Abstract
Quantification of human immunodeficiency virus type 1 (HIV-1) RNA as a measure of viral load has greatly improved the monitoring of therapies for infected individuals. With the significant reductions in viral load now observed in individuals treated with highly active anti-retroviral therapy (HAART), viral load assays have been adapted to achieve greater sensitivity. Two commercially available ultrasensitive assays, the Bayer Quantiplex HIV-1 bDNA version 3.0 (bDNA 3.0) assay and the Roche Amplicor HIV-1 Monitor Ultrasensitive version 1.5 (Amplicor 1.5) assay, are now being used to monitor HIV-1-infected individuals. Both of these ultrasensitive assays have a reported lower limit of 50 HIV-1 RNA copies/ml and were developed from corresponding older generation assays with lower limits of 400 to 500 copies/ml. However, the comparability of viral load data generated by these ultrasensitive assays and the relative costs of labor, disposables, and biohazardous wastes were not determined in most cases. In this study, we used matched clinical plasma samples to compare the quantification of the newer bDNA 3.0 assay with that of the older bDNA 2.0 assay and to compare the quantification and costs of the bDNA 3.0 assay and the Amplicor 1.5 assay. We found that quantification by the bDNA 3.0 assay was approximately twofold higher than that by the bDNA 2.0 assay and was highly correlated to that by the Amplicor 1.5 assay. Moreover, cost analysis based on labor, disposables, and biohazardous wastes showed significant savings with the bDNA 3.0 assay as compared to the costs of the Amplicor 1.5 assay.
Management of human immunodeficiency virus type 1 (HIV-1)-infected individuals has greatly improved with the introduction of quantitative viral load testing and highly active anti-retroviral therapy (HAART). With successful HAART, viral load measurements have dropped below the limit of detection of previously available commercial assays (<400 to <500 HIV-1 RNA copies/ml) (3, 5, 10, 12, 18). This degree of reduction in HIV-1 viral load, and the general consensus that HAART therapy should aim to suppress HIV-1 replication as fully as possible in order to attain durable virologic responses, has prompted the need for even more sensitive viral load quantification assays (2). Consequently, a number of manufacturers, including Bayer Diagnostics (formerly Chiron Diagnostics, Emeryville, Calif.) and Roche Molecular Systems, Inc. (Somerville, N.J.), have adapted their existing viral load assays to permit a lower limit of detection of 50 HIV-1 RNA copies/ml.
Most HIV-1-infected patients are now monitored by ultrasensitive viral load assays. However, it is unclear whether ultrasensitive assays, which use different technologies, generate comparable quantitative values. The comparability of viral load data is an important issue for effective management of HIV-1-infected individuals since in many cases, with no apparent choice, patient samples may be switched from one assay to another at any given time as a result of insurance coverage or test accessibility. This poses a significant problem since studies have shown that the Quantiplex HIV-1 bDNA version 2.0 (bDNA 2.0) assay and the Amplicor HIV-1 Monitor Standard version 1.0 (Amplicor 1.0) assay do not generate comparable quantification results (1, 7, 9, 11). Another factor to consider in comparing the ultrasensitive viral load assays is assay cost. In addition to the cost of assay reagents, costs associated with labor, disposables, and biohazardous waste should be considered when evaluating ultrasensitive viral load assays for routine use in the clinical laboratory.
This study compared HIV-1 viral load quantification and associated costs of two commercially available ultrasensitive HIV-1 viral load assays—Quantiplex HIV-1 bDNA version 3.0 assay (bDNA 3.0; Bayer Diagnostics) and Amplicor HIV-1 Monitor Ultrasensitive version 1.5 assay (Amplicor 1.5; Roche Molecular Systems, Inc.). Using clinical plasma samples, we evaluated the correlation and mean difference in HIV-1 RNA copy number between the older and newer versions of the Quantiplex HIV-RNA assay, bDNA 2.0 and bDNA 3.0, respectively, and between bDNA 3.0 and Amplicor 1.5. We also conducted a cost analysis based on labor, disposables, biohazardous wastes, and assay components between bDNA 3.0 and Amplicor 1.5.
MATERIALS AND METHODS
Clinical plasma samples.
Excess uncharacterized plasma from HIV-1-infected subjects remaining after routine testing by bDNA 3.0 (or by bDNA 2.0 until July 1998) at the San Francisco General Hospital (SFGH) Clinical Laboratory was used. Plasma from EDTA-anticoagulated whole blood was processed and frozen to −80°C within 6 h following blood draw. For routine testing, frozen plasma samples (approximately 5-ml aliquots) were transferred from −80°C to cold tap water for 10 to 15 min until thawed and then transferred to wet ice. Plasma samples were maintained on wet ice for approximately 45 min during preparation of aliquots for viral load testing. The excess plasma was returned to −80°C until viral load testing was successfully completed. Excess plasma samples then were transferred to the University of California, San Francisco (UCSF) Clinical Microbiology Research Laboratory and stored at −80°C for further analysis.
A total of 1,220 plasma samples, previously tested with bDNA 2.0 at the SFGH Clinical Laboratory, were retested with bDNA 3.0 at both the UCSF Clinical Microbiology Research Laboratory and the Bayer Reference Testing Laboratory (BRTL; Emeryville, Calif.). These 1,220 samples were selected to cover a wide viral load range (determined with bDNA 2.0) as follows: 1,035 samples at <500 copies/ml, 64 at 500 to 10,000 copies/ml, 57 at 10,001 to 100,000 copies/ml), and 64 at >100,000 copies/ml.
A total of 159 additional uncharacterized clinical plasma samples, previously tested with bDNA 3.0 by the SFGH Clinical Laboratory, were tested again with bDNA 3.0 and the Amplicor 1.5 at the UCSF Clinical Microbiology Research Laboratory. These samples were randomly selected from approximately 500 banked samples that previously were collected over 2 weeks and covered a wide viral load range (determined with bDNA 3.0) as follows: 48 samples at <50 copies/ml, 46 at 50 to 1,000 copies/ml, 33 at 1,001 to 10,000 copies/ml, and 32 at 10,001 to 72,942 copies/ml. These clinical samples were concurrently dispensed as 1.0- and 0.5-ml aliquots in 1.5-ml Sarstedt microcentrifuge tubes for bDNA 3.0 and Amplicor 1.5, respectively, and returned to −80°C. In this way, samples for testing with both assays were handled in the same manner and subjected to the same number of freeze thaw cycles (no more than three).
