Abstract
Physical signals provided by the extracellular matrix (ECM) are key microenvironmental parameters for the fate decision of hematopoietic stem and progenitor cells (HSPC) in bone marrow. Insights into cell-ECM interactions are critical for advancing HSC-based tissue engineering. Herein, we employed collagen hydrogels and collagen-alginate hydrogels of defined stiffness to study the behaviors of hematopoietic progenitor cells (HPCs). Three-dimensional (3D) collagen hydrogels with a stiffness of 45 Pa were found to promote HPC maintenance and colony formation of monocyte/macrophage progenitors. Using single-cell RNA sequencing (scRNA-seq), we also characterized the comprehensive transcriptional profiles of cells randomly selected from two-dimensional (2D) and 3D hydrogels. A distinct maturation trajectory from HPCs into macrophages within the 3D microenvironment was revealed by these results. 3D-derived macrophages expressed high levels of various cytokines and chemokines, such as Saa3, Cxcl2, Socs3 and Tnf. Furthermore, enhanced communication between 3D-macrophages and other hematopoietic clusters based on ligand-repair interactions was demonstrated through bioinformatic analyses. Our research underlines the regulatory role of matrix-dimensionality in HPC differentiation and therefore probably be applied to the generation of specialized macrophages.
Keywords: Hematopoietic progenitor cells, Collagen, Matrix-dimensionality, Macrophages
Graphical abstract
Highlights
-
•
3D collagen hydrogels influenced the maintenance of hematopoietic progenitor cells.
-
•
3D matrices modulated the lineage specification of hematopoietic progenitors and promoted the formation of CFU-M colonies.
-
•
Single-cell RNA sequencing identified a cluster of specialized macrophages within a 3D microenvironment.
1. Introduction
Adult hematopoiesis in mammals is guaranteed by hematopoietic stem cells (HSCs), which have the properties of self-renewal and multipotency. They also give rise to progenitor cells that become committed to the main marrow cell lines. Unfortunately, the reduced cell viability and elusory fate decision of isolated HSCs ex vivo impedes their medical use in stem cell therapies for hematological disorders. In vivo, the bone marrow (BM) niche appears to be uniquely adapted to support the functions of HSCs and hematopoietic progenitor cells (HPCs), thus balancing the enormous production requirements for blood cells and a variety of immune cells [1]. The niche microenvironment nourishes and modulates these cell behaviors. Much of our current understanding of HSC-niche interactions is limited to the chemical signals such as the function of extracellular matrix (ECM) components and the cell adhesion molecules secreted by niche cell populations. Yet, the complexity of the niche network underscores the necessity to consider other regulatory signals that may influence hematopoiesis development or determine whether HSCs live in a quiescent stage or egress and circulate away from their niche [2].
Although HSCs are non-anchor dependent cells, an increasing number of biomaterial based studies have demonstrated that HSCs may “feel” or sense the physical diversity of their surrounding environments in vitro [3]. For example, in 2011, John E J Rasko et al. proposed that mechanical signals from substrates, such as matrix elasticity, impacted the expansion of hematopoietic stem and progenitor cells (HSPCs) [4]. In 2012, Brendan A. C. Harley showed that collagen substrate dimensionality (two-dimensional (2D) vs. three-dimensional (3D)) affected HSPC viability and morphology, as did 2D polyacrylamide (PA) substrates with different rigidities ranging from 0.71 to 196 kPa [5]. A simultaneous study performed by Joachim P. Spatz confirmed the importance of matrix elasticity on HSC adhesion and migration. Human CD34+ HPCs on harder PEGDA hydrogels (>38 kPa) showed faster migration and shorter pauses than those on softer gels (≤20 kPa), indicating the mechanosensitivity of HPC behaviors [6]. Next, Brendan A. C. Harley revealed how the stiffness and ECM ligands integrated together to alter the early fate determination of HSCs. Fibronectin-coated 44 kPa PA substrates resembling the osteoid region within the marrow cavity in particular promoted the proliferation of myeloid lineages, while laminin-coated 3.7 kPa gels resembling the sinusoid region favored erythroid lineage specification [7]. Subsequent study further corroborated the mediated role of ECM stiffness during megakaryocyte differentiation and proplatelet production [8,9]. These findings suggest that niche physical parameters are independent or interdependent (with chemical elements) signals that can be received by HSCPs. Heterogeneous stiffness throughout the BM niche also emerges from the heterogeneous distribution of ECM structural proteins. While PA substrates are able to recapitulate mechanical cues of rigid marrow regions and have been utilized for investigating the behaviors of HPCs near the bone surface (>30 kPa) and perivascular regions (1–3 kPa) [7], effort is still required to define whether HPCs can be retained ex vivo on substrates with compliant stiffness, such as the stiffness of adipose-rich marrow regions that exhibit a Young's modulus lower than 1 kPa [10,11].
Another noteworthy biophysical regulator is spatial confinement. In terms of the physiological relevance of experiments performed in vitro, monolayers of 2D cultured cells only just receive polarized mechanical cues from the ventral surface, whereas 3D encapsulation strategies appropriately enhance cell-ECM contacts and mimic the real physical situation of niche cells in native tissues [12,13]. Direct cell-cell interactions (juxtacrine signals) existing in 2D platforms depend on the activation of membrane-bound cell adhesion molecules. Within a 3D microenvironment, thin filopodial protrusions from HSCs have been shown to extend into the surrounding gel [5], and indirect cell-cell communications between HSCs and niche cells (paracrine signals) or autocatalytic effects from HSCs themselves (autocrine signals) are dominantly modulated by diffusible biomolecules [14]. Accordingly, we hypothesized that 3D encapsulated hematopoietic progenitors lead to distinct fate events when compared to cells in 2D monolayer cultures or conventional fluid cultures.
In this study, we simulated the BM adipose tissue microenvironment using collagen and collagen-alginate hydrogel scaffolds, and assessed their impact on HSC fate decisions. We further utilized single-cell RNA sequencing (scRNA-seq) to characterize the alternations in the cell transcriptome as a result of changes in matrix-dimensionality. Our findings emphasize the importance of physical factors in HPC fate regulation. scRNA-seq served as a powerful tool for revealing the HPC fate choices driven by mechanical confinement.
2. Materials and methods
2.1. HPC isolation
Animal experimental procedures were performed with the approval of Lab Animal Ethics & Welfare committee of the Northwestern Polytechnical University. Wide-type female C57BL6 mice between 5 and 8 weeks of age were obtained from Laboratory Animal Center of the Xi'an Jiaotong University. Primary BM cell suspension was isolated by crushing and filtering the mice femurs and tibias marrow tissue through a 40-μm cell strainer. BM mononuclear cells were separated by Ficoll-Hypaque gradient centrifugation (density 1.084 g/ml, GE healthcare). Lin-c-kit+ HPCs were collected using magnetic activated cell sorting technology (MACS, Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's instructions. The purity of the sorted cells was assessed by flow cytometry. Freshly isolated cells were either immediately analyzed or kept in IMDM medium supplemented with 10%FBS (Gibco), 50 ng/ml rm SCF, 20 ng/mL rm TPO and 20 ng/mL rm Flt-3L (all from PeproTech, Rocky Hill, NJ) for hydrogel culture.
2.2. Fabrication of hydrogel culture systems
Hydrogels were prepared using rat tail derived type I collagen (Corning, NY, USA) and alginate sodium (Sigma-Aldrich, MO, USA). All reagents were per-cooled at 4 °C and placed on ice to prevent any premature gelation upon mixing. Gelling procedure was carried out in accordance with the manufacturer's instructions. In brief, collagen stocks were diluted with culture medium to 3 mg/ml and buffered to a pH 7.4 with 1 N NaOH and PBS buffer. For 2D systems, the neutral collagen solutions were delivered into tissue culture plates (TCPs) for completely polymerization at 37 °C prior to cell seeding. 3D hydrogel scaffolds were prepared by adding the cell suspension to the neutral collagen stock in lieu of the equivalent volume of cell medium alone. For 3D collagen-alginate composites, collagen was mixed with alginate to create a final pre-gel solution of 0.1% alginate and 3 mg/ml collagen. Cell suspensions were added to alginate-collagen solutions at the same seeding density as 2D. In the same manner, the mixtures were given 1 h at 37 °C, 5% CO2 to trigger the formation of collagen fibril. Collagen-alginate gels were then treated with CaCl2 (11 mg/ml) for 20 min to induce ionotropic alginate gelation. The blended gels were finally washed with PBS twice before medium supplementing. Encapsulated cells in 3D collagen gels were harvest by digestion using type IV collagenase and sterile sodium citrate solution before subsequent analysis. Similarly, cells grown in gel surface were treated with equal collagenase.
2.3. Mechanical measurements of hydrogels
Nano-indentation is a technology to probe the micro-mechanical properties of soft biomaterials [15]. The elastic moduli of collagen and collagen-alginate hydrogels were quantified via Piuma Nanoindenter by the following parameters: Probe Stiffness (N/m) was 0.048; Probe Radius (um) was 9.5; Speed (um/second) was 5; Indenter Type was sphere. The Young's Modulus of each test was calculated using a Hertzian model on 80% of the data on the loading section of the load-indentation data curve.
2.4. Swelling and porosity characterization of hydrogels
The swelling behavior and water absorption of hydrogels were measured by immersing lyophilized gels in PBS buffer (pH = 7.4) at 37 °C, and then measuring their weights after various incubation times with an analytical balance until a constant weight was observed. The swelling ratio (SR) was defined using equation (1):
| SR= (wt - w0) / w0, | (1) |
where wt is the constant weight of a hydrogel after equilibrium swelling and w0 is the weight of a dry hydrogel [16].
The microstructures of lyophilized hydrogels were imaged after being sputtered with platinum by scanning electron microscopy (SEM, Tescan, Czech Republic) at 10 kV. Image J 1.52v software was used to analyze the pore structure characteristics of hydrogels using the obtained SEM images [17,18]. The sum of the areas of the pores was divided by the total exposed sample areas to obtain the percent porosity.
