Abstract
Amyotrophic lateral sclerosis (ALS) has historically posed unique challenges for gene-therapy-based approaches, due to a paucity of therapeutic targets as well as the difficulty of accessing both the brain and spinal cord. Recent advances in our understanding of disease mechanism and ALS genetics, however, have combined with tremendous strides in CNS targeting, gene delivery, and gene editing and knockdown techniques to open new horizons of therapeutic possibility. Gene therapy clinical trials are currently underway for ALS patients with SOD1 mutations, C9orf72 hexanucleotide repeat expansions, ATXN2 trinucleotide expansions, and FUS mutations, as well as sporadic disease without known genetic cause. In this review, we provide an in-depth exploration of the state of ALS-directed gene therapy, including antisense oligonucleotides, RNA interference, CRISPR, adeno-associated virus (AAV)-mediated trophic support, and antibody-based methods. We discuss how each of these approaches has been implemented across known genetic causes as well as sporadic ALS, reviewing preclinical studies as well as completed and ongoing human clinical trials. We highlight the transformative potential of these evolving technologies as the gene therapy field advances toward a true disease-modifying treatment for this devastating illness.
Keywords: ALS, amyotrophic lateral sclerosis, gene therapy, AAV, ASO, RNAi, CRISPR, gene delivery, clinical trial
In this review, Davidson and colleague provide an in-depth review of the preclinical and clinical gene therapy advances made in treatment of both inherited and sporadic amyotrophic lateral sclerosis (ALS). They discuss antisense oligonucleotides, RNA interference, CRISPR, AAV-mediated trophic support, and antibody-based methods, highlighting ongoing challenges and anticipated future directions.
Introduction
Amyotrophic lateral sclerosis (ALS) is a uniformly fatal disease characterized by degeneration of upper and lower motor neurons, leading to progressive paralysis, respiratory failure, and death in 2–5 years. It is also the most common adult motor neuron disease, with a prevalence of 5 per 100,0001 and a lifetime risk of 1:400–1:800.2 The search for disease-modifying therapies has long been hampered by a poor understanding of disease mechanism. In fact, out of >80 human clinical trials,3 only riluzole4,5 and edaravone6, 7, 8 have slowed progression, both of them modestly and by targeting non-specific factors such as excitotoxicity or oxidative stress. Recent advances in our understanding of ALS pathophysiology, including the discovery of many of its genetic underpinnings, have enabled the development of increasingly targeted therapies, with today’s clinical trials holding growing promise and hope for efficacious treatments.
A pathologic hallmark of ALS, affecting over 97% of patients, is ubiquitinated cytoplasmic inclusions consisting largely of Tar-DNA binding protein of 43 kDA (TDP-43).9,10 TDP-43 pathology is also found in 50% of frontotemporal dementia (FTD) patients and is seen in nearly all ALS-FTD spectrum cases.9, 10, 11, 12, 13 Normally found in the nucleus, TDP-43 broadly mediates transcription, translation, and splicing,14, 15, 16, 17 interacting with ~30% of the transcriptome.14 Its cytoplasmic mislocalization is thought to be detrimental on two fronts: (1) toxicity of cytoplasmic aggregates,18,19 and (2) loss of normal nuclear function.20, 21, 22, 23, 24 In motor neurons from ALS patients, TDP-43 associates with cytoplasmic stress granules (SGs),25,26 conglomerates of proteins and RNA that become maladaptive in disease. SGs additionally bind a host of nuclear import, export, and transcription and translation factors, resulting in loss of nucleocytoplasmic shuttling and eventual cell death.25, 26, 27 This intracellular cascade provides several therapeutic targets aimed at pathologic elements common to most ALS cases.14
Another approach, and one particularly amenable to gene therapy techniques, is targeting genetic mutations known to cause ALS. Ten percent of ALS can be classified as familial, with ~70% of familial cases explained by known gene mutations.28 The first ALS-associated gene, superoxide dismutase-1 (SOD1),29 was described in 1993 and accounts for 12%–20% of familial and 1%–2% of sporadic ALS.28,29 For the decade and a half following its discovery, this remained the only major known heritable cause and therefore was the basis of all preclinical ALS models.30,31 However, unlike the 97% of cases described above, SOD1-ALS patients do not demonstrate TDP-43 pathology at autopsy, raising concern that SOD1 animal models similarly do not represent most ALS pathology, and studies conducted in these models may not broadly translate.30,31 Indeed, most early studies failed in human clinical trials.32,33 It was not until 2008 that a second contributor to heritable ALS, TARDBP—which encodes TDP-43—was discovered.34,35 TARDBP mutations account for 4% of familial and 1% of sporadic cases, and the discovery of both this direct genetic cause and the identification of TDP-43 in motor neuron inclusions yielded additional mouse models.30,31,36,37 A year later, mutations in the fused in sarcoma (FUS) gene were discovered, accounting for another 4% of familial ALS,38,39 although, like SOD1, FUS patients lack TDP-43 pathology. In 2010, intermediate-length CAG trinucleotide repeat expansions in the ataxin-2 (ATXN2) gene were found to be associated with up to 4.7% of all ALS.40 The most significant genetic discovery to date has been that of C9Orf72 in 2011;41,42 G4C2 hexanucleotide repeat expansions in this gene have been found in 40% of familial and 8% of sporadic cases, or 11% of all ALS,28,43 and some mouse models recapitulate ALS pathophysiology.44,45 More than 20 additional ALS-associated genes together account for an additional 12%–15% of familial ALS,28,46 leaving 25%–30% of familial cases unexplained. Aside from SOD1 and FUS, TDP-43 pathology is seen across all mutations, with nearly all known ALS-associated genes also being identified in FTD or acting as FTD phenotype modifiers, SOD1 being a notable exception.10
Gene therapy involves the delivery of genetic material to cells in order to introduce functional copies of dysfunctional genes, introduce trophic factors and other disease-modifying genes, or silence harmful gene expression using antisense oligonucleotides (ASOs), RNA interference (RNAi), or gene-editing technology such as CRISPR. In order to target the central nervous system (CNS), this material can be delivered naked, as is often the case for ASOs; by viral vectors including adeno-associated virus (AAV) and others; or by physical or chemical systems such as nanoparticles.2,47 Methods to access the CNS across the blood-brain barrier include intravenous (i.v.), intracerebroventricular (i.c.v.), intrathecal (i.t.), or intraparenchymal delivery. An ALS-specific challenge is the need to reach both the motor cortex and the spinal cord.
Viral vectors have the advantage of conferring sustained expression of genetic material in transduced cells after a single treatment. Among these, AAV has emerged as the lead vector for CNS delivery, based on its ability to transduce terminally differentiated cells and to establish nuclear episomes without incurring the risk of insertional mutagenesis.48,49 Equally advantageous is the selective tropism of its many different serotypes, both naturally occurring and modified.47,50 Thus far, the serotypes that have dominated ALS preclinical studies and clinical trials are the naturally occurring AAV9 and AAVrh10, due to their ability to target motor neurons.47,51,52 In particular, the discovery that self-complementary AAV9 (scAAV9) vectors cross the blood-brain barrier51 led to their use in treatment of spinal muscular atrophy (SMA), an inherited lower motor neuron disorder in which patients lack a functioning copy of the SMN1 gene. i.v. delivery of scAAV9 encoding SMN1 rescued SMA mouse models,53, 54, 55 and subsequent human clinical trials showed dramatic efficacy in SMA patients,56 leading to US Food and Drug Administration (FDA) approval of Zolgensma in 2019. This served as an important proof of principle for ALS therapeutics, which also must target lower motor neurons.49 Recently, novel technologies have used guided evolution to create novel AAV serotypes with increased CNS tropism and expression while de-targeting peripheral tissues,57, 58, 59 granting the potential to broaden the efficacy and increase the specificity of future AAV-mediated therapeutics. Of note, lentiviral approaches have also been used with some success in preclinical models of ALS,49,60 but their translatability is limited by their small area of transduction, low viral titers, and broad tropism, as well as their integration into the host genome with associated risk of mutagenesis.47,61 For this reason, the majority of preclinical ALS lentiviral studies, reviewed elsewhere,49 have focused on delivery to muscle or ex vivo gene transfer,49,62 including one approach combining both strategies62 that recently completed a phase I/II clinical trial (ClinicalTrials.gov: NCT02943850).
In this manuscript, we provide an overview of the most promising gene-therapy-based approaches for ALS to date, including ASO therapy, AAV-mediated gene silencing, and AAV-mediated gene delivery. We focus primarily on recent studies and review how these strategies have been applied in preclinical models and human clinical trials for each of the most common familial forms of ALS, as well as for sporadic disease.
ASOs
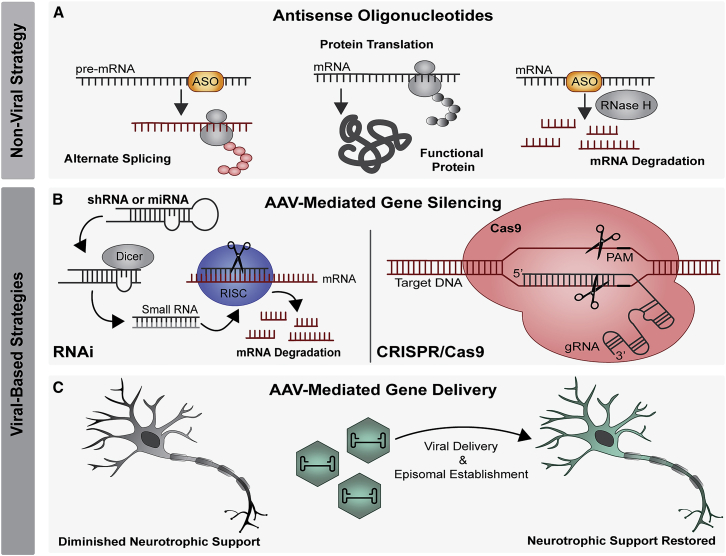
ASOs are single-stranded, 8- to 50-base sequences of synthetic oligonucleotides that can be designed to complement target mRNAs for RNase H enzyme-mediated target degradation or designed against primary transcripts to induce alternative splicing (Figure 1A).63, 64, 65 Recently, various modifications have been made to increase ASO stability, protect against nuclease degradation, improve cellular uptake, recruit RNase H, and reduce immunogenicity.64 Although they do not cross the blood-brain barrier, ASOs achieve widespread CNS distribution after i.t. delivery,63,65 making them an important tool in the arsenal against ALS. Indeed, ASOs were successfully developed to treat SMA, providing an important proof-of-concept for ALS. For SMA therapy, delivery of a splice-altering ASO resulting in the conversion of SMN2 to SMN1 showed efficacy in mouse models of SMA as well as distribution to affected areas of the spinal cord in nonhuman primates (NHPs).66 This was followed by clinical trials showing marked improvements in strength and longevity in children with SMA,67, 68, 69 leading to FDA approval of nusinersen in 2016 and its current widespread use in the clinic.
Figure 1.
Summary of gene therapy strategies
(A) Non-viral strategies include using ASOs to induce alternate splicing or RNase H-mediated degradation. (B and C) Viral strategies include (B) AAV-mediated gene silencing, through RNA interference or CRISPR-Cas9 or (C) AAV-mediated gene delivery including neurotrophic factors. AAV, adeno-associated virus; ASO, antisense oligonucleotide; Cas, CRISPR-associated system; miRNA, microRNA; PAM, protospacer adjacent motif; RISC, RNA-induced silencing complex; RNAi, RNA interference; shRNA, small hairpin RNA.
