DNA nanotubes can cross blood-brain tumor barrier and enhance survival with no toxicity when used for the local delivery of DOX.
Abstract
Effective treatment of glioblastoma remains a daunting challenge. One of the major hurdles in the development of therapeutics is their inability to cross the blood-brain tumor barrier (BBTB). Local delivery is an alternative approach that can still suffer from toxicity in the absence of target selectivity. Here, we show that nanotubes formed from self-assembly of ssDNA-amphiphiles are stable in serum and nucleases. After bilateral brain injections, nanotubes show preferential retention by tumors compared to normal brain and are taken up by glioblastoma cells through scavenger receptor binding and macropinocytosis. After intravenous injection, they cross the BBTB and internalize in glioblastoma cells. In a minimal residual disease model, local delivery of doxorubicin showed signs of toxicity in the spleen and liver. In contrast, delivery of doxorubicin by the nanotubes resulted in no systemic toxicity and enhanced mouse survival. Our results demonstrate that ssDNA nanotubes are a promising drug delivery vehicle to glioblastoma.
INTRODUCTION
Glioblastoma (GBM) is the most malignant and aggressive type of primary brain tumor. Despite multimodal treatment through tumor resection, radiation, and chemotherapy, long-term survival remains low with a high rate of recurrence, a median survival of 15 to 18 months, and less than 5% of patients estimated to be alive 5 years after diagnosis (1, 2). Therefore, novel therapies for the treatment of GBM are desperately needed. One of the major hurdles in the development of therapeutics is the inability of agents to cross the blood-brain barrier (BBB). Although the BBB is disrupted during tumor progression in high-grade gliomas [known as the blood-brain tumor barrier (BBTB)], its permeability is heterogeneous in GBM, leading to inconsistent responses to systemically delivered therapeutics (3, 4).
Delivery of anticancer therapies to GBM using nanoparticles has shown promise (5–8); however, nanoparticle accumulation in GBM tumors after systemic administration remains very low, limiting their clinical applicability (9, 10). Thus, other strategies have been investigated to enhance the delivery to GBM using methods that do not rely on the circulatory system and circumvent the BBB/BBTB, such as BBB disruption, intranasal delivery, direct injections of therapeutics, convection-enhanced delivery, and use of implantable controlled-release polymer systems (11, 12). Local delivery to the brain limits the potential for neurotoxicity as delivered doses do not need to be as high as those needed for systemic delivery. However, intratumoral delivery of drugs such as doxorubicin (DOX) can still result in systemic toxicity (13). Therefore, even with local intratumoral delivery of chemotherapeutics, there is a critical need to design targeting strategies that localize therapeutics to specific cell types (14, 15).
DNA nanotechnology is a growing field that allows for the design of nanoparticles where DNA is the building block. DNA origami (16) and DNA tile assembly (17) are two methods that allow for the assembly of specific nanoparticle structures composed solely of DNA molecules, and both techniques have been used to create DNA nanotubes (NT) that can be delivered to cells in vitro and in vivo (18–22). It was shown, for example, that NT made from tile assembly are internalized by immune cells, human cervical cancer cells, and nasopharyngeal epidermal carcinoma cells when functionalized with either CpG sequences or folic acid (18–20). However, their clinical applicability has been limited because of processing complexities and poor in vivo stability (23). DNA origami and DNA tile assembly require precise ions, solvent, and thermal conditions to allow for DNA annealing and assembly, and often, computer-aided design is required to determine the DNA sequences needed to achieve certain structures. DNA NT made of DNA tiles also degrade in the presence of serum and endonucleases (18, 19).
We previously reported on an alternative approach to forming DNA NT through the spontaneous self-assembly of single-stranded DNA (ssDNA)–amphiphiles (24, 25). The amphiphiles consist of three building blocks: an ssDNA hydrophilic headgroup, a dialkyl hydrophobic tail, and an alkyl spacer between the headgroup and tail. The ssDNA-amphiphiles self-assemble in aqueous solution into ellipsoidal micelles and bilayer nanotapes that can progress into hollow NT (24). We have previously demonstrated that the architecture of the ssDNA-amphiphiles can be used to manage the dimensions of the NT, where the NT diameter can be controlled by the hydrophobic tail length, and the presence of intermolecular G-quadruplexes in the ssDNA headgroup dictates the NT length (25). As the dialkyl tail length increases from (C16)2 to (C20)2, the diameter of the NT increases from approximately 30 to 55 nm (25). Coarse-grained molecular dynamics simulations revealed an increase in area per molecule within the bilayer as tail length increases, resulting in wider nanotapes that could transition into NT with larger diameters. ssDNA-amphiphiles with guanine (G)–rich sequences that form G-quadruplexes can assemble into ellipsoidal micelles and short NT with submicrometer lengths (25). Sequences with a total of 10, 25, and 40 nucleotides (nt) were examined (25). Ellipsoidal micelles are the first assembled nanostructures from the ssDNA-amphiphiles (24). Therefore, the micelles have to release the amphiphiles that can attach to the nanotapes and allow for the nanotape growth. However, micelles with G-rich sequences, where ssDNA headgroups can form intermolecular stacked G-tetrad planes, are more stable (26). Thus, we hypothesized that the amphiphiles may be less prone to leaving the micelles and assembling in the nanotapes, which may explain the formation of shorter NT (25). The short NT are stable in different electrolytes (KCl, NaCl, CaCl2, and MgCl2) or buffers [acetate buffer (pH 5.0) and carbonate-bicarbonate buffer (pH 9.0)] after overnight exposure (25). In this study, we synthesized ssDNA-amphiphiles that can form short NT, consisting of a 10-nt G-rich ssDNA headgroup that is attached to a (C16)2 tail through a C12 spacer. An ssDNA with 10-nt is used as it is the smallest G-rich sequence that was tested and shown to form short NT (25).
We evaluated the internalization of the NT in GL261 cells and normal astrocytes and investigated their uptake mechanism via trafficking and colocalization experiments. For the in vivo studies, we examined their stability in serum and nucleases via gel electrophoresis and transmission electron microscopy (TEM). Fluorescence and confocal microscopy were used to evaluate the presence of NT in tumoral and normal brain hemispheres after bilateral brain injections. Their biodistribution after tail vein injection was evaluated via micropositron emission tomography/computed tomography (μPET/CT) and ex vivo measurements of radioactivity. The ability of the NT to cross the BBTB was further investigated via confocal microscopy and flow cytometry. Last, in a minimal residual disease mouse model of orthotopic GBM, we delivered via Alzet micro-osmotic pumps DOX and DOX intercalated in the NT, and we measured survival by Kaplan-Meier plots and systemic toxicity via histopathological analysis. The findings of this work suggest that the ssDNA NT are a promising approach to targeting GBM and can be used for the delivery of therapeutics, such as DOX, with minimal to no systemic toxicity.
RESULTS AND DISCUSSION
Synthesis and characterization of ssDNA-amphiphile NTs
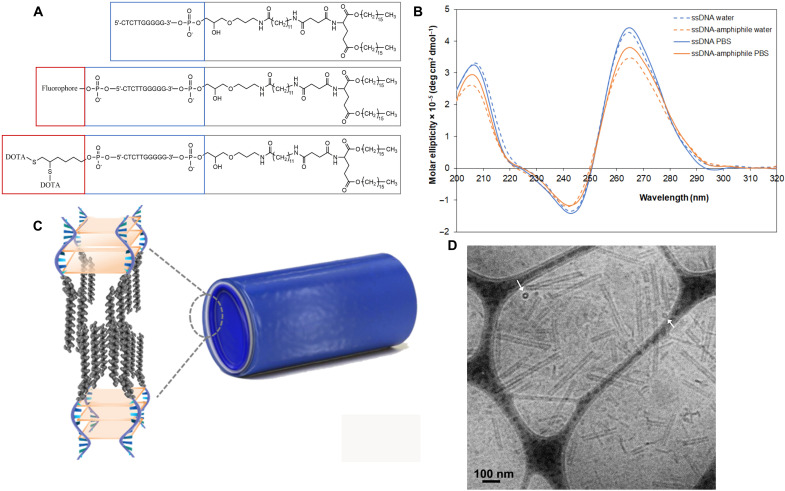
Synthesis steps for all ssDNA-amphiphiles are shown in fig. S1. Successful synthesis was verified using liquid chromatography–mass spectrometry (LC-MS) (table S1). Figure 1A shows the chemical structure of the 10-nt ssDNA-amphiphile, with and without a fluorophore and chelating agent. The secondary structure of the free ssDNA and ssDNA-amphiphiles was investigated by circular dichroism (CD). Parallel stranded G-quadruplex structures have a CD spectrum exhibiting strong positive peaks around 260 to 265 nm and 210 nm and a smaller negative peak around 245 nm (27, 28). They are stabilized by small cations that fit within the G-quartet structure but can also be formed in pure water (24, 29). The CD spectra of the free 10-nt ssDNA and ssDNA-amphiphiles in Milli-Q water and phosphate-buffered saline (PBS) have maxima at 206 and 265 nm and a minimum at 243 nm (Fig. 1B), which is characteristic of a parallel G-quadruplex structure. Conjugation of the 10-nt ssDNA to a hydrophobic tail and self-assembly did not alter its secondary structure. The 10-nt ssDNA-amphiphiles form weakly ellipsoidal micelles and hollow NT in water (Fig. 1C), as demonstrated before via cryogenic TEM (cryo-TEM) and small angle x-ray scattering (24, 25). In this study, we separated the NT from the micelles using size exclusion chromatography. The NT were 238 ± 122 nm long, with a diameter of 35 ± 4 nm and a bilayer wall thickness of 8 ± 2 nm (n = 100), as evaluated by cryo-TEM images (Fig. 1D). The white arrows in Fig. 1D point to short NT that are viewed end-on, demonstrating the hollow nature of the ssDNA-amphiphile NT, as previously shown (24, 25).
Fig. 1. Ten nucleotide ssDNA-amphiphiles form parallel G-quadruplexes and self-assemble into hollow nanotubes.
(A) Chemical structures of the 10-nt ssDNA-amphiphiles. (B) CD spectra in Milli-Q water and PBS of the free 10-nt ssDNA and the ssDNA-amphiphiles. (C) Schematic representation of the nanotubes and the ssDNA-amphiphiles in the nanotubes, not drawn to scale. ssDNA is shown in blue, and the hydrophobic tails are shown in gray. The light orange represents G-quartet planes. (D) Cryo-TEM image of ssDNA nanotubes. The white arrows point to nanotubes that are viewed end-on, demonstrating their hollow nature.
Preferential GL261 uptake of ssDNA NTs via scavenger receptors and macropinocytosis
We studied the interaction between the 10-nt ssDNA-amphiphile NT with GL261 mouse GBM cells and C8-D1A healthy mouse astrocytes. NT labeled with the fluorophore hexachlorofluorescein (HEX), containing 20 mole percent (mol %) HEX-labeled ssDNA-amphiphiles, were incubated with the cells for 24 hours at 37°C, and confocal microscopy was used to qualitatively determine the extent of cell internalization [Fig. 2A and movies S1 and S2 with three-dimensional (3D) reconstructions of 2D confocal image stacks]. After incubation for 24 hours, the NT showed strong internalization into the GL261 cells, but only showed minimal surface binding and no internalization into the C8-D1A cells. Internalization for the NT in GL261 cells after 3-hour incubation at 37°C was also verified (fig. S2).
