Abstract
Background: Every year, nearly 170,000 people die from bladder cancer worldwide. A major problem after transurethral resection of bladder tumor is that 40–80% of the tumors recur. Ferroptosis is a type of regulatory necrosis mediated by iron-catalyzed, excessive oxidation of polyunsaturated fatty acids. Increasing the sensitivity of tumor cells to ferroptosis is a potential treatment option for cancer. Establishing a diagnostic and prognostic model based on ferroptosis-related genes may provide guidance for the precise treatment of bladder cancer.
Methods: We downloaded mRNA data in Bladder Cancer from The Cancer Genome Atlas and analyzed differentially expressed genes based on and extract ferroptosis-related genes. We identified relevant pathways and annotate the functions of ferroptosis-related DEGs using Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis and Gene Ontology functions. On the website of Search Tool for Retrieving Interacting Genes database (STRING), we downloaded the protein-protein interactions of DEGs, which were drawn by the Cytoscape software. Then the Cox regression analysis were performed so that the prognostic value of ferroptosis-related genes and survival time are combined to identify survival- and ferroptosis-related genes and establish a prognostic formula. Survival analysis and receiver operating characteristic curvevalidation were then performed. Risk curves and nomograms were generated for both groups to predict survival. Finally, RT-qPCR was applied to analyze gene expression.
Results: Eight ferroptosis-related genes with prognostic value (ISCU, NFE2L2, MAFG, ZEB1, VDAC2, TXNIP, SCD, and JDP2) were identified. With clinical data, we established a prognostic model to provide promising diagnostic and prognostic information of bladder cancer based on the eight ferroptosis-related genes. RT-qPCR revealed the genes that were differentially expressed between normal and cancer tissues.
Conclusion: This study found that the ferroptosis-related genes is associated with bladder cancer, which may serve as new target for the treatment of bladder cancer.
Keywords: bladder cancer, ferroptosis, prognostic model construction, survival analysis, mRNA
Introduction
The mortality of Bladder cancer ranks 13th among cancers (Antoni et al., 2017). To make a diagnosis and give treatment of bladder cancer, prognosis and risk assessment are important. The TNM staging system has been widely applied to predict the prognosis in cancer patients (Kluth et al., 2015). In recent years, the diagnostic or prognostic prediction model established by combining molecular markers and staging systems has improved the diagnosis and treatment ability of bladder cancer (Chen et al., 2020a; Yang et al., 2020). However, most molecular markers are derived from differential gene analysis, and the function of genes has not been further studied. Therefore, it is necessary to explore new functional biomarkers.
Ferroptosis is a new type of programmed cell death that is iron-dependent (Latunde-Dada, 2017; Wang et al., 2020b; Riegman et al., 2020). Ferroptosis is associated with many important physiological processes such as mitochondrial function, lipid metabolism and oxidative stress, so that induction of ferroptosis play a pivotal role in inhibiting the progression of tumor (Torti and Torti, 2020). There are increasing indications that ferroptosis can produce a marked effect in tumor treatment, and several studies have focused on cancer therapies that rely on ferroptosis (Conrad et al., 2020; Wu et al., 2020; Zuo et al., 2020). Wang et al. identified branched-chain amino acid aminotransferase 2 (BCAT2) as a novel inhibitor of ferroptosis using CRISPR/Cas9-based assays in hepatoma cells (Wang et al., 2020a). Li et al. (2019) constructed CaO2 and Fe3O4 nanoparticles and utilized cascade reaction-mediated ferroptosis to synergize with immune regulation in cancer treatment (Li and Rong, 2020). In a related bladder cancer study, Chen et al. (2020b) found that the quinazolinyl-arylurea derivatives can induce ferroptosis based on the structural modification of sorafenib. Therefore, genes associated with ferroptosis may predict the survival of patients with bladder cancer and new personalized therapeutic targets.
We collected mRNA expression data and corresponding clinical data from BLCA-TCGA and analyzed the biological functions of the ferroptosis-related DEGs. We then constructed a prognostic 8-gene signature associated with ferroptosis, which was validated by survival analysis and risk assessment. In addition, we performed biological functions analysis to investigate the potential mechanism of ferroptosis-related genes. These results will help to afford new insights for the prognosis and treatment of bladder cancer.
Materials and Methods
Data Source
The mRNA data and clinical data of bladder cancer patients were obtained from the Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). Clinical features of bladder urothelial carcinoma patients (BLCA) from the TCGA database are shown in Table 1. The ferroptosis-related genes were then downloaded on the FerrDb database (http://www.zhounan.org/ferrdb)(Zhou and Bao, 2020). A list of related genes is provided in the Supplementary Material (Supplementary Table S1). Combined with the transcriptome sequencing map of bladder cancer, ferroptosis-related genes of bladder cancer were obtained. All data were publicly available online.
TABLE 1.
Clinical features of BLCA patients (n = 412) from the TCGA database.
| Variables | Patients, n (%) |
|---|---|
| Sex | |
| Male | 108 (26.21%) |
| Female | 304 (37.79%) |
| Age (year) | |
| ≤65 | 162 (39.32%) |
| >65 | 250 (60.68%) |
| TNM stage | |
| I | 2 (00.49%) |
| II | 131 (31.80%) |
| III | 141 (34.22%) |
| IV | 136 (33.01%) |
| unknown | 2 (00.49%) |
| T stage | |
| T0 | 1 (00.24%) |
| T1 | 3 (00.73%) |
| T2 | 120 (29.13%) |
| T3 | 196 (47.57%) |
| T4 | 59 (14.32%) |
| TX | 1 (00.24%) |
| unknown | 32 (7.77%) |
| N stage | |
| N0 | 239 (58.01%) |
| N1 | 47 (11.41%) |
| N2 | 76 (18.45%) |
| N3 | 8 (1.94%) |
| NX | 36 (8.74%) |
| unknown | 6 (1.46%) |
| M stage | |
| M0 | 196 (47.57%) |
| M1 | 11 (2.67%) |
| MX | 202 (49.03%) |
| Unknown | 3 (0.73%) |
| Grade | |
| High Grade | 388 (94.017%) |
| Low Grade | 21 (5.10%) |
| Unknown | 3 (0.73%) |
Data Processing of Ferroptosis-Related Differentially Expressed Genes
Using the R software, we analyzed ferroptosis-related differentially expressed genes (DEGs) to identify differences between tumor and sample groups. The difference is significant when | log2Foldchange | > 1 and false discovery rate (FDR) < 0.05.
