Abstract
Glioma is a heterogeneous cellular environment in which immune cells play critical roles in tumor progression. Myeloid-derived suppressor cells (MDSCs) contribute to the formation of the immunosuppressive microenvironment of glioma; however, how glioma cells interact with MDSCs and how this interaction affects the function of other immune cells are unclear. Glioma cells can systemically communicate with immune cells via the secretion of exosomes, which contain microRNAs (miRNAs). Leveraging miRNA sequencing of exosomes, we identified enrichment of miR-1246 in glioma-derived exosomes and exosomes isolated from the cerebrospinal fluid (CSF) of glioma patients. We demonstrated that miR-1246 drives the differentiation and activation of MDSCs in a dual specificity phosphatase 3 (DUSP3)/extracellular signal‑regulated kinase (ERK)-dependent manner. In addition, postoperative CSF exosomal miR-1246 expression was found to be associated with the glioma recurrence rate. Hypoxia, a well-recognized feature of the glioblastoma microenvironment, increased miR-1246 levels in glioma-derived exosomes by enhancing miR-1246 transcription and selective packaging via upregulation of POU class 5 homeobox 1 (POU5F1) and heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1). Importantly, we identified a mechanism of 2-methoxyestradiol, a microtubule inhibitor currently undergoing clinical trials for glioblastoma. 2-Methoxyestradiol suppresses MDSC activation by inhibiting hypoxia-driven exosomal miR-1246 expression in glioma cells and PD-L1 expression in MDSCs.
Keywords: glioma, exosomes, cerebrospinal fluid, miR-1246, MDSC, 2-methoxyestradiol
Graphical abstract
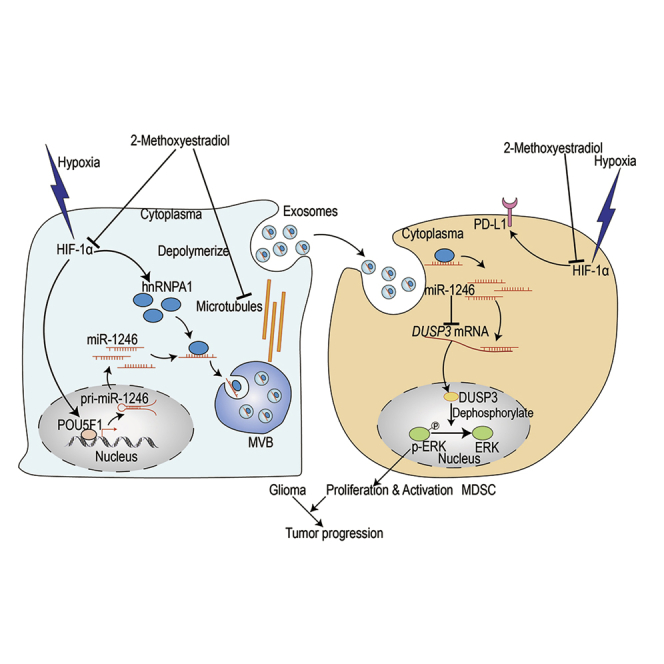
Gang Li and colleagues provide evidence that glioma patient cerebrospinal fluid-, plasma-, and glioma cell-derived exosomal miR-1246 promotes the differentiation and immunosuppressive function of myeloid-derived suppressor cells (MDSCs) and 2-methoxyestradiol suppresses MDSC activation by inhibiting hypoxia-driven exosomal miR-1246 expression in glioma cells and PD-L1 expression in MDSCs.
Introduction
Gliomas represent 40% of brain tumors.1 Glioblastoma is the most common type of glioma2 and the deadliest glioma, with a median survival of only 14 months despite recent advances in therapeutic strategies. Glioblastoma is resistant to conventional therapy because of its complex immunosuppressive microenvironment. Myeloid-derived suppressor cells (MDSCs) are a population of immature myeloid cells that accumulate in glioma patients. MDSCs suppress the function of T cells to interfere with normal immune responses and contribute to glioma progression.3 The accumulation of MDSCs in glioma patients causes systemic immunosuppression and impairs the effectiveness of immunotherapies.4 Therefore, we focused on the differentiation and activation of glioma-associated monocytic MDSCs (M-MDSCs) and aimed to target M-MDSCs to improve immunotherapy.
MDSCs can be differentiated from myeloid progenitors in the bone marrow or from peripheral monocytes/neutrophils.4, 5, 6 Factors in the plasma may affect the differentiation and function of peripheral and circulating MDSCs. In the central nervous system, cerebrospinal fluid (CSF) can interact directly with the brain tumor microenvironment7 and can potentially drive the differentiation and suppressive function of glioma-infiltrating MDSCs. In glioma patients, the differentiation of MDSCs can begin prior to their migration into glioma,8 suggesting a long-distance interaction between glioma cells and MDSCs.
Exosomes are small vesicles (50–150 nm in size) secreted by most cell types, including cancer cells.9 Exosome donor cells (e.g., glioma cells) can transport various forms of RNAs and proteins through multiple types of body fluids, such as extracellular fluid, CSF, and plasma, to target cells.10 We propose that glioma-derived exosomes are present in the CSF, where exosomes are taken up by local glioma-infiltrating MDSCs, and in the peripheral circulatory system, where exosomes interact with circulating MDSCs in the blood or with MDSC progenitors in the bone marrow. Because almost all cells produce exosomes, the composition of blood exosomes is complex, and plasma exosomal microRNA (miRNA) sequencing in glioma patients may not be sensitive enough to detect glioma-derived exosomes.11 Given the proximity of CSF to brain tumors, CSF may allow for a better detection of glioma-derived exosomes via exosomal miRNA sequencing. In this study, we uncovered that exosomes isolated from glioma patient plasma, CSF, and glioma cells enhanced the differentiation of human monocytes into MDSCs. By performing miRNA sequencing in exosomes isolated from glioma patient CSF and gliomas, we identified that miR-1246 is enriched in glioma-derived exosomes and mediates the differentiation and immunosuppressive function of M-MDSCs via the dual specificity phosphatase 3 (DUSP3)/ERK pathway. In addition, CSF exosomal miR-1246 expression is associated with recurrence in glioma patients.
Hypoxia is a well-recognized feature of the glioblastoma microenvironment and uniquely establishes an immunosuppressive environment by affecting both glioma cells and immune cells.12,13 We found that hypoxia not only locally influences glioma-associated MDSCs by upregulating MDSC PD-L1 expression but also remotely influences the functional propagation of M-MDSCs in the peripheral blood by altering the miRNA profile of glioma-derived exosomes. We determined that hypoxia promotes the enrichment of miR-1246 in glioma-derived exosomes by increasing miR-1246 transcription and selective exosomal sorting through upregulating POU class 5 homeobox 1 (POU5F1) and heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1). Finally, we revealed an anti-glioma mechanism of 2-methoxyestradiol (2-ME2) via blocking hypoxia-mediated M-MDSC functional propagation.
Results
Glioma patient CSF-, plasma- and glioma cell-isolated exosomes promote the differentiation and immunosuppressive function of MDSCs
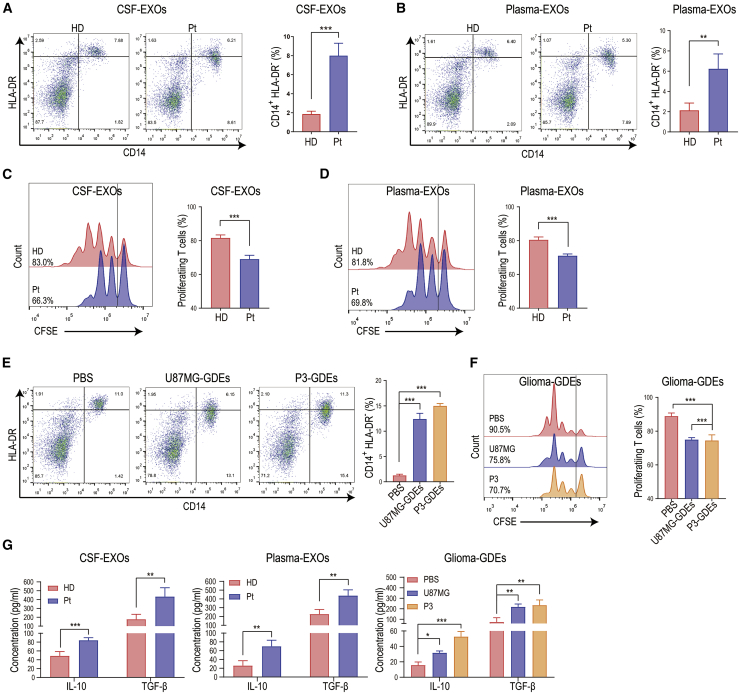
To study the effect of body fluid exosomes from glioma patients on MDSCs, we isolated exosomes from CSF, blood plasma, and glioma cell (U87MG and P3 cells) culture medium (Figures S1A–S1C). Isolated exosomes were used to stimulate human peripheral blood monocytes isolated from healthy donors (Figure S1D). Exosomes from glioma patient CSF and plasma significantly promoted differentiation of monocytes into CD14+HLA-DR− M-MDSCs compared to exosomes isolated from healthy donor CSF and plasma (Figures 1A and 1B). The expression of CD11b and CD33 on the above CD14+HLA-DR− cells was also validated (Figure S1E). We next examined the immunosuppressive function of glioma patient CSF and plasma exosome-induced M-MDSCs. Since the primary immunosuppressive function of MDSCs in the tumor microenvironment is to inhibit the proliferation of CD8+ T lymphocytes, we co-cultured glioma patient exosome-induced M-MDSCs with human T cells and examined T cell proliferation. Compared to healthy donor CSF and plasma exosome-induced M-MDSCs, glioma patient body fluid exosome-induced MDSCs had a higher propensity to inhibit T cell proliferation (Figures 1C and 1D). Glioma cell (U87MG and P3) exosomes also promoted the differentiation of monocytes into CD14+HLA-DR− M-MDSCs compared to the control (Figure 1E). MDSCs induced by glioma cell exosomes suppressed the proliferation of CD8+ T cells (Figure 1F). In addition, glioma patient CSF, plasma, and glioma cell exosome-induced MDSCs secreted higher levels of interleukin (IL)-10 and transforming growth factor (TGF)-β than MDSCs induced by healthy donor CSF- and plasma-derived exosomes (Figure 1G).
