Abstract
The devastating impact of the ongoing coronavirus disease 2019 (COVID-19) on public health, caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has made targeting the COVID-19 pandemic a top priority in medical research and pharmaceutical development. Surveillance of SARS-CoV-2 mutations is essential for the comprehension of SARS-CoV-2 variant diversity and their impact on virulence and pathogenicity. The SARS-CoV-2 open reading frame 10 (ORF10) protein interacts with multiple human proteins CUL2, ELOB, ELOC, MAP7D1, PPT1, RBX1, THTPA, TIMM8B, and ZYG11B expressed in lung tissue. Mutations and co-occurring mutations in the emerging SARS-CoV-2 ORF10 variants are expected to impact the severity of the virus and its associated consequences. In this article, we highlight 128 single mutations and 35 co-occurring mutations in the unique SARS-CoV-2 ORF10 variants. The possible predicted effects of these mutations and co-occurring mutations on the secondary structure of ORF10 variants and host protein interactomes are presented. The findings highlight the possible effects of mutations and co-occurring mutations on the emerging 140 ORF10 unique variants from secondary structure and intrinsic protein disorder perspectives.
Keywords: SARS-CoV-2, ORF10, Co-occurring mutations, Intrinsic protein disorder, And ubiquitin ligase complex
1. Introduction
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) continues the pandemic spread of coronavirus disease 2019 (COVID-19), with over 227 million people confirmed infected and at least 4.66 million deaths worldwide [1], [2]. In 2021, in almost every region of the SARS-CoV-2 genome, several mutations compared to the wild-type SARS-CoV-2 (NC 045512) were discovered [3], [4], [5]. Like other RNA viruses, the SARS-CoV-2 is constantly evolving by mutations and new variants with different characteristics are emerging [6], [7], [8], [9], [10], [11], [12]. Detection and mutation surveillance of SARS-CoV-2 is of utmost priority to investigate the origin and to combat the virus [13]. To date, no method can rapidly diagnose multiple viral infections and determine variants in a high-throughput manner [14]. SARS-CoV-2 is one of the largest RNA viruses with a genome of approximately 29Kb, which includes eleven open reading frames (ORFs) [15], [16], [17], [18]. These ORFs (1a and 1b) possess two polypeptides that are translated into sixteen non-structural proteins (NSP1–16) [19], [20], [21]. The main non-structural proteins (NSP) include RNA-dependent RNA polymerase (RdRp or NSP12) and a 3′-5′ exonuclease [22]. RNA viruses typically show a high mutation rate ranging from 10−6 to 10−4 [23], [24]. Notably, SARS-CoV-2 possesses the 3′-5′ exonuclease capable of correcting mistakes during replication [25]. While surveilling mutations to comprehend the genetic diversity across various SARS-CoV-2 variants, it is also important to decipher whether the increase in mutation frequency is because of the natural selection, and to determine the possible consequences for SARS-CoV-2 fitness, such as increased infectivity and pathogenicity, or due to adaptation, thereby becoming drug-resistant or possessing ability to evade the immune system [26], [27]. Non-synonymous mutations of various SARS-CoV-2 proteins have been reported [28], [29], [30], [31].
The ORF proteins are dispensable for viral growth in vitro, and might play important roles within the environment of the infected host [32]. The SARS-CoV-2 open reading frame 10 (ORF10) protein shows no sequence similarity with other known coronavirus proteins [33], [34]. The SARS-CoV-2 ORF10, a putative 38-amino acid viral protein encoded in the 3′ accessory region of the genome, is a highly ordered, hydrophobic, and thermally stable protein, which contains at least one transmembrane region [34], [35]. The ORF10 binds to components of a Cullin-2-RING-ligase (CRL2) complex containing Cullin-2, RBX1, Elongin B, Elongin C, and ZYG11B (CRL2ZY G11B) [36], [37], [38]. Earlier, it has been reported that the extreme N terminus of ORF10 contains a methionine-glycine-tyrosine motif, which would presumably aid ORF10 to be recruited into the CRL2ZY G11B ubiquitin ligase complex [37]. It was further confirmed that interaction between ORF10 and CRL2ZY G11B is not relevant for SARS-CoV-2 infection in vitro [37], [39]. There is no evidence of ORF10 regulating or being regulated by CRL2ZY G11B [37]. On the other hand, the ORF10 protein in some SARS-CoV-2 variants, resulted in non-attenuation of disease and maintained transmissibility [40]. Furthermore, ORF10 is not essential for viral infection and replication while encoding a truncated protein that is neutrally evolving, through positive selection [39], [41].
In this study, we report 128 single mutations and 35 co-occurring mutations in the unique SARS-CoV-2 ORF10 variants. This report illuminates potential effects due to mutations and co-occurring mutations in the emerging ORF10 variants from the secondary structure and intrinsic protein disorder perspectives.
2. Materials and methods
2.1. Data
A total of 202,968 SARS-CoV-2 ORF10 amino acid sequences were retrieved from the National Center for Biotechnology Information (NCBI) database on June 29, 2021. Note that none of these ORF10 sequences contained any ambiguous characters. Among the 202,968 SARS-CoV-2 ORF10 sequences only 140 were unique and contained amino acid substitutions that made them different from the ORF10 sequence found in the original hCoV-19/Wuhan/WIV04/2019 strain. Furthermore, SARS-CoV-2 ORF10 protein sequences from the GISAID database were used for finding the co-occurring mutations in the CoVal database.
2.2. Methods
2.2.1. Transmembrane topology and secondary structure prediction
Prediction of a transmembrane protein topology of a given protein is one of the classical issues in bioinformatics. The Phobius program was used to predict transmembrane topology for SARS-CoV-2 ORF10 protein variants [42], [43].
The secondary structure of the SARS-CoV-2 ORF10 protein was predicted using the JPred 4 webserver [44]. In addition to protein secondary structure JPred was used to predict solvent accessibility and coiled-coil regions. The following keys were used in the secondary structure prediction of ORF10 variants [44].
-
•
Shades of red: The more red a position is, the higher the level of conservation of chemical properties of the amino acids.
-
•
Jnetpred: Final secondary structure prediction for a query.
-
•
Jnet 25: Jnet prediction of burial, less than 25% solvent accessibility.
-
•
Jnet 5: Jnet prediction of burial, less than 5% exposure.
-
•
Jnet 0: Jnet prediction of burial, 0% exposure.
-
•
Jnetconf: Jnet reliability of prediction accuracy ranges from 0 to 9, bigger is better.
2.2.2. Intrinsic disorder analysis
All SARS-CoV-2 ORF10 variants were subjected to the per-residue disorder analysis, with PONDR-VSL2 algorithm [45]. This tool showed good performance on proteins containing both structure and disorder, and is considered as an accurate standalone disorder predictor [46], [47], [48]. Predisposition scores for the per-residual conditions are 0 to 1, where 0 indicates residues entirely arranged, and 1 indicates residues completely disordered. Residues with predicted disorder scores between 0.25 and 0.5 were considered as moderately disordered, residues with disorder scores between 0.1 and 0.25 were considered flexible, whereas residues with the values of the predicted disorder scores higher than 0.5 were considered disordered.
2.2.3. Analysis of sequence variation
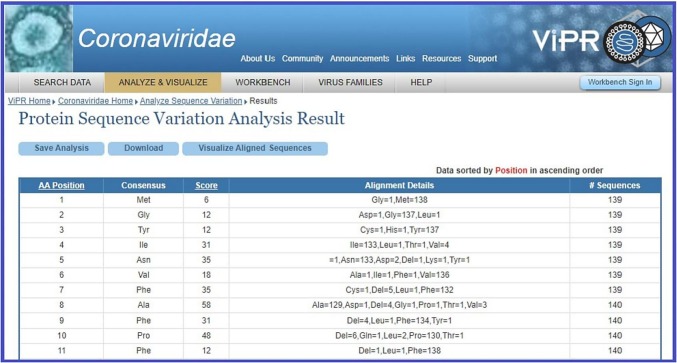
Single mutations in all the 140 unique ORF10 proteins were determined using the Virus Pathogen Resource ViPR by uploading a Fasta file of ORF10 sequences [49]. A snapshot of the ORF10 sequence variations is presented in Fig. 1 .
Fig. 1.
A snapshot of ViPR database showing the ORF10 sequence variations and consensus amino acid residues.
Furthermore, the predicted effect on pathogenicity of all the mutations was analyzed with PredictSNP and PhD-SNP [50], [51]. Note that PredictSNP web server makes a consensus based on other prediction tools such as MAPP, PolyPhen-1 and PolyPhen-2, SIFT, SNAP, and PANTHER. Therefore, the degree of accuracy is expected to be high. The pathogenicity score (PredictSNP consensus prediction) was calculated using the following equation:
where N is the number of integrated tools, δi represents the overall prediction (+1 for the deleterious prediction, -1 for the neutral prediction) and S i expresses the transformed confidence scores [52].
In addition, co-occurring of mutations in ORF10 proteins were also detected by the CoVal database.
A quantitative measure was defined to localize the co-occurring mutations in the ORF10 protein in a given geo-location with regards to other geo-locations worldwide.
Localization across countries: For each co-occurring mutation in the ORF10 protein, localization is defined as
where, NGm and TGm denote the number of SARS-CoV-2 genomes with these specific simultaneous mutations (m) in a geo-location, and the total number of SARS-CoV-2 genomes with this mutation (m) worldwide, respectively. It varies from 0 to 1. The normalized factor 0 denotes uniformly spreading of the mutations across various geo-locations, whereas 1 denotes the detection of the mutation in a single geo-location (discussed in CoVal database).
2.2.4. Frequency distribution of amino acids and clustering
The frequency distribution of each amino acid present in the ORF10 sequence was determined using standard bioinformatics routine in Matlab [53]. For each ORF10 sequence, a twenty-dimensional frequency-vector considering the frequency of standard twenty amino acids can be obtained. The distance (Euclidean metric) between any two pairs of frequency vectors was calculated for each pair of ORF10 sequences. By having the distance matrix, a set of clusters was obtained applying the well-known K-means clustering method using the standard routine in Matlab-2021a [53], [54].
3. Results
3.1. Continent-wise unique ORF10 variants and mutations
Continent-wise unique variations of ORF10 proteins and their mutations with predicted effects are presented in the following subsections.
3.1.1. ORF10 variants and transmembrane topology of SARS-CoV-2 ORF10
Continent-wise distribution of the total 140 unique ORF10 variants is presented in Table 1 . The highest frequency of unique ORF10 variants was found in North America among the total of 140 SARS-CoV-2 ORF10 variants.
Table 1.
Unique mutations in the unique ORF10 variants across six continents. # denotes “number”.
| Africa | Asia | Europe | North America | Oceania | South America | |
|---|---|---|---|---|---|---|
| # of total ORF10 | 1183 | 3393 | 1033 | 186,572 | 10,290 | 497 |
| # of unique ORF10 | 15 | 21 | 7 | 132 | 15 | 6 |
| % of unique ORF10 | 1.27 | 0.62 | 0.68 | 0.07 | 0.15 | 1.21 |
| # of residue positions of mutations in ORF10 | 12 | 15 | 6 | 37 | 12 | 5 |
| % of total number of residues | 31.6 | 39.5 | 15.8 | 97.4 | 31.6 | 13.2 |
Continent-wise, a decreasing order percentage of unique ORF10 variants was Africa>South America>Europe>Asia>Oceania>North America (Table 1).
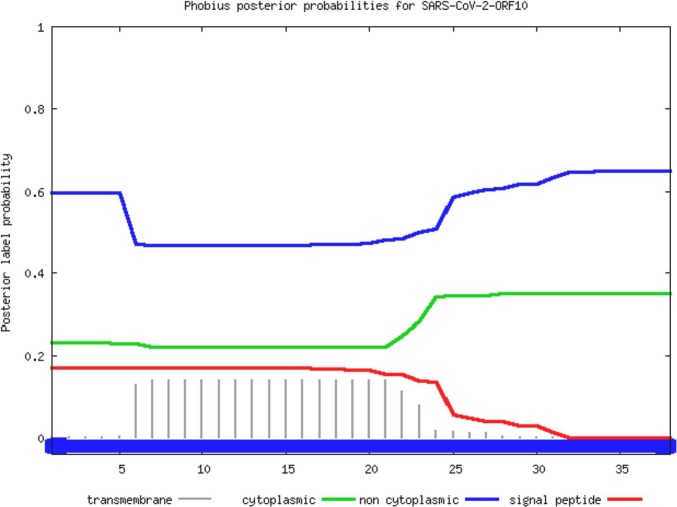
In addition, the transmembrane topology and signal peptides for the wild-type SARS-CoV-2 ORF10 (YP 009725255) protein were predicted using the Phobius webserver. The associated posterior probability for the topology prediction is presented in Fig. 2 .
