The vasculature is a multifunctional organ critical for life; In addition to facilitating the distribution of oxygen and nutrients across tissues of the body, blood vessels play a key role in the active transport of immune cells, metabolites, and therapeutics, and in the regulation of blood pressure and the maintenance of normal body temperature. Given the diverse biological functions of the vasculature, it is not surprising that the disruption or maladaptation of the vasculature is a critical step in the progression of many diseases. While some diseases afflict the vasculature itself such as coronary artery disease1 and Sturge-Weber Syndrome2, other diseases such as cancer co-opt blood vessels to meet growing metabolic needs. As such, the vasculature has long been considered as a therapeutic target3–5 and a major contributor to tissue identity and function. Unfortunately, the vasculature is rarely included in in vitro disease models, perhaps contributing to the slowdown in approval of new drugs, despite our increased understanding of the genotypic and phenotypic basis of disease. Indeed, recent data show that the Food and Drug Administration (FDA) approves a mere 13.8% of all drug candidates.6 This pervasive clinical failure of therapeutics emphasizes the need for improved, physiologically-relevant in vitro models of human disease for drug screening, and we make the argument that incorporating the vasculature into these models will be essential.
Current preclinical studies rely heavily on 2D in vitro models or murine in vivo models, both of which, for different reasons, fail to recapitulate vascularized human tissues and thus the behavior of drugs in the body. To address the prevailing dissonance between preclinical models and human studies, researchers have sought alternative human cell-based in vitro platforms not only to assess novel therapeutics, but also to interrogate disease biology. There has been a growing focus on 3D culture technologies, for example, such as stem cell-derived organoids7,8 and patient-derived spheroids,9 as these better maintain in vivo functions in culture owing to the 3D conformation they adopt, allowing for more physiologic cell-cell and cell-extracellular matrix interactions. Unfortunately, the potential of these 3D culture techniques to yield preclinical data with a higher predictive value compared to 2D studies has yet to materialize, mostly due to issues surrounding biological and disease relevance, maintenance of tissue identity, and scalability.7 As such, none of these 3D in vitro models has, to our knowledge, been ubiquitously adopted in preclinical studies by the pharmaceutical industry or in academia, although strategies focused on developing the capacity to vascularize organoids is growing.10
Microphysiological System Platforms: A systematic approach to identify key cellular and molecular determinants driving human physiology, pathophysiology and clinical pharmacology.
Recently, microphysiological system (MPS, also known as, organ-on-a-chip or tissue chip) platforms have emerged with great promise to improve the predictive capacity of preclinical modeling and bridge the translational gap. While platforms vary in design, MPS are bioengineered in vitro environments containing human cells that aim to recapitulate key functional units of tissues and organs. As recently defined by the National Center for Advancing Translational Sciences (NCATS) at the National Institutes of Health (NIH), MPS platforms have three critical and defining characteristics: “the 3D nature and arrangements of the tissues on the platforms; the presence and integration of multiple cell types to reflect a more physiological balance of cells; and the presence of biomechanical forces relevant to the tissue being modeled.”11 The rapid development and maturation of MPS models have been made possible by the adoption of several biomedical technologies developed over the last decade including advanced cell sourcing options, genome editing, and microfluidic techniques. In addition, the advent of single cell RNA sequencing has allowed unparalleled insights into how cells interact with each other in complex tissues. As a result, MPS are uniquely positioned to become physiologically and functionally relevant models that recapitulate several organ micro-features (multicellular composition and architecture, native ECM constituents etc.), including the ability to emulate biomechanical forces, such as flow-derived shear stress.
Much of the MPS literature published thus far has focused on demonstrating the physiological relevance of individual platforms and the proof-of-concept experiments relating to the capacity of the platforms to replace preclinical 2D culture methods. Increasing evidence is emerging detailing how these platforms not only have the capacity to work synergistically with murine models, but also how they provide an independent research tool for interrogating biological phenomena. Utilizing these human-derived systems, diverse biological advances have been made, from resolving aspects of the neuroinflammation and endothelial-to-mesenchymal transition (EndoMT) pathways, to elucidating the role of diseased endothelium in Hutchinson-Gilford progeria syndrome (HGPS).12–14 Thus, MPS platforms hold great promise for both basic science and clinical breakthroughs.
The Vascular Niche and MPS Platforms: Recapitulating developmental and regenerative programs to improve model organ fidelity and function.
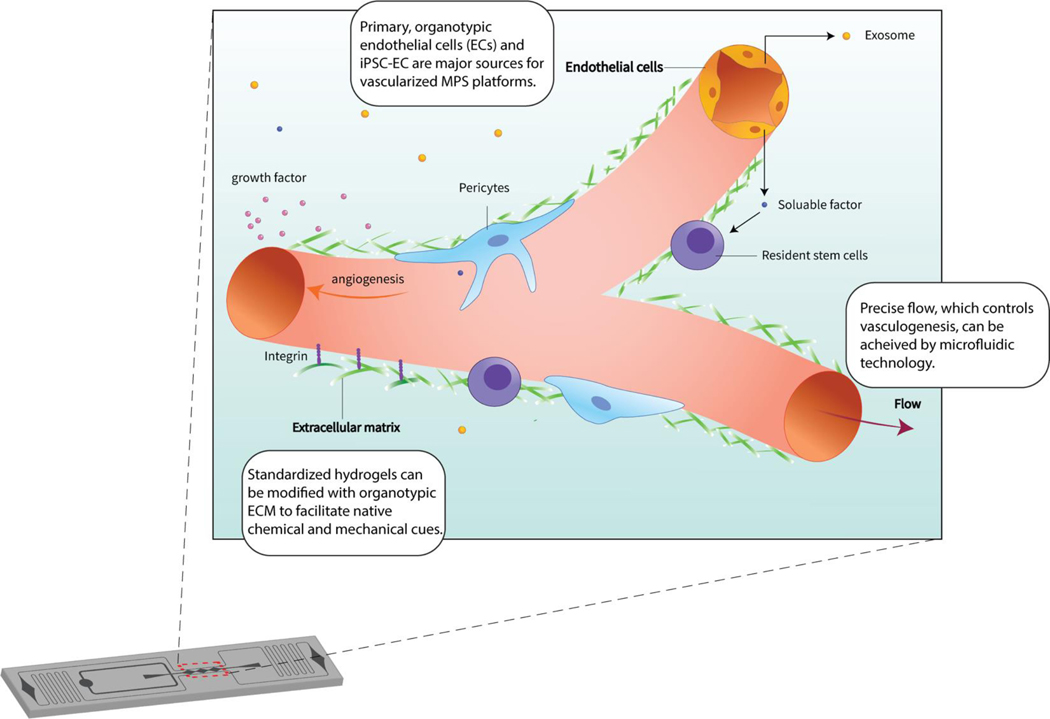
A shared goal amongst many developers is the incorporation of a vascular compartment into the next iteration of MPS platforms. Inclusion of the vasculature is considered an important developmental target as blood vessels not only pervade the body’s tissues providing a thoroughfare for blood borne solutes and cells, but also plays a critical role in organogenesis and organ identity. Indeed, increasing evidence is accumulating regarding the extent to which the unique molecular signatures and functional properties of tissue specific endothelial cells (ECs, the single layer of cells lining the blood vessel lumen) contribute to organ differentiation, identity and function.15,16 As such, the incorporation of a vascular compartment is not only pertinent to the delivery of drugs and nutrients in an in vivo-like fashion to the MPS parenchyma, but also to the development of tissue identity itself. In other words, the vasculature and its niche are essential components of generating physiologically-relevant tissues. Here, we define the vascular niche as the microenvironment proximal to organotypic microvasculature whereby distinct, endothelial extrinsic cues (soluble factors, exosomes, ECM deposition, etc.) actively influence organ regeneration, patterning, and homeostasis often orchestrated with or executed by a resident stem cell population (Fig. 1). Accordingly, current and the next generation of MPS platforms that incorporate a vascular component leverage several developmental and functional characteristics of the endothelium that are critical for the creation of more physiologically relevant models.
Fig 1. Core Engineering Strategies of Vascularized MPS.
Microfluidic technology precisely tunes the microenvironment, including flow dynamics and chemical gradients. By integrating considerations such as endothelial cell sourcing, extracellular matrix components and shear control into the MPS, the platform generates a unique in vitro microenvironment with the capacity to support a resident stem cell population.
This review will discuss published platforms and present considerations for next generation MPS platforms looking to incorporate this critical constituent. First, we discuss EC sourcing, including predominant cell types currently used in the field. Next, we discuss EC mechanosensing and mechanotransduction and discuss options for extracellular matrix that supports not only the vasculature, but also contributes to organ identity. This is followed by a discussion on endothelial shear stress and interstitial flow and how this affects endothelial homeostasis and vascular network formation. Finally, we discuss how establishing the vasculature in next-generation vascularized MPS platforms will take advantage of unique organogenic processes governed by ECs, and how incorporation of a vascular niche will establish a unique human cell-derived in vitro model for stem cell development.
Endothelial Cell Sourcing for MPS Applications
For decades, Human Umbilical Vein Endothelial Cells (HUVECs) have served as the primary in vitro model for studying the physiological and pathological processes of vasculature. As HUVECs are easy to source, culture and manipulate for experimental purposes, it is unsurprising the vast majority of MPS platforms with a vascular component utilize these primary ECs. Recently, transcriptomic developments have detailed the extent to which distinct organ-specific EC expression patterns drive organotypic development and function, supporting the long-standing supposition that the vasculature is remarkably diverse.17 Based on the idea the endothelium is genetically tailored on a per-organ basis that aids overall function, the question arises whether HUVECs, widely considered as a mature EC with a phenotype unique to its resident tissue, best serve organ-specific vascularized MPS models. In this section, we review EC sourcing and outline the benefits, detriments and various characteristics of ECs available for integration into MPS platforms.
Primary Endothelial Cells
Due to their widespread availability and relative ease of use, primary ECs dominate the study of vascular biology, and this has proven a huge advantage as they show superior physiological relevance compared to immortalized lines that rarely if ever retain the ability to make lumenized vessels in culture. Thus, the use of HUVECs and other primary ECs cells, such HMVECs (Human Microvascular ECs), along with Endothelial Colony Forming Cell-derived ECs (ECFC-ECs) is common. The long-term use of primary ECs in the field and subsequent breadth of validation studies demonstrating the ability of primary EC to recapitulate critical aspects of vascular physiology ex vivo has benefited development of vascularized MPS platforms immensely. HUVECs, for instance express many important endothelial junctional proteins (e.g. ZO-1, Claudin-5),18 adhesion molecules (e.g. VE-Cadherin, PECAM-1, ICAM‐1, VCAM-1)18,19 and selectins (e.g. E-Selectin and P-selectin).19,20 HUVECs also store, within Weibel-Palade bodies, von Willebrand factor (vWF), a key player in hemostasis and a well-characterized EC marker.21 Critically, HUVECs retain the capacity to form three-dimensional tubular structures ex vivo with a marked angiogenic,22 leukocyte trafficking, 23 and Nitric Oxide (NO)24 production potential. Thus, as a well characterized, general model of the endothelium, it is not surprising HUVECs have been incorporated into several vascular compartments of organ-specific MPS platforms (Table 1).
Table 1.
