obesity-Associated FTO. Nat. Chem. Biol. 7, 885–887. 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S., Zhang X., Miao Y., Liang P., Zhu K., She Y., et al. (2018). m6A RNA Modification Controls Autophagy through Upregulating ULK1 Protein abundanceA RNA Modification Controls Autophagy through Upregulating ULK1 Protein Abundance. Cell Res 28, 955–957. 10.1038/s41422-018-0069-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthiya R., Khandelia P. (2020). m6A RNA Methylation: Ramifications for Gene Expression and Human Health. Mol. Biotechnol. 62, 467–484. 10.1007/s12033-020-00269-5 [DOI] [PubMed] [Google Scholar]
- Kasowitz S. D., Ma J., Anderson S. J., Leu N. A., Xu Y., Gregory B. D., et al. (2018). Nuclear m6A Reader YTHDC1 Regulates Alternative Polyadenylation and Splicing during Mouse Oocyte Development. Plos Genet. 14, e1007412. 10.1371/journal.pgen.1007412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuckles P., Lence T., Haussmann I. U., Jacob D., Kreim N., Carl S. H., et al. (2018). Zc3h13/Flacc Is Required for Adenosine Methylation by Bridging the mRNA-Binding Factor Rbm15/Spenito to the m6A Machinery Component Wtap/Fl(2)d. Genes Dev. 32, 415–429. 10.1101/gad.309146.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer J., Rao H., Hackert P., Sloan K. E., Höbartner C., Bohnsack M. T. (2018). The m6A Reader Protein YTHDC2 Interacts with the Small Ribosomal Subunit and the 5′-3′ Exoribonuclease XRN1. Rna 24, 1339–1350. 10.1261/rna.064238.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang F., Singh R. K., Pei Y., Zhang S., Sun K., Robertson E. S. (2019). EBV Epitranscriptome Reprogramming by METTL14 Is Critical for Viral-Associated Tumorigenesis. Plos Pathog. 15, e1007796. 10.1371/journal.ppat.1007796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lence T., Paolantoni C., Worpenberg L., Roignant J.-Y. (2019). Mechanistic Insights into m6A RNA Enzymes. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1862, 222–229. 10.1016/j.bbagrm.2018.10.014 [DOI] [PubMed] [Google Scholar]
- Li H.-B., Tong J., Zhu S., Batista P. J., Duffy E. E., Zhao J., et al. (2017). m6A mRNA Methylation Controls T Cell Homeostasis by Targeting the IL-7/STAT5/SOCS pathwaysA mRNA Methylation Controls T Cell Homeostasis by Targeting the IL-7/STAT5/SOCS Pathways. Nature 548, 338–342. 10.1038/nature23450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Wu H., Wang Q., Ning S., Xu S., Pang D. (2021). Dual Effects of N6-Methyladenosine on Cancer Progression and Immunotherapy. Mol. Ther. - Nucleic Acids 24, 25–39. 10.1016/j.omtn.2021.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.-J., Fan Y.-G., Leng R.-X., Pan H.-F., Ye D.-Q. (2018). Potential Link between M 6 A Modification and Systemic Lupus Erythematosus. Mol. Immunol. 93, 55–63. 10.1016/j.molimm.2017.11.009 [DOI] [PubMed] [Google Scholar]
- Li L., Zang L., Zhang F., Chen J., Shen H., Shu L., et al. (2017). Fat Mass and Obesity-Associated (FTO) Protein Regulates Adult Neurogenesis. Hum. Mol. Genet. 26, 2398–2411. 10.1093/hmg/ddx128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Jiang Y., Sun X., Wu Y., Chen Z. (2021). METTL3 Is Required for Maintaining β-cell Function. Metabolism 116, 154702. 10.1016/j.metabol.2021.154702 [DOI] [PubMed] [Google Scholar]
