Abstract
Salvia hispanica L. (Chia) seeds are good source of proteins with diverse health benefits. The seed protein was extracted through alkaline solubilisation followed by acid precipitation to separate fibres and are digested sequentially by pepsin and pancreatin. Enzyme-substrate ratio, temperature and contact time had high impact on degree of hydrolysis affecting their chelating ability. Maximum degree of hydrolysis (14.06%) and maximum copper chelation (74.98%) was obtained at 4% w/w enzyme-substrate ratio at 37 °C for 4 h. Copper chelating enzymatic hydrolysate was isolated by HiTrap chelating column and purified further by rpHPLC. Out of nine fractions obtained by rpHPLC the sixth fraction with 93.09 ± 0.16% of copper chelating activity and 82.91 ± 0.52% of antioxidant activity was further characterized as Copper chelating Chia Protein Hydrolysate (CCPH). Ultraviolet spectroscopy and fluorescence spectroscopic studies revealed the interaction of the major chelating sites of the CCPH with the copper divalent ion. The purified CCPH was subjected to LC-MS/ESI-TOF analysis from which six major intense peaks obtained with m/z value ranging from 0.4 kDa to 2.5 kDa were identified and sequenced using Mascot database. The functional behaviour and the binding capacity of these peptides were analysed by their amino acid composition. The CCPH was stable in a simulated gastric condition and its chelating ability remained unaltered. These results explored an informative bioactive peptides with varied activity and one valuable among is the copper chelating with antioxidant property. Furthermore, these Chia seed protein hydrolysates can be useful as dietary supplements to enhance mineral bioavailability.
Keywords: Chia seed, Protein hydrolysate, Copper chelating peptides
Graphical abstract
Highlights
-
•
Optimization of Pepsin-Pancreatin hydrolysis of Salvia hispanica L. (Chia) seed protein.
-
•
Purification of Copper Chelating Protein Hydrolysate (CCPH) using affinity chromatography and rpHPLC.
-
•
The structure-function analysis of purified CCPH by UV/Vis and Fluorescence spectroscopic analysis and mass determination by LC-MS/ESI-TOF.
-
•
In vitro gastro-intestinal simulation studies of CCPH indicated its structural stability with constant chelating property.
-
•
Hence CCPH can be a promising, safe alternative of metal salts for mineral fortification.
1. Introduction
Protein hydrolysates and derived peptides are known to possess several biological activities, such as antioxidant, anti-inflammatory, anti-hypertensive, and mineral chelation. Bioactive peptides have varied effects on organism's health and chelation of transient divalent metal ions like iron, calcium, copper, zinc etc., is one of their proposed mechanisms of action. Now a day's food industries are showing great interest over functional foods which have paved research in this field focusing mainly on natural products that are rich in bioactive compound and one such bioactive compound is peptides/proteins. Among various bioactivities of proteins/peptides, the ability to bind with metals, hence acting as chelating agent has high significance. Although copper and iron are essential co-factors for various enzymes, their excess can have unfavourable pro-oxidant effects in vivo, by the formation of reactive oxygen species. They also aid oxidative reactions in food which in succession has negative influence on the flavour, texture, nutritive value and shelf life of food products. Hence, chelating peptides make up a powerful tool for preventing these oxidative processes both in foods and in vivo. Peptide-metal complexes are more stable than metal in its free form under gastrointestinal conditions (Fernandes et al., 2019). Also, chelating of metal ions prevents the formation of insoluble metal complexes with other food components, which is a major cause of decrease in its bioavailability. This also indicates that peptide-metal complex may have a significant role in improving the nutritional quality of foods, thus, enhancing the health of an individual. The production of protein isolates is proposed as a method to reduce anti-nutritional and toxic factors. These protein isolates can be further processed to obtain protein hydrolysates (PH) which have better functional and nutritional properties (Gilani et al., 2005). These protein hydrolysates are also sources of bioactive peptides that may show beneficial biological effects. The chelating phosphopeptides procured from milk proteins have been best characterized and their positive impact on in vivo and in vitro absorption of minerals such as zinc, calcium or iron has been reported (Kim et al., 2007; Miao et al., 2019; Walters et al., 2018).
Salvia hispanica L. (Chia) is an herbaceous plant belonging to the order Lamiales, family Lamiaceae, subfamily Nepetoideae and genus Salvia (Grancieri et al., 2019). This edible plant is native to south-eastern Mexico and gaining popularity in recent years for its properties. Chia seeds are rich in fibers, polyunsaturated fatty acids (PUFAs) and antioxidants that help slow cellular aging. The PUFAs contribute to lower cholesterol and triglycerides in humans. Amino acid analysis in the seed revealed a high content of sulphur, essential and non-essential amino acids, but a poor content of tryptophan and lysine. The amino acid content, particularly of tyrosine, cysteine, and methionine, makes this protein a suitable substrate for the production of antioxidant peptides, released by enzymatic hydrolysis (Chim-Chi et al., 2017). There has been constant search for substances with biological activity (such as beneficial effects on health and preventing deterioration of food) obtained from natural sources to be included as foods or additives. Therefore this study aims to isolate and characterize the bioactive protein hydrolysate/peptides obtained by enzymatic hydrolysis of Chia seed protein and could be used as an active ingredient in formulation of functional foods. Further, pepsin and pancreatin are the common gastro intestinal proteases present in the human body and probably the physiological digestion of the Chia seed protein may evolve various bioactive peptides with beneficial effects in vivo.
2. Materials and methods
The chia seeds used for the study were collected from a local market in Mysuru, Karnataka, India. The proteases, Pepsin and Pancreatin were procured from MERCK (SIGMA-ALDRICH). HiTrap chelating HP immobilized metal affinity chromatography columns were procured from GE Healthcare Biosciences. All the chemicals used were of analytical grade and solvents were of HPLC grade.
