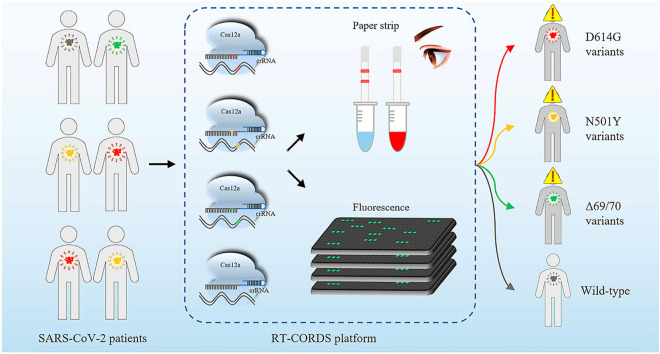
Abstract
The increasing prevalence of SARS-CoV-2 variants with spike mutations has raised concerns owing to higher transmission rates, disease severity, and escape from neutralizing antibodies. Rapid and accurate detection of SARS-CoV-2 variants provides crucial information concerning the outbreaks of SARS-CoV-2 variants and possible lines of transmission. This information is vital for infection prevention and control. We used a Cas12a-based RT-PCR combined with CRISPR on-site rapid detection system (RT-CORDS) platform to detect the key mutations in SARS-CoV-2 variants, such as 69/70 deletion, N501Y, and D614G. We used type-specific CRISPR RNAs (crRNAs) to identify wild-type (crRNA-W) and mutant (crRNA-M) sequences of SARS-CoV-2. We successfully differentiated mutant variants from wild-type SARS-CoV-2 with a sensitivity of 10−17 M (approximately 6 copies/μL). The assay took just 10 min with the Cas12a/crRNA reaction after a simple RT-PCR using a fluorescence reporting system. In addition, a sensitivity of 10−16 M could be achieved when lateral flow strips were used as readouts. The accuracy of RT-CORDS for SARS-CoV-2 variant detection was 100% consistent with the sequencing data. In conclusion, using the RT-CORDS platform, we accurately, sensitively, specifically, and rapidly detected SARS-CoV-2 variants. This method may be used in clinical diagnosis.
Keywords: CRISPR/Cas12, RT-CORDS, D614G, N501Y, 69/70 deletion, SARS-CoV-2 variants
Graphical abstract
1. Introduction
A number of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants have emerged since its first outbreak in 2019 (Sanyaolu et al., 2021). Some of these variants (termed as Alpha/Beta/Gamma/Delta), which have been reported to influence viral transmissibility, disease severity, diagnostics, and vaccine efficacy, were defined as the variants of concern (VOC) by the World Health Organization (WHO) (World Health Organization, 2021). The emergence of VOC has reported in >190 countries and regions, and >68% of global samples are positive for VOC (Table S1).
Because the spike protein mediates SARS-CoV-2 entry into the host and is also a significant target of neutralizing antibodies and vaccines, some key mutations of VOC occurring in the spike protein (e.g., D614G, N501Y and 69/70 deletion) have garnered significant scientific attention (Harvey et al., 2021; Salvatori et al., 2020). D614G, one of the key mutations, is a nonsynonymous amino acid change in the spike protein due to a 23,403 A>G single-base substitution in the SARS-CoV-2 reference sequence; the mutation was first identified in early 2020 (Korber et al., 2020). It is found in four SARS-CoV-2 VOC: B.1.1.7, P1, P1.351, and B.1.617.2. Patients with G614 SARS-CoV-2 show higher viral loads and 61% higher mortality than those with D614-SARS-CoV-2 (Davies et al. 2021a, 2021b; Korber et al., 2020; Plante et al., 2020). In addition, D614G variants have been demonstrated to increase the transmissibility in in vitro human cell models (from 1.3 to 7.7 folds) and in vivo animal models. The D614G mutation may alter proteolytic cleavage and further increase S1 shedding, or it could enhance the binding affinity between RBD and AEC2 (Daniloski et al., 2021; Hou et al., 2020; Plante et al., 2020; Wang et al., 2021a; Zhang et al., 2020).
N501Y is also a nonsynonymous amino acid change due to 23,063 A>T substitution in the SARS-CoV-2 reference sequence. This mutation is identified in various VOC such as P1, B.1.1.7, and B.1.351 lineages. Clinical data indicate that N501Y-positive VOC significantly increase the risks of hospitalization (59% higher), intensive care unit admission (105% higher), and death (61% higher) (Fisman and Tuite, 2021). Recent evidence indicates that the N501Y mutation enhances the affinity to bind with the host angiotensin-converting enzyme 2 (ACE2) receptor by up to 7-fold more than that of the wild-type (WT) (Ali et al., 2021; Laffeber et al., 2021; Li et al., 2021b; Liu et al., 2021) and also enhances viral transmission both in vivo (1.0–5.3 fold) and in vitro (1.3–5.4 fold) (Liu et al., 2021). Furthermore, the N501Y mutation exhibits poorer binding potential for 82.7% of the common alleles of the major histocompatibility complex than N501 and might escape immune defenses (Castro et al., 2021). The 69/70 deletion results from 204 to 209del in the S gene and has been found in both B.1.1.7 and B.1.526.1 variants. This small deletion is associated with the S-gene target failure of a three-target reverse transcription polymerase chain reaction (RT-PCR) assay (Bal et al., 2021). Recently, a 2-fold increase in pseudovirus infectivity of 69/70 deletion was observed in several cell models of ACE2 expression (Meng et al., 2021a). Therefore, owing to the prominent impacts on viral transmissibility and disease severity, SARS-CoV-2 VOC or other variants with key mutations should be monitored and identified for enhanced epidemic control and improved clinical therapy.
Until now, sequencing has been the gold standard for identifying SARS-CoV-2 variant mutations. This method is accurate but time-consuming and costly and must be performed off-site. Moreover, quantitative RT-PCR (RT-qPCR) has been developed as a gold standard for detection of SARS-CoV-2 infection, identification of SARS-CoV-2 mutations with mutation-matched primers or probes, and identification through amplification and melting curve analyses (Aoki et al., 2021; Bedotto et al., 2021; Vega-Magaña et al., 2021; Zelyas et al., 2021). However, primer/probe mismatches in a qPCR may only slightly alter these curves (Boyle et al., 2009; Stadhouders et al., 2010; Süß et al., 2009). Thus, the PCR-based identification of single-base mutations is nonspecific and not sufficiently reliable. In addition, performing quantitative RT-PCR assays requires professional operations and bulky instruments.
Once Cas12a/crRNA bound to an activator (ssDNA or dsDNA) that has complementary base-pairing to the guide crRNA, the trans-acting cleavage of Cas12a is initiated and then capable of nonspecific cleavage of ssDNA (single-stranded DNA) (Chen et al., 2018). When combined with a nonspecific ssDNA reporter, Cas12a has been used to detect nucleic acids reliably (Bai et al., 2019; Ding et al., 2021; Gong et al. 2021a, 2021b; Ma et al., 2021; Wang et al., 2021b). By introducing an additive mismatch in crRNA, the CRISPR/Cas12 system can even be used for single-nucleotide polymorphism (SNP) genotyping with single-base specificity (Chen et al., 2021; Huang et al., 2021; Lee et al., 2020; Lee Yu et al., 2021; Meng et al., 2021b). The identification of a single base makes Cas12 an ideal approach for mutation detection.
Therefore, we assessed the detection of SARS-CoV-2 mutations of concern (69/70 deletion, N501Y, and D614G) using an improved Cas12a-based CRISPR on-site rapid detection system (CORDS) platform—a rapid, sensitive, and specific on-site biosensing system developed in our laboratory. We designed and screened mutant (MT)-specific crRNA (crRNA-M). Cas12/crRNA-M could specifically and rapidly differentiate SARS-CoV-2 mutations compared with WT-specific crRNA (crRNA-W). The sensitivity was 10−17 M (6 copies/μL) with a fluorescence reporting system and up to 10−16 M (60 copies/μL) using a lateral flow strip reporting system. The accuracy of RT-CORDS is 100% consistent with that of the sequencing method. These findings indicate that the improved RT-CORDS platform is a powerful tool for monitoring the key mutations in VOC and other SARS-CoV-2 variants.