Endothelial Cell Sourcing for Vascularized MPS Platforms
| Primary ECs: Human Umbilical Vein Endothelial Cell (HUVEC) | |||||
|---|---|---|---|---|---|
| Organ / Tissue | Vascularized MPS Study Focus | Vascular Network Formation Strategy | Lumenization? | Vascular barrier function assayed? | Ref. |
| Liver | Evaluation of anticancer bioactivity in the TME and hepatotoxicity in the liver | Pre-Patterned | No | - | [189] |
| Various Cancers | Endothelial regulation of chemotherapeutic transport | Self-Assembled | Yes | Yes | [102] |
| Ovarian Cancer | Platelet extravasation through the endothelium into tumor microenvironment | Pre-Patterned | Yes | Yes | [190] |
| Bone Marrow | Bone marrow pathophysiology | Pre-Patterned | Yes | No | [103] |
| Vasculature | Endothelial-to-mesenchymal transition axis | Self-Assembled | Yes | Yes | [13] |
| Various Cancers | Tumor heterogeneity and its influence on vasculature formation | Self-Assembled | Yes | No | [191] |
| Various Cancers | Tumor spheroid induced angiogenesis | Self-Assembled | No | - | [192] |
| Myocardia | Endothelialized-myocardium platform for cardiovascular toxicity evaluation | Bioprinted | Yes | No | [123] |
| Lung | Pulmonary toxicity of nanoparticles | Pre-Patterned | No | Yes | [193] |
| Lung | Anti-fibrotic drug nintedanib and its effect on vascular remodeling | Self-Assembled | Yes | Yes | [194] |
| Bone Marrow | Perivascular bone niche to study metastatic colonization of the bone | Self-Assembled | Yes | Yes | [136] |
| Lung | Pathophysiology of pulmonary thrombosis and advance drug development | Pre-Patterned | Yes | Yes | [195] |
| Retina | Outer blood-retinal barrier model | Self-Assembled | Yes | Yes | [196] |
| Glioma | Bioprinted glioblastoma tumors derived from patient-derived tumor cells | Bioprinted | No | - | [124] |
| Vasculature | Effects of ambient fine particulate matter on the vasculature | Self-Assembled | Yes | Yes | [197] |
| Primary ECs: Organotypic Endothelial Cells | |||||
| Kidney | Tubular–vascular exchange of solutes akin to native kidney tissue. | Pre-Patterned | Yes | Yes | [27] |
| Brain | Contributions of individual cell types of the blood brain barrier (BBB) to inflammatory stimuli | Pre-Patterned | Yes | Yes | [12] |
| Liver | Continuous zonated liver model for diseases modeling and ADME/TOX | Pre-Patterned | No | Yes | [198] |
| Kidney | Constructing a functional kidney glomerular-capillary-wall | Pre-Patterned | Yes | Yes | [199] |
| Vasculature | Endothelial barrier dysfunction associated inflammatory and hematological diseases | Pre-Patterned | Yes | Yes | [200] |
| Brain | Neurovascular microfluidic bioreactor for modeling of BBB function and testing of drug toxicity and permeability | Pre-Patterned | Yes | Yes | [201] |
| Primary ECs: Endothelial Colony Forming Cell derived Endothelial Cells (ECFC-ECs) | |||||
| Vasculature | Vascular inflammation and thrombosis | Pre-Patterned | Yes | Yes | [202] |
| Vasculature / Cancer | Reproducible vascularized micro-organs and tumors | Self-Assembled | Yes | Yes | [100,203] |
| Vasculature | Spatial and temporal control of oxygen tensions characteristic of in vivo biology | Self-Assembled | Yes | Yes | [204] |
| Vasculature | Large-scale perfusable microvascular networks | Self-Assembled | Yes | Yes | [205] |
| Bone Marrow | Hematopoietic stem/progenitor cell culture in parallel perivascular and endosteal niche | Self-Assembled | Yes | Yes | [101] |
| Colorectal Cancer | Capturing tumor heterogeneity, vascular disruption and TME interactions | Self-Assembled | Yes | Yes | [206] |
| Induced Pluripotent Stem Cells derived Endothelial Cells (iPSC-ECs) | |||||
| Vasculature | Developing a CDH5-mCherry reporter iPS cell line amenable to form stable, perfusable microvessels | Self-Assembled | Yes | Yes | [52] |
| Progeria Syndrome | Elucidating diseased endothelium’s role in Hutchinson-Gilford progeria syndrome | Pre-Patterned | Yes | No | [14] |
| Pancreas | Microvessel-pancreatic islet interactions | Self-Assembled | Yes | Yes | [51] |
With the advent of advanced transcriptomic technologies, our understanding of the extent to which organ-specific EC gene clusters establish a functionally diverse endothelium has become increasingly resolved, offering new perspectives on the relevance of incorporating HUVECs into organotypic MPS. Marcu et al., for instance, investigated single-donor EC’s isolated from four major organs—the heart, lung, liver, and kidneys— and demonstrated that these organ-specific ECs have unique gene expression patterns, and show distinct barrier properties, angiogenic potential, and metabolic rates.16 These isolated organ-specific ECs also have the capacity to support in vitro organ development and parenchyma function. When co-cultured with isolated fetal liver ECs, for example, hepatocytes exhibited markedly improved survival and supportive function in terms of albumin production. As evidence accumulates supporting the extent to which specialized ECs contribute to organ identity and function,25 it is apt to consider integrating organotypic ECs into vascularized MPS over more universal models of the endothelium to best recapitulate the in vivo environment and organ function.
Indeed, several vascularized MPS devices that utilize organ-specific ECs see marked physiologic improvement. In comparison to HUVEC-derived in vitro microvasculature, Uwamori et al. demonstrated vascular networks generated from human brain microvascular ECs (BMECs) retain a significantly lower permeability coefficient.26 Not surprisingly, this barrier-like phenotype was characterized by maintenance of tight junction protein expression, a genetic hallmark of blood-brain barrier endothelium. Thus, it may be most important to utilize organotypic ECs in MPS platforms where the microenvironment, including the endothelium, facilitates robust organ function, which may include filtration, absorption, secretion and/or selective permeability. Lin et al., for example, created a 3D vascularized renal proximal tubule MPS composed of renal cortex epithelium and glomerular microvascular ECs (GMECs) that exhibits active reabsorption via tubular–vascular exchange of solutes.27 Importantly, GMECs retain a phenotype within the MPS device reminiscent of its in vivo state including CD31 localization at cell junctions, vWF and tight junction expression, and glycocalyx deposition on the luminal surface. With a microenvironment incorporating organotypic vasculature, this model not only exhibits selective reabsorption and transport of solutes but can also facilitate investigative studies of endothelium–epithelium cross-talk under homeostatic and disease conditions.
While the prospect of utilizing organ-specific ECs within vascularized MPS has advantages based on the potential to facilitate improved organ identity and function, in many cases access to primary, organotypic ECs may not be possible. As most available tissues are acquired from post-operation waste, biopsies for diagnostic purposes or post-mortem tissues, it would be unreasonable to suggest all MPS must incorporate organ specific ECs to be considered as physiologically relevant. While EC may well take on the “correct” phenotype if the surrounding tissue is physiologic enough to provide the appropriate cues, several strategies and technologies can be employed to engineer primary cells in higher abundance to recapitulate an organotypic phenotype, either by genetic manipulation or chemical treatment. Short-term expression of ETV2, a developmental EC transcription factor, for instance, “resets” HUVECs to a malleable progenitor state able to adopt tissue specific characteristics.28 Additionally, all-trans retinoic acid (ATRA) has been utilized to induce a blood-brain barrier phenotype in ECs including ECFC-ECs,29 a progenitor-like EC commonly isolated from umbilical cord or peripheral blood that expresses markers of the three endothelial differentiation lineages (venous, arterial, and lymphatic).30,31 Increasingly, ECFC-ECs, with the capacity to adopt tissue-specific EC phenotypes, are growing in popularity with MPS platform developers for their relative ease of accessibility and heightened proliferative and vasculogenic capacity (Table 1).30,32
With the expansion of single cell RNA sequencing technologies, we can begin to understand how well the transcriptomes of generalized endothelium maps onto those of organotypic ECs to determine how well suited the former can be utilized in organ specific models. Lukowski et al. demonstrated that differentiated murine ECs originating from the aorta, for instance, have a transcriptomic profile with significant overlap to the HUVEC transcriptome.33 The idea organotypic ECs share some transcriptomic basis with those from another organ has also been supported by recent findings; while the transcriptome of ECs derived from the brain and liver presented unique identities, ECs from other tissues (adipose, heart, and aorta) shared more significant transcriptomic similarities.25 It is currently unclear to what extent functional EC properties are intrinsic, acquired during early differentiation or reprogrammed by the local environment. Thus, in some cases incorporating cells that are more mature (i.e., HUVEC, arterial EC) may be appropriate, while in others, a cell with more plasticity (i.e., ECFC-EC) might be a better choice. Indeed, MPS models may be the ideal environment in which to study these very questions.
Induced Pluripotent Stem Cell (iPSC)-derived Endothelial Cells
While incorporating primary ECs within MPS platforms is perhaps the most straightforward sourcing strategy to faithfully recapitulate the endothelium ex vivo, variable tissue availability, relatively poor regeneration capacity and patient-to-patient variability tends to compromise model reproducibility. The advancement of iPSC technologies can remedy these difficulties as a potential infinitely renewable and reproducible cell source. Generated by reprogramming somatic cells via forced transcription factor expression, iPSCs have the capacity to differentiate into any lineage of the three germ layers.34 This unprecedented opportunity to generate ECs including those with vessel- and organ-specific properties has spurred great developments within vascular biology, benefitting developmental,35,36 disease37,38 and therapeutic modeling39–41 alike.
iPSC-derived EC (iPSC-EC) differentiation strategies have been extensively described.42,43 Briefly, most protocols direct iPSC-EC differentiation through a mesoderm linage and then introduce endothelial specification by the addition of vascular endothelial growth factor (VEGF). Additional components such as miR-21 and transforming growth factor beta (TGF-β) amongst others can be added to increase differentiation efficiency.44 While these chemically defined differentiation protocols are the most frequent approaches, iPSC-ECs can also be generated through the overexpression of endothelial development transcription factor ETV2.45 Regardless of differentiation approach, pure iPSC-EC populations are captured by fluorescence-activated cell sorting (FACS) or magnetic-activated sorting (MACS) based on expression of the endothelial marker PECAM1 (CD31), and lack of hematopoietic markers, such as CD45.46
Like their primary EC counterparts, iPSC-EC recapitulate genetic hallmarks of the endothelium, including PECAM-1 (CD31), VE-Cadherin (CD144), VEGFR2 (KDR), and vWF expression.47 Multiple sources also report iPSC-EC exhibit typical cobblestone morphology and basic endothelial functions including limited “tube” formation on Matrigel,48 hematological and inflammatory responses,49,50 and NO production.36 Critically, reports show iPSC-ECs are capable of undergoing vasculogenic and angiogenic processes to form 3D conduits and vascular networks, and these have been incorporated into a handful of vascularized MPS platforms (Table 1). Unfortunately, widespread integration of iPSC-EC has been hampered by the limitations of current-generation EC differentiation techniques. Most protocols, for example, generate iPSC-ECs exhibiting deficiencies in capillary morphogenesis; in comparison to primary ECs, iPSC-ECs often make fragmented or inconsistent vascular networks.51,52 This suggests current iPSC-ECs protocols produce EC populations with a limited vasculogenic potential, most likely due to limited endothelial commitment and a more immature phenotype. While the molecular basis for endothelial tube formation is increasingly understood,53,54 the critical genetic characteristics or biophysical cues necessary for iPSC-ECs to obtain and maintain a vasculogenic capacity have yet to be determined.
Patient-derived iPSCs can also be generated, presenting a new frontier in personalized medicine. The development of iPSC-derived diseased endothelium is a particularly interesting point of vascularized MPS development as dysfunctional vasculature is a key driver in many diseases.55 Hereditary Hemorrhagic Telangiectasia (HHT), for example, is associated with loss-of-function mutations of Endoglin (ENG) or Activin A receptor like type 1 (ACVRL1/ALK1) and is characterized by vascular lesions, including telangiectasias and arteriovenous malformations (AVMs).56 Patient-derived iPSC-ECs harboring genetic determinants of disease, such as those driving HHT, can not only be utilized to evaluate genetic mutations as causative factors, but also be manipulated via genomic editing to revert mutations and generate isogenic controls. For example, Zhou et al. generated iPSC-ECs from an HHT patient with a novel ENG missense mutation. After subcellular analysis, the authors demonstrated mutant ENG sequesters within the endoplasmic reticulum, affecting downstream of BMP signaling, a modulator of vascular homeostasis and remodeling. Following autologous mutant correction, partial BMP signaling was restored.57 In a similar vein, Gu et al. corrected BMPR2 mutations in iPSC-ECs derived from patients with familial pulmonary arterial hypertension (FPAH) and confirmed that the aberrant gene is causal for the abnormal phenotype.58 Upon confirmation that these isogenic lines retain a vasculogenic capacity to form networks in vitro, the paired cell types can be integrated into MPS platforms, not only to reveal diseased endothelium genotype-phenotype associations, but also to facilitate predictive personalized medicine applications that take into account the patient’s unique genetic background.
iPSC-ECs have the potential to become widely utilized within vascularized MPS platforms as they model critical aspects of the endothelium and alleviate primary EC accessibility challenges. While there has been considerable progress on subtype- and patient-specific iPSC-ECs derivation, including protocols to generate arterial and venous-like ECs,36 several areas of optimization remain including maximizing efficiency, decreasing differentiation time, and generating bona fide organotypic endothelial subtypes. Moreover, iPSC-ECs as currently generated, often do not exhibit the same vasculogenic potential as either HUVECs or ECFC-EC as noted above, nor are they a homogeneous population; Paik et al. uncovered 4 transcriptionally unique subpopulations of iPSC-EC.59 This heterogeneity likely plagues most iPSC-EC populations. Protocols that address these issues, however, lack uniformity, and the absence of stringent genetic and functional assessments prevents meaningful cross-comparison analysis between research groups. The problem of increasingly divergent differentiation protocols utilizing variable quality control measures was recently exemplified in a transcriptomic study of iPSC-BMECs that revealed how a widely-adopted iPSC-to-brain EC differentiation protocol60–62 actually generates a homogenous epithelial cell population.63 Papers that have used these cells in the formation of blood-brain barrier models will need to be re-evaluated.