2.1. Seed flour preparation and protein extraction
Chia seeds were soaked in 0.5M sodium bicarbonate solution (1:10 w/v) for 24 h with constant agitation using magnetic stirrer. The soaked seeds are centrifuged at 11500g for 30 min to remove mucilage. The seeds obtained in the pellet are air dried and remaining mucilage is removed manually. Demucilaged seeds are further ground into fine powder. Chia seed flour (CSF) is defatted in Soxhlett chamber for 8 h using n-hexane at 65 °C. Defatted flour is air dried to remove traces of n-hexane and stored at 4 °C until use.
Protein extraction was done according to Yang et al. (2014) with slight modification. Defatted CSF was dispersed in water (1:10 w/v) and the pH was adjusted to 8.5 using 1M NaOH solution. Solution was kept on stirrer for 1 h at 500 rpm at room temperature followed by centrifugation at 8820g for 20 min at 4 °C. This was repeated twice and the supernatants obtained were collected and the pH was adjusted to 3.8 with 0.1M HCl to precipitate protein. The protein pellet obtained was dispersed in water (1:1 w/v) and dialysed at 4 °C for 48 h (1 kDa cut off). Protein slurry was lyophilized and stored at −20 °C.
2.2. Digestion of protein isolates
Freeze-dried chia seed protein isolates were digested at three different temperature (30 °C, 37 °C and 45 °C) along with three different enzyme concentration (2%, 4% and 8% w/w) at three different time intervals (2hrs, 4hrs, 6hrs and 8hrs). The pH was kept constant at 2.0 and 7.0 for protein digestion using pepsin and pancreatin respectively. Optimum temperature, enzyme substrate concentration and the incubation time were standardized for digestion of protein isolates. Pepsin is an important enzyme produced in the human gut during food digestion. It has endopeptidase activity and it is most efficient in cleaving peptide bonds between hydrophobic and aromatic amino acids, such as Phe, Tyr, and Trp. Pancreatin includes proteases, such as trypsin or chymotrypsin which are released by the pancreas in the small bowel. Trypsin is an endopeptidase that is very efficient in cleaving peptide bonds between basic amino acids, such as Arg and Lys. Chymotrypsin is an endopeptidase which is efficient in cleaving peptide bonds between aromatic amino acids, such as Tyr, Phe, and Trp.
Protein hydrolysates were prepared according to Meg et al. (2007) with modifications. Freeze-dried protein isolate was dissolved in 0.1M KCl–HCl buffer of pH 2.0. Proteins were digested at 37 °C using pepsin (4% w/w) for 4 h. Reaction was terminated by keeping the solution on boiling water bath for 10 min and the pH was neutralized to 7.0 by adding 2N NaOH and further hydrolysed with pancreatin (4% w/w) for 4 h at 37 °C. Enzyme was inactivated by heating in boiling water bath for 10 min. The suspension obtained was centrifuged at 11227g for 30 min at 4 °C. Supernatant were collected and stored at −20 °C.
2.3. Protein estimation and degree of hydrolysis of chia protein
Protein content in protein isolate and hydrolysates were determined by Bradford's method. Degree of hydrolysis of Chia protein was quantified according to Silvestre et al. (2013). Church's solution was freshly prepared by adding 25 ml of 100 mM borax, 2.5 ml of 20% w/w SDS, 40 mg of O-phthaldialdehyde dissolved in 1 ml of methanol and 100 μL of β-mercaptoethanol and volume made up to 50 ml using deionized water. 2 ml of Church's reagent was added to 50 μL of protein hydrolysate and incubated for 2 min at room temperature. Absorbance was taken at 340 nm. The peptide content was quantified using casein tryptone as standard (Church et al., 1983) and protein isolate as control. Degree of hydrolysis (DH) was measured using the formula;
| DH (%) = (ABS x 1934 x d)/c |
Where ABS is the absorbance of samples, d is the dilution factor and c is the protein concentration of the sample (g L–1).
2.4. Purification of copper chelating chia protein hydrolysate by HiTrap cationic exchange chromatography and preparative rpHPLC
Copper chelating protein hydrolysates were isolated using HiTrap™ metal chelating HP column. Column was pre-washed with 5 column volume (CV) of distilled water. The column was then charged with copper using 1 CV of 0.1M solution (in water) of copper salt. Column is then washed with 5 CV of distilled water to remove unbound ions. Blank run was carried out using 5 CV of binding buffer (0.5M sodium chloride and 5 mM imidazole in 20 mM tris buffer of pH 7.0) followed by 5 CV elution buffer (0.5M sodium chloride and 0.5M imidazole in 20 mM tris buffer of pH 7.0) and concluding with 5 CV binding buffer. The sample of 1 CV is then loaded followed by 10 CV of binding buffer and 25 CV of elution buffer (in a linear gradient of 0–60%), again 5 CV of elution buffer (linear gradient up to 100%). Column is then washed with 4 CV of 100% elution buffer and equilibrated with 5 CV of binding buffer. After purification, column was stripped using 5 CV binding buffer containing 50 mM EDTA. Column was again washed with distilled water. The column is finally washed with 5 CV 20% ethanol and stored at 4 °C.
The Hi-Trap column purified copper chelating protein hydrolysates were further separated and purified using preparative rpHPLC. Preparative rpHPLC was performed using Waters system provided with waters 2545 quaternary gradient module, waters 2707 Auto sampler, waters 2998 photodiode array detector and waters fraction collector III. The HiTrap purified copper chelating protein hydrolysate of 8 ml (10 mg/ml) was injected into the column (waters 10 × 250mm semi-preparative column) and was eluted with a linear gradient of 0–80% solvent A (acetonitrile with 0.1% trifluoroacetic acid) and solvent B (water with 0.1% trifluoroacetic acid) for 70 min maintaining the constant flow rate of 7.0 ml/min at 20 °C. Analytical rpHPLC was carried out in Agilent 1260 Infinity Quaternary equipped with G1311 B/C Quaternary pump, G1329B Auto sampler, G1330B Thermostat and G4212B VWD HPLC system equipped with Eclipse plus C18 column (4.6 × 150mm I.D., 5 μm particle) with the same solvent conditions as mentioned above to check the homogeneity of the purified CCPH fraction.
