Abstract
Interaction between the dietary fiber and the gut microbes can regulate host bile acid metabolism. This study sought to explore the effects of guar gum combined with pregelatinized waxy maize starch (GCW) in a gestation diet on reproductive performance, gut microbiota composition, and bile acid homeostasis of sows. A total of 61 large white sows were randomly grouped into the control (n = 33) and 2% GCW (n = 28) groups during gestation. GCW diet increased birth-weight of piglets, and decreased the percentage of intrauterine growth restriction (IUGR) piglets. In addition, dietary GCW reduced gut microbial diversity and modulated gut microbial composition in sows on day 109 of gestation. The relative abundance of bile salt hydrolase (BSH) gene-encoding bacteria, Lactobacillus and Bacteroides decreased after GCW administration, whereas no significant difference was observed in the fecal level of total glycine-conjugated and taurine-conjugated bile acids between the two groups. Dietary GCW increased the relative abundance of Ruminococcaceae (one of few taxa comprising 7α-dehydroxylating bacteria), which was associated with elevated fecal deoxycholic acid (DCA) in the GCW group. GCW administration lowered the concentrations of plasma total bile acid (TBA) and 7α-hydroxy-4-cholesten-3-one (C4) (reflecting lower hepatic bile acid synthesis) at day 90 and day 109 of gestation compared with the control diet. Furthermore, the levels of plasma glycoursodeoxycholic acid (GUDCA), tauroursodeoxycholic acid (TUDCA) and glycohyocholic acid (GHCA) were lower in the GCW group compared with the control group. Spearman correlation analysis showed alterations in the composition of the gut microbiota by GCW treatment was associated with improved bile acid homeostasis and reproductive performance of sows. In conclusion, GCW-induced improves bile acid homeostasis during gestation which may enhance reproductive performance of sows.
Keywords: soluble fiber, gut microbiota, bile acid, reproductive performance, sow
Introduction
Increased maternal total bile acid (TBA) of serum is implicated in dysregulation of bile acid homeostasis, which is associated with fetal distress (1), unexplained stillbirth (2), spontaneous preterm labor (3), neonatal low birth weight (4), and intrauterine fetal pig death (5). Notably, elevated maternal serum TBA levels are associated with accumulation of toxic bile acids (e.g., LCA) in placenta (6). In addition, bile acids can be transported across the placenta causing adverse fetal outcome in intrahepatic cholestasis of pregnancy (ICP) patients (7).
Synthesis and metabolism of bile acids is tightly regulated by gut microbiota (8). Alterations in gut microbiota composition and function can affect host bile acid metabolism (9, 10). Dietary fiber may serve as a platform for interaction between gut microbiota and bile acids and thus improve production of secondary bile acids (11). Secondary bile acids (e.g., glycoursodeoxycholic acid) influence host bile acids metabolism via the modulation of intestinal farnesoid X receptor (FXR) signaling (12). Diets high in whole grains increases the plasma levels of taurocholic acid (TCA) and glycocholic acid (GCA), which are implicated in activation of nuclear FXR target genes thus modulating bile acid metabolism (13). Soluble dietary fibers (such as wheat arabinoxylan and oat β-glucan) reduce the levels of circulating bile acids in swine model (14, 15). Notably, increase in serum bile acid levels is inhibited by oral guar gum treatment in women with ICP (16, 17). Our previous studies report that konjac flour supplementation during gestation alters gut microbiota composition in sows (18). Moreover, supplementing diet with 0.8% soluble dietary fiber can reduce incidence of intrauterine growth restriction (IUGR) piglets and improve intra-litter uniformity in replacement gilts (19). However, previous studies have not explored the mechanism by which soluble dietary fiber enhances reproductive performance in sows.
This study explored the impacts of dietary GCW intake on reproductive performance and gut microbiota composition and bile acid homeostasis of sows during gestation. The hypothesis of the study was that dietary GCW intake in gestation perturbs the gut microbiota accompanied with changes in bile acid composition in feces, and in turn induces intestinal FXR signaling, which may reduce TBA levels in plasma.
Materials and Methods
Animal use and care protocol used in this study was approved by Institutional Animal Care and Use Committee of Huazhong Agricultural University. The ethical approval number of this study is HZAUSW-2016-023.
Animals, Diets, and Sample Collection
GCW used in this experiment was obtained by mixing 85.7% pregelatinized waxy maize starch (Hangzhou, China) with 14.3% guar gum (Yunzhou, China). A total of 61 multiparous sows (large white, parity 2–6) were randomly grouped into two dietary treatments using a complete block design. During gestation, sows were fed with corresponding experimental diets: the control group was fed with basal diet (control, n = 33) whereas the GCW group was fed with basal diet supplemented with 2% GCW to replace rice bran meal (GCW, n = 28). All experimental diets were formulated to meet nutrient requirements of gestating sows according to recommendations by NRC (20). Treatments were isonitrogenous and isoenergetic diets. Diet compositions and nutrient levels are presented in Supplementary Table 1. Animals were fed twice a day (07:00 and 14:30 h). Blood samples (6~8 mL) were collected from the marginal ear vein of sow with disposable vacuum blood collection tubes 4 h after the morning meal (7~10 sows per group) on days 30, 60, 90, 109 of gestation (G30, G60, G90, and G109). Plasma was extracted by centrifugation of blood samples at 3,000 × g for 5 min at 4°C and stored at −80°C until analysis. Fresh feces were collected on day 109 of gestation and stored in a −80°C freezer. All sows did not present any signs of disease before sampling.
Measurement of Plasma TBA, ALT, and AST
Plasma TBA, alanine aminotransferase (ALT), and alanine aminotransferase (AST), were determined through standard routine procedures using a Mindray BS-240 automatic biochemical analyzer (Mindray biomedical electronic co. LTD, Shenzhen, China).
Measurement of Plasma FGF19 and C4
Concentrations of fibroblast growth factor 19 (FGF19) and 7α-hydroxy-4-cholesten-3-one (C4) in plasma were determined using porcine enzyme-linked immunosorbent assay kits (Nanjing Camilo biological engineering co. LTD, Nanjing, China), following the manufacturer's instructions.
