Abstract
The application of scaffolding materials is believed to hold enormous potential for tissue regeneration. Despite the widespread application and rapid advance of several tissue-engineered scaffolds such as natural and synthetic polymer-based scaffolds, they have limited repair capacity due to the difficulties in overcoming the immunogenicity, simulating in-vivo microenvironment, and performing mechanical or biochemical properties similar to native organs/tissues. Fortunately, the emergence of decellularized extracellular matrix (dECM) scaffolds provides an attractive way to overcome these hurdles, which mimic an optimal non-immune environment with native three-dimensional structures and various bioactive components. The consequent cell-seeded construct based on dECM scaffolds, especially stem cell-recellularized construct, is considered an ideal choice for regenerating functional organs/tissues. Herein, we review recent developments in dECM scaffolds and put forward perspectives accordingly, with particular focus on the concept and fabrication of decellularized scaffolds, as well as the application of decellularized scaffolds and their combinations with stem cells (recellularized scaffolds) in tissue engineering, including skin, bone, nerve, heart, along with lung, liver and kidney.
Keywords: Decellularization, ECM, 3D scaffolds, Tissue regeneration, Recellularization
Graphical abstract
Highlight
-
•
dECM scaffolds for tissue regeneration are primarily introduced according to the development and fabrication of decellularized scaffolds, as well as the application of dECM scaffolds and their constructs combined with stem cells in various tissue engineering.
-
•
Limitations, challenges, and perspectives of dECM for accelerating development of tissue regeneration are discussed, respectively.
1. Introduction of decellularized extracellular matrix scaffolds
Along with the rapid development of tissue engineering in the past decades, biological scaffolds have attracted significant interest in this field due to their great biocompatibility and bioactivity, and moderate mechanical performances for supporting cells. Among various types of scaffolds, decellularized extracellular matrix (dECM) scaffolds, which refer to biomaterials formed by human or animal organs/tissues with the removal of immunogenic cellular components via decellularized technologies, are under the spotlight [1]. dECM scaffolds mainly consist of extracellular matrix (ECM), which is a three-dimensional (3D) framework containing extracellular macromolecules such as collagen, elastin, fibronectin, laminin, and matricellular proteins, as listed in Table 1 [[2], [3], [4], [5]]. Meanwhile, the physicochemical signals and biological performance of dECM can be remained after decellularization, which provides a substrate for mechanical supporting and a biological 3D carrier for subsequent cell seeding [6,7]. Therefore, such recellularized dECM can be implanted into patients for tissue engineering, including restoration of damaged organs, regeneration of endogenous tissues, and replacement of missing organs [2,8].
Table 1.
List of main protein components in dECM.
| Protein | Distributions | Function | Reference |
|---|---|---|---|
| Collagen | The skeletal systems and most soft tissues | Provide tensile strength, connect the framework of tissue/organ, affect cell types disposition | [2,9,10] |
| Elastin | Most soft tissues, such as vasculature and muscle | Provide elasticity, adjust mechanical properties, increase hemocompatibility | [2,9,10] |
| Fibronectin | Most organs/tissues | Regulate cell behavior and function, guide branching morphogenesis in the development of complicated structures | [2,8,10] |
| Laminin | Cartilage, muscle | Promote cell adhesion and migration, provide bioactive sites for cross-linking | [2,8] |
| Matricellular Proteins | Vasculature | Contain binding sites for ECM structural proteins and cell surface receptors, and modulate activities of specific growth factors | [9] |
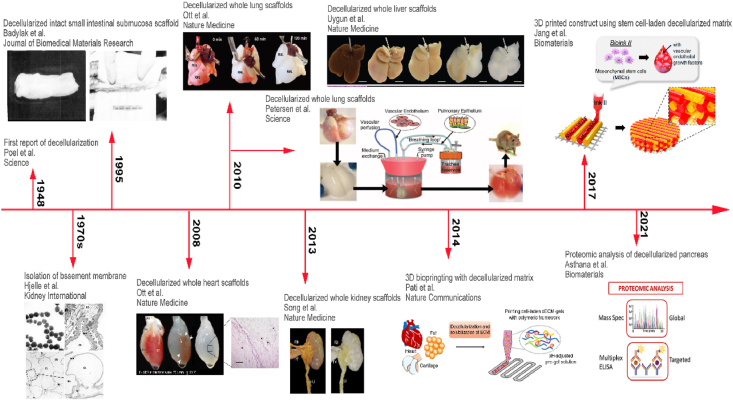
Since the conception of decellularization was introduced, researchers have carried out numerous studies and explorations on the application of dECM scaffolds, as Fig. 1 shows the timeline of dECM. The first exploration of decellularization was achieved by Poel in 1948 [11]. After that, a few studies had reported decellularization technologies in the 1970s [12]. In 1995, Badylak's team reported acellular scaffolds of small intestinal submucosa (SIS) for Achilles tendon repair [13]. Since then, various 3D biological scaffolds derived from decellularized tissues or whole organs have been harvested and applied in a broad range of tissue engineerings, such as dermal tissue repair, heart valves replacement, and vascular tissue regeneration [1,2]. A tremendous milestone of decellularized whole organs was reported by Ott in 2008, where a cadaveric ECM scaffold derived from rat heart was fabricated via perfusion decellularization [14]. From then on, many research groups demonstrated the feasibility of obtaining acellular scaffolds from a variety of organs. In 2010, Ott, as well as Petersen, reported bioartificial lungs using decellularization process and verified that the acellular lung could participate in gas exchange [15,16]. Similarly, Uygun obtained a transplantable liver scaffold via perfusion decellularization and demonstrated the recellularized grafts could normally support albumin secretion, urea synthesis and cytochrome P450 expression [17]. In 2013, Song et al. introduced the orthotopic transplantation of decellularized kidneys in rodents and showed their functions of urine production and metabolites clearance, demonstrating the possibility and feasibility of fabricating dECM bioscaffolds with complex physiological functions [18]. Moreover, with the steady improvement of decellularization technology, dECM scaffolds have been regarded as promising biomaterials to be solubilized into bioinks for 3D-bioprinting since the dECM could transform from a pre-gel fluid to hydrogel [[19], [20], [21], [22]]. In 2017, Jang used stem cell-laden heart tissue-derived dECM bioinks in 3D cell printing, in which cell-to-cell interactions and differentiation capability of stem cells are improved, and thus the pre-vascularized construct provided a cardiac-like microenvironment and resulted in beneficial effects on cardiac repair [23]. Recently, researchers are paying more attention to the in-depth profiling of the proteome of decellularized ECM, in order to provide guidelines for comprehensively defining the ECM atlas and thus designing improved dECM scaffolds [24]. In conclusion, the emergence and development of dECM scaffolds open a new avenue for human-organ/tissue-derived biomaterials in tissue engineering [2].
Fig. 1.
Timeline of decellularized extracellular matrix (dECM) scaffolds. Crude decellularization technique emerged in 1948 [11], but the production of tissue-specific ECM was first reported in the 1970s [12]. Intact acellular small intestinal submucosa matrices were produced for Achilles tendon repair in 1995 [13]. Whole rat decellularized hearts were first introduced in 2008 [14], and decellularized lungs and acellular livers were fabricated in 2010, respectively [[15], [16], [17]]. Orthotopic transplantation of the decellularized kidney was operated in 2013 [18]. dECM hydrogels were involved in 3D-bioprinting from 2014 [22], followed by a 3D-printed construct using stem cell-laden dECM bioinks in 2017 [23]. Recently, various proteomics methods have been developed to analyze dECM materials in 2021 [24].
2. Classification and fabrication of dECM scaffolds
2.1. Classification of dECM scaffolds
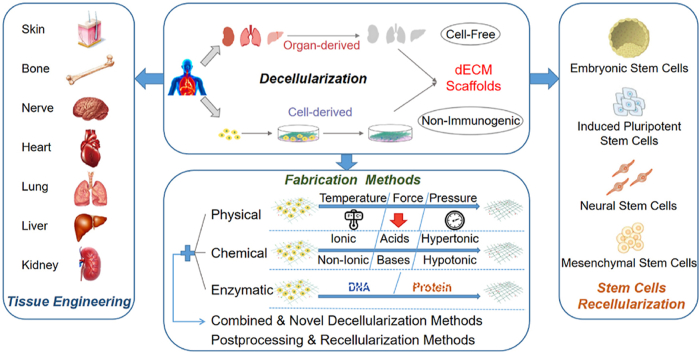
Decellularization is a method of eliminating cellular components from organs/tissues/cells to generate a structural ECM template and maintain a biomimetic ECM microenvironment [25]. According to the origins of ECM, dECM scaffolds are classified into three categories, including autogenous dECM, allogeneic dECM and xenogeneic dECM. Since autogenous dECM scaffolds face the situation of tissue limitations and surgical complications, most dECM scaffolds are from allogeneic or xenogeneic donor tissues; however, donor site morbidity, architecture and mass composition differences, and immunogenicity problems caused by incomplete decellularization may exist in allogeneic/xenogeneic dECM [26,27]. According to the sources of ECM, dECM scaffolds can also be classified into two main categories, including organ/tissue-derived dECM scaffolds and cell-derived dECM scaffolds, as Fig. 2 shows.
Fig. 2.
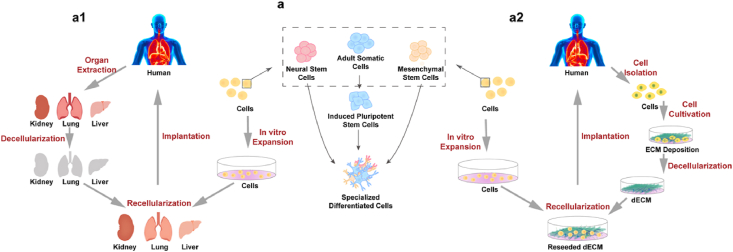
Classification of dECM scaffolds. (a1) Organ/Tissue-derived dECM as a decellularized scaffold for tissue engineering. Human-derived organs/tissues undergo decellularization progress to obtain dECM scaffolds. Then cells are extracted, expanded and then seeded onto the dECM scaffolds to generate recellularized grafts for organ/tissue bioengineering. (a2) Cell-derived dECM as a decellularized scaffold for tissue engineering. Cell-deposited extracellular matrix (ECM) undergoes decellularization progress to obtain dECM scaffolds. Cells from other sources are recellularized onto the dECM scaffolds to generate bioengineered grafts for tissue engineering. (a) Recellularized stem cells and their classification.
2.1.1. Organ/tissue(O/T)-derived dECM scaffolds
O/T-derived dECM scaffolds refer to the scaffolds derived from specific organs or tissues, which possess the natural 3D architecture from the whole organ or tissue by eliminating immunogenic cellular components and preserving non-immunogenic ECM (Fig. 2a1). These organs/tissues go through decellularization process, and the resulted acellular ECM scaffolds can be reseeded with specific cells to generate tissue-engineered grafts. O/T-derived dECM scaffolds serve as reservoirs for site-specific bioactive molecules and cell-matrix interactions, offering numerous advantages for tissue regeneration. First, since the native ECM contains memory factors and cues, O/T-derived dECM retains distinct tissue-specific memory that can drive tissue-specific differentiation [25,28]. Second, O/T-derived dECM partly remains the sophisticated ECM structure and interior architecture, such as the morphology of pores and the alignment of collagen fibers, which might significantly regulate cell adhesion, proliferation and differentiation [29]. Furthermore, their mechanical properties and microenvironment conditions are more similar to the native ECM than cell-deposited ECM discussed below.
2.1.2. Cell-derived dECM scaffolds
Cell-derived dECM scaffolds are alternative scaffolds to O/T-derived dECM scaffolds to provide a requisite niche. When cells are cultured in vitro, they can secrete cell-specific ECM, which can be decellularized subsequently to be a substratum for yielding large-quantity cells (Fig. 2a2) [25,30]. The acellular ECM, named cell-derived dECM, can be recellularized to form another excellent decellularized graft. These dECM scaffolds have attracted considerable interest since they possess several unique benefits. First, compared to the sophisticated fabrication of O/T-derived dECM scaffolds, acquiring cell-derived dECM scaffolds is relatively more exercisable to achieve, especially for obtaining dECM from certain progenitors or stem cells. Second, cell-derived ECM scaffolds can reduce the risk of pathogen transfer caused by allogenic ECM and eliminate adverse host immune responses induced by xenogeneic ECM [[31], [32], [33]]. Furthermore, in-vitro cultured cell-derived dECM can be more operable to modify and graft on other surfaces of biomaterials such as hydroxyapatite and biphasic calcium phosphate [[34], [35], [36]].