2.5. Flow cytometry
A FACSCalibur system (BD Biosciences) was used for cell phenotype analysis. Cell suspensions were incubated in PBS at 4 °C with combinations of appropriate antibodies. HPCs were stained with a cocktail of FITC-conjugated lineage (Lin) antibodies (CD3 (17A2), CD45R (B220), CD11b, TER-119 (TER-119), Ly-G6 (Gr-1)), PE-conjugated c-Kit (CD117), and APC-conjugated Sca-1. BM myeloid cells and lymphoid cells were stained with a FITC-anti CD11b monoclonal antibody (M1/70) and a PE-anti CD19 monoclonal antibody (1D3) [19]. All antibodies were purchased from eBioscience or Biolegend. FACS data were analyzed using FlowJo software Version 7.6.1 (TreeStar).
2.6. Cell viability assays
Cell viability was estimated using an Annexin V/FITC apoptosis kit (Dojindo, Japan). Harvested cells were stained with 5 μl of FITC conjugated annexin V and 5 μl of propidium iodide (PI) for 15 min at the room temperature. Doubly stained cells were counted using flow cytometry. Meanwhile, a Cell Counting Kit-8 (CCK-8) (Dojindo, Japan) kit was used to estimate the cell proliferative activity after hydrogel culture. Cell medium mixed with CCK-8 solution was incubated for a fixed length of time, and the relevant absorbance at 450 nm was tested with a microplate reader.
2.7. Colony-forming unit assays
To compare the multi-lineage potential of HPCs from different culture conditions, after accurate cell counting, cells were harvested and resuspended in methylcellulose medium (M3434, stem cell technology, Canada) and incubated at 37 °C for 7 days. Different colonies were enumerated using an inverted microscope, according to the examples of the manufacturer's guideline.
2.8. Droplet-based scRNA-sequencing
scRNA-seq was performed based on the 10 × Genomics single-cell RNA sequencing platform (10 × Genomics, Pleasanton, CA, USA). Briefly, cells were washed immediately after the hydrogel digestion. Live cells were captured by MACS with a Dead Cell Removal Kit (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the recommended protocol. Cell viability (>95%) were determined twice using a hemocytometer (TC20, Bio-Rad, Hercules, CA, USA). Droplet-based single-cell partitioning and complementary DNA libraries were prepared using a Chromium Single Cell 3′ GEM, Library & Gel Bead Kitq v3 (10 × Genomics); cDNA was purified using a SPRIselect Reagent Kit (Beckman Coulter). The constructed libraries were sequenced on an Illumina HiSeq X Ten sequencer (Illumina, San Diego, CA, USA) and pair-ended 150 bp (PE150) reads were generated for downstream analysis.
2.9. Single-cell data analysis
The Cell Ranger Single-Cell Software Suite (https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome) was used to align reads, generate feature-barcode matrices, and estimate raw gene counts. The STAR software was used for alignment (with reference genome mm10/GRCm38) of the cDNA insert sequence. Downstream analyses including quality filtering, normalization, dimensional reduction, cell clustering and the identification of differentially expressed genes (DEGs) were performed with the R package Seurat (version 3.2.0). Briefly, cells with expression of <500 or >6000 genes or a mitochondrial gene expression >5% of the total gene expression were removed. After filtering unwanted cells, the dataset was normalized by the total expression, multiplied by a scale factor of 10,000, and log-transformed the result. Downstream unsupervised clustering and UMAP analysis were performed based on the statistically significant principal components with the Seurat FindClusters function (resolution = 0.8). Marker genes for each cluster were determined with the Wilcoxon rank-sum test by the Seurat FindAllMarkers function (genes with Log Fold Change threshold above 0.25 were identified as DEG, top 10 DEGs are shown in Supplementary Table S1). For the cell-type identification, we annotated clusters on the basis of DEGs in each cluster using marker genes in Mouse Cell Atlas (http://bis.zju.edu.cn/MCA/index.html) and Cell Marker Database (http://bio-bigdata.hrbmu.edu.cn/CellMarker/).
Gene Ontology (GO, http://geneontology.org/) enrichments as well as Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.kegg.jp/) pathway annotations were analyzed using positive markers of target clusters, through the corresponding databases of Carlson M (2019). org.Mm.eg.db, and STRINGI, respectively. GO and KEGG terms with a P value < 0.05 were considered to be significantly enriched.
To analyze the dynamic trajectory development of HPCs, we used package Monocle 2.0 and Monocle 3.0 to organize cells in pseudo-developmental timeline, followed by optional statistical tests to find genes which varied in expression over those trajectories. The intercellular communication between defined clusters were systematically inferred and visualized using the Celltalker package, which is characterized by a comprehensive signaling molecule interaction database including multimeric ligand-receptor pairs, soluble agonists and antagonists, as well as stimulatory and inhibitory co-ligands and co-receptors.
To identify the transcription factors (TFs) activity of interested hematopoietic subpopulations, the SCENIC was performed in R to reconstruct gene-regulatory networks (GRNs) and identify stable cell states based on cis-regulatory cues with default settings. SCENIC workflow was run as described in the website of SCENIC: http://aertslab.org/#scenic. First, TF-gene co-expression modules were reconstructed with GENIE3 in a data-driven manner. Subsequently, modules were trimmed by RcisTarget analysis and genes in the respective TF binding motifs were enriched. These significant gene regulatory networks are termed as regulons. Once the regulons were obtained, AUCell activity of each regulon across individual cell was evaluated and a binary regulon activity matrix was obtained.
2.10. Quantitative RT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen) and reversely transcribed into cDNA using GoScript™ Reverse Transcriptase (Promega). Real-time PCR was performed using iQ™ SYBR® Green Supermix (Bio-rad). Gapdh gene expression was used as a standard. The gene expression values were calculated via the 2−ΔΔCt method.
2.11. Statistical analysis
The data are expressed as mean and standard error of the mean (SEM). Statistical significance between data groups (defined as P < 0.05) was determined using unpaired t-test method from GraphPad Prism 5.
3. Results
3.1. Cell viability inside different 3D collagen matrices
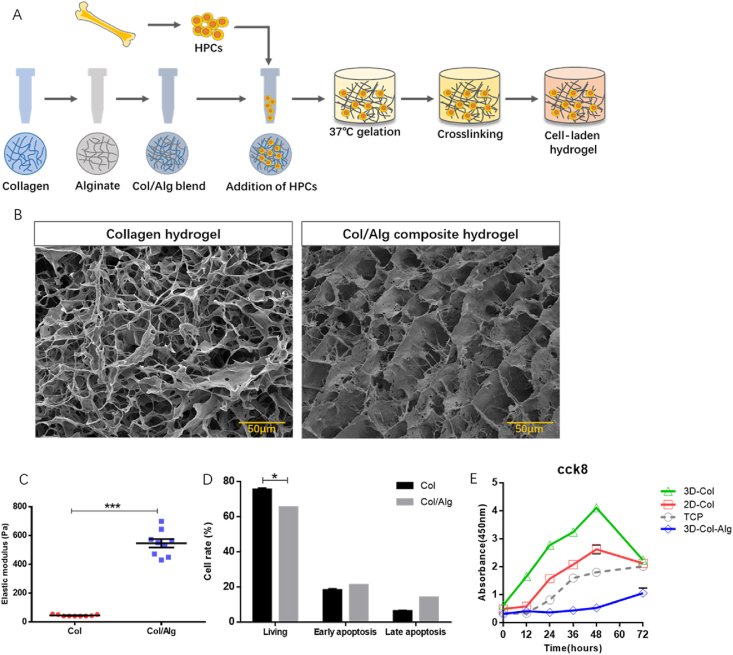
The elastic moduli measurements of collagen hydrogels were taken once gels were completely polymerized. The hydrogels used in this experiment exhibited significantly different in terms of their modulus of elasticity (Fig. 1A–C). The average stiffness of collagen hydrogels was 45.12 ± 2.0 Pa, whereas the collagen-alginate hydrogels exhibited a mean stiffness of 546.5 ± 29.47 Pa. The elastic stiffness of both gels mimicked the natural stiffness of the ECM in BM adipose tissue. The microstructural properties of collagen matrices changed simultaneously with the addition of alginate. The cumulative pore area and the average pore sizes are shown in Fig. S1. Introduction of alginate increased the aperture diameters of collagen matrices, but no significant changes were revealed in swelling tests.
Fig. 1.
Cell viability evaluation of different cell culture matrices.
To evaluate the cellular compatibility of these cell culture substrates, cells from 3D hydrogel matrices were obtained and stained. The percentages of viable cells, early apoptotic cells and late apoptotic cells were measured by flow cytometry (Fig. 1D). Collagen gels displayed a relatively higher frequency of viable cells, whereas collagen-alginate gels had more apoptotic cells. The cell viability remained greater than 60% after encapsulation culturing within both of the fabricated substrates. Meanwhile, proliferation capacity evaluation of cells from hydrogel systems was performed (Fig. 1E). Trypan blue dye exclusion was used to ensure that the live cell numbers were consistent for different groups. CCK8 assays demonstrated that collagen hydrogels had a significant effect on cell proliferation activity compared to normal fluid conventional culture on TCP. 3D collagen encapsulation culture had the most relative absorbance at 450 nm, suggesting the strongest cell activity. Compared to cells that were conventionally cultured, collagen systems increased the cell proliferation capacity, but the cell proliferation seemed to be reduced after culture within composite hydrogels.
(A) Schematic of scaffolds of collagen-alginate formation. (B) Internal architecture of collagen matrices (left) and collagen-alginate matrices (right). (C) The elastic properties of collagen and collagen-alginate hydrogels (n = 9 recipients per group from three separate experiments). (D) Cells cultured in 3D substrates of different rigidities were harvested, counted, and analyzed to compare cell viabilities by flow cytometry. Collagen-alginate matrices increased early apoptosis of HPCs. (E) Cell proliferation abilities of HPCs cultured on top and inside of collagen hydrogels. HPCs (Hematopoietic progenitor cells). Col (Collagen). Col/Alg (collagen-alginate). TCP (tissue culture plate). Three biological replicates are represented in (D) and (E). *P < 0.05, ***P < 0.001.