Quantification of Baij, Bsh1, and Bsh2 Gene by Real-Time PCR
Total fecal microbial DNA was extracted using TIANamp Stool DNA Kit (Tiangen Biotech co. LTD, Beijing, China), following the manufacturer's instructions. Expression levels of bacterial bile acid metabolism-related genes (baij, bsh1, and bsh2) were determined by quantitative real-time PCR (Q-PCR). Expression levels of baij, bsh1, and bsh2 genes were normalized using 16s rDNA. The reaction system and conditions used for Q-PCR were as described in a previous study (21). Primer sequences used in this study are presented in Supplementary Table 2.
Bile Acid Analysis
All bile acid standards were purchased from Olchemim Ltd. (Olomouc, Czech Republic) and Sigma (St. Louis, MO, USA). For plasma samples, 200 μL methanol was added to 50 μL plasma-spiked with 10 μL internal standard (IS), vortexed, and centrifuged at 12,000 r/min for 10 min. For feces samples, 20 mg of feces were spiked with 10 μL IS and mixed with 200 μL methanol, vortexed, and centrifuged at 12,000 r/min for 10 min. The supernatant were evaporated to dryness, reconstituted in 100 μL 50% aqueous methanol (V/V) for further LC-MS analysis.
Bile acids were quantified using ultra-performance liquid chromatography (UPLC, Shim-pack UFLC SHIMADZU CBM30A system) coupled to a tandem mass spectrometry system (MS/MS, Applied Biosystems 6500 QTRAP). Chromatographic separations were performed with an ACQUITY UPLC HSS C18 column (1.8 μm, 2.1 mm 100 mm, Waters Corp., Milford, MA, USA). The mobile phase was water (1,000 mL) + formicacid (100 μL) + ammonium acetate (5 mM; A) and formicacid (100 μL) + acetonitrile (1,000 mL; B). The gradient conditions were as follows: 0–0.5 min (5% B), 0.5–4.5 min (40–50% B), 4.5–7.5 min (50–80% B), 7.5–10 min (80–95% B), and 10–12 min (5% B). The total analysis time was 12 min and the injection volume was 3 μL. The column temperature was maintained at 40°C with the flow rate set at 0.35 mL/min. The mass spectrometer was operated with an electrospray ionization interface in negative ionization (ESI–) mode. The UPLC-MS raw data were collected using Multiple Reaction Monitoring (MRM) mode. The qualitative analysis of mass spectrometry data was performed based on the self-built database MWDB (Metware Biotechnology Co., Ltd., Wuhan, China), and the quantitative analysis were performed using the Multiquant 3.0 software (Sciex, Framingham, MA, USA).
Analysis of Gut Microbiota
Total bacterial DNA was extracted from fecal samples using a QIAamp DNA Stool Mini Kit (Qiagen, Germany) following the manufacturer's protocol. Bacterial 16S rRNA gene V3-V4 region of each sample was then amplified using 16S rRNA universal primers. PCR products were mixed then sequenced on an Illumina MiSeq platform (2 × 250 bp) (Illumina, United States). Microbiome bioinformatics were performed with QIIME2 2019.4 (22) with slight modification according to the official tutorials. Briefly, raw sequence data were demultiplexed using the demux plugin following by primers cutting with cutadapt plugin (23). Sequences were then quality filtered, denoised, merged, and chimera removed using the DADA2 plugin (24). Non-singleton amplicon sequence variants (ASVs) were aligned with mafft (25) and used to construct a phylogeny with fasttree2 (26). Alpha-diversity values of the samples were determined based on the Observed species, Chao1, Shannon, and Simpson index. Beta-diversity measures were calculated using weighted-UniFrac distance. Linear discriminant analysis (LDA) effect size analysis (LEfSe) was used to identify key gut microbiota of the two groups (p < 0.05, Wilcoxon rank-sum test; log LDA > 2).
Statistical Analysis
Data were analyzed with SPSS (SPSS 23.0, SPSS Inc., Chicago, IL). Categorical variables were analyzed using χ2-tests. Normally distributed data were analyzed using student's unpaired t-test; whereas data that was not normally distributed was analyzed using non-parametric Mann Whitney U-test. Spearman correlation analyses were performed using Prism GraphPad 8.0.1 (GraphPad Software, San Diego, CA, United States). P < 0.05 were considered significant.
Results
Reproductive Performance
Effects of GCW in the gestation diet on reproductive performance of sows are presented in Table 1. GCW showed no significant effect on total number of born piglets and number of alive piglets. GCW significantly increased birth weight of piglets compared with the weight of piglets in the control group. Moreover, the percentage of IUGR piglets (< 800 g) in the total alive piglets for the GCW group was 0.91%, which was significantly lower compared with that of the control group (3.89%). However, no significant differences were observed for the number of piglets with birth weights ranging between 800–1,000, 1,000–1,200, 1,200–1,400, and more than 1,400 g between the two groups.
Table 1.
Effects of dietary supplementation with GCW during gestation on reproductive performance of sowsa.
| Parameter | CON | GCW | P-value |
|---|---|---|---|
| No. of litters | 33 | 28 | |
| Litter size, No/litter | |||
| Total born | 12.97 ± 2.59 | 12.07 ± 2.65 | 0.187 |
| Born alive | 12.45 ± 2.53 | 11.82 ± 2.56 | 0.337 |
| Birth weight of litters, kg | 18.36 ± 3.91 | 18.04 ± 3.61 | 0.747 |
| Birth weight of piglets, kg | 1.47 ± 0.33 | 1.53 ± 0.32 | 0.030 |
| Birth weight distribution of piglets, % b | |||
| <800 g, % | 3.89 (16) | 0.91 (3) | 0.010 |
| 800–1,000 g, % | 4.62 (19) | 5.13 (17) | 0.746 |
| 1,000–1,200 g, % | 8.76 (36) | 10.88 (36) | 0.333 |
| 1,200–1,400 g, % | 18.98 (78) | 19.64 (65) | 0.821 |
| >1,400 g, % | 63.75 (262) | 63.44 (210) | 0.932 |
Data on fetus and birth weight distribution of piglets are expressed as proportion of total born piglets in both groups (actual number in the group); other data are presented as mean ± SD.
The birth weight distribution of piglets refers to the ratio of piglets in each weight range to the total live-born piglets in both groups.