2.2. Fabrication of dECM scaffolds
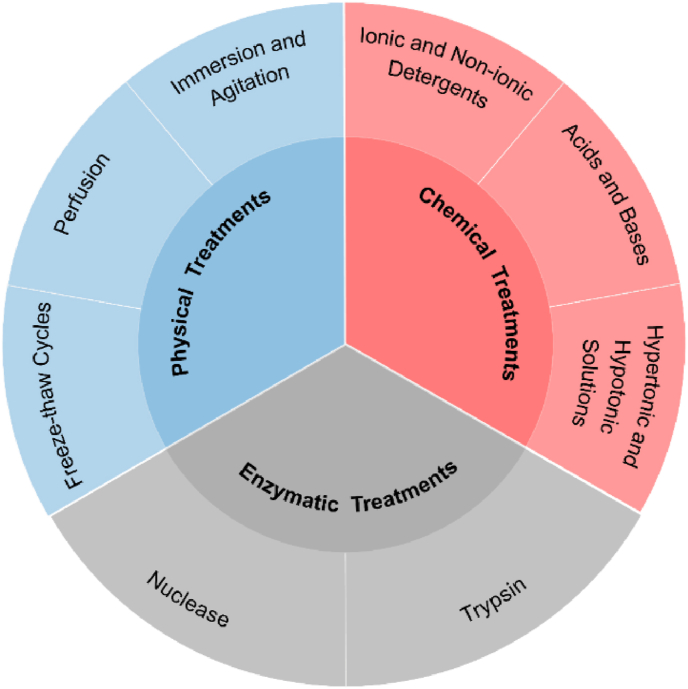
Ideal decellularization refers to the complete removal of cellular components with remaining the original architecture and composition, biochemical and mechanical properties of the native ECM [37,38]. To date, there is no methodological gold standard of decellularization, which is largely dependent on multiple factors of the source tissue, such as species and ages, anatomical location and sizes [39,40]; however, researchers have developed a wide variety of decellularization protocols, including physical, chemical, enzymatic treatments and a combination of these approaches (Fig. 3).
Fig. 3.
Decellularization methods: physical treatments, chemical treatments, and enzymatic treatments. The typical processes for each treatment are cataloged.
2.2.1. Physical treatments
Physical treatments modulate physical characteristics, such as temperature, force and pressure, to facilitate rinsing of detergent solutions, disruption of cell membranes, and removal of cellular contents. Particularly, freeze-thaw cycles, immersion and agitation, and perfusion are widely used for decellularization. Herein, various physical treatments, and their mechanisms and disadvantages are summarized in Table 2.
Table 2.
List of physical treatments used for decellularization.
| Method | Mechanism | Disadvantages | Reference |
|---|---|---|---|
| Freeze-thaw cycles | Disrupt cell membranes and cause cell lysis by forming intracellular ice crystals | Disrupt ECM microstructure by ice crystals. | [42,43,[72], [73], [74], [75]] |
| Require subsequent treatments for the removal of cellular contents | |||
| Immersion and agitation | Rupture tissues and cells, and isolate cells from basement membranes | Alter ECM architecture. | [42,50,65,66,70,72] |
| Need optimal standards for mechanical force and exposure time | |||
| Perfusion | Remove cells from ECM and help the removal of cellular components and debris within the organ's natural vasculature | Cannot process tissues without innate vasculature. | [14,[49], [50], [51], [52],57,[76], [77], [78], [79], [80], [81]] |
| Sophisticated to operate the perfusion devices | |||
| Scraping | Mechanically remove cells from ECM surface | Damage underlying basement membranes. | [82] |
| Need standards for controlling mechanical force | |||
| Sonication | Generate acoustic cavitation bubbles, induce shear stress effect, and thus rupture the cell membrane; Ease and assist the penetration of agents by emitting the vibration, and help cellular debris removal | High power or longer duration of sonication could disrupt the main structural fibers and produce adverse effects on the vascular tissues | [[83], [84], [85]] |
| Supercritical fluids | Facilitate chemical exposure, and lead to cell removal | Not yet widely used | [[86], [87], [88]] |
| Pressure gradient/Convective flow | Burst cells, and aid in the delivery of solutions to force cellular components and soluble proteins out of tissues | Make solutions difficult to penetrate ECM due to the constantly high pressure | [[89], [90], [91], [92], [93]] |
| Pressurization | Result in cell lysis | Denature the proteins, and disrupt the ECM structure by formed ice crystals. | [51,94,95] |
| Expensive to operate | |||
| Electroporation | Cause micropore formation in cell membranes and lead to cell lysis | Hard to process large-sized tissues. | [[96], [97], [98], [99]] |
| Used solvents can be toxic |
2.2.1.1. Freeze-thaw cycles
Freeze-thaw cycles refer to a repetitive decellularization process that involves freeze-drying in nitrogen and subsequent thawing in buffer solution. Freeze-thaw procedures are operated to disintegrate cell membranes, promote cell lysis and detach cells from the ECM network by forming intracellular ice crystals. Freeze-thaw cycles have been used for both O/T-derived ECM and cell-derived ECM due to its improved decellularization efficiency, reduced residual chemicals and cytotoxicity, and well-preserved biochemical composition, biomechanical performance and tissue structures [41,42]. The decellularization efficiency is mainly determined by its cooling/thawing rate, the setpoint temperature, processing time, and repetitive cycles [43]. Freeze-thaw cycles is typically combined with chemical or enzymatic methods to form a porous structure, rendering ECM susceptible to subsequent chemical treatments [43,44]. However, without chemical or enzymatic agents, cyclic freeze-thawing could also achieve satisfactory results [34,36,45]. For instance, repetitive freeze-thawing procedures could deplete all native cells from peripheral nerve sheath, while basal lamina maintains integrity and flexibility [46].
2.2.1.2. Perfusion
Perfusion is a typical decellularized route for larger, thicker tissues or whole organs, which refers to cannulating organs/tissues and subsequently establishing a channel for circulating agents through their intrinsic vascular system [47]. The efficiency of perfusion is primarily determined by perfusion route (artery or portal vein route), infusion mode (antegrade or retrograde), perfusion paraments (pressure, temperature, and flow rate), types and concentrations of perfusate, and the dimension of organ/tissue [[48], [49], [50], [51], [52], [53]]. This method has attracted great attention because it intensely improves the decellularization efficiency by rapid accessing to the whole organ and homogeneous perfusion through the native vasculature [[54], [55], [56]]. Second, after perfusion, the intact and intricate vascular network, the vital construction of the whole organ, is maintained, including portal vein, artery inflow, and venous outflow, which is essential for the transportation of nutrition and oxygen within organs/tissues [5,57,58]. However, perfusion requires additional hardware and complex setup for flow control, such as an infusion/withdrawal syringe pump and pressure transducer [[59], [60], [61]].
2.2.1.3. Immersion and agitation
Compared with perfusion treatment, immersion and agitation is a more appropriate method for small, fragile and thin sections of organs as well as tissues without innate vascular structures [[62], [63], [64]]. Immersion and agitation refers to the method of submerging tissues into decellularization solutions with constant mechanical agitation, whose efficiency depends on different paraments, including agitation intensity, decellularization agent and tissue dimension [62,65]. After immersing tissues in the agents, agitation facilitates the rupture of cells, the detachment of cells from basement membranes, and the removal of cellular components. As an optimal physical way to decellularize, the utilization of immersion and agitation owns plenty of advantages. First, using dynamic immersion and agitation can attain a more homogeneous detergent exposure than static decellularization, and achieve a better decellularization effect with less exposure time to aggressive decellularization agents [60,66]. Second, this method does not significantly affect the ECM surface structure, collagen structure and integrity, mechanical strength, and GAG content [56,[66], [67], [68], [69]]. Third, compared to whole organ perfusion, this method is a more accessible and easily executed procedure [56]. However, compared with perfusion, using this method may cause more damages to tissues due to the limited diffusion of chemicals by agitation [70,71].
2.2.2. Chemical treatments
Chemical treatments use detergents and chemical agents to disrupt cellular bonds and remove cellular components. An overview of chemical treatments and their typical solutions, mechanisms, and disadvantages is briefly summarized in Table 3. In particular, detergents, acids and bases, hypertonic and hypotonic solutions are discussed in the following sections.
Table 3.
List of chemical treatments used for decellularization.
| Method | Typical Agents | Mechanism | Disadvantages | Reference |
|---|---|---|---|---|
| Ionic detergents | SDS, | Solubilize cytoplasmic membranes, nucleic membranes and nucleic acids, and dissociate deoxyribonucleic acid (DNA) from protein | Damage ECM structure and collagen integrity. | [36,50,51,58,114,115] |
| SDC, | Reduce the contents of glycosaminoglycan (GAG) and growth factors | |||
| Triton X-200 | ||||
| Non-ionic detergents | Triton X-100 | Solubilize cell membranes and dissociate DNA from proteins (especially suitable for cell-derived dECM) without disrupting protein-protein interactions | Alter the ECM architecture. | [43,50,51,57,58,107,108,115,116] |
| Reduce laminins/fibronectin content. | ||||
| Remove cells incompletely | ||||
| Zwitterionic detergents | CHAPS, | Solubilize cell membranes and dissociate DNA from proteins by disrupting DNA-protein, lipid-lipid, lipid-protein interactions | Disrupt and dissociate proteins in the ECM | [100,[117], [118], [119], [120]] |
| SB-10, | ||||
| SB-16 | ||||
| Acids | Peracetic acid, | Disrupt cell membranes, solubilize cytoplasmic organelles, induce cell lysis, denature proteins and catalyze hydrolytic degradation of biomolecules | Damage ECM architecture. | [76,105,114,121] |
| Hydrochloric acid | Affect intracellular molecules such as GAG | |||
| Bases | Ammonium hydroxide | Solubilize cytoplasmic components, disrupt nucleic acids and catalyze hydrolytic degradation of biomolecules | Influence GAG content, collagen and growth factors. | [57,61,105] |
| Weaken mechanical properties | ||||
| Hypertonic solutions | Sodium chloride | Rupture cell membranes and lyse cells by osmotic shock | Difficult to remove DNA remnants | [42,118] |
| Hypotonic solutions | Tris-HCl | Induce cell lysis, and detach DNA from proteins | Difficult to remove DNA remnants | [116,118,122] |
| Organic solvents | Ethanol, Methanol, Acetones | Extract the lipid of the ECM and cause tissue dehydration, leading to cell membranes solubilization and cell lysis | Cross-link and precipitate proteins, including collagen. | [35,56,73,[123], [124], [125]] |
| Chelators | EDTA, EGTA | Dissociate cells and ECM by binding divalent metal ions necessary for cell adhesion | Disrupt protein-protein interactions, and denature proteins in ECM | [75,118,119,125] |
2.2.2.1. Ionic and non-ionic detergents
Detergents, including ionic, non-ionic and zwitterionic detergents, are soluble amphiphiles that can disrupt the hydrophobic-hydrophilic interactions among molecules. They effectively remove immunogenic cellular components by solubilizing cell membranes and dissociating DNA from proteins, but strong detergents can have adverse effects on ECM by disrupting and dissociating proteins. Ionic detergent is an effective chemical treatment for decellularization, which solubilizes nuclear and cytoplasmic membranes by disrupting lipid-lipid, lipid-protein, deoxyribonucleic acid (DNA)-protein and protein-protein interactions, leading to the elimination of cells and genetic contents. The most common ionic detergent are sodium dodecyl sulfate (SDS), sodium deoxycholate (SDC), sodium lauryl sulfate, sodium lauryl glutamate, sodium lauryl ester sulfate and Triton X-200 [[100], [101], [102]]. It is verified that SDS treatment can thoroughly remove the native cells as well as genetic materials [29,74]. The concentration of SDS plays a critical role in the decellularization process. The higher concentration of SDS results in lower remained DNA content and reduced mechanical strength in the dECM scaffolds, while the lower concentration of SDS leads to more retention of collagen, less denaturation of ECM proteins and increased cytotoxic effects with more cellular remnants [29,60,61,65,103]. However, due to interrupting protein-protein interactions, ionic detergents have a harmful impact on ECM structures and bioactive components [48,51,61,103,104]. Besides, remaining SDS is difficult to remove due to its strong interactions with ECM proteins, which may subsequently induce cytotoxicity to recellularized cells [41,[104], [105], [106]]. Compared to ionic detergents, non-ionic detergents provide a gentler treatment to solubilize cell membranes and dissociate DNA from proteins by breaking up interactions of lipid-lipid, lipid-protein, DNA-protein, but leaving protein-protein interactions conserved. Because of the incapacity to denature proteins, non-ionic detergents can clear off cellular contents without damaging the structure and orientation of collagen [42,48,61,107]. However, because non-ionic detergents are milder and gentler, they are insufficient to remove nuclei and DNA [51,58,108]. Therefore, these detergents always require the assistance of other solutions or physical treatments to ensure the complete removal of cellular components [43,48,61].