Fig. 2.
Posterior probability plot for prediction of transmembrane topology and signal peptides for the wild-type SARS-CoV-2 ORF10.
From the posterior probability distributions, it was observed that the SARS-CoV-2 ORF10 protein was entirely non-cytoplasmic (Fig. 2).
3.1.2. Unique ORF10 variants and their single and co-occurring mutations
The amino acid residue positions of SARS-CoV-2 ORF10 single-mutations on each continent are listed in Table 2 .
Table 2.
Continent-wise amino acid positions of single-mutations in SARS-CoV-2 ORF10 unique variants.
| Continent-wise residue positions of single-mutations | |||||
|---|---|---|---|---|---|
| Africa | Asia | Europe | North America | Oceania | South America |
| 3 | 6 | 3 | 1–38 (except 18th) | 3 | 3 |
| 5 | 7 | 6 | 4 | 19 | |
| 7 | 8 | 13 | 8 | 23 | |
| 10 | 9 | 23 | 10 | 28 | |
| 23 | 10 | 30 | 17 | 35 | |
| 24 | 13 | 31 | 22 | ||
| 28 | 14 | 23 | |||
| 30 | 22 | 24 | |||
| 31 | 23 | 28 | |||
| 33 | 24 | 30 | |||
| 37 | 30 | 37 | |||
| 38 | 31 | 38 | |||
| 33 | |||||
| 35 | |||||
| 37 | |||||
It was noticed that except for the residue in position 18, all amino acids at each position from 1 to 38 possessed point-missense mutations (Table 2). The common residue was S23, where two mutations S23F and S23P were found across all six continents. A total of eight common residue positions with mutations were observed in Asia and Africa, whereas several common residual mutation positions were associated with Asia and Oceania.
The predicted effects of pathogenicity for each mutation are listed in Table 3 . Our data revealed that among a total of 128 mutations, 72 were neutral, and 56 deleterious with regards to pathogenicity.
Table 3.
The predicted effect of ORF10 single mutations on pathogenicity.
| Predicted effect of mutations | |||||||
|---|---|---|---|---|---|---|---|
| Percentage of pathogenicity |
Percentage of pathogenicity |
||||||
| Mutation | PredictSNP | PhD-SNP | Predicted type | Mutation | PredictSNP | PhD-SNP | Predicted type |
| V33A | 83% | 66% | Neutral | N22H | 83% | 51% | Neutral |
| V33D | 87% | 77% | Deleterious | N22I | 87% | 61% | Deleterious |
| V33F | 83% | 45% | Neutral | N22K | 87% | 77% | Deleterious |
| V33I | 83% | 78% | Neutral | N22S | 83% | 68% | Neutral |
| D31G | 87% | 73% | Deleterious | N22T | 87% | 61% | Deleterious |
| D31H | 87% | 58% | Deleterious | N22Y | 87% | 73% | Deleterious |
| D31N | 87% | 68% | Deleterious | I4L | 83% | 78% | Neutral |
| D31V | 87% | 82% | Deleterious | I4T | 83% | 55% | Neutral |
| D31Y | 87% | 86% | Deleterious | I4V | 83% | 83% | Neutral |
| T38A | 83% | 78% | Neutral | R20G | 87% | 58% | Deleterious |
| T38I | 83% | 55% | Neutral | R20I | 83% | 68% | Neutral |
| L37F | 83% | 83% | Neutral | R20K | 83% | 68% | Neutral |
| L37H | 83% | 58% | Neutral | R20T | 83% | 51% | Neutral |
| L37I | 83% | 83% | Neutral | M21I | 83% | 51% | Neutral |
| L37P | 87% | 61% | Deleterious | M21K | 87% | 82% | Deleterious |
| L37R | 87% | 61% | Deleterious | M21L | 87% | 58% | Deleterious |
| Y3C | 83% | 58% | Neutral | M21T | 87% | 73% | Deleterious |
| Y3H | 83% | 72% | Neutral | M21V | 83% | 58% | Neutral |
| N5D | 83% | 66% | Neutral | N25D | 83% | 45% | Neutral |
| N5K | 83% | 55% | Neutral | N25G | 87% | 59% | Deleterious |
| N5S | 83% | 66% | Neutral | N25S | 83% | 55% | Neutral |
| N5Y | 87% | 68% | Deleterious | Y26C | 87% | 68% | Deleterious |
| R24C | 87% | 61% | Deleterious | Y26F | 83% | 68% | Neutral |
| R24H | 83% | 51% | Neutral | Y26H | 83% | 58% | Neutral |
| R24L | 87% | 61% | Deleterious | I27K | 87% | 86% | Deleterious |
| R24S | 83% | 58% | Neutral | I27L | 83% | 51% | Neutral |
| S23F | 87% | 58% | Deleterious | I27M | 87% | 58% | Deleterious |
| S23P | 83% | 51% | Neutral | I27R | 87% | 86% | Deleterious |
| P10L | 87% | 86% | Deleterious | I27T | 87% | 77% | Deleterious |
| P10Q | 7% | 73% | Deleterious | I27V | 83% | 66% | Neutral |
| P10S | 83% | 51% | Neutral | D31G | 87% | 73% | Deleterious |
| P10T | 87% | 68% | Deleterious | D31H | 87% | 58% | Deleterious |
| F7C | 87% | 59% | Deleterious | D31N | 87% | 68% | Deleterious |
| F7L | 83% | 55% | Neutral | D31V | 87% | 82% | Deleterious |
| F7S | 83% | 55% | Neutral | D31Y | 87% | 86% | Deleterious |
| V6A | 83% | 78% | Neutral | M1G | 83% | 72% | Neutral |
| V6F | 83% | 55% | Neutral | G2D | 83% | 68% | Neutral |
| V6I | 83% | 78% | Neutral | G2L | 83% | 58% | Neutral |
| Y14C | 87% | 68% | Deleterious | F11L | 87% | 73% | Deleterious |
| Y14F | 83% | 72% | Neutral | F11S | 87% | 61% | Deleterious |
| Y14H | 83% | 55% | Neutral | T12A | 83% | 58% | Neutral |
| A28P | 87% | 73% | Deleterious | T12M | 87% | 73% | Deleterious |
| A28S | 83% | 51% | Neutral | S15C | 83% | 51% | Neutral |
| A28V | 83% | 55% | Neutral | S15G | 83% | 58% | Neutral |
| V30A | 83% | 55% | Neutral | L16P | 87% | 86% | Deleterious |
| V30I | 83% | 78% | Neutral | L17F | 83% | 72% | Neutral |
| V30L | 83% | 51% | Neutral | L17P | 87% | 86% | Deleterious |
| F35C | 87% | 59% | Deleterious | C19F | 87% | 59% | Deleterious |
| F35S | 83% | 51% | Neutral | S23F | 87% | 58% | Deleterious |
| A8D | 87% | 77% | Deleterious | S23P | 83% | 51% | Neutral |
| A8G | 83% | 68% | Neutral | Q29H | 83% | 55% | Neutral |
| A8P | 87% | 73% | Deleterious | Q29L | 87% | 61% | Deleterious |
| A8S | 83% | 55% | Neutral | Q29R | 87% | 58% | Deleterious |
| A8V | 83% | 58% | Neutral | V32A | 83% | 66% | Neutral |
| F9L | 87% | 61% | Deleterious | V32I | 83% | 78% | Neutral |
| F9S | 87% | 58% | Deleterious | V32L | 83% | 55% | Neutral |
| F9Y | 83% | 51% | Neutral | N34D | 83% | 66% | Neutral |
| I13L | 83% | 55% | Neutral | N34Y | 87% | 73% | Deleterious |
| I13M | 83% | 51% | Neutral | N36K | 83% | 51% | Neutral |
| I13T | 87% | 61% | Deleterious | N36S | 83% | 72% | Neutral |
| I13V | 83% | 68% | Neutral | T38A | 83% | 78% | Neutral |
| N22D | 83% | 58% | Neutral | T38I | 83% | 55% | Neutral |
| N22F | 83% | 55% | Neutral | ||||
Among a total of 37 residue positions with single mutations, the residue positions 1, 2, 11, 12, 15, 16, 20, 21, 25, 26, 27, 29, 32, 34, and 36 were unique in North America. Among all these residue positions, only the mutations at positions 11 and 16 in ORF10 variants in North America were predicted to be deleterious.
Furthermore, co-occurring mutations in the SARS-CoV-2 ORF10 in some geo-locations were listed in Table 4 . The highest number of simultaneous mutations (at 14 positions among the total of 38 amino acid residues) in ORF10 was noticed in a SARS-CoV-2 variant from Russia on March 21, 2020. Interestingly, no report of this kind of co-occurring mutation globally, hence denoted by localized index 1.
Table 4.
Co-occurring mutations in the ORF10 variants in various geo-locations.
| Mutations | Frequency | Date first collected | Localization across countries |
|---|---|---|---|
| USA | |||
| M1K; G2A; Y3D; I4G; N5L; V6Y; F7K; A8R | 9 | 16-01-2021 | 0.75 |
| P10S; L37F | 9 | 30-11-2020 | 1 |
| P10L; R24C | 2 | 17-03-2021 | 1 |
| R24L; A28V | 2 | 01-04-2021 | 1 |
| C19F; A28V | 1 | 24-03-2021 | 1 |
| I4V; N5D | 1 | 07-04-2021 | 1 |
| L37F; T38I | 1 | 09-12-2020 | 0.5 |
| M1G; G2L | 1 | 18-03-2020 | 0.4 |
| M1Q; Y3R; I4W; N5A; V6I; A8T | 1 | 23-02-2021 | 0.5 |
| M1R; G2W; Y3A | 1 | 30-03-2021 | 0.5 |
| M1R; G2W; Y3A; N5T; V6F | 1 | 22-07-2020 | 1 |
| M21I; R24C | 1 | 06-04-2021 | 1 |
| N36C; T38I | 1 | 08-03-2021 | 1 |
| S23F; I27K | 1 | 02-01-2021 | 1 |
| UK | |||
| M1G; G2L | 2 | 01-05-2020 | 0.4 |
| Q29I; V30F; D31N; V32C; N34T; F35L; N36G; L37R | 1 | 29-04-2020 | 1 |
| R20I; A28V | 1 | 21-05-2020 | 1 |
| T12M; V30L | 1 | 18-01-2021 | 0.5 |
| India | |||
| A8L; F9I | 1 | 09-03-2021 | 1 |
| M1Q; Y3R; I4W; N5A; V6I; A8T | 1 | 11-02-2021 | 0.5 |
| P10S; F11V | 1 | 23-04-2021 | 1 |
| South Africa | |||
| R24C; V30L | 1 | 06-01-2021 | 0.4 |
| Spain | |||
| V30L; T38I | 20 | 20-01-2021 | 0.95 |
| P10S; V30L | 13 | 20-01-2021 | 0.39 |
| V30L; D31N | 8 | 12-02-2021 | 0.8 |
| V30L; L37F | 4 | 18-01-2021 | 0.54 |
| M1L; G2R; Y3P; I4K; N5L; V6M; F7Q | 3 | 11-03-2020 | 1 |
| R24C; V30L | 2 | 01-03-2021 | 0.4 |
| S23F; V30L | 1 | 20-01-2021 | 0.38 |
| Germany | |||
| P10S; V30L | 15 | 24-02-2021 | 0.39 |
| V30L; D31H | 4 | 11-03-2021 | 1 |
| L16P; V30L | 3 | 16-03-2021 | 1 |
| P10S; S23F | 3 | 28-03-2021 | 1 |
| F7V; A8T | 1 | 10-04-2021 | 1 |
| L17I; R20I; V30L | 1 | 2021-01 | 1 |
| L37F; T38I | 1 | 06-04-2021 | 0.5 |
| S23F; V30L | 1 | 2020-12 | 0.38 |
| T12M; V30L | 1 | 24-03-2021 | 0.5 |
| Greece | |||
| I13M; V30L | 1 | 26-02-2021 | 1 |
| Mexico | |||
| M1K; G2A; Y3D; I4G; N5L; V6Y; F7K; A8R | 1 | 09-04-2021 | 0.75 |
| P10S; F35S | 1 | 21-03-2021 | 1 |
| Russia | |||
| R20T; M21R; S23P; R24A; Y26N; I27A; Q29I; D31L; V32Q; V33L; N34P; F35Q; N36G; L37T | 1 | 21-03-2020 | 1 |
Several co-occurring mutations in SARS-CoV-2 ORF10 variants have been reported in the USA, UK, India, South Africa, Spain, Germany, Greece, Mexico, and Russia. The most co-occurring mutations (V30L, T38I, first reported on January 20, 2021) were found in 20 infected patients in Spain. In the US the first detected and reported co-occurring mutations were M1K, G2A, Y3D, I4G, N5L, V6Y, F7K, and A8R on January 16, 2021 from 9 infected patients, and later on April 9 the first case was reported in Germany (Table 4). It is worth knowing that no single mutation at the residue positions 1 and 2 have been reported, and none of the co-occurring mutations among M1K, G2A, Y3D, I4G, N5L, V6Y, F7K, and A8R were reported as a single mutation in the ORF10 variants. Double co-occurring mutations P10S; V30l were reported in Spain on January 20, 2021, and on February 24, 2021, in Germany. Surprisingly, the pathogenic effects were predicted to be neutral (Table 3). It was noticed that V30L, one of the most common mutations in ORF10, was co-occurring with most of the other co-occurring mutations. Furthermore it was noticed that many mutations such as L17I, A8T, V6M, N34P, and F35Q did not appear as a single mutation, they appeared as one of the co-occurring mutations. It appears that co-occurring mutations in the SARS-CoV-2 ORF10 variants is an emerging trend.