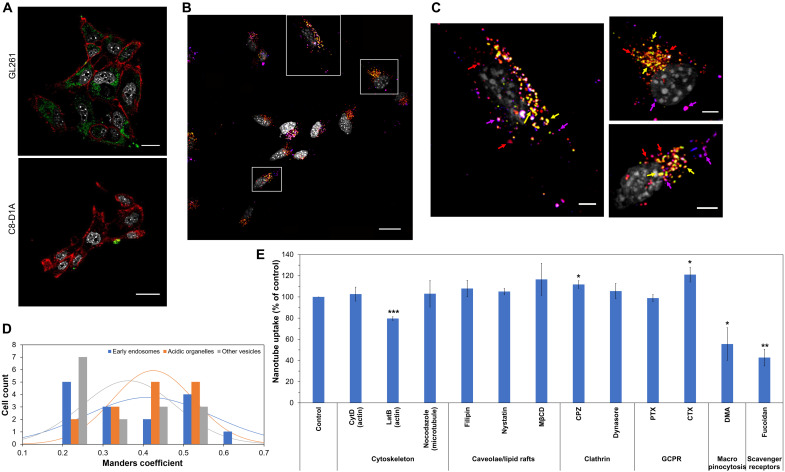
Fig. 2. Preferential uptake of nanotubes by GL261 GBM cells via the macropinocytosis pathway and scavenger receptors.
(A) Confocal microscopy of HEX-labeled nanotubes (green) after incubation with GL261 GBM cells and C8-D1A normal astrocytes for 24 hours at 37°C. Nuclei are shown in gray and cell membranes in red. Scale bars, 20 μm. (B) Maximum-intensity projection of a confocal microscopy image of HEX-labeled nanotubes (red) after incubation with GL261 cells for 24 hours at 37°C. Nuclei are shown in gray, early endosomes are shown in blue, and acidic organelles are shown in green. Colocalization of nanotubes with early endosomes is shown in magenta and with acidic organelles in yellow. Scale bar, 20 μm. (C) Zoom-in images of the square areas shown in (B). Magenta, yellow, and red arrows point to select areas of nanotube colocalization with early endosomes, acidic organelles, and other vesicles that were not labeled, respectively. Scale bars, 5 μm. (D) Manders coefficient values quantify the different levels of colocalization between nanotubes and early endosomes, acidic organelles, and vesicles not labeled (n = 15). (E) GL261 association of nanotubes in the presence of different endocytosis inhibitors. Data are shown as means ± SD (n = 3). Statistical significance with that of the control (incubation with nanotubes in the absence of inhibitors) was determined using a two-sided unpaired t test; *P < 0.05, **P < 0.01, and ***P < 0.005. There was no significant statistical difference for all other treatments (P > 0.05).
The intracellular fate of the NT was further determined by incubating GL261 cells with the NT for 24 hours and staining for early endosomes (shown in blue) and acidic organelles, such as late endosomes and lysosomes (shown in green). Results showed that after 24 hours, the NT (shown in red) were colocalized with early endosomes and acidic organelles, as indicated by the magenta and yellow color observed in the images, respectively (Fig. 2, B and C, and movie S3 with 3D reconstructions of 2D confocal image stacks). In addition, the NT (red dots) were also located in the cytosol, not associated with early endosomes or acidic organelles, suggesting that the NT may also be localized in nonacidic or moderately acidic vesicles. Calculation of the Manders coefficient (Fig. 2D) verified these observations. The Manders coefficient is a fraction between 0 and 1 that quantifies the overlap of two signals in an image (30). The closer the coefficient is to 1, the stronger the colocalization between the two signals. Here, the Manders coefficients were calculated with respect to the nanoparticle signal, showing what percentage of the nanoparticle signal overlapped with either the early endosomes, or the acidic organelles, or the vesicles that were not labeled.
To reveal the internalization mechanism of the NT, an endocytosis inhibition experiment was performed where the uptake of the ssDNA NT was evaluated in GL261 cells in the presence of different endocytosis inhibitors that did not induce any toxicity to the cells (Fig. 2E and fig. S3). The inhibitors used can be classified into six major groups based on their effects in cells: cytoskeleton [cytochalasin D (CytD) disrupts actin microfilaments, latrunculin B (LatB) inhibits actin polymerization, and nocodazole disrupts microtubule assembly], caveolae/lipid rafts [filipin, nystatin, and methyl-β-cyclodextrin (MβCD) inhibit caveolae and lipid raft internalization through depletion of cholesterol from the cell membrane by forming inclusion complexes with cholesterol], clathrin [chlorpromazine (CPZ) prevents the assembly and disassembly of clathrin lattices on cell surfaces, and dynasore is an inhibitor of dynamin that participates in clathrin-mediated endocytosis], G protein–coupled receptors (GPCR) [pertussis toxin (PTX) is an inhibitor of Gαi protein, and cholera toxin (CTX) is an inhibitor of Gαt/s proteins], macropinocytosis [5-(N,N-dimethyl)-amiloride hydrochloride (DMA) inhibits Na+/H+ exchanger activity], and scavenger receptors (fucoidan is a polysaccharide that binds to various types of scavenger receptors) (31, 32). The cellular uptake of the NT was inhibited on average by 44% in the presence of DMA and 57% in the presence of fucoidan. In addition, treatment with LatB decreased the internalization of the ssDNA NT by 20%, consistent with the finding that macropinocytosis is involved in the internalization of the NT, because macropinocytosis depends on actin polymerization. In contrast, neither caveolae/lipid rafts, nor clathrin, nor GPCR-associated pathway inhibitors decreased NT internalization. Scavenger receptors are known to bind a wide variety of negatively charged macromolecules and to induce their internalization using different endocytic pathways (33). Class A scavenger receptors have been shown to uptake spherical polyvalent oligonucleotide-functionalized gold nanoparticles via a lipid raft–dependent, caveolae-mediated pathway, where, after 24 hours, the nanoparticles showed moderate colocalization with early endosomes, strong colocalization with late endosomes, and no appreciable colocalization with lysosomes (34–36). Class A scavenger receptors have also been implicated in the uptake of nanocomplexes composed of the cell-penetrating peptide PepFect14 and splice-correcting oligonucleotides through macropinocytosis and caveolin-dependent endocytosis, where, after 4 hours, complexes resided mainly in neutral or slightly acidic vesicles (37). Numerous studies have demonstrated elevated expression of scavenger receptors in different cancers, including glioma (38), thus potentially explaining the enhanced internalization in GL261 cells compared to C8-D1A cells. Macropinocytosis forms large macropinosomes (0.2 to 5 μm) that can internalize our NT, is highly up-regulated in cancer cells compared to normal cells, and has been shown to function as a unique mechanism for transporting extracellular proteins as a form of extracellular energy into cancer cells such as GBM (39). Our results show that the ssDNA NT do not internalize into C8-D1A normal astrocytes and further suggest that they bind to scavenger receptors on GL261 cells and internalize through macropinocytosis, thus providing a strategy to target the macropinocytosis pathway and provide a therapeutic approach to treat GBM.
NT stability and in vivo tumor targeting
A common limitation of ssDNA-based nanoparticles is their stability when delivered in vivo (23). The main degradation pathways are through desorption of ssDNA by serum proteins and degradation by nucleases, where direct cleavage of ssDNA at an internal site by endonucleases or removal of nucleotides at the terminus by exonucleases is a possibility (40). The stability of the NT in different serum and nuclease concentrations was investigated using gel electrophoresis and TEM. The NT were exposed to PBS, 10 and 50% (v/v) fetal bovine serum (FBS) in PBS (Fig. 3A). After incubation at 37°C for 24 hours, it was found that there was no change in the electrophoretic mobility of the NT when incubated with 10% FBS, suggesting no change to the NT structure. At 50% FBS concentration, there was band smear at a higher molecular weight. The decrease in the electrophoretic mobility of the NT suggests a possible adsorption of serum proteins onto the surface of the NT. However, no degradation products were observed after incubation with any serum solutions as indicated by the lack of distinct bands with higher mobility than the control sample. The NT were also tested for their stability after exposure to varying concentrations of endonuclease deoxyribonuclease I (DNase I) and exonuclease III. (Fig. 3A). After incubation with nuclease concentrations between 0 and 5 U/ml for 24 hours at 37°C, it was found that there was no degradation of the ssDNA NT when exposed to either DNase I or exonuclease III. The NT were also stable after exposure to T5 exonuclease at 5 U/ml for 24 hours at 37°C (Fig. 3A). The presence of NT was also verified via TEM after their exposure to 50% FBS, DNase I (5 U/ml), exonuclease III (5 U/ml), and T5 exonuclease (5 U/ml) (fig. S4). The stability of the ssDNA NT in different conditions is therefore promising for their in vivo use. The average concentration of circulating DNase I in healthy human patients is 0.356 ± 0.410 U/ml, while the circulating concentration of DNase I in human patients with GBM is 0.045 ± 0.007 U/ml (41, 42). Therefore, even at much higher than physiologically relevant concentrations of DNase, the ssDNA-amphiphile NT show no degradation. No degradation was observed for the exonuclease III, as the 3′ terminus of the amphiphiles is conjugated to the dialkyl tail, preventing exonuclease III from binding to the amphiphile. It has been shown that 5′ exonucleases from serum do not play a major role in DNA breakdown even when the 5′ terminus is exposed (40), and our results verified that the NT were stable in the presence of T5 exonuclease. The high local concentration of the ssDNA headgroups in the self-assembled NT likely prevents interaction with nucleases, thus preventing degradation by internal or external cleavage, in agreement with previous work that showed steric hindrance through dense ssDNA packing in a micelle corona limits accessibility of nucleases (43, 44). The stability of our ssDNA-amphiphile NT is in sharp contrast to other DNA nanostructures formed through tiles that were stable for up to 8 hours in cell medium without serum at 37°C, but complete degradation was observed after 2-hour incubation at 37°C in pure serum (18, 19). In DNase I (0.5 U/ml) at 37°C, no degradation of these tubes formed from DNA tiles was observed for 2 hours, while at higher DNase I concentrations, the tubes survived for up to 15 min (18).
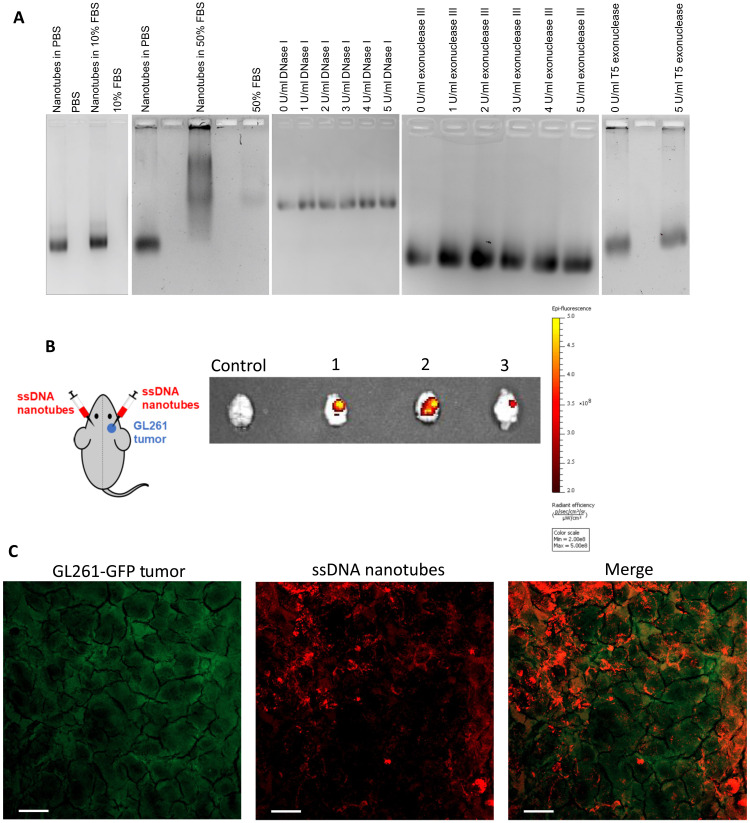
Fig. 3. Nanotube stability and preferential uptake by the tumor.
