Abstract
Background:
Estimated glomerular filtration rate (eGFR), albuminuria and serum uric acid (SUA) are markers of kidney function that have been associated with cognitive ability. However, whether these associations are causal is unclear.
Methods:
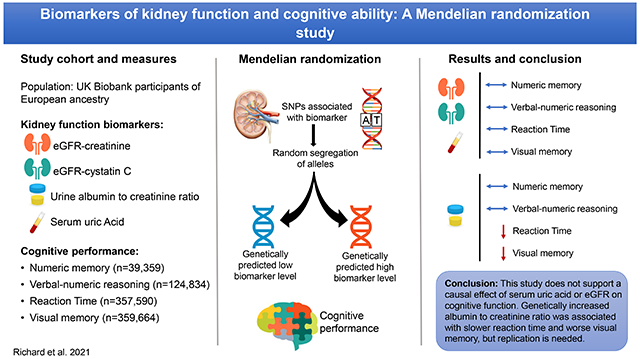
We performed one-sample Mendelian randomization (MR) to estimate the effects of kidney function markers on cognitive performance using data from the UK Biobank. Polygenic scores for SUA, urine albumin to creatinine ratio (ACR), estimated glomerular filtration rate based on serum creatinine (eGFRcre) and serum cystatin C (eGFRcys) were used as instrumental variables, and cognitive function outcomes included a test of verbal-numeric reasoning and reaction time.
Results:
We found no evidence of a causal effect of genetically determined SUA, eGFRcre or eGFRcys on cognitive function outcomes. There was no association between a polygenic score for ACR and verbal-numeric reasoning or numeric memory. However, there was suggestive evidence of a relationship between genetically increased ACR and slower reaction time and worse visual memory. ACR was no longer significantly associated with visual memory in analyses using an unweighted polygenic score and in analyses stratified by sex and age category. Pleiotropy adjusted estimates were directionally consistent with those of the principal analysis but overlapped with the null.
Conclusions:
This MR study does not support causal effects of SUA, eGFRcre or eGFRcys on cognitive performance. Genetically increased ACR was associated with slower processing speed and visual memory, but results need confirmation in independent samples.
Keywords: albuminuria, glomerular filtration rate, uric acid, cognition, Mendelian randomization
Graphical Abstract
1. Introduction
Dementia imposes significant societal and economic burdens. It is a leading cause of disability and was estimated to cost the equivalent to 1.1% of global gross domestic product in 2015 (1). As no effective therapeutic treatment is currently available, identification and mitigation of modifiable risk factors remain of central importance. Chronic kidney disease (CKD) has emerged as a possible risk indicator for cognitive impairment (2,3). Individuals living with chronic kidney disease experience higher rates of cognitive impairment and dementia compared to healthy adults (4). Observational studies of associations between biomarkers of kidney function and cognition suggest that this risk may extend to individuals with only mild kidney impairment (5,6). Most common of these biomarkers, estimated glomerular filtration rate based on serum creatinine (eGFRcre) has been associated with cognitive performance (4,6–8) but studies have been conflicting (2). Though less studied, cystatin C based GFR estimates (GFRcys) have shown stronger associations with cognitive performance compared to creatinine-based measurements (9,10). An increased urine albumin to creatinine ratio (ACR) or albuminuria, which is highly indicative of vascular dysfunction has been associated with higher odds of cognitive impairment and dementia (2,11). However, whether this reflects a causal effect of albuminuria independent of concomitant cardiovascular disease is unclear. Higher serum uric acid (SUA) levels are correlated with diabetes, cardiovascular and kidney disease (12), but associations with cognitive ability are inconsistent (13–15). Somewhat paradoxically, case-control and cross-sectional studies have reported lower levels of SUA in individuals with Alzheimer’s disease compared to those with normal cognition (15–17). This finding has been attributed to a potential neuroprotective role of SUA through its antioxidant properties (18).
However, observational studies are susceptible to confounding and reverse causation and are therefore not appropriate for inferring causation. For example, prior associations may have been confounded by environmental factors such as socio-economic status or comorbid disease. Reverse causation whereby the state of dementia leads to alterations in biomarker concentrations may also explain some of the observed inconsistencies in past studies, particularly with respect to the relationship between serum uric acid and Alzheimer’s disease as individuals with Alzheimer’s may change their eating habits (19).
The Mendelian randomization approach attempts to provide evidence of a causal association using genetic variants as instrumental variables for the exposure of interest. Analogous to randomization in clinical trials, the random assortment of alleles during meiosis allows confounding factors to be distributed evenly across genotypes. Furthermore, genotype at conception confers a lifelong increase or decrease in the exposure of interest minimizing the effects of reverse causation. The validity of the instrumental variable in Mendelian randomization relies on three key assumptions: 1. the genetic variant is strongly associated with the exposure, 2. the variant is not associated with confounders of exposure-outcome association, and 3. the variant-outcome association is explained only through the effect of the exposure of interest. The second and third assumptions can be violated in the presence of pleiotropy, where a genetic variant is associated with factors on a different causal pathway (20). However, sensitivity analyses have been developed to address pleiotropic effects.
Individual-level data from the UK Biobank (UKBB) and summary data from previous genome-wide association studies (GWAS) were used to construct polygenic scores for multiple markers of kidney function including SUA, eGFRcre, eGFRcys or ACR. We then performed a one-sample MR using these scores as instrumental variables to test for causal associations between each kidney function biomarker and cognitive performance.
2. Materials and Methods
2.1. Study population
The UKBB is a prospective cohort that enrolled 502,617 participants aged 40-73 years from across the United Kingdom between 2006 and 2010. Details of enrollment procedures have been previously described (21). At the baseline assessment, participants completed a detailed, computerized questionnaire including a wide range of information pertaining to lifestyle and health characteristics. Participants completed a battery of cognitive function tests via touchscreen interface at this time. Blood and urine samples for the full cohort were collected and stored for biochemical tests and genotyping. Ethical approval for data collection was received from the North-West Multi-centre Research Ethics Committee and the research was carried out in accordance with the Declaration of Helsinki of the World Medical Association and approved by the University of California San Diego Institutional Review Board. Written informed consent was obtained for all participants.