3.2. Intrinsic disorder regions of SARS-CoV-2 ORF10 variants
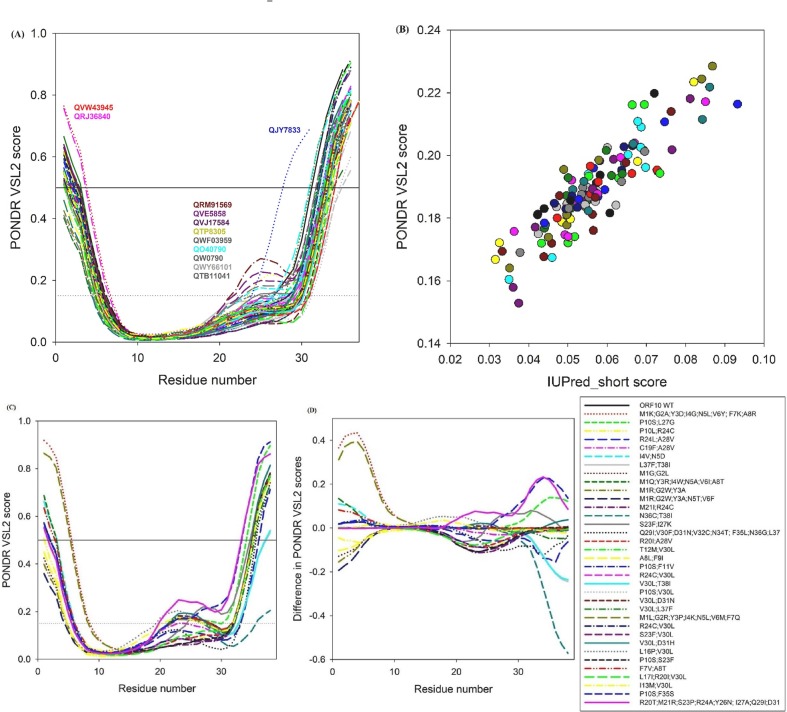
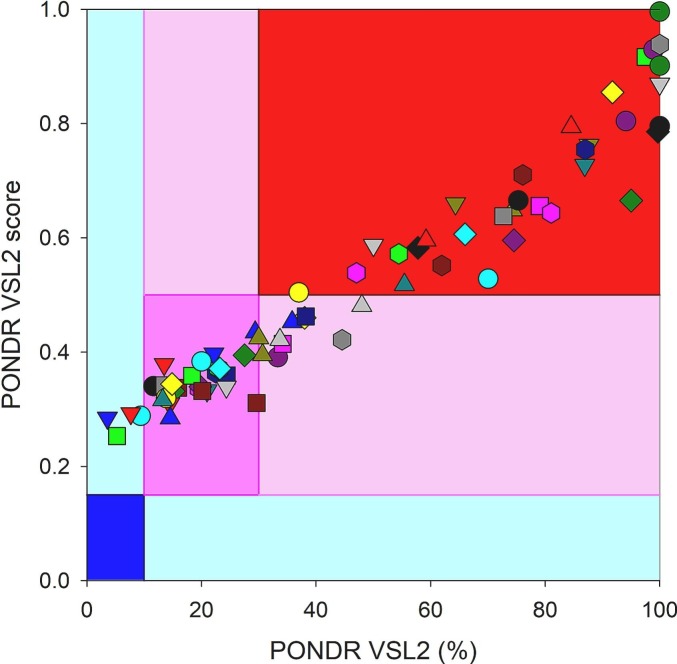
The per-residue disorder profiles for 138 unique SARS-CoV-2 ORF10 variants (variants are too short for disorder analysis) and 35 co-occurring mutations are presented in Fig. 3 . Fig. 3A shows that the intrinsic disorder predisposition of ORF10 is noticeably affected by single mutations. Mutations present the largest effects at the N- and C-terminal regions, where the disorder predisposition can vary from 0.4 to 0.8 and from 0.55 to 0.9 for the N- and C-termini, respectively, and for a region centered at residue 25, where the disorder score can change from 0.059 to 0.27. Fig. 3B compares the outputs of PONDR-VSL2 for 138 unique ORF10 variants with the results generated by one of the most conservative predictors of intrinsic disorder, and IUPred short [55].
Fig. 3.
(A): Per-residue disorder profiles for 138 SARS-CoV-2 ORF10 variants, (B): correlation between the outputs of two commonly used disorder predictors, (C): disorder propensity is changed in ORF10 variants with co-occurring mutations, and (D): difference spectra calculated by subtracting of the per-residue disorder scores of wild type ORF10 from the per-residue disorder scores of corresponding mutants.
Here, we investigate the effects of point mutations on the overall disorder score for the entire protein. This analysis showed that although the disorder score values generated by IUPred short are noticeably smaller than the corresponding PONDR-VSL2 data, the scales of changes introduced by single mutations in intrinsic disorder predisposition of ORF10 as evaluated by IUPred (from 0.03 to 0.095) are comparable to that generated by PONDR VSL2 (from 0.15 to 0.23). Furthermore, both predictors mostly agree on the direction of the changes introduced in the global disorder propensity of this protein by single mutations. Fig. 3C shows how disorder propensity is changed in ORF10 variants with co-occurring mutations. Noticeably, the effects of co-occurring mutants are qualitatively similar to those of single mutations, as most variability is observed at the disorder predisposition of N- and C-terminal regions and a region centered at residue 25. However, in the case of co-occurring mutants, scales of changes at the terminal regions are noticeably larger (the disorder predisposition can vary from 0.35 to 0.92 and from 0.2 to 0.9 for the N- and C-termini, respectively). Finally, to simplify comparison of the effect of co-occurring mutations on the disorder predisposition of ORF10, Fig. 3D shows “difference spectra” calculated by subtracting of the per-residue disorder scores of wild type ORF10 from the per-residue disorder scores of corresponding mutants. In this plot, intensities of the resulting “bands” reflect the magnitude of changes, whereas their sign, reflects the mutation-induced increase or decrease (positive or negative values, respectively) in local disorder propensity. Fig. 3D shows that most of the co-occurring mutations increase the disorder predisposition of the N-terminal region of ORF10, whereas many such mutations decrease the disorder propensity of the region centered at residue 25, and most of the mutations do not affect disorder predisposition of the C-terminal region.
Since ORF10 is not an enzyme, the accumulating point and co-occurring mutations are likely to affect some other functions of this protein, e.g., its capability to be engaged in protein-protein interactions. In fact, as discussed in the following section, accumulated data suggested the presence of multiple host partner proteins for ORF10. In fact, Fig. 3 shows that ORF10 is a short hybrid protein containing disordered tails and a more ordered central region; it is likely that this protein is using both ordered and disordered segments for its interactions.
3.3. Interactability of ORF10 and intrinsic disorder status of its host partners
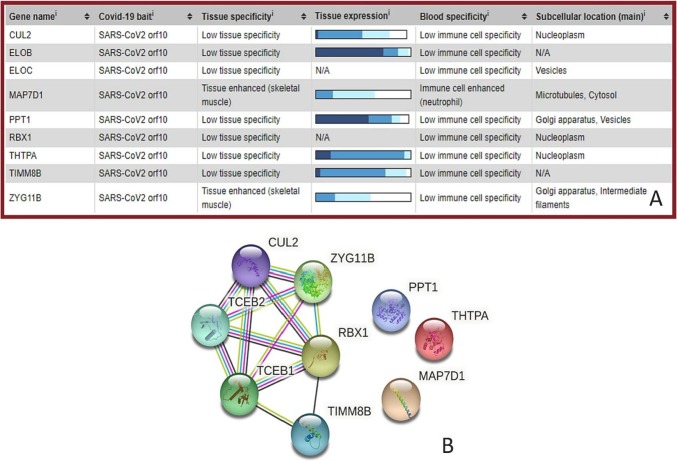
The SARS-CoV-2 ORF10 protein was found to interact with few of the human proteins such as CUL2, ELOB, ELOC, MAP7D1, PPT1, RBX1, THTPA, TIMM8B, and ZYG11B [37]. A detailed summary of the tissue and cellular expression patterns of SARS-CoV-2 ORF10 interacting human proteins, based on transcriptomics and antibody-based proteomics, are presented in Fig. 5. The ORF10 of SARS-CoV-2 interacts with members of the Cullin ubiquitin ligase CUL2ZY G11B complex. Interestingly, among the genes, ZYG11B scored the highest with regards to ORF10 interactome, which confirmed a direct interaction between ORF10 and ZYG11B [36].
Fig. 5.
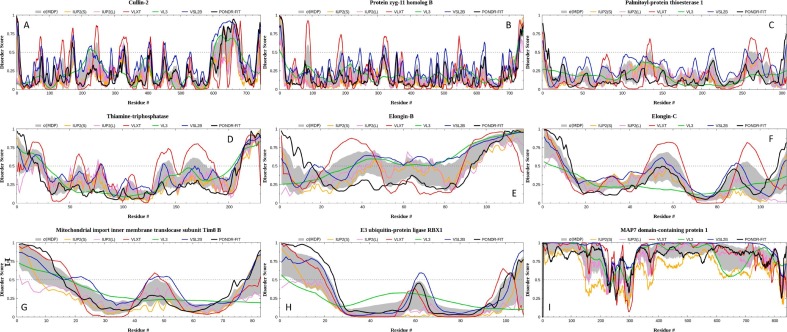
Intrinsic disorder predisposition of human proteins interacting with ORF10 (A: CUL2; B: ZYG11B; C: PPT1; D: THTPA; E: ELOB; F: ELOC; G: TIMM8B; H: RBX1; and I: MAD7D1) analyzed by six per-residue predictors, PONDR-VLXT, PONDR-VSL2, PONDR-VL3, PONDR-FIT, and the IUPred2A computational platform that allows identification of either short or long regions of intrinsic disorder, IUPred-L and IUPred-S [56], [57], [58], [59], [60]. The outputs of the evaluation of the per-residue disorder propensity by these tools are represented as real numbers between 1 (ideal prediction of disorder) and 0 (ideal prediction of order). A threshold of ≥0.5 was used to identify disordered residues and regions in query proteins. For each query protein in this study, the predicted percentage of intrinsic disorder (PPID) was calculated based on the outputs of per-residue disorder predictors. Here, PPID in a query protein represents a percent of residues with disorder scores exceeding 0.5.
The SARS-CoV-2 ORF10 interacting with human proteins CUL2, ELOB, PPT1, THTPA, and TIMM8B were ex- pressed in various tissues, including the lung. SARS-CoV-2 ORF10 contains an α-helical region (amino acid residue positions 3 to 20), which may interact and form a complex with CUL2ZY G11B [36]. It was furthermore reported that by forming a complex with CUL2ZY G11B, ORF10 hijacks it for ubiquitination and degradation of restriction factors, or alternatively, may bind to the N-terminal glycine in ORF10 to target it for degradation [36], [37].