Endothelial Cell Sourcing Outlook
To optimally recapitulate the endothelial compartment, integrating ECs into MPS systems relies on fit-for-use modeling decisions based on the unique MPS microenvironment, the capacity to generate or harvest organ-specific ECs and the hypothesis in question. Currently, HUVECs dominate MPS development. With the progression of transcriptomic analysis detailing the extent of endothelial diversity and the improvement of iPSC technologies, it is likely the next generation of MPS devices will primarily utilize organotypic and iPSC-ECs to improve organ fidelity. Indeed, advanced iPSC protocols are maturing, including recent approaches to generate specific endothelial lineages from iPSC derived from ECFC-ECs.64 This approach suggests iPSC-ECs may retain an endothelial epigenetic memory and maintain an endothelial phenotype better than ECs generated from fibroblast-derived iPSC. Ultimately, the expanding use of iPSC technology for the generation of EC, and other cells, calls for the generation of standardized protocols with stringent, agreed-upon functional and genetic validation.
Endothelial Extracellular Matrix: Vasculogenesis, Homeostasis, and Repair
The ECM is a multifunctional biological scaffold that not only provides physical support, but also contributes to tissue homeostasis via its surface topology, surface ligand landscape and composition.65 This non-cellular meshwork is composed of a multitude of proteins and proteoglycans and delivers critical biochemical and biomechanical cues to resident cells that are essential for organogenesis, homeostasis, angiogenesis and tissue repair.66 Thus, it is no surprise the ECM has long been used as a substrate to promote functional tissue reconstruction in clinical regenerative medicine, including esophageal repair,67 cartilage and bone regeneration,68 and musculotendinous tissue repair.69 These methods to restore tissue functionality involve the delivery of constructive cues and modulation of the microenvironment through ECM signaling. Accordingly, understanding the crosstalk between cells and their surrounding ECM will offer new strategies on how to fine-tune gene expression and cell phenotype within MPS platforms. A more robust and functionally relevant model can be achieved by taking advantage of the distinct ECM-cell receptor pairings that trigger intracellular signal events and cellular responses, including pro-proliferative and migratory EC phenotypes.70–72 Moreover, studies demonstrate ECM composition (the scaffolding proteins and embedded growth factors) has a high degree of tissue and organ specificity, which plays a critical role in tissue and organ morphogenesis.73 From this perspective, understanding how ECM influences EC behavior and organ identity is key to generating physiologically relevant and organ-specific vascularized MPS. In the following paragraphs, we briefly discuss how ECs sense and respond to chemical and mechanical cues of the ECM, what MPS scaffolding substrates are currently utilized and offer suggestions on incorporating organ-specific ECM in next-generation MPS platforms.
Endothelial Cells and Mechanosensing
The ECM is composed of various biopolymers and specialized macromolecules, including collagen, elastin, adhesion proteins (e.g., laminins and fibronectin), hyaluronans, and proteoglycans.74 Together, these constituents work in concert to support tissues and organs; while collagen and elastin act as a scaffold providing mechanical support to resident cells, hyaluronans and proteoglycans coordinate to fill the empty spaces between fibrous structures and promote water retention within the tissue.74 Mechanosensing describes the ability of cells to sense and integrate mechanical cues from their matrix-based microenvironment. Receptor-mediated mechanosensing is initiated via the ligation of a distinct set of cellular surface receptors and ECM pairings. This coupling results in the transmission and activation of intracellular signaling cascades including those involved in cell cycle control, migration, and matrix remodeling. The integrin family, which represents the principal cellular receptors cells use to attach to the ECM, for example, consists of eighteen α-subunits and eight β-subunits, which join into 24 combinations.75 This subunit diversity allows for the high degree of integrin-ECM specificity necessary for determining behavior of distinct cellular subsets.
The instructive role of ECM in endothelial homeostasis and angiogenesis
While its scaffolding function suggests a rigid-like structure, the ECM is a malleable and highly dynamic network. The constant remodeling of the ECM governed by deposition-degradation dynamics actively influences endothelial proliferative and migratory processes necessary for angiogenesis. Immobilized within the matrix of many tissues, for instance, are angiogenic cytokines, including vascular endothelial growth factor (VEGF), insulin like growth factor (IGF), fibroblast growth factor (FGF), transforming growth factor-β1 (TGF-β1) and hepatocyte growth factor (HGF). Upon chemical modification or enzymatic degradation of the ECM, previously sequestered factors generate instructive chemical gradients, including those necessary to induce pro-proliferative and chemotactic phenotypes. Small-molecular-weight peptides derived from degraded ECM also play a role in the attraction of ECs to remodeling sites during physiological processes such as organogenesis and wound healing.76 Furthermore, mechanical cues generated by the ECM topography can also influence vascular remodeling in a process known as haptotaxis.77 Haptotaxis refers to EC movement in response to gradients of ECM components that are governed by ECM-integrin interactions. HMVECs, for example, show haptotactic migration in response to gradients of fibronectin.78
Current ECM Considerations for Vascularized MPS Platforms
Indeed, the ECM not only provides a scaffold essential for maintaining the organization of vasculature, but also the anchorage necessary for endothelial morphogenesis, proliferation, migration, survival, and homeostasis.73,77 Unsurprisingly, ECs in 3D cultures retain more faithful genotypic and phenotypic identities ex vivo than do the same cells grown in 2D.79 While the geometric intricacies of vascularized MPS vary, capillary-like structures are primarily generated in vitro via three engineering strategies – self-assembled, pre-patterned, or 3D-bioprinted vasculature – with each method owning a particular set of strengths and weaknesses. Self-assembled vascularized MPS platforms, for example, recapitulate de novo vasculature formation in vitro and are thus a strong model of vascular biology. Souring ECs that maintain a strong vasculogenic potential ex vivo, however, is required, and in general, a hierarchical vasculature is hard to create. Conversely, pre-patterned and 3D-bioprinted vascularized MPS devices form fully perfusable vasculature with predefined dimensions and controlled configurations, but vessel size is largely limited by the resolution and size of the mold or nozzle, resulting in lumen diameters of 100μm or more.10,80. In addition, in many cases these vessels cannot, or do not, remodel in response to external cues. Regarding ECM selection for each vascularized MPS engineering technique, while pre-patterned vasculature MPS platforms focus on utilizing ECM micro-architectures to adhere ECs onto inert biomaterials, self-assembled and bioprinted MPS platforms seek to provide mechanical support for de novo vascular formation. Thus, scaffolding selection and subsequent deposition techniques necessary to generate ECM-laden platforms are largely dependent on MPS microfabrication approach.
Self-Assembled Vascularized MPS Platforms
Self-assembled vascularized MPS platforms encompass those that utilize the intrinsic capacity of ECs to form capillary-like structures within biocompatible matrices (Fig. 2A).81–83 This requires a malleable microenvironment that mechanically supports EC engraftment and subsequent lumenization. While several matricellular proteins, including SPARC, β-IgH3 and IGFBP7, have been shown to be critical for lumen formation,84 matrix contributions during capillary morphogenesis remains less resolved. Much of our current understanding regarding matrix-lumen dynamics derives from earlier studies in non-perfusable 3D culture that established the basic parameters for self-assembly of vessel-like structures, including a bead-based angiogenic sprouting model22,85 and a model of human capillary tube formation embedded within hydrogels.86
Fig 2. Strategies to build vascularized MPS.
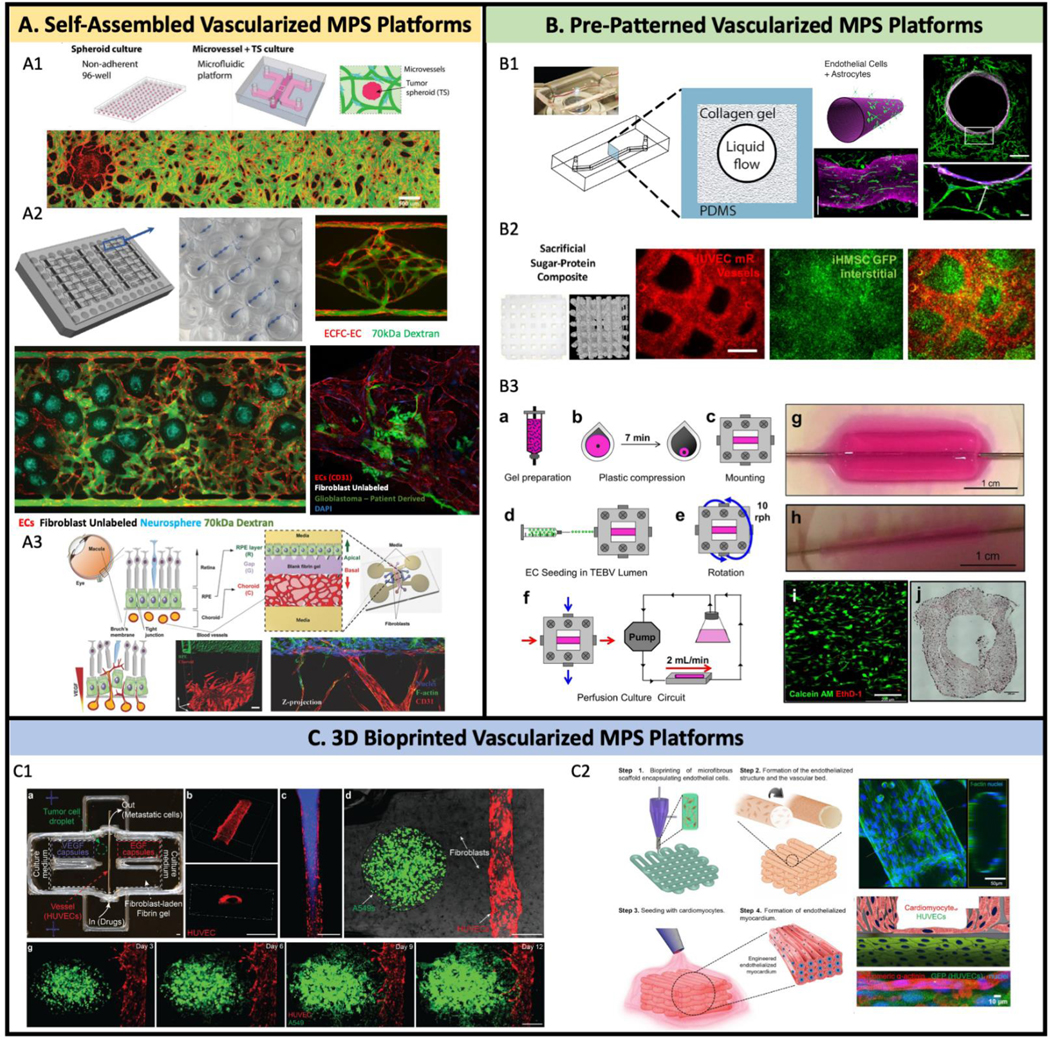
(A) Self-assembled in vitro vascular network on MPS platforms. (A1) 3D vascularized tumor on-chip was built by co-culture 3D tumor spheroid, EC, and fibroblast into fibrin gel. (A2) 70kDa FITC dextran flowing through the upper “arteriole”, through the capillary network, and out through the low pressure “venule” at the bottom. Additional cell types can be seeded into the VMO as an in vitro model of other organs, vascular diseases, and cancer types and is amenable to the growth of neurospheres. Patient derived glioblastoma in the VMO tends to migrate towards and along the vasculature, suggesting the presence of a “preferred” vascular niche. (A3) A organotypic eye-on-a-chip model mimics the pathogenesis of choroidal neovascularization (CNV) in the retinal pigment epithelium (RPE) choroid complex in vitro. (B) Pre-pattern vascularized MPS platforms. (B1) A 3D model of the human blood-brain barrier (BBB) within a cylindrical collagen lumen structure. (B2) Micron-resolution vascular features made by sugar-protein based sacrificial structure. (B3) Tissue engineered human blood vessels (TEBV) with human neonatal dermal fibroblasts (hNDFs) embedded in collagen gel. Coronary artery disease endothelial progenitor cells (EPCs) are seeded into TEBV lumen. (C) 3D bioprinted vascularized MPS platforms. (C1) Vascularized tumor models are built by 3D printing techniques to mimic cancer dissemination. (C2) Heart-on-chip with bioprinting endothelialized myocardium.