3.2. The effects of different collagen matrices on hematopoietic cell maintenance ex vivo
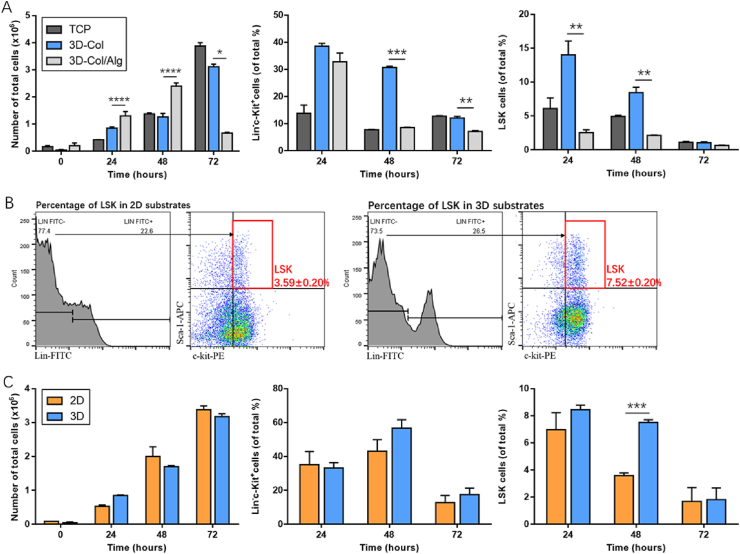
Although the role of microenvironmental mechanical properties, such as stiffness in determining the behaviors of HSCs has been discussed by others, the effect of softer rigidity on cell behavior requires attention because of the spatio-temporal diversity of niche components [20]. To explore the effect of physiologically relevant rigidity remodeling of adipose-rich region stiffness (<1 kPa) on the maintenance of HPCs, we used collagen substrates and collagen-alginate composites with a modulus of 45.12 Pa and 546.5 Pa, respectively, to model 3D-niche environments for HPCs in vitro. For up to three days, we compared the percentages of Lin-c-Kit+ cells and LSK cells at each time point across these substrates with different compliances (Fig. 2A). Although there was a rapid increase in total cell numbers, it seemed that the cells cultured in vitro had difficulty retaining the Lin-c-Kit+ phenotype (Fig. S2). By the end of the three-day culture period, the percentages of Lin-c-Kit+ cells were lower than 20%, and a dramatic decline in cell numbers inside collagen-alginate scaffolds was observed. Within 24 h of culture, both 3D hydrogel systems had higher Lin-c-Kit+ percentages than traditional fluid culture, but the percentages of LSK cells in composite matrices were lower. In the two-day period, the percentages of Lin-c-Kit+ cells from collagen-alginate scaffolds showed a dramatic decrease, whereas collagen hydrogels maintained the percentages of Lin-c-Kit+ cells at approximate 30% in the present experiment, indicating the superior effect of collagen hydrogels at 45.12 Pa for preserving hematopoietic progenitor phenotype. However, changes in the pore geometry induced by alginate suggested that these values should not be directly compared by stiffness. The increased efficiency of collagen matrices for HPC maintenance possibly resulted from their stiffness and overall microstructure.
Fig. 2.
Impact of different hydrogel matrices on HPC maintenance.
CCK8 assays had revealed that collagen hydrogels accelerated cell proliferation; therefore, we selected collagen hydrogels to compare the percentages of HPCs seeded on top of (2D) or encapsulated inside (3D) gels, and confirm the sustain abilities of different matrix-dimensionality on HPC maintenance. The histogram given in Fig. 2C shows that there were no significant differences in terms of the percentages of Lin-c-Kit+ cells within three days of culture between the two groups. As for LSK cells, the frequencies were found to directly correlate to the hydrogel dimensionality. 2D culture decreased the LSK percentages from 6.99 ± 1.24% to 3.59 ± 0.20% after 24 h, whereas encapsulation culture showed a significantly higher number at 7.52 ± 0.20%. This result suggested that altering substrate dimensionality affected the HPC fraction, and the 3D system was favorable for maintaining HPC phenotypes up to two days ex vivo.
(A) Number of total cells, percentages of Lin-c-kit+ cells and LSK cells inside collagen scaffolds and collagen-alginate scaffolds were analyzed after 24 h, 48 h, and 72 h of culture. Although compared to normal liquid culture, 3D collagen-alginate constructs retained higher Lin-c-kit+ phenotypes after 24 h, the percentages of LSK were lower than TCP controls and collagen gels with softer stiffness. (B) Representative results from flow cytometry showed higher LSK cell proportions in HPCs cultured on 3D collagen gels at 48 h. Hematopoietic progenitor populations were identified as Lin-c-kit+. Mice HSPC fractions were denoted using Lin−Sca-1+c-kit+ (LSK). Cells in red boxes showed the percentages of double positive cells (Sca-1+c-kit+) in gated lineage negative cells. Representative profiles of isotype controls are shown in Fig. S3 (C) Comparative analysis of total cell number, percentages of Lin-c-kit+ cells and LSK cells on surface of collagen and cultured inside the same gel types. Before 72 h, 3D environment had the competitive ability to retain HPC phenotypes. LSK (Lin-c-kit+sca-1+). 24 h (24 h), 48 h (48 h) and 72 h (72 h). Data are mean ± SEM from three separate experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
3.3. 3D matrices modulate the lineage specification of hematopoietic cells
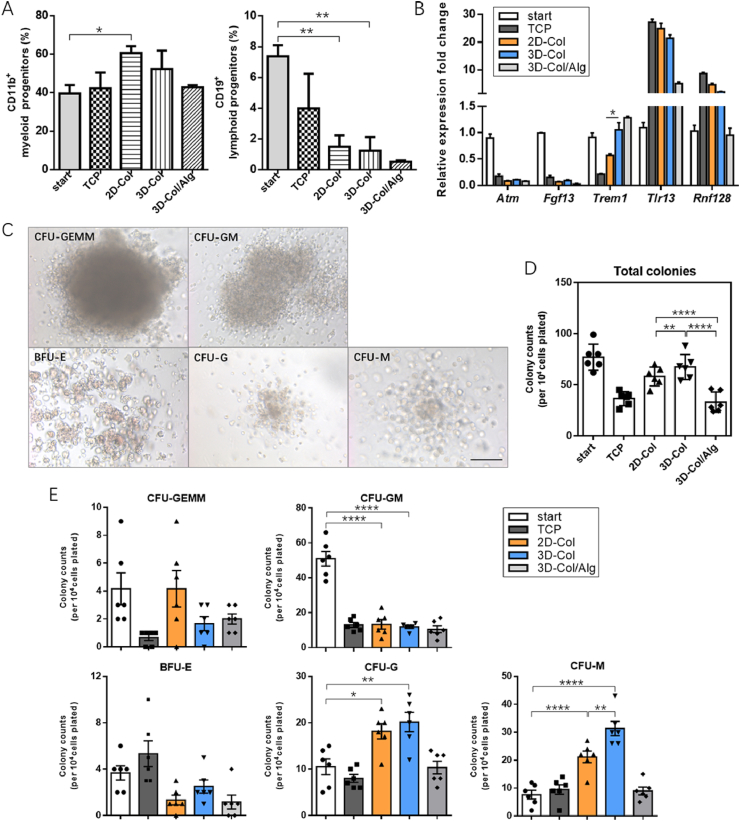
To investigate the changes in the cell lineage decisions as a result of matrix physical features, the cells from different culture systems were stained with CD11b and CD19 antibodies, the corresponding antigens of which are mostly expressed in myeloid-lineage cells and lymphoid-lineage cells respectively. The evaluation of flow cytometry data revealed all of the culture methods tended to increase the myeloid differentiation of HPCs, and among of these, cells on 2D hydrogel surfaces had the highest myeloid proportion. Meanwhile, the percentages of CD19 reflected HPCs failed to retain the differentiative capacity of lymphoid cells when cultured ex vivo irrespective of culture conditions. Hydrogel systems were not conducive to the maintenance of lymphoid-lineage cells when compared to traditional culture systems. Notably, HPCs cultured inside stiffer 3D collagen composites were found to have an evident decline in lymphoid percentage (Fig. 3A). Subsequently, we compared the expression of myeloid and lymphoid fingerprint genes between HPCs cultured on top of or inside collagen hydrogels (Fig. 3B). There was a significant increase in the fold change of Trem1 in 3D cultured HPCs, a gene that plays a pivotal role in mature monocyte development of myeloid progenitors [21].
Fig. 3.
Changes in the myeloid and lymphoid lineage potential of HPCs in 2D culture and 3D matrices.
The impact on the determination of lineage specification was next confirmed through colony forming assays (Fig. 3C). The types of colony-forming unit (CFU) colonies observed corresponded to the different stages of lineage commitment generated from hematopoietic cells. Culturing in vitro impaired the total colony formation ability of HPCs, mainly because of the relatively decreased colony counts for granulocyte and macrophage progenitor cells (CFU-GM) (Fig. 3D and E). However, there were relatively more colonies after culture on collagen systems regardless of the initial HPCs. Increased numbers of total colonies were found for cells after 3D embedded culture, in contrast to 2D culture (Fig. 3D). Compared to TCP controls, hydrogels seemingly reduced the numbers of erythroid progenitor cells (BFU-E) (Fig. 3E). We noted that collagen substrates enhanced the potency of HPCs to produce CFU-macrophage (CFU-M) and CFU-granulocyte (CFU-G) colonies compared to uncultured HPCs and HPCs after TCP culture, although no significant quantity variance was found for early multi-potential progenitor cells (CFU-GEMM), as well as granulocyte and macrophage progenitor cells (CFU-granulocyte, macrophage (CFU-GM)). We then compared the CFU-M formation of HPCs in 2D and 3D matrices, and a conspicuous elevated colony number was found for cells derived from 3D culture, indicating that 3D matrices might have promote the macrophage differentiation (Fig. 3E). Therefore, we hypothesized that a selective modulated effect from matrix-dimensionality impacted late myeloid differentiation events for HPCs.
