Abstract
Lung cancer is the leading cause of cancer‐related death worldwide due to diagnosis in the advanced stage and drug resistance in the subsequent treatments. Development of novel diagnostic and therapeutic methods is urged to improve the disease outcome. Exosomes are nano‐sized vehicles which transport different types of biomolecules intercellularly, including DNA, RNA and proteins, and are implicated in cross‐talk between cells and their surrounding microenvironment. Tumor‐derived exosomes (TEXs) have been revealed to strongly influence the tumor microenvironment, antitumor immunoregulatory activities, tumor progression and metastasis. Potential of TEXs as biomarkers for lung cancer diagnosis, prognosis and treatment prediction is supported by numerous studies. Moreover, exosomes have been proposed to be promising drug carriers. Here, we review the mechanisms of exosomal formation and uptake, the functions of exosomes in carcinogenesis, and potential clinical utility of exosomes as biomarkers, tumor vaccine and drug delivery vehicles in the diagnosis and therapeutics of lung cancer.
Keywords: biomarker, diagnosis, exosome, lung cancer, tumor‐derived exosome
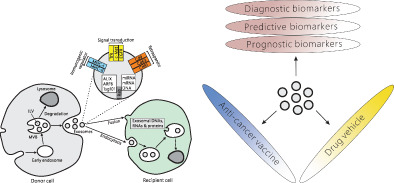
Formation and secretion of exosomes by donor cells and uptake of exosomal contents by recipient cells. Diverse applications of tumor‐derived exosomes in lung cancer management.
INTRODUCTION
Lung cancer, which accounts for 11.6% of all cancer cases and 18.4% of cancer related mortalities worldwide, represents a serious public health problem. 1 Non‐small cell lung cancer (NSCLC), consisting of adenocarcinoma, squamous cell carcinoma and large cell carcinoma, is the predominant histological subtype of lung cancer encompassing more than 80% of total cases. 2 Chemotherapy is currently the primary therapy for advanced lung cancer. Targeted therapies with EGFR‐, ALK‐, BRAF‐ or MET‐inhibitors and immunotherapies with PD‐L1, PD1 or CTLA4 antibody have also been developed in NSCLC treatment. 3 While a number of potential biomarkers have been explored, none are recommended for lung cancer screening, which accounts for the current situation whereby only 15% of lung cancer patients are diagnosed at an early stage. 4 Improvement of lung cancer outcome calls for the development of biomarkers for lung cancer management.
Exosomes are a type of extracellular vehicle (EV) of endosomal origin ranging from 30 to 150 nm in diameter which are secreted by most cells and found in various body fluids, such as plasma, saliva, urine, and ascites. 5 Despite the diversity in size and type of body fluid, all exosomes contain a subgroup of membrane proteins, including TSG101, ALIX and CD63, owing to their common endosomal origin.
When first discovered in 1983, 6 , 7 exosomes were regarded as garbage cans for unwanted materials from the cell of origin. Accumulative studies have subsequently illustrated that exosomes are capable of conducting intracellular communication by transporting DNA, RNA, and proteins, which in turn affects the physiological condition of the recipient cell. Tumor‐derived exosomes (TEX) can remold the tumor microenvironment to favor tumor progression and metastasis, for example, by transporting oncoproteins K‐RAS and MET or oncogenic miRNAs to surrounding healthy cells, 8 or by initiating a premetastatic niche and guiding tumor cells to prospective metastatic spots mediated by exosomal integrins. 9 , 10 , 11 TEXs can also serve as tumor vaccine to induce immune response against tumor by affecting activities of natural killer (NK) cells. 12 Moreover, the ability of exosomes to transfer biomolecules and drugs to recipient cells makes them promising drug delivery vehicles, 13 and exosomal nucleic acids (miRNA, mRNA, DNA) and proteins have shown potential in serving as diagnostic, prognostic, and predictive biomarkers for various cancers.
In the current review, we first portray recent studies on the mechanisms of formation and uptake of exosomes. We summarize tumor‐promoting functions of exosomes in correlation with crosstalk between cancer cells and tumor microenvironment. Finally, we discuss the clinical potential of exosomes as biomarkers, tumor vaccine and drug carrier in lung cancer diagnosis and therapeutics.
FORMATION OF EXOSOMES
Exosomes are generated by inward budding of the membrane of a type of endosomes called multivesicular bodies (MVBs) to form intraluminal vesicles (ILVs) 14 , 15 followed by release of the ILVs into the extracellular space upon fusion of the MVBs with the cellular membrane 16 (Figure 1). Alternatively, MVBs can fuse with the lysosome, which results in the degradation of ILVs‐containing MVBs and their contents, and may therefore modulate the secretion of exosomes. 17 Biogenesis of exosomes starts with the formation of ILVs within MVBs. The endosomal membrane is first restructured and gets enriched with tetraspanins (such as CD9 and CD63) which play a crucial role in exosomal formation. 18 The endosomal sorting complexes required for transport (ESCRTs) are then anchored to the site of ILV formation. 15 , 19 ESCRT I/II initiate and drive the inward budding of endosomal membrane and ESCRT III terminates this course, respectively. 20 , 21 Recruitment of ESCRT I/II to the cytoplasmic side of the early endosomal membrane is stimulated by a variety of factors, such as hepatocyte growth factor‐regulated tyrosine kinase substrate (HRS), the ubiquitination of the cytosolic tail of endocytosed proteins, phosphatidylinositol 3‐phosphate (PIP3), and curved membrane topology. 22 , 23 , 24 , 25 , 26 , 27 Despite the predominant role of the ESCRT pathway in exosomal biogenesis, Stuffers and colleagues revealed that, depending on the cell type, depletion of the ESCRTs did not block the formation of MVBs, indicative of the existence of ESCRT‐independent mechanisms of exosomal biogenesis in parallel to the ESCRT pathway. 28 In a recent study, Baietti et al. have shown that an alternative pathway, the syndecan‐syntenin‐ALIX pathway which includes heparanase, syndecan heparan sulfate proteoglycans, ADP ribosylation factor 6 (ARF6), phospholipase D2 (PLD2) and syntenin, is involved in mediating exosomal biogenesis. 14 Release of exosomes into the extracellular milieu is initiated by fusion of the MVB membrane with the plasma membrane, a process revealed to involve a variety of mechanisms and facilitated by a couple of Rab GTPases, including RAB11 and RAB35, or RAB27A/B. 29 , 30 , 31
FIGURE 1.

Biogenesis and secretion of exosomes by donor cells and uptake of exosomal contents by recipient cells. Exosomes originate by membrane invagination of the multivesicular bodies (MVBs) to form intraluminal vesicles (ILVs). MVBs fuse either with lysosomes or with the cellular membrane, leading to degradation of the MVBs or secretion of the exosomes into the extracellular space. Exosomes merge with the recipient cell either by fusion of the exosomal membrane with the cellular membrane or via the endocytosis pathway, leading to the discharge of exosomal contents into the cytosol of the recipient cell. Components generally found in the exosome include miRNA, mRNA, DNA and proteins such as tetraspanins (CD9, CD63, and CD81) involved in exosomal biogenesis, RAB GTPases (RAB11, RAB27A/B and RAB35) involved in exosomal secretion, ALIX, ARF6 and TSG101 in the endosome‐related pathways, and proteins involved in the signal transduction and antigen presentation (such as TfR, c‐Met, EGFR, MHC I/II). Abbreviations: miRNA, microRNA; ARF, ADP‐ribosylation factor; TSG101, tumor susceptibility gene 101; MHC, major histocompatibility complex; EGFR, epidermal growth factor receptor; TfR, transferrin receptor
EXOSOMAL CONTENTS AND THEIR SELECTIVE LOADING INTO EXOSOMES
Exosomes contain various cytoplasmic proteins, lipids, and genetic materials including DNA, mRNA, and non‐coding RNAs from the cell of origin (Figure 1). These data along with the purification procedures used to isolate the exosomes have been assembled into four publicly accessible databases: Exocarta, 32 EVpedia, 33 Vesiclepedia 34 and exoRBase. 35 Extensive research has revealed that exosomal contents change with different cell types and physiological conditions.Proteins involved in exosomal biogenesis, including different types of tetraspanins (for example, CD9, CD63, and CD81), in exosomal release (such as RAB27A and RAB11) and in the endosome‐related pathways (for example, ESCRT components, ALIX, ARF6, and TSG101, etc.), are universally present in exosomes. In addition, endosomal trans‐membrane proteins and proteins involved in signal transduction and antigen presentation (such as TfR, LAMP1, EGFR, MHC I/II), are also commonly found in exosomes. In contrast, resident proteins of the endoplasmic reticulum, Golgi, and nucleus are barely present in exosomes. 36 A large amount of RNAs, including mRNAs, rRNAs, tRNA, miRNAs, long non‐coding RNAs (lncRNAs) and circular RNAs, have been characterized in exosomes by next‐generation sequencing (NGS) and other techniques. 37 , 38 , 39 , 40 The majority of exosomal RNAs are no longer than 200 nucleotides and only a minority can reach up to 4 kb. 41 After exosomes are released from the donor cell, the exosomal membrane is believed to protect the RNAs in the exosomes from RNase digestion in the extracellular environment. 42 Despite that, exosomes share similar lipid composition with the donor cells, sphingomyelin, cholesterol, ganglioside GM3, phosphatidylserine, and ceramide were revealed to be enriched in exosomes, whereas phosphatidylcholine and diacyl‐glycerol are reduced in exosomes comparing to the cell of origin. 43 , 44 Studies have indicated that phosphatidylserine enrichment in exosomal membrane may promote their uptake into recipient cells via the internalization pathway. 45 , 46 Sphingolipid ceramide has been found to be required for the formation of ILVs by facilitating the inward budding of MVB membrane through its cone‐shaped structure. 46 In accordance with this observation, ceramide and its derivatives were detected to be abundant in exosomes. 47 , 48 However, inactivation of neutral sphingomyelinase (nSMase), a protein which produces ceramide, does not inhibit MVB formation or exosomal release in some cell types. Different molecular machineries, therefore, may be involved in exosomal formation in different cell types and related to the difference in the exosomal contents.Indeed, exosomes released via RAB11 and RAB35 were enriched with flotillin and cell‐specific proteins including Wnt, PLP and the transferrin receptor (TfR), 44 while exosomes released via RAB27A/B were enriched with late endosomal resident proteins, such as CD63, ALIX, and TSG101. 30 , 49 Yang and colleagues indicated proteins can be loaded into vesicles by associating with the plasma membrane as an oligomeric complex. 50 More efforts are required to unveil molecular mechanisms involved in the selective loading of exosomal proteins.Studies have shown that exosomal RNA profiles, to some extent, were different from the cytosolic counterparts of the cells of origin, indicating selective recruitment of RNA cargos into the exosmes. 51 , 52 , 53 Various molecular mechanisms have been unveiled in selective loading of miRNAs into the exosome. An interaction between a four nucleotide (GGAG) motif enriched in exsomal miRNAs and the ribonucleoprotein hnRNPA2B1 has been proposed to sort these miRNAs into MVBs. 54 In addition, 3′‐uridylation of miRNAs may contribute to recruitment of the miRNAs into exosomes while 3′‐adenylation can block exosomal loading of the miRNAs. 55
UPTAKE OF EXOSOMES
Apart from components on the exosomal membrane, such as EGFRvIII, Notch1 and Rheb, which can trigger signaling in the recipient cell by ligand‐receptor interactions without exosomal merge, functionality of exosomal contents necessitate their discharge into the recipient cell to work. 56 , 57 Indeed, a number of studies have observed the transfer of exosomal RNAs into the recipient cell, both in vitro and in vivo. 58 , 59 , 60 Exosomes released from donor cells are either taken up by neighboring cells or travel through the circulation system followed by merging into cells at distance. Merging of exosomes into the recipient cell can be accomplished either by fusion of the exosomal membrane with the plasma membrane, resulting in release of exosomal contents into the cytosol, or via the cellular endocytosis pathway 61 , 62 (Figure 1). By the endocytosis pathway, exosomes are first encompassed in the endosomal compartments followed by fusion of the exosomal membrane with the endosomal membrane to escape their destination for lysosomal degradation, leading to discharge of exosomal cargos into the cytosol of the recipient cell. 63 Spatiotemporal tracking by fluorescence microscopy has shown that exosomes in the medium were first docked onto plasma membrane and diffused slowly in the cytoplasm followed by switching to a rapid and directed movement mode, indicative of active trafficking along actin filaments or microtubules. 64 The mechanisms of exosomal cargo discharge from the endosome require further investigation.