Banked and characterized serial plasma samples were obtained from 24 individuals from the Options study at UCSF (F. M. Hecht, M. A. Chesney, M. P. Busch, B. D. Rawal, S. I. Staprans, and J. O. Kahn, Abstr. 5th Conf. Retrovir. Opportun. Infect., abstr. 582, 1998; F. M. Hecht, B. D. Rawal, J. O. Kahn, M. Swanson, M. A. Chesney, J. A. Levy, and P. Busch, Abstr. 6th Conf. Retrovir. Opportun. Infect., abstr. 178, 1999). These individuals had been recently infected with HIV-1 (as documented by a positive HIV-1 RNA and a negative or indeterminate HIV-1 antibody test or documented HIV-1 antibody seroconversion within 6 months) and initiated on HAART. The serial samples, which previously had been tested with bDNA 2.0, included two baseline samples (collected immediately prior to HAART) as well as samples collected at 4- to 8-week intervals from each subject. Stored specimens were retested with bDNA 3.0 at the UCSF Clinical Microbiology Research Laboratory.
Committee on human research.
This study was performed in accordance with the guidelines of the UCSF committee on human research. In order to preserve patient confidentiality, specimens were unlinked to information such as date, patient identifier, patient name, and health center identification.
Study sites and technical training.
All comparative analyses between bDNA 2.0 and bDNA 3.0 were performed at the SFGH Clinical Laboratory and the BRTL. All viral load results and cost analyses (labor, disposables, and biohazardous waste) for comparison between bDNA 3.0 and Amplicor 1.5 were generated at the UCSF Clinical Microbiology Research Laboratory at San Francisco General Hospital.
One operator performed all viral load determinations for the study comparing bDNA 3.0 to Amplicor 1.5. He successfully completed all training requirements by Roche Diagnostics (Branchburg, N.J.) and Bayer Diagnostics (on site) and was issued certificates of training from both companies. He then was retrained at the study evaluation site at San Francisco General Hospital.
Amplicor 1.5 assay.
The Amplicor 1.5 assay is based on reverse transcriptase (RT)-PCR, with a reported dynamic range from 50 to 75,000 HIV-1 RNA copies/ml (15). This assay has been shown to be equivalent to the Amplicor HIV-1 Monitor version 1.0 assay (Amplicor 1.0) (17), which has been approved by the United States Federal Drug Administration (FDA). The Amplicor 1.5 assay was performed according to manufacturer's instructions, and all samples were run singly.
Samples were processed for the Amplicor 1.5 assay following a three-step work flow: (i) specimen preparation (including centrifugation and aspiration of clarified plasma, viral lysis, RNA precipitation, RNA washing, and suspension of purified RNA in buffer), (ii) amplification, and (iii) detection. According to the training provided by Roche Diagnostics (Branchburg, N.J.), the Amplicor 1.5 assay (like the Amplicor 1.0 assay) required four separated areas dedicated to sample processing (HIV-1 concentration, RNA extraction and purification, and combination of RNA and master mix solution), master mix preparation (RT-PCR reagent preparation), amplification (thermal cycling and reaction termination), and detection (plate hybridization, plate incubation, plate wash, detection). The Amplicor 1.5 assay setup was strictly unidirectional, and the operator was required to change gloves and gown when entering each work area.
bDNA 2.0 and 3.0 assays.
The bDNA 2.0 and 3.0 assays are based on branched DNA (bDNA) signal amplification technology. Both assays utilize synthetic oligonucleotide probes (capture probes, target probes, preamplifier probes, amplifier probes, and label probes) to generate signal amplification through hybridization. In the earlier version, bDNA 2.0, nonspecific hybridization between these probes increased background noise, which in turn limited the sensitivity of the assay. In the present version, bDNA 3.0, the nonnatural bases isocytidine (isoC) and isoguanidine (isoG) are incorporated in all probes except those that hybridize to the target HIV-1 RNA (capture probes and target probes). Since isoC and isoG do not hybridize with natural bases, the nonspecific hybridization between probes containing these nonnatural bases is greatly reduced. With the incorporation of isoC and isoG to reduce background noise and the development of a new target probe set and optimized buffer conditions to increase signal generation capacity, the bDNA 3.0 assay achieves a lower detection limit of 50 copies/ml (4). The bDNA 2.0 and 3.0 assays were performed according to manufacturer's instructions. All samples for bDNA 2.0 were run in duplicate, whereas all samples for bDNA 3.0 were run singly (4, 8). The second-generation bDNA 2.0 assay is performed manually and has a dynamic range of 500 to 800,000 HIV-1 RNA copies/ml (8, 16). The bDNA 3.0 assay was used in conjunction with the semi-automated Quantiplex 340 system and has a dynamic range of 50 to 500,000 HIV-1 RNA copies/ml (4).
Samples were processed for the bDNA 3.0 assay following a three-step work flow: (i) specimen preparation (including centrifugation and aspiration of clarified plasma), (ii) hybridization, and (iii) detection. The bDNA 3.0 assay required two work areas—one area for sample processing (HIV-1 concentration and viral lysis) and one area for hybridization and detection (heating block for hybridization and Quantiplex 340 for additional hybridization steps, washing, and detection). This assay allowed for bidirectional movement during assay performance, and the operator could wear the same gown and gloves between both sites for all but the final detection step, which required a new set of gloves.
Comparative cost analysis.
For comparative analysis of costs of disposables and labor, the routine assay procedures were followed for both the bDNA 3.0 and the Amplicor 1.5 assays, as described in the package inserts. Costs for disposables were based on list prices as of October 1998 from various vendors in the United States. No discounted pricing was used. For the assessment of labor, each procedure was timed from the start to the end. Except for the bDNA 3.0 overnight incubation and the Amplicor 1.5 thermal cycler step, all other “hands off” intervals that required the presence of the operator (i.e., short incubation steps) were included in this analysis. Since technical steps within each procedure were staggered, there was little hands off time to permit the operator to attend to unrelated projects.