(A) The fractions of CD11b+ myeloid cells (left) and CD19+ lymphoid cells (right) on collagen and collagen-alginate were tested. Data are mean ± SEM from three separate experiments. (B) HPCs from different systems were collected, and gene markers indicating myeloid and lymphoid differentiation of hematopoietic cells were tested. Data are mean ± SEM from three separate experiments. (C) The typical appearance of different CFU-colonies, including CFU-GEMM, CFU-GM, in addition to more mature BFU-E, CFU-G, and CFU-M colonies. Scale bar: 50 μm. (D) CFU-assays reflecting HSC lineage specification were influenced by matrix-dimensionality. After 48 h of culture, HPCs inside 3D collagen gels generated the largest number of colonies (n = 6 recipients per group from three separate experiments). (E) Collagen gels increased CFU-G and CFU-M numbers, regardless of 2D or 3D culture. When compared to 2D, 3D encapsulation significantly increased the number of CFU-M colonies. Compared to TCP controls, no significant differences could be observed for collagen-alginate cultured HPCs (n = 6 from three independent experiments). start (freshly isolated Lin-c-kit+ cells). CFU (colony-forming unit). CFU-GEMM (colony-forming unit-granulocyte, erythrocyte, macrophage, megakaryocyte). CFU-GM (colony-forming unit-granulocyte, macrophage). CFU-M (colony-forming unit-macrophage). CFU-G (colony-forming unit-granulocyte). BFU-E (burst-forming unit-erythroid). *P < 0.05, **P < 0.01, ****P < 0.0001.
3.4. Heterogeneity of differentiated hemopoietic cells within collagen matrices at single-cell resolution
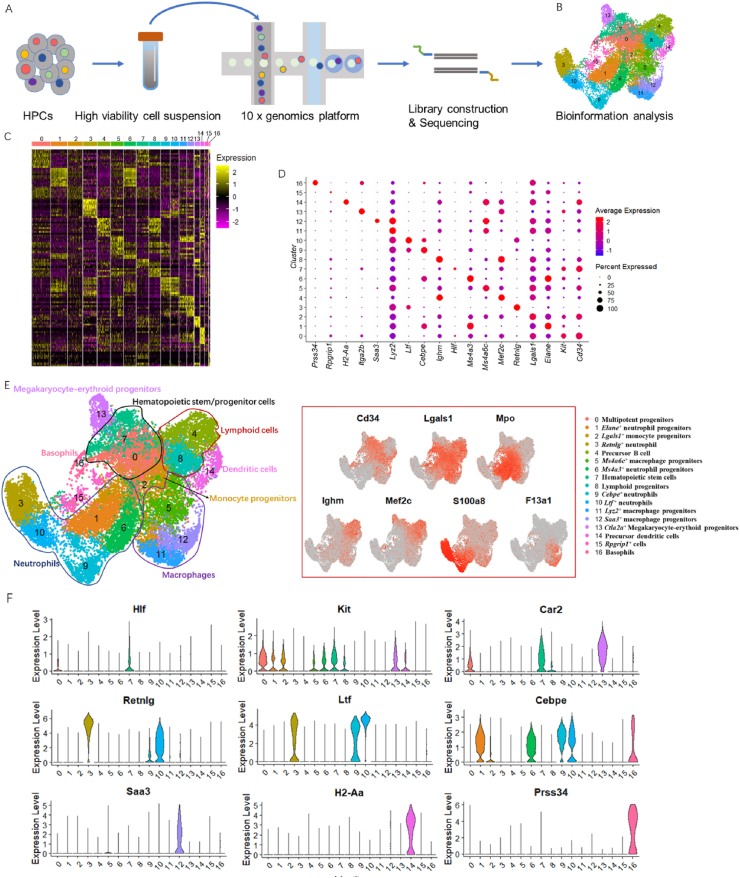
To comprehensively identify cell specification commitment and thoroughly explore the distinct role of matrix-dimensionality on HPC transcription dynamics, we applied scRNA-seq (10x Genomics Chromium, Fig. 4A) on cells from 2D and 3D collagen gels after two days of culture. We randomly selected approximately 10,000 single live cells for each system and then obtained single-cell transcriptomics data using the CellRanger package. After quality control and cell filtering, we ultimately achieved a large dataset of 22,775 signature genes across all cells, consisting of 9677 cells from gel surface and 9365 gel-encapsulated cells. Through Seurat classification, a total of 17 clusters were generated and visualized by uniform manifold approximation and projection (UMAP) plots, as shown in Fig. 4B and C. We subsequently annotated each cluster type depending on the DEGs across different clusters (Table S1). All clustering information and their corresponding markers are shown in Fig. 4, including an HSC subpopulation, a multipotent progenitor subpopulation, downstream progenitor subpopulations and mature cell subpopulations.
Fig. 4.
Single-cell resolution transcriptomic atlas of HPCs reveals the extent of cellular heterogeneity in HPCs on collagen hydrogel scaffolds.
Since the continuous differentiation landscapes have little or no discrete differentiation stages and smooth transitions across cells [22], except for some markers that are precisely expressed in certain mature lineage, many gene markers were found to be concurrently expressed in different hematopoietic clusters (Fig. 4D–F). The combined expression of the HSC fingerprint gene Hlf and the stem cell marker Ifitm1 allowed us to distinguish a cluster of HSCs from progenitor clusters expressing Cd34 and Kit [23]. Clustering by the high expression of Car2 and Itga2b was identified as megakaryocyte-erythroid progenitors. Using Lgals1 and Mpo as markers, we were able to identify monocyte clusters, including monocyte progenitors expressing Kit and Cd34, diverse granulocytes (marked by S100a8 and S100a9) and macrophages (marked by F13a1 and Psap). Neutrophil categories could be further divided by their respective highly expressed DEGs, such as Elane, Retnlg, Ms4a3, Cebpe, and Ltf. Macrophage signatures likewise could be divided into three subpopulations by the expression of Ms4a6c, Lyz2 and Saa3. The specific expression of Prss34 marked a cluster of basophils, while we were able to identify a cluster of dendritic cells (DCs) by unique DCs markers, such as H2-Aa and H2-Eb1. Moreover, we also identified lymphoid clusters marked by Ighm, including a precursor B cell cluster marked by expressing Mef2c and Cd 74.
(A) Schematic workflow of 10x Genomics analysis for our experimental strategy. (B) Uniform manifold approximation and projection (UMAP) analysis for visualizing sequencing data with reduced-dimensionality. Each point represents a cell, with color distinguishing clusters. Unbiased Seurat clustering defined a total of 17 cell clusters. (C) Gene expression heatmap showing the top 10 significantly differential expressed genes for each cell cluster defined in Fig. 4B. Yellow corresponds to high expression level; purple and black correspond to low expression level. Gene lists can be found in Table S1. Rows represent individual genes found to be selectively upregulated in individual clusters; columns represent individual cells, ordered by Seurat clusters. Margin color bars highlight cell clusters matching the classified hematopoietic cells dataset. (D) Dot plot profile of select cluster-specific markers across discriminative clusters. The scale and color of circles indicate the cell percent and expression level of each cluster. (E) Cell type assignment for each cluster module. Middle featured UMAP plots represent confirmation and gene symbols defining several major cell types containing hematopoietic progenitors (cluster 0, 7, marked by Cd34), neutrophil cells (cluster 1,3,6,9,10, marked by Lgals1, Mpo, and S100a8), lymphoid cells (cluster 4, 8 marked by Ighm and Mef2c) and macrophages (cluster 5, 11,12, marked by F13a1). In detail, cell clusters are all labeled by identified cell type (right) according to (F) violin plots of marker genes that were enriched in specific clusters.
3.5. Single-cell profiling characterizes a distinct macrophage cluster in response to 3D microenvironments
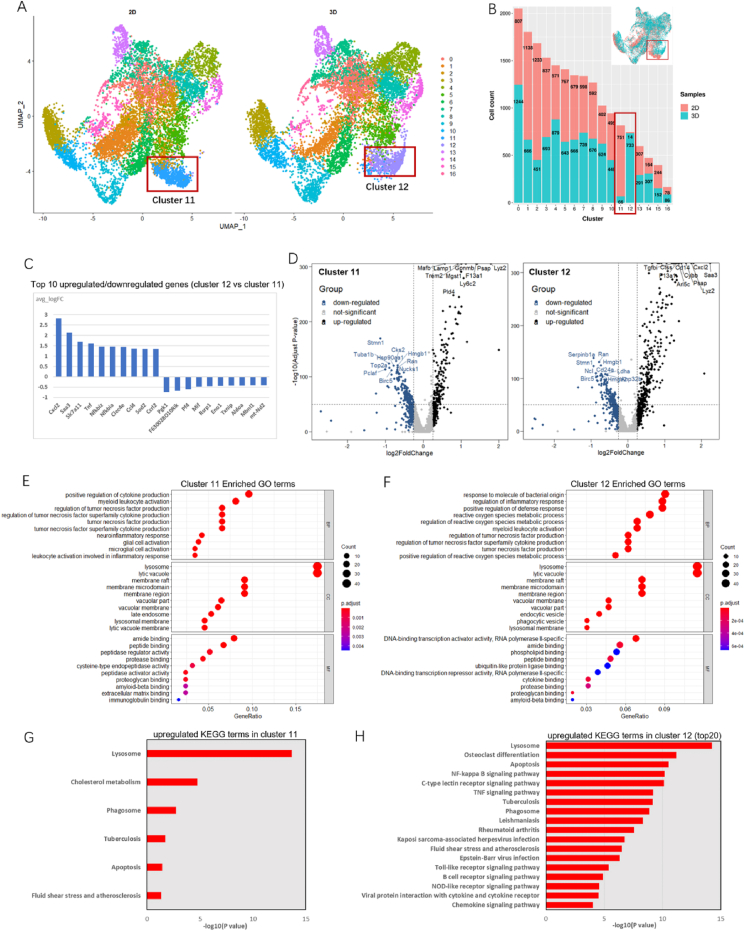
On the basis of cell classification types, we sought to define similarities and differences in the transcriptomic landscapes between 2D and 3D cell samples. Separate UMAP plots (Fig. 5A) revealed notable alternations in macrophage clusters. We analyzed the cell counts of each cluster, as shown in Fig. 5B. The HSC cluster contained 598 cells from 2D samples and 739 cells from 3D samples, and the (MPP) cluster had the most cells, including 807 2D cells and 1244 3D cells. These data suggested that 3D cultured cells possess a higher frequency of HSPCs, and this phenomenon was consistent with the consequences obtained by the previous cell phenotype identifications (Fig. 2C). We examined differences in the ratio of 2D/3D cells for cluster 11 and cluster 12, representing distinct macrophage types with their cell-type-specific gene expression profiles. A 751/817 (87.6%) cell ratio for cluster 11 belonged to the 2D samples, whereas 733/747 (98.1%) of cells derived from 3D samples were found in cluster 12, with an almost complete absence of 2D cells. Despite several macrophage markers such as Lyz2, Psap and F13a1 being detected in both clusters, cluster 12 macrophages were determined to have more upregulated DEGs, including Saa3, Cxcl2, Slc7a11, Tnf, and other genes, as shown in Fig. 5C and D.
