Abstract
The product of rat gene 33 was identified as an ErbB-2-interacting protein in a two-hybrid screen employing the ErbB-2 juxtamembrane and kinase domains as bait. This interaction was reproduced in vitro with a glutathione S-transferase fusion protein spanning positions 282 to 395 of the 459-residue gene 33 protein. Activation of ErbB-2 catalytic function was required for ErbB-2–gene 33 physical interaction in living cells, whereas ErbB-2 autophosphorylation was dispensable. Expression of gene 33 protein was absent in growth-arrested NIH 3T3 fibroblasts but was induced within 60 to 90 min of serum stimulation or activation of the ErbB-2 kinase and decreased sharply upon entry into S phase. New differentiation factor stimulation of mitogen-deprived mammary epithelial cells also caused accumulation of gene 33 protein, which could be found in a complex with ErbB-2. Overexpression of gene 33 protein in mouse fibroblasts inhibited (i) cell proliferation driven by ErbB-2 but not by serum, (ii) cell transformation induced by ErbB-2 but not by Ras or Src, and (iii) sustained activation of ERK 1 and 2 by ErbB-2 but not by serum. The gene 33 protein may convey inhibitory signals downstream to ErbB-2 by virtue of its association with SH3-containing proteins, including GRB-2, which was found to associate with gene 33 protein in living cells. These data indicate that the gene 33 protein is a feedback inhibitor of ErbB-2 mitogenic function and a suppressor of ErbB-2 oncogenic activity. We propose that the gene 33 protein be renamed with the acronym RALT (receptor-associated late transducer).
Protein-protein interactions play a crucial role in the regulation of signal transduction pathways activated by receptor tyrosine kinases (RTKs) (58). SH2 (Src homology 2) and PTB (phosphotyrosine [PTyr] binding) domains recognize PTyr residues in the context of specific peptide sequences and can therefore bind to autophosphorylated receptors or to tyrosine-phosphorylated RTK substrates (58, 74). Modules based on PTyr-independent molecular recognition such as EH, PDZ, SH3, and WW domains (58, 74) are also involved in signaling downstream to activated RTKs. In general, protein-protein interaction modules are found both in polypeptides possessing intrinsic catalytic properties and in adapter-scaffold proteins. In the former case protein-protein interactions may modulate the function of a given enzyme by simply regulating its subcellular distribution or by allosteric activation (58). Adapter-scaffold proteins, on the other hand, are essentially made up of protein-protein interaction domains that allow for the assembly of multiprotein complexes in which the functions of different enzymes are integrated both spatially and temporally (57).
Upon ligand activation, RTKs target not only positive effectors but also enzymes involved in negative regulation of receptor signaling, such as tyrosine phosphatases (39), the Ras GTPase-activating protein (15), and c-Cbl (8, 37, 44). Adapter proteins such as Slap (67) and the SOCS gene family products (55) are also implicated in negative regulation of signaling by both receptor and nonreceptor tyrosine kinases.
It is likely that coordinated activation of positive- and negative-effector pathways tunes the magnitude and duration of signals evoked by RTK activity. The fact that quantitative differences in the outputs of RTK signals can have a dramatic impact on the execution of distinct cellular programs is demonstrated by the finding that graded ERK activation by Torso leads to different cell fates in embryonic termini of Drosophila melanogaster (26). In a remarkable analogy, RTKs are able to instruct mammalian cells to either proliferate or differentiate depending on the temporal profile of ERK 1 and 2 activity that they induce (48). Furthermore, it is becoming increasingly clear that the execution of developmental programs initiated by RTKs requires the function of inhibitory signaling molecules, which are expressed in the context of transcriptional responses activated by the RTKs themselves (60). Hence, the study of the regulatory circuitry which grades signaling by RTKs in time and space is receiving increasing attention.
The ErbB-2 receptor is a member of the epidermal growth factor (EGF) receptor (EGFR) subfamily of RTKs, whose direct ligand is still to be identified. The available evidence indicates that ErbB-2 acts as a coreceptor for a number of ErbB ligands in the context of heterodimers with EGFR, ErbB-3, and ErbB-4 (1, 66). It is thought that this network of combinatorial receptor interactions provides for signal diversification, as ErbB heterodimers have signaling competence distinct from that of homodimers (56). Remarkably, ErbB-2 plays a fundamental role in this process, since it emerges as the favorite partner for each of the other ErbB receptors and therefore dictates the hierarchy of ligand-driven heterodimerization among members of the ErbB family (30, 76). The relevance of ErbB-2 function in mammalian organisms is underscored by the severe developmental defects observed in ErbB-2 nullizygous mice (2, 7, 22, 41, 52). Furthermore, unabated ErbB-2 function leads to cell transformation in different model systems (21) and is associated with a poor clinical outcome in about 30% of human breast and ovary carcinomas (21).
We have searched for novel regulators of ErbB-2 signaling using the ErbB-2 juxtamembrane and tyrosine kinase domains as bait in a yeast two-hybrid screen. We now report the identification of the previously described gene 33 protein (13, 42) as an inhibitor of ErbB-2 mitogenic signaling.
MATERIALS AND METHODS
Two-hybrid system.
A fragment of the erbB-2 cDNA spanning nucleotides (nt) 2197 to 2646 (amino acids [aa] 682 to 832) was obtained by PCR amplification and cloned in the pBTM116 vector (78) to generate the LexA–ErbB-2 Δ832 fusion. The SacII site at position 2589 of the erbB-2 cDNA was used to generate the LexA–ErbB-2 bait by subcloning a SacII-MluI fragment from the LTR–ErbB-2 Δ1050 vector (18) in the SacII-SalI sites of the LexA–ErbB-2 Δ832 clone (SalI and MluI overhangs were rendered blunt with Klenow polymerase). A stop codon corresponding to position 715 of the ErbB-2 product was generated by cloning an adapter oligonucleotide in the LexA–ErbB-2 bait at the BamHI site located at position 2289 of the erbB-2 cDNA. A mouse embryo cDNA library expressed by the pVP16 vector (78) was transformed in L40 cells containing the LexA–ErbB-2 bait; growth, selection, and screening of yeast transformants have been described previously (78). pVP16-Raf and pLexA-RasV12 (78) were used as positive controls for the His+/LacZ+ phenotype in L40 cells, and LexA-lamin was used as a control for nonspecific interactions of clone 52.
Expression of recombinant gene 33 proteins in Escherichia coli and mammalian cells.
The coding sequence of the rat gene 33 cDNA (nt 264 to 1664) (13) was amplified by reverse transcription-PCR using total RNA extracted from rat liver as the source of the template, sequenced, and cloned in the pcDNA3 vector (Invitrogen). Retroviral vectors expressing RALT and green fluorescent protein (GFP) were generated by cloning the gene 33 cDNA in the PINCO plasmid (32); in this vector expression of RALT cDNA is driven by promoter-enhancer sequences of the Moloney murine leukemia virus 5′ long terminal repeat, whereas GFP expression is controlled by an internal cytomegalovirus promoter. Fragments of RALT were expressed in E. coli as fusion products with glutathione S-transferase (GST) using pGEX vectors (Pharmacia). The GST-RALT 1-262 and GST-RALT 263-459 fusion proteins were generated using the EcoRV site at position 1063 of the RALT cDNA in order to clone EcoRI-EcoRV and EcoRV-SalI fragments, respectively, from pcDNA3-RALT into pGEX 4T1. A GST-clone 52 fusion was generated by cloning a BamHI-EcoRI cDNA insert from the pVP16-c152 vector into pGEX-3X. GST fusion proteins were expressed in the BL21 E. coli strain and purified as described previously (73).
Antiserum generation and immunochemical procedures.
The S1 antiserum was generated by immunizing rabbits with purified GST-clone 52 fusion protein. Purified anti-RALT immunoglobulin G fractions were obtained by sequential affinity chromatography of S1 sera onto GST and GST-clone 52 resins. The 19C5/4 monoclonal antibody (MAb) was obtained by immunizing BALB/c mice with purified GST-RALT 263-459 fusion protein. Conditioned supernatants were obtained from subclones of hybridomas and were shown to recognize a single band in Western blots of quiescent PINCO-RALT cells. Such reactivity was absent in control lysates of PINCO cells. Immunoprecipitation, Western blotting, and subcellular fractionation procedures have been described previously (65). Polyclonal antibodies to PTyr and phospholipase C-γ (PLC-γ) (UBI) and to GRB-2 (Transduction Laboratories) and rabbit polyclonal antisera to SHC (Transduction Laboratories) and to the ErbB-2 kinase domain (18) were used as previously described (65). The anti-cyclin D1 MAb 72-13G and polyclonal anti-ERK 1 and anti-ERK 2 antiserum (Santa Cruz Biotechnology) and anti-P-ERK antibodies (New England Biolabs) were used at 0.2 μg/ml; affinity-purified S1 antibodies were used at 1 μg/ml. Anti-EGFR MAb (Ab1; Oncogene Science), anti-ErbB-2 MAb W6-100 (65), and affinity-purified S1 were used at 2 to 3 μg per mg of cell lysate for immunoprecipitation. Polyclonal anti-RET (where RET stands for rearranged during transfection) and anti-EGFR antibodies (Santa Cruz Biotechnology) were used in Western blotting procedures at 1 μg/ml. Procedures for binding assays with GST proteins have been described previously (65). Assays for binding to PTyr-agarose (Sigma) were like those for binding to glutathione-agarose (65). For blot overlay assays, recombinant RALT proteins were labeled with biotin as described previously (65) and used at 5 μg/ml to probe nitrocellulose filters containing recombinant GST-SH3 domains. Incubation and washing procedures were like those used for Western blots. RALT proteins captured by the filter were detected by horseradish peroxidase (HRP)-conjugated streptavidin (Pierce) (0.1 μg/ml) followed by enhanced chemiluminescence (ECL).
Cell culture and gene transfer procedures.
NIH-ErbB-2, NIH-ErbB-2 Δ1050, NIH-ErbB-2 5F, and NIH-EGFR/ErbB-2 cells were derived by transfection of NIH 3T3 fibroblasts (18, 71). To induce quiescence, subconfluent monolayers of NIH 3T3 and NIH-EGFR/ErbB-2 cells were cultured in mitogen-free medium (MFM; a 1:1 mixture of Dulbecco's minimal essential medium and Ham's F-12 medium containing 0.2% [vol/vol] serum, 10−7 M Na2SeO3, and 2 μg of transferrin/ml) for 24 to 36 h. For transient-transfection experiments, 293 cells were transfected with plasmid DNAs using a calcium phosphate procedure (12). For flow cytometry studies, cells were resuspended in phosphate-buffered saline (PBS) containing 0.1% Triton X-100, 20 μg of RNase A/ml, and 50 μg of propidium iodide/ml and analyzed with an Epics XL cytometer (Coulter).
Cell proliferation and cell transformation assays.
Phoenix packaging cells (32) were transfected with PINCO and PINCO-RALT plasmid DNAs, selected with puromycin (1 μg/ml) and used as a source of recombinant retrovirus stocks. For cell growth assays, NIH-EGFR/ErbB-2 cells were seeded at 5 × 103 cells/well in 24-well plates coated with gelatin. After 16 h, medium was replaced with conditioned medium obtained from PINCO and PINCO-RALT packaging cell lines; this procedure was repeated after 12 h. Following two rounds of infection, cells were washed with PBS and either MFM or MFM containing EGF (UBI) or serum was added. Cells were cultured under these conditions for 40 h and pulsed with 1 μCi of [methyl-3H]thymidine (Amersham)/ml for 3 h before being harvested. Incorporated radioactivity was measured as described previously (72). To measure a single round of synchronous DNA replication, cells were cultured in serum-containing medium for 16 h after retroviral infection, switched to MFM for 24 h to induce quiescence, and then challenged with mitogens for 24 h before being harvested; [methyl-3H]thymidine was added to the culture medium for the last 12 h.