FUNCTIONALITY OF EXOSOMES
Abundant research has illustrated that tumor‐derived exosomes (TEXs) play important roles in regulation of cell proliferation, migration and invasion. TEXs regulate tumor progression and metastasis by modulating local immune response, 65 , 66 epithelial‐mesenchymal transition (EMT), 67 angiogenesis 68 and drug resistence. 69 Various molecular mechanisms have been unveiled to be involved in the modulation of tumor progression and metastasis conducted by TEX contents, such as proteins and microRNAs.The immune response significantly affects cancer outcomes. 70 As important roles of EVs being revealed in regulation of inflammatory reactions in different inflammatory diseases including lung inflammation and injury, 71 , 72 TEXs can trigger the release of cytokines/chemokines from immune cells which result in the stimulation of anti‐tumor immune reactions or in a systemic immunosuppression. 73 TEXs have been reported to suppress CD8+ T cells‐induced anti‐tumor immune activities stimulated by tumor‐specific antigens, resulting in promotion of tumor growth. 74 Exosomes derived from exhausted CD8+ T cells could be uptake by non‐exhausted CD8+ T cells and subsequently impaired the anticancer function of normal CD8+ T cells. 75 Rather than suppress tumor growth by undermining cancer cells, the affected immune system promotes tumor progression by supporting the chronic inflammation and suppressing antitumor immunity. 76 Epithelial–mesenchymal transition (EMT) is a process by which epithelial cells acquire mesenchymal cell properties, which enables the cells to be invasive and migrate to distant sites leading to metastasis and tumor progression. 77 , 78 The importance of EMT in lung cancer has also been illuminated in various studies. 79 , 80 Serum TEXs isolated from patients with late stage lung cancer can induce EMT in recipient human bronchial epithelial cells. 81 These TEXs contain high levels of vimentin, which is a member of the type III intermediate filament protein family and a marker for EMT. The correlation between vimentin expression level and metastasis and invasion ability has been observed in lung cancer 81 , 82 , 83 and many other cancers, including prostate, colorectal and gastric cancers. Angiogenesis, a process regulated by various mechanisms and angiogenic factors, is crucial for tumor progression and metastasis. Hypoxia, a hallmark of the tumor microenvironment, has been reported to cause enhanced TEX production and a change in their content which enables TEXs to induce angiogenesis. 84 , 85 , 86 Among the exosomal contents, miRNAs are most studied. MiRNAs are a type of short noncoding RNAs which can mediate paracrine and endocrine effects by post‐transcriptionally modulating gene expression and cellular function in the recipient cells. 87 Specifically, TEX production and the level of exosomal miR‐23a were detected to be increased during hypoxia‐induced angiogenesis in CL1‐5 lung adenocarcinoma cells. Uptake of TEX‐associated miR‐23a, in turn, leads to targeting of prolyl hydroxylase 1 and 2 (PHD1 and 2), the accumulation of hypoxia‐inducible factor (HIF)‐1α, and the boost of angiogenesis. 85 Studies have demonstrated that hepatocellular carcinoma (HCC) cell‐derived exosomal miRNA‐21 could convert hepatocyte stellate cells to cancer‐associated fibroblasts and thus promote tumor progression by secreting angiogenic cytokines selected, 88 while metastatic breast cancer cells secrete miR‐105 to boost cell migration by down‐regulating expression of the tight junction protein ZO‐1. 89 Additionally, studies have identified exosomal proteins that may play crucial roles in the recipient cells. 90 EVs of tumor origin induced tumor angiogenesis by transporting proangiogenic peptides (for example, EGFRvIII) to the surrounding endothelial cells of microvessels, leading to activation of transforming signal pathways and regulation of the expression levels of vascular endothelial growth factor (VEGF). 91 In recent years, functional research on exosomal dsDNAs, 92 lncRNA 93 and circRNA 94 has also greatlyincreased.Formation of a premetastatic niche is the primary step required for metastasis, and is initiated through various mechanisms that promote a series of events beginning with vascular leakage which facilitates colonization of CTCs to the premetastatic site. 95 , 96 Exosomes released by breast cancer play an important role in promoting breast cancer bone metastasis, which is associated with the formation of a premetastatic niche via transferring miR‐21 to osteoclasts. 97 Hypoxia‐induced exosomal miR‐135a‐5p could initiate LATS2‐YAP‐MMP7 axis to form a premetastatic niche, and eventually promote the occurrence of CRC liver metastasis. 98 MiR‐25‐3p, a metastasis‐promoting miRNA of colorectal cancer (CRC), can be transferred from CRC to endothelial cells via exosomes, and promotes premetastatic niche formation by inducing vascular permeability and angiogenesis. 99 TLR3 in lung epithelial cells can be activated by the small RNA content of TEXs, and subsequently stimulate chemokine secretion and neutrophil recruitment to the lung, which together promote the niche formation and tumor lung metastasis. 100 Tumor exosome integrins also play important roles in preparing the premetastatic niche. A different tumor exosomal integrin subtype has been linked to specific organ metastasis and exosomal integrins have been suggested to be used for predicting organ‐specific metastasis. 10 TEXs are also involved in drug resistance in cancer. Recent studies indicated that exosomal delivery of functional P‐glycoprotein and multidrug resistance associated protein‐1 (MRP‐1) from drug resistant cancer cells led to acquired multidrug resistance by drug sensitive cancer cells. 101 , 102 Tumor‐associated macrophage‐derived mir21 can be transferred to the gastric cancer cells, where it suppresses cell apoptosis and enhances activation of PI3K/AKT signaling pathway by downregulation of PTEN, thus confer cisplatin resistance in gastric cancer. 103 In lung cancer, hypoxia‐induced exosomal PKM2 reprogrammed CAFs to create an acidic microenvironment promoting NSCLC cells proliferation and transmitted cisplatin‐resistance to sensitive NSCLC cells, led to cisplatin resistance in vitro and in vivo. 104 Therefore, exosomes play an important role in drug resistance by transfer of biomolecules to affect the characteristics of receptor cells or microenvironment, and exosomes may be used as drug delivery vehicles to tumor drugs or gene therapy. 105 Accordingly, exosomal miRNAs and mRNAs may predict drug resistance and help improve treatment options.Epigenetic modification plays an important role in tumor occurrence and development. TEX signaling participates in the adjustment of epigenetic modification. Exosome‐derived ncRNAs may serve as potential drivers of epigenetic reprogramming of cancer stem cells. 106 In NSCLC cell lines (A549 and H1299), exosome‐transmitted UFC1 promote progression by inhibiting PTEN expression via EZH2‐mediated epigenetic silencing. 107 Normal human gastric epithelial GES‐1 Cells absorbed gastric cancer cells released exosomal lncHEIH can upregulate EZH2 expression, which inhibited the expression of the tumor suppressor GSDME by methylation of the GSDME promoter, thus promoting the malignant transformation of normal gastric cells. 108 SNHG9, a papillary thyroid cancer cell exosome‐enriched lncRNA, inhibits cell autophagy and promotes cell apoptosis of normal thyroid epithelial cell. 109 LINC00470 in exosomes from glioma patients inhibiting autophagy and enhancing the proliferation of glioma cells by regulating WEE1 expression and activation of the PI3K/AKT/mTOR pathway.
