Abstract
The performance of a new, rapid, easy-to-perform assay based on neuraminidase enzyme activity for detection of influenza virus types A and B was compared to detection by culture, indirect immunofluorescence, and enzyme immunoassay in 479 nasal wash specimens from children with respiratory infections. Compared to isolation of influenza virus by culture, the neuraminidase assay had a sensitivity of 70.1%, specificity of 92.4%, positive predictive value of 76.3%, and negative predictive value of 89.9%. There was a higher sensitivity for the detection of influenza A virus (76.4%) than for influenza B virus (40.9%). Indirect immunofluorescence showed a sensitivity of 59.8% and specificity of 97% compared to culture isolation for detection of influenza A and B viruses. Enzyme immunoassay showed a sensitivity of 89.7% and specificity of 98.1% for the detection of influenza A alone. The quality of the nasal wash specimen had a significant effect on the detection of influenza virus by all of the assays. A strong response of the neuraminidase assay was more likely to represent a culture-confirmed influenza infection. This new rapid neuraminidase assay was useful for the detection of influenza A and B viruses in nasal wash specimens.
Influenza viruses are a significant cause of morbidity and hospitalizations in children, especially young infants and those with chronic diseases (6, 8, 11, 22, 25). The standard method of diagnosis for influenza infection is isolation of the virus by culture from respiratory secretions, which may take several days. Tests for rapid diagnosis of influenza A and B virus by direct or indirect immunofluorescence assay on exfoliated nasopharyngeal cells have shown variable sensitivity (40 to 100%) and specificity (86 to 99%) (9, 16, 19, 23, 24, 26). Influenza A virus may also be detected rapidly in nasal wash secretions by enzyme immunoassay (EIA), but this test does not detect influenza B virus (9, 13). The rapid detection of both types of influenza infections would allow appropriate antiviral therapy and is particularly important, since agents active against both influenza A and B are now available (12, 17, 21). Rapid detection also may decrease the use of antibiotics in patients with respiratory tract infections (20, 27).
A new rapid diagnostic kit based on the detection of neuraminidase specifically produced by influenza viruses (Zstat Flu; ZymeTx, Oklahoma, Okla.) recently has been approved for the rapid diagnosis of influenza A and B infections in throat swab specimens by the Food and Drug Administration (FDA) (18). While the manufacturer recommends testing in throat swab specimens, preliminary studies using this kit showed similar performance using nasal wash specimens, which are frequently used in children for diagnosis of other viral respiratory infections, such as respiratory syncytial virus (9, 18). The ability to use one sample for multiple tests is advantageous to the patient and to health care professionals.
The purposes of this study were to describe the performance of a new neuraminidase detection assay for the detection of influenza A and B viruses in nasal wash specimens and to compare it to established diagnostic modalities commonly used in pediatric patients.
MATERIALS AND METHODS
Patient samples.
All nasal wash and nasal aspirate specimens submitted between 30 December 1998 and 20 March 1999, during weekday, daytime shifts, to the Diagnostic Virology Laboratory, Texas Children's Hospital, Houston, for detection of respiratory viruses were included in this study. Nasal wash and aspirates were performed by respiratory therapists according to a standardized procedure. One to three milliliters of normal saline was instilled into the patient's nares. A sterile suction catheter was introduced approximately 2.5 cm into the nasopharynx, suction was applied, and secretions were collected in a sterile mucous trap. Samples were prospectively processed for rapid detection of influenza and for viral culture.
Laboratory methods.
Three different rapid detection assays were evaluated. The first detection assay was a newly FDA-approved neuraminidase detection assay (Zstat Flu; ZymeTx) and was performed according to the manufacturer's instructions. This test detects both influenza A and B viruses but does not distinguish between the two viruses. Briefly, nasal wash specimens (1 ml) were mixed with 200 μl of concentrated buffered solution. The reconstituted substrate vials were inverted several times for mixing and then placed in an electric heating mantle at 41°C for 30 min. Next, 0.5 ml of alkaline solution was added to stop the reaction, and the solution was transferred into a collecting device. After the solution drained, the collecting device's membrane color turned blue for a positive reaction. A negative reaction was noted when the membrane color remained white. Results were also categorized as weak positive when the coloration was light and strong positive when the membrane was strongly colored. The test required approximately 30 min to perform. The only equipment required was a specially designed heating block, provided by the manufacturer. Initially, the assay was performed by one technician (B.C.), who had received instructions from the manufacturer, scored 100% proficiency in two separate proficiency tests of 15 coded samples, and was experienced with this test (18). During the time period of conduction of the study, 130 additional samples were tested by other technicians trained by B.C. in the use of the test.
