Abstract
Opposite-sex social relationships are important predictors of fitness in many animals, including several group-living mammals. Consequently, understanding sources of variance in the tendency to form opposite-sex relationships is important for understanding social evolution. Genetic contributions are of particular interest due to their importance in long-term evolutionary change, but little is known about genetic effects on male–female relationships in social mammals, especially outside of the mating context. Here, we investigate the effects of genetic ancestry on male–female affiliative behaviour in a hybrid zone between the yellow baboon, Papio cynocephalus, and the anubis baboon, Papio anubis, in a population in which male–female social bonds are known predictors of life span. We place our analysis within the context of other social and demographic predictors of affiliative behaviour in baboons. Genetic ancestry was the most consistent predictor of opposite-sex affiliative behaviour we observed, with the exception of strong effects of dominance rank. Our results show that increased anubis genetic ancestry is associated with a subtle, but significantly higher, probability of opposite-sex affiliative behaviour, in both males and females. Additionally, pairs of anubis-like males and anubis-like females were the most likely to socially affiliate, resulting in moderate assortativity in grooming and proximity behaviour as a function of genetic ancestry. Our findings indicate that opposite-sex affiliative behaviour partially diverged during baboon evolution to differentiate yellow and anubis baboons, despite overall similarities in their social structures and mating systems. Furthermore, they suggest that affiliative behaviour may simultaneously promote and constrain baboon admixture, through additive and assortative effects of ancestry, respectively.
Keywords: baboon, genetic ancestry, grooming, hybrid zone, opposite-sex social bond, Papio anubis, Papio cynocephalus
Social relationships, both within and between sexes, are ubiquitous features in the lives of social mammals. Affiliative interactions among members of the same sex are positively associated with fertility or survival in a number of social mammal species, including group-living primates, equids, cetaceans and rodents (e.g. Cameron et al., 2009; Ellis et al., 2019; Frère, Krützen, Mann, Connor et al. 2010; Schülke et al., 2010; Silk et al., 2009; Silk et al., 2010; Weidt et al., 2008). Opposite-sex affiliative bonds can also have important consequences. In monogamous species, strong social bonds between sexual partners predict shorter interbirth intervals, increased offspring number and improved offspring survival, potentially due to improved coordination between partners in caring for young, obtaining resources or defence against predators (e.g. Black, 2001; Griggio & Hoi, 2011; Ribble, 1992; Sánchez-Macouzet et al., 2014). Furthermore, in some group-living primates, females compete for access to males outside of mating contexts, suggesting that social bonds with males are themselves an important resource (Archie et al., 2014; Baniel et al, 2016, 2018; Cheney et al., 2012; Haunhorst et al., 2019; Lemasson et al., 2008; Palombit et al., 2001; Seyfarth, 1978). In support of this idea, females in several cercopithecine monkey species benefit from opposite-sex social bonds via enhanced survival, care for their offspring and protection from harassment (Archie et al., 2014; Baniel et al., 2016; Haunhorst et al., 2017; Kulik et al., 2012; Lemasson et al., 2008; Moscovice et al., 2009; Nguyen et al., 2009; Palombit et al., 1997; Seyfarth, 1978; Silk et al., 2020; Weingrill, 2000). Males of these species may also benefit from social bonds with females. For example, baboon males who form strong social bonds with females tend to live longer than those who do not (Campos et al., 2020). Males may also benefit by gaining mating opportunities (although the evidence for this benefit is mixed), opportunities to care for their offspring or access to infants that can be exploited for social gain (Ménard et al., 2001; Packer, 1979b; Smuts, 1985; van Schaik & Paul, 1996; Whitten, 1987).
While a number of studies have investigated the sources of variance in same-sex affiliative relationships in group-living mammals (Best et al., 2014; Frère, Krützen, Mann, Watson-Capps, et al., 2010; Langergraber et al., 2009; Mitani, 2009; Möller et al., 2001; Seyfarth, 1976; Seyfarth et al., 2014; Silk, Alberts, et al., 2006, Silk, Altmann, et al., 2006; Smith et al., 2006; Widdig et al., 2001), we know comparably less about the sources of variance in opposite-sex relationships, especially outside the mating context. Addressing this gap is important for understanding the evolution of heterosexual bonds. In particular, if the tendency to form opposite-sex social bonds is affected by genotype, then it has the potential to evolve in response to natural selection. Strong evidence for genetic effects comes from interspecific comparisons between pair-bonded and multiply mating species. For example, comparisons between the monogamous prairie vole, Microtus ochrogaster, and other, closely related promiscuous voles have identified genetic divergence in the pathways that regulate arginine vasopressin, oxytocin and dopamine signalling, which in turn influences pair-bonding behaviour (Young et al., 1996; Young et al., 1999; Young, Waymire, et al., 1997; Young, Winslow, et al., 1997; reviewed in: Carter & Perkeybile, 2018; Johnson & Young, 2015; Sadino & Donaldson, 2018; Young et al., 2011). These pathway differences may in part be due to differences in the distribution and densities of hormone receptors in the brain, suggesting one important mechanism through which variation in opposite-sex social relationships evolves (Insel et al., 1994; Insel & Shapiro, 1992; Smeltzer et al., 2006). Research in other pair-bonded rodents, primates, fish, frogs and birds has placed these findings in a broader context, indicating that these and other pathways (e.g. Young et al., 2019) consistently influence pair bonding across divergent species, although they may do so in a species-specific manner (reviewed in: Carter & Perkeybile, 2018; Fischer, Nowicki, et al., 2019; Johnson & Young, 2015).
Despite these important discoveries in pair-bonded species, little is known about genetic influences on opposite-sex social bonding in group-living animals, including the degree to which genotype contributes to differences between species with similar social and mating systems. Here, we investigate the association between genetic ancestry and male–female affiliative behaviour in a well-studied natural primate population, the baboons of Kenya’s Amboseli basin (Alberts, 2018; Alberts & Altmann, 2012). Baboons (genus Papio) began speciating ~1.4 million years ago, and today, the six extant species occupy distinct geographical ranges across Africa (Rogers et al., 2019). Most species of baboons, including those in Amboseli, live in multimale, multifemale social groups in which multiple individuals of both sexes mate and form social bonds (Fischer. Higham, et al., 2019). Amboseli lies in a hybrid zone between two such species, the yellow baboon, Papio cynocephalus, and the anubis baboon, Papio anubis (also known as the olive baboon) (Alberts & Altmann, 2001; Samuels & Altmann, 1986; Tung et al., 2008; Wall et al., 2016). While yellow baboons contribute the majority of genetic ancestry in this population, the range of admixture we observe – a continuous distribution from animals that are almost entirely yellow to those that are almost entirely anubis – gives us the opportunity to examine potential genetic ancestry effects on opposite-sex affiliative relationships. Complementary data on social and demographic variables for the same individuals allow us to place these effects in the context of other, environmental sources of variance.
Genetic Ancestry Effects on Male–Female Interactions in Hybrid Zones
The Amboseli baboon hybrid zone provides a ‘natural laboratory’ for understanding the relationship between genetic ancestry and affiliative behaviour because it allows individuals with varyingly admixed genomes to be observed in a shared environment (Hewitt, 1988). In turn, studying social behaviour in hybrid zones can shed light onto hybrid zone dynamics, as most clearly illustrated in cases where ancestry influences mating behaviour. In such cases, assortative mating by ancestry limits gene flow and can reinforce species boundaries, whereas ancestry-related mating advantages can lead to asymmetric gene flow and range expansion (e.g. Baldassarre et al., 2014; Baldassarre & Webster, 2013; Kronforst et al., 2006; Mavárez et al., 2006).
Ancestry effects on mating behaviour have also been detected in both the yellow baboon–anubis baboon hybrid zone in Amboseli and in an anubis baboon–hamadryas baboon hybrid zone (Papio anubis × Papio hamadryas) in Ethiopia. In Amboseli, anubis-like males are more likely to obtain consortships (extended mate-guarding associations between an adult male and an adult female in oestrus, during which most conceptions occur), and male–female pairs with similar genetic ancestry are more likely to consort than pairs with different ancestry (Tung et al., 2012). In the Ethiopian hybrid zone, ancestry affects both male mating strategy and how females respond to males, and nonoestrous females assortatively groom phenotypically similar males in at least some demographic contexts (Bergman & Beehner, 2003). However, significant gaps remain in our understanding of how genetic ancestry affects male–female affiliation outside of the mating context (but see Bergman et al. 2008 for an analysis of male interest in nonoestrous females). If such effects occur, genetic ancestry effects on male–female social relationships may be more important than indicated by analyses of mating behaviour alone. Specifically, because opposite-sex social affiliation also predicts life span in the Amboseli population (Archie et al., 2014; Campos et al., 2020), ancestry effects on this trait may secondarily affect how long individuals live and who they co-reside with, thus influencing the genetic composition of subsequent generations.
Goals of This Study
Here, we evaluated the extent to which genetic ancestry predicts the formation of male–female social relationships in baboons. We focused specifically on male–female affiliative behaviour in nonmating contexts (i.e. periods when females were pregnant or lactating and not sexually cycling) because social relationships in these contexts are not driven by immediate sexual interactions. Using two multivariate models (one for grooming behaviour and one for proximity behaviour), we simultaneously tested for (1) the additive effects of male and female individual characteristics, including genetic ancestry, on the probability of affiliative social behaviour between males and females, and (2) characteristics defined by the pair, including ancestry-related assortativity. In the same model, we also tested two additional hypotheses: (3) that opposite-sex affiliation depends on female reproductive state (i.e. pregnancy or lactation), based on evidence that the stability of male–female relationships varies across baboon species as a function of female reproductive state (Baniel et al., 2016; Fischer et al., 2017; Goffe et al., 2016; Nguyen et al., 2009; Städele et al., 2019; Weingrill, 2000); and (4) that opposite-sex affiliation depends on group demography, based on findings that male–female interactions in baboons and other primates also depend on group composition (Archie et al., 2014; Bergman & Beehner, 2003; Rosenbaum et al., 2016; Tung et al., 2012).
METHODS
Study Subjects
Study subjects were adult baboons from an intensively studied wild population inhabiting the Amboseli ecosystem of southern Kenya (Alberts, 2018; Alberts & Altmann, 2012). This population consists of multigeneration hybrids (Tung et al., 2008). Estimates from whole genome resequencing data indicate that while most baboons in Amboseli are predominantly yellow, admixture is pervasive (Vilgalys et al., 2021; Wall et al., 2016). Phenotypically, study subjects represent the full continuum, from animals that appear fully yellow to animals that are markedly anubis-like, with multiple representatives at all points along the continuum (Alberts & Altmann, 2001; Tung et al., 2008). This natural hybrid population is situated within a narrow hybrid zone that likely extends along the geographical boundary between yellow baboon and anubis baboon distributions in East Africa (Charpentier et al., 2012).
Members of the Amboseli baboon study population are individually recognized based on physical appearance and are monitored on a near-daily basis by trained observers who record demographic data (e.g. group membership, births, deaths, immigration, emigration) and behavioural data (e.g. social interactions, mating, travelling, resting, feeding). Importantly, the continuum of hybrid phenotypes has been present in Amboseli for many decades (Samuels & Altmann,1986; Tung et al., 2008), meaning that anubis-like animals are not rare and/or unusually distinct to observers; our observational protocols, described below, also guard against nonrandom behavioural sampling. Study subjects were parous adult females (because parous females are strongly preferred over nulliparous females as mates by adult males of most primate species: Anderson, 1986; Gesquiere et al., 2007) and adult males that had achieved a social dominance rank among other adult males in their group (Appendix, Table A1). Overall, we considered members of 12 social groups that were studied between November 1999 and December 2015. We restricted the data set to include only males and females for whom estimates of genetic ancestry, genetic diversity and genetic relatedness between individuals in male–female pairs could be calculated from previously generated microsatellite data (Buchan et al., 2003; Tung et al., 2008, 2012). The resulting sample contained 136 females and 160 males, who together formed 3468 unique male–female dyads across the grooming and proximity data sets. These study subjects make up the vast majority of adult males (87%) and adult females (97%) in our population that otherwise met our inclusion criteria for this study (see Affiliative Social Behaviour below). Note that because we attempt to genotype every individual in our study groups, our genotyping protocol is unbiased with respect to ancestry.
Affiliative Social Behaviour
Grooming and maintenance of close spatial proximity (hereafter, proximity) are affiliative behaviours important to establishing, maintaining and strengthening social bonds in nonhuman primates (Cords, 1997, 2012; Palombit et al., 1997; Silk et al., 2013). Although male–female grooming and proximity events were moderately correlated in our data set (Pearson’s correlation: r26,243 = 0.222, P < 10−15), we analysed grooming and proximity separately because grooming measures explained a small proportion of the variance in the proximity data (i.e. r2 = 0.049). Almost all of the grooming data (99.8%) were collected during systematic monitoring of the population, following a sampling protocol in which observers move in a predetermined random order throughout the group. We refer to this data collection approach as ‘representative interaction sampling’. In this sampling approach, observers record all grooming interactions in their line of sight, while simultaneously conducting random-order focal animal sampling on adult females. Therefore, observers rarely remain in the same location in the group for more than 10 min (the duration of each sample), which avoids biases due to uneven sampling of subjects. Additionally, members of baboon social groups in the Amboseli population are frequently spread out across a relatively large area, so there is no single central position in the group (Alberts et al., 2020; Archie et al., 2014). A potential concern with representative interaction sampling is that, because each observation day involves the same number of observers, regardless of group size, fewer grooming events per individual will inevitably be observed in larger groups. We control for this bias using a measure of observer effort (see Observer effort below). Additional grooming data, and all proximity data, were collected during random-order focal animal sampling on adult females, during which the identity of the nearest adult male within 5 m and grooming activity, if any, were recorded once per minute for the duration of each 10 min sample (Alberts et al., 2020).
For each co-resident opposite-sex dyad, we determined whether they were observed grooming or in proximity at least once during a given 2-month interval (see also Appendix, Additional Methods). We chose 2-month intervals as our unit for analysis to maximize comparability with an earlier analysis of ancestry effects on mating behaviour (Tung et al., 2012) and to facilitate unambiguous assignment of reproductive state to females (during longer intervals, females often switch between reproductive states, and in 2-month periods they do so as well but less frequently). We note that our 0/1 measure of grooming and proximity thus captures the probability that male–female affiliative behaviour occurs, and not the strength of a dyad’s social bond. Specifically, even though bonds are formed via affiliative behaviour, accurate assessment of bond strength in this system generally requires ~1 year of observational data (Silk, Altmann, et al., 2006), a period that frequently overlaps multiple female reproductive states. We collected a mean ± SD of 63.3 ± 39.7 min (range 0–255) and 66.2 ± 37.5 min (range 8–255) of focal observation data per female per 2-month interval, for the grooming and proximity data sets, respectively.