Fig. 5.
Distinct landscape of gene expression features in macrophage clusters between 2D and 3D cells.
Single-cell hematopoietic landscapes highlight the differentiated journeys toward different hematopoietic lineages in native bone marrow [24]. To demonstrate that cluster 11 and cluster 12 macrophages were indeed differentiated in vitro, we compared the expression of those macrophage-related DEGs in the single-cell transcriptional template of uncultured Lin-c-kit+ cells, which was provided by Dahlin's study [24]. Fig. S4 and Fig. S5 demonstrated that the expression of these genes was specific to our dataset, and thus illustrated that these two clusters of macrophages were generated from HPCs after culture on collagen gels.
Using their respective up-regulated DEGs, we performed GO enrichment analysis to gain insight into the cell-type related cell functional states of these clusters. There were indeed several top-level GO terms differently enriched. DEGs from cluster 11 macrophages relating to the three main categories of GO function were primarily enriched in “positive regulation of cytokine production”, “lysosome” and “amide binding” (Fig. 5E). In contrast, DEGs from cluster 12 macrophages were associated with GO terms including “response to molecule of bacterial origin”, “lysosome”, and “DNA-binding transcription activator activity, RNA polymerase II-specific” (Fig. 5F). Moreover, KEGG analysis revealed that the DEGs from cluster 11 were involved in 6 KEGG pathways, among which “Lysosome” contained the most DEGs (23 DEGs) (Fig. 5G). Conversely, the upregulated genes of cluster 12 cells were related to more pathways, including the KEGG pathways that were found in cluster 11. In total, 29 DEGs from cluster 12 were involved in “Lysosome”, and pathways such as “Osteoclast differentiation”, “NF-kappa B signaling pathway”, “C-type lectin receptor signaling pathway”, “TNF signaling pathway”, and “Leishmaniasis” were also associated with cluster 12 macrophages, as displayed in Fig. 5H.
(A) UMAP dimensional reduction displaying relative position of 2D cells (left) and 3D cells (right). Obvious differences were found in macrophage subsets cluster 11 and cluster 12. (B) Bar plot of the distributions of the number of successfully profiled single cells in each cell cluster sorted from 2D or 3D. Cluster 11 contained 751 2D cells and 66 3D cells, while cluster 12 contained 14 2D cells and 733 3D cells. (C) Top 10 differentially expressed genes from cluster 12 compared to cluster 11. (D) Volcano plots of cell type-specific genes differentially expressed in cluster 11 and cluster 12 cells. (E and F) Top “biological process (BP)”, “molecular function (MF)”, and “cellular component (CC)” Gene Ontology (GO) terms enriched for upregulated DEGs of (E) cluster 11 and (F) cluster 12. (G and H) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis for genes up-regulated in (G) cluster 11 and (H) cluster 12 (top 20).
3.6. Distinct transcriptional trajectory of macrophage lineage guided by a 3D microenvironment
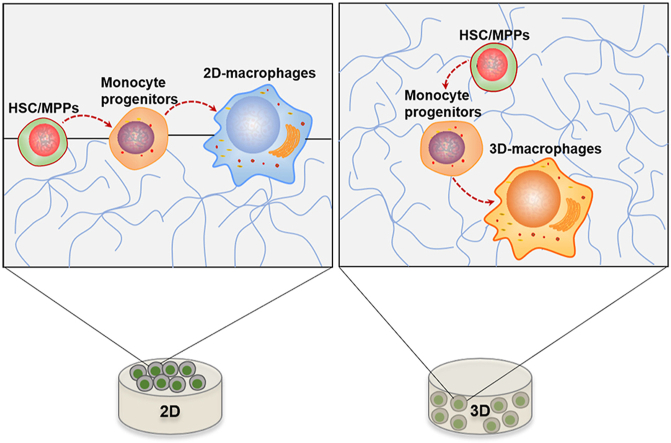
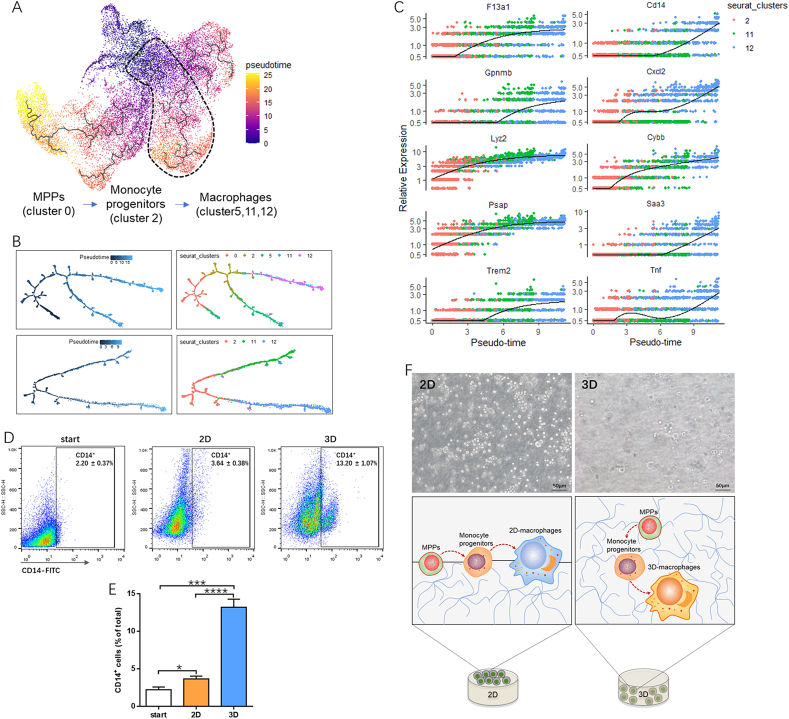
Theoretically, HSCs will differentiate into monocyte progenitors, and macrophages are generated from their upstream monocytes. Our dataset was classified into 17 clusters containing a cluster of monocyte progenitors (cluster 2) and 3 clusters of macrophages (cluster 5/11/12), and the molecular features among macrophage cluster 11 and 12 were revealed by several preceding analyses. To investigate the maturation trajectories symbolizing the cell type traverse from progenitor to mature lineage, especially to macrophage lineage, pseudotime trajectories were established. Integrated pseudotime in Fig. 6A indicated the developmental hierarchy between these 17 clusters, reflecting progenitor clusters emerging earlier than mature clusters during HSC specification. Then the developmental cascades in macrophage related populations, including clusters 0, 2, 5, 11, and 12, were extracted and visualized to reflect the hierarchical relationship from multipotent progenitor to mature macrophage. As expected, cluster 0 was demonstrated to be the highest “source” cell state in our MPP-macrophage specification line, while cluster 2 served as a likely intermediary progenitor state. Clusters 5, 11, and 12 occupied at a relative later stage and shared common upriver progenitors (Fig. 6B). In particular, clusters 11 and 12 were located on the same branch as cluster 2, indicating the parallel pseudotime ordering during macrophage differentiation. mRNA levels of genes that distinguish macrophage subtypes were next scrutinized with respect to their pseudotime coordinates. Along with the maturation of monocytes, the expression of macrophage markers such as F13a1, Lyz2, and Psap increased gradually. Genes (Saa3, Cd14, Cxcl2, and Tnf) that were highly expressed in cluster 12 macrophages showed trajectory-dependent patterns, and increased along with cluster 12 macrophage maturation (Fig. 6C). In addition, consistent with the gene expression of Cd14, CD14+ cells were significantly increased in 3D cultured cells compared to the fleshly sorted cells (start) (Fig. 6D). Taken together, on the basis of temporal extrapolation results and marker gene expression, cluster 11 and cluster 12 were identified as contemporaneous macrophages. Therefore, we could conclude that a transferred macrophage lineage trajectory was a result of matrix-dimensionality. Differentiating monocytes appeared to be skewed preferentially toward cluster 12 macrophages in 3D collagen matrices (3D-macrophages), while 2D matrices tended to produce the cluster 11 type macrophages (2D-macrophages, Fig. 6E).
Fig. 6.
Dynamic pseudotime analysis reveals two different macrophage clusters that occupy similar developmental hierarchies.