The second rapid detection test for influenza viruses was antigen detection by indirect immunofluorescence assay (IFA) (Microscan; Bartels Viral Respiratory Screening and Identification, Baxter Laboratories, West Sacramento, Calif.) that also detects influenza A and B viruses. Samples were processed according to previously described procedures (16). This test required about 4 h to perform and required a fluorescence microscope and a senior technician experienced in the reading of immunofluorescence slides. Specimens were divided into three categories according to the cellularity observed during IFA testing. When <20 cells per slide were observed, the specimen was considered inadequate for IFA analysis, since a positive test could be missed due to the low number of cells examined. Specimens with >20 cells per slide were qualitatively categorized as having sparse or abundant cellularity if there were less or more than 3 to 5 cells per high-power field, respectively. Specimens with >20 cells per slide were considered positive when at least 1 cell showed immunofluorescence staining.
The third rapid detection test for influenza virus was EIA (Directigen FluA; Becton-Dickinson, Cockeysville, Md.). This test detected only influenza A virus and was performed according to the manufacturer's instructions (26). It required 30 min to perform and required no special equipment and was easily performed by all technicians.
Viral cultures also were performed in all samples according to standard virologic technique (15). Samples were inoculated into human foreskin fibroblast, rhesus monkey kidney, and human lung carcinoma (A549) cell culture monolayers. Cultures were visually inspected under light microscopy daily for 14 days for evidence of viral cytopathic effect. Hemadsorption with a 0.4% suspension of guinea pig red blood cells to rhesus monkey kidney cell culture tubes was performed on days 2, 7, and 14 of incubation. Respiratory viruses isolated in culture were identified by immunofluorescence assay (Microscan; Bartels Viral Respiratory Screening and Identification, Baxter Laboratories), herpes simplex virus and cytomegalovirus were confirmed by immunofluorescence (Syva Microtrak; Syva Company, Palo Alto, Calif.), and picornaviruses were identified as rhinovirus or enterovirus by acid lability (pH 3) testing. The mean time for initial identification of viral effect by hemadsorption for influenza virus-positive cultures was 3.6 (±2.3) days.
A viral nucleic acid assay, utilizing a previously described reverse transcription-PCR (RT-PCR) assay was performed in apparently false-positive samples for which a positive neuraminidase detection assay result was found but the viral culture result was negative (1, 2). Extraction of viral RNA from clinical specimens was performed by a modification of the method described by Boom et al. (4). Briefly, 50 μl of the specimen was added to a reaction vessel containing 900 μl of L6 buffer and size-fractionated silica. The L6 buffer was prepared by dissolving 120 g of GuSCN in 100 ml of 0.1 M Tris hydrochloride, pH 6.4; subsequently, 22 ml of 0.2 M EDTA solution was added to the solution, the pH was adjusted with NaOH to pH 8.0, and 2.6 g of Triton X-100 was added to the solution. After the solution was homogenized by vortexing for 5 s, it was centrifuged to pellet the silica-nucleic acid complex. The RNA was purified by washing the pellet with L2 buffer (120 g of GuSCN dissolved in 100 ml of 0.1 M Tris hydrochloride [pH 6.4]), 70% ethanol, and acetone. Purified RNA was eluted from the dried silica in TE buffer (10 mM Tris hydrochloride [pH 8.0] and 1 mM EDTA) and assayed for influenza A virus by RT-PCR using the primers FAM1 and FAM2 and for influenza B virus using the primers B1 and B2B (1, 2). Positive results by RT-PCR were identified by hybridization using virus-specific, digoxigenin-labeled probes.
Statistical analysis.
Sensitivity, specificity, and positive and negative predictive values were calculated from two-by-two contingency tables. Continuous variables were compared using the independent sample t test. Categorical variables were analyzed using Fisher's exact test or chi-squared test as appropriate. All comparisons were done using two-tailed tests, and a P value of <0.05 was considered significant.