Gut Microbiota Analysis
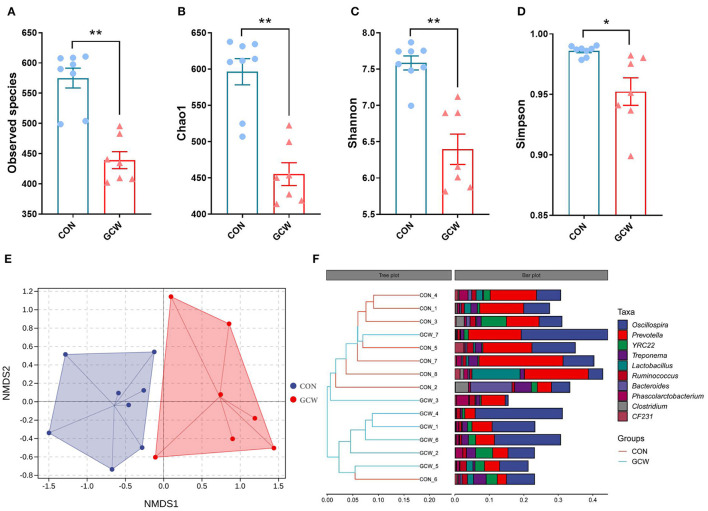
To explore the composition of gut microbiota for the two groups on day 109 of gestation, 16S ribosomal RNA gene (V3-V4 gene regions) sequencing was conducted. Analysis of Alpha-diversity indexes showed that the number of observed species, Chao1, Shannon, and Simpson index were significantly lower in the GCW group compared with the control group (Figures 1A–D). Nonmetric multidimensional scaling (NMDS) analysis for beta density showed that the control group was separately clustered from the GCW group (Figure 1E). Hierarchical Cluster Analysis (HCA) based on the weighted-UniFrac distance showed that control samples clustered together whereas GCW samples clustered together (Figure 1F). This finding validated the results obtained by NMDS analysis.
Figure 1.
Alpha and beta diversity of gut microbiota. The number of Observed species (A), Chao 1 (B), Shannon (C), and Simpson index (D) between control (n = 8) and GCW (n = 7) groups. (E) Non-metric multidimensional scaling (NMDS) plot. (F) Dendrogram of hierarchical clustering analysis. Data are presented as means ± SEM. *p < 0.05, **p < 0.01.
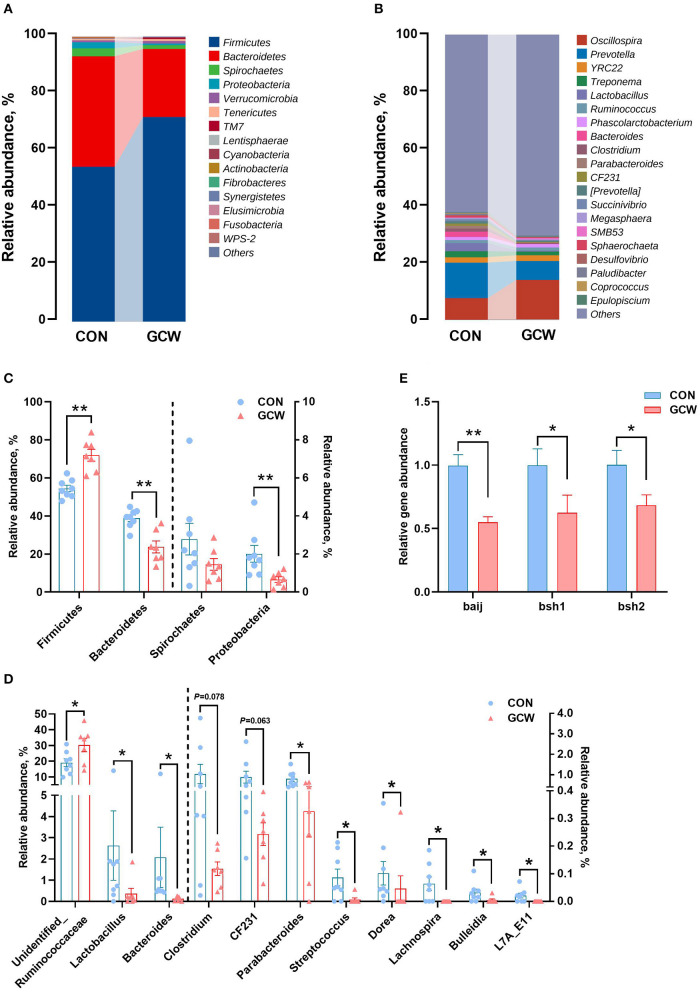
Firmicutes, Bacteroidetes, Spirochaetes, and Proteobacteria were the four most dominant phyla (Figure 2A). At the genus level, Oscillospira, Prevotella, YRC22, and Treponema were the four most abundant genera (Figure 2B). The GCW group showed higher abundances of Firmicutes and lower abundances of Bacteroidetes and Proteobacteria compared with the control group (Figure 2C). The GCW group had lower abundances of Lactobacillus, Bacteroides, Parabacteroides, Streptococcus, Dorea, Lachnospira, Bulleidia, and L7A_E11 and greater abundances of Unidentified_Ruminococcaceae compared with the control group (Figure 2D). In addition, GCW treatment tended to reduce the relative abundance of Clostridium and CF231 compared with the levels in the control group (Figure 2D). At the family level, the GCW group showed higher abundances of Ruminococcaceae and lower abundances of Lactobacillaceae, Bacteroidaceae, and RF16 compared with the levels in the control group (Supplementary Figure 1). Notably, dietary GCW supplementation significantly reduced expression level of baij gene for Clostridium scinden, bsh1 gene for Lactobacillus plantarum, and bsh2 gene for Bacteroides ovatus compared with the levels in the control group (Figure 2E). LEfSe analysis showed similar changes in composition of gut microbiota at the phyla, family, and genus taxonomic levels (Supplementary Figure 2).
Figure 2.
Gut microbiota composition profiles. Bar plot showing relative abundance of microbiota at the phylum (A) and genus (B) levels for the control and GCW groups. Different levels of bacteria at the phylum (C) and genus (D) levels between control (n = 8) and GCW (n = 7) groups. (E) Relative abundance of genes involved in bile acid 7α-dehydroxylation (baij) and bile salt hydrolysis (bsh1 and bsh2) between control (n = 15) and GCW (n = 14) groups. Data are presented as means ± SEM. *p < 0.05, **p < 0.01.