- Li X., Tang J., Huang W., Wang F., Li P., Qin C., et al. (2017). The M6A Methyltransferase METTL3: Acting as a Tumor Suppressor in Renal Cell Carcinoma. Oncotarget 8, 96103–96116. 10.18632/oncotarget.21726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang J., Huang C., Shen M., Zhan H., Xu K. (2020). RNA N6-Methyladenosine: a Promising Molecular Target in Metabolic Diseases. Cell Biosci 10, 19. 10.1186/s13578-020-00385-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang Q., Cui G., Zhao F., Tian X., Sun B.-F., et al. (2020). m6A Regulates Liver Metabolic Disorders and Hepatogenous DiabetesA Regulates Liver Metabolic Disorders and Hepatogenous Diabetes. Genomics, Proteomics & Bioinformatics 18, 371–383. 10.1016/j.gpb.2020.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Niu Y., Wan A., Chen D., Liang H., Chen X., et al. (2020). RNA M6 A Methylation Regulates Sorafenib Resistance in Liver Cancer through FOXO3-Mediated Autophagy. Embo j 39, e103181. 10.15252/embj.2019103181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Yang Z., Li R., Wu Y., Chi M., Gao S., et al. (2021). Potential Roles of N6-Methyladenosine (m6A) in Immune Cells. J. Transl Med. 19, 251. 10.1186/s12967-021-02918-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zhang X., Chen K., Cheng Y., Liu S., Xia M., et al. (2019). CCR7 Chemokine Receptor-Inducible Lnc-Dpf3 Restrains Dendritic Cell Migration by Inhibiting HIF-1α-Mediated Glycolysis. Immunity 50, 600–615. e15. 10.1016/j.immuni.2019.01.021 [DOI] [PubMed] [Google Scholar]
- Liu N., Dai Q., Zheng G., He C., Parisien M., Pan T. (2015). N6-methyladenosine-dependent RNA Structural Switches Regulate RNA-Protein Interactions. Nature 518, 560–564. 10.1038/nature14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Zhou K. I., Parisien M., Dai Q., Diatchenko L., Pan T. (2017). N 6-methyladenosine Alters RNA Structure to Regulate Binding of a Low-Complexity Protein. Nucleic Acids Res. 45, 6051–6063. 10.1093/nar/gkx141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu Z., Tang H., Shen Y., Gong Z., Xie N., et al. (2019). The N6-Methyladenosine (m6A)-Forming Enzyme METTL3 Facilitates M1 Macrophage Polarization through the Methylation of STAT1 mRNA. Am. J. Physiology-Cell Physiol. 317, C762–c775. 10.1152/ajpcell.00212.2019 [DOI] [PubMed] [Google Scholar]
- Lu T. X., Zheng Z., Zhang L., Sun H.-L., Bissonnette M., Huang H., et al. (2020). A New Model of Spontaneous Colitis in Mice Induced by Deletion of an RNA m6A Methyltransferase Component METTL14 in T Cells. Cell Mol. Gastroenterol. Hepatol. 10, 747–761. 10.1016/j.jcmgh.2020.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q., Fu B., Zhang L., Guo Y., Huang Z., Li J. (2020). Decreased Peripheral Blood ALKBH5 Correlates with Markers of Autoimmune Response in Systemic Lupus Erythematosus. Dis. Markers 2020, 8193895. 10.1155/2020/8193895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q., Gao Y., Zhang L., Rao J., Guo Y., Huang Z., et al. (2020). Decreased ALKBH5, FTO, and YTHDF2 in Peripheral Blood Are as Risk Factors for Rheumatoid Arthritis. Biomed. Res. Int. 2020, 5735279. 10.1155/2020/5735279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q., Rao J., Zhang L., Fu B., Guo Y., Huang Z., et al. (2020). The Study of METTL14, ALKBH5, and YTHDF2 in Peripheral Blood Mononuclear Cells from Systemic Lupus Erythematosus. Mol. Genet. Genomic Med. 8, e1298. 10.1002/mgg3.1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J., Zhang Y., Gao S., Zhang C., Chen Y., Li W., et al. (2018). Endothelial-specific m6A Modulates Mouse Hematopoietic Stem and Progenitor Cell Development via Notch Signaling. Cell Res 28, 249–252. 10.1038/cr.2017.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Chang M., Lv H., Zhang Z.