(A) Stability of nanotubes in different concentrations of serum, DNase I, exonuclease III, and T5 exonuclease. (B) Schematic of bilateral intracranial injections of nanotubes to a mouse with GL261 tumor only on the right hemisphere of the brain. NIR fluorescence images of mouse brains excised at different time points: mouse 1 at 45 min, mouse 2 at 70 min, and mouse 3 at 105 min. Control mouse had a GL261 tumor but did not receive intracranial nanotube injections. The radiant efficiency of NIR fluorescently labeled nanotubes is shown with a heat map. (C) Mice bearing orthotopic tumors of GL261 cells expressing GFP (GL261-GFP) were intracranially injected with HEX-labeled nanotubes. Mice were euthanized 3 hours after nanotube injection, and tumor tissues were removed and processed. Maximum-intensity projection of a confocal microscopy image shows colocalization of GL261 cells (green) with the nanotubes (red). Scale bars, 20 μm.
After observing strong cell internalization of the NT after incubation with GL261 cells in vitro and verifying their stability, the retention of NT in a more clinically applicable system was tested by direct intracranial injection of NT into an orthotopic GBM mouse model. GL261 tumors were grown in the right hemisphere of mouse brains, and IRDye 800CW–labeled NT were intracranially injected into both hemispheres (Fig. 3B). The mice were euthanized at different time points, and their brains were excised and imaged for near-infrared (NIR) fluorescence. A GL261 tumor–bearing mouse did not receive any NT injections as a control. The average ratio of radiant efficiency between the tumor-bearing hemisphere and healthy hemisphere, after both hemispheres received NT injections, was 2.25 ± 0.07 (n = 3), while the ratio for the control mouse was 1.02. Observed differences in NIR fluorescence between healthy and tumor-bearing hemispheres were a result of differential retention of the NT by the two regions despite both sides receiving an equivalent volume of IRDye 800CW–labeled NT. Additional imaging of brain slices showed that this observation was consistent throughout different brain sections (fig. S5), indicating that only the tumor-bearing hemisphere retained the ssDNA-amphiphile NT. To examine the type of cells that were targeted by the ssDNA NT in the tumor, fluorescently labeled NT were injected intracranially into mice bearing GL261 tumors, where the GL261 cells were either unlabeled or expressed green fluorescent protein (GL261-GFP). Results showed that 3 hours after NT injection, the NT were internalized by GL261 cells (Fig. 3C) and tumor-associated macrophages (TAMs) (fig. S6).
Biodistribution of systemically delivered NT in orthotopic GL261 tumor–bearing mice
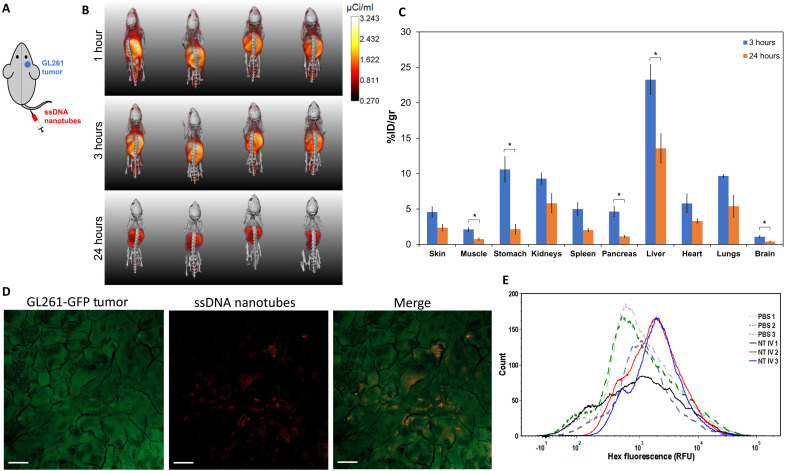
NT labeled with 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA) were used to chelate 64Cu for the biodistribution experiments. DOTA labeling has been shown to result in minimal changes to the size and surface charge of nanoparticles (45, 46). TEM verified the presence of DOTA-labeled NT (fig. S7). The DOTA-labeled amphiphiles were present in approximately 1850× molar excess of 64Cu, ensuring that all available copper was chelated by the NT (44). Mice bearing orthotopic GL261 tumors on the right hemisphere were injected with NT through their lateral tail vein (Fig. 4A) and imaged with μPET/CT at 1, 3, and 24 hours after injection (Fig. 4B). Figure 4B shows that the liver is the organ with the highest radioactivity at every time point, and some activity is also shown on the head of the mice. To accurately evaluate biodistribution of the NT, mice were euthanized at either 3 or 24 hours. Excised organs were weighed and measured for radioactivity to evaluate NT biodistribution at each time point. The activity of each organ was adjusted for the half-life decay of 64Cu and expressed as percentage of injected dose per gram of tissue (%ID/g), shown in Fig. 4C. All organs measured showed a decrease in radioactivity between 3 and 24 hours (Fig. 4C), and the organ with the highest accumulation at both time points was the liver, consistent with the blood clearance profiles of many types of nanoparticles (47). Brain accumulation at 3 hours was 1.08 ± 0.16 %ID/g and at 24 hours was 0.40 ± 0.07 %ID/g. This brain accumulation is higher than literature reports of other negatively charged nanoparticles, such as liposomes and spherical micelles, where tumor accumulation after 24 hours varied from 0.04 to 0.14 %ID/g in orthotopic GBM animal models (48, 49).
Fig. 4. Nanotube biodistribution and BBTB crossing.
(A) Schematic of intravenous injection of nanotubes to a mouse with GL261 tumor on the right hemisphere of the brain. (B) Full body volumetric 3D reconstruction of μPET/CT mice imaged at 1, 3, and 24 hours after intravenous injection of 64Cu-radiolabeled nanotubes. The μPET intensity scale bar for all images has units of microcuries per milliliter, and the intensity of the μPET signal was not adjusted for the half-life of 64Cu. (C) Ex vivo biodistribution of 64Cu-labeled nanotubes at 3 or 24 hours after intravenous injection to mice bearing GL261 orthotopic tumors. Radioactivity and weights of different organs were measured, and data were adjusted for the 12.7 hours half-life of 64Cu and plotted as means ± SEM (n = 3 to 4). Statistical significance was determined using a two-sided unpaired t test; *P < 0.05. There was no significant statistical difference for pairs without brackets (P > 0.05). (D and E) Mice bearing orthotopic tumors of GL261 cells expressing GFP (GL261-GFP) were injected intravenously with HEX-labeled nanotubes. Mice were euthanized 6 hours after nanotube injection, and tumor tissues were removed and processed for confocal microscopy (D) and flow cytometry (E). (D) Maximum-intensity projection of a confocal microscopy image showing colocalization of GL261-GFP cells (green) with the HEX-labeled nanotubes (red). Scale bars, 20 μm. (E) Flow cytometry plots from GL261-GFP tumors from mice that were intravenously injected with PBS or HEX-labeled nanotubes (NT IV).
To evaluate whether the NT were on the tumor-bearing hemisphere after systemic administration, maximum-intensity projections of the heads of each mouse from the μPET/CT images at 1-, 3-, and 24-hour time points were examined (fig. S8A). These images are tail view projections, viewing the head of the mouse looking from the tail, through the head, and out through the nose of the mouse. μPET profile intensity plots relative to the left edge of the mouse cranium suggest that there may be preferential NT accumulation in the tumor-bearing right hemisphere (fig. S8B). To further elucidate whether the NT could cross the BBTB, the colocalization of GL261 cells and HEX-labeled NT was evaluated after intravenous injection of the NT into mice bearing orthotopic tumors of GL261 cells expressing GFP (GL261-GFP). Tumor tissues were removed 6 hours after NT injection and evaluated via confocal microscopy and flow cytometry. Confocal images (Fig. 4D) demonstrated that the NT were internalized by the GL261-GFP cells. In addition, NT associated with the GL261-GFP cells had a mean fluorescence intensity increase of 2.6-fold compared to control mice that did not receive NT injections, as evaluated via flow cytometry (Fig. 4E). Together, these data suggest that the NT can cross the BBTB after intravenous injection and associate with the GBM cells.
Treatment of GBM with NT intercalating DOX
We examined the ability of the NT to deliver a therapeutic load, such as DOX. DOX has been shown to intercalate into the double-stranded region of ssDNA stem-loop or G-quadruplex structures, thus forming physical complexes with the ssDNA sequences through noncovalent intercalations (50, 51). The retention of DOX by the NT was investigated by dialyzing a sample of the NT that intercalated DOX against PBS at 37°C for 6 weeks. There is an initial burst release in the first day, where 37 ± 1% of DOX has been released, followed by a slower sustained release, with 56 ± 4% of DOX released after 1 week, and 72 ± 2% after 6 weeks (fig. S9). The effect of NT, DOX, and DOX intercalated in the NT (NT-DOX) on the viability of the GL261 cells was also assessed (fig. S10). The empty ssDNA NT were shown to have no effect on cell viability. However, there was a significant improvement in GBM cell killing activity through delivery of NT-DOX (31 ± 6%) compared to free DOX (47 ± 6%).