2.2. Genotyping
The UKBB study was genotyped on the Affymetrix (now part of ThermoFisher Scientific) UK BiLEVE Axiom array (n=49,950 participants) or the similar UKBB Axiom array (n=438,427). To facilitate use of the UKBB resource by the research community, genotyping, quality control (QC) and genotype imputation were performed centrally by the primary UKBB investigators (22). Genotype imputation is a statistical technique that leverages directly genotyped variants and a reference panel to infer ungenotyped variants. Prior to imputation, genetic data from two arrays was combined and a QC procedure performed. Post quality control, genetic data is available for 488,377 subjects on 805,426 genetic markers and 92,693,895 imputed variants. We carried out the following additional quality control and filtering steps. Individuals with the following characteristics were excluded: extreme heterozygosity or missingness (n=968), individuals with sex chromosome aneuploidy (n=651), individuals whose reported sex did not match genetically inferred sex (n=186), and individuals with high levels of cryptic relatedness (n=73). Principal components were then calculated for the remaining 486,387 participants using 1000 Genomes as the reference population (23). We used the “aberrant” clustering package in R (24) with a lambda parameter of 8.2 to determine the European ancestry cluster. Subjects with self-report of non-British or non-European ancestry included in European ancestry cluster were excluded, resulting in, 454,488 participants with European ancestry. To avoid inflation in test statistics due to inclusion of related individuals, we used a custom script that implements a greedy algorithm to determine the unrelated subset. Relatedness was first determined by UKBB using identity by State (IBS). The algorithm sequentially breaks related pairs to retain only unrelated individuals while preferentially maximizing the number of individuals with a user defined characteristic. In this study we chose to maximize those with available verbal-numeric reasoning scores. We excluded those with approximately second degree or closer relatedness (pi-hat =0.0625, n= 69,378 removed). After additionally excluding those who had withdrawn consent at the time of this study, pregnant women (n=119), individuals with probable type 1 diabetes (n=1670) and participants missing data on kidney function exposures or cognitive test scores there remained 39,359, 124,834, 357,590 and 359,664 participants for analyses of numeric memory, verbal-numeric reasoning, reaction time, and visual memory, respectively.
2.3. Kidney function biomarkers
At the initial assessment (2006-2010), blood and spot urine samples were collected and analyzed at a centralized laboratory. Sampling, handling, and quality control of biochemical measures have been described in detail previously (25). Briefly, serum creatinine, urine creatinine and urine albumin were measured on a Beckman Coulter AU5800 instrument. An enzymatic, IDMS-traceable method was used to measure serum and urine creatinine. Urine albumin was quantified using an immune-turbidimetric method (Randox laboratories) with a lower limit of detection of 6.7 mg/L. Measurements below the lower limit of detection were set to 6.7 mg/L as done previously (26). Serum cystatin C was measured using an Immuno-turbidimetric assay on a Siemens ADVIA 1800 instrument. Estimated GFR was calculated using creatinine (eGFRcre) or cystatin C (eGFRcys) by the CKD-EPI equation as described previously (27,28). SUA was measured by uricase PAP analysis on a Beckman Coulter AU5800.
2.4. Cognitive function
Cognitive function was assessed using self-administered, computerized tests designed specifically for the UKBB (29). The verbal-numeric reasoning, reaction time, visual memory test, and numeric memory test were used in this analysis and are described below:
Verbal-numeric reasoning:
The verbal-numeric reasoning test, originally labelled the ‘fluid intelligence’ test, (Field ID 20016) included 13 logic/reasoning-type questions. The score was the number of questions answered correctly within a two-minute time limit. The Cronbach alpha coefficient for this test has been previously reported elsewhere as 0.62 (30). This test was added part-way through the initial assessment period and therefore was not administered to all participants.
Reaction time:
The reaction time test is similar to the card game “Snap”. Participants were shown a series of card pairs with symbols on them and were instructed to press a large red button as quickly as possible when the cards matched (Field ID 20023). The score was the mean time, in milliseconds (ms), to press the button across all trials with a matching pair.
Visual memory:
The “pairs-matching” test was used to assess episodic visual memory in the UKBB (Field ID 399). Participants were briefly shown the positions of six card pairs and were then asked to match them from memory in as few attempts as possible. The score on this test was the number of errors made. Pairs matching scores were log(x+1) transformed for analyses as done in prior studies (31,32).
Numeric memory:
To assess numeric short-term memory (Field ID 4282) participants were asked to recall a 2-digit number that was shown on the screen. The number became 1-digit longer each time they remembered correctly up to a maximum of 12 digits. The score for analysis was the maximum digit length recalled correctly. This test was removed from the baseline assessment before recruitment concluded and therefore is available for a subset of participants.
2.5. Covariates
Years of education were estimated by mapping each of the educational qualifications reported by UKBB participants to categories defined in the 1997 International Standard Classification of Education (ISCED) and imputing the number of years of schooling as described by Okbay et al. (33). Hypertension was defined as systolic blood pressure ≥140 mmHg or diastolic blood pressure ≥90 mmHg, self-report of a past hypertension diagnosis or use of antihypertensive medications.