-W., Zhang W., He X., et al. (2018). RNA m6A Methylation Participates in Regulation of Postnatal Development of the Mouse Cerebellum. Genome Biol. 19, 68. 10.1186/s13059-018-1435-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Wang X., Cai J., Dai Q., Natchiar S. K., Lv R., et al. (2019). N6-Methyladenosine Methyltransferase ZCCHC4 Mediates Ribosomal RNA Methylation. Nat. Chem. Biol. 15, 88–94. 10.1038/s41589-018-0184-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. z., Yang F., Zhou C. c., Liu F., Yuan J. h., Wang F., et al. (2017). METTL14 Suppresses the Metastatic Potential of Hepatocellular Carcinoma by Modulating N 6 ‐methyladenosine‐dependent Primary MicroRNA Processing. Hepatology 65, 529–543. 10.1002/hep.28885 [DOI] [PubMed] [Google Scholar]
- Mauer J., Sindelar M., Despic V., Guez T., Hawley B. R., Vasseur J.-J., et al. (2019). FTO Controls Reversible m6Am RNA Methylation during snRNA Biogenesis. Nat. Chem. Biol. 15, 340–347. 10.1038/s41589-019-0231-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McComb S., Thiriot A., Akache B., Krishnan L., Stark F. (2019). Introduction to the Immune System. Methods Mol. Biol. 2024, 1–24. 10.1007/978-1-4939-9597-4_1 [DOI] [PubMed] [Google Scholar]
- Men L., Sun J., Luo G., Ren D. (2019). Acute Deletion of METTL14 in β-Cells of Adult Mice Results in Glucose Intolerance. Endocrinology 160, 2388–2394. 10.1210/en.2019-00350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. D., Patil D. P., Zhou J., Zinoviev A., Skabkin M. A., Elemento O., et al. (2015). 5′ UTR m6A Promotes Cap-independent Translation. Cell 163, 999–1010. 10.1016/j.cell.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. D., Saletore Y., Zumbo P., Elemento O., Mason C. E., Jaffrey S. R. (2012). Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3′ UTRs and Near Stop Codons. Cell 149, 1635–1646. 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo X.-B., Lei S.-F., Qian Q.-Y., Guo Y.-F., Zhang Y.-H., Zhang H. (2019). Integrative Analysis Revealed Potential Causal Genetic and Epigenetic Factors for Multiple Sclerosis. J. Neurol. 266, 2699–2709. 10.1007/s00415-019-09476-w [DOI] [PubMed] [Google Scholar]
- Mohammad M., Hassanpour M., Tsai V., Li H., Ruitenberg M., Booth D., et al. (2012). Dendritic Cells and Multiple Sclerosis: Disease, Tolerance and Therapy. Ijms 14, 547–562. 10.3390/ijms14010547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Műzes G., Molnár B., Sipos F. (2012). Regulatory T Cells in Inflammatory Bowel Diseases and Colorectal Cancer. World J. Gastroenterol. 18, 5688–5694. 10.3748/wjg.v18.i40.5688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W., Yao S., Zhou Y., Liu Y., Huang P., Zhou A., et al. (2019). Long Noncoding RNA GAS5 Inhibits Progression of Colorectal Cancer by Interacting with and Triggering YAP Phosphorylation and Degradation and Is Negatively Regulated by the m6A Reader YTHDF3. Mol. Cancer 18, 143. 10.1186/s12943-019-1079-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y., Zhao X., Wu Y.-S., Li M.-M., Wang X.-J., Yang Y.-G. (2013). N6-methyl-adenosine (m6A) in RNA: an Old Modification with a Novel Epigenetic Function. Genomics, Proteomics & Bioinformatics 11, 8–17. 10.1016/j.gpb.2012.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohl K., Tenbrock K. (2015). Regulatory T Cells in Systemic Lupus Erythematosus. Eur. J. Immunol. 45, 344–355. 10.1002/eji.201344280 [DOI] [PubMed] [Google Scholar]