RESULTS
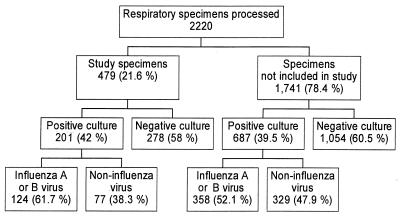
There were 2,220 specimens processed by the Diagnostic Virology Laboratory for respiratory viral culture during the study period. A total of 479 specimens were tested by the neuraminidase assay performed by a single technician (B.C.) during daylight, weekday hours, and constitute the study samples. Figure 1 shows results from viral cultures for all specimens processed during the study period. There were 130 specimens that had the neuraminidase assay performed by other technicians and are not included in the study sample.
FIG. 1.
Results of respiratory viral cultures performed at the Diagnostic Virology Laboratory during the study period (1 January 1999 to 20 March 1999). The numbers are the numbers of specimens in the different categories.
The 479 study samples were obtained from patients with a mean age of 3.8 years (range, 7 days to 27 years). Of the 479 patients, 293 (61.1%) were male and 186 (38.8%) were female. A total of 423 (88.3%) of these samples were also processed for IFA and 417 (87%) had an EIA for influenza A detection performed. All three rapid tests were performed on 374 (78%) specimens. Of the 479 study specimens, 201 (41.9%) had a virus isolated (102 influenza virus type A, 22 influenza virus type B, 32 rhinovirus, 17 parainfluenza virus, 17 respiratory syncytial virus, 6 adenovirus, 7 cytomegalovirus, 2 enterovirus, and 1 herpes simplex virus; in 5 specimens, a dual infection was detected).
Table 1 shows the performance of all three rapid detection assays compared to viral isolation by culture. There were 114 samples positive by the neuraminidase detection assay; the sensitivity was 70.1%, and the specificity was 92.4%. The sensitivity for detection of influenza A virus alone was 76.4%, and for influenza B virus alone, it was 40.9%. When tests categorized only as strong positive were analyzed as positive, the specificity for detection of influenza A and B virus was higher (95.8%), with a lower sensitivity (64.2%). When performance of 130 additional tests carried out by other technicians (not B.C.) less experienced with the kit was analyzed, lower sensitivity (45.7%) and specificity (76.2%) for detection of all influenza viruses compared to culture were observed.
TABLE 1.
Performance of rapid diagnostic tests for the detection of influenza virus infection in nasal wash samples compared to viral culture
| Test | n | Sensitivity (%) | Specificity (%) | Predictive value (%)
|
|
|---|---|---|---|---|---|
| Positive | Negative | ||||
| Neuraminidase assay | 479 | 70.1 | 92.4 | 76.3 | 89.9 |
| IFA | 321a | 59.8 | 97 | 88.1 | 86.6 |
| EIA for influenza A virus | |||||
| Detection of influenza A virus | 417 | 89.7 | 98.1 | 93.5 | 96.9 |
| Detection of influenza A or B virus | 417 | 74.3 | 98 | 93.5 | 90.7 |
A total of 423 samples were tested, but 102 were found to have inadequate numbers of cells.
Of the 479 specimens, 423 also were tested by IFA. Microscopic analysis showed that 102 (24.1%) samples were inadequate for IFA (<20 cells), while 60 had sparse cellularity, and 261 had abundant cells. Of the 321 specimens adequate for IFA, there were 48 (14.9%) positive for influenza A virus and 11 (3.4%) positive for influenza B virus. The sensitivity and specificity of IFA compared to viral culture for detection of any influenza virus was 59.8 and 97%, respectively. Influenza A EIA was performed in 417 of the 479 specimens. There were 93 (22.3%) positive and 324 (77.6%) negative test results, providing a sensitivity of 89.7% and specificity of 98.1% for detection of influenza A virus by EIA.
There was a decreased ability to recover viruses by culture in specimens that were considered to have sparse or inadequate cellularity during IFA compared to those with abundant cellularity (Table 2). However, when isolation of influenza viruses alone was compared between the groups, the difference did not attain statistical significance. There was no difference in the mean time for a culture to become positive between the three different IFA cellularity groups (mean times of 4.5, 4, and 4.2 days). The sensitivity for detection of influenza A or B viruses in specimens with abundant cellularity was 77% and the specificity of 89.4%, compared to sensitivity of 57.1% and specificity of 96.8% for the combination of the groups with sparse and inadequate cellularity. Sample cellularity also adversely influenced the performance of the IFA and EIA rapid tests.