2.6. Statistical analysis
2.6.1. Polygenic scores and MR analysis
Single nucleotide polymorphisms (SNPs) included in the SUA, eGFRcre and eGFRcys polygenic scores were identified from GWAS meta-analyses of 288,649, 567,460 and 32,861 participants of European ancestry, respectively (26,34,35). We used European ancestry specific summary statistics from a meta-analysis (n=547,361) carried out by Teumer et al. to derive the polygenic score for ACR (55). All summary statistics were downloaded from the data page maintained by the CKDGen consortium (ckdgen.imbi.uni-freiburg.de/). It should be noted that UKBB participants made up the largest proportion of individuals included in the Teumer et al. meta-analysis which may to contribute to the winner’s curse phenomenon (36). However, prior GWAS were underpowered and would be unlikely to yield a polygenic score with adequate instrument strength (37). For each biomarker we constructed two polygenic scores based on SNPs that passed a p-value threshold p<5 ×10−8 or p<1 ×10−5 in prior GWAS. SNPs were pruned based on the 1000 Genomes data with an R2 <0.1 and a 500kb clumping window to find the SNP with the with the lowest p-value for each clump. The variance explained by the polygenic scores was calculated as the adjusted R2 from the association of each score with the biomarker adjusted for age, sex and the first 10 principal components (PCs) of population structure minus the adjusted R2 from the regression with age, sex and the 10 PCs only.
We performed mendelian randomization analyses through two-stage least squares (2SLS) regression using the ivreg command from the AER package in R (38). In 2SLS the exposure of interest is regressed on the polygenic score, and the outcome is regressed on the predicted values of the exposure and the residuals from the first regression. All 2SLS models were adjusted for age, sex, and the first 10 PCs. ACR was log-transformed for normality before analysis. The instrument strength of each polygenic score was assessed using the F-statistic from the first regression where an F-statistic less than 10 suggests a weak instrument.
2.6.2. Sensitivity analysis
Modification by sex or age was assessed by performing 2SLS stratified by sex and by the median age in the UKBB (<58 years vs 58 years or older). Potential confounding or mediation by hypertension status was assessed by adjusting for this factor in 2SLS models. Similar adjustment for years of schooling was carried out to address confounding due to differences in education. We repeated 2SLS analyses using an unweighted allele score for ACR to minimize bias from the use of internally derived weights (39). Although, 2SLS is the standard method for MR in one-sample settings it does not address the problem of pleiotropy which violates the assumption that the genetic variant-outcome association is explained only through the effect of the exposure of interest. Alternatively, methods to address pleiotropy have been developed for two-sample MR where the effect estimates for the SNP-exposure and SNP-outcome are gleaned from independent study populations (40). Using the MendelianRandomization package in R (41), we applied two methods, MRegger (42) and weighted-median regression (43) to account for the potential effects of pleiotropic SNPs in the ACR polygenic score. First, the β-estimates for the associations between each SNP with ACR and with cognitive performance on reaction time and visual memory tasks were obtained, and inverse-variance weighted fixed effects meta-analysis (IVW) was used to derive the MR estimate to approximate the 2SLS estimate (44). The MR-Egger method was used to estimate the causal effect as the slope from the weighted regression and the average pleiotropic effect as the intercept. If the intercept from the MR-Egger analysis is not equal to zero, this indicates directional pleiotropy. Weighted median regression was then used to estimate the causal effect assuming at least 50% of the SNPs in the polygenic score are valid and that there is not directional pleiotropy.
3. Results
Characteristics of the study population are shown in Table 1. The mean age was 56.7 years and 54% were female. On average, participants had a mean verbal-numeric reasoning score of 6.18 (standard deviation (SD)=2.11), a mean reaction time of 555ms (SD=113), a mean visual memory score of 4.10 errors (SD=3.27), and a mean numeric memory score of 6.74 digits (SD=1.32). Mean eGFR was slightly lower when estimated using cystatin C rather than creatinine (mean (SD)=88.31 (15.95) ml/min for eGFRcys and 90.66 (13.10) ml/min for eGFRcre). SUA was higher in men compared to women (mean (SD)=5.96 (1.20) mg/dl vs 4.54 (1.10) mg/dl), but the median ACR was lower in men (median (IQR)=0.86 (0.83) mg/mmol in men and 1.37 (1.33) mg/mmol in women).
Table 1.
Characteristics of the study population overall and according to sex: the UK Biobank
| All Participants | Female | Male | |
|---|---|---|---|
|
| |||
| n=359,664 | n=193,807 | n=165,857 | |
| Age (years) | 56.71 (8.01) | 56.52 (7.91) | 56.94 (8.11) |
| Smoking status | |||
| Current | 36,934 (10.3) | 17,028 (8.8) | 19,906 (12.0) |
| Never | 194,805 (54.2) | 113,933 (58.8) | 80,872 (48.8) |
| Past | 127,925 (35.6) | 62,846 (32.4) | 65,079 (39.2) |
| Some university education | 203,753 (56.7) | 105,864 (54.6) | 97,889 (59.0) |
| Alcohol drinking status | |||
| Current | 336,449 (93.5) | 178,672 (92.2) | 157,777 (95.1) |
| Never | 10,995 (3.1) | 8,236 (4.2) | 2,759 (1.7) |
| Past | 12,220 (3.4) | 6,899 (3.6) | 5,321 (3.2) |
| Body mass index (kg/m2) | 27.34 (4.74) | 26.95 (5.12) | 27.81 (4.21) |
| LDL-c (mmol/L) | 3.57 (0.87) | 3.64 (0.87) | 3.49 (0.86) |
| Triglycerides (mmol/L) | 1.75 (1.02) | 1.55 (0.85) | 1.98 (1.14) |
| Hypertension | 198,572 (55.2) | 93,941 (48.5) | 104,631 (63.1) |
| Type II diabetes | 17,622 (4.9) | 6,400 (3.3) | 11,222 (6.8) |
| Coronary artery disease | 12,803 (3.6) | 2,744 (1.4) | 10,059 (6.1) |
| History of stroke | 5,842 (1.6) | 2,411 (1.2) | 3,431 (2.1) |
| Heart failure | 1,011 (0.3) | 258 (0.1) | 753 (0.5) |
| Cholesterol-lowering medication | 60,496 (16.8) | 23,420 (12.1) | 37,076 (22.4) |
| Antihypertensive medication | 72,545 (20.2) | 32,659 (16.9) | 39,886 (24.0) |
| ACR (mg/mmol) | 1.11 (1 .17) | 1.37 (1.33) | 0.86 (0.83) |
| SUA (mg/dl) | 5.20 (1.35) | 4.54 (1.10) | 5.96 (1.20) |
| GFRcre (ml/min) | 90.66 (13.10) | 90.79 (13.11) | 90.51 (13.09) |
| GFRcys (ml/min) | 88.31 (15.95) | 88.68 (15.71) | 87.87 (16.22) |
| Verbal-numeric reasoning score | 6.18 (2.11) | 6.06 (2.03) | 6.31 (2.18) |
| Reaction time (ms) | 555.19 (113.12) | 563.19 (113.53) | 545.83 (111.92) |
| Visual memory (errors) | 4.10 (3.27) | 4.11 (3.19) | 4.09 (3.36) |
| Numeric memory score | 6.74 (1.32) | 6.63 (1.31) | 6.86 (1.32) |
Abbreviations: ACR, albumin-to-creatinine ratio; eGFRcre, creatinine-based estimated glomerular filtration rate; eGFRcys, cystatin C-based estimated glomerular filtration rate; SUA, serum uric acid; ACR shown as median (IQR). Other values shown as n (%) for categorical variables and mean (SD) for continuous variables. All characteristics are significantly different by sex except eGFRcre (p-value=0.53) and visual memory (p-value=0.27).