2.5. DPPH free radical scavenging assay
DPPH method was carried out according to the Zhang et al. (2020) with little modifications. The fractions of CCPH and standard ascorbic acid were prepared in 0.1 mg/mL and 1.0 mg/mL with methanol as preliminary screening concentration, respectively. The reaction mixture contained 5 μl of test samples and 95 μl of DPPH in methanol. These reaction mixtures were taken in 96 well plates and were vigorously shaken and left for 20 min in the dark at room temperature. The absorbance was then measured at 515 nm. Percentage radical scavenging activity was determined by comparison with a methanol treated control. All reactions were carried out with three replications. Inhibition of DPPH radical inhibition was calculated as follows:
| DPPH Radical Scavenging (%) = [(ABC – ABS)/ABC] x 100 |
Where, ABC is the absorbance of the control (methanol treated) and ABS is the absorbance of the sample (CCPH and standard).
2.6. Copper chelating activity
Cu2+ chelating activity was determined using the pyrocatechol violet reagent according to Saiga et al. (2003). Hydrolysates (50 μg) were added to 96-well plates containing 250 μL of 50 mM sodium acetate buffer pH 6.0, 6.25 μL of 4 mM pyrocatechol violet and Cu (1 μg, CuSO4). EDTA (50 μg) and ascorbic acid (50 μg) were used as a positive and negative control respectively. Absorbance at 632 nm was determined after incubation for 1 min at room temperature. Copper chelating activity was calculated using the formula;
| Chelating activity (%) = (1 – ABS at 632 nm/ABC at 632 nm) × 100 |
Where ABS is the absorbance of samples and ABC is the absorbance of control.
2.7. Characterization of CCPH
2.7.1. Protein hydrolysate and metal interaction studies by fluorescence spectroscopy and ultraviolet spectroscopy
Fluorescence spectroscopy analysis was done according to Cai et al. (2016) with little modification. This analysis was used to understand the conformational changes of the CCPH with copper ions using Hitachi F-4600 fluorescence spectrophotometer. The excitation wavelength was set to 285 nm and the emission wavelengths between 250 and 400 nm was recorded. The slit width for excitation and emission was 10 and 20 nm respectively. For determinations, 20 μg/ml of CCPH solution was prepared and the pH was adjusted to 6.0. Then CuSO4 Solution was added constantly after every 10 min with the concentration ranging from 1 mM to 7 mM and the fluorescence emission spectra were recorded.
The ultraviolet spectroscopy analysis was done according to Lin et al. (2015) with little modifications. The ultraviolet spectra of copper chelating CCPH were monitored over the wavelength range of 190 nm–400 nm using ultraviolet spectrophotometer (U-2900, Hitachi). For determinations, 20 μg/ml of CCPH solution was prepared and the pH was adjusted to 6.0. Then 0, 0.5, 1.0, 1.0, 1.0 and 1.0 μl of 2M CuSO4 were added constantly after every 10 min and the UV spectra were recorded.
2.7.2. LC-MS/ESI-TOF of rpHPLC purified CCPH fraction
The purified CCPH was analysed further by LC-MS/ESI-TOF. The analysis was carried out using Acquity UPLC model system from Waters equipped with auto-sampler gradient pump and Dual Wavelength Detector (DAD). The column used was Accucore C18 with a length of 50 mm having inner diameter of 4.6 mm and particle size of 2.6 μm. 10 μl of CCPH fraction was injected into the column having water, acetonitrile and 0.1% formic acid gradient as mobile phase. A constant flow rate of 1 ml/min was maintained for a run time of 10 min. UPLC system was coupled with Q-TOF mass spectrometer of XevoG2-XS model from Waters. The monoisotopic mass were calculated and the data was processed by using Mascot database search engine. The mascot search parameters were set as; 1% of the false-positive discovery rate (FDR) of peptide identification, 1 missed cleavage site, trypsin enzyme, the peptide mass tolerance of ±100 ppm, oxidation as variable modifications. The peptide with maximum Mascot scores of at least 20 was considered (Tu et al., 2020).
2.7.3. Stability of CCPH-Copper complex under simulated gastric condition
The CCPH-copper complex was prepared according to the methodology described by Fernandes et al. (2019). 10 μg of copper sulphate was mixed with 8.0 ml of 50 mM sodium phosphate buffer of pH 6.0. To this 2.0 ml of sample containing 60 μg of CCPH was added. The mixture was incubated under stirring at room temperature for 1 h maintaining the constant pH 6.0. Subsequently the mixture was centrifuged for 20 min at 2800g. The supernatant was lyophilized and stored at room temperature.
To evaluate the stability of CCPH-copper complex under simulated gastric conditions, the methodology described by Fernandes et al. (2019) was employed. In these tests, 2.0 mg of CCPH-copper complex was mixed with 1.9 ml of gastric fluid (NaCl 35 mM, pH 2.0). The mixture was incubated for 15 min at 37 °C and then, 100 μl of pepsin solution (1.0 mg/ml) was added and was incubated under shaking water bath for 1 h at 37 °C. Subsequently, the reaction mixture was adjusted to pH 6.0 for the CCPH-copper complex. The samples were then centrifuged for 20 min at 2800g and the content of unbound copper in the supernatant was determined by colorimetric method using pyrocatechol violet as described by Saiga et al. (2003).
2.8. Statistical analysis
For each analysis, determinations were made in triplicate and the mean and standard deviation was reported.