(A) Comprehensive developmental trajectories of HPCs cultured on/in collagen hydrogels at single-cell resolution reconstructed by monocle 3, spanning from early progenitor stage (purple and blue) to terminal differentiation stage (orange and yellow). (B) Clusters of cells containing MPPs (cluster 0), monocytes (cluster 2) and macrophages (clusters 5,11, and 12) were subsequently sorted for unbiased dynamic lineage analysis generating hypothetical developmental relationships by monocle2 algorithms. Cells are ordered in pseudotime. Cluster 5, 11, and 12 represent individual macrophage linage bifurcation in Monocle. (C) Pseudotime kinetics of cluster-defining genes along macrophage lineages (from monocyte progenitor to cluster 11 and 12). Lines from F13a1, Gpnmb, Lyz2, Psap, Trem2, and Cybb show the smoothed expression over pseudotime in macrophage lineage, and Cd14, Cxcl2, Saa3 and Tnf were transiently upregulated in cluster 12 macrophages. (D and E) Flow cytometry analysis of percentages of CD14+ cells on surface of collagen and cultured inside the same gel types. Data are mean ± SEM from three separate experiments (n = 3 recipients in start group; n = 5 recipients in 2D and 3D groups). *P < 0.05, ***P < 0.001, ****P < 0.0001. (F) Dimensionality-dependent developmental trajectories of macrophage lineages. A 3D environment alters the direction of monocyte progenitors and produced cluster 12 macrophages, instead of cluster 11 macrophages, which were formed on 2D matrices. MPPs (Multipotent progenitors).
3.7. Extensive cell communications between 3D-macrophages and other hematopoietic subpopulations
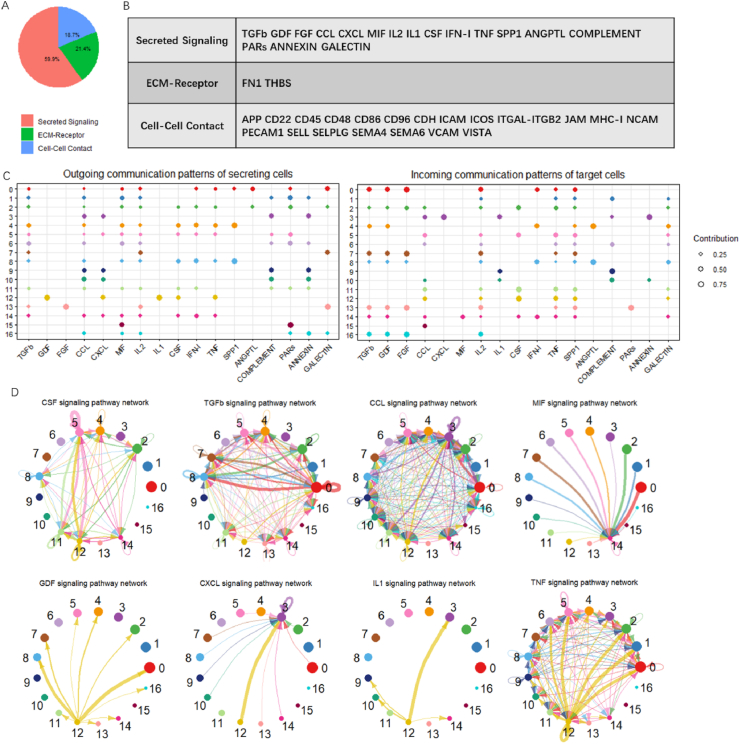
To infer the global cell communications across 17 cell clusters, the Cellchat package was adopted used due to its available repository of mouse ligand-receptor interactions. We were able to quantitatively analyze the dominant signal inputs and outputs among these clusters [25]. Our Seurat clusters were input into Cellchat to create complex intercellular communication networks from the three aspects of "Secreted Signaling", "ECM-Receptor", and "Cell-Cell Contact" (Fig. 7A). Hematopoietic subsets connected and interacted to each other by ligand-receptor pairs (Supplementary Table S2). A total of 41 cell-cell interactions between different hematopoietic populations were examined, containing 17 “Secreted Signaling”, 2 “ECM-Receptor”, and 22 “Cell-cell Contact” pathways (as listed at Fig. 7B and C and Supplementary Fig. S6 A and B). On the basis of “Secreted Signaling”, macrophage cluster 5, 11 and 12 showed similar incoming patterns, such as CCL, IL2, CSF, TNF, SPP1, and GALECTIN, implying similar receptors exist in different macrophage subsets enable cells to receive the same signals from other cell subsets (Fig. 7C and D). However, divergence was noted in the outgoing communication patterns. Cluster 11 and 5 macrophages had low expression of ligands involved in secreting signaling such as TGFb, CCL, and MIF pathways. Cluster 12 macrophages, in contrast, secreted distinct ligands related to GDF, CXCL, IL1 and TNF pathways.
Fig. 7.
Analysis of cell-cell communications between hematopoietic subsets.
Notably, 3D-macrophages highly expressed Cxcl2, which stimulated the mobilization and accelerated the release of BM neutrophils during inflammatory reactions [26]. As expected, CXCL and IL1 signaling networks reflected the intense communications of cluster 12 macrophages and cluster 3 neutrophils (Fig. 7D), which were linked by the ligand-receptor complexes of Cxcl2/Cxcr2 (in an autocrine and paracrine manner) and Il1b/Il1r2 (in a paracrine manner). TNF networks showed that the receptors Tnfrsf1a and Tnfrsf1b interacted with TNF, which contributed to the close relationship of 3D-macrophages and surrounding groups (clusters 0, 1, 2, 5, and 6) including monocyte progenitors and neutrophils. Moreover, the ligand Gdf15 interacted with the receptor Tnfrsf1b in lymphoid clusters (clusters 4 and 8), allowing the cross talk in the GDF signaling network between 3D-macrophages and lymphoid cells. This could inhibit macrophage-treated neutrophil recruitment [27]. In summary, high levels of DEGs in 3D-macrophages might enhance the secretion of ligands and thereby strengthen their communication with other cell groups within 3D environment. Some of these 3D-enhanced signaling pathways might connect with each other and contribute to the same physiological processes.
(A) The ligand-receptor interaction database Cellchat contains 2021 validated interactions, including 59.9% of secreting interactions, 21.4% of ECM-receptor interactions, and 18.7% of cell-cell contact interactions. (B) Enrichment in Cellchat signaling based on over-expression of ligands and receptors for each cell cluster. (C) Overview of incoming and outgoing signals (Secreted Signaling) for each cell group. The colored cell node size is proportional to the contribution score computed by leveraging pattern recognition approaches. Outgoing signaling, defined as treating cells as sources and incoming signaling, defined as treating cells as targets, were then calculated for cells from each cluster. (D) Representative circle plots showing globe connectomes across the 17 clusters predict intensive relationships between macrophages from cluster 12 and other subpopulations. Vertex (colored cell node) size is proportional to the contribution score in (C), while the thickness of each line is proportional to the sum of the weights between two nodes. The directions of arrows provide quick insight into the autocrine vs. paracrine manner for signaling in a given cell type. Similarities and differences in overall signaling structure between the cell nodes were readily observable.
3.8. Potential TFs underlying the regulation across 3D specialized macrophages
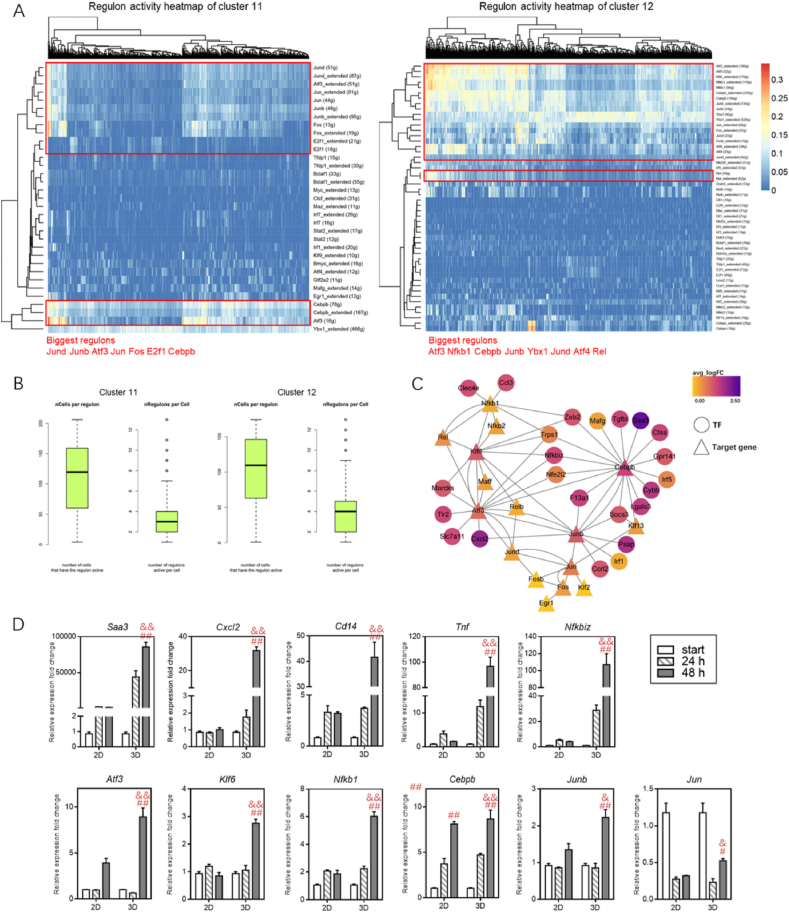
We next applied the SCENIC method to clarify the activities of candidate TF combinations related to our identified 2D/3D-macrophage clusters. The 33 biggest (not extended) regulons representing the TF-gene co-expression modules were found in 2D-macrophages, and the 50 biggest regulons were highly expressed in 3D-macrophages (Fig. 8A and B). Correspondingly, more TFs with their recognition motifs were significantly enriched in their co-expressed gene sets in 3D-macrophages (Fig. S7A). Comparative analysis of the AUC activity dynamics of selected common regulons with regard to the HSC-MPP-monocytes-macrophage axis was performed (Fig. S7B). The activities of Cebpb and Atf3 increased following the development of macrophage differentiation in the 3D microenvironment. Regulons such as Jun and Jund were also highly expressed in 3D-HSCs and 3D-macrophages. Combined with upregulated cluster-DEGs, TF-target regulatory networks specific to 3D-macrophages were constructed (Fig. 8C), and time-dependent increase in mRNA levels for dominating TFs and DEGs were validated on the aggregate level by statistical significance (Fig. 8D). Straightforward relationships of TFs such as Atf3, Cebpb, Junb and Klf6 were observed. Analysis of TF regulons further illustrated the high transcriptome activity of macrophages induced by 3D matrices in vitro.
