Abstract
In order to achieve more sensitive and specific results for the rapid diagnosis of tuberculosis, we have developed a new method, named balanced heminested PCR, which avoids the inconvenience of asymmetric amplification and has the advantages of single-tube heminested PCR. This was achieved by replacing the outer primer that participates in both rounds of amplification in the standard heminested technique by another primer containing the sequence of the inner primer attached at its 5′ end. When both techniques were tested for the IS6110 target of Mycobacterium tuberculosis complex in 80 smear-negative culture-positive sputum samples and 60 control samples, the results showed 100% specificity for both techniques and sensitivities of 60 and 75% for heminested PCR and balanced heminested PCR, respectively (P = 0.02). In conclusion, the balanced heminested technique shows a higher sensitivity than that of the standard heminested, and it could be applied to any PCR by attaching the inner primer at the 5′ end of the opposite outer primer. Thus, the balanced heminested technique provides a target for the inner primer in both strands, avoiding asymmetric amplification and thereby resulting in a more efficient amplification, and, in practice, a higher sensitivity without loss of specificity and with a minimum risk of cross-contamination.
Tuberculosis (TB) is one of the most widespread, lethal infectious diseases affecting humans (23). Laboratory diagnosis is commonly based on culture and staining for acid-fast bacilli (AFB). The latter is the most rapid and economic method for detecting mycobacteria. However, given that half of the new cases of TB are smear negative, many diagnoses can not be confirmed at the time of presentation (3). This leads to delays in initiating appropriate treatment and/or the use of invasive procedures to firmly establish the diagnosis. Contrary to the general idea that AFB smear-negative patients do not contribute significantly to the spread of infection, Behr et al. found that up to 27% of recently acquired disease in San Francisco, California, was transmitted from smear-negative cases (2). Thus, for nucleic acid-based amplification techniques to be useful in TB control programs, sensitivity and specificity must be improved when the AFB smear is negative (1, 9).
Nevertheless, for achieving the best results it is necessary to optimize and combine good protocols for extraction, amplification, and detection of nucleic acids (17, 19). In this study, we focused on improving the amplification step. Thus, compared with conventional single-step PCR, nested amplification can enhance sensitivity approximately 1,000 fold but with a high risk of contamination (14). In order to completely eliminate this risk, single-tube nested PCR with a uracil-N-glycosylase (UNG)-dUTP system (15) can be performed but without the possible advantages of adding fresh enzyme or diluting inhibitors (22). Nonetheless, when the design and number of primers that can be used are limited due to the sequence of the target (the TB genome has a 65.5% G+C content [8] and it is difficult to find good primers in some regions), incompatibilities between primers and/or a large number of additional products could make the performance of a heminested PCR (HN) in one tube appropriate. However, the addition of primers at different concentrations results in an asymmetric amplification which makes the reaction less efficient.
In order to increase the yield of the reaction and to improve the sensitivity of detection of the Mycobacterium tuberculosis complex in smear-negative specimens, we developed a single-tube balanced HN PCR (B-HN) which avoids asymmetric amplification. This was achieved by replacing the original outer primer by another primer which also contained the sequence of the opposite inner primer attached at the 5′ end.
Here, for the first time we describe this modification, which overcomes some of the disadvantages of the HN technique.
MATERIALS AND METHODS
Clinical specimens.
All clinical specimens were processed in the Microbiology Laboratory of the Hospital Clínic (Barcelona, Spain). Eighty sputum samples of good quality belonging to 80 human immunodeficiency virus-negative patients with pulmonary TB were studied to analyze sensitivity. All samples were positive for M. tuberculosis in culture but were AFB smear negative.
Additionally, 60 culture- and stain-negative samples from human immunodeficiency virus-negative control patients were included: saliva samples from 40 healthy young people (student volunteers) who were tuberculin skin test negative and sputum samples from 20 chronic obstructive lung disease patients admitted for an acute episode without clinical signs, radiological lesions, or a history of TB.