3. Results and discussion
3.1. Enzymatic hydrolysis and peptide quantification
Chia seeds are rich in dietary fibres and proteins. The total protein content of the Chia seeds was found to be 10.2% whereas 12.62% was reported by Chim-chi in 2017. This difference may be due to geographical variation and also due to loss of protein during removal of the mucilage from the seeds. Enzymatic hydrolysis (pepsin followed by pancreatin) was performed at different temperature (30 °C, 37 °C and 45 °C) and enzyme substrate ratio (2%, 4% and 8% w/w) for varied time intervals from 2hrs to 8hrs. Maximum degree of hydrolysis (14.06%) and highest copper chelating activity (74.98 ± 0.03%) was achieved with 4% w/w enzyme substrate ratio and incubation period of 4 h at 37 °C (Fig. 1). Protein hydrolysis with pepsin followed by pancreatin each for 4 h showed 14.06% degree of hydrolysis which increased to 21.1% further when the incubation time was increased upto 6 h, but the copper chelating ability was decreased. This standardized condition of hydrolysis was followed for in vitro simulated gastric digestion to study the stability of the purified Chia seed Copper Chelating Protein Hydrolysate (CCPH). Protein digestion was done using pepsin followed by pancreatin as they are secreted in the stomach and small bowel during food digestion. Pepsin cleaves peptide bonds to the N-terminal side of cyclic amino acid residues and helps in generating hydrolysates that can withstand acidic pH (pH 2.0) of stomach. Pancreatin is a combination of digestive enzymes such as trypsin and chymotrypsin that specifically cleaves peptides to the C-terminal of lysine, arginine and aromatic amino acids, this helps in generating wide variety of hydrolysates with various bioactivities. It has been reported that the different proteases in pancreatin extract extends the protein digestion process over a long time. These protein hydrolysates generated using these proteases resemble those produced in the organism during the digestion of chia seed protein. These bioactive hydrolysates can be easily absorbed intact and also favour metal transport and absorption. It was also noted that the degree of hydrolysis increased with increase in hydrolysis time but this showed decreased copper chelating activity. Among four different time intervals of hydrolysis (2hrs, 4hrs, 6hrs and 8hrs) conducted, maximum chelation was at 4 h of contact time.
Fig. 1.
Degree of hydrolysis (DH) and percentage copper chelation (PCC) of chia seed protein at different temperature and enzyme concentrations.
3.2. Isolation and purification of CCPH by HiTrap and rpHPLC
Copper binding Chia seed protein hydrolysates were isolated from pepsin-pancreatin hydrolysed chia seed protein through HiTrap chelating column charged with Cu2+ ions. The isolated PHs were further purified by preparative rpHPLC where nine distinct peaks were obtained and are eluted separately (Fig. 2A.). Out of nine fractions collected the sixth fraction (CCPH) eluted at the retention time of 43.58 min showed 93.09 ± 0.16% of maximum copper binding potential than the other eight fractions obtained. Homogeneity of the purified CCPH was confirmed by the presence of single peak at 42.53 min of retention time by analytical rpHPLC (Fig. 2B.). Structure-function relationship of the purified CCPH fraction was analysed by Ultraviolet and fluorescence spectroscopy. The molecular mass and amino acid composition were analysed through LC-MS/ESI-TOF data.
Fig. 2.
Analytical rpHPLC elution profile of (A) pepsin-pancreatin hydrolysed chia seed protein (CPH) and (B) purified copper chelating chia protein hydrolysate (CCPH).
3.3. DPPH free radical scavenging assay
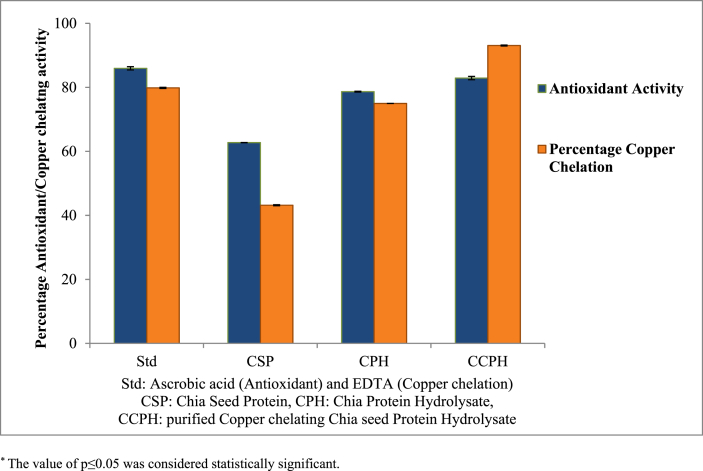
As shown in Fig. 3, the antioxidant activity of chia seed protein increased from 62.74 ± 0.04% to 78.67 ± 0.17% with hydrolysis, further increase was observed after purification of CCPH through HiTrap column followed by rpHPLC. The purified CCPH obtained have the ability to scavenge the DPPH radical (82.91 ± 0.52%) which is almost equivalent to that of standard ascorbic acid (85.95 ± 0.50%) taken as positive control. Antioxidant activity seen in CCPH obtained from pepsin-pancreatin hydrolysis is almost equal to the antioxidant activity of chia PH obtained from alcalase-flavourzyme hydrolysis (86.16%) as discussed by Chim-Chi et al. (2017). The CCPH obtained from pepsin-pancreatin hydrolysis also have chelating ability with copper which was not reported in alcalase-flavourzyme hydrolysed peptides.
Fig. 3.
Percentage antioxidant activity and chelation of copper ions by chia seed protein and its hydrolysates.