SOD1
Mutations in SOD1 are believed to cause ALS through toxic gain of function caused by aggregation of misfolded SOD1 protein.30,70 Although more than 150 ALS-associated SOD1 mutations have been described, the G93A mutation, rare in humans, is the most commonly studied in preclinical models, as there exists a readily available SOD1G93A mouse71,72 and rat.73 In a key preclinical study, a 20-mer ASO targeting SOD1 was delivered via continuous infusion in the lateral ventricle of rats and the lumbar spine of NHPs and demonstrated significant and widespread ASO concentrations throughout the brain and spinal cord with deep tissue penetration.74 In SOD1G93A rats treated pre-symptomatically, there was substantial SOD1 mRNA and protein knockdown in the brain and spinal cord that was associated with slowed disease progression and extended survival.74 This led to a first-in-human phase I clinical trial (ClinicalTrials.gov: NCT01041222), where ASO ISIS 333611 was delivered via a single, 11.5-h i.t. infusion to patients with SOD1-associated ALS.75 While the study demonstrated safety and tolerability as well as establishing the use of cerebrospinal fluid (CSF) SOD1 as a pharmacokinetic marker, there were no reductions in CSF SOD1 protein at the conservative concentrations used (maximum dose, 3 mg). A subsequent trial (ClinicalTrials.gov: NCT02623699) delivered the ASO, now named BIIB067/Tofersen (IONIS-SOD1Rx), via serial lumbar i.t. injections over 12 weeks, with patients cohorted into dosing groups ranging from 20–100 mg.76 The trial demonstrated safety at these higher doses, as well as reductions in CSF SOD1 protein of 33% in the highest-dose group. Exploratory analyses showed promising reductions in rate of decline as measured by the revised ALS functional rating scale (ALSFRS-R) in the high-dose group, especially among fast progressors.76 Safety and efficacy are currently being evaluated in a phase 3, randomized, double-blind, placebo-controlled trial (ClinicalTrials.gov: NCT02623699) and its extension study (ClinicalTrials.gov: NCT03070119). In addition to the traditional delivery of naked ASOs described in these studies, another group reported a novel approach using AAV to achieve targeted and sustained ASO expression. AAVrh10 expressing antisense sequences targeting SOD1 embedded in a U7 small-nuclear RNA was delivered both i.v. and i.c.v. to SOD1G93A mice, at birth or in pre-symptomatic adulthood, leading to nonsense-mediated SOD1 decay and markedly increased survival, strength, and weight in both age groups.77
C9orf72
Although the full mechanisms by which C9orf72 hexanucleotide repeat expansions cause ALS are unknown, studies suggest a prominent role for a toxic gain of function, mediated in part by the G4C2 repeat RNA and in part by dipeptide repeat proteins produced by repeat-associated non-ATG (RAN) translation.78,79 The first ASO-based strategies focused on reducing this toxicity in induced pluripotent stem cells (iPSCs) derived from humans with C9orf72 expansions. Three such studies published in tandem in 2013 found that ASO-mediated reduction of the C9orf72 transcript—whether through binding within the repeat expansion or within the surrounding regions—reduced nuclear RNA foci in C9orf72-associated ALS.78,80,81 Two of the groups found additionally that ASO treatment reversed aberrant gene expression and reduced susceptibility to excitotoxicity in iPSC-differentiated neurons,78,81 while the third group demonstrated persistence of aberrant RNA expression after sense-strand targeting and evidence suggesting the importance of simultaneously targeting the antisense strand.80 This group also performed the first in vivo studies, demonstrating sustained efficacy and tolerability of knocking down C9orf72 RNA in wild-type mice for up to 18 weeks after a single injection of ASO into the lateral ventricle.80 In a subsequent study, the authors used a single i.c.v. injection to deliver ASOs targeting G4C2 repeat-containing sense-strand RNAs to pre-symptomatic adult mice expressing up to 450 repeats. They demonstrated sustained reduction in RNA foci and dipeptide repeat proteins in the cortex and spinal cord, as well as amelioration of cognitive deficits.82 This work has ultimately led to a phase I clinical trial of the ASO BIIB078 (ClinicalTrials.gov: NCT03626012) for C9orf72-ALS patients.
ATXN2
Although long trinucleotide repeat expansions in ATXN2 have been known to cause spinocerebellar ataxia type 2,83, 84, 85, 86 intermediate-length expansions were discovered in 2010 to be a relatively common cause of heritable ALS.40,87, 88, 89 Ataxin-2 serves diverse functions in the cell, including RNA processing and receptor endocytosis, but its critical functions in the formation of stress granules27,90 and induction of aberrant TDP-43 cleavage by caspase 391,92 are of particular relevance in ALS. In 2018 it was demonstrated that a host of nucleocytoplasmic transport factors are bound in ataxin-2-containing SGs upon induction of stress in HEK293T cells. Further, delivery of ASOs targeting ataxin-2 to neuronal-differentiated iPSCs from C9orf72-ALS patients reversed cytoplasmic mislocalization of nuclear proteins.27 Seminal work in vivo used a rapidly progressive TDP-43 ALS mouse model to demonstrate that a one-time delivery of Atxn2-ASO to the lateral ventricle at birth resulted in sustained, marked reduction of Atxn2 mRNA as well as prolonged survival and improved gait.93 While this strategy can be of benefit to patients with ATXN2-ALS, importantly, it also offers a therapeutic avenue for the broader ALS population, as TDP-43 localization to ATXN2-dependent stress granules is a shared pathologic endpoint.25 A phase I clinical trial of the ASO BIIB105 (ClinicalTrials.gov: NCT04494256) is currently underway, enrolling patients both with and without CAG repeat expansions in ATXN2.
FUS
FUS is the most commonly mutated gene found in juvenile and pediatric ALS, with the p.P525L mutation causing a particularly aggressive and early-onset form94,95 through a toxic gain-of-function mechanism. In 2019, Ionis Pharmaceutical, in conjunction with Columbia Medical Center, developed an ASO targeting this mutation via i.t. delivery and obtained FDA approval for experimental use in a young woman, for whom the therapy, jacifusen, is named.96 Jaci has since passed away, but jacifusen has been used to treat three additional FUS-ALS patients through the FDA’s compassionate use protocol, and 8 more patients are approved for investigational treatment with funding from the ALS association and Project ALS.95,97,98
AAV-mediated gene silencing
AAV-mediated silencing has the advantage over ASOs of eliminating the need for readministration, which is of use when targeting both the brain and spinal cord. AAVs are frequently used to deliver small noncoding RNAs to achieve RNAi, a naturally occurring process in which double-stranded RNA molecules regulate the expression of mRNA through homologous base pairing and subsequent cleavage via the RNAi-induced silencing complex (RISC; Figure 1B).2,99 Two types of small noncoding RNAs, small hairpin RNA (shRNA) and artificial microRNA (miRNA), are in increasingly widespread use as a therapeutic strategy against neurodegenerative disease, particularly miRNA due to its more favorable safety profile.100,101 Further refinement has been achieved using bioinformatic tools to minimize passenger strand loading102 and optimize promoter, serotype, and dosing choices,99 leading to greatly decreased off-target effects. The first AAV-RNAi human clinical trial for a neurodegenerative disease (ClinicalTrials.gov: NCT04120493) is currently in phase I/II and uses AAV5 to deliver a miRNA targeting Huntingtin to treat Huntington’s disease.2,103 This study will provide important safety and feasibility data for the ALS field.
An alternative approach to treat gain-of-function disorders is gene editing, which can be achieved using CRISPR-associated (Cas) systems, the most widely used of which is the RNA-guided Cas9 endonuclease104 (Figure 1B). CRISPR-Cas9 enables specific targeting of genomic sequences and induction of double-stranded DNA breaks, causing frameshift-inducing base insertions or deletions (indels) with diverse applications,105 although its use in the ALS field is relatively nascent.106 Additional CRISPR-based systems have been developed that have transformative potential.107, 108, 109, 110 These include CRISPR-interference (CRISPRi), which uses a catalytically dead Cas9 to repress DNA without the potential mutagenic effects associated with cleavage;111 the Cas13 family, which targets RNA for degradation112,113 and variants of which are small enough for AAV packaging;114 and prime editing, which is a method of introducing specific genomic insertions or deletions without inducing double-stranded breaks.115 The application of these technologies to the ALS field is highly anticipated as they are further developed.
SOD1
As the first-identified form of ALS, SOD1 has been the target of the majority of AAV-RNAi-based approaches, with increasingly promising results. The first AAV9 studies delivering shRNA against SOD1 compared i.v. administration in SOD1G93A mice at differing ages and found a 39% increase in survival after P1-injection that decreased substantially with age and disease progression;116 they noted increasingly preferential glial targeting over neurons with later injections.116,117 They also found markedly increased survival in the slower-progressing SOD1G37R mouse model even when treated after disease onset, and they demonstrated robust SOD1 mRNA knockdown throughout the spinal cord in NHPs after lumbar i.t. injection.116 Another group used bilateral direct cortical injection of AAV9-shRNA targeting SOD1 in presymptomatic adult SOD1G93A rats and found that upper motor SOD1 suppression alone was able to preserve motor function and increase survival by 20 days.118 A 2016 study used AAV9 to deliver an artificial miRNA against SOD1 (mi-SOD1) to the ventricles in neonatal SOD1G93A mice. The authors found a 50% extension in survival with significant delay in hindlimb paralysis, as well as improvements across multiple histopathologic parameters including number of spinal motor neurons.119 Notably, neonatal treatment has limited translatability, as most patients present in adulthood after symptom onset, and the authors therefore conducted a separate study using rAAVRh10 to deliver mi-SOD1 systemically to adult SOD1G93A mice. They found substantially delayed disease onset and 21% extended survival, with preserved strength and respiratory function.120 They then tested the vector in marmosets via i.t. injection and found significant lowering of SOD1 in lower motor neurons throughout the spinal cord without short-term toxicity.120 Subsequent studies of rAAVRh10.mi-SOD1 in macaques, delivered i.t. with the head down at 30 degrees, demonstrated robust silencing at the mRNA level in laser-captured motor neurons throughout the spinal cord that was proportionate to the strength of the promoter used.121 The authors demonstrated a low off-targeting profile and found no adverse events up to 92 days post-infusion, paving the way for a human study published in July 2020. In that study, two patients with SOD1-ALS were treated with i.t. infusion of rAAVRh10.mi-SOD1. The first patient had transient improvement in strength in his right leg and slightly lowered CSF SOD1 levels without other signs of benefit, and his course was complicated by a severe meningoradiculitis with sensory symptoms. These sensory symptoms were not seen in the second patient, who was treated with immunosuppression at the time of infusion. At autopsy, SOD1 levels were lower in the spinal cord of patient 1 when compared to those in untreated SOD1 patients, and neurons were depleted in bilateral dorsal root ganglia (DRG) with a T-lymphocytic infiltrate of nerve roots. Patient 2 maintained stable strength and vital capacity throughout the 12-month period reported.122
The DRG toxicity seen in the above clinical trial has also been seen with AAV9 when administered systemically to neonatal pigs and juvenile primates,123 although the NHPs did not show clinical signs of toxicity. Further meta-analyses of CSF and systemic administration across 33 NHP studies (a total of 256 NHPs) showed DRG toxicity in 83% of CSF-treated and 32% of systemic-treated NHPs that was independent of serotype, sex, promoter, or transgene; importantly, however, pathology was dose dependent and overall mild, and no clinical sequelae were seen.124 Interesting alternative approaches to avoid DRG toxicity have been tried with varying degrees of success. For instance, the AAV-Rh10-mediated miSOD1 treatment described above also resulted in 50-day survival prolongation after direct injection of the tongue and intrapleural space of adult presymptomatic SOD1G93A mice,125 areas in which weakness is the lead cause of death in ALS patients. Recent studies successfully used DRG-specific miRNAs to downregulate transgene expression in these cells while preserving intended CNS transduction, an efficient detargeting strategy that can be employed across any AAV-based therapeutic.126 Finally, one notable study used a novel subpial delivery method, administering AAV9-shRNA to the narrow space between the spinal cord parenchyma and the innermost layer of the meninges. The authors achieved long-term suppression of motor neuron disease in SOD1G37R mice treated pre-symptomatically as well as potent blocking of disease progression in mice treated after disease onset. They then used this delivery method in adult pigs and NHPs and found homogeneous and robust transgene expression throughout the spinal cord, including motor neurons.127 Although technically challenging, the demonstration of feasibility in these larger animal models speaks well to the promise of subpial AAV delivery in humans, who share comparable spinal anatomy and size.