Bile Acid Homeostasis-Related Parameters in Plasma
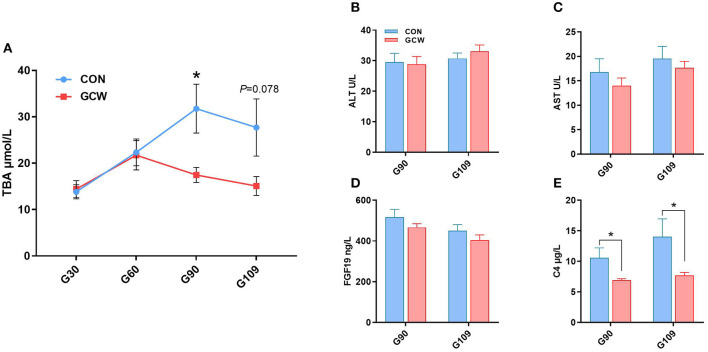
To explore the effect of GCW on bile acid homeostasis, levels of several related plasma biological parameters were determined. Plasma TBA levels in the GCW group on day 90 of gestation were significantly reduced by 45.04% compared with the levels in the control group (Figure 3A). Moreover, GCW treatment tended to decrease plasma TBA level on day 109 of gestation compared with the level in the control group (Figure 3A). However, the two groups showed no significant difference in the levels of plasma ALT, AST, and FGF19 during late gestation (days 90 and 109) (Figures 3B–D). Notably, GCW diet significantly reduced plasma C4 concentration during late gestation compared with the control diet (Figure 3E).
Figure 3.
(A) Dynamic change of maternal peripheral plasma TBA between the two groups across gestation (n = 7–10/group). Plasma concentrations of ALT (B), AST (C), FGF19 (D), and C4 (E) between the two groups during late gestation (n = 8–10/group). Data are presented as means ± SEM. *p < 0.05.
Plasma Bile Acids Composition
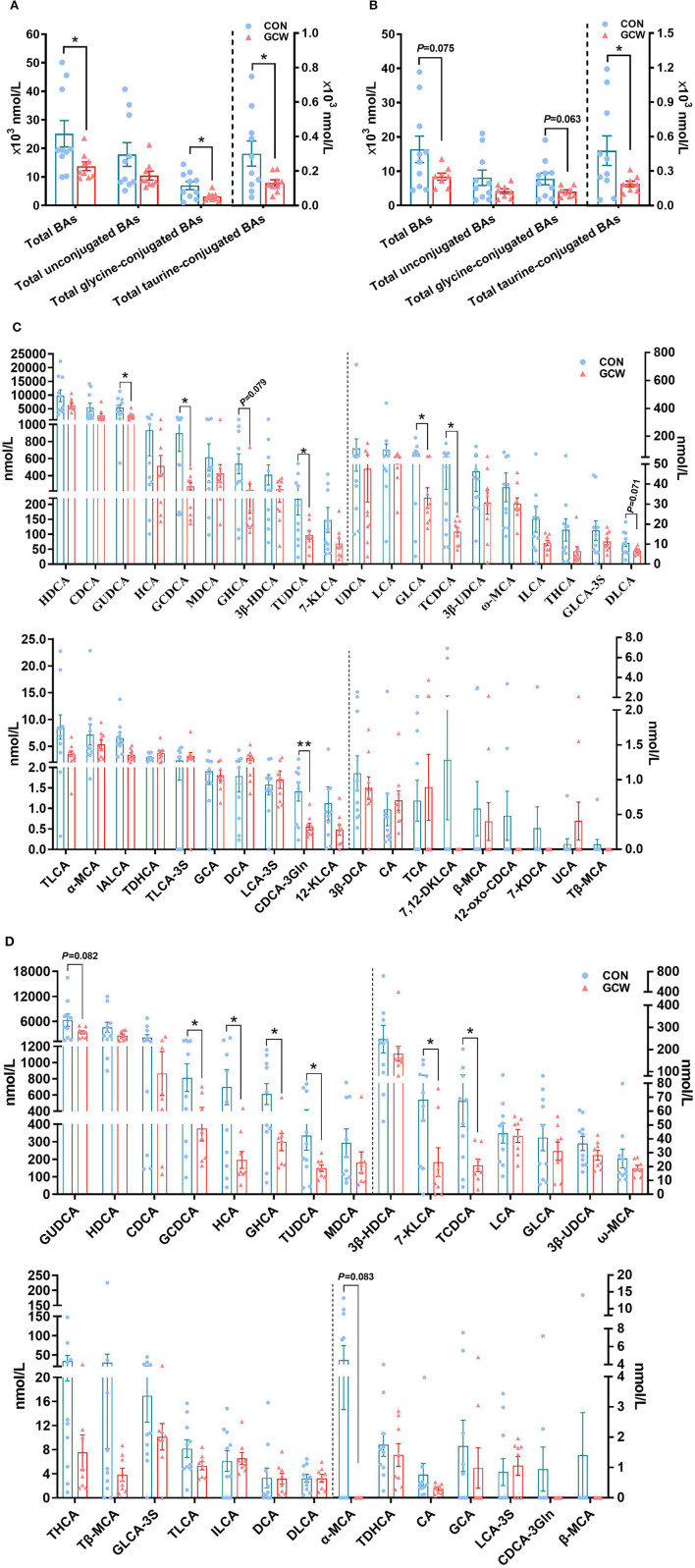
To further characterize the effect of GCW feeding on bile acid homeostasis of sows, we analyzed bile acids composition in plasma samples obtained from control and GCW groups. Most plasma bile acids were unconjugated, followed by glycine-conjugated bile acids and taurine-conjugated bile acids (Figures 4A,B). Furthermore, analysis of individual bile acids showed high abundance of HDCA and CDCA and GUDCA bile acids in plasma (Figures 4C,D). In addition, the concentrations of plasma total bile acids, total glycine-conjugated bile acids and total taurine-conjugated bile acids in the GCW group on day 90 of gestation were significantly lower compared with the levels in the control group (Figure 4A). Moreover, concentrations of total taurine-conjugated bile acids were significantly reduced by GCW treatment, and level of total bile acids and total glycine-conjugated bile acids tended to decrease on day 109 of gestation in the GCW group compared with the control group (Figure 4B). Plasma levels of GUDCA, GCDCA, TUDCA, GLCA, TCDCA, and CDCA-3Gln were significantly decreased in GCW group on day 90 of gestation compared with the control group (Figure 4C). In addition, GCW intervention induced lower levels of GCDCA, HCA, GHCA, 7-KLCA, and TCDCA in plasma of sows on day 109 of gestation (Figure 4D).