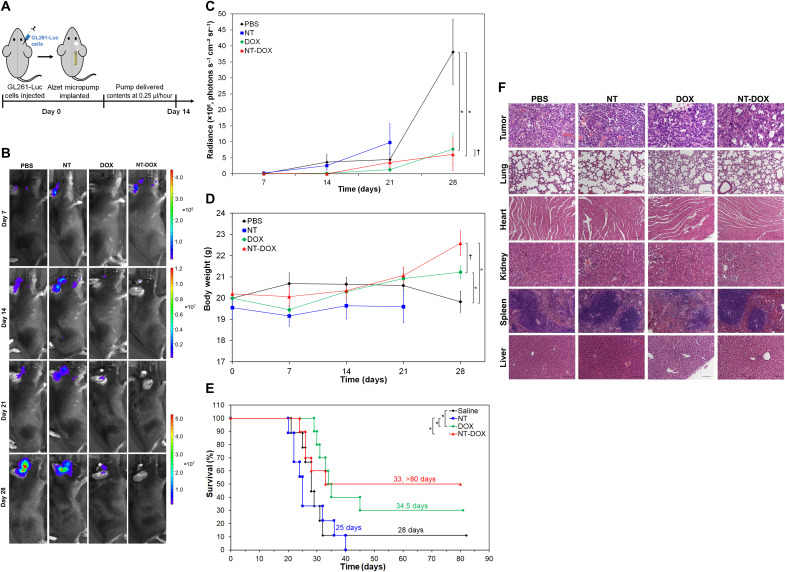
On the basis of the in vitro results, we wanted to further evaluate the NT as a delivery vehicle for DOX in a minimal residual disease model of orthotopic GBM. On the day of the surgery, 3 × 104 GL261-Luc cells (GL261 cells expressing luciferase) were injected to the right side of the brain to simulate remaining residual tumor cells after tumor resection. Immediately following intracranial injection of the cells, the micro-osmotic Alzet pump was implanted subcutaneously, and the cannula, connected to the pump through a catheter, was lowered into the brain though the same burr hole used to inject the cells (Fig. 5A). The pump delivered different treatments over the course of 14 days at a rate of 0.25 μl/hour. The Alzet pumps were loaded with either PBS, 70 μM DOX (0.2 mg DOX/kg mouse), NT at 95 μM total ssDNA-amphiphiles, or NT-DOX at the same concentrations of DOX and amphiphiles. Mice were imaged weekly for 4 weeks (Fig. 5B). After 28 days from the day of surgery, analysis of the bioluminescence signal from the brain showed that the mice that received PBS had a significant increase in the size of the tumor compared to mice that received DOX or NT-DOX (Fig. 5C). Mice treated with DOX or NT-DOX demonstrated an 80 and 84% decrease in tumor signal, respectively, compared to the PBS control group. There was no significant statistical difference in the tumor signal between the mice that received DOX and NT-DOX. We also measured the changes in body weight for all groups (Fig. 5D) and found that mice treated with PBS lost on average 1% body weight after 28 days, whereas mice treated with DOX gained 6% body weight, and mice treated with NT-DOX gained on average 12% body weight during the same time. The effective inhibition of tumor growth by the DOX and NT-DOX treatments correlated with an increase in animal survival (Fig. 5E). The median survival time of mice was 28 days for the PBS group. The median survival time of mice that were treated with NT was 25 days. In contrast, mice that received free DOX for 14 days had a significant increase in their median survival (34.5 days, 3 of 10 mice survived for >80 days). For the group that received NT-DOX for 14 days, the survival curve is horizontal at 50% survival. The median survival time was 33 days, and 5 of the 10 mice survived for >80 days. While survival differences between the NT-DOX and DOX groups were not statistically significant due to the size of the groups, the increase in survival for the NT-DOX group is notable. The NT-DOX treatment, where half of the mice survived for >80 days, also shows an advantage compared to other local treatments. For example, intracranial delivery of DOX loaded in mesoporous silica nanoparticles 3 days after GBM cell implantation resulted in a median survival of 36.5 days with no survivors after 40 days (52). Similarly, intracranial delivery of DOX through a biodegradable polymer wafer that was implanted 5 days after GBM cell implantation resulted in a median survival of 34 or 45 days, depending on DOX loading, with no survivors after 45 days (53).
Fig. 5. Local brain delivery of DOX and nanotubes intercalating DOX.
(A) Preparation and treatment schedule of mice. The right side of the brain was injected with 3 × 104 GL261-Luc cells on day 0. Immediately after that, a micro-osmotic Alzet pump was implanted subcutaneously, and the cannula, connected to the pump through a catheter, was lowered into the same burr hole used to inject the cells. The pumps were loaded with either PBS, 70 μM DOX (0.2 mg DOX/kg mouse), nanotubes (NT) at 95 μM ssDNA-amphiphiles, or NT-DOX at the same concentrations of DOX and amphiphiles and delivered their content in about 14 days at a pumping rate of 0.25 μl/hour. (B) Representative bioluminescence images of mice at different time points. Scale bars are shown on the side. (C) Quantification of tumor bioluminescence values in the different treatment groups over time. (D) Body weight of mice in different groups during treatment. In (C) and (D), data are shown as means ± SEM (n = 9 to 10). For the NT group, data are not reported on day 28 as only three mice were alive. Statistical significance on day 28 was determined using a two-sided unpaired t test; †P > 0.05 and *P < 0.05. (E) Survival curves corresponding to the different treatment groups (n = 9 to 10). Statistical significance was determined using a two-sided log-rank test; *P < 0.05. There was no significant statistical difference for pairs without brackets (P > 0.05). (F) Representative images of hematoxylin and eosin staining of tumors and other organs from mice that received different treatments. Images were taken with 20× objective lens. Scale bars, 100 μm.
The advantage of the NT-DOX group compared to DOX was shown after histological examination of different organs. Histological analysis of the brain, liver, spleen, lungs, kidneys, and heart tissues of mice was performed at the end of the experiment. From the mice that were examined and were long-term survivors at the end of the experiment on day 82, no tumors were observed in their brains (fig. S11A). For the rest of the mice, tumors showed several histological features characteristic of high-grade gliomas, including infiltrative growth and brain invasion, focal necrosis (ranging from <5 to 10%), multinucleation, marked cell anaplasia with high nuclear-to-cytoplasmic ratio, and frequent abnormal mitoses (Fig. 5F and fig. S11B). There were few and inconsistent differences between the morphology of the tumors in the different groups. Compared to the PBS control, lung, kidney, and heart tissues of mice treated with DOX, NT, or NT-DOX showed no significant findings (Fig. 5F). However, the spleen of mice treated with DOX showed diffuse, moderate to marked depletion of the spleen white pulp associated with zonal lymphocytic apoptosis and necrosis. Moderate, diffuse excessive accumulation of iron deposits (hemosiderosis) was observed in the red pulp (Fig. 5F). In addition, their liver exhibited multifocal areas of hepatocyte degeneration and necrosis occasionally associated with neutrophilic and lymphohistiocytic infiltrate, multifocal individual hepatocyte necrosis/apoptosis, and diffuse glycogen depletion (Fig. 5F). DOX has been shown to trigger splenic marginal depletion of the spleen white pulp (54, 55) and liver focal necrosis (56, 57). In contrast, there were no significant findings in spleen and liver tissues of mice treated with NT or NT-DOX, thus demonstrating no microscopic toxicity to mice. Therefore, selective delivery of DOX with ssDNA NT is effective in limiting systemic toxicity associated with DOX, a property that can be advantageous in cases where repeated DOX administration is necessary.
Moving forward, further studies are needed to evaluate treatment of GBM and systemic toxicity after administration of NT-DOX and DOX via tail vein injection. When DOX, either free or encapsulated in nanoparticles, is administered via tail vein injection to mice and rats with orthotopic GBM tumors, typical DOX doses vary from 1.5 mg/kg every 3 to 4 days to 5 mg/kg once weekly (58–60). Considering that nanoparticle accumulation 24 hours after injection varies from 0.04 to 0.14 %ID/g brain in orthotopic GBM animal models (48, 49), a weekly 5 mg DOX/kg mouse injection will result in a maximum accumulation of 140 ng DOX/g brain 24 hours after injection. In sharp contrast, a weekly DOX injection of 5 mg DOX/kg mouse via our NT-DOX, with an accumulation of 0.40 ± 0.07 %ID/g brain 24 hours after injection, will result on average in 400 ng DOX/g brain, with potentially limited systemic toxicity from DOX.
We have previously demonstrated that amphiphiles with G-rich ssDNA sequences can form short NT, whereas in the absence of guanines, the NT are micrometers in length (25). In addition, changes in the length of the hydrophobic tail result in changes in the diameter of the NT (25). Therefore, future studies can examine the effect of NT length and diameter in the delivery of different drugs to GBM and other cancers. Moreover, investigations on the state of NT upon cellular internalization (intact versus fragmented) could shed light on the mechanism of their cellular uptake. Future explorations could also include coassembly of the ssDNA-amphiphiles with aptamer-amphiphiles or formation of the NT only from aptamer-amphiphiles that will allow for the targeting of specific receptors. In addition, the presence of the bilayer with a hydrophobic core allows for the encapsulation of hydrophobic compounds, and therefore, the NT can be used for the delivery of drug combinations that will either have a synergistic effect or chemosensitize cancer cells to different treatments.
Collectively, our work demonstrates the promising therapeutic advantages of ssDNA NT intercalated with DOX as an anticancer agent, as it increased the number of long-term survivors and minimized toxicity to healthy organs. NT bind to scavenger receptors and are internalized by GBM cells through macropinocytosis. Both mechanisms are highly active in GBM cells compared to normal cells. Benefits of this technology include stability in nucleases and serum, ability to cross the BBTB and accumulate in the brain, preferential retention by tumors compared to normal brain, and ability to load and deliver chemotherapy drugs such as DOX. NT formed through the self-assembly of ssDNA-amphiphiles may have potential for translation as a drug delivery vehicle to GBM tumors.
MATERIALS AND METHODS
Materials
All materials were purchased from Sigma-Aldrich and used without further purification or modification unless otherwise stated. Buffers include high-performance liquid chromatography (HPLC) buffer A (100 mM hexafluoroisopropanol and 15 mM triethylamine in Milli-Q water), HPLC buffer B (100 mM hexafluoroisopropanol and 15 mM triethylamine in methanol), TEAA buffer (50% molar basis triethylamine, 50% molar basis glacial acetic acid, pH 7.0), Cu-TBTA [10 mM copper(II)-tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine in 55% dimethyl sulfoxide (DMSO), 45% Milli-Q water], 1× PBS [137 mM sodium chloride, 2.7 mM potassium chloride, 10 mM disodium phosphate, 1.8 mM monopotassium phosphate in Milli-Q water (pH 7.4)], and 1× TAE [40 mM tris(hydroxymethyl)aminomethane, 20 mM acetic acid, 1 mM ethylenediaminetetraacetic acid].
Synthesis of ssDNA-amphiphiles
ssDNA sequences were purchased from Integrated DNA Technologies (IDT) (Coralville, IA) with a 3′-amino modifier and an optional 5′ HEX [538/555 nm excitation (ex)/emission (em)] or hexynyl (alkyne) group. A 10-nt sequence (5′-CTCTTGGGGG-AmMo-3′) was used in this study. ssDNA was precipitated in Milli-Q water using 100 mM cetyl trimethylammonium bromide (CTAB) and centrifuged for 15 min at 16,100g, followed by removal of the liquid and drying of the precipitate under an air stream to remove any excess water. The dried precipitated ssDNA was then resuspended in 90%/10% (v/v) mixture of dimethylformamide (DMF) and DMSO at 500 μM. The C16 dialkyl tail with the C12 hydrocarbon spacer was synthesized as described elsewhere (28), added in 10 times molar excess, and reacted for 16 hours at 65°C. The solution was concentrated by drying in a vacuum oven until approximately 50 μl in volume. The reaction product with the ssDNA-amphiphile and unreacted ssDNA was precipitated by a lithium perchlorate precipitation, where 1 ml of lithium perchlorate in acetone (2.5% w/v) was added and the solution was mixed until homogeneous, followed by the addition of 100 μl of Milli-Q water and placed in a −20°C freezer for 15 min. The precipitate was centrifuged for 15 min at 16,100g and rehydrated with 1 ml of Milli-Q water and filtered through a 0.45-μm polyether sulfone filter (GE Healthcare, Chicago, IL). The filtered ssDNA-amphiphiles were separated from unreacted ssDNA using HPLC with HPLC buffer A and HPLC buffer B over 30 min. ssDNA-amphiphiles were then dried under an air stream to approximately 150 μl, precipitated with 1 ml of lithium perchlorate in acetone to remove HPLC buffer components, and rehydrated at 500 μM in Milli-Q water for storage at −20°C. DOTA-labeled ssDNA-amphiphiles were synthesized as previously described (44). The molecular weight of ssDNA-amphiphiles was verified by LC-MS as described previously (44). For the synthesis of IRDye 800CW–labeled amphiphiles, ssDNA-amphiphiles with a 5′-alkyne modification were mixed in 50% Milli-Q water and 50% DMSO to a final concentration of 100 μM. TEAA buffer was added to a concentration of 200 mM, Cu-TBTA buffer was added to a concentration of 1 mM, ascorbic acid was added to a concentration of 2 mM, and IRDye 800CW Azide (778/794 nm ex/em) (Licor, Lincoln, NE) was added in five times molar excess of the ssDNA-amphiphiles. The solution was mixed and left overnight in the dark at room temperature, followed by a lithium perchlorate in acetone precipitation to remove excess buffer components. The dye-labeled ssDNA-amphiphiles were rehydrated at 500 μM in Milli-Q water for storage at −20°C.