All samples were decontaminated by a standard N-acetyl-l-cysteine–NaOH procedure (12). The resulting pellet was resuspended in 2 ml of phosphate-buffered saline (140 mM NaCl, 2.6 mM KCl, 10.1 mM Na2HPO4, 1.7 mM KH2PO4 [pH 7.4]). Auramine staining and Lowenstein-Jensen cultures were performed. The remaining pellet was frozen at −20°C until PCR processing was carried out.
PCR assay. (i) Sample preparation.
Aliquots of 500 μl were inactivated by heating at 95°C during 30 min and concentrated by centrifugation at 13,000 × g for 15 min. The pellet was resuspended in 300 μl of 10× Tris-EDTA solution (100 mM Tris-HCl, 10 mM EDTA [pH 8.0]) with 2 mg of lysozyme per ml and incubated at 37°C for 1 h. Proteinase K and sodium dodecyl sulfate were added to final concentrations of 250 μg/ml and 1% (wt/vol), respectively, and incubated at 43°C for 1 h. The suspension was extracted twice with phenol-chloroform-isoamyl alcohol (25:24:1, vol:vol:vol) and twice with chloroform-isoamyl alcohol (24:1, vol:vol). The pellet was treated with 2 volumes of 100% ethanol and 0.2 M NaCl, stored overnight at −20°C, then washed with 70% ethanol, dried, and resuspended in 100 μl of distilled water. Twenty microliters was used for PCR amplification. Both HN and B-HN used the same extract.
(ii) HN.
HN was based on the nested PCR technique described by Kennedy et al. and Wilson et al. (10, 22), but some modifications were performed in order to improve the sensitivity and specificity of the reaction. For this reason, the primer Tb670 was suppressed (J. González, A. García, J. Almeda, et al., Abstr. VIIth Reunión del Grupo Espan. de Micobacteríol., 1996; J. González, J. Almeda, A. García, et al., Abstr. XVIIIth Congr. Eur. Soc. Mycobacteriol., 1997).
Amplification was performed in 0.5-ml PCR tubes with a total reaction volume of 50 μl by using a 480 thermal cycler (Perkin-Elmer). Each reaction tube contained 2.5 U of Taq DNA polymerase (Gibco BRL); 0.5 U of UNG (Boehringer Mannheim); 200 μM (each) dATP, dCTP, and dGTP; 600 μM dUTP (Boehringer Mannheim); 1× final buffer (20 mM Tris-HCl, 50 mM KCl [pH 8.4]); 2 mM MgCl2, and 20 μl of sample. The primers used were 100 nM Tb850 (5′-TAGGCGTCGGTGACAAAGGCCACG-3′), 1 μM Tb505 (5′-ACGACCACATCAACC-3′), and 10 nM Tb294 (5′-GGACAACGCCGAATTGCGAAGGGC-3′).
All the primers and reagents were added at the beginning of the reaction, therefore not requiring opening of the tube to add the nested primer. These primers belong to the insertion sequence IS6110 that is present several times in M. tuberculosis complex genomes (21). To date, all strains studied in our area have IS6110 copies, thereby validating the use of this target for this study (P. Coll, personal communication).
(iii) B-HN.
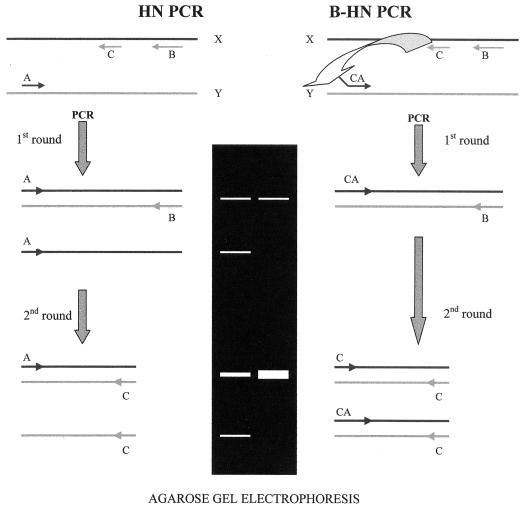
The reaction mixture and conditions were identical to those described above, but instead of the Tb850, the primer Tb505-850 (5′-ACGACCACATCAACCTAGGCGTCGGTGACAAAGGCCACG-3′) was used. Tb505-850 consists of the sum of the sequences of the primers Tb505 and Tb850. We also tested primer Tb505-850 at two different concentrations: 100 nM and 10 nM. The comparison between HN and B-HN is shown in Fig. 1.