We excluded data from periods in which behavioural monitoring was inconsistent or when social groups were too unstable (i.e. social groups were fissioning or fusing) to unambiguously determine an individual’s group membership. We also excluded all data from the 2009 hydrological year (1 November 2008–31 October 2009), which included the most severe drought documented in the Amboseli basin in more than 40 years (Okello et al., 2016; Tuqa et al., 2014). Omitting data from 2009 ensured that effects from this rare and extreme event, which altered patterns of female fertility and reproductive states, did not influence our results (Fitzpatrick et al., 2014; Lea et al., 2015).
Predictor Variables
We investigated the relationship between genetic ancestry and opposite-sex affiliative social behaviour using the following predictors, motivated in part by known predictors of mating behavior in this population (Tung et al., 2012) (see Appendix, Table A2–A3 for correlations among all predictor variables).
Genetic ancestry
Genetic estimates of hybridity (i.e. the proportion of each individual’s genome estimated to be from anubis ancestry) were included for females (hf) and males (hm). These estimates were based on genotypes at up to 13 highly polymorphic microsatellite markers and average ancestry assignments produced using the Bayesian clustering algorithm STRUCTURE 2.3.4 (Falush et al., 2003; Pritchard et al., 2000; see Tung et al., 2008; mean ± SD typed loci per individual = 12.40 ± 1.10). These assignments range continuously from 0 to 1, where 0 corresponds to unadmixed yellow baboon ancestry and 1 corresponds to unadmixed anubis baboon ancestry. These estimates are strongly correlated with recent genome-wide ancestry estimates (Pearson’s correlation: r21 =0.717, N = 23 individuals that overlapped between data sets, P = 1.17 × 10−4) (Wall et al., 2016); however, because genome-wide estimates are available for only a subset of the population, we used the microsatellite-based estimates here. Individuals in our study vary in their genetic ancestry from 0.03 to 0.92 (mean ± SD: 0.31 ± 0.28; see Appendix, Table A1 for genetic ancestry summary statistics, stratified by social group).
Assortative genetic ancestry index
To test the possibility that males and females of similar genetic ancestry are more likely to socially affiliate, we calculated a pairwise assortative genetic ancestry index, b, as a function of the genetic ancestry estimates of the female and male (hf and hm, respectively), paralleling the approach used in Tung et al.’s (2012) pairwise assortative mating index, a:
This index ranges from 0 to 1: high values indicate highly assortative male–female pairs (i.e. individuals in the pair both have low or high genetic ancestry estimates) and low values indicate highly disassortative male–female pairs (i.e. individuals in the pair have different genetic ancestry estimates). Intermediate values indicate male–female pairs where both individuals are of intermediate ancestry.
Heterozygosity
High genetic diversity is sometimes thought to be a measure of genetic quality (Kempenaers, 2007). Because it is relevant to mate choice (Kempenaers, 2007) and potentially social partner choice, we therefore included a measure of genetic diversity for both males and females using up to 14 highly polymorphic microsatellite markers (mean ± SD typed loci per individual = 13.13 ± 1.22; 13 of these markers were also used to assign genetic ancestry scores). We estimated individual genetic diversity by dividing the number of heterozygous loci by the number of genotyped loci for each individual (following Charpentier et al., 2008). Importantly, there is no overall effect of species identity (i.e. yellow or anubis) on genetic diversity using these markers (Charpentier et al., 2012). The correlation between genetic diversity and genetic ancestry is moderate within each sex (Pearsons correlation for both grooming and proximity: r = 0.31, P < 10−4 for males; r < 0.26, P < 0.01 for females).
Relatedness
Because social affiliation may be affected by kinship, we included an estimate of genetic relatedness for each male–female dyad using the method of Queller and Goodnight (1989). These estimates, based on the same genotype data used to estimate heterozygosity, were calculated using the function ‘coancestry’ with the estimator ‘quellergt’ in the R package ‘related’ (v.1.0; Pew et al., 2015; Wang, 2011).
Social dominance rank
Social dominance rank can enhance access to valuable resources, including desirable social partners (e.g. Archie et al., 2014; Baniel et al., 2016; Haunhorst et al., 2019; Lemasson et al., 2008; Palombit et al., 2001). We therefore modelled female rank, male rank and the interaction between female and male ranks as additional fixed effects in the models. Female and male ranks were assigned separately for each sex, on a monthly basis, based on the outcomes of dyadic agonisms between all pairs of individuals in the same group (Alberts et al., 2020; Alberts & Gordon, 2018). We represented rank using an ordinal approach, where the highest-ranking individual holds rank 1 and lower-ranking individuals occupy ranks of successively higher numbers. Since female and male ranks were assigned on a monthly basis and our time window for analyses of grooming and proximity interactions spanned a 2-month period, we used the average of each individual’s rank across both months for each 2-month interval. Note that although higher-ranking males and anubis-like males are more likely to gain consortships in this population (Tung et al., 2012), genetic ancestry is not correlated with dominance rank (P > 0.35 for both grooming and proximity data sets; see also Franz et al., 2015).
Age
Female age may also affect a female’s social interactions. To account for possible age-related effects, we modelled a linear effect of female age, averaged across each 2-month analysis window (i.e. her age at the start of the second month), as a continuous predictor variable in our models. We also included a transformed measure of female age that reflects the relationship between female age and conception probability in this population, where the highest conception probability occurs at ~14 years of age (Beehner et al., 2006). Following Tung et al. (2012), we calculated female-transformed age, at, as a function of au, the untransformed female age:
This transformation assigns 0 for the value of at at 14 years, the age at which conception probabilities are highest; values of at become increasingly negative with distance from age 14. For 90.4% (123 out of 136) of the females in the data set, birth dates were known to within a few days. For the other females in the data set, birth dates were estimated to within 6 months (i.e. ±3 months’ error). Male age was not included in any models since it is tightly correlated with rank in male baboons (Alberts et al., 2003) and its effect on mating and social behaviour is likely to be linked to rank (Silk et al., 2020; Tung et al., 2012).
Group composition
To incorporate group level demographic effects on social behaviour, we included the number of adult females and the number of adult males in the social group of a male–female pair in both models (averaged across each 2-month analysis period).
Reproductive state
Because female reproductive state affects the stability of male–female bonds in other baboon species (Baniel et al., 2016; Weingrill, 2000), we also included female reproductive state as a categorical variable in our models. To capture opposite-sex affiliation outside the context of mating, we excluded all data points in which the female member of a potential pair was cycling. Thus, reproductive state was either pregnant or lactating, both of which meant that the female was not actively mating and could not conceive. Pregnancy and lactation were coded as −1 and 1, respectively, which avoided numerical instability that occurred if we used a 0/1 encoding (see Appendix, Additional Methods). To test whether the effects of female reproductive state on male–female social affiliation depended on genetic ancestry, we also modelled an interaction between female reproductive state and female genetic ancestry and a separate interaction between female reproductive state and male genetic ancestry.
Pair co-residency
The number of days that a male and female were observed in the same social group may influence both their tendency to affiliate and our ability to detect interactions between them. We therefore included the total number of days in each 2-month interval that a male and female were censused in the same group as a model covariate.
Observer effort
The number of field observers and the amount of time spent conducting behavioural observations for each study group was consistent across all study groups regardless of their size (Appendix, Fig. A1). Consequently, the probability of observing grooming or proximity events could vary as a function of social group size, because an observer watching a small group is likely to capture a larger fraction of interactions in a given period than that same observer watching a much larger group (note that had we measured the ‘proportion of time’ a dyad was in proximity – conditional on the female being observed – instead of as a binary event, observer effort would not be expected to affect the outcome). Thus, we calculated observer effort and included it as a covariate in both models. Observer effort was estimated as the average number of minutes of focal sample data collected per adult female per social group in a given 2-month interval (see Appendix, Additional Methods).
Statistical Analyses
Grooming and proximity behaviour were modelled as binary events and analysed separately using binomial mixed effects models. Each row of data corresponded to a unique, co-resident female–male dyad in a given 2-month interval and was assigned a value of ‘1’ if the dyad was observed grooming or in proximity at least once during the 2-month interval and a ‘0’ if they were not. We used 2 months as our time interval because our resolution for grooming and proximity behaviour is relatively coarse on a month-to-month basis, even after excluding months in which observer effort was low (see Appendix, Additional Methods).
We retained all 2-month intervals in which focal females groomed or were in proximity with any candidate male social partner at least once, except for 2-month intervals in which females transitioned between reproductive states and 2-month intervals in which the average number of adult males in the social group was less than two. We also excluded any male in a female’s 2-month interval if he was only present for 1 of the 2 months and excluded all data for females and males who were observed for less than 8 months because sparse data makes it difficult to estimate individual level random effects. The final grooming data set included 127 unique females and 160 unique males across 1866 female 2-month intervals (17 356 female–male pair–interval combinations), and the final proximity data set included 131 unique females and 160 unique males across 2338 female 2-month intervals (21 130 total female–male pair–interval combinations).
We ran binomial mixed effects models using the function glmmTMB (family = ‘binomial’) in the R package glmmTMB (v.1.0.0; Brooks et al., 2017), using a logit link:
where yij is a 0/1 value indicating whether male–female dyad i was observed grooming or in proximity during a 2-month interval j. yij is drawn from a binomial distribution, where the probability of grooming or proximity (pij) is modelled as the function of the logit-transformed sum of (1) the intercept, β0; (2) the fixed effects (Xij β) of male genetic ancestry, female genetic ancestry, the assortative genetic ancestry index for that pair, male heterozygosity, female heterozygosity, genetic relatedness between individuals in that pair, male dominance rank, female dominance rank, female age, transformed female age, the number of adult females in the social group, the number of adult males in the social group, female reproductive state (pregnant or lactating), the interaction between female reproductive state and female genetic ancestry, the interaction between female reproductive state and male genetic ancestry, the interaction between male and female dominance ranks, pair co-residency and observer effort (Xij represents all of these data using standard matrix notation and β refers to the vector of all fixed effect estimates); and (3) the random effects of male identity, mi, and female identity, fi. εij represents model error.
To assess statistical significance, we used permutation tests to account for unequal representation of individuals in the data set and predictors that did not follow standard parametric distributions. We followed the procedure of Tung et al. (2012), who conducted a similar analysis on mating behaviour. Specifically, we first computed, for each female–interval combination, the proportion of dyads where an event (grooming or proximity) occurred. These values are estimates of the probability of grooming or proximity with any male, per female–interval combination. These probabilities were then permuted across all female 2-month intervals, and randomized response variables (0/1) were generated by drawing from a binomial distribution with pij equal to the permuted grooming or proximity probability for each female–interval. This approach preserves the structure of the predictor variables (including correlations between predictors), the number of times each individual is represented in the data set and the distribution of grooming or proximity events for each female–interval. We then fitted the model used to analyse the real data to the permuted data set and calculated a P value for each predictor variable based on the number of times that the absolute value of the effect size estimated from the permuted data sets was greater than the absolute value of the effect size estimated from the observed data set, across 1000 permutations. All analyses were run in R (v.3.6.1; R Core Team, 2019).
Ethical Note
The research in this study was approved by the Institutional Animal Care and Use Committee (IACUC) at Duke University (no. A273–17-12) and adhered to the laws and guidelines of the Kenyan government. The majority of our data collection was entirely noninvasive. The animals are completely habituated to the presence of observers and show no signs of stress associated with observation. Observers further reduce animal stress by maintaining at least a 5 m distance from habituated study subjects during observational sampling, and faecal samples for genotyping were not collected until all animals moved away from where the sample was deposited. The animals experience no stress during faecal sample collection. For the minority of samples for which microsatellite genotypes were obtained from blood, samples were collected using a minimally invasive protocol on animals that had been briefly anaesthetized (Lea et al., 2018; Sapolsky & Altmann, 1991; Tung et al., 2011). Stress is reduced by darting only when animals are not looking towards the observer, by keeping the period of anaesthetization brief (usually less than 1 h), by ensuring that the animals are kept in a quiet, undisturbed area in the shade during recovery and by returning them to their social groups as quickly as possible (usually within 4 h of anaesthetization). The darting protocol is overseen by a veterinarian and includes animal welfare oversight. In all cases, study subjects returned quickly to their social group and showed no signs of adverse reactions to the sampling.
RESULTS
Individual Characteristics: Genetic Ancestry and Dominance Rank Predict Opposite-sex Affiliative Social Behaviour in Males and Females
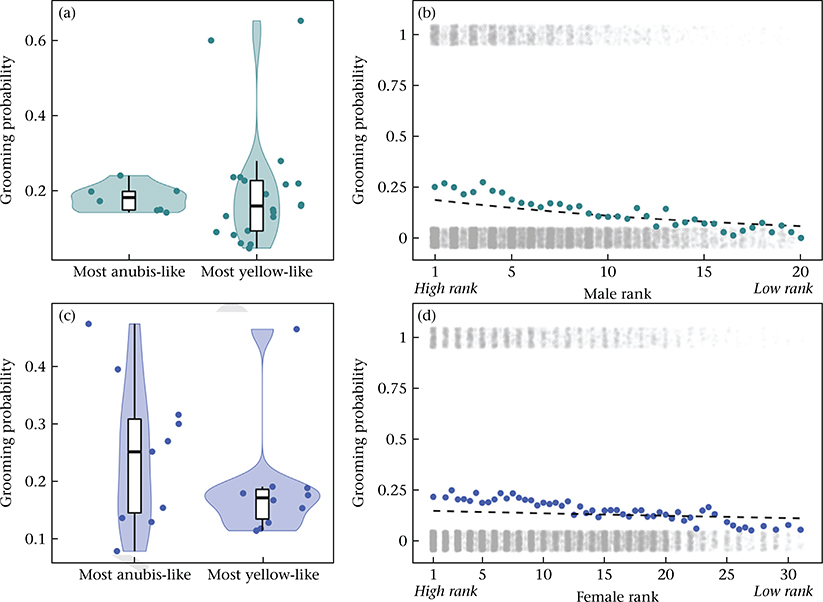
Our models identified two male characteristics that consistently predicted opposite-sex grooming and proximity behaviour (Tables 1–2). Specifically, grooming and proximity were more likely to occur if the male in the dyad had more anubis ancestry (grooming: β = 0.429, P < 0.001; Table 1, Fig. 1a; proximity: β = 0.270, P < 0.001; Table 2, Appendix, Fig. A2a) and was higher ranking (grooming: β = −0.096, P < 0.001; Table 1, Fig. 1b; proximity: β = −0.047, P < 0.001; Table 2, Appendix, Fig. A2b). Male heterozygosity was not significantly associated with either grooming or proximity behaviour.
Table 1.