3.4. Copper chelating activity
Hydrolysis of chia seed protein resulted in an increase in copper chelating activity as shown in Fig. 3. Thus, protein hydrolysis has led to increased Cu2+ chelating activity, this may be due to increased carboxylic group which promotes the conversion of proteins into anionic form that facilitates chelation of Cu2+ and increases the exposure of metal chelating amino acid residues. Protein hydrolysis with pepsin and pancreatin also increased iron chelating activity although the maximum iron chelating activity corresponding to the hydrolysates produced by treatment with pepsin and pancreatin was almost 3 times lower than the maximum copper chelating activity showed by the CCPH. Peng et al. (2010) have also noted that whey protein hydrolysates have a far weaker Fe2+ chelating capacity than Cu2+ chelating capacity. Kong & Xiong. (2006) mentioned that this behaviour may be related to the higher coordination number of Fe2+ thus, it requires more chelators than in Cu2+. There was notable decrease in copper binding ability of protein hydrolysates (from 74.98% to 14.2%) with increase in hydrolysis time from 4 to 6 h for both pepsin and pancreatin. The increase in hydrolysis time produces very low molecular weight peptides which have reduced copper trapping ability that in-turn decreases their copper chelation potential (Chim-Chi et al., 2017). The purified fraction of CCPH showed higher copper chelating activity of 93.09% in comparison to non-purified PH (74.98%), it may be due to some protein hydrolysates in the mixture that bind to the metal binding sites hence blocking their exposure to metal ions.
3.5. Structural characterization of CCPH
3.5.1. Fluorescence spectroscopy analysis
The pepsin pancreatin protein hydrolysates showed an endogenous fluorescence at 350 nm and the purified CCPH showed endogenous fluorescence at 310 nm (Fig. 4A.) which reduced with the increase in Cu2+ concentration. However, when the concentration reached 7.0 mM, no further changes were noted. This suggests that changes in the fluorescence were observed when copper chelated with protein hydrolysates and excess of free copper ions has no impact on fluorescence intensity. The decrease in the intensity of endogenous fluorescence with the increase in copper ion concentration shows chelating activity. Similar result has been reported by Wu et al. (2012) that fluorescence quenching was observed when calcium ions combined with calcium-chelating peptide. Two most known fluorescence quenching mechanisms are dynamic quenching and static quenching (Li et al., 2020). According to Shrivastava and Nair (2004), the energy transfer from a donor (aromatic amino acid) to an acceptor (metal complex) results in fluorescence quenching. This endogenous fluorescence quenching is of static quenching mechanism type (Crouse et al., 2014). It is defined as a process of quenching matter and fluorescent material by producing non-luminescent material at its wavelength (Lian et al., 2021). Dynamic quenching only effects the fluorophore in excited state without altering its UV–visible spectra whereas static quenching causes distress in fluorophore absorption spectrum due to the formation of ground-state complex (Makarska-Bialokoz, 2017). In our work, the fluorescence peak decreased with the addition of copper salt solution, which indicates the formation of non-fluorescent peptide-metal complex which is an example of static quenching mechanism.
Fig. 4.
Structural analysis of purified copper chelating CCPH using (A) Fluorescence quenching at wavelength 310 nm (B) UV spectra with different CuSO4 concentrations over the wavelength range from 190 to 400 nm.
3.5.2. Ultraviolet spectroscopy analysis
As shown in the Fig. 4B, the UV absorption spectra of CCPH- Cu2+ chelate showed distinct difference from that of protein hydrolysate, which suggests the formation of new substance with the interaction of protein hydrolysate with copper ions. Protein hydrolysate showed maximum UV absorption peak at about 216 nm. With the increase in copper ion concentration, the absorbance of the maximum absorption peak increased gradually from 1.762 to 1.867, showing hyperchromic effect and red shift phenomenon. According to Cai et al. (2016), the chelating process generates polarizing changes during the binding of copper ions with the ligands which is due to the presence of the chromophore groups (-C O, –COOH) and auxochrome groups (-OH, –NH2).
3.5.3. LC-MS/ESI-TOF analysis of purified CCPH fraction
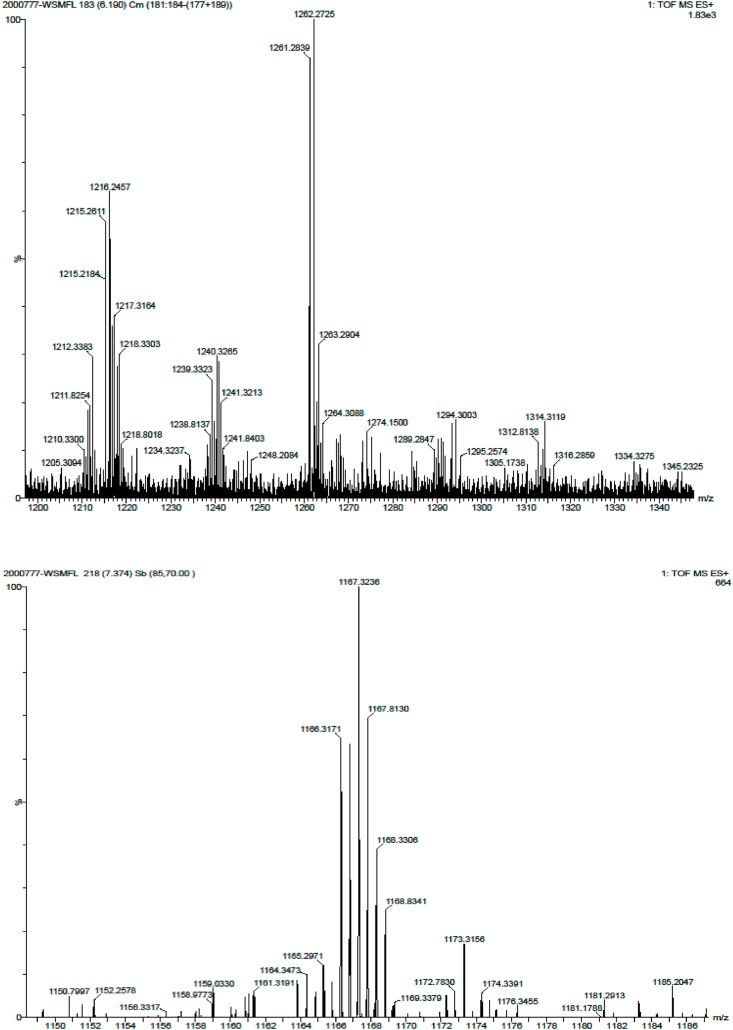