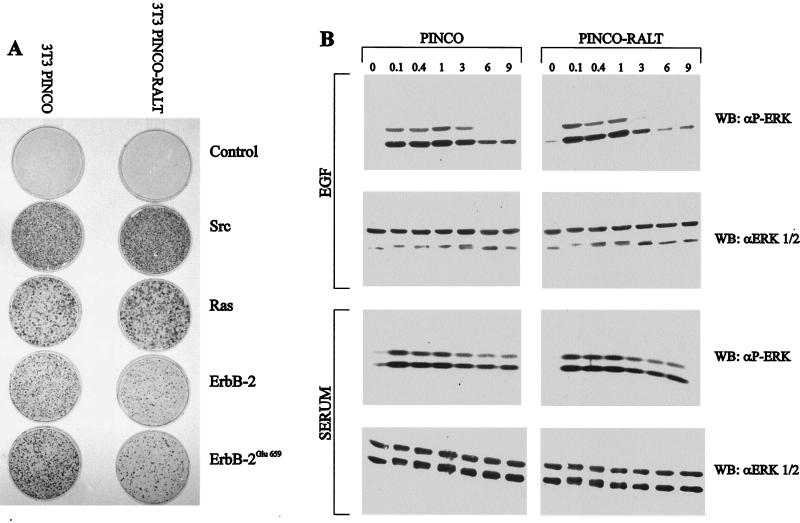
For cell transformation studies, NIH 3T3 cells transduced with either PINCO or PINCO-RALT retroviruses were used as targets for infection with replication-defective retrovirus stocks generated by transient transfection of Bosc23 cells (59) with LTR-2, LTR-erbB-2 (17), LTR-erbB-2Glu659 (70), or zip-v-ras (20); stocks of v-src were obtained as described previously (23). Foci were scored after 10 to 12 days by staining dishes with methylene blue.
Northern blots.
Total cellular RNA was extracted from cells using Trizol reagent (Life Technologies), according to the manufacturer's instructions. RNA samples were size fractionated by agarose electrophoresis, transferred onto nitrocellulose filters, and processed for Northern hybridization. cDNA probes were labeled with [32P]dCTP using a random priming procedure (Ready-To-Go; Amersham Pharmacia Biotech) as indicated by the manufacturer. Inhibition of mRNA or protein synthesis was obtained by adding to the culture medium actinomycin D (1 μg/ml) or cycloheximide (10 μg/ml), respectively.
Immunofluorescence and confocal microscopy analysis.
Cultures of PINCO-RALT cells in 35-mm-diameter plastic dishes were brought to quiescence by a 24-h serum deprivation and then challenged with 10% newborn calf serum or 10 ng of EGF/ml for different lengths of time. Cells were fixed in 4% (vol/vol) paraformaldehyde in PBS at 20°C for 10 min and permeabilized with 0.25% (vol/vol) Triton X-100 in PBS for 10 min. Incubation with anti-RALT 19C5/4 MAb was carried out at 4°C for 16 h. After being washed with PBS, dishes were incubated with affinity-purified goat anti-mouse antibody (Jackson Immunoresearch) at 4 μg/ml to enhance the RALT signal along with diluted M6 rabbit antiserum against the ErbB-2 COOH terminus for 1 h at 20°C. After being washed with PBS, dishes were incubated with tetramethyl rhodamine isocyanate-conjugated donkey anti-goat serum and Cy5-conjugated donkey anti-rabbit serum (Jackson Immunoresearch) for 1 h. After a final washing, nuclei were counterstained with Hoechst 33258 (Calbiochem) and mounted in Gelvatol. Samples were routinely examined with a Zeiss microscope equipped with ×40 and ×50 water immersion objectives. Confocal analysis was carried out with a Leica TCS-NT system equipped with 40× (1.00 to 0.5) and 100× (1.3 to 0.6) oil immersion lenses and an acousto-optical tunable filter (AOTF) to correct for channel cross talk.
RESULTS
Identification of the gene 33 protein as a protein interacting with the kinase domain of ErbB-2.
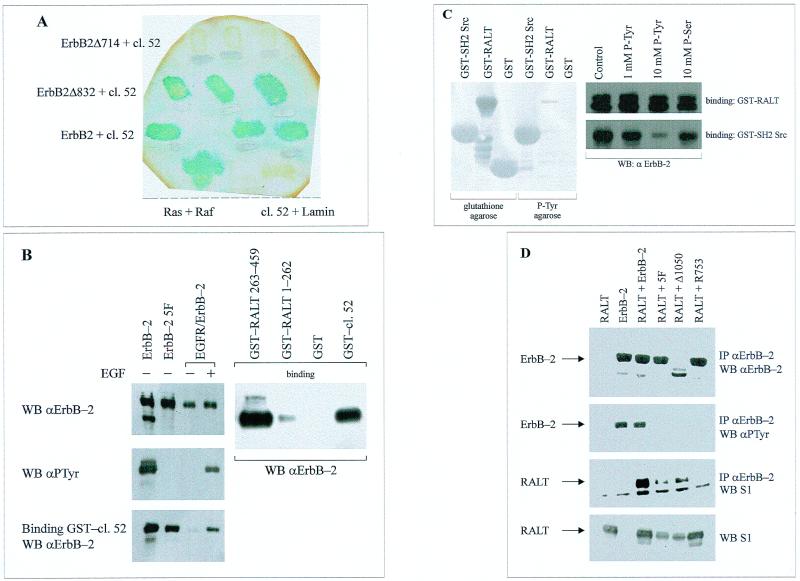
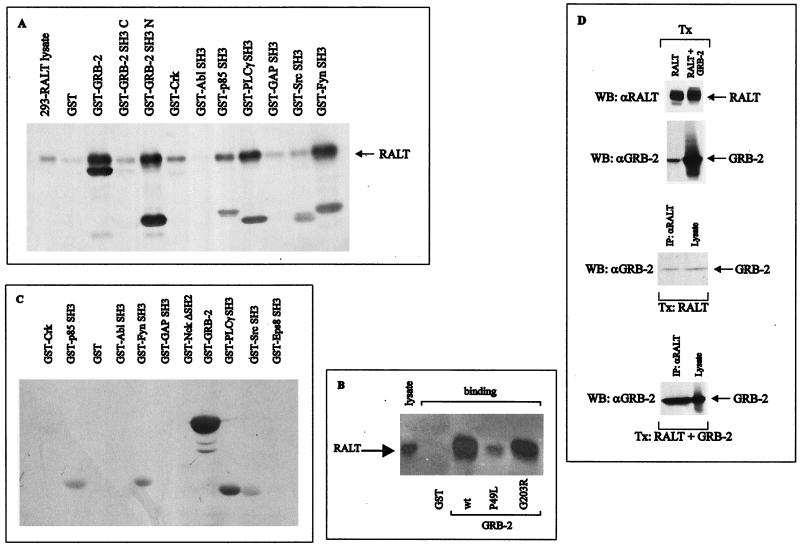
A LexA–ErbB-2 bait (aa 682 to 1049 of ErbB-2) lacking the major ErbB-2 autophosphorylation sites (47, 71) was used to screen a murine embryo cDNA library expressed as fusion products with the VP16 activation domain (78). One cDNA clone (clone 52) generated a fusion protein which showed strong interaction with the LexA–ErbB-2 bait but not with a LexA-lamin fusion protein (Fig. 1A). The strength of the ErbB-2–clone 52 interaction was comparable to that of Ras-Raf complexes (Fig. 1A). Truncation of the COOH-terminal half of the ErbB-2 bait (residues 833 to 1049 of gp185ErbB-2) did not impair the ErbB-2–clone 52 interaction. However, deletion of aa 715 to 1049 (ErbB-2 Δ714), leaving intact only the ErbB-2 juxtamembrane region, abolished the ErbB-2–clone 52 interaction (Fig. 1A).
FIG. 1.
Physical interaction of RALT with ErbB-2. (A) Physical interactions between the indicated LexA–ErbB-2 baits and VP16 fusions were detected in colonies of yeast transformants by assaying β-galactosidase activity on replica filters. Three independent colonies expressing VP16-clone 52 (cl. 52) and ErbB-2 baits were tested. The Ras-Raf interaction was used as positive control for β-galactosidase detection, whereas LexA-lamin was coexpressed with VP16-clone 52 as a negative control. (B) Lysates from NIH–ErbB-2, NIH–ErbB-2 5F, and NIH-EGFR/ErbB-2 transfectants were analyzed for receptor expression (top left blot) and receptor PTyr content (middle blot) by immunoblot analysis; EGFR–ErbB-2 receptors were activated by stimulation with 50 ng of EGF/ml for 5 min at 37°C. Solubilized ErbB-2 and EGFR–ErbB-2 receptors were tested for their ability to bind to the indicated GST fusion proteins immobilized onto glutathione-agarose beads (bottom left and top right blots). Proteins bound to agarose beads were immunoblotted with anti-ErbB-2 antibodies and detected with ECL reaction. WB, Western blot. (C) (Left) Bacterial lysates containing recombinant GST-SH2 Src, GST-clone 52 (GST-RALT), and GST were incubated with either glutathione-agarose or PTyr-agarose beads for 30 min at 4°C. After being washed, bound proteins were resolved by SDS-PAGE, transferred onto a nitrocellulose filter, and stained with Ponceau's red (blotting with anti-GST antibodies gave identical results; not shown). (Right) GST-clone 52 (GST-RALT) and GST-SH2 Src proteins were bound to glutathione-agarose beads and incubated for 30 min with either buffer alone (control) or the indicated phosphoamino acids. Resins were then assayed for the ability to bind to ErbB-2 solubilized from NIH–ErbB-2 transfectants, as described for panel B. P-Ser, phosphoserine (D) 293 cells were transiently transfected with the indicated expression vectors for RALT and/or various ErbB-2 alleles. Anti-ErbB-2 immunoprecipitates (IP) were analyzed by immunoblotting for receptor content (anti-ErbB-2 blot), receptor autophosphorylation (anti-PTyr blot), and the presence of coprecipitated RALT (S1 blot); expression of transfected RALT in each sample was assessed by Western blot analysis of 50 μg of total cell protein with affinity-purified S1 antibodies (bottom). Primary antibodies were detected with HRP-labeled secondary antibodies and the ECL reaction.
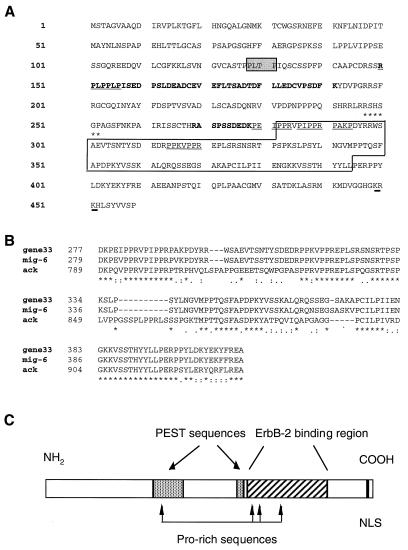
The clone 52 insert encodes 114 aa, which have 99% identity with residues 282 to 395 of the predicted product of rat gene 33 (13, 42) and 97% identity with residues 284 to 395 of the human Mig-6 protein (80). We conclude that clone 52 contains a partial cDNA clone corresponding to the murine orthologue of the widely expressed rat gene 33 (51) and, most likely, of the human mig-6 gene. Both gene 33 (13, 42) and mig-6 (80) have been described as mitogen-induced genes, and their products are 81% identical (87% overall homology) (80).
The 459-aa sequence of the gene 33 protein (Fig. 2A) does not have significant homologies with other proteins in the database with the exception of the noncatalytic portion of ACK, a nonreceptor type tyrosine kinase (46) (see Discussion). However, it reveals previously unnoticed features (shared with Mig-6) which make this polypeptide a candidate for a signal transducer (Fig. 2A): (i) several Pro-rich sequences, which may serve as recognition motifs for SH3 domain-containing proteins (49); (ii) two blocks of PEST sequences (aa 150 to 191 with a PEST score of 6.7 and aa 269 to 278 with a PEST score of 14.35) often found in labile proteins involved in cell cycle regulation (64); (iii) a potential nuclear localization signal (KRKH) at positions 449 to 452; (iv) a PLTP consensus sequence for phosphorylation by ERK 1 and ERK 2 (28); (v) a potential binding site for 14-3-3 proteins (82); and (vi) several potential sites of phosphorylation by protein kinase (PKC) (81), protein kinase A (PKA) (27), and casein kinase II (CKII) (62). As we propose to rename the gene 33 protein with the acronym RALT (see Discussion), the two designations are used interchangeably hereafter.
FIG. 2.
Structural features of RALT. (A) The 459-amino-acid sequence of RALT is shown. The sequence corresponding to that of the clone 52 insert is boxed; PEST sequences (as scored by the PEST-FIND program) are in boldface; candidate SH3 binding sequences are underlined, as is the nuclear localization signal (NLS) KRKH. Grey box, PLTP consensus sequence for phosphorylation by ERK 1 and ERK 2; asterisks, a putative binding sequence for 14-3-3 proteins. Note the presence of several consensus sequences for Thr or Ser phosphorylation by PKA (R/K2-X-S/T), PKC (S/T-X-R/K), and CKII (S/T-X2-D/E). (B) Alignment of the RALT homology region of human ACK (ack) with RALT (gene33) and Mig-6 (mig-6) homologous sequences, as defined by the CLUSTAL W program; asterisks, double dots, and single dots, identities and conserved and semiconserved substitutions, respectively. (C) Schematic representation of RALT as a multidomain protein.