2.2.2.2. Acids and bases
Acids and bases are the decellularization methods based on catalyzing the hydrolytic degradation of biomolecules, cytoplasmic components and nucleic acids [109]. Similar to detergents, they can disrupt ECM constituents and structures.Acid compounds donate hydrogen ion H+ or form a covalent bond with an electron pair to catalyze hydrolytic degradation, among which peracetic acid (PAA), hydrochloric acid and acetic acid are typically utilized for decellularization [38]. After PAA treatment, cytoplasmic organelles are solubilized and nucleic acids are disrupted. In addition, PAA performs germicidal activity against a broad spectrum of bacteria, viruses and fungi [110]. However, acids also damage ECM microarchitecture, reduce collagen content and weaken tissue strength [37]. Therefore, proper acids and optimized concentrations should be selected. PAA at the concentration of 0.1% is regarded as an ideal treatment for thin tissues, since it has minimal effects on ECM structures and components [111]. Compared to acid compounds, alkaline could release hydroxide OH− and react with acids to form salts, and the most common bases for decellularization are ammonium hydroxide, sodium hydroxide and sodium sulfide [37]. Bases decellularize tissues by denaturing chromosomal DNA and inducing cellular lysis. Especially alkaline solutions with a pH higher than 11 can efficiently remove cellular remnants, since DNA is susceptible to denature [109]. However, alkaline can affect ECM structure, change mechanical and viscoelastic properties, reduce GAG content, and even remove growth factors within tissues if exposed for a long time. Besides, alkaline solutions with pH 10–12 may greatly damage collagen fibers, fibronectin and GAGs, as well as elicit severe host responses and form fibrotic tissues [109].
2.2.2.3. Hypertonic and hypotonic solutions
Hypertonic/Hypotonic solution treatment respectively refers to using solutions with a higher/lower solute concentration than that in cells to remove cellular components out of cells. Due to the osmotic effects, they decellularize tissues through inducing cell lysis, cell dehydration and cell death. In addition to offering a pressure condition, it is verified that the removal of protein can be achieved by hypertonic solutions, and the removal of nuclei and DNA can occur in hypotonic solutions, which could explain the reason that they are regarded as chemical routes for decellularization [112]. Besides, since these solutions are more operable to be washed out of tissues, the possibility of inducing cellular toxicity from the remained solution is minimized [42]. Moreover, hypertonic/hypotonic solutions of saline (e.g., sodium chloride) can effectively remove other residual chemicals and cellular leftovers from ECM [72]. Further, hypertonic solutions could maintain basal lamina structures and tissue functionality [112]. For example, NaCl treatment in PBS shows a negligible effect on the transparency of corneas compared to the treatment of SDS or nuclease [67]. However, cell lysis in hypotonic solutions may cause the dispersion of antigens in tissues and result in tissue swelling [112]. In addition, despite hypertonic/hypotonic solutions can aid in cell death, they cannot achieve complete elimination of cellular residues unless combined with other detergents or enzymes [113]. For instance, the hypertonic-DNase wash strategy results in a more effective removal (>95%) of DNA content than the hypertonic-hypotonic wash method (<95%) [112].
2.2.3. Enzymatic treatments
Enzymatic treatments provide high specificity for the removal of cell components and undesirable ECM constituents by breaking specific chains within cellular fragments or cell-matrix adhesions. The typical enzymatic treatments, along with their mechanism and disadvantages, are shown in Table 4. In particular, nuclease and trypsin are discussed in the following sections.
Table 4.
List of enzymatic treatments used for decellularization.
| Method | Mechanism | Disadvantages | Reference |
|---|---|---|---|
| Nuclease | Catalyze the hydrolysis of the interior or terminal bonds of RNA and DNA, and aid in the complete removal of residual nucleic acids | Induce severe distortion of ECM structure. | [67,100,101,117,126,128,129,134] |
| Hard to be removed incompletely, and can impede recellularization and transplantation | |||
| Trypsin | Cleave peptide bonds on the carboxyl-side of arginine and lysine | Hard to achieve sufficient decellularization. | [130,133,[135], [136], [137], [138], [139], [140]] |
| Need lengthy incubation time. | |||
| Disrupt elastin and collagen structure | |||
| Dispase | Cleave specific peptides (collagen Ⅳ and fibronectin) in the basement membranes | Damage basement membranes and ECM | [67] |
| Lipase | Catalyze the hydrolysis of lipids and aids in delipidation | Hard to remove all lipids | [141,142] |
| Phospholipase | Hydrolyze phospholipid components of cells and solubilizes cells | Significantly reduce GAG content | [143,144] |
2.2.3.1. Nuclease
Nucleases catalyze the hydrolysis of deoxyribonucleotide and ribonucleotide chains, and therefore aid in cell removal. Deoxyribonuclease (DNase), as a typical nuclease, has been commonly used for decellularization, due to its powerful specificity to facilitate the removal of DNA content while retaining proteins by cleaving nucleic acid sequences [126]. DNase is generally used after detergents that increase internal spacing and porosity, making DNase easier and quicker to infiltrate into tissues. The penetrated DNase can remove residual nucleic acids and cellular debris, as well as assist in the washing out of detergents [67,100,127]. DNase with detergents is shown to be essential for the complete removal of cellular materials as >95% DNA removal is achieved after DNase treatment [61,101,112,120,128]. However, the long-standing processing time of nucleases can alter ECM structure and decrease mechanical stability, and result in the loss of ECM constituents such as GAG, laminin and collagen IV [67,100,120]. Furthermore, multi-stepped and extensive washing should be performed because the remaining enzymatic products may invoke an immune response and impede subsequent recellularization and implantation [129].
2.2.3.2. Trypsin
Trypsin is another commonly used enzyme for decellularization processes. Its mechanism relies on the cleavage of the peptide bonds on the carbon side of carboxyl-side of arginine and lysine, resulting in the separation of cellular components from ECM. Similar to DNase, trypsin has been widely used since it could effectively decellularize ECM without inducing a cytotoxic effect [75,130]. However, trypsin can cleave proteins within the ECM, possibly leading to the damage of preserved ECM and alteration of mechanical stability [38]. Therefore, proper concentrations and exposure time are recommended [[131], [132], [133]]. For instance, Lin clarified that the selection of proper trypsin concentrations (<0.03–0.5%) and exposure times (<12 h) are mandatory for the decellularization of the porcine coronary artery [133]. Ramm demonstrated that the trypsin treatment of heart valves should be limited to less than 90 min, which showed effective removal of DNA and N-linked glycans, and excellent mechanical stability [131].
2.2.4. Combinations of physical, chemical and enzyme treatments
These different methods mentioned above, physical, chemical and enzymatic treatments, have their own advantages and disadvantages. They all can remove cellular components to a certain degree but also do harm to the composition, structure and properties of the remaining ECM. In general, to minimize the adverse effect on ECM and maximize the removal of cellular contents, the use of any of these treatments alone is insufficient; thus these methods should be combined [51,145,146]. For instance, decellularization was more effective when using the combination of chemical treatments (Triton X-100 and hypotonic solution) and physical method (freeze-thaw cycles), leaving only roughly 1% residual nuclei and 20% residual DNA, whereas samples that had not undergone additional freeze-thaw cycles contained approximately 20% residual nuclei and 40% residual DNA [108]. Furthermore, Shafiq optimized the decellularization protocols for auricular cartilage with the combination of chemical (SDC), physical (freeze-thaw cycles), and enzymatic (DNase and Trypsin) routes. They claimed that detergent/enzymatic method was one of the most effective processes, and their optimized methods could mimic a biophysical and physiological niche similar to human auricular cartilage ECM. This is because the usage of DNase and SDC could effectively decellularize cartilage, freeze-thaw could minimize the duration of exposure to detergent/enzymatic forms, and trypsin could accelerate decellularization by removing cells while maintaining collagen and biomechanical properties [75]. It is noted that the choices of decellularization methods, depending on but are not limited to tissue thickness, cellular density and scaffold size. A proper method for specific tissue or organ may weaken decellularized effects or even induce adverse effects on other organs or tissues. For example, studies have proved the superiority of agitation in 1% SDS to agitation in 1% Triton X-100 for decellularization of human testicular tissue due to the strong ability of SDS to penetrate thick tissues [147]. However, it is observed that tissues following the agitation in 1% SDS protocol showed shrinkage and rigidity, indicating that it is not suitable for the decellularization of a force-resistant matrix such as dense and fibrous tendons [42].
2.2.5. Recently emerged treatments of decellularization methods
In addition to the above classical methods, several novel treatments, such as vacuum-assisted decellularization and apoptosis-assisted decellularization, are recently emerged. Vacuum-assisted decellularization refers to using negative pressure to accelerate the processing of decellularization [[148], [149], [150], [151]]. The method is highly effective when combined with physical, chemical or enzyme treatment; however, the prolonged vacuum time may increase the pore size and destruct the collagen fiber bundles, and thus decrease the mechanical strength of specific tissues, such as pericardium [152,153]. Apoptosis-assisted decellularization removes cellular components via apoptosis-inducing agents such as camptothecin and rotenone, and is demonstrated to be promising for complex structures like nerve tissues or vasculature [112,154,155]. However, due to the complicated mechanism, these decellularization-assisted novel methods have not been applied extensively, remaining a research field worthy of explorations.
2.3. Postprocessing of dECM scaffolds: sterilization, cross-linking and modification
Sterilization. In order to remove the toxic components and improve the biocompatibility of the dECM scaffolds, decellularized scaffolds should be post-processed through sterilization and disinfection. The ideal sterilization or disinfection of the acellular scaffolds can effectively remove microorganisms without inducing toxicity. However, the sterilization often results in the changes in the physical and chemical properties, such as causing cross-linking of collagen matrix and breaking of macromolecular chains. Irradiation is a physical sterilization method, mainly including gamma rays produced by a 60Co device and electron beams produced by electron accelerators. Irradiation can directly destroy the nucleic acid, protein and enzyme in the microorganism, due to its strong penetration capability. However, irradiation could lead to changes in physical properties, chemical properties, and biological compatibility of dECM [156,157]. Ethylene oxide (EO) sterilization is a relatively mature sterilization method, which can make the amino and carboxyl groups in microbial protein and nucleic acid alkylation, and inactivate the macromolecules [158]. EO can be performed at room temperature and does not cause damage to materials, but it produces toxic residues after sterilization [159,160]. Antibiotic disinfection can significantly inhibit the growth of bacteria by destroying bacterial cell wall and inhibiting protein synthesis and DNA synthesis, and have a negligible effect on the structure of acellular scaffold. But it is basically ineffective on viruses and spores, and besides, the antimicrobial spectrum of each antibiotic is limited [161,162]. Peracetic acid (PAA) is a chemical detergent, but it can also achieve a bactericidal effect under certain conditions. PAA could destroy the cell wall of the microorganism and affect the enzyme system in the microorganism, and its decomposition products are non-toxic [163]. However, the strong oxidation of PAA will alter the physical and chemical properties of the scaffolds [164,165].