The tumor microenvironment (TME) is a highly heterogeneous system incorporating cancer cells, endothelial cells, fibroblasts, adipocytes, mesenchymal stem cells, immunocyte and extracellular matrix. Tumor cells are closely connected with immune and stromal cells in TME and interact to form an environment of chronic inflammation and immunosuppression. Tumor‐associated macrophages (TAMs) are macrophages derived from peripheral blood monocytes recruited into solid tumor tissue microenvironment. Increased infiltration of tumor‐associated macrophages (TAMs) is observed in most cancer tissues compared with paracancer or normal tissues. 110 , 111 TAMs lose their killing ability and acquire an inhibitory phenotype, which promotes tumor development. Generally, macrophages differentiate into two main phenotypes: classically activated (M1) and alternatively activated (M2) 112 .TEXs can polarize M1 113 or M2 macrophage 114 and consequently inhibit or promote tumor metastasis. Exosomes from M2 macrophage promote the development of cancer. 115 Cancer‐associated fibroblasts (CAFs) are activated fibroblasts in tumor tissues. Extensive evidence suggests that CAFs are involved in stimulating cancer cell proliferation and progression. 116 TEXs can activate fibroblasts and promote CAF conversion. In cervical cancer, tumor‐secreted exosomal Wnt2B activates fibroblasts and promotes CAF conversion to promote cervical cancer progression. 117 Tumor‐secreted exosomal lncRNA POU3F3 promotes cisplatin resistance in ESCC by inducing fibroblast differentiation into CAFs. 118
EXOSOMES AS DISEASE BIOMARKERS
Abundant evidence has shown that cells from individuals with diseases and healthy subjects secreted exosomes containing different proteins and RNAs into the circulation and body fluid, which makes exosomes applicable for liquid biopsy as potential diagnostic biomarkers. 119 , 120 Melo and coworkers found that exosomal proteoglycan glypican‐1 (GP1) was specifically present in the serum of patients with pancreatic cancer with high sensitivity and exosomal GP1 level is highly positively correlated with the tumor burden. Correspondingly, exosomal GP1 level is also correlated with survival of pre‐ and post‐surgical patients, indicating exosomal GP1 ideal to serve as diagnostic and prognostic biomarkers for pancreatic cancer. 121 Exosomal cytoskeleton‐associated protein 4 (CKAP4) was secreted by pancreatic ductal adenocarcinoma (PDAC) cells and was highly detected in pancreatic tumor‐bearing xenografted mice and patients with PDAC, whereas CKAP4 was barely detectable in normal mice and postoperative patients, suggests that CKAP4 secreted in exosomes may represent a biomarker for PDAC. 122 In addition, exosomes released by tumor tissues are enriched with miRNAs and exosomal miRNAs may be explored as potential biomarkers for early diagnosis of cancers. 123 , 124 A number of clinical studies on exosomes as diagnostic biomarkers of cancer are ongoing.Potential of exosomes as biomarkers for noncancer diseases have been investigated as well. The expression of exosomal miR‐331‐5p and miR‐505 were significantly higher in patients with Parkinson's disease (PD) compared with healthy controls with the ROC curve 0.849 and 0.898, respectively, which suggests that exosomal miRNAs could potentially act as biomarkers for PD. 125 Exosomes collected from bronchoalveolar lavage fluid from patients with asthma and healthy subjects contain different miRNA contents, exhibiting potential to serve as diagnostic biomarker of asthma. 126
EXOSOMAL miRNAs AS DIAGNOSTIC, PREDICTIVE AND PROGNOSTIC BIOMARKERS FOR LUNG CANCER
Studies have illustrated that the composition of exosomal miRNAs differs among NSCLC subtypes and may serve as biomarkers for diagnosis, therapeutics and prognosis of NSCLC 127 , 128 (Figure 2).As early as in 2009, Rabinowits et al. 129 described a set of 12 miRNAs (including miR‐17‐3p, miR‐21, miR‐106a, miR‐146, miR‐155, miR‐199, miR‐192, miR‐203, miR‐205, miR‐210, miR‐212 and miR‐214) isolated from serum exosomes of NSCLC patients while not from those of healthy subjects, suggesting that exosomal miRNAs could be used for NSCLC diagnosis. In another study, 746 exosome‐derived miRNAs were globally screened in lung adenocarcinoma (LAC) patients, lung granuloma patients and healthy controls. 2 miRNA panels consisting of 4 miRNAs (miR‐378a, miR‐379, miR‐139‐5p and miR‐200‐5p) and 6 miRNAs (miR‐151a‐5p, miR‐30a‐3p, miR‐200b‐5p, miR‐629, miR‐100 and miR‐154‐3p), respectively, were explored for screening of LAC against healthy subjects and patients with benign lung nodules with high sensitivity (96% and 97.5%) but probably insufficient specificity (60% and 72%). 130 More recently, an miRNA panel containing let‐7b‐5p, let‐7e‐5p, miR‐23a‐3p, and miR‐486‐5p were developed for NSCLC diagnosis with sensitivity of 80.25%, specificity of 92.31% and AUC value of 0.899, respectively. In the meanwhile, levels of miR‐181b‐5p and miR‐361b‐5p, and of miR‐10b‐5p and miR‐320b were indicated to identify lung adenocarcinoma (LAC) and lung squamous cell carcinoma (LSCC), respectively, with sensitivity of 80.65% and 83.33%, specificity of 91.67% and 90.32%, and AUC value of 0.936 and 0.911, respectively. 131 Exosomal miRNAs have also been illustrated in prediction of treatment response of lung cancer patients. Two studies showed that upregulation of miR‐96, and miR‐208‐a in NSCLC promoted tumor growth and resistance to radiotherapy, indicative of potential novel therapeutic targets. 132 , 133 Level of exosomal miR‐146a‐5p was demonstrated to predict sensitivity of NSCLC to the cisplatin therapy through regulating autophagy pathway. 134 Similarly, exosomal miR‐100‐5p was shown to be downregulated in cisplatin‐resistant A549 (A549/DDP) cells in comparison with that in wild‐type A549, and A549/DDP‐derived exosomes were capable of endowing cisplatin resistance to wild‐type A549 cells mediated by mTOR pathway. 135 In a recent study, Poroyko et al. 136 analyzed exosomal miRNAs from the serum of NSCLC, SCLC and healthy controls by shotgun sequencing and identified 17 differentially displayed miRNAs between lung cancer patients and healthy subjects. A set of exosomal miRNAs were differentially displayed in the patient cohort before and after chemotherapy, indicative of the potential of exosomal miRNA profiling in disease subtyping and treatment efficacy evaluation of lung cancer. exosomal could be a noninvasive diagnostic and prognostic marker of radioresistant NSCLC For patients who received tyrosine kinase receptor inhibitors, exosomal miRNAs has also shown the predictive effect in drug response. MiR‐184 and miR‐3913‐5p derived from exosomes in the peripheral blood of NSCLC patients could be used as biomarkers to indicate osimertinib resistanceLevels of miR‐21 and miR‐4257 were found to be significantly increased in relapsing NSCLC patients after surgery and predict low disease free survival. 137 Low level of miR‐146a‐5p in serum exosomes indicated poor progression free survival of NSCLC patients. 134 Higher level of plasma exosomal miR‐451a from NSCLC patients of stages I, II or III before surgery were detected in those with recurrence and poor disease‐free and overall survival, indicative of the potential of exosomal miR‐451a serving as the prognostic biomarker for NSCLC patients. 138 In another study, 83 tumor‐related miRNAs in serum exosomes were screened and nine miRNAs were detected to be differentially present in exosomes of NSCLC patients. Among these nine miRNAs, miR‐23b‐3p, miR‐10b‐5p and miR‐21‐5p were upregulated in NSCLC compared to healthy subjects, and higher levels of the three miRNAs predicted low overall survival of the patients. 139
FIGURE 2.

Potential applications of exosomes in lung cancer management. Tumor‐derived exosomes can serve as promising biomarkers for diagnosis, prognosis and treatment efficacy of lung cancer. Exosomes could also act as drug delivery vehicles and anticancer vaccine in lung cancer therapy
EXOSOMAL PROTEINS AS DIAGNOSTIC AND PROGNOSTIC BIOMARKERS FOR LUNG CANCER
Recent studies by proteomic analysis have detected a number of lung cancer exosome‐enriched proteins that are involved either in biogenesis, transport and fusion of exosomes or play important roles in tumor metastasis, angiogenesis and immunoregulation. 140 These exosomal proteins can reflect the donor cells and pathological state of disease, which make them potential biomarkers for diagnosis and prognosis of lung cancer (Figure 2).Quantitative proteomic analyses of exosomal proteins in NSCLC cells and normal bronchial epithelial cells have identified NSCLC exosome‐enriched proteins involved in cell signaling, cell adhesion and extracellular matrix remodeling. Levels of EGFR and SRC as well as their downstream effectors GRB2 and RALA, and MET, RAC1 and KRAS proteins were detected to be upregulated in NSCLC exosomes. 141 In contrast to similar plasma EGFR levels between lung cancer patients and normal subjects, remarkable higher level of exosomal EGFR was observed in lung cancer patients in comparison to normal subjects 142 with 80% of serum exosomes from NSCLC being identified to be EGFR positive. 143 These studies indicated exosomal EGFR as potential diagnostic biomarker for lung cancer. Jakobsen and coworkers used an extracellular vesicle array containing 37 antibodies against lung cancer‐related proteins to capture and phenotype serum exosomes of NSCLC patients. A combined 30‐marker model was explored to distinguish NSCLC with sensitivity of 75%, specificity of 76% and diagnostic accuracy of 75.3%. 144 Another study of 581 patients by the same team indicated three markers CD151, CD171, and tetraspanin 8 as the strongest separators of patients with cancer of all histological subtypes versus patients without cancer. 145 Recently, Wang et al. 146 detected the differential expression protein in exosomes of distant metastatic and nonmetastatic NSCLC patients identified by multidimensional liquid chromatography and mass spectrometry analysis and found lipopolysaccharide‐binding proteins (LBP) in the exosomes to be well distinguished between patients with metastatic and patients with nonmetastatic NSCLC. The area under the curve (AUC) was 0.803 with a sensitivity of 83.1% and a specificity of 67%, suggesting exosome LBP might be promising and effective candidates of metastatic NSCLC. Further research by the same team found that a combination of AHSG, ECM1, and carcinoembryonic antigen improved the diagnostic potential of NSCLC with the diagnostic values AUC of 0.938 for NSCLC and 0.911 for early stage NSCLC versus healthy individuals, further suggesting the potential diagnostic value of serum exosome proteins. 147 Sandfeld‐Paulsen et al. performed proteomic analyses of 49 exosomal membrane bound proteins from a cohort of 276 NSCLC patients and identified nine exosomal membrane bound proteins to be potential prognostic biomarker of NSCLC. Specifically, high levels of EGFR, NYESO‐1 and PLAP are indicative of poor prognosis of NSCLC patients. 148 Recently, saliva has emerged as a novel medium for cancer detection as its collection is simple and noninvasive. 149 Exosomes released by cells or organs could also be detected in saliva. 150 In a xenografted mouse model of human lung cancer, salivary exosome‐like microvesicles were found to carry tumor cell‐specific mRNA and protein from blood to saliva. 151 Salivary exosomal proteins were systematically quantitatively compared by LC‐MS/MS between lung cancer patients and normal subjects, and 150 proteins in salivary exosomes were identified to be dysregulated in lung cancer, among which 25 proteins were from remote organs and five were lung‐associated proteins. These studies indicated that salivary exosomal proteins could also be explored as a diagnostic biomarker of lung cancer. 152 Exosome‐based detection of EGFR mutation in plasma from NSCLC patients has also achieved favorable diagnostic results. 153 Detection of the T790M mutation on exosomal cfDNA achieved 92% sensitivity and 89% specificity using tumor biopsy results as gold standard. 154 In another study, the sensitivity was 98% for detection of activating EGFR mutations and 90% for EGFR T790M based on exosomal cfDNA. 155 Recently, an exosome‐focused translational research for afatinib (EXTRA) study has been carried out to identify a novel predictive biomarker and a resistance marker for patients who received afatinib treantment. 156 All these studies demonstrate that exosomal EGFR mutation detection might be used as diagnostic and predictive biomarker for EGFR‐TKI treatment and help to avoid unnecessary tumor biopsies.