2. Materials and methods
2.1. Materials
LbCas12a, HiScribe T7 High-Yield RNA Synthesis Kit, RNA Cleanup Kit, NEBNext Q5 Hot Start HiFi PCR Master Mix, NEBuffer2.1 were ordered from NEB (Beijing, China). DNaseI, recombination RNase inhibitor (RRI), PrimeSTAR max were ordered from Takara Bio (Beijing, China). T-vectors, Pfu DNA polymerase were ordered from TIANGEN Biotechnology (Beijing, China). One-Step RT-PCR Kit was ordered from Vazyme Biotech (Nanjing, China). Gel Extraction Kit was ordered from Omega Biotech (Shanghai, China). Lateral flow strips were purchased from Magigen Biotech (Guangzhou, China). Oligonucleotides were synthesized by GENEWIZ (Jiangsu, China). S gene DNA targets were synthesized by Generay Biotech (Shanghai, China). Nucleic acid was quantified using Thermo Fisher Nanodrop 1000 Spectrophotom. Fluorescence signals were recorded with Tecan's Spark 20M.
2.2. Design and transcription of crRNAs
We designed the crRNAs for mutation identification based on a previous study (Bai et al., 2019). In brief, an 18-nt sequence following the protospacer adjacent motif (PAM; TTTV), which covers the mutation, was selected as the crRNA. In addition, as a single-base mutation of D614G and N501Y, an additional mismatch was introduced in the seed region of their crRNA-W and crRNA-M to enhance the differentiability between the WT and MT variants (Table S2). For the 69/70 target, TTC was selected as the PAM and no additional mismatch was introduced to the crRNA-W/M because this mutation was small deletion instead of single-base substitution (Table S2).
crRNA transcription templates were prepared using 60-nt oligos containing a T7 promoter, scaffold (Table S3). After oligo-F and oligo-R annealing, the annealed products were ligated to T-vectors to obtain pGM-T-crRNA plasmids. Finally, transcription templates were obtained through PCR amplification with Pfu DNA polymerase from pGM-T-crRNA and were purified using a Gel Extraction Kit.
In vitro transcription of crRNAs used a HiScribe T7 High-Yield RNA Synthesis Kit. Reactions were performed in 20 μL volume at 37 °C for 16 h following the manufacturer's instruction for short RNA transcripts. Finally, the crRNA transcripts were treated and purified with an RNA Cleanup Kit.
2.3. Preparation of WT and MT SARS-CoV-2 DNA and RNA targets
The DNA targets of WT S gene and MT SARS-CoV-2 (covering D614G, N501Y and 69/70 deletion sites) were synthesized (Table S4) and then cloned into pUC57 plasmid.
RNA targets were prepared through in vitro transcription with a HiScribe T7 High-Yield RNA Synthesis Kit. Briefly, RNA transcription templates containing a T7 promoter were amplified from pUC57-WT-DNA and pUC57-MT-DNA with specific primers (Table S2) using the NEBNext Q5 Hot Start HiFi PCR Master Mix according to manufacturer's instruction, followed by purification using a Gel Extraction Kit. In vitro transcription was performed in 20 μL reaction volume, according to a standard RNA synthesis protocol, at 37 °C for 16 h. Finally, RNA transcripts were treated with DNaseI to remove transcription templates, purified with an RNA Cleanup Kit, and quantified using Thermo Fisher Nanodrop 1000 Spectrophotom.
2.4. In vitro cleavage assays
Briefly, LbCas12a-mediated target cleavage assays were performed in 20 μL reaction volume with 50 nM LbCas12a protein, 100 nM crRNA, 1× NEBuffer2.1, 20 U recombination RNase inhibitor (RRI), 5 nM linear pUC57-WT-DNA or pUC57-MT-DNA plasmids, and nuclease-free water. LbCas12a was preincubated with crRNA and RRI in NEBuffer2.1 at room temperature for 10 min to form ribonucleoproteins. Target DNA was then added, and the reaction mixture was incubated at 37 °C for 1 or 2 h. Finally, cleaved products were verified using gel electrophoresis.
To investigate the identification of mutation through CRISPR/Cas12a trans-cleavage of fluorescence reporter, the in vitro trans-cleavage assays were performed in 20 μL reaction volume with 50 nM LbCas12a protein, 100 nM crRNA, 1 × NEBuffer2.1, 20 U recombination RNase inhibitor (RRI), 1 nM pUC57-WT-DNA or pUC57-MT-DNA plasmids, and nuclease-free water. The reaction mixtures were then quickly transferred to a 384-well fluorescence plate reader and incubated at 37 °C. Fluorescence signals were collected every 5 min at an excitation wavelength of 485 nm and an emission wavelength of 535 nm.
2.5. Modified CORDS assay
Bioinformatic alignment of multiple SARS-CoV-2 sequences downloaded from GISAID was performed to identify specific and relatively conserved sequences adjacent to mutations. These sequences were used to design PCR/RT-PCR primers (Table S2).
The modified CORDS fluorescence assay comprises two steps, PCR amplification and Cas12a sensing. PCR reactions were performed in 50 μL reaction volume containing 20 μL of DNA, 4 μL of 10 μM primer F/R mix, 25 μL of 2 × PrimeSTAR max, and nuclease-free water. Thermal cycling was as follows: 3 min at 98 °C for initial denaturation; 30 cycles of 10 s at 98 °C for denaturation, 10 s at 55 °C for annealing, and 5 s at 72 °C for extension; and 5 min at 72 °C for the final extension. For Cas12a reaction, FAM- and BHQ-labeled 12-nt ssDNA reporter (FQ-ssDNA) (FAM-NNNNNNNNNNNN-BHQ) for reporting the collateral cleavage of Cas12a was synthesized. The fluorescence reporting assay was performed following a detection protocol. The whole 20 μL reaction volume contained 50 nM LbCas12a, 100 nM crRNA, and 20 U RNase inhibitor, 1 × NEBuffer2.1, 10 μL PCR product, 500 nM FQ-ssDNA, and nuclease-free water at room temperature. The reaction mixture was then quickly transferred to a 384-well fluorescence plate reader and incubated at 37 °C. Fluorescence signals were detected using Tecan's Spark 20M multimode microplate reader at an excitation wavelength of 485 nm and an emission wavelength of 535 nm. Fluorescence assay kinetics assay were assessed with signals collected every 2.5 or 5 min.
The PCR reaction for the CORDS lateral flow strip assay was performed as described earlier in this section. Digoxin- and biotin-labeled 14-nt ssDNA reporter (DB-ssDNA) (Digoxin-NNNNNNNNNNNNNN-Biotin) was used to report Cas12a collateral activity in the lateral flow strip reporting assay. Briefly, a Cas12a paper strip assay was performed in 40 μL reaction volume containing nuclease-free water, 1× NEBuffer2.1, 40 U RNase inhibitor, 50 nM LbCas12a, 100 nM crRNA, 2 nM DB-ssDNA, and 20 μL PCR product. The reaction mixture was then incubated at 37 °C for 60 min. Finally, lateral flow strips were inserted into the reaction mixture and incubated at room temperature for 5 min for use as readouts.
2.6. RT-CORDS assay
The RT-CORDS fluorescence assay comprises an RT-PCR reaction for amplification and a Cas12a reaction for sensing. A One-Step RT-PCR Kit was used. In brief, 50 μL reaction volume contains 18.5 μL of RNA sample, 4 μL of 10 μM primer F/R mix, 25 μL of 2× One Step mix and 2.5 μL of One Step Enzyme mix. The amplification program was as follows: 30 min at 50 °C for reverse transcription; 3 min at 94 °C for initial denaturation; 30 cycles of 30 s at 94 °C for denaturation, 30 s at 55 °C for annealing, and 30 s at 72 °C for extension; and 5 min at 72 °C for the final extension. The Cas12a reaction was performed using a method similar to that used for the CORDS fluorescence assay, except the RT-PCR product substituted the PCR product.
The RT-PCR reaction for the RT-CORDS lateral strip assay was performed as described earlier in this section. The Cas12a reaction was performed using a method similar to that used for the CORDS lateral flow strip assay, except the RT-PCR product substituted the PCR product.
2.7. SARS-CoV-2 variant detection
RNA samples of 18 SARS-CoV-2 variants were collected from Guangdong Provincial Center for Disease Control and Prevention (CDC) and treated in strict accordance with the WHO-recommended procedure and were used in the RT-CORDS assay to detect real SARS-CoV-2 variants. All samples with N501Y, D614G, and 69/70 deletion mutations were detected. RT-PCR products of the samples were also sequenced for further validation.
2.8. Statistical analysis
All the replicate experiments in this study consisted of three repeats. Uncertainties in mean values are provided as standard errors. Statistical analyses were performed using GraphPad Prism. Statistical significance was assessed using 2-way analysis of variance and multiple comparisons. Significance was considered at *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; ns indicates no significance.