Circular dichroism
ssDNA-amphiphile solutions or pure ssDNA were diluted to 35 μM in Milli-Q water or PBS and transferred to a 0.1-cm path length cuvette. CD spectra from 320 to 200 nm were collected using an AVIV 420 CD Spectrometer (Center for Molecular Biophysics, Johns Hopkins University) using a 1-nm step size with an averaging time of 5 s and a settling time of 0.333 s. The background spectrum from the Milli-Q water or PBS was subtracted, and the raw ellipticity values were converted to molar ellipticity. Data were smoothed using the Savitzky-Golay filter function (sgolayfilt) on MATLAB using an order of 3 and a frame length of 11.
NT preparation
ssDNA NT containing different amphiphiles (unlabeled mixed with fluorescently labeled or DOTA labeled) were created by combining the desired amphiphiles at the correct ratio in Milli-Q water. One volume equivalent of DMSO was added to the mixtures so the final DMSO concentration was 50% (v/v). The mixtures were then stirred for 4 hours, during which Milli-Q water was slowly added to the mixtures until the final DMSO concentration at 4 hours was 10% (v/v). Mixtures were dialyzed overnight using a Tube-O-DIALYZER Medi 1K molecular weight cut off (MWCO) dialysis membrane (G-Biosciences, St. Louis, MO) to remove excess DMSO and dried under an air stream to 500 μM to prepare for nanoparticle separation. NT were separated from micelles using size exclusion chromatography on an Äkta fast protein liquid chromatography (Amersham Biosciences, Piscataway, NJ). A C10/20 Column (GE Healthcare, Chicago, IL) loaded with Sepharose CL-4B chromatography matrix was used to separate the nanoparticles. ssDNA-amphiphile mixtures (500 μM) were loaded at 500 μl per run onto the column and separated using Milli-Q water as a buffer. Fractions were collected on the basis of UV absorbance of the eluent and dried under an air stream to 500 μM.
Cryo-TEM and room temperature TEM
Lacey formvar/carbon 200 mesh copper grids were purchased from Ted Pella Inc. (Redding, CA) and glow discharged for 1 min to make the grids more hydrophilic. ssDNA-amphiphiles (500 μM; 4.5 μl) in Milli-Q water were deposited onto the grid and vitrified in liquid ethane using a Vitrobot (Vitrobot parameters: blot time, 5 s; wait time, 3 s; relax time, 3 s; offset, 0; humidity, 95%; 25°C). The grids were transferred to and kept under liquid nitrogen until imaged on a Tecnai G2 Spirit TWIN 20-120 kV/LaB6 TEM operated at an accelerating voltage of 120 kV using an Eagle 2k charge-coupled device (CCD) camera at the University of Minnesota Characterization Facility.
For the room temperature TEM experiments, 5 μl of ssDNA-amphiphiles was used at a concentration of 50 μM for the DOTA-labeled amphiphiles, 47 μM for the nuclease stability, and 27 μM for the serum stability experiment. Pure carbon 300 mesh copper grids (Ted Pella Inc.) were glow discharged in a GloQube glow discharge system (Quorum). Samples were then deposited on the grid. After 5 min, the excess sample was wicked off with filter paper. The grids were then imaged in a FEI Talos 200SC FEG TEM with a CETA scintillated 4k CMOS camera at the Integrated Imaging Center at the Johns Hopkins University.
Cell culture
GL261 mouse GBM cells [originally from the National Institutes of Health (NIH)] or C8-D1A normal mouse astrocytes (American Type Culture Collection, Manassas, VA) were cultured at 37°C and 5% CO2 using Dulbecco’s modified Eagle medium (DMEM) (Thermo Fisher Scientific, Rockford, IL) supplemented with 10% (v/v) FBS (Thermo Fisher Scientific, Rockford, IL) and penicillin (100 U/ml) and streptomycin (0.1 mg/ml). Cells were passaged when they reached 80% confluence by treatment with TrypLE Express Cell dissociation agent (Thermo Fisher Scientific, Rockford, IL).
NT cell internalization and organelle colocalization via confocal microscopy
ssDNA-amphiphile NT containing 20 mol % HEX-labeled ssDNA-amphiphiles were prepared at 250 μM in PBS. Two hundred thousand GL261 or C8-D1A cells were deposited onto glass coverslips within wells of a 24-well plate and allowed to adhere and proliferate for 24 hours. The next day, medium was replaced with 500 μl of fresh medium, and nanoparticles were added to a final concentration of 12.5 μM. After 24 hours, the medium containing NT was removed, and the cells were washed once with PBS. The cells were then stained simultaneously for their nuclei and membranes using Hoechst 33342 (Thermo Fisher Scientific, Rockford, IL) at 0.92 μg/ml and Wheat Germ Agglutinin Alexa Fluor 647 (Thermo Fisher Scientific, Rockford, IL) at 5.0 μg/ml, respectively, for 7 min at 37°C. The cells were then washed once with PBS and fixed using 4% paraformaldehyde (PFA) in PBS for 10 min at room temperature and then washed twice with PBS to remove any remaining PFA. Cells were mounted onto glass slides using ProLong Diamond Antifade Mountant (Thermo Fisher Scientific, Rockford, IL) and imaged with an Olympus FluoView FV1000 BX2 Upright Confocal Microscope (University Imaging Centers, University of Minnesota). Image analysis was performed using ImageJ software. Organelle colocalization was performed in the same manner, but the medium replenishment after 24 hours used 1 ml instead of 500 μl. Early endosomes were stained by adding 10 μl of CellLight Early Endosomes-GFP Bacman 2.0 (Thermo Fisher Scientific, Rockford, IL) for a final concentration of 10 viral particles per well. The CellLight solution was added at the same time as the HEX-labeled ssDNA-amphiphiles, which had a final concentration of 12.5 μM. Two hours before the completion of the 24-hour incubation, LysoTracker Deep Red (Thermo Fisher Scientific, Rockford, IL) was added to the wells at a final concentration of 200 nM. At the end of the 24-hour incubation, the medium containing nanoparticles was removed, and the cells were washed once with PBS. The nuclei were then stained using Hoechst 33342 at 0.92 μg/ml for 10 min at 37°C, washed once with PBS, fixed with 4% PFA in PBS for 10 min at room temperature, washed twice with PBS, and mounted onto glass slides using ProLong Diamond Antifade Mountant. The cells were then imaged with an Olympus FluoView FV1000 BX2 Upright Confocal Microscope, with image analysis performed in ImageJ software. Manders coefficients were calculated by drawing a region of interest (ROI) around each cell excluding the nuclei and using ImageJ’s Coloc2 plugin. The coefficients reported here are the percent of nanoparticle signal that overlapped with either early endosomes or acidic organelles. Percent nanoparticle signal in vesicles that were not labeled for each cell was calculated by using ImageJ’s Measure tool to sum the total pixel intensity of nanoparticle signal that did not co-occur with either endosomes or acidic organelles, and dividing by the total nanoparticle signal pixel intensity. Corresponding normal curves were graphed for each histogram using the mean and standard deviation of the dataset of interest and scaling the area under the curve to equal the area under the histogram.
Inhibition of endocytosis
Two hundred thousand GL261 cells were plated per well in 12-well plates and incubated at 37°C and 5% CO2 overnight. The following day, medium was replaced with fresh medium, and inhibitor stock solutions were diluted and delivered to the cells at targeted concentrations: CytD (1.25 μg/ml), 10 μM LatB, nocodazole (15 μg/ml), filipin (5 μg/ml), nystatin (2.5 μg/ml), MβCD (1.32 mg/ml), CPZ (2.5 μg/ml), dynasore (12.5 μg/ml), PTX (0.2 μg/ml), CTX (2 μg/ml), DMA (30 μg/ml), and fucoidan (500 μg/ml). Cells with no inhibitor were used as positive controls. Plates were gently shaken by hand to ensure even distribution of inhibitors and placed in the incubator for 30 min. After that, 5 nmol of NT containing 20 mol % HEX-labeled ssDNA-amphiphiles was added to each well, and plates were gently shaken by hand and placed in the incubator for 3 hours. Cells were washed twice with 1 ml of PBS, trypsin-EDTA at 37°C was added to each well, and the contents of each well were individually mixed via up and down pipetting and transferred to 1.5-ml centrifuge tubes. Cells were washed twice with PBS, reconstituted in 500 μl of PBS, and transferred to flow cytometry tubes. Five microliters of propidium iodide (PI) (10 μg/ml) solution was added to each tube and vortexed. The cells were run on a BD FACSCanto flow cytometer (Integrated Imaging Center, Johns Hopkins University) and were examined for PI, and HEX fluorescence via excitation at 488 nm with a 585/42 filter. Cells were gated by PI staining to select for live cells and by scattering (forward scatter channel A and side scatter channel A) to select for single cells. To evaluate cytotoxicity of inhibitors, 10,000 GL261 cells were plated in 96-well plates and incubated overnight in a 37°C 5% CO2 incubator. The next day, medium was replaced with fresh formulated medium, and inhibitor stock solutions were diluted and delivered to the cells at concentrations mentioned above. Plates were gently shaken by hand and placed in the incubator for 3 hours. Cell viability was measured using CellTiter-Glo 2.0 assay (Promega, Madison, WI) following the manufacturer’s protocol. Luminescence was recorded using a Synergy H1 microplate reader (Biotek, Winooski, VT), and cell viability was normalized to untreated cells.
NT serum and nuclease stability evaluated via gel electrophoresis
NT containing 20 mol % HEX-labeled ssDNA-amphiphiles prepared in Milli-Q water were used for each of these experiments, except for the T5 exonuclease and 50% FBS gels for which unlabeled ssDNA-amphiphiles were used. For the serum stability experiment, NT were mixed into three separate conditions all at a 12.5 μM final ssDNA-amphiphile concentration. The three solutions used were 1× PBS, 10% (v/v) FBS–90% (v/v) 1× PBS to mimic in vitro conditions, and 50% (v/v) FBS–50% (v/v) 1× PBS to mimic in vivo conditions. For the nuclease stability experiment, NT were mixed with DNase I, exonuclease III (Thermo Fisher Scientific, Rockford, IL), or T5 Exonuclease (New England Biolabs, Ipswich, MA). Nuclease concentrations were tested between 0 and 5 U/ml in 1× reaction buffer provided by each kit. NT were at a 12.5 μM ssDNA-amphiphile final concentration. All NT-serum and NT-nuclease solutions were incubated at 37°C for 24 hours combined with 6X Orange DNA Loading Dye (Thermo Scientific, Rockford, IL), were run on 2% agarose gels (2% agarose in 1× TAE buffer) at 120 V for 35 min, and imaged using a ChemiDoc MP Imaging System (Bio-Rad, Hercules, CA). For those experiments using unlabeled NT, the DNA was stained using Diamond Nucleic Acid Dye (Promega, Madison, WI) for 30 min before imaging.