Functional and Pathway Enrichment
Kyoto Encyclopedia of Genes and Genomes (KEGG) is a pathway-related database that systematically analyzes gene function and links genomic information and functional information. Gene Ontology (GO) is an international standardized classification system for gene functions that divides the functions of genes into three parts: cellular component (CC), molecular function (MF), and biological process (BP) (Yu et al., 2012). Bar graphs and bubbles were drawn by GO and KEGG analysis of ferroptosis-related DEGs. The difference is significant when p values < 0.05.
Protein-Protein Interaction Network
We analyzed the PPI network of ferroptosis-related DEGs in STRING (https://string-db.org/). Subsequently, the PPI network was analyzed and visualized by Cytoscape 3.7.1, and the most relevant subnetwork module was obtained using Cytoscape plug-in MCODE.
Identification of Prognostic Ferroptosis-Related Differentially Expressed Genes
We performed Kaplan-Meier survival analysis to assess the effect of ferroptosis-related genes on overall survival (OS) in bladder cancer patients. Then we conducted Cox regression. Prognostic ferroptosis-related genes (p < 0.05) were screened out. We also retained the ferroptosis-related DEGs if | log2Foldchange | >1 and FDR <0.05. The intersection of the two results in prognostic ferroptosis-related DEGs in BLCA.
Construction and Analysis of Prognostic Models
We analyzed the prognostic value and survival time of ferroptosis-related DEGs using Cox regression analysis to identify ferroptosis-related genes associated with survival and generated forest maps. Univariate Cox regression analysis was conducted to identify the OS-related core genes, and genes with p < 0.1 were utilized for the subsequent multivariate Cox regression analysis. Thereafter, samples from TCGA were randomly divided into two groups in a 1:1 ratio to construct the best prognostic model, including a training group (n = 197) and a testing group (n = 196). We identified eight survival-related ferroptosis-related genes were by this model and obtained the correlation coefficients of each gene. , where “Exp” represents the gene expression, and “coef” represents the coefficient score from multivariate Cox analysis. The risk score of each patient was calculated based on this model. Then, we analyzed the survival analysis, receiver operating characteristic curve (ROC) and risk curves of each group. Furthermore, by univariate and multivariate analysis, a nomogram was generated to provide patient survival information.
Preparation of Bladder Cancer Clinical Tissue Samples and Adjacent Tissue Samples
Throughout the study, we obtained informed consent from each patient for the collection and analysis of tissue samples, which was approved by the Union Hospital, Tongji Medical College, and Huazhong University of Science and Technology ethics review boards. We immediately frozen and stored the tissue at −80°C after extracting it.
Total Ribo Nucleic Acid Extraction, Reverse Transcription, and Real-Time Quantitative-Polymerace Chain Reaction Analysis
We extracted total RNA from tissue samples using Trizol reagent and used the nanodrop2000 to assess RNA integrity. We used reverse transcription reagents to react with extracted RNA to produce cDNA complementary to the single strand of RNA, which was then quantified in real time using the SYBR Green PCR kit. Each gene’s cycle threshold (CT) was recorded. The relative mean value of each gene’s expression was calculated, and the 2-∆∆CT method was used to determine whether the differences between groups were statistically significant, and the mean value was used as the final experimental result for replicate wells. All procedures were carried out in accordance with the manufacturer’s instructions.
Statistical Analysis
The predictive significance of ferroptosis associated genes and other clinical characteristics was determined by using univariate and multivariate Cox regression. The effect of this model was assessed by using Kaplan Meier curves and the log rank test method to validate the difference in survival time between different risk groups based on the ferroptosis prognostic model. The effectiveness of this ferroptosis prognostic model to predict the prognosis of bladder cancer patients was assessed using a receiver operating characteristic curve (ROC). p values less than 0.05 were deemed statistically significant in two-sided statistical testing. The graphing software graphpad prism version 8.0.1 and the R language version 4.0.2 were used in this investigation. The R packages mainly used in this paper include limma, pheatmap, survival, iGraph, reshape2, glmnet, clusterprofiler, org. hs.eg.db, enrichplot, ggplot2.
Results
Identification of Differentially Expressed Ferroptosis-Related Genes
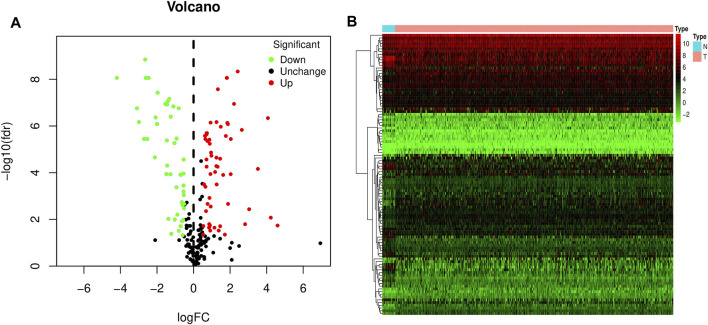
The mRNA expression levels of all ferroptosis-related genes were compared between 19 control bladder tissues and 412 bladder cancer tissues in the TCGA-BLCA dataset. 103 ferroptosis-related DEGs were obtained, including 57 upregulated ferroptosis-related genes and 46 downregulated ferroptosis-related genes, and plotted volcano and heat maps (Figure 1).