The first study using CRISPR in ALS was published in 2017. The authors used a modified AAV9 to deliver Staphylococcus aureus-derived Cas9 (SaCas9) and a single-guide RNA (sgRNA) targeting the hSOD1 gene via the facial vein to neonatal SOD1G93A mice.128 They found concentrated SaCas9 expression throughout the ventral horn cells of the spinal cord, with >2.5-fold decrease in SOD1 protein levels. Treated mice showed preserved motor neurons with improved motor function, a 37% delay in disease onset, and a 25% increase in survival, with a low off-target indel rate across spinal cord transgenes.128 Another group used AAV9-SaCas9-sgRNA to treat neonatal SOD1G93A mice via i.c.v. injection and also found reduced SOD1 in anterior horn cells. These studies also reported an increase in motor neurons and greatly improved motor function, as well as a notable 54.6% increase in survival, with a low off-target indel rate in predicted off-target sequences.129 In a subsequent study by the first group, an intein-mediated trans-splicing system was devised for in vivo single-base editing of SOD1, in order to minimize the potential mutagenic outcomes of double-stranded DNA breaks. The authors used dual AAV9 vectors to deliver this system i.t. to pre-symptomatic adult SOD1G93A mice and found a 40% reduction in SOD1 inclusions accompanied by improved neuromuscular function and muscle bulk, as well as 11% increase in survival,130 despite astrocyte-predominant targeting. These CRISPR-based strategies hold great promise as refinement continues.
C9of72
Due to the challenges of targeting intronic, GC-rich repeat expansions131 and achieving knockdown in the nucleus, where C9orf72-mediated RNA foci are predominantly found, RNAi-based approaches for C9orf72 have been challenging compared to SOD1. In 2015, a group was able to engineer duplex RNAs to overcome these hurdles and effectively target both sense and antisense G4C2 expansions in ALS-patient-derived fibroblast cells.132 By introducing mismatches in key regions, they destabilized the parent duplex to allow entry into the RISC complex, with resultant 40%–60% reduction in nuclear RNA foci from both strands. The authors subsequently engineered single-stranded silencing RNAs (ss-siRNAs) that function like ASOs but mediate degradation through RNAi, and found that these achieved even more potent inhibition of sense and antisense G4C2 repeat expansions in human mutant C9orf72 fibroblast cells.133 The biotech company uniQure developed a different bidirectional miRNA-based approach, using concatenated miRNA hairpins targeting C9orf72 (miC), and achieved 50% reduction of sense and antisense nuclear RNA foci in (G4C2)44-expressing cells. They further incorporated these miC constructs into AAV5 and effectively silenced C9orf72 in iPSC-derived neurons from an FTD patient.134 In a subsequent study, they used miC constructs to reduce both nuclear and cytoplasmic C9orf72 mRNA in FTD patient-derived iPSCs.135 They then delivered their AAV5-miC constructs to the striatum of adult transgenic C9orf72_3 line 112 mice, a strain harboring several tandem copies of human C9orf72 that does not develop neurodegeneration but exhibits RNA foci and poly(GP) protein. They found 20%–40% reduction of C9orf72 mRNA and sense intronic transcripts in transduced regions, although the mice were not followed long enough to determine poly(GP) reduction.135
There have been few CRISPR-mediated studies for C9orf72, but a recent study of note used CRISPR-Cas9 to generate deletions in the promoter region of iPSCs derived from patients harboring C9orf72 expansions. They found that these deletions reduced the expression of the C9orf72 variant containing the repeat expansion, which in turn nearly eliminated production of all dipeptide repeats and markedly reduced axonal degeneration.136 Another recent study identified Ku80 as a DNA repair protein that is overactivated in in Drosophila harboring G4C2 expansions and in C9orf72-ALS patient-derived iPSCs, leading to premature apoptosis. The authors used CRISPR-Cas9 to reduce Ku80 and found reductions in pro-apoptotic pathways; they also found decreased nuclear RNA foci after direct CRISPR-mediated deletion of the G4C2 repeat expansion.137 These are promising approaches for future in vivo studies.
ATXN2
To date there are no published RNAi or CRISPR-based approaches targeting ATXN2. However, the potential for this approach can be inferred from related studies in the spinocerebellar ataxia field and from the ASO-mediated reduction of Atxn2 in mouse models of SCA2138 and ALS93. Viral delivery of RNAi-based approaches has demonstrated success in mouse models of SCA1,139,140 SCA3,141,142 and SCA7,143 while a CRISPR-based approach in patient-derived iPSCs has shown early promise for SCA3.144 Additional ASO, RNAi, and CRISPR-based approaches for spinocerebellar ataxia are reviewed extensively elsewhere145 and demonstrate the feasibility and promise of this approach if adapted to motor neuron-targeting AAV delivery methods.
AAV-mediated gene delivery
The most long-established application of gene therapy is delivery of a therapeutic transgene. Unlike in the case of Zolgensma for SMA, however, no ALS-causing mutations have been found to have an exclusive loss-of function mechanism, and so therapeutic transgene delivery has focused on neurotrophic factors to provide support for degenerating neurons (Figure 1C). Neurotrophins are typically secreted proteins involved in neuronal growth or survival, and several neurotrophins have been found to decline over time in ALS patients and animal models,49 suggesting a potential role in prolonging neuronal health. While nonspecific and unlikely to be curative, these methods are attractive for their potential to prolong survival across all forms of ALS. Non-gene-therapy-mediated growth factor studies32,146 have unfortunately not been successful in clinical trials,147 which is felt to be in large part due to inadequate dosing to affected areas of the nervous system. For this reason, gene therapy approaches have been pursued by numerous groups in an effort to improve CNS delivery and dosing. Early gene therapy studies for neurotrophic support purposes have been extensively reviewed elsewhere.47,49 Here, we focus on recent AAV-mediated work, most of which was conducted in SOD1G93A mice. We also discuss AAV-mediated delivery of non-trophic factors, including neuromuscular junction modulators and aggregation-directed therapies.
Intramuscular delivery of insulin-like growth factor 1 (IGF1) using AAV9 was shown in two separate studies to preserve spinal motor neurons and modestly extend lifespan in SOD1G93A mice treated pre-symptomatically or in early post-symptomatic adulthood.148,149 Similar results were obtained using systemic AAV9-IGF1 delivery.150 In a different approach, i.c.v. delivery of AAV4 was used to target ependymal cells for IGF1 secretion into the CSF in early symptomatic SOD1G93A mice. The authors found improved motor function and modestly improved survival,151 effects that were slightly greater with delivery of AAV4-mediated vascular endothelial growth factor (VEGF), which has been hypothesized to act in the same pathway as IGF1.152 Delivery of both AAV4-IGF1 and AAV4-VEGF did not provide further benefit.151 VEGF has been adapted for ALS therapy at both the gene therapy and protein levels.153 Using i.t. delivery of AAV9, one group demonstrated that VEGF expression in adult symptomatic SOD1G93A mice improved motor function and extended survival.154 When AAV1 and AAV9 were used to deliver VEGF to a feline model of lower motor neuron disease via i.c.v., i.v., or intra-cisterna magna (i.c.m.) injection, only i.c.m. delivery resulted in sustained, high levels of VEGF expression in the spinal cord; however, there was no therapeutic benefit.155
Although early studies of AAV-mediated glial-derived neurotrophic factor (GDNF) delivery showed promise,156,157 effects were modest and seen only after intramuscular delivery. A 2017 study of a more translatable, systemic AAV9-GDNF delivery in pre-symptomatic SOD1G93A rats showed modest improvement in strength and delay of forelimb paralysis. However, there was no extension in survival, and adverse side effects were seen, including decreases in working memory, activity levels, and weight .158 Another neurotrophic factor, granulocyte-colony stimulating factor (G-CSF), was found to preserve motor units and extend survival by 10% when delivered to adult pre-symptomatic SOD1G93A mice via direct intraspinal injection of AAV1.159 Hepatocyte growth factor (HGF) has also been evaluated, using i.t. AAV1160 or intramuscular AAV6161 to treat adult pre-symptomatic SOD1G93A mice, with modest improvements noted in strength and survival using both delivery methods.
Factors that act at the neuromuscular junction have also been assessed, including intramuscular delivery of AAV1-neuregulin to promote collateral sprouting and electrophysiologic function in adult symptomatic SOD1G93A mice. However, there were no effects on strength or survival.162 Systemic administration of AAV9 delivering the gene encoding the neuromuscular junction protein DOK7 preserved the neuromuscular junction, improved locomotion, and modestly increased lifespan in early symptomatic SOD1G93A mice, without affecting motor neuron counts.163 D-amino acid oxidase (DAO) is thought to modulate excitotoxicity, and i.t. delivery of AAV9-DAO to early symptomatic SOD1G93A mice resulted in motor neuron preservation and a modest survival extension.164
Factors with direct effects on misfolded ALS-associated proteins are also being studied. For example, the cytosolic chaperone, macrophage migration inhibitory factor (MIF), inhibits mutant SOD1 misfolding.165 AAV9-mediated expression of MIF after direct intraspinal delivery to neonatal SOD1G93A mice or the slower-progressing loxSOD1G37R mouse model significantly reduced misfolded SOD1 levels in the spinal cord of the SOD1G93A mice and improved strength and survival in both models.166 i.t. delivery of AAV1 expressing a monoclonal antibody targeting misfolded SOD1 to pre-symptomatic adult SOD1G93A mice resulted in a notable 28% extension of lifespan accompanied by reduced misfolded spinal SOD1 and attenuated neuronal stress signals and gliosis.167 A similar strategy was tested in symptomatic TDP-43G348C mice, which manifest cytoplasmic accumulation of TDP-43 as well as memory impairment. AAV9-mediated cortical delivery of an antibody targeting an aggregation-prone region of TDP-43 reduced TDP-43 proteinopathy, cognitive impairment, motor defects, and neuroinflammation.168
Conclusions and perspectives
The ability to provide targeted, sustained treatment gives gene therapy the potential to permanently alter the therapeutic landscape for diseases of the CNS, as it has already done in the case of SMA. ALS in particular is in dire need of impactful, disease-modifying approaches capable of reaching some of the body’s most protected regions, the cerebral cortex and the anterior horn cells of the spinal cord, which can increasingly be achieved through minimally invasive means by harnessing the tremendous potential of gene therapy. Once delivered to their target, these therapies can act through knockdown-mediated amelioration of gain-of-function mechanisms or through delivery of protective agents, enabling approaches ranging from targeting a specific ALS-causing mutation to modifying a common pathologic endpoint across sporadic cases. As we continue to hone our delivery methods, vector specificity, and cargo efficacy, there is great hope that truly impactful treatments for ALS will emerge in the near future.