3.1. MR analysis
F-statistics for the eight polygenic scores ranged from 552 to 10,004 which suggests that they were not weak instruments. The number of SNPs included in each polygenic score, the variance explained by the score and the corresponding effect estimates from the 2SLS regression are shown in Table 2. We detected no evidence for a significant causal effect of SUA, eGFRcre, or eGFRcys on performance on any of the cognitive tasks (all p-values ≥0.06). There was no apparent effect of ACR on verbal-numeric reasoning or numeric memory score, however increased ACR as predicted by the 293-SNP score was significantly associated with slower reaction-time scores (β (95% confidence interval [CI])) for 1 SD logACR=4.93 (1.60 to 8.26) ms, p=0.004). The association was slightly weaker using the 76-SNP score (β (95%CI) =4.82 (0.95 to 8.68), p=0.01). Genetically increased ACR was also significantly associated with a higher percent of visual memory errors (β (95% CI)= 2.06(0.07 to 4.09)%, p=0.04), but only using the 293-SNP score.
Table 2.
Results from two-stage least squares MR analyses for the association of kidney function biomarkers with cognitive performance
| P-value cutoff | SNPs (n) | Variance explained | Verbal-numeric reasoning score |
Reaction time (ms) |
Visual memory (% errors) |
Numeric memory score |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β | 95%CI | p | β | 95%CI | p | β | 95%CI | p | β | 95%CI | p | |||
| SUA (mg/dl) | ||||||||||||||
| 1×10−5 | 693 | 5.90% | −0.02 | −0.06, 0.02 | 0.26 | 0.39 | −0.72, 1.51 | 0.48 | −0.65 | −1.51, 0.22 | 0.14 | −0.03 | −0.07, 0.01 | 0.13 |
| 5×10−8 | 297 | 5.40% | −0.03 | −0.07, 0.01 | 0.13 | 0.24 | −0.91, 1.39 | 0.68 | −0.51 | −1.41, 0.4 | 0.27 | −0.04 | −0.08, 0.01 | 0.10 |
| eGFRcre (10ml/min) | ||||||||||||||
| 1×10−5 | 1120 | 4.50% | 0.02 | −0.02, 0.06 | 0.25 | −0.65 | −1.94, 0.64 | 0.31 | 0.71 | −0.04, 1.47 | 0.06 | 0.01 | −0.04, 0.05 | 0.74 |
| 5×10−8 | 453 | 4.00% | 0.03 | −0.01, 0.07 | 0.15 | −0.58 | −1.95, 0.79 | 0.40 | 0.74 | −0.06, 1.54 | 0.07 | −0.03 | −0.07, 0.02 | 0.30 |
| eGFRcys (10ml/min) | ||||||||||||||
| 1×10−5 | 16 | 3.40% | 0.007 | −0.01, 0.24 | 0.72 | −0.47 | −1.68, 0.74 | 0.43 | 0.11 | −0.6, 0.82 | 0.76 | −0.03 | −0.08, 0.01 | 0.13 |
| 5×10−8 | 4 | 3.00% | 0.006 | −0.01, 0.25 | 0.77 | −0.21 | −1.87, 1.45 | 0.79 | 0.26 | −0.44, 0.96 | 0.46 | −0.02 | −0.06, 0.02 | 0.29 |
| Log ACR (1 SD) | ||||||||||||||
| 1×10−5 | 293 | 1.10% | −0.03 | −0.17, 0.12 | 0.72 | 4.93 | 1.56, 8.30 | 0.004 | 2.06 | 0.07, 4.09 | 0.04 | −0.08 | −0.21, 0.06 | 0.26 |
| 5×10−8 | 76 | 0.60% | −0.03 | −0.17, 0.11 | 0.69 | 4.82 | 0.92, 8.72 | 0.01 | 2.64 | −0.06, 5.42 | 0.055 | −0.01 | −0.20, 0.18 | 0.89 |
Abbreviations: ACR, albumin-to-creatinine ratio; CI, confidence interval; eGFRcre, creatinine-based estimated glomerular filtration rate; eGFRcys, cystatin C-based estimated glomerular filtration rate; SUA, serum uric acid 1 SD logACR = 0.74 log(mg/mmol). For visual memory test scores, percent errors are the exponentiated beta coefficients (exp(β)−1) ×100).