Cross-linking. To enhance the mechanical strength, and ensure a stable 3D network structure, decellularized scaffolds are always undergone a cross-linking treatment. Since decellularization often has adverse effects on the biomechanical properties of ECM, physical and chemical crosslinking methods are usually used to preserve the 3D structure of dECM and improve the mechanical properties of scaffolds [166]. Glutaraldehyde is a commonly used chemical crosslinker for collagen-based materials, and Yu cross-linked decellularized kidney scaffold with glutaraldehyde perfusion; but its cytotoxicity may affect subsequent studies [167,168]. Alternatively, glyoxal and genipin were used as crosslinkers with reduced cytotoxicity [169,170]. Interestingly, researchers have found that cross-linking approaches could assist in promoting cell adhesion and differentiation. For example, Xing used EDC crosslinking combined with chemical extraction to conduct tissue decellularization, which could promote the adhesion and differentiation of mesenchymal stem cells [171].
Modification. To increase the pore density and improve cell infiltration, surface modification methods, such as laser, solvent casting and particle leaching, and electrospinning modification, are developed [172]. Laser modification, employing a laser beam to selectively sinter the material surface and producing a large number of minute pores at a high density, was demonstrated to generate micropores on tissue surfaces and thus speed up the decellularized processing. Take the cartilage for an example, laser modification facilitates migration and dedifferentiation of reseeded chondrocytes, and also maintains the mechanical strength of native tissues [[173], [174], [175]]. Solvent-casting/salt-leaching, leaving uniform salt solvent evaporated and forming a sponge-like porous structure in water, was shown to provide an optimal template for cell infiltration and nutrient transfer [172,176]. Furthermore, coating of inorganic polymers and peptide modification have also been applied as ECM modifications recently [177,178]. Attempts have been made by covalently coupling bioactive peptides to the inner surface of dECM, which showed improved biofunction performance such as cell adhesion, proliferation and differentiation [[179], [180], [181]]. Besides, ECM-derived peptides with signaling domains, a type of ECM fragments, have also been used to modify the other biomaterials and enhance their functions by connecting with receptors on the cell membrane surface [[182], [183], [184]].
2.4. Recellularization of dECM scaffolds
Recellularization is defined as renewed repopulation of acellular ECM scaffolds with specific cell types or stem cells, to form an engineered construct with specific structures and functions [185]. Stem cells, the undifferentiated cells that can differentiate into specific lineages and self-renew by dividing themselves to produce more stem cells, have been widely used for recellularization [186,187]. They are generally grouped into pluripotent stem cells and multipotent stem cells (Fig. 2a). Pluripotent stem cells, including embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), can be theoretically differentiated into almost all kinds of somatic cells while remaining their indefinite growth; multipotent stem cells, such as mesenchymal stem cells (MSCs), can be found in varieties of tissues such as bone marrow, adipose tissues and central nervous system (CNS) [186]. Embryonic stem cells (ESCs), isolated from embryos maintaining the unique capability of self-renewal and multi-lineage differentiation, have attracted much interest for organ/tissue regeneration [188]. Thus, the combination of ESCs and dECM scaffolds is widely studied in various tissue repairs, such as cardiac tissues, cornea and kidneys [[189], [190], [191], [192], [193]]. However, due to moral issues and ethical controversy, researches and translational medicine related to ESCs are strictly regulated, or even prohibited. Another pluripotent stem cells, iPSCs, reprogramed from somatic cells by introducing a set of transcription factors, can achieve similar gene expression profiles of ESCs and obtain unlimited capacity of proliferation and differentiation [194,195]. However, some researches indicate that the blunted phenotype of iPSC-derived cell lineages can limit their overall cellular potency compared with primary cells [196,197]. Interestingly, studies reveal that 3D dECM scaffolds can improve the phenotype and viability of iPSC-derived cells [196]. Applications of iPSC-derived cells with dECM scaffolds can be found in various tissue engineerings such as heart, kidney, lung and pancreas [[198], [199], [200]]. Ott's group combined human decellularized cardiac matrix with human iPSC-derived cardiomyocytes, and built functional 3D cardiac tissue constructs with force-generating human myocardial tissue, which showed feasible methods for repair and replacement of myocardium [201]. MSCs, an important population of multipotent stem cells with less immune system-mediated rejection, can differentiate into numerous cell types, showing notable performances in the field of tissue engineerings such as dermal wound healing, tendon repair, cardiomyogenesis and bone regeneration [186,[202], [203], [204], [205], [206], [207], [208], [209]]. In recent years, the decellularized matrix has been shown as proper bioscaffolds for MSCs in the application of tissue repairing, since its native biophysical and biochemical properties can induce the site-specific differentiation of MSCs and further coordinate with cells to repair organs/tissues [209]. For instance, Brouki Milan harvested decellularized dermal matrix (DDM) from human skin and repopulated it with human umbilical cord perivascular cells (HUCPVCs), a type of MSCs that could accelerate the early wound healing of the skin. According to experimental results of an accelerated wound closure and a promoted wound maturity, the HUCPVCs-loaded DDM scaffolds showed positive effects on full-thickness wound healing, such as diabetic wound models [204]. To sum up, recellularization not only depends on the variety of cell types, but also relies on the recellularization methods, the cell density, culture conditions, etc.
Although these cell-based strategies have been widely applied to tissue repair and regeneration, these methods have some drawbacks, such as cell contamination, short transplant life, increased disease transmission, and microvascular obstruction [210]. The cell-free strategy aims at overcoming the above drawbacks and repairing damaged tissues/organs by recruiting endogenous stem cells to the damaged site. A successful acellular strategy requires appropriate stimulation and recruitment factors to active and accumulate cells at the injury site, followed by cellular proliferation and differentiation, and tissue repairing and regeneration [211]. For example, studies showed that the meniscus acellular composite scaffold had great biomechanics and biocompatibility, and could promote the regeneration of meniscus [212]. However, the key technical problem in the cell-free strategy is searching the appropriate stimulation and recruitment factors to recruit sufficient populations of endogenous stem cells.
3. dECM scaffolds for tissue engineering
Up to now, since organ/tissue transplantation is concerned for the risk of donor shortage and health risks such as diseases and pathogenicity transmission, dECM scaffolds with critical components and native structures are investigated as new alternatives for organ/tissue regeneration. In the following sections, we mainly focus on dECM scaffolds for skin, bone, nerve, heart along with lung, liver, and kidney.
3.1. dECM scaffolds for skin tissue engineering
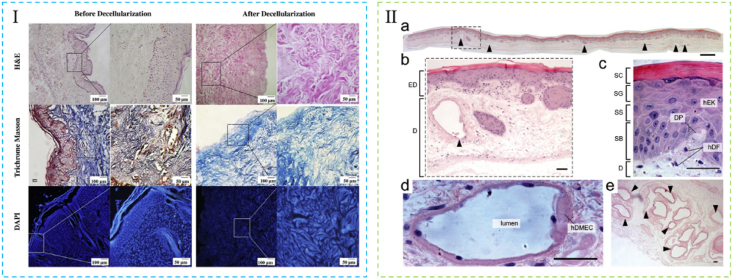
Skin, the largest organ in the human body, acts as a barrier against the external environment and toxins. Loss of skin integrity can result in acutely physiologic imbalance and subsequently further injuries [213]. dECM scaffolds could repair and regenerate skin tissues, since they preserve physical signals such as the porous dermal bilayer architecture, vessel structure, adhesive force and elasticity, and bioactive molecules such as endogenous growth factors [[214], [215], [216]]. For example, the maintained intact basement membrane zone was demonstrated to facilitate the adhesion of keratinocytes, and the preserved dermal structure with papillary and reticular dermis is verified to promote the growth of angiogenic cells [217,218]. In addition, the release of basic fibroblast growth factors from dECM is shown to reduce wound contraction, leading to less scar formation [215]. Interestingly, once autologous skin is regenerated, the degradation products of dECM show negligible toxicity and even possess the ability to induce the formation of tube-like structures and the regeneration of skin [219]. It should be noted that skin is a bilayer structure comprising avascular epidermal and vascular dermal layers [220]. Brouki Milan reported a decellularized dermal matrix (DDM) scaffold, and the structure of DDM and its biomechanical strength were relatively similar to those of fresh human dermis. After reseeding DDM scaffolds with human umbilical cord perivascular cells (HUCPVCs), the construct significantly enhanced cutaneous wound healing by performing an accelerating wound closure rate, faster re-epithelization and decreased collagen deposition (Fig. 4I) [204]. Further, Groeber employed a decellularized porcine jejunum to fabricate a bilayer skin alternative, and proved that native vascular networks, topography and collagen contents were preserved (Fig. 4II). By seeding human dermal microvascular endothelial cells into the vascular system, and inoculating dermal fibroblasts and human epidermal keratinocytes onto the surface of dECM scaffolds, a human vascularized skin equivalent composed of an epidermal and a vascular dermal layer was generated, proved to have the potential for skin grafting [221].
Fig. 4.
dECM for skin tissue engineering. (Ⅰ) Histological assessment of human dermis before and after decellularization. Reproduced from Ref. [204]. (Ⅱ) H&E stained cross-section of the vascularized skin equivalent (vSE). (a) Overview of the vSE. (b) Magnification of the area indicated in (a). (c) Detailed view of the epidermal layer. (d) Detailed image of a vascular structure that has been reseeded with human dermal microvascular endothelial cells (hDMEC). (e) Image of the vasculature in efferent vessels. Reproduced from Ref. [221].
To date, diabetic foot ulcers (DPUs), a full-thickness skin loss, has been investigated and treated by using dECM scaffolds [222,223]. Angiogenesis, essential for the transportation of oxygen and nutrients to the wound site, is one of the major issues for DPUs treatment. Although there are plenty of treatments such as extensive necrotic tissue debridement, biological dressings and hyperbaric oxygen therapy, they all cannot completely resolve the healing problem [224]. Recently, dECM scaffolds with a high capacity of neovascularization have been used for DPU due to their preserved vasculature and endogenous angiogenesis factor [[225], [226], [227]]. It is shown that dECM-based scaffolds can accelerate wound healing of DPUs by affecting granulation tissue formation, epithelial regeneration and pro-angiogenesis activity, and may reduce scar formation by shortening the inflammatory phase [204]. Nowadays, there are several acellular products successfully transferring into clinical application, such as AlloDerm® regenerative tissue matrix and Oasis® wound matrix [228]. However, some procedures need to be optimized and the criteria of these products should be enhanced to perfect the skin alternatives emerging in the market. For example, the reconstitution of adnexal structures such as hair follicles and sweat glands has not been shown in clinical products, which is a main criterion for skin formation. Fortunately, some studies have reported that decellularized human placenta-derived ECM could modulate the healing of full-thickness wounds accompanied by hair follicle formation [215,229].