Figure 1.
Glioma patient body fluid-derived exosomes exhibited strong MDSC induction potency
(A and B) Exosomes (EXOs) isolated from glioma patient (Pt) CSF (A) and plasma (B) increased the differentiation of peripheral blood monocytes to CD14+HLA-DR− cells (M-MDSCs) compared to EXOs isolated from healthy donors (HD) (n = 4 for each group). (C and D) M-MDSCs were induced by stimulating CD14+ cells with glioma patient CSF exosomes or plasma exosomes. CFSE-labeled CD14-depleted PBMCs were co-cultured with induced M-MDSCs for 72 h (n = 4 for each group). After 72 h of co-culture, CD8+ T cells among the above CD14-depleted PBMCs were labeled with anti-CD8 antibody and gated through flow cytometry. CD8+ T cell proliferation was evaluated by measuring the CFSE dilution signal. More diluted signal indicates more proliferation. (E) Glioma-derived exosome (GDE) (from U87MG and P3 cells) stimulation enhanced the differentiation of CD14+HLA-DR− cells among PBMCs (n = 3 for each group). (F) CFSE-labeled CD14-depleted PBMCs were co-cultured with M-MDSCs induced by GDEs (isolated from U87MG and P3 cells) (n = 3 for each group). CD8+ T cell proliferation was evaluated by CFSE dilution using flow cytometry after 72 h of co-culture. (G) Glioma patient CSF (n = 4)-, plasma (n = 4)-, and glioma cell (n = 3)-EXOs induced M-MDSCs secrete increased levels of IL-10 and TGF-β compared to healthy donor CSF-EXO (n = 4)-, plasma-EXO (n = 4)-, and PBS (n = 3)-induced M-MDSCs. The data are presented as the mean ± SD; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Glioma patient CSF and plasma exosomal miR-1246 promotes MDSC activation
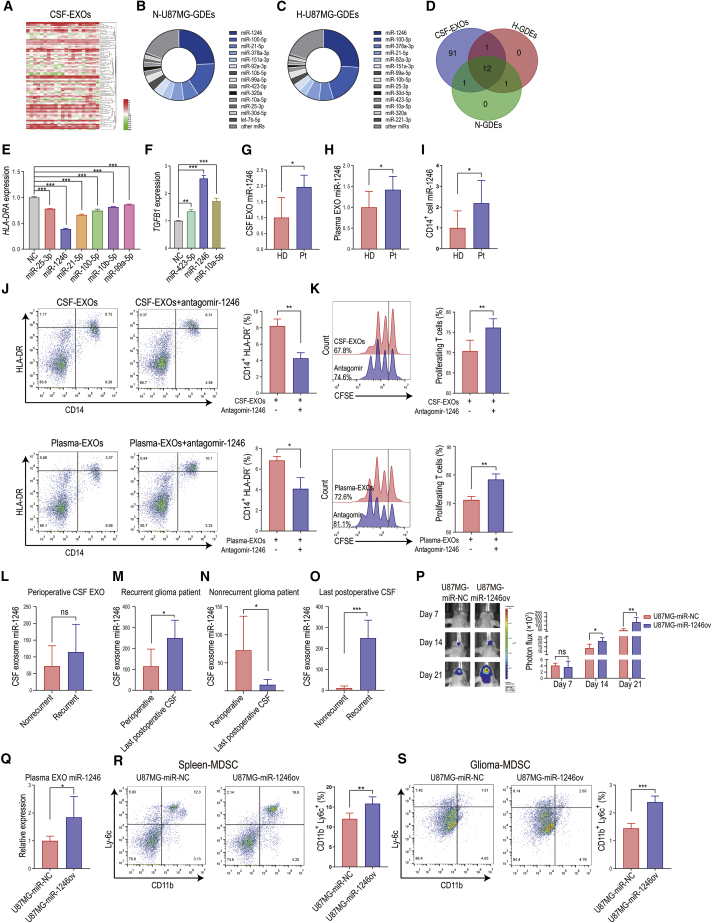
Almost all cells secrete exosomes, and exosomes are abundant in the blood. The source of plasma exosomes in cancer patients is complex, and the number of exosomes is ∼5 × 109 vesicles per milliliter of plasma.14 CSF interacts directly with the glioma microenvironment and is less complex in exosome composition than plasma, which allows for more straightforward detection. We performed miRNA sequencing of CSF exosomes collected perioperatively from 21 glioma patients (National Genomics Data Center [GSA no. CRA002339]) (Figure 2A; Table S1). We also performed miRNA sequencing of U87MG human glioma cell-derived exosomes (normoxia or hypoxia conditioned, SRA no. SRP123461). Overall, 105 miRNAs were highly (transcripts per million [TPM] > 500) expressed in glioma patient CSF exosomes. In both normoxia- and hypoxia-conditioned glioma-derived exosomes (N-GDEs and H-GDEs, respectively), the top 14 expressing miRNAs accounted for >80% of total miRNAs (Figures 2B and 2C). All the top 14 miRNAs were upregulated in H-GDEs compared to N-GDEs (Figure S2A). Other miRNAs each accounted for <1% of total miRNAs. Twelve miRNAs were highly expressed in both CSF exosomes and GDEs (Figure 2D). We overexpressed the miRNAs in human CD14+ monocytes and examined HLA-DRA and TGFB1 expression. miR-1246, miR-25-3p, miR-21-5p, miR-100-5p, miR-10b-5p, and miR-99a-5p reduced HLA-DRA expression, whereas miR-1246, miR-423-5p, and miR-10a-5p enhanced TGFB1 gene expression (Figures 2E and 2F). miR-1246 exhibited the strongest induction of MDSCs and MDSC-mediated immunosuppressive effects. miR-1246 was also the most highly expressed miRNA in GDEs and accounted for 24.4% of miRNAs in N-GDEs and 26.0% of miRNAs in H-GDEs (Figures 2B and 2C).
Figure 2.
Glioma patient CSF- and tumor cell-derived exosome miRNA sequencing identified miR-1246 as the key miRNA mediating the functional expansion of M-MDSCs
(A) Heatmap of differential CSF-exosome miRNA expression in 21 patients with gliomas of different grades. (B and C) Distribution of the top 14 enriched glioma exosomal miRNAs (higher than 1% of total miRNA) in normoxia- and hypoxia-conditioned glioma-derived exosomes (N-GDEs and H-GDEs, respectively). (D) Twelve miRNAs were shared among the miRNAs in all glioma patient CSF EXOs, N-GDEs, and H-GDEs. (E) Among the 12 miRNAs, miR-1246, miR-25-3p, miR-21-5p, miR-100-5p, miR-10b-5p, and miR-99a-5p overexpression in CD14+ cells reduced HLA-DRA expression. miR-1246 overexpression exhibited the strongest MDSC induction potency in CD14+ cells. (F) miR-1246, miR-423-5p, and miR-10a-5p increased gene expression of the immunosuppressive cytokine TGFB1. miR-1246 increased TGFB1 levels to the highest among all miRNAs tested. (G and H) The expression of miR-1246 was 1.96- and 1.42-fold higher in the EXOs of CSF (n = 5) and plasma (n = 7) collected from glioma patients than in those collected from healthy people. (I) miR-1246 expression increased 2.19-fold in CD14+ cells isolated from glioma patients (n = 7) compared to those isolated from healthy donors (n = 7). (J) Antagomir-1246 treatment impaired glioma patient CSF and plasma EXO-induced MDSC differentiation (n = 4 for each group, paired two-tailed Student’s t tests). (K) M-MDSCs obtained by stimulating CD14+ cells with glioma patient CSF EXOs or plasma EXOs were transfected with antagomir-1246 during EXOs stimulation. CFSE-labeled CD14-depleted PBMCs were co-cultured with M-MDSCs for 72 h. After 72 h of co-culture, CD8+ T cells among the above CD14-depleted PBMCs were labeled with anti-CD8 antibody and gated through flow cytometry. CD8+ T cell proliferation was evaluated by measuring the CFSE dilution signal (n = 4 for each group, paired two-tailed Student’s t tests). (L) No difference in EXO miR-1246 expression was identified in perioperative CSF collected from recurrent (n = 6) and nonrecurrent (n = 5) glioma patients. (M) In recurrent glioma patients, miR-1246 expression was significantly higher in the last postoperative CSF EXO sample than in the perioperative CSF EXO sample (n = 6). (N) In nonrecurrent glioma patients, EXO miR-1246 expression in the last postoperative CSF sample was reduced by 84.9% compared to that in perioperative CSF sample (n = 5). (O) miR-1246 expression in the last postoperative CSF sample from recurrent glioma patients (n = 6) was 22.9-fold higher than that in the postoperative CSF sample from nonrecurrent glioma patients (n = 5). (P) Bioluminescence images and luminescence quantification in mice implanted with control U87MG cells (U87MG-miR-NC) and miR-1246-overexpressing U87MG cells (U87MG-miR-1246ov) (n = 5 for each group). (Q) Plasma EXO miR-1246 expression was measured in glioma-bearing mice with quantitative real-time PCR (n = 5 for each group). (R and S) The percentage of CD11b+Ly6c+ M-MDSCs in spleens and tumors (n = 5 for each group). The data are presented as the mean ± SD; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗ p < 0.001.
To determine whether miR-1246 is the key miRNA in CSF and plasma exosome-mediated monocyte differentiation into MDSCs, we first compared body fluid exosomal miR-1246 expression in glioma patients and healthy donors. miR-1246 expression was 1.96 and 1.42 times higher in glioma patient CSF and plasma exosomes than in healthy donors (Figures 2G and 2H). CD14+ cells in glioma patients also expressed increased levels of miR-1246 compared to those in healthy donors (Figure 2I). We next transfected peripheral blood mononuclear cells (PBMCs) with miR-1246 antagomir and stimulated them with CSF and plasma exosomes. The miR-1246 antagomir impaired glioma patient CSF and plasma exosome-induced MDSC differentiation (Figure 2J) and immunosuppressive function (Figure 2K). These findings suggest that exosomal miR-1246 in glioma patient body fluid enhances monocyte differentiation into MDSCs and MDSC activation.
Among the 21 glioma patients whose CSF exosomes we sequenced above, complete prognosis information was available for 11 patients, among whom 6 patients developed recurrent gliomas (Tables S2 and S3). We performed miRNA sequencing around every 3 months postoperatively in these patients. No significant difference in perioperative CSF exosomal miR-1246 expression was found between recurrent and nonrecurrent glioma patients (Figure 2L). However, exosomal miR-1246 was 2.2 times higher in the last postoperative CSF sample than in the perioperative CSF sample in recurrent glioma patients (Figure 2M). Exosomal miR-1246 was 84.9% lower in the last postoperative CSF sample compared to the perioperative CSF sample in nonrecurrent glioma patients (Figure 2N). The levels of miR-1246 in the last postoperative CSF sample from recurrent glioma patients was 22.9 times higher than that in nonrecurrent glioma patients (Figure 2O), suggesting that CSF exosomal miR-1246 is associated with glioma presence and can potentially be used as a biomarker to detect tumor recurrence after surgery.
Next, we employed a glioma xenograft mouse model to determine the function of GDE miR-1246 in vivo. We implanted miR-NC (control group)- and miR-1246-overexpressing U87MG cells into the brains of nude mice (Figure 2P) and measured plasma exosomal miR-1246 expression. Since miR-1246 is not expressed by mouse cells15, 16, 17 (Figure S2B), all exosomal miR-1246 identified in the plasma originated from human U87MG glioma cells. The plasma exosomes from mice bearing miR-1246-overexpressing U87MG glioma cells contained significantly higher levels of miR-1246 than those from the control U87MG group (Figure 2Q). In addition, a higher percentage of CD11b+Ly6c+ mouse M-MDSCs in the spleen and glioma was observed in mice bearing miR-1246-overexpressing U87MG cells than in the control U87MG group (Figures 2R and 2S).
GDE miR-1246 potentiates M-MDSCs by activating the DUSP3/ERK pathway
To directly visualize the transportation of glioma cell-derived exosomal miR-1246 to MDSCs, GDEs isolated from U87MG/P3 cells transiently transfected with Cy3-tagged miR-1246 were used to treat monocytes for 48 h. Fluorescently labeled miR-1246 was observed in monocytes, demonstrating that miR-1246 was transferred from glioma cells to human monocytes via exosomes (Figure 3A). Similarly, the expression of miR-1246 was increased in monocytes after stimulation with different types of exosomes (Figure S2C). When we pretreated the cells with 100 nM antagomir-1246 before exosome treatment, the expression level of miR-1246 failed to increase after exosome treatment (Figure S2C).
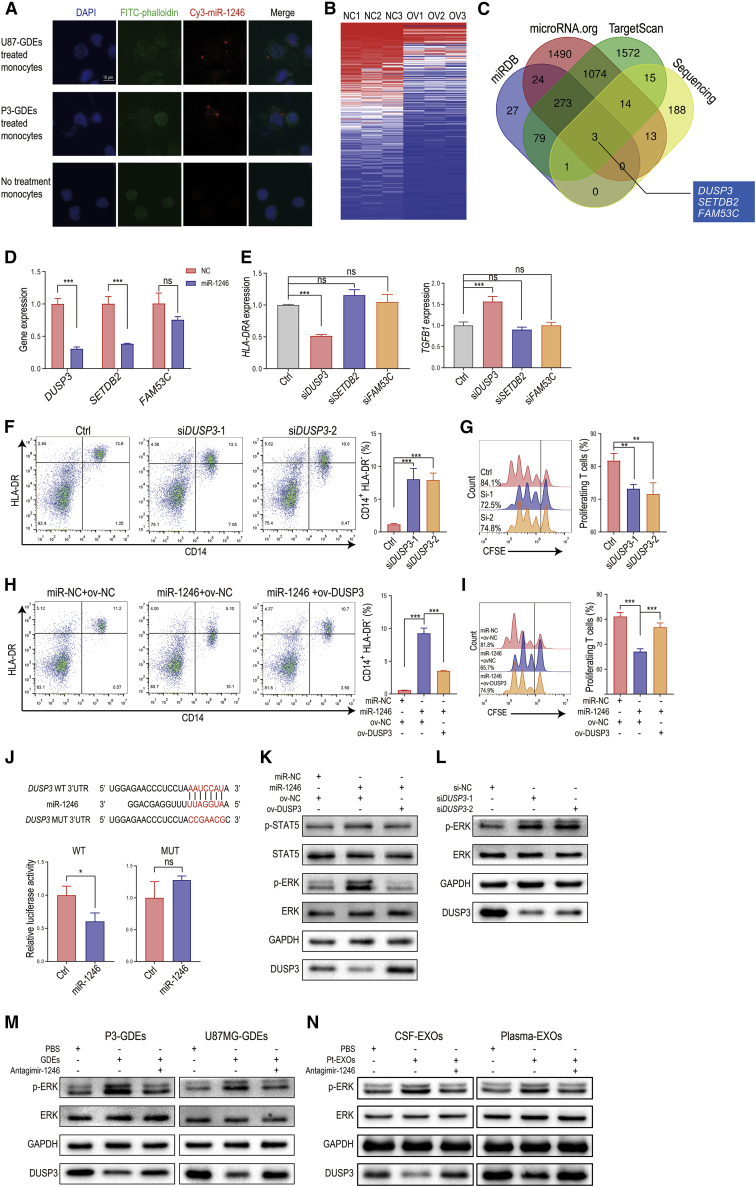
Figure 3.
miR-1246 in GDEs promotes MDSC differentiation and function through the DUSP3/ERK pathway
(A) GDEs derived from Cy3-tagged miR-1246-expressing glioma cells were used to treat human monocytes for 48 h. The monocytes were observed with confocal microscopy. Scale bar, 10 μm. (B) Heatmap of RNA sequencing results for miR-1246-overexpressing and control U937 cell lines. (C) Venn diagram identifying the overlap of miR-1246 target genes identified with the miRDB, microRNA.org, and TargetScan databases and the miR-1246 overexpression sequencing results. (D) DUSP3, SETDB2, and FAM53C expression was studied in miR-1246-overexpressing human CD14+ monocytes. (E) HLA-DRA and TGFB1 expression levels were measured after DUSP3, SETDB2, or FAM53C knockdown in monocytes. (F) The percentage of CD14+HLA-DR- MDSCs among DUSP3 knockdown PBMCs was measured by flow cytometry. (G) MDSCs with DUSP3 knockdown and control MDSCs were co-cultured with human CD8+ T cells. The proliferation of CD8+ T cells was measured. (H and I) The differentiation and function of MDSCs induced by miR-NC or miR-1246 mimics and nonsense sequence or DUSP3 overexpression plasmids. (J) Schematic diagram of the predicted wild-type binding site between miR-1246 and the DUSP3 3′ UTR and the mutated sequences. (K and L) Activation of the ERK and STAT5 pathways was studied after miR-1246 overexpression and DUSP3 knockdown. (M) GDE stimulation increased the phosphorylation of ERK, and this process was blocked by antagomir-1246. (N) Glioma patient CSF- and plasma-isolated exosomes activate the DUSP3/ERK pathway, and this effect was blocked by antagomir-1246. The data are presented as the mean ± SD; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Considering the significant individual variations, we first chose the U937 human monocyte cell line to perform RNA sequencing and investigated the downstream pathways of miR-1246, which would be helpful in controlling variables and enhancing repeatability. We overexpressed miR-NC and miR-1246 in U937 human monocytic cell lines and performed RNA sequencing in the treated cells (SRA no. SRP216625) (Figure 3B). Genes (p < 0.05) downregulated in the miR-1246 group relative to the miR-NC group were filtered by overlap with predicted miR-1246 targets (determined with the online bioinformatic tools TargetScan, miRDB, and microRNA.org) (Figure 3C). DUSP3, SETDB2, and FAM53C were identified as potential targets of miR-1246. We further validated that their expression in CD14+ monocytes was downregulated after miR-1246 overexpression (Figure 3D). To determine whether these genes are the key downstream molecules in miR-1246-mediated MDSC activation, we used small interfering RNAs (siRNAs) (si-DUSP3, si-SETDB2, and si-FAM53C) to knock down these 3 genes in human CD14+ cells and measured HLA-DRA and TGFB1 gene expression (Figure 3E). Only DUSP3 knockdown reduced HLA-DRA expression and increased TGFB1 expression in human CD14+ cells. We further showed that DUSP3 knockdown promoted monocyte differentiation into MDSCs (Figure 3F) and inhibited T cell proliferation (Figure 3G). In addition, DUSP3 overexpression reversed the effect caused by miR-1246 in MDSCs (Figures 3H and 3I).
To test whether miR-1246 directly targets DUSP3, 3′ UTR seed sequence mutations and 3′ UTR luciferase assays were conducted in human CD14+ cells. DUSP3 luciferase activity in miR-1246-transfected cells was decreased to ∼60% of the control group (Figure 3J), indicating that DUSP3 is a direct target of miR-1246. Among the potential DUSP3 downstream molecules signal transducer and activator of transcription 5 (STAT5), ERK1/2, c-Jun N-terminal kinase 1/2, p38, epidermal growth factor receptor, and ErbB2,18 only the STAT5 and ERK1/2 pathways have been reported to modulate MDSC activation.19,20 We overexpressed miR-1246 in human CD14+ cells and measured STAT5 and ERK1/2 activation. Only phosphorylation of ERK1/2, not STAT5, was elevated by miR-1246 and could be reversed by overexpression of DUSP3 (Figure 3K). DUSP3 knockdown with 2 different siRNAs (si-DUSP3) also increased ERK1/2 phosphorylation (Figure 3L). In addition, the ERK1/2 pathway was activated when we incubated human CD14+ monocytes with P3/U87MG GDEs. GDE-induced MDSC activation was blocked by transfection with antagomir-1246 (Figure 3M). Similarly, we treated human CD14+ monocytes with glioma patient CSF- and plasma-isolated exosomes. Glioma patient-derived exosomes activated the DUSP3/ERK pathway, and this effect was blocked by antagomir-1246 (Figure 3N).