Figure 4.
Plasma bile acid composition between the two groups at day 90 and day 109 of gestation. Plasma total bile acids, total unconjugated bile acids, total glycine-conjugated, and total taurine-conjugated bile acids at day 90 (A) and day 109 (B) of gestation. Plasma individual bile acids at day 90 (C) and day 109 (D) of gestation. Data are presented as means ± SEM (n = 8–10/group). *p < 0.05, **p < 0.01.
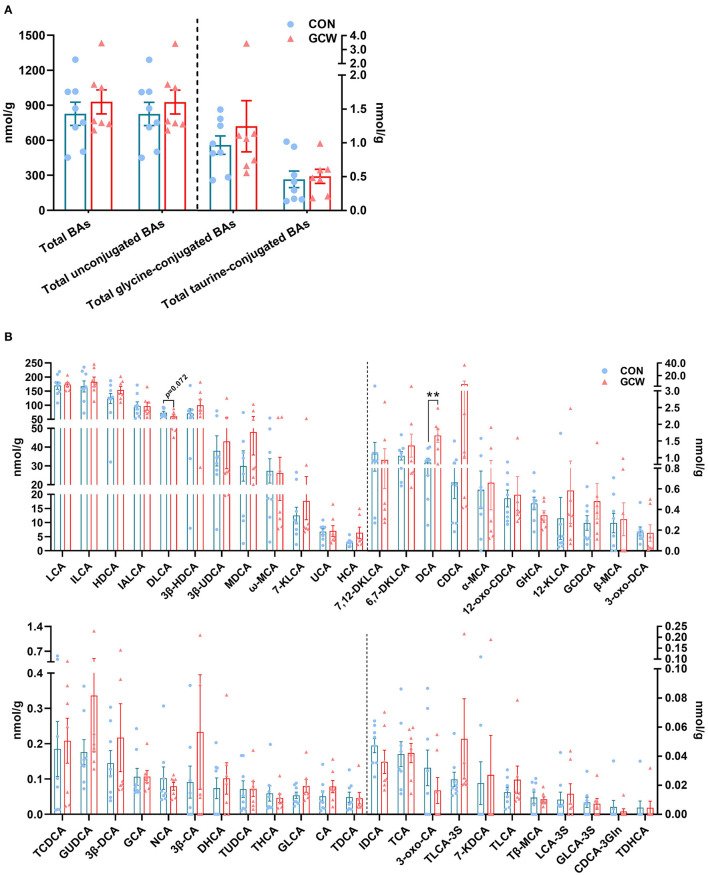
Fecal Bile Acids Composition
Alterations in gut microbial flora induced by GCW treatment affected fecal bile acids composition. Predominant bile acids in fecal samples were unconjugated, followed by glycine-conjugated bile acids and taurine-conjugated bile acids (Figure 5A). Notably, LCA and ILCA and HDCA and IALCA were the most abundant fecal bile acids (Figure 5B). GCW had no significant effect on fecal total bile acids, total unconjugated bile acids, total glycine-conjugated bile acids or total taurine-conjugated bile acids (Figure 5A). In addition, GCW treatment upregulated the levels of fecal HDCA, 3β-HDCA, MDCA, 7-KLCA, HCA, DCA, and CDCA, with higher levels observed for DCA (Figure 5B). However, the levels of fecal DLCA decreased in GCW group compared with the control (Figure 5B).
Figure 5.
Fecal bile acid composition between the two groups on day 109 of gestation. (A) Fecal total bile acids, total unconjugated bile acids, total glycine-conjugated bile acids, and total taurine-conjugated bile acids. (B) Fecal individual bile acids. Data are presented as means ± SEM (n = 8 sows in the control group and 7 sows in the GCW group). **p < 0.01.
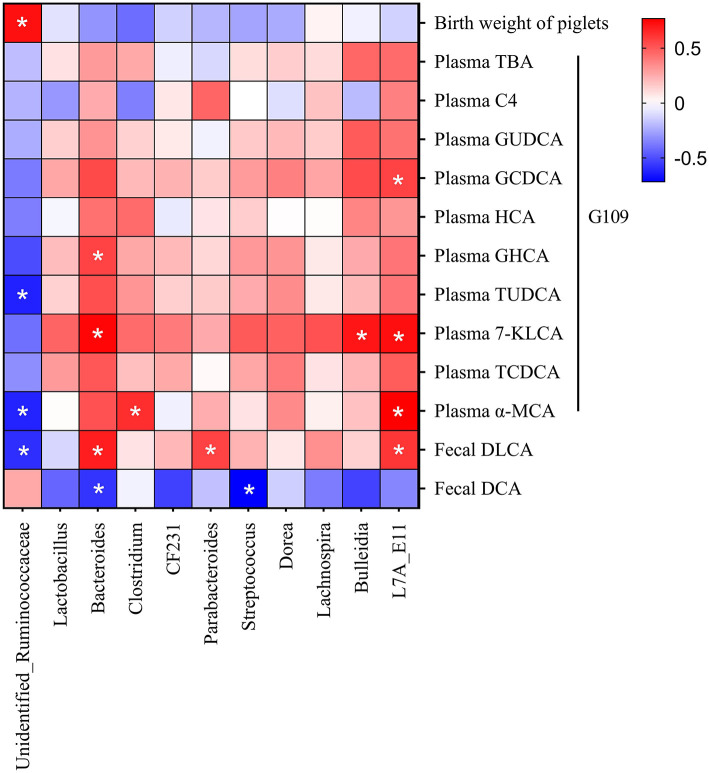
Correlation Between Microbiome and Parameters of Bile Acid Metabolism and Reproductive Performance of Sows
Spearman correlation analysis was conducted to explore the potential link between microbiome and parameters of bile acid metabolism and reproductive performance of sows (Figure 6). Unidentified_Ruminococcaceae was positively correlated with birth weight of piglets, meanwhile negatively correlated with plasma TUDCA, plasma α-MCA, and fecal DLCA. Bacteroides was negatively correlated with fecal DCA, but positively correlated with plasma GHCA, plasma 7-KLCA and fecal DLCA. Additionally, Clostridium was positively correlated with plasma α-MCA, Parabacteroides was positively correlated with fecal DLCA, and Streptococcus was negatively correlated with fecal DCA.