(A1) Adapted from (102), with permission of John Wiley and Sons.
(A3) Adapted from (196), with permission of John Wiley and Sons.
(B1) Adapted from (12), with permission of PLOS.
(B2) Adapted from (111), with permission of Elsevier.
(B3) Adapted from (207).
(C1) Adapted from (125), with permission of John Wiley and Sons.
(C2) Adapted from (123), with permission of Elsevier.
Hydrogels mimic ECM microstructures with the capacity to retain water and facilitate nutrient and metabolite transport. Critically, hydrogels have several tunable characteristics including permeability and viscoelasticity, while advanced temperature-87 and light88-responsive hydrogels permit dynamic cell culture. As vasculogenic and angiogenic processes are necessary to form capillary-like networks in vitro, promoting specific EC-ECM interactions is especially critical for vascularized MPS platforms that are not pre-patterned. Thus, to support self-assembled vascularized MPS platforms, a composite matrix of hydrogel supplemented with additional ECM proteins is commonly used.
Collagen I hydrogels
Collagen I, an ECM protein that naturally assembles into a fibrous hydrogel, is abundantly present in many tissues. Several vascularized MPS platforms incorporate purified collagen I,89–92 as this hydrogel is amenable to changes in stiffness via alterations in concentration93 or gelation temperature94 and promotes endothelial survival and angiogenic processes.95 When type I collagen binds α1β1 or α2β1 integrins (amongst many), the mitogen-activated protein (MAP) kinase cascade is trigged, which promotes EC viability and angiogenesis.96,97 Thus, collagen I not only promotes cell association within MPS platforms and its biomaterials, but also mimics the physiological cues supporting ECs and vascular network formation.
Besides EC-collagen signaling cascades that benefit angiogenesis, the ability to inject collagen I directly into microfluidic channels makes it useful for MPS applications. As a thermo- and pH-responsive polymer, collagen I is stable in acidic solutions at low temperatures and is capable of crosslinking below 37 ºC and at neutral pH.98 Because of these properties, it is easy to control collagen crosslinking after injection without the need for a catalyst. Moreover, tuning the concentration of collagen can optimize the growth of different types of cells. Adriani et al. presented a co-culture microfluidic platform, incorporating neurons, astrocytes, and cerebral ECs, to create a blood-brain barrier model.99 To optimize the hydrogel for each cell type, they tuned the concentration of collagen I and found that increasing the concentration to 7 mg/ml was optimal for astrocytes, while still avoiding gel contraction, whereas 2.5 mg/mL was most suitable for neuron survival.
Fibrin hydrogels
Fibrin gel, another popular hydrogel employed in MPS platforms, exhibits excellent biocompatibility, promotes cell attachment, and can be remodeled by ECs.100–103 Fibrinogen (the precursor to fibrin) self-assembles into a polymer network upon mixing with thrombin, which constitutes the basis of blood clotting. Thus, fibrin gels can “cure” in situ and is a suitable material to inject suspended cells into microfluidic channels.104 Furthermore, this substrate can enhance angiogenic processes in vitro.105 Roy et al., for example, investigated the effects of platelet-rich fibrin matrix (PRFM) on EC behavior and showed that PRFM releases several growth factors, including VEGF-A and TGF-β, both of which are involved in endothelial remodeling and neovascularization.106 Additionally, to tune the matrix’s mechanical properties closer to the stiffness of the organ in vivo, fibrin can be mixed with different ECM components and inorganic minerals; Ahn et al. developed a microfluidic model that encapsulates the 3D bone tumor microenvironment.107 Since bone retains inorganic mineral hydroxyapatite (HA), which confers high stiffness and toughness properties, HA nanoparticles were added to the ECM to recapitulate the physiological microenvironment in vitro.
Pre-Patterned Vascularized MPS Platforms
Pre-patterned vasculature relies on preformed microfluidic channels to generate a lumenized vascular structure (Fig. 2B).108,109 Typically, several microfluidic channels are arrayed in parallel connected by a porous membrane that delineates different organ compartments (vascular, stromal, parenchymal, etc.). More complex vascular networks can be formed using sacrificial templates made of sugar or alginate.110,111 In most cases, to form a vessel-like structure these channels need to be coated with matrix to promote EC adhesion. This is accomplished by flushing the channels with relevant ECM proteins.
Laminin, Fibronectin and Collagen Coating
Options in EC coating techniques have been extensively described.112,113 Briefly, endothelial basement membrane proteins, such as laminin, collagen, or fibronectin are used to coat the surface and provide the native substrate necessary to achieve stable anchoring of ECs. Laminin, a cross shaped glycoprotein, is a major basement membrane component and a common EC in vitro substrate. Besides its structural role, laminins facilitate vascular tissue morphogenesis and homeostasis by regulating tissue architecture and adhesion-migration dynamics.114 Importantly, the use of laminin substrates is beneficial in vascularized MPS platforms that incorporate progenitor-like or iPSC-ECs, as it has been shown to promote commitment towards endothelial lineages.115 Fibronectin, another glycoprotein, is also a commonly used EC substrate coating.116,117 It benefits EC growth by promoting VEGF-induced differentiation118 and is efficiently absorbed onto PDMS,119a common MPS biomaterial. Finally, collagen is a suitable ECM coating for EC attachment and broadly employed for biomedical engineering purposes.120 As micropatterning studies uncover the extent to which cellular topographies regulate morphology, migration and mortality117,121 strategies to leverage collagen’s capacity to finely guide endothelial alignment have been employed. At increasing flow rates of injection, for example, collagen fibrils align parallel to experienced shear; ECs seeded on the aligned matrix migrate along the direction of fiber alignment, suggesting that the orientation of ECM regulates EC morphology.122 This technique for producing oriented collagen fibers can be applied to pre-patterned vascularized MPS system by controlling the device geometry and flow conditions of collagen necessary for effective EC attachment.
3D Bioprinted Vascularized MPS Platforms
3D bioprinted vascularized MPS platforms include those where tissues are generated layer-by-layer from a single-cell suspension (Fig. 2C).123–125 In recent years, 3D printing has emerged as an important biomedical tool. A key advantage of 3D printing is the ability to have precise control within a 3D space to generate complex geometrical features of vascular tissue in vitro. Cui et al., for instance, utilized thermal inkjet printing technology to fabricate micro-sized vascular channels. Before printing, HMVECs were premixed with thrombin, which serves as the “bio-ink,” and printed onto a fibrinogen substrate. Thrombin then catalyzes fibrin gel polymerization, thereby minimizing structure deformation after printing. The authors found cells aligned themselves within the fibrin channel and proliferated to form a confluent lining.126
As the base bio-ink material, an ideal hydrogel should show the following properties: printability, mechanical strength, and post printing gelation. Bio-ink printability depends on several parameters such as viscosity of the solution, surface tension of the bio-ink, and the ability to initiate crosslinking. If the bio-ink is highly viscous, for example, the 3D printed structures will have higher stability, but this requires higher extruding pressure during printing.127 As individual printed filaments require enough strength and stiffness to maintain structural integrity after printing, a balance needs to be attained between printability and printed structure stability. Finally, hydrogel gelation is an important aspect in preserving the shape of bioprinted structures and minimizing structure collapse.128 As 3D printing is a relatively young technology, the majority of research developed thus far focuses on fine tuning broader characteristics of bio-inks including printability, biocompatibility and ease of use; bio-ink specifics for optimal EC culture have yet to be developed.
The Promise of Tissue-Specific ECM in Next-Generation Vascularized MPS Platforms
Increasing evidence shows that tissue-specific ECM can promote site-appropriate differentiation of stem cells and maintain the appropriate phenotype in vitro.129,130 Sellaro et al., for example, cultured sinusoidal ECs (SECs) on different substrates, including ECM derived from the liver (L-ECM), bladder (UBM-ECM), and small intestine submucosa (SIS-ECM)131 and showed that while SECs cultured on L-ECM maintain a differentiated phenotype for a minimum of three days, those cultured on UBM-ECM and SIS-ECM showed signs of dedifferentiation after 24 hours. These results verify tissue-specific ECM scaffolding optimally supports a unique population of cells in vivo and in vitro and is currently an underutilized within organotypic MPS platforms. Finally, tissue-specific ECM is proposed to have distinct tissue development roles through influencing cell differentiation, variation in pore diameter,132,133 the surface texture, and the rigidity and elasticity134,135 of the tissues.
As noted, most vascularized MPS platforms currently utilize standardized (non-tissue-specific) ECM coatings and hydrogels to promote endothelial attachment and maintain a vasculogenic potential. Only more recently have developers applied organ-specific ECM within MPS platforms harvested from decellularized tissues. Marturano-Kruik et al., for example, incorporated ECs and bone marrow mesenchymal stem cells (MSCs) within a decellularized 3D bone matrix embedded microfluidic platform to create a perivascular niche-on-a-chip.136 The authors demonstrate bone marrow MSCs undergo phenotypical transition toward mural cell lineages and support the formation of capillary-like structures. Furthermore, ECM derived from diseased organs can also provide informative cues that support pathogenesis. Romero-López et al. compared the protein composition and stiffness of decellularized ECM from normal and colon tumor tissue.137 After evaluating vascular network formation and tumor growth in each of the reconstituted matrices, the authors concluded that the tumor-derived matrix enhanced tumor cell proliferation and altered vascular network formation.
While decellularized ECM constitutes a perfect source of biological matrix possessing the organotypic molecular and mechanical properties necessary for tissue support, there are several obstacles to its universal application within MPS devices, such as limited tissue biopsy access, heterogeneity, and challenges associated with cell removal versus preservation of ECM composition.138,139 Thus, we believe that modifying the EC hydrogel scaffold or coating to mimic the important properties from the tissue in vivo, such as ECM composition, stiffness, and cell attachment is a more practical way to achieve tissue-specific ECM characteristics while current methods to purify organotypic ECM is currently inefficient.
Endothelial Cells under Flow Conditions Sense Shear
Under physiological conditions, the endothelium is exposed to various mechanical and hemodynamic forces due to interstitial and pulsatile blood flow. While radial forces caused by intravascular pressures are derived from the former, axial shear forces caused by friction are driven by the latter. Force and shear vary spatially and temporally throughout the vasculature depending on vessel diameter, wall thickness, geometry and pre- or post-capillaries. For instance, in straighter sections of blood vessels flow is laminar and largely unidirectional; at regions of blood vessel curvature, flow is more likely to be disturbed with stagnant dead volumes. Moreover, straighter vessels experience greater wall shear stresses than curved vasculature.140,141 Importantly, however, at the small diameters used in MPS systems the Reynolds number is low and the flow can safely be assumed to be linear. Existing in such a highly dynamic microenvironment, ECs integrate this collection of biomechanical cues through a suite of luminal transmembrane receptors, cell-cell junctions and cell-basement membrane interactions mediated by integrins. These signals modulate a variety of cellular responses including morphogenesis, cell function and gene expression.142,143 Increasingly, flow shear stress has been shown to regulate vascular network formation, remodeling, and stabilization.144 Thus, in order to create a physiological environment for vessel formation and maintenance in vitro, precisely controlling flow dynamics within proper ranges is key to maintaining normal EC function within MPS devices. In this section we explore the interaction between flow shear and EC barrier junction and the regulation of EC homeostasis by shear.
Mechanotransduction on Endothelial Barrier Junctions
The most common mechanical force ECs experience derives from fluid flow. ECs utilize membrane bound mechanotransducers, such as G protein-coupled receptors, glycocalyx and endothelial-specific junctional complexes to convert mechanical forces into biological responses in a process called mechanotransduction.145 The Piezo1 ion channel, a newly discovered shear stress mechanosensor, senses pulsatile blood flow and triggers Ca2+ signaling, for example, which can result in the reorganization of ECs towards the polarity of the applied force.146 Additionally, ECs maintain specialized cell-cell contacts via VE-cadherin-based adheren junctions to retain vascular integrity in a force-filled environment.147
Effects of Flow Shear on Endothelial Phenotype and Homeostasis
Subjecting ECs to unidirectional or disturbed shear stress patterns has been shown to significantly alter cell morphology and phenotype.148 To demonstrate this phenomenon, Song et al. designed a microfluidic model of angiogenic sprouting and studied the interplay of shear stress, transverse interstitial flow, and VEGF gradients in mediating sprouting morphogenesis.149 In this model, liquid perfuses two adjacent channels lined with ECs sandwiching a collagen filled chamber that allows vessel sprouting. By controlling the flow configuration in the two-channel device, the direction and flow profile of interstitial flow across the collagen chamber could be determined, and VEGF gradients, which govern EC morphogenesis, could be established. In line with previous observations, they found that endothelial tip cells orientate against the direction of interstitial flow, and likely, the VEGF gradient.