Fig. 4B represents the STRING-generated network of the host proteins interacting with ORF10 and demonstrates that CUL2, ZYG11B, RBX1, ELOB/TCEB1, ELOC/TCEB2, and TIMM8B form a tightly linked cluster, whereas PPT1, THTPA, and MAP7D1 are not involved in interaction with other host proteins considered here [56]. Intrinsic disorder profiles for these proteins, ranging from 83 (TIMM88) to 841 residues (MAD7D1), are presented in Fig. 5. These profiles generated a set of commonly used disorder predictors and show that ORF10 interacting proteins possess rather different levels of intrinsic disorder, with some of them being mostly ordered (e.g., PPT1, which is an expected feature of an enzyme), and others (e.g., MAP7D1) being highly disordered. In fact, based on their increasing disorder content evaluated by PONDR-VSL2 as a percent of predicted intrinsically disordered residues (PPIDR), these proteins can be arranged into the following sequence: PPT1 (3.59%) < ZYG11B (9.41%) < THTPA (22.17%) < CUL2 (22.5%) < ELOC/TCEB2 (23.21%) < RBX1 (29.63%) < TIMM8B (44.58%) < ELOB/TCEB1 (74.58%) < MAD7D1 (98.93%).
Fig. 4.
A: A snapshot summary of the tissue and cellular expression patterns of SARS-CoV-2 ORF10 interacting human proteins, based on transcriptomics and antibody-based proteomics. B: Inter-set protein-protein interaction network of human proteins interacting with ORF10 generated by Search Tool for the Retrieval of Interacting Genes; STRING, http://string-db.org/. STRING produces a network of protein- protein interactions based on predicted and experimentally-validated information on the interaction partners of a protein of interest [56]. In the corresponding network, the nodes correspond to proteins, whereas the edges show predicted or known functional associations. Seven types of evidence are used to build the corresponding network, where they are indicated by the differently colored lines: a green line represents neighborhood evidence; a red line – the presence of fusion evidence; a purple line – experimental evidence; a blue line – co-occurring evidence; a light blue line – database evidence; a yellow line – text mining evidence; and a black line – co-expression evidence [56]. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
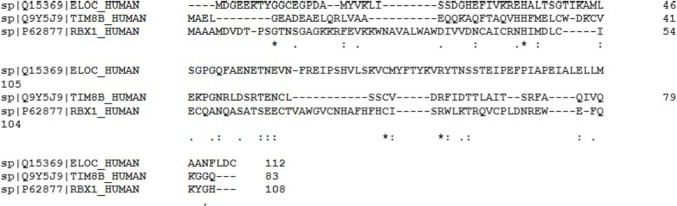
Therefore, based on the accepted classification, proteins are considered as mostly ordered, moderately disordered and highly disordered if their PPDR < 10%, 10 ≤ PPDR < 30%, and highly disordered PPDR ≥ 30%, respectively, PPT1 and ZYG11B are highly ordered, THTPA, CUL2, and ELOC/TCEB2 are moderately disordered, and RBX1, TIMM8B, ELOB/TCEB1, and MAD7D1 are highly disordered [61]. Curiously, Fig. 5 shows close similarity of the disorder profiles of TIMM88 and RBX1, and to a latter degree, ELOB/TCEB1 despite the fact that these three proteins are characterized by relatively low sequence identity (ranging from 11.69 to 25.64%) (Fig. 6 ):
Fig. 6.
Sequence alignment among ELOC, TIM8B, and RBX1 human proteins.
We further looked at the presence of potential disorder-based binding sites (molecular recognition features, MoRFs; i.e., disordered regions that gain ordered structure at interaction with binding partners) in these nine proteins using the ANCHOR algorithm [62]. This analysis revealed that more than half of these proteins contains MoRFs: ZYG11B (residues 716-721), CUL2 (residues 679-688), ELOB (residues 1-6 and 75-96), THTPA (residues 1-6 and 193-200), and MAD7D1 (residues 1-22, 37-80, 88-107, 115-125, 132-142, 149-163, 232-237, 251-268, 291-315, 336-345, 355-386, 398-406, 408-414, 427-491, 500-550,565-608, 635-642, 713-719, 737-762, 772-799, and 814-832), suggesting the role of intrinsic disorder in their functionality. This analysis indicates that at least some of these MoRFs can be involved in interaction with ORF10. Obviously, subsequent experimental analysis is needed to verify this interesting hypothesis.
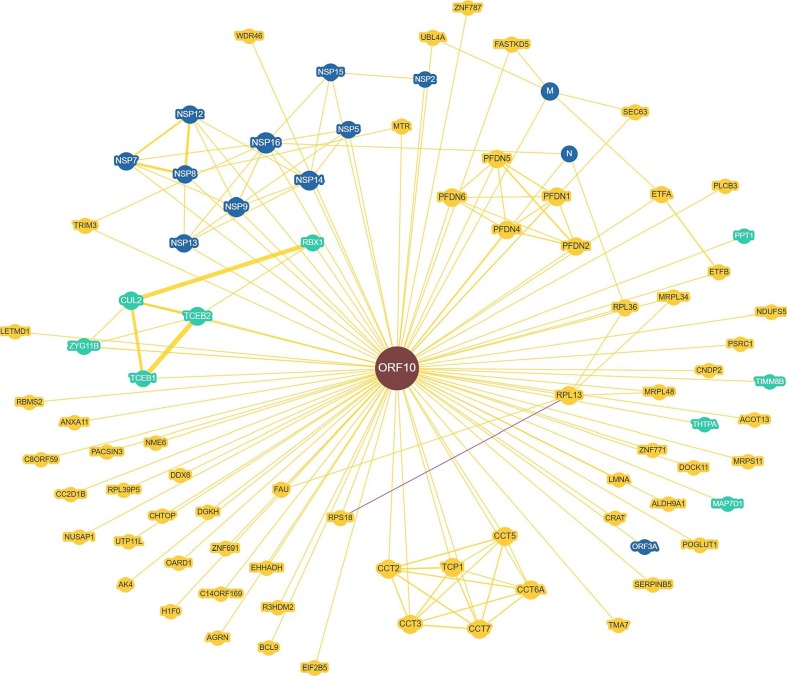
To further extend the interactability analysis of ORF10, we used a Biological General Repository for Interaction Datasets BioGRID platform, which is a comprehensively curated biomedical interaction repository containing 2,133,831 protein and genetic interactions, 29,417 chemical interactions, and 1,128,339 post translational modifications from major model organism species [63]. One of the recent BioGRID activities is a COVID-19 coronavirus curation project, where 15,645 non-redundant interactions are described for 32 SARS-CoV-2 proteins. Fig. 7 represents a protein-protein inter- action (PPI) network generated by BioGRID for ORF10. This network 72 contains 72 host proteins (including 9 proteins discussed in the previous sections) and 12 SARS-CoV-2 proteins. These data indicate that despite its small size, ORF10 is a highly promiscuous protein interacting with multiple host proteins and potentially affecting a multitude of functional pathways in the host. It is also likely that the unique mutations found in the natural variants of this protein can affect its interactability and associated multifunctionality, generating highly heterogeneous outputs (Fig. 8).
Fig. 7.
ORF10-centered protein-protein interaction network generated by BioGRID. This network includes both human and SARS-CoV-2 proteins engaged in interaction with ORF10 (dark red circle in the middle of the plot). Nine human proteins discussed in the previous section are shown in green, whereas the remaining human proteins are shown by yellow symbols. Blue symbols correspond to proteins from SARS-CoV-2. Yellow lines represent interactions with physical evidence, purple lines show association with genetic and physical evidence. Node size reflects the number of edges to/from that node, with the larger node possessing more edges attached to it. Similarly, the edge thickness serves as a reflection of the number of unique curated interactions supporting the association, with thicker lines representing edges with more unique curated interactions supporting its existence. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
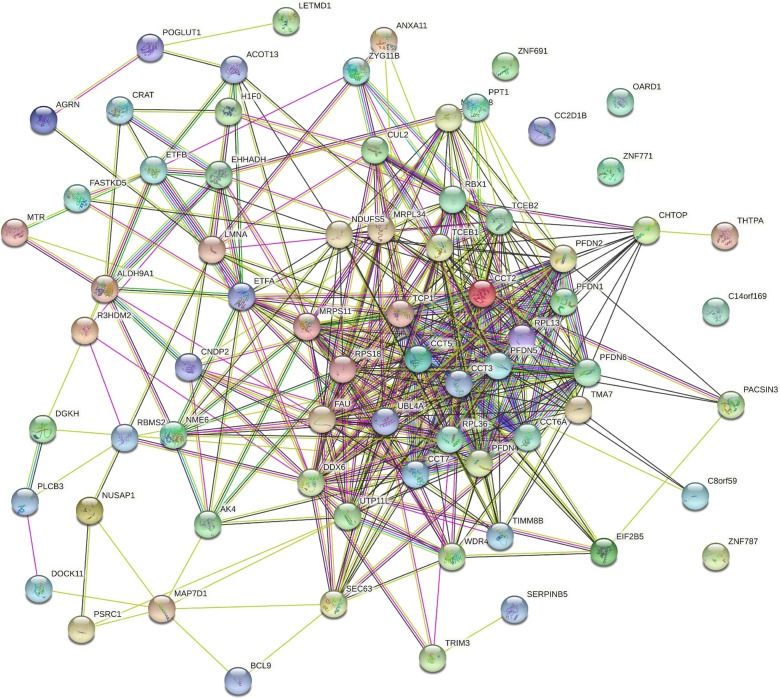
Fig. 8.
STRING-based analysis of the inter-set interactivity of 72 human proteins interacting with SARS-CoV-2 ORF10 using the low confidence level of 0.15. This confidence level was selected to ensure maximal inclusion of host proteins into the resulting PPI.
Analysis of this network in terms of the significantly enriched Gene Ontology terms (GO-terms) indicated that the molecular functions of its proteins include unfolded protein binding, structural constituent of ribosome, and RNA binding. Furthermore, the 37 significantly enriched GO-terms corresponding to biological processes include positive regulation of establishment of protein localization to telomere (GO:1904851), positive regulation of protein localization to Cajal body (GO:1904871), positive regulation of telomerase RNA localization to Cajal body (GO:1904874), positive regulation of telomere maintenance via telomerase (GO:0032212), binding of sperm to zona pellucida (GO:0007339), toxin transport (GO:1901998), regulation of telomere maintenance (GO:0032204), fatty acid beta-oxidation (GO:0006635), regulation of protein localization to nucleus (GO:1900180), SRP-dependent cotranslational protein targeting to membrane (GO:0006614), protein folding (GO:0006457), regulation of transcription from RNA polymerase II promoter in response to hypoxia (GO:0061418), positive regulation of chromosome organization (GO:2001252), translational initiation (GO:0006413), protein stabilization (GO:0050821), cellular response to hypoxia (GO:0071456), peptide biosynthetic process (GO:0043043), regulation of chromosome organization (GO:0033044), translation (GO:0006412), regulation of protein stability (GO:0031647), protein targeting (GO:0006605), regulation of DNA metabolic process (GO:0051052), amide biosynthetic process (GO:0043604), establishment of protein localization to organelle (GO:0072594), positive regulation of organelle organization (GO:0010638), cellular amide metabolic process (GO:0043603), protein localization to organelle (GO:0033365), cellular macromolecule catabolic process (GO:0044265), organonitrogen compound biosynthetic process (GO:1901566), positive regulation of cellular component organization (GO:0051130), macromolecule catabolic process (GO:0009057), cellular nitrogen compound biosynthetic process (GO:0044271), organic substance catabolic process (GO:1901575), cellular catabolic process (GO:0044248), cellular nitrogen compound metabolic process (GO:0034641), cellular metabolic process (GO:0044237), and cellular process (GO:0009987). It is tempting therefore to hypothesize that at least part of these functions and processes will be affected by ORF10 via its binding to specific host proteins.