Hypoxia-induced glioma exosomal miR-1246 promotes MDSC activation
Hypoxia is a common feature of the glioma microenvironment. According to our GDE sequencing data, miR-1246 was upregulated 3.4-fold in H-GDEs compared to N-GDEs (Figure S2A). In addition, hypoxia increased miR-1246 levels in GDEs from the four glioma cell lines analyzed (Figure S2D), suggesting that hypoxia-induced GDE miR-1246 expression can lead to miR-1246 enrichment in glioma patient body fluid exosomes.
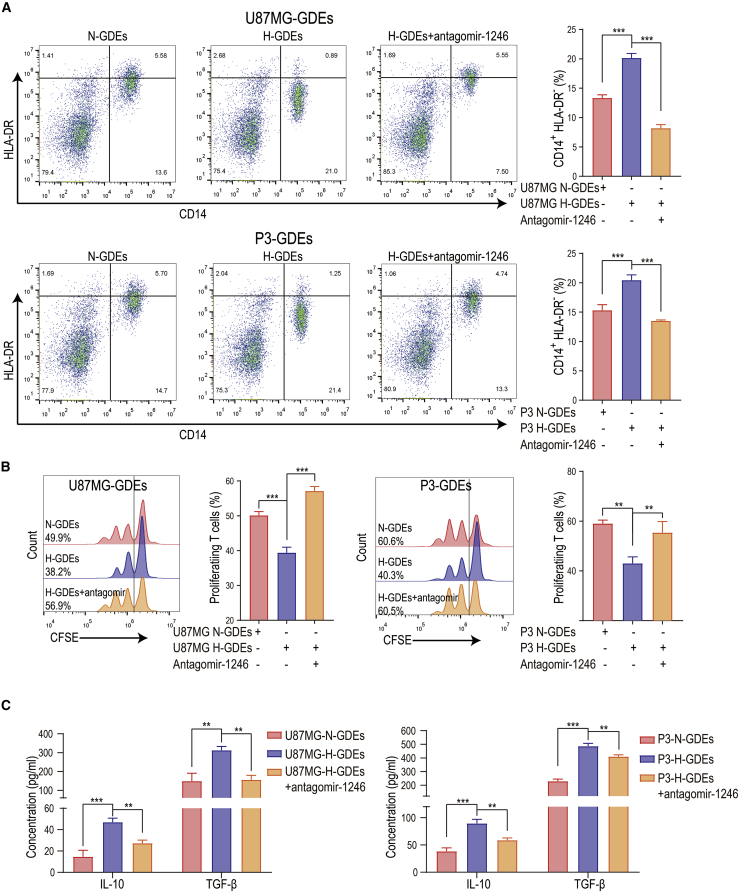
We next sought to determine the MDSC induction effect of N-GDEs and H-GDEs by incubating PBMCs from healthy donors with N-GDEs and H-GDEs. H-GDEs exhibited a stronger MDSC induction ability than N-GDEs (Figure 4A). H-GDE-induced MDSCs also exhibited a stronger ability to suppress CD8+ T cell proliferation and to produce IL-10 and TGF-β than N-GDE-induced MDSCs (Figures 4B and 4C). Consistent with the role of miR-1246 in glioma patient CSF and plasma exosomes, antagomir-1246 rescued the effect induced by H-GDEs (Figures 4A–4C), indicating that hypoxia-inducible GDE miR-1246 mediates the H-GDE-induced MDSC functional propagation.
Figure 4.
Hypoxia-induced exosomal miR-1246 upregulation mediates the functional differentiation of MDSCs
(A) The differentiation of MDSCs was measured after U87MG and P3 N-GDE and H-GDE stimulation. Antagomir-1246 impaired GDE-induced MDSC differentiation. (B) GDE-induced MDSCs were co-cultured with human peripheral CD8+ T cells from healthy volunteers at a 1:2 ratio. Three days later, CD8+ T cell proliferation was evaluated via flow cytometry by monitoring CFSE dilution. One representative experiment is shown. (C) IL-10 and TGF-β production was measured in N-GDE-, H-GDE-, and H-GDE+antagomir-1246-treated MDSCs. The data are presented as the mean ± SD; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
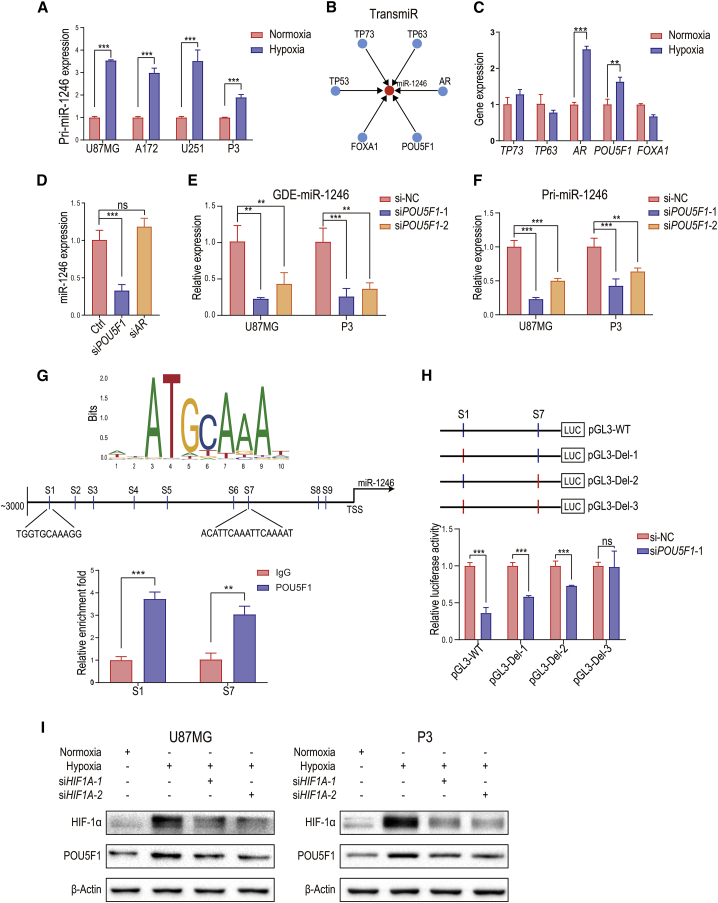
Hypoxia induces POU5F1 to promote miR-1246 transcription
The biogenesis of miRNA involves three steps. First, primary miRNA (pri-miRNA) is transcribed in the nucleus. Second, pri-miRNAs are processed by DGCR8 and Drosha to become precursor miRNAs (pre-miRNAs). Finally, the pre-miRNA is exported to the cytoplasm and cleaved by Dicer to become mature miRNA. To test whether hypoxia promotes miR-1246 transcription, we first determined whether hypoxia upregulates pri-miR-1246 levels in U87MG, A172, U251, and P3 human glioma cell lines (Figure 5A). To identify the mechanism by which hypoxia drives pri-miR-1246 transcription, we used TransmiR (http://www.cuilab.cn/transmir) to identify pri-miR-1246 transcription factor candidates: TP73, TP63, AR, POU5F1, FOXA1, and TP53 (Figure 5B). Because U251 and P3 human glioma cell lines harbor TP53 mutations, TP53 was excluded from subsequent analyses. We examined TP73, TP63, AR, POU5F1, and FOXA1 gene expression in hypoxia-conditioned U87MG cells and found that only POU5F1 and AR were upregulated after hypoxia stimulation (Figure 5C). Whereas AR knockdown in hypoxia-stimulated glioma cells did not affect miR-1246 expression, POU5F1 knockdown significantly reduced the miR-1246 level in hypoxia-conditioned glioma cells and H-GDEs (Figures 5D and 5E; Figures S3A and S3B). The expression of pri-miR-1246 was also decreased after POU5F1 knockdown (Figure 5F).
Figure 5.
Hypoxia-induced POU5F1 promotes miR-1246 transcription
(A) Levels of pri-miR-1246 in normoxia- and hypoxia-conditioned glioma cells. (B) TransmiR (http://www.cuilab.cn/transmir) prediction of miR-1246 transcription factors. (C) Levels of TP73, TP63, AR, POU5F1, and FOXA1 were measured in normoxia- and hypoxia-conditioned U87MG cells. (D) The expression of miR-1246 was measured in POU5F1 and AR knockdown hypoxia-stimulated glioma cells. (E) The expression of miR-1246 was examined in control and POU5F1 knockdown hypoxia-conditioned GDEs. (F) The expression of pri-miR-1246 was examined in control and POU5F1 knockdown hypoxia-conditioned glioma cells. (G) Computational prediction of POU5F1 binding sites (S1–S9) in the miR-1246 regulatory region and confirmation of POU5F1 binding to candidate miR-1246 sites via ChIP-qPCR analysis in hypoxia-conditioned U87MG cells. (H) Dual-luciferase reporter assay demonstrating the luciferase activity of the wild-type, S1 deletion, S7 deletion, and S1+S7 deletion miR-1246 promoter in U87MG cells transfected with si-NC or si-POU5F1 (n = 4 for each group). (I) POU5F1 protein levels were measured in normoxia- and hypoxia-conditioned U87MG and P3 cells. HIF1A knockdown inhibited the hypoxia-induced POU5F1 expression. The data are presented as the mean ± SD; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Nine POU5F1 binding sites were predicted in the upstream region (−1 to −3,000) of miR-1246 (Figure 5G; predicted by JASPAR [http://jaspar.genereg.net]). To determine which of the 9 sites are involved in the POU5F1-miR-1246 interaction, we performed a chromatin immunoprecipitation (ChIP) assay. DNA fragments were collected from hypoxia-conditioned U87MG cells and immunoprecipitated with anti-POU5F1 antibody and the IgG control. We observed high binding affinity of endogenous POU5F1 to miR-1246 in U87MG cells at two predicted sites (site 1 and site 7, Figure 5G), suggesting that POU5F1 directly binds to the miR-1246 promoter region to increase miR-1246 expression. Next, we engineered the wild-type and three mutant (S1 deletion, S7 deletion, and S1+S7 deletion) promoters to examine their binding ability to POU5F1. POU5F1 increased luciferase expression in wild-type, S1 deletion, and S7 deletion but not the S1+S7 deletion conditions (Figure 5H), indicating that POU5F1 promotes pri-miR-1246 transcription by binding to site 1 and site 7 on the miR-1246 promoter. In addition, POU5F1 expression was upregulated in hypoxia-conditioned U87MG and P3 cells. Cells mainly respond to hypoxia by activating hypoxia-inducible factors (HIFs). We found that HIF1A knockdown inhibited hypoxia-mediated POU5F1 expression, indicating that hypoxia drives POU5F1 expression via HIF-1α (Figure 5I).