Fig. 8.
SCENIC reveals significant regulons activated in 2D- and 3D-macrophages.
(A) Heatmap showing the activity of each regulon in individual cells of cluster 11and 12. After RcisTarget identification, 33 and 50 regulons with only direct targets are significantly enriched in cluster 11 and 12, respectively, which are subsequently scored by AUCell function in SCENIC. (B) Comparative boxplots showing cluster 12 has relative more cells that have active regulons as well as more activated regulons per cell. (C) Gene regulatory networks inferred for putative key transcription factors (TFs) that motivate upregulated DEGs in cluster 12 macrophages, based on TF-gene correlations identified by SCENIC enrichment analysis. (D) Expression levels of select upregulated genes and TFs were validated by quantitative real-time PCR analysis (data are mean ± SEM from three separate experiments). &P < 0.01, &&P < 0.0001 with respect to 2D at 48 h; #P < 0.01, ##P < 0.0001 with respect to start group.
4. Discussion
As one of the abundant ECM components, type I collagen has been engineered for hydrogels to observe and quantify the effects of mechanics on cellular responses [28]. In the present work, collagen hydrogels and collagen-alginate hydrogels were applied for comparing the maintenance and lineage specification of HPCs on substrates with different physical properties. Crosslinking of sodium alginate strengthened the mechanical characteristics of primary collagen hydrogels (from 45.12 Pa to 546.5 Pa) while changing the microstructure accordingly. The decreased cell viability, flow cytometric HPC frequencies and potency to generate CFU colonies suggested that HPC viability was susceptible to external physical signals and probably failed to endure the enforced stiffness or altered pore geometry in 3D conditions.
3D cell-matrix models with diverse stiffnesses are more representative of the actual niche microenvironment. The superiority of 3D collagen hydrogels, with their soft mechanical stiffness, on cell viability and HPC phenotype maintenance was also presented here. 3D collagen encapsulated HPCs had increased CFU-M forming abilities. Even though a direct link between in vitro matrix biophysical properties and early HSC fate specification events was demonstrated by Harley's study [7], it is interesting to speculate that there is an influence of culture space constraints on the late developmental stage toward macrophage lineages as demonstrated by our study. In reality, HPCs can compensate for the constraints of a collagen gel in an autocrine/paracrine fashion [14], but the specific mechanism by which encapsulated HPCs sense physical signals and change the direction of differentiation needs additional study [3].
Studies have shown that HSCs are capable of promoting the expansion of human CD34+ hematopoietic cells, and promoting the number of myeloid colony-forming units (CFU–C) [29]. Furthermore, a large number of cytokines or chemokines are overexpressed in these cells, including IL-8, MIP-α, CXCL2, TREM1 and TNF- α as well as many TFs such as FOSB, JUN, and ATF3, suggesting that complex cell-cell and cell-ECM interactions orchestrate the proliferation and mobilization of HSCs [29]. However, in these experimental settings, using fluid suspension cultivation as a control, rather than a collagen basal surface, was perhaps insufficient to counteract the chemical effect caused by the presence of collagen I. In contrast, gene expression tested by bulk sequencing failed to identity the reaction of individual cell groups to external stimulation or cellular heterogeneity during proliferation or differentiation of HPCs. By means of scRNA-seq, we generated a comprehensive single-cell compendium of heterogenetic hematopoietic populations and their hierarchical models in 2D- and 3D-micoenvironments. ScRNA-seq, which has been recognized as a promising technological advancement for stem cell research, permits the dissection of cellular heterogeneity in an unbiased manner (with no need of a priori knowledge of a cell population) and allow us to discover novel intrinsic characteristics of stem cell populations that may have been masked by traditional evaluation methods [22,30]. ScRNA-seq converts static differentiation landscapes into a dynamic cell flux model and yielded a more holistic understanding of HPCs differentiation events when they were cultured in hydrogels that provide physical regulatory cues. This provides evidence of the altered cell development trajectory induced by 3D matrices.
3D collagen scaffold slices have been reported to modulate the lineage commitment of DCs and contribute to the production of regulatory DCs from BM cells. A 3D microenvironment prevents antigen uptake by DCs, but these cells possess immunoregulatory functions, according to the up-regulated expression of immunosuppressive molecules [31]. Intriguingly, our collagen encapsulation scaffolds resulted in a highly specialized population of macrophages in the absences of Lin+ cells, instead of DCs, suggesting that mature BM Lin+ cells could be basic mediators for 3D-DCs lineage.
The diverse macrophage subpopulations derived from 2D/3D cultivated HPCs are profoundly dissectible in terms of their single-cell transcriptome landscape. The 3D-macrophages were naturally generated on collagen 3D matrices. Compared with BM native macrophage subsets and macrophages that are induced by macrophage colony-stimulating factor (M-CSF or CSF1) in vitro, there are possible differences in functional plasticity. This remains to be determined. The majority of molecules implicated in the TFs-target network of 3D-macrophages play roles in inflammatory reactions, macrophage polarization and other cell functions. Some are overexpressed genes that have been previously found in collagen cultured human HPCs. For example, the transcription factor Atf3, acts as a regulator of T cells to balance mucosal immunity [32], control mitochondrial homeostasis by suppressing the mRNA synthesis of PINK1 [33], and join in the JNK signaling pathway to mediate the inhibitory action of Tnf-α on osteoblast differentiation [34]. Tnf, Cxcl2, Nfkbiz and Jun belong to the NF-kappa B and TNF signaling pathways, which are not only extensively activated in inflammatory reactions and macrophage polarization, but also are positively associated with hematopoietic regeneration and myeloid differentiation [35]. Therefore, we can conclude that certain homologous functions of these genes control the fate determination of HPCs in 3D matrices. Except for the role of the chemokine Cxcl2 produced by macrophages for neutrophil migration that has been mentioned previously, our analysis on the communications of cell clusters intuitively illustrates the interactions between 3D-macrophages and other clusters, and such cooperation may occur within a 3D microenvironment rather a 2D. Unlike other genes, Saa3, which was uniquely expressed in 3D-macrophages, had a more than 20-fold increase in 3D cells at the 24 h time point. Thus, we can regard Saa3 as an early marker for such transformations in macrophage types. In recent years, Saa3 has attracted attention as a novel mediator in multiple positive/negative feedbacks between diverse niche functional cells. For instance, Saa3 acts on the T cell signaling pathways and directly stimulates the cell differentiation program of Th17 cells [36]. Saa3 is also an inhibitory element expressed by RANKL-treated BM macrophages to block PTH-stimulated osteoblastic differentiation [37]. Furthermore, 3D matrices may slightly suppress the expression of Pgk1, Eno1 and Pf4. Inhibition of Pgk1 and Eno1 has been deemed as an effective therapeutic mechanism to disrupt macrophage-promoted tumor growth [38,39]. The chemokine gene Pf4 (Cxcl4), which is mainly expressed by the megakaryocytic lineage, has been deemed to be an inhibitor of myeloid progenitors and macrophage phagocytosis [40,41]. The low expression of Pf4 indicated the undamaged macrophage phagocytic capacity of 3D-macrophages. Taken together, the data we present here demonstrated that HPCs were capable of sensitively reacting to space constraints and this prompts us to further study the biological characteristics of 3D-macrophages.
CRediT authorship contribution statement
Pan Zhang: Conceptualization, Methodology, Investigation, Writing – original draft, Writing – review & editing. Linmu Xu: Methodology, Investigation. Jingsong Gao: Conceptualization. Guangkui Xu: Investigation, Validation. Yanping Song: Conceptualization, Methodology, Investigation. Guang Li: Conceptualization, Methodology. Jingjing Ren: Conceptualization, Methodology. Yunjie Zhang: Conceptualization, Methodology. Cheng Yang: Methodology, Data curation. Yu Zhang: Methodology, Data curation. Ruiheng Xie: Data curation, Visualization. Nu Zhang: Resources, Language modification. Hui Yang: Writing – review & editing, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by grants from National Natural Science Foundation of China (Nos. 12002285, 11672227, 61927810), Natural Science Basic Research Plan in Shaanxi Province of China (Nos. 2018JQ1033, 2020JZ-11) and Innovation Foundation for Doctor Dissertation of Northwestern Polytechnical University (No. CX202058).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2021.08.032.
Contributor Information
Nu Zhang, Email: zhangnu@nwpu.edu.cn.