Acknowledgments
D.A.A. acknowledges funding from the US National Institute of Neurological Disorders and Stroke (NS114106). B.L.D is supported by NS09435 and NS111671 and the CHOP Research Institute. We thank Ashley Robbins for her assistance in generating figures for this review and Lauren Elman and Colin Quinn for their critical reading of the manuscript.
Author contributions
D.A.A. and B.L.D. planned the manuscript. D.A.A. wrote and coordinated the draft and designed the figure and Table 1. D.A.A. and B.L.D. reviewed and revised the manuscript.
Table 1.
Gene therapy-based therapeutic strategies for ALS
| Gene therapy | Date | Model | Route | Outcome | Citation |
|---|---|---|---|---|---|
| Antisense oligonucleotides | |||||
| SOD1 | 2006 | rat; NHP SOD1G93A | i.c.v. (rat), i.t. (NHP) | increased survival; good biodistribution in NHP | Smith et al.74 |
| 2013 | human SOD1-ALS phase I | i.t. | safe; no benefit | Miller et al.75 | |
| 2017 | mouse SOD1G93A | i.c.v. + i.v. | increased survival, strength, and weight | Biferi et al.77 | |
| 2020 | human SOD1-ALS phase I/II | i.t. | safe; reduced CSF SOD1 | Miller et al.76 | |
| current | human SOD1-ALS phase III | i.t. | TBD | ClinicalTrials.gov: NCT02623699 | |
| C9orf72 | 2013 | human iPSC C9-ALS | reduced nuclear RNA foci and excitotoxicity; reversed aberrant gene expression | Donnelly et al.78 | |
| 2013 | human iPSC C9-ALS | reduced nuclear RNA foci and excitotoxicity; reversed aberrant gene expression | Sareen et al.81 | ||
| 2013 | human iPSC C9-ALS; mouse (WT) | reduced nuclear RNA foci; persistent aberrant expression; safe in mice | Lagier-Tourenne et al.80 | ||
| 2016 | mouse C9(450) | i.c.v. | sustained reduction in nuclear RNA foci and DPRs in motor neurons; cognitive benefit | Jiang et al.82 | |
| current | human C9-ALS phase I | i.t. | TBD | ClinicalTrials.gov: NCT03626012 | |
| ATXN2 | 2017 | mouse hTDP-43 | i.c.v. | reduced Atxn2 mRNA; increased survival; improved gait | Becker et al.93 |
| 2018 | human iPSC C9-ALS | reduced nuclear RNA foci and excitotoxicity; reversed aberrant gene expression | Zhang et al.27 | ||
| Current | human ATXN2-ALS & sporadic ALS phase I | i.t. | TBD | ClinicalTrials.gov: NCT04494256 | |
| FUS |
2019 | human FUS-ALS | i.t. | patient passed; further clinical data not reported | Arnold96 |
| Current | human FUS-ALS | i.t. | TBD | Figueiredo97 | |
| AAV-mediated gene silencing | |||||
| AAV-RNAi | |||||
| SOD1 | 2013 | mouse SOD1G93A & SOD1G37R; NHP | i.v. (mice), i.t. (NHP) | increased survival, decreasing with age of administration; reduced SOD1 mRNA in NHP spinal cord | Foust et al.116 |
| 2014 | rat SOD1G93A | cortex | delayed disease onset and increased survival; preserved motor function | Thomsen et al.118 | |
| 2016 | mouse SOD1G93A | i.c.v. | increased survival; delayed paralysis; increase in motor neurons | Stoica et al.119 | |
| 2016 | mouse SOD1G93A; NHP | i.v. (mice), i.t. (NHP) | delayed onset; increased survival; preserved motor and respiratory function in mice; reduced SOD1 mRNA in NHP spinal cord | Borel et al.120 | |
| 2018 | NHP | i.t. | reduced SOD1 mRNA in spinal cord | Borel et al.121 | |
| 2019 | mouse SOD1G93A | tongue, subplural | increased survival; lowered SOD1 mRNA in muscle, tongue, and diaphragm | Keeler et al.125 | |
| 2020 | human SOD1-ALS phase I | i.t. | possible clinical stabilization in 1 patient; lowered spinal cord SOD1 in second, with DRG toxicity and no clear clinical benefit | Mueller et al.122 | |
| 2020 | mouse SOD1G37R; pig; NHP | lumbar subpial | stopped disease progression in mice; strong LMN targeting in large animals | Bravo-Hernandez et al.127 | |
| C9orf72 |
2015 | human iPSC C9-ALS | marked reduction in nuclear foci | Hu et al.132 | |
| 2017 | human iPSC C9-ALS | marked reduction in nuclear foci | Hu et al.133 | ||
| 2019 | (G4C2)44 cells; human iPSC C9-FTD | reduced nuclear foci; silenced C9orf72 in iPSC | Martier et al.134 | ||
| 2019 | human iPSC C9-FTD; mouse C9orf72_3 | striatum | reduced nuclear foci in iPSCs, reduced C9orf72 mRNA in mouse striatum | Martier et al.135 | |
| AAV-CRISPR | |||||
| SOD 1 | 2017 | mouse SOD1G93A | i.v. | increased survival and strength with decreased SOD1 protein in spinal cord | Gaj et al.128 |
| 2020 | mouse SOD1G93A | i.c.v. | increased survival and strength with decreased SOD1 protein in spinal cord | Duan et al.129 | |
| 2020 | mouse SOD1G93A | i.t. | increased survival and strength with decreased SOD1 inclusions in spinal cord | Lim et al.130 | |
| C9orf72 |
2019 | Drosophila C9; human iPSC C9-ALS | reduced apoptotic pathway activation; reduced nuclear foci | Lopez-Gonzalez et al.137 | |
| 2020 | human iPSC C9-ALS | reduced C9orf72 expression; near elimination of dipeptide repeats; reduced axonal degeneration | Krishnan et al.136 | ||
| AAV-mediated gene delivery | |||||
| Neurotrophic support | |||||
| IGF | 2010 | mouse SOD1G93A | i.c.v. | improved motor function and increased survival | Dodge et al.151 |
| 2016 | mouse SOD1G93A | i.m. | preserved motor neurons; increased survival | Allodi et al.148 | |
| 2018 | mouse SOD1G93A | i.m. | preserved motor neurons; increased survival | Lin et al.149 | |
| 2018 | mouse SOD1G93A | i.v. | preserved motor neurons; increased survival | Wang et al.150 | |
| VEGF | 2010 | mouse SOD1G93A | i.c.v. | improved motor function and increased survival | Dodge et al.151 |
| 2013 | feline Lix1−/− (LMN disease) | i.c.v., i.v., or i.c.m. | only i.c.m. delivery resulted in sustained VEGF in spinal cord, without therapeutic benefit | Bucher et al.155 | |
| 2016 | mouse SOD1G93A | i.t. | improved motor function and increased survival | Wang et al.154 | |
| GDNF | 2017 | rat SOD1G93A | i.v. | improved strength, but no effect on survival; worsened cognitive function, decreased activity | Thomsen et al.158 |
| G-CSF | 2011 | mouse SOD1G93A | intra-spinal | preserved motor units; increased survival | Henriques et al.159 |
| HGF |
2019 | mouse SOD1G93A | i.t. | improved motor function and increased survival | Lee et al.160 |
| 2019 | mouse SOD1G93A | i.t. | improved motor function and increased survival | Lee et al.161 | |
| Neuromuscular junction modulators | |||||
| Neuregulin | 2016 | mouse SOD1G93A | i.m. | no effect on strength or survival | Mancuso et al.162 |
| DOK7 | 2017 | mouse SOD1G93A | i.v. | preserved neuromuscular junction, improved locomotion, increased survival; no effect on motor neuron counts | Miyoshi et al.163 |
| DAO | 2017 | mouse SOD1G93A | i.t. | preserved motor neurons, increased survival | Wang et al.164 |
| Targeting misfolded protein | |||||
| MIF | 2019 | mouse SOD1G93A + loxSOD1G37R | intra-spinal | improved strength and increased survival; reduced misfolded SOD1 in SOD1G93A mice | Leyton-Jaimes et al.166 |
| SOD1 Ab | 2014 | mouse SOD1G93A | i.t. | increased survival, reduced misfolded SOD1, reduced neuronal stress | Patel et al.167 |
| TDP-43 Ab | 2019 | mouse TDP-43G348C | cortical | reduced TDP-43 proteinopathy, cognitive impairment, motor defects & neuroinflammation | Pozzi et al.168 |
CSF, cerebrospinal fluid; i.c.m., intra-cisterna magna; i.c.v., intracerebroventricular; i.m., intramuscular; i.t., intrathecal; i.v., intravenous; NHP, nonhuman primate; WT, wild type.
Declaration of interests
B.L.D. is a founder of Spark Therapeutics and Spirovant Sciences and is on the SAB of Patch Bio, Resilience, Saliogen Therapeutics, Panorama Medicines, Homology Medicines, and Spirovant Sciences.
Contributor Information
Defne A. Amado, Email: defne@pennmedicine.upenn.edu.
Beverly L. Davidson, Email: davidsonbl@email.chop.edu.