Analysis of the interaction between RALT and ErbB-2 in vitro and in intact cells.
A GST-clone 52 fusion protein readily bound constitutively active gp185ErbB-2 (Fig. 1B) solubilized from NIH–ErbB-2 transfectants (17), whereas no ErbB-2 binding to GST beads (Fig. 1B) and GST-clone 52 beads incubated with NIH 3T3 lysates (data not shown) was observed. The region of RALT spanning residues 282 to 395 (i.e., those corresponding to the polypeptide encoded by the clone 52 cDNA insert) is the only one involved in ErbB-2 recognition, since a longer GST fusion protein (RALT 263-459) bound to gp185ErbB-2 as efficiently as GST-clone 52 and since specific binding to gp185ErbB-2 was not observed with a GST-RALT 1-262 recombinant protein (Fig. 1B).
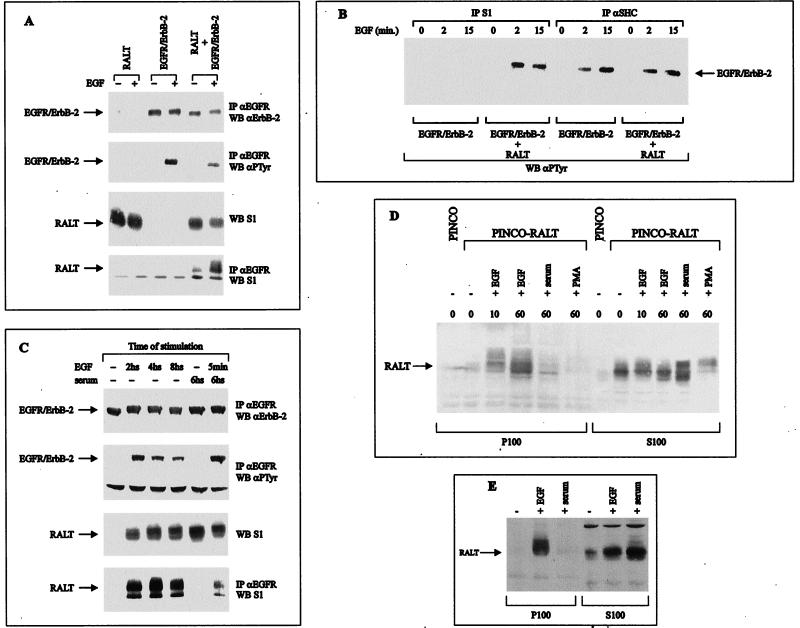
Transient expression of gene 33 cDNA in 293 cells produced a protein resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as a 53- to 54-kDa doublet, which was recognized in immunoblot analysis by affinity-purified antibodies raised against either GST-clone 52 (S1) (Fig. 1D, bottom) or a synthetic peptide spanning positions 443 to 455 of RALT (S3; data not shown). This immunoreactivity is in agreement with the predicted molecular weight of RALT and was absent in 293 cells transfected with the empty parental expression vector. The doublet is likely to reflect different extents of Ser and Thr phosphorylation of RALT, as predicted by the presence of consensus sequences potentially targeted by PKA, PKC, CKII, and ERKs. Indeed, we have found that in living cells 32P incorporation into RALT is dramatically increased by activation of PKC, PKA, and ErbB-2 (data not shown). Anti-ErbB-2 immunoprecipitates from lysates of 293 cells cotransfected with expression vectors for gp185ErbB-2 and RALT contained the 53- to 54-kDa gene 33 polypeptide; this was observed only upon coexpression of ErbB-2 and RALT (Fig. 1D). Complex formation between RALT and ErbB-2 in 293 cells was not affected by the 5F mutation (Fig. 1D), which inactivates major ErbB-2 autophosphorylation sites (71); this was confirmed in in vitro binding assays (Fig. 1B). Likewise, the Δ1050 mutant, which lacks the ErbB-2 COOH tail (18), bound RALT efficiently in intact cells (Fig. 1D) and in vitro (data not shown). That tyrosine phosphorylation does not tag ErbB-2 for molecular recognition by RALT was confirmed by the in vitro experiments whose results are shown in Fig. 1C. Soluble PTyr, unlike phosphoserine, effectively competed out in a dose-dependent fashion the ability of GST-SH2 Src to bind to ErbB-2, whereas it had no effect on the interaction of GST-clone 52 with ErbB-2 (Fig. 1C, right). Furthermore PTyr-agarose efficiently bound GST-SH2 Src but neither GST nor GST-clone 52 (Fig. 1C, left). All three recombinant proteins, however, were bound by glutathione-agarose with comparable efficiencies, as expected (Fig. 1C). Coimmunoprecipitation of RALT with ErbB-2 was abolished by the R753 mutation (Fig. 1D), which ablates tyrosine kinase activity of gp185ErbB-2 (71). The requirement for ErbB-2 catalytic activation in the generation of RALT–ErbB-2 complexes in intact cells was further investigated in 293 cells stably expressing RALT and the EGFR–ErbB-2 recombinant receptor (which is silent unless activated by EGF [24]). Anti-RALT immunoreactivity could be recovered only in antireceptor immunoprecipitates from EGF-stimulated cells and was contingent on EGFR–ErbB-2 and RALT coexpression (Fig. 3A). This was confirmed by the in vitro binding assay shown in Fig. 1B.
FIG. 3.
Activation of the ErbB-2 kinase drives formation of RALT–ErbB-2 complexes and causes relocation of RALT to the membrane compartment. (A) 293 cells were transfected with expression vectors for RALT and/or EGFR–ErbB-2, as indicated. Lysates were prepared either before or after stimulation with EGF (50 ng/ml) for 5 min at 37°C. Antireceptor immunoprecipitates (IP) were analyzed for EGFR–ErbB-2 content (anti-ErbB-2 blot), EGFR–ErbB-2 autophosphorylation (anti-PTyr blot), and the presence of coprecipitating RALT (S1 blot, bottom). Expression of transfected RALT was assessed by Western blot (WB) analysis of 50 μg of total cell protein with S1 antibodies. (B) 293 cells expressing either EGFR–ErbB-2 or EGFR–ErbB-2 and RALT were stimulated for 2 or 15 min with 50 ng of EGF/ml at 37°C; lysates were subjected to immunoprecipitation with antibodies against RALT or SHC followed by Western blot analysis with anti-PTyr antibodies. The indicated bands correspond to tyrosine-phosphorylated EGFR–ErbB-2 molecules, as also proved by their reactivity with anti-ErbB-2 antibodies and their absence in IP prepared with lysates of 293 RALT cells (data not shown). (C) Expression of endogenous RALT protein was induced in quiescent NIH-EGFR/ErbB-2 cells by stimulation with 10 ng of EGF/ml or 10% serum for the indicated times. Antireceptor IP (prepared with a MAb against the extracellular domain of human EGFR) were analyzed by immunoblotting with anti-ErbB-2, anti-PTyr, and S1 antibodies. Expression of RALT was monitored by immunoblotting 50 μg of cell lysate with S1 antibodies. (D) NIH-EGFR/ErbB-2 transfectants infected with PINCO or PINCO-RALT retroviruses (see the legend for Fig. 8 for details) were serum starved for 24 h; cells were lysed either before (−) or after (+) stimulation with 10 ng of EGF/ml, 10% serum, or 50 ng of PMA/ml for the indicated times (minutes). Cytosolic (S100) and membrane fractions (P100) were prepared and analyzed by immunoblotting with S1 antibodies. Under these conditions the S1 antibody detects only ectopically expressed RALT; similar results were obtained with anti-RALT MAb 19C5/4 (not shown). (E) Expression of endogenous RALT protein was induced in quiescent NIH-EGFR/ErbB-2 cells by a 3-h stimulation with 10 ng of EGF/ml or 10% serum; cytosolic and membrane fractions were analyzed by immunoblotting with S1 antibodies. In all panels primary antibodies were detected by incubation with HRP-labeled secondary antibodies and the ECL reaction.
Activation of the ErbB-2 kinase regulates expression of RALT in growth-arrested NIH 3T3 cells.
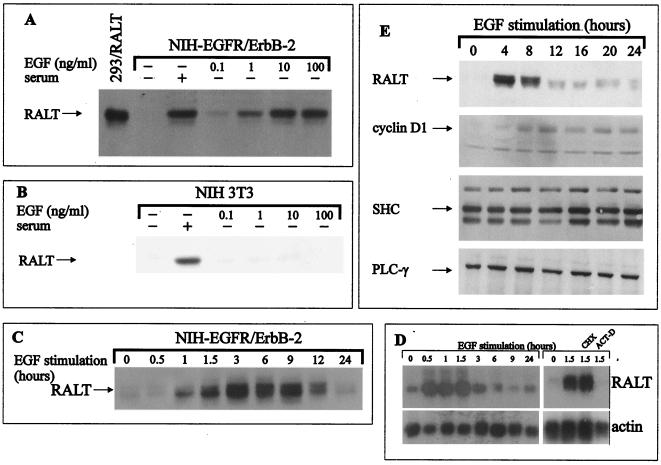
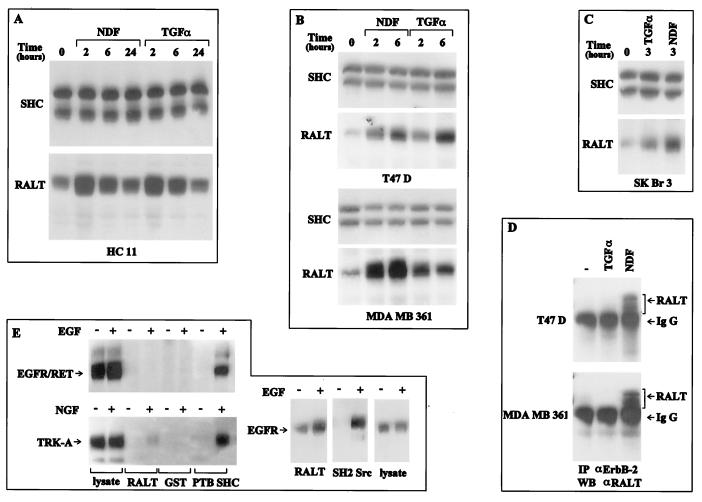
Expression of gene 33 and mig-6 mRNAs is not detected in growth-arrested fibroblasts but is induced by serum stimulation (50, 51, 80). Consistently, expression of RALT was absent in growth-arrested NIH-EGFR/ErbB-2 and NIH 3T3 fibroblasts (Fig. 4A and B), whereas it was readily detectable in cycling cells (Fig. 6B) and quiescent fibroblasts stimulated with serum for 3 h (Fig. 4A and B).
FIG. 4.
Expression of RALT is regulated during the cell cycle. Quiescent NIH-EGFR/ErbB-2 (A) and NIH 3T3 cells (B) were lysed either before (−) or after (+) stimulation for 3 h at 37°C with 10% serum or EGF at the indicated concentrations. Samples were normalized for protein content and analyzed by immunoblotting with S1 affinity-purified antibodies. A lysate of 293 and RALT transfectants was used as control for S1 reactivity in panel A. (C) Quiescent EGFR/ErbB-2 cells were stimulated at 37°C for the indicated times with 10 ng of EGF/ml. Lysates were normalized for protein content and analyzed by Western blotting with S1 antibodies. (D) NIH-EGFR/ErbB-2 cells were made quiescent and then challenged with EGF as in panel C. Total RNA from each sample was subjected to Northern hybridization with a RALT cDNA probe and exposed for autoradiography (top). The same filter was subsequently stripped of radioactivity and hybridized to a human β-actin probe (bottom). When indicated, actinomycin D (ACT-D) or cycloheximide (CHX) was added to medium along with EGF to inhibit de novo mRNA or protein synthesis, respectively. (E) Quiescent EGFR/ErbB-2 transfectants were lysed either before or after stimulation with 10 ng of EGF/ml for the indicated times; lysates were analyzed by blotting with the indicated antibodies. Detection of primary antibodies in Western blots shown in panels A to C and E was with the ECL reaction.
FIG. 6.