Results from a multivariate logistic regression model predicting grooming behaviour
| Predictor variablea | Effect estimate | P b | Effect directionc | |
|---|---|---|---|---|
|
| ||||
| Intercept | −1.474 | 0.339 | – | |
| Genetic effects | Female genetic ancestry | 0.513 | <0.001 | More anubis ancestry in females → ↑ Pr(groom) |
| Male genetic ancestry | 0.429 | <0.001 | More anubis ancestry in males → ↑ Pr(groom) | |
| Assortative genetic ancestry index (b) | 0.646 | <0.001 | Females and males of similar genetic ancestry → ↑ Pr(groom) | |
| Female heterozygosity | −0.249 | 0.406 | – | |
| Male heterozygosity | 0.228 | 0.189 | – | |
| Genetic relatedness | −0.090 | 0.489 | – | |
| Rank effects | Female ordinal rank | −0.027 | <0.001 | Higher ranking females → ↑ Pr(groom) |
| Male ordinal rank | −0.096 | <0.001 | Higher ranking males → ↑ Pr(groom) | |
| Female ordinal rank × male ordinal rank | 0.003 | <0.001 | Females and males of similar rank → ↑ Pr(groom) | |
| Age effects | Female age (au) | −0.007 | 0.332 | – |
| Female age transformed (at) | −0.192 | 0.562 | – | |
| Demographic effects | Adult females in group | −0.039 | <0.001 | More adult females in group → ↑ Pr(groom) |
| Adult males in group | −0.030 | 0.018 | More adult males in group → ↑ Pr(groom) | |
| Reproductive state effects | Reproductive state (pregnant = −1, lactating = +1) | −0.067 | 0.165 | – |
| Reproductive state (as above) × female genetic ancestry | 0.086 | 0.412 | – | |
| Reproductive state (as above) × male genetic ancestry | 0.004 | 0.949 | – | |
| Co-residency effects | Pair co-residency | 0.047 | <0.001 | Longer co-residency → ↑ Pr(groom) |
| Observer effects | Observer effort | 0.002 | 0.425 | – |
All variables included in this table were fitted as fixed effects in the multivariate logistic regression model. Male and female identity were fitted as random effects.
Predictor variables for which P < 0.01 are bolded and P < 0.05 are italicized.
Pr(groom) = probability of observing grooming between a given male–female dyad at least once during a 2-month interval.
Table 2.
Results from a multivariate logistic regression model predicting proximity behaviour
| Predictor variablea | Effect estimate | P b | Effect directionc | |
|---|---|---|---|---|
|
| ||||
| Intercept | −1.584 | 0.008 | – | |
| Genetic effects | Female genetic ancestry | 0.270 | 0.022 | More anubis ancestry in females → ↑ Pr(prox) |
| Male genetic ancestry | 0.270 | <0.001 | More anubis ancestry in males → ↑ Pr(prox) | |
| Assortative genetic ancestry index (b) | 0.303 | <0.001 | Females and males of similar genetic ancestry → ↑ Pr(prox) | |
| Female heterozygosity | −0.120 | 0.654 | – | |
| Male heterozygosity | 0.042 | 0.745 | – | |
| Genetic relatedness | −0.041 | 0.693 | – | |
| Rank effects | Female ordinal rank | −0.024 | <0.001 | Higher ranking females → ↑ Pr(prox) |
| Male ordinal rank | −0.047 | <0.001 | Higher ranking males → ↑ Pr(prox) | |
| Female ordinal rank × male ordinal rank | 0.001 | 0.011 | Females and males of similar rank → ↑ Pr(prox) | |
| Age effects | Female age (au) | −0.004 | 0.644 | – |
| Female age transformed (at) | −0.028 | 0.916 | – | |
| Demographic effects | Adult females in group | −0.018 | 0.073 | – |
| Adult males in group | −0.036 | 0.005 | More adult males in group → ↑ Pr(prox) | |
| Reproductive state effects | Reproductive state (pregnant = −1, lactating = +1) | 0.056 | 0.187 | – |
| Reproductive state (as above) × female genetic ancestry | 0.021 | 0.851 | – | |
| Reproductive state (as above) × male genetic ancestry | −0.059 | 0.269 | – | |
| Co-residency effects | Pair co-residency | 0.037 | <0.001 | Longer co-residency → ↑ Pr(prox) |
| Observer effects | Observer effort | 0.027 | <0.001 | Greater observer effort → ↑ Pr(prox) |
All variables included in this table were fitted as fixed effects in the multivariate logistic regression model. Male and female identity were fitted as random effects.
Predictor variables for which P < 0.01 are bolded and P < 0.05 are italicized.
Pr(prox) = probability of observing a given male–female dyad in proximity at least once during a 2-month interval.
Figure 1.
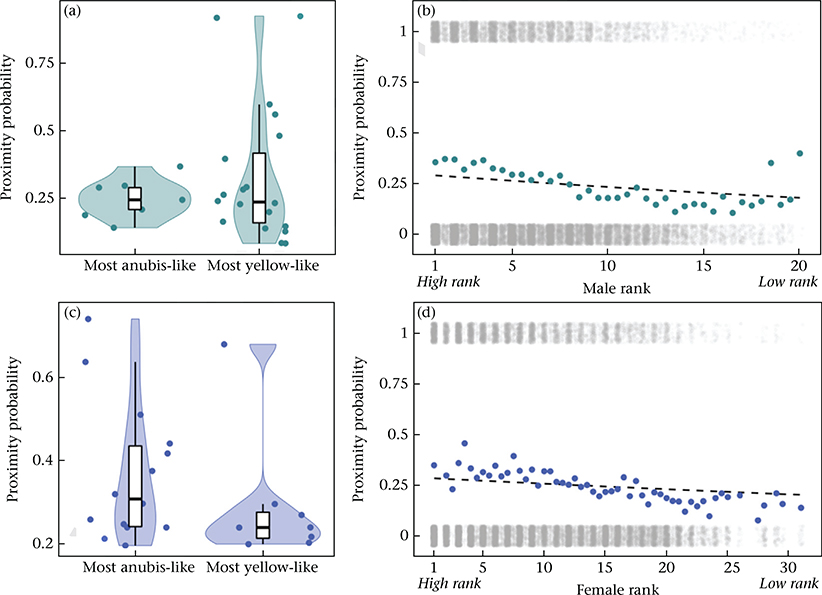
The influence of genetic ancestry and dominance rank on the tendency to groom with an opposite-sex partner. (a) The probability of grooming among co-resident opposite-sex pairs, per 2-month interval, for the most anubis-like males (above the 90th percentile for male genetic ancestry in the data set, >83.6% anubis ancestry, N = 8 males) and the most yellow-like males (below the 10th percentile for male genetic ancestry in the data set, <4.8% anubis ancestry, N = 21 males). Probabilities are shown here based on the data without adjustment for other covariates. (b) The probability of grooming among co-resident opposite-sex pairs, per 2-month interval, as a function of male dominance rank. Coloured dots show probabilities based on counts of grooming occurrences, without adjustment for other covariates (as in (a)), and the dashed line shows the predicted relationship based on model estimates, assuming average values for all other covariates (see Appendix, Additional Methods). Grey dots show the presence (y = 1) or absence (y = 0) of grooming behaviour for all 17 356 female–male pair–interval combinations, as a function of male dominance rank (dots are jittered vertically for visibility). Noninteger values correspond to individuals that changed ranks during a 2-month interval in the data set. (c) As in (a), for the most anubis-like females (above the 90th percentile for female genetic ancestry in the data set, >76.0% anubis ancestry, N = 11 females) and the most yellow-like females (below the 10th percentile for female genetic ancestry in the data set, <3.5% anubis ancestry, N = 10 females). (d) As in (b), with the probability of grooming shown as a function of female dominance rank.
Similar patterns were observed for females, although the effect of female rank was weaker than that of male rank. Grooming and proximity were more likely to occur if the female in a dyad had more anubis ancestry (grooming: β = 0.513, P < 0.001; Table 1, Fig. 1c; proximity: β = 0.270, P = 0.022; Table 2, Appendix, Fig. A2c) and was higher ranking (grooming: β = −0.027, P < 0.001; Table 1, Fig. 1d; proximity: β = −0.024, P < 0.001; Table 2, Fig. A2d). Female reproductive state, female age, transformed female age and female heterozygosity did not significantly affect grooming or proximity behaviour, nor did the interaction between female reproductive state and female genetic ancestry.
Dyad Level Characteristics: Traits of Both Partners Predict the Propensity to Affiliate with the Opposite Sex
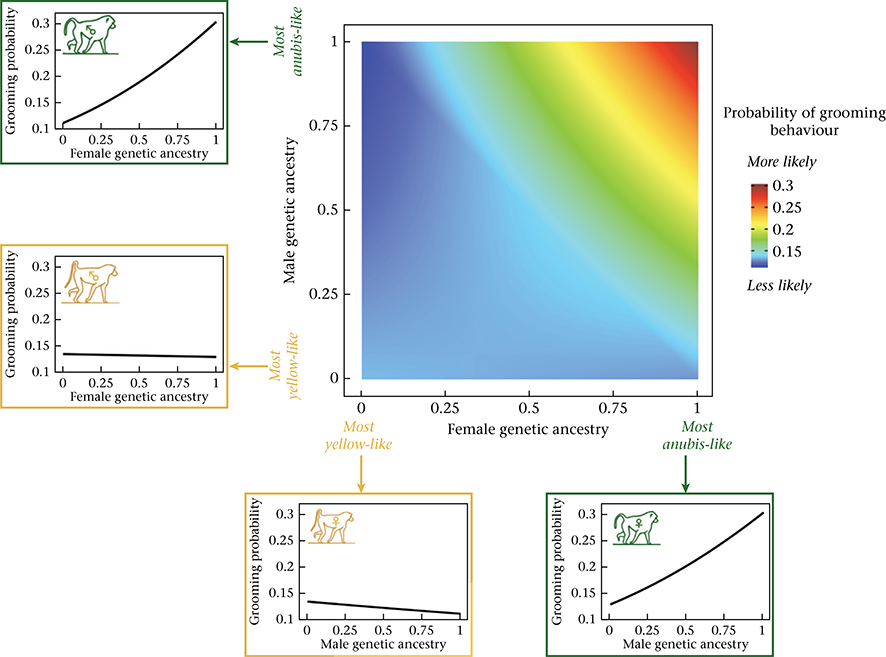
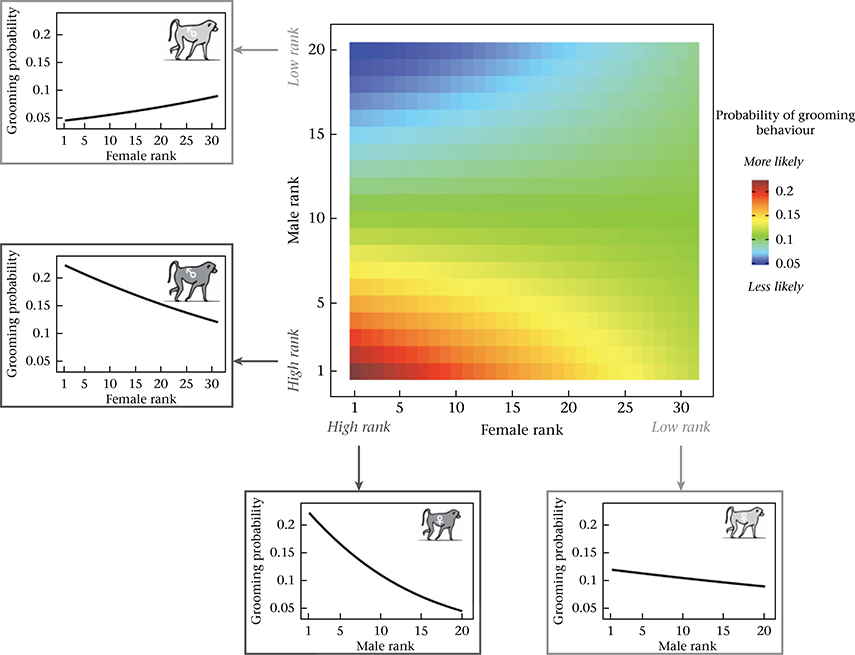
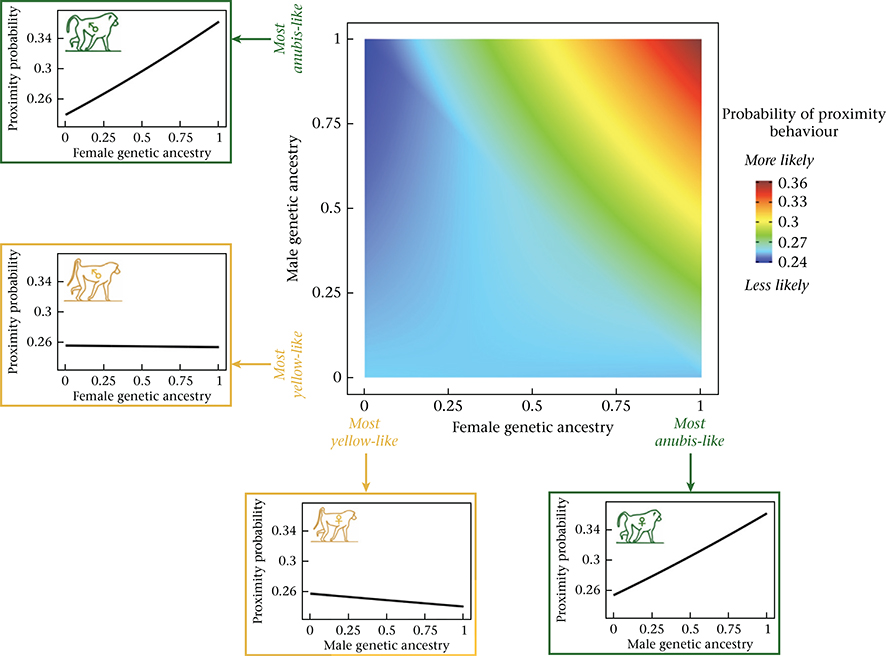
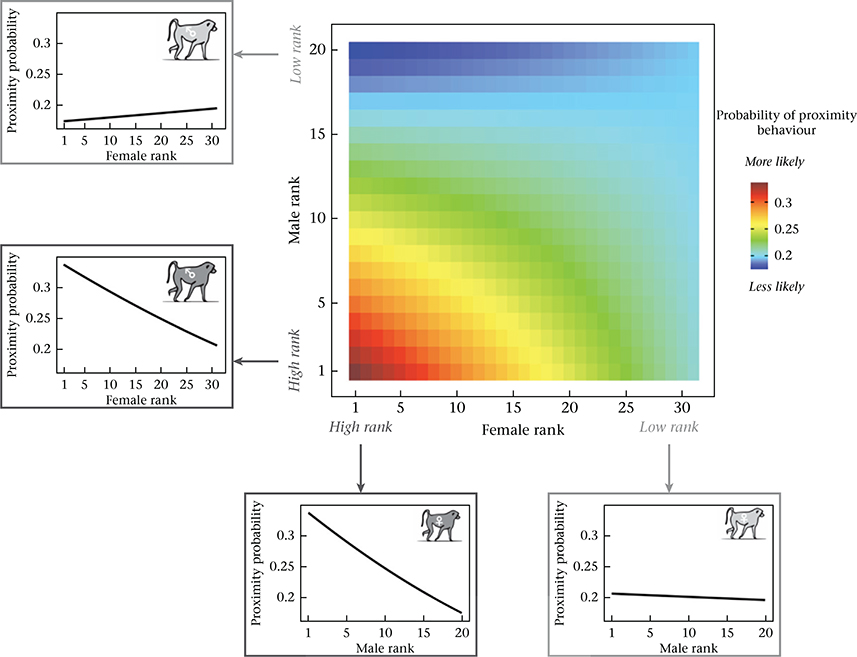
In addition to individual level effects, we found that the combined characteristics of the female and male in each dyad predicted the probability of grooming and proximity. First, affiliative interactions were assortative with respect to genetic ancestry: they were more likely to occur when both partners were of similar genetic ancestry (i.e. both anubis-like or both yellow-like) and less likely to occur if they were of different genetic ancestry (grooming: β = 0.646, P < 0.001; Table 1, Fig. 2; proximity: β = 0.303, P < 0.001; Table 2, Appendix, Fig. A3). Overall, the probability of grooming and proximity was highest for pairs where both partners were anubis-like. Affiliative interactions were also assortative with respect to dominance rank: if both partners were high ranking, the probability of affiliative interaction was higher than explained by the separate, additive effects of high male rank and high female rank alone (grooming: β = 0.003, P < 0.001; Table 1, Fig. 3; proximity: β = 0.001, P = 0.011; Table 2, Appendix, Fig. A4). The effects of ancestry-based assortativity and rank-based assortativity are likely to be independent, as the assortative genetic ancestry index we used here was only weakly correlated with the product of male and female rank (absolute value of Pearson’s correlation: r < 0.07, P < 3.7 × 10−13 for both grooming and proximity data sets). The correlation between rank and genetic ancestry was similarly weak within each sex (absolute value of Pearson’s correlation: r < 0.03 for unique male rank–genetic ancestry combinations, P > 0.35 for both grooming and proximity; r < 0.08 for unique female rank–genetic ancestry combinations, P = 0.37 for grooming and P = 0.04 for proximity).