References
- 1.Oskarsson B., Gendron T.F., Staff N.P. Amyotrophic Lateral Sclerosis: An Update for 2018. Mayo Clin. Proc. 2018;93:1617–1628. doi: 10.1016/j.mayocp.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Martier R., Konstantinova P. Gene Therapy for Neurodegenerative Diseases: Slowing Down the Ticking Clock. Front. Neurosci. 2020;14:580179. doi: 10.3389/fnins.2020.580179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J.J. Overview of current and emerging therapies for amytrophic lateral sclerosis. Am. J. Manag. Care. 2020;26(9, Suppl):S191–S197. doi: 10.37765/ajmc.2020.88483. [DOI] [PubMed] [Google Scholar]
- 4.Bensimon G., Lacomblez L., Meininger V., ALS/Riluzole Study Group A controlled trial of riluzole in amyotrophic lateral sclerosis. N. Engl. J. Med. 1994;330:585–591. doi: 10.1056/NEJM199403033300901. [DOI] [PubMed] [Google Scholar]
- 5.Bellingham M.C. A review of the neural mechanisms of action and clinical efficiency of riluzole in treating amyotrophic lateral sclerosis: what have we learned in the last decade? CNS Neurosci. Ther. 2011;17:4–31. doi: 10.1111/j.1755-5949.2009.00116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abe K., Itoyama Y., Sobue G., Tsuji S., Aoki M., Doyu M., Hamada C., Kondo K., Yoneoka T., Akimot M., et al. Confirmatory double-blind, parallel-group, placebo-controlled study of efficacy and safety of edaravone (MCI-186) in amyotrophic lateral sclerosis patients Amyotroph. Lateral Scler. Frontotemporal Degener. 2014;15:610–617. doi: 10.3109/21678421.2014.959024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Writing Group on Behalf of the Edaravone (MCI-186) ALS 17 Study Group Exploratory double-blind, parallel-group, placebo-controlled extension study of edaravone (MCI-186) in amyotrophic lateral sclerosis Amyotroph. Lateral Scler. Frontotemporal Degener. 2017;18(Suppl 1):20–31. doi: 10.1080/21678421.2017.1362000. [DOI] [PubMed] [Google Scholar]
- 8.Yoshino H., Kimura A. Investigation of the therapeutic effects of edaravone, a free radical scavenger, on amyotrophic lateral sclerosis (Phase II study) Amyotroph. Lateral Scler. 2006;7:241–245. doi: 10.1080/17482960600881870. [DOI] [PubMed] [Google Scholar]
- 9.Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., et al. Ubiquitinated TDP-43 in Frontotemporal Lobar Degeneration and Amyotrophic Lateral Sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 10.Baradaran-Heravi Y., Van Broeckhoven C., van der Zee J. Stress granule mediated protein aggregation and underlying gene defects in the FTD-ALS spectrum. Neurobiol. Dis. 2020;134:104639. doi: 10.1016/j.nbd.2019.104639. [DOI] [PubMed] [Google Scholar]
- 11.Mackenzie I.R.A., Rademakers R. The molecular genetics and neuropathology of frontotemporal lobar degeneration: recent developments. Neurogenetics. 2007;8:237–248. doi: 10.1007/s10048-007-0102-4. [DOI] [PubMed] [Google Scholar]
- 12.Johnson J.K., Diehl J., Mendez M.F., Neuhaus J., Shapira J.S., Forman M., Chute D.J., Roberson E.D., Pace-Savitsky C., Neumann M., et al. Frontotemporal lobar degeneration: demographic characteristics of 353 patients. Arch. Neurol. 2005;62:925–930. doi: 10.1001/archneur.62.6.925. [DOI] [PubMed] [Google Scholar]
- 13.Josephs K.A., Holton J.L., Rossor M.N., Godbolt A.K., Ozawa T., Strand K., Khan N., Al-Sarraj S., Revesz T. Frontotemporal lobar degeneration and ubiquitin immunohistochemistry. Neuropathol. Appl. Neurobiol. 2004;30:369–373. doi: 10.1111/j.1365-2990.2003.00545.x. [DOI] [PubMed] [Google Scholar]
- 14.Guo L., Shorter J. Biology and Pathobiology of TDP-43 and Emergent Therapeutic Strategies. Cold Spring Harb. Perspect. Med. 2017;7:a024554. doi: 10.1101/cshperspect.a024554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freibaum B.D., Chitta R.K., High A.A., Taylor J.P. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J. Proteome Res. 2010;9:1104–1120. doi: 10.1021/pr901076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ling S.C., Polymenidou M., Cleveland D.W. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sephton C.F., Cenik C., Kucukural A., Dammer E.B., Cenik B., Han Y., Dewey C.M., Roth F.P., Herz J., Peng J., et al. Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J. Biol. Chem. 2011;286:1204–1215. doi: 10.1074/jbc.M110.190884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barmada S.J., Skibinski G., Korb E., Rao E.J., Wu J.Y., Finkbeiner S. Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J. Neurosci. 2010;30:639–649. doi: 10.1523/JNEUROSCI.4988-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu E.Y., Cali C.P., Lee E.B. RNA metabolism in neurodegenerative disease. Dis. Model. Mech. 2017;10:509–518. doi: 10.1242/dmm.028613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vanden Broeck L., Callaerts P., Dermaut B. TDP-43-mediated neurodegeneration: towards a loss-of-function hypothesis? Trends Mol. Med. 2014;20:66–71. doi: 10.1016/j.molmed.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 21.Melamed Z., López-Erauskin J., Baughn M.W., Zhang O., Drenner K., Sun Y., Freyermuth F., McMahon M.A., Beccari M.S., Artates J.W., et al. Premature polyadenylation-mediated loss of stathmin-2 is a hallmark of TDP-43-dependent neurodegeneration. Nat. Neurosci. 2019;22:180–190. doi: 10.1038/s41593-018-0293-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cascella R., Capitini C., Fani G., Dobson C.M., Cecchi C., Chiti F. Quantification of the Relative Contributions of Loss-of-function and Gain-of-function Mechanisms in TAR DNA-binding Protein 43 (TDP-43) Proteinopathies. J. Biol. Chem. 2016;291:19437–19448. doi: 10.1074/jbc.M116.737726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boeynaems S., Bogaert E., Van Damme P., Van Den Bosch L. Inside out: the role of nucleocytoplasmic transport in ALS and FTLD. Acta Neuropathol. 2016;132:159–173. doi: 10.1007/s00401-016-1586-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y.-J., Xu Y.F., Cook C., Gendron T.F., Roettges P., Link C.D., Lin W.L., Tong J., Castanedes-Casey M., Ash P., et al. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc. Natl. Acad. Sci. USA. 2009;106:7607–7612. doi: 10.1073/pnas.0900688106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu-Yesucevitz L., Bilgutay A., Zhang Y.J., Vanderweyde T., Citro A., Mehta T., Zaarur N., McKee A., Bowser R., Sherman M., et al. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS ONE. 2010;5:e13250. doi: 10.1371/journal.pone.0013250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y.R., King O.D., Shorter J., Gitler A.D. Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 2013;201:361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang K., Daigle J.G., Cunningham K.M., Coyne A.N., Ruan K., Grima J.C., Bowen K.E., Wadhwa H., Yang P., Rigo F., et al. Stress Granule Assembly Disrupts Nucleocytoplasmic Transport. Cell. 2018;173:958–971.e17. doi: 10.1016/j.cell.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chia R., Chiò A., Traynor B.J. Novel genes associated with amyotrophic lateral sclerosis: diagnostic and clinical implications. Lancet Neurol. 2018;17:94–102. doi: 10.1016/S1474-4422(17)30401-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosen D.R., Siddique T., Patterson D., Figlewicz D.A., Sapp P., Hentati A., Donaldson D., Goto J., O’Regan J.P., Deng H.X., et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 30.Philips T., Rothstein J.D. Rodent Models of Amyotrophic Lateral Sclerosis. Curr. Protoc. Pharmacol. 2015;69:5.67.1–5.67.21. doi: 10.1002/0471141755.ph0567s69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joyce P.I., Fratta P., Fisher E.M.C., Acevedo-Arozena A. SOD1 and TDP-43 animal models of amyotrophic lateral sclerosis: recent advances in understanding disease toward the development of clinical treatments. Mamm. Genome. 2011;22:420–448. doi: 10.1007/s00335-011-9339-1. [DOI] [PubMed] [Google Scholar]
- 32.Mitsumoto H., Brooks B.R., Silani V. Clinical trials in amyotrophic lateral sclerosis: why so many negative trials and how can trials be improved? Lancet Neurol. 2014;13:1127–1138. doi: 10.1016/S1474-4422(14)70129-2. [DOI] [PubMed] [Google Scholar]
- 33.Rothstein J.D. Of mice and men: reconciling preclinical ALS mouse studies and human clinical trials. Ann. Neurol. 2003;53:423–426. doi: 10.1002/ana.10561. [DOI] [PubMed] [Google Scholar]
- 34.Sreedharan J., Blair I.P., Tripathi V.B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J.C., Williams K.L., Buratti E., et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rutherford N.J., Zhang Y.J., Baker M., Gass J.M., Finch N.A., Xu Y.F., Stewart H., Kelley B.J., Kuntz K., Crook R.J., et al. Novel mutations in TARDBP (TDP-43) in patients with familial amyotrophic lateral sclerosis. PLoS Genet. 2008;4:e1000193. doi: 10.1371/journal.pgen.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janssens J., Wils H., Kleinberger G., Joris G., Cuijt I., Ceuterick-de Groote C., Van Broeckhoven C., Kumar-Singh S. Overexpression of ALS-associated p.M337V human TDP-43 in mice worsens disease features compared to wild-type human TDP-43 mice. Mol. Neurobiol. 2013;48:22–35. doi: 10.1007/s12035-013-8427-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wils H., Kleinberger G., Janssens J., Pereson S., Joris G., Cuijt I., Smits V., Ceuterick-de Groote C., Van Broeckhoven C., Kumar-Singh S. TDP-43 transgenic mice develop spastic paralysis and neuronal inclusions characteristic of ALS and frontotemporal lobar degeneration. Proc. Natl. Acad. Sci. USA. 2010;107:3858–3863. doi: 10.1073/pnas.0912417107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwiatkowski T.J., Jr., Bosco D.A., Leclerc A.L., Tamrazian E., Vanderburg C.R., Russ C., Davis A., Gilchrist J., Kasarskis E.J., Munsat T., et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 39.Vance C., Rogelj B., Hortobágyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elden A.C., Kim H.J., Hart M.P., Chen-Plotkin A.S., Johnson B.S., Fang X., Armakola M., Geser F., Greene R., Lu M.M., et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renton A.E., Majounie E., Waite A., Simón-Sánchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H., van Swieten J.C., Myllykangas L., et al. ITALSGEN Consortium A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J., et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Majounie E., Renton A.E., Mok K., Dopper E.G., Waite A., Rollinson S., Chiò A., Restagno G., Nicolaou N., Simon-Sanchez J., et al. Chromosome 9-ALS/FTD Consortium. French research network on FTLD/FTLD/ALS. ITALSGEN Consortium Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11:323–330. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y., Pattamatta A., Zu T., Reid T., Bardhi O., Borchelt D.R., Yachnis A.T., Ranum L.P. C9orf72 BAC Mouse Model with Motor Deficits and Neurodegenerative Features of ALS/FTD. Neuron. 2016;90:521–534. doi: 10.1016/j.neuron.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 45.Batra R., Lee C.W. Mouse Models of C9orf72 Hexanucleotide Repeat Expansion in Amyotrophic Lateral Sclerosis/ Frontotemporal Dementia. Front. Cell. Neurosci. 2017;11:196. doi: 10.3389/fncel.2017.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Renton A.E., Chiò A., Traynor B.J. State of play in amyotrophic lateral sclerosis genetics. Nat. Neurosci. 