3.2. dECM scaffolds for bone tissue engineering
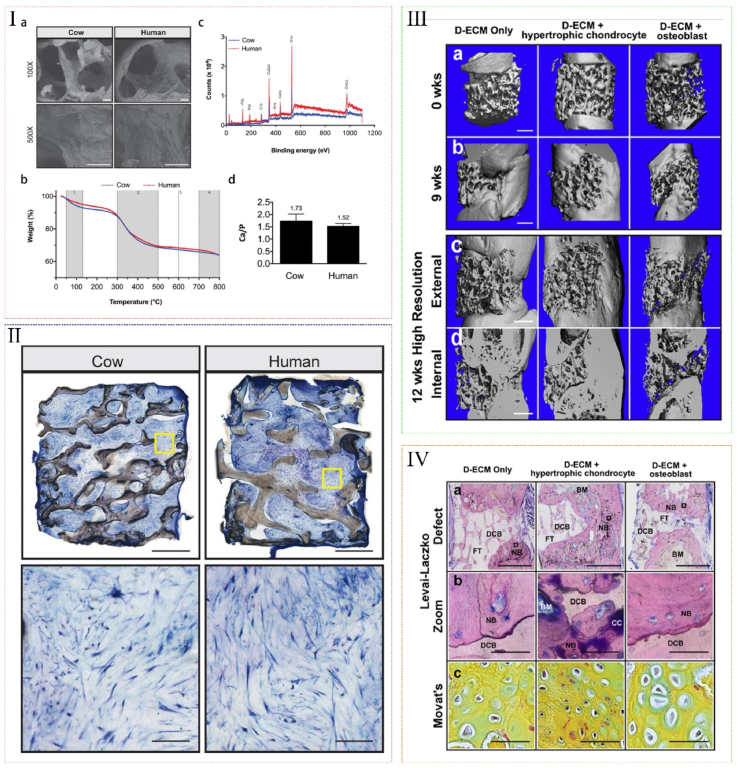
Bone is a dynamic tissue with the innate capacity to heal and regenerate itself following injury [230,231]. However, large defects may cause reversible impacts and require external interventions for osteogenesis [232]. Recently, decellularized matrices represent alternative scaffolds for bone regeneration due to their biomimetic environment supporting histogenesis, including their structure features, bioactive molecules and trace elements. First, dECM scaffolds preserve the macro-structural features like geometry and highly porous structure, and the microstructural features like surface roughness and micropores, which greatly improve the osteoinductivity [[233], [234], [235]]. Second, rigid matrices mimic the cross-linked collagen structure of osteoid so that stem cells on stiff substrates tend to differentiate into osteogenic lineages [236,237]. Third, dECM scaffolds preserve immunomodulatory cytokines, such as transforming growth factor-β, bone morphogenic proteins and bFGFs, which regulate the pro-inflammatory response [203,[238], [239], [240], [241], [242]]. In addition, the trace elements preserved in dECM, such as magnesium and strontium, play a crucial role in maintaining the integrity and cellular activity of bone [231,243]. To date, decellularized bone scaffolds are mainly derived from livestock animals due to their accessibility and similarity with human bone [233,235,237]. Sladkova compared decellularized cow and human cadaveric bone scaffolds with the same decellularized protocol. The results showed that the scaffolds displayed similar porous and trabecular structure, while interspecies differences existed in cortical bone parameters such as composition and density (Fig. 5Ⅰ). Then the scaffolds were then seeded with human induced pluripotent stem cells (iPSC)-derived mesenchymal progenitor cells, and the constructs were subsequently cultured in osteogenic inducing medium and later transformed to perfusion bioreactors. After 5 weeks’ cultivation, samples displayed a similar extent of tissue formation, with the cells embedded and distributed uniformly throughout the scaffolds (Fig. 5Ⅱ). In conclusion, despite minor differences in architecture and mass composition, it was reported that dECM scaffolds from both human and animal sources could support cell viability and the formation of mineralized tissues [244].
Fig. 5.
dECM for bone tissue engineering. (Ⅰ) Characterization of decellularized cow and human bone scaffolds. (a) SEM, (b) Thermogravimetric analysis, (c) X-ray photoelectron spectroscopy analysis and (d) Ca/P ratio of decellularized cow and human bone scaffolds. (Ⅱ) Histological analysis of tissue formation on decellularized cow and human bone scaffolds after cultured in perfusion bioreactors with mesenchymal progenitor cells for 5 weeks. (Ⅰ) and (Ⅱ) reproduced from Ref. [244]. (Ⅲ) (a–b) Bridging of critically sized femoral defects represented by 3D μCT reconstructions of the rat femur at 0 and 9 weeks after post-implantation with acellular scaffolds, hypertrophic chondrocyte and osteoblast grafts. (c–d) Internal and external regions are shown at 12 weeks. (Ⅳ) Defect regeneration and bone formation are shown at 12 weeks after post-implantation. (a) Hard bone histology using the Levai-Laczko stain. (b) Magnified views allowed detection of calcified cartilage. (c) At the location of new bone formation, a cartilage anlage characteristic of endochondral ossification was present (green staining in Movat's pentachrome sections). Reproduced from Ref. [230].
Endochondral ossification, as an innate self-heal mode of the long bone, has been designed to repair large segmental defects with the utilization of biomaterials in the past few years. It is verified that tissue-engineered bone grafts based on dECM scaffolds could regenerate bone through endochondral ossification [231,[245], [246], [247], [248]]. Bernhard et al. prepared bone decellularized scaffolds, and applied them for the treatment of orthotopic, critical-size defects in rat femur. The scaffolds were infused with adipose stem cells, and cultured in chondrogenic medium for 2 weeks and subsequently in hypertrophic medium for 3 weeks to form hypertrophic chondrocyte-seeded constructs. Then constructs showed endochondral-like characteristics, and exhibited excellent bone regenerative features after implantation, including enhanced bone deposition, fast integration with adjacent native bone, bridging of femoral defects, and regenerative milieu established within the defect space (Fig. 5III-IV) [230]. Therefore, dECM scaffolds provide an osteoinductive microenvironment for the differentiation of chondrocytes, the maturation of hypertrophic chondrocytes, and the osteogenic activity of tissues, which offer a promising method to improve the repair effect of the long bone defects. However, there still exist further steps for the dECM scaffolds applied in the bone tissue engineering. For example, decalcification of dECM should be added because demineralized matrices remain mineral components and facilitate abundant osteogenesis-related factors exposure [249]. Moreover, osteoconductive inorganic materials, such as nanohydroxyapatite, should be embedded to dECM scaffolds to provide inorganic cues to osteoblasts for their adhesion, proliferation, and differentiation capacity towards osteogenic lineages [250].
3.3. dECM scaffolds for nerve tissue engineering
Injuries of the nervous system are one of the most devastating conditions for patients, while the recovery of nerve injuries remains a challenging problem due to the intrinsically low neuron regeneration capacity, especially for the central nervous system (CNS) injuries. Recently, stem cell therapy is a promising strategy for nerve injuries, but cells would be washed out of the lesion and their migration would be hampered by glial scars, or they may be attacked by inflammatory factors and oxygen free radicals [251]. In dECM scaffolds, on the one hand, the native architecture is preserved, guiding cell migration and directing axonal trajectories [[252], [253], [254], [255]]. On the other hand, the retained ECM components, such as GAGs and proteoglycans, are shown to affect neural stem cells (NSCs) proliferation and regulate the synapse formation. The preserved neuro-supportive molecules could stimulate neurons to modulate their phenotype from a transmitting state to a regenerative state [256,257]. Moreover, during dECM scaffolds proteolysis degradation, the discharged peptide motifs may increase vascular permeability, induce angiogenesis, and modulate the innate immune responses [[258], [259], [260], [261]]. Since CNS tissues have a more limited physiological capability to spontaneously regenerate after structural disruptions than the peripheral nervous system (PNS), the research of dECM scaffolds used for CNS has received relatively scarce attention [255,262]. Crapo applied a decellularization method for different CNS tissues, including optic nerve, spinal cord, and brain (Fig. 6Ⅰ). The resulting matrix was proved to be sufficiently decellularized, and bioactive molecules, including myelin, laminin, neuro-inductive bFGF, and the neurotrophin nerve growth factors, were retained (Fig. 6Ⅱ-Ⅲ). The resultant scaffolds could regulate behaviors of neural-like cells (PC12), including their proliferation, migration and neural differentiation [263]. Besides, they have found that CNS ECM versus non-CNS ECM shown tissue-specific functionality for PC12 cells. Interestingly, dECM could be solubilized into a liquid phase, and subsequently form a hydrogel phase [264]. Such hydrogel scaffolds are widely investigated due to the advantages of their injectability and in situ polymerization, which delivers the hydrogels through a minimally invasive method and enhances astrocyte/axon interactions through direct integration with host tissue [[264], [265], [266]]. For instance, DeQuach decellularized porcine brains and turned them into a liquid form by using enzymatic digestion. After decellularization, the absence of intact nuclei indicated the removal of cellular contents, while critical protein components such as sulfated GAGs, multiple collagen isoforms, laminin and proteoglycan perlecan of native brain ECM were preserved (Fig. 6ⅠV). After being seeded with iPSC-derived neurons, the brain matrix was proved to support the culture and maturation of neurons in vitro, and formed a nanofibrous scaffold in vivo (Fig. 6V) [267].
Fig. 6.
dECM for nerve tissue engineering. (Ⅰ) Biologic scaffolds derived from porcine central nervous system (CNS) tissues. (a) Native optic nerve tissue (left) and optic nerve ECM (right). (b) Native spinal cord tissue (left) and spinal cord ECM (right). (c) Native brain tissue (left) and brain ECM (right). (Ⅱ) Characterization of residual deoxyribonucleic acid (DNA) in CNS ECM scaffolds. (a) native optic nerve tissue (b) optic nerve ECM. (c) native spinal cord tissue, (d) spinal cord ECM, (e) native brain tissue, (f) brain ECM. (Ⅲ) Protein content of CNS ECM scaffolds. (a) native optic nerve tissue, (b) optic nerve ECM, (c) native spinal cord tissue, (d) spinal cord ECM, (e) native brain tissue, and (f) brain ECM. (g) native optic nerve, (h) optic nerve ECM, (j) native spinal cord, (k) spinal cord ECM, (l) native brain, and (m) brain ECM. Reproduced from Ref. [263]. (Ⅳ) H&E-stained sections of porcine brain matrix (a) decellularized brain matrix (b). (Ⅴ) Brain matrix material was loaded into a syringe (a) and injected subcutaneously, whereupon the injected material self-assembled into a gel (b). Reproduced from Ref. [267].
Spinal cord injury (SCI), caused by the formation of glial scars, loss of vast amounts of neurons and insufficient expression of neurotrophic factors, along with sensory or motor deficits, devastating neurological disorders and cognitive impairments in patients, is a tough task to be conquered [268]. Reactive microgliosis and heightened inflammatory responses are the top issues of SCI, which form a cycle to chronically activate the microglial cell inflammatory responses and self-propels neurotoxicity [269]. Interestingly, it is verified that dECM scaffolds can cause a significant decrease in the ratio of M1 to M2 macrophage phenotype, shifting from primarily pro-inflammatory to primarily anti-inflammatory in CNS, and thus increase the survival of seeded cells and limit deleterious effects [265]. Meanwhile, the tissue-engineered constructs based on dECM also greatly enhance CNS functional recovery by axon regeneration along with the remyelination of axons [251,255]. Not only for SCI, dECM also displays extraordinary regeneration capacity for brain nerve injuries, especially for traumatic brain injury (TBI) [262,[270], [271], [272]]. For example, Wu demonstrated that brain dECM offered long-lasting structural and functional protection against traumatic brain injury-induced neurological deficits by improving neurobehavioral function, and mitigating glial scar formation and pro-inflammatory microglial responses [273]. Sood proved that brain ECM could enhance functional tissue-specific differentiation of human-induced NSC into healthy neurons and astrocytes, and inhibit reactive astrogliosis by downregulation of chondroitin sulfate proteoglycan [274]. Although numerous studies have been done for PNS and CNS repair, improvements are still required urgently to form a more biomimetic scaffold. First, for weak and readily disintegrated CNS tissues like brain tissues, optimal decellularization methods should be established to retain mechanical properties and avoid tissue loss [275]. Meanwhile, the chemical crosslinking process may enhance the elastic modulus, compressive strength and stiffness of dECM, and thus affects neural differentiation of seeded cells and alters the proportion of neuronal and glial cells [265,266,276]. Second, since the regeneration effects are significantly affected by the remaining growth-inhibiting molecules (e.g., interleukin-α and tumor necrosis factor-α, which exacerbate secondary tissue damage and the formation of an inhibitory glial scar), the final composition after decellularization should be balanced by removing those growth-inhibiting molecules and keeping growth-improving factors [277,278].