FIG. 1.
Comparison between HN and B-HN. Primers A and CA hybridize to the Y strand. Primers B and C hybridize to the X strand. (Left) During the first round of HN, only outer primers anneal. The inner primer can not hybridize due to its low Tm. Primer A is more concentrated than B because it must also participate during the second round. This results in asymmetric amplification, since more strand X is synthesized by primer A. Primer B runs out due to its low concentration. During the second round, the annealing temperature is lower than in the first round, and the inner primer can hybridize. There are more X strands and a higher concentration of primer C, thereby helping the synthesis of strand Y which is produced in large amounts. This results in additional bands when electrophoresis is performed. (Right) During the first round of B-HN, only outer primers anneal. The inner primer can not hybridize due to its low Tm. The concentration of primer CA is equal to that of B, since it could be replaced by C during the second round. This results in a symmetric amplification. (Primer CA can also be more concentrated than B, which is not represented in the drawing; in this case, the first round is similar to HN and produces asymmetric amplification, but this is balanced during the second round). Primer B runs out due to its low concentration. Primer CA provides the target for primer C in both strands. During the second round, the annealing temperature is lower than in the first round, and the inner primer can hybridize. There is a higher concentration of primer C, thereby helping the synthesis of strand Y. C can then anneal at both strands, depending on the need to balance the output of both strands. Primer C will only anneal with the Y strand if primer CA places the target before it. This results in a more efficient reaction because single-stranded bands are not produced.
(iv) PCR conditions.
The conditions were the same for both methods. After 15 min at 25°C to allow UNG to work, the temperature was raised to 94°C for 5 min to deactivate the enzyme. The first stage of amplification involved 30 cycles of denaturation at 94°C for 45 s, with primer annealing and extension carried out in one step at 72°C for 1.5 min. The second stage included 30 cycles of denaturation at 94°C for 45 s, primer annealing at 55°C for 1 min, and extension at 72°C for 30 s, after which the reaction mixture was held at 72°C in a soak file until storage at −20°C.
(v) Product detection.
Twenty microliters of the amplified product was electrophoresed on a 2% (wt/vol) agarose gel stained with 0.5 μg of ethidium bromide per ml and visualized by UV transillumination. The presence of a 369-bp band for HN and a 384-bp band for B-HN indicated successful amplification of the IS6110 target. These bands were indistinguishable on the gel.
Determination of method sensitivity.
To determine the theoretical sensitivity of both techniques, 10-fold serial dilutions of one McFarland standard density equivalent (∼3 · 108 CFU/ml) were performed with a clinical strain of M. tuberculosis followed by amplification. DNA was prepared by boiling the organisms (11). The concentration was calculated by measuring the absorbance at 260 nm (1 A260 U = 50 μg of double-stranded DNA per ml) and taking 5 fg of DNA as one mycobacterium equivalent (8).
Statistical methods.
The chi-square test was used to analyze the results. Confidence intervals were calculated for 95%.
RESULTS
Balanced amplification.
Theoretically when both primers, Tb505-850 and Tb294, were at the same concentration (10 nM), asymmetric amplification during the first and second stage was avoided. When the Tb505-850 concentration was increased to 100 nM, this asymmetric amplification occurred during the first stage but not in the second. Nonetheless, the effect of asymmetric amplification on the overall sensitivity and specificity of the reaction during the first stage was minimal; thus, we decided to run all experiments using 100 nM of Tb505-850 in order to have conditions identical to those of HN.
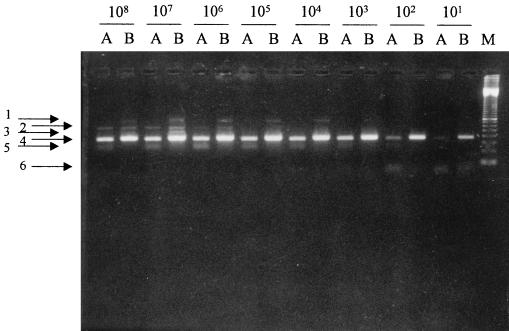
Serial dilutions.