RALT binds to SH3 domains. (A) Lysates of 293-RALT transfectants were incubated with the indicated recombinant GST fusion proteins immobilized onto glutathione-agarose beads. Proteins bound to resins were analyzed by immunoblotting with anti-RALT antibodies and the ECL reaction. Lysate in the left lane represents 5% of the input lysate in the binding reaction. (B) GST–GRB-2 fusions expressed from either wt GRB-2 cDNA or cDNA encoding the indicated GRB-2 mutant proteins were purified onto glutathione-agarose beads in similar amounts and used as affinity reagents for the capture of RALT solubilized from 293 transfectants; lysate in the left lane corresponds to 5% input in the binding reaction. Proteins bound to resin were subjected to immunoblot analysis with S1 anti-RALT antibodies. (C) Each of the indicated recombinant proteins (2 μg; purified by affinity chromatography on glutathione-agarose resin) was run on SDS-PAGE gel and transferred to nitrocellulose filters. After the proteins were stained with Ponceau's red to ensure that equal amounts of protein were present in each lane, independent filters were probed with biotin-labeled GST–RALT 1-262 or GST–RALT 263-459; detection was with HRP-conjugated streptavidin followed by the ECL reaction. No specific reactivity was observed with RALT 1-262, and therefore only the filter probed with GST–RALT 263-459 is shown. (D) Lysates from 293 cells transfected with RALT or RALT and GRB-2 expression vectors were analyzed for RALT and GRB-2 expression by immunoblotting (upper two panels). Anti-RALT immunoprecipitates from these lysates were blotted with anti-GRB-2 antibodies along with lysates representing 10% of the input protein in the immunoprecipitation reaction (lower two panels). ECL exposure of the third panel from top was 90 s, while that of the bottom panel was 45 s. WB, Western blotting; Tx, transfection.
Activation of the ErbB-2 kinase by EGF induces a mitogenic response in NIH 3T3 fibroblasts expressing an EGFR–ErbB-2 chimera (24). As shown in Fig. 4C, expression of RALT protein could be detected in growth-arrested NIH-EGFR/ErbB-2 cells within 1 h of EGF stimulation, peaked between 3 and 9 h, and declined thereafter; this effect was dose dependent (Fig. 4A). EGF stimulation of parental NIH 3T3 cells induced neither progression to S phase (not shown) nor detectable expression of RALT (Fig. 4B).
Induction of RALT protein expression upon EGF stimulation of EGFR/ErbB-2 transfectants followed transcriptional activation of gene 33, as indicated by Northern blot experiments shown in Fig. 4D. RALT mRNA expression was almost undetectable in quiescent cells but was promptly induced by EGF stimulation, peaking after 60 to 90 min of EGF triggering and returning to baseline by 6 h. The kinetics of RALT mRNA induction is reminiscent of that of immediate-early genes. Indeed, induction of RALT mRNA by EGF was unaffected by treatment of cells with the protein synthesis inhibitor cycloheximide, whereas actinomycin D abolished RALT mRNA accumulation induced by EGF stimulation (Fig. 4D).
Endogenous RALT protein expressed in EGFR/ErbB-2 transfectants could be found in a complex with the ErbB-2 kinase (Fig. 3C). Serum-starved NIH-EGFR/ErbB-2 fibroblasts were either left untreated or stimulated with mitogens; EGFR–ErbB-2 molecules were immunoprecipitated, and immune complexes were probed with S1 antibodies. RALT could be coimmunoprecipitated with EGFR–ErbB-2 only in EGF-stimulated samples, whereas no RALT–ErbB-2 complexes were detected in serum-stimulated cells. However a brief exposure to EGF was sufficient to activate the ErbB-2 kinase in serum-induced cells and to produce coimmunoprecipitation between EGFR–ErbB-2 and RALT (Fig. 3C, bottom right).
Next, we investigated the relationship between the expression of RALT protein and the kinetics of progression of quiescent EGFR/ErbB-2 transfectants into S phase. In the experiment shown in Fig. 4E, EGFR/ErbB-2 transfectants, made quiescent by serum deprivation, entered S phase 10 to 12 h after EGF stimulation and reached a peak of DNA synthesis between 16 and 20 h; by 24 h most of the cycling cells had completed DNA duplication and accumulated in G2/M (data not shown). Under these conditions cyclin D1 expression was faint after 4 h of EGF stimulation, peaked between 8 and 12 h, and declined thereafter. RALT expression was undetectable in quiescent cells, peaked between the 4- and 8-h time points, and dropped significantly upon progression into S phase. In contrast SHC and PLC-γ expression did not change over the 24-h interval examined (Fig. 4E). Similar kinetics was observed when cells were stimulated with serum (not shown). Thus, mitogenic stimulation of quiescent murine fibroblasts induces expression of RALT protein, which is maintained throughout most of the G1 phase of the cell cycle and which declines sharply upon transition into S phase.
Regulation of RALT in normal and malignant breast epithelial cells.
We next addressed whether expression of RALT protein is also regulated by ErbB-2 signaling in cells in which ErbB-2 is naturally expressed as a coreceptor for new differentiation factor (NDF) (1, 66). We also assayed whether RALT expression is controlled by transforming growth factor alpha (TGF-α).
The murine HC11 cell line is derived from normal mammary gland epithelium and is known to respond mitogenically to ErbB ligands such as TGF-α and NDF (4). Stimulation of quiescent HC11 cultures with either TGF-α or NDF-β1 induced a robust increase of RALT expression, as assessed by immunoblot analysis of samples stimulated with these mitogens for 2 and 6 h. RALT immunoreactivity returned to baseline levels by 24 h (Fig. 5A). By contrast, no changes in the expression of the 46- and 52-kDa SHC isoforms were detected in the same samples (Fig. 5A).
FIG. 5.
Specificity of regulation of RALT expression and RALT–ErbB-2 interaction. Subconfluent cultures of HC11 normal mammary epithelial cells (A) and T47D and MDA-MB 361 (B) and SK-Br3 (C) breast carcinoma cell lines were kept in MFM for 36 h and then either lysed or stimulated for the indicated lengths of time with NDF-β1 or TGF-α (each at 20 ng/ml). Equal amounts of total cell protein were analyzed by immunoblotting with anti-RALT antibodies. Filters were subsequently stripped and reprobed with anti-SHC antibodies. Note that the ECL reaction mixtures corresponding to the anti-RALT blots of T47D, MDA-MB 361, and SK-Br3 samples were exposed for the same times, thus allowing for a fair comparison of relative levels of expression of RALT. (D) T47D and MDA-MB 361 cultures were kept in MFM for 36 h and then stimulated for 3 h with the indicated ligands (each at 20 ng/ml). Lysates were subjected to immunoprecipitation (IP) with the anti-ErbB-2 MAb and immunoblotting with the anti-RALT antibody. The band below RALT in each lane corresponds to the MAb heavy chain. WB, Western blotting; IgG, immunoglobulin G. (E) Quiescent monolayers of NIH 3T3 transfectants expressing EGFR, TRK-A, or the chimeric EGFR-RET receptor were stimulated with either carrier or the indicated ligand for 5 min at 37°C. After lysis, equal amounts of cellular protein were assayed in binding reactions with the indicated GST fusion proteins immobilized onto glutathione-agarose beads. Precipitated proteins were analyzed by immunoblotting with the indicated antireceptor antibodies. Antireceptor immunoreactivity in 5% of the input lysate in corresponding binding reactions is shown.
We extended our analysis to the T47D, MDA-MB 361, and SK-Br3 breast carcinoma cell lines, which display increasing levels of expression of ErbB-2 (40). NDF stimulation of these cell lines prominently activates ErbB-2–ErbB-3 heterodimers, while TGF-α binds to EGFR and possibly transmodulates ErbB-2 (4). Stimulation of T47D, MDA-MB 361, and SK-Br3 cells with either NDF-β1 or TGF-α led to increased expression of RALT protein, whereas anti-SHC immunoreactivity was not affected by either treatment (Fig. 5B and C). Interestingly, serum stimulation did not induce RALT expression in these epithelial cell lines, at variance with what we observed in murine fibroblasts (not shown). Noticeably, stimulation of RALT expression by NDF in these breast tumor cell lines did not require abnormal levels of ErbB-2 receptors: T47D cells express quasinormal levels of ErbB-2 and yet NDF induced as much RALT protein in these cells as in the SK-Br3 cell line, which contains an amplified ErbB-2 gene (40). We conclude that in normal and malignant breast epithelial cells RALT expression is controlled by signals propagated by either EGFR (upon TGF-α stimulation) or heterodimers containing ErbB-2 (upon NDF stimulation). Our data do not rule out, however, that transmodulation of ErbB-2 by TGF-α-activated EGFR contributes to the induction of RALT expression in breast epithelial cells.
Anti-ErbB-2 immunoprecipitates prepared from T47D and MDA-MB 361 cells stimulated with NDF-β1 contained anti-RALT immunoreactivity (Fig. 5D). This immunoreactivity was absent in anti-ErbB-2 immunoprecipitates prepared from either carrier-stimulated cultures or TGF-α-treated cells (Fig. 5D). In breast tumor cells which do not overexpress EGFR, such as T47D and MDA-MB 361, TGF-α-driven ErbB-1–ErbB-2 heterodimers are not detected in coimmunoprecipitation assays (53); therefore our results indicate that in cells derived from breast epithelium RALT is recruited by ErbB-2 in the context of NDF-driven ErbB-2–ErbB-3 heterodimers.
Finally, we assessed whether RTKs bearing different degrees of structural homology to ErbB-2 were able to interact with RALT in vitro. GST-clone 52 fusion protein interacted with EGFR solubilized from NIH-EGFR transfectants (Fig. 5E). This binding was dependent to some extent on EGF stimulation. At variance with EGFR, ErbB-2, and the EGFR–ErbB-2 chimera, neither TRK-A, the high-affinity NGF receptor, nor the EGFR-RET chimera (69) was able to bind to GST-clone 52 (Fig. 5E). In control experiments, however, both TRK-A and EGFR-RET bound efficiently to GST-SHC PTB (58) upon ligand stimulation (Fig. 5E).
RALT binds to SH3 domains.
The presence of several Pro-rich sequences in RALT suggests that it may complex with proteins containing SH3 domains (49). This hypothesis was tested by assaying the ability of recombinant SH3 domains to interact with RALT protein solubilized from 293 transfectants. Poor binding or no binding at all was detected with GST–c-Crk and SH3 domains from Abl, Ras GAP, Src (Fig. 6A), Eps 8, and Nck (data not shown; Fig. 6C). A significant fraction of RALT protein bound to GST fusion proteins containing the SH3 domain of the p85 subunit of phosphatidylinositol 3-kinase, Fyn, and PLC-γ1; GST–GRB-2 bound RALT with the highest affinity, and this interaction was accounted for entirely by the NH2-terminal SH3 domain of GRB-2 (Fig. 6A). Consistently, the P49L substitution in the GRB-2 NH2 SH3 module (68), which is homologous to a loss-of-function mutation in Caenorhabditis elegans Sem-5 (14), caused a dramatic reduction in the GRB-2–RALT interaction (Fig. 6B). On the other hand the G203R mutation (68), which disrupts the function of the COOH SH3 domains of Sem-5 and GRB-2 (14), had no effect on GRB-2 binding to RALT (Fig. 6B).
Binding to recombinant SH3 domains was direct and entirely confined to the RALT COOH-terminal half, as demonstrated by blot overlay assays (Fig. 6C). Purified GST-RALT 1-262 and GST-RALT 263-459 proteins were labeled with biotin and used to probe nitrocellulose filters containing a battery of recombinant SH3 domains expressed as GST fusions. Soluble RALT 263-459 interacted with filter-bound GST–GRB-2 and also with SH3 domains from p85, Fyn, PLC-γ, and Src, albeit less efficiently than with GRB-2 (Fig. 6C). GST itself and other SH3 domains did not react with RALT 263-459. Soluble RALT 1-262 did not bind to any of the recombinant proteins tested in Fig. 6C. Ponceau's red staining of the blots shown in Fig. 6A to C indicated that the inputs of recombinant proteins in all lanes were comparable (not shown). Collectively these experiments indicate that RALT binds with the highest affinity to the NH2-terminal SH3 domain of GRB-2 and to a weaker extent to the PLC-γ, p85, Src, and Fyn SH3 modules. The RALT–GRB-2 interaction was also detected in intact cells. No anti-GRB-2 immunoreactivity was found in anti-RALT immunoprecipitates prepared from 293 cells transfected with the GRB-2 expression vector (not shown), due to very low expression of endogenous RALT in 293 cells (see Fig. 1D and 3A). Anti-RALT immunoprecipitates from 293 cells expressing ectopic RALT contained GRB-2 (Fig. 6D, third panel from top). Anti-GRB-2 immunoreactivity in anti-RALT immunoprecipitates was markedly increased by concomitant transfection of RALT and GRB-2 expression vectors, as shown in the bottom panel of Fig. 6D.
Spatial regulation of RALT by activation of the ErbB-2 kinase.