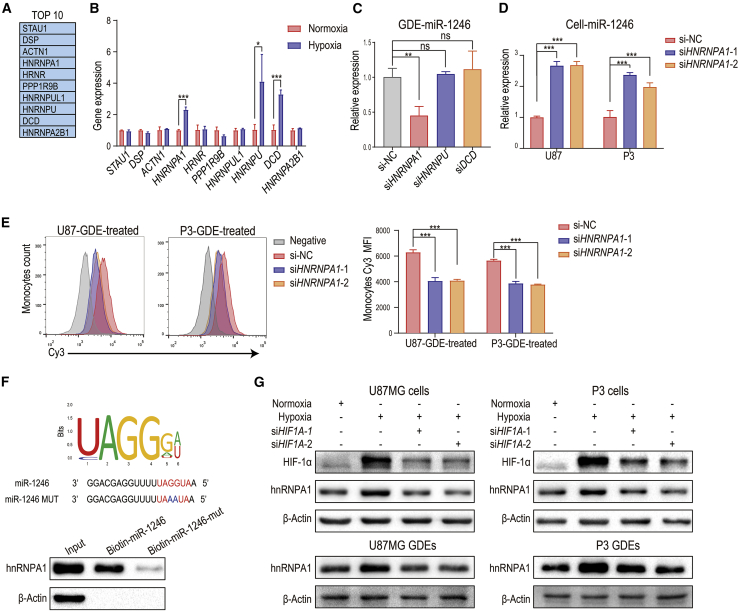
Hypoxia-induced hnRNPA1 selectively packages miR-1246 into glioma exosomes
To investigate the sorting of miR-1246 into exosomes, we analyzed a published proteomic dataset from an RNA:protein pull-down assay using biotinylated miR-1246.21 The top 10 proteins identified are shown in Figure 6A. We measured the RNA levels of these 10 genes in normoxia- and hypoxia-conditioned U87MG cells and found that hypoxia upregulated HNRNPA1, HNRNPU, and DCD expression (Figure 6B). Knockdown of HNRNPA1, but not HNRNPU and DCD, in glioma cells reduced exosomal miR-1246 levels (Figure 6C). Furthermore, HNRNPA1 knockdown reduced miR-1246 expression in hypoxia-conditioned GDEs and increased intracellular miR-1246 levels in hypoxia-conditioned glioma cells (Figures 6C and 6D; Figures S3C and S3D). HNRNPA1 knockdown had no effect on glioma pri-miR-1246 expression, suggesting that hnRNPA1 does not regulate miR-1246 transcription (Figure S3E). In addition, the amount of miR-1246 transferred from glioma to MDSCs via exosomes was reduced when hypoxia-conditioned glioma cells were transfected with si-HNRNPA1 (Figure 6E).
Figure 6.
Hypoxia-inducible hnRNPA1 selectively packages miR-1246 into exosomes
(A) In the pull-down assay,21 biotinylated miR-1246 was pulled down with protein lysates from wild-type (WT) HCT116 cells. Proteins pulled down with miR-1246 were analyzed with mass spectrometry. The top 10 identified proteins are displayed in the table. (B) The expression of the top 10 proteins in the pull-down assay was examined by quantitative real-time PCR in hypoxia- and normoxia-stimulated U87MG glioma cells. (C) The expression of miR-1246 in hypoxia-stimulated HNRNPA1, HNRNPU, and DCD knockdown and control U87MG GDEs was examined. (D) The expression of miR-1246 was examined in control and HNRNPA1 knockdown hypoxia-stimulated glioma cells. (E) Monocytes were treated with GDEs derived from Cy3-miR-1246-overexpressing U87 and P3 cells transfected with NC or si-HNRNPA1. The mean Cy3 fluorescence intensity of treated monocytes was measured. (F) The interaction between hnRNPA1 and miR-1246 was validated by pull-down assays. The binding capacity of hnRNPA1 to miR-1246 was impaired when UAGGUA was mutated to UAAAUA. (G) The expression of hnRNPA1 was studied in normoxia- and hypoxia-stimulated U87MG and P3 cells and GDEs. HIF1A knockdown impaired hypoxia-induced hnRNPA1 expression. The data are presented as the mean ± SD; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
We next predicted the potential hnRNPA1 binding sequence in miR-1246, using an online database (RBPDB, http://rbpdb.ccbr.utoronto.ca/; threshold = 0.7),22 and identified a potential site “UAGGUA,” the mutation of which interrupted the interaction between hnRNPA1 and miR-1246 (Figure 6F). We further showed that hypoxia induces hnRNPA1 expression in U87MG and P3 cell lines and their exosomes. To determine whether hypoxia drives exosomal miR-1246 packaging through HIF-1α, we knocked down HIF1A expression in U87MG/P3 cells with siRNA (si-HIF1A). Compared to the control group, hypoxia did not upregulate hnRNPA1 expression in glioma cells and GDEs in the si-HIF1A group, indicating that hypoxia mediates hnRNPA1 expression via HIF-1α (Figure 6G).
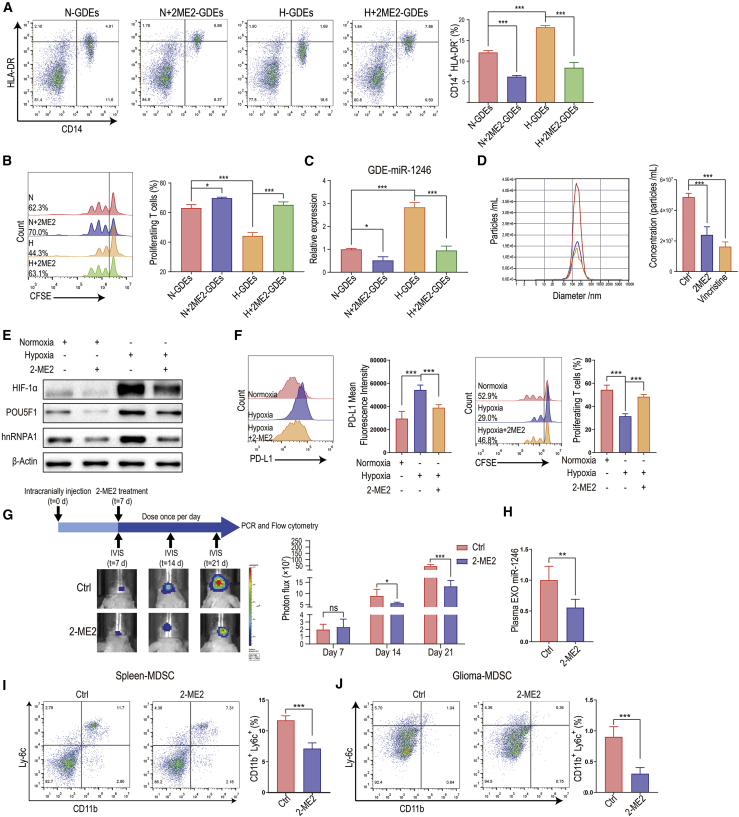
2-Methoxyestradiol inhibits glioma exosome production and exosomal miR-1246 expression
2-ME2 is a natural estrogen metabolite that exhibits antiproliferative and antiangiogenic activity by inducing apoptosis and inhibiting HIF-1α and tumor microtubules.23 2-ME2 inhibits the cellular accumulation of HIF-1α under both normoxic and hypoxic conditions.24 The efficacy of 2-ME2 in treating recurrent glioma has been demonstrated in early-phase clinical trials (Kirkpatrick et al., 2007, Am. Soc. Clin. Oncol., abstract). Since our study demonstrated that hypoxia-mediated GDE miR-1246 expression is HIF-1α dependent and it has been reported that the microtubule network is required for transport of exosomes to the plasma membrane (Granger et al.;25 Pederson et al., 2021, Am. Assoc. Cancer Res., abstract), we sought to determine whether 2-ME2 inhibits GDE miR-1246 expression and the subsequent functional propagation of MDSCs.
We first incubated normoxic and hypoxic glioma cells with 2-ME2 (50 μM, MedChemExpress, USA). Exosomes were isolated from glioma cell culture medium after 2-ME2 treatment and used to stimulate PBMCs. Compared to H-GDEs, 2-ME2-treated H-GDEs resulted in less monocyte differentiation into MDSCs with weaker MDSC-mediated T cell suppression (Figures 7A and 7B). Moreover, 2-ME2 reduced miR-1246 expression in H-GDEs and the release of GDEs from glioma cells (Figures 7C and 7D). Vincristine can also induce depolymerization of microtubules and was used as a positive control. Consistent with our findings that GDE miR-1246 expression is mediated by HIF-1α-dependent pathways (POU5F1 and hnRNPA1), 2-ME2 inhibited the hypoxia-mediated POU5F1 and hnRNPA1 upregulation (Figure 7E). It has been reported that hypoxia increases PD-L1 expression in MDSCs in a HIF-1α-dependent manner.13 Activated MDSCs express high levels of PD-L1, which interacts with PD-1 on T cells to induce T cell exhaustion. We found that 2-ME2 treatment reduced hypoxia-mediated PD-L1 expression in MDSCs and reversed the T cell exhaustion effect (Figure 7F).