3.2. Sensitivity analyses
We found similar null associations between SUA, eGFRcre, and eGFRcys with cognitive performance when 2SLS analyses was stratified by sex or age. ACR was not associated with verbal-numeric reasoning or numeric memory in stratified MR analyses. Associations between genetically increased ACR and slower reaction time were significant in men and women (β (95% CI)= 5.69 (0.42 to 10.96), p=0.03 and 4.27 (0.06 to 8.48), p=0.048, respectively). The association between ACR and reaction time was slightly stronger in individuals younger than 58 years compared to those who were older (β (95% CI)=6.02 (1.37 to 10.67), p=0.01 vs 4.46 (0.05 to 8.87), p=0.047, respectively). Associations between the 293-SNP ACR score and visual memory where no longer significant when analyses were stratified into subgroups. However, similar to the trends observed for the stratified associations between ACR and reaction time, visual memory effect estimates for a one standard deviation increase in log ACR were slightly larger in men compared women (β (95% CI)= 2.32 (−0.26 to 4.96), p=0.08 and 1.78 (−1.29 to 4.94), p=0.26, respectively) and in individuals younger than 58 years compared to those who were older (β (95% CI)= 2.44 (−0.46 to 5.42), p=0.10 vs 1.72 (−1.06 to 4.51), p=0.22, respectively). For 2SLS analyses, there was minimal difference in effect estimates when hypertension or education were added to the models with the exception that the association between ACR and reaction time was slightly attenuated after accounting for hypertension status (β (95% CI)= 4.59 (1.19-7.99), p=0.008).
The results of 2SLS, IVW, MREgger and weighted median regressions for the association between the 293-SNP ACR polygenic score and reaction time and visual memory are shown in Figures 1A and 1B. Genetically increased ACR estimated using the unweighted polygenic score was significantly associated with slower reaction time scores (β (95% CI)= 5.84 (2.08 to 9.60), p=0.001), but was no longer associated with visual memory (β (95% CI)=1.71 (−0.45 to 3.87), p=0.11). The results of the analyses that control for pleiotropy (MREgger and weighted median regression) were directionally consistent with that of the main analysis, but not statistically significant. However, the precision of the estimates was much lower for these methods as they demand high statistical power. The MREgger regression did not indicate the presence of directional pleiotropy (β (95% CI)=0.01 (−0.73 to 0.95), p=0.79 for reaction time intercept and 0.005 (−0.09 to 0.10), p=0.61 for visual memory intercept).
Figure 1.
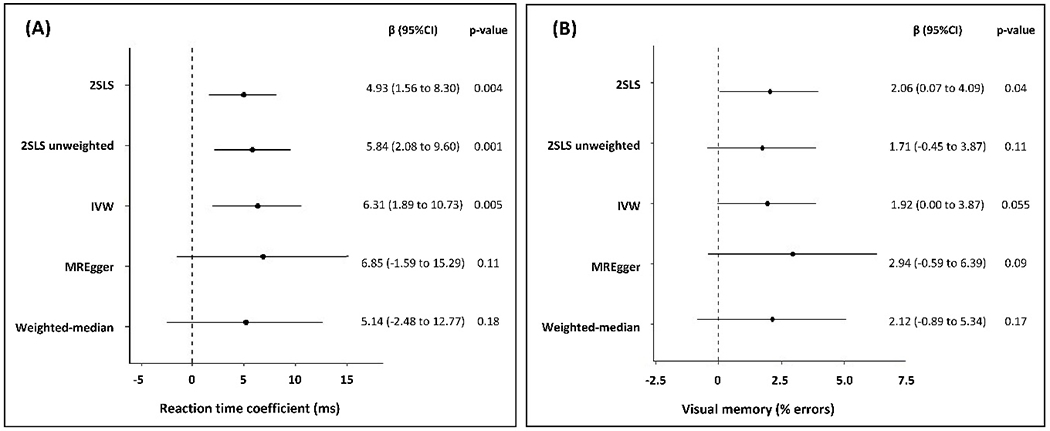
Mendelian randomization estimates for the effect of 1-standard deviation (SD) increase in genetically determined logACR on (A) reaction time (ms) and (B) visual memory (% errors).
Abbreviations: CI, confidence interval; IVW, inverse-variance weighted. 2SLS, Two-stage least-squares
4. Discussion
In this study, we used MR analyses to investigate the potential causal associations of four markers of kidney function and cognitive ability in a large population-based cohort of European descent. Genetically increased ACR was associated with reaction time, a measure of processing speed. Our study did not provide evidence to support a causal effect of genetically determined serum uric acid, eGFRcre or eGFRcys levels on cognitive performance despite the associations observed in observational studies (2,3,14,15).
In a recent two-sample MR using summary GWAS data, Efstathiadou et al. found no association between genetically increased SUA and global cognitive function in participants of the UKBB and the Cognitive Genomics (COGENT) consortium (n= 110,347) (45). We found a similar null association in a one-sample setting which is not affected by heterogeneity between the populations used to obtain the SNP-exposure and SNP-outcome association statistics. Furthermore, we extend these results to a tests of reaction time and memory. This conflicts with past studies that found lower levels of SUA in Alzheimer’s disease cases versus controls (15,16,46). However, the results of this study along with those of prospective studies that show an inverse association between SUA and cognitive performance (13,47) suggest that it is unlikely that increasing SUA would benefit cognition. It is important to note that the 2SLS regression assumes a linear relationship between the exposure of interest and the outcome and therefore may not capture threshold effects. We therefore could not fully characterize the potential effects of the hyperuricemic state on cognitive ability.