- Ottaviani C., Nasorri F., Bedini C., de Pità O., Girolomoni G., Cavani A. (2006). CD56brightCD16- NK Cells Accumulate in Psoriatic Skin in Response to CXCL10 and CCL5 and Exacerbate Skin Inflammation. Eur. J. Immunol. 36, 118–128. 10.1002/eji.200535243 [DOI] [PubMed] [Google Scholar]
- Palm A.-K. E., Friedrich H. C., Mezger A., Salomonsson M., Myers L. K., Kleinau S. (2015). Function and Regulation of Self-Reactive Marginal Zone B Cells in Autoimmune Arthritis. Cell Mol Immunol 12, 493–504. 10.1038/cmi.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil D. P., Chen C.-K., Pickering B. F., Chow A., Jackson C., Guttman M., et al. (2016). m6A RNA Methylation Promotes XIST-Mediated Transcriptional repressionA RNA Methylation Promotes XIST-Mediated Transcriptional Repression. Nature 537, 369–373. 10.1038/nature19342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton K. E., Chen B., Liu K., Hunter O. V., Xie Y., Tu B. P., et al. (2017). The U6 snRNA M 6 A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 169, 824–835. e14. 10.1016/j.cell.2017.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping X.-L., Sun B.-F., Wang L., Xiao W., Yang X., Wang W.-J., et al. (2014). Mammalian WTAP Is a Regulatory Subunit of the RNA N6-Methyladenosine Methyltransferase. Cell Res 24, 177–189. 10.1038/cr.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto R., Vågbø C. B., Jakobsson M. E., Kim Y., Baltissen M. P., O’Donohue M.-F., et al. (2020). The Human Methyltransferase ZCCHC4 Catalyses N6-Methyladenosine Modification of 28S Ribosomal RNA. Nucleic Acids Res. 48, 830–846. 10.1093/nar/gkz1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin S. J. S., Bloom M. S., Robinson W. H. (2019). B Cell Checkpoints in Autoimmune Rheumatic Diseases. Nat. Rev. Rheumatol. 15, 303–315. 10.1038/s41584-019-0211-0 [DOI] [PubMed] [Google Scholar]
- Růžička K., Zhang M., Campilho A., Bodi Z., Kashif M., Saleh M., et al. (2017). Identification of Factors Required for M(6) A mRNA Methylation in Arabidopsis Reveals a Role for the Conserved E3 Ubiquitin Ligase HAKAI. New Phytol. 215, 157–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatino J. J., Jr., Pröbstel A.-K., Zamvil S. S. (2019). B Cells in Autoimmune and Neurodegenerative central Nervous System Diseases. Nat. Rev. Neurosci. 20, 728–745. 10.1038/s41583-019-0233-2 [DOI] [PubMed] [Google Scholar]
- Sakaguchi S., Yamaguchi T., Nomura T., Ono M. (2008). Regulatory T Cells and Immune Tolerance. Cell 133, 775–787. 10.1016/j.cell.2008.05.009 [DOI] [PubMed] [Google Scholar]
- Shao F., Zheng P., Yu D., Zhou Z., Jia L. (2020). Follicular Helper T Cells in Type 1 Diabetes. FASEB j. 34, 30–40. 10.1096/fj.201901637r [DOI] [PubMed] [Google Scholar]
- Shapouri-Moghaddam A., Mohammadian S., Vazini H., Taghadosi M., Esmaeili S. A., Mardani F., et al. (2018). Macrophage Plasticity, Polarization, and Function in Health and Disease. J. Cell Physiol 233, 6425–6440. 10.1002/jcp.26429 [DOI] [PubMed] [Google Scholar]
- Shi F.-D., Ljunggren H.-G., La Cava A., Van Kaer L. (2011). Organ-specific Features of Natural Killer Cells. Nat. Rev. Immunol. 11, 658–671. 10.1038/nri3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Wang X., Lu Z., Zhao B. S., Ma H., Hsu P. J., et al. (2017). YTHDF3 Facilitates Translation and Decay of N6-Methyladenosine-Modified RNA. Cell Res 27, 315–328. 10.1038/cr.2017.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Wei J., He C. (2019). Where, when, and How: Context-dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 74, 640–650. 10.1016/j.molcel.2019.