RALT may complex with ErbB-2 because it is either a substrate of the ErbB-2 kinase or a regulator of ErbB-2 catalytic function. RALT phosphorylation on tyrosine residues was not detected upon ligand activation of EGFR–ErbB-2 (data not shown). This is consistent with the observation that there are no tyrosine residues in the RALT sequence embedded in a context canonically suitable for catalytic recognition by tyrosine kinases. Expression of ectopic RALT altered neither the constitutive level of tyrosine phosphorylation of gp185ErbB-2 in 293 cells (Fig. 1D) nor the rate of ligand-dependent autophosphorylation of EGFR–ErbB-2 in 293 and NIH 3T3 transfectants (data not shown). Finally, EGFR–ErbB-2 molecules precipitated by anti-RALT antibodies were readily labeled by anti-PTyr antibodies (i.e., they belonged to the pool of activated receptors) at a stoichiometry comparable to that of receptors coprecipitated by anti-SHC antibodies (Fig. 3B shows an anti-PTyr blot of anti-SHC and anti-RALT immunoprecipitations containing similar levels of EGFR–ErbB-2). These data argue against RALT being either an ErbB-2 substrate or a negative regulator of ErbB-2 intrinsic kinase function.
As RALT protein lacks membrane-targeting sequences, we surmised that RALT resides in the cytosol and may transit to cellular membranes upon interaction with ErbB-2. Such a behavior would be consistent with RALT being an adapter protein able to relocate cytosolic effectors containing SH3 domains (Fig. 6) via its conditional interaction with ErbB-2 (Fig. 1 and 3).
Immunoblotting analysis of cytosolic and membrane fractions prepared from quiescent NIH-EGFR/ErbB-2 cells expressing ectopic RALT (PINCO-RALT cells; see below for more details) indicated that RALT was localized exclusively in the cytosol. Upon EGF stimulation a rapid and sustained relocation of RALT to the membrane fraction was observed, whereas serum and phorbol myristate acetate (PMA) were ineffective (Fig. 3D). Likewise, when synthesis of endogenous RALT was induced in NIH-EGFR/ErbB-2 cells by a 3-h stimulation with either serum or EGF, RALT relocation to cell membranes was observed only in the EGF-treated sample (Fig. 3E).
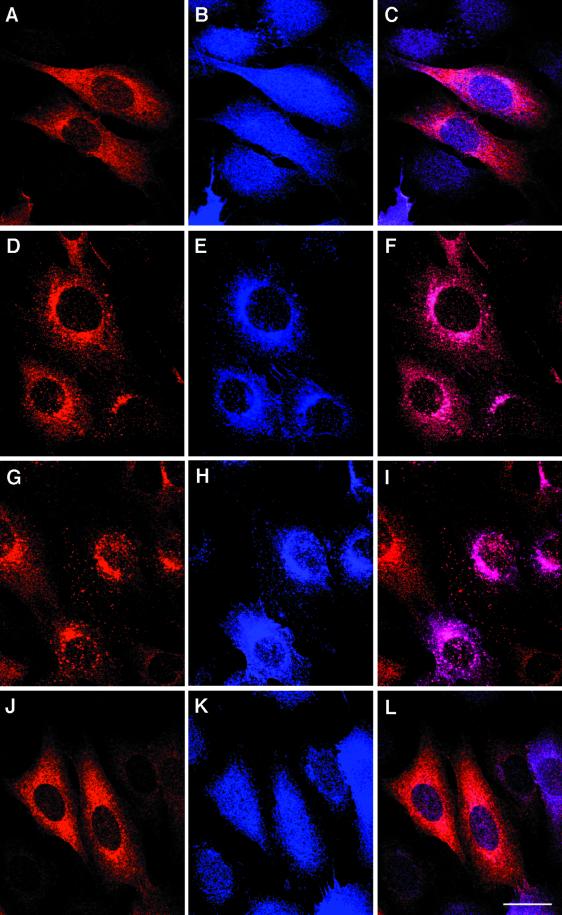
We also performed immunocytochemistry experiments to obtain structural information on the subcellular localization of RALT. As the S1 antibody performed poorly in immunofluorescence assays, we used PINCO-RALT cells and detected ectopic RALT by a MAb specific for the rat RALT species (see Materials and Methods). The use of PINCO-RALT cells also allowed us to study RALT location in a cell not subjected to mitogenic stimuli necessary to induce synthesis of endogenous RALT. In mitogen-deprived PINCO-RALT cells, staining for ectopic RALT was finely granular and diffuse throughout the cytoplasm (Fig. 7A). Upon EGF treatment at 37°C, RALT staining accumulated in a time-dependent fashion to the perinuclear region and yielded a coarsely granular pattern indicative of RALT being relocated to membranous structures (Fig. 7D and G). In EGF-stimulated cells, EGFR–ErbB-2 molecules relocated from the plasma membrane (Fig. 7B) to intracellular structures (Fig. 7E and H), most likely identifiable as endosomes (45). Merging of images indicated extensive colocalization of RALT and EGFR–ErbB-2 in EGF-treated cells (Fig. 7F and I). On the other hand, serum stimulation of PINCO-RALT cells for 60 min did not cause relocation of either RALT or EGFR–ErbB-2 molecules (Fig. 7J to L). Similar results were obtained when cells were stimulated with serum for either longer or shorter lengths of time (data not shown).
FIG. 7.
Immunofluorescence imaging of RALT in EGFR/ErbB-2 transfectants. Subconfluent monolayers of PINCO-RALT cells (see legend for Fig. 8A and B for details) were kept in MFM for 24 h. Dishes were processed for immunocytochemistry either before (A to C) or after (D to I) stimulation with EGF (10 ng/ml) for 15 (D to F) and 60 min (G to I). Samples shown in panels J to L were stimulated with 10% serum for 60 min. Samples shown in panels A, D, G, and J were stained with anti-RALT MAb 19C5/4; those shown in panels B, E, H, and K were stained with anti-ErbB-2 M6 antiserum. (C, F, I, L) Merged images. Bar (L), 20 μm.
Collectively, the experiments discussed above prove that activation of ErbB-2 kinase is necessary to relocate RALT from cytosol to membrane compartments and therefore suggest that spatial control of RALT is implemented by the ErbB-2–RALT physical interaction.
Biological analysis of RALT in mitogenic signaling activated by ErbB-2.
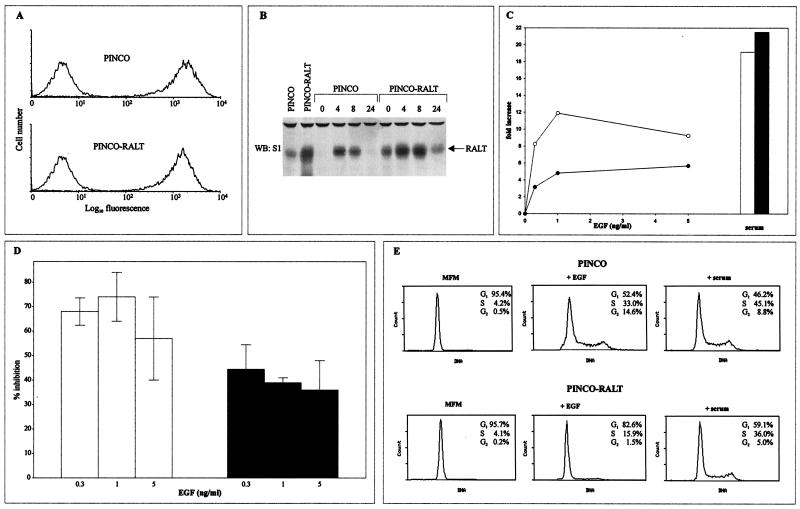
We addressed the possible function of RALT in ErbB-2 mitogenic signaling by overexpressing RALT in NIH-EGFR/ErbB-2 cells. The PINCO vector (32) was used to generate high-titer recombinant retrovirus stocks expressing either GFP or both GFP and RALT. Infection efficiencies of target NIH-EGFR/ErbB-2 cells (hereafter designated PINCO and PINCO-RALT cells, respectively) were routinely greater than 95%, as determined by flow cytometry assessment of GFP-positive cells (Fig. 8A). In cycling cells, ectopic RALT was overexpressed about threefold over the levels of the endogenous protein (Fig. 8B). In quiescent PINCO-RALT cells, ectopic RALT was detected at levels comparable to those of the endogenous protein expressed by quiescent PINCO cells after 4 h of EGF stimulation (Fig. 8B). Expression of ectopic RALT was high throughout G1 phase. For biological experiments, PINCO and PINCO-RALT cells were switched to MFM supplemented with either serum or EGF soon after infection. DNA synthesis was assessed after 40 h. EGF-treated PINCO cells showed a dose-dependent increase of thymidine incorporation over control cultures kept in MFM; optimal responses were obtained at 1 ng/ml EGF (Fig. 8C). By contrast, EGF-treated PINCO-RALT cells grew poorly at suboptimal to optimal EGF concentrations (Fig. 8C), their thymidine incorporation being reduced by 68.3 and 74.8% at 0.3 and 1 ng/ml, respectively (Fig. 8D). At high EGF concentrations (5 ng/ml) the RALT inhibitory effect was partially alleviated (57.2% average inhibition; Fig. 8D), and indeed optimal responses of PINCO-RALT cells were routinely obtained at 5 ng of EGF/ml. RALT overexpression was not toxic per se, since the viabilities of PINCO and PINCO-RALT cells in MFM and EGF-containing media were comparable and PINCO-RALT cells grew as efficiently as PINCO cells in media containing either serum (Fig. 8C) or PMA (data not shown).
FIG. 8.
RALT overexpression inhibits ErbB-2 mitogenic activity. (A) NIH-EGFR/ErbB-2 cells were infected with replication-defective PINCO and PINCO-RALT retroviruses; transduction efficiency was monitored by flow cytometry assessment of GFP-positive cells. Background fluorescence of parental cells is indicated by the left peak in each graph. (B) RALT expression in PINCO and PINCO-RALT cells was assessed by immunoblot analysis of lysates prepared either from cycling (left two lanes) or from quiescent cells stimulated for the indicated times (hours) with 5 ng of EGF/ml. WB, Western blotting. (C) NIH-EGFR/ErbB-2 cells were plated in 24-well plates, subjected to two rounds of infection with PINCO and PINCO-RALT viruses, and then switched to either MFM or MFM supplemented with serum or EGF. DNA synthesis was assessed after 40 h by pulse-labeling quadruplicate wells with [3H]thymidine. Data are expressed as fold increases of thymidine incorporation in mitogen-treated wells over that in mitogen-free wells. A typical dose-response analysis for EGF stimulation is shown. Open circles, PINCO; solid circles, PINCO-RALT. Bars at right, mitogenic response to 5% serum. Open bar, PINCO; solid bar, PINCO-RALT. (D) Open bars, average inhibition of ErbB-2-driven DNA synthesis in PINCO-RALT cells grown in EGF-supplemented MFM compared to that in PINCO cells; data were obtained from five experiments performed as detailed in the legend to panel C; solid bars, average inhibition of DNA synthesis in growth-arrested PINCO-RALT cells stimulated to enter S phase by EGF compared to that in PINCO cells. Assays were performed in quadruplicate, and data were obtained from four experiments. Error bars, standard deviations. (E) After infection, PINCO and PINCO-RALT cells were grown for 40 h either in MFM or MFM supplemented with 5 ng of EGF/ml or 5% serum. Distribution of cells in the cell cycle was assessed by staining with propidium iodide and analysis with an Epics flow cytometer. Percentages of cells in G1, S, and G2 phases are indicated.
PINCO cells grown for 40 h in either serum or EGF-containing media showed a profile of cell cycle distribution typical of exponentially growing cells and similar to that of PINCO-RALT cells growing in serum-supplemented media (note that the slightly reduced growth fraction of PINCO-RALT cells in serum reported in Fig. 8E was not observed in other experiments). PINCO-RALT cells cultured in EGF-containing media, however, accumulated in G0/G1 (Fig. 8E). Hence, RALT overexpression appears to antagonize ErbB-2-driven cell proliferation by inducing G1 arrest. This conclusion was reinforced by experiments in which we measured the influence of RALT overexpression on the ability of quiescent EGFR/ErbB-2 transfectants to undergo a synchronous round of DNA replication. RALT overexpression exerted a robust inhibitory effect on EGF-driven DNA synthesis (Fig. 8D), whereas comparable responses to serum stimulation were observed in PINCO and PINCO-RALT cells (not shown).
Since cell transformation experiments accommodate the complexity of chronic deregulation of mitogenic signals, we assayed whether RALT overexpression was also able to antagonize ErbB-2 function under these stringent conditions. NIH 3T3-PINCO and NIH 3T3 PINCO-RALT fibroblasts were superinfected with replication-defective recombinant retroviruses expressing v-src, v-ras, erbB-2, and erbB-2Glu659. Foci of transformed cells were scored after 10 to 12 days (Fig. 9A). RALT overexpression did not affect focus formation by v-src and v-ras, whereas cell transformation by retroviruses expressing either wild-type (wt) erbB-2 or the gain-of-function erbB-2Glu659 mutant (70) was significantly inhibited (Fig. 9A).
FIG. 9.