3.4. dECM scaffolds for cardiac tissue engineering
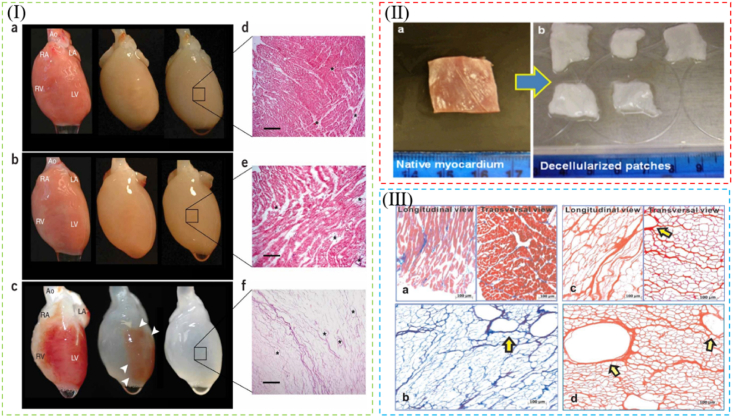
Heart failure remains a burgeoning public health problem worldwide and can cause an irreversible impact on patients [279]. To date, dECM scaffolds that mimic native cardiac environment are demonstrated as a promising material for heart failure [280,281]. First, the 3D structure, vasculature tree of native heart, and cardiac-specific functionalities are preserved after decellularization [81,282,283]. Especially, perfusion-decellularized whole heart scaffolds even retain the chamber geometry and valve competency, which could act as templates to generate synchronously beating heart tissues subsequently [14,284,285]. The maintained cardiac vascular networks could be vital to ensure sufficient blood supply for oxygen and nutrients [286,287]. In addition, the preserved properties make scaffolds resist the strength during the continuous contraction/relaxation of the heart, leading to the stabilization of the infarcted region [288]. Second, dECM scaffolds retain alignments such as topography, which is shown to regulate angiogenic growth factors and guide anisotropic microvascular progression, important for cardiac homeostasis and remodeling [284,286]. Third, dECM maintained bioactive cues like ErbB receptors, are demonstrated to have extracellular ligands to induce cardiomyocyte proliferation [23,280,289]. Furthermore, biomimetic culture in dECM scaffolds facilitates cardiac gene expression, and generates myocardial ECM with contractile function and electrical conductivity [285,290,291]. It should be noted that heart muscle, unlike heart valves or blood vessels, has rare alternatives, so the creation of myocardial tissue is the most challenging goal for cardiac tissue engineering, on which the following contents concentrate [292]. For cardiac tissue engineering, the most studied dECM scaffolds are decellularized whole heart scaffolds [81,287,293]. Ott first reported an optimal decellularized rat whole heart scaffold (Fig. 7I). The decellularized heart was shown to be a complex, biocompatible cardiac ECM scaffold with a perfusable vascular tree, patent valves and a four-chamber-geometry template. After repopulating decellularized hearts with neonatal cardia cells or aortic endothelial cells and subsequently culturing the construct in simulated physiological conditions, the nascent pump function of cardiac tissues was regenerated [14]. In addition to decellularized whole heart scaffolds, dECM patches have also been widely developed [23,285,286,290]. Cardiac patches can be as effective as the whole heart in repairing the damaged myocardium, with the advantage of exercisable implantation [294]. Wang created cardiac dECM patches (Fig. 7IⅠ). After decellularization, the 3D structure and vasculature templates (vessel-like structure) were preserved (Fig. 7IⅡ). By reseeding the decellularized patches, bone marrow mononuclear cells within the scaffolds showed good cell viability, proliferation and differentiation. The mechanical properties of cardiac tissues were recovering with the culture time increasing, and tissues performed angiogenesis potential and cardiac regeneration [295].
Fig. 7.
(Ⅰ) Perfusion decellularization of whole heart scaffold. Retrograde perfusion of rat heart using polyethylene glycol (a), Triton X-100 (b) or sodium dodecyl sulfate (SDS) (c) over 12 h. Corresponding H&E staining of thin sections from LV of rat hearts perfused with polyethylene glycol (d) or Triton X-100 (e). (f) H&E staining of a thin section of SDS-treated heart showing no intact cells or nuclei. Reproduced from Ref. [14]. (Ⅱ) Morphology of (a) native myocardium and (b) decellularized myocardial scaffold after decellularization treatment of 2.5 weeks. (Ⅲ) Mason's trichrome staining for (a) native myocardium and (b) decellularized myocardial scaffold.H&E staining of (c) the longitudinal and transversal views of the acellular scaffold, and (d) vasculature templates in the decellularized myocardial scaffold. Reproduced from Ref. [295].
Myocardial infarction (MI), a typical ischemic heart disease, referring to the situation that the blockage of a coronary artery reduces the blood supply to the heart muscle, makes the lack of oxygen in the affected tissue, leading to the cardiomyocyte death and tissue necrosis [296]. It is of great importance to prevent the pathological remodeling process and repair the infarcted tissue after MI, which may lead to the loss of normal contractility and functionality, since fibroblasts and endothelial cells can replace the impacted area with tough, rigid, collagenous and fibrotic scar tissue. It has been shown that the application of dECM scaffolds for MI could prevent adverse remodeling, reduce the scar expansion, and protect the local area from stiffening. In addition, heart dECM scaffolds could promote revascularization of the injured region, regulate cardiomyocyte proliferation and differentiation, and induce cardiac ECM remodeling [23,280,289,297]. Although dECM scaffolds have shown great promise in cardiac tissue repair and regeneration, some issues still need to be addressed. Especially, considering the importance of adequate blood supply for myocardial tissues, optimal decellularization methods should be further developed to attain more intact cardiac vascular networks. Moreover, the timing of bioscaffold transplantation and experimental endpoints are not standardized, so paraments of decellularization, recellularization and in-vitro culture cannot be compared and fully optimized. For example, some groups identified that the usage of decellularized heart ECM showed a great effect on the angiogenesis and myocardial regeneration effect after only 2 weeks of MI; however, others observed the influence of dECM on cardiac tissue after 4 or even 6 weeks of MI [280,283,297].
3.5. dECM scaffolds for whole-organ tissue engineering
The regeneration of organs/tissues has gradually focused their attention on dECM scaffolds over the past few years, which started from the restoration of simpler tissues (such as skin, bone, nerve and heart, discussed in Section 1, 2, 3), to the reconstruction of more complex whole organs, such as lung, liver and kidney. Since organ transplantation is largely limited by donor shortage, and the highly intricate vascular networks and architecture make it difficult for artificial organs and scaffolds to integrate with vessels and other systems [298]. For these solid organs, whole organ-derived dECM scaffolds have unique advantages, which can offer 3D organ matrices with preserved intact macroscopic 3D architecture, ultrastructure, vascular network and ECM components. Especially, the retained circulatory network of native organs can allow the constructs to be connected to blood torrent. Taking these into account, here we mainly overview decellularized whole organ scaffolds of these solid organs for tissue engineering, including lung, liver and kidney.
3.5.1. dECM scaffolds for lung tissue engineering
Respiratory disease is the third leading cause of death worldwide, however, lung transplantation is facing the situation of severe donor lung shortage [299]. Interestingly, dECM scaffolds remain spatial architecture, intact alveolar structure, thin basement membrane and hierarchical branching geometry, which offer a high surface area for gas exchange. Besides, they also preserve physiological mechanical stresses. Moreover, the maintained ECM signals and surface topography cues have synergistic effects on regulating pulmonary cell functions [118,[299], [300], [301], [302]]. Especially, pre-existing arterial and venous vascular tree and bronchial tree networks can be preserved as a framework for the reconstruction of vasculature and the lung organ [49,303,304]. In 2010, Ott created a bioartificial lung through decellularization, which provided acellular vasculature, airways, alveoli, and preserved alveolar septation, alveolar surface area, and ECM proteins (Fig. 8I). After seeding with lung epithelial and endothelial cells, the cell-seeded constructs were perfused and ventilated in a bioreactor to simulate the physiological microenvironment of lung. It was shown that the recellularized lung could perform gas exchange both in vitro and in vivo [15]. To date, the decellularized lung matrix has excellent potential for the treatment of pulmonary diseases, such as chronic obstructive pulmonary disease [305].
Fig. 8.
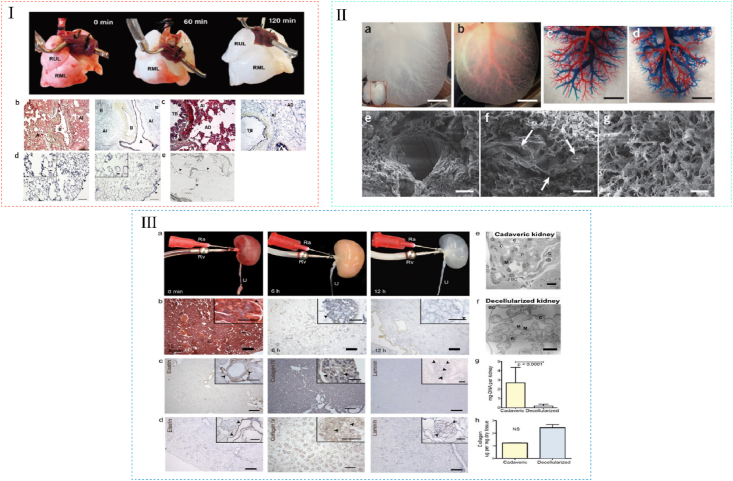
dECM for lung, liver and kidney tissue engineering. (Ⅰ) Perfusion decellularization of whole rat lungs. (a) Freshly isolated lung (left), after 60 min of SDS perfusion (middle), and after 120 min of SDS perfusion (right). (b,c) Corresponding Movat's pentachrome staining of thin sections from parenchyma of native (left panels) and decellularized (right panels) lung. (d) Corresponding Verhoeff's elastic-tissue staining of thin sections from parenchyma of native (left) and decellularized (right) lung. (e) TEM image of decellularized lung. Reproduced from Ref. [15]. (II) DLM retains intact lobular structure and vascular bed. (a) Representative photograph of decellularized left lateral and median lobes of rat liver. (b) The vascular tree, after perfusion with Allura Red AC dye. (c,d) Corrosion cast model of left lobe of a normal liver (c) and the DLM (d). (e–g) SEM images of a vessel (e), a section featuring bile duct-like small vessels (arrows) (f), ECM within the parenchyma (g), with hepatocyte-size free spaces. Reproduced from Ref. [17]. (III) Perfusion decellularization of whole rat kidneys. (a) Freshly isolated kidney (left), after 6 h of SDS perfusion (middle), and after 12 h of SDS perfusion (right). (b) Corresponding Movat's pentachrome stained sections of rat kidney during perfusion decellularization. (c) Representative immunohistochemical stains of cadaveric rat kidney sections showing distribution of elastin, collagen IV and laminin. (d) Corresponding sections of decellularized rat kidney tissue after immunohistochemical staining for elastin, collagen IV and laminin. (e) TEM of a cadaveric rat glomerulus showing capillaries (C), mesangial matrix (M) and podocytes (P) surrounded by Bowman's capsule (BC). (f) TEM of decellularized rat glomerulus exhibiting cellularity in decellularized kidneys. (g, h) Biochemical quantification of DNA and total collagen in cadaveric and decellularized rat kidney tissue. Reproduced from Ref. [18].
3.5.2. dECM scaffolds for hepatic tissue engineering
Liver is the largest solid organ in the body and plays an essential role in detoxification, protein synthesis and bile production, and thus liver disease has raised awareness of the public [306,307]. dECM scaffolds have provided a new way for hepatic tissue engineering, since they can preserve liver-specific ECM components, liver functional characteristics, the biliary tree with adequate branching, and vascular networks such as the portal vein, hepatic artery and hepatic vein [57,[308], [309], [310], [311], [312]]. In 2010, Uygun first reported the production of a transplantable liver graft. The decellularized liver matrix preserved 3D architecture, liver ECM, and microvascular network (Fig. 8II). Recellularized grafts could support liver-specific functions, including albumin secretion, urea synthesis and cytochrome P450 expression in vitro, and support hepatocyte survival and function with minimal ischemic damage in vivo [17]. From then on, the development of dECM scaffolds for hepatic tissue engineering has paid more attention to optimizing decellularization methods for reducing the decellularization time and attaining more intact vascular networks [51,57,309]. Owing to their relatively intact structure and functions, dECM scaffold is a potential treatment for liver diseases, including acute liver injury and chronic liver damage [306].
3.5.3. dECM scaffolds for kidney tissue engineering
Kidney is the most important organ for water homeostasis and waste excretion, and its unit consists of a glomerulus for ultrafiltration and a tubular apparatus for reabsorption [198,313]. To date, dECM scaffolds provide a therapy for patients with renal diseases, which preserve both composition and structure of renal ECM, as well as kidney-specific functions such as filtration, secretion and reabsorption [298,314,315]. Particularly, preserved renal microvasculatures, such as glomeruli and peritubular capillaries, are crucial for vascular patency [[316], [317], [318]]. In 2013, Song reported the first experimental orthotopic transplantation of a bioengineered kidney in rodents. They created decellularized whole-organ scaffolds with intact and perfusable vascular, glomerular and tubular compartments (Fig. 8III). After repopulating these scaffolds with renal endothelial and epithelial cells and perfusing the constructs in the bioreactor, the grafts showed in vitro capacity to clear metabolites, to reabsorb electrolytes and to generate concentrated urine, and produced urine via ureteral conduit after transplantation in vivo [18]. From then on, more researchers have made progress in decreasing decellularization time, increasing the vasculature integrity and maintaining kidney-specific growth factors [58,115,318,319]. To date, dECM scaffolds are proved to be attractive alternatives for the chronic kidney disease, end-stage renal disease [313,314,318].