Serial dilutions were performed to determine the end points of the techniques (Fig. 2). Both methods detected as few as 10 bacillus equivalents, although bands were stronger with B-HN, indicating a more efficient reaction. Asymmetric amplification yielded a lower band that was absent in B-HN.
FIG. 2.
Comparison of both methods in ten-fold serial dilutions of M. tuberculosis. A, HN; B, B-HN; M, 100-bp ladder marker. 1, bands of unknown origin, present only in B-HN, which may be due to artifacts produced by the tailed primer. This does not affect the final sensitivity or specificity. 2, bands corresponding to double-stranded DNA produced during first-round amplification. They are only present in concentrated dilutions. 3, bands corresponding to single-stranded DNA produced during first-round amplification. 4, bands corresponding to the desired double-stranded final product. 5, bands due to asymmetric amplification corresponding to single-stranded DNA. These are only present in HN samples. This detracts from sensitivity to the desired final product. 6, bands corresponding to dimer primers. They are only present in low dilutions.
Patient samples.
Of the 80 patients with pulmonary TB, 65 had unilateral infiltrated radiological lesions, and the other 15 had cavitated and/or bilateral radiological lesions. Forty-five samples were positive by both HN and B-HN, 3 samples were positive for HN but negative for B-HN, 15 samples were positive for B-HN but negative for HN, and 17 samples were negative for both. All control samples were negative by both techniques. The overall sensitivity was 60% for HN and 75% for B-HN (P = 0.02) (Table 1). Table 2 shows the results according to the radiological lesions displayed by the patient group.
TABLE 1.
Results of both PCR methods
| Study group | No. of samples | No. (%) of samples positive bya:
|
|
|---|---|---|---|
| HN | B-HN | ||
| TB patients | 80 | 48 (60)∗ | 60 (75)∗ |
| Controls | 80 | 0 (100) | 0 (100) |
∗, significant difference (P = 0.02).
TABLE 2.
Sensitivity of both PCR methods by radiological statusa
| Radiological lesion | No. of patients | No. (%) of samples showing sensitivity byb:
|
|
|---|---|---|---|
| HN | B-HN | ||
| Unilateral infiltrate | 65 | 37 (57)∗ | 49 (75.5)∗ |
| Cavitated and/or bilateral | 15 | 11 (73) | 11 (73) |
Eighty patients with positive cultures but negative sputum samples by AFB staining.
∗, significant difference (P = 0.02).
DISCUSSION
For smear-negative specimens, most published studies report a sensitivity of around 60% or even less, depending on the number of samples and experiment conditions (3, 4, 7, 9, 13, 16, 18, 20, 24). This low sensitivity for smear-negative specimens shows that current amplification assays may be unsuitable in replacing cultures for the diagnosis of tuberculosis.
Our objective was to improve the sensitivity of these tests, especially with smear-negative samples. For this reason, we developed an HN method. The use of a single tube diminishes the possibility of contamination and increases the sensitivity and specificity compared to a standard PCR. Besides preventing contamination by previously amplified PCR products, the addition of UNG has an effect similar to a “hot start” (6), since it degrades any elongated product initially and the reaction begins hot when UNG has been inactivated. The high annealing temperatures avoid nonspecific amplifications (5).
The four different-size bands observed by Wilson et al. after agarose gel electrophoresis (22) are due to the four possible combinations allowed between the four primers used in the reaction. In the HN protocol, four bands can also be observed, but in this case two are combinations between primers belonging to the first and second amplification product, and the other two are due to the asymmetric amplification. When B-HN is performed, these latter two bands can be avoided, leading to better interpretation of the results and greater band intensity.