The Amplicor 1.5 assay was performed in one run with 14 plates per run (yielding 169 tests, or 159 reportable results). Costs of disposables for the Amplicor 1.5 assay were determined for each of the three workflow steps. Costs of sample preparation disposables were calculated for the following: 182 aerosolized 200-μl tips ($80/960), 672 aerosolized 1,000-μl tips ($80/720), 672 fine-tip transfer pipets ($57/500), 14 5-ml pipets ($64/200), 14 10-ml pipets ($67/200), 168 1.5-ml screw-cap tubes ($235/1000), 14 50-ml tubes ($78/200), 154 ml of reagent-grade ethanol ($90/4 liters), 101 ml of 2-propanol ($67/4 liters), and 12 pairs of gloves ($65/50 pair). Costs for disposables for amplification were calculated for the following: 350 aerosolized 200-μl tips ($80/960), 14 25-μl Eppendorf dispensers ($112/100), 2 25-ml reagent reservoirs ($58/100), 168 caps ($75/2,400), 168 reaction tubes ($195/2,000), and 12 pairs of gloves ($65/50 pair). Costs for disposables for detection were calculated for the following: 1,008 aerosolized 200-μl tips ($80/960), 56 25-ml reagent reservoirs ($58/100), 14 5-ml pipets ($64/200), 14 10-ml pipets ($67/200), 14 50-ml tubes ($78/200), and 7 pairs of gloves ($65/50 pair).
The bDNA 3.0 assay was performed either as two runs of one plate per run (yielding 192 tests, or 168 reportable results) or as one run of two plates per run (yielding 192 tests, or 168 reportable results). Costs of disposables for the bDNA 3.0 assay were determined for each of these two scenarios according to the three workflow steps. For the bDNA 3.0 assay performed in two runs of one plate per run, costs of sample preparation disposables were calculated for the following: 192 aerosolized 1,000-μl tips ($90/800), 200 nonaerosolized 200-μl tips ($61/960), 2 25-μl Eppendorf dispensers ($112/100), 192 1.5-ml screw-cap tubes ($235/1,000), and 8 pairs of gloves ($65/50 pair). Costs of disposables for amplification (bDNA 3.0 in two runs, one plate per run) were calculated for the following: 388 aerosolized 200-μl tips ($90/960), 2 2-ml pipets ($125/500), 2 10-ml pipets ($67/200), 2 50-ml tubes ($78/200), and 4 pairs of gloves ($65/50 pair). Costs of disposables for detection (bDNA 3.0 in two runs, one plate per run) were calculated for the following: 102 aerosolized 200-μl tips ($90/960), 2 aerosolized 1000-μl tips ($90/800), 2 100-μl Eppendorf dispensers ($100.5/100), 8 55-ml reagent reservoirs ($96/80), 8 10-ml pipets ($67/200), 8 50-ml tubes ($78/200), and 8 pairs of gloves ($65/50 pair).
For the bDNA 3.0 assay performed in one run of two plates per run, costs of sample preparation disposables were calculated for the following: 192 aerosolized 1,000-μl tips ($90/800), 200 nonaerosolized 200-μl tips ($61/960), 1 25-μl Eppendorf dispenser ($112/100), 192 1.5-ml screw-cap tubes ($235/1,000), and 8 pairs of gloves ($65/50 pair). Costs of disposables for amplification (bDNA in one run, two plates per run) were calculated for the following: 388 aerosolized 200-μl tips ($90/960), 1 2-ml pipet ($125/500), 1 10-ml pipet ($67/200), 1 50-ml tube ($78/200), and 4 pairs of gloves ($65/50 pair). Costs of disposables for detection (bDNA in one run, two plates per run) were calculated for the following: 51 aerosolized 200-μl tips ($90/960), 1 aerosolized 1,000-μl tips ($90/800), 1 100-μl Eppendorf dispenser ($110.50/100), 4 55-ml reagent reservoirs ($96/80), 4 10-ml pipets ($67/200), 4 50-ml tubes ($78/200), and 4 pairs of gloves ($65/50 pair).
Statistical analysis.
For the purpose of analysis, samples reported to be below the level of detection for a particular assay were assigned HIV-1 RNA copy number values of one-half of the cutoff level for that assay. Similarly, samples reported to be above the dynamic range of an assay were assigned copy number values of one plus the highest value of the dynamic range of the assay. In comparative analyses, the lower cutoff value of the two assays was used for both sets of data. Correlation and linear regression analyses were performed on log10-transformed HIV-1 RNA copy numbers. All analyses were carried out with the SAS Statistical Analyses package (SAS Institute, Cary, N.C.).
For the correlation analyses, both the Pearson correlation coefficient and the nonparametric Spearman rank correlation coefficient, along with their respective 95% confidence intervals, were calculated. Standard statistical methods were used to test for significant correlation (6). Linear regression models were created with least-squares linear regression on log10-transformed HIV-1 RNA copy numbers with the SAS regression procedure, allowing for an intercept term. Significance of the overall model was assessed with the F statistic, and individual terms in the model were tested with the t statistic.
RESULTS
Quantification comparison: bDNA 2.0 versus bDNA 3.0.
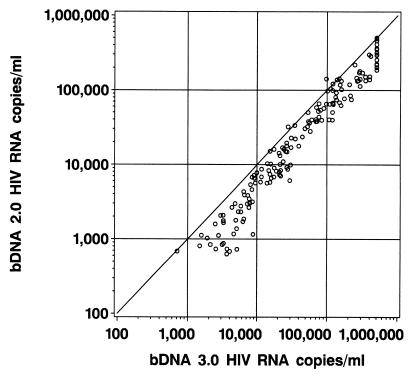
The agreement between quantification values by the bDNA 2.0 and bDNA 3.0 assays is shown in Fig. 1. The quantification values obtained with these assays were significantly correlated with one another, as indicated by the Pearson correlation coefficient (r value) of 0.982 (95% confidence interval, 0.976 to 0.986; P < 0.0001).
FIG. 1.
Results of HIV-1 RNA quantification of 185 samples by the older bDNA 2.0 assay and the more sensitive bDNA 3.0 assay. The solid line represents the identity line, where all determinations should fall if a perfect correlation between the two assays was achieved.