Figure 7.
2-ME2 impairs glioma cell exosome production and exosomal miR-1246 expression
(A) Normoxic and hypoxic exosomes were isolated from glioma cell culture medium after 2-ME2 treatment and used to stimulate PBMCs. (B) MDSCs induced by the 4 types of GDEs in (A) were co-cultured with human CD8+ T cells. The proliferation of CD8+ T cells was measured by flow cytometry. (C) The expression of miR-1246 in the 4 types of GDEs in (A) was measured by quantitative real-time PCR. (D) The U87MG GDE concentration was measured with ZetaView after control (DMSO), 2-ME2, and vincristine (50 nM, MedChemExpress) treatment. (E) The expression of hnRNPA1 and POU5F1 in normoxic and hypoxic U87MG cells treated with 2-ME2 was measured by immunoblotting. (F) PD-L1 expression and T cell suppression assay of MDSCs isolated from GBM patient peripheral blood treated with hypoxia and 2-ME2 (n = 5 for each group). (G) Bioluminescence images and the corresponding quantification of U87MG glioma in mice treated with control (DMSO) or 2-ME2 (n = 5 for each group). (H) Plasma exosomal miR-1246 levels were measured with quantitative real-time PCR (n = 5 for each group). (I and J) The percentage of Cd11b+Ly6c+ M-MDSCs in the spleen and tumors of glioma-bearing mice was measured (n = 5 for each group). The data are presented as the mean ± SD; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
We also studied the effect of 2-ME2 on GDE miR-1246 in a glioma xenograft mouse model (Figure 7G). Plasma exosomes, splenocytes, and glioma xenografts were collected from mice treated with DMSO (vehicle) or 2-ME2 (200 mg/kg/day). 2-ME2 reduced exosomal miR-1246 levels and the number of splenic and tumor-infiltrating MDSCs (Figures 7H–7J; Figure S4A). Our previous research showed that cytochalasin D, an actin polymerization inhibitor, impairs exosome uptake by MDSCs.26 Since 2-ME2 inhibits microtubule function, it may impair exosome uptake by MDSCs. To confirm whether 2-ME2 inhibits the uptake of glioma exosomes by MDSCs, we incubated 2-ME2-treated MDSCs with PKH67-labeled exosomes for 24 h and found that exosome uptake was decreased compared to that in the control (DMSO) group (Figure S4B). The transport of RNAs to exosomes likely depends on cytoskeletal components. Whether 2-ME2 impairs miR-1246 exosomal packaging in glioma cells requires further investigation.
Collectively, 2-ME2 interferes with hypoxia-induced MDSC differentiation and immune suppressive function in 3 ways: First, 2-ME2 inhibits hypoxia-induced miR-1246 transcription and exosomal packaging in glioma cells. Second, 2-ME2 reduces exosome production by glioma cells by interfering with microtubules. Finally, 2-ME2 blocks hypoxia-induced MDSC PD-L1 expression.
Discussion
Here we studied the MDSC induction potential of glioma patient-derived body fluids from both the central nervous system (CSF) and blood (plasma). We found that exosomes from both the CSF and blood of glioma patients can induce differentiation and activation of MDSCs. Glioma-derived exosomes are present in the CSF, where exosomes could be taken up by local glioma-infiltrating MDSCs, and in the peripheral circulatory system, where exosomes could interact with circulating MDSCs in the blood or with MDSC progenitors in the bone marrow. Importantly, CSF is in close proximity to glioma and serves as an ideal reservoir to identify exosome biomarkers to predict the prognosis of glioma patients and to study how tumor-secreted exosomes shape the immunosuppressive function of local MDSCs.7
In this study, we showed that human GDEs are enriched with miR-1246, miR-151a-3p, and miR-320a, which are not expressed by mice and therefore were not identified in our previous studies using mouse GDEs.26,27 Higher expression of exosomal miR-1246 is associated with pathological grade, distant metastasis, and poor prognosis in various cancer types.28, 29, 30, 31 The roles of miR-1246 in modulating MDSC function and induction in cancer have not been reported. Here, we uncovered that miR-1246 present in glioma patient CSF and plasma exosomes promotes the differentiation of monocytes into MDSCs, as well as the immunosuppressive function of MDSCs. In addition, glioma local hypoxia promotes monocyte differentiation and MDSC function by enriching miR-1246 in GDEs and directly enhancing the immunosuppressive function of MDSCs.
We found that hypoxia induces the enrichment of miR-1246 in GDEs by increasing the production of mature miR-1246 and selective exosomal packaging of miR-1246. To identify the protein that interacts with mature miR-1246 and modulates its packaging, we analyzed the published miR-1246 binding partner dataset21 and determined that hnRNPA1 selectively binds miR-1246. To exclude the possibility that hnRNPA1 promotes transcription and splicing of miR-1246, we knocked down HNRNPA1 expression in U87MG and P3 cell lines and examined pri-miR-1246 expression in glioma cells and miR-1246 expression in their GDEs. Only GDE miR-1246 was reduced after HNRNPA1 knockdown, whereas the cellular pri-miR-1246 levels were not changed, supporting our hypothesis that hnRNPA1 only promotes the packaging of miR-1246 into GDEs. In addition, we found that the expression of miR-1246 in glioma cells was increased, which could be caused by reduced exosomal miR-1246 packaging and the subsequent intracellular accumulation of miR-1246. This finding is intriguing because researchers could inhibit the expression of specific sorting proteins of some tumor suppressor miRNAs, which would cause accumulation of tumor suppressor miRNAs in cells and inhibit tumor progression, potentially providing an avenue of treating brain tumors.
Glioblastoma is resistant to conventional chemotherapy partially because of the complex immune microenvironment. Thus, it is essential to identify therapeutic agents that target the immunosuppressive environment of glioblastoma. 2-ME2, a natural metabolite of estradiol, has been reported to inhibit tumor growth and angiogenesis by inhibiting HIF activity and microtubules.24 This drug showed potent anti-glioblastoma activity in preclinical models, and multiple studies have highlighted its potential as an antitumor agent.32,33 2-ME2 has completed phase I/II clinical trials for recurrent glioblastoma (Kirkpatrick et al., 2007, Am. Soc. Clin. Oncol., abstract). We identified an antiglioma mechanism by which 2-ME2 blocks hypoxia-induced MDSC functional proliferation at three levels. First, 2-ME2 blocks HIF-1α-induced exosomal miR-1246 transportation from glioma cells to M-MDSCs. Second, 2-ME2 impairs glioma-derived exosome release by interfering with microtubules. Third, 2-ME2 directly inhibits hypoxia-induced PD-L1 expression in M-MDSCs, which reduces the T cell suppressive effect of M-MDSCs.
In summary, the glioma patient body fluid exosomal miR-1246-induced MDSC functional expansion elucidated in this study highlights how local tumor hypoxia contributes to the formation of an immunosuppressive tumor microenvironment. Our study identified miR-1246 as a potential miRNA biomarker for monitoring glioma recurrence. These findings offer opportunities to manage glioma patients by targeting the tumor immune microenvironment.
Materials and methods
Patient sample collection
The glioma patient CSF samples for exosome miRNA sequencing were isolated from 21 glioma patients who were admitted to Qilu Hospital of Shandong University from November 2017 to October 2019. The glioma patient CSF and plasma samples used to stimulate human monocytes were isolated from 4 glioblastoma multiforme (GBM) patients admitted to Qilu Hospital of Shandong University from November 2017 to August 2019. The four normal CSF samples used for stimulating human monocytes were obtained from shunt procedures for normal pressure hydrocephalus (NPH) patients in the Department of Neurosurgery of Qilu Hospital of Shandong University. All glioma patients had received surgical resection of gliomas. The study was approved by the Clinical Research Ethics Committee of Qilu Hospital of Shandong University, and written informed consent was obtained from all patients.
Cell culture
The human glioma cell lines U87MG, U251, and A172 were purchased from the Chinese Academy of Sciences Cell Bank and cultured in DMEM containing 10% fetal bovine serum (FBS). The human primary glioblastoma cell line P3 was a kind gift from Dr. Rolf Bjerkvig, University of Bergen, and cultured in neurobasal medium (Gibco, USA) supplemented with GlutaMAX (2 mM), B-27 supplement (1×), and basic fibroblast growth factor and epidermal growth factor (20 ng/mL). Normoxia- or hypoxia-treated cells were established by culturing cells in incubators containing 21% or 1% O2 for 48 h. All cell lines were validated by short tandem repeat profiling.
Exosome isolation from plasma, CSF, and cell culture supernatant
All exosome isolation steps were conducted according to the minimal information for studies of extracellular vesicles (MISEV) guidelines.34 CSF and plasma samples from glioma patients and healthy volunteers in Qilu Hospital of Shandong University were collected and stored in liquid nitrogen. Plasma was diluted 1:1 with 1× PBS before centrifugation. Culture supernatant was collected from glioma cells cultured under normoxic or hypoxic conditions in DMEM supplemented with 10% exosome-depleted FBS for 48 h. Plasma, CSF, and glioma cell culture medium was centrifuged at 2,000 × g for 30 min and 12,000 × g for 45 min and filtered through a 0.22-μm filter to remove cell debris and large vesicles. The collected supernatant was centrifuged at 110,000 × g for 70 min. After the supernatant was removed, the pellet was washed with PBS and centrifuged at 110,000 × g for 70 min. Exosomes were collected and resuspended in 20–50 μL of PBS for further study.