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., Zheng Y., Luo S., Li X., Zhang Y., Meng X., et al. (2021). METTL3 Promotes Activation and Inflammation of FLSs through the NF-κB Signaling Pathway in Rheumatoid Arthritis. Front. Med. 8, 607585. 10.3389/fmed.2021.607585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima H., Matsumoto M., Ishigami Y., Ebina M., Muto A., Sato Y., et al. (2017). S-adenosylmethionine Synthesis Is Regulated by Selective N6-Adenosine Methylation and mRNA Degradation Involving METTL16 and YTHDC1. Cell Rep. 21, 3354–3363. 10.1016/j.celrep.2017.11.092 [DOI] [PubMed] [Google Scholar]
- Shimoda S., Harada K., Niiro H., Shirabe K., Taketomi A., Maehara Y., et al. (2011). Interaction between Toll-like Receptors and Natural Killer Cells in the Destruction of Bile Ducts in Primary Biliary Cirrhosis. Hepatology 53, 1270–1281. 10.1002/hep.24194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R.-h., Zhao J., Gao C.-q., Qin Q., Zhang J.-a. (2021). Inclusion of ALKBH5 as a Candidate Gene for the Susceptibility of Autoimmune Thyroid Disease. Adv. Med. Sci. 66, 351–358. 10.1016/j.advms.2021.07.006 [DOI] [PubMed] [Google Scholar]
- Souness J. E., Stouffer J. E., de Sanchez V. C. (1982). Effect of N6-Methyladenosine on Fat-Cell Glucose Metabolism. Biochem. Pharmacol. 31, 3961–3971. 10.1016/0006-2952(82)90642-6 [DOI] [PubMed] [Google Scholar]
- Tong J., Cao G., Zhang T., Sefik E., Amezcua Vesely M. C., Broughton J. P., et al. (2018). m6A mRNA Methylation Sustains Treg Suppressive functionsA mRNA Methylation Sustains Treg Suppressive Functions. Cell Res 28, 253–256. 10.1038/cr.2018.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J., Flavell R. A., Li H.-B. (2018). RNA m6A Modification and its Function in Diseases. Front. Med. 12, 481–489. 10.1007/s11684-018-0654-8 [DOI] [PubMed] [Google Scholar]
- Tong J., Wang X., Liu Y., Ren X., Wang A., Chen Z., et al. (2021). Pooled CRISPR Screening Identifies M(6)A as a Positive Regulator of Macrophage Activation. Sci. Adv. 7. 10.1126/sciadv.abd4742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu L. P., Pickering B. F., Cheng Y., Zaccara S., Nguyen D., Minuesa G., et al. (2017). The N6-Methyladenosine (m6A)-Forming Enzyme METTL3 Controls Myeloid Differentiation of normal Hematopoietic and Leukemia Cells. Nat. Med. 23, 1369–1376. 10.1038/nm.4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahren-Herlenius M., Dörner T. (2013). Immunopathogenic Mechanisms of Systemic Autoimmune Disease. The Lancet 382, 819–831. 10.1016/s0140-6736(13)60954-x [DOI] [PubMed] [Google Scholar]
- Wan Y. Y. (2010). Regulatory T Cells: Immune Suppression and beyond. Cell Mol Immunol 7, 204–210. 10.1038/cmi.2010.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.-X., Cui G.-S., Liu X., Xu K., Wang M., Zhang X.-X., et al. (2018). METTL3-mediated m6A Modification Is Required for Cerebellar Development. Plos Biol. 16, e2004880. 10.1371/journal.pbio.2004880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Hu X., Huang M., Liu J., Gu Y., Ma L., et al. (2019). Mettl3-mediated mRNA m6A Methylation Promotes Dendritic Cell Activation. Nat. Commun. 10, 1898. 10.1038/s41467-019-09903-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Yan S., Lu H., Wang S., Xu D. (2019). METTL3 Attenuates LPS-Induced Inflammatory Response in Macrophages via NF-κB Signaling Pathway. Mediators Inflamm. 2019, 3120391. 10.1155/2019/3120391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Chen L., Qiang P. (2021). The Role of IGF2BP2, an m6A Reader Gene, in Human Metabolic Diseases and Cancers. Cancer Cell Int 21, 99. 10.1186/s12935-021-01799-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Doxtader K. A., Nam Y. (2016). Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol. Cell 63, 306–317. 10.1016/j.molcel.2016.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Chen C., Ding Q., Zhao Y., Wang Z., Chen J., et al. (2020). METTL3-mediated m6A Modification of HDGF mRNA Promotes Gastric Cancer Progression and Has Prognostic Significance. Gut 69, 1193–1205. 10.1136/gutjnl-2019-319639 [DOI] [PubMed] [Google Scholar]