Our results show that B-HN is more sensitive than standard HN, allowing the diagnosis to be advanced in 75% of the cases in which the smear was negative without waiting for culture results, and this fact is more evident in patients without cavitated lesions, who in our geographical area represent around 80% of the cases with pulmonary involvement. As indicated by the American Thoracic Society (1), we also recommend the use of these tests in conjunction with the available clinical data on the patient. In conclusion, the B-HN method is more sensitive than the HN technique. It does not decrease the specificity of the reaction. It can be applied to any PCR without further manipulation by attaching the sequence of the inner primer at the 5′ end of the opposite outer primer. B-HN provides a target to the inner primer in both strands, resulting in a more efficient reaction which, in practice, means a higher sensitivity. This modification also has additional advantages over current amplification protocols when further manipulation of the PCR products is required, such as cloning or sequence capture, since one restriction enzyme will cut both ends of the product and more strands will be captured or labelled by the primers.
ACKNOWLEDGMENTS
This work was supported by Fondo de Investigaciones Sanitarias de la Seguridad Social (FIS) grants 96/0028-01 and 98/1282 from the Ministerio de Salud, Madrid, Spain, and Sociedad Española de Neumología y Cirugía Torácica (SEPAR) grant 96/444. Albert García-Quintanilla was granted a predoctoral fellowship from the Departament de Microbiologia i Parasitologia Sanitàries, Divisió Ciències de la Salut, Universitat de Barcelona, Spain.
We thank Julià González, Rosa Monté, and Dolors Ricart for their assistance in supplying samples.
REFERENCES
- 1.American Thoracic Society Workshop. Rapid diagnostic tests for tuberculosis. What is the appropriate use? Am J Respir Crit Care Med. 1997;155:1804–1814. doi: 10.1164/ajrccm.155.5.9154896. [DOI] [PubMed] [Google Scholar]
- 2.Behr M A, Warren S A, Salamon H, Hopewell P C, Ponce de Leon A, Daley C L, Small P M. Transmission of Mycobacterium tuberculosis from patients smear-negative for acid-fast bacilli. Lancet. 1999;353:444–449. doi: 10.1016/s0140-6736(98)03406-0. [DOI] [PubMed] [Google Scholar]
- 3.Bennedsen J, Thomsen V O, Pfyffer G E, Funke G, Feldmann K, Beneke A, et al. Utility of PCR in diagnosing pulmonary tuberculosis. J Clin Microbiol. 1996;34:1407–1411. doi: 10.1128/jcm.34.6.1407-1411.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brugiere O, Vokurka M, Lecossier D, Mangiapan G, Amrane A, Milleron B, Mayaud C, Cadranel J, Hance A J. Diagnosis of smear-negative pulmonary tuberculosis using sequence capture polymerase chain reaction. Am J Respir Crit Care Med. 1997;155:1478–1481. doi: 10.1164/ajrccm.155.4.9105098. [DOI] [PubMed] [Google Scholar]
- 5.Chan C M, Yuen K Y, Chan K S, Yam W C, Yim K H M, Ng W F, Ng M H. Single-tube nested PCR in the diagnosis of tuberculosis. J Clin Pathol. 1996;49:290–294. doi: 10.1136/jcp.49.4.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chou Q, Russell M, Birch D E, Raymond J, Block W. Prevention of pre-PCR mis-priming and primer dimerization improves low-copy number amplifications. Nucleic Acids Res. 1992;20:1717–1723. doi: 10.1093/nar/20.7.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarridge J E, III, Shawar R M, Shinnick T M, Plikaytis B B. Large-scale use of polymerase chain reaction for detection of Mycobacterium tuberculosis in a routine mycobacteriology laboratory. J Clin Microbiol. 1993;31:2049–2056. doi: 10.1128/jcm.31.8.2049-2056.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 9.Doern G V. Diagnostic mycobacteriology: where are we today? J Clin Microbiol. 1996;34:1873–1876. doi: 10.1128/jcm.34.8.1873-1876.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kennedy N, Gillespie S H, Saruni A O S, Kisyombe G, McNerney R, Ngowi F I, Wilson S. Polymerase chain reaction for assessing treatment response in patients with pulmonary tuberculosis. J Infect Dis. 1994;170:713–716. doi: 10.1093/infdis/170.3.713. [DOI] [PubMed] [Google Scholar]