RALT overexpression inhibits ErbB-2-driven cell transformation and late ERK activation. (A) NIH 3T3 fibroblasts were infected with either PINCO or PINCO-RALT retroviruses; >95% of target cells were transduced as indicated by GFP expression. PINCO or PINCO-RALT NIH 3T3 cells were superinfected with the indicated replication-defective recombinant retroviruses. Dishes were stained with methylene blue after 10 to 12 days. (B) NIH-EGFR/ErbB-2 fibroblasts transduced with PINCO or PINCO-RALT retroviruses were made quiescent and then challenged with either 10% serum or 5 ng of EGF/ml for the indicated times (hours). Lysates were analyzed by immunoblotting with anti-ERK 1 and 2 and anti-P-ERK antibodies, followed by the ECL reaction.
RALT regulates the kinetics of ERK 1 and 2 activation by ErbB-2.
Sustained activation of ERK 1 and 2 is crucial to ErbB-2 mitogenic and transforming function (5) and is a major consequence of ErbB-2 recruitment into heterodimers between ErbB family members (31). We therefore assessed whether RALT overexpression alters the profile of activation of ERK 1 and 2 by ErbB-2. Quiescent PINCO or PINCO-RALT cells were stimulated with either EGF or serum for different times. Activated ERK 1 and 2 molecules were detected by Western blot analysis of cell lysates with antibodies against phosphorylated ERK 1 and 2 (29). Serum activated ERK 1 and 2 with comparable strengths and durations in PINCO and PINCO-RALT cells (Fig. 9B). EGF-dependent activation of the ErbB-2 kinase also produced rapid and sustained activation of ERKs in PINCO cells. RALT overexpression did not affect the initial activation of ERKs by ErbB-2, whereas late ERK 1 and 2 activation (>1 h) was significantly reduced by RALT overexpression (Fig. 9B), despite comparable levels of ERK 1 and 2 (Fig. 9B) and receptor expression (data not shown).
DISCUSSION
Rat gene 33 (13, 42) and its likely human orthologue, mig-6 (80), were previously cDNA cloned as genes whose expression is induced in quiescent cells by mitogenic stimulation. However, those studies did not provide clues to either the structure or the function of the gene products. We report here that the gene 33 protein is a modular protein consisting of two blocks of PEST sequences in its NH2-terminal half, in addition to functional SH3 binding motifs and an ErbB-2 binding domain in its COOH-terminal portion (Fig. 2C). We also identify the gene 33 protein as a signal regulator able to complex with the kinase domain of the ErbB-2 receptor and to antagonize mitogenic signals propagated by gp185ErbB-2. Expression of the gene 33 protein and formation of its complexes with ErbB-2 occur in G1 phase at a stage when biochemical responses which immediately follow RTK triggering have already attained their peak. Hence we propose to rename the gene 33 protein RALT (receptor-associated late transducer).
Physical interaction between RALT and gp185ErbB-2.
Unlike most of the RTK signal transducers identified so far, RALT contains neither an SH2 nor a PTB domain and binds to ErbB-2 in a PTyr-independent fashion via a region between aa 282 and 396 (Fig. 1). The minimal EBR (ErbB-2 binding region) of RALT can be narrowed down to a 59-aa stretch which spans positions 313 to 372 (our unpublished data). The EBR is located within a region of homology of RALT and Mig-6 with the noncatalytic COOH-terminal portion of ACK (Fig. 2B), a nonreceptor tyrosine kinase also containing a CRIB domain and an SH3 module (46). RALT and ACK are 80.6% homologous (53.7% identical) in this 134-aa stretch, which we propose to name RALT homology region (RHR). It has been suggested that the portion of ACK containing the RHR is involved in ACK regulation (46, 83). In light of our findings we propose that it may bind via its EBR-like module to the catalytic domain of RTKs (ErbB receptors would be likely candidates) or, alternatively, to the kinase domain of ACK itself. Given their conservation, it is likely that RHR sequences which flank the EBR module may regulate a function(s) integrated with that of the EBR itself. Remarkably, all of the SH3 binding motifs present in the RALT COOH-terminal half are conserved in the RHR of ACK; thus, it appears that EBR modules cosegregated with adjacent SH3 binding motifs during evolution (see below for further discussion).
Integrity of aa 715 to 832 of ErbB-2 is required for the RALT–ErbB-2 interaction to occur (Fig. 1A). This sequence spans the NH2-terminal lobe of the ErbB-2 kinase domain, as defined by the molecular model of the ErbB-2 catalytic domain (54). Our observation lends support to the contention that this region is relevant for signal diversification (72) and expands the functional relevance of the ErbB-2 kinase domain.
RALT–ErbB-2 complex formation is contingent on receptor activation. In principle, signaling by ErbB-2 could induce rapid posttranslational modifications of RALT, which would allow RALT itself to be bound by ErbB-2. This is unlikely, however, as the RALT–ErbB-2 interaction can be reconstituted in vitro using E. coli-expressed RALT fragments. It is more plausible that receptor activation unmasks a cryptic RALT binding site in the ErbB-2 kinase domain. This may be caused by ErbB-2 phosphorylation on Ser and Thr residues, which is enhanced by receptor signaling, or by the precise rotational coupling of ErbB-2 monomers which is required for productive dimerization of ErbB-2 (9, 36).
Despite its interaction with the ErbB-2 kinase domain, RALT is neither a substrate of gp185ErbB-2 nor a direct regulator of ErbB-2 catalytic activity. Rather, RALT's interaction with ErbB-2 apparently allows it to be translocated from cytosol to the membrane compartment. Spatial regulation of RALT may be pertinent to its functioning as an adapter/scaffold protein (see below) and its ability to inhibit cell proliferation. Indeed, in PINCO-RALT cells stimulated with serum or PMA, we observed neither RALT relocation to the membrane compartment nor inhibition of proliferation.
RALT is an inhibitor of ErbB-2 mitogenic function and transforming activity.
Overexpression of RALT in NIH 3T3 fibroblasts inhibited ErbB-2-dependent cell proliferation and caused cells to accumulate in the G0/G1 phase of the cell cycle. Ectopic expression of RALT was also capable of antagonizing the transforming activity of overexpressed gp185ErbB-2. RALT, however, does not seem to behave as a general inhibitor of mitogenic signaling and cell transformation. The oncogenic activity of Ras and Src were not affected by ectopic RALT, nor did we observe adverse effects of RALT overexpression on cell proliferation driven by serum. The available data indicate that the function of RALT in the context of mitogenic signaling may be restricted to receptors of the ErbB family, as its overexpression in the same cellular context inhibits cell proliferation driven by EGFR, ErbB-4, and several ErbB heterodimers, but not by platelet-derived growth factor receptor (PDGF-R) and fibroblast growth factor receptor 1 (FGF-R1) (our unpublished data). The promiscuity of RALT within the ErbB family is not surprising, since ErbB receptors and their ligands form an integrated signaling network driven by combinatorial homodimeric and heterodimeric receptor interactions (1, 66). Biochemically, this correlates with the finding that EGFR binds in vitro to RALT.
An apparent paradox, however, resides in the observation that mitogens such as serum, PMA, basic FGF, and PDGF induce RALT expression in NIH 3T3 cells and yet ectopic expression of RALT does not antagonize their mitogenic programs (Fig. 4 and 8) (our unpublished observations). One possibility is that RALT is involved in the control of cellular functions other than ErbB-2-driven proliferation. Alternatively, RALT function may be required in the context of a wide range of mitogenic stimuli, due to the fact that ErbB family members are cross-activated by a number of receptors, including integrins (10), interleukin-6 receptor (63), and G protein-coupled receptors (GPCRs) (33). RALT may also provide a safeguard mechanism, which allows cells stimulated by non-ErbB ligands to resist perturbation by concomitant and inappropriate activation of ubiquitously expressed ErbB receptors. In either of the last two scenarios, transcriptional activation of RALT could be envisaged, at least in fibroblasts, as a stereotyped genetic response devoted to specific regulation of ErbB signaling.
RALT can be categorized as a cell-autonomous feedback inhibitor of gp185ErbB-2, whose physiological function is that of tuning the strength and duration of ErbB-2 mitogenic signals. This may be particularly relevant in light of (i) the hierarchical privilege enjoyed by gp185ErbB-2 in the assembly of heterodimers between members of the ErbB family (30) and (ii) the sustained signaling activity afforded to heterodimers containing ErbB-2 by their decelerated rates of ligand dissociation (38) and internalization (3, 43). Given the relevance of ErbB-2 function in development (2, 7, 22, 41, 52) and the expression of RALT in embryonic tissues, it is also possible that fine regulation of ErbB-2 signaling by RALT plays a role in morphogenic processes. Indeed, developmental studies have indicated that transcriptionally activated inhibitors of Drosophila EGFs, and FGF receptor, such as Kekkon 1 (25) and Sprouty (11), act in a cell-autonomous fashion to restrain RTK function, in order that patterning can be induced properly (60).
Structural determinants of RALT implicated in signaling downstream to the ErbB-2 receptor.
How does RALT exert its regulatory function on ErbB-2 signaling? Neither steady-state expression nor catalytic activation of gp185ErbB-2 and EGFR–ErbB-2 seems to be altered by RALT overexpression. Furthermore, overexpression of RALT inhibited with comparable efficiencies both wt ErbB-2 and ErbB-2Glu659, two receptor species endowed with markedly different rates of internalization (45). In addition, we did not observe significant consequences of RALT overexpression on the rate and kinetics of ligand-induced EGFR–ErbB-2 down-regulation (our unpublished observations). These experiments indicate that RALT is unlikely to inhibit ErbB-2 mitogenic function by altering receptor trafficking and suggest that RALT may function as a bona fide signal transducer which affects mitogenic pathways downstream to ErbB-2. Its ability to bind to SH3 domains and its property of being spatially regulated by virtue of interaction with gp185ErbB-2 are consistent with RALT behaving as an adapter/scaffold protein which links activated ErbB-2 receptors to effectors containing SH3 domains. This may be a fundamental function of RALT since, as discussed above, its EBR and SH3 binding motifs appear to be evolutionarily linked and possibly functionally connected to each other. The NH2-terminal SH3 domain of GRB-2 bound RALT in vitro with the highest affinity, and RALT–GRB-2 association was detected in intact cells. Although it has not been determined as yet whether the RALT–GRB-2 interaction is relevant to the growth-suppressive function of RALT, we note an intriguing analogy between the ErbB-2–RALT–GRB-2 interaction and the c-Kit/SOCS-1/GRB-2 pathway. SOCS1 is a scaffold protein whose expression is induced in the hematopoietic lineage by c-Kit signaling. SOCS1 binds in turn to c-Kit in a ligand-dependent fashion, recruits GRB-2 by binding to GRB-2 SH3 domains, and inhibits mitogenic signaling by the Kit receptor without affecting its intrinsic catalytic activity (16).
Temporal regulation of RALT expression.
In quiescent NIH 3T3 cells, ralt mRNA is induced by mitogens with kinetics similar to that of immediate-early genes (this study; 51). Therefore RALT belongs to an expanding class of mammalian regulators of tyrosine kinase signaling, whose expression is transcriptionally controlled by receptor activation (6, 16, 55, 60, 75).
The presence of PEST sequences in the RALT protein suggests that RALT may be labile. Consistently, proteasome inhibitors induce accumulation of RALT protein (our unpublished observations). These findings indicate that accumulation of RALT is governed at several levels, including gene transcription and protein stability. Such a biochemical lability may provide RALT with the property to rapidly fluctuate in G1 and timely tune ErbB-2 signaling. The eventual loss of RALT expression upon entry into S phase is likely needed to avoid a priori inhibition of ErbB-2 mitogenic signaling in cycling cells which reenter G1.
The temporal profile of ERK 1 and 2 activation by ErbB-2 is shortened in cells overexpressing RALT. Although the molecular mechanisms linking RALT overexpression to inhibition of ERK activity have not been detailed, this finding is relevant for two reasons. Given the importance of ERK 1 and 2 activity in ErbB-2 mitogenic signaling (5), we suggest that the inhibitory function of RALT is funneled, at least in part, via down-regulation of this pathway. The kinetic experiments shown in Fig. 9B also indicate that overexpressed RALT intercepts ErbB-2-activated pathways only in a temporal window in which endogenous RALT is expressed. Hence, the phenotype observed upon RALT overexpression (at least at the levels obtained in our experimental conditions) seems to be caused by a gene dosage effect, rather than by the temporally inappropriate expression of RALT.
Spatial determinants of RALT function.