The distribution of differences between bDNA 2.0 and bDNA 3.0 results over the range of samples tested is shown in Table 1. Overall, HIV-1 RNA copy numbers were 0.239 log10 higher for bDNA 3.0 as compared to those of the bDNA 2.0 assay; however, this difference varied over the range of samples tested. For example, samples with fewer than 10,000 HIV RNA copies/ml by bDNA 3.0 showed a significantly higher shift from bDNA 2.0 results than did samples with >10,000 HIV RNA copies/ml. Table 1 also shows the results of the linear regression of the log10-transformed copy numbers with bDNA 3.0 results as the dependent variable and bDNA 2.0 results as the independent variable (i.e., prediction of bDNA 3.0 results from bDNA 2.0 results). This analysis indicated a strong linear relationship between the results given by the two assays over the range of samples tested (r = 0.98). Within the assay ranges tested, the <10,000 HIV-1 RNA copies/ml range showed a weaker relationship between assay values than did the two ranges above 10,000 HIV-1 RNA copies/ml (r = 0.91 and r = 0.90, respectively).
TABLE 1.
Relationship between log10-transformed quantification values obtained with the bDNA 2.0 and bDNA 3.0 assaysa
| Assay range (HIV-1 RNA copies/ml) | n | Correlation (95% CI) | Mean log10 difference (95% CI) | P value |
|---|---|---|---|---|
| 499–1,000,000 | 185 | 0.982 (0.976–0.986) | 0.239 (0.212–0.266) | <0.0001 |
| 499–10,000 | 45 | 0.750 (0.585–0.855) | 0.367 (0.306–0.429) | <0.0001 |
| 10,001–100,000 | 69 | 0.908 (0.850–0.944) | 0.246 (0.209–0.285) | <0.0001 |
| 100,000–1,000,000 | 80 | 0.899 (0.846–0.934) | 0.161 (0.125–0.196) | <0.0001 |
CI, confidence interval. P values are for correlations.
To further assess differences in quantification values for samples tested with bDNA 2.0, an additional 1,035 clinical samples below the detection limit of the bDNA 2.0 assay (<500 HIV-1 RNA copies/ml) were retested with the bDNA 3.0 assay at the BRTL. As shown in Table 2, approximately 80% of samples initially reported by bDNA 2.0 to have <500 copies/ml were confirmed to have <500 copies/ml by bDNA 3.0 (50.7% had <50 copies/ml, 29.6% had 51 to 500 copies/ml). The remaining samples were quantified above 500 copies/ml by bDNA 3.0 (9.8% had 501 to 1,000 copies/ml, 8.6% had 1,001 to 2,000 copies/ml, 1.2% had 2,001 to 3,000 copies/ml, 0.1% had >3,001 copies/ml).
TABLE 2.
bDNA 3.0 assay results for 1,035 clinical plasma samples previously quantified as <500 copies/ml by bDNA 2.0
| bDNA 3.0 results (copies/ml) | n | % of total | Cumulative % |
|---|---|---|---|
| <50 | 524 | 50.7 | 50.7 |
| 51–500 | 305 | 29.6 | 80.3 |
| 501–1,000 | 100 | 9.8 | 90.1 |
| 1,001–2,000 | 88 | 8.6 | 98.7 |
| 2,001–3,000 | 12 | 1.2 | 99.9 |
| >3,001 | 1 | 0.1 | 100 |
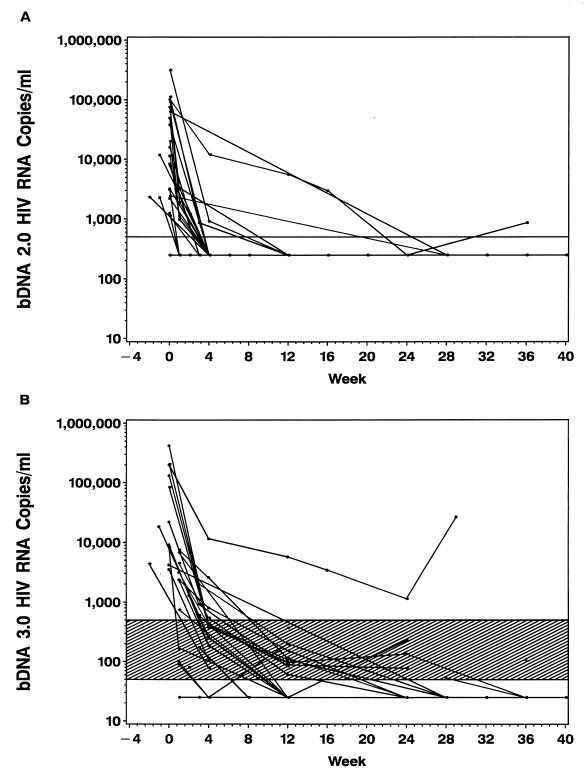
A comparison of bDNA 2.0 and bDNA 3.0 results for serial samples from 24 individuals newly infected with HIV-1 is shown in Fig. 2. When serial samples were tested with bDNA 2.0, the average time for viral load to be reduced to nondetectable levels (<500 copies/ml) was approximately 4 weeks after the initiation of HAART. However, when matched samples were retested with the more sensitive bDNA 3.0 assay, additional viral load data were generated. Using the bDNA 3.0 assay, the average time for the viral load to be reduced to nondetectable levels (<50 copies/ml) was approximately 24 weeks after the initiation of HAART.
FIG. 2.
Viral load results for 24 HIV-infected patients monitored for up to 36 weeks as measured with bDNA 2.0 (A) and bDNA 3.0 (B). The horizontal lines indicate the lower limits of detection for the bDNA 2.0 and bDNA 3.0 assays at 500 and 50 copies/ml, respectively. Data points falling below the limit of detection are plotted at half the limit of detection for each assay. The shaded area indicates the difference between the detection limits of the bDNA 2.0 and bDNA 3.0 assays.
Quantification comparison: bDNA 3.0 versus Amplicor 1.5.