3. Results
3.1. The principle of RT-CORDS based biosensor for SARS-CoV-2 mutation detection
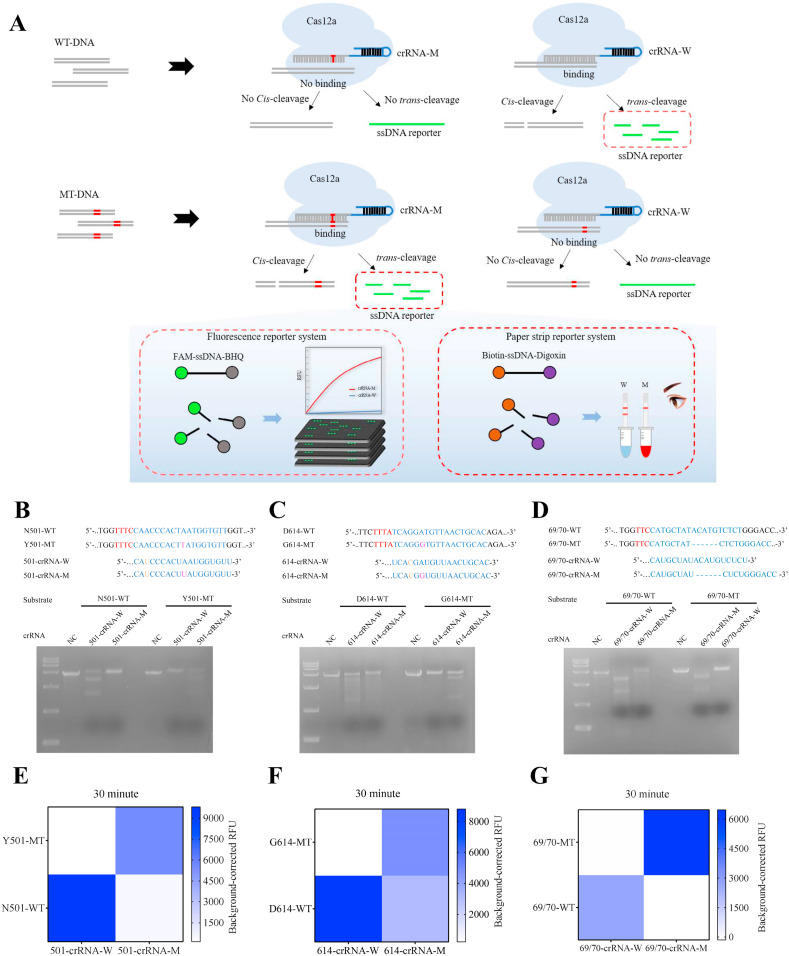
The principle of the biosensor was depicted in Fig. 1 A. Mutant specific Cas12a/crRNA-M and wild-type specific Cas12a/crRNA-W were designed to differentiate MT from WT sample. The crRNAs for single-base mutation were designed by introducing a transition mismatch in the seed region to enhance specificity, such as U-T mismatch in N501Y and C–C mismatch in D614G detection (Fig. 1B and C). Once Cas12a/crRNA-M bound to mutant target, the trans-acting cleavage of Cas12a is initiated and then capable of nonspecific cleavage of ssDNA reporter (FAM-ssDNA-BHQ or Biotin-ssDNA-Digoxin). Finally, the signal of mutant target is detected by fluorescence or paper strip (Fig. 1A).
Fig. 1.
Specific identification of SARS-CoV-2 mutations using CRISPR/Cas12a cis-cleavage and trans-cleavage by MT-specific crRNA-M and WT-specific crRNA-W. (A) Schematic representation of Cas12a/crRNA-W and Cas12a/crRNA-M to differentiate MT-DNA from WT-DNA. (B, C, D) in vitro cleavage assay of N501Y, D614G and 69/70 deletion targets using Cas12a/crRNA-W and Cas12a-crRNA-M. WT: wild-type DNA; MT: mutant DNA; NC: negative control. (E, F, G) Identification of N501Y, D614G and 69/70 deletion targets through CRISPR/Cas12a trans-cleavage of the fluorescence reporter.
3.2. Highly specific SARS-CoV-2 mutation cleavage by modified crRNA
The SARS-CoV-2 reference genome sequence was downloaded from the National Center for Biotechnology Information and mutation information for SARS-CoV-2 variants was obtained from GISAID (https://www.gisaid.org/). Because CRISPR/Cas12a has been reported to differentiate SNPs with single-base resolution (Chen et al., 2021; Lee Yu et al., 2021; Li et al., 2018), we used Cas12a to identify SARS-CoV-2 N501Y, D614G, and 69/70 deletion mutations; these are the key mutations of SARS-CoV-2 VOC (Table S1). These mutations are associated with transmissibility and clinical characteristics of SARS-CoV-2 variants (Hou et al., 2020; Laffeber et al., 2021; Meng et al., 2021a). We synthesized crRNA-W and crRNA-M to target WT and MT sequences for each mutation and performed in vitro cleavage assays to verify the ability of the CRISPR/Cas12a system to identify SARS-CoV-2 mutations (Table S1, Fig. 1A–D).
We designed 501-crRNA-W (N501-WT-specific crRNA) and 501-crRNA-M (Y501-MT-specific crRNA) to detect N501Y mutation (Table S2). The kinetic profiling of single and double mismatches between the crRNA and target (Jones et al., 2021), suggested the introduction of a skillful mismatch to the seed region of both 501-crRNA-W and 501-crRNA-M that make MTs easier to differentiate from WTs. As expected, we found that N501-WT substrate was significantly cleaved in the presence of 501-crRNA-W, but no cleavage was observed in the presence of 501-crRNA-M. Likewise, the 501-MT substrate was cleaved significantly by 501-crRNA-M compared with 501-crRNA-W (Fig. 1B). The abovementioned findings indicate that 501-crRNA-W and 501-crRNA-M can identify N501Y MT variants specifically.
We also introduced a skillful mismatch into the seed region of 614-crRNA-W and 614-crRNA-M for D614G. The targets were respectively recognized and cleaved specifically and efficiently by 614-crRNA-W and 614-crRNA-M (Fig. 1C). However, no canonical PAM TTTV motif was present adjacent to the 69/70 mutation site. We selected TTC as the PAM for LbCas12a recognition (Table S2); LbCas12a can also cleave targets with this PAM (Yamano et al., 2017). We designed 69/70-crRNA-W and 69/70-crRNA-M to specifically target 69/70-WT and 69/70-MT without introducing any artificial mismatch. 69/70-WT substrate was only cleaved by 69/70-crRNA-W, and 69/70-MT substrate was cleaved by only 69/70-crRNA-M (Fig. 1D). We conclude that the specific crRNA-W and crRNA-M that we designed can successfully and specifically identify N501Y, D614G, and 69/70 deletion mutations in SARS-CoV-2 variants.
3.3. CORDS can accurately and rapidly differentiate the DNA targets of SARS-CoV-2 mutations
We have previously established a CORDS platform for in vitro virus nucleic acid detection (Bai et al., 2019). For mutation identification, we also introduced fluorescence probes, 12-nt ssDNAs labeled with 5′ -FAM and 3′-BHQ, to report the collateral activity of Cas12a. We assessed the specificity of mutation discrimination by integrating a fluorescence reporter system. Fluorescence increased solely when 501-crRNA-M was used to detect Y501-MT substrate (Fig. 1E, S1A, and S1B). The same results were observed when the substrate was N501-WT, 69/70-WT, or 69/70-MT (Fig. 1F and G, and S1C–S1F). The G614-MT substrate could only activate Cas12a/614-crRNA-M but not Cas12a/614-crRNA-W. D614-MT can be slightly activated by Cas12a/614-crRNA-W (Fig. 1F and S1D). However, significant differences in fluorescence intensities between 614-crRNA-W and 614-crRNA-M were noted, and this slight activation had no impact on D614G discrimination. Furthermore, kinetic data showed that 30 min is enough time to differentiate the MT variants from the WT (Fig. 1E–G).