FIGURE 1.
Volcano and heat map of differentially expressed ferroptosis-related genes in bladder cancer. (A) Volcano map. (B) Heat map. Red dots represent upregulated genes, and green dots represent downregulated genes.
Functional Enrichment Analyses of Differentially Expressed Ferroptosis-Related Genes
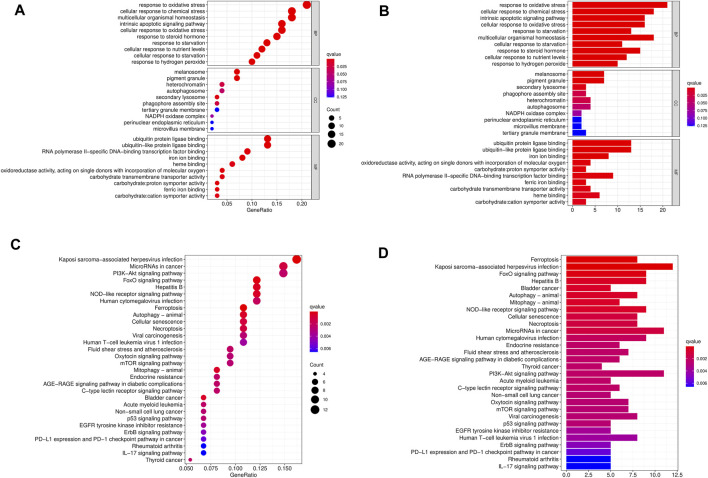
To study the biological functions of 103 differentially expressed ferroptosis-related genes, GO function and KEGG pathway enrichment were performed, and bar plots and bubbles were plotted. As shown in Figure 2, the top 10 most relevant genes were selected. GO results indicate that differentially expressed ferroptosis-related genes are involved in some important biological processes, such as response to stimuli, maintenance of cell homeostasis, cell autophagy, apoptosis, and a variety of iron-related functions. KEGG pathway enrichment analysis indicated that 103 genes were mainly involved in PD-L1/PD-1 pathway in cancer, multiple signaling pathways related to cell differentiation, tumorigenesis and development, and autophagy-related pathways.
FIGURE 2.
GO and KEGG enrichment pathways of differentially expressed ferroptosis-related genes. The dot graph (A) and bar graph (B) of GO and the dot graph (C) and bar graph (D) of KEGG are shown. The size of the dot indicates the count, and the color indicates the Q value.
Protein-Protein Interaction Network Construction
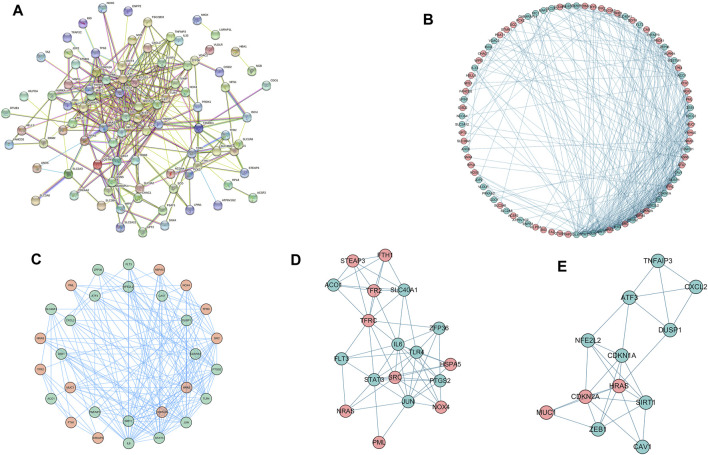
We used STRING tools to predict the protein interactions among the differentially expressed ferroptosis-related genes. We found 101 nodes and 348 edges in the PPI network. Low degree values were associated with smaller node sizes, while lower co-expression values were associated with smaller edge sizes. Thereafter, a network diagram of 102 genes was drawn using the Cytoscape software, as shown in Figure 3A. In addition, two key subnets were extracted and visualized using the MCODE plug-in, as shown in Figure 3B. Two most significant MCODE modules visualization and two most significant MCODE components form the PPI network as shown in Figures 3C-E. The results of KEGG pathway enrichment and GO function on the genes of the two subnetworks were shown in Table 2 and Table 3.
FIGURE 3.
Differentially expressed ferroptosis-related genes were used to construct protein-protein interaction networks and subnetworks. (A) PPI interaction network map obtained from the STRING database. (B) Genes of the interacting PPI network visualized by Cytoscape. Red nodes represent upregulated genes, while blue nodes represent downregulated genes. (C) Two most significant MCODE modules visualization. (D,E) Two most significant MCODE components form the PPI network.
TABLE 2.
The GO function enrichment analysis of the two most significant MCODE components.