- 11.Kocagoz T, Yilmaz E, Ozkara S, Kocagoz S, Hayran M, Sachedeva M, Chambers H F. Detection of Mycobacterium tuberculosis in sputum samples by polymerase chain reaction using a simplified procedure. J Clin Microbiol. 1993;31:1435–1438. doi: 10.1128/jcm.31.6.1435-1438.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kubica G P W, Dye E, Cohn M L, Middlebrook G. Sputum digestion and decontamination with N-acetyl-l-cysteine-sodium hydroxide for culture of mycobacteria. Am Rev Respir Dis. 1963;87:775–779. doi: 10.1164/arrd.1963.87.5.775. [DOI] [PubMed] [Google Scholar]
- 13.Lebrun L, Mathieu D, Saulnier C, Nordmann P. Limits of commercial molecular tests for diagnosis of pulmonary tuberculosis. Eur Respir J. 1997;10:1874–1876. doi: 10.1183/09031936.97.10081874. [DOI] [PubMed] [Google Scholar]
- 14.Liu P Y-F, Shi Z-Y, Lau Y-J, Hu B-S. Rapid diagnosis of tuberculous meningitis by a simplified nested amplification protocol. Neurology. 1994;44:1161–1164. doi: 10.1212/wnl.44.6.1161. [DOI] [PubMed] [Google Scholar]
- 15.Longo M C, Berninger M S, Hartley J L. DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene. 1990;93:125–128. doi: 10.1016/0378-1119(90)90145-h. [DOI] [PubMed] [Google Scholar]
- 16.Nolte F S, Metchock B, McGowan J E, Jr, Edwards A, Kwumabua O, Thurmond C, et al. Direct detection of Mycobacterium tuberculosis in sputum by polymerase chain reaction and DNA hybridization. J Clin Microbiol. 1993;31:1777–1782. doi: 10.1128/jcm.31.7.1777-1782.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noordhoek G T, Kolk A H J, Bjune G, Catty D, Dale J W, Fine P E M, Godfrey-Faussett P, Cho S-N, Shinnick T, Svenson S B, Wilson S, van Embden J D A. Sensitivity and specificity of PCR for detection of Mycobacterium tuberculosis: a blind comparison study among seven laboratories. J Clin Microbiol. 1994;32:277–284. doi: 10.1128/jcm.32.2.277-284.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pierre C, Lecossier D, Boussougant Y, Bocart D, Joly V, Yeni P, et al. Use of a reamplification protocol improves sensitivity of detection of Mycobacterium tuberculosis in clinical samples by amplification of DNA. J Clin Microbiol. 1991;29:712–717. doi: 10.1128/jcm.29.4.712-717.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roth A, Schaberg T, Mauch H. Molecular diagnosis of tuberculosis: current clinical validity and future perspectives. Eur Respir J. 1997;10:1877–1891. doi: 10.1183/09031936.97.10081877. [DOI] [PubMed] [Google Scholar]
- 20.Shawar R M, El Zaatari F A K, Nataraj A, Clarridge J E. Detection of Mycobacterium tuberculosis in clinical samples by two-step polymerase chain reaction and nonisotopic hybridization methods. J Clin Microbiol. 1993;31:61–65. doi: 10.1128/jcm.31.1.61-65.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thierry D, Cave M D, Eisenach K D, Crawford J T, Bates J H, Gicquel B, et al. IS6110, an IS-like element of Mycobacterium tuberculosis complex. Nucleic Acids Res. 1990;18:188. doi: 10.1093/nar/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson S M, McNerney R, Nye P M, Godfrey-Faussett P D, Stoker N G, Voller A. Progress toward a simplified polymerase chain reaction and its application to diagnosis of tuberculosis. J Clin Microbiol. 1993;31:776–782. doi: 10.1128/jcm.31.4.776-782.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.World Health Organization. WHO report on the tuberculosis epidemic 1997. Geneva, Switzerland: WHO; 1998. [Google Scholar]
- 24.Yuen K, Yam W C, Wong L P, Seto W H. Comparison of two automated DNA amplification systems with a manual one-tube nested PCR assay for diagnosis of pulmonary tuberculosis. J Clin Microbiol. 1997;35:1385–1389. doi: 10.1128/jcm.35.6.1385-1389.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]