Characterization of exosomes
CSF-, plasma-, and glioma cell-derived exosomes were first loaded onto carbon-coated electron microscopy grids (with membranes) and incubated for 10 min. Next, excess exosomes were removed, and one drop of 3% glutaraldehyde was placed on the grids for 5 min. The grids were washed with distilled water for 2 min, and a total of five washes were performed. The grids were then incubated with 4% uranyl acetate solution for 10 min and 1% methylcellulose solution for 5 min. Finally, the grids were dried and observed with a TEM 1011 electron microscope at 80 kV (JEOL-1200EX). Exosome particle size and concentration were analyzed with a ZetaView system (Particle Metrix, Germany).
Exosome uptake assay
Exosomes were labeled with PKH67 (Sigma-Aldrich, USA) according to the manufacturer’s protocol. PKH lipophilic dyes are fluorescent, and their aliphatic domains intercalate into lipid bilayers, such that exosomes stained with PKH67 can be visualized via fluorescence microscopy. The PKH67-labeled exosomes were incubated with monocytes. Once the PKH67-labeled exosomes are internalized by monocytes, the fluorescence signal from PKH67 can be observed within the recipient cells. Briefly, exosomes were reconstituted in 50 μL of PBS before 1 mL of Diluent C was added. Four microliters of PKH67 dye was added to 1 mL of Diluent C before being added to the exosomes. The samples were mixed gently for 4 min. To neutralize the excess dye, the PKH67-labeled exosome solution was mixed with 3 mL of 0.5% BSA and centrifuged at 100,000 × g for 1 h. The exosome pellet was resuspended in PBS and added to the culture medium of human monocytes. The monocytes were then fixed and examined under a fluorescence microscope.
Peripheral blood mononuclear cell and monocyte isolation
PBMCs were isolated from the venous blood of healthy donors and patients with lymphocyte separation medium (LTS1077, TBD, China). Blood was separated with standard density gradient centrifugation (30 min at 500 × g at 21°C), and the PBMC layer was carefully transferred to another tube. CD14+ monocytes were separated from PBMCs with CD14 MicroBeads (Miltenyi Biotec, Germany) according to the manufacturer’s protocol. Purity (>95%) was confirmed by flow cytometry.
MDSC induction
PBMCs (5 × 105) from healthy donors were cultured in 12-well plates with 0.5 mL of exosome-depleted RPMI 1640 complete medium. CSF-, plasma-, or glioma cell-derived exosomes (10 μg) were added to the culture medium and co-cultured with PBMCs for 72 h. In the miR-1246 validation experiment, human PBMCs were transfected with antagomir-1246 for 6 h before 72 h of exosome stimulation. PBMCs were examined via flow cytometry after the co-culture.
Transfection with miRNA mimics, antagomir, siRNA, and lentivirus
The miRNA mimics, control mimics, antagomir-1246, and siRNA (si-DUSP3, si-HIF1A, si-POU5F1, and si-HNRNPA1) were purchased from GenePharma (China). The miR-1246 overexpression lentivirus and negative control lentivirus were synthesized by GeneChem (Shanghai, China). All the sequence information is listed in Table S4. Monocytes were transfected with the following 12 miRNAs: miR-1246, miR-100-5p, miR-21-5p, miR-378a-3p, miR-151a-3p, miR-99a-5p, miR-92a-3p, miR-10b-5p, miR-25-3p, miR-30d-5p, miR-10a-5p, and miR-423-5p. Briefly, 5 × 105 monocytes were cultured in a 12-well plate with 0.2 mL of RPMI 1640 medium containing FBS. HiPerFect Transfection Reagent (QIAGEN, Germany) (6 μL) was mixed with 100 nM mimics or siRNA in 200 μL of RPMI 1640 without FBS for 10 min, and the mixture was added to the cultured monocytes. 4 h later, 400 μL of medium containing FBS was added. Monocytes were collected for subsequent experiments 2 days later.
Flow cytometry
For the MDSC differentiation assay, PBMCs were stained with anti-CD14-APC (eBioscience, USA) and anti-HLA-DR-PE (eBioscience) antibodies. For the MDSC PD-L1 assay, HLA-DR negative cells were separated from GBM patient PBMCs with anti-HLA-DR MicroBeads (Miltenyi Biotec) and stained with anti-CD14-APC and anti-PD-L1-PerCP-eFluor 710 antibodies (eBioscience). Flow cytometry was performed with a BD Accuri C6 flow cytometer, and data were analyzed with FlowJo V10 software.
T cell suppression assay
CD14-depleted PBMCs were labeled with 2.5 μM carboxyfluorescein diacetate succinimidyl ester (CFSE) and stimulated with coated anti-CD3 (1 μg/mL, eBioscience) and soluble anti-CD28 (1 μg/mL, eBioscience) antibodies. Exosome-induced monocytes or miR-1246 mimics/si-DUSP3-transfected monocytes were cultured with these cells at a ratio of 1:2 in a U-bottom 96-well plate. Three days later, the cells were stained with anti-CD8-APC antibody and analyzed for CFSE dilution.
Cytokine production
The concentrations of the suppressive cytokines IL-10 and TGF-β in the culture medium of exosome-induced monocytes were measured with ELISA kits (Proteintech, USA) according to the manufacturer’s protocol.
RNA extraction and quantitative real-time PCR
Exosomal RNA was extracted with a SeraMir Exosome RNA Extraction Kit (System Biosciences, USA), and cellular RNA was extracted with TRIzol (Invitrogen, USA) as previously described.26 Reverse transcription was performed with a ReverTra Ace qPCR RT kit (FSQ-101, Toyobo, Japan). Quantitative PCR was conducted with TB Green Premix Ex Taq (Takara, Japan) on a Roche Light Cycler 480. U6 was used as endogenous control. The primers used are listed in Table S5.
Western blotting
Protein was extracted from cells and exosomes with RIPA lysis buffer containing protease inhibitor cocktail (Sigma-Aldrich). Antibodies against the following proteins were used: β-Actin (8457, Cell Signaling Technology, USA), GAPDH (5174, Cell Signaling Technology), Calnexin (2679, Cell Signaling Technology), TSG101 (ab125011, Abcam, UK), CD9 (EXOAB-CD9A-1, System Biosciences), POU5F1 (11263-1-AP, Proteintech), hnRNPA1 (sc-32301, Santa Cruz Biotechnology), HIF-1α (36169S, Cell Signaling Technology), ERK (16443-1-AP, Proteintech), p-ERK (9101S, Cell Signaling Technology), p-STAT5 (4322T, Cell Signaling Technology), STAT5 (94205S, Cell Signaling Technology), and DUSP3 (ab125077, Abcam).
Chromatin immunoprecipitation assay
ChIP assays were performed with an EZ-Magna ChIP A/G Chromatin Immunoprecipitation Kit (17-10086, Millipore, USA) and anti-POU5F1 antibody (ab181557, Abcam). Immunoprecipitated DNA fragments were quantified with qPCR. The primers used are listed in Table S6.
miRNA target prediction and luciferase reporter assay
The online miRNA prediction tools miRDB (http://mirdb.org/), TargetScan (http://www.targetscan.org/vert_72/), and microRNA.org (http://www.microrna.org) were used to predict targets of miR-1246. The reporter genes containing pGL3-DUSP3-wt and pGL3-DUSP3-mut were synthesized by Bio-Asia (Jinan, China). Monocytes were transfected with the wild-type/mutant luciferase reporter and miR-1246 mimics. Two days later, a dual-luciferase reporter assay kit (Promega, USA) was used to perform luciferase assays.
Biotin miRNA pull-down assay
Cytoplasmic extracts were isolated from U87MG cells with a PARIS Kit (AM1921, Invitrogen, USA) and incubated with 100 pmol of biotinylated miR-1246 for 4 h at 4°C. Washed streptavidin magnetic beads (20164, Thermo Scientific, USA) were added and incubated with the extracts for another 4 h at 4°C. Beads were washed 5 times and mixed with loading buffer, followed by western blot analysis.
Animal studies
Luciferase-labeled glioma U87MG (miR-NC and miR-1246 overexpressing) cells (106/mouse) were injected into the brains of nude mice. Seven days after glioma implantation, 2-ME2 was injected intraperitoneally (200 mg/kg/day for 14 days). Bioluminescence was analyzed with a Spectrum In Vivo Imaging System (IVIS) (PerkinElmer). Blood, glioma tissue, and the spleen were harvested for further analysis after the last injection. All procedures that involved mice were approved by the Animal Care and Use Committee of Qilu Hospital of Shandong University.
Exosomal miRNA transportation assay
Cy3-labeled miR-1246 was purchased from GenePharma. GDEs were isolated from the supernatant of U87MG and P3 cells transfected with Cy3-labeled miR-1246 and used to treat monocytes. Two days later, the monocytes were collected and stained with DAPI and fluorescein isothiocyanate (FITC)-phalloidin.
Statistical analyses
Data analyses were performed with GraphPad Prism software. Unpaired two-tailed Student’s t tests were used to determine differences between two groups. Data for multiple comparisons were analyzed with one-way analysis of variance (ANOVA). Fisher’s exact tests were used to compare differences in age, sex, WHO grade, histology, and surgical methods between recurrent and nonrecurrent glioma patients. Statistical significance was set at p < 0.05. All data are presented as the mean values with standard deviation. All experiments were replicated at least three times with similar results.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81874083; 82072776; 82072775; 81702468; 81802966; 81902540), Natural Science Foundation of Shandong Province of China (Nos. ZR2019BH057; ZR2020QH174), Key Clinical Research project of Clinical Research Center of Shandong University (2020SDUCRCA011), and Taishan Pandeng Scholar Program of Shandong Province (No. tspd20210322).
Author contributions
W.Q., X.G., H.X., and G.L. conceived and designed the project. W.Q., X.G., B.L., Y.Q., and S. Zhao performed experiments. W.Q., X.G., Q.G., and S. Zhang analyzed the data. X.G. and W.Q. wrote the manuscript. Z.C., M.Q., S.W., Z.Z., and Z.P. provided reagents and materials. J.W., R.Z., and L.D. gave intellectual input. H.X. and G.L. supervised the study.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2021.06.023.
Contributor Information
Hao Xue, Email: xuehao@sdu.edu.cn.