Animals
C57BL/6J mice were purchased from the Jackson Laboratories (Bar Harbor, ME). Studies were approved either by the Institutional Animal Care and Use Committee of the University of Minnesota or the Johns Hopkins University Animal Care and Use Committee in accordance with the NIH’s Guide for the Care and Use of Laboratory Animals.
Bilateral intracranial injections of NT to orthotopic GL261 tumor–bearing mice and whole-brain imaging
Four mice were placed into deep anesthesia using an intraperitoneal injection of ketamine (100 mg/kg; Vedco, St. Joseph, MO) and xylazine (10 mg/kg; Akorn Animal Health, Lake Forest, IL). Buprenorphine (0.03 mg/ml, intramuscularly) was administered, the mouse head was sterilized, and a 1-cm incision was made along the scalp. Thirty thousand GL261 cells in sterile PBS were implanted into the right-side striatum of the mice using a murine stereotaxic system (Stoelting Co., Wood Dale, IL). NT containing 20 mol % IRDye 800CW–labeled ssDNA-amphiphiles were prepared in PBS. Three of the mice were anesthetized with ketamine (100 mg/kg) and xylazine (10 mg/kg), and 2 μl of the NT solution (2 nmol of ssDNA-amphiphiles) was injected bilaterally into both the left (normal side) and right (tumor side) striatum, 14 days after tumor cell implantation. At 45 to 105 min after NT injections, mice were decapitated, and brains were taken and fixed in 4% PFA overnight. Mouse brains were imaged using an In Vivo Imaging System (IVIS) (University Imaging Centers, University of Minnesota) with 780/820 nm ex/em settings. Following direct imaging, the mouse brains were dehydrated in 30% sucrose in PBS and embedded with Tissue-Tek optimal cutting temperature (O.C.T.) cryo-compound (Sacura, Torrance, CA). The brains were then frozen at −80°C, and 10-μm sections were cut using a Leica cryostat (Wetzlar, Germany), mounted onto charged Superfrost Plus glass slides (Thermo Fisher Scientific, Rockford, IL), and stored at −20°C until imaging. Mounted slices were imaged using a Nikon Eclipse TE2000-U inverted wide-field fluorescence microscope (University Imaging Centers, University of Minnesota). Image analysis was performed using ImageJ software.
Evaluation of NT association with tumors after intracranial or intravenous injections to orthotopic GL261 tumor–bearing mice
To establish the xenograft, GFP-labeled GL261 cells were dissociated into single-cell suspensions and stereotactically injected into the brains of 12-week-old mice (50,000 cells per injection). Three weeks after cell injection, 20 mol % HEX-labeled NT were administered to the mice via intracranial or intravenous injection. For the intracranial route, 1 nmol of ssDNA-amphiphiles was dissolved in 2 μl of PBS, or 2 μl of PBS was injected for each mouse. For the intravenous route, 30 nmol of ssDNA-amphiphiles was dissolved in 200 μl of PBS, or 200 μl of PBS was injected for each mouse. The mice were euthanized 3 hours after intracranial injection, or 6 hours after intravenous injection of NT, and tumor tissues were removed. For confocal microscopy, resected tumor tissues were immediately snap frozen and later cryosectioned axially into 30-μm slices using a Leica CM 1905 cryostat. Mounted slices were imaged on a Zeiss LSM700 confocal microscope (Integrated Imaging Center at the Johns Hopkins University). Settings were optimized to avoid background fluorescence using untreated brain slices. Zen software was used to process the obtained images.
Tumors evaluated via flow cytometry were rinsed with PBS, transferred to a petri dish, and mechanically disaggregated to slurry consistency with fine scissors. DMEM-F12 medium (1 to 2 ml; Thermo Fisher Scientific, Rockford, IL) was added to tumor slurry, followed by repeatedly pipetting up and down with a 1-ml pipette tip to break down visible aggregates. Dissociated tumor samples were then pipetted up and down with a 200-μl pipette tip, transferred to a 15-ml tube, and centrifuged at 300g for 5 min. Cell pellets were resuspended in 2 ml of DMEM-F12 medium and filtered through a cell strainer (70 μm). Cells were diluted with PBS and fixed with 4% PFA. Fixed samples in PBS were subsequently run on a BD FACSCanto flow cytometer (Integrated Imaging Center, Johns Hopkins University).
For evaluation of NT association with TAMs, 8-week-old mice were intracranially inoculated with 100,000 GL261 cells. On day 14 after tumor inoculation, 20 mol % HEX-labeled NT (1 nmol of ssDNA-amphiphiles dissolved in 2 μl of PBS) were administered intratumorally to a depth of 2 mm into the original burr hole for tumor inoculation. Brains were collected 3 hours after NT injection and immediately placed in 10% formalin solution overnight, followed by a daily sucrose gradient (10, 20, and then 30% sucrose in PBS) to wash out the formalin. Fixed brains were then flash frozen on dry ice and cryosectioned axially into 30-μm slices using a Leica CM 1905 cryostat. Brains were stained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize cell nuclei and Iba1 primary antibody at 1:200 (Wako Pure Chemical Corporation, Tokyo, Japan) to visualize macrophages. Briefly, brain slices were blocked with tris-buffered saline (Corning, Corning, NY) supplemented with 0.1% Triton X-100, 1% bovine serum albumin, and 5% normal goat serum (Thermo Fisher Scientific, Waltham, MA) for 4 hours, followed by incubation with unconjugated primary antibodies overnight at 4°C. Then, slices were washed and incubated with goat anti-rabbit 488 secondary antibody (Invitrogen, Carlsbad, CA) for 2 hours at room temperature. Last, slices were incubated with DAPI nuclear stain for 15 min, mounted in fluorescence mounting medium (Agilent Technologies, Santa Clara, CA), and sealed and imaged using a Zeiss LSM710 confocal microscope. Settings were optimized to avoid background fluorescence using untreated brain slices. Zen software was used to process the obtained images.
μPET/CT imaging of orthotopic GL261 tumor–bearing mice
Tumors were prepared in the same manner as for the bilateral intracranial injections of NT. Fourteen days after GL261 implantation, NT containing 5 mol % DOTA-labeled ssDNA-amphiphiles were mixed with 64CuCl2 (Department of Medical Physics, University of Wisconsin, WI). The dried 64CuCl2 salt was dissolved in 100 mM sodium acetate in Milli-Q water (pH 6) at 2 μCi/μl. The NT at 250 μM were diluted to 150 μM in 2× PBS and then mixed with the 64Cu solution (1:1 v/v), giving final concentrations of 75 μM ssDNA-amphiphile and 1 μCi/μl 64Cu in 1× PBS. The mixture was incubated at 37°C for 1 hour to allow for chelation of the radioisotope by the DOTA moieties and to preheat the solution before injection.
Mice were placed under a heat lamp before injections to dilate the veins in their tails. The tails were wiped with ethanol swabs to clean them before injection, and 200 μl of the 64Cu-labeled NT solution was injected into the lateral tail veins of the mice. The final solution injected contained approximately 0.8 pmol of 64Cu and 1.5 nmol of DOTA-labeled ssDNA-amphiphiles, approximately 1850 times molar excess of DOTA to 64Cu, which has been shown to entirely chelate all available copper (44). The radioactivity and time of measurement for each individual syringe were measured immediately before and after tail vein injections. Fifteen minutes before each imaging time point (1, 3, and 24 hours after injection), mice were anesthetized using 3% isofluorane in oxygen at 0.8 liter/min flow. μPET and CT scans were taken on a Siemens Inveon μPET/CT scanner (University Imaging Centers, University of Minnesota). After the 1-hour imaging time point, mice were placed under a heat lamp until they regained consciousness. After either the 3-hour or the 24-hour time point, mice were euthanized for ex vivo organ radioactivity measurements; if mice were not euthanized after the 3-hour time point, then they were placed under a heat lamp until they regained consciousness.
Images from the μPET/CT scans were saved as DICOM files and cropped to separate each individual mouse. This was done using a custom interactive tool written for MevisLab (www.mevislab.de). Care was taken to maintain the coordinate system and the calibrated radiological values contained in the original DICOM files. From these separated images, volumetric 3D renderings of each mouse were created for the whole mouse body. ImageJ was used to create the maximum-intensity projections of the head of each mouse and to plot the μPET intensity profiles as a function of distance across the head of the mouse, starting from the left hemisphere.
Ex vivo biodistribution analysis
At 3 or 24 hours after injections, mice were euthanized to collect organs for the biodistribution measurements. Organs were excised and weighed to determine their mass. The radioactivity of each organ [kilo counts per minute (kcpm)] was recorded using a scintillator and converted to microcuries using a calibration curve. The radiation values for each organ were then adjusted for the decay half-life of 64Cu (12.7 hours). The total injected dose was calculated by measuring the decay-adjusted radiation in the syringe before the injection and subtracting the decay-adjusted radiation in the syringe after injection. In addition, the decay-adjusted radiation in each mouse’s tail at the time of euthanasia was subtracted because of the possibility of missing the vein during injection, thereby limiting the amount of 64Cu systemically delivered. Organ radioactivity was scaled to the normalized injected dose and then scaled by the mass of the organ. Data were plotted as percent injected dose per gram of tissue (%ID/g).
Preparation of NT-DOX and DOX release
DOX–hydrogen chloride dissolved in water at 1 mg/ml was combined on an equimolar basis with ssDNA-amphiphiles in water at 500 μM. DMSO was added to the solution until the final DMSO concentration was 50% (v/v). The solution was stirred for 2 hours. Over four additional hours, water was slowly added until the final concentration was 90% water and 10% DMSO (v/v) at the end of the 4-hour period. The mixture was dialyzed overnight in a Tube-O-DIALYZER Medi 1K MWCO dialysis membrane (G-Biosciences, St. Louis, MO) to remove the DMSO. NT intercalating DOX were separated from micelles intercalating DOX as described above under NT preparation. DNA concentration was calculated through the absorbance of light at 260 nm. However, DOX also absorbs light at this wavelength. Therefore, the absorbance of mixtures of ssDNA and DOX was measured at both 260 and 488 nm, the maximum absorbance wavelengths for DNA and DOX, respectively. The extinction coefficient of the ssDNA at 260 nm was provided by IDT as 89,300 cm−1 M−1 and assumed to remain the same after the attachment of the hydrophobic tail. The extinction coefficient of the ssDNA at 488 nm was calculated by measuring the absorbance of a known amount of ssDNA at both 260 and 488 nm, providing an extinction coefficient at 488 nm of 135 cm−1 M−1. Several known concentrations of DOX were prepared by weighing out solid DOX and suspending in known volumes of Milli-Q water. The absorbance for each DOX sample was measured at both 260 and 488 nm, allowing for the calculation of the extinction coefficients for DOX as 14,715 and 10,200 cm−1 M−1, respectively. With all four extinction coefficients and the absorbance measurements at both 260 and 488 nm, the concentration of ssDNA-amphiphiles and DOX was calculated by solving the two coupled linear equations. It was assumed that the absorbance of the NT and DOX was additive with no interacting terms.