Figure 2.
The combined effect of genetic ancestry characteristics of females and males on the probability of grooming. The central heatmap shows the probability of grooming behaviour as a function of female genetic ancestry (X axis) and male genetic ancestry (Y axis), based on model estimates assuming average values for all other covariates (see Appendix, Additional Methods). Assortative affiliative behaviour is reflected by the increased probability of yellow-like females grooming with yellow-like males, relative to anubis-like males, and anubis-like females grooming with anubis-like males, relative to yellow-like males. The probability of grooming is highest for pairs where both partners are anubis-like. Line graphs surrounding the heatmap show model predictions for the probability of grooming behaviour for males (left) and females (bottom) at the two extremes of genetic ancestry, as a function of the genetic ancestry of potential opposite-sex social partners.
Figure 3.
The combined effect of rank characteristics of females and males on the probability of grooming. The central heatmap shows the probability of grooming behaviour as a function of female dominance rank (X axis) and male dominance rank (Y axis), based on model estimates assuming average values for all other covariates (see Appendix, Additional Methods). The probability of grooming is highest for pairs where both partners are high ranking. Line graphs surrounding the heatmap show model predictions for the probability of grooming behaviour for males (left) and females (bottom) at the extremes of the rank distribution, as a function of the dominance rank of potential opposite-sex social partners.
Genetic relatedness did not predict either grooming or proximity behaviour within dyads (grooming: Table 1; proximity: Table 2), in contrast to the effects of relatedness on mating behaviour, where relatives are less likely to mate (consistent with inbreeding avoidance in baboons: Alberts & Altmann, 1995; Packer, 1979a; Tung et al., 2012). In other words, opposite-sex kin were neither more likely nor less likely to socially affiliate than opposite-sex nonkin. Additionally, male–female affiliation did not depend on the interaction between female reproductive state and male genetic ancestry (grooming: Table 1; proximity: Table 2).
Group Demography Influences Grooming and Proximity Behaviour, and Observer Effort Affects Ascertainment of These Behaviours
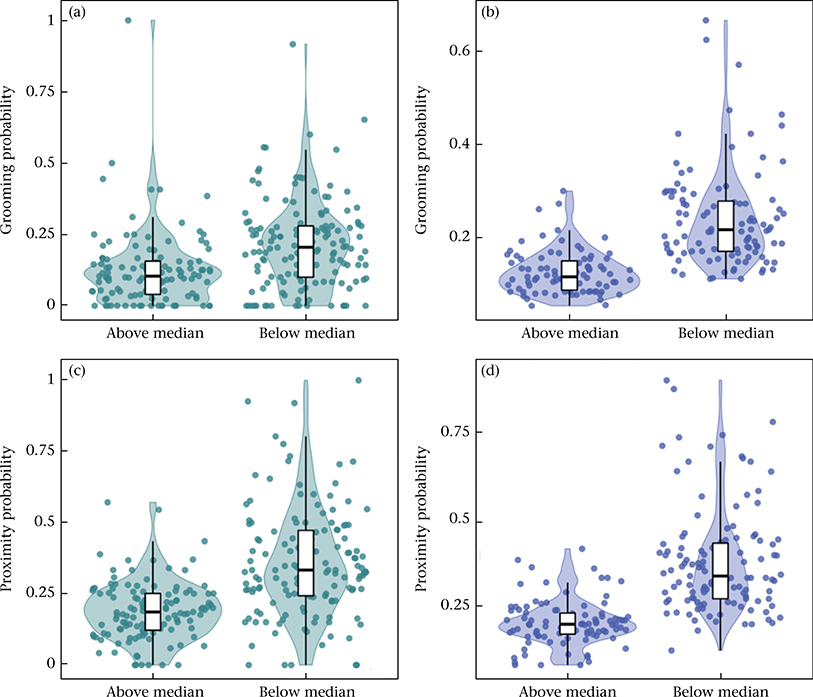
In addition to individual and dyad level effects, we found that aspects of group demography also influence male–female affiliative behaviour. The probability of grooming was lower for all dyads when the social group contained more adult males (β = −0.030, P = 0.018; Table 1, Appendix, Fig. A5a–b) and more adult females (β = −0.039, P < 0.001; Table 1, Appendix, Fig. A6a–b). Similarly, the probability of proximity was lower for all dyads when the social group contained more adult males (β = −0.036, P = 0.005; Table 2, Appendix, Fig. A5c–d), but not more adult females (β = −0.018, P = 0.073; Table 2, Appendix, Fig. A6c–d).
Finally, the probability of grooming and proximity behaviour was higher for all dyads the more days they were observed together in the same group (grooming: β = 0.047, P < 0.001; Table 1; proximity: β = 0.037, P < 0.001; Table 2). The probability of observing a given male–female dyad in proximity, but not of observing them grooming at least once, also increased with greater observer effort (grooming: β = 0.002, P = 0.425; Table 1; proximity: β = 0.027, P < 0.001; Table 2).
DISCUSSION
Our results show that opposite-sex affiliative relationships are predicted by genetic ancestry in a natural baboon hybrid zone. Genetic ancestry effects are of particular interest because opposite-sex relationships must have a partial genetic basis in order to respond to natural selection. Additionally, genetic ancestry-associated differences provide prima facie evidence that this trait has evolved in the past. Specifically, in the Amboseli baboons, genetic ancestry acts alongside the effects of dominance rank and group demography to predict grooming and proximity behaviour between adult males and adult females outside the mating context. These effects are not only detectable as a function of the individual characteristics of males and females, but also as a function of the properties of each opposite-sex pair. Although more anubis-like males and females were more likely to affiliate with the opposite sex regardless of their partner’s ancestry, pairs of anubis-like males and anubis-like females were the most likely to be observed grooming or in close proximity (Fig. 2, Appendix, Fig. A3). Our findings thus suggest that the tendency to engage in opposite-sex affiliative behaviour partially diverged during baboon evolution to differentiate yellow and anubis baboons. We note that while we tested for the effects of genetic ancestry in this study, not genotype per se, baboons in Amboseli inherit anubis ancestry from both maternal and paternal lines (Tung et al., 2008). Our data set also contains many multigeneration hybrids, such that genetic ancestry estimates vary continuously between mostly yellow to mostly anubis. Thus, the signature of genetic ancestry reported here likely arises from ancestry-associated differences in genotype, as opposed to ancestry-associated maternal or environmental effects on social preference.
These results add to previous evidence that male–female social bonds vary across baboon species (Baniel et al., 2016; Fischer et al., 2017; Goffe et al., 2016; Nguyen et al., 2009; Städele et al., 2019; Weingrill, 2000). For instance, male–female social relationships in chacma baboons, Papio ursinus, are short-lived and occur primarily when females have dependent infants, whereas in Guinea baboons, Papio papio, close male–female social relationships commonly last for several years (Baniel et al., 2016; Fischer et al., 2017; Goffe et al., 2016). Yellow and anubis baboons, which have similar social organization and mating systems (multimale, multifemale groups with male-biased dispersal and polygynandry), are thought to fall between these two extremes, such that male–female relationships can sometimes, although not always, be long-lasting (Nguyen et al., 2009; Smuts, 1985; Städele et al., 2019). The identification of genetic ancestry effects in this study thus suggests that subtle differences in the nature of opposite-sex social relationships can evolve even between species that are otherwise quite similar in their social systems and behavioural repertoires.
Our results also support an emerging perspective that behavioural variation within the genus Papio is relatively continuous (Fischer, Higham, et al., 2019). If so, the commonly used distinction between multilevel societies (e.g. as in Guinea and hamadryas baboons) and single level societies (e.g. as in yellow, anubis, chacma and Kinda baboons, Papio kindae) is not sufficient to capture behavioural diversity in baboons. Our finding that anubis-like individuals more readily affiliate with opposite-sex social partners than yellow-like individuals also suggests that anubis baboons share behavioural similarities with other ‘northern clade’ baboons (Guinea and hamadryas) that may reflect their shared genetic history (Jolly, 2020; Rogers et al., 2019). Interestingly, while more anubis-like baboons in our study were relatively more social with the opposite sex compared to yellow-like baboons, anubis baboons are comparably less social with opposite-sex partners than hamadryas baboons in the anubis–hamadryas Ethiopian hybrid zone (Bergman & Beehner, 2004). This again highlights that both substantial, discrete differences (e.g. between Guinea baboons and chacma baboons) and subtle, continuous variation (e.g. between the yellow baboons and anubis baboons in our study) characterize social behaviour in the baboon genus (Fischer, Higham, et al., 2019).
Several lines of evidence also support the relevance of genetic ancestry effects on opposite-sex affiliative behaviour to current variation in fitness. First, opposite-sex social relationships predict longevity in the Amboseli baboons (Archie et al., 2014; Campos et al., 2020), and longevity is an important contributor to lifetime reproductive success in both male and female baboons, as well as in other long-lived vertebrates (Alberts et al., 2006; Clutton-Brock, 1988; Lawler, 2007; McDonald, 1993; McLean et al., 2019; Newton, 1989; Wroblewski et al., 2009). Second, male–female social bonds can also lead to other reproductive gains, including offspring care that may improve survival (Anderson, 1992; Buchan et al., 2003; Busse & Hamilton, 1981; Huchard et al., 2013; Moscovice et al., 2009; Nguyen et al., 2009; Silk et al., 2020). Indeed, our findings that affiliative behaviour was less common for any given dyad in large groups and that both male and female rank predicted social interactions suggest that male–female social bonds are an important and limited social resource for both sexes (Archie et al., 2014; Baniel et al., 2016; Haunhorst et al., 2019; Lemasson et al., 2008; Palombit et al., 2001; Seyfarth, 1976; Städele et al., 2019). This interpretation agrees with reports in chacma baboons that pregnant and lactating females direct aggression towards cycling females that are mate-guarded by and copulate with a shared male social partner (Baniel et al., 2018). Together with ancestry-related differences in affiliative behaviour, our observations indicate that opposite-sex affiliative behaviour has not only evolved in baboons in the past, but may also be the target of selection in the Amboseli population today.
The long-term ramifications of our findings for the stability or resolution of the hybrid zone remain somewhat unclear. If strong opposite-sex social bonds are fitness enhancing, more anubis-like ancestry should be favoured in Amboseli. However, assortative social preferences (both in mating and nonmating contexts) can also act as a barrier to admixture and could reduce the rate of anubis expansion. Along with previous findings in Amboseli (Charpentier et al., 2008; Franz et al., 2015; Tung et al., 2012), our results thus suggest that genetic ancestry is associated with a range of selectively relevant behavioural and life-history traits that do not universally point towards either anubis range expansion or to behaviourally mediated reproductive isolation. Furthermore, any effect of genetic ancestry in baboons must necessarily be filtered through the effect of dominance rank, which is the most robust predictor of male mating behaviour and, based on the results of this study, also a major contributor to opposite-sex affiliative behaviour (Tung et al., 2012). Finally, the effect of assortativity necessarily depends on the characteristics of available social partners. The interplay between genetic ancestry effects on an individual level and at the dyadic level will therefore be dynamic across populations and over time. The complexity of these co-acting factors may help explain why yellow baboons and anubis baboons remain phenotypically and genetically distinct, even though genomic analyses indicate repeated bouts of gene flow between yellow baboons and anubis baboons over hundreds to thousands of generations (Rogers et al., 2019;Wall et al., 2016). More broadly, our results suggest that simple behavioural barriers to admixture, such as wing pattern-based mate choice in butterflies or vibrational-based courtship signals in treehoppers (Jiggins et al., 2001; Rodríguez et al., 2004), are unlikely to occur in socially complex animals like baboons. Future work using social network analysis, which could help reveal how genetic ancestry structures the topology and connectivity of social relationships in whole groups, may help shed light on this question.
Finally, our study suggests both parallels and differences between opposite-sex social interactions within versus outside the context of mating. Specifically, effects of female age and kinship were weak or undetectable in our analysis of male–female affiliative behaviour outside the mating context. These results contrast with our previous results for mating: male baboons are less likely to mate with females in older age classes (Tung et al., 2012), and baboons and other primates avoid mating with relatives (Alberts & Altmann, 1995; Godoy et al., 2016; Packer, 1979a; Tung et al., 2012; Walker et al., 2017; Widdig et al., 2017), probably as a behavioural strategy that minimizes inbreeding depression (Robinson et al., 2019). However, in both settings, higher-ranking and more anubis-like males are more likely to interact with females, and assortativity by genetic ancestry and dominance rank characterizes both mating and male–female affiliative behaviour (Tung et al., 2012). These parallels could be partially explained if mating success also leads males to affiliate with the mothers of their joint offspring, after those offspring are born. If so, anubis-like males who are successful at gaining consortships would also be involved in more female-directed affiliative behaviour. Indeed, evidence from several studies of baboons suggests that males frequently form affiliative relationships with the mothers of their genetic offspring (Huchard et al., 2010; Moscovice et al., 2010; Nguyen et al., 2009; Silk et al., 2020; Städele et al., 2019). In contrast, opposite-sex social affiliation is not a strong predictor of future mating events (Huchard et al., 2010; Moscovice et al., 2010; Nguyen et al., 2009; Silk et al., 2020; Städele et al., 2019), so differences in male–female behaviour in mating versus nonmating contexts may arise because the benefits of opposite-sex social bonds differ from the benefits of mating. Female choice and female–female competition may also be more important in predicting grooming and proximity than mating behaviour. For instance, whereas only one or a few females experience oestrus at any given time (Bercovitch,1983; Bulger, 1993; Levy et al., 2020), all adult females are available as, and may actively be searching out, grooming partners. Notably, grooming relationships in baboons are more often initiated and maintained by females than by males, whereas males primarily bear the costs of mate guarding in a mating context (Alberts et al., 1996; Nguyen et al., 2009; Packer, 1979b; Palombit et al., 1997; but see; Weyher et al., 2014). Thus, sexual and social preferences, and the extent to which they are expressed by males versus females, are likely to vary across different types of opposite-sex relationships – a distinction reminiscent of differences between social monogamy and genetic monogamy in pair-bonded birds and mammals (Carter & Perkeybile, 2018; Gowaty, 1996).