In addition to modulating the morphology and migration of ECs, shear stress also regulates EC homeostasis. Several studies have reported that shear stress induced by blood flow triggers essential signaling pathways that regulate endothelial homeostasis, such as those leading to generation of reactive oxygen species (ROS) and nitric oxide (NO).150,151 To demonstrate this effect, Chen et al. targeted SIRT1, an important modulator in cardiovascular function in health and disease and found that overexpression of SIRT1 prevented endothelial senescence, whereas inhibition of SIRT1 caused premature senescence.152 The authors found that, compared to ECs cultured under static conditions, laminar flow at 12 dyn/cm2 increased EC SIRT1 expression, thereby increasing NO bioavailability.
Flow Shear Regulates Vascular Morphogenesis
Vascular morphogenesis is a dynamic process regulated by shear stress. As the magnitude of flow changes, thereby impacting the levels of shear that ECs experience, vessels modify their diameter proportionally to restore the initial shear stress level.153 This suggests that ECs maintain flow shear stress within a desired range. Baeyens et al. introduced the flow shear stress set-point theory, which stipulates that vessel deviation from a set point induces readjustment of vessel diameters.154 More specifically, flow shear stress at the set point maintains vessel quiescence, while shear higher or lower than the set point triggers vessel remodeling. Thus, microfluidic platforms that can precisely regulate the flow dynamic field, including flow rate, shear, flow direction, and Péclet number (Pe) are a powerful tool to investigate the role of physiological mass transport during vasculogenesis. Hsu et al.,144 for example, designed three microfluidic flow conditions: high interstitial flow (Pe>10) in the transverse and the longitudinal flow direction, an intermediate state that is close to normal living tissue, and diffusion-dominant flow (Pe<0.1) and reported that vasculogenesis can be induced under high interstitial flow and diffusion dominant flow, but not under intermediate state (normal) flow.
Controlling Dynamic Flow Fields in Microfluidic Platforms
MPS platforms benefit from microfluidic-based technology that enables the use of small culture medium volumes, control over chemical and physical cues in the microenvironment, and the ability to perform relatively high throughput experiments. Fluid flow can be driven in MPS platforms via several techniques. One of the simplest and most common methods utilizes hydrostatic pressure: two liquid reservoirs with different liquid heights generate passive flow from the higher reservoir to the lower. This simple method is used extensively as no external pumps are required. However, flow rate decay occurs over time as the pressure driven by the hydrostatic head diminishes.100 As a result, regular supplement of liquid is needed to maintain liquid levels. To solve the issue of passive pumps, Wang et al. proposed a siphon-based micropump system with autofill function.155 Alternatively, rocker-based platforms can be used to periodically reverse the direction of flow, thereby maintaining a reasonably uniform flow rate.156,157 Finally, active mechanical pumps, such as syringe pumps with fine tuning flow capabilities, enable stable flow rate over time.83 Syringe pumps, however, are bulky and not suitable at scale.
The Vascular Niche and MPS Organ-Specific Identity
The vasculature was long described as a passive conduit for delivering oxygen and nutrients, aiding blood coagulation, and facilitating the transportation of immune cells. Only more recently has the vasculature been appreciated for its genotypic and functional diversity, including the unique microenvironment it generates, and its contributions to organ development, homeostasis and regeneration. Indeed, the milieu proximal to organotypic microvasculature (i.e. the vascular niche) harbors distinct endothelial extrinsic cues affecting cellular differentiation, survival and proliferation.158–160 These instructive interactions between the vasculature and its resident cells was first hinted at by the finding that stem cells often reside close, to and interact with, capillary beds.161,162 The influence of the vascular niche on stem cells, parenchymal cells and disease progression is becoming of increasing interest to investigators in multiple fields. In this section, we briefly describe the vascular niche, its role in organogenesis, homeostasis, repair, and disease progression, and how incorporating a vascular compartment can strengthen the physiological relevance of the next generation of MPS devices.
The Instructive Role of the Vascular Niche
While the transcriptional specification of the vascular plexus163 and its anatomical development164,165 are reasonably well understood, the molecular basis of organotypic specialization is an emerging field. Our current understanding of organotypic vascular development largely relies on seminal studies of endothelial specialization in the liver,166 pancreas,167 and brain.168 Capillaries permeate all developing tissues and maturation of organotypic endothelium occurs synergistically with its resident organ. In other words, the endothelium assumes a distinct genotypic profile induced by the parenchyma and in return parenchymal differentiation is influenced by secreted, and likely transmembrane, endothelial-derived signals. It is not yet known whether this reciprocal developmental relationship extrapolates to all organs. While the molecular underpinnings of EC specialization remain unclear, the active influence of ECs on proximal tissue is better understood.
Organogenesis, Homeostasis and Repair
On a per organ basis, the vascular niche performs distinctive and versatile functions in response to local needs. The endothelium imparts an instructive influence on its microenvironment utilizing soluble (e.g. angiocrine factors)169 and insoluble (e.g. ECM-cell and cell-cell) interactions. This influence begins during development and prior to the maturation of the circulatory system as the endothelium extensively orchestrates organogenesis and organ patterning.170 In the case of several developing organs, including the liver166 and pancreas,167 embryological work demonstrates nascent vasculature is not only informative, but critical for organogenic processes. Following development, the best articulated function of the vascular niche is its homeostatic role in directing stem cell proliferation, self-renewal, and differentiation.171–173 The niche bathes stem cells in stoichiometrically relevant concentrations of angiocrine factors that control a quiescent versus pro-proliferative/differentiation phenotype. Moreover, EC-derived ECM also plays a key role in niche signaling as it not only contributes to the polarization of cells, influencing the symmetric and asymmetric cell division of stem cells, but also provides a basement membrane to plastic cells unable to deposit scaffolding proteins.174,175 Finally, the vascular niche is implicated in repair processes as a local modulator of regeneration, supporting the expansion of stem and progenitor cells176 and controlling fibrosis.177 Accordingly, the functional capacity of the vasculature is multifaceted and it can be seen as a broad gatekeeper of organogenic and homeostatic processes.
Disease Progression
An essential step in the pathophysiology of many diseases includes remodeling of the endothelium and maladaptation of the vascular niche. Many cancers, for instance, co-op or recruit the vasculature.178 This capacity not only allows the cancer to grow beyond its diffusion limit, but also provides access to a formative niche with the ability to modulate tumor progression, migration, and metastasis. 169,179 Moreover, the cancer stem cell (CSC) hypothesis stipulates this rare population is responsible for the initiation, recurrence and therapeutic resistance of many malignant tumors.180 In a parallel fashion to non-malignant stem cells, the vascular niche appears to fortify the apex cell of the tumor differentiation hierarchy and support neoplasms.181 It is thus unsurprising the vascular niche is heavily considered as a therapeutic target.182–184
The Vascular Niche and MPS Platforms
Vascularized MPS Platforms Mimic Organogenesis
Organogenesis requires the integration of signals from several parallel inputs including chemical, mechanical, and cell-cell interactions, characteristics of which vascularized MPS models support and maintain. The proximity of the endothelium to developing tissue during organogenesis suggests that ECs may play an essential role during organ development and maturation. Arguably, the formation of on-chip microtissues utilizes several developmental pathways to reform a tissue from a single cell suspension. Incorporation of EC-guided morphogenic and maturation processes can be utilized in tissue development ex vivo and do not require the use of organotypic185 or even species-specific186 endothelia. For example, when human embryonic stem cell-derived pancreatic progenitors cells are co-cultured with HUVECs or rat heart microvascular endothelial cells (rHMVECs), improved maturation was observed.186 Thus, the regulatory function of the vasculature and its niche have the potential to induce marked improvements in the physiological capacity of all MPS platforms.
Stem Cell Culture and Vascularized MPS
While in vivo models have helped identify cells residing in vascular niches, manipulating 2D in vitro “microenvironments”, which have fewer degrees of freedom compared to 3D tissues, remains challenging if we want to study the contribution of niche components to surrounding tissues. Thus, vascularized MPS platforms recapitulating the 3D functional unit of organs are uniquely positioned to not only benefit from the instructive nature of the endothelium to improve the model’s fidelity, but also to elucidate mechanisms underlying spatial and temporal control of the vascular niche. Recently, a handful of vascular niche-related MPS platforms have been published, many of which demonstrate the capacity to maintain a stem cell population for an extensive time period, validate progenitor cells egression, and observe lineage maturation.101,136,187,188 The majority of current niche-focused MPS platforms recapitulate the bone marrow microenvironment, as the complex, multi-niche interactions amongst hematopoietic, osteoblast and vascular cell populations to facilitate hematopoiesis are almost impossible to create with 2D culture strategies. Glaser et al. created a bone marrow MPS platform that retains perivascular and endosteal niches in a dual chamber design.101 This MPS demonstrates the capability to maintain hematopoietic stem/progenitor cells for two weeks in vitro in either niche, track the differentiation of resident stem cells into the myeloid and erythroid lineages, and witness the intravasation of immature neutrophils into the adjacent microfluidic lines. Impressively, the platform incorporates an additional chamber used to study the trafficking of malignant cells into the adjacent perivascular and endosteal niches, providing insight into cancers that preferentially metastasize to different bone marrow niches. Thus, MPS platforms have the capacity to support multiple microenvironments and maintain proximal communication of the niches for the purposes of clinical and basic science research.
Conclusion
Although developments in MPS technology have emerged rapidly with several strategies to incorporate vessel conduits, including self-assembled, pre-patterned, and 3D bioprinted methodologies, barriers to translationally bridge in vitro platforms and clinical results remain. Ideally, a fully functional, organotypic vascular network, rather than a passive conduit, is needed to develop superior physiologically relevant MPS platforms. The vasculature has been proven to not only support nutrient delivery and metabolic processes, but also to play a vital role in organogenesis through unique molecular and phenotypic signatures. These complex functions need to be captured in MPS models so that the target tissue behaves in vitro as it does in vivo. This is equally true in patient-derived organoids where maintenance of in vivo structure and function is the goal. If vasculature is not incorporated into these tissues, they will rapidly lose important maintenance cues, and as they grow will also become hypoxic, further shifting gene expression away from that present in vivo.10 The vascular niche also maintains stem cell populations. While a small handful of MPS platforms have successfully incorporated organotypic vasculature and its niche, the universal adoption of organ-specific vasculature within MPS platforms is unlikely to materialize soon, primarily due to sourcing issues. As a result, researchers must build MPS platforms that balance several engineering choices to get as close to an organotypic phenotype as possible.
In this review, we have discussed current developments in vascularized MPS platforms and future perspectives with regard to establishing organotypic models. EC sourcing is a critical consideration as it contributes to the tissues genotypic and phenotypic capacities and improves the fidelity of the vascularized MPS to the model organ. HUVECs, thus far, are the most common EC source as they are easy to manipulate and culture for experimental purposes. As iPSC technology matures, iPSC-derived ECs with the capacity to maintain organ and patient-specific properties are likely to become increasingly utilized to promote organotypic characteristics. Furthermore, a clearer understanding is developing of the crucial role ECM plays as an indispensable factor for organogenesis. ECM not only provides the mechanical support to maintain vascular organization, but also supplies the chemical cues necessary for EC homeostasis and tissue organogenesis. Besides utilizing the standardized hydrogels as ECM in MPS, decellularized 3D matrix from tissue in vivo and the modification of archetypal ECM will be required to promote functional tissue reconstruction, provide constructive cues, and generate a vascular niche. Flow-derived shear also plays a key role in vascular network formation and EC function in MPS platforms. Microfluidic technology is a powerful tool when designing MPS platforms since it can precisely tune the microenvironment, including flow dynamics and chemical gradients. By integrating considerations such as EC source, ECM and shear control into the MPS, the system will generate a unique microenvironment that benefits organ development and homeostasis. We envision that integration of biological substrates, microfluidic techniques and stem cell biology strategies will generate vascularized MPS platforms that more accurately model the complexities of specific tissues in health and disease.
Acknowledgements
This work has been supported by U54 CA217378, R01 CA180122, and UH3 TR-000481. C.C.W.H. receives support from the Chao Family Comprehensive Cancer Center (CFCCC) through an NCI Center Grant [P30A062203].