2014;17:17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cappella M., Ciotti C., Cohen-Tannoudji M., Biferi M.G. Gene Therapy for ALS-A Perspective. Int. J. Mol. Sci. 2019;20:4388. doi: 10.3390/ijms20184388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murlidharan G., Samulski R.J., Asokan A. Biology of adeno-associated viral vectors in the central nervous system. Front. Mol. Neurosci. 2014;7:76. doi: 10.3389/fnmol.2014.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tosolini A.P., Sleigh J.N. Motor Neuron Gene Therapy: Lessons from Spinal Muscular Atrophy for Amyotrophic Lateral Sclerosis. Front. Mol. Neurosci. 2017;10:405. doi: 10.3389/fnmol.2017.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Colella P., Ronzitti G., Mingozzi F. Emerging Issues in AAV-Mediated In Vivo Gene Therapy. Mol. Ther. Methods Clin. Dev. 2017;8:87–104. doi: 10.1016/j.omtm.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duque S., Joussemet B., Riviere C., Marais T., Dubreil L., Douar A.M., Fyfe J., Moullier P., Colle M.A., Barkats M. Intravenous administration of self-complementary AAV9 enables transgene delivery to adult motor neurons. Mol. Ther. 2009;17:1187–1196. doi: 10.1038/mt.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanguy Y., Biferi M.G., Besse A., Astord S., Cohen-Tannoudji M., Marais T., Barkats M. Systemic AAVrh10 provides higher transgene expression than AAV9 in the brain and the spinal cord of neonatal mice. Front. Mol. Neurosci. 2015;8:36. doi: 10.3389/fnmol.2015.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Foust K.D., Wang X., McGovern V.L., Braun L., Bevan A.K., Haidet A.M., Le T.T., Morales P.R., Rich M.M., Burghes A.H., Kaspar B.K. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat. Biotechnol. 2010;28:271–274. doi: 10.1038/nbt.1610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Dominguez E., Marais T., Chatauret N., Benkhelifa-Ziyyat S., Duque S., Ravassard P., Carcenac R., Astord S., Pereira de Moura A., Voit T., Barkats M. Intravenous scAAV9 delivery of a codon-optimized SMN1 sequence rescues SMA mice. Hum. Mol. Genet. 2011;20:681–693. doi: 10.1093/hmg/ddq514. [DOI] [PubMed] [Google Scholar]
- 55.Valori C.F., Ning K., Wyles M., Mead R.J., Grierson A.J., Shaw P.J., Azzouz M. Systemic delivery of scAAV9 expressing SMN prolongs survival in a model of spinal muscular atrophy. Sci. Transl. Med. 2010;2:35ra42. doi: 10.1126/scitranslmed.3000830. [DOI] [PubMed] [Google Scholar]
- 56.Mendell J.R., Al-Zaidy S., Shell R., Arnold W.D., Rodino-Klapac L.R., Prior T.W., Lowes L., Alfano L., Berry K., Church K., et al. Single-Dose Gene-Replacement Therapy for Spinal Muscular Atrophy. N. Engl. J. Med. 2017;377:1713–1722. doi: 10.1056/NEJMoa1706198. [DOI] [PubMed] [Google Scholar]
- 57.Deverman B.E., Pravdo P.L., Simpson B.P., Kumar S.R., Chan K.Y., Banerjee A., Wu W.L., Yang B., Huber N., Pasca S.P., Gradinaru V. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 2016;34:204–209. doi: 10.1038/nbt.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chan K.Y., Jang M.J., Yoo B.B., Greenbaum A., Ravi N., Wu W.L., Sánchez-Guardado L., Lois C., Mazmanian S.K., Deverman B.E., Gradinaru V. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 2017;20:1172–1179. doi: 10.1038/nn.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Challis R.C., Ravindra Kumar S., Chan K.Y., Challis C., Beadle K., Jang M.J., Kim H.M., Rajendran P.S., Tompkins J.D., Shivkumar K., et al. Systemic AAV vectors for widespread and targeted gene delivery in rodents. Nat. Protoc. 2019;14:379–414. doi: 10.1038/s41596-018-0097-3. [DOI] [PubMed] [Google Scholar]
- 60.Azzouz M. Gene Therapy for ALS: progress and prospects. Biochim. Biophys. Acta. 2006;1762:1122–1127. doi: 10.1016/j.bbadis.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 61.Choudhury S.R., Hudry E., Maguire C.A., Sena-Esteves M., Breakefield X.O., Grandi P. Viral vectors for therapy of neurologic diseases. Neuropharmacology. 2017;120:63–80. doi: 10.1016/j.neuropharm.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suzuki M., Svendsen C.N. Ex Vivo Gene Therapy Using Human Mesenchymal Stem Cells to Deliver Growth Factors in the Skeletal Muscle of a Familial ALS Rat Model. Methods Mol. Biol. 2016;1382:325–336. doi: 10.1007/978-1-4939-3271-9_24. [DOI] [PubMed] [Google Scholar]
- 63.Ly C.V., Miller T.M. Emerging antisense oligonucleotide and viral therapies for amyotrophic lateral sclerosis. Curr. Opin. Neurol. 2018;31:648–654. doi: 10.1097/WCO.0000000000000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schoch K.M., Miller T.M. Antisense Oligonucleotides: Translation from Mouse Models to Human Neurodegenerative Diseases. Neuron. 2017;94:1056–1070. doi: 10.1016/j.neuron.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bennett C.F., Swayze E.E. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 66.Passini M.A., Bu J., Richards A.M., Kinnecom C., Sardi S.P., Stanek L.M., Hua Y., Rigo F., Matson J., Hung G., et al. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci. Transl. Med. 2011;3:72ra18. doi: 10.1126/scitranslmed.3001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chiriboga C.A., Swoboda K.J., Darras B.T., Iannaccone S.T., Montes J., De Vivo D.C., Norris D.A., Bennett C.F., Bishop K.M. Results from a phase 1 study of nusinersen (ISIS-SMN(Rx)) in children with spinal muscular atrophy. Neurology. 2016;86:890–897. doi: 10.1212/WNL.0000000000002445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Finkel R.S., Chiriboga C.A., Vajsar J., Day J.W., Montes J., De Vivo D.C., Yamashita M., Rigo F., Hung G., Schneider E., et al. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet. 2016;388:3017–3026. doi: 10.1016/S0140-6736(16)31408-8. [DOI] [PubMed] [Google Scholar]
- 69.Finkel R.S., Mercuri E., Darras B.T., Connolly A.M., Kuntz N.L., Kirschner J., Chiriboga C.A., Saito K., Servais L., Tizzano E., et al. ENDEAR Study Group Nusinersen versus Sham Control in Infantile-Onset Spinal Muscular Atrophy. N. Engl. J. Med. 2017;377:1723–1732. doi: 10.1056/NEJMoa1702752. [DOI] [PubMed] [Google Scholar]
- 70.Ekhtiari Bidhendi E., Bergh J., Zetterström P., Forsberg K., Pakkenberg B., Andersen P.M., Marklund S.L., Brännström T. Mutant superoxide dismutase aggregates from human spinal cord transmit amyotrophic lateral sclerosis. Acta Neuropathol. 2018;136:939–953. doi: 10.1007/s00401-018-1915-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gurney M.E., Pu H., Chiu A.Y., Dal Canto M.C., Polchow C.Y., Alexander D.D., Caliendo J., Hentati A., Kwon Y.W., Deng H.X., et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 72.Ripps M.E., Huntley G.W., Hof P.R., Morrison J.H., Gordon J.W. Transgenic mice expressing an altered murine superoxide dismutase gene provide an animal model of amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA. 1995;92:689–693. doi: 10.1073/pnas.92.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nagai M., Aoki M., Miyoshi I., Kato M., Pasinelli P., Kasai N., Brown R.H., Jr., Itoyama Y. Rats expressing human cytosolic copper-zinc superoxide dismutase transgenes with amyotrophic lateral sclerosis: associated mutations develop motor neuron disease. J. Neurosci. 2001;21:9246–9254. doi: 10.1523/JNEUROSCI.21-23-09246.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smith R.A., Miller T.M., Yamanaka K., Monia B.P., Condon T.P., Hung G., Lobsiger C.S., Ward C.M., McAlonis-Downes M., Wei H., et al. Antisense oligonucleotide therapy for neurodegenerative disease. J. Clin. Invest. 2006;116:2290–2296. doi: 10.1172/JCI25424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miller T.M., Pestronk A., David W., Rothstein J., Simpson E., Appel S.H., Andres P.L., Mahoney K., Allred P., Alexander K., et al. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomised, first-in-man study. Lancet Neurol. 2013;12:435–442. doi: 10.1016/S1474-4422(13)70061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miller T., Cudkowicz M., Shaw P.J., Andersen P.M., Atassi N., Bucelli R.C., Genge A., Glass J., Ladha S., Ludolph A.L., et al. Phase 1-2 Trial of Antisense Oligonucleotide Tofersen for SOD1 ALS. N. Engl. J. Med. 2020;383:109–119. doi: 10.1056/NEJMoa2003715. [DOI] [PubMed] [Google Scholar]
- 77.Biferi M.G., Cohen-Tannoudji M., Cappelletto A., Giroux B., Roda M., Astord S., Marais T., Bos C., Voit T., Ferry A., Barkats M. A New AAV10-U7-Mediated Gene Therapy Prolongs Survival and Restores Function in an ALS Mouse Model. Mol. Ther. 2017;25:2038–2052. doi: 10.1016/j.ymthe.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Donnelly C.J., Zhang P.W., Pham J.T., Haeusler A.R., Mistry N.A., Vidensky S., Daley E.L., Poth E.M., Hoover B., Fines D.M., et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Balendra R., Isaacs A.M. C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat. Rev. Neurol. 2018;14:544–558. doi: 10.1038/s41582-018-0047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lagier-Tourenne C., Baughn M., Rigo F., Sun S., Liu P., Li H.R., Jiang J., Watt A.T., Chun S., Katz M., et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc. Natl. Acad. Sci. USA. 2013;110:E4530–E4539. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sareen D., O’Rourke J.G., Meera P., Muhammad A.K., Grant S., Simpkinson M., Bell S., Carmona S., Ornelas L., Sahabian A., et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci. Transl. Med. 2013;5:208ra149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jiang J., Zhu Q., Gendron T.F., Saberi S., McAlonis-Downes M., Seelman A., Stauffer J.E., Jafar-Nejad P., Drenner K., Schulte D., et al. Gain of Toxicity from ALS/FTD-Linked Repeat Expansions in C9ORF72 Is Alleviated by Antisense Oligonucleotides Targeting GGGGCC-Containing RNAs. Neuron. 2016;90:535–550. doi: 10.1016/j.neuron.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Imbert G., Saudou F., Yvert G., Devys D., Trottier Y., Garnier J.M., Weber C., Mandel J.L., Cancel G., Abbas N., et al. Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats. Nat. Genet. 1996;14:285–291. doi: 10.1038/ng1196-285. [DOI] [PubMed] [Google Scholar]
- 84.Pulst S.M., Nechiporuk A., Nechiporuk T., Gispert S., Chen X.N., Lopes-Cendes I., Pearlman S., Starkman S., Orozco-Diaz G., Lunkes A., et al. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat. Genet. 1996;14:269–276. doi: 10.1038/ng1196-269. [DOI] [PubMed] [Google Scholar]
- 85.Sanpei K., Takano H., Igarashi S., Sato T., Oyake M., Sasaki H., Wakisaka A., Tashiro K., Ishida Y., Ikeuchi T., et al. Identification of the spinocerebellar ataxia type 2 gene using a direct identification of repeat expansion and cloning technique, DIRECT. Nat. Genet. 1996;14:277–284. doi: 10.1038/ng1196-277. [DOI] [PubMed] [Google Scholar]
- 86.Fernandez M., McClain M.E., Martinez R.A., Snow K., Lipe H., Ravits J., Bird T.D., La Spada A.R. Late-onset SCA2: 33 CAG repeats are sufficient to cause disease. Neurology. 2000;55:569–572. doi: 10.1212/wnl.55.4.569. [DOI] [PubMed] [Google Scholar]