NT-DOX mixtures (200 μl) with DOX (75 μg/ml) and 76 μM ssDNA-amphiphiles on average in PBS were placed in a D-Tube Dialyzer Midi, MWCO 3.5 kDa. The dialysis tube was placed in a beaker with 100 ml of PBS at 37°C. At several time points during the dialysis, small samples were taken out, and the absorbance at 260 and 488 nm was measured to determine the DOX concentration.
Cell viability
The effect of DOX, NT, and NT-DOX on cell viability was assessed using the CellTiter-Glo 2.0 assay. Ten thousand GL261 cells were deposited into black 96-well tissue culture treated plates with 100 μl of medium and allowed to adhere for 24 hours at 37°C. The next day, medium was removed, 95 μl of fresh medium was added, and 5 μl of each test sample dissolved in Milli-Q water was added: water (control), NT at 5 to 6.4 μM ssDNA-amphiphiles, free DOX at 5 μg/ml, or NT-DOX at the same DOX and amphiphile concentrations. The samples were incubated with cells for 12 hours at 37°C, followed by a single wash with PBS. Fresh medium (100 μl) was added, and cells were incubated at 37°C for an additional 36 hours. Cells were allowed to equilibrate to room temperature, while the CellTiter-Glo 2.0 solution was placed in a room temperature water bath. The CellTiter-Glo 2.0 solution (100 μl) was added to each well of cells simultaneously, and the entire plate was placed on an orbital shaker for 2 min and then allowed to rest for 10 min. The luminescence signal of each well was measured, and the luminescence of each group was normalized to the luminescence of the untreated cells.
Intracranial delivery of NT intercalating DOX and bioluminescence imaging of mice
GL261-Luc cells (originally from NIH) were transfected to express luciferase by the Ohlfest Lab at the University of Minnesota. Glioma medium consisted of DMEM high glucose and l-glutamine (Genesee Scientific, 25-500), supplemented with 10% FBS, 1% penicillin-streptomycin (HyClone, SV30010), and 1% MEM NEAA (Gibco, 1140-050). Medium was changed every other day, and cells were passaged when reaching 80% confluence using TrypLE. Before transplantation, cells were washed three times with PBS followed by trypsinization for 5 min at 37°C followed by inactivation of the trypsin and centrifugation. The resulting pellet was resuspended in cold Hanks’ balanced salt solution (HBSS; Life Technologies) for counting using a hemocytometer. The cells were centrifuged a second time and resuspended at a concentration of roughly 1 × 104 cells per microliter of cold HBSS. The final cell solution was counted, and viability was assessed using Trypan Blue exclusion. The final cell count was calculated as the total number of viable cells per microliter.
Eight-week-old mice were used for this study. Animals were first anesthetized with isoflurane oxygen mixture, and then, the head of the animal was shaved and treated with betadine. Following mounting in a stereotaxic frame, a single midline incision was made along the scalp and skin retracted to expose bregma. A 10-μl Hamilton syringe was loaded with the cell solution. A small burr hole was drilled in the skull above the injection site in the right hemisphere (from bregma: anterior, 1.0 mm; and lateral, 1.5 mm). The needle was slowly inserted into the brain (3.1 mm ventral to the pia mater in mice), and 1 × 104 viable GL261 cells were injected at a speed of 0.5 μl/min. Following injection, the needle remained in place for 1 min. The injection needle was raised 0.1 mm and again 0.2 mm from the initial injection site, and the injection was repeated with 1 × 104 cells injected at each site for a total of 3 × 104 viable cells across three sites. At the conclusion of the last injection, the needle remained in place for 3 min before being slowly withdrawn. Immediately following intracranial injection of GL261-Luc cells, hemostats were then inserted into the incision site and used to create a subcutaneous pouch immediately posterior to the scapula of the mouse by which the micro-osmotic pump (Alzet 1002) was inserted with the catheter tubing connected to the cannula (Alzet brain infusion kit 3) extending through the incision site. The cannula was slowly lowered into the brain though the same burr hole using a cannula holder (Alzet cannula holder 1) to sit 3 mm below the skull. Cannulas were fixed to the skull of mice using Loctite 454, and then, the cannula guide was removed using bone shears. The incision was then closed using 4-0 absorbable sutures, and mice were transferred to a heated recovery cage until fully sternal, at which point, mice were singly housed and returned to colony rooms. The pumps were loaded with either PBS, 70 μM DOX (0.2 mg DOX/kg mouse), NT at 95 μM ssDNA-amphiphiles, or NT-DOX at the same concentrations of DOX and amphiphiles. Mice were monitored twice daily for signs of advanced tumor progression. Mice were imaged weekly for 4 weeks after tumor implantation. The substrate d-luciferin (Thermo Fisher Scientific) was administered via intraperitoneal injection at 150 μg/g body weight in 200 μl of PBS. The mice were then placed onto the warmed stage inside the imaging chamber with continuous exposure to 1 to 1.5% isoflurane in oxygen at 1 liter/min. Bioluminescence images were acquired using the IVIS 1000 system (Xenogen) equipped with a highly sensitive cooled CCD camera, 10 to 15 min after d-luciferin administration. Images were analyzed by using the Living Image software (Xenogen). ROIs were defined in the brain, which were held constant across all images. The photon counts within each ROI were quantified. For visualization purposes, the bioluminescence image and the corresponding white light surface image were fused into a transparent pseudocolor overlay. When a mouse reached a moribund state, the animal was deeply anesthetized via ketamine overdose (100 mg/kg, intraperitoneally) and perfused with ice-cold PBS followed by 4% PFA. The brain, heart, lung, kidneys, and spleen were removed from the animals and stored in PFA overnight at 4°C and then transferred to 70% ethanol.
Histopathological analysis
Following perfusion and fixation with 10% neutral buffered formalin, tissues were processed into paraffin blocks using standard histology techniques, sectioned at a thickness of 4 μm, stained with hematoxylin and eosin, and evaluated using light microscopy by two American College of Veterinary Pathologists (ACVP) board-certified pathologists (A.-F.T. and M.G.O.). All work was done at the Masonic Cancer Center Comparative Pathology Shared Resource laboratory at the University of Minnesota.
Statistical analysis
Statistical differences were determined using unpaired two-tailed Student’s t tests or one-way analysis of variance (ANOVA) with Tukey’s post hoc test. Survival Kaplan-Meier curves were constructed and compared using a two-sided log-rank test. Statistical analyses were performed using Excel (Microsoft) and the Real Statistics Excel Resource Pack.
Acknowledgments
We would like to acknowledge H. Bui at the Integrated Imaging Center for assistance with some of the flow cytometry studies. We also acknowledge use of the University of Minnesota Imaging Centers and the following facilities at the Johns Hopkins University: NMR Core Facility, Center for Molecular Biophysics, and Integrated Imaging Center.
Funding: This work was partially supported by a grant to E.K. and W.C.L. through the University of Minnesota’s MnDRIVE program, and the Whiting School of Engineering of the Johns Hopkins University (E.K.). Parts of this work were carried out in the University of Minnesota Characterization Facility, which receives partial support from the NSF through the MRSEC program.
Author contributions: E.K. conceived the idea, designed the study, supervised the project, and contributed to data analysis. M.A.H., H.K., Z.S., M.L.S., A.T.C., M.R.C., A.-F.T., T.P., K.L., and B.X. performed the experiments and contributed to data analysis. L.L. contributed to data analysis. C.C.C., M.G.O., R.M.K., and W.C.L. contributed to the design of experiments. E.K. wrote the manuscript with contributions and feedback from all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
Table S1
Legends for movies S1 to S3
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S3
REFERENCES AND NOTES
- 1.Cantrell J. N., Waddle M. R., Rotman M., Peterson J. L., Ruiz-Garcia H., Heckman M. G., Quinones-Hinojosa A., Rosenfeld S. S., Brown P. D., Trifiletti D. M., Progress toward long-term survivors of glioblastoma. Mayo Clin. Proc. 94, 1278–1286 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Poon M. T. C., Sudlow C. L. M., Figueroa J. D., Brennan P. M., Longer-term (≥2 years) survival in patients with glioblastoma in population-based studies pre- and post-2005: A systematic review and meta-analysis. Sci. Rep. 10, 11622 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arvanitis C. D., Ferraro G. B., Jain R. K., The blood-brain barrier and blood-tumour barrier in brain tumours and metastases. Nat. Rev. Cancer 20, 26–41 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarkaria J. N., Hu L. S., Parney I. F., Pafundi D. H., Brinkmann D. H., Laack N. N., Giannini C., Burns T. C., Kizilbash S. H., Laramy J. K., Swanson K. R., Kaufmann T. J., Brown P. D., Agar N. Y. R., Galanis E., Buckner J. C., Elmquist W. F., Is the blood–brain barrier really disrupted in all glioblastomas? A critical assessment of existing clinical data. Neuro Oncol. 20, 184–191 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liaw K., Zhang F., Mangraviti A., Kannan S., Tyler B., Kannan R. M., Dendrimer size effects on the selective brain tumor targeting in orthotopic tumor models upon systemic administration. Bioeng. Transl. Med. 5, e10160 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolhar P., Anselmo A. C., Gupta V., Pant K., Prabhakarpandian B., Ruoslahti E., Mitragotri S., Using shape effects to target antibody-coated nanoparticles to lung and brain endothelium. Proc. Natl. Acad. Sci. U.S.A. 110, 10753–10758 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lam F. C., Morton S. W., Wyckoff J., Han T. L. V., Hwang M. K., Maffa A., Balkanska-Sinclair E., Yaffe M. B., Floyd S. R., Hammond P. T., Enhanced efficacy of combined temozolomide and bromodomain inhibitor therapy for gliomas using targeted nanoparticles. Nat. Commun. 9, 1991 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitchell M. J., Billingsley M. M., Haley R. M., Wechsler M. E., Peppas N. A., Langer R., Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 20, 101–124 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jain K. K., A critical overview of targeted therapies for glioblastoma. Front. Oncol. 8, 419 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai Q., Wilhelm S., Ding D., Syed A. M., Sindhwani S., Zhang Y., Chen Y. Y., MacMillan P., Chan W. C. W., Quantifying the ligand-coated nanoparticle delivery to cancer cells in solid tumors. ACS Nano 12, 8423–8435 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Hendricks B. K., Cohen-Gadol A. A., Miller J. C., Novel delivery methods bypassing the blood-brain and blood-tumor barriers. Neurosurg. Focus 38, E10 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Chaichana K. L., Pinheiro L., Brem H., Delivery of local therapeutics to the brain: Working toward advancing treatment for malignant gliomas. Ther. Deliv. 6, 353–369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu W., Chen H., Shan F., Zhou J., Sun X., Zhang L., Gong T., A novel doxorubicin-loaded in situ forming gel based high concentration of phospholipid for intratumoral drug delivery. Mol. Pharm. 