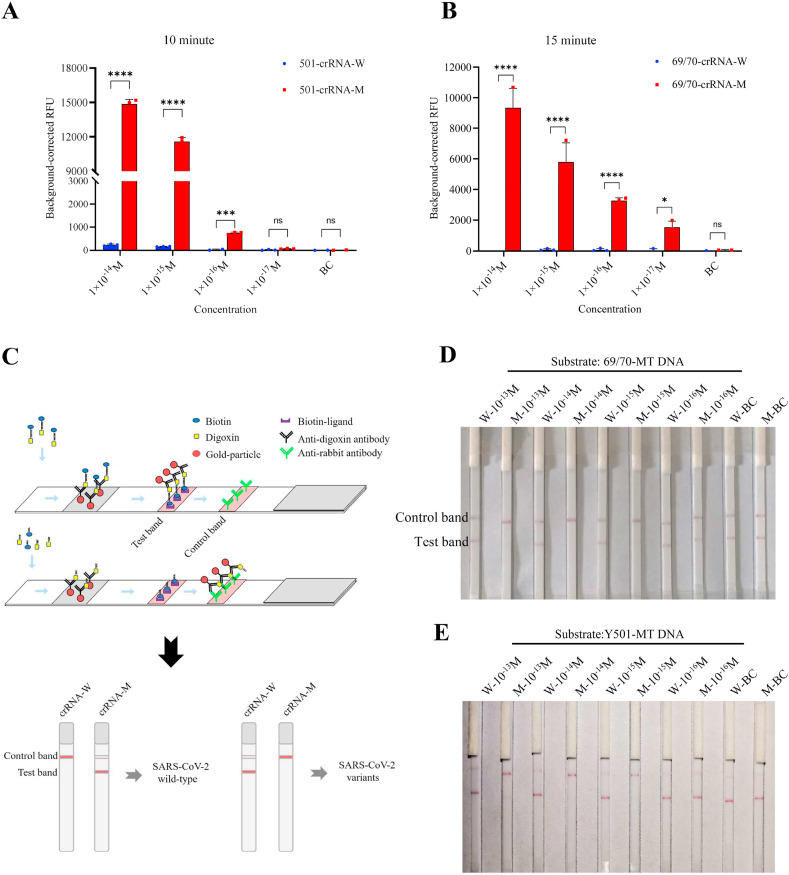
After validating the Cas12a fluorescence assay to differentiate mutations, we introduced PCR amplification to improve the sensitivity of mutation identification. The limit of detection (LOD) can reach 10−16 M when Y501-MT or N501-WT amplicons serve as substrates (Fig. 2A, S2, S4A, and S4B). The 69/70 deletion can be identified even when substrate is present at 10−17 M using TTC as the PAM (Fig. 2B, and S3). Thus, we developed CORDS fluorescence reporting system that rapidly detects mutations from SARS-CoV-2 variants, with high sensitivity and specificity.
Fig. 2.
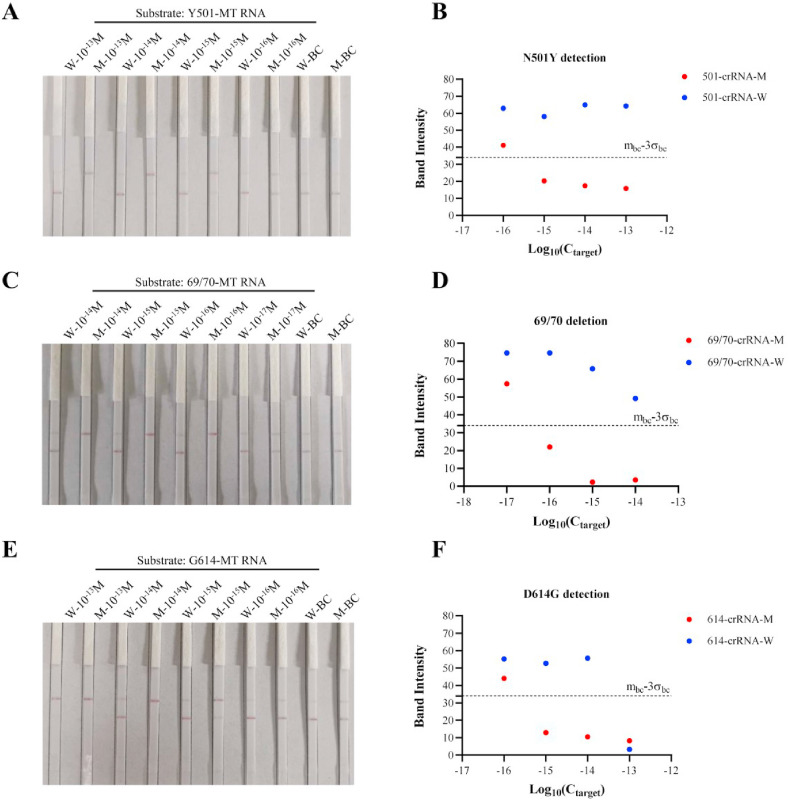
Detection of N501Y and 69/70 deletion mutations in SARS-CoV-2 using the CORDS platform. (A) Sensitivity of N501Y detection using the CORDS fluorescence reporting assay after 10 min of Cas12a/crRNA sensing time. (B) Sensitivity of 69/70 deletion identification using the CORDS fluorescence reporting assay after 15 min of Cas12a/crRNA sensing time. (C) Schematic representation of Cas12a lateral flow strips to differentiate mutation-containing SARS-CoV-2 variants. (D, E) Sensitivity of 69/70 deletion and N501Y mutation detection using the CORDS strips assay. W: crRNA-W; M: crRNA-M; BC: no target. In all figures error bars represent the means ± standard deviation (SD) from replicates (n = 3).
The system was made more convenient with reduced dependence of instruments when we combined CORDS with lateral flow strips as a substitute for the fluorescence intensity readout (Fig. 2 C). In this assay, 5′-digoxin and 3′-biotin-labeled 14-nt ssDNA and lateral flow strips were used to report the collateral activity of Cas12a (Fig. 2C). Similar to the fluorescence reporting assay, we detected serial concentrations of substrate to determine the sensitivity of the CORDS lateral flow strips reporting system. Results indicated that LOD can reached 10−15 M with Y501-MT or N501-WT as the substrate (Fig. 2D and S4C). This sensitivity can also be achieved for 69/70-MT as the substrate (Fig. 2E). CORDS paper strips system can rapidly detect SARS-CoV-2 variants mutations, with high sensitivity and without the need for a signal detection instrument.
The established CORDS fluorescence and paper strip systems can be used for rapid and sensitive detection of SARS-CoV-2 mutations with DNA targets reversely transcribed from SARS-CoV-2 RNA.
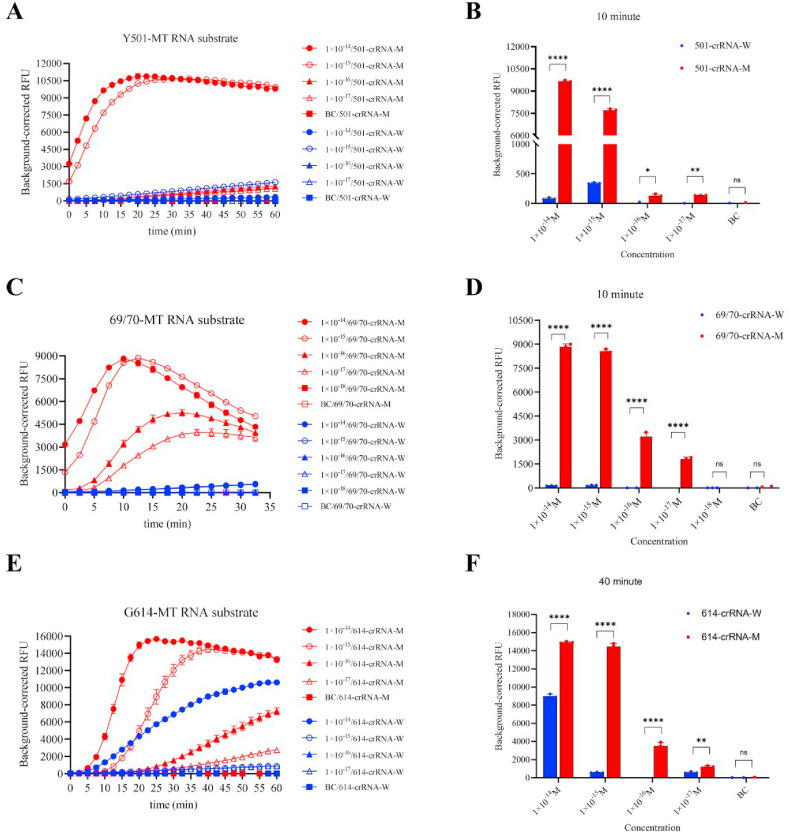
3.4. RT-CORDS can sensitively and accurately detect the synthetic RNA targets of SARS-CoV-2 mutations
In RT-CORDS, RT-PCR was introduced for RNA reverse transcription and amplification instead of isothermal amplification because the latter is always unstable when combined with reverse transcription (Fig. S5). LOD reached 10−17 M with Y501-MT RNA as substrate (Fig. 3 A and B). The sensitivity of the RT-CORDS fluorescence reporting system was higher than that of the CORDS system, perhaps because of the different amplification ability of the DNA polymerase. The sensitivity of RT-CORDS for N501Y detection was 10−15 M, the same sensitivity as that of CORDS, when lateral flow strips were used as final read out (Fig. 2, Fig. 4 B). RT-CORDS fluorescence assay for 69/70 deletion identification showed a sensitivity of 10−17 M, the same value as that achieved for N501Y (Fig. 3C and D). The LOD for RT-CORDS lateral flow strip was 10−16 M (Fig. 4C and D), higher than observed for N501Y detection and in a previous study (Bai et al., 2019). The sensitivity for D614G detection was 10−17 M for RT-CORDS fluorescence and 10−15 M for RT-CORDS paper strip reporting systems (Fig. 3, Fig. 4F). Increased fluorescence and test band disappearance were observed for 614-crRNA-W at 10−14 M and 10−13 M for G614-MT RNA in the fluorescence reporting and lateral flow strip systems. D614G detection is thus more suitable for samples when SARS-CoV-2 viral load is <10−14 M (approximately 6000 copies/μL), and appropriate dilution of samples should be considered for higher viral loads (Table S5). Notably, kinetic analyses showed N501Y and 69/70 deletion can be identified in as small duration as 5–10 min, indicating a shorter reaction time for mutation tracking (Figs. S6 and S7). However, 40 min is the most advisable for D614G detection (Fig. S8).