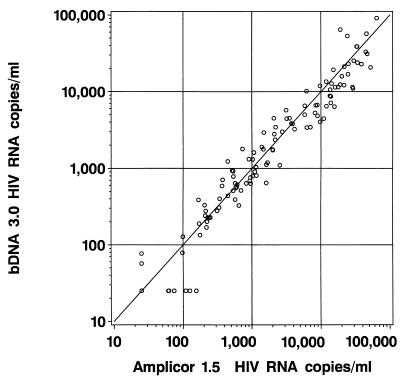
The agreement between quantification values measured by the bDNA 3.0 and Amplicor 1.5 assays is shown in Fig. 3. The quantification values obtained with these assays were significantly correlated with one another, as indicated by the Pearson correlation coefficient of 0.984 (95% confidence interval, 0.978 to 0.984; P < 0.0001).
FIG. 3.
Results of HIV-1 RNA quantification of 159 samples by the bDNA 3.0 assay based on bDNA signal amplification technology and the PCR-based Amplicor 1.5 assay. The solid line represents the identity line, where all determinations should fall if a perfect correlation between the two assays was achieved.
The correlation and mean difference between bDNA 3.0 and Amplicor 1.5 results are shown in Table 3. For samples above 10,000 HIV-1 RNA copies/ml, the two assays were approximately 90% correlated, and the mean difference in log10 copy number between the two assays was significantly different from zero. For samples with 10,000 or fewer HIV-1 RNA copies/ml, the correlation between bDNA 3.0 and Amplicor 1.5 results was less strong (approximately 75% correlated), and the mean difference in log10 copy number between the two assays was not significantly different from zero.
TABLE 3.
Relationship between log10-transformed quantification values obtained with the bDNA 3.0 and Amplicor 1.5 assaysa
| Assay range (HIV-1 RNA copies/ml) | n | Correlation (95% CI) | Mean log10 difference (95% CI) | P value |
|---|---|---|---|---|
| 0–100,000 | 159 | 0.984 (0.978–0.988) | −0.0365 (−0.067–−0.005) | <0.0001 |
| 0–1,000 | 91 | 0.953 (0.930–0.969) | −0.0032 (−0.071–0.007) | <0.0001 |
| 1,001–10,000 | 40 | 0.853 (0.738–0.920) | −0.034 (−0.104–0.035) | <0.0001 |
| 10,001–100,000 | 28 | 0.643 (0.355–0.819) | −0.055 (−0.135–0.078) | 0.0001 |
Assay range was determined with the bDNA 3.0 assay. CI, confidence interval. P values are for the correlations.
When the results of bDNA 3.0 and Amplicor 1.5 were dichotomized at 50 HIV-1 RNA copies/ml, there was excellent agreement between the two assays for which samples contained less than 50 copies/ml and those that contained more than 50 copies/ml or more (Table 4). Of the 159 samples tested, the two assays disagreed on only eight—two samples reported by Amplicor 1.5 as having <50 copies/ml were reported by bDNA 3.0 as having >50 copies/ml, and six samples reported by Amplicor 1.5 as having >50 copies/ml were reported by bDNA 3.0 as having <50 copies/ml. Moreover, relatively low viral levels were measured for the discrepant samples. The six samples reported by bDNA 3.0 as having <50 copies/ml were quantified by Amplicor 1.5 at 61, 62, 74, 109, 125, and 155 copies/ml, while the two samples reported by Amplicor 1.5 as having <50 copies/ml were quantified by bDNA 3.0 at 57 and 77 copies/ml.
TABLE 4.
Comparison of bDNA 3.0 and Amplicor 1.5 assay results dichotomized at 50 HIV-1 RNA copies/ml
| bDNA 3.0 assay results | Amplicor 1.5 assay results
|
Total | |
|---|---|---|---|
| <50 copies/ml | >50 copies/ml | ||
| <50 copies/ml | 45 | 6a | 51 |
| >50 copies/ml | 2b | 106 | 108 |
| Total | 47 | 112 | 159 |
These 6 samples were quantified by Amplicor 1.5 at 61, 62, 74, 109, 125, and 155 copies/ml.
These 2 samples were quantified by bDNA 3.0 at 57 and 77 copies/ml.
Assay component cost comparison: bDNA 3.0 versus Amplicor 1.5.
The cost of assay components also were compared for bDNA 3.0 and Amplicor 1.5. As of November 1999, the list price for Amplicor 1.5 was $2,500 per kit (U.S. pricing; Roche Diagnostics). Each Amplicor kit contains two 96-well plates that are designed to test 21 clinical samples and 3 controls, for a total of 24 tests per kit. As such, the cost of assay components for Amplicor 1.5 was $119.00 per clinical sample, or $104.17 per test (which includes controls). By comparison, the list price for bDNA 3.0 was $10,500 per kit (U.S. pricing; Bayer Diagnostics). Each bDNA 3.0 kit contains one 96-well plate that is designed to test 84 clinical samples, 9 standards, and 3 controls, for a total of 96 tests per kit. As such, the cost of assay components for bDNA 3.0 was $125.00 per clinical sample, or $109.38 per test (which includes standards and controls). Hence, the cost of assay components is approximately 5% higher for bDNA 3.0 than for Amplicor 1.5. These calculations assume that all wells are used for each assay run and thus indicate the lowest per-sample or per-test costs for assay components. If fewer samples are included in each run (i.e., not all wells are used), then the cost of assay components would be higher. Also, it is important to note that these calculations are based on the U.S. list prices for each of the kits and that any reduced pricing that may be offered by the manufacturers would affect the per-sample or per-test cost of assay components.
Overall cost comparison: bDNA 3.0 versus Amplicor 1.5.
The overall costs for the Amplicor 1.5 and bDNA 3.0 are shown in Table 5. Costs for Amplicor 1.5 were evaluated for the assay performed as a single run consisting of 14 plates (total of 168 tests: 159 clinical samples and 9 controls). Costs for bDNA 3.0 were evaluated both for the assay performed in a single run consisting of two plates run simultaneously (total of 192 tests: 168 clinical samples and 24 controls) as well as a two separate runs consisting of one plate per run (total of 192 tests: 168 clinical samples and 24 controls). In Table 5, the cost of the assay kits were determined according to the U.S. list price (as of November 1999), although kit prices may be reduced according to discounted pricing offered by the manufacturer. Labor costs are noted in terms of time (minutes) since different sites may have different pay schedules for technicians.