11, 3378–3385 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Cao J., Huang D., Peppas N. A., Advanced engineered nanoparticulate platforms to address key biological barriers for delivering chemotherapeutic agents to target sites. Adv. Drug Deliv. Rev. 167, 170–188 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Vance D., Martin J., Patke S., Kane R. S., The design of polyvalent scaffolds for targeted delivery. Adv. Drug Deliv. Rev. 61, 931–939 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Rothemund P. W. K., Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Yan H., Park S. H., Finkelstein G., Reif J. H., LaBean T. H., DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science 301, 1882–1884 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Sellner S., Kocabey S., Nekolla K., Krombach F., Liedl T., Rehberg M., DNA nanotubes as intracellular delivery vehicles in vivo. Biomaterials 53, 453–463 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Kocabey S., Meinl H., MacPherson I. S., Cassinelli V., Manetto A., Rothenfusser S., Liedl T., Lichtenegger F. S., Cellular uptake of tile-assembled DNA nanotubes. Nanomaterials 5, 47–60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ko S., Liu H., Chen Y., Mao C., DNA nanotubes as combinatorial vehicles for cellular delivery. Biomacromolecules 9, 3039–3043 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y.-X., Shaw A., Zeng X., Benson E., Nyström A. M., Högberg B., DNA origami delivery system for cancer therapy with tunable release properties. ACS Nano 6, 8684–8691 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Hamblin G. D., Carneiro K. M. M., Fakhoury J. F., Bujold K. E., Sleiman H. F., Rolling circle amplification-templated DNA nanotubes show increased stability and cell penetration ability. J. Am. Chem. Soc. 134, 2888–2891 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Stephanopoulos N., Strategies for stabilizing DNA nanostructures to biological conditions. Chembiochem 20, 2191–2197 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Pearce T. R., Kokkoli E., DNA nanotubes and helical nanotapes via self-assembly of ssDNA-amphiphiles. Soft Matter 11, 109–117 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Kuang H., Gartner T. E. III, de Mello M. D., Guo J., Zuo X., Tsapatsis M., Jayaraman A., Kokkoli E., ssDNA-amphiphile architecture used to control dimensions of DNA nanotubes. Nanoscale 11, 19850–19861 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Wilner S. E., Sparks S. E., Cowburn D., Girvin M. E., Levy M., Controlling lipid micelle stability using oligonucleotide headgroups. J. Am. Chem. Soc. 137, 2171–2174 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearce T. R., Waybrant B., Kokkoli E., The role of spacers on the self-assembly of DNA aptamer-amphiphiles into micelles and nanotapes. Chem. Commun. 50, 210–212 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Waybrant B., Pearce T. R., Kokkoli E., Effect of polyethylene glycol, alkyl, and oligonucleotide spacers on the binding, secondary structure, and self-assembly of fractalkine binding FKN-S2 aptamer-amphiphiles. Langmuir 30, 7465–7474 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Choi E. W., Nayak L. V., Bates P. J., Cancer-selective antiproliferative activity is a general property of some G-rich oligodeoxynucleotides. Nucleic Acids Res. 38, 1623–1635 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manders E. M. M., Verbeek F. J., Aten J. A., Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 169, 375–382 (1993). [DOI] [PubMed] [Google Scholar]
- 31.Zhang L. W., Monteiro-Riviere N. A., Mechanisms of quantum dot nanoparticle cellular uptake. Toxicol. Sci. 110, 138–155 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Contreras J., Xie J., Chen Y. J., Pei H., Zhang G., Fraser C. L., Hamm-Alvarez S. F., Intracellular uptake and trafficking of difluoroboron dibenzoylmethane-polylactide nanoparticles in HeLa cells. ACS Nano 4, 2735–2747 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy J. E., Tedbury P. R., Homer-Vanniasinkam S., Walker J. H., Ponnambalam S., Biochemistry and cell biology of mammalian scavenger receptors. Atherosclerosis 182, 1–15 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Patel P. C., Giljohann D. A., Daniel W. L., Zheng D., Prigodich A. E., Mirkin C. A., Scavenger receptors mediate cellular uptake of polyvalent oligonucleotide-functionalized gold nanoparticles. Bioconjug. Chem. 21, 2250–2256 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu X. A., Choi C. H. J., Zhang C., Hao L., Mirkin C. A., Intracellular fate of spherical nucleic acid nanoparticle conjugates. J. Am. Chem. Soc. 136, 7726–7733 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi C. H. J., Hao L., Narayan S. P., Auyeung E., Mirkin C. A., Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates. Proc. Natl. Acad. Sci. U.S.A. 110, 7625–7630 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Juks C., Padari K., Margus H., Kriiska A., Etverk I., Arukuusk P., Koppel K., Ezzat K., Langel Ü., Pooga M., The role of endocytosis in the uptake and intracellular trafficking of PepFect14-nucleic acid nanocomplexes via class A scavenger receptors. Biochim. Biophys. Acta 1848, 3205–3216 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Yu X., Guo C., Fisher P. B., Subjeck J. R., Wang X.-Y., Scavenger receptors: Emerging roles in cancer biology and immunology. Adv. Cancer Res. 128, 309–364 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang J.-L., Jiang G., Song Q.-X., Gu X., Hu M., Wang X.-L., Song H.-H., Chen L.-P., Lin Y.-Y., Jiang D., Chen J., Feng J.-F., Qiu Y.-M., Jiang J.-Y., Jiang X.-G., Chen H.-Z., Gao X.-L., Lipoprotein-biomimetic nanostructure enables efficient targeting delivery of siRNA to Ras-activated glioblastoma cells via macropinocytosis. Nat. Commun. 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zagorovskya K., Chou L. Y. T., Chan W. C. W., Controlling DNA–nanoparticle serum interactions. Proc. Natl. Acad. Sci. U.S.A. 113, 13600–13605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamkovich S. N., Cherepanova A. V., Kolesnikova E. V., Rykova E. Y., Pyshnyi D. V., Vlassov V. V., Laktionov P. P., Circulating DNA and DNase activity in human blood. Ann. N. Y. Acad. Sci. 1075, 191–196 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Al-Chalabi K. A. M., Mahmood Al-Abachi S. Z., Enzymatic changes in serum of benign and malignant brain tumors patients. J. Edu. Sci. 21, 19–30 (2008). [Google Scholar]
- 43.Rush A. M., Thompson M. P., Tatro E. T., Gianneschi N. C., Nuclease-resistant DNA via high-density packing in polymeric micellar nanoparticle coronas. ACS Nano 7, 1379–1387 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harris M. A., Pearce T. R., Pengo T., Kuang H., Forster C., Kokkoli E., Aptamer micelles targeting fractalkine-expressing cancer cells in vitro and in vivo. Nanomedicine: NBM 14, 85–96 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Bartlett D. W., Davis M. E., Physicochemical and biological characterization of targeted, nucleic acid-containing nanoparticles. Bioconjug. Chem. 18, 456–468 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schluep T., Hwang J., Hildebrandt I. J., Czernin J., Choi C. H., Alabi C. A., Mack B. C., Davis M. E., Pharmacokinetics and tumor dynamics of the nanoparticle IT-101 from PET imaging and tumor histological measurements. Proc. Natl. Acad. Sci. U.S.A. 106, 11394–11399 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alexis F., Pridgen E., Molnar L. K., Farokhzad O. C., Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol. Pharm. 5, 505–515 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seo J. W., Ang J., Mahakian L. M., Tam S., Fite B., Ingham E. S., Beyer J., Forsayeth J., Bankiewicz K. S., Xu T., Ferrara K. W., Self-assembled 20-nm 64Cu-micelles enhance accumulation in rat glioblastoma. J. Control. Release 220, 51–60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kadari A., Pooja D., Gora R. H., Gudem S., Kolapalli V. R. M., Kulhari H., Sistla R., Design of multifunctional peptide collaborated and docetaxel loaded lipid nanoparticles for antiglioma therapy. Eur. J. Pharm. Biopharm. 132, 168–179 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Manet I., Manoli F., Zambelli B., Andreano G., Masi A., Cellai L., Monti S., Affinity of the anthracycline antitumor drugs Doxorubicin and Sabarubicin for human telomeric G-quadruplex structures. Phys. Chem. Chem. Phys. 13, 540–551 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Kuang H., Schneiderman Z., Shabana A. M., Russo G. C., Guo J., Wirtz D., Kokkoli E., Effect of an alkyl spacer on the morphology and internalization of MUC1 aptamer-naphthalimide amphiphiles for targeting and imaging triple negative breast cancer cells. Bioeng. Transl. Med. e10194 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng Y., Morshed R., Cheng S. H., Tobias A., Auffinger B., Wainwright D. A., Zhang L., Yunis C., Han Y., Chen C. T., Lo L. W., Aboody K. S., Ahmed A. U., Lesniak M. S., Nanoparticle-programmed self-destructive neural stem cells for glioblastoma targeting and therapy. Small 9, 4123–4129 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lesniak M. S., Upadhyay U., Goodwin R., Tyler B., Brem H., Local delivery of doxorubicin for the treatment of malignant brain tumors in rats. Anticancer Res 25, 3825–3831 (2005). [PMC free article] [PubMed] [Google Scholar]
- 54.Blomme E. A. G., Ciurlionis R., Marsh K. C., Waring J. F., Yang Y., Evaluation of the effects of serial phlebotomy on the transcriptome of major tissues and on the response to toxicants in rats. Toxicol. Lett. 176, 138–148 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Jadapalli J. K., Wright G. W., Kain V., Sherwani M. A., Sonkar R., Yusuf N., Halade G. V., Doxorubicin triggers splenic contraction and irreversible dysregulation of COX and LOX that alters the inflammation-resolution program in the myocardium. Am. J. Physiol. Heart Circ. Physiol. 315, H1091–h1100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.El-Sayyad H. I., Ismail M. F., Shalaby F. M., Abou-El-Magd R. F., Gaur R. L., Fernando A., Raj M. H. G., Ouhtit A., Histopathological effects of cisplatin, doxorubicin and 5-flurouracil (5-FU) on the liver of male albino rats. Int. J. Biol. Sci. 5, 466–473 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martinel Lamas D. J., Nicoud M. B., Sterle H. A., Carabajal E., Tesan F., Perazzo J. C., Cremaschi G. A., Rivera E. S., Medina V. A., Selective cytoprotective effect of histamine on doxorubicin-induced hepatic and cardiac toxicity in animal models. Cell Death Discov. 1, 15059–15059 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo L., Fan L., Pang Z., Ren J., Ren Y., Li J., Chen J., Wen Z., Jiang X., TRAIL and doxorubicin combination enhances anti-glioblastoma effect based on passive tumor targeting of liposomes. J. Control. Release 154, 93–102 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Gaillard P. J., Appeldoorn C. C., Dorland R., van Kregten J., Manca F., Vugts D. J., Windhorst B., van Dongen G. A., de Vries H. E., Maussang D., van Tellingen O., Pharmacokinetics, brain delivery, and efficacy in brain tumor-bearing mice of glutathione pegylated liposomal doxorubicin (2B3-101). PLOS ONE 9, e82331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Steiniger S. C., Kreuter J., Khalansky A. S., Skidan I. N., Bobruskin A. I., Smirnova Z. S., Severin S. E., Uhl R., Kock M., Geiger K. D., Gelperina S. E., Chemotherapy of glioblastoma in rats using doxorubicin-loaded nanoparticles. Int. J. Cancer 109, 759–767 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Table S1
Legends for movies S1 to S3
Movies S1 to S3