Footnotes
Competing Interests
MLE and YHC declare no competing interests. CCWH and APL have equity interests in Aracari Biosciences, Inc, which is commercializing the microfluidic devices described in this paper (Figure 2). The terms of this arrangement have been reviewed and approved by the University of California, Irvine in accordance with its conflict-of-interest policies.
Reference
- 1.Malakar AK, Choudhury D, Halder B, Paul P, Uddin A. and Chakraborty S, J Cell Physiol, 2019, 234, 16812–16823. [DOI] [PubMed] [Google Scholar]
- 2.Higueros E, Roe E, Granell E. and Baselga E, Actas Dermosifiliogr, 2017, 108, 407–417. [DOI] [PubMed] [Google Scholar]
- 3.Luxán G. and Dimmeler S, Cardiovasc Res, 2021, cvab047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaaf MB, Garg AD and Agostinis P, Cell Death Dis, 2018, 9, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Govindpani K, McNamara LG, Smith NR, Vinnakota C, Waldvogel HJ, Faull RL and Kwakowsky A, J Clin Med, 2019, 8, 651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong CH, Siah KW and Lo AW, Biostatistics, 2019, 20, 273–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schutgens F. and Clevers H, Annu Rev Pathol, 2020, 15, 211–234. [DOI] [PubMed] [Google Scholar]
- 8.McCauley HA and Wells JM, Development, 2017, 144, 958–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon S-J, Lee D, Gopal S, Ku A, Moon H. and Dordick JS, MEDICAL DEVICES & SENSORS, 2020, 3, e10067. [Google Scholar]
- 10.Zhang S, Wan Z. and Kamm RD, Lab on a Chip, 2021, 21, 473–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Low LA, Mummery C, Berridge BR, Austin CP and Tagle DA, Nat Rev Drug Discov, 2021, 20, 345–361. [DOI] [PubMed] [Google Scholar]
- 12.Herland A, van der Meer AD, FitzGerald EA, Park T-E, Sleeboom JJF and Ingber DE, PLOS ONE, 2016, 11, e0150360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hultgren NW, Fang JS, Ziegler ME, Ramirez RN, Phan DTT, Hatch MMS, Welch-Reardon KM, Paniagua AE, Kim LS, Shon NN, Williams DS, Mortazavi A. and Hughes CCW, Nat Commun, 2020, 11, 5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atchison L, Abutaleb NO, Snyder-Mounts E, Gete Y, Ladha A, Ribar T, Cao K. and Truskey GA, Stem Cell Reports, 2020, 14, 325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nolan DJ, Ginsberg M, Israely E, Palikuqi B, Poulos MG, James D, Ding B-S, Schachterle W, Liu Y, Rosenwaks Z, Butler JM, Xiang J, Rafii A, Shido K, Rabbany SY, Elemento O. and Rafii S, Dev Cell, 2013, 26, 204–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcu R, Choi YJ, Xue J, Fortin CL, Wang Y, Nagao RJ, Xu J, MacDonald JW, Bammler TK, Murry CE, Muczynski K, Stevens KR, Himmelfarb J, Schwartz SM and Zheng Y, iScience, 2018, 4, 20–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Potente M. and Mäkinen T, Nat Rev Mol Cell Biol, 2017, 18, 477–494. [DOI] [PubMed] [Google Scholar]
- 18.Kluger MS, Clark PR, Tellides G, Gerke V. and Pober JS, Arterioscler Thromb Vasc Biol, 2013, 33, 489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madugundu AK, Na CH, Nirujogi RS, Renuse S, Kim KP, Burns KH, Wilks C, Langmead B, Ellis SE, Collado‐Torres L, Halushka MK, Kim M. and Pandey A, Proteomics, 2019, 19, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lorenzon P, Vecile E, Nardon E, Ferrero E, Harlan JM, Tedesco F. and Dobrina A, J Cell Biol, 1998, 142, 1381–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Müller AM, Hermanns MI, Skrzynski C, Nesslinger M, Müller K-M and Kirkpatrick CJ, Exp Mol Pathol, 2002, 72, 221–229. [DOI] [PubMed] [Google Scholar]
- 22.Nakatsu MN, Sainson RCA, Aoto JN, Taylor KL, Aitkenhead M, Pérez-del-Pulgar S, Carpenter PM and Hughes CCW, Microvasc Res, 2003, 66, 102–112. [DOI] [PubMed] [Google Scholar]
- 23.Jaczewska J, Abdulreda MH, Yau CY, Schmitt MM, Schubert I, Berggren P-O, Weber C, Koenen RR, Moy VT and Wojcikiewicz EP, J Leukoc Biol, 2014, 95, 265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papapetropoulos A, García-Cardeña G, Madri JA and Sessa WC, J Clin Invest, 1997, 100, 3131–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paik DT, Tian L, Williams IM, Rhee S, Zhang H, Liu C, Mishra R, Wu SM, Red-Horse K. and Wu JC, Circulation, 2020, 142, 1848–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uwamori H, Ono Y, Yamashita T, Arai K. and Sudo R, Microvasc Res, 2019, 122, 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin NYC, Homan KA, Robinson SS, Kolesky DB, Duarte N, Moisan A. and Lewis JA, Proc Natl Acad Sci U S A, 2019, 116, 5399–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palikuqi B, Nguyen D-HT, Li G, Schreiner R, Pellegata AF, Liu Y, Redmond D, Geng F, Lin Y, Gómez-Salinero JM, Yokoyama M, Zumbo P, Zhang T, Kunar B, Witherspoon M, Han T, Tedeschi AM, Scottoni F, Lipkin SM, Dow L, Elemento O, Xiang JZ, Shido K, Spence JR, Zhou QJ, Schwartz RE, De Coppi P, Rabbany SY and Rafii S, Nature, 2020, 585, 426–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizee MR, Wooldrik D, Lakeman KAM, van het Hof B, Drexhage JAR, Geerts D, Bugiani M, Aronica E, Mebius RE, Prat A, de Vries HE and Reijerkerk A, J Neurosci, 2013, 33, 1660–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paschalaki KE and Randi AM, Front Med (Lausanne), 2018, 5, 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kutikhin AG, Tupikin AE, Matveeva VG, Shishkova DK, Antonova LV, Kabilov MR and Velikanova EA, Cells, 2020, 9, 876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banno K. and Yoder MC, Pediatr Res, 2018, 83, 283–290. [DOI] [PubMed] [Google Scholar]
- 33.Lukowski SW, Patel J, Andersen SB, Sim S-L, Wong HY, Tay J, Winkler I, Powell JE and Khosrotehrani K, Cell Reports, 2019, 27, 2748–2758.e3. [DOI] [PubMed] [Google Scholar]
- 34.Shi Y, Inoue H, Wu JC and Yamanaka S, Nat Rev Drug Discov, 2017, 16, 115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Protze SI, Lee JH and Keller GM, Cell Stem Cell, 2019, 25, 311–327. [DOI] [PubMed] [Google Scholar]
- 36.Rosa S, Praça C, Pitrez PR, Gouveia PJ, Aranguren XL, Ricotti L. and Ferreira LS, Sci Rep, 2019, 9, 3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Songstad AE, Wiley LA, Duong K, Kaalberg E, Flamme-Wiese MJ, Cranston CM, Riker MJ, Levasseur D, Stone EM, Mullins RF and Tucker BA, Invest Ophthalmol Vis Sci, 2015, 56, 8258–8267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kachamakova-Trojanowska N, Stepniewski J. and Dulak J, Cells, 2019, 8, 11:1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olgasi C, Talmon M, Merlin S, Cucci A, Richaud-Patin Y, Ranaldo G, Colangelo D, Di Scipio F, Berta GN, Borsotti C, Valeri F, Faraldi F, Prat M, Messina M, Schinco P, Lombardo A, Raya A. and Follenzi A, Stem Cell Reports, 2018, 11, 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho H, Macklin BL, Lin Y-Y, Zhou L, Lai MJ, Lee G, Gerecht S. and Duh EJ, JCI Insight, 2020, 5, e131828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abaci HE, Guo Z, Coffman A, Gillette B, Lee W-H, Sia SK and Christiano AM, Adv Healthc Mater, 2016, 5, 1800–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoder MC, Curr Opin Hematol, 2015, 22, 252–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu M, He J, Zhang C, Xu J. and Wang Y, Stem Cell Research & Therapy, 2019, 10, 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di Bernardini E, Campagnolo P, Margariti A, Zampetaki A, Karamariti E, Hu Y. and Xu Q, J Biol Chem, 2014, 289, 3383–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morita R, Suzuki M, Kasahara H, Shimizu N, Shichita T, Sekiya T, Kimura A, Sasaki K, Yasukawa H. and Yoshimura A, PNAS, 2015, 112, 160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Beijnum JR, Rousch M, Castermans K, van der Linden E. and Griffioen AW, Nat Protoc, 2008, 3, 1085–1091. [DOI] [PubMed] [Google Scholar]
- 47.Jang S, Collin de l’Hortet A. and Soto-Gutierrez A, Am J Pathol, 2019, 189, 502–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belair DG, Whisler JA, Valdez J, Velazquez J, Molenda JA, Vickerman V, Lewis R, Daigh C, Hansen TD, Mann DA, Thomson JA, Griffith LG, Kamm RD, Schwartz MP and Murphy WL, Stem Cell Rev Rep, 2015, 11, 511–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Focosi D, Amabile G, Di Ruscio A, Quaranta P, Tenen DG and Pistello M, Blood Cancer J, 2014, 4, e211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L, Xiang M, Liu Y, Sun N, Lu M, Shi Y, Wang X, Meng D, Chen S. and Qin J, Biomicrofluidics, 2016, 10, 014106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rambøl MH, Han E. and Niklason LE, Tissue Eng Part A, 2020, 26, 556–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kurokawa YK, Yin RT, Shang MR, Shirure VS, Moya ML and George SC, Tissue Eng Part C Methods, 2017, 23, 474–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davis GE, Stratman AN, Sacharidou A. and Koh W, Int Rev Cell Mol Biol, 2011, 288, 101–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Naito H, Iba T. and Takakura N, Int Immunol, 2020, 32, 295–305. [DOI] [PubMed] [Google Scholar]
- 55.Rajendran P, Rengarajan T, Thangavel J, Nishigaki Y, Sakthisekaran D, Sethi G. and Nishigaki I, Int J Biol Sci, 2013, 9, 1057–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kritharis A, Al-Samkari H. and Kuter DJ, Haematologica, 2018, 103, 1433–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou F, Zhao X, Liu X, Liu Y, Ma F, Liu B. and Yang J, Pulm Circ, 2020, 10, 4: 2045894019885357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gu M, Shao N-Y, Sa S, Li D, Termglinchan V, Ameen M, Karakikes I, Sosa G, Grubert F, Lee J, Cao A, Taylor S, Ma Y, Zhao Z, Chappell J, Hamid R, Austin ED, Gold JD, Wu JC, Snyder MP and Rabinovitch M, Cell Stem Cell, 2017, 20, 490–504.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paik DT, Tian L, Lee J, Sayed N, Chen IY, Rhee S, Rhee J-W, Kim Y, Wirka RC, Buikema JW, Wu SM, Red-Horse K, Quertermous T. and Wu JC, Circ Res, 2018, 123, 443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lippmann ES, Azarin SM, Kay JE, Nessler RA, Wilson HK, Al-Ahmad A, Palecek SP and Shusta EV, Nat Biotechnol, 2012, 30, 783–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park T-E, Mustafaoglu N, Herland A, Hasselkus R, Mannix R, FitzGerald EA, Prantil-Baun R, Watters A, Henry O, Benz M, Sanchez H, McCrea HJ, Goumnerova LC, Song HW, Palecek SP, Shusta E. and Ingber DE, Nat Commun, 2019, 10, 2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vatine GD, Barrile R, Workman MJ, Sances S, Barriga BK, Rahnama M, Barthakur S, Kasendra M, Lucchesi C, Kerns J, Wen N, Spivia WR, Chen Z, Van Eyk J. and Svendsen CN, Cell Stem Cell, 2019, 24, 995–1005.e6. [DOI] [PubMed] [Google Scholar]