TABLE 5.
Cost comparison for Amplicor 1.5 versus bDNA 3.0
| Cost category | Amplicor 1.5 (1 run, 14 plates, 168 tests [159 reportable results])
|
bDNA 3.0 (2 runs, 2 plates, 192 tests [168 reportable results])
|
bDNA 3.0 (1 run, 2 plates, 192 tests [168 reportable results])
|
|||
|---|---|---|---|---|---|---|
| All tests | Per test | All tests | Per test | All tests | Per test | |
| Assay components | $17,500a | $104.17 | $21,000b | $109.38 | $21,000b | $109.38 |
| Disposables | ||||||
| Specimen preparation | $241.30 | $1.44 | $92.07 | $0.48 | $90.95 | $0.47 |
| Amplification/hybridization | $83.24 | $0.50 | $43.54 | $0.23 | $42.56 | $0.22 |
| Detection | $140.21 | $0.83 | $37.80 | $0.20 | $18.90 | $0.10 |
| Total all steps | $464.75 | $2.77 | $173.41 | $0.91 | $152.41 | $0.79 |
| Labor | ||||||
| Specimen preparation | 975 min | 5.8 min | 300 min | 1.6 min | 285 min | 1.5 min |
| Amplification/hybridization | 77 min | 0.5 min | 340 min | 1.8 min | 210 min | 1.1 min |
| Detection | 540 min | 3.2 min | 470 min | 2.5 min | 255 min | 1.3 min |
| Total all steps | 1,592 min | 9.5 min | 1,110 min | 5.8 min | 750 min | 3.9 min |
| Biohazardous waste (total all steps) | 30 cm3 | 0.18 cm3 | 16 cm3 | 0.08 cm3 | 15 cm3 | 0.08 cm3 |
Seven kits (24 tests per kit) at U.S. list price of $2,500 per kit.
Two kits (96 tests per kit) at U.S. list price of $10,500 per kit.
The costs of disposables and labor for the two assays were compared for each of the three work flow steps: sample preparation, amplification/hybridization, and detection. The highest costs of disposables (not including assay components) were incurred in the sample preparation step for both assays. Labor time was highest during the sample preparation step for Amplicor 1.5. Labor time for the bDNA 3.0 assay was fairly evenly distributed throughout all three work flow steps when the assay was performed in a single run with two plates, but was somewhat higher for the detection step when the assay was performed in two runs at one plate per run.
The lowest costs (not including assay components) were incurred with bDNA 3.0. Per-test costs for the bDNA 3.0 assay performed in two runs were significantly less (63% lower costs for disposables, 30% less labor, 50% less biohazardous waste) than those for Amplicor 1.5. A further overall reduction in disposables, labor, and biohazard waste for the bDNA 3.0 assay to less than half that of Amplicor 1.5 was realized when the bDNA 3.0 assay was performed in a single run (two plates run simultaneously, containing 24 controls and 168 clinical samples).
DISCUSSION
With the introduction of newer, more effective therapies such as HAART that can greatly reduce viral load to below 400 to 500 copies/ml, more laboratories are using ultrasensitive assays with lower detection limits of 50 copies/ml to monitor HIV-1-infected individuals. Since it is not uncommon for patients to have longitudinal viral load data from more than one assay or from earlier versions of the same assay, it is important to understand whether these ultrasensitive assays yield comparable results. Concerns about the comparability of viral load data have been supported by earlier studies. For example, in the Stadi trial, in which the combination of stavudine and didanosine was clinically evaluated, viral quantification obtained with the Amplicor 1.0 assay (standard and ultrasensitive) was significantly higher than that obtained with the bDNA 2.0 assay (14). Other studies also have showed discordance in viral load quantification by the Amplicor 1.0 assay as compared to that of the bDNA 2.0 assay (7, 9, 11). The observed discordance between the results of different assays has led to the recommendation that patients be monitored with a single manufacturer's assay (7, 14). Although use of an external standard can negate differences in the copy number estimates made by different assays (1), external standards are not routinely used in clinical practice. Therefore, it is important to understand the comparability of viral load data generated by the different assays in order to avoid complications in patient monitoring should assays be changed. Moreover, it is important to document the comparative costs of the different assays in order to give laboratories the information needed to select the most cost-effective system. Not only should the cost of assay components be considered but also the costs incurred for disposables, labor, and biohazardous waste. Assay cost considerations are particularly important in these times in which laboratories may be required to cut costs as a result of lower operating budgets and lower reimbursements.
In this study, we compared the quantification and costs of the most recent versions of commercially available ultrasensitive viral load assays—Amplicor 1.5 and bDNA 3.0. The two manufacturers, Roche Diagnostics and Bayer Diagnostics, respectively, have used different approaches in adapting their assays to address the current clinical need for more sensitive viral load quantification. The older Amplicor 1.0 assay (including both the standard and ultrasensitive extraction procedures) has been approved by the FDA and thus is the “gold standard” assay by which all other HIV-1 RNA assays must favorably compare in terms of quantification. The newer Amplicor 1.5 assay incorporates primer pairs designed to detect all HIV-1 clades (17) and has replaced version 1.0 in most labs worldwide, except for those in North America. Studies have shown that the Amplicor HIV-1 Monitor versions 1.5 and 1.0 assays generate comparable quantification on matched clinical samples using the standard and ultrasensitive procedures (17), and as such fall in line with the FDA-approved product. In order to achieve enhanced sensitivity with the Amplicor assay, Roche Diagnostics modified the specimen preparation procedure to allow greater input of RNA from 10-fold more plasma. Whereas the standard procedure for the Amplicor 1.0 or 1.5 assay requires direct lysis of 0.2 ml of plasma and resuspension of RNA in 0.4 ml of buffer, the ultrasensitive procedure for these assays requires ultracentrifugation of 0.5 ml of plasma, lysis of the viral pellet, and resuspension of RNA in 0.1 ml of buffer. With the incorporation of this high-speed centrifugation step to concentrate the virus, the lower detection limit of the Amplicor assay drops from 400 to 50 copies/ml (15). By contrast, the sensitivity of the bDNA assay was improved by modifying the chemistry of the assay. In addition to developing a new target probe set and optimizing buffer conditions to increase signal generation capacity, the nonnatural bases isoC and isoG were incorporated into the binding regions of probes to reduce nonspecific hybridization and thus lower background noise. With these modifications to the assay chemistry, the bDNA 3.0 assay achieves a lower detection limit of 50 copies/ml (4).