4. Conclusions and outlooks
Over the past few decades, along with the emergency and development of a vast number of decellularization techniques, researches on dECM bioscaffolds have evolved from initial attempts of decellularized simple tissue scaffolds to successful preparation of decellularized whole organ scaffolds. Via physical treatments, chemical treatments and/or enzymatic treatments, dECM scaffolds can ensure the removal of immunogenic cellular components and the retention of significant compositions and native structures, which offer a microenvironment for tissue repair, restoration and regeneration. To date, decellularized scaffolds along with stem cell technologies play an essential role in the areas of tissue engineering, and provide clinically promising therapies for diseases such as skin wound healing, bone defects, nerve injuries, heart diseases, as well as coverage on whole-organ tissue regeneration related to lung, liver and kidney. Compared with other engineered biological scaffolds, dECM scaffolds attract more attention due to their outstanding advantages for tissue engineering. First, the removal of immunogenic cellular components reduces the potential adverse immune responses after implantation. Second, the 3D architecture of dECM is retained from original organs/tissues after decellularization, which provides biomimetic scaffolds with stable physical structures and signals for subsequent cell-cell interactions and cell-ECM interactions [7,22,320]. Third, compared to synthetic polymers, dECM scaffolds provide a better surface for cell attachment, owing to the remaining biological and biochemical components like cell adhesion ligands on the surface [2,321].
Based on these advantages, dECM-based products have recently emerged as an attractive substrate for clinical application. For example, AlloDerm® (BioHorizons) was applied to skin regeneration. After the materials were implanted, scar quality and skin functions in the implantation area were significantly improved [322]. Oasis® (Smith & Nephew) is another acellular product for skin repair, derived from acellular porcine SIS, primarily used for chronic wound treatment [323]. Products used for tendon and ligament injuries include GraftJacket® (Wright Medical) and Allopatch HD™ (MTF Sports Medicine), are prepared by decellularization of human dermis ECM. Particularly, GraftJacket® has been approved for clinical use by FDA in 2014. CardioCel® (Admedus IHS Inc.) is a heart valve product based on the acellular pericardium, which performs great biocompatible and facilitates the proliferation and differentiation of stem cells [324]. The application of these products has in turn promoted the development of decellularization technologies. However, it is notable that decellularization process could bring certain damages to the original structure and properties of ECM, and induce immunogenicity with residual cellular components [320]. A clinical application of Synergraft® valves, the decellularized porcine heart valves introduced in Europe in 2001, caused three of four patient's death because of the incomplete removal of cell remnants in decellularized products [325].
Therefore, during the explorations and clinical transformation of dECM scaffolds, there expose several challenges (Fig. 9), including (i) more sensible detection methods should be developed to test if there remain residual toxic products in dECM; (ii) decellularized protocols should be optimized to minimize toxicity by either reducing toxic and hard-to-remove decellularized agents, or choosing treatments that assist in removing those agents; (iii) the degradation speed of dECM scaffolds should be consistent with the process of bone remodeling, which affects the time for cell ingrowth, and an improper speed can result in transplantation failure [202,326]; (iv) the stricter selection of organ/tissue sources should be adopted due to the risks of transplantation, such as disease transmission risks of non-autogenous dECM, and pigmentation disorders derived from decellularized skin ECM; (v) the standardized quality control of dECM should be established according to the ECM source or the organ/tissue variety for further in vivo follow-up studies; (vi) the forms of dECM scaffolds should be adjusted as required. For example, dECM can be switched into particles for better portability, hydrogels for minimally invasive injectability, or films on other surfaces for enhanced regenerative capacity; (vii) to achieve functional regeneration of organs/tissues, reconstruction of vascular networks and adnexal structures should also be focused on, so besides the ECM structures and composition, these vital architectures should also be maintained; (viii) the immunomodulatory of dECM and their degradation products, induced by the remaining physiological motifs and bioactive receptors, has been proved, though the exact mechanism in still unclear and needs further investigation [[327], [328], [329]]; (ix) the optimization for recellularization and cultivation within dECM scaffolds should be further developed, since they all affect the functional integration of dECM scaffolds with the host and clinical success of tissue-engineered constructs; (x) surface modification can be added to dECM to provide biological cues that assist in tissue regeneration, such as mechanical modification and loading with factors or other bioactive molecules; (xi) the timing of biomaterial transplantation and experimental endpoints are needed to be standardized, facilitating the comparison of results between different study groups. These lingering challenges can be offered as the catalysts for future clinical applications once they can be further settled in detail.
Fig. 9.
Prospects and challenges of dECM applied in tissue engineering.
In conclusion, despite the shortcomings mentioned above, dECM is acknowledged as an ideal choice for immune-compatible organ/tissue alternatives. Besides, as a platform, dECM could provide a biomimetic microenvironment for integrating with stem cells and other bioactive materials, which is a promising inspiration for the design of future bioscaffolds as well as an optimized solution to problems in tissue engineering and regenerative therapies.
Declaration of competing interest
We declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted, which entitled “Decellularized Extracellular Matrix Scaffolds: Recent Trends and Emerging Strategies in Tissue Engineering” by Xuewei Zhang, Xi Chen*, Hua Hong, Rubei Hu, Jiashang Liu and Changsheng Liu*.
Acknowledgments
This work was primarily leveraged by National Natural Science Foundation of China for Innovative Research Groups (No. 51621002), National Key R&D Program of China (No. 2018YFC1105700), and the Fundamental Research Funds for the Central Universities (No. JKVD12001002, JKD01211612). Any opinions, findings, conclusions, or recommendations expressed herein are those of the author(s).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Contributor Information
Xi Chen, Email: chenxi@ecust.edu.cn.
Changsheng Liu, Email: liucs@ecust.edu.cn.
References
- 1.Hinderer S., Layland S.L., Schenke-Layland K. Adv. Drug Deliv. Rev. 2016;97:260–269. doi: 10.1016/j.addr.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Chen F.M., Liu X.H. Prog. Polym. Sci. 2016;53:86–168. doi: 10.1016/j.progpolymsci.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu M., Zeng X., Ma C., Yi H., Ali Z., Mou X.B., Li S., Deng Y., He N.Y. Bone Res. 2017;5 doi: 10.1038/boneres.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourgine P.E., Gaudiello E., Pippenger B., Jaquiery C., Klein T., Pigeot S., Todorov A., Jr., Feliciano S., Banfi A., Martin I. Adv. Funct. Mater. 2017;27 [Google Scholar]
- 5.Duisit J., Amiel H., Wuthrich T., Taddeo A., Dedriche A., Destoop V., Pardoen T., Bouzin C., Joris V., Magee D., Vogelin E., Harriman D., Dessy C., Orlando G., Behets C., Rieben R., Gianello P., Lengele B. Acta Biomater. 2018;73:339–354. doi: 10.1016/j.actbio.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 6.Hoshiba T. Materials. 2019;12:1311. doi: 10.3390/ma12081311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhandayuthapani B., Yoshida Y., Maekawa T., Kumar D.S. Int. J. Polym. Sci. 2011 doi: 10.1155/2011/290602. 290602. [DOI] [Google Scholar]
- 8.Song J.J., Ott H.C. Trends Mol. Med. 2011;17:424–432. doi: 10.1016/j.molmed.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Ramaswamy A.K., Vorp D.A., Weinbaum J.S. Front. Cardiovasc. Med. 2019;6 doi: 10.3389/fcvm.2019.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catoira M.C., Fusaro L., Di Francesco D., Ramella M., Boccafoschi F. J. Mater. Sci. Mater. Med. 2019;30 doi: 10.1007/s10856-019-6318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poel W.E. Science. 1948;108:390. doi: 10.1126/science.108.2806.390-a. [DOI] [PubMed] [Google Scholar]
- 12.Hjelle J.T., Carlson E.C., Brendel K., Meezan E. Kidney Int. 1979;15:20–32. doi: 10.1038/ki.1979.3. [DOI] [PubMed] [Google Scholar]
- 13.Badylak S.F., Tullius R., Kokini K., Shelbourne K.D., Klootwyk T., Voytik S.L., Kraine M.R., Simmons C. J. Biomed. Mater. Res. 1995;29:977–985. doi: 10.1002/jbm.820290809. [DOI] [PubMed] [Google Scholar]
- 14.Ott H.C., Matthiesen T.S., Goh S.-K., Black L.D., Kren S.M., Netoff T.I., Taylor D.A. Nat. Med. 2008;14:213–221. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- 15.Ott H.C., Clippinger B., Conrad C., Schuetz C., Pomerantseva I., Ikonomou L., Kotton D., Vacanti J.P. Nat. Med. 2010;16:927–933. doi: 10.1038/nm.2193. [DOI] [PubMed] [Google Scholar]
- 16.Petersen T.H., Calle E.A., Zhao L.P., Lee E.J., Gui L.Q., Raredon M.B., Gavrilov K., Yi T., Zhuang Z.W., Breuer C., Herzog E., Niklason L.E. Science. 2010;329:538–541. doi: 10.1126/science.1189345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uygun B.E., Soto-Gutierrez A., Yagi H., Izamis M.L., Guzzardi M.A., Shulman C., Milwid J., Kobayashi N., Tilles A., Berthiaume F., Hertl M., Nahmias Y., Yarmush M.L., Uygun K. Nat. Med. 2010;16:814–820. doi: 10.1038/nm.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song J.J., Guyette J.P., Gilpin S.E., Gonzalez G., Vacanti J.P., Ott H.C. Nat. Med. 2013;19:646–651. doi: 10.1038/nm.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saldin L.T., Cramer M.C., Velankar S.S., White L.J., Badylak S.F. Acta Biomater. 2017;49:1–15. doi: 10.1016/j.actbio.2016.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mandrycky C., Wang Z.J., Kim K., Kim D.H. Biotechnol. Adv. 2016;34:422–434. doi: 10.1016/j.biotechadv.2015.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garreta E., Oria R., Tarantino C., Pla-Roca M., Prado P., Fernández-Avilés F., Campistol J.M., Samitier J., Montserrat N. Mater. Today. 2017;20:166–178. [Google Scholar]