- 63.Lu TM, Houghton S, Magdeldin T, Durán JGB, Minotti AP, Snead A, Sproul A, Nguyen D-HT, Xiang J, Fine HA, Rosenwaks Z, Studer L, Rafii S, Agalliu D, Redmond D. and Lis R, Proc Natl Acad Sci U S A, 2021, 118, e2016950118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu S, Zhao M-T, Jahanbani F, Shao N-Y, Lee WH, Chen H, Snyder MP and Wu JC, JCI Insight, 2016, 1, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ford AJ and Rajagopalan P, WIREs Nanomedicine and Nanobiotechnology, 2018, 10, e1503. [DOI] [PubMed] [Google Scholar]
- 66.Streuli C, Curr Opin Cell Biol, 1999, 11, 634–640. [DOI] [PubMed] [Google Scholar]
- 67.Londono R, Jobe BA, Hoppo T. and Badylak SF, World J Gastroenterol, 2012, 18, 6894–6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Benders KEM, van Weeren PR, Badylak SF, Saris DBF, Dhert WJA and Malda J, Trends Biotechnol, 2013, 31, 169–176. [DOI] [PubMed] [Google Scholar]
- 69.Turner NJ and Badylak SF, Eur Cell Mater, 2013, 25, 130–143. [DOI] [PubMed] [Google Scholar]
- 70.James BD and Allen JB, ACS Biomater. Sci. Eng, 2018, 4, 3818–3842. [DOI] [PubMed] [Google Scholar]
- 71.Charbonier FW, Zamani M. and Huang NF, Adv Biosyst, 2019, 3, 2: e1800252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hoffmann GA, Wong JY and Smith ML, Biochemistry, 2019, 58, 4710–4720. [DOI] [PubMed] [Google Scholar]
- 73.Badylak SF, Semin Cell Dev Biol, 2002, 13, 377–383. [DOI] [PubMed] [Google Scholar]
- 74.Frantz C, Stewart KM and Weaver VM, J Cell Sci, 2010, 123, 4195–4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim S-H, Turnbull J. and Guimond S, J Endocrinol, 2011, 209, 139–151. [DOI] [PubMed] [Google Scholar]
- 76.Li F, Li W, Johnson S, Ingram D, Yoder M. and Badylak S, Endothelium, 2004, 11, 199–206. [DOI] [PubMed] [Google Scholar]
- 77.Davis GE and Senger DR, Circ Res, 2005, 97, 1093–1107. [DOI] [PubMed] [Google Scholar]
- 78.Smith JT, Elkin JT and Reichert WM, Exp Cell Res, 2006, 312, 2424–2432. [DOI] [PubMed] [Google Scholar]
- 79.Baker BM and Chen CS, J Cell Sci, 2012, 125, 3015–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee S, Ko J, Park D, Lee S-R, Chung M, Lee Y. and Li Jeon N, Lab on a Chip, 2018, 18, 2686–2709. [DOI] [PubMed] [Google Scholar]
- 81.Sobrino A, Phan DTT, Datta R, Wang X, Hachey SJ, Romero-López M, Gratton E, Lee AP, George SC and Hughes CCW, Sci Rep, 2016, 6, 31589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jeon JS, Bersini S, Whisler JA, Chen MB, Dubini G, Charest JL, Moretti M and Kamm RD, Integr Biol (Camb), 2014, 6, 555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Paek J, Park SE, Lu Q, Park K-T, Cho M, Oh JM, Kwon KW, Yi Y-S, Song JW, Edelstein HI, Ishibashi J, Yang W, Myerson JW, Kiseleva RY, Aprelev P, Hood ED, Stambolian D, Seale P, Muzykantov VR and Huh D, ACS Nano, 2019, 13, 7627–7643. [DOI] [PubMed] [Google Scholar]
- 84.Newman AC, Nakatsu MN, Chou W, Gershon PD and Hughes CCW, Mol Biol Cell, 2011, 22, 3791–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nakatsu MN, Sainson RCA, Pérez-del-Pulgar S, Aoto JN, Aitkenhead M, Taylor KL, Carpenter PM and Hughes CCW, Lab Invest, 2003, 83, 1873–1885. [DOI] [PubMed] [Google Scholar]
- 86.Bell SE, Mavila A, Salazar R, Bayless KJ, Kanagala S, Maxwell SA and Davis GE, J Cell Sci, 2001, 114, 2755–2773. [DOI] [PubMed] [Google Scholar]
- 87.Mahboubian A, Vllasaliu D, Dorkoosh FA and Stolnik S, Biomacromolecules, 2020, 21, 4737–4746. [DOI] [PubMed] [Google Scholar]
- 88.Zheng Y, Han MKL, Jiang Q, Li B, Feng J and del Campo A, Mater. Horiz, 2020, 7, 111–116. [Google Scholar]
- 89.de Graaf MNS, Cochrane A, van den Hil FE, Buijsman W, van der Meer AD, van den Berg A, Mummery CL and Orlova VV, APL Bioeng, 2019, 3, 026105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jia L, Han F, Yang H, Turnbull G, Wang J, Clarke J, Shu W, Guo M and Li B, Adv Healthc Mater, 2019, 8, e1900435. [DOI] [PubMed] [Google Scholar]
- 91.van Duinen V, Zhu D, Ramakers C, van Zonneveld AJ, Vulto P and Hankemeier T, Angiogenesis, 2019, 22, 157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mathur T, Abhay Singh K, Pandian NKR, Tsai S-H, Hein TW, Gaharwar AK, Flanagan JM and Jain A, Lab on a Chip, 2019, 19, 2500–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Miron-Mendoza M, Seemann J and Grinnell F, Biomaterials, 2010, 31, 6425–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang Y, Motte S and Kaufman LJ, Biomaterials, 2010, 31, 5678–5688. [DOI] [PubMed] [Google Scholar]
- 95.Mongiat M, Andreuzzi E, Tarticchio G and Paulitti A, Int J Mol Sci, 2016, 17. 11:1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Perruzzi CA, de Fougerolles AR, Koteliansky VE, Whelan MC, Westlin WF and Senger DR, J Invest Dermatol, 2003, 120, 1100–1109. [DOI] [PubMed] [Google Scholar]
- 97.Whelan MC and Senger DR, J Biol Chem, 2003, 278, 327–334. [DOI] [PubMed] [Google Scholar]
- 98.Williams BR, Gelman RA, Poppke DC and Piez KA, J Biol Chem, 1978, 253, 6578–6585. [PubMed] [Google Scholar]
- 99.Adriani G, Ma D, Pavesi A, Kamm RD and Goh ELK, Lab Chip, 2017, 17, 448–459. [DOI] [PubMed] [Google Scholar]
- 100.Phan DTT, Wang X, Craver BM, Sobrino A, Zhao D, Chen JC, Lee LYN, George SC, Lee AP and Hughes CCW, Lab Chip, 2017, 17, 511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Glaser DE, Curtis MB, Sariano PA, Rollins ZA, Shergill BS, Anand A, Deely AM, Shirure VS, Anderson L, Lowen JM, Ng NR, Weilbaecher K, Link DC and George SC, bioRxiv, 2020, 2020.04.17.039339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Haase K, Offeddu GS, Gillrie MR and Kamm RD, Advanced Functional Materials, 2020, 30, 2002444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chou DB, Frismantas V, Milton Y, David R, Pop-damkov P, Ferguson D, Macdonald A, Bölükbaşı ÖV, Joyce CE, Teixeira LSM, Rech A, Jiang A, Calamari E, Jalili-firoozinezhad S, Furlong BA, Sullivan LRO, Ng CF, Choe Y, Marquez S, Myers KC, Weinberg OK, Hasserjian RP, Ewart L and Ingber DE, Nat. Biomed. Eng, 2020, 4, 394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li Y, Meng H, Liu Y and Lee BP, ScientificWorldJournal, 2015, 2015, 685690. [Google Scholar]
- 105.Morin KT and Tranquillo RT, Exp Cell Res, 2013, 319, 2409–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Roy S, Driggs J, Elgharably H, Biswas S, Findley M, Khanna S, Gnyawali U, Bergdall VK and Sen CK, Wound Repair Regen, 2011, 19, 753–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ahn J, Lim J, Jusoh N, Lee J, Park T-E, Kim Y, Kim J and Jeon NL, Front Bioeng Biotechnol, 2019, 7, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zervantonakis IK, Hughes-Alford SK, Charest JL, Condeelis JS, Gertler FB and Kamm RD, Proc Natl Acad Sci U S A, 2012, 109, 13515–13520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, Kermani P, Hempstead B, Fischbach-Teschl C, López JA and Stroock AD, Proc Natl Acad Sci U S A, 2012, 109, 9342–9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang X-Y, Jin Z-H, Gan B-W, Lv S-W, Xie M and Huang W-H, Lab Chip, 2014, 14, 2709–2716. [DOI] [PubMed] [Google Scholar]
- 111.Eltaher HM, Abukunna FE, Ruiz-Cantu L, Stone Z, Yang J and Dixon JE, Acta Biomater, 2020, 113, 339–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kubota Y, Kleinman HK, Martin GR and Lawley TJ, J Cell Biol, 1988, 107, 1589–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Akther F, Yakob SB, Nguyen N-T and Ta HT, Biosensors (Basel), 2020, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hamill KJ, Kligys K, Hopkinson SB and Jones JCR, J Cell Sci, 2009, 122, 4409–4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ohta R, Niwa A, Taniguchi Y, Suzuki NM, Toga J, Yagi E, Saiki N, Nishinaka-Arai Y, Okada C, Watanabe A, Nakahata T, Sekiguchi K and Saito MK, Sci Rep, 2016, 6, 35680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dickinson LE, Moura ME and Gerecht S, Soft Matter, 2010, 6, 5109–5119. [Google Scholar]
- 117.Chen CS, Mrksich M, Huang S, Whitesides GM and Ingber DE, Science, 1997, 276, 1425–1428. [DOI] [PubMed] [Google Scholar]
- 118.Wijelath ES, Rahman S, Murray J, Patel Y, Savidge G and Sobel M, J Vasc Surg, 2004, 39, 655–660. [DOI] [PubMed] [Google Scholar]
- 119.Toworfe GK, Composto RJ, Adams CS, Shapiro IM and Ducheyne P, J Biomed Mater Res A, 2004, 71, 449–461. [DOI] [PubMed] [Google Scholar]
- 120.Lee CH, Singla A and Lee Y, Int J Pharm, 2001, 221, 1–22. [DOI] [PubMed] [Google Scholar]
- 121.Anderson DEJ and Hinds MT, Ann Biomed Eng, 2011, 39, 2329–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lee P, Lin R, Moon J and Lee LP, Biomed Microdevices, 2006, 8, 35–41. [DOI] [PubMed] [Google Scholar]
- 123.Zhang YS, Arneri A, Bersini S, Shin S-R, Zhu K, Goli-Malekabadi Z, Aleman J, Colosi C, Busignani F, Dell’Erba V, Bishop C, Shupe T, Demarchi D, Moretti M, Rasponi M, Dokmeci MR, Atala A and Khademhosseini A, Biomaterials, 2016, 110, 45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yi H-G, Jeong YH, Kim Y, Choi Y-J, Moon HE, Park SH, Kang KS, Bae M, Jang J, Youn H, Paek SH and Cho D-W, Nat Biomed Eng, 2019, 3, 509–519. [DOI] [PubMed] [Google Scholar]
- 125.Meng F, Meyer Carolyn. M., Joung D, Vallera Daniel. A., McAlpine Michael. C. and Panoskaltsis-Mortari A, Adv Mater, 2019, 31, e1806899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cui X and Boland T, Biomaterials, 2009, 30, 6221–6227. [DOI] [PubMed] [Google Scholar]
- 127.Tirella A, Orsini A, Vozzi G and Ahluwalia A, Biofabrication, 2009, 1, 045002. [DOI] [PubMed] [Google Scholar]
- 128.Malda J, Visser J, Melchels FP, Jüngst T, Hennink WE, Dhert WJA, Groll J and Hutmacher DW, Advanced Materials, 2013, 25, 5011–5028. [DOI] [PubMed] [Google Scholar]
- 129.Zeng D, Ou D-B, Wei T, Ding L, Liu X-T, Hu X-L, Li X and Zheng Q-S, BMC Cell Biol, 2013, 14, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen X-D, Birth Defects Res C Embryo Today, 2010, 90, 45–54. [DOI] [PubMed] [Google Scholar]
- 131.Sellaro TL, Ravindra AK, Stolz DB and Badylak SF, Tissue Eng, 2007, 13, 2301–2310. [DOI] [PubMed] [Google Scholar]
- 132.Lien S-M, Ko L-Y and Huang T-J, Acta Biomater, 2009, 5, 670–679. [DOI] [PubMed] [Google Scholar]
- 133.Woodfield TBF, Van Blitterswijk CA, De Wijn J, Sims TJ, Hollander AP and Riesle J, Tissue Eng, 2005, 11, 1297–1311. [DOI] [PubMed] [Google Scholar]
- 134.Engler AJ, Carag-Krieger C, Johnson CP, Raab M, Tang H-Y, Speicher DW, Sanger JW, Sanger JM and Discher DE, J Cell Sci, 2008, 121, 3794–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Engler AJ, Sen S, Sweeney HL and Discher DE, Cell, 2006, 126, 677–689. [DOI] [PubMed] [Google Scholar]