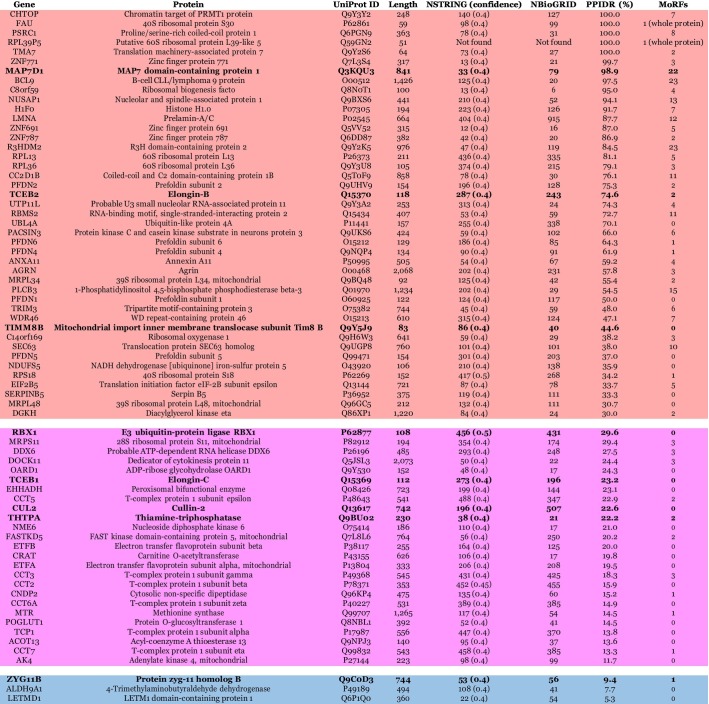
One should keep in mind that each of these human proteins interacting with ORF10 is a promiscuous binder itself. This is illustrated by Table 5 that provides some basic information about these host proteins. It is seen that with almost no exception, these proteins are engaged in multiple interactions, often serving as hubs of corresponding PPI networks. As high binding promiscuity is often associated with the presence of intrinsic disorder in a query of proteins, we also analyzed intrinsic disorder predispositions of these host proteins. Corresponding data are shown in Table 5 and Fig. 9. Based on the accepted practice, per-protein features derived from the corresponding disorder profiles can be used to classify proteins as mostly ordered, moderately or highly disordered. This analysis revealed that only four proteins are expected to be mostly ordered, 43 are highly disordered and 25 are moderately disordered. Furthermore, most of the 72 human proteins contain MoRFs (see Table 5), and all of these proteins can undergo extensive post translational modifications (PTMs, data not shown). This is not surprising, as many enzymatically-catalyzed PTMs are commonly found within the intrinsically disordered or flexible regions [64], [65], [66]. These observations indicate that these host proteins utilize their intrinsic disorder for interaction with specific partners (including ORF10) and for modulation of these interactions via PTMs.
Table 5.
Interactability and intrinsic disorder of human proteins interacting with ORF10. Proteins are arranged by their intrinsic disorder content and lines are colored in red, pink and blue to show highly disordered, moderately disordered and highly ordered proteins, respectively.
Fig. 9.
Intrinsic disorder predisposition of 72 human proteins interacting with SARS-CoV-2 ORF10 protein based on their mean disorder scores and percent of predicted disordered residues as evaluated by PONDR(R) VSL2 algorithm. Large values of each parameter indicate increasing disorder. Color blocks indicate regions which are mostly ordered (blue and light blue), moderately disordered (pink and light pink), or mostly disordered (red). If the two parameters agree, the corresponding part of the background is dark (blue or pink), whereas light blue and light pink reflect areas in which only one of these criteria applies. It is noteworthy that only four human proteins that interact with ORF10 are mostly (located within the light blue area). The remaining 68 proteins are either moderately or highly disordered. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. Predicting secondary structures of ORF10 protein variants with co-occurring mutations
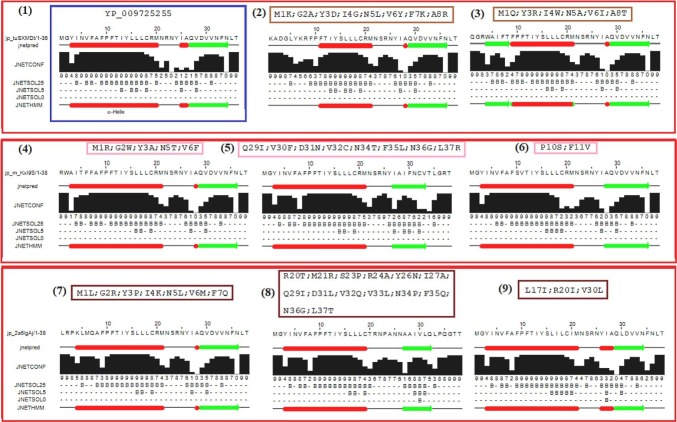
The secondary structures of nine ORF10 variants including the wild-type ORF10 (YP 009725255) with several co-occurring mutations were predicted (Fig. 10). The α-helix secondary structural amino acid (aa) residue positions 3 to 20 were invariant for the six ORF variants with co-occurring mutations as mentioned in panels (4) to (9) of Fig. 10, although the reliability indices were changed for the α-helix region [36].
Fig. 10.
Secondary structure predispositions of nine ORF10 variants including the wild ORF10 (YP 009725255) with co-occurring mutations as predicted by the JPred 4 webserver [44]. Red and green stretches correspond to the predicted α-helices and β-strands, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Furthermore, it was observed that the α-helix amino acid residue positions (3-20) were switched to 11-21 and 9-21 due to the co-occurring mutations M1K, G2A, Y3D, I4G, N5L, V6Y, F7K, A8R, and M1Q, Y3R, I4W, N5A, V6I, A8T in two different ORF10 variants, respectively. Therefore, due to these two sets of co-occurring mutations, SARS-CoV-2 ORF10 interactions with the CUL2ZY G11B complex might be affected. Due to the amino acid residue positional changes in the secondary structure in the ORF10 variants (other than the reference ORF10 (YP 009725255)), these ORF10 variants with human proteins other than the CUL2ZY G11B complex are likely to be affected. Mutations at position 1 affect the methionine initiation codon, for which replacement only valine has been shown to support translation of protein. As for the ORF10 mutations methionine at position 1 is replaced by arginine (M1R), glutamine (M1Q), leucine (M1L), and lysine (M1K), respectively, the mutants should not code for protein and therefore possess no secondary structure.
3.5. Amino acid frequency distribution across the unique ORF10 variants and associated clusters
The frequency of each amino acid in the 140 unique ORF10 sequences is presented in Table 6, Table 7 . Also, Fig. 11(A) shows the amino acid frequency vector of each unique ORF10 sequence. It was noticed that glutamic acid (E) and tryptophan (W) were absent in each ORF10 variant. Among 140 unique ORF10 variants, lysine (K) was present with a single frequency only in seven North American ORF10 variants (QTD22916.1, QLA48060.1, QWY66101.1, QVO40425.1, QTP28305.1, QSL79091.1, and QRM91569.1).
Table 6.
Frequency distribution of amino acids over the 140 unique SARS-CoV-2 ORF10 variants (from QKM75696.1 to QWN58574.1).
| ORF10 | A | R | N | D | C | Q | E | G | H | I | L | K | M | F | P | S | T | W | Y | V |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| QKM75696.1 | 2 | 2 | 4 | 1 | 1 | 1 | 0 | 0 | 0 | 2 | 4 | 0 | 1 | 3 | 1 | 2 | 2 | 0 | 2 | 3 |
| QKG88643.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 5 | 0 | 1 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QVJ13930.1 | 2 | 2 | 5 | 2 | 1 | 1 | 0 | 0 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QWE67724.1 | 2 | 2 | 5 | 1 | 2 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 2 | 4 |
| QRJ36840.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 1 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 2 | 4 |
| QQY03084.1 | 2 | 2 | 4 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QRG22086.1 | 2 | 2 | 4 | 2 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QTD22916.1 | 2 | 2 | 4 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 1 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QUL69971.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 2 | 0 | 2 | 2 | 0 | 3 | 3 |
| QWB85197.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 5 | 1 | 2 | 2 | 0 | 3 | 3 |
| QWJ83116.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 4 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 3 |
| QSE09446.1 | 2 | 2 | 5 | 1 | 2 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 3 | 1 | 2 | 2 | 0 | 3 | 4 |
| QWZ00470.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 5 | 0 | 2 | 4 | 0 | 2 | 2 | 0 | 3 | 4 |
| QTP26076.1 | 3 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 1 | 0 | 3 | 4 |
| QWK62875.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 3 | 4 | 1 | 2 | 1 | 0 | 3 | 4 |
| QVG57396.1 | 2 | 2 | 5 | 1 | 2 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 2 | 4 |
| QRA60944.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 5 | 1 | 2 | 2 | 0 | 2 | 4 |
| QWE68295.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 1 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 2 | 4 |
| QWT58729.1 | 2 | 2 | 5 | 1 | 2 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 1 | 2 | 0 | 3 | 4 |
| QWM42669.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 2 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 1 | 2 | 0 | 3 | 4 |
| QWS64226.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 3 | 0 | 2 | 5 | 1 | 2 | 2 | 0 | 3 | 4 |
| QVJ47956.1 | 2 | 1 | 5 | 1 | 1 | 1 | 0 | 2 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QVW78101.1 | 2 | 1 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 4 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QLA48060.1 | 2 | 1 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 1 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QVO98764.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 4 | 4 | 0 | 1 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QWY66101.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 1 | 1 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QWQ05246.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 5 | 0 | 1 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QTS35265.1 | 2 | 2 | 4 | 2 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QVP24786.1 | 2 | 2 | 4 | 1 | 1 | 1 | 0 | 1 | 1 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QVO85840.1 | 2 | 2 | 4 | 1 | 1 | 1 | 0 | 1 | 0 | 4 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QVO40425.1 | 2 | 2 | 4 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 1 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QWU52456.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 5 | 1 | 1 | 2 | 0 | 3 | 4 |
| QTP28305.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 2 | 4 | 1 | 2 | 5 | 1 | 1 | 2 | 0 | 3 | 4 |
| QWS07290.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 2 | 1 | 2 | 0 | 3 | 4 |
| QWF07009.1 | 2 | 1 | 5 | 1 | 2 | 1 | 0 | 1 | 0 | 3 | 3 | 0 | 2 | 5 | 1 | 2 | 2 | 0 | 3 | 4 |
| QWY19801.1 | 2 | 1 | 5 | 1 | 2 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QWX30181.1 | 2 | 1 | 5 | 1 | 1 | 1 | 0 | 1 | 1 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QWY95666.1 | 2 | 1 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 5 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QVU00656.1 | 2 | 2 | 4 | 2 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QTA53643.1 | 3 | 2 | 4 | 1 | 1 | 1 | 0 | 2 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 3 |
| QUG14309.1 | 2 | 2 | 4 | 1 | 1 | 1 | 0 | 2 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QWY70751.1 | 2 | 2 | 5 | 1 | 2 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 2 | 4 |
| QSJ35636.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 5 | 1 | 2 | 2 | 0 | 2 | 4 |
| QWF03959.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 1 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 2 | 4 |
| BCY15724.1 | 2 | 2 | 3 | 0 | 1 | 0 | 0 | 1 | 0 | 3 | 3 | 0 | 2 | 3 | 1 | 2 | 1 | 0 | 3 | 1 |
| QWT72678.1 | 2 | 2 | 5 | 1 | 1 | 0 | 0 | 1 | 1 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QUM42028.1 | 2 | 2 | 5 | 1 | 1 | 0 | 0 | 1 | 0 | 3 | 5 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QUX49292.1 | 3 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 3 |
| QWK69365.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 4 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 3 |
| QTA74333.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 5 | 1 | 2 | 2 | 0 | 3 | 3 |
| QWY54619.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 5 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 3 |
| QUV26065.1 | 3 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 3 |
| QWF05003.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 4 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 3 |
| QUP00476.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 5 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 3 |
| QTJ93574.1 | 3 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 3 |
| QWN49685.1 | 2 | 2 | 5 | 2 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 3 |
| QVO91006.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 5 | 1 | 2 | 2 | 0 | 3 | 3 |
| QVV18442.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 4 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 3 |
| QJY78233.1 | 2 | 2 | 3 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 3 | 0 | 2 | 3 | 1 | 2 | 1 | 0 | 3 | 4 |
| QVL90897.1 | 2 | 2 | 4 | 2 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QOH29638.1 | 2 | 2 | 5 | 1 | 2 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 3 | 1 | 2 | 2 | 0 | 3 | 4 |
| QWC74916.1 | 2 | 2 | 4 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QSL79091.1 | 2 | 2 | 4 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 1 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| BCX23983.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 3 | 0 | 2 | 4 | 1 | 2 | 1 | 0 | 3 | 4 |
| QWY94400.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 3 | 0 | 2 | 5 | 1 | 2 | 2 | 0 | 3 | 4 |
| QSE25736.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 1 | 3 | 3 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QWN58574.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 4 | 3 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
Table 7.