EXOSOMES IN LUNG CANCER THERAPY
The overall patient outcomes of lung cancer therapy remain unsatisfactory. Improvement of efficacy necessitates the exploration of novel therapeutic approaches for lung cancer. Immunotherapy by targeting PD‐1 and PD‐L1, which negatively regulate T cell activation, represents a novel treatment approach for broad range of cancers, including lung cancer. Exosomes are involved in the regulation of inflammatory signals in the tumor microenvironment and therefore affect the immunotherapeutic efficacy in lung cancer. 70 As a result, inhibition of TEX release or of integrin‐mediated TEX uptake by blocking integrins may restrain the development of an amicable tumor microenvironment, which leads to the repression of tumor progression. 157 , 158 In addition, the exosome represents a potential delivery tool of biological molecules and drugs (Figure 2). In comparison to other drug delivery tools developed, including nanoparticles and liposomes, 159 exosomes display numerous advantages, such as less toxicity, low immunogenicity, targeting specific recipient cells mediated by ligands and peptides on the exosomal membrane, and the ability to transport across the blood brain barrier. 160 Natural compounds can be loaded into exosomes during exosomal biogenesis process or through in vitro incubation, transfection or electroporation of purified exosomes. 161 Docetaxel (DTX), the first‐line of the antitumor agent used to treat NSCLC, was selected payload into exosome by electroporation, compared to the free DTX, exosomes significantly increased the cellular uptake in vitro evaluation and showed better targeting to tumor tissue in the mice. 162 In another research, engineered targeting tLyp‐1 exosomes had high transfection efficiency into lung cancer and cancer stem cells and were able to knockdown the target gene of cancer cells and to reduce the stemness of cancer stem cells. 163 All these researches suggest that exosomes might offer a promising gene delivery platform for future cancer therapy.Exosomes have been explored as anticancer vaccine based on the consideration that they contain tumor‐specific antigens 164 (Figure 2). Yaddanapudi et al. 165 recently reported that vaccination with GM‐CSF positive but not GM‐CSF negative exosomes from murine embryonic stem cells (ESCs) delayed or prevented tumorigenesis in mice by boosting tumor‐specific responses and Th1 cytokine reactions of the tumor‐infiltrating lymphocytes, suggesting potential roles of GM‐CSF positive human ESCs in cancer‐preventing vaccination of susceptible individuals. In a clinical study, MAGE peptides loaded exosomes derived from autologous dendritic cells (DEXs) of NSCLC patients were evaluated for their safety and efficacy as tumor vaccine. 166 Enhanced immune responses with NK activities and T cell response against MAGE peptides were observed and long term disease stability were achieved in some patients. In another clinical study, clinical benefits of cancer antigens‐loaded IFN‐g‐DEXs were evaluated in NSCLC patients without disease progression after chemotherapy. 167 Augmented antitumor function of NK cells induced by DEXs has been established in advanced NSCLC patients with defective NKp30 expression. These studies reveal potential applications of dendritic cell‐derived exosomes as anticancer vaccine.
DISCUSSION
Crosstalk between cancer cells and the tumor microenvironment mediated by tumor derived exosomes has been established to play an important role in tumor progression and metastasis. However, little is known about how TEXs interact with tissues in distance. Further investigation is necessary to understand the function of TEXs in tumor progression and metastasis, which will facilitate the development of novel cancer therapeutic approaches.Exosomes appear to be desirable biomarkers for prognosis, diagnosis and treatment prediction of lung cancer on account of exosomal cargos mirroring tissue expression patterns, the noninvasiveness of liquid biopsy which makes consecutive surveillance possible, and the stability of genetic materials safeguarded by the exosomal membrane. There has been extensive research on exosomal miRNAs and proteins as potential diagnostic biomarker for lung cancer. These studies suggest that miRNA and protein panels, instead of single miRNAs and proteins, present more clinical values. The outcomes from different studies, despite being inconsistent, do partially overlap. In consideration of the variations in study subjects, exosome origins and exosome isolation approaches, production of comparable outcomes necessitates larger‐scale investigation with unified patient classification and exosome isolation methods.Moreover, possible therapeutic applications of exosomes could open up new avenues in lung cancer treatment. The potential of exosomes to serve as drug delivery vehicles have been proposed. Exosomes derived from dendritic cells serving as cancer vaccine have been proven to be promising in lung cancer therapy. While vaccination of DEXs has been illustrated to enhance NK cell activity in some lung cancer patients, little is known about the application of exosomes in adjuvant therapy of lung cancer or as drug delivery tool. Further clinical studies are warranted to confirm the capacity of exosomes in lung cancer management.In conclusion, exosomes are a new exciting field of research which have opened a new window to the diagnosis, prognosis and treatment of lung cancer. Although a great number of inspiring findings and potential applications for exosomes have been published, methods for exosome isolation remain to be standardized. Future investigations should be conducted on approaches to manage the biogenesis, cargo loading, release, and interaction of lung cancer exosomes so as to better understand their molecular mechanisms and develop novel therapeutics with maximal on‐target efficiency.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
ACKNOWLEDGMENTS
This work was supported by the Key Research and Development Program of Hunan Province, China (2018SK21215).
Xia Z, Qing B, Wang W, Gu L, Chen H, Yuan Y. Formation, contents, functions of exosomes and their potential in lung cancer diagnostics and therapeutics. Thorac Cancer. 2021;12:3088–3100. 10.1111/1759-7714.14217
Funding information Key Research and Development Program of Hunan Province of China, Grant/Award Number: 2018SK21215
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2. Oser MG, Niederst MJ, Sequist LV, Engelman JA. Transformation from non‐small‐cell lung cancer to small‐cell lung cancer: molecular drivers and cells of origin. Lancet Oncol. 2015;16(4):e165–72. 10.1016/S1470-2045(14)71180-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ettinger DS, Wood DE, Aisner DL, Akerley W, Bauman JR, Bharat A, et al. NCCN guidelines insights: non‐small cell lung cancer, Version 2.2021. J Natl Compr Canc Netw. 2021;19(3):254–66. 10.6004/jnccn.2021.0013 [DOI] [PubMed] [Google Scholar]
- 4. Jacobsen MM, Silverstein SC, Quinn M, Waterston LB, Thomas CA, Benneyan JC, et al. Timeliness of access to lung cancer diagnosis and treatment: a scoping literature review. Lung Cancer. 2017;112:156–64. 10.1016/j.lungcan.2017.08.011 [DOI] [PubMed] [Google Scholar]
- 5. Whiteside TL. Tumor‐derived exosomes and their role in cancer progression. Adv Clin Chem. 2016;74:103–41. 10.1016/bs.acc.2015.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harding C, Heuser J, Stahl P. Receptor‐mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol. 1983;97(2):329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33(3):967–78. [DOI] [PubMed] [Google Scholar]
- 8. Sun T, Kalionis B, Lv G, Xia S, Gao W. Role of exosomal noncoding RNAs in lung carcinogenesis. Biomed Res Int. 2015;2015:125807. 10.1155/2015/125807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang Y, Wang XF. A niche role for cancer exosomes in metastasis. Nat Cell Biol. 2015;17(6):709–11. 10.1038/ncb3181 [DOI] [PubMed] [Google Scholar]
- 10. Hoshino A, Costa‐Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–35. 10.1038/nature15756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang M, Zhao X, Huang F, Wang L, Huang J, Gong Z, et al. Exosomal proteins: key players mediating pre‑metastatic niche formation and clinical implications (Review). Int J Oncol. 2021;58(4):4. 10.3892/ijo.2021.5184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li Q, Huang Q, Huyan T, Wang Y, Huang Q, Shi J. Bifacial effects of engineering tumour cell‐derived exosomes on human natural killer cells. Exp Cell Res. 2018;363(2):141–50. 10.1016/j.yexcr.2017.12.005 [DOI] [PubMed] [Google Scholar]
- 13. Gong C, Zhang X, Shi M, Li F, Wang S, Wang Y, et al. Tumor exosomes reprogrammed by low pH are efficient targeting vehicles for smart drug delivery and personalized therapy against their homologous tumor. Adv Sci (Weinh). 2021;8(10):2002787. 10.1002/advs.202002787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, et al. Syndecan‐syntenin‐ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14(7):677–85. 10.1038/ncb2502 [DOI] [PubMed] [Google Scholar]