Gang Li, Email: dr.ligang@sdu.edu.cn.
Supplemental information
References
- 1.Turner J.D., Williamson R., Almefty K.K., Nakaji P., Porter R., Tse V., Kalani M.Y. The many roles of microRNAs in brain tumor biology. Neurosurg. Focus. 2010;28:E3. doi: 10.3171/2009.10.FOCUS09207. [DOI] [PubMed] [Google Scholar]
- 2.Thakkar J.P., Dolecek T.A., Horbinski C., Ostrom Q.T., Lightner D.D., Barnholtz-Sloan J.S., Villano J.L. Epidemiologic and molecular prognostic review of glioblastoma. Cancer Epidemiol. Biomarkers Prev. 2014;23:1985–1996. doi: 10.1158/1055-9965.EPI-14-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raychaudhuri B., Rayman P., Ireland J., Ko J., Rini B., Borden E.C., Garcia J., Vogelbaum M.A., Finke J. Myeloid-derived suppressor cell accumulation and function in patients with newly diagnosed glioblastoma. Neuro-oncol. 2011;13:591–599. doi: 10.1093/neuonc/nor042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodrigues J.C., Gonzalez G.C., Zhang L., Ibrahim G., Kelly J.J., Gustafson M.P., Lin Y., Dietz A.B., Forsyth P.A., Yong V.W., Parney I.F. Normal human monocytes exposed to glioma cells acquire myeloid-derived suppressor cell-like properties. Neuro-oncol. 2010;12:351–365. doi: 10.1093/neuonc/nop023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tcyganov E., Mastio J., Chen E., Gabrilovich D.I. Plasticity of myeloid-derived suppressor cells in cancer. Curr. Opin. Immunol. 2018;51:76–82. doi: 10.1016/j.coi.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao Y., Poschke I., Wennerberg E., Pico de Coaña Y., Egyhazi Brage S., Schultz I., Hansson J., Masucci G., Lundqvist A., Kiessling R. Melanoma-educated CD14+ cells acquire a myeloid-derived suppressor cell phenotype through COX-2-dependent mechanisms. Cancer Res. 2013;73:3877–3887. doi: 10.1158/0008-5472.CAN-12-4115. [DOI] [PubMed] [Google Scholar]
- 7.Shen F., Zhang Y., Yao Y., Hua W., Zhang H.S., Wu J.S., Zhong P., Zhou L.F. Proteomic analysis of cerebrospinal fluid: toward the identification of biomarkers for gliomas. Neurosurg. Rev. 2014;37:367–380. doi: 10.1007/s10143-014-0539-5. discussion 380. [DOI] [PubMed] [Google Scholar]
- 8.Kumar V., Patel S., Tcyganov E., Gabrilovich D.I. The Nature of Myeloid-Derived Suppressor Cells in the Tumor Microenvironment. Trends Immunol. 2016;37:208–220. doi: 10.1016/j.it.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Théry C., Zitvogel L., Amigorena S. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 10.Kalluri R., LeBleu V.S. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noerholm M., Balaj L., Limperg T., Salehi A., Zhu L.D., Hochberg F.H., Breakefield X.O., Carter B.S., Skog J. RNA expression patterns in serum microvesicles from patients with glioblastoma multiforme and controls. BMC Cancer. 2012;12:22. doi: 10.1186/1471-2407-12-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiu D.K., Tse A.P., Xu I.M., Di Cui J., Lai R.K., Li L.L., Koh H.Y., Tsang F.H., Wei L.L., Wong C.M., et al. Hypoxia inducible factor HIF-1 promotes myeloid-derived suppressor cells accumulation through ENTPD2/CD39L1 in hepatocellular carcinoma. Nat. Commun. 2017;8:517. doi: 10.1038/s41467-017-00530-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noman M.Z., Desantis G., Janji B., Hasmim M., Karray S., Dessen P., Bronte V., Chouaib S. PD-L1 is a novel direct target of HIF-1α, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. J. Exp. Med. 2014;211:781–790. doi: 10.1084/jem.20131916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Notarangelo M., Zucal C., Modelska A., Pesce I., Scarduelli G., Potrich C., Lunelli L., Pederzolli C., Pavan P., la Marca G., et al. Ultrasensitive detection of cancer biomarkers by nickel-based isolation of polydisperse extracellular vesicles from blood. EBioMedicine. 2019;43:114–126. doi: 10.1016/j.ebiom.2019.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y., Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozomara A., Birgaoanu M., Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pavic K., Duan G., Köhn M. VHR/DUSP3 phosphatase: structure, function and regulation. FEBS J. 2015;282:1871–1890. doi: 10.1111/febs.13263. [DOI] [PubMed] [Google Scholar]
- 19.Chalmin F., Ladoire S., Mignot G., Vincent J., Bruchard M., Remy-Martin J.P., Boireau W., Rouleau A., Simon B., Lanneau D., et al. Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J. Clin. Invest. 2010;120:457–471. doi: 10.1172/JCI40483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ko J.S., Rayman P., Ireland J., Swaidani S., Li G., Bunting K.D., Rini B., Finke J.H., Cohen P.A. Direct and differential suppression of myeloid-derived suppressor cell subsets by sunitinib is compartmentally constrained. Cancer Res. 2010;70:3526–3536. doi: 10.1158/0008-5472.CAN-09-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooks T., Pateras I.S., Jenkins L.M., Patel K.M., Robles A.I., Morris J., Forshew T., Appella E., Gorgoulis V.G., Harris C.C. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat. Commun. 2018;9:771. doi: 10.1038/s41467-018-03224-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin X., Guo H., Wang X., Zhu X., Yan M., Wang X., Xu Q., Shi J., Lu E., Chen W., Zhang J. Exosomal miR-196a derived from cancer-associated fibroblasts confers cisplatin resistance in head and neck cancer through targeting CDKN1B and ING5. Genome Biol. 2019;20:12. doi: 10.1186/s13059-018-1604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar B.S., Raghuvanshi D.S., Hasanain M., Alam S., Sarkar J., Mitra K., Khan F., Negi A.S. Recent Advances in chemistry and pharmacology of 2-methoxyestradiol: An anticancer investigational drug. Steroids. 2016;110:9–34. doi: 10.1016/j.steroids.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 24.Mabjeesh N.J., Escuin D., LaVallee T.M., Pribluda V.S., Swartz G.M., Johnson M.S., Willard M.T., Zhong H., Simons J.W., Giannakakou P. 2ME2 inhibits tumor growth and angiogenesis by disrupting microtubules and dysregulating HIF. Cancer Cell. 2003;3:363–375. doi: 10.1016/s1535-6108(03)00077-1. [DOI] [PubMed] [Google Scholar]
- 25.Granger E., McNee G., Allan V., Woodman P. The role of the cytoskeleton and molecular motors in endosomal dynamics. Semin. Cell Dev. Biol. 2014;31:20–29. doi: 10.1016/j.semcdb.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo X., Qiu W., Liu Q., Qian M., Wang S., Zhang Z., Gao X., Chen Z., Xue H., Li G. Immunosuppressive effects of hypoxia-induced glioma exosomes through myeloid-derived suppressor cells via the miR-10a/Rora and miR-21/Pten Pathways. Oncogene. 2018;37:4239–4259. doi: 10.1038/s41388-018-0261-9. [DOI] [PubMed] [Google Scholar]
- 27.Guo X., Qiu W., Wang J., Liu Q., Qian M., Wang S., Zhang Z., Gao X., Chen Z., Guo Q., et al. Glioma exosomes mediate the expansion and function of myeloid-derived suppressor cells through microRNA-29a/Hbp1 and microRNA-92a/Prkar1a pathways. Int. J. Cancer. 2019;144:3111–3126. doi: 10.1002/ijc.32052. [DOI] [PubMed] [Google Scholar]
- 28.Bhagirath D., Yang T.L., Bucay N., Sekhon K., Majid S., Shahryari V., Dahiya R., Tanaka Y., Saini S. microRNA-1246 Is an Exosomal Biomarker for Aggressive Prostate Cancer. Cancer Res. 2018;78:1833–1844. doi: 10.1158/0008-5472.CAN-17-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X.J., Ren Z.J., Tang J.H., Yu Q. Exosomal MicroRNA MiR-1246 Promotes Cell Proliferation, Invasion and Drug Resistance by Targeting CCNG2 in Breast Cancer. Cell. Physiol. Biochem. 2017;44:1741–1748. doi: 10.1159/000485780. [DOI] [PubMed] [Google Scholar]
- 30.Sakha S., Muramatsu T., Ueda K., Inazawa J. Exosomal microRNA miR-1246 induces cell motility and invasion through the regulation of DENND2D in oral squamous cell carcinoma. Sci. Rep. 2016;6:38750. doi: 10.1038/srep38750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhai L.Y., Li M.X., Pan W.L., Chen Y., Li M.M., Pang J.X., Zheng L., Chen J.X., Duan W.J. In Situ Detection of Plasma Exosomal MicroRNA-1246 for Breast Cancer Diagnostics by a Au Nanoflare Probe. ACS Appl. Mater. Interfaces. 2018;10:39478–39486. doi: 10.1021/acsami.8b12725. [DOI] [PubMed] [Google Scholar]
- 32.Kang S.H., Cho H.T., Devi S., Zhang Z., Escuin D., Liang Z., Mao H., Brat D.J., Olson J.J., Simons J.W., et al. Antitumor effect of 2-methoxyestradiol in a rat orthotopic brain tumor model. Cancer Res. 2006;66:11991–11997. doi: 10.1158/0008-5472.CAN-06-1320. [DOI] [PubMed] [Google Scholar]
- 33.Muh C.R., Joshi S., Singh A.R., Kesari S., Durden D.L., Makale M.T. PTEN status mediates 2ME2 anti-tumor efficacy in preclinical glioblastoma models: role of HIF1α suppression. J. Neurooncol. 2014;116:89–97. doi: 10.1007/s11060-013-1283-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Théry C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin-Smith G.K., et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.