- 22.Pati F., Jang J., Ha D.-H., Won Kim S., Rhie J.-W., Shim J.-H., Kim D.-H., Cho D.-W. Nat. Commun. 2014;5:3935. doi: 10.1038/ncomms4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jang J., Park H.-J., Kim S.-W., Kim H., Park J.Y., Na S.J., Kim H.J., Park M.N., Choi S.H., Park S.H., Kim S.W., Kwon S.-M., Kim P.-J., Cho D.-W. Biomaterials. 2017;112:264–274. doi: 10.1016/j.biomaterials.2016.10.026. [DOI] [PubMed] [Google Scholar]
- 24.Asthana A., Tamburrini R., Chaimov D., Gazia C., Walker S.J., Van Dyke M., Tomei A., Lablanche S., Robertson J., Opara E.C., Soker S., Orlando G. Biomaterials. 2021;270:120613. doi: 10.1016/j.biomaterials.2020.120613. [DOI] [PubMed] [Google Scholar]
- 25.Rana D., Zreiqat H., Benkirane-Jessel N., Ramakrishna S., Ramalingam M. J. Tissue Eng. Regen. Med. 2017;11:942–965. doi: 10.1002/term.2061. [DOI] [PubMed] [Google Scholar]
- 26.Porzionato A., Stocco E., Barbon S., Grandi F., Macchi V., De Caro R. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19124117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.K L., C N., M G., A L., G T., M A., Z M., G K., K Z., C Z. Med. Case Rep. 2018 04. [Google Scholar]
- 28.Ullah I., Busch J.F., Rabien A., Erguen B., Stamm C., Knosalla C., Hippenstiel S., Reinke P., Kurtz A. Adv. Sci. 2020;7 doi: 10.1002/advs.201901198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Safari F., Fani N., Eglin D., Alini M., Stoddart M.J., Eslaminejad M.B. J. Biomed. Mater. Res. 2019;107:1793–1802. doi: 10.1002/jbm.a.36698. [DOI] [PubMed] [Google Scholar]
- 30.Cheng C.W., Solorio L.D., Alsberg E. Biotechnol. Adv. 2014;32:462–484. doi: 10.1016/j.biotechadv.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma D., Ferguson M., Zhao F. In: Caballero D., Kundu S.C., Reis R.L., editors. vol. 156. Academic Press; 2020. pp. 3–13. (Methods in Cell Biology). [Google Scholar]
- 32.Assunção M., Dehghan-Baniani D., Yiu C.H.K., Später T., Beyer S., Blocki A. Front. Bioeng. Biotechnol. 2020;8 doi: 10.3389/fbioe.2020.602009. 602009-602009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoshiba T. J. Mater. Chem. B. 2017;5:4322–4331. doi: 10.1039/c7tb00074j. [DOI] [PubMed] [Google Scholar]
- 34.Kumar A., Nune K.C., Misra R.D.K. J. Biomed. Mater. Res. 2016;104:2751–2763. doi: 10.1002/jbm.a.35809. [DOI] [PubMed] [Google Scholar]
- 35.Kumar A., Nune K.C., Misra R.D.K. J. Biomed. Mater. Res. 2016;104:1343–1351. doi: 10.1002/jbm.a.35664. [DOI] [PubMed] [Google Scholar]
- 36.Kim B., Ventura R., Lee B.-T. J. Tissue Eng. Regen. Med. 2018;12:E1256–E1267. doi: 10.1002/term.2529. [DOI] [PubMed] [Google Scholar]
- 37.Gupta S.K., Mishra N.C., Dhasmana A. Methods Mol. Biol. 2018;1577:1–10. doi: 10.1007/7651_2017_34. [DOI] [PubMed] [Google Scholar]
- 38.Paulo Zambon J., Atala A., Yoo J.J. Methods (San Diego, Calif.) 2019;171:3–10. doi: 10.1016/j.ymeth.2019.09.016. [DOI] [PubMed] [Google Scholar]
- 39.Philips C., Campos F., Roosens A., Sánchez-Quevedo M.D.C., Declercq H., Carriel V. Ann. Biomed. Eng. 2018;46:1921–1937. doi: 10.1007/s10439-018-2082-y. [DOI] [PubMed] [Google Scholar]
- 40.Mendibil U., Ruiz-Hernandez R., Retegi-Carrion S., Garcia-Urquia N., Olalde-Graells B., Abarrategi A. Int. J. Mol. Sci. 2020;21:5447. doi: 10.3390/ijms21155447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu X., Cai Y., Xia C., Wu H., Li Q., Xu Z., Lu F. Artif. Organs. 2019;43:1162–1169. doi: 10.1111/aor.13519. [DOI] [PubMed] [Google Scholar]
- 42.Aeberhard P.-A., Grognuz A., Peneveyre C., McCallin S., Hirt-Burri N., Antons J., Pioletti D., Raffoul W., Applegate L.A. Artif. Organs. 2019;44:E161–E171. doi: 10.1111/aor.13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roth S.P., Glauche S.M., Plenge A., Erbe I., Heller S., Burk J. BMC Biotechnol. 2017;17:13. doi: 10.1186/s12896-017-0329-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng J., Wang C., Gu Y. Bio Med. Mater. Eng. 2019;30:191–205. doi: 10.3233/BME-191044. [DOI] [PubMed] [Google Scholar]
- 45.Dang Y., Waxman S., Wang C., Jensen A., Loewen R.T., Bilonick R.A., Loewen N.A. PeerJ. 2017;5:e3629. doi: 10.7717/peerj.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plant C.D., Plant G.W. Methods Mol. Biol. 2018;1739:455–466. doi: 10.1007/978-1-4939-7649-2_30. [DOI] [PubMed] [Google Scholar]
- 47.Goh S.-K., Bertera S., Vaidya V., Dumpe S., Barner S., Mathew S., Banerjee I. Technology. 2018;6:118–134. [Google Scholar]
- 48.Nara S., Chameettachal S., Midha S., Murab S., Ghosh S. RSC Adv. 2016;6:2225–2240. [Google Scholar]
- 49.Wang Z., Wang Z., Yu Q., Xi H., Weng J., Du X., Chen D., Ma J., Mei J., Chen C. J. Biomed. Mater. Res. 2016;104:2567–2575. doi: 10.1002/jbm.a.35794. [DOI] [PubMed] [Google Scholar]
- 50.Struecker B., Butter A., Hillebrandt K., Polenz D., Reutzel-Selke A., Tang P., Lippert S., Leder A., Rohn S., Geisel D., Denecke T., Aliyev K., Joehrens K., Raschzok N., Neuhaus P., Pratschke J., Sauer I.M. J. Tissue Eng. Regen. Med. 2017;11:531–541. doi: 10.1002/term.1948. [DOI] [PubMed] [Google Scholar]
- 51.Willemse J., Verstegen M.M.A., Vermeulen A., Schurink I.J., Roest H.P., van der Laan L.J.W., de Jonge J. Mater. Sci. Eng. C-Mater. Biol. Appl. 2020;108 doi: 10.1016/j.msec.2019.110200. [DOI] [PubMed] [Google Scholar]
- 52.Mesina M., Mindrila I., Mesina C., Obleaga C.V., Istratoaie O. J. Mind Med. Sci. 2019;6:137–142. [Google Scholar]
- 53.Elebring E., Kuna V.K., Kvarnstroem N., Sumitran-Holgersson S. J. Tissue Eng. 2017;8 doi: 10.1177/2041731417738145. 2041731417738145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seetapun D., Ross J.J. Surgery. 2017;161:1474–1478. doi: 10.1016/j.surg.2016.09.041. [DOI] [PubMed] [Google Scholar]
- 55.Park S.M., Yang S., Rye S.-M., Choi S.W. Biomed. Eng. Online. 2018;17:15. doi: 10.1186/s12938-018-0445-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simsa R., Vila X.M., Salzer E., Teuschl A., Jenndahl L., Bergh N., Fogelstrand P. PloS One. 2019;14 doi: 10.1371/journal.pone.0220743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Verstegen M.M.A., Willemse J., van den Hoek S., Kremers G.-J., Luider T.M., van Huizen N.A., Willemssen F.E.J.A., Metselaar H.J., Ijzermans J.N.M., van der Laan L.J.W., de Jonge J. Stem Cell. Dev. 2017;26:1304–1315. doi: 10.1089/scd.2017.0095. [DOI] [PubMed] [Google Scholar]
- 58.Kajbafzadeh A.-M., Khorramirouz R., Nabavizadeh B., Seyedian S.-S.L., Akbarzadeh A., Heidari R., Masoumi A., Azizi B., Beigi R.S.H. Mater. Sci. Eng. C-Mater. Biol. Appl. 2019;98:392–400. doi: 10.1016/j.msec.2019.01.018. [DOI] [PubMed] [Google Scholar]
- 59.Schilling B.K., Lamberti K.K., Snowden M.J., Baker J.S., Byrd K., Komatsu C., Solari M.G., Marra K.G. Tissue Eng. 2019;26:253–264. doi: 10.1089/ten.tea.2019.0209. [DOI] [PubMed] [Google Scholar]
- 60.Matuska A.M., McFetridge P.S. J. Biomed. Mater. Res. B Appl. Biomater. 2018;106:1858–1868. doi: 10.1002/jbm.b.33997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Farag A., Hashimi S.M., Vaquette C., Volpato F.Z., Hutmacher D.W., Ivanovski S. Arch. Oral Biol. 2018;88:67–76. doi: 10.1016/j.archoralbio.2018.01.014. [DOI] [PubMed] [Google Scholar]
- 62.Duisit J., Amiel H., Orlando G., Dedriche A., Behets C., Gianello P., Lengele B. Plast. Reconstr. Surg. 2018;141:95–103. doi: 10.1097/PRS.0000000000003910. [DOI] [PubMed] [Google Scholar]
- 63.Schiavo Matias G.d.S., Rigoglio N.N., Oliveira Carreira A.C., Romagnolli P., Nunes Barreto R.d.S., Mess A.M., Miglino M.A., Fratini P. Cells Tissues Organs. 2018;205:217–225. doi: 10.1159/000492466. [DOI] [PubMed] [Google Scholar]
- 64.Alshaikh A.B., Padma A.M., Dehlin M., Akouri R., Song M.J., Brannstrom M., Hellstrom M. J. Ovarian Res. 2019;12:58. doi: 10.1186/s13048-019-0531-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.My Thi Ngoc N., Ha Le Bao T. Cells Tissues Organs. 2018;206:296–307. doi: 10.1159/000501807. [DOI] [PubMed] [Google Scholar]
- 66.Tchoukalova Y.D., Hintze J.M., Hayden R.E., Lott D.G. Int. J. Artif. Organs. 2017;41:100–107. doi: 10.5301/ijao.5000648. [DOI] [PubMed] [Google Scholar]
- 67.Wilson S.L., Sidney L.E., Dunphy S.E., Dua H.S., Hopkinson A. Curr. Eye Res. 2016;41:769–782. doi: 10.3109/02713683.2015.1062114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Monibi F.A., Bozynski C.C., Kuroki K., Stoker A.M., Pfeiffer F.M., Sherman S.L., Cook J.L. Tissue Eng. C Methods. 2016;22:1059–1070. doi: 10.1089/ten.TEC.2016.0276. [DOI] [PubMed] [Google Scholar]
- 69.Qu J., Van Hogezand R.M., Zhao C., Kuo B.J., Carlsen B.T. Ann. Plast. Surg. 2015;75:112–116. doi: 10.1097/SAP.0000000000000157. [DOI] [PubMed] [Google Scholar]
- 70.Guimaraes A.B., Correia A.T., Alves B.P., Da Silva R.S., Martins J.K., Pego-Fernandes P.M., Xavier N.S., Dolhnikoff M., Cardoso P.F.G. Transplant. Proc. 2019;51:1611–1613. doi: 10.1016/j.transproceed.2019.01.042. [DOI] [PubMed] [Google Scholar]
- 71.Zhang J., Hu Z.Q., Turner N.J., Teng S.F., Cheng W.Y., Zhou H.Y., Zhang L., Hu H.W., Wang Q., Badylak S.F. Biomaterials. 2016;89:114–126. doi: 10.1016/j.biomaterials.2016.02.040. [DOI] [PubMed] [Google Scholar]
- 72.Roth S.P., Erbe I., Burk J. In: Decellularized Scaffolds and Organogenesis: Methods and Protocols. Turksen K., editor. Springer New York; New York, NY: 2018. pp. 227–237. [DOI] [Google Scholar]
- 73.Song M., Liu Y., Hui L. Mol. Med. Rep. 2018;17:138–146. doi: 10.3892/mmr.2017.7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khosroshahi A.F., Rad J.S., Kheirjou R., Roshangar B., Rashtbar M., Salehi R., Ranjkesh M.R., Roshangar L. J. Cell. Physiol. 2020;235:1556–1567. doi: 10.1002/jcp.29074. [DOI] [PubMed] [Google Scholar]
- 75.Rahman S., Griffin M., Naik A., Szarko M., Butler P.E.M. Sci. Rep. 2018;8:3097. doi: 10.1038/s41598-018-20592-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pan J., Yan S., Gao J.-j., Wang Y.-y., Lu Z.-j., Cui C.-w., Zhang Y.-h., Wang Y., Meng X.-q., Zhou L., Xie H.-y., Zheng J., Zheng M.H., Zheng S.-s. Int. J. Biochem. Cell Biol. 2016;80:124–131. doi: 10.1016/j.biocel.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 77.Watanabe M., Yano K., Okawa K., Yamashita T., Tajima K., Sawada K., Yagi H., Kitagawa Y., Tanishita K., Sudo R. Acta Biomater. 2019;95:307–318. doi: 10.1016/j.actbio.2018.12.042. [DOI] [PubMed] [Google Scholar]
- 78.Kojima H., Yasuchika K., Fukumitsu K., Ishii T., Ogiso S., Miyauchi Y., Yamaoka R., Kawai T., Katayama H., Yoshitoshi-Uebayashi E.