Fig. 3.
Sensitivity of SARS-CoV-2 mutant RNA identification using the RT-CORDS fluorescence reporting system. (A, B) Sensitivity of synthetic Y501-MT RNA detection using the RT-CORDS fluorescence assay. (A) Fluorescence kinetics of FQ-ssDNA reporter transcleaved by Cas12a using various Y501-MT RNA concentrations. (B) Background-corrected RFU of Y501-MT detection using various Y501-MT RNA concentrations at 10 min. (C, D) Sensitivity of synthetic 69/70-MT RNA detection using the RT-CORDS fluorescence assay. (E, F) Sensitivity of synthetic G614-MT RNA detection using the RT-CORDS fluorescence assay. In all figures error bars represent the means ± standard deviation (SD) from replicates (n = 3).
Fig. 4.
Sensitivity of SARS-CoV-2 mutant RNA identification using the RT-CORDS lateral flow strip reporting system. (A, B) Sensitivity of N501Y detection using the RT-CORDS lateral flow strips system. (C, D) Sensitivity of 69/70 deletion detection using the RT-CORDS lateral flow strips system. (E, F) Sensitivity of D614G detection using the RT-CORDS lateral flow strips system. (A, C, E) Visual readout of the paper strips. (B, D, F) Mean gray values of the test band at different target concentrations. The dashed line mbc-3σbc indicates the positive cutoff.
3.5. RT-CORDS ensures 100% accurate detection of SARS-CoV-2 variants
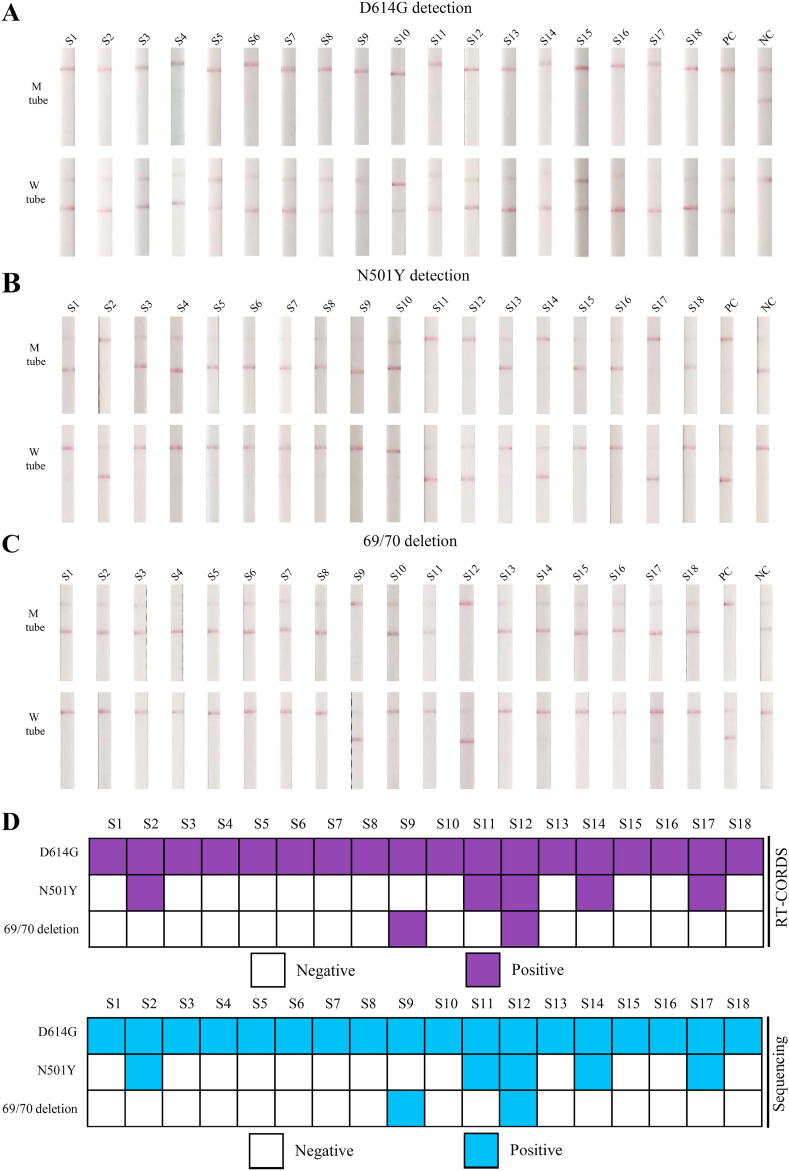
In total, 18 SARS-CoV-2 variant RNA samples were tested using the RT-CORDS paper strip system. All samples carried the D614G mutation (Fig. 5 A). Furthermore, five samples carrying the N501Y mutation and two samples carrying the 69/70 deletion were identified (Fig. 5B–D). Results were 100% consistent with the sequencing data (Fig. 5 and S9–S11).
Fig. 5.
Detection of SARS-CoV-2 variants using RT-CORDS. (A) D614G detection of SARS-CoV-2 variants using the RT-CORDS paper strip assay. (B) N501Y detection of SARS-CoV-2 variants samples using the RT-CORDS paper strip assay. (C) 69/70 deletion detection of SARS-CoV-2 variants samples using the RT-CORDS paper strip assay. (D) Summary of the detection of three SARS-CoV-2 variant mutations using RT-CORDS and sequencing.
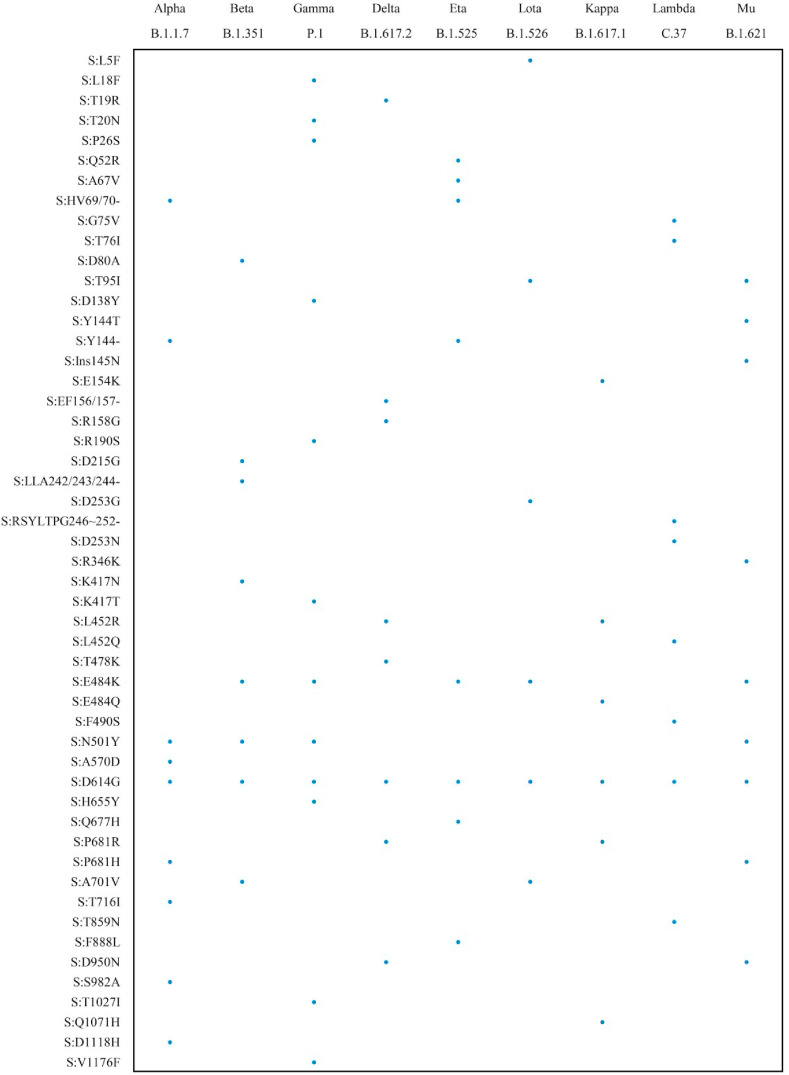
We simplified the RT-CORDS procedure for storage and use through the lyophilized paper strip system (Fig. S12A). Lyophilization did not alter the LOD for mutation identification (Fig. S12B). Besides, according to our accelerated stability test, the lyophilization of Cas12a mix work well after storage at 4 °C for 1 week without loss of sensitivity (Fig. S12C). RT-CORDS is a rapid, robust, and accurate method for the identification of SARS-CoV-2 N501Y, D614G, and 69/70 deletion mutations and may be used for additional tracking of spike mutations through a simple process in clinical diagnostics (Fig. 6 and S13).