This study showed a statistically significant correlation between the bDNA 3.0 and the Amplicor 1.5 assays over a wide range of viral load (50 to 75,000 copies/ml) when tested with matched samples. Also, this study showed that the lower limit of detection was equivalent for both assays when tested on matched clinical samples. Discordant samples from both assays had viral loads within the lower end of the dynamic range (61 to 155 copies/ml) and were well within the reported coefficient of variation for the assays. Assay performance in this study was within acceptable limits since the standard deviations for the operator, the assays, and the environment in which this study was conducted were within assay specifications (within threefold, or less than a 0.5-log difference) based upon controls. Moreover, the performance of the operator was evaluated and approved by both companies by using certified test panels so as to rule out potential bias against either assay. Hence, viral load data generated by either assay yielded comparable results.
The close agreement between the more recent versions of the Amplicor and bDNA assays observed in this study, as compared to the findings of earlier studies of older assay versions (7, 11) may be explained, at least in part, by quantification differences between the 2.0 versus 3.0 versions of the bDNA assay. We found a two- to sevenfold difference in quantification between bDNA 2.0 and 3.0 for samples containing above 500 HIV-1 RNA copies/ml, with the greatest difference occurring at the lower end of the bDNA 2.0 dynamic range (<10,000 HIV-1 RNA copies/ml). It is perhaps not surprising that this anomaly was not noted earlier, since bDNA 2.0 was introduced before the introduction of HAART and thus prior to the significant reductions in viral load now commonly observed. Indeed, discrepancies between bDNA 2.0 and Amplicor 1.0 were noticed only after viral levels in the majority of HAART-treated individuals began to decline. Our study also showed that the use of a more sensitive viral load assay in monitoring serial samples from recently infected individuals beginning HAART provided at least an additional 20 weeks of detectable viral load as compared to the older, less sensitive assay. This added viral load information improves the management of infected individuals since it provides critical information on the efficacy of HAART and helps to rapidly identify the emergence of viral breakthrough in the absence of genotypic or phenotypic analysis.
Given the comparability of assay results, other factors, such as assay components, labor, disposables, and biohazardous waste, may influence the selection of a viral load assay in a clinical setting. Based on U.S. list prices as of November 1999, the per-test cost for the bDNA 3.0 assay was approximately 5% higher than that of the Amplicor 1.5 assay. However, our study demonstrated a significant savings in labor (∼30 to 53%), disposables (∼63 to 67%), and biohazardous waste (∼50%) for the bDNA 3.0 assay as compared to costs of the Amplicor 1.5 assay. It is important to consider that although manufacturers may offer various discounted price plans for kits based on the number of kits purchased, these discounts may not offset the additional hidden costs incurred for labor, disposables, and biohazardous waste. Discounts in the prices of disposables would lead to additional cost savings, but these discounts would be realized for both assays, and the relative difference in the cost of disposables must still be taken into account. The time involved in labor also differed between the two assays. The bDNA 3.0 assay required fewer steps and involved less repeat pipetting and lower complexity (i.e., RNA extraction) than the Amplicor 1.5 assay. The most significant difference in labor between these assays was the sample preparation step—only one manipulation, lysis of the pellet, is required for the bDNA 3.0 assay, whereas Amplicor 1.5 requires four manipulations—one for the viral pellet and three for the RNA pellet. The complexity of the sample preparation step for the Amplicor 1.5 assay adds significantly to the cost of disposables and labor, although automation of this step should significantly reduce the labor component. The other steps (i.e., amplification/hybridization and detection) for bDNA 3.0 and Amplicor 1.5 are easily performed and require relatively minimal labor. Use of the fully automated COBAS system for Amplicor 1.5 will simplify the amplification/hybridization steps. Thus, in selecting the most cost-effective approach, laboratories must consider not only the price of assay components but also all of the costs incurred in HIV-1 viral load testing.
In addition to the costs associated with viral load testing described in this study (assay components, labor, disposables, and biohazardous waste), HIV-1 viral load testing laboratories need to consider the dynamic ranges of the assays for additional potential costs savings. For example, the two specimen preparation procedures of the Amplicor 1.0 and 1.5 assays allow for dynamic ranges of 50 to 75,000 copies/ml and 400 to 750,000 copies/ml for the ultrasensitive and standard procedures, respectively. However, additional costs for viral load testing may result from the need to reflex from one specimen preparation procedure to another. This issue was addressed in a recent study (13) in which it was proposed that, depending on the virologic response, the standard procedure, the ultrasensitive procedure, or a combination of both procedures could be used as the most cost-effective strategy for the Amplicor 1.0 assay. Whereas two extraction procedures are needed to realize the full dynamic range of the Amplicor assay, this approach is not necessary with the bDNA 3.0 assay since the dynamic range of this assay is from 50 to 500,000 copies/ml.
Different individuals working in the management of HIV-1 infection may be impacted differently by the findings of this study on the comparability of viral load data and the relative costs of the bDNA 3.0 and Amplicor 1.5 assays. For the health care worker, the correlation between the quantification values of the bDNA 3.0 and Amplicor 1.5 assays may provide more flexibility in monitoring patients should patients be switched from one assay to another since longitudinal viral load data collected with either or both assays should still be meaningful. For the laboratory director, the availability of two commercial ultrasensitive assays that are comparable allows greater flexibility in selecting the most cost-effective approach for viral load testing. It is essential that comparative studies continue to be performed as new assays for the management of HIV-1 infection are introduced. Moreover, such studies should be supported and encouraged by the manufacturers. As such, any claims can rapidly be confirmed by independent groups in a timely manner.
ACKNOWLEDGMENTS
We thank Roche Molecular Systems, Roche Diagnostics, and Bayer Diagnostics for providing kits and disposables.