TABLE 2.
Effect of quality of nasal wash specimen on the ability to isolate viruses and detect influenza virus by rapid diagnosis or by culturea
| Group | n | Culture positive (any virus) | Culture positive (influenza virus) | NAb positive | IFA positive | EIA positivec |
|---|---|---|---|---|---|---|
| 1 (abundant cellsd) | 261 | 50.2% | 28.4% | 29.5% | 21.5% | 26.3% |
| 2 (sparse cellse) | 60 | 31.7% | 21.7% | 18.3% | 5% | 14% |
| 3 (inadequate cellsf) | 102 | 26.5% | 21.6% | 12.7% | 0% | 19.5% |
| P values | ||||||
| Between groups 1 and 2 | P = 0.01 | NSg | NS | P = 0.003 | P = 0.04 | |
| Between groups 1 and 3 | P < 0.001 | NS | P = 0.001 | P < 0.001 | NS | |
| Between groups 2 and 3 | NS | NS | NS | P < 0.001 | NS |
Includes 423 specimens in which IFA was performed.
NA, neuraminidase assay.
EIA, enzyme immunoassay for influenza A virus. Includes only 395 specimens for which both EIA and IFA were performed.
Abundant cells, >20 cells per well with >3 to 5 cells per high-power field.
Sparse cells, >20 cells per well with <3 to 5 cells per high-power field.
Inadequate cells, <20 cells per well.
NS, difference not statistically significant.
There were 27 samples for which there was a positive neuraminidase test result, but no virus was recovered (Table 3). In six of the specimens, influenza virus RNA was detected by RT-PCR (influenza A virus detected in five and influenza B virus detected in one). In addition, 5 of the 27 samples were positive by EIA and/or IFA, providing further corroboration that the neuraminidase assay results represented true-positive results in these samples.
TABLE 3.
Results of additional tests for 27 nasal wash specimens that tested positive by neuraminidase detection assay but tested negative for influenza virus by culture
| n | Neuraminidase assay | IFA | Influenza A EIA | RT-PCR |
|---|---|---|---|---|
| 2 | Positive | Influenza A | Positive | Influenza A |
| 1 | Positive | Influenza A | Positive | Negative |
| 2 | Positive | Influenza A | Negative | Influenza A |
| 1 | Positive | Negative | Negative | Influenza A |
| 1 | Positive | Negative | Negative | Influenza B |
| 20 | Positive | Negative | Negative | Negative |
When the performance of the neuraminidase assay was analyzed again, considering all positive results obtained from any additional diagnostic tests as an influenza infection, the specificity of the neuraminidase assay increased to 94.2%, with a positive predictive value of 82.5%, while the sensitivity and negative predictive value were 68.6 and 88.2%, respectively.
There were 37 samples negative by the neuraminidase assay from which influenza virus was grown (24 samples for influenza A virus and 13 for influenza B virus). Of the 37 samples, 36 were tested by EIA and 14 were positive. A total of 24 samples were tested by IFA; 18 had abundant cellularity, while 6 and 9 had sparse and inadequate cellularity, respectively. In six samples, the IFA result was positive. Influenza virus strains isolated from these 37 samples (false-negative result by the neuraminidase assay) were tested directly with the neuraminidase assay to confirm the affinity of the substrate for the neuraminidase enzyme produced by each strain, and all strains tested positive.
DISCUSSION
Traditional rapid diagnostic tests for influenza virus, such as EIA and IFA, rely on antigen-antibody reactions to detect the presence of the virus in clinical samples. Detection of influenza virus by a neuraminidase assay is a novel approach in rapid diagnosis. Neuraminidase is an enzyme produced by both influenza A and B viruses (10), so this enzyme assay detects both influenza A and B viruses but cannot distinguish between them. Although other viruses, including parainfluenza virus and the paramyxovirus causing mumps, and some bacteria (including Streptococcus pneumoniae, group B streptococcus, Streptococcus viridans, and Pseudomonas sp.) produce molecules with enzymatic activities similar to that of neuraminidase (3, 7, 14), a previous study did not find any cross-reactivity when this neuraminidase detection assay was tested against pure cultures of any of these viruses or bacteria (18).