Fig. 6.
Detectable mutations which have been reported in spike protein of VOC and VOI by RT-CORDS. Sequences around mutations containing TTTV or TTV PAM sequence were detectable by this method.
4. Discussion
DNA sequencing is the gold-standard for SARS-CoV-2 variant tracking as it can provide complete information regarding the genome sequence. However, sequencing is time-consuming, expensive and inconvenient for general laboratories. Therefore, we established the RT-CORDS for SARS-CoV-2 variant tracking, which is cost-effective, rapid, specific, and sensitive. The system does not require any special laboratory equipment.
The strategy of designing mutant specific crRNA in RT-CORDS was efficient and efficacious, as well as its broad applicability for almost all SARS-CoV-2 mutations. Among those existing similar sensors, Meng et al. and Huang et al. both introduced single-base mismatch covering all possible nucleotide substitutions in the position −2/3 to +2/3 around the mutant site, then the optimized crRNA was determined by an extensive and laborious screening process (Huang et al., 2021; Meng et al., 2021b). However, the specificity of crRNA binding is closely associated with the seed region of the crRNA and it is more efficient to introduce a single-base mismatch at the seed region (Chen et al., 2018). Here we introduced the single-base mismatch just at the seed region of crRNA according to the kinetic mechanism of CRISPR/Cas12a (Jones et al., 2021). And the results indicated that all the designed specific crRNAs worked well for different types of mutation, yet without laborious screening process (Fig. 1B–G).
High accuracy is the key critical benefit of the RT-CORDS. By introducing a skillful point mutation at the seed region of crRNA, the RT-CORDS can detect the SARS-CoV-2 single-base mutations (N501Y and D614G) with high specificity. RT-CORDS can also accurately identify a small deletion mutation in SARS-CoV-2 (69/70 deletion) with specific crRNAs without introducing a point mutation. In this study, 18 SARS-CoV-2 variants were detected, and all three mutations identified by RT-CORDS were perfectly consistent with the sequencing data, indicating 100% accuracy (Fig. 5). Compared with similar CRISPR-based methods, the accuracy in this study was higher than that of a Cas9-based N501Y detection method (accuracy: 86%) (Kumar et al., 2021). Meng et al. also utilized a symRNA-Cas12a detection method to distinguish the D614G mutant samples correctly, but merely 5 mutant samples involved (Meng et al., 2021b). Conversely, the accuracy of the most frequently used RT-qPCR for SARS-CoV-2 Marseille-4 variant detection, a point mutation in ORF1, with a specific probe is 93% (Bedotto et al., 2021). Furthermore, the ability of RT-qPCR to differentiate mutations distinguishment is influenced by adjacent mutations, resulting in decreased accuracy (Boudet et al., 2021; Zelyas et al., 2021). This issue illustrates a limitation of RT-qPCR for variant screening.
The RT-CORDS differentiates mutations in SARS-CoV-2 variants accurately and exhibits high sensitivity for all three SARS-CoV-2 mutation targets. RT-CORDS with fluorescence showed LODs for all three mutations reached 10−17 M (approximately 6 copies/μL) (Fig. 3, Table S5), which was higher than that of the Cas13-based N501Y detection system (100 copies/μL) (de Puig et al., 2021). Similar sensitivity was reported in Meng et al. and Huang et al., but RT-CORDS took less reaction time (only 10 min) after Cas12a added to amplicons because of the more specific and efficient crRNAs we used (Huang et al., 2021; Meng et al., 2021b). Furthermore, RT-CORDS based paper strip system also can distinguish the SARS-CoV-2 mutation efficiently with naked eyes, which is not studied in those existing sensors. The LOD of RT-CORDS with paper strip were up to 10−15 M for N501Y and D614G detection and 10−16 M for 69/70 detection (Fig. 4, Table S5). The LOD of RT-CORDS for SARS-CoV-2 variant tracking was consistent with our previous study, which detected ASFV using CORDS (Bai et al., 2019). Thus, CORDS detection is robust and reproducible. Clinical data reveal that viral loads of SARS-CoV-2 in throat swab and sputum samples peak around 5–6 days after symptom onset and range from 10⁴ to 10⁷ copies per mL; even higher numbers have been reported for the variants (Fajnzylber et al., 2020; Luo et al., 2021; Pan et al., 2020). RT-CORDS is, therefore, sufficiently sensitive to facilitate the early screening of SARS-CoV-2 variants.
The narrow range of targets makes the PAM sequences a key obstacle for designing an efficient crRNA for mutation detection. We used TTC as an alternative PAM for 69/70 deletion. No TTTV sequence was present near the 69/70 site. Although several studies have reported a lower affinity of TTV for Cas12a loading, the LOD for the 69/70 deletion was the same as that for canonical PAM, indicating that TTV can also be efficient for activating Cas12a system sensitively (Fig. 2, Fig. 3, Fig. 4C) (Yamano et al., 2017). Sequences adjacent to the S gene mutations of SARS-CoV-2 VOC and VOI revealed TTTV and TTV near all mutations, indicating that these mutations are targetable and can be tracked using RT-CORDS (Fig. 6 ). Conversely, RAY, a Cas9-based SARS-CoV-2 variant tracking system, only targets a subset of mutations reported in SARS-CoV-2 variants, because no NGG PAM sequence exists near the mutation targets (Kumar et al., 2021). Hence, RT-CORDS is widely applicable for mutation tracking.
Despite the single-base resolution of Cas12, the kinetics of CRISPR enzyme for molecular diagnosis recently revealed that realistic LODs for Cas13/Cas12-based detection system were only 10 pM to 100 fM, regardless of the number of targets (Ramachandran and Santiago, 2021). Thus, signal amplification is essential either upstream or downstream of CRISPR reactions to achieve high sensitivity (Chen et al., 2020; Li et al., 2021a; Qiao et al., 2021). Isothermal amplification such as RT-RPA or RT-LAMP is a common choice for Cas12 based point-of-care testing (Broughton et al., 2020; Meng et al., 2021b). However, isothermal amplification usually exhibits low efficiency and stability in mutation detection as optimal primer design is limited within the narrow range of mutant targets. Thus, we turned to conventional and stable RT-PCR before Cas12 sensing. Conveniently, RT-PCR is also much cheaper and more available, which facilitate its use in conventional laboratory conditions.
5. Conclusions
In summary, we report that the RT-CORDS is an accurate and sensitive system for identifying single-base mutations (N501Y and D614G) and a small deletion (69/70 deletion) mutation in SARS-CoV-2 variants using CRISPR/Cas12a. This system is easy to assemble for SARS-CoV-2 mutation screening in conventional laboratory conditions. This characteristic will be helpful for monitoring the VOC and allow the rapid tracing of epidemic trends. Although RT-CORDS still relies on target amplification as many POCT methods, the high specificity and accuracy of this system make it an ideal choice for high throughput SARS-CoV-2 variant screening.
CRediT authorship contribution statement
Changsheng He: Writing – original daft, designed and performed experiments and wrote the manuscript. Cailing Lin: Writing – original daft, designed and performed experiments and wrote the manuscript.. Guosheng Mo: Writing – original daft, designed and performed experiments and wrote the manuscript. Binbin Xi: Formal analysis, performed bioinformatic analysis of sequences. An′an Li: performed variant RNA samples assay. Dongchao Huang: Formal analysis, performed bioinformatic analysis of sequences. Yanbin Wan: Formal analysis, analyzed the data. Feng Chen: Formal analysis, analyzed the data. Yufeng Liang: performed variant RNA samples assay. Qingxia Zuo: Formal analysis, analyzed the data. Wanqing Xu: made figures. Dongyan Feng: made figures. Guanting Zhang: performed variant RNA samples assay. Changwen Ke: advised on experimental design. Hongli Du: Supervision, Writing – original daft, Funding acquisition, conceived and designed the study, supervised the study, wrote manuscript and acquired the research funds. Lizhen Huang: Supervision, Writing – original daft, Funding acquisition, conceived and designed the study, supervised the study, wrote manuscript and acquired the research funds.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Three patents have been filed through South China University of Technology related to this work.