| Ontology | ID | Description | P value | Adjusted P Value | Count |
|---|---|---|---|---|---|
| Subnetwork 1 | |||||
| BP | GO:0055072 | iron ion homeostasis | 9.84E-11 | 1.59E-07 | 6 |
| BP | GO:0055076 | transition metal ion homeostasis | 2.02E-09 | 1.63E-06 | 6 |
| BP | GO:0006879 | cellular iron ion homeostasis | 3.08E-09 | 1.66E-06 | 5 |
| CC | GO:0005788 | endoplasmic reticulum lumen | 0.000152 | 0.015066 | 4 |
| CC | GO:0010008 | endosome membrane | 0.000803 | 0.039742 | 4 |
| MF | GO:0035258 | steroid hormone receptor binding | 0.000105 | 0.003937 | 3 |
| MF | GO:0035259 | glucocorticoid receptor binding | 0.000116 | 0.003937 | 2 |
| MF | GO:0061629 | RNA polymerase II-specific DNA-binding transcription factor binding | 0.000151 | 0.003937 | 4 |
| Subnetwork 2 | |||||
| BP | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 1.76E-10 | 2.35E-07 | 6 |
| BP | GO:0001933 | negative regulation of protein phosphorylation | 2.31E-09 | 1.41E-06 | 7 |
| BP | GO:0006469 | negative regulation of protein kinase activity | 3.24E-09 | 1.41E-06 | 6 |
| CC | GO:0000792 | heterochromatin | 0.000944 | 0.038697 | 2 |
| MF | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity | 2.77E-05 | 0.002883 | 2 |
| MF | GO:0051019 | mitogen-activated protein kinase binding | 0.000136 | 0.00706 | 2 |
| MF | GO:0030291 | protein serine/threonine kinase inhibitor activity | 0.000207 | 0.007167 | 2 |
TABLE 3.
The KEGG function enrichment analysis of the two most significant MCODE components.
| ID | Description | P value | Adjusted P Value | Count |
|---|---|---|---|---|
| Subnetwork 1 | ||||
| hsa05167 | Kaposi sarcoma-associated herpesvirus infection | 6.34E-08 | 9.9E-06 | 7 |
| hsa05161 | Hepatitis B | 6.12E-07 | 4.78E-05 | 6 |
| hsa04216 | Ferroptosis | 1.3E-06 | 5.49E-05 | 4 |
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | 1.45E-06 | 5.49E-05 | 5 |
| hsa04625 | C-type lectin receptor signaling pathway | 1.76E-06 | 5.49E-05 | 5 |
| Subnetwork 2 | ||||
| hsa05219 | Bladder cancer | 1.42E-05 | 0.000968 | 3 |
| hsa05206 | MicroRNAs in cancer | 1.74E-05 | 0.000968 | 5 |
| hsa04218 | Cellular senescence | 2.57E-05 | 0.000968 | 4 |
Construction and Analysis of Prognostic Signature
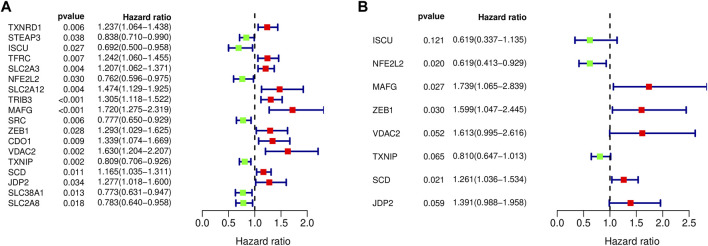
103 differentially expressed ferroptosis-related genes interacting with survival time were analyzed to identify the prognostic value of by Cox regression; 18 survival-related genes were obtained, and forest maps were generated (Figure 4A). The samples from TCGA were randomly divided into a training set (n = 197) and test set (n = 196) according to a 1:1 ratio. Eight survival-related genes were screened out through the training set and are used to construct the best prognostic signature, and the correlation coefficients for each gene were obtained. These eight genes were: iron-sulfur cluster assembly enzymes (ISCU), nuclear factor, erythroid 2 like 2 (NFE2L2), MAF bZIP transcription factor G (MAFG), zinc finger E-box-binding protein 1 (ZEB1), voltage-dependent anion channel 2 (VDAC2), thioredoxin-interacting protein (TXNIP), stearoyl-CoA desaturase (SCD), and Jun dimerization protein 2 (JDP2) (Figure 4B). Among them, ISCU, NFE2L2, and TXNIP are classified as low-risk genes, while ISCU, MAFG, ZEB1, VDAC2, SCD, JDP2 are classified as high-risk genes. In addition, details of the prognostic signature are listed in Table 4.
FIGURE 4.
Forest maps of differentially expressed ferroptosis-related genes. (A) Eighteen prognostically related ferroptosis-related genes are shown in the forest map, with red denoting high-risk genes and green denoting low-risk genes. (B) Eight ferroptosis-related genes were obtained by constructing the prognostic model shown in the forest map.
TABLE 4.
Detailed information on ferroptosis-related genes for constructing the prognostic signature.