The performance of the neuraminidase assay compared to isolation of influenza virus in culture showed a sensitivity of 70.1% and specificity of 92.4%, with both false-positive and false-negative results encountered. Although viral culture is considered the “gold standard” for virologic confirmation of influenza infections, a single specimen might have a false-negative viral culture result. For example, Younkin et al. found the initial specimen obtained for viral culture positive in only 83% of 47 cases of culture-confirmed influenza infections (28). Reasons for the inability to isolate the virus by culture include inadequate specimen collection or handling, low virus titer, and inactivation of the virus by the immune system. The ability to detect influenza virus by other methods in 25.9% of the apparently false-positive specimens supports the probability that these results represented true infections. When these specimens were considered truly positive, the specificity of the neuraminidase assay increased slightly to 94.2%, with a positive predictive value of 82.5%. Based on these findings and the lack of cross-reactivity between pure cultures of viruses or bacteria and this neuraminidase assay, we conclude that this assay is specific for the detection of influenza virus in nasal wash specimens.
It is unclear why 37 specimens yielded an apparently false-negative neuraminidase assay result. A possible explanation for this finding is decreased affinity of the viral neuraminidase enzyme for the assay substrate, possibly caused by mutation in the neuraminidase gene. We explored this possibility by directly testing the influenza virus strains isolated from these specimens with the neuraminidase assay. Since all of the strains tested were neuraminidase positive, decreased affinity of the enzyme for the substrate does not account for the false-negative results. An alternative theoretical explanation is the presence of specific antibodies against the neuraminidase, produced by the host as a result of immunity against a current or past influenza infection, that could have decreased the enzymatic activity found in the specimen. Also, a higher viral load may be required for the neuraminidase assay to give a positive result compared to that of isolation by culture, and specimens with low or inadequate cellularity may have had lower viral titers contributing to the discrepant results.
An important variable that appears to affect the results is the quality of the sample submitted for testing. Specimens considered inadequate for IFA testing due to poor cellularity were significantly less likely to grow a virus in culture or to give a positive result by neuraminidase testing. In fact, when results from specimens with abundant cells were analyzed separately, the sensitivity of the neuraminidase assay compared to culture was 77% compared to 57.1% for the groups with sparse and inadequate cellularity. It is therefore possible that centrifugation of the sample and testing of the cellular pellet may enhance sensitivity of this assay.
The results in this study show decreased sensitivity and higher specificity to those observed in our preliminary study in throat and nasal wash specimens, where these values were 96 and 77%, respectively (18). In our prior study, most positive results by culture and the neuraminidase assay were found in throat swab specimens. The ability of a diagnostic test to detect respiratory viruses may vary, depending on the type of sample and anatomic sites (5). Differences in site-specific viral replication, antigen expression, or immune response may account for some of this variability.
An important observation was the lower sensitivity for detection of influenza B virus than for influenza A virus by the neuraminidase assay. A potential advantage of this test compared to detection of influenza A virus by EIA is that it can also detect influenza B virus. However, since sensitivity for detection of influenza B appears to be low, this advantage will depend on the relative contribution of influenza B virus to a particular influenza epidemic. The affinity to the substrate of neuraminidase produced by influenza B strains may be lower than that of strains of influenza A, but we did not find objective evidence of this. Modification of the assay may lead to increased affinity and sensitivity in detection of influenza B strains.
A surprising finding in the study was the variability in the performance of the test when technical personnel not involved with the study performed the neuraminidase assay. Although the test is easy to perform, reading of results, particularly weakly positive test results, may vary between persons. We observed that positive results varied in intensity, presumably according to the amount of enzyme present in the sample, with tests that showed a strong response demonstrating higher specificity than those with a weak response.
Our observations suggest that this novel neuraminidase assay for the detection of influenza viruses in nasal wash specimens is very specific and moderately sensitive and that the sensitivity depends upon the adequacy of the sample, experience of the technician, and the influenza virus type. Its overall performance is comparable to that of IFA, a procedure commonly used in many diagnostic laboratories for the rapid diagnosis of viral respiratory infections, whereas the EIA demonstrated a better performance profile for the detection of influenza A virus alone. The neuraminidase assay currently may offer an advantage over EIA for detection of influenza A virus in seasons when influenza B infections are prevalent. In addition, since antiviral neuraminidase inhibitors that are effective against influenza A and B virus are now available to clinicians, the neuraminidase assay will have the advantage of offering rapid diagnosis of both types of influenza virus.