Acknowledgments
This work was supported by the National Natural Science Foundation of China [grant number 31871292]; Fundamental Research Funds for the Central Universities [grant number 2019MS089] from South China University of Technology; Natural Science Foundation of Guangdong Province of China [grant number 2019A1515010619]; Medical Scientific Research Foundation of Guangdong Province of China [grant number A2019432]; The National Key R&D Program of China [grant number 2018YFC0910201].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113857.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ali F., Kasry A., Amin M. The new SARS-CoV-2 strain shows a stronger binding affinity to ACE2 due to N501Y mutant. Med. Drug Discov. 2021;10:100086. doi: 10.1016/j.medidd.2021.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki A., Mori Y., Okamoto Y., Jinno H. Development of a genotyping platform for SARS-CoV-2 variants using high-resolution melting analysis. J. Infect. Chemother. 2021;27(9):1336–1341. doi: 10.1016/j.jiac.2021.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J., Lin H., Li H., Zhou Y., Liu J., Zhong G., Wu L., Jiang W., Du H., Yang J., Xie Q., Huang L. Cas12a-Based on-site and rapid nucleic acid detection of African swine fever. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.02830. 2830-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bal A., Destras G., Gaymard A., Stefic K., Marlet J., Eymieux S., Regue H., Semanas Q., D'Aubarede C., Billaud G., Laurent F., Gonzalez C., Mekki Y., Valette M., Bouscambert M., Gaudy-Graffin C., Lina B., Morfin F., Josset L., Group t.C.-D.H.S. Two-step strategy for the identification of SARS-CoV-2 variant of concern 202012/01 and other variants with spike deletion H69–V70, France, August to December 2020. Euro Surveill. 2021;26(3):2100008. doi: 10.2807/1560-7917.ES.2021.26.3.2100008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedotto M., Fournier P.E., Houhamdi L., Levasseur A., Delerce J., Pinault L., Padane A., Chamieh A., Tissot-Dupont H., Brouqui P., Sokhna C., Azar E., Saile R., Mboup S., Bitam I., Colson P., Raoult D. Implementation of an in-house real-time reverse transcription-PCR assay for the rapid detection of the SARS-CoV-2 Marseille-4 variant. J. Clin. Virol. 2021;139:104814. doi: 10.1016/j.jcv.2021.104814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudet A., Stephan R., Bravo S., Sasso M., Lavigne J.P. Limitation of screening of different variants of SARS-CoV-2 by RT-PCR. Diagnostics. 2021;11(7):1241. doi: 10.3390/diagnostics11071241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle B., Dallaire N., MacKay J. Evaluation of the impact of single nucleotide polymorphisms and primer mismatches on quantitative PCR. BMC Biotechnol. 2009;9(1):75. doi: 10.1186/1472-6750-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro A., Carter H., Zanetti M. Potential global impact of the N501Y mutation on MHC-II presentation and immune escape. bioRxiv. 2021:429431. 2021.02.02. [Google Scholar]
- Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Mei Y., Jiang X. Universal and high-fidelity DNA single nucleotide polymorphism detection based on a CRISPR/Cas12a biochip. Chem. Sci. 2021;12:4455–4462. doi: 10.1039/d0sc05717g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Shi Y., Chen Y., Yang Z., Wu H., Zhou Z., Li J., Ping J., He L., Shen H., Chen Z., Wu J., Yu Y., Zhang Y., Chen H. Contamination-free visual detection of SARS-CoV-2 with CRISPR/Cas12a: a promising method in the point-of-care detection. Biosens. Bioelectron. 2020;169:112642. doi: 10.1016/j.bios.2020.112642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniloski Z., Jordan T.X., Ilmain J.K., Guo X., Bhabha G., tenOever B.R., Sanjana N.E. The Spike D614G mutation increases SARS-CoV-2 infection of multiple human cell types. Elife. 2021;10 doi: 10.7554/eLife.65365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A.B., Russell T.W., Tully D.C., Washburne A.D., Wenseleers T., Gimma A., Waites W., Wong K.L.M., Van Zandvoort K., Silverman J.D., CMMID COVID-19 Working Group. Diaz-Ordaz K., Keogh R., Eggo R.M., Funk S., Jit M., Atkins K.E., Edmunds W.J. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372(6538) doi: 10.1126/science.abg3055. COVID-19 Genomics UK (COG-UK) Consortium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Jarvis C.I., CMMID COVID-19 Working Group. Edmunds W.J., Jewell N.P., Diaz-Ordaz K., Keogh R.H. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021;593(7858):270–274. doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Puig H., Lee R.A., Najjar D., Tan X., Soekensen L.R., Angenent-Mari N.M., Donghia N.M., Weckman N.E., Ory A., Ng C.F., Nguyen P.Q., Mao A.S., Ferrante T.C., Lansberry G., Sallum H., Niemi J., Collins J.J. Minimally instrumented SHERLOCK (miSHERLOCK) for CRISPR-based point-of-care diagnosis of SARS-CoV-2 and emerging variants. Sci. Adv. 2021;7(32) doi: 10.1126/sciadv.abh2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R., Long J., Yuan M., Zheng X., Shen Y., Jin Y., Yang H., Li H., Chen S., Duan G. CRISPR/Cas12-Based ultra-sensitive and specific point-of-care detection of HBV. Int. J. Mol. Sci. 2021;22(9):4842. doi: 10.3390/ijms22094842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajnzylber J., Regan J., Coxen K., Corry H., Wong C., Rosenthal A., Worrall D., Giguel F., Piechocka-Trocha A., Atyeo C., Fischinger S., Chan A., Flaherty K.T., Hall K., Dougan M., Ryan E.T., Gillespie E., Chishti R., Li Y., Jilg N., Hanidziar D., Baron R.M., Baden L., Tsibris A.M., Armstrong K.A., Kuritzkes D.R., Alter G., Walker B.D., Yu X., Li J.Z. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat. Commun. 2020;11(1):5493. doi: 10.1038/s41467-020-19057-5. The Massachusetts Consortium for Pathogen Readiness. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisman D.N., Tuite A.R. Progressive increase in virulence of novel SARS-CoV-2 variants in Ontario, Canada. medRxiv. 2021 doi: 10.1503/cmaj.211248. 2021.07.05.21260050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S., Li J., Pan W., Li N., Tang B. Duplex-specific nuclease-assisted CRISPR-Cas12a strategy for MicroRNA detection using a personal Glucose meter. Anal. Chem. 2021;93(30):10719–10726. doi: 10.1021/acs.analchem.1c02478. [DOI] [PubMed] [Google Scholar]
- Gong S., Zhang S., Lu F., Pan W., Li N., Tang B. CRISPR/Cas-Based in vitro diagnostic platforms for Cancer biomarker detection. Anal. Chem. 2021;93(35):11899–11909. doi: 10.1021/acs.analchem.1c02533. [DOI] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J., Robertson D.L. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. COVID-19 Genomics UK (COG-UK) Consortium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., Leist S.R., Schäfer A., Nakajima N., Takahashi K., Lee R.E., Mascenik T.M., Graham R., Edwards C.E., Tse L.V., Okuda K., Markmann A.J., Bartelt L., de Silva A., Margolis D.M., Boucher R.C., Randell S.H., Suzuki T., Gralinski L.E., Kawaoka Y., Baric R.S. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370(6523):1464. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Zhang F., Zhu K., Lin W., Ma W. dsmCRISPR: dual synthetic mismatches CRISPR/Cas12a-based detection of SARS-CoV-2 D614G mutation. Virus Res. 2021;304:198530. doi: 10.1016/j.virusres.2021.198530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S.K., Hawkins J.A., Johnson N.V., Jung C., Hu K., Rybarski J.R., Chen J.S., Doudna J.A., Press W.H., Finkelstein I.J. Massively parallel kinetic profiling of natural and engineered CRISPR nucleases. Nat. Biotechnol. 2021;39(1):84–93. doi: 10.1038/s41587-020-0646-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Sheffield COVID-19 Genomics Group. McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Gulati S., Ansari A.H., Phutela R., Acharya S., Azhar M., Murthy J., Kathpalia P., Kanakan A., Maurya R., Vasudevan J.S., Murali A., Pandey R., Maiti S., Chakraborty D. FnCas9 based CRISPR diagnostic for rapid and accurate detection of major SARS-CoV-2 variants on a paper strip. Elife. 2021;10 doi: 10.7554/eLife.67130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffeber C., de Koning K., Kanaar R., Lebbink J.H.G. Experimental evidence for enhanced receptor binding by rapidly spreading SARS-CoV-2 variants. J. Mol. Biol. 2021;433(15) doi: 10.1016/j.jmb.2021.167058. 167058-167058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., Park Y.H., Jin Y.B., Kim S.U., Hur J.K. CRISPR diagnosis and therapeutics with single base pair precision. Trends Mol. Med. 2020;26(3):337–350. doi: 10.1016/j.molmed.2019.09.008. [DOI] [PubMed] [Google Scholar]