To our knowledge, this is the first study to examine the association between eGFR and cognitive ability using the MR approach. Our finding of a null association between genetically determined eGFR and cognitive performance is consistent with a recent meta-analysis of observational studies by Deckers et al. that showed no significant differences in cognitive impairment according to eGFRcre (2). The authors described substantial study heterogeneity which could reflect differential distributions or treatment of confounding factors between populations. Taken together, the results of this study and that of Deckers et al. suggest that past significant associations between eGFRcre and cognitive function may have been affected by residual confounding. However, because eGFR often has a nonlinear association with outcomes, we may have not been able to detect any threshold effects.
In contrast with observational studies that support a positive association between eGFRcys and cognitive performance (9,48), genetically determined eGFRcys did not predict cognitive ability in this study. A large proportion (85%) of the total variance in eGFRcys that was explained by the polygenic score is attributable to one SNP (rs1158167) which is found near the cystatin C precursor gene family (CST3, CST4, CST9) and explains 2.7% of the variance of serum cystatin C in the UKBB. It is likely that this SNP reflects cystatin C expression rather than renal filtration. Although this suggests cystatin C concentrations do not causally affect cognition, a better understanding of the genetic underpinnings of eGFRcys independent of cystatin C expression may be necessary to draw further conclusions.
Our finding that genetically determined ACR was associated with reaction time and memory is consistent with prior observational studies (9,49–51). While the mechanism for this association is not known, it may be mediated through increased blood pressure and cardiovascular risk. Using bidirectional MR, Haas et al. suggested that high blood pressure contributes to albuminuria which in turn leads to further increased blood pressure in a feed-forward loop (52). Indeed, we found that the association between ACR and reaction time was slightly weaker when hypertension status was accounted for lending support to a potential mediating role of blood pressure. Cognition may also be affected due to the consequences of kidney damage including anemia, hyperparathyroidism, acidosis, hyperhomocysteinemia, inflammation, and exposure to uremic toxins accumulation (53).
Key strengths of this study were the large sample size and access to individual level data for one-sample MR allowing for stratification by sex and age group. This method also avoids the problem of sample heterogeneity that affects two-sample MR. In addition, the extensive biochemistry measurements of the UKBB data allowed for the investigation of multiple measures of kidney function. Some limitations of our study should also be noted. First, the ACR polygenic score was constructed using weights from GWAS that included UKBB participants. This can exacerbate weak instrument bias which can overestimate the exposure-outcome association in one-sample MR studies (54). However, we also found a significant positive association with reaction time when an unweighted polygenic score was used. ACR was not as robustly associated with visual memory, therefore these results should be interpreted cautiously but do provide motivation for replication in other large independent cohorts. Furthermore, our analysis may not be able to detect nonlinear associations. Future studies should consider using polygenic scores for binary kidney function traits in this context. However, results from adequately powered GWAS will be required. Our analysis was restricted to participants of European ancestry, so our results may not be generalizable to other ethnic groups. In addition, the cognitive tests in the UKBB were developed to be administered on a large scale and without supervision and may therefore not be highly sensitive to cognitive differences. However, the tests used showed substantial correlation with previously validated tests in an independent sample of individuals (31). Finally, we did not examine causal associations with specific cognitive diagnoses. However, at the time of this study there are likely insufficient numbers of participants with dementia (n=1888) and Alzheimer’s disease (n=707) to provide adequate power for MR analyses. With full integration of primary care data, case detection will likely increase as will the actual prevalence of these “hard outcomes” as the cohort ages allowing for the examination of these associations in future work.
5. Conclusions
In conclusion, our MR analyses do not support a causal effect of SUA, eGFRcre or eGFRcys on cognitive function. A polygenic score for ACR was associated with reaction time, a measure of processing speed, and less robustly with a task of visual memory. Replication in independent cohorts is needed.
Highlights.
Markers of kidney function have been associated with cognitive performance.
Mendelian randomization was used to assess causality using data from the UK Biobank.
We found no causal effect of uric acid or glomerular filtration rate on cognition.
Genetically predicted urine albumin was associated with slower reaction time.
Acknowledgements
We are grateful to the UK Biobank participants for taking part in the study and to the UK Biobank for providing access to the resource.
Funding
This work was supported by the National Heart, Lung, and Blood Institute grants R00HL122515. Computing resources were supported by National Institute of General Medical Sciences P41-GM103504.
Abbreviations:
- ACR
albumin-to-creatinine ratio
- eGFRcre
creatinine-based estimated glomerular filtration rate
- eGFRcys
cystatin C-based estimated glomerular filtration rate
- MR
Mendelian randomization
- SUA
serum uric acid
- 2SLS
two-stage least squares
Footnotes
Declarations of interest: none
Ethics statement
Ethical approval for UKBB data collection was received from the North-West Multi-centre Research Ethics Committee and the research was carried out in accordance with the Declaration of Helsinki of the World Medical Association. Written informed consent was obtained for all participants. This analysis of UKBB data was conducted in compliance with the University of California San Diego Institutional Review Board.
Competing interests
The authors declare that they have no other competing interests.
Data availability
The UK Biobank resource is available to bona fide researchers for health-related research in the public interest. All researchers who wish to access the research resource must register with UK Biobank by completing the registration form in the Access Management System (AMS – https://bbams.ndph.ox.ac.uk/ams/).
References
- 1.World Heath Organization Dementia Fact Sheet. Available from: https://www.who.int/news-room/fact-sheets/detail/dementia (accessed 11 Jul 2020).