- 15. Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, et al. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci. 2013;126(Pt 24):5553–65. 10.1242/jcs.128868 [DOI] [PubMed] [Google Scholar]
- 16. Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nat Rev Mol Cell Biol. 2009;10(9):597–608. 10.1038/nrm2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Villarroya‐Beltri C, Baixauli F, Mittelbrunn M, Fernández‐Delgado I, Torralba D, Moreno‐Gonzalo O, et al. ISGylation controls exosome secretion by promoting lysosomal degradation of MVB proteins. Nat Commun. 2016;7:13588. 10.1038/ncomms13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pols MS, Klumperman J. Trafficking and function of the tetraspanin CD63. Exp Cell Res. 2009;315(9):1584–92. 10.1016/j.yexcr.2008.09.020 [DOI] [PubMed] [Google Scholar]
- 19. Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464(7290):864–9. 10.1038/nature08849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Babst M, Katzmann DJ, Estepa‐Sabal EJ, Meerloo T, Emr SD. Escrt‐III: an endosome‐associated heterooligomeric protein complex required for mvb sorting. Dev Cell. 2002;3(2):271–82. [DOI] [PubMed] [Google Scholar]
- 21. Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21(1):77–91. 10.1016/j.devcel.2011.05.015 [DOI] [PubMed] [Google Scholar]
- 22. Fernandez‐Borja M, Wubbolts R, Calafat J, Janssen H, Divecha N, Dusseljee S, et al. Multivesicular body morphogenesis requires phosphatidyl‐inositol 3‐kinase activity. Curr Biol. 1999;9(1):55–8. [DOI] [PubMed] [Google Scholar]
- 23. Katzmann DJ, Babst M, Emr SD. Ubiquitin‐dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT‐I. Cell. 2001;106(2):145–55. [DOI] [PubMed] [Google Scholar]
- 24. Bache KG, Brech A, Mehlum A, Stenmark H. Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J Cell Biol. 2003;162(3):435–42. 10.1083/jcb.200302131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Razi M, Futter CE. Distinct roles for Tsg101 and Hrs in multivesicular body formation and inward vesiculation. Mol Biol Cell. 2006;17(8):3469–83. 10.1091/mbc.e05-11-1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shields SB, Oestreich AJ, Winistorfer S, Nguyen D, Payne JA, Katzmann DJ, et al. ESCRT ubiquitin‐binding domains function cooperatively during MVB cargo sorting. J Cell Biol. 2009;185(2):213–24. 10.1083/jcb.200811130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tamai K, Tanaka N, Nakano T, Kakazu E, Kondo Y, Inoue J, et al. Exosome secretion of dendritic cells is regulated by Hrs, an ESCRT‐0 protein. Biochem Biophys Res Commun. 2010;399(3):384–90. 10.1016/j.bbrc.2010.07.083 [DOI] [PubMed] [Google Scholar]
- 28. Stuffers S, Sem Wegner C, Stenmark H, Brech A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic. 2009;10(7):925–37. 10.1111/j.1600-0854.2009.00920.x [DOI] [PubMed] [Google Scholar]
- 29. Hsu C, Morohashi Y, Yoshimura S, Manrique‐Hoyos N, Jung SY, Lauterbach MA, et al. Regulation of exosome secretion by Rab35 and its GTPase‐activating proteins TBC1D10A‐C. J Cell Biol. 2010;189(2):223–32. 10.1083/jcb.200911018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ostrowski M, Carmo NB, Krumeich S, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12(1):19–30. 10.1038/ncb2000 [DOI] [PubMed] [Google Scholar]
- 31. Blanc L, Vidal M. New insights into the function of Rab GTPases in the context of exosomal secretion. Small GTPases. 2018;9(1–2):95–106. 10.1080/21541248.2016.1264352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Keerthikumar S, Chisanga D, Ariyaratne D, al Saffar H, Anand S, Zhao K, et al. ExoCarta: a web‐based compendium of exosomal cargo. J Mol Biol. 2016;428(4):688–92. 10.1016/j.jmb.2015.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim DK, Lee J, Kim SR, Choi DS, Yoon YJ, Kim JH, et al. EVpedia: a community web portal for extracellular vesicles research. Bioinformatics. 2015;31(6):933–9. 10.1093/bioinformatics/btu741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pathan M, Fonseka P, Chitti SV, Kang T, Sanwlani R, Van Deun J, et al. Vesiclepedia 2019: a compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res. 2019;47(D1):D516–9. 10.1093/nar/gky1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li S, Li Y, Chen B, Zhao J, Yu S, Tang Y, et al. exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018;46(D1):D106–12. 10.1093/nar/gkx891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thery C, Boussac M, Veron P, et al. Proteomic analysis of dendritic cell‐derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166(12):7309–18. [DOI] [PubMed] [Google Scholar]
- 37. Geis‐Asteggiante L, Belew AT, Clements VK, Edwards NJ, Ostrand‐Rosenberg S, el‐Sayed NM, et al. Differential content of proteins, mRNAs, and miRNAs suggests that MDSC and their exosomes may mediate distinct immune suppressive functions. J Proteome Res. 2018;17(1):486–98. 10.1021/acs.jproteome.7b00646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lasser C, Shelke GV, Yeri A, et al. Two distinct extracellular RNA signatures released by a single cell type identified by microarray and next‐generation sequencing. RNA Biol. 2017;14(1):58–72. 10.1080/15476286.2016.1249092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–4. 10.1038/cr.2015.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu H, Wang Q, Zhong H, Li L, Zhang Q, Huang Q, et al. Differentially expressed microRNAs in exosomes of patients with breast cancer revealed by nextgeneration sequencing. Oncol Rep. 2020;43(1):240–50. 10.3892/or.2019.7401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Batagov AO, Kurochkin IV. Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3′‐untranslated regions. Biol Direct. 2013;8:12. 10.1186/1745-6150-8-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7(3):e30679. 10.1371/journal.pone.0030679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Llorente A, Skotland T, Sylvanne T, et al. Molecular lipidomics of exosomes released by PC‐3 prostate cancer cells. Biochim Biophys Acta. 2013;1831(7):1302–9. [DOI] [PubMed] [Google Scholar]
- 44. Laulagnier K, Grand D, Dujardin A, Hamdi S, Vincent‐Schneider H, Lankar D, et al. PLD2 is enriched on exosomes and its activity is correlated to the release of exosomes. FEBS Lett. 2004;572(1–3):11–4. 10.1016/j.febslet.2004.06.082 [DOI] [PubMed] [Google Scholar]
- 45. Fitzner D, Schnaars M, van Rossum D, Krishnamoorthy G, Dibaj P, Bakhti M, et al. Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J Cell Sci. 2011;124(Pt 3):447–58. 10.1242/jcs.074088 [DOI] [PubMed] [Google Scholar]
- 46. Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319(5867):1244–7. 10.1126/science.1153124 [DOI] [PubMed] [Google Scholar]
- 47. Wubbolts R, Leckie RS, Veenhuizen PT, et al. Proteomic and biochemical analyses of human B cell‐derived exosomes. Potential implications for their function and multivesicular body formation. J Biol Chem. 2003;278(13):10963–72. 10.1074/jbc.M207550200 [DOI] [PubMed] [Google Scholar]
- 48. Brouwers JF, Aalberts M, Jansen JW, et al. Distinct lipid compositions of two types of human prostasomes. Proteomics. 2013;13(10‐11):1660–6. 10.1002/pmic.201200348 [DOI] [PubMed] [Google Scholar]
- 49. Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10(8):513–25. 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- 50. Yang JM, Gould SJ. The cis‐acting signals that target proteins to exosomes and microvesicles. Biochem Soc Trans. 2013;41(1):277–82. 10.1042/BST20120275 [DOI] [PubMed] [Google Scholar]
- 51. Huang Q, Yang J, Zheng J, Hsueh C, Guo Y, Zhou L. Characterization of selective exosomal microRNA expression profile derived from laryngeal squamous cell carcinoma detected by next generation sequencing. Oncol Rep. 2018;40(5):2584–94. 10.3892/or.2018.6672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Preusser C, Hung LH, Schneider T, et al. Selective release of circRNAs in platelet‐derived extracellular vesicles. J Extracell Vesicles. 2018;7(1):1424473. 10.1080/20013078.2018.1424473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nolte‐'t Hoen EN, Buermans HP, Waasdorp M, Stoorvogel W, Wauben MH, t Hoen PA. Deep sequencing of RNA from immune cell‐derived vesicles uncovers the selective incorporation of small non‐coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012;40(18):9272–85. 10.1093/nar/gks658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Villarroya‐Beltri C, Gutierrez‐Vazquez C, Sanchez‐Cabo F, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. 10.1038/ncomms3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Koppers‐Lalic D, Hackenberg M, Bijnsdorp IV, Van Eijndhoven MAJ, Sadek P, Sie D, et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014;8(6):1649–58. 10.1016/j.celrep.2014.08.027 [DOI] [PubMed] [Google Scholar]
- 56. Al‐Nedawi K, Meehan B, Micallef J, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10(5):619–24. 10.1038/ncb1725 [DOI] [PubMed] [Google Scholar]
- 57. Patel B, Patel J, Cho JH, Manne S, Bonala S, Henske E, et al. Exosomes mediate the acquisition of the disease phenotypes by cells with normal genome in tuberous sclerosis complex. Oncogene. 2016;35(23):3027–36. 10.1038/onc.2015.358 [DOI] [PubMed] [Google Scholar]