Figure 6.
Correlation between microbiome and parameters of bile acid metabolism, and reproductive performance of sows. Red indicates positive correction, and blue indicates negative correction. *p < 0.05.
Discussion
Previous studies have explored interaction between dietary fiber, bile acids, and the gut microbiota (11). However, the effects of dietary soluble fiber on bile acid homeostasis during gestation have not been fully explored. Bile acids bind to the nuclear receptor FXR, which is primarily expressed in hepatocytes and distal ileal enterocytes, and thus regulate their own biosynthesis and transport (27). A recent study on pregnant women and mice reported reduced ileal FXR-mediated suppression of bile acid synthesis in the liver, resulting in hypercholanemia (28). In this study, dietary GCW treatment reduced plasma C4 level (reflecting lower hepatic bile acid synthesis). GCW-fed sows showed lower plasma primary bile acids, such as GCDCA, GHCA, and TCDCA, implying that GCW reduced hepatic bile acid synthesis. Bile acids such as HCA species (e.g., HCA and HDCA), GUDCA and TUDCA are antagonists of FXR (12, 29). Furthermore, plasma levels of GUDCA, TUDCA and GHCA were lower in the GCW group, which inhibited FXR signaling activity thus increasing hepatic bile acid synthesis. Notably, the levels of plasma conjugated bile acids were lower in the GCW group than in controls, which implies that conjugated bile acids were harmful to fetal pigs.
Guar gum binds to bile acids in the intestinal lumen and enhances their excretion in feces (30), which, in turn, lowers TBA concentration in plasma. However, concentration of fecal total bile acids was not significantly different between the two groups, which could be due to differences in fiber composition. The fecal total bile acid excretion was not evaluated in this, which is a potential limitation to our study. An increased intestinal permeability was detected in ICP patients during pregnancy, which may enhance the absorption of bacterial endotoxin (31). Furthermore, the disorder of bile acid metabolism in the late stage of pregnant mice could be induced by intraperitoneal injection of lipo-polysaccharide (LPS) (32). Notably, dietary GCW treatment reduced intestinal permeability and plasma endotoxin level (date not shown) and increased relative abundances of butyrate-producing bacteria (Ruminococcaceae) on day 109 of gestation. Also, GCW-fed sows showed higher plasma and fecal butyrate levels on day 109 of gestation (date not shown), which led to a strengthened gut barrier function. Therefore, the present study did not exclude that dietary GCW treatment reduced plasma TBA levels via downregulating plasma endotoxin levels of sows during gestation.
Previous studies report that dietary fiber modulates intestinal microbiota composition (33). Decreased microbiome diversity is a feature of dysbiosis (34). In this study, dietary GCW treatment reduced microbiome alpha diversity, possibly by selectively inhibiting the growth of harmful microorganisms (such as several members of the phyla Proteobacteria), which was consistent with findings from prior studies (35, 36). In the large intestine, gut microbiota-derived bile salt hydrolases (BSH) deconjugates host-derived conjugated primary bile acids to produce free primary bile acids, taurine and glycine (9). Metagenomic analyses show presence of BSH genes in Gram-positive bacteria genera (e.g., Lactobacillus, Bifidobacterium, Enterococcus, Clostridium, and Listeria) and Gram-negative bacteria genera (e.g., Bacteroides) (37). Upon deconjugation, the hydroxyl group at the C7 position of bile acids is then removed by 7α-dehydroxylating bacteria (e.g., Clostridium and Eubacterium), thus converting primary bile acids to secondary bile acids (38). In this study, dietary soluble fiber treatment increased the relative abundance of Ruminococcaceae, which was correlated with increase in the level of fecal DCA. Notably, members of Ruminococcaceae are can carry out 7α-dehydroxylation resulting in formation of secondary bile acids (39, 40). The bsh1 and bsh2 genes participated in converting conjugated bile acids to unconjugated bile acids (41). The baij gene could participate in the process of converting primary bile acids into secondary bile acids (42). Microbiota-derived bile acids, LCA and DCA and their taurine and glycine conjugated forms have high affinities for G protein Bile acid-activated receptor (GPBAR)-1, also known as TGR5 (43). Furthermore, the relative abundance of BSH-encoding bacteria (including Lactobacillus and Bacteroides) were lower in the GCW group compared with the levels in the control group. However, the concentration of fecal total glycine-conjugated and taurine-conjugated bile acids were not significantly different between two groups. Notably, the abundance of bile acid-metabolizing intestinal bacteria coincided with the expression levels of bacterial bile acid metabolism-related genes.