- 136.Marturano-Kruik A, Nava MM, Yeager K, Chramiec A, Hao L, Robinson S, Guo E, Raimondi MT and Vunjak-Novakovic G, Proc Natl Acad Sci U S A, 2018, 115, 1256–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Romero-López M, Trinh AL, Sobrino A, Hatch MMS, Keating MT, Fimbres C, Lewis DE, Gershon PD, Botvinick EL, Digman M, Lowengrub JS and Hughes CCW, Biomaterials, 2017, 116, 118–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Badylak SF, Taylor D and Uygun K, Annu Rev Biomed Eng, 2011, 13, 27–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Scarritt ME, Pashos NC and Bunnell BA, Front Bioeng Biotechnol, 2015, 3, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cybulsky MI and Marsden PA, Arterioscler Thromb Vasc Biol, 2014, 34, 1806–1808. [DOI] [PubMed] [Google Scholar]
- 141.Chiu J-J and Chien S, Physiol Rev, 2011, 91, 327–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nishimura T and Takeichi M, Curr Top Dev Biol, 2009, 89, 33–54. [DOI] [PubMed] [Google Scholar]
- 143.Maruthamuthu V, Aratyn-Schaus Y and Gardel ML, Curr Opin Cell Biol, 2010, 22, 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hsu Y-H, Moya ML, Hughes CCW, George SC and Lee AP, Lab Chip, 2013, 13, 2990–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hahn C and Schwartz MA, Nat Rev Mol Cell Biol, 2009, 10, 53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li J, Hou B, Tumova S, Muraki K, Bruns A, Ludlow MJ, Sedo A, Hyman AJ, McKeown L, Young RS, Yuldasheva NY, Majeed Y, Wilson LA, Rode B, Bailey MA, Kim HR, Fu Z, Carter DA, Bilton J, Imrie H, Ajuh P, Dear TN, Cubbon RM, Kearney MT, Prasad RK, Evans PC, Ainscough JF and Beech DJ, Nature, 2014, 515, 279–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dejana E, Orsenigo F and Lampugnani MG, J Cell Sci, 2008, 121, 2115–2122. [DOI] [PubMed] [Google Scholar]
- 148.Chien S, Ann Biomed Eng, 2008, 36, 554–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Song JW and Munn LL, Proc Natl Acad Sci U S A, 2011, 108, 15342–15347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Matlung HL, Bakker ENTP and VanBavel E, Antioxid Redox Signal, 2009, 11, 1699–1709. [DOI] [PubMed] [Google Scholar]
- 151.Ding Z, Liu S, Wang X, Deng X, Fan Y, Sun C, Wang Y and Mehta JL, Antioxid Redox Signal, 2015, 22, 760–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chen Z, Peng I-C, Cui X, Li Y-S, Chien S and Shyy JY-J, Proc Natl Acad Sci U S A, 2010, 107, 10268–10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tuttle JL, Nachreiner RD, Bhuller AS, Condict KW, Connors BA, Herring BP, Dalsing MC and Unthank JL, Am J Physiol Heart Circ Physiol, 2001, 281, H1380–1389. [DOI] [PubMed] [Google Scholar]
- 154.Baeyens N, Nicoli S, Coon BG, Ross TD, Van den Dries K, Han J, Lauridsen HM, Mejean CO, Eichmann A, Thomas J-L, Humphrey JD and Schwartz MA, Elife, 2015, 4, e04645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang X, Zhao D, Phan DTT, Liu J, Chen X, Yang B, Hughes CCW, Zhang W and Lee AP, Lab Chip, 2018, 18, 2167–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang YI and Shuler ML, Lab Chip, 2018, 18, 2563–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Esch MB, Prot J-M, Wang YI, Miller P, Llamas-Vidales JR, Naughton BA, Applegate DR and Shuler ML, Lab Chip, 2015, 15, 2269–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rafii S, Butler JM and Ding B-S, Nature, 2016, 529, 316–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Baruah J and Wary KK, Front Cell Dev Biol, 2020, 7, 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gattazzo F, Urciuolo A and Bonaldo P, Biochim Biophys Acta, 2014, 1840, 2506–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Shen Q, Goderie SK, Jin L, Karanth N, Sun Y, Abramova N, Vincent P, Pumiglia K and Temple S, Science, 2004, 304, 1338–1340. [DOI] [PubMed] [Google Scholar]
- 162.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C and Morrison SJ, Cell, 2005, 121, 1109–1121. [DOI] [PubMed] [Google Scholar]
- 163.Sunita P-H and Pa D, Int J Dev Biol, 2011, 55, 353–363.21732275 [Google Scholar]
- 164.Ono S, Egawa G and Kabashima K, Inflamm Regen, 2017, 37, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.dela Paz NG and D’Amore PA, Cell Tissue Res, 2009, 335, 5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Matsumoto K, Yoshitomi H, Rossant J and Zaret KS, Science, 2001, 294, 559–563. [DOI] [PubMed] [Google Scholar]
- 167.Lammert E, Cleaver O and Melton D, Science, 2001, 294, 564–567. [DOI] [PubMed] [Google Scholar]
- 168.Bautch VL and James JM, Cell Adh Migr, 2009, 3, 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Butler JM, Kobayashi H and Rafii S, Nat Rev Cancer, 2010, 10, 138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Crivellato E, Nico B and Ribatti D, J Anat, 2007, 211, 415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sugimura R, Int. J. Hematol, 2018, 107, 642–645. [DOI] [PubMed] [Google Scholar]
- 172.He N, Zhang L, Cui J and Li Z, Bone Marrow Res, 2014, 2014, 128436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Licht T and Keshet E, Mech Dev, 2015, 138 Pt 1, 56–62. [DOI] [PubMed] [Google Scholar]
- 174.Witjas FMR, van den Berg BM, van den Berg CW, Engelse MA and Rabelink TJ, Stem Cells Transl Med, 2019, 8, 375–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Nikolova G, Strilic B and Lammert E, Trends Cell Biol, 2007, 17, 19–25. [DOI] [PubMed] [Google Scholar]
- 176.Zhao L, Zhang R, Su F, Dai L, Wang J, Cui J, Huang W and Zhang S, Oxid Med Cell Longev, 2020, 2020, 7865395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Winkler M, Staniczek T, Kürschner SW, Schmid CD, Schönhaber H, Cordero J, Kessler L, Mathes A, Sticht C, Neßling M, Uvarovskii A, Anders S, Zhang X-J, von Figura G, Hartmann D, Mogler C, Dobreva G, Schledzewski K, Géraud C, Koch P-S and Goerdt S, J Hepatol, 2021, 74, 380–393. [DOI] [PubMed] [Google Scholar]
- 178.Forster JC, Harriss-Phillips WM, Douglass MJ and Bezak E, Hypoxia (Auckl), 2017, 5, 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ghajar CM, Peinado H, Mori H, Matei IR, Evason KJ, Brazier H, Almeida D, Koller A, Hajjar KA, Stainier DYR, Chen EI, Lyden D and Bissell MJ, Nat Cell Biol, 2013, 15, 807–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Moharil RB, Dive A, Khandekar S and Bodhade A, J Oral Maxillofac Pathol, 2017, 21, 463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Cao Z, Scandura JM, Inghirami GG, Shido K, Ding B-S and Rafii S, Cancer Cell, 2017, 31, 110–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Singhal M and Augustin HG, Cancer Res, 2020, 80, 659–662. [DOI] [PubMed] [Google Scholar]
- 183.Mammoto A and Mammoto T, Front Bioeng Biotechnol, 2019, 7, 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Behrmann L, Wellbrock J and Fiedler W, Expert Opin Ther Targets, 2020, 24, 451–462. [DOI] [PubMed] [Google Scholar]
- 185.Kao D-I, Lacko LA, Ding B-S, Huang C, Phung K, Gu G, Rafii S, Stuhlmann H and Chen S, Stem Cell Reports, 2015, 4, 181–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Jaramillo M and Banerjee I, J Vis Exp, 2012, 3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Nelson MR, Ghoshal D, Mejías JC, Rubio DF, Keith E and Roy K, bioRxiv, 2019, 2019.12.15.876813. [Google Scholar]
- 188.Ma C, Witkowski MT, Harris J, Dolgalev I, Sreeram S, Qian W, Tong J, Chen X, Aifantis I and Chen W, Sci Adv, 2020, 6, eaba5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hou Y, Ai X, Zhao L, Gao Z, Wang Y, Lu Y, Tu P and Jiang Y, Lab Chip, 2020, 20, 2482–2494. [DOI] [PubMed] [Google Scholar]
- 190.Saha B, Mathur T, Handley KF, Hu W, Afshar-Kharghan V, Sood AK and Jain A, Blood Adv, 2020, 4, 3329–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lee S, Lim J, Yu J, Ahn J, Lee Y and Jeon NL, Lab Chip, 2019, 19, 2071–2080. [DOI] [PubMed] [Google Scholar]
- 192.Ko J, Ahn J, Kim S, Lee Y, Lee J, Park D and Li Jeon N, Lab on a Chip, 2019, 19, 2822–2833. [DOI] [PubMed] [Google Scholar]
- 193.Zhang M, Xu C, Jiang L and Qin J, Toxicol Res (Camb), 2018, 7, 1048–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zeinali S, Bichsel CA, Hobi N, Funke M, Marti TM, Schmid RA, Guenat OT and Geiser T, Angiogenesis, 2018, 21, 861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Jain A, Barrile R, van der Meer AD, Mammoto A, Mammoto T, De Ceunynck K, Aisiku O, Otieno MA, Louden CS, Hamilton GA, Flaumenhaft R and Ingber DE, Clin Pharmacol Ther, 2018, 103, 332–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Chung M, Lee S, Lee BJ, Son K, Jeon NL and Kim JH, Advanced Healthcare Materials, 2018, 7, 1700028. [DOI] [PubMed] [Google Scholar]
- 197.Li Y, Pi Q-M, Wang P-C, Liu L-J, Han Z-G, Shao Y, Zhai Y, Zuo Z-Y, Gong Z-Y, Yang X and Wu Y, RSC Adv., 2017, 7, 56108–56116. [Google Scholar]
- 198.Li X, George SM, Vernetti L, Gough AH and Taylor DL, Lab Chip, 2018, 18, 2614–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Musah S, Mammoto A, Ferrante TC, Jeanty SSF, Hirano-Kobayashi M, Mammoto T, Roberts K, Chung S, Novak R, Ingram M, Fatanat-Didar T, Koshy S, Weaver JC, Church GM and Ingber DE, Nat Biomed Eng, 2017, 1, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Qiu Y, Ahn B, Sakurai Y, Hansen CE, Tran R, Mimche PN, Mannino RG, Ciciliano JC, Lamb TJ, Joiner CH, Ofori-Acquah SF and Lam WA, Nat Biomed Eng, 2018, 2, 453–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Brown JA, Pensabene V, Markov DA, Allwardt V, Neely MD, Shi M, Britt CM, Hoilett OS, Yang Q, Brewer BM, Samson PC, McCawley LJ, May JM, Webb DJ, Li D, Bowman AB, Reiserer RS and Wikswo JP, Biomicrofluidics, 2015, 9, 054124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Mathur T, Singh KA, R Pandian NK, Tsai S-H, Hein TW, Gaharwar AK, Flanagan JM and Jain A, Lab Chip, 2019, 19, 2500–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Liu Y, Sakolish C, Chen Z, Phan DTT, Bender RHF, Hughes CCW and Rusyn I, Toxicology, 2020, 445, 152601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lam SF, Shirure VS, Chu YE, Soetikno AG and George SC, PLoS One, 2018, 13, e0209574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Yue T, Zhao D, Phan DTT, Wang X, Park JJ, Biviji Z, Hughes CCW and Lee AP, Microsyst Nanoeng, 2021, 7, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Hachey SJ, Movsesyan S, Nguyen QH, Burton-Sojo G, Tankazyan A, Wu J, Hoang T, Zhao D, Wang S, Hatch MM, Celaya E, Gomez S, Chen GT, Davis RT, Nee K, Pervolarakis N, Lawson DA, Kessenbrock K, Lee AP, Lowengrub J, Waterman ML and Hughes CCW, Lab Chip, 2021, 21, 1333–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Fernandez CE, Yen RW, Perez SM, Bedell HW, Povsic TJ, Reichert WM and Truskey GA, Sci Rep, 2016, 6, 21579. [DOI] [PMC free article] [PubMed] [Google Scholar]