Data Availability
Data on grooming behaviour and proximity behaviour and all predictor variables tested in our models are available in the Duke Research Data Repository at https://doi.org/10.7924/r4kp82d1z. R code for recreating the analyses and figures in the manuscript is available at https://github.com/ArielleF/Genetic-Ancestry-Social-Affiliation.
Acknowledgments
We gratefully acknowledge the support of the U.S. National Science Foundation (NSF) and the U.S. National Institutes of Health (NIH) for the majority of the data represented here, currently through NSF IOS 1456832, NIH R01AG053308, R01AG053330, R01HD088558 and P01AG031719. We also thank the North Carolina Biotechnology Center for support for high-performance computing resources (2016-IDG-1013). A.S.F. was supported by NSF GRFP (DGE 1644868) and NIH T32GM007754; E.M.M was supported by NSF IOS 1501971. We thank Duke University, Princeton University and the University of Notre Dame for financial and logistical support. In Kenya, our research was approved by the Kenya Wildlife Service (KWS), the National Environment Management Authority (NEMA) and the National Council for Science, Technology, and Innovation (NACOSTI). We also thank the University of Nairobi, Institute of Primate Research, the National Museums of Kenya, the members of the Amboseli-Longido pastoralist communities, the Enduimet Wildlife Management Area, Ker & Downey Safaris, Air Kenya and Safarilink for their cooperation and assistance in the field. Particular thanks go to the Amboseli Baboon Research Project field team (R. S. Mututua, S. Sayialel, J. K. Warutere, I. L. Siodi, G. Marinka, B. Oyath) and camp staff, to T. Wango and V. Oudu for their untiring assistance in Nairobi and to Jeanne Altmann for her fundamental contributions to the Amboseli baboon research. The baboon project database, Babase, was designed and programmed by K. Pinc and is expertly managed by N. H. Learn and J. B. Gordon. Finally, we thank current and previous members of the Alberts and Tung labs, especially N. Snyder-Mackler, A. J. Lea and C. R. Campbell, and two anonymous referees for their helpful feedback. For a complete set of acknowledgments of funding sources, logistical assistance and data collection and management, please visit http://amboselibaboons.nd.edu/acknowledgements/. Any opinions, findings and conclusions expressed in this material are those of the authors and do not necessarily reflect the views of our funding bodies.
APPENDIX
Additional Methods
Estimating observer effort
All baboon study groups are observed by the same number of field observers for roughly the same amount of time, regardless of group size. As a consequence, the individual level density of our grooming and proximity data varies by social group (i.e. there are differences among groups in observer effort per individual baboon). To account for these differences, we estimated observer effort using focal animal sampling data from adult females. Each focal sample typically consists of 10 point samples, collected once per minute, for 10 min (Alberts et al., 2020; Archie et al., 2014). We summed the total number of point samples collected for adult females per social group across each female’s 2-month interval and then divided this value by the total number of adult females present in the social group during that same time period. We used this value as a measure of observer effort and included it as a fixed effect covariate in our models.
Filtering of grooming and proximity data
In the main text, we report results for grooming and proximity behaviour using 2-month time intervals. Initially, we attempted to measure grooming and proximity behaviour on a monthly basis. However, on a monthly basis, grooming and proximity were not observed in 93.2% and 87.2% of rows (N = 62 195 total rows, where a row represents a male–female dyad that could potentially be observed grooming or in proximity), respectively. While some of these ‘0’ value rows reflect a true absence of male–female interactions, observations in a single month may be too sparse to accurately reflect whether a given male–female dyad interacted. Indeed, even after omitting social group-month combinations with low observer effort (i.e. less than an average of 20 point samples per female per social group-month), grooming and proximity were still not observed in 92.1% and 83.9% of the data set, respectively. Thus, we concluded that our resolution of grooming and proximity behaviour was too coarse to measure on a monthly basis.
To address this concern, we expanded our time window from one month to two months and excluded any male in a female’s 2-month interval if he was only present for one of the two months. This decision reduced the percentage of rows where grooming and proximity did not occur (87.9% and 76.9% of rows, respectively, N = 26 245 total rows). Because we were most interested in the predictors of opposite-sex affiliative behaviour, conditioned on it actually occurring, we also filtered out any 2-month interval where the focal female did not interact with any male social partner for the entire interval (31.7% and 17.1% of intervals for the grooming and proximity data sets, respectively). We performed this filtering separately for the grooming and proximity data sets, thus producing two separate data sets for downstream analysis. We also excluded all data for females and males who were observed for less than 8 months. If these filters resulted in no observations of grooming or proximity for a given female interval, we also removed that interval (and repeated this procedure until all filtering criteria were met). Finally, we did not consider 2-month intervals in which females transitioned between reproductive states (pregnant or lactating) and 2-month intervals in which there were fewer than two adult males available for a female to interact with.
Encoding female reproductive state
In the main text, we report results for grooming and proximity behaviour from models where female reproductive state was coded as −1 for pregnancy and 1 for lactation. This deviates from the usual arbitrary coding of binary states as 0/1. We made this decision because we found that, with the 0/1 encoding, our beta and P value estimates for some model parameters depended on whether we assigned pregnancy to the 0 state versus lactation. Using the −1/1 alternative encoding eliminated this issue, produced consistent results across R packages (glmmTMB (version 1.0.0; Brooks et al., 2017) versus lme4 (version 1.1.23; Bates et al., 2015)), and also qualitatively matched our estimates using a fixed effects-only model (using the function glm from base R). Hence, we report the results using the −1/1 encoding in the main text.
Visualizing model effects
For a subset of figures, we plotted the probability of grooming and proximity behaviour as a function of a varying predictor of interest and model estimates assuming average values for all other covariates (Figs 1b, 1d, 2, 3, Appendix, Figs A2b, A2d, A3, A4). For example, to calculate the probability of grooming and proximity as a function of male dominance rank, we modelled an ‘average’ male (apart from his rank) interacting with an ‘average’ female in an ‘average’ demographic environment. We then estimated how variation in male rank is predicted to affect the probability of grooming or proximity using R’s predict function. For predictors that reflect the combined characteristics of the male–female pair, we used the same approach, but extended it to vary the characteristic of interest for both the male and the female, while holding the other predictor variables constant at their average value.
Table A1.
Summary of social groups, study subjects and observation periods
| Group ID | Femalesa | Malesa | Genetic ancestryb | Months in grooming data setc |
Months in proximity data setc |
Observation periodd | Observation monthse | ||
|---|---|---|---|---|---|---|---|---|---|
| Per female | Per male | Per female | Per male | ||||||
|
| |||||||||
| 1.1 | 33 | 48 | 0.20 ± 0.24 (0.03–0.87) | 21.88 ± 13.60 | 28.02 ± 23.40 | 30.06 ± 18.76 | 28.04 ± 23.38 | November 1999–September 2010 | 119 |
| 1.11 | 17 | 23 | 0.28 ± 0.26 (0.03–0.80) | 14.53 ± 8.19 | 20.52 ± 18.07 | 21.25 ± 6.81 | 21.09 ± 18.54 | May 2011–December 2015 | 56 |
| 1.12 | 5 | 9 | 0.16 ± 0.20 (0.03–0.72) | 10.40 ± 1.67 | 9.22 ± 5.83 | 12.00 ± 2.00 | 9.22 ± 5.83 | May 2011–November 2012 | 19 |
| 1.21 | 12 | 24 | 0.26 ± 0.29 (0.03–0.88) | 17.78 ± 10.74 | 13.25 ± 13.60 | 19.27 ± 12.78 | 13.46 ± 14.04 | November 1999–September 2008 | 72 |
| 1.211 | 8 | 13 | 0.32 ± 0.26 (0.04–0.89) | 16.50 ± 7.07 | 19.31 ± 10.55 | 19.75 ± 7.59 | 19.38 ± 10.52 | August 2012–December 2015 | 41 |
| 1.22 | 25 | 35 | 0.36 ± 0.28 (0.04–0.88) | 31.84 ± 18.97 | 29.80 ± 26.77 | 38.08 ± 21.60 | 30.29 ± 27.19 | November 1999–November 2012 | 145 |
| 1.221 | 8 | 9 | 0.38 ± 0.29 (0.04–0.90) | 6.33 ± 2.34 | 9.56 ± 5.10 | 11.25 ± 1.83 | 11.56 ± 4.82 | June 2014–December 2015 | 19 |
| 1.222 | 11 | 9 | 0.39 ± 0.30 (0.04–0.81) | 7.27 ± 2.57 | 12.22 ± 7.03 | 8.91 ± 2.74 | 12.89 ± 7.37 | June 2014–December 2015 | 19 |
| 2.1 | 20 | 44 | 0.26 ± 0.25 (0.03–0.82) | 27.50 ± 20.01 | 24.23 ± 19.73 | 32.50 ± 24.96 | 24.68 ± 19.89 | November 1999–March 2011 | 124 |
| 2.11 | 12 | 21 | 0.33 ± 0.27 (0.04–0.92) | 15.80 ± 7.08 | 17.67 ± 13.44 | 20.17 ± 5.75 | 18.14 ± 13.64 | June 2011–December 2015 | 55 |
| 2.12 | 8 | 10 | 0.32 ± 0.28 (0.03–0.82) | 6.33 ± 1.97 | 9.40 ± 6.98 | 8.75 ± 3.01 | 9.90 ± 7.36 | June 2011–November 2012 | 18 |
| 2.2 | 31 | 48 | 0.43 ± 0.30 (0.04–0.92) | 25.42 ± 13.54 | 29.27 ± 26.48 | 31.19 ± 15.62 | 29.52 ± 26.66 | November 1999–September 2011 | 121 |
Number of unique individuals included in this data set, per unique social group. Individuals may be represented in more than one social group because of social group fissions and fusions and/or secondary dispersal by males.
Mean ± SD (minimum–maximum).
Mean ± SD. Due to our filtering criteria, individuals had to be in each data set for at least 8 months, but could be counted as members of different social groups across those months.
The observation starting and ending month and year for each social group across both grooming and proximity data sets. Some observation periods span time periods in which behavioural monitoring was inconsistent or when social groups were too unstable (i.e. social groups were fissioning or fusing) to unambiguously determine an individual’s group membership. For our analysis, we excluded these time periods as well as the 2009 hydrological year (1 November 2008–31 October 2009). We also excluded months when the number of adult males in the social group was less than two.
The number of unique observation months for each social group included across the grooming and proximity data sets.
Table A2.
Pearson’s product-moment correlation (r) between predictor variables in the final grooming data set
| Genetic ancestry |
Assortative genetic ancestry index | Heterozygosity |
Genetic relatedness | Dominance rank |
Female age | Adults in social group |
Reproductive state | Pair co-residency | Observer effort | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Male | Female | Male | Female | Male | Female | Male | |||||||
|
| |||||||||||||
| Genetic ancestry | |||||||||||||
| Female | 0.098 | −0.391 | 0.282 | 0.070 | −0.086 | 0.027 | 0.028 | −0.009 | 0.189 | 0.110 | 0.020 | −0.053 | −0.054 |
| Male | −0.612 | 0.048 | 0.292 | −0.158 | 0.047 | −0.016 | 0.036 | 0.115 | 0.108 | −0.002 | 0.042 | −0.062 | |
| Assortative genetic ancestry index | −0.131 | −0.159 | 0.244 | −0.035 | −0.069 | −0.001 | −0.090 | −0.082 | 0.001 | −0.044 | 0.017 | ||
| Heterozygosity | |||||||||||||
| Female | 0.044 | −0.091 | 0.185 | 0.067 | −0.015 | 0.205 | 0.151 | 0.005 | −0.034 | −0.152 | |||
| Male | −0.086 | 0.020 | 0.010 | 0.009 | 0.056 | 0.077 | −0.013 | 0.000 | −0.042 | ||||
| Genetic relatedness | 0.011 | −0.045 | −0.005 | −0.030 | −0.029 | −0.002 | 0.033 | 0.010 | |||||
| Dominance rank | |||||||||||||
| Female | 0.167 | −0.007 | 0.396 | 0.335 | 0.026 | 0.039 | −0.255 | ||||||
| Male | 0.014 | 0.399 | 0.505 | −0.028 | −0.033 | −0.220 | |||||||
| Female age | 0.025 | 0.021 | 0.068 | 0.076 | −0.064 | ||||||||
| Adults in social group | |||||||||||||
| Female | 0.804 | 0.018 | −0.029 | −0.614 | |||||||||
| Male | −0.050 | 0.039 | −0.436 | ||||||||||
| Reproductive state | −0.027 | 0.017 | |||||||||||
| Pair co-residency | 0.026 | ||||||||||||
Predictor variables for which P < 0.01 are bolded and P < 0.05 are italicized.
Table A3.
Pearson’s product-moment correlation (r) between predictor variables in the final proximity data set
| Genetic ancestry |
Assortative genetic ancestry index | Heterozygosity |
Genetic relatedness | Dominance rank |
Female age | Adults in social group |
Reproductive state | Pair co-residency | Observer effort | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Male | Female | Male | Female | Male | Female | Male | |||||||
|
| |||||||||||||
| Genetic ancestry | |||||||||||||
| Female | 0.095 | −0.390 | 0.253 | 0.071 | −0.086 | −0.008 | 0.021 | 0.001 | 0.160 | 0.098 | 0.011 | −0.032 | −0.040 |
| Male | −0.655 | 0.050 | 0.272 | −0.164 | 0.032 | −0.021 | 0.039 | 0.102 | 0.090 | −0.001 | 0.053 | −0.052 | |
| Assortative genetic ancestry index | −0.124 | −0.163 | 0.237 | −0.016 | −0.056 | −0.008 | −0.068 | −0.060 | 0.001 | −0.056 | 0.011 | ||
| Heterozygosity | |||||||||||||
| Female | 0.045 | −0.086 | 0.190 | 0.045 | −0.027 | 0.189 | 0.116 | −0.004 | −0.028 | −0.148 | |||
| Male | −0.087 | 0.016 | 0.000 | 0.015 | 0.056 | 0.074 | −0.009 | 0.003 | −0.045 | ||||
| Genetic relatedness | 0.020 | −0.041 | 0.002 | −0.017 | −0.015 | 0.002 | 0.036 | 0.001 | |||||
| Dominance rank | |||||||||||||
| Female | 0.150 | 0.013 | 0.378 | 0.304 | 0.066 | 0.029 | −0.253 | ||||||
| Male | 0.050 | 0.375 | 0.491 | −0.026 | −0.037 | −0.222 | |||||||
| Female age | 0.089 | 0.095 | 0.070 | 0.077 | −0.101 | ||||||||
| Adults in social group | |||||||||||||
| Female | 0.781 | 0.016 | −0.028 | −0.637 | |||||||||
| Male | −0.051 | 0.035 | −0.451 | ||||||||||
| Reproductive state | −0.016 | 0.002 | |||||||||||
| Pair co-residency | 0.022 | ||||||||||||
Predictor variables for which P < 0.01 are bolded and P < 0.05 are italicized.