| Id | ENSG_ID | Chromosome | Coef | HR |
|---|---|---|---|---|
| ISCU | ENSG00000136003 | 12q23.3 | −0.48 | 0.619 (0.337−1.135) |
| NFE2L2 | ENSG00000116044 | 2q31.2 | −0.48 | 0.619 (0.413−0.929) |
| MAFG | ENSG00000197063 | 17q25.3 | 0.55 | 1.739 (1.065−2.839) |
| ZEB1 | ENSG00000148516 | 10p11.22 | 0.47 | 1.599 (1.047−2.445) |
| VDAC2 | ENSG00000165637 | 10q22.2 | 0.48 | 1.613 (0.995−2.616) |
| TXNIP | ENSG00000265972 | 1q21.1 | −0.21 | 0.810 (0.647−1.013) |
| SCD | ENSG00000099194 | 10q24.31 | 0.23 | 1.261 (1.036−1.534) |
| JDP2 | ENSG00000140044 | 14q24.3 | 0.33 | 1.391 (0.988−1.958) |
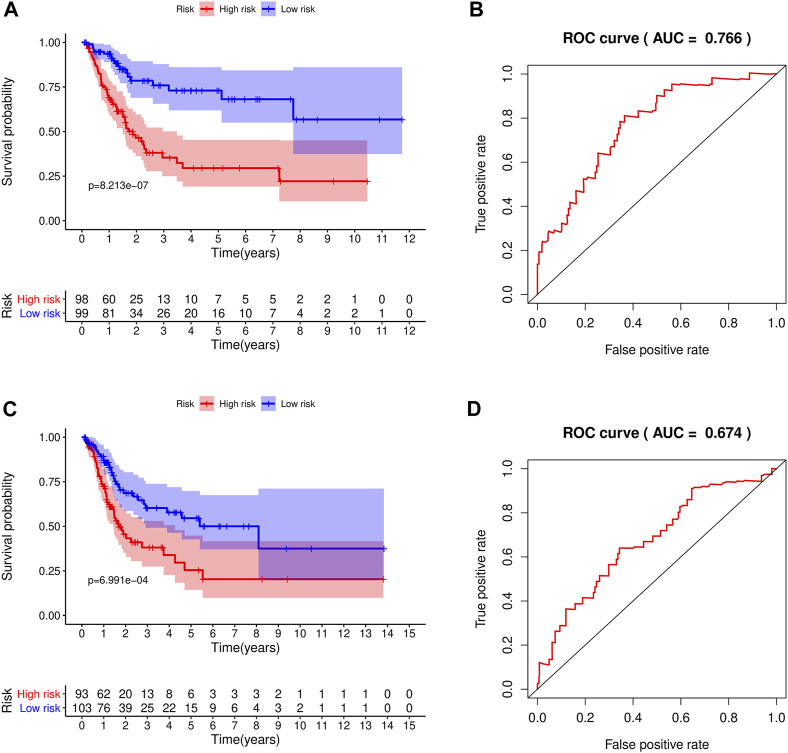
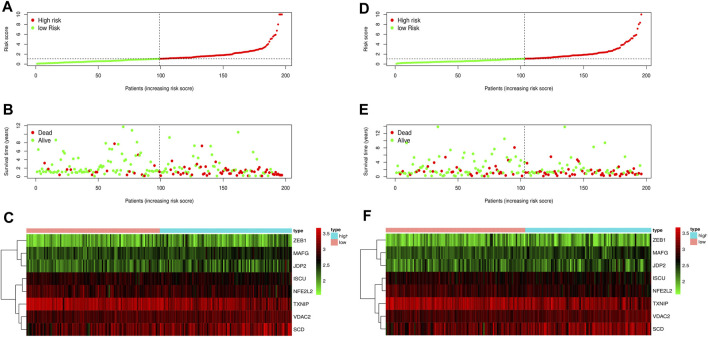
We obtained a risk score for each patient and obtained the median of the risk scores for all patients according to the risk formula. The risk formula is We classified patients below the median as a low-risk group and those above the median as a high-risk group. The patients were divided into high-risk and low-risk groups in the training and test groups, respectively, according to the risk score (Figure 5A). In the training group, there were 98 patients in the high-risk group and 99 patients in the low-risk group. In the test set, there were 93 patients in the high-risk group and 103 patients in the low-risk group. As shown in the results, patients with high-risk scores had shorter survival times than those with low-risk scores. The ROC curve of survival prediction showed satisfactory performance (Figure 5B). The AUC (Area Under Curve) value in the training set was 0.766, and that in the test set was 0.674 (Figure 6). The closer the AUC value is to 1.0, the more authentic the detection method is. The same analysis was also performed in the test group with consistent results (Figure 5C,D).
FIGURE 5.
The constructed prognostic model was used for survival analysis of the training set and test set, and ROC was analyzed. (A) Survival analysis curve of the training set; red indicates patients in the high-risk group, blue denotes patients in the low-risk group. (B) ROC curve of the training set. (C) Survival analysis curve of the test set. (D) ROC curve of the test set.
FIGURE 6.
Risk curve of the training set and test set. (A) Risk score distribution of the training set. (B) Survival state distribution of training set. (C) Heat maps of eight ferroptosis-related genes in the training set. (D) Distribution of risk scores in the test set. (E) Survival state distribution of the test set. (F) Heat maps of eight ferroptosis-related genes in the test set.
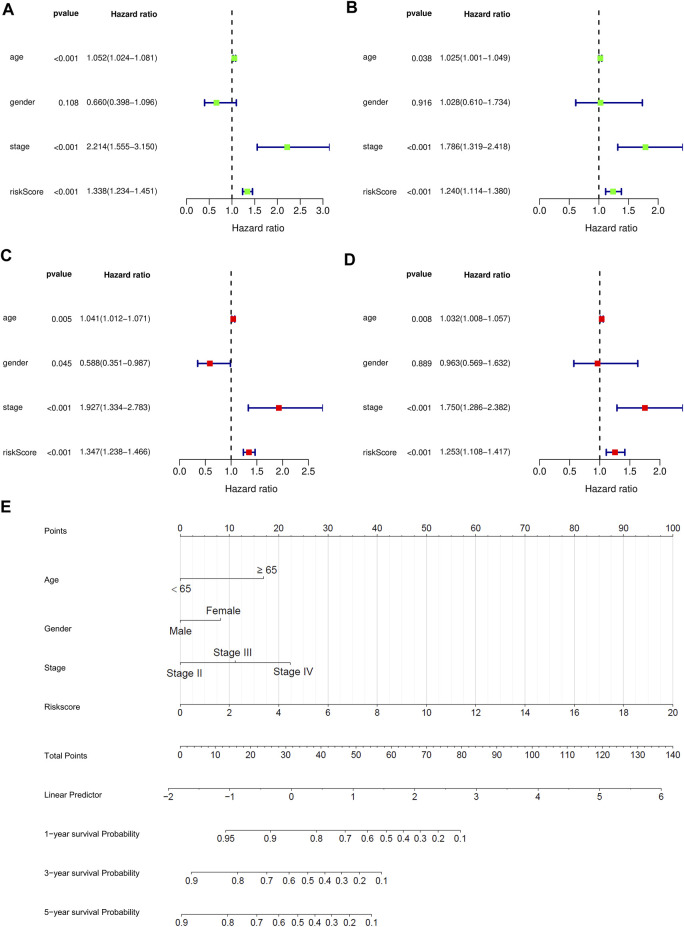
We performed univariate and multivariate independent prognostic analysis. Age, tumor stage, and the risk score could be used as independent prognostic factors for survival of bladder cancer patients in the training and validation sets (p < 0.05) in the univariate independent prognostic analysis. Sex, age, tumor stage, and the risk score were independent prognostic factors of bladder cancer (p < 0.05) in the multivariate independent prognostic analysis. For the test set, age was not found to be a prognostic factor (p = 0.889) (Figures 7A-D). All the prognostic factors were included in the nomograms (Figure 7E).