Conclusion
The findings of this study show that dietary GCW intervention can enhance reproductive performance probably by maintaining bile acid homeostasis in sows during gestation. Moreover, dietary GCW treatment increased levels of fecal DCA by regulating gut microbiota in sows.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA771933.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee of Huazhong Agricultural University. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
JP conceived and designed the experiments and wrote and revised the manuscript. XW performed the experiments, analyzed the data, and wrote part of the manuscript. SY and CC performed the experiments and took part in the data analysis. CX analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Foundation of China (31772609) and Hubei Province Science and Technology Innovation Major Project (2019ABA081).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
Abbreviations
- TBA
total bile acid
- FGF19
fibroblast growth factor 19
- C4
7α-hydroxy-4-cholesten-3-one
- ALT
alanine aminotransferas
- AST
alanine aminotransferase
- FXR
farnesoid X receptor
- TGR5
Takeda G protein-coupled receptor
- TNF-α
tumor necrosis factor-α
- BSH
bile salt hydrolase
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- 12-KLCA
12-ketolithocholic acid
- 12-oxo-CDCA
12-oxochenodeoxycholic acid
- 3-oxo-CA
3-oxocholic acid
- 3-oxo-DCA
3-oxodeoxycholic acid
- 3β-CA
3β-cholic acid
- 3β-DCA
3β-deoxycholic acid
- 3β-HDCA
β-hyodeoxycholic acid
- 3β-UDCA
3β-ursodeoxycholic acid
- 6, 7-DKLCA, 6
7-diketolithocholic acid
- 7, 12-DKLCA, 7
12-diketolithocholic acid
- 7-KDCA
7-ketodeoxycholic acid
- 7-KLCA
7-ketolithocholic acid
- CA
cholic acid
- CDCA-3Gln
chenodeoxycholic acid-3-β-D-glucuronide
- CDCA
chenodeoxycholic acid
- DCA
deoxycholic acid
- DHCA
dehydrocholic acid
- DLCA
dehydrolithocholic acid
- GCA
glycocholic acid
- GCDCA
glycochenodeoxycholic acid
- GHCA
glycohyocholic acid
- GLCA
glycolithocholic acid
- GLCA-3S
glycolithocholic acid-3-sulfate
- GUDCA
glycoursodeoxycholic acid
- HCA
hyocholic acid
- HDCA
hyodeoxycholic acid
- IALCA
isoallolithocholic acid
- IDCA
isodeoxycholic acid
- ILCA
isolithocholic acid
- LCA
lithocholic acid
- LCA-3S
lithocholic acid-3-sulfate
- MDCA
murideoxycholic acid
- NCA
norcholic acid
- TCA
Taurocholic acid
- TCDCA
taurochenodeoxycholic acid
- TDCA
taurodeoxycholic acid
- TDHCA
taurodehydrocholic acid
- THCA
taurohyocholic acid
- TLCA
taurolithocholic acid
- TLCA-3S
taurolithocholic acid-3-sulfate
- TUDCA
tauroursodeoxycholic acid
- Tβ-MCA
tauro-β-muricholic acid
- UCA
ursocholic acid
- α-MCA
α-muricholic acid
- β-MCA
β-muricholic acid
- ω-MCA
ω-muricholic acid.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.756910/full#supplementary-material
References
- 1.Joutsiniemi T, Timonen S, Leino R, Palo P, Ekblad U. Ursodeoxycholic acid in the treatment of intrahepatic cholestasis of pregnancy: a randomized controlled trial. Arch Gynecol Obstetr. (2014) 289:541–7. 10.1007/s00404-013-2995-5 [DOI] [PubMed] [Google Scholar]
- 2.Ovadia C, Seed PT, Sklavounos A, Geenes V, Di Ilio C, Chambers J, et al. Association of adverse perinatal outcomes of intrahepatic cholestasis of pregnancy with biochemical markers: results of aggregate and individual patient data meta-analyses. Lancet. (2019) 393:899–909. 10.1016/S0140-6736(18)31877-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.You S, Cui A.-M., Hashmi SF, Zhang X, Nadolny C, et al. Dysregulation of bile acids increases the risk for preterm birth in pregnant women. Nat Commun. (2020) 11:2111. 10.1038/s41467-020-15923-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li L, Chen W, Ma L, Liu ZB, Lu X, Gao XX, et al. Continuous association of total bile acid levels with the risk of small for gestational age infants. Sci Rep. (2020) 10:9257. 10.1038/s41598-020-66138-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang P, Zhong H, Song Y, Yuan P, Li Y, Lin S, et al. Targeted metabolomics analysis of maternal-placental-fetal metabolism in pregnant swine reveals links in fetal bile acid homeostasis and sulfation capacity. Am J Physiol Gastrointestinal Liver Physiol. (2019) 317:G8–16. 10.1152/ajpgi.00056.2019 [DOI] [PubMed] [Google Scholar]
- 6.Esti ú MC, Monte MJ, Rivas L, Moirón M, Gomez-Rodriguez L, Rodriguez-Bravo T, et al. Effect of ursodeoxycholic acid treatment on the altered progesterone and bile acid homeostasis in the mother-placenta-foetus trio during cholestasis of pregnancy. Br J Clin Pharmacol. (2015) 79:316–29. 10.1111/bcp.12480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brouwers L, Koster MP, Page-Christiaens GC, Kemperman H, Boon J, Evers IM, et al. Intrahepatic cholestasis of pregnancy: maternal and fetal outcomes associated with elevated bile acid levels. Am J Obstet Gynecol. (2015) 212:100.e1–7. e107. 10.1016/j.ajog.2014.07.026 [DOI] [PubMed] [Google Scholar]
- 8.Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall H-U, Bamberg K, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. (2013) 17:225–35. 10.1016/j.cmet.2013.01.003 [DOI] [PubMed] [Google Scholar]
- 9.Wahlström A, Sayin SI, Marschall H-U, Bäckhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. (2016) 24:41–50. 10.1016/j.cmet.2016.05.005 [DOI] [PubMed] [Google Scholar]
- 10.Winston JA, Theriot CM. Diversification of host bile acids by members of the gut microbiota. Gut Microb. (2020) 11:158–71. 10.1080/19490976.2019.1674124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh J, Metrani R, Shivanagoudra SR, Jayaprakasha GK, Patil BS. Review on bile acids: effects of the gut microbiome, interactions with dietary fiber, and alterations in the bioaccessibility of bioactive compounds. J Agric Food Chem. (2019) 67:9124–38. 10.1021/acs.jafc.8b07306 [DOI] [PubMed] [Google Scholar]
- 12.Sun L, Xie C, Wang G, Wu Y, Wu Q, Wang X, et al. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat Med. (2018) 24:1919–29. 10.1038/s41591-018-0222-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ginos BN, Navarro SL, Schwarz Y, Gu H, Wang D, Randolph TW, et al. Circulating bile acids in healthy adults respond differently to a dietary pattern characterized by whole grains, legumes and fruits and vegetables compared to a diet high in refined grains and added sugars: a randomized, controlled, crossover feeding study. Metabolism. (2018) 83:197–204. 10.1016/j.metabol.2018.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunness P, Michiels J, Vanhaecke L, De Smet S, Kravchuk O, Van de Meene A, et al. Reduction in circulating bile acid and restricted diffusion across the intestinal epithelium are associated with a decrease in blood cholesterol in the presence of oat β-glucan. FASEB J. (2016) 30:4227–38. 10.1096/fj.201600465R [DOI] [PubMed] [Google Scholar]