Frequency distribution of amino acids over the 140 unique SARS-CoV-2 ORF10 variants (from QTI75440.1 to QWY55420.1).
| ORF10 | A | R | N | D | C | Q | E | G | H | I | L | K | M | F | P | S | T | W | Y | V |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| QTI75440.1 | 3 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 1 | 0 | 3 | 4 |
| QWS53172.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 4 | 4 | 0 | 2 | 4 | 1 | 2 | 1 | 0 | 3 | 4 |
| YP 009725255.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QWY95092.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 3 | 0 | 2 | 4 | 2 | 2 | 2 | 0 | 3 | 4 |
| QWQ66744.1 | 2 | 3 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 3 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QSW62483.1 | 2 | 2 | 4 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 3 | 2 | 0 | 3 | 4 |
| QVU28280.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 5 | 0 | 2 | 3 | 1 | 2 | 2 | 0 | 3 | 4 |
| QWW53635.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 3 | 1 | 3 | 2 | 0 | 3 | 4 |
| QQX05795.1 | 2 | 2 | 4 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 4 | 4 |
| QVE28736.1 | 2 | 2 | 5 | 0 | 1 | 1 | 0 | 2 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QTC84700.1 | 2 | 2 | 5 | 0 | 1 | 1 | 0 | 1 | 1 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QWN49673.1 | 2 | 2 | 6 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 3 |
| QUA36764.1 | 2 | 2 | 6 | 0 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QSW42096.1 | 2 | 2 | 5 | 0 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 5 |
| QWY22460.1 | 2 | 2 | 5 | 0 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 4 | 4 |
| QUF19963.1 | 2 | 3 | 5 | 1 | 1 | 0 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QTB11041.1 | 1 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 2 | 2 | 2 | 0 | 3 | 4 |
| QWX09518.1 | 1 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 3 | 2 | 0 | 3 | 4 |
| QWU01215.1 | 1 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 5 |
| QRM91569.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 2 | 4 | 1 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QRX03618.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 2 | 5 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QSO40790.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 2 | 4 | 0 | 3 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QVE25858.1 | 2 | 3 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 2 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QVJ17584.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 2 | 4 | 0 | 2 | 4 | 1 | 2 | 3 | 0 | 3 | 4 |
| QVO95200.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 2 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 5 |
| QVE30392.1 | 2 | 2 | 4 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 3 | 2 | 0 | 3 | 4 |
| QTC19517.1 | 2 | 1 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 3 | 2 | 0 | 3 | 4 |
| QVJ37366.1 | 2 | 2 | 4 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 3 | 2 | 0 | 3 | 4 |
| QWQ76823.1 | 2 | 2 | 4 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 3 | 0 | 3 | 4 |
| QVV08801.1 | 2 | 2 | 4 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 4 | 4 |
| QUI12106.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 1 | 4 | 1 | 2 | 3 | 0 | 3 | 4 |
| QWU53472.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 1 | 4 | 1 | 2 | 2 | 0 | 3 | 5 |
| QTS24551.1 | 2 | 1 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 3 | 0 | 3 | 4 |
| QNV50343.1 | 2 | 2 | 5 | 1 | 0 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 5 | 1 | 2 | 2 | 0 | 3 | 4 |
| QUB17908.1 | 1 | 2 | 5 | 1 | 0 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 5 | 1 | 2 | 2 | 0 | 3 | 5 |
| QVL64016.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 3 | 0 | 2 | 4 | 2 | 2 | 2 | 0 | 3 | 4 |
| QWU68360.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 3 | 0 | 2 | 4 | 2 | 2 | 2 | 0 | 3 | 4 |
| QTO29824.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 2 | 5 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QWU51246.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 2 | 4 | 0 | 3 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QVX69392.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 2 | 4 | 0 | 2 | 4 | 1 | 2 | 3 | 0 | 3 | 4 |
| QWO21857.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 2 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 5 |
| QTK02152.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 5 | 0 | 2 | 3 | 1 | 2 | 2 | 0 | 3 | 4 |
| QWY72735.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 3 | 1 | 3 | 2 | 0 | 3 | 4 |
| QVH90751.1 | 2 | 2 | 5 | 1 | 1 | 2 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 0 | 2 | 2 | 0 | 3 | 4 |
| QVX36355.1 | 2 | 1 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 5 | 0 | 2 | 4 | 0 | 3 | 2 | 0 | 3 | 4 |
| QWY95104.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 3 | 0 | 2 | 5 | 0 | 3 | 2 | 0 | 3 | 4 |
| QWY18545.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 0 | 3 | 2 | 0 | 3 | 4 |
| QVL15727.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 0 | 2 | 3 | 0 | 3 | 4 |
| QTO07027.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 3 | 0 | 2 | 2 | 0 | 3 | 4 |
| BCX25240.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 5 | 0 | 2 | 3 | 1 | 2 | 2 | 0 | 3 | 4 |
| QWB65585.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 3 | 1 | 3 | 2 | 0 | 3 | 4 |
| QTW57386.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 3 | 1 | 2 | 2 | 0 | 4 | 4 |
| QWW38212.1 | 1 | 2 | 5 | 2 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QSE30156.1 | 1 | 2 | 5 | 1 | 1 | 1 | 0 | 2 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QUF17645.1 | 1 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 2 | 2 | 2 | 0 | 3 | 4 |
| QUF20717.1 | 1 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 3 | 1 | 2 | 2 | 0 | 3 | 4 |
| QWB83606.1 | 1 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 3 | 2 | 0 | 3 | 4 |
| QPF60767.1 | 1 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 3 | 0 | 3 | 4 |
| QVR42407.1 | 1 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 2 | 0 | 2 | 2 | 0 | 3 | 4 |
| QUA32182.1 | 1 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 5 | 0 | 2 | 4 | 0 | 2 | 2 | 0 | 3 | 5 |
| QRG41735.1 | 1 | 1 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 5 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 5 |
| QWT73590.1 | 1 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 5 |
| QWK62266.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 5 | 0 | 2 | 3 | 1 | 2 | 2 | 0 | 3 | 4 |
| QVM67662.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 4 | 4 | 0 | 2 | 3 | 1 | 3 | 1 | 0 | 3 | 4 |
| QQE14148.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 3 | 1 | 3 | 2 | 0 | 3 | 4 |
| QVU00728.1 | 2 | 2 | 4 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 3 | 2 | 0 | 3 | 4 |
| QUA79573.1 | 2 | 2 | 4 | 1 | 1 | 1 | 0 | 1 | 0 | 3 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 4 | 4 |
| QWT65692.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 2 | 5 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 4 |
| QVW43945.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 2 | 4 | 0 | 2 | 4 | 1 | 2 | 3 | 0 | 3 | 4 |
| QUM21266.1 | 2 | 2 | 4 | 2 | 1 | 1 | 0 | 1 | 0 | 2 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 5 |
| QTX06599.1 | 2 | 1 | 5 | 1 | 2 | 1 | 0 | 1 | 0 | 2 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 5 |
| QVM14069.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 2 | 3 | 0 | 2 | 5 | 1 | 2 | 2 | 0 | 3 | 5 |
| QWY55420.1 | 2 | 2 | 5 | 1 | 1 | 1 | 0 | 1 | 0 | 2 | 4 | 0 | 2 | 4 | 1 | 2 | 2 | 0 | 3 | 5 |
Fig. 11.
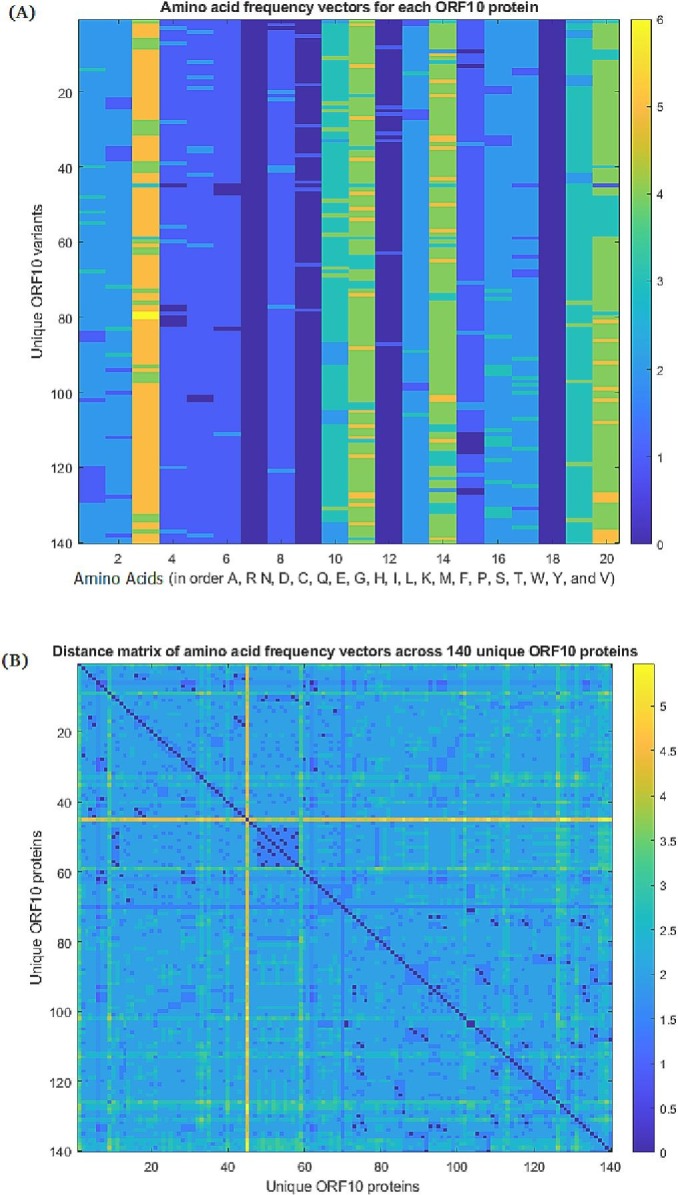
(A): Frequency distribution of amino acids across the unique 140 ORF10 proteins, and (B) pairwise distance matrix of frequency vectors for each ORF10 protein
It was noticed that asparagine (N) was present in the ORF10 sequences with the highest frequencies ranging from 4 to 6 (Fig. 11(A)). Also, the frequencies of leucine (L), phenylalanine (F), and valine (V) in each ORF10 variant were dominant as compared to that of other amino acids.
For each pair of frequency vectors associated with ORF10 sequences, distances were enumerated (Fig. 11(B)).
The most distant frequency vector was detected for the SARS-CoV-2 ORF10 BCY15724.1 (Japan, collected in November, 2020). Based on frequency-vector similarity (Euclidean distance-wise nearness), unique 140 ORF10 proteins were clustered using the K-means clustering method (Table 8 ).
Table 8.
K-means clustering (clusters: 20) of the unique ORF10 variants based on amino acid frequency distribution.