FIGURE 7.
Independent prognostic analysis and 1 -, 2 -, and 3-years survival prediction were performed for bladder cancer patients in the training and testing sets. (A) Univariate prognostic analysis of training set. (B) Multivariate prognostic analysis of training set. (C) Univariate prognostic analysis of test set. (D) Multivariate prognostic analysis of test set. (E) The nomogram was used to predict 1-year, 2-years, and 3-years survival rates of bladder cancer patients in the training set.
Finally, the nomogram of these eight prognostic genes was drawn in the training set. The RNA expression of eight genes was used as a parameter to draw the dotted lines in the nomogram and the total score can predict the survival rates.
Validation of Ferroptosis-Related Genes
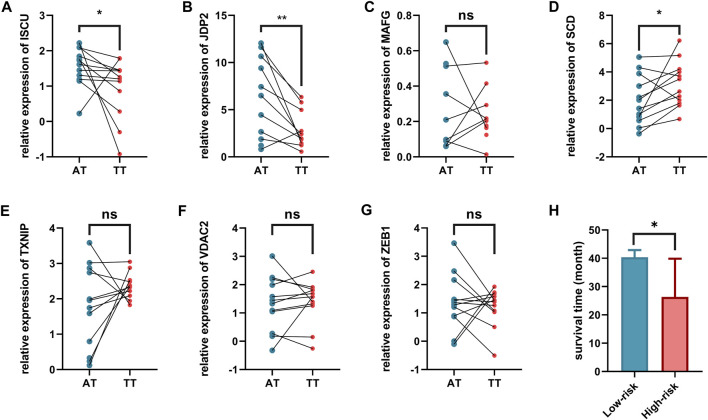
We further validated the expression of these eight genes in 12 pairs of clinical sample tissues (Figure 8). The results showed that the expression levels of ISCU and JDP2 were lower in tumor samples and higher in paracancerous tissues. In clinical sample tissues, the expression of SCD was exactly the opposite. Although the p-value was not less than 0.05, we found that MAFG with VDAC2 presented higher expression in paracancerous tissues than in cancer tissues, and TXNIP presented lower expression in paracancerous tissues than in cancer tissues. The low abundance of nfe2l2 expression in tissues precludes its detection by qPCR. Because the number of clinical samples was small and the plotting of survival curves was difficult, we divided the patients into high-risk and low-risk patients by the risk model derived in the previous study and found a statistical difference in survival time between these two groups. Low-risk patients had a longer survival time than high-risk patients (Figure 8H).
FIGURE 8.
Validation of expression levels of eight ferroptosis related genes in tumor tissues. and adjacent normal tissues. (A) ISCU, (B) JDP2, (C) MAFG, (D) SCD, (E) TXNIP, (F) VDAC2, (G) ZEB1, (H) Comparison of survival time among patients with different risk characteristics. AT, adjacent normal tissues. TT, tumor tissues.
Discussion
Bladder cancer has a certain recurrence rate after operation. Therefore, it is meaningful to find new diagnostic biomarkers or therapeutic targets for bladder cancer. Ferroptosis is associated with many physiological and pathological processes, such as tumor suppression, neuronal degeneration, antiviral immune response, and ischemia-reperfusion injury. From the perspective of cancer therapy, we hope to eliminate cancer cells by promoting ferroptosis or inhibiting ferroptosis in healthy cells. However, the specific mechanism of ferroptosis remains to be investigated in bladder cancer.
We integrated mRNA data from TCGA database and identified 103 ferroptosis-related DEGs, including 57 upregulated and 46 downregulated ferroptosis-related genes. In addition, through functional analysis, we found that ferroptosis-related DEGs were involved in apoptosis and immune checkpoint pathways. We then established a prognostic model based on eight ferroptosis-related genes to predict the accurate survival for patients with bladder cancer based on these results. By combining important clinical data and risk scores, a nomogram was constructed to predict survival rates.