Figure A1.
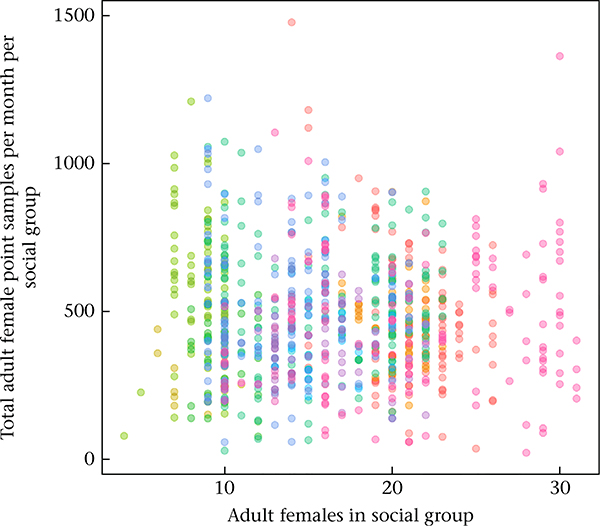
The total effort invested in behavioural observations across study groups of different sizes. Each dot represents a unique social group–month combination, coloured by social group (N = 12 social groups, N = 812 total group-months from the initial, monthly based data set). There is no relationship between the number of adult females present in a social group and the total number of point samples recorded for adult females per social group (10 point samples are collected per focal sample, if the sample is complete; linear model estimate for the effect of number of adult females on total number of point samples: β = −1.615, P = 0.221). The result is that observer effort per group does not vary by group size, but observer effort per individual baboon does. Hence, the probability of observing whether a given male–female dyad groomed or were in proximity at least once in a given time interval is smaller for large groups than for small groups. To address this problem, we included a measure of observer effort (total adult female point samples per social group/total adult females present in social group) in our models.
Figure A2.
The influence of genetic ancestry and dominance rank on the tendency to be in proximity to an opposite-sex partner. (a) The probability of being in proximity among co-resident opposite-sex pairs, per 2-month interval, for the most anubis-like males (above the 90th percentile for male genetic ancestry in the data set, >81.5% anubis ancestry, N = 9 males) and the most yellow-like males (below the 10th percentile for male genetic ancestry in the data set, <4.8% anubis ancestry, N = 20 males). Probabilities are shown here based on the data without adjustment for other covariates (note that, although not visually apparent in the 1/0 proximity data as summarized here, more anubis-like males are more likely to be observed in proximity with female partners in the full model: β = 0.270, P < 0.001, Table 2). (b) The probability of being in proximity among co-resident opposite-sex pairs, per 2-month interval, as a function of male dominance rank. Coloured dots show probabilities based on counts of proximity occurrences, without adjustment for other covariates (as in (a)), and the dashed line shows the predicted relationship based on model estimates, assuming average values for all other covariates (see Appendix, Additional Methods). Grey dots show the presence (y = 1) or absence (y = 0) of proximity behaviour for all 21 130 female–male pair–interval combinations, as a function of male dominance rank (dots are jittered vertically for visibility). Noninteger values correspond to individuals that changed ranks during a 2-month interval in the data set. (c) As in (a), for the most anubis-like females (above the 90th percentile for female genetic ancestry in the data set, >74.1% anubis ancestry, N = 14 females) and the most yellow-like females (below the 10th percentile for female genetic ancestry in the data set, <3.2% anubis ancestry, N = 8 females). (d) As in (b), with the probability of being in proximity shown as a function of female dominance rank.
Figure A3.
The combined effect of genetic ancestry characteristics of females and males on the probability of being in proximity. The central heatmap shows the probability of proximity behaviour as a function of female genetic ancestry (X axis) and male genetic ancestry (Y axis), based on model estimates assuming average values for all other covariates (see Appendix, Additional Methods). Assortative affiliative behaviour is reflected by the increased probability of yellow-like females being in proximity with yellow-like males, relative to anubis-like males, and anubis-like females being in proximity with anubis-like males, relative to yellow-like males. The probability of being in proximity is highest for pairs where both partners are anubis-like. Line graphs surrounding the heatmap show model predictions for the probability of proximity behaviour for males (left) and females (bottom) at the two extremes of genetic ancestry, as a function of the genetic ancestry of potential opposite-sex social partners.
Figure A4.
The combined effect of rank characteristics of females and males on the probability of being in proximity. The central heatmap shows the probability of proximity behaviour as a function of female dominance rank (X axis) and male dominance rank (Y axis), based on model estimates assuming average values for all other covariates (see Appendix, Additional Methods). The probability of being in proximity is highest for pairs where both partners are high ranking. Line graphs surrounding the heatmap show model predictions for the probability of proximity behaviour for males (left) and females (bottom) at the extremes of the rank distribution, as a function of the dominance rank of potential opposite-sex social partners.
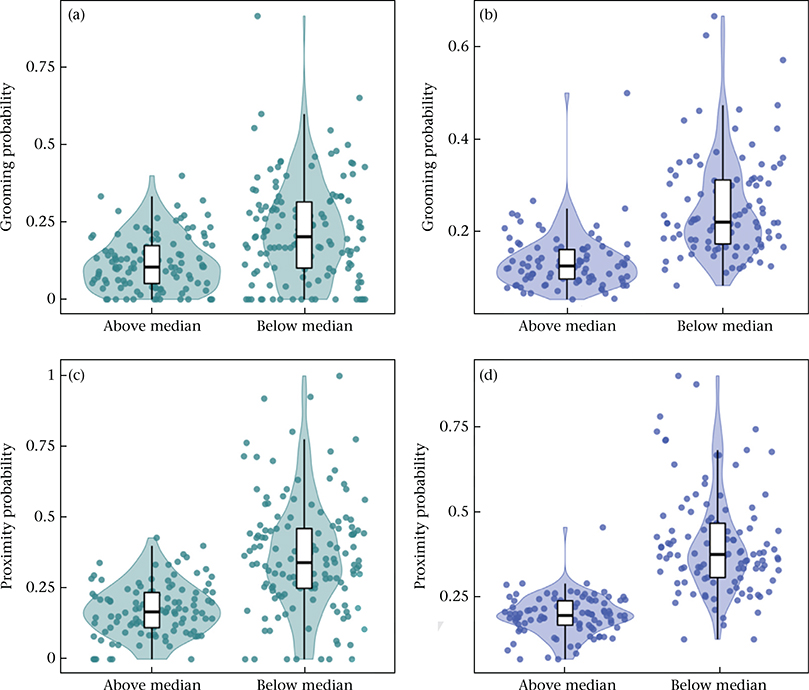
Figure A5.
The influence of the number of adult males present in a social group on grooming and proximity behaviour. (a, b) The probability of grooming among co-resident opposite-sex pairs, per 2-month interval, for each male (a) and female (b) in social groups with greater or less than the median number of co-resident males in the sample (N = 12 males). Probabilities are shown here based on the data without adjustment for other covariates. (c, d) As in (a, b), for proximity behaviour (median = 11.5 males).
Figure A6.
The influence of the number of adult females present in a social group on grooming and proximity behaviour. (a, b) The probability of grooming among co-resident opposite-sex pairs, per 2-month interval, for each male (a) and female (b) in social groups with greater or less than the median number of co-resident females in the sample (N = 20 females). Probabilities are shown here based on the data without adjustment for other covariates. (c, d) As in (a, b), for proximity behaviour (median = 20 females). Note that in the full model, the apparent difference observable for proximity was not significant after taking into account other model covariates and random effects (β = −0.018, P = 0.073; Table 2).
References
- Alberts SC (2018). Social influences on survival and reproduction: Insights from a long-term study of wild baboons. Journal of Animal Ecology, 88(1), 47–66. 10.1111/1365-2656.12887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts SC, & Altmann J (1995). Balancing costs and opportunities: Dispersal in male baboons. American Naturalist, 145(2), 279–306. 10.1086/285740 [DOI] [Google Scholar]
- Alberts SC, & Altmann J (2001). Immigration and hybridization patterns of yellow and anubis baboons in and around Amboseli, Kenya. American Journal of Primatology, 53(4), 139–154. 10.1002/ajp.1 [DOI] [PubMed] [Google Scholar]
- Alberts SC, & Altmann J (2012). The Amboseli Baboon Research Project: 40 years of continuity and change. In Kappeler PM, & Watts DP (Eds.), Long-term field studies of primates (pp. 261–287). Berlin, Germany: Springer-Verlag. [Google Scholar]
- Alberts SC, Altmann J, & Wilson ML (1996). Mate guarding constrains foraging activity of male baboons. Animal Behaviour, 51(6), 1269–1277. 10.1006/anbe.1996.0131 [DOI] [Google Scholar]
- Alberts SC, Archie EA, Altmann J, & Tung J (2020). Monitoring guide for the Amboseli Baboon Research Project: Protocols for long-term monitoring and data collection. Retrieved from https://amboselibaboons.nd.edu/assets/384683/abrp_monitoring_guide_9april2020.pdf.
- Alberts SC, Buchan JC, & Altmann J (2006). Sexual selection in wild baboons: From mating opportunities to paternity success. Animal Behaviour, 72(5), 1177–1196. 10.1016/j.anbehav.2006.05.001 [DOI] [Google Scholar]
- Alberts SC, & Gordon JB (2018). Protocols for data management. Retrieved from https://papio.biology.duke.edu/babasewiki/DataManagement?action=AttachFile&do=view&target=DukeþDataþManagementþProtocol.pdf.
- Alberts SC, Watts HE, & Altmann J (2003). Queuing and queue-jumping: Long-term patterns of reproductive skew in male savannah baboons, Papio cynocephalus. Animal Behaviour, 65(4), 821–840. 10.1006/anbe.2003.2106 [DOI] [Google Scholar]
- Anderson CM (1986). Female age: Male preference and reproductive success in primates. International Journal of Primatology, 7(3), 305–326. 10.1007/bf02736394 [DOI] [Google Scholar]
- Anderson CM (1992). Male investment under changing conditions among chacma baboons at Suikerbosrand. American Journal of Physical Anthropology, 87(4), 479–496. 10.1002/ajpa.1330870408 [DOI] [PubMed] [Google Scholar]
- Archie EA, Tung J, Clark M, Altmann J, & Alberts SC (2014). Social affiliation matters: Both same-sex and opposite-sex relationships predict survival in wild female baboons. Proceedings of the Royal Society B: Biological Sciences, 281(1793), 20141261. 10.1098/rspb.2014.1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldassarre DT, & Webster MS (2013). Experimental evidence that extra-pair mating drives asymmetrical introgression of a sexual trait. Proceedings of the Royal Society B: Biological Sciences, 280(1771), 20132175. 10.1098/rspb.2013.2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldassarre DT, White TA, Karubian J, & Webster MS (2014). Genomic and morphological analysis of a semipermeable avian hybrid zone suggests asymmetrical introgression of a sexual signal. Evolution, 68(9), 2644–2657. 10.1111/evo.12457 [DOI] [PubMed] [Google Scholar]
- Baniel A, Cowlishaw G, & Huchard E (2016). Stability and strength of male–female associations in a promiscuous primate society. Behavioral Ecology and Sociobiology, 70(5), 761–775. 10.1007/s00265-016-2100-8 [DOI] [Google Scholar]
- Baniel A, Cowlishaw G, & Huchard E (2018). Jealous females? Female competition and reproductive suppression in a wild promiscuous primate. Proceedings of the Royal Society B: Biological Sciences, 285(1886). 10.1098/rspb.2018.1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, & Walker S (2015). Fitting linear mixed-effects models using lme4. Journal of Statistical Software, 67(1), 1–48. 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- Beehner JC, Onderdonk DA, Alberts SC, & Altmann J (2006). The ecology of conception and pregnancy failure in wild baboons. Behavioral Ecology, 17(5), 741–750. 10.1093/beheco/arl006 [DOI] [Google Scholar]
- Bercovitch FB (1983). Time budgets and consortships in olive baboons (Papio anubis). Folia Primatologica, 41(3–4), 180–190. 10.1159/000156130 [DOI] [Google Scholar]
- Bergman TJ, & Beehner JC (2003). Hybrid zones and sexual selection: Insights from the Awash baboon hybrid zone (Papio hamadryas anubis × P.h. hamadryas). In Jones CB, & American Society of Primatologists (Eds.), Sexual selection and reproductive competition in primates: New perspectives and directions (pp. 503–537). Norman, OK: American Society of Primatologists. [Google Scholar]
- Bergman TJ, & Beehner JC (2004). Social system of a hybrid baboon group (Papio anubis × P. hamadryas). International Journal of Primatology, 25(6), 1313–1330. [Google Scholar]
- Bergman TJ, Phillips-Conroy JE, & Jolly CJ (2008). Behavioral variation and reproductive success of male baboons (Papio anubis × Papio hamadryas) in a hybrid social group. American Journal of Primatology, 70(2), 136–147. 10.1002/ajp.20467 [DOI] [PubMed] [Google Scholar]
- Best EC, Dwyer RG, Seddon JM, & Goldizen AW (2014). Associations are more strongly correlated with space use than kinship in female eastern grey kangaroos. Animal Behaviour, 89, 1–10. 10.1016/j.anbehav.2013.12.011 [DOI] [Google Scholar]
- Black JM (2001). Fitness consequences of long-term pair bonds in barnacle geese: Monogamy in the extreme. Behavioral Ecology, 12(5), 640–645. 10.1093/beheco/12.5.640 [DOI] [Google Scholar]
- Brooks ME, Kristensen K, van Benthem KJ, Magnusson A, Berg CW, Nielsen A, et al. (2017). glmmTMB balances speed and flexibility among packages for zero-inflated generalized linear mixed modeling. R Journal, 9(2), 378–400. 10.32614/Rj-2017-066 [DOI] [Google Scholar]
- Buchan JC, Alberts SC, Silk JB, & Altmann J (2003). True paternal care in a multi-male primate society. Nature, 425(6954), 179–181. 10.1038/nature01866 [DOI] [PubMed] [Google Scholar]
- Bulger JB (1993). Dominance rank and access to estrous females in male savanna baboons. Behaviour, 127(1–2), 67–103. [DOI] [Google Scholar]
- Busse C, & Hamilton WJ (1981). Infant carrying by male chacma baboons. Science, 212(4500), 1281–1283. 10.1126/science.212.4500.1281 [DOI] [PubMed] [Google Scholar]
- Cameron EZ, Setsaas TH, & Linklater WL (2009). Social bonds between unrelated females increase reproductive success in feral horses. Proceedings of the National Academy of Sciences of the United States of America, 106(33), 13850–13853. 10.1073/pnas.0900639106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos FA, Villavicencio F, Archie EA, Colchero F, & Alberts SC (2020). Social bonds, social status and survival in wild baboons: A tale of two sexes. Philosophical Transactions of the Royal Society B, 375(1811), 20190621. 10.1098/rstb.2019.0621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CS, & Perkeybile AM (2018). The monogamy paradox: What do love and sex have to do with it? Frontiers in Ecology and Evolution, 6, 202. 10.3389/fevo.2018.00202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier MJ, Fontaine MC, Cherel E, Renoult JP, Jenkins T, Benoit L, Barthès N, Alberts SC, & Tung J (2012). Genetic structure in a dynamic baboon hybrid zone corroborates behavioural observations in a hybrid population. Molecular Ecology, 21(3), 715–731. 10.1111/j.1365-294X.2011.05302.x [DOI] [PubMed] [Google Scholar]