| ORF10 | Cluster | ORF10 | Cluster | ORF10 | Cluster | ORF10 | Cluster |
|---|---|---|---|---|---|---|---|
| QKM75696.1 | 8 | QVW78101.1 | 11 | BCY15724.1 | 17 | QWN58574.1 | 15 |
| QKG88643.1 | 1 | QLA48060.1 | 1 | QWT72678.1 | 16 | QTI75440.1 | 16 |
| QVJ13930.1 | 1 | QVO98764.1 | 11 | QUM42028.1 | 1 | QWS53172.1 | 11 |
| QWE67724.1 | 7 | QWY66101.1 | 1 | QUX49292.1 | 14 | YP 009725255.1 | 5 |
| QRJ36840.1 | 7 | QWQ05246.1 | 1 | QWK69365.1 | 11 | QWY95092.1 | 15 |
| QQY03084.1 | 5 | QTS35265.1 | 20 | QTA74333.1 | 3 | QWQ66744.1 | 15 |
| QRG22086.1 | 20 | QVP24786.1 | 20 | QWY54619.1 | 14 | QSW62483.1 | 5 |
| QTD22916.1 | 20 | QVO85840.1 | 20 | QUV26065.1 | 14 | QVU28280.1 | 6 |
| QUL69971.1 | 18 | QVO40425.1 | 20 | QWF05003.1 | 11 | QWW53635.1 | 19 |
| QWB85197.1 | 3 | QWU52456.1 | 3 | QUP00476.1 | 14 | QQX05795.1 | 20 |
| QWJ83116.1 | 11 | QTP28305.1 | 10 | QTJ93574.1 | 14 | QVE28736.1 | 16 |
| QSE09446.1 | 6 | QWS07290.1 | 16 | QWN49685.1 | 14 | QTC84700.1 | 16 |
| QWZ00470.1 | 1 | QWF07009.1 | 10 | QVO91006.1 | 3 | QWN49673.1 | 14 |
| QTP26076.1 | 16 | QWY19801.1 | 1 | QVV18442.1 | 11 | QUA36764.1 | 16 |
| QWK62875.1 | 16 | QWX30181.1 | 1 | QJY78233.1 | 8 | QSW42096.1 | 4 |
| QVG57396.1 | 7 | QWY95666.1 | 1 | QVL90897.1 | 20 | QWY22460.1 | 16 |
| QRA60944.1 | 3 | QVU00656.1 | 20 | QOH29638.1 | 6 | QUF19963.1 | 16 |
| QWE68295.1 | 7 | QTA53643.1 | 16 | QWC74916.1 | 5 | QTB11041.1 | 4 |
| QWT58729.1 | 16 | QUG14309.1 | 20 | QSL79091.1 | 20 | QWX09518.1 | 4 |
| QWM42669.1 | 16 | QWY70751.1 | 7 | BCX23983.1 | 15 | QWU01215.1 | 4 |
| QWS64226.1 | 3 | QSJ35636.1 | 3 | QWY94400.1 | 3 | QRM91569.1 | 9 |
| QVJ47956.1 | 1 | QWF03959.1 | 7 | QSE25736.1 | 15 | QRX03618.1 | 12 |
| ORF10 | Cluster | ORF10 | Cluster | ORF10 | Cluster | ORF10 | Cluster |
|---|---|---|---|---|---|---|---|
| QSO40790.1 | 9 | QVH90751.1 | 1 | QVU00728.1 | 5 | QWU68360.1 | 15 |
| QVE25858.1 | 9 | QVX36355.1 | 2 | QUA79573.1 | 20 | QTO29824.1 | 12 |
| QVJ17584.1 | 9 | QWY95104.1 | 10 | QWT65692.1 | 12 | QWU51246.1 | 9 |
| QVO95200.1 | 9 | QWY18545.1 | 1 | QVW43945.1 | 9 | QVX69392.1 | 9 |
| QVE30392.1 | 5 | QVL15727.1 | 1 | QUM21266.1 | 10 | QWO21857.1 | 9 |
| QTC19517.1 | 1 | QTO07027.1 | 6 | QTX06599.1 | 10 | QTK02152.1 | 6 |
| QVJ37366.1 | 5 | BCX25240.1 | 6 | QVM14069.1 | 10 | QWY72735.1 | 19 |
| QWQ76823.1 | 20 | QWB65585.1 | 19 | QWY55420.1 | 9 | ||
| QVV08801.1 | 20 | QTW57386.1 | 6 | QVR42407.1 | 18 | ||
| QUI12106.1 | 1 | QWW38212.1 | 4 | QUA32182.1 | 2 | ||
| QWU53472.1 | 4 | QSE30156.1 | 4 | QRG41735.1 | 2 | ||
| QTS24551.1 | 1 | QUF17645.1 | 4 | QWT73590.1 | 4 | ||
| QNV50343.1 | 3 | QUF20717.1 | 6 | QWK62266.1 | 6 | ||
| QUB17908.1 | 2 | QWB83606.1 | 4 | QVM67662.1 | 13 | ||
| QVL64016.1 | 15 | QPF60767.1 | 4 | QQE14148.1 | 19 |
Based on amino acid compositions in each ORF10 sequence, unique ORF10 proteins were clustered into 20 different clusters. Cluster-1 contains the highest number (17) of ORF10 variants (which fall in the US), whereas clusters 13 and 17 contain only one ORF10 variant each. The ORF10 proteins BCY15724 (collected from Japan) and QVM67662 (USA: Florida) belong to cluster 13 and 17, respectively (Table 9 ).
Table 9.
Frequency of ORF10 variants in each cluster.
| Cluster number | Frequency | Cluster number | Frequency | Cluster number | Frequency |
|---|---|---|---|---|---|
| 1 | 17 | 5 | 7 | 19 | 4 |
| 20 | 14 | 11 | 7 | 12 | 3 |
| 16 | 13 | 14 | 7 | 8 | 2 |
| 4 | 11 | 15 | 7 | 18 | 2 |
| 9 | 10 | 7 | 6 | 13 | 1 |
| 3 | 9 | 10 | 6 | 17 | 1 |
| 6 | 9 | 2 | 4 |
Clearly, twenty different clusters with different frequencies of ORF10 variants showed wide variations of ORF10 sequences based on amino acid compositions.
4. Discussion
In this study, a total of 140 unique SARS-CoV-2 ORF10 sequences were observed among 202968 ORF10 sequences obtained from the NCBI database. Remarkably, most unique ORF10 variants were first reported in North America only (Table 1). Consequently, the highest number of point mutations and co-occurring mutations were found in North America. Continent-wise Africa had the highest percentage (1.27%) of unique variants of ORF10 sequences. Furthermore, it was observed that all amino acids of ORF10 except for residue 18, possessed single-missense mutations (Table 2). The unique ORF10 variants from Asia, Africa, and Oceania had several point-mutations at common residue positions (Table 2).
We noticed that in North America, ORF10 mutations only at the amino acid residue positions 11 (F11S, F11L) and 16 (L16P) were deleterious. A significant percentage of non-synonymous mutations, 53 among the total of 128 mutations were deleterious, which may alter the intensity of the interactome between ORF10 and host proteins. The 22 unique co-occurring mutations were observed in different geo-locations (Table 4). It was noticed that co-occurring mutations in ORF10 variants became an emerging trend, which varied slightly in 2020, and inevitably in future ORF10 variants with co-occurring mutations might be transmitted to other geo-locations while restriction on international travel are lifted. Furthermore, it was observed that the effects of co-occurring mutants are qualitatively similar to those of single mutations, as most variability is at the disorder predisposition of N- and C-terminal regions and a region centered at residue 25. However, in co-occurring mutants, scales of changes at the terminal regions are noticeably larger. Most of the co-occurring mutations increase the disorder predisposition of the N-terminal region of ORF10, whereas many such mutations decrease the disorder propensity of the region centered at residue 25. Consequently, these mutations may affect the secondary structure and associated functions of the ORF10 variants. In addition, amino acid residue positions of the secondary structures (especially the N-terminal α-helix regions) of several ORF10 variants changed, and therefore interaction of those ORF10 variants with the human protein CUL2ZY G11B complex are most likely altered.
5. Conclusions
Unique variations in SARS-CoV-2 ORF10 proteins are an emerging trend across different continents due to the appearance of various single point and co-occurring mutations. The highest percentages of unique ORF10 variants were found in Africa, though the highest frequency of unique ORF10 variants was found in North America. Among the possible explanations for this phenomenon are: a) (world largest number of the sequenced SARS-CoV-2 isolates originates from North America); b) the existence of the co-infection between the different SARS-CoV-2 strains in North America (which is almost non-existent in the other parts of the world, with the noticeable exception for England) [67]; c) highly diversified population, which is traditionally separated into ethnically, racially, and culturally different enclaves; d) implementation of very different approaches for controlling the COVID-19 spread by local administrations potentially generating conditions for the locally diversified evolution of SARS-CoV-2.
It was further observed that the growth rate of emerging non-synonymous mutations in ORF10 proteins is increasing non-linearly, which is certainly alarming with regards to stability or instability of emerging SARS-CoV-2 variants. Due to significant deleterious mutations, expression of the SARS-CoV-2 ORF10 proteins might get altered, a phenomenon, which will affect functional virus-host protein-protein interactions. Also, it was reported that the secondary structure α helix regions of the ORF10 variants was changed due to co-occurring mutations. Consequently, virulence/pathogenicity of SARS-CoV-2 may get influenced directly or indirectly and therefore, continuous surveillance of mutations and their associated effects is necessary.
CRediT authorship contribution statement
Sk. Sarif Hassan: Conceptualization; Formal analysis; Investigation; Methodology; Research design; Supervision; Validation; Visualization; Writing – original draft; Writing – review & editing
Kenneth Lundstrom: Data curation; Formal analysis; Investigation; Writing – original draft; Writing – review & editing
Angel Serrano-Aroca: Data curation; Formal analysis; Investigation; Writing – review & editing
Parise Adadi: Data curation; Formal analysis; Investigation; Writing – review & editing
Alaa A. A. Aljabali: Data curation; Formal analysis; Investigation; Writing – review & editing
Elrashdy M. Redwan: Data curation; Formal analysis; Investigation; Writing – review & editing
Amos Lal: Data curation; Formal analysis; Investigation; Writing – review & editing
Ramesh Kandimalla: Data curation; Formal analysis; Investigation; Writing – review & editing
Tarek Mohamed Abd El-Aziz: Data curation; Formal analysis; Investigation; Writing – review & editing
Pabitra Pal Choudhury: Data curation; Formal analysis; Investigation; Writing – review & editing
Gajendra Kumar Azad: Data curation; Formal analysis; Investigation; Writing – review & editing
Samendra P Sherchan: Data curation; Formal analysis; Investigation; Writing – review & editing
Gaurav Chauhan: Data curation; Formal analysis; Investigation; Writing – review & editing
Murtaza Tambuwala: Data curation; Formal analysis; Investigation; Writing – review & editing
Kazuo Takayama: Data curation; Formal analysis; Investigation; Writing – review & editing
Debmalya Barh: Data curation; Formal analysis; Investigation; Writing – review & editing
Giorgio Palu: Data curation; Formal analysis; Investigation; Writing – review & editing
Pallab Basu: Data curation; Formal analysis; Investigation; Writing – review & editing
Vladimir N. Uversky: Conceptualization; Formal analysis; Investigation; Methodology; Research design; Supervision; Validation; Visualization; Writing – original draft; Writing – review & editing
All authors read the final article and approved it for submission.
Acknowledgements
We gratefully acknowledge the authors from the originating laboratories responsible for obtaining the specimens and the Submitting laboratories where genetic sequence data were generated and shared via the NCBI and GISAID Initiatives, on which this research is based.
Data availability
Data will be made available on request.