ACKNOWLEDGMENTS
The neuraminidase assay kits were provided by ZymeTx.
We thank all the staff at the Diagnostic Virology Laboratory for their participation in this study.
REFERENCES
- 1.Atmar R L, Baxter B D, Dominguez E A, Taber L H. Comparison of reverse transcription-PCR with tissue culture and other rapid diagnostic assays for detection of type A influenza virus. J Clin Microbiol. 1996;34:2604–2606. doi: 10.1128/jcm.34.10.2604-2606.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atmar R L, Baxter B D. Typing and subtyping clinical isolates of influenza virus using reverse transcription-polymerase chain reaction. Clin Diagn Virol. 1996;7:77–84. doi: 10.1016/s0928-0197(96)00254-1. [DOI] [PubMed] [Google Scholar]
- 3.Beighton D, Whiley R A. Sialidase activity of the “Streptococcus milleri group” and other viridans group streptococci. J Clin Microbiol. 1990;28:1431–1433. doi: 10.1128/jcm.28.6.1431-1433.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Correa A G, Allen C, Taber L H, Couch R B. Program and abstracts of the 32nd Infectious Diseases Society of America Annual Meeting. Washington, D.C.: Infectious Diseases Society of America; 1994. Comparison of clinical specimens in the rapid diagnosis of respiratory syncytial virus infection in infants, abstr. 327; p. 56A. [Google Scholar]
- 6.Dagan R, Hall C B. Influenza A virus infection imitating bacterial sepsis in early infancy. Pediatr Infect Dis. 1984;3:218–220. doi: 10.1097/00006454-198405000-00007. [DOI] [PubMed] [Google Scholar]
- 7.Davis L, Baig M M, Ayoub E M. Properties of extracellular neuraminidase produced by group A streptococcus. Infect Immun. 1979;24:780–786. doi: 10.1128/iai.24.3.780-786.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denny F W, Clyde W A., Jr Acute lower respiratory tract infections in nonhospitalized children. J Pediatr. 1986;108:635–646. doi: 10.1016/s0022-3476(86)81034-4. [DOI] [PubMed] [Google Scholar]
- 9.Dominguez E A, Taber L H, Couch R B. Comparison of rapid diagnostic techniques for respiratory syncytial and influenza A virus respiratory infections in young children. J Clin Microbiol. 1993;31:2286–2290. doi: 10.1128/jcm.31.9.2286-2290.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drzeniek R. Viral and bacterial neuraminidases. Curr Top Microbiol Immunol. 1972;59:35–74. doi: 10.1007/978-3-642-65444-2_2. [DOI] [PubMed] [Google Scholar]
- 11.Glezen W P, Paredes A, Taber L H. Influenza in children. Relationship to other respiratory agents. JAMA. 1980;243:1345–1349. doi: 10.1001/jama.243.13.1345. [DOI] [PubMed] [Google Scholar]
- 12.Hayden F G, Osterhaus A D M E, Treanor J J, Fleming D M, Aoki F Y, Nicholson K G, Bohnen A M, Hirst H M, Keene O, Wightman K. Efficacy and safety of the neuraminidase inhibitor zanamivir in the treatment of influenza virus infections. N Engl J Med. 1997;337:874–880. doi: 10.1056/NEJM199709253371302. [DOI] [PubMed] [Google Scholar]
- 13.Johnston S L G, Bloy H. Evaluation of a rapid enzyme immunoassay for detection of influenza A virus. J Clin Microbiol. 1993;31:142–143. doi: 10.1128/jcm.31.1.142-143.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee L T, Howe C. Pneumococcal neuraminidase. J Bacteriol. 1966;91:1418–1426. doi: 10.1128/jb.91.4.1418-1426.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lennette E H, Lennette D A, Lennette E T. Diagnostic procedures for viral, rickettsial and chlamydial infections. 7th ed. Washington, D.C.: American Public Health Association; 1995. [Google Scholar]