- 58. Koppers‐Lalic D, Hogenboom MM, Middeldorp JM, Pegtel DM. Virus‐modified exosomes for targeted RNA delivery; a new approach in nanomedicine. Adv Drug Deliv Rev. 2013;65(3):348–56. 10.1016/j.addr.2012.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lai CP, Kim EY, Badr CE, Weissleder R, Mempel TR, Tannous BA, et al. Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat Commun. 2015;6:7029. 10.1038/ncomms8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ridder K, Keller S, Dams M, Rupp AK, Schlaudraff J, Del Turco D, et al. Extracellular vesicle‐mediated transfer of genetic information between the hematopoietic system and the brain in response to inflammation. PLoS Biol. 2014;12(6):e1001874. 10.1371/journal.pbio.1001874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. O'Donoghue EJ, Krachler AM. Mechanisms of outer membrane vesicle entry into host cells. Cell Microbiol. 2016;18(11):1508–17. 10.1111/cmi.12655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014;3(1). 10.3402/jev.v3.24641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Heusermann W, Hean J, Trojer D, Steib E, Von Bueren S, Graff‐Meyer A, et al. Exosomes surf on filopodia to enter cells at endocytic hot spots, traffic within endosomes, and are targeted to the ER. J Cell Biol. 2016;213(2):173–84. 10.1083/jcb.201506084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tian T, Zhu YL, Hu FH, Wang YY, Huang NP, Xiao ZD. Dynamics of exosome internalization and trafficking. J Cell Physiol. 2013;228(7):1487–95. 10.1002/jcp.24304 [DOI] [PubMed] [Google Scholar]
- 65. Miyazaki T, Ikeda K, Sato W, Horie‐Inoue K, Inoue S. Extracellular vesicle‐mediated EBAG9 transfer from cancer cells to tumor microenvironment promotes immune escape and tumor progression. Oncogenesis. 2018;7(1):7. 10.1038/s41389-017-0022-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shen Y, Guo D, Weng L, Wang S, Ma Z, Yang Y, et al. Tumor‐derived exosomes educate dendritic cells to promote tumor metastasis via HSP72/HSP105‐TLR2/TLR4 pathway. Onco Targets Ther. 2017;6(12):e1362527. 10.1080/2162402X.2017.1362527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sun X, Lin F, Sun W, Zhu W, Fang D, Luo L, et al. Exosome‐transmitted miRNA‐335‐5p promotes colorectal cancer invasion and metastasis by facilitating EMT via targeting RASA1. Mol Ther Nucleic Acids. 2021;24:164–74. 10.1016/j.omtn.2021.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Behera J, Kumar A, Voor MJ, Tyagi N. Exosomal lncRNA‐H19 promotes osteogenesis and angiogenesis through mediating Angpt1/Tie2‐NO signaling in CBS‐heterozygous mice. Theranostics. 2021;11(16):7715–34. 10.7150/thno.58410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li S, Yi M, Dong B, Jiao Y, Luo S, Wu K. The roles of exosomes in cancer drug resistance and its therapeutic application. Clin Transl Med. 2020;10(8):e257. 10.1002/ctm2.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Whiteside TL. Exosomes in cancer: another mechanism of tumor‐induced immune suppression. Adv Exp Med Biol. 2017;1036:81–9. 10.1007/978-3-319-67577-0_6 [DOI] [PubMed] [Google Scholar]
- 71. Lee H, Abston E, Zhang D, Rai A, Jin Y. Extracellular vesicle: an emerging mediator of intercellular crosstalk in lung inflammation and injury. Front Immunol. 2018;9:924. 10.3389/fimmu.2018.00924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Peng Q, Zhang J, Zhou G. Circulating exosomes regulate T‐cell mediated inflammatory response in oral lichen planus. J Oral Pathol Med. 2018;48:143–50. 10.1111/jop.12804 [DOI] [PubMed] [Google Scholar]
- 73. Altevogt P, Bretz NP, Ridinger J, Utikal J, Umansky V. Novel insights into exosome‐induced, tumor‐associated inflammation and immunomodulation. Semin Cancer Biol. 2014;28:51–7. 10.1016/j.semcancer.2014.04.008 [DOI] [PubMed] [Google Scholar]
- 74. Maybruck BT, Pfannenstiel LW, Diaz‐Montero M, Gastman BR. Tumor‐derived exosomes induce CD8+ T cell suppressors. J Immunother Cancer. 2017;5:65. 10.1186/s40425-017-0269-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang X, Shen H, He Q, Tian W, Xia A, Lu XJ. Exosomes derived from exhausted CD8+ T cells impaired the anticancer function of normal CD8+ T cells. J Med Genet. 2019;56(1):29–31. 10.1136/jmedgenet-2018-105439 [DOI] [PubMed] [Google Scholar]
- 76. Kadota T, Yoshioka Y, Fujita Y, Kuwano K, Ochiya T. Extracellular vesicles in lung cancer‐From bench to bedside. Semin Cell Dev Biol. 2017;67:39–47. 10.1016/j.semcdb.2017.03.001 [DOI] [PubMed] [Google Scholar]
- 77. Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat Rev Cancer. 2018;18(2):128–34. 10.1038/nrc.2017.118 [DOI] [PubMed] [Google Scholar]
- 78. Derynck R, Weinberg RA. EMT and cancer: more than meets the eye. Dev Cell. 2019;49(3):313–6. 10.1016/j.devcel.2019.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Mahmood MQ, Ward C, Muller HK, Sohal SS, Walters EH. Epithelial mesenchymal transition (EMT) and non‐small cell lung cancer (NSCLC): a mutual association with airway disease. Med Oncol. 2017;34(3):45. 10.1007/s12032-017-0900-y [DOI] [PubMed] [Google Scholar]
- 80. Chockley PJ, Chen J, Chen G, Beer DG, Standiford TJ, Keshamouni VG. Epithelial‐mesenchymal transition leads to NK cell‐mediated metastasis‐specific immunosurveillance in lung cancer. J Clin Invest. 2018;128(4):1384–96. 10.1172/JCI97611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Rahman MA, Barger JF, Lovat F, Gao M, Otterson GA, Nana‐Sinkam P. Lung cancer exosomes as drivers of epithelial mesenchymal transition. Oncotarget. 2016;7(34):54852–66. 10.18632/oncotarget.10243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ye Z, Zhang X, Luo Y, Li S, Huang L, Li Z, et al. Prognostic values of vimentin expression and its clinicopathological significance in non‐small cell lung cancer: a meta‐analysis of observational studies with 4118 cases. PLoS One. 2016;11(9):e0163162. 10.1371/journal.pone.0163162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tadokoro A, Kanaji N, Liu D, Yokomise H, Haba R, Ishii T, et al. Vimentin regulates invasiveness and is a poor prognostic marker in non‐small cell lung cancer. Anticancer Res. 2016;36(4):1545–51. [PubMed] [Google Scholar]
- 84. Shao C, Yang F, Miao S, Liu W, Wang C, Shu Y, et al. Role of hypoxia‐induced exosomes in tumor biology. Mol Cancer. 2018;17(1):120. 10.1186/s12943-018-0869-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hsu YL, Hung JY, Chang WA, Lin YS, Pan YC, Tsai PH, et al. Hypoxic lung cancer‐secreted exosomal miR‐23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO‐1. Oncogene. 2017;36(34):4929–42. 10.1038/onc.2017.105 [DOI] [PubMed] [Google Scholar]
- 86. Matsuura Y, Wada H, Eguchi H, Gotoh K, Kobayashi S, Kinoshita M, et al. Exosomal miR‐155 derived from hepatocellular carcinoma cells under hypoxia promotes angiogenesis in endothelial cells. Dig Dis Sci. 2019;64(3):792–802. 10.1007/s10620-018-5380-1 [DOI] [PubMed] [Google Scholar]
- 87. Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153(3):516–9. 10.1016/j.cell.2013.04.003 [DOI] [PubMed] [Google Scholar]
- 88. Zhou Y, Ren H, Dai B, Li J, Shang L, Huang J, et al. Hepatocellular carcinoma‐derived exosomal miRNA‐21 contributes to tumor progression by converting hepatocyte stellate cells to cancer‐associated fibroblasts. J Exp Clin Cancer Res. 2018;37(1):324. 10.1186/s13046-018-0965-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, et al. Cancer‐secreted miR‐105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25(4):501–15. 10.1016/j.ccr.2014.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Silva J, Garcia V, Rodriguez M, Compte M, Cisneros E, Veguillas P, et al. Analysis of exosome release and its prognostic value in human colorectal cancer. Genes Chromosomes Cancer. 2012;51(4):409–18. [DOI] [PubMed] [Google Scholar]
- 91. Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–6. 10.1038/ncb1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Qu X, Li Q, Yang J, Zhao H, Wang F, Zhang F, et al. Double‐stranded DNA in exosomes of malignant pleural effusions as a novel DNA source for EGFR mutation detection in lung adenocarcinoma. Front Oncol. 2019;9:931. 10.3389/fonc.2019.00931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Zheng W, Ji D, Zhou Y, Yu L, Huang P, Zheng Y, et al. Exosomal non‐coding RNAs in hepatobiliary cancer: a rising star. Mol Cancer Ther. 2021;20:1777–88. 10.1158/1535-7163.MCT-21-0363 [DOI] [PubMed] [Google Scholar]
- 94. Yang B, Teng F, Chang L, et al. Tumor‐derived exosomal circRNA_102481 contributes to EGFR‐TKIs resistance via the miR‐30a‐5p/ROR1 axis in non‐small cell lung cancer. Aging (Albany NY). 2021;13(9):13264–86. 10.18632/aging.203011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Liu Y, Cao X. Characteristics and Significance of the Pre‐metastatic Niche. Cancer Cell. 2016;30(5):668–81. 10.1016/j.ccell.2016.09.011 [DOI] [PubMed] [Google Scholar]
- 96. Peinado H, Zhang H, Matei IR, Costa‐Silva B, Hoshino A, Rodrigues G, et al. Pre‐metastatic niches: organ‐specific homes for metastases. Nat Rev Cancer. 2017;17(5):302–17. 10.1038/nrc.2017.6 [DOI] [PubMed] [Google Scholar]