References
- 1.Worldometer COVID live update. https://www.worldometers.info/coronavirus/
- 2.Johns-Hopkin-University Coronavirus Resource Center. https://coronavirus.jhu.edu/map.html
- 3.Laha S., Chakraborty J., Das S., Manna S.K., Biswas S., Chatterjee R. Characterizations of SARS-CoV-2 mutational profile, spike protein stability and viral transmission. Infect. Genet. Evol. 2020;85 doi: 10.1016/j.meegid.2020.104445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim J.-S., Jang J.-H., Kim J.-M., Chung Y.-S., Yoo C.-K., Han M.-G. Genome-wide identification and characterization of point mutations in the SARS-CoV-2 genome. Osong Public Health Res. Perspect. 2020;11(3):101. doi: 10.24171/j.phrp.2020.11.3.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weber S., Ramirez C.M., Weiser B., Burger H., Doerfler W. Sars-cov-2 worldwide replication drives rapid rise and selection of mutations across the viral genome: a time-course study–potential challenge for vaccines and therapies. EMBO Mol. Med. 2021;13(6) doi: 10.15252/emmm.202114062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krause P.R., Fleming T.R., Longini I.M., Peto R., Briand S., Heymann D.L., Beral V., Snape M.D., Rees H., Ropero A.-M., et al. SARS-CoV-2 variants and vaccines. N. Engl. J. Med. 2021;385(2):179–186. doi: 10.1056/NEJMsr2105280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Touret F., Luciani L., Baronti C., Cochin M., Driouich J.-S., Gilles M., Thirion L., Nougairede A., de Lamballerie X. Replicative fitness of a SARS-CoV-2 20i/501y. v1 variant from lineage b. 1.1. 7 in human reconstituted bronchial epithelium. Mbio. 2021;12(4) doi: 10.1128/mBio.00850-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Padhi A.K., Tripathi T. Can SARS-CoV-2 accumulate mutations in the s-protein to increase pathogenicity? ACS Pharmacol. Transl. Sci. 2020;3(5):1023–1026. doi: 10.1021/acsptsci.0c00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen A.T., Altschuler K., Zhan S.H., Chan Y.A., Deverman B.E. COVID-19 cg enables SARS-CoV-2 mutation and lineage tracking by locations and dates of interest. eLife. 2021;10 doi: 10.7554/eLife.63409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Padhi A.K., Shukla R., Saudagar P., Tripathi T. High-throughput rational design of the remdesivir binding site in the RDRP of SARS-CoV-2: implications for potential resistance. Iscience. 2021;24(1) doi: 10.1016/j.isci.2020.101992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desai S., Rashmi S., Rane A., Dharavath B., Sawant A., Dutt A. An integrated approach to determine the abundance, mutation rate and phylogeny of the SARS-CoV-2 genome. Brief. Bioinform. 2021;22(2):1065–1075. doi: 10.1093/bib/bbaa437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Padhi A.K., Dandapat J., Saudagar P., Uversky V.N., Tripathi T. Interface-based design of the favipiravir-binding site in SARS-CoV-2 RNA-dependent RNA polymerase reveals mutations conferring resistance to chain termination. FEBS Lett. 2021;595(18):2366–2382. doi: 10.1002/1873-3468.14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maurano M.T., Ramaswami S., Zappile P., Dimartino D., Boytard L., Ribeiro-dos Santos A.M., Vulpescu N.A., Westby G., Shen G., Feng X., et al. Sequencing identifies multiple early introductions of SARS-CoV-2 to the New York city region. Genome Res. 2020;30(12):1781–1788. doi: 10.1101/gr.266676.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bi C., Ramos-Mandujano G., Tian Y., Hala S., Xu J., Mfarrej S., Esteban C.R., Delicado E.N., Alofi F.S., Khogeer A., et al. Simultaneous detection and mutation surveillance of SARS-CoV-2 and multiple respiratory viruses by rapid field-deployable sequencing. Med. 2021;2(6):689–700.e4. doi: 10.1016/j.medj.2021.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Satarker S., Nampoothiri M. Structural proteins in severe acute respiratory syndrome coronavirus-2. Arch. Med. Res. 2020;51(6):482–491. doi: 10.1016/j.arcmed.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang D., Jiang A., Feng J., Li G., Guo D., Sajid M., Wu K., Zhang Q., Ponty Y., Will S., et al. The SARS-CoV-2 subgenome landscape and its novel regulatory features. Mol. Cell. 2021;81(10):2135–2147. doi: 10.1016/j.molcel.2021.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasibhatla S.M., Kinikar M., Limaye S., Kale M.M., Kulkarni-Kale U. Understanding evolution of SARS-CoV-2: a perspective from analysis of genetic diversity of RDRP gene. J. Med. Virol. 2020;92(10):1932–1937. doi: 10.1002/jmv.25909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michel C.J., Mayer C., Poch O., Thompson J.D. Characterization of accessory genes in coronavirus genomes. Virol. J. 2020;17(1):1–13. doi: 10.1186/s12985-020-01402-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silva S.J.R.d., Silva C.T.A.d., Mendes R.P.G., Pena L. Role of nonstructural proteins in the pathogenesis of SARS-CoV-2. 92 (9) 2020. pp. 1427–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584(7819):154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 21.Clark L.K., Green T.J., Petit C.M. Structure of nonstructural protein 1 from SARS-CoV-2. J. Virol. 2021;95(4):e02019–e02020. doi: 10.1128/JVI.02019-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shannon A., Le N.T.-T., Selisko B., Eydoux C., Alvarez K., Guillemot J.-C., Decroly E., Peersen O., Ferron F., Canard B. Remdesivir and SARS-CoV-2: structural requirements at both Nsp12 RDRP and Nsp14 exonuclease active-sites. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018;16(8) doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elena S.F., Sanjuán R. Adaptive value of high mutation rates of RNA viruses: separating causes from consequences. J. Virol. 2005;79(18):11555–11558. doi: 10.1128/JVI.79.18.11555-11558.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan L., Yang Y., Li M., Zhang Y., Zheng L., Ge J., Huang Y.C., Liu Z., Wang T., Gao S., et al. Coupling of n7-methyltransferase and 3`-5` exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading. Cell. 2021;184(13):3474–3485. doi: 10.1016/j.cell.2021.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J., Wang R., Wang M., Wei G.-W. Mutations strengthened SARS-CoV-2 infectivity. J. Mol. Biol. 2020;432(19):5212–5226. doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eskier D., Karaku¨lah G., Suner A., Oktay Y. RDRP mutations are associated with SARS-CoV-2 genome evolution. PeerJ. 2020;8:e9587. doi: 10.7717/peerj.9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta A.M., Chakrabarti J., Mandal S. Non-synonymous mutations of SARS-CoV-2 leads epitope loss and segregates its variants. Microbes Infect. 2020;22(10):598–607. doi: 10.1016/j.micinf.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hassan S.S., Choudhury P.P., Roy B. SARS-CoV-2 envelope protein: non-synonymous mutations and its consequences. Genomics. 2020;112(6):3890–3892. doi: 10.1016/j.ygeno.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Issa E., Merhi G., Panossian B., Salloum T., Tokajian S. SARS-CoV-2 and Orf3a: nonsynonymous mutations, functional domains, and viral pathogenesis. mSystems. 2020;5(3) doi: 10.1128/mSystems.00266-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seyran M., Pizzol D., Adadi P., El-Aziz T.M., Hassan S.S., Soares A., Kandimalla R., Lundstrom K., Tambuwala M., Aljabali A.A., et al. Questions concerning the proximal origin of SARS-CoV-2. J. Med. Virol. 2021;93(3):1204. doi: 10.1002/jmv.26478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narayanan K., Huang C., Makino S. Sars coronavirus accessory proteins. Virus Res. 2008;133(1):113–121. doi: 10.1016/j.virusres.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hassan S.S., Attrish D., Ghosh S., Choudhury P.P., Uversky V.N., Aljabali A.A., Lundstrom K., Uhal B.D., Rezaei N., Seyran M., et al. Notable sequence homology of the orf10 protein introspects the architecture of SARS-CoV-2. Int. J. Biol. Macromol. 2021;181:801–809. doi: 10.1016/j.ijbiomac.2021.03.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuster N.A. 2021. Characterization and structural prediction of the putative orf10 protein in SARS-CoV-2, bioRxiv. 2020–10. [Google Scholar]
- 35.Altincekic N., Korn S.M., Qureshi N.S., Dujardin M., Ninot-Pedrosa M., Abele R., Abi Saad M.J., Alfano C., Almeida F.C., Alshamleh I., et al. Large-scale recombinant production of the SARS-CoV-2 proteome for high-throughput and structural biology applications. Front. Mol. Biosci. 2021;8:89. doi: 10.3389/fmolb.2021.653148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A sars-cov-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mena E.L., Donahue C.J., Vaites L.P., Li J., Rona G., O’Leary C., Lignitto L., Miwatani-Minter B., Paulo J.A., Dhabaria A. Orf10–cullin-2–zyg11b complex is not required for SARS-CoV-2 infection. Proc. Natl. Acad. Sci. 2021;118(17) doi: 10.1073/pnas.2023157118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J., Guo M., Tian X., Wang X., Yang X., Wu P., Liu C., Xiao Z., Qu Y., Yin Y., et al. Virus-host interactome and proteomic survey reveal potential virulence factors influencing SARS-CoV-2 pathogenesis. Med. 2021;2(1):99–112. doi: 10.1016/j.medj.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pancer K., Milewska A., Owczarek K., Dabrowska A., Kowalski M., Łabaj P.P., Branicki W., Sanak M., Pyrc K. The SARS-CoV-2 Orf10 is not essential in vitro or in vivo in humans. PLoS Pathog. 2020;16(12) doi: 10.1371/journal.ppat.1008959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang D.-M., Lin F.-C., Tsai P.-H., Chien Y., Wang M.-L., Yang Y.-P., Chang T.-J. Pandemic analysis of infection and death correlated with genomic open reading frame 10 mutation in severe acute respiratory syndrome coronavirus 2 victims. J. Chin. Med. Assoc. 2021;84(5):478–484. doi: 10.1097/JCMA.0000000000000542. [DOI] [PubMed] [Google Scholar]
- 41.Cagliani R., Forni D., Clerici M., Sironi M. Coding potential and sequence conservation of SARS-CoV-2 and related animal viruses. Infect. Genet. Evol. 2020;83 doi: 10.1016/j.meegid.2020.104353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.K¨all L., Krogh A., Sonnhammer E.L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004;338((5):1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 43.Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R., Potter S.C., Finn R.D., et al. The embl-ebi search and sequence analysis tools apis in 2019. Nucleic Acids Res. 2019;47(W1):W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drozdetskiy A., Cole C., Procter J., Barton G.J. Jpred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015;43(W1):W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Obradovic Z., Peng K., Vucetic S., Radivojac P., Dunker A.K. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins. 2005;61(S7):176–182. doi: 10.1002/prot.20735. [DOI] [PubMed] [Google Scholar]
- 46.Meng F., Uversky V.N., Kurgan L. Comprehensive review of methods for prediction of intrinsic disorder and its molecular functions. Cell. Mol. Life Sci. 2017;74(17):3069–3090. doi: 10.1007/s00018-017-2555-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng Z.-L., Kurgan L. Comprehensive comparative assessment of in-silico predictors of disordered regions. Curr. Protein Pept. Sci. 2012;13(1):6–18. doi: 10.2174/138920312799277938. [DOI] [PubMed] [Google Scholar]
- 48.Fan X., Kurgan L. Accurate prediction of disorder in protein chains with a comprehensive and empirically designed consensus. J. Biomol. Struct. Dyn. 2014;32(3):448–464. doi: 10.1080/07391102.2013.775969. [DOI] [PubMed] [Google Scholar]
- 49.Pickett B.E., Sadat E.L., Zhang Y., Noronha J.M., Squires R.B., Hunt V., Liu M., Kumar S., Zaremba S., Gu Z., et al. Vipr: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012;40(D1):D593–D598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bendl J., Stourac J., Salanda O., Pavelka A., Wieben E.D., Zendulka J., Brezovsky J., Damborsky J. Predictsnp: robust and accurate consensus classifier for prediction of disease-related mutations, PLoS computational biology 10 (1) (2014) e1003440.E. Capriotti, P. Fariselli, Phd-snpg: a webserver and lightweight tool for scoring single nucleotide variants. Nucleic Acids Res. 2017;45(W1):W247–W252. doi: 10.1093/nar/gkx369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henson R., Cetto L. Encyclopedia of Genetics, Genomics, Proteomics And Bioinformatics. 2004. The Matlab bioinformatics toolbox. [Google Scholar]
- 52.Bendl J., Stourac J., Salanda O., Pavelka A., Wieben E.D., Zendulka J., Brezovsky J., Damborsky J. PredictSNP: robust and accurate consensus classifier for prediction of disease-related mutations. PLoS Comput. Biol. 2014;10(1) doi: 10.1371/journal.pcbi.1003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hassan S.S., Aljabali A.A., Panda P.K., Ghosh S., Attrish D., Choudhury P.P., Seyran M., Pizzol D., Adadi P., El-Aziz T.M.Abd. A unique view of SARS-CoV-2 through the lens of orf8 protein. Comput. Biol. Med. 2021;133 doi: 10.1016/j.compbiomed.2021.104380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hassan S.S., Choudhury P.P., Roy B., Jana S.S. Missense mutations in SARS-CoV-2 genomes from Indian patients. Genomics. 2020;112(6):4622–4627. doi: 10.1016/j.ygeno.2020.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mészáros B., Erdos G., Dosztányi Z. IUPred2a: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018;46(W1):W329–W337. doi: 10.1093/nar/gky384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2010;39(suppl 1):D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Romero P., Obradovic Z., Li X., Garner E.C., Brown C.J., Dunker A.K. Sequence complexity of disordered protein. Proteins. 2001;42(1):38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 58.Peng K., Vucetic S., Radivojac P., Brown C.J., Dunker A.K., Obradovic Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J. Bioinforma. Comput. Biol. 2005;3(01):35–60. doi: 10.1142/s0219720005000886. [DOI] [PubMed] [Google Scholar]
- 59.Peng K., Radivojac P., Vucetic S., Dunker A.K., Obradovic Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinforma. 2006;7(1):1–17. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xue B., Dunbrack R.L., Williams R.W., Dunker A.K., Uversky V.N. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta Protein Proteomics. 2010;1804(4):996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rajagopalan K., Mooney S.M., Parekh N., Getzenberg R.H., Kulkarni P. A majority of the cancer/testis antigens are intrinsically disordered proteins. J. Cell. Biochem. 2011;112(11):3256–3267. doi: 10.1002/jcb.23252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mészáros B., Simon I., Dosztányi Z. Prediction of protein binding regions in disordered proteins. PLoS Comput. Biol. 2009;5(5) doi: 10.1371/journal.pcbi.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oughtred R., Stark C., Breitkreutz B.-J., Rust J., Boucher L., Chang C., Kolas N., O’Donnell L., Leung G., McAdam R., et al. The biogrid interaction database: 2019 update. Nucleic Acids Res. 2019;47(D1):D529–D541. doi: 10.1093/nar/gky1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Iakoucheva L.M., Radivojac P., Brown C.J., O’Connor T.R., Sikes J.G., Obradovic Z., Dunker A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32(3):1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pejaver V., Hsu W.-L., Xin F., Dunker A.K., Uversky V.N., Radivojac P. The structural and functional signatures of proteins that undergo multiple events of post-translational modification. Protein Sci. 2014;23(8):1077–1093. doi: 10.1002/pro.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Darling A.L., Uversky V.N. Intrinsic disorder and posttranslational modifications: the darker side of the biological dark matter. Front. Genet. 2018;9:158. doi: 10.3389/fgene.2018.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haddad D., John S.E., Mohammad A., Hammad M.M., Hebbar P., Channanath A., Nizam R., Al-Qabandi S., Al Madhoun A., Alshukry A., Ali H., Thanaraj T.A., Al-Mulla F. SARS-CoV-2: possible recombination and emergence of potentially more virulent strains. PLoS One. 2021;16(5) doi: 10.1371/journal.pone.0251368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.