The CCPH fraction obtained after purification was subjected to LC-MS/ESI-TOF analysis. From the data obtained monoisotopic masses were calculated and the amino acid sequences of six major ions with m/z value Fig. 5 ranging from 0.4 kDa to 2.5 kDa were identified using Mascot database search (Matrix Science) as shown in Table 1. Knowing the amino acid composition can help in the better understanding of functional behaviour of the CCPH. Amino acid composition shows that the side group such as amino and carboxyl of the acidic amino acids (Glu and Asp) and basic amino acids (Lys, His and Arg) are involved in chelating copper ions. Involvement of these side groups are reported by Aderinola et al. (2018). The Imidazole ring of histidine plays a major role in metal binding and antioxidant activity of peptides. Studies have shown that higher concentration of hydrophobic amino acids and the presence of negatively charged amino acids play major role in promoting their biological activities such as antioxidant and metal chelation. The MS data have shown that the CCPH is rich in hydrophobic amino acids such as Gly, Val, Leu, Ile, Pro and Met (23.8%) and negatively charged amino acids such as Asp and Glu (12.69%). It is also known that metal ions such as Cu2+, Zn2+ and Ni2+ bind with oxygen-rich groups like phosphate group (serine) and carboxyl groups (Glu and Asp) and nitrogen-rich group such as imidazole group (His) (Zhu et al., 2015; Sovago et al., 2012). The copper binding ability of CCPH is supported by the N-imidazole of His and N-amide of Lys which are known to act as electron donors (Vo et al., 2020; Radomska et al., 1990; Mendola et al., 2012). The amino acid sequence (Table 1) identified from the peptides of mass 2.5 kDa (AAATAMHSTALAGQSLVKPVNELSR) and 0.43 kDa (TVSK) have been reported to be found in proteins having binding and acetylation properties. Based on their homology it has been reported to be a part of major light harvesting chlorophyll-a/b-binding protein in the photosystem II light harvesting complex. The peptide sequence of mass 0.48 kDa (GGCCSG) is reported to be found in protein possessing GTPase activity. Generation of high metal affinity is observed in the presence of two negatively charged amino acids such as Asp and Glu having carboxyl side chain (Kallay et al., 2005). The CCPH not only contains amino acids for metal binding but also shows high contents of essential amino acids. The high essential amino acid content such as Thr, His, Lys, Val, Leu, Ile and Met (30.13%) is associated with their high nutritional values. These observations also indicate that the peptide of 2.332 kDa (ATCDCEHDYELNEEIGMCCR) has high copper binding ability due to the high content of amino acids with both oxygen-rich and nitrogen-rich side chains in comparison to other five identified sequences. The purified CCPH fraction is also rich in Cys and Met which are sulphur-containing amino acids that enhance copper binding capacity of the protein hydrolysate through free electrons of S atoms in their sulfhydryl and thioether group respectively (Vo et al., 2020; Sovago et al., 2012). Besides the amino acid sequence there are many factors such as length of peptide sequence/molecular weight and steric structure of a peptide sequence that may influence the copper binding ability of a peptide. Hence, further investigation is necessary to explore these factors.
Fig. 5.
Q-TOF mass spectrometer analysis of Hi-Trap and rpHPLC purified CCPH fraction showing six major ions with m/z value of 0.562 kDa, 0.508 kDa, 0.433 kDa, 0.482 kDa, 2.522 kDa and 2.332 kDa respectively.
Table 1.
Molecular weight along with the sequence identified using Mascot database search tool for CCPH.
| Molecular Weight (in kDa) | Molecular Weight (in kDa) | Isoelectric Point | Net Chargea (at pH 7.0) | Hydrophobic Ratio (%)b |
|---|---|---|---|---|
| 0.562 | CERR | 9.2 | +0.9 | 0 |
| 0.508 | KPHK | 10.69 | +2.1 | 25 |
| 0.433 | TVSK | 9.82 | +1.0 | 25 |
| 0.482 | GGCCSG | 2.75 | -0.1 | 0 |
| 2.522 | AAATAMHSTALAGQSLVKPVNELSR | 10.18 | +1.1 | 52 |
| 2.332 | ATCDCEHDYELNEEIGMCCR | 3.69 | -5.2 | 20 |
Net charge was calculated based on negatively charged amino acid (E and D) and positively charged amino acids (R, H and K) in the peptide sequence using the online tool https://pepcalc.com/.
Calculated the percentage of hydrophobic residues (I, V, L, F, C, M, A, W) in the peptide sequence using the online tool https://peptide2.com/N_peptide_hydrophobicity_hydrophilicity.php.
3.6. Stability of CCPH-copper complex to gastric conditions
Absorption of metal ions is generally regulated to avoid the accumulation of ions. One of the main barriers for the uptake of metal ion is the maintenance of the ion in digestive tract. The main point of control is the stomach due to the combined action of the enzyme pepsin and the extremely acidic pH of this region. Gastric stability of the chelates during passage through the gastrointestinal tract is a major step to initially ensure the absorption of copper from the body. The results shown in Table 2, suggested that the chelating ability of CCPH remained unaltered at gastric conditions, thus indicating that the CCPH-copper complex is resistant to gastro intestinal conditions.
Table 2.
Stability of purified CCPH- Cu2+ complex in simulated In Vitro gastric conditions.
| Sample Conditions | Percentage Copper Chelationa (%) | |
|---|---|---|
| CCPH – Cu2+ Complex |
Initial | 94.72 ± 0.45 |
| NaCl pH 2.0 | 93.58 ± 0.07 | |
| pH 2.0 with pepsin | 93.34 ± 0.08 |
The value of p ≤ 0.05 was considered statistically significant.