- Wang X., Zhang J., Wang Y. (2019). Long Noncoding RNA GAS5-AS1 Suppresses Growth and Metastasis of Cervical Cancer by Increasing GAS5 Stability. Am. J. Transl Res. 11, 4909–4921. [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wu R., Liu Y., Zhao Y., Bi Z., Yao Y., et al. (2020). m6A mRNA Methylation Controls Autophagy and Adipogenesis by Targeting Atg5 and Atg7A mRNA Methylation Controls Autophagy and Adipogenesis by Targeting Atg5 and Atg7. Autophagy 16, 1221–1235. 10.1080/15548627.2019.1659617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhao B. S., Roundtree I. A., Lu Z., Han D., Ma H., et al. (2015). N6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161, 1388–1399. 10.1016/j.cell.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-N., Jin H.-Z. (2020). Transcriptome-Wide m6A Methylation in Skin Lesions from Patients with Psoriasis Vulgaris. Front. Cell Dev. Biol. 8, 591629. 10.3389/fcell.2020.591629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li Y., Toth J. I., Petroski M. D., Zhang Z., Zhao J. C. (2014). N6-methyladenosine Modification Destabilizes Developmental Regulators in Embryonic Stem Cells. Nat. Cell Biol 16, 191–198. 10.1038/ncb2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. N., Yu C. Y., Jin H. Z. (2020). RNA N6-Methyladenosine Modifications and the Immune Response. J. Immunol. Res. 2020, 6327614. 10.1155/2020/6327614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardowska A. (2021). m6A RNA Methylation in Systemic Autoimmune Diseases-A New Target for Epigenetic-Based Therapy? Pharmaceuticals (Basel) 14. 10.3390/ph14030218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J., Lv R., Ma H., Shen H., He C., Wang J., et al. (2018). Zc3h13 Regulates Nuclear RNA m6A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cell 69, 1028–1038. 10.1016/j.molcel.2018.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worbs T., Hammerschmidt S. I., Förster R. (2017). Dendritic Cell Migration in Health and Disease. Nat. Rev. Immunol. 17, 30–48. 10.1038/nri.2016.116 [DOI] [PubMed] [Google Scholar]
- Wu J., Frazier K., Zhang J., Gan Z., Wang T., Zhong X. (2020). Emerging Role of M6 A RNA Methylation in Nutritional Physiology and Metabolism. Obes. Rev. 21, 10.1111/obr.12942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Feng J., Jiang D., Zhou X., Jiang Q., Cai M., et al. (2017). AMPK Regulates Lipid Accumulation in Skeletal Muscle Cells through FTO-dependent Demethylation of N6-Methyladenosine. Sci. Rep. 7, 41606. 10.1038/srep41606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Yang X., Chen Z., Tian L., Jiang G., Chen F., et al. (2019). m6A-induced lncRNA RP11 Triggers the Dissemination of Colorectal Cancer Cells via Upregulation of Zeb1A-Induced lncRNA RP11 Triggers the Dissemination of Colorectal Cancer Cells via Upregulation of Zeb1. Mol. Cancer 18, 87. 10.1186/s12943-019-1014-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., Adhikari S., Dahal U., Chen Y.-S., Hao Y.-J., Sun B.-F., et al. (2016). Nuclear M 6 A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 61, 507–519. 10.1016/j.molcel.2016.01.012 [DOI] [PubMed] [Google Scholar]