Hui Yang, Email: kittyyh@nwpu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Morrison S.J., Scadden D.T. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi J.S., Mahadik B.P., Harley B.A. Engineering the hematopoietic stem cell niche: frontiers in biomaterial science. Biotechnol. J. 2015;10:1529–1545. doi: 10.1002/biot.201400758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee-Thedieck C., Spatz J.P. Biophysical regulation of hematopoietic stem cells. Biomater Sci. 2014;2:1548–1561. doi: 10.1039/c4bm00128a. [DOI] [PubMed] [Google Scholar]
- 4.Holst J., Watson S., Lord M.S., Eamegdool S.S., Bax D.V., Nivison-Smith L.B., et al. Substrate elasticity provides mechanical signals for the expansion of hemopoietic stem and progenitor cells. Nat. Biotechnol. 2010;28:1123–1128. doi: 10.1038/nbt.1687. [DOI] [PubMed] [Google Scholar]
- 5.Choi J.S., Harley B.A. The combined influence of substrate elasticity and ligand density on the viability and biophysical properties of hematopoietic stem and progenitor cells. Biomaterials. 2012;33:4460–4468. doi: 10.1016/j.biomaterials.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Lee-Thedieck C., Rauch N., Fiammengo R., Klein G., Spatz J.P. Impact of substrate elasticity on human hematopoietic stem and progenitor cell adhesion and motility. J. Cell Sci. 2012;125:3765. doi: 10.1242/jcs.095596. [DOI] [PubMed] [Google Scholar]
- 7.Choi J.S., Harley B.A.C. Marrow-inspired matrix cues rapidly affect early fate decisions of hematopoietic stem and progenitor cells. Science advances. 2017;3 doi: 10.1126/sciadv.1600455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ward C.M., Ravid K. Matrix mechanosensation in the erythroid and megakaryocytic lineages. Cells. 2020;9:894. doi: 10.3390/cells9040894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aguilar A., Pertuy F., Eckly A., Strassel C., Collin D., Gachet C., et al. Importance of environmental stiffness for megakaryocyte differentiation and proplatelet formation. Blood. 2016;128:2022–2032. doi: 10.1182/blood-2016-02-699959. [DOI] [PubMed] [Google Scholar]
- 10.Discher D.E., Mooney D.J., Zandstra P.W. Growth factors, matrices, and forces combine and control stem cells. Science (New York, NY) 2009;324:1673–1677. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel P.N., Smith C.K., Patrick C.W., Jr. Rheological and recovery properties of poly(ethylene glycol) diacrylate hydrogels and human adipose tissue. J. Biomed. Mater. Res. 2005;73:313–319. doi: 10.1002/jbm.a.30291. [DOI] [PubMed] [Google Scholar]
- 12.Knight E., Przyborski S. Advances in 3D cell culture technologies enabling tissue-like structures to be created in vitro. J. Anat. 2015;227:746–756. doi: 10.1111/joa.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gvaramia D., Müller E., Müller K., Atallah P., Tsurkan M., Freudenberg U., et al. Combined influence of biophysical and biochemical cues on maintenance and proliferation of hematopoietic stem cells. Biomaterials. 2017;138:108–117. doi: 10.1016/j.biomaterials.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 14.Mahadik B.P., Bharadwaj N.A., Ewoldt R.H., Harley B.A. Regulating dynamic signaling between hematopoietic stem cells and niche cells via a hydrogel matrix. Biomaterials. 2017;125:54–64. doi: 10.1016/j.biomaterials.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mattei G., Gruca G., Rijnveld N., Ahluwalia A. The nano-epsilon dot method for strain rate viscoelastic characterisation of soft biomaterials by spherical nano-indentation. Journal of the mechanical behavior of biomedical materials. 2015;50:150–159. doi: 10.1016/j.jmbbm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 16.Bajpai M., Bajpai S.K., Jyotishi P. Water absorption and moisture permeation properties of chitosan/poly(acrylamide-co-itaconic acid) IPC films. Int. J. Biol. Macromol. 2016;84:1–9. doi: 10.1016/j.ijbiomac.2015.11.088. [DOI] [PubMed] [Google Scholar]
- 17.Spiller K.L., Laurencin S.J., Charlton D., Maher S.A., Lowman A.M. Superporous hydrogels for cartilage repair: evaluation of the morphological and mechanical properties. Acta Biomater. 2008;4:17–25. doi: 10.1016/j.actbio.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Lee P.F., Bai Y., Smith R.L., Bayless K.J., Yeh A.T. Angiogenic responses are enhanced in mechanically and microscopically characterized, microbial transglutaminase crosslinked collagen matrices with increased stiffness. Acta Biomater. 2013;9:7178–7190. doi: 10.1016/j.actbio.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X., Zhao J., Gu C., Cui Y., Dai Y., Song G., et al. Med23 serves as a gatekeeper of the myeloid potential of hematopoietic stem cells. Nat. Commun. 2018;9:3746. doi: 10.1038/s41467-018-06282-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bello A.B., Park H., Lee S.H. Current approaches in biomaterial-based hematopoietic stem cell niches. Acta Biomater. 2018;72:1–15. doi: 10.1016/j.actbio.2018.03.028. [DOI] [PubMed] [Google Scholar]
- 21.Gingras M.C., Lapillonne H., Margolin J.F. TREM-1, MDL-1, and DAP12 expression is associated with a mature stage of myeloid development. Mol. Immunol. 2022;38:817–824. doi: 10.1016/s0161-5890(02)00004-4. [DOI] [PubMed] [Google Scholar]
- 22.Watcham S., Kucinski I., Gottgens B. New insights into hematopoietic differentiation landscapes from single-cell RNA sequencing. Blood. 2019;133:1415–1426. doi: 10.1182/blood-2018-08-835355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giladi A., Paul F., Herzog Y., Lubling Y., Weiner A., Yofe I., et al. Single-cell characterization of haematopoietic progenitors and their trajectories in homeostasis and perturbed haematopoiesis. Nat. Cell Biol. 2018;20(7):836–846. doi: 10.1038/s41556-018-0121-4. [DOI] [PubMed] [Google Scholar]
- 24.Dahlin J.S., Hamey F.K., Pijuan-Sala B., Shepherd M., Lau W.W.Y., Nestorowa S., et al. A single-cell hematopoietic landscape resolves 8 lineage trajectories and defects in Kit mutant mice. Blood. 2018;131:e1–e11. doi: 10.1182/blood-2017-12-821413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin S., Guerrero-Juarez C.F., Zhang L., Chang I., Ramos R., Kuan C.-H., et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021;12:1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burdon P.C., Martin C., Rankin S.M. The CXC chemokine MIP-2 stimulates neutrophil mobilization from the rat bone marrow in a CD49d-dependent manner. Blood. 2005;105:2543–2548. doi: 10.1182/blood-2004-08-3193. [DOI] [PubMed] [Google Scholar]
- 27.Artz A., Butz S., Vestweber D. GDF-15 inhibits integrin activation and mouse neutrophil recruitment through the ALK-5/TGF-βRII heterodimer. Blood. 2016;128:529–541. doi: 10.1182/blood-2016-01-696617. [DOI] [PubMed] [Google Scholar]
- 28.Plant A.L., Bhadriraju K., Spurlin T.A., Elliott J.T. Cell response to matrix mechanics: focus on collagen. Biochim. Biophys. Acta. 2009;1793:893–902. doi: 10.1016/j.bbamcr.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 29.Oswald J., Steudel C., Salchert K., Joergensen B., Thiede C., Ehninger G., et al. Gene-expression profiling of CD34+ hematopoietic cells expanded in a collagen I matrix. Stem Cell. 2006;24(3):494–500. doi: 10.1634/stemcells.2005-0276. [DOI] [PubMed] [Google Scholar]
- 30.Wen L., Tang F. Single-cell sequencing in stem cell biology. Genome Biol. 2016;17:71. doi: 10.1186/s13059-016-0941-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang Y., Wang B., Zhao Y., Xiao Z., Li J., Cui Y., et al. Collagen scaffold microenvironments modulate cell lineage commitment for differentiation of bone marrow cells into regulatory dendritic cells. Sci. Rep. 2017;7:42049. doi: 10.1038/srep42049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao Y., Yang Q., Deng H., Tang J., Hu J., Liu H., et al. Transcriptional factor ATF3 protects against colitis by regulating follicular helper T cells in Peyer's patches. Proc. Natl. Acad. Sci. Unit. States Am. 2019;116:6286–6291. doi: 10.1073/pnas.1818164116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bueno M., Brands J., Voltz L., Fiedler K., Mays B., St Croix C., et al. ATF3 represses PINK1 gene transcription in lung epithelial cells to control mitochondrial homeostasis. Aging Cell. 2018;17 doi: 10.1111/acel.12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeong B.C. ATF3 mediates the inhibitory action of TNF-α on osteoblast differentiation through the JNK signaling pathway. BBRC (Biochem. Biophys. Res. Commun.) 2018;499:696–701. doi: 10.1016/j.bbrc.2018.03.214. [DOI] [PubMed] [Google Scholar]
- 35.Yamashita M., Passegué E. TNF-α coordinates hematopoietic stem cell survival and myeloid regeneration. Cell stem cell. 2019;25:357–372. doi: 10.1016/j.stem.2019.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J.Y., Hall J.A., Kroehling L., Wu L., Najar T., Nguyen H.H., et al. Serum amyloid A proteins induce pathogenic Th17 cells and promote inflammatory disease. Cell. 2020;180:79–91. doi: 10.1016/j.cell.2019.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Estus T.L., Choudhary S., Pilbeam C.C. Prostaglandin-mediated inhibition of PTH-stimulated β-catenin signaling in osteoblasts by bone marrow macrophages. Bone. 2016;85:123–130. doi: 10.1016/j.bone.2016.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cappello P., Curcio C., Mandili G., Roux C., Bulfamante S., Novelli F. Next generation immunotherapy for pancreatic cancer: DNA vaccination is seeking new combo partners. Cancers. 2018;10:51. doi: 10.3390/cancers10020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y., Yu G., Chu H., Wang X., Xiong L., Cai G., et al. Macrophage-associated PGK1 phosphorylation promotes aerobic glycolysis and tumorigenesis. Molecular cell. 2018;71:201–215. doi: 10.1016/j.molcel.2018.06.023. [DOI] [PubMed] [Google Scholar]
- 40.Lindsey M.L., Jung M., Yabluchanskiy A., Cannon P.L., Iyer R.P., Flynn E.R., et al. Exogenous CXCL4 infusion inhibits macrophage phagocytosis by limiting CD36 signalling to enhance post-myocardial infarction cardiac dilation and mortality. Cardiovasc. Res. 2019;115:395–408. doi: 10.1093/cvr/cvy211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hérault A., Binnewies M., Leong S., Calero-Nieto F.J., Zhang S.Y., Kang Y.A., et al. Myeloid progenitor cluster formation drives emergency and leukaemic myelopoiesis. Nature. 2017;544:53–58. doi: 10.1038/nature21693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.