- Charpentier MJ, Tung J, Altmann J, & Alberts SC (2008). Age at maturity in wild baboons: Genetic, environmental and demographic influences. Molecular Ecology, 17(8), 2026–2040. 10.1111/j.1365-294X.2008.03724.x [DOI] [PubMed] [Google Scholar]
- Cheney DL, Silk JB, & Seyfarth RM (2012). Evidence for intra-sexual selection in wild female baboons. Animal Behaviour, 84(1), 21–27. 10.1016/j.anbehav.2012.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutton-Brock TH (1988). Reproductive success: Studies of individual variation in contrasting breeding systems. Chicago, IL: University of Chicago Press. [Google Scholar]
- Cords M (1997). Friendships, alliances, reciprocity and repair. In Whiten A, & Byrne RW (Eds.), Machiavellian intelligence II: Extensions and evaluations (pp. 24–49). Cambridge, U.K.: Cambridge University Press. [Google Scholar]
- Cords M (2012). The behavior, ecology, and social evolution of cercopithecine monkeys. In Mitani JC, Call J, Kappeler PM, Palombit RA, & Silk JB (Eds.), The evolution of primate societies (pp. 91–112). Chicago, IL: The University of Chicago Press. [Google Scholar]
- Ellis S, Snyder-Mackler N, Ruiz-Lambides A, Platt ML, & Brent LJN (2019). Deconstructing sociality: The types of social connections that predict longevity in a group-living primate. Proceedings of the Royal Society B: Biological Sciences, 286(1917), 20191991. 10.1098/rspb.2019.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, & Pritchard JK (2003). Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics, 164(4), 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J, Higham JP, Alberts SC, Barrett L, Beehner JC, Bergman TJ, Carter AJ, Collins A, Elton S, Fagot J, Ferreira da Silva MJ, Hammerschmidt K, Henzi P, Jolly CJ, Knauf S, Kopp GH, Rogers J, Roos C, Seyfarth RM,…Zinner D (2019). Insights into the evolution of social systems and species from baboon studies. Elife, 8, Article e50989. 10.7554/eLife.50989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J, Kopp GH, Dal Pesco F, Goffe A, Hammerschmidt K, Kalbitzer U, Klapproth M, Maciej P, Ndao I, Patzelt A, & Zinner D (2017). Charting the neglected West: The social system of Guinea baboons. American Journal of Physical Anthropology, 162(Suppl. 63), 15–31. 10.1002/ajpa.23144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer EK, Nowicki JP, & O’Connell LA (2019). Evolution of affiliation: Patterns of convergence from genomes to behaviour. Philosophical Transactions of the Royal Society B: Biological Sciences, 374(1777), 20180242. 10.1098/rstb.2018.0242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick CL, Altmann J, & Alberts SC (2014). Sources of variance in a female fertility signal: Exaggerated estrous swellings in a natural population of baboons. Behavioral Ecology and Sociobiology, 68(7), 1109–1122. 10.1007/s00265-014-1722-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz M, McLean E, Tung J, Altmann J, & Alberts SC (2015). Self-organizing dominance hierarchies in a wild primate population. Proceedings of the Royal Society B: Biological Sciences, 282(1814), 20151512. 10.1098/rspb.2015.1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frère CH, Krützen M, Mann J, Connor RC, Bejder L, & Sherwin WB (2010a). Social and genetic interactions drive fitness variation in a free-living dolphin population. Proceedings of the National Academy of Sciences of the United States of America, 107(46), 19949–19954. 10.1073/pnas.1007997107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frère CH, Krützen M, Mann J, Watson-Capps JJ, Tsai YJ, Patterson EM, Connor R, Bejder L, & Sherwin WB (2010b). Home range overlap, matrilineal and biparental kinship drive female associations in bottlenose dolphins. Animal Behaviour, 80(3), 481–486. 10.1016/j.anbehav.2010.06.007 [DOI] [Google Scholar]
- Gesquiere LR, Wango EO, Alberts SC, & Altmann J (2007). Mechanisms of sexual selection: Sexual swellings and estrogen concentrations as fertility indicators and cues for male consort decisions in wild baboons. Hormones and Behavior, 51(1), 114–125. 10.1016/j.yhbeh.2006.08.010 [DOI] [PubMed] [Google Scholar]
- Godoy I, Vigilant L, & Perry SE (2016). Inbreeding risk, avoidance and costs in a group-living primate, Cebus capucinus. Behavioral Ecology and Sociobiology, 70(9), 1601–1611. 10.1007/s00265-016-2168-1 [DOI] [Google Scholar]
- Goffe AS, Zinner D, & Fischer J (2016). Sex and friendship in a multilevel society: Behavioural patterns and associations between female and male Guinea baboons. Behavioral Ecology and Sociobiology, 70, 323–336. 10.1007/s00265-015-2050-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowaty PA (1996). Battle of the sexes and origins of monogamy. In Black JM (Ed.), Partnerships in birds: The study of monogamy (pp. 21–52). Oxford, U.K.: Oxford University Press. [Google Scholar]
- Griggio M, & Hoi H (2011). An experiment on the function of the long-term pair bond period in the socially monogamous bearded reedling. Animal Behaviour, 82(6), 1329–1335. 10.1016/j.anbehav.2011.09.016 [DOI] [Google Scholar]
- Haunhorst CB, Fürtbauer I, Schülke O, & Ostner J (2019). Female macaques compete for ‘power’ and ‘commitment’ in their male partners. Evolution and Human Behavior, 41(2), 117–125. 10.1016/j.evolhumbehav.2019.11.001 [DOI] [Google Scholar]
- Haunhorst CB, Heesen M, Ostner J, & Schülke O (2017). Social bonds with males lower the costs of competition for wild female Assamese macaques. Animal Behaviour, 125, 51–60. 10.1016/j.anbehav.2017.01.008 [DOI] [Google Scholar]
- Hewitt GM (1988). Hybrid zones: Natural laboratories for evolutionary studies. Trends in Ecology & Evolution, 3(7), 158–167. 10.1016/0169-5347(88)90033-X [DOI] [PubMed] [Google Scholar]
- Huchard E, Alvergne A, Féjan D, Knapp LA, Cowlishaw G, & Raymond M (2010). More than friends? Behavioural and genetic aspects of heterosexual associations in wild chacma baboons. Behavioral Ecology and Sociobiology, 64(5), 769–781. 10.1007/s00265-009-0894-3 [DOI] [Google Scholar]
- Huchard E, Charpentier MJ, Marshall H, King AJ, Knapp LA, & Cowlishaw G (2013). Paternal effects on access to resources in a promiscuous primate society. Behavioral Ecology, 24(1), 229–236. 10.1093/beheco/ars158 [DOI] [Google Scholar]
- Insel TR, & Shapiro LE (1992). Oxytocin receptor distribution reflects social organization in monogamous and polygamous voles. Proceedings of the National Academy of Sciences of the United States of America, 89(13), 5981–5985. 10.1073/pnas.89.13.5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel TR, Wang ZX, & Ferris CF (1994). Patterns of brain vasopressin receptor distribution associated with social organization in microtine rodents. Journal of Neuroscience, 14(9), 5381–5392. 10.1523/jneurosci.14-09-05381.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins CD, Naisbit RE, Coe RL, & Mallet J (2001). Reproductive isolation caused by colour pattern mimicry. Nature, 411(6835), 302–305. 10.1038/35077075 [DOI] [PubMed] [Google Scholar]
- Johnson ZV, & Young LJ (2015). Neurobiological mechanisms of social attachment and pair bonding. Current Opinion in Behavioral Sciences, 3, 38–44. 10.1016/j.cobeha.2015.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly CJ (2020). Philopatry at the frontier: A demographically driven scenario for the evolution of multilevel societies in baboons (Papio). Journal of Human Evolution, 146, 102819. 10.1016/j.jhevol.2020.102819 [DOI] [PubMed] [Google Scholar]
- Kempenaers B (2007). Mate choice and genetic quality: A review of the heterozygosity theory. Advances in the Study of Behavior, 37, 189–278. 10.1016/S0065-3454(07)37005-8 [DOI] [Google Scholar]
- Kronforst MR, Young LG, Kapan DD, McNeely C, O’Neill RJ, & Gilbert LE (2006). Linkage of butterfly mate preference and wing color preference cue at the genomic location of wingless. Proceedings of the National Academy of Sciences of the United States of America, 103(17), 6575–6580. 10.1073/pnas.0509685103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulik L, Muniz L, Mundry R, & Widdig A (2012). Patterns of interventions and the effect of coalitions and sociality on male fitness. Molecular Ecology, 21(3), 699–714. 10.1111/j.1365-294X.2011.05250.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langergraber K, Mitani J, & Vigilant L (2009). Kinship and social bonds in female chimpanzees (Pan troglodytes). American Journal of Primatology, 71(10), 840–851. 10.1002/ajp.20711 [DOI] [PubMed] [Google Scholar]
- Lawler RR (2007). Fitness and extra-group reproduction in male Verreaux’s sifaka: An analysis of reproductive success from 1989–1999. American Journal of Physical Anthropology, 132(2), 267–277. 10.1002/ajpa.20507 [DOI] [PubMed] [Google Scholar]
- Lea AJ, Akinyi MY, Nyakundi R, Mareri P, Nyundo F, Kariuki T, Alberts SC, Archie EA, & Tung J (2018). Dominance rank-associated gene expression is widespread, sex-specific, and a precursor to high social status in wild male baboons. Proceedings of the National Academy of Sciences of the United States of America, 115(52), E12163–E12171. 10.1073/pnas.1811967115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea AJ, Altmann J, Alberts SC, & Tung J (2015). Developmental constraints in a wild primate. American Naturalist, 185(6), 809–821. 10.1086/681016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemasson A, Palombit RA, & Jubin R (2008). Friendships between males and lactating females in a free-ranging group of olive baboons (Papio hamadryas anubis): Evidence from playback experiments. Behavioral Ecology and Sociobiology, 62(6), 1027–1035. 10.1007/s00265-007-0530-z [DOI] [Google Scholar]
- Levy EJ, Zipple MN, McLean E, Campos FA, Dasari M, Fogel AS, Franz M, Gesquiere LR, Gordon JB, Grieneisen L, Habig B, Jansen DJ, Learn NH, Weibel CJ, Altmann J, Alberts SC, & Archie EA (2020). A comparison of dominance rank metrics reveals multiple competitive landscapes in an animal society. Proceedings of the Royal Society B: Biological Sciences, 287(1934), 20201013. 10.1098/rspb.2020.1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavárez J, Salazar CA, Bermingham E, Salcedo C, Jiggins CD, & Linares M (2006). Speciation by hybridization in Heliconius butterflies. Nature, 441(7095), 868–871. 10.1038/nature04738 [DOI] [PubMed] [Google Scholar]
- McDonald DB (1993). Demographic consequences of sexual selection in the long-tailed manakin. Behavioral Ecology, 4(4), 297–309. 10.1093/beheco/4.4.297 [DOI] [Google Scholar]
- McLean EM, Archie EA, & Alberts SC (2019). Lifetime fitness in wild female baboons: Trade-offs and individual heterogeneity in quality. American Naturalist, 194(6), 745–759. 10.1086/705810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard N, von Segesser F, Scheffrahn W, Pastorini J, Vallet D, Gaci B, Martin RD, & Gautier-Hion A (2001). Is male-infant caretaking related to paternity and/or mating activities in wild Barbary macaques (Macaca sylvanus)? Comptes Rendus de l’Academie des Sciences - Series III: Sciences de la Vie, 324(7), 601–610. 10.1016/S0764-4469(01)01339-7 [DOI] [PubMed] [Google Scholar]
- Mitani JC (2009). Male chimpanzees form enduring and equitable social bonds. Animal Behaviour, 77(3), 633–640. 10.1016/j.anbehav.2008.11.021 [DOI] [Google Scholar]
- Möller LM, Beheregaray LB, Harcourt RG, & Krützen M (2001). Alliance membership and kinship in wild male bottlenose dolphins (Tursiops aduncus) of southeastern Australia. Proceedings of the Royal Society B: Biological Sciences, 268(1479), 1941–1947. 10.1098/rspb.2001.1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscovice LR, Di Fiore A, Crockford C, Kitchen DM, Wittig R, Seyfarth RM, & Cheney DL (2010). Hedging their bets? Male and female chacma baboons form friendships based on likelihood of paternity. Animal Behaviour, 79(5), 1007–1015. 10.1016/j.anbehav.2010.01.013 [DOI] [Google Scholar]
- Moscovice LR, Heesen M, Di Fiore A, Seyfarth RM, & Cheney DL (2009). Paternity alone does not predict long-term investment in juveniles by male baboons. Behavioral Ecology and Sociobiology, 63(10), 1471–1482. 10.1007/s00265-009-0781-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton I (1989). Lifetime reproduction in birds. London, U.K.: Academic Press. [Google Scholar]
- Nguyen N, Van Horn RC, Alberts SC, & Altmann J (2009). ‘Friendships’ between new mothers and adult males: Adaptive benefits and determinants in wild baboons (Papio cynocephalus). Behavioral Ecology and Sociobiology, 63(9), 1331–1344. 10.1007/s00265-009-0786-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okello MM, Kenana L, Maliti H, Kiringe JW, Kanga E, Warinwa F, Bakari S, Ndambuki S, Massawe E, Sitati N, Kimutai D, Mwita M, Gichohi N, Muteti D, Ngoru B, & Mwangi P (2016). Population density of elephants and other key large herbivores in the Amboseli ecosystem of Kenya in relation to droughts. Journal of Arid Environments, 135, 64–74. 10.1016/j.jaridenv.2016.08.012 [DOI] [Google Scholar]
- Packer C (1979a). Inter-troop transfer and inbreeding avoidance in Papio anubis. Animal Behaviour, 27(1), 1–36. 10.1016/0003-3472(79)90126-x [DOI] [PubMed] [Google Scholar]