- Lee Yu H., Cao Y., Lu X., Hsing I.M. Detection of rare variant alleles using the AsCas12a double-stranded DNA trans-cleavage activity. Biosens. Bioelectron. 2021;189:113382. doi: 10.1016/j.bios.2021.113382. [DOI] [PubMed] [Google Scholar]
- Li F., Ye Q., Chen M., Zhou B., Zhang J., Pang R., Xue L., Wang J., Zeng H., Wu S., Zhang Y., Ding Y., Wu Q. An ultrasensitive CRISPR/Cas12a based electrochemical biosensor for Listeria monocytogenes detection. Biosens. Bioelectron. 2021;179:113073. doi: 10.1016/j.bios.2021.113073. [DOI] [PubMed] [Google Scholar]
- Li Q., Nie J., Wu J., Zhang L., Ding R., Wang H., Zhang Y., Li T., Liu S., Zhang M., Zhao C., Liu H., Nie L., Qin H., Wang M., Lu Q., Li X., Liu J., Liang H., Shi Y., Shen Y., Xie L., Zhang L., Qu X., Xu W., Huang W., Wang Y. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell. 2021;184(9):2362–2371. doi: 10.1016/j.cell.2021.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.Y., Cheng Q.X., Wang J.M., Li X.Y., Zhang Z.L., Gao S., Cao R.B., Zhao G.P., Wang J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018;4:20. doi: 10.1038/s41421-018-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu J., Plante K.S., Plante J.A., Xie X., Zhang X., Ku Z., An Z., Scharton D., Schindewolf C., Menachery V.D., Shi P.Y., Weaver S.C. The N501Y spike substitution enhances SARS-CoV-2 transmission. bioRxiv. 2021:434499. doi: 10.1038/s41586-021-04245-0. 2021.03.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C.H., Morris C.P., Sachithanandham J., Amadi A., Gaston D., Li M., Swanson N.J., Schwartz M., Klein E.Y., Pekosz A., Mostafa H.H. Infection with the SARS-CoV-2 delta variant is associated with higher infectious virus loads compared to the Alpha variant in both unvaccinated and vaccinated individuals. medRxiv. 2021 doi: 10.1093/cid/ciab986. 2021.08.15.21262077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Yin L., Li X., Chen S., Peng L., Liu G., Ye S., Zhang W., Man S. A smartphone-based visual biosensor for CRISPR-Cas powered SARS-CoV-2 diagnostics. Biosens. Bioelectron. 2021;195:113646. doi: 10.1016/j.bios.2021.113646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng B., Kemp S.A., Papa G., Datir R., Ferreira I., Marelli S., Harvey W.T., Lytras S., Mohamed A., Gallo G., Thakur N., Collier D.A., Mlcochova P., Duncan L.M., Carabelli A.M., Kenyon J.C., Lever A.M., De Marco A., Saliba C., Culap K., Cameroni E., Matheson N.J., Piccoli L., Corti D., James L.C., Robertson D.L., Bailey D., Gupta R.K. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep. 2021;35(13):109292. doi: 10.1016/j.celrep.2021.109292. COVID-19 Genomics UK (COG-UK) Consortium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q., Wang X., Wang Y., Dang L., Liu X., Ma X., Chi T., Wang X., Zhao Q., Yang G., Liu M., Huang X., Ma P. Detection of the SARS-CoV-2 D614G mutation using engineered Cas12a guide RNA. Biotechnol. J. 2021;16(6) doi: 10.1002/biot.202100040. [DOI] [PubMed] [Google Scholar]
- Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20(4):411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., Mirchandani D., Scharton D., Bilello J.P., Ku Z., An Z., Kalveram B., Freiberg A.N., Menachery V.D., Xie X., Plante K.S., Weaver S.C., Shi P.Y. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2020;592(7852):116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao B., Xu J., Yin W., Xin W., Ma L., Qiao J., Liu Y. “Aptamer-locker” DNA coupling with CRISPR/Cas12a-guided biosensing for high-efficiency melamine analysis. Biosens. Bioelectron. 2021;183:113233. doi: 10.1016/j.bios.2021.113233. [DOI] [PubMed] [Google Scholar]
- Ramachandran A., Santiago J.G. CRISPR enzyme kinetics for molecular diagnostics. Anal. Chem. 2021;93(20):7456–7464. doi: 10.1021/acs.analchem.1c00525. [DOI] [PubMed] [Google Scholar]
- Salvatori G., Luberto L., Maffei M., Aurisicchio L., Roscilli G., Palombo F., Marra E. SARS-CoV-2 SPIKE PROTEIN: an optimal immunological target for vaccines. J. Transl. Med. 2020;18(1):222. doi: 10.1186/s12967-020-02392-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyaolu A., Okorie C., Marinkovic A., Haider N., Abbasi A.F., Jaferi U., Prakash S., Balendra V. The emerging SARS-CoV-2 variants of concern. Therapeut. Adv. Infect. Dis. 2021;8:1–10. doi: 10.1177/20499361211024372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadhouders R., Pas S.D., Anber J., Voermans J., Mes T.H.M., Schutten M. The effect of primer-template mismatches on the detection and quantification of nucleic acids using the 5' nuclease assay. J. Mol. Diagn. 2010;12(1):109–117. doi: 10.2353/jmoldx.2010.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Süß B., Flekna G., Wagner M., Hein I. Studying the effect of single mismatches in primer and probe binding regions on amplification curves and quantification in real-time PCR. J. Microbiol. Methods. 2009;76(3):316–319. doi: 10.1016/j.mimet.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Vega-Magaña N., Sánchez-Sánchez R., Hernández-Bello J., Venancio-Landeros A.A., Peña-Rodríguez M., Vega-Zepeda R.A., Galindo-Ornelas B., Díaz-Sánchez M., García-Chagollán M., Macedo-Ojeda G., García-González O.P., Muñoz-Valle J.F. RT-qPCR Assays for Rapid Detection of the N501Y, 69-70del, K417N, and E484K SARS-CoV-2 Mutations: a Screening Strategy to Identify Variants With Clinical Impact. Front. Cell. Infect. Microbiol. 2021;11(434):672562. doi: 10.3389/fcimb.2021.672562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Zheng Y., Niu Z., Jiang X., Sun Q. The virological impacts of SARS-CoV-2 D614G mutation. J. Mol. Cell Biol. 2021 doi: 10.1093/jmcb/mjab045. mjab045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Qian C., Pang Y., Li M., Yang Y., Ma H., Zhao M., Qian F., Yu H., Liu Z., Ni T., Zheng Y., Wang Y. opvCRISPR: one-pot visual RT-LAMP-CRISPR platform for SARS-cov-2 detection. Biosens. Bioelectron. 2021;172:112766. doi: 10.1016/j.bios.2020.112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2021. Coronavirus Disease 2019 (COVID-19) Weekly Epidemiological Edition 60.https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---5-october-2021 URL: [Google Scholar]
- Yamano T., Zetsche B., Ishitani R., Zhang F., Nishimasu H., Nureki O. Structural basis for the canonical and non-canonical PAM recognition by CRISPR-Cpf1. Mol. Cell. 2017;67(4):633–645. doi: 10.1016/j.molcel.2017.06.035. e633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelyas N., Pabbaraju K., Croxen M.A., Lynch T., Buss E., Murphy S.A., Shokoples S., Wong A., Kanji J.N., Tipples G. Precision response to the rise of the SARS-CoV-2 B.1.1.7 variant of concern by combining novel PCR assays and genome sequencing for rapid variant detection and surveillance. Microbiol. Spectr. 2021;9(1) doi: 10.1128/spectrum.00315-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jackson C.B., Mou H., Ojha A., Peng H., Quinlan B.D., Rangarajan E.S., Pan A., Vanderheiden A., Suthar M.S., Li W., Izard T., Rader C., Farzan M., Choe H. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020;11(1):6013. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.