Our sincere thanks to Lynette Sawyer at the Bayer Reference Testing Laboratory for providing HIV-1 viral load results on clinical samples used for this study. We also greatly thank Linda Wuestehube for editorial assistance.
REFERENCES
- 1.Brambilla D, Leung S, Lew J, Todd J, Herman S, Cronin M, Shapiro D E, Bremer J, Hanson C, Hillyer G V, McSherry G D, Sperling R S, Coombs R W, Reichelderfer P S. Absolute copy number and relative change in determinations of human immunodeficiency virus type 1 RNA in plasma: effect of an external standard on kit comparisons. J Clin Microbiol. 1998;36:311–314. doi: 10.1128/jcm.36.1.311-314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carpenter C C, Fischl M A, Hammer S M, Hirsch M S, Jacobsen D M, Katzenstein D A, Montaner J S, Richman D D, Saag M S, Schooley R T, Thompson M A, Vella S, Yeni P G, Volberding P A. Antiretroviral therapy for HIV infection in 1998: updated recommendations of the International AIDS Society-USA Panel. JAMA. 1998;280:78–86. doi: 10.1001/jama.280.1.78. [DOI] [PubMed] [Google Scholar]
- 3.Chun T W, Stuyver L, Mizell S B, Ehler L A, Mican J A, Baseler M, Lloyd A L, Nowak M A, Fauci A S. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins M L, Irvine B, Tyner D, Fine E, Zayati C, Chang C-A, Horn T, Ahle D, Detmer J, Shen L-P, Kolberg J, Bushnell S, Urdea M S, Ho D D. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 1997;25:2979–2984. doi: 10.1093/nar/25.15.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finzi D, Hermankova M, Pierson T, Carruth L M, Buck C, Chaisson R E, Quinn T C, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho D D, Richman D D, Siliciano R F. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 6.Fleiss J L. Statistical methods for rates and proportions. xviii. New York, N.Y: Wiley; 1981. p. 321. [Google Scholar]
- 7.Hodara V, Monticelli A, Pampuro S, Salomón H, Jauregui Rueda H, Libonatti O. HIV-1 viral load: comparative evaluation of three commercially available assays in Argentina. Acta Physiol Pharmacol Ther Latinoam. 1998;48:107–113. [PubMed] [Google Scholar]
- 8.Kern D, Collins M, Fultz T, Detmer J, Hamren S, Peterkin J J, Sheridan P, Urdea M, White R, Yeghiazarian T, Todd J. An enhanced-sensitivity branched-DNA assay for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1996;34:3196–3202. doi: 10.1128/jcm.34.12.3196-3202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin H J, Pedneault L, Hollinger F B. Intra-assay performance characteristics of five assays for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1998;36:835–839. doi: 10.1128/jcm.36.3.835-839.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Natarajan V, Bosche M, Metcalf J A, Ward D J, Lane H C, Kovacs J A. HIV-1 replication in patients with undetectable plasma virus receiving HAART. Lancet. 1999;353:119–120. doi: 10.1016/s0140-6736(05)76156-0. [DOI] [PubMed] [Google Scholar]
- 11.Nolte F S, Boysza J, Thurmond C, Clark W S, Lennox J L. Clinical comparison of an enhanced-sensitivity branched-DNA assay and reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1998;36:716–720. doi: 10.1128/jcm.36.3.716-720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Notermans D W, Jurriaans S, de Wolf F, Foudraine N A, de Jong J J, Cavert W, Schuwirth C M, Kauffmann R H, Meenhorst P L, McDade H, Goodwin C, Leonard J M, Goudsmit J, Danner S A. Decrease of HIV-1 RNA levels in lymphoid tissue and peripheral blood during treatment with ritonavir, lamivudine and zidovudine. Ritonavir/3TC/ZDV Study Group. AIDS. 1998;12:167–173. doi: 10.1097/00002030-199802000-00006. [DOI] [PubMed] [Google Scholar]
- 13.Raboud J M, Seminari E, Rae S L, Harrigan P R, Hogg R S, Conway B, Sherlock C, Schechter M T, O'Shaughnessy M V, Montaner J S G. Comparison of costs of strategies for measuring levels of human immunodeficiency virus type 1 RNA in plasma by using Amplicor and Ultra Direct assays. J Clin Microbiol. 1998;36:3369–3371. doi: 10.1128/jcm.36.11.3369-3371.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Segondy M, Izopet J, Pellegrin I, Montes B, Dumon B, Pasquier C, Peeters M, Fleury H J, Puel J, Reynes J. Comparison of the Quantiplex HIV-1 RNA 2.0 assay with the Amplicor HIV-1 Monitor 1.0 assay for quantitation of levels of human immunodeficiency virus type 1 RNA in plasma of patients receiving stavudine-didanosine combination therapy. J Clin Microbiol. 1998;36:3392–3395. doi: 10.1128/jcm.36.11.3392-3395.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun R, Ku J, Jayakar H, Kuo J C, Brambilla D, Herman S, Rosenstraus M, Spadoro J. Ultrasensitive reverse transcription-PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1998;36:2964–2969. doi: 10.1128/jcm.36.10.2964-2969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Todd J, Pachl C, White R, Yeghiazarian T, Johnson P, Taylor B, Holdoniy M, Kern D, Hamren S, Chernoff D, Urdea M. Performance characteristics for the quantitation of plasma HIV-1 RNA using branched DNA signal amplification technology. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;10:S35–S44. [PubMed] [Google Scholar]
- 17.Triques K, Coste J, Perret J L, Segarra C, Mpoudi E, Reynes J, Delaporte E, Butcher A, Dreyer K, Herman S, Spadoro J, Peeters M. Efficiencies of four versions of the AMPLICOR HIV-1 MONITOR test for quantification of different subtypes of human immunodeficiency virus type 1. J Clin Microbiol. 1999;37:110–116. doi: 10.1128/jcm.37.1.110-116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vandamme A M, Van Vaerenbergh K, De Clercq E. Anti-human immunodeficiency virus drug combination strategies. Antivir Chem Chemother. 1998;9:187–203. doi: 10.1177/095632029800900301. [DOI] [PubMed] [Google Scholar]