- 2.Deckers K, Camerino I, van Boxtel MPJ, Verhey FRJ, Irving K, Brayne C, et al. Dementia risk in renal dysfunction. Neurology. 2017. January 10;88(2):198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Etgen T Kidney disease as a determinant of cognitive decline and dementia. Alzheimers Res Ther. 2015;7(1):29–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Etgen T, Chonchol M, Frstl H, Sander D. Chronic kidney disease and cognitive impairment: A systematic review and meta-analysis. Am J Nephrol. 2012;35(5):474–82. [DOI] [PubMed] [Google Scholar]
- 5.Hailpern SM, Melamed ML, Cohen HW, Hostetter TH. Moderate Chronic Kidney Disease and Cognitive Function in Adults 20 to 59 Years of Age: Third National Health and Nutrition Examination Survey (NHANES III). J Am Soc Nephrol. 2007. June;18(7):2205–13. [DOI] [PubMed] [Google Scholar]
- 6.Seliger SL, Wendell CR, Waldstein SR, Ferrucci L, Zonderman AB. Renal Function and Long-Term Decline in Cognitive Function: The Baltimore Longitudinal Study of Aging. Am J Nephrol. 2015;41(4–5):305–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zammit AR, Katz MJ, Lai JY, Zimmerman ME, Bitzer M, Lipton RB. Association between Renal Function and Cognitive Ability Domains in the Einstein Aging Study: A Cross-Sectional Analysis. J Gerontol-Ser Biol Sci Med Sci. 2015;70(6):764–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elias MF, Dore GA, Davey A. Kidney disease and cognitive function. Contrib Nephrol. 2013;179:42–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martens RJH, Kooman JP, Stehouwer CDA, Dagnelie PC, van der Kallen CJH, Koster A, et al. Estimated GFR, Albuminuria, and Cognitive Performance: The Maastricht Study. Am J Kidney Dis. 2017. February 1;69(2):179–91. [DOI] [PubMed] [Google Scholar]
- 10.Wei Y, Wei YK, Zhu J. Early markers of kidney dysfunction and cognitive impairment among older adults. J Neurol Sci. 2017;375:209–14. [DOI] [PubMed] [Google Scholar]
- 11.Georgakis MK, Dimitriou NG, Karalexi MA, Mihas C, Nasothimiou EG, Tousoulis D, et al. Albuminuria in Association with Cognitive Function and Dementia: A Systematic Review and Meta-Analysis. J Am Geriatr Soc. 2017;65(6):1190–8. [DOI] [PubMed] [Google Scholar]
- 12.Feig DI, Kang D-H, Johnson RJ. Uric Acid and Cardiovascular Risk. N Engl J Med. 2008. October 23;359(17):1811–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Latourte A, Soumaré A, Bardin T, Perez-ruiz F, Debette S, Richette P. Uric acid and incident dementia over 12 years of follow-up: a population-based cohort study. 2017;1–8. [DOI] [PubMed] [Google Scholar]
- 14.Euser SM, Hofman A, Westendorp RGJ, Breteler MMB. Serum uric acid and cognitive function and dementia. Brain J Neurol. 2009. February;132(Pt 2):377–82. [DOI] [PubMed] [Google Scholar]
- 15.Al-Khateeb E, Althaher A, Al-Khateeb M, Al-Musawi H, Azzouqah O, Al-Shweiki S, et al. Relation between uric acid and Alzheimer’s disease in elderly Jordanians. J Alzheimers Dis. 2015;44(3):859–65. [DOI] [PubMed] [Google Scholar]
- 16.Du N, Xu D, Hou X, Song X, Liu C, Chen Y, et al. Inverse Association Between Serum Uric Acid Levels and Alzheimer’s Disease Risk. Mol Neurobiol. 2016;53(4):2594–9. [DOI] [PubMed] [Google Scholar]
- 17.Khan AA, Quinn TJ, Hewitt J, Fan Y, Dawson J. Serum uric acid level and association with cognitive impairment and dementia: systematic review and meta-analysis. Age Dordr Neth. 2016;38(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Giorgi A, Fabbian F, Pala M, Tiseo R, Parisi C, Misurati E, et al. Uric acid: Friend or foe? Uric acid and cognitive function “gout kills more wise men than simple.” Eur Rev Med Pharmacol Sci. 2015;19(4):640–6. [PubMed] [Google Scholar]
- 19.Fostinelli S, De Amicis R, Leone A, Giustizieri V, Binetti G, Bertoli S, et al. Eating Behavior in Aging and Dementia: The Need for a Comprehensive Assessment. Front Nutr. 2020;7:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawlor DA, Harbord RM, Sterne JAC, Timpson N, Smith GD. Mendelian randomization: Using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27(8):1133–63. [DOI] [PubMed] [Google Scholar]
- 21.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLOS Med. 2015. March;12(3):e1001779–e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature, 562 (7726) (2018. October), pp. 203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Auton A, Abecasis GR, Altshuler DM, Durbin RM, Abecasis GR, Bentley DR, et al. A global reference for human genetic variation. Nature. 2015. October;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bellenguez C, Strange A, Freeman C, Donnelly P, Spencer CCA. A robust clustering algorithm for identifying problematic samples in genome-wide association studies. Bioinformatics. 2012. January 1;28(1):134–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elliott P, Peakman TC, UK Biobank. The UK Biobank sample handling and storage protocol for the collection, processing and archiving of human blood and urine. Int J Epidemiol. 2008. April;37(2):234–44. [DOI] [PubMed] [Google Scholar]
- 26.German Chronic Kidney Disease Study, Lifelines Cohort Study, V. A. Million Veteran Program, Tin A, Marten J, Halperin Kuhns VL, et al. Target genes, variants, tissues and transcriptional pathways influencing human serum urate levels. Nat Genet. 2019. October;51(10):1459–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levey AS, Stevens LA, Schmid CH, Zhang Y (Lucy), Castro AF, Feldman HI, et al. A New Equation to Estimate Glomerular Filtration Rate. Ann Intern Med. 2009. May 5;150(9):604–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inker LA, Schmid CH, Tighiouart H, Eckfeldt JH, Feldman HI, Greene T, et al. Estimating Glomerular Filtration Rate from Serum Creatinine and Cystatin C. N Engl J Med. 2012. July 5;367(1):20–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cullen B, Nicholl BI, Mackay DF, Martin D, Ul-Haq Z, McIntosh A, et al. Cognitive function and lifetime features of depression and bipolar disorder in a large population sample: Cross-sectional study of 143,828 UK Biobank participants. Eur Psychiatry. 2015. November;30(8):950–8. [DOI] [PubMed] [Google Scholar]