Immunofluorescence imaging indicated that RALT colocalizes with EGFR–ErbB-2 in what appeared to be endosomal structures (Fig. 7). Although it is entirely possible that some RALT molecules may interact with ErbB-2 at the plasma membrane, our data suggest that the endosomal compartment may be a privileged site for RALT–ErbB-2 complexes. It is noteworthy that RALT protein expression is detected in different cell types after prolonged ErbB-2 or EGFR activation, at a time when a significant fraction of receptors has undergone internalization. A growing body of literature indicates that this pool of internalized receptors has signaling competence and may contribute to spatial regulation of signaling complexes activated by RTKs (19, 34, 35, 77). For instance, EGFR internalization is coupled to Ras activation (34), whereas PLC-γ molecules bound to internalized EGFR do not promote phosphatidylinositol hydrolysis, despite being efficiently phosphorylated on tyrosine residues by EGFR itself (35). Consistently, internalization was shown to be required for efficient ERK activation by EGFR, under conditions in which EGFR triggering depended on either direct ligand stimulation (77) or transmodulation by GPCRs (61). An attractive hypothesis could therefore hold that RALT molecules bound to internalized gp185ErbB-2 are involved in tuning ERK activation carried out by trafficking ErbB complexes.
Concluding remarks.
We have shown that the triggering of ErbB-2 induces expression of RALT, which in turn may function as a feedback inhibitor of ErbB-2 mitogenic activity. Signals of different strengths and durations are generated by ligand-receptor complexes of the ErbB family as a function of their stability at the plasma membrane and their relative propensities for either being degraded or recycled back to the cell surface once internalized (79). This study adds a novel level of complexity to the network which modulates in time the output of ErbB mitogenic signals. Given the pathogenic role of deranged ErbB-2 activity in human cancer (1, 66), it will be of interest to determine whether there is as a selection for genetic or epigenetic disruption of RALT expression and/or function in human tumors.
ACKNOWLEDGMENTS
We are grateful to S. Hollenberg who made available vectors, cDNA library, yeast strains, and protocols for the two-hybrid system. P. P. Di Fiore, T. Miki, A. Hall, S. Gutkind, M. Cippitelli, M. Sudol, B. Mayer, A. Sacchi, M. Santoro, S. Strano, and P. G. Pelicci are acknowledged for reagents. We also thank R. Fraioli and C. Full for technical assistance, G. Casale and F. Delprete for photographic work, and M. V. Sarcone and P. Franke for typing and editing the manuscript.
C.P. was the recipient of an AIRC fellowship. This work was supported by grants awarded to O.S. by AIRC, EC, and the Italy-USA Program; S.A. and P.B. were supported by AIRC and CNR (PF-Biotecnologie).
L.F. and C.P. contributed equally to this work.
O.S. thanks A.Z. for her supportive presence.
Footnotes
Dedicated to the cherished memory of Stefania and Raffaele.
REFERENCES
- 1.Alroy I, Yarden Y. The ErbB signaling network in embryogenesis and oncogenesis: signal diversification through combinatorial ligand-receptor interactions. FEBS Lett. 1997;410:83–86. doi: 10.1016/s0014-5793(97)00412-2. [DOI] [PubMed] [Google Scholar]
- 2.Anton E S, Marchionni M A, Lee K F, Rakic P. Role of GGF/neuregulin signaling in interactions between migrating neurons and radial glia in the developing cerebral cortex. Development. 1997;124:3501–3510. doi: 10.1242/dev.124.18.3501. [DOI] [PubMed] [Google Scholar]
- 3.Baulida J, Kraus M H, Alimandi M, Di Fiore P P, Carpenter G. All ErbB receptors other than the epidermal growth factor receptor are endocytosis impaired. J Biol Chem. 1996;271:5251–5257. doi: 10.1074/jbc.271.9.5251. [DOI] [PubMed] [Google Scholar]
- 4.Beerli R R, Hynes N E. Epidermal growth factor-related peptides activate distinct subsets of ErbB receptors and differ in their biological activities. J Biol Chem. 1996;271:6071–6076. doi: 10.1074/jbc.271.11.6071. [DOI] [PubMed] [Google Scholar]
- 5.Ben Levy R, Paterson H F, Marshall C J, Yarden Y. A single autophosphorylation site confers oncogenicity to the Neu/ErbB-2 receptor and enables coupling to the MAP kinase pathway. EMBO J. 1994;13:3302–3311. doi: 10.1002/j.1460-2075.1994.tb06632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourette R P, Arnaud S, Myles G M, Blanchet J P, Rohrschneider L R, Mouchiroud G. Mona, a novel hematopoietic-specific adaptor interacting with the macrophage colony-stimulating factor receptor, is implicated in monocyte/macrophage development. EMBO J. 1998;17:7273–7281. doi: 10.1093/emboj/17.24.7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Britsch S, Li L, Kirchhoff S, Theuring F, Brinkmann V, Birchmeier C, Riethmacher D. The ErbB2 and ErbB3 receptors and their ligand, neuregulin-1, are essential for development of the sympathetic nervous system. Genes Dev. 1998;12:1825–1836. doi: 10.1101/gad.12.12.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broome M A, Galisteo M L, Schlessinger J, Courtneidge S A. The proto-oncogene c-Cbl is a negative regulator of DNA synthesis initiated by both receptor and cytoplasmic tyrosine kinases. Oncogene. 1999;18:2908–2912. doi: 10.1038/sj.onc.1202873. [DOI] [PubMed] [Google Scholar]
- 9.Burke C L, Stern D F. Activation of Neu (ErbB-2) mediated by disulfide bond-induced dimerization reveals a receptor tyrosine kinase dimer interface. Mol Cell Biol. 1998;18:5371–5379. doi: 10.1128/mcb.18.9.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carpenter G. Employment of the epidermal growth factor receptor in growth factor-independent signaling pathways. J Cell Biol. 1999;146:697–702. doi: 10.1083/jcb.146.4.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casci T, Vinos J, Freeman M. Sprouty, an intracellular inhibitor of Ras signaling. Cell. 1999;96:655–665. doi: 10.1016/s0092-8674(00)80576-0. [DOI] [PubMed] [Google Scholar]
- 12.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chrapkiewicz N B, Davis C M, Chu D T, Caldwell C M, Granner D K. Rat gene 33: analysis of its structure, messenger RNA and basal promoter activity. Nucleic Acids Res. 1989;17:6651–6667. doi: 10.1093/nar/17.16.6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark S G, Stern M J, Horvitz H R. C. elegans cell-signalling gene sem-5 encodes a protein with SH2 and SH3 domains. Nature. 1992;356:340–344. doi: 10.1038/356340a0. [DOI] [PubMed] [Google Scholar]
- 15.Cleghon V, Feldmann P, Ghiglione C, Copeland T D, Perrimon N, Hughes D A, Morrison D K. Opposing actions of CSW and RasGAP modulate the strength of Torso RTK signaling in the Drosophila terminal pathway. Mol Cell. 1998;2:719–727. doi: 10.1016/s1097-2765(00)80287-7. [DOI] [PubMed] [Google Scholar]
- 16.De Sepulveda P, Okkenhaug K, Rose J L, Hawley R G, Dubreuil P, Rottapel R. Socs1 binds to multiple signalling proteins and suppresses steel factor-dependent proliferation. EMBO J. 1999;18:904–915. doi: 10.1093/emboj/18.4.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Fiore P P, Pierce J H, Kraus M H, Segatto O, King C R, Aaronson S A. erbB-2 is a potent oncogene when overexpressed in NIH/3T3 cells. Science. 1987;237:178–182. doi: 10.1126/science.2885917. [DOI] [PubMed] [Google Scholar]
- 18.Di Fiore P P, Segatto O, Lonardo F, Fazioli F, Pierce J H, Aaronson S A. The carboxy-terminal domains of erbB-2 and epidermal growth factor receptor exert different regulatory effects on intrinsic receptor tyrosine kinase function and transforming activity. Mol Cell Biol. 1990;10:2749–2756. doi: 10.1128/mcb.10.6.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Guglielmo G M, Baass P C, Ou W J, Posner B I, Bergeron J J. Compartmentalization of SHC, GRB2 and mSOS, and hyperphosphorylation of Raf-1 by EGF but not insulin in liver parenchyma. EMBO J. 1994;13:4269–4277. doi: 10.1002/j.1460-2075.1994.tb06747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dotto G P, Parada L F, Weinberg R A. Specific growth response of ras-transformed embryo fibroblasts to tumour promoters. Nature. 1985;318:472–475. doi: 10.1038/318472a0. [DOI] [PubMed] [Google Scholar]
- 21.Dougall W C, Qian X, Peterson N C, Miller M J, Samanta A, Greene M I. The neu-oncogene: signal transduction pathways, transformation mechanisms and evolving therapies. Oncogene. 1994;9:2109–2123. [PubMed] [Google Scholar]
- 22.Erickson S L, O'Shea K S, Ghaboosi N, Loverro L, Frantz G, Bauer M, Lu L H, Moore M W. ErbB3 is required for normal cerebellar and cardiac development: a comparison with ErbB2- and heregulin-deficient mice. Development. 1997;124:4999–5011. doi: 10.1242/dev.124.24.4999. [DOI] [PubMed] [Google Scholar]
- 23.Falcone G, Provenzano C, Alema S, Tato F. Transformation of NIH3T3 cells by Rous sarcoma virus occurs with high efficiency in the absence of proviral rearrangements or amplification. Oncogene. 1992;7:1913–1920. [PubMed] [Google Scholar]
- 24.Fazioli F, Kim U-H, Rhee S G, Molloy C J, Segatto O, Di Fiore P P. The erbB-2 mitogenic signaling pathway: tyrosine phosphorylation of phospholipase C-γ and GTPase-activating protein does not correlate with erbB-2 mitogenic potency. Mol Cell Biol. 1991;11:2040–2048. doi: 10.1128/mcb.11.4.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghiglione C, Carraway III K L, Amundadottir L T, Boswell R E, Perrimon N, Duffy J B. The transmembrane molecule kekkon 1 acts in a feedback loop to negatively regulate the activity of the Drosophila EGF receptor during oogenesis. Cell. 1999;96:847–856. doi: 10.1016/s0092-8674(00)80594-2. [DOI] [PubMed] [Google Scholar]
- 26.Ghiglione C, Perrimon N, Perkins L A. Quantitative variations in the level of MAPK activity control patterning of the embryonic termini in Drosophila. Dev Biol. 1999;205:181–193. doi: 10.1006/dbio.1998.9102. [DOI] [PubMed] [Google Scholar]
- 27.Glass D B, el Maghrabi M R, Pilkis S J. Synthetic peptides corresponding to the site phosphorylated in 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase as substrates of cyclic nucleotide-dependent protein kinases. J Biol Chem. 1986;261:2987–2993. [PubMed] [Google Scholar]
- 28.Gonzalez F A, Raden D L, Davis R J. Identification of substrate recognition determinants for human ERK1 and ERK2 protein kinases. J Biol Chem. 1991;266:22159–22163. [PubMed] [Google Scholar]
- 29.Grammer T C, Blenis J. Evidence for MEK-independent pathways regulating the prolonged activation of the ERK-MAP kinases. Oncogene. 1997;14:1635–1642. doi: 10.1038/sj.onc.1201000. [DOI] [PubMed] [Google Scholar]
- 30.Graus-Porta D, Beerli R R, Daly J M, Hynes N E. ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J. 1997;16:1647–1655. doi: 10.1093/emboj/16.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graus-Porta D, Beerli R R, Hynes N E. Single-chain antibody-mediated intracellular retention of ErbB-2 impairs Neu differentiation factor and epidermal growth factor signaling. Mol Cell Biol. 1995;15:1182–1191. doi: 10.1128/mcb.15.3.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grignani F, Kinsella T, Mencarelli A, Valtieri M, Riganelli D, Grignani F, Lanfrancone L, Peschle C, Nolan G P, Pelicci P G. High-efficiency gene transfer and selection of human hematopoietic progenitor cells with a hybrid EBV/retroviral vector expressing the green fluorescence protein. Cancer Res. 1998;58:14–19. [PubMed] [Google Scholar]
- 33.Hackel P O, Zwick E, Prenzel N, Ullrich A. Epidermal growth factor receptors: critical mediators of multiple receptor pathways. Curr Opin Cell Biol. 1999;11:184–189. doi: 10.1016/s0955-0674(99)80024-6. [DOI] [PubMed] [Google Scholar]