- 15.Gunness P, Williams BA, Gerrits WJ, Bird AR, Kravchuk O, Gidley MJ. Circulating triglycerides and bile acids are reduced by a soluble wheat arabinoxylan via modulation of bile concentration and lipid digestion rates in a pig model. Mol Nutr Food Res. (2016) 60:642–51. 10.1002/mnfr.201500686 [DOI] [PubMed] [Google Scholar]
- 16.Gylling H, Riikonen S, Nikkil ä K, Savonius H, Miettinen TA. Oral guar gum treatment of intrahepatic cholestasis and pruritus in pregnant women: effects on serum cholestanol and other non-cholesterol sterols. Europ J Clin Invest. (1998) 28:359–63. 10.1046/j.1365-2362.1998.00282.x [DOI] [PubMed] [Google Scholar]
- 17.Riikonen S, Savonius H, Gylling H, Nikkil ä K, Tuomi A.-M., et al. Oral guar gum, a gel-forming dietary fiber relieves pruritus in intrahepatic cholestasis of pregnancy. Acta Obstetr Gynecol Scand. (2000) 79:260–4. 10.1034/j.1600-0412.2000.079004260.x [DOI] [PubMed] [Google Scholar]
- 18.Tan C, Wei H, Ao J, Long G, Peng J. Inclusion of konjac flour in the gestation diet changes the gut microbiota, alleviates oxidative stress, and improves insulin sensitivity in sows. Appl Environ Microbiol. (2016) 82:5899–909. 10.1128/AEM.01374-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhuo Y, Shi X, Lv G, Hua L, Zhou P, Che L, et al. Beneficial effects of dietary soluble fiber supplementation in replacement gilts: pubertal onset and subsequent performance. Anim Reproduct Sci. (2017) 186:11–20. 10.1016/j.anireprosci.2017.08.007 [DOI] [PubMed] [Google Scholar]
- 20.National Research Council (NRC) (2012). Nutrient Requirements of Swine. [Google Scholar]
- 21.Xiang Q, Wu X, Pan Y, Wang L, Guo Y, Cui C, et al. Early intervention using fecal microbiota transplantation combined with probiotics influence the growth performance, diarrhea, and intestinal barrier function of piglets. Appl Sci. (2020) 10:568. 10.3390/app10020568 [DOI] [Google Scholar]
- 22.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. (2019) 37:852–857. 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. (2011) 17:10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 24.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Mehods. (2016) 13:581–3. 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katoh K, Misawa K, Kuma Ki, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. (2002) 30:3059–66. 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evolut. (2009) 26:1641–50. 10.1093/molbev/msp077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Aguiar Vallim TQ, Tarling EJ, Edwards PA. Pleiotropic roles of bile acids in metabolism. Cell Metab. (2013) 17:657–69. 10.1016/j.cmet.2013.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ovadia C, Perdones-Montero A, Spagou K, Smith A, Sarafian MH, Gomez-Romero M, et al. Enhanced microbial bile acid deconjugation and impaired ileal uptake in pregnancy repress intestinal regulation of bile acid synthesis. Hepatology. (2019) 70:276–93. 10.1002/hep.30661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng X, Chen T, Jiang R, Zhao A, Wu Q, Kuang J, et al. Hyocholic acid species improve glucose homeostasis through a distinct TGR5 and FXR signaling mechanism. Cell Metab. (2020) 33:791–803. 10.2139/ssrn.3528695 [DOI] [PubMed] [Google Scholar]
- 30.Turner P, Tuomilehto J, Happonen P, La Ville A, Shaikh M, Lewis B. Metabolic studies on the hypolipidaemic effect of guar gum. Atherosclerosis. (1990) 81:145–50. 10.1016/0021-9150(90)90021-A [DOI] [PubMed] [Google Scholar]
- 31.Reyes H, Zapata R, Hernández I, Gotteland M, Sandoval L, Jirón MI, et al. Is a leaky gut involved in the pathogenesis of intrahepatic cholestasis of pregnancy? Hepatology. (2006) 43:715–22. 10.1002/hep.21099 [DOI] [PubMed] [Google Scholar]
- 32.Zhang C, Gan Y, Lv JW, Qin MQ, Hu WR, Liu ZB, et al. The protective effect of obeticholic acid on lipopolysaccharide-induced disorder of maternal bile acid metabolism in pregnant mice. Int Immunopharmacol. (2020) 83:106442. 10.1016/j.intimp.2020.106442 [DOI] [PubMed] [Google Scholar]
- 33.Sanders ME, Merenstein DJ, Reid G, Gibson GR, Rastall RA. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat Rev Gastroenterol Hepatol. (2019) 16:605–616. 10.1038/s41575-019-0173-3 [DOI] [PubMed] [Google Scholar]
- 34.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. (2013) 500:541–6. 10.1038/nature12506 [DOI] [PubMed] [Google Scholar]
- 35.Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. (2018) 359:1151–6. 10.1126/science.aao5774 [DOI] [PubMed] [Google Scholar]
- 36.Zou J, Chassaing B, Singh V, Pellizzon M, Ricci M, Fythe MD, et al. Fiber-mediated nourishment of gut microbiota protects against diet-induced obesity by restoring IL-22-mediated colonic health. Cell Host Microb. (2018) 23:41–53. e44. 10.1016/j.chom.2017.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gérard P. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens. (2014) 3:14–24. 10.3390/pathogens3010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Devlin AS, Fischbach MA. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat Chem Biol. (2015) 11:685–90. 10.1038/nchembio.1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. (2013) 58:949–55. 10.1016/j.jhep.2013.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sinha SR, Haileselassie Y, Nguyen LP, Tropini C, Wang M, Becker LS, et al. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microb. (2020) 27:659–70.e655. 10.1016/j.chom.2020.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duary RK, Batish VK, Grover S. Relative gene expression of bile salt hydrolase and surface proteins in two putative indigenous Lactobacillus plantarum strains under in vitro gut conditions. Mol Biol Rep. (2012) 39:2541–52. 10.1007/s11033-011-1006-9 [DOI] [PubMed] [Google Scholar]
- 42.Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. (2013) 499:97–101. 10.1038/nature12347 [DOI] [PubMed] [Google Scholar]
- 43.Jia W, Xie G, Jia W. Bile acid–microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol. (2018) 15:111. 10.1038/nrgastro.2017.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA771933.