Ferroptosis-related genes in this study included ISCU, NFE2L2, MAFG, ZEB1, VDAC2, TXNIP, SCD, and JDP2. These genes have been studied in metabolic processes or tumor development. ISCU is an important scaffold protein and can assemble iron and sulfur into iron-sulfur clusters These clusters participate in the maturation of [2Fe-2S] and [4Fe-4S] proteins in the mitochondria and cytoplasm and the regulation of iron metabolism (Duan et al., 2009). As an important redox center, iron-sulfur clusters participated in many physiological functions such especially in metabolism (Xu and Møller, 2011). NFE2L2 is transcription factor, which regulate genes that contain antioxidant response elements in their promoters. NFE2L2 plays a vital role in the treatment of neurodegenerative diseases and regulation of ferroptosis (Liu et al., 2020; Song and Long, 2020). MAFG is a small MAF protein (Liu et al., 2018). MAFG-mediated cisplatin resistance lies in lower reactive oxygen species production; therefore, MAFG may be a potential therapeutic target (Vera-Puente et al., 2018). ZEB1 is a transcriptional repressor. Studies have shown that ZEB1 is abnormally expressed in many liver diseases, including hepatocellular carcinoma (Li et al., 2019). Studies have found that ZEB1 overexpression increases sensitivity to ferroptosis (Lee et al., 2020). VDAC2 is a member of the voltage-dependent anion channel pore-forming protein family, which is thought to play an important role in the metabolite diffusion process on the outer mitochondrial membrane. Moreover, it can participate in the mitochondrial apoptotic pathway by regulating the activity of BCL2-antagonist/killer 1 protein. TXNIP is a thioredoxin-binding protein, which participates in cellular redox signaling and plays an important role in oxidative stress. TXNIP has been found to be involved in the autophagy and ferroptosis of a class of protein disulfide isomerase inhibitors in the treatment of glioblastoma (Kyani et al., 2018). SCD encodes enzymes involved in fatty acid biosynthesis. In one study, SCD expression in bladder cancer patients was found to be significantly associated with poor prognosis. Loss-of-function experiments further revealed that inhibiting SCD expression can reduce cell proliferation and invasion (Piao et al., 2019). JDP2, a member of the bZIP transcription factor family and acts as a broad AP-1 inhibitory protein. JDP2 plays an important role in inhibiting AP-1-driven biological processes, such as cell cycle regulation, cell differentiation, apoptosis, and tumorigenesis (Chen et al., 2017). In hepatocellular carcinoma, miR-501 can regulate JDP2 to promote tumor cell development (Yu et al., 2019). In conclusion, these eight genes are mainly involved in iron metabolism, lipid metabolism, and amino acid metabolism.
In general, we investigated the biological function and prognostic value of ferroptosis-related genes and established a new prognostic model based that are closely related to the overall survival of bladder cancer patients.
The main limitation of this study is the small number of clinical samples used for validation, which requires further investigation. And we need to experimentally validate the hypothesis. Also, more validation research is needed to explore the mechanism of ferroptosis-related genes in bladder cancer. In spite of all its limitations, our results suggest that prognostic markers based on ferroptosis-related genes may be a reliable predictive tool for the prognosis of bladder cancer patients.
Data Availability Statement
The data analyzed in this study were obtained from publicly available datasets. The mRNA data and clinical can be found here: The Cancer Genome Atlas (https://portal.gdc.cancer.gov/). The ferroptosis-related genes are downloaded from the FerrDb database (http://www.zhounan.org/ferrdb).
Author Contributions
KY and FW conceived and designed the study. KY and YR analyzed the data. CW and XT revised the images. KY and JL drafted the manuscript. FW, YX and XZ revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Improvement Project for Theranostic ability on Difficulty Miscellaneous disease (Tumor) and the Science and Technology Innovation Fostering Foundation of Zhongnan Hospital of Wuhan University (ZLYNXM202008, Nos. znpy2019050, znpy2019004, znpy2018027), This work was also funded by Medical Top-talented youth development project of Hubei Province, and by Medical talented youth development project in Health Commission of Hubei Province (No. WJ2019Q049).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.631152/full#supplementary-material
References
- Antoni S., Ferlay J., Soerjomataram I., Znaor A., Jemal A., Bray F. (2017). Bladder Cancer Incidence and Mortality: A Global Overview and Recent Trends. Eur. Urol. 71 (1), 96–108. 10.1016/j.eururo.2016.06.010 [DOI] [PubMed] [Google Scholar]
- Chen C., Liu X., Jiang J., Li S., Wang G., Ju L., et al. (2020a). Matrix Metalloproteinase 11 Is a Potential Biomarker in Bladder Cancer Diagnosis and Prognosis. Ott Vol. 13, 9059–9069. 10.2147/ott.S243452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.-N., Li T., Cheng L., Qin T.-S., Sun Y.-X., Chen C.-T., et al. (2020b). Synthesis and In Vitro Anti-bladder Cancer Activity Evaluation of Quinazolinyl-Arylurea Derivatives. Eur. J. Med. Chem. 205, 112661. 10.1016/j.ejmech.2020.112661 [DOI] [PubMed] [Google Scholar]
- Chen Y.-L., Chan S.-H., Lin P.-Y., Chu P.-Y. (2017). The Expression of a Tumor Suppressor Gene JDP2 and its Prognostic Value in Hepatocellular Carcinoma Patients. Hum. Pathol. 63, 212–216. 10.1016/j.humpath.2017.03.003 [DOI] [PubMed] [Google Scholar]
- Conrad M., Lorenz S. M., Proneth B. (2021). Targeting Ferroptosis: New Hope for As-Yet-Incurable Diseases. Trends Mol. Med. 27, 113–122. 10.1016/j.molmed.2020.08.010 [DOI] [PubMed] [Google Scholar]