4. Conclusion
In summary, protein hydrolysate contains different combination of peptides having various functional properties. Therefore, the bioactive peptide of interest has to be separated from the pool of peptide mixture present in the protein hydrolysate to increase their functional capabilities (Budseekoad et al., 2018). The simpler spatial structure and more exposure of metal ion binding sites by low molecular weight protein hydrolysates/peptides increases their metal chelating ability (Wang et al., 2014; Zhang et al., 2021) Hence, a specific CCPH fraction showing strong copper chelating ability from pepsin-pancreatin hydrolysed chia seed protein was purified. The chelating activity, structural characteristics and gastro-intestinal stability was investigated. The results indicated that copper ions could form bonds with carboxyl oxygen atoms and amino nitrogen atoms which lead to a new and stable CCPH-copper chelate complex formation. The CCPH- copper chelate complex was found to be resistant to simulated gastric digestion and remained active, which makes it more available for intestinal absorption. The stability of CCPH-copper chelate was greater than CuSO4, which in turn suggests that the purified CCPH has the potential to be used as functional nutraceutical additive.
Declaration of competing interest
The authors declare that they have no competing financial interests and personal relationship that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported by key project funded by Life Science Research Board- Defence Research and Development organization (LSRB-DRDO), India (LSRB-325).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crfs.2021.11.007.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Aderinola T.A., Fagbemi T.N., Enujiugha V.N., Alashi A.M., Aluko R.E. Amino acid composition and antioxidant properties of Moringa oleifera seed protein isolate and enzymatic hydrolysates. Heliyon. 2018;4(10) doi: 10.1016/j.heliyon.2018.e00877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budseekoad S., Yupanqui C.T., Sirinupong N., Alashi A.M., Aluko R.E., Youravong W. Structural and functional characterization of calcium and iron-binding peptides from mung bean protein hydrolysate. J. Funct. Foods. 2018;49:333–341. doi: 10.1016/j.jff.2018.07.041. [DOI] [Google Scholar]
- Cai X., Lin J., Wang S. Novel peptide with specific calcium-binding capacity from schizochytrium sp. protein hydrolysates and calcium bioavailability in caco-2 cells. Mar. Drugs. 2016;15(1):3. doi: 10.3390/md15010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chim-Chi Y., Gallegos-Tintore S., Jimenez-Martínez C., Davila-Ortiz G., Chel-Guerrero L. Antioxidant capacity of Mexican chia (Salvia hispanica L.) protein hydrolyzates. J. Food Meas. Char. 2017;12(1):323–331. doi: 10.1007/s11694-017-9644-9. [DOI] [Google Scholar]
- Church F.C., Swaisgood H.E., Porter D.H., Catignani G.L. Spectrophotometric assay using o-phthaldialdehyde for determination of proteolysis in milk and isolated milk proteins. J. Dairy Sci. 1983;66(6):1219–1227. doi: 10.3168/jds.S0022-0302(83)81926-2. [DOI] [Google Scholar]
- Crouse H.F., Potoma J., Nejrabi F., Snyder D.L., Chohan B.S., Basu S. Quenching of tryptophan fluorescence in various proteins by a series of small nickel complexes. Dalton Trans. 2014;41:2720–2731. doi: 10.1039/C2DT12169G. [DOI] [PubMed] [Google Scholar]
- Fernandes K.F., Ribeiro J.V.V., Batista K.A. Potential iron and copper chelating activity of naturally occurring peptides and protein fractions from common bean (Phaseolus vulgaris) Int. J. Biochem. Physiol. 2019;4(3):161. doi: 10.23880/ijbp-16000161. [DOI] [Google Scholar]
- Gilani G.S., Cockell K.A., Sepehr E. Effects of antinutritional factors on protein digestibility and amino acid availability in foods. J. AOAC Int. 2005;88(3):967–987. doi: 10.1093/jaoac/88.3.967. [DOI] [PubMed] [Google Scholar]
- Grancieri M., Martino H.S.D., Gonzalez de Mejia E. Chia seed (Salvia hispanica L.) as a source of proteins and bioactive peptides with health benefits: a review. Compr. Rev. Food Sci. Food Saf. 2019;18(2):480–499. doi: 10.1111/1541-4337.12423. [DOI] [PubMed] [Google Scholar]
- Kallay C., Varnagy K., Micera G., Sanna D., Sovago I. Copper(II) complexes of oligopeptides containing aspartyl and glutamyl residues. Potentiometric and spectroscopic studies. J. Inorg. Biochem. 2005;99(7):1514–1525. doi: 10.1016/j.jinorgbio.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Kim S.B., Seo I.S., Khan M.A., Ki K.S., Nam M.S., Kim H.S. Separation of iron-binding protein from whey through enzymatic hydrolysis. Int. Dairy J. 2007;17:625–631. doi: 10.1016/j.idairyj.2006.09.001. [DOI] [Google Scholar]
- Kong B., Xiong Y.L. Antioxidant activity of zein hydrolysates in a liposome system and the possible mode of action. J. Agric. Food Chem. 2006;54(16):6059–6068. doi: 10.1021/jf060632q. [DOI] [PubMed] [Google Scholar]
- Li J., Song Y., Vogt R.D., Liu Y., Luo J., Li T. Bioavailability and cytotoxicity of cerium- (IV), copper- (II), and zinc oxide nanoparticles to human intestinal and liver cells through food. Sci. Total Environ. 2020;702 doi: 10.1016/j.scitotenv.2019.134700. [DOI] [PubMed] [Google Scholar]