- Packer C (1979b). Male dominance and reproductive activity in Papio anubis. Animal Behaviour, 27(1), 37–45. 10.1016/0003-3472(79)90127-1 [DOI] [PubMed] [Google Scholar]
- Palombit RA, Cheney DL, & Seyfarth RM (2001). Female–female competition for male ‘friends’ in wild chacma baboons (Papio cynocephalus ursinus). Animal Behaviour, 61(6), 1159–1171. 10.1006/anbe.2000.1690 [DOI] [Google Scholar]
- Palombit RA, Seyfarth RM, & Cheney DL (1997). The adaptive value of ‘friendships’ to female baboons: Experimental and observational evidence. Animal Behaviour, 54(3), 599–614. 10.1006/anbe.1996.0457 [DOI] [PubMed] [Google Scholar]
- Pew J, Wang J, Muir P, & Frasier T (2015). Related: An R package for analyzing pairwise relatedness data based on codominant molecular markers (R package Version 1.0). https://github.com/timothyfrasier/related. [DOI] [PubMed]
- Pritchard JK, Stephens M, & Donnelly P (2000). Inference of population structure using multilocus genotype data. Genetics, 155(2), 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queller DC, & Goodnight KF (1989). Estimating relatedness using genetic-markers. Evolution, 43(2), 258–275. 10.2307/2409206 [DOI] [PubMed] [Google Scholar]
- R Core Team. (2019). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. https://www.R-project.org/. [Google Scholar]
- Ribble DO (1992). Lifetime reproductive success and its correlates in the monogamous rodent, Peromyscus californicus. Journal of Animal Ecology, 61(2), 457–468. 10.2307/5336 [DOI] [Google Scholar]
- Robinson JA, Belsare S, Birnbaum S, Newman DE, Chan J, Glenn JP, Ferguson B, Cox LA, & Wall JD (2019). Analysis of 100 high-coverage genomes from a pedigreed captive baboon colony. Genome Research, 29(5), 848–856. 10.1101/gr.247122.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez RL, Sullivan LE, & Cocroft RB (2004). Vibrational communication and reproductive isolation in the Enchenopa binotata species complex of treehoppers (Hemiptera: Membracidae). Evolution, 58(3), 571–578. 10.1111/j.0014-3820.2004.tb01679.x [DOI] [PubMed] [Google Scholar]
- Rogers J, Raveendran M, Harris RA, Mailund T, Leppala K, Athanasiadis G, Schierup MH, Cheng J, Munch K, Walker JA, Konkel MK, Jordan V, Steely CJ, Beckstrom TO, Bergey C, Burrell A, Schrempf D, Noll A, Kothe M, … Baboon Genome Analysis, Consortium. (2019). The comparative genomics and complex population history of Papio baboons. Science Advances, 5(1), Article eaau6947. 10.1126/sciadv.aau6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum S, Maldonado-Chaparro AA, & Stoinski TS (2016). Group structure predicts variation in proximity relationships between male–female and male–infant pairs of mountain gorillas (Gorilla beringei beringei). Primates, 57(1), 17–28. 10.1007/s10329-015-0490-2 [DOI] [PubMed] [Google Scholar]
- Sadino JM, & Donaldson ZR (2018). Prairie voles as a model for understanding the genetic and epigenetic regulation of attachment behaviors. ACS Chemical Neuroscience, 9(8), 1939–1950. 10.1021/acschemneuro.7b00475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels A, & Altmann J (1986). Immigration of a Papio anubis male into a group of Papio cynocephalus baboons and evidence for an anubis–cynocephalus hybrid zone in Amboseli, Kenya. International Journal of Primatology, 7(2), 131–138. [Google Scholar]
- Sánchez-Macouzet O, Rodríguez C, & Drummond H (2014). Better stay together: Pair bond duration increases individual fitness independent of age-related variation. Proceedings of the Royal Society B: Biological Sciences, 281(1786), 20132843. 10.1098/rspb.2013.2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapolsky RM, & Altmann J (1991). Incidence of hypercortisolism and dexamethasone resistance increases with age among wild baboons. Biological Psychiatry, 30(10), 1008–1016. 10.1016/0006-3223(91)90121-2 [DOI] [PubMed] [Google Scholar]
- Schülke O, Bhagavatula J, Vigilant L, & Ostner J (2010). Social bonds enhance reproductive success in male macaques. Current Biology, 20(24), 2207–2210. 10.1016/j.cub.2010.10.058 [DOI] [PubMed] [Google Scholar]
- Seyfarth RM (1976). Social relationships among adult female baboons. Animal Behaviour, 24(4), 917–938. 10.1016/s0003-3472(76)80022-x [DOI] [PubMed] [Google Scholar]
- Seyfarth RM (1978). Social relationships among adult male and female baboons. II. Behaviour throughout the female reproductive cycle. Behaviour, 64, 227–247. [Google Scholar]
- Seyfarth RM, Silk JB, & Cheney DL (2014). Social bonds in female baboons: The interaction between personality, kinship and rank. Animal Behaviour, 87, 23–29. 10.1016/j.anbehav.2013.10.008 [DOI] [Google Scholar]
- Silk JB, Alberts SC, & Altmann J (2006). Social relationships among adult female baboons (Papio cynocephalus) II. Variation in the quality and stability of social bonds. Behavioral Ecology and Sociobiology, 61(2), 197–204. 10.1007/s00265-006-0250-9 [DOI] [Google Scholar]
- Silk JB, Altmann J, & Alberts SC (2006). Social relationships among adult female baboons (Papio cynocephalus) I. Variation in the strength of social bonds. Behavioral Ecology and Sociobiology, 61(2), 183–195. 10.1007/s00265-006-0249-2 [DOI] [Google Scholar]
- Silk JB, Beehner JC, Bergman TJ, Crockford C, Engh AL, Moscovice LR, Wittig RM, Seyfarth RM, & Cheney DL (2009). The benefits of social capital: Close social bonds among female baboons enhance offspring survival. Proceedings of the Royal Society B: Biological Sciences, 276(1670), 3099–3104. 10.1098/rspb.2009.0681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silk JB, Beehner JC, Bergman TJ, Crockford C, Engh AL, Moscovice LR, et al. (2010). Strong and consistent social bonds enhance the longevity of female baboons. Current Biology, 20(15), 1359–1361. 10.1016/j.cub.2010.05.067 [DOI] [PubMed] [Google Scholar]
- Silk JB, Cheney D, & Seyfarth R (2013). A practical guide to the study of social relationships. Evolutionary Anthropology, 22(5), 213–225. 10.1002/evan.21367 [DOI] [PubMed] [Google Scholar]
- Silk JB, Städele V, Roberts EK, Vigilant L, & Strum SC (2020). Shifts in male reproductive tactics over the life course in a polygynandrous mammal. Current Biology, 30(9), 1716–1720.e3. 10.1016/j.cub.2020.02.013 [DOI] [PubMed] [Google Scholar]
- Smeltzer MD, Curtis JT, Aragona BJ, & Wang Z (2006). Dopamine, oxytocin, and vasopressin receptor binding in the medial prefrontal cortex of monogamous and promiscuous voles. Neuroscience Letters, 394(2), 146–151. 10.1016/j.neulet.2005.10.019 [DOI] [PubMed] [Google Scholar]
- Smith JE, Memenis SK, & Holekamp KE (2006). Rank-related partner choice in the fission–fusion society of the spotted hyena (Crocuta crocuta). Behavioral Ecology and Sociobiology, 61(5), 753–765. 10.1007/s00265-006-0305-y [DOI] [Google Scholar]
- Smuts BB (1985). Sex and friendship in baboons. New York, NY: Aldine. [Google Scholar]
- Städele V, Roberts ER, Barrett BJ, Strum SC, Vigilant L, & Silk JB (2019). Male–female relationships in olive baboons (Papio anubis): Parenting or mating effort? Journal of Human Evolution, 127, 81–92. 10.1016/j.jhevol.2018.09.003 [DOI] [PubMed] [Google Scholar]
- Tung J, Akinyi MY, Mutura S, Altmann J, Wray GA, & Alberts SC (2011). Allele-specific gene expression in a wild nonhuman primate population. Molecular Ecology, 20(4), 725–739. 10.1111/j.1365-294X.2010.04970.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung J, Charpentier MJ, Garfield DA, Altmann J, & Alberts SC (2008). Genetic evidence reveals temporal change in hybridization patterns in a wild baboon population. Molecular Ecology, 17(8), 1998–2011. 10.1111/j.1365-294X.2008.03723.x [DOI] [PubMed] [Google Scholar]
- Tung J, Charpentier MJ, Mukherjee S, Altmann J, & Alberts SC (2012). Genetic effects on mating success and partner choice in a social mammal. American Naturalist, 180(1), 113–129. 10.1086/665993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuqa JH, Funston P, Musyoki C, Ojwang GO, Gichuki NN, Bauer H, Tamis W, Dolrenry S, Van’t Zelfde M, de Snoo GR, & de Iongh HH (2014). Impact of severe climate variability on lion home range and movement patterns in the Amboseli ecosystem, Kenya. Global Ecology and Conservation, 2, 1–10. 10.1016/j.gecco.2014.07.006 [DOI] [Google Scholar]
- van Schaik CP, & Paul A (1996). Male care in primates: Does it ever reflect paternity? Evolutionary Anthropology, 5, 152–156. [Google Scholar]
- Vilgalys TP, Fogel AS, Anderson JA, Kim SY, Voyles TN, Robinson JA, Wall JD, Archie EA, Alberts SC, & Tung J (2021). Selection against admixture and gene regulatory divergence in a natural primate hybrid zone. Manuscript in preparation. [Google Scholar]
- Walker KK, Rudicell RS, Li Y, Hahn BH, Wroblewski E, & Pusey AE (2017). Chimpanzees breed with genetically dissimilar mates. Royal Society Open Science, 4(1), 160422. 10.1098/rsos.160422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Schlebusch SA, Alberts SC, Cox LA, Snyder-Mackler N, Nevonen KA, Carbone L, & Tung J (2016). Genomewide ancestry and divergence patterns from low-coverage sequencing data reveal a complex history of admixture in wild baboons. Molecular Ecology, 25(14), 3469–3483. 10.1111/mec.13684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J (2011). COANCESTRY: A program for simulating, estimating and analysing relatedness and inbreeding coefficients. Molecular Ecology Resources, 11(1), 141–145. 10.1111/j.1755-0998.2010.02885.x [DOI] [PubMed] [Google Scholar]
- Weidt A, Hofmann SE, & König B (2008). Not only mate choice matters: Fitness consequences of social partner choice in female house mice. Animal Behaviour, 75(3), 801–808. 10.1016/j.anbehav.2007.06.017 [DOI] [Google Scholar]
- Weingrill T (2000). Infanticide and the value of male–female relationships in mountain chacma baboons. Behaviour, 137(3), 337–359. 10.1163/156853900502114 [DOI] [Google Scholar]
- Weyher AH, Phillips-Conroy JE, Fourrier MS, & Jolly CJ (2014). Male-driven grooming bouts in mixed-sex dyads of Kinda baboons (Papio kindae). Folia Primatologica, 85(3), 178–191. 10.1159/000362544 [DOI] [PubMed] [Google Scholar]
- Whitten PL (1987). Infants and adult males. In Smuts BB, Cheney DL, Seyfarth RM, Wrangham RW, & Struhsaker TT (Eds.), Primate societies (pp. 343–357). Chicago, IL: The University of Chicago Press. [Google Scholar]
- Widdig A, Muniz L, Minkner M, Barth Y, Bley S, Ruiz-Lambides A, Junge O, Mundry R, & Kulik L (2017). Low incidence of inbreeding in a long-lived primate population isolated for 75 years. Behavioral Ecology and Sociobiology, 71(1), 18. 10.1007/s00265-016-2236-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widdig A, Nürnberg P, Krawczak M, Streich WJ, & Bercovitch FB (2001). Paternal relatedness and age proximity regulate social relationships among adult female rhesus macaques. Proceedings of the National Academy of Sciences of the United States of America, 98(24), 13769–13773. 10.1073/pnas.241210198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewski EE, Murray CM, Keele BF, Schumacher-Stankey JC, Hahn BH, & Pusey AE (2009). Male dominance rank and reproductive success in chimpanzees, Pan troglodytes schweinfurthii. Animal Behaviour, 77(4), 873–885. 10.1016/j.anbehav.2008.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RL, Ferkin MH, Ockendon-Powell NF, Orr VN, Phelps SM, Pogany A, Richards-Zawacki CL, Summers K, Szekely T, Trainor BC, Urrutia AO, Zachar G, O’Connell LA, & Hofmann HA (2019). Conserved transcriptomic profiles underpin monogamy across vertebrates. Proceedings of the National Academy of Sciences of the United States of America, 116(4), 1331–1336. 10.1073/pnas.1813775116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KA, Gobrogge KL, Liu Y, & Wang Z (2011). The neurobiology of pair bonding: Insights from a socially monogamous rodent. Frontiers in Neuroendocrinology, 32(1), 53–69. 10.1016/j.yfrne.2010.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LJ, Huot B, Nilsen R, Wang Z, & Insel TR (1996). Species differences in central oxytocin receptor gene expression: Comparative analysis of promoter sequences. Journal of Neuroendocrinology, 8(10), 777–783. 10.1046/j.1365-2826.1996.05188.x [DOI] [PubMed] [Google Scholar]
- Young LJ, Nilsen R, Waymire KG, MacGregor GR, & Insel TR (1999). Increased affiliative response to vasopressin in mice expressing the V1a receptor from a monogamous vole. Nature, 400(6746), 766–768. 10.1038/23475 [DOI] [PubMed] [Google Scholar]
- Young LJ, Waymire KG, Nilsen R, Macgregor GR, Wang Z, & Insel TR (1997). The 5’ flanking region of the monogamous prairie vole oxytocin receptor gene directs tissue-specific expression in transgenic mice. Annals of the New York Academy of Sciences, 807(1), 514–517. 10.1111/j.1749-6632.1997.tb51955.x [DOI] [PubMed] [Google Scholar]
- Young LJ, Winslow JT, Nilsen R, & Insel TR (1997). Species differences in V1a receptor gene expression in monogamous and nonmonogamous voles: Behavioral consequences. Behavioral Neuroscience, 111(3), 599–605. 10.1037//0735-7044.111.3.599 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data on grooming behaviour and proximity behaviour and all predictor variables tested in our models are available in the Duke Research Data Repository at https://doi.org/10.7924/r4kp82d1z. R code for recreating the analyses and figures in the manuscript is available at https://github.com/ArielleF/Genetic-Ancestry-Social-Affiliation.