- Xie L. J., Yang X. T., Wang R. L., Cheng H. P., Li Z. Y., Liu L., et al. (2019). Identification of Flavin Mononucleotide as a Cell‐Active Artificial N 6 ‐Methyladenosine RNA Demethylase. Angew. Chem. Int. Ed. 58, 5028–5032. 10.1002/anie.201900901 [DOI] [PubMed] [Google Scholar]
- Yao Y., Yang Y., Guo W., Xu L., You M., Zhang Y.-C., et al. (2021). METTL3-dependent m6A Modification Programs T Follicular Helper Cell Differentiation. Nat. Commun. 12, 1333. 10.1038/s41467-021-21594-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K.-J., Ringeling F. R., Vissers C., Jacob F., Pokrass M., Jimenez-Cyrus D., et al. (2017). Temporal Control of Mammalian Cortical Neurogenesis by m6A Methylation. Cell 171, 877–889. 10.1016/j.cell.2017.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y., Liu J., Cui X., Cao J., Luo G., Zhang Z., et al. (2018). VIRMA Mediates Preferential m6A mRNA Methylation in 3′UTR and Near Stop Codon and Associates with Alternative Polyadenylation. Cell Discov 4, 10. 10.1038/s41421-018-0019-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccara S., Ries R. J., Jaffrey S. R. (2019). Reading, Writing and Erasing mRNA Methylation. Nat. Rev. Mol. Cell Biol 20, 608–624. 10.1038/s41580-019-0168-5 [DOI] [PubMed] [Google Scholar]
- Zhang C., Chen Y., Sun B., Wang L., Yang Y., Ma D., et al. (2017). m6A Modulates Haematopoietic Stem and Progenitor Cell Specification. Nature 549, 273–276. 10.1038/nature23883 [DOI] [PubMed] [Google Scholar]
- Zhang C., Fu J., Zhou Y. (2019). A Review in Research Progress Concerning m6A Methylation and Immunoregulation. Front. Immunol. 10, 922. 10.3389/fimmu.2019.00922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Zhao B. S., Zhou A., Lin K., Zheng S., Lu Z., et al. (2017). m 6 A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation ProgramA Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 31, 591–606. e6. 10.1016/j.ccell.2017.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Yang Y., Sun B.-F., Shi Y., Yang X., Xiao W., et al. (2014). FTO-dependent Demethylation of N6-Methyladenosine Regulates mRNA Splicing and Is Required for Adipogenesis. Cell Res 24, 1403–1419. 10.1038/cr.2014.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G., Dahl J. A., Niu Y., Fedorcsak P., Huang C.-M., Li C. J., et al. (2013). ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Mol. Cell 49, 18–29. 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q., Hou J., Zhou Y., Li Z., Cao X. (2017). The RNA Helicase DDX46 Inhibits Innate Immunity by Entrapping m6A-Demethylated Antiviral Transcripts in the Nucleus. Nat. Immunol. 18, 1094–1103. 10.1038/ni.3830 [DOI] [PubMed] [Google Scholar]
- Zheng Z., Zhang L., Cui X.-L., Yu X., Hsu P. J., Lyu R., et al. (2020). Control of Early B Cell Development by the RNA N6-Methyladenosine Methylation. Cell Rep. 31, 107819. 10.1016/j.celrep.2020.107819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Zhang X., Hu J. (2021). m6A Demethylase ALKBH5 Controls CD4+ T Cell Pathogenicity and Promotes Autoimmunity. Sci. Adv. 7 (25), eabg0470. 10.1126/sciadv.abg0470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Zhao Y., Zou L., Zhang D., Aki D., Liu Y.-C. (2019). The E3 Ligase VHL Promotes Follicular Helper T Cell Differentiation via Glycolytic-Epigenetic Control. J. Exp. Med. 216, 1664–1681. 10.1084/jem.20190337 [DOI] [PMC free article] [PubMed] [Google Scholar]