- 97. Yuan X, Qian N, Ling S, Li Y, Sun W, Li J, et al. Breast cancer exosomes contribute to pre‐metastatic niche formation and promote bone metastasis of tumor cells. Theranostics. 2021;11(3):1429–45. 10.7150/thno.45351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sun H, Meng Q, Shi C, Yang H, Li X, Wu S, et al. Hypoxia‐inducible exosomes facilitate liver‐tropic pre‐metastatic niche in colorectal cancer. Hepatology. 2021;74(5):2633–51. 10.1002/hep.32009 [DOI] [PubMed] [Google Scholar]
- 99. Zeng Z, Li Y, Pan Y, Lan X, Song F, Sun J, et al. Cancer‐derived exosomal miR‐25‐3p promotes pre‐metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun. 2018;9(1):5395. 10.1038/s41467-018-07810-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Liu Y, Gu Y, Han Y, Zhang Q, Jiang Z, Zhang X, et al. Tumor exosomal RNAs promote lung pre‐metastatic Niche formation by activating alveolar epithelial TLR3 to recruit neutrophils. Cancer Cell. 2016;30(2):243–56. 10.1016/j.ccell.2016.06.021 [DOI] [PubMed] [Google Scholar]
- 101. Ji R, Zhang B, Zhang X, Xue J, Yuan X, Yan Y, et al. Exosomes derived from human mesenchymal stem cells confer drug resistance in gastric cancer. Cell Cycle. 2015;14(15):2473–83. 10.1080/15384101.2015.1005530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Lopes‐Rodrigues V, Di Luca A, Sousa D, et al. Multidrug resistant tumour cells shed more microvesicle‐like EVs and less exosomes than their drug‐sensitive counterpart cells. Biochim Biophys Acta. 2016;1860(3):618–27. 10.1016/j.bbagen.2015.12.011 [DOI] [PubMed] [Google Scholar]
- 103. Zheng P, Chen L, Yuan X, Luo Q, Liu Y, Xie G, et al. Exosomal transfer of tumor‐associated macrophage‐derived miR‐21 confers cisplatin resistance in gastric cancer cells. J Exp Clin Cancer Res. 2017;36(1):53. 10.1186/s13046-017-0528-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wang D, Zhao C, Xu F, Zhang A, Jin M, Zhang K, et al. Cisplatin‐resistant NSCLC cells induced by hypoxia transmit resistance to sensitive cells through exosomal PKM2. Theranostics. 2021;11(6):2860–75. 10.7150/thno.51797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Zhang HD, Jiang LH, Hou JC, Zhong SL, Zhu LP, Wang DD, et al. Exosome: a novel mediator in drug resistance of cancer cells. Epigenomics. 2018;10(11):1499–509. 10.2217/epi-2017-0151 [DOI] [PubMed] [Google Scholar]
- 106. Jayaseelan VP, Arumugam P. Exosome‐derived ncRNAs as potential drivers of epigenetic reprogramming of cancer stem cells. Epigenomics. 2021;13(18):1439–41. 10.2217/epi-2021-0139 [DOI] [PubMed] [Google Scholar]
- 107. Zang X, Gu J, Zhang J, Shi H, Hou S, Xu X, et al. Exosome‐transmitted lncRNA UFC1 promotes non‐small‐cell lung cancer progression by EZH2‐mediated epigenetic silencing of PTEN expression. Cell Death Dis. 2020;11(4):215. 10.1038/s41419-020-2409-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Lu Y, Hou K, Li M, Wu X, Yuan S. Exosome‐delivered LncHEIH promotes gastric cancer progression by upregulating EZH2 and stimulating methylation of the GSDME promoter. Front Cell Dev Biol. 2020;8:571297. 10.3389/fcell.2020.571297 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 109. Wen D, Liu WL, Lu ZW, Cao YM, Ji QH, Wei WJ. SNHG9, a papillary thyroid cancer cell exosome‐enriched lncRNA, inhibits cell autophagy and promotes cell apoptosis of normal thyroid epithelial cell Nthy‐ori‐3 through YBOX3/P21 pathway. Front Oncol. 2021;11:647034. 10.3389/fonc.2021.647034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Huang YK, Wang M, Sun Y, di Costanzo N, Mitchell C, Achuthan A, et al. Macrophage spatial heterogeneity in gastric cancer defined by multiplex immunohistochemistry. Nat Commun. 2019;10(1):3928. 10.1038/s41467-019-11788-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Franklin RA, Liao W, Sarkar A, Kim MV, Bivona MR, Liu K, et al. The cellular and molecular origin of tumor‐associated macrophages. Science. 2014;344(6186):921–5. 10.1126/science.1252510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Yang M, McKay D, Pollard JW, Lewis CE. Diverse functions of macrophages in different tumor microenvironments. Cancer Res. 2018;78(19):5492–503. 10.1158/0008-5472.CAN-18-1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Dong H, Xie C, Jiang Y, Li K, Lin Y, Pang X, et al. Tumor‐derived exosomal protein tyrosine phosphatase receptor Type O polarizes macrophage to suppress breast tumor cell invasion and migration. Front Cell Dev Biol. 2021;9:703537. 10.3389/fcell.2021.703537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Chen J, Zhang K, Zhi Y, Wu Y, Chen B, Bai J, et al. Tumor‐derived exosomal miR‐19b‐3p facilitates M2 macrophage polarization and exosomal LINC00273 secretion to promote lung adenocarcinoma metastasis via Hippo pathway. Clin Transl Med. 2021;11(9):e478. 10.1002/ctm2.478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Zhang S, Li D, Zhao M, Yang F, Sang C, Yan C, et al. Exosomal miR‐183‐5p shuttled by M2 polarized tumor‐associated macrophage promotes the development of colon cancer via targeting THEM4 mediated PI3K/AKT and NF‐kappaB pathways. Front Oncol. 2021;11:672684. 10.3389/fonc.2021.672684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16(9):582–98. 10.1038/nrc.2016.73 [DOI] [PubMed] [Google Scholar]
- 117. Liang LJ, Yang Y, Wei WF, Wu XG, Yan RM, Zhou CF, et al. Tumor‐secreted exosomal Wnt2B activates fibroblasts to promote cervical cancer progression. Oncogenesis. 2021;10(3):30. 10.1038/s41389-021-00319-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Tong Y, Yang L, Yu C, Zhu W, Zhou X, Xiong Y, et al. Tumor‐secreted exosomal lncRNA POU3F3 promotes cisplatin resistance in ESCC by inducing fibroblast differentiation into CAFs. Mol Ther Oncolytics. 2020;18:1–13. 10.1016/j.omto.2020.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Luo R, Liu M, Yang Q, Cheng H, Yang H, Li M, et al. Emerging diagnostic potential of tumor‐derived exosomes. J Cancer. 2021;12(16):5035–45. 10.7150/jca.59391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Verdi J, Ketabchi N, Noorbakhsh N, Saleh M, Ebrahimi‐Barough S, Seyhoun I, et al. Development and clinical application of tumor‐derived exosomes in patients with cancer. Curr Stem Cell Res Ther. 2021;16. 10.2174/1574888X16666210622123942 [DOI] [PubMed] [Google Scholar]
- 121. Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, et al. Glypican‐1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523(7559):177–82. 10.1038/nature14581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Kimura H, Yamamoto H, Harada T, Fumoto K, Osugi Y, Sada R, et al. CKAP4, a DKK1 receptor, is a biomarker in exosomes derived from pancreatic cancer and a molecular target for therapy. Clin Cancer Res. 2019;25(6):1936–47. 10.1158/1078-0432.CCR-18-2124 [DOI] [PubMed] [Google Scholar]
- 123. Zhou L, Wang W, Wang F, Yang S, Hu J, Lu B, et al. Plasma‐derived exosomal miR‐15a‐5p as a promising diagnostic biomarker for early detection of endometrial carcinoma. Mol Cancer. 2021;20(1):57. 10.1186/s12943-021-01352-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Lu X, Lu J, Wang S, Zhang Y, Ding Y, Shen X, et al. Circulating serum exosomal miR‐92a‐3p as a novel biomarker for early diagnosis of gastric cancer. Future Oncol. 2021;17(8):907–19. 10.2217/fon-2020-0792 [DOI] [PubMed] [Google Scholar]
- 125. Yao YF, Qu MW, Li GC, Zhang FB, Rui HC. Circulating exosomal miRNAs as diagnostic biomarkers in Parkinson's disease. Eur Rev Med Pharmacol Sci. 2018;22(16):5278–83. 10.26355/eurrev_201808_15727 [DOI] [PubMed] [Google Scholar]
- 126. Levanen B, Bhakta NR, Torregrosa Paredes P, et al. Altered microRNA profiles in bronchoalveolar lavage fluid exosomes in asthmatic patients. J Allergy Clin Immunol. 2013;131(3):894–903. 10.1016/j.jaci.2012.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Wu J, Shen Z. Exosomal miRNAs as biomarkers for diagnostic and prognostic in lung cancer. Cancer Med. 2020;9(19):6909–22. 10.1002/cam4.3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Smolarz M, Widlak P. Serum exosomes and their miRNA Load‐A potential biomarker of lung cancer. Cancers (Basel). 2021;13(6):1373. 10.3390/cancers13061373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Rabinowits G, Gercel‐Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10(1):42–6. 10.3816/CLC.2009.n.006 [DOI] [PubMed] [Google Scholar]
- 130. Cazzoli R, Buttitta F, Di Nicola M, et al. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol. 2013;8(9):1156–62. 10.1097/JTO.0b013e318299ac32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Jin X, Chen Y, Chen H, Fei S, Chen D, Cai X, et al. Evaluation of tumor‐derived exosomal mirna as potential diagnostic biomarkers for early‐stage non‐small cell lung cancer using next‐generation sequencing. Clin Cancer Res. 2017;23(17):5311–9. 10.1158/1078-0432.CCR-17-0577 [DOI] [PubMed] [Google Scholar]