- Duan X., Yang J., Ren B., Tan G., Ding H. (2009). Reactivity of Nitric Oxide with the [4Fe-4S] Cluster of Dihydroxyacid Dehydratase from Escherichia coli . Biochem. J. 417 (3), 783–789. 10.1042/bj20081423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluth L. A., Black P. C., Bochner B. H., Catto J., Lerner S. P., Stenzl A., et al. (2015). Prognostic and Prediction Tools in Bladder Cancer: A Comprehensive Review of the Literature. Eur. Urol. 68 (2), 238–253. 10.1016/j.eururo.2015.01.032 [DOI] [PubMed] [Google Scholar]
- Kyani A., Tamura S., Yang S., Shergalis A., Samanta S., Kuang Y., et al. (2018). Discovery and Mechanistic Elucidation of a Class of Protein Disulfide Isomerase Inhibitors for the Treatment of Glioblastoma. ChemMedChem 13 (2), 164–177. 10.1002/cmdc.201700629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latunde-Dada G. O. (2017). Ferroptosis: Role of Lipid Peroxidation, Iron and Ferritinophagy. Biochim. Biophys. Acta (Bba) - Gen. Subjects 1861 (8), 1893–1900. 10.1016/j.bbagen.2017.05.019 [DOI] [PubMed] [Google Scholar]
- Lee J., You J. H., Kim M.-S., Roh J.-L. (2020). Epigenetic Reprogramming of Epithelial-Mesenchymal Transition Promotes Ferroptosis of Head and Neck Cancer. Redox Biol. 37, 101697. 10.1016/j.redox.2020.101697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.-y., Yang C.-c., Yang J.-f., Li H.-d., Zhang B.-y., Zhou H., et al. (2019). ZEB1 Regulates the Activation of Hepatic Stellate Cells through Wnt/β-Catenin Signaling Pathway. Eur. J. Pharmacol. 865, 172787. 10.1016/j.ejphar.2019.172787 [DOI] [PubMed] [Google Scholar]
- Li Z., Rong L. (2020). Cascade Reaction-Mediated Efficient Ferroptosis Synergizes with Immunomodulation for High-Performance Cancer Therapy. Biomater. Sci. 8, 6272–6285. 10.1039/d0bm01168a [DOI] [PubMed] [Google Scholar]
- Liu T., Yang H., Fan W., Tu J., Li T. W. H., Wang J., et al. (2018). Mechanisms of MAFG Dysregulation in Cholestatic Liver Injury and Development of Liver Cancer. Gastroenterology 155 (2), 557–571.e514. 10.1053/j.gastro.2018.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Lv X., Song E., Song Y. (2020). Fostered Nrf2 Expression Antagonizes Iron Overload and Glutathione Depletion to Promote Resistance of Neuron-like Cells to Ferroptosis. Toxicol. Appl. Pharmacol. 407, 115241. 10.1016/j.taap.2020.115241 [DOI] [PubMed] [Google Scholar]
- Piao C., Cui X., Zhan B., Li J., Li Z., Li Z., et al. (2019). Inhibition of Stearoyl CoA Desaturase-1 Activity Suppresses Tumour Progression and Improves Prognosis in Human Bladder Cancer. J. Cel Mol Med 23 (3), 2064–2076. 10.1111/jcmm.14114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegman M., Sagie L., Galed C., Levin T., Steinberg N., Dixon S. J., et al. (2020). Ferroptosis Occurs through an Osmotic Mechanism and Propagates Independently of Cell Rupture. Nat. Cel Biol 22 (9), 1042–1048. 10.1038/s41556-020-0565-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Long D. (2020). Nrf2 and Ferroptosis: A New Research Direction for Neurodegenerative Diseases. Front. Neurosci. 14, 267. 10.3389/fnins.2020.00267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torti S. V., Torti F. M. (2020). Iron and Cancer: 2020 Vision. Cancer Res. 80, 5435–5448. 10.1158/0008-5472.Can-20-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera-Puente O., Rodriguez-Antolin C., Salgado-Figueroa A., Michalska P., Pernia O., Reid B. M., et al. (2018). MAFG Is a Potential Therapeutic Target to Restore Chemosensitivity in Cisplatin-Resistant Cancer Cells by Increasing Reactive Oxygen Species. Translational Res. 200, 1–17. 10.1016/j.trsl.2018.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Zhang Z., Tsai H.-i., Liu Y., Gao J., Wang M., et al. (2020a). Branched-chain Amino Acid Aminotransferase 2 Regulates Ferroptotic Cell Death in Cancer Cells. Cell Death Differ 28, 1222–1236. 10.1038/s41418-020-00644-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wei Z., Pan K., Li J., Chen Q. (2020b). The Function and Mechanism of Ferroptosis in Cancer. Apoptosis 25, 786–798. 10.1007/s10495-020-01638-w [DOI] [PubMed] [Google Scholar]
- Wu Y., Yu C., Luo M., Cen C., Qiu J., Zhang S., et al. (2020). Ferroptosis in Cancer Treatment: Another Way to Rome. Front. Oncol. 10, 571127. 10.3389/fonc.2020.571127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X. M., Møller S. G. (2011). Iron-sulfur Clusters: Biogenesis, Molecular Mechanisms, and Their Functional Significance. Antioxid. Redox Signaling 15 (1), 271–307. 10.1089/ars.2010.3259 [DOI] [PubMed] [Google Scholar]
- Yang J.-L., Wang C. C. N., Cai J.-H., Chou C.-Y., Lin Y.-C., Hung C.-C. (2020). Identification of GSN and LAMC2 as Key Prognostic Genes of Bladder Cancer by Integrated Bioinformatics Analysis. Cancers 12 (7), 1809. 10.3390/cancers12071809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.-G., Han Y., He Q.-Y. (2012). clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS: A J. Integr. Biol. 16 (5), 284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W., Deng W., Zhao Q., Zhuang H., Zhang C., Jian Z. (2019). miR-501 Acts as an Independent Prognostic Factor that Promotes the Epithelial-Mesenchymal Transition through Targeting JDP2 in Hepatocellular Carcinoma. Hum. Cel 32 (3), 343–351. 10.1007/s13577-019-00243-7 [DOI] [PubMed] [Google Scholar]
- Zhou N., Bao J. (20202020). FerrDb: A Manually Curated Resource for Regulators and Markers of Ferroptosis and Ferroptosis-Disease Associations, 2020. Oxford: Database. 10.1093/database/baaa021 FerrDb: a Manually Curated Resource for Regulators and Markers of Ferroptosis and Ferroptosis-Disease Associations [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo S., Yu J., Pan H., Lu L. (2020). Novel Insights on Targeting Ferroptosis in Cancer Therapy. Biomark Res. 8, 50. 10.1186/s40364-020-00229-w [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data analyzed in this study were obtained from publicly available datasets. The mRNA data and clinical can be found here: The Cancer Genome Atlas (https://portal.gdc.cancer.gov/). The ferroptosis-related genes are downloaded from the FerrDb database (http://www.zhounan.org/ferrdb).