- 87.Yu Z., Zhu Y., Chen-Plotkin A.S., Clay-Falcone D., McCluskey L., Elman L., Kalb R.G., Trojanowski J.Q., Lee V.M., Van Deerlin V.M., et al. PolyQ repeat expansions in ATXN2 associated with ALS are CAA interrupted repeats. PLoS ONE. 2011;6:e17951. doi: 10.1371/journal.pone.0017951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee T., Li Y.R., Ingre C., Weber M., Grehl T., Gredal O., de Carvalho M., Meyer T., Tysnes O.B., Auburger G., et al. Ataxin-2 intermediate-length polyglutamine expansions in European ALS patients. Hum. Mol. Genet. 2011;20:1697–1700. doi: 10.1093/hmg/ddr045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Van Damme P., Veldink J.H., van Blitterswijk M., Corveleyn A., van Vught P.W., Thijs V., Dubois B., Matthijs G., van den Berg L.H., Robberecht W. Expanded ATXN2 CAG repeat size in ALS identifies genetic overlap between ALS and SCA2. Neurology. 2011;76:2066–2072. doi: 10.1212/WNL.0b013e31821f445b. [DOI] [PubMed] [Google Scholar]
- 90.Nonhoff U., Ralser M., Welzel F., Piccini I., Balzereit D., Yaspo M.L., Lehrach H., Krobitsch S. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol. Biol. Cell. 2007;18:1385–1396. doi: 10.1091/mbc.E06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hart M.P., Gitler A.D. ALS-associated ataxin 2 polyQ expansions enhance stress-induced caspase 3 activation and increase TDP-43 pathological modifications. J. Neurosci. 2012;32:9133–9142. doi: 10.1523/JNEUROSCI.0996-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van den Heuvel D.M.A., Harschnitz O., van den Berg L.H., Pasterkamp R.J. Taking a risk: a therapeutic focus on ataxin-2 in amyotrophic lateral sclerosis? Trends Mol. Med. 2014;20:25–35. doi: 10.1016/j.molmed.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 93.Becker L.A., Huang B., Bieri G., Ma R., Knowles D.A., Jafar-Nejad P., Messing J., Kim H.J., Soriano A., Auburger G., et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature. 2017;544:367–371. doi: 10.1038/nature22038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Conte A., Lattante S., Zollino M., Marangi G., Luigetti M., Del Grande A., Servidei S., Trombetta F., Sabatelli M. P525L FUS mutation is consistently associated with a severe form of juvenile amyotrophic lateral sclerosis. Neuromuscul. Disord. 2012;22:73–75. doi: 10.1016/j.nmd.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 95.Cappella M., Pradat P.-F., Querin G., Biferi M.G. Beyond the Traditional Clinical Trials for Amyotrophic Lateral Sclerosis and The Future Impact of Gene Therapy. J. Neuromuscul. Dis. 2021;8:25–38. doi: 10.3233/JND-200531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Arnold C. Tailored treatment for ALS poised to move ahead. Nat. Med. 2019 doi: 10.1038/d41591-019-00013-w. Published online May 30, 2019. [DOI] [PubMed] [Google Scholar]
- 97.Figueiredo M. 2020. Collaboration Funds Experimental Therapy for Rare FUS-ALS.https://alsnewstoday.com/news-posts/2020/03/16/jacifusen-collaboration-funds-experimental-therapy-for-patients-with-rare-fus-als/ [Google Scholar]
- 98.ALS Association. 2021. The ALS Association and Project ALS to Fund Columbia University Drug Trial for Patients with Rare Genetic Forms of ALS.http://web.alsa.org/site/PageNavigator/blog_021420.html [Google Scholar]
- 99.Borel F., Kay M.A., Mueller C. Recombinant AAV as a platform for translating the therapeutic potential of RNA interference. Mol. Ther. 2014;22:692–701. doi: 10.1038/mt.2013.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Boudreau R.L., Martins I., Davidson B.L. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol. Ther. 2009;17:169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McBride J.L., Boudreau R.L., Harper S.Q., Staber P.D., Monteys A.M., Martins I., Gilmore B.L., Burstein H., Peluso R.W., Polisky B., et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc. Natl. Acad. Sci. USA. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Boudreau R.L., Spengler R.M., Hylock R.H., Kusenda B.J., Davis H.A., Eichmann D.A., Davidson B.L. siSPOTR: a tool for designing highly specific and potent siRNAs for human and mouse. Nucleic Acids Res. 2013;41:e9. doi: 10.1093/nar/gks797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Evers M.M., Miniarikova J., Juhas S., Vallès A., Bohuslavova B., Juhasova J., Skalnikova H.K., Vodicka P., Valekova I., Brouwers C., et al. AAV5-miHTT Gene Therapy Demonstrates Broad Distribution and Strong Human Mutant Huntingtin Lowering in a Huntington’s Disease Minipig Model. Mol. Ther. 2018;26:2163–2177. doi: 10.1016/j.ymthe.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yun Y., Ha Y. CRISPR/Cas9-Mediated Gene Correction to Understand ALS. Int. J. Mol. Sci. 2020;21:3801. doi: 10.3390/ijms21113801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Anzalone A.V., Koblan L.W., Liu D.R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020;38:824–844. doi: 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- 108.Burmistrz M., Krakowski K., Krawczyk-Balska A. RNA-Targeting CRISPR-Cas Systems and Their Applications. Int. J. Mol. Sci. 2020;21:1122. doi: 10.3390/ijms21031122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Doudna J.A. The promise and challenge of therapeutic genome editing. Nature. 2020;578:229–236. doi: 10.1038/s41586-020-1978-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang D., Zhang F., Gao G. CRISPR-Based Therapeutic Genome Editing: Strategies and In Vivo Delivery by AAV Vectors. Cell. 2020;181:136–150. doi: 10.1016/j.cell.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B., Shmakov S., Makarova K.S., Semenova E., Minakhin L., et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Abudayyeh O.O., Gootenberg J.S., Essletzbichler P., Han S., Joung J., Belanto J.J., Verdine V., Cox D.B.T., Kellner M.J., Regev A., et al. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Konermann S., Lotfy P., Brideau N.J., Oki J., Shokhirev M.N., Hsu P.D. Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell. 2018;173:665–676.e14. doi: 10.1016/j.cell.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M., Chen P.J., Wilson C., Newby G.A., Raguram A., Liu D.R. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Foust K.D., Salazar D.L., Likhite S., Ferraiuolo L., Ditsworth D., Ilieva H., Meyer K., Schmelzer L., Braun L., Cleveland D.W., Kaspar B.K. Therapeutic AAV9-mediated suppression of mutant SOD1 slows disease progression and extends survival in models of inherited ALS. Mol. Ther. 2013;21:2148–2159. doi: 10.1038/mt.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Foust K.D., Nurre E., Montgomery C.L., Hernandez A., Chan C.M., Kaspar B.K. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat. Biotechnol. 2009;27:59–65. doi: 10.1038/nbt.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Thomsen G.M., Gowing G., Latter J., Chen M., Vit J.P., Staggenborg K., Avalos P., Alkaslasi M., Ferraiuolo L., Likhite S., et al. Delayed disease onset and extended survival in the SOD1G93A rat model of amyotrophic lateral sclerosis after suppression of mutant SOD1 in the motor cortex. J. Neurosci. 2014;34:15587–15600. doi: 10.1523/JNEUROSCI.2037-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stoica L., Todeasa S.H., Cabrera G.T., Salameh J.S., ElMallah M.K., Mueller C., Brown R.H., Jr., Sena-Esteves M. Adeno-associated virus-delivered artificial microRNA extends survival and delays paralysis in an amyotrophic lateral sclerosis mouse model. Ann. Neurol. 2016;79:687–700. doi: 10.1002/ana.24618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Borel F., Gernoux G., Cardozo B., Metterville J.P., Toro Cabrera G.C., Song L., Su Q., Gao G.P., Elmallah M.K., Brown R.H., Jr., Mueller C. Therapeutic rAAVrh10 Mediated SOD1 Silencing in Adult SOD1(G93A) Mice and Nonhuman Primates. Hum. Gene Ther. 2016;27:19–31. doi: 10.1089/hum.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Borel F., Gernoux G., Sun H., Stock R., Blackwood M., Brown R.H., Jr., Mueller C. Safe and effective superoxide dismutase 1 silencing using artificial microRNA in macaques. Sci. Transl. Med. 2018;10:eaau6414. doi: 10.1126/scitranslmed.aau6414. [DOI] [PubMed] [Google Scholar]
- 122.Mueller C., Berry J.D., McKenna-Yasek D.M., Gernoux G., Owegi M.A., Pothier L.M., Douthwright C.L., Gelevski D., Luppino S.D., Blackwood M., et al. SOD1 Suppression with Adeno-Associated Virus and MicroRNA in Familial ALS. N. Engl. J. Med. 2020;383:151–158. doi: 10.1056/NEJMoa2005056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hinderer C., Katz N., Buza E.L., Dyer C., Goode T., Bell P., Richman L.K., Wilson J.M. Severe Toxicity in Nonhuman Primates and Piglets Following High-Dose Intravenous Administration of an Adeno-Associated Virus Vector Expressing Human SMN. Hum. Gene Ther. 2018;29:285–298. doi: 10.1089/hum.2018.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hordeaux J., Buza E.L., Dyer C., Goode T., Mitchell T.W., Richman L., Denton N., Hinderer C., Katz N., Schmid R., et al. Adeno-Associated Virus-Induced Dorsal Root Ganglion Pathology. Hum. Gene Ther. 2020;31:808–818. doi: 10.1089/hum.2020.167. [DOI] [PubMed] [Google Scholar]
- 125.Keeler A.M., Zieger M., Semple C., Pucci L., Veinbachs A., Brown R.H., Jr., Mueller C., ElMallah M.K. Intralingual and Intrapleural AAV Gene Therapy Prolongs Survival in a SOD1 ALS Mouse Model. Mol. Ther. Methods Clin. Dev. 2019;17:246–257. doi: 10.1016/j.omtm.2019.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hordeaux J., Buza E.L., Jeffrey B., Song C., Jahan T., Yuan Y., Zhu Y., Bell P., Li M., Chichester J.A., et al. MicroRNA-mediated inhibition of transgene expression reduces dorsal root ganglion toxicity by AAV vectors in primates. Sci. Transl. Med. 2020;12:eaba9188. doi: 10.1126/scitranslmed.aba9188. [DOI] [PubMed] [Google Scholar]
- 127.Bravo-Hernandez M., Tadokoro T., Navarro M.R., Platoshyn O., Kobayashi Y., Marsala S., Miyanohara A., Jushas S., Juhasova J., Skalnikova H., et al. Spinal subpial delivery of AAV9 enables widespread gene silencing and blocks motoneuron degeneration in ALS. Nat. Med. 2020;26:118–130. doi: 10.1038/s41591-019-0674-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gaj T., Ojala D.S., Ekman F.K., Byrne L.C., Limsirichai P., Schaffer D.V. In vivo genome editing improves motor function and extends survival in a mouse model of ALS. Sci. Adv. 2017;3:eaar3952. doi: 10.1126/sciadv.aar3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Duan W., Guo M., Yi L., Liu Y., Li Z., Ma Y., Zhang G., Liu Y., Bu H., Song X., et al. The deletion of mutant SOD1 via CRISPR/Cas9/sgRNA prolongs survival in an amyotrophic lateral sclerosis mouse model. Gene Ther. 2020;27:157–169. doi: 10.1038/s41434-019-0116-1. [DOI] [PubMed] [Google Scholar]
- 130.Lim C.K.W., Gapinske M., Brooks A.K., Woods W.S., Powell J.E., Zeballos C M.A., Winter J., Perez-Pinera P., Gaj T. Treatment of a Mouse Model of ALS by In Vivo Base Editing. Mol. Ther. 2020;28:1177–1189. doi: 10.1016/j.ymthe.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Petri S., Meister G. SsiRNA design principles and off-target effects. Methods Mol. Biol. 2013;986:59–71. doi: 10.1007/978-1-62703-311-4_4. [DOI] [PubMed] [Google Scholar]