- 16.Leonardi G P, Leib H, Birkhead G S, Smith C, Costello P, Conron W. Comparison of rapid detection methods for influenza A virus and their value in health-care management of institutionalized geriatric patients. J Clin Microbiol. 1994;32:70–74. doi: 10.1128/jcm.32.1.70-74.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendel D B, Tai C Y, Escarpe P A, Li W, Sidwell R W, Huffman J H, Sweet C, Jakeman K J, Merson J, Lacy S A, Lew W, Williams M A, Zhang L, Chen M S, Bischofberger N, Kim C U. Oral administration of a prodrug of the influenza virus neuraminidase inhibitor GS 4071 protects mice and ferrets against influenza infection. Antimicrob Agents Chemother. 1998;42:640–646. doi: 10.1128/aac.42.3.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noyola D E, Paredes A J, Clark B, Demmler G J. Evaluation of a neuraminidase detection assay for the rapid detection of influenza A and B virus in children. Pediatr Dev Pathol. 2000;3:162–167. doi: 10.1007/s100240050020. [DOI] [PubMed] [Google Scholar]
- 19.Rabalais G, Stout G, Waldeyer S. Rapid detection of influenza B virus in respiratory secretions by immunofluorescence during an epidemic. Diagn Microbiol Infect Dis. 1992;15:35–37. doi: 10.1016/0732-8893(92)90054-w. [DOI] [PubMed] [Google Scholar]
- 20.Serwint J R, Miller R M. Why diagnose influenza infections in hospitalized pediatric patients? Pediatr Infect Dis J. 1993;12:200–204. doi: 10.1097/00006454-199303000-00005. [DOI] [PubMed] [Google Scholar]
- 21.Sidwell R W, Huffman J H, Barnard D L, Bailey K W, Wong M, Morrison A, Syndergaard T, Kim C U. Inhibition of influenza virus infections in mice by GS4104, an orally effective influenza virus neuraminidase inhibitor. Antiviral Res. 1998;37:107–120. doi: 10.1016/s0166-3542(97)00065-x. [DOI] [PubMed] [Google Scholar]
- 22.Sugaya N, Nerome K, Ishida M, Nerome R, Nagae M, Takeuchi Y, Osano M. Impact of influenza virus infection as a cause of pediatric hospitalization. J Infect Dis. 1992;165:373–375. doi: 10.1093/infdis/165.2.373. [DOI] [PubMed] [Google Scholar]
- 23.Takimoto S, Grandien M, Ishida M A, Pereira M S, Paiva T M, Ishimaru T, Makita E M, Martinez C H O. Comparison of enzyme-linked immunosorbent assay, indirect immunofluorescence assay, and virus isolation for detection of respiratory viruses in nasopharyngeal secretions. J Clin Microbiol. 1991;29:470–474. doi: 10.1128/jcm.29.3.470-474.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Todd S J, Minnich L, Waner J L. Comparison of rapid immunofluorescence procedure with Testpack RSV and Directigen FLU-A for diagnosis of respiratory syncytial virus and influenza A virus. J Clin Microbiol. 1995;33:1650–1651. doi: 10.1128/jcm.33.6.1650-1651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Troendle J F, Demmler G J, Glezen W P, Finegold M, Romano M J. Fatal influenza B virus pneumonia in pediatric patients. Pediatr Infect Dis J. 1992;11:117–121. doi: 10.1097/00006454-199202000-00011. [DOI] [PubMed] [Google Scholar]
- 26.Waner J L, Todd S J, Shalaby H, Murphy P, Wall L V. Comparison of Directigen FLU-A with viral isolation and direct immunofluorescence for the rapid detection and identification of influenza A virus. J Clin Microbiol. 1991;29:479–482. doi: 10.1128/jcm.29.3.479-482.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woo P C Y, Chiu S S, Seto W, Peiris M. Cost-effectiveness of rapid diagnosis of viral respiratory tract infections in pediatric patients. J Clin Microbiol. 1997;35:1579–1581. doi: 10.1128/jcm.35.6.1579-1581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Younkin S W, Betts R F, Roth F K, Douglas R G., Jr Reduction in fever and symptoms in young adults with influenza A/Brazil/78 H1N1 infection after treatment with aspirin or amantadine. Antimicrob Agents Chemother. 1983;23:577–582. doi: 10.1128/aac.23.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]