- 34.Haugh J M, Huang A C, Wiley H S, Wells A, Lauffenburger D A. Internalized epidermal growth factor receptors participate in the activation of p21(ras) in fibroblasts. J Biol Chem. 1999;274:34350–34360. doi: 10.1074/jbc.274.48.34350. [DOI] [PubMed] [Google Scholar]
- 35.Haugh J M, Schooler K, Wells A, Wiley H S, Lauffenburger D A. Effect of epidermal growth factor receptor internalization on regulation of the phospholipase C-gamma 1 signaling pathway. J Biol Chem. 1999;274:8958–8965. doi: 10.1074/jbc.274.13.8958. [DOI] [PubMed] [Google Scholar]
- 36.Jiang G, Hunter T. Receptor signaling: when dimerization is not enough. Curr Biol. 1999;9:R568–R571. doi: 10.1016/s0960-9822(99)80357-1. [DOI] [PubMed] [Google Scholar]
- 37.Joazeiro C A, Wing S S, Huang H, Leverson J D, Hunter T, Liu Y C. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 38.Karunagaran D, Tzahar E, Beerli R R, Chen X, Graus-Porta D, Ratzkin B J, Seger R, Hynes N E, Yarden Y. ErbB-2 is a common auxiliary subunit of NDF and EGF receptors: implications for breast cancer. EMBO J. 1996;15:254–264. [PMC free article] [PubMed] [Google Scholar]
- 39.Klingmuller U, Lorenz U, Cantley L C, Neel B G, Lodish H F. Specific recruitment of SH-PTP1 to the erythropoietin receptor causes inactivation of JAK2 and termination of proliferative signals. Cell. 1995;80:729–738. doi: 10.1016/0092-8674(95)90351-8. [DOI] [PubMed] [Google Scholar]
- 40.Kraus M H, Popescu N C, Amsbaugh S C, King C R. Overexpression of the EGF receptor-related proto-oncogene erbB-2 in human mammary tumor cell lines by different molecular mechanisms. EMBO J. 1987;6:605–610. doi: 10.1002/j.1460-2075.1987.tb04797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee K F, Simon H, Chen H, Bates B, Hung M C, Hauser C. Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature. 1995;378:394–398. doi: 10.1038/378394a0. [DOI] [PubMed] [Google Scholar]
- 42.Lee K L, Makkinje A, Ch'Ang L Y, Kenney F T. Molecular cloning and analysis of full-length cDNAs cognate to a rat gene under multihormonal control. Arch Biochem Biophys. 1989;269:106–113. doi: 10.1016/0003-9861(89)90091-x. . (Erratum, 276:554, 1990.) [DOI] [PubMed] [Google Scholar]
- 43.Lenferink A E, Pinkas-Kramarski R, van de Poll M L, van Vugt M J, Klapper L N, Tzahar E, Waterman H, Sela M, van Zoelen E J, Yarden Y. Differential endocytic routing of homo- and hetero-dimeric ErbB tyrosine kinases confers signaling superiority to receptor heterodimers. EMBO J. 1998;17:3385–3397. doi: 10.1093/emboj/17.12.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon W Y, Beguinot L, Geiger B, Yarden Y. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lotti L V, Di Lazzaro C, Zompetta C, Frati L, Torrisi M R. Surface distribution and internalization of erbB-2 proteins. Exp Cell Res. 1992;202:274–280. doi: 10.1016/0014-4827(92)90075-j. [DOI] [PubMed] [Google Scholar]
- 46.Manser E, Leung T, Salihuddin H, Tan L, Lim L. A non-receptor tyrosine kinase that inhibits the GTPase activity of p21cdc42. Nature. 1993;363:364–367. doi: 10.1038/363364a0. [DOI] [PubMed] [Google Scholar]
- 47.Margolis B L, Lax I, Kris R, Dombalagian M, Honegger A M, Howk R, Givol D, Ullrich A, Schlessinger J. All autophosphorylation sites of epidermal growth factor (EGF) receptor and HER2/neu are located in their carboxyl-terminal tails. Identification of a novel site in EGF receptor. J Biol Chem. 1989;264:10667–10671. [PubMed] [Google Scholar]
- 48.Marshall C J. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 49.Mayer B J, Eck M J. SH3 domains. Minding your p's and q's. Curr Biol. 1995;5:364–367. doi: 10.1016/s0960-9822(95)00073-x. [DOI] [PubMed] [Google Scholar]
- 50.Mohn K L, Laz T M, Hsu J C, Melby A E, Bravo R, Taub R. The immediate-early growth response in regenerating liver and insulin-stimulated H-35 cells: comparison with serum-stimulated 3T3 cells and identification of 41 novel immediate-early genes. Mol Cell Biol. 1991;11:381–390. doi: 10.1128/mcb.11.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mohn K L, Laz T M, Melby A E, Taub R. Immediate-early gene expression differs between regenerating liver, insulin-stimulated H-35 cells, and mitogen-stimulated Balb/c 3T3 cells. Liver-specific induction patterns of gene 33, phosphoenolpyruvate carboxykinase, and the jun, fos, and egr families. J Biol Chem. 1990;265:21914–21921. [PubMed] [Google Scholar]
- 52.Morris J K, Lin W, Hauser C, Marchuk Y, Getman D, Lee K F. Rescue of the cardiac defect in ErbB2 mutant mice reveals essential roles of ErbB2 in peripheral nervous system development. Neuron. 1999;23:273–283. doi: 10.1016/s0896-6273(00)80779-5. [DOI] [PubMed] [Google Scholar]
- 53.Muller W J, Arteaga C L, Muthuswamy S K, Siegel P M, Webster M A, Cardiff R D, Meise K S, Li F, Halter S A, Coffey R J. Synergistic interaction of the Neu proto-oncogene product and transforming growth factor alpha in the mammary epithelium of transgenic mice. Mol Cell Biol. 1996;16:5726–5736. doi: 10.1128/mcb.16.10.5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murali R, Brennan P J, Kieber-Emmons T, Greene M I. Structural analysis of p185c-neu and epidermal growth factor receptor tyrosine kinases: oligomerization of kinase domains. Proc Natl Acad Sci USA. 1996;93:6252–6257. doi: 10.1073/pnas.93.13.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nicholson S E, Willson T A, Farley A, Starr R, Zhang J G, Baca M, Alexander W S, Metcalf D, Hilton D J, Nicola N A. Mutational analyses of the SOCS proteins suggest a dual domain requirement but distinct mechanisms for inhibition of LIF and IL-6 signal transduction. EMBO J. 1999;18:375–385. doi: 10.1093/emboj/18.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olayioye M A, Graus-Porta D, Beerli R R, Rohrer J, Gay B, Hynes N E. ErbB-1 and ErbB-2 acquire distinct signaling properties dependent upon their dimerization partner. Mol Cell Biol. 1998;18:5042–5051. doi: 10.1128/mcb.18.9.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pawson T, Nash P. Protein-protein interactions define specificity in signal transduction. Genes Dev. 2000;14:1027–1047. [PubMed] [Google Scholar]
- 58.Pawson T, Scott J D. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 59.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perrimon N, McMahon A P. Negative feedback mechanisms and their roles during pattern formation. Cell. 1999;97:13–16. doi: 10.1016/s0092-8674(00)80710-2. [DOI] [PubMed] [Google Scholar]
- 61.Pierce K L, Maudsley S, Daaka Y, Luttrell L M, Lefkowitz R J. Role of endocytosis in the activation of the extracellular signal-regulated kinase cascade by sequestering and nonsequestering G protein-coupled receptors. Proc Natl Acad Sci USA. 2000;97:1489–1494. doi: 10.1073/pnas.97.4.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pinna L A. Casein kinase 2: an ‘eminence grise’ in cellular regulation? Biochim Biophys Acta. 1990;1054:267–284. doi: 10.1016/0167-4889(90)90098-x. [DOI] [PubMed] [Google Scholar]
- 63.Qiu Y, Ravi L, Kung H J. Requirement of ErbB2 for signalling by interleukin-6 in prostate carcinoma cells. Nature. 1998;393:83–85. doi: 10.1038/30012. [DOI] [PubMed] [Google Scholar]
- 64.Rechsteiner M, Rogers S W. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 65.Ricci A, Lanfrancone L, Chiari R, Belardo G, Pertica C, Natali P G, Pelicci P G, Segatto O. Analysis of protein-protein interactions involved in the activation of the Shc/Grb-2 pathway by the ErbB-2 kinase. Oncogene. 1995;11:1519–1529. [PubMed] [Google Scholar]
- 66.Riese D J, Stern D F. Specificity within the EGF family/ErbB receptor family signaling network. Bioessays. 1998;20:41–48. doi: 10.1002/(SICI)1521-1878(199801)20:1<41::AID-BIES7>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 67.Roche S, Alonso G, Kazlauskas A, Dixit V M, Courtneidge S A, Pandey A. Src-like adaptor protein (Slap) is a negative regulator of mitogenesis. Curr Biol. 1998;8:975–978. doi: 10.1016/s0960-9822(98)70400-2. [DOI] [PubMed] [Google Scholar]
- 68.Rozakis-Adcock M, Fernley R, Wade J, Pawson T, Bowtell D. The SH2 and SH3 domains of mammalian Grb2 couple the EGF receptor to the Ras activator mSos1. Nature. 1993;363:83–85. doi: 10.1038/363083a0. [DOI] [PubMed] [Google Scholar]
- 69.Santoro M, Wong W T, Aroca P, Santos E, Matoskova B, Grieco M, Fusco A, Di Fiore P P. An epidermal growth factor receptor/ret chimera generates mitogenic and transforming signals: evidence for a ret-specific signaling pathway. Mol Cell Biol. 1994;14:663–675. doi: 10.1128/mcb.14.1.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Segatto O, King C R, Pierce J H, Di Fiore P P, Aaronson S A. Different structural alterations upregulate in vitro tyrosine kinase activity and transforming potency of the erbB-2 gene. Mol Cell Biol. 1988;8:5570–5574. doi: 10.1128/mcb.8.12.5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Segatto O, Lonardo F, Pierce J H, Bottaro D P, Di Fiore P P. The role of autophosphorylation in modulation of erbB-2 transforming function. New Biol. 1990;2:187–195. [PubMed] [Google Scholar]
- 72.Segatto O, Lonardo F, Wexler D, Fazioli F, Pierce J H, Bottaro D P, White M F, Di Fiore P P. The juxtamembrane regions of the epidermal growth factor receptor and gp185erbB-2 determine the specificity of signal transduction. Mol Cell Biol. 1991;11:3191–3202. doi: 10.1128/mcb.11.6.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Segatto O, Pelicci G, Giuli S, Digiesi G, Di Fiore P P, McGlade J, Pawson T, Pelicci P G. Shc products are substrates of erbB-2 kinase. Oncogene. 1993;8:2105–2112. [PubMed] [Google Scholar]
- 74.Sudol M. From Src homology domains to other signaling modules: proposal of the ‘protein recognition code.’. Oncogene. 1998;17:1469–1474. doi: 10.1038/sj.onc.1202182. [DOI] [PubMed] [Google Scholar]
- 75.Sun H, Charles C H, Lau L F, Tonks N K. MKP-1 (3CH134), an immediate early gene product, is a dual specificity phosphatase that dephosphorylates MAP kinase in vivo. Cell. 1993;75:487–493. doi: 10.1016/0092-8674(93)90383-2. [DOI] [PubMed] [Google Scholar]
- 76.Tzahar E, Waterman H, Chen X, Levkowitz G, Karunagaran D, Lavi S, Ratzkin B J, Yarden Y. A hierarchical network of interreceptor interactions determines signal transduction by Neu differentiation factor/neuregulin and epidermal growth factor. Mol Cell Biol. 1996;16:5276–5287. doi: 10.1128/mcb.16.10.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vieira A V, Lamaze C, Schmid S L. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science. 1996;274:2086–2089. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- 78.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 79.Waterman H, Sabanai I, Geiger B, Yarden Y. Alternative intracellular routing of ErbB receptors may determine signaling potency. J Biol Chem. 1998;273:13819–13827. doi: 10.1074/jbc.273.22.13819. [DOI] [PubMed] [Google Scholar]
- 80.Wick M, Burger C, Funk M, Muller R. Identification of a novel mitogen-inducible gene (mig-6): regulation during G1 progression and differentiation. Exp Cell Res. 1995;219:527–535. doi: 10.1006/excr.1995.1261. [DOI] [PubMed] [Google Scholar]
- 81.Woodgett J R, Gould K L, Hunter T. Substrate specificity of protein kinase C. Use of synthetic peptides corresponding to physiological sites as probes for substrate recognition requirements. Eur J Biochem. 1986;161:177–184. doi: 10.1111/j.1432-1033.1986.tb10139.x. [DOI] [PubMed] [Google Scholar]
- 82.Yaffe M B, Rittinger K, Volinia S, Caron P R, Aitken A, Leffers H, Gamblin S J, Smerdon S J, Cantley L C. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 83.Yang W, Cerione R A. Cloning and characterization of a novel Cdc42-associated tyrosine kinase, ACK-2, from bovine brain. J Biol Chem. 1997;272:24819–24824. doi: 10.1074/jbc.272.40.24819. [DOI] [PubMed] [Google Scholar]