- 132. Zheng Q, Ding H, Wang L, Yan Y, Wan Y, Yi Y, et al. Circulating exosomal miR‐96 as a novel biomarker for radioresistant non‐small‐cell lung cancer. J Oncol. 2021;2021:5893981. 10.1155/2021/5893981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Tang Y, Cui Y, Li Z, Jiao Z, Zhang Y, He Y, et al. Radiation‐induced miR‐208a increases the proliferation and radioresistance by targeting p21 in human lung cancer cells. J Exp Clin Cancer Res. 2016;35:7. 10.1186/s13046-016-0285-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Yuwen DL, Sheng BB, Liu J, Wenyu W, Shu YQ. MiR‐146a‐5p level in serum exosomes predicts therapeutic effect of cisplatin in non‐small cell lung cancer. Eur Rev Med Pharmacol Sci. 2017;21(11):2650–8. [PubMed] [Google Scholar]
- 135. Qin X, Yu S, Zhou L, Shi M, Hu Y, Xu X, et al. Cisplatin‐resistant lung cancer cell‐derived exosomes increase cisplatin resistance of recipient cells in exosomal miR‐100‐5p‐dependent manner. Int J Nanomedicine. 2017;12:3721–33. 10.2147/IJN.S131516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Poroyko V, Mirzapoiazova T, Nam A, et al. Exosomal miRNAs species in the blood of small cell and non‐small cell lung cancer patients. Oncotarget. 2018;9(28):19793–806. 10.18632/oncotarget.24857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Dejima H, Iinuma H, Kanaoka R, Matsutani N, Kawamura M. Exosomal microRNA in plasma as a non‐invasive biomarker for the recurrence of non‐small cell lung cancer. Oncol Lett. 2017;13(3):1256–63. 10.3892/ol.2017.5569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Kanaoka R, Iinuma H, Dejima H, Sakai T, Uehara H, Matsutani N, et al. Usefulness of Plasma Exosomal MicroRNA‐451a as a Noninvasive Biomarker for Early Prediction of Recurrence and Prognosis of Non‐Small Cell Lung Cancer. Oncology. 2018;94(5):311–23. 10.1159/000487006 [DOI] [PubMed] [Google Scholar]
- 139. Liu Q, Yu Z, Yuan S, et al. Circulating exosomal microRNAs as prognostic biomarkers for non‐small‐cell lung cancer. Oncotarget. 2017;8(8):13048–58. 10.18632/oncotarget.14369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Zhou L, Lv T, Zhang Q, Zhu Q, Zhan P, Zhu S, et al. The biology, function and clinical implications of exosomes in lung cancer. Cancer Lett. 2017;407:84–92. 10.1016/j.canlet.2017.08.003 [DOI] [PubMed] [Google Scholar]
- 141. Clark DJ, Fondrie WE, Yang A, Mao L. Triple SILAC quantitative proteomic analysis reveals differential abundance of cell signaling proteins between normal and lung cancer‐derived exosomes. J Proteomics. 2016;133:161–9. 10.1016/j.jprot.2015.12.023 [DOI] [PubMed] [Google Scholar]
- 142. Yamashita T, Kamada H, Kanasaki S, Maeda Y, Nagano K, Abe Y, et al. Epidermal growth factor receptor localized to exosome membranes as a possible biomarker for lung cancer diagnosis. Pharmazie. 2013;68(12):969–73. [PubMed] [Google Scholar]
- 143. Huang SH, Li Y, Zhang J, Rong J, Ye S. Epidermal growth factor receptor‐containing exosomes induce tumor‐specific regulatory T cells. Cancer Invest. 2013;31(5):330–5. 10.3109/07357907.2013.789905 [DOI] [PubMed] [Google Scholar]
- 144. Jakobsen KR, Paulsen BS, Baek R, Varming K, Sorensen BS, Jorgensen MM. Exosomal proteins as potential diagnostic markers in advanced non‐small cell lung carcinoma. J Extracell Vesicles. 2015;4:26659. 10.3402/jev.v4.26659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Sandfeld‐Paulsen B, Jakobsen KR, Baek R, et al. Exosomal Proteins as Diagnostic Biomarkers in Lung Cancer. J Thorac Oncol. 2016;11(10):1701–10. 10.1016/j.jtho.2016.05.034 [DOI] [PubMed] [Google Scholar]
- 146. Wang N, Song X, Liu L, Niu L, Wang X, Song X, et al. Circulating exosomes contain protein biomarkers of metastatic non‐small‐cell lung cancer. Cancer Sci. 2018;109(5):1701–9. 10.1111/cas.13581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Niu L, Song X, Wang N, Xue L, Song X, Xie L. Tumor‐derived exosomal proteins as diagnostic biomarkers in non‐small cell lung cancer. Cancer Sci. 2018;110:433–42. 10.1111/cas.13862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Sandfeld‐Paulsen B, Aggerholm‐Pedersen N, Baek R, Jakobsen KR, Meldgaard P, Folkersen BH, et al. Exosomal proteins as prognostic biomarkers in non‐small cell lung cancer. Mol Oncol. 2016;10(10):1595–602. 10.1016/j.molonc.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Eftekhari A, Hasanzadeh M, Sharifi S, Dizaj SM, Khalilov R, Ahmadian E. Bioassay of saliva proteins: the best alternative for conventional methods in non‐invasive diagnosis of cancer. Int J Biol Macromol. 2019;124:1246–55. 10.1016/j.ijbiomac.2018.11.277 [DOI] [PubMed] [Google Scholar]
- 150. Sharma S, Rasool HI, Palanisamy V, Mathisen C, Schmidt M, Wong DT, et al. Structural‐mechanical characterization of nanoparticle exosomes in human saliva, using correlative AFM, FESEM, and force spectroscopy. ACS Nano. 2010;4(4):1921–6. 10.1021/nn901824n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Yang J, Wei F, Schafer C, Wong DT. Detection of tumor cell‐specific mRNA and protein in exosome‐like microvesicles from blood and saliva. PLoS One. 2014;9(11):e110641. 10.1371/journal.pone.0110641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Sun Y, Huo C, Qiao Z, Shang Z, Uzzaman A, Liu S, et al. Comparative proteomic analysis of exosomes and microvesicles in human saliva for lung cancer. J Proteome Res. 2018;17(3):1101–7. 10.1021/acs.jproteome.7b00770 [DOI] [PubMed] [Google Scholar]
- 153. Jouida A, McCarthy C, Fabre A, Keane MP. Exosomes: a new perspective in EGFR‐mutated lung cancer. Cancer Metastasis Rev. 2021;40(2):589–601. 10.1007/s10555-021-09962-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Castellanos‐Rizaldos E, Grimm DG, Tadigotla V, Hurley J, Healy J, Neal PL, et al. Exosome‐based detection of EGFR T790M in plasma from non‐small cell lung cancer patients. Clin Cancer Res. 2018;24(12):2944–50. 10.1158/1078-0432.CCR-17-3369 [DOI] [PubMed] [Google Scholar]
- 155. Krug AK, Enderle D, Karlovich C, Priewasser T, Bentink S, Spiel A, et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann Oncol. 2018;29(3):700–6. 10.1093/annonc/mdx765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Okuma Y, Morikawa K, Tanaka H, Yokoyama T, Itani H, Horiuchi K, et al. Prospective exosome‐focused translational research for afatinib study of non‐small cell lung cancer patients expressing EGFR (EXTRA study). Thorac Cancer. 2019;10(2):395–400. 10.1111/1759-7714.12923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Wu YJ, Muldoon LL, Gahramanov S, Kraemer DF, Marshall DJ, Neuwelt EA. Targeting alphaV‐integrins decreased metastasis and increased survival in a nude rat breast cancer brain metastasis model. J Neurooncol. 2012;110(1):27–36. 10.1007/s11060-012-0942-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Inamdar S, Nitiyanandan R, Rege K. Emerging applications of exosomes in cancer therapeutics and diagnostics. Bioeng Transl Med. 2017;2(1):70–80. 10.1002/btm2.10059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Kotmakci M, Bozok Cetintas V. Extracellular vesicles as natural nanosized delivery systems for small‐molecule drugs and genetic material: steps towards the future nanomedicines. J Pharm Pharm Sci. 2015;18(3):396–413. [DOI] [PubMed] [Google Scholar]
- 160. Johnsen KB, Gudbergsson JM, Skov MN, Pilgaard L, Moos T, Duroux M. A comprehensive overview of exosomes as drug delivery vehicles ‐ endogenous nanocarriers for targeted cancer therapy. Biochim Biophys Acta. 2014;1846(1):75–87. 10.1016/j.bbcan.2014.04.005 [DOI] [PubMed] [Google Scholar]
- 161. Farooqi AA, Desai NN, Qureshi MZ, Librelotto DRN, Gasparri ML, Bishayee A, et al. Exosome biogenesis, bioactivities and functions as new delivery systems of natural compounds. Biotechnol Adv. 2018;36(1):328–34. 10.1016/j.biotechadv.2017.12.010 [DOI] [PubMed] [Google Scholar]
- 162. Wang Y, Guo M, Lin D, Liang D, Zhao L, Zhao R, et al. Docetaxel‐loaded exosomes for targeting non‐small cell lung cancer: preparation and evaluation in vitro and in vivo. Drug Deliv. 2021;28(1):1510–23. 10.1080/10717544.2021.1951894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Bai J, Duan J, Liu R, du Y, Luo Q, Cui Y, et al. Engineered targeting tLyp‐1 exosomes as gene therapy vectors for efficient delivery of siRNA into lung cancer cells. Asian J Pharm Sci. 2020;15(4):461–71. 10.1016/j.ajps.2019.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Santos P, Almeida F. Exosome‐based vaccines: history, current state, and clinical trials. Front Immunol. 2021;12:711565. 10.3389/fimmu.2021.711565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Yaddanapudi K, Meng S, Whitt AG, al Rayyan N, Richie J, Tu A, et al. Exosomes from GM‐CSF expressing embryonic stem cells are an effective prophylactic vaccine for cancer prevention. Onco Targets Ther. 2019;8(3):1561119. 10.1080/2162402X.2018.1561119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Morse MA, Garst J, Osada T, Khan S, Hobeika A, Clay TM, et al. A phase I study of dexosome immunotherapy in patients with advanced non‐small cell lung cancer. J Transl Med. 2005;3(1):9. 10.1186/1479-5876-3-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Besse B, Charrier M, Lapierre V, Dansin E, Lantz O, Planchard D, et al. Dendritic cell‐derived exosomes as maintenance immunotherapy after first line chemotherapy in NSCLC. Onco Targets Ther. 2016;5(4):e1071008. 10.1080/2162402X.2015.1071008 [DOI] [PMC free article] [PubMed] [Google Scholar]