- 30.Hagenaars SP, Harris SE, Davies G, Hill WD, Liewald DCM, Ritchie SJ, et al. Shared genetic aetiology between cognitive functions and physical and mental health in UK Biobank (N =112 151) and 24 GWAS consortia . Mol Psychiatry. 2016. November;21(11):1624–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fawns-Ritchie C, Deary IJ. Reliability and validity of the UK Biobank cognitive tests. PLOS ONE. 2020. April 20;15(4):e0231627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cornelis MC, Wang Y, Holland T, Agarwal P, Weintraub S, Morris MC. Age and cognitive decline in the UK Biobank. PloS One. 2019;14(3):e0213948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okbay A, Beauchamp JP, Fontana MA, Lee JJ, Pers TH, Rietveld CA, et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016. May 11;533(7604):539–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wuttke M, Li Y, Li M, Sieber KB, Feitosa MF, Gorski M, et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. 2019;42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li M, Li Y, Weeks O, Mijatovic V, Teumer A, Huffman JE, et al. SOS2 and ACP1 Loci Identified through Large-Scale Exome Chip Analysis Regulate Kidney Development and Function. J Am Soc Nephrol. 2017. March;28(3):981–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burgess S, Thompson SG. Use of allele scores as instrumental variables for Mendelian randomization. Int J Epidemiol. 2013. August;42(4):1134–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teumer A, Tin A, Sorice R, Gorski M, Yeo NC, Chu AY, et al. Genome-wide association studies identify genetic loci associated with Albuminuria in diabetes. Diabetes. 2016;65(3):803–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleiber C, Zeileis A (2019). AER: Applied Econometrics with R. R package version 1.2–7. Available at: https://CRAN.R-project.org/package=AER. [Google Scholar]
- 39.Burgess S, Thompson SG. Use of allele scores as instrumental variables for Mendelian randomization. Int J Epidemiol. 2013. August 1;42(4):1134–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burgess S, Davey Smith G, Davies NM, Dudbridge F, Gill D, Glymour MM, et al. Guidelines for performing Mendelian randomization investigations. Wellcome Open Res. 2020. April 28;4:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017. 01;46(6):1734–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015. April;44(2):512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016. May;40(4):304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013. November;37(7):658–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anthoula Efstathiadou, Dipender Gill, Frances McGrane, Terence Quinn, Jesse Dawson. Genetically Determined Uric Acid and the Risk of Cardiovascular and Neurovascular Diseases: A Mendelian Randomization Study of Outcomes Investigated in Randomized Trials. J Am Heart Assoc. 2019. September 3;8(17):e012738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim T, Pae C, Yoon S, Jang W, Lee NJ, Kim J, et al. Decreased plasma antioxidants in patients with Alzheimer ‘ s disease. 2006;(June 2005):344–8. [DOI] [PubMed] [Google Scholar]
- 47.Alam AB, Wu A, Power MC, West NA, Alonso A. Associations of serum uric acid with incident dementia and cognitive decline in the ARIC-NCS cohort. J Neurol Sci. 2020. July 15;414:116866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheppach JB, Coresh J, Wu A, Gottesman RF, Mosley TH, Knopman DS, et al. Albuminuria and Estimated GFR as Risk Factors for Dementia in Midlife and Older Age: Findings From the ARIC Study. Am J Kidney Dis. 2020. December 1;76(6):775–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ekblad LL, Toppala S, Johansson JK, Koskinen S, Sundvall J, Rinne JO, et al. Albuminuria and Microalbuminuria as Predictors of Cognitive Performance in a General Population: An 11-Year Follow-Up Study. J Alzheimers Dis. 2018. February 20;62(2):635–48. [DOI] [PubMed] [Google Scholar]
- 50.Sacre JW, Magliano DJ, Zimmet PZ, Polkinghorne KR, Chadban SJ, Anstey KJ, et al. Associations of Chronic Kidney Disease Markers with Cognitive Function: A 12-Year Follow-Up Study. Anstey K, Peters R, editors. J Alzheimers Dis. 2019. August 13;70(s1):S19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiménez-Balado J, Riba-Llena I, Pizarro J, Palasí A, Penalba A, Ramírez C, et al. Kidney function changes and their relation with the progression of cerebral small vessel disease and cognitive decline. J Neurol Sci. 2020. February;409:116635. [DOI] [PubMed] [Google Scholar]
- 52.Haas ME, Aragam KG, Emdin CA, Bick AG, Hemani G, Davey Smith G, et al. Genetic Association of Albuminuria with Cardiometabolic Disease and Blood Pressure. Am J Hum Genet. 2018. October 4;103(4):461–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bugnicourt J-M, Godefroy O, Chillon J-M, Choukroun G, Massy ZA. Cognitive Disorders and Dementia in CKD: The Neglected Kidney-Brain Axis. J Am Soc Nephrol. 2013;24(3):353–63. [DOI] [PubMed] [Google Scholar]
- 54.Zheng J, Baird D, Borges M-C, Bowden J, Hemani G, Haycock P, et al. Recent Developments in Mendelian Randomization Studies. Curr Epidemiol Rep. 2017;4(4):330–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teumer A, Li Y, Ghasemi S, Prins B, Wuttke M, Hermle T, et al. Genome-wide association meta-analyses and fine-mapping elucidate pathways influencing albuminuria. Nature communications, 10 (1) (2019), Article 4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The UK Biobank resource is available to bona fide researchers for health-related research in the public interest. All researchers who wish to access the research resource must register with UK Biobank by completing the registration form in the Access Management System (AMS – https://bbams.ndph.ox.ac.uk/ams/).