- Lian J., Yang Y., Qiu W., Huang L., Wang C., Chen Q., Ke Q., Wang Q. Fluorescent characteristics and metal binding properties of different molecular weight fractions in stratified extracellular polymeric substances of activated sludge. Separations. 2021;8:120–140. doi: 10.3390/separations8080120. [DOI] [Google Scholar]
- Lin J.P., Cai X.X., Tang M.R., Wang S.Y. Preparation and evaluation of the chelating nanocomposite fabricated with marine algae Schizochytrium sp. Protein hydrolysate and calcium. J. Agric. Food Chem. 2015;63(44):9704–9714. doi: 10.1021/acs.jafc.5b04001. [DOI] [PubMed] [Google Scholar]
- Makarska-Bialokoz Magdalena. Interactions of hemin with bovine serum albumin and human haemoglobin: a fluorescence quenching study. Spectrochim. Acta Mol. Biomol. Spectrosc. 2017;193:23–32. doi: 10.1016/j.saa.2017.11.063. [DOI] [PubMed] [Google Scholar]
- Meg C., Pedroche A.J., Yust M.M., Calle J.G.A.N., Alaiz M., Milla F. Affinity purification of copper chelating peptides from chickpea protein hydrolysates. J. Agric. Food Chem. 2007;55(10):3949–3954. doi: 10.1021/jf063401s. [DOI] [PubMed] [Google Scholar]
- Mendola D.L., Magrì A., Santoro A.M., Nicoletti V.G., Rizzarelli E. Copper(II) interaction with peptide fragments of histidine–proline-rich glycoprotein: speciation, stability and binding details. J. Inorg. Biochem. 2012;111:59–69. doi: 10.1016/j.jinorgbio.2012.02.027. [DOI] [PubMed] [Google Scholar]
- Miao J., Liao W., Pan Z., Wang Q., Duan S., Xiao S., Yang Z., Cao Y. Isolation and identification of iron-chelating peptides from casein hydrolysates. Food Funct. 2019;10(5):2372–2381. doi: 10.1039/C8FO02414F. [DOI] [PubMed] [Google Scholar]
- Peng X., Kong B., Xia X., Liu Q. Reducing and radical scavenging activities of whey protein hydrolysates prepared with Alcalase. Int. Dairy J. 2010;20(5) doi: 10.1016/j.idairyj.2009.11.019. 0–365. [DOI] [Google Scholar]
- Radomska B., Sovago I., Kiss T. Tyrosinate and lysinate as bridging residues in copper(II) dipeptide complexes. J. Chem. Soc., Dalton Trans. 1990;(1):289–292. doi: 10.1039/DT9900000289. [DOI] [Google Scholar]
- Saiga A., Tanabe S., Nishimura T. Antioxidant activity of peptides obtained from porcine myofibrillar proteins by protease treatment. J. Agric. Food Chem. 2003;51(12):3661–3667. doi: 10.1021/jf021156g. [DOI] [PubMed] [Google Scholar]
- Shrivastava H.Y., Nair B.U. Fluorescence resonance energy transfer from tryptophan to a chromium (III) complex accompanied by non-specific cleavage of albumin: a step forward towards the development of a novel photoprotease. J. Inorg. Chem. 2004;98:991–994. doi: 10.1016/j.jinorgbio.2004.02.014. [DOI] [PubMed] [Google Scholar]
- Silvestre M.P.C., Morais H.A., Silva V.D.M., Silva M.R. Degree of hydrolysis and peptide profile of whey proteins using pancreatin hydrolysis of whey protein. Nutrire. 2013;38(3):278–290. doi: 10.4322/nutrire.2013.026. [DOI] [Google Scholar]
- Sovago I., Kallay C., Varnagy K. Peptides as complexing agents: factors influencing the structure and thermodynamic stability of peptide complexes. Coord. Chem. Rev. 2012;256(19–20):2225–2233. doi: 10.1016/j.ccr.2012.02.026. [DOI] [Google Scholar]
- Tu M., Qiao X., Wang C., Liu H., Cheng S., Xu Z., Du M. In vitro and in silico analysis of dual-function peptides derived from casein hydrolysate. Food Sci. Hum. Wellness. 2020;10(1):32–37. doi: 10.1016/j.fshw.2020.08.014. [DOI] [Google Scholar]
- Vo T.D.L., Pham K.T., Doan K.T. Identification of copper-binding peptides and investigation of functional properties of acetes japonicus proteolysate. Waste and Biomass Valorization. 2020;12:1565–1579. doi: 10.1007/s12649-020-01112-3. [DOI] [Google Scholar]
- Walters M., Esfandi R., Tsopmo A. Potential of food hydrolyzed proteins and peptides to chelate iron or calcium and enhance their absorption. Foods. 2018;7(10):172. doi: 10.3390/foods7100172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Wang C., Li B., Li H.L. Zn(II) chelating with peptides found in sesame protein hydrolysates: identification of the binding sites of complexes. Food Chem. 2014;165(15):594–602. doi: 10.1016/j.foodchem.2014.05.146. [DOI] [PubMed] [Google Scholar]
- Wu H., Liu Z., Zhao Y., Zeng M. Enzymatic preparation and characterization of iron-chelating peptides from anchovy (Engraulis japonicas) muscle protein. Food Res. Int. 2012;48(2):435–441. doi: 10.1016/j.foodres.2012.04.013. [DOI] [Google Scholar]
- Yang N., Li Y.M., Zhang K., Jiao R., Ma K.Y., Zhang R., Chen Z.Y. Hypocholesterolemic activity of buckwheat flour is mediated by increasing sterol excretion and down-regulation of intestinal NPC1L1 and ACAT2. J. Funct. Foods. 2014;6:311–318. doi: 10.1016/j.jff.2013.10.020. [DOI] [Google Scholar]
- Zhang J., Zhou L., Cui L., Liu Z., Wei J., Kang W. Antioxidant and α-glucosidase inhibitiory activity of Cercis chinensis flowers. Food Sci. Hum. Wellness. 2020;9(4):313–319. doi: 10.1016/j.fshw.2020.04.003. [DOI] [Google Scholar]
- Zhang Y., Ding X., Li M. Preparation, characterization and in vitro stability of iron-chelating peptides from mung beans. Food Chem. 2021;349 doi: 10.1016/j.foodchem.2021.129101. [DOI] [PubMed] [Google Scholar]
- Zhu K.X., Wang X.P., Guo X.N. Isolation and characterization of zinc-chelating peptides from wheat germ protein hydrolysates. J. Funct. Foods. 2015;12:23–32. doi: 10.1016/j.jff.2014.10.030. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.