- 132.Hu J., Liu J., Li L., Gagnon K.T., Corey D.R. Engineering Duplex RNAs for Challenging Targets: Recognition of GGGGCC/CCCCGG Repeats at the ALS/FTD C9orf72 Locus. Chem. Biol. 2015;22:1505–1511. doi: 10.1016/j.chembiol.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hu J., Rigo F., Prakash T.P., Corey D.R. Recognition of c9orf72 Mutant RNA by Single-Stranded Silencing RNAs. Nucleic Acid Ther. 2017;27:87–94. doi: 10.1089/nat.2016.0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Martier R., Liefhebber J.M., Miniarikova J., van der Zon T., Snapper J., Kolder I., Petry H., van Deventer S.J., Evers M.M., Konstantinova P. Artificial MicroRNAs Targeting C9orf72 Can Reduce Accumulation of Intra-nuclear Transcripts in ALS and FTD Patients. Mol. Ther. Nucleic Acids. 2019;14:593–608. doi: 10.1016/j.omtn.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Martier R., Liefhebber J.M., García-Osta A., Miniarikova J., Cuadrado-Tejedor M., Espelosin M., Ursua S., Petry H., van Deventer S.J., Evers M.M., Konstantinova P. Targeting RNA-Mediated Toxicity in C9orf72 ALS and/or FTD by RNAi-Based Gene Therapy. Mol. Ther. Nucleic Acids. 2019;16:26–37. doi: 10.1016/j.omtn.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Krishnan G., Zhang Y., Gu Y., Kankel M.W., Gao F.B., Almeida S. CRISPR deletion of the C9ORF72 promoter in ALS/FTD patient motor neurons abolishes production of dipeptide repeat proteins and rescues neurodegeneration. Acta Neuropathol. 2020;140:81–84. doi: 10.1007/s00401-020-02154-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lopez-Gonzalez R., Yang D., Pribadi M., Kim T.S., Krishnan G., Choi S.Y., Lee S., Coppola G., Gao F.B. Partial inhibition of the overactivated Ku80-dependent DNA repair pathway rescues neurodegeneration in C9ORF72-ALS/FTD. Proc. Natl. Acad. Sci. USA. 2019;116:9628–9633. doi: 10.1073/pnas.1901313116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Scoles D.R., Meera P., Schneider M., Paul S., Dansithong W., Figueroa K.P., et al. Antisense oligonucleotide therapy for spinocerebellar ataxia type 2. Nature. 2017;544:362–366. doi: 10.1038/nature22044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Keiser M.S., Boudreau R.L., Davidson B.L. Broad therapeutic benefit after RNAi expression vector delivery to deep cerebellar nuclei: implications for spinocerebellar ataxia type 1 therapy. Mol. Ther. 2014;22:588–595. doi: 10.1038/mt.2013.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Keiser M.S., Monteys A.M., Corbau R., Gonzalez-Alegre P., Davidson B.L. RNAi prevents and reverses phenotypes induced by mutant human ataxin-1. Ann. Neurol. 2016;80:754–765. doi: 10.1002/ana.24789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nóbrega C., Nascimento-Ferreira I., Onofre I., Albuquerque D., Hirai H., Déglon N., de Almeida L.P. Silencing mutant ataxin-3 rescues motor deficits and neuropathology in Machado-Joseph disease transgenic mice. PLoS ONE. 2013;8:e52396. doi: 10.1371/journal.pone.0052396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nóbrega C., Nascimento-Ferreira I., Onofre I., Albuquerque D., Déglon N., de Almeida L.P. RNA interference mitigates motor and neuropathological deficits in a cerebellar mouse model of Machado-Joseph disease. PLoS ONE. 2014;9:e100086. doi: 10.1371/journal.pone.0100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ramachandran P.S., Boudreau R.L., Schaefer K.A., La Spada A.R., Davidson B.L. Nonallele specific silencing of ataxin-7 improves disease phenotypes in a mouse model of SCA7. Mol. Ther. 2014;22:1635–1642. doi: 10.1038/mt.2014.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ouyang S., Xie Y., Xiong Z., Yang Y., Xian Y., Ou Z., Song B., Chen Y., Xie Y., Li H., Sun X. CRISPR/Cas9-Targeted Deletion of Polyglutamine in Spinocerebellar Ataxia Type 3-Derived Induced Pluripotent Stem Cells. Stem Cells Dev. 2018;27:756–770. doi: 10.1089/scd.2017.0209. [DOI] [PubMed] [Google Scholar]
- 145.Buijsen R.A.M., Toonen L.J.A., Gardiner S.L., van Roon-Mom W.M.C. Genetics, Mechanisms, and Therapeutic Progress in Polyglutamine Spinocerebellar Ataxias. Neurotherapeutics. 2019;16:263–286. doi: 10.1007/s13311-018-00696-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Petrov D., Mansfield C., Moussy A., Hermine O. ALS Clinical Trials Review: 20 Years of Failure. Are We Any Closer to Registering a New Treatment? Front. Aging Neurosci. 2017;9:68. doi: 10.3389/fnagi.2017.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bartus R.T., Johnson E.M., Jr. Clinical tests of neurotrophic factors for human neurodegenerative diseases, part 1: Where have we been and what have we learned? Neurobiol. Dis. 2017;97:156–168. doi: 10.1016/j.nbd.2016.03.027. [DOI] [PubMed] [Google Scholar]
- 148.Allodi I., Comley L., Nichterwitz S., Nizzardo M., Simone C., Benitez J.A., Cao M., Corti S., Hedlund E. Differential neuronal vulnerability identifies IGF-2 as a protective factor in ALS. Sci. Rep. 2016;6:25960. doi: 10.1038/srep25960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lin H., Hu H., Duan W., Liu Y., Tan G., Li Z., Liu Y., Deng B., Song X., Wang W., et al. Intramuscular Delivery of scAAV9-hIGF1 Prolongs Survival in the hSOD1 G93A ALS Mouse Model via Upregulation of D-Amino Acid Oxidase. Mol. Neurobiol. 2018;55:682–695. doi: 10.1007/s12035-016-0335-z. [DOI] [PubMed] [Google Scholar]
- 150.Wang W., Wen D., Duan W., Yin J., Cui C., Wang Y., Li Z., Liu Y., Li C. Systemic administration of scAAV9-IGF1 extends survival in SOD1G93A ALS mice via inhibiting p38 MAPK and the JNK-mediated apoptosis pathway. Brain Res. Bull. 2018;139:203–210. doi: 10.1016/j.brainresbull.2018.02.015. [DOI] [PubMed] [Google Scholar]
- 151.Dodge J.C., Treleaven C.M., Fidler J.A., Hester M., Haidet A., Handy C., Rao M., Eagle A., Matthews J.C., Taksir T.V., et al. AAV4-mediated expression of IGF-1 and VEGF within cellular components of the ventricular system improves survival outcome in familial ALS mice. Mol. Ther. 2010;18:2075–2084. doi: 10.1038/mt.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li Z., Duan W., Cui C., Liu Y., Li C., Liu Y. AAV9-IGF1 protects TDP-25 cells from apoptosis and oxidative stress partly via up-regulating the expression of VEGF in vitro. Neurosci. Lett. 2017;640:123–129. doi: 10.1016/j.neulet.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 153.Keifer O.P., Jr., O’Connor D.M., Boulis N.M. Gene and protein therapies utilizing VEGF for ALS. Pharmacol. Ther. 2014;141:261–271. doi: 10.1016/j.pharmthera.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang Y., Duan W., Wang W., Wen D., Liu Y., Liu Y., Li Z., Hu H., Lin H., Cui C., et al. scAAV9-VEGF prolongs the survival of transgenic ALS mice by promoting activation of M2 microglia and the PI3K/Akt pathway. Brain Res. 2016;1648:1–10. doi: 10.1016/j.brainres.2016.06.043. [DOI] [PubMed] [Google Scholar]
- 155.Bucher T., Colle M.A., Wakeling E., Dubreil L., Fyfe J., Briot-Nivard D., Maquigneau M., Raoul S., Cherel Y., Astord S., et al. scAAV9 intracisternal delivery results in efficient gene transfer to the central nervous system of a feline model of motor neuron disease. Hum. Gene Ther. 2013;24:670–682. doi: 10.1089/hum.2012.218. [DOI] [PubMed] [Google Scholar]
- 156.Wang L.-J., Lu Y.Y., Muramatsu S., Ikeguchi K., Fujimoto K., Okada T., Mizukami H., Matsushita T., Hanazono Y., Kume A., et al. Neuroprotective effects of glial cell line-derived neurotrophic factor mediated by an adeno-associated virus vector in a transgenic animal model of amyotrophic lateral sclerosis. J. Neurosci. 2002;22:6920–6928. doi: 10.1523/JNEUROSCI.22-16-06920.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Li W., Brakefield D., Pan Y., Hunter D., Myckatyn T.M., Parsadanian A. Muscle-derived but not centrally derived transgene GDNF is neuroprotective in G93A-SOD1 mouse model of ALS. Exp. Neurol. 2007;203:457–471. doi: 10.1016/j.expneurol.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 158.Thomsen G.M., Alkaslasi M., Vit J.P., Lawless G., Godoy M., Gowing G., Shelest O., Svendsen C.N. Systemic injection of AAV9-GDNF provides modest functional improvements in the SOD1G93A ALS rat but has adverse side effects. Gene Ther. 2017;24:245–252. doi: 10.1038/gt.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Henriques A., Pitzer C., Dittgen T., Klugmann M., Dupuis L., Schneider A. CNS-targeted viral delivery of G-CSF in an animal model for ALS: improved efficacy and preservation of the neuromuscular unit. Mol. Ther. 2011;19:284–292. doi: 10.1038/mt.2010.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lee S.H., Kim S., Lee N., Lee J., Yu S.S., Kim J.H., Kim S. Intrathecal delivery of recombinant AAV1 encoding hepatocyte growth factor improves motor functions and protects neuromuscular system in the nerve crush and SOD1-G93A transgenic mouse models. Acta Neuropathol. Commun. 2019;7:96. doi: 10.1186/s40478-019-0737-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lee S.H., Lee N., Kim S., Lee J., Choi W., Yu S.S., Kim J.H., Kim S. Intramuscular delivery of HGF-expressing recombinant AAV improves muscle integrity and alleviates neurological symptoms in the nerve crush and SOD1-G93A transgenic mouse models. Biochem. Biophys. Res. Commun. 2019;517:452–457. doi: 10.1016/j.bbrc.2019.07.105. [DOI] [PubMed] [Google Scholar]
- 162.Mancuso R., Martínez-Muriana A., Leiva T., Gregorio D., Ariza L., Morell M., Esteban-Pérez J., García-Redondo A., Calvo A.C., Atencia-Cibreiro G., et al. Neuregulin-1 promotes functional improvement by enhancing collateral sprouting in SOD1(G93A) ALS mice and after partial muscle denervation. Neurobiol. Dis. 2016;95:168–178. doi: 10.1016/j.nbd.2016.07.023. [DOI] [PubMed] [Google Scholar]
- 163.Miyoshi S., Tezuka T., Arimura S., Tomono T., Okada T., Yamanashi Y. DOK7 gene therapy enhances motor activity and life span in ALS model mice. EMBO Mol. Med. 2017;9:880–889. doi: 10.15252/emmm.201607298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wang W., Duan W., Wang Y., Wen D., Liu Y., Li Z., Hu H., Cui H., Cui C., Lin H., Li C. Intrathecal Delivery of ssAAV9-DAO Extends Survival in SOD1 G93A ALS Mice. Neurochem. Res. 2017;42:986–996. doi: 10.1007/s11064-016-2131-6. [DOI] [PubMed] [Google Scholar]
- 165.Israelson A., Ditsworth D., Sun S., Song S., Liang J., Hruska-Plochan M., McAlonis-Downes M., Abu-Hamad S., Zoltsman G., Shani T., et al. Macrophage migration inhibitory factor as a chaperone inhibiting accumulation of misfolded SOD1. Neuron. 2015;86:218–232. doi: 10.1016/j.neuron.2015.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Leyton-Jaimes M.F., Kahn J., Israelson A. AAV2/9-mediated overexpression of MIF inhibits SOD1 misfolding, delays disease onset, and extends survival in mouse models of ALS. Proc. Natl. Acad. Sci. USA. 2019;116:14755–14760. doi: 10.1073/pnas.1904665116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Patel P., Kriz J., Gravel M., Soucy G., Bareil C., Gravel C., Julien J.P. Adeno-associated virus-mediated delivery of a recombinant single-chain antibody against misfolded superoxide dismutase for treatment of amyotrophic lateral sclerosis. Mol. Ther. 2014;22:498–510. doi: 10.1038/mt.2013.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Pozzi S., Thammisetty S.S., Codron P., Rahimian R., Plourde K.V., Soucy G., Bareil C., Phaneuf D., Kriz J., Gravel C., Julien J.P. Virus-mediated delivery of antibody targeting TAR DNA-binding protein-43 mitigates associated neuropathology. J. Clin. Invest. 2019;129:1581–1595. doi: 10.1172/JCI123931. [DOI] [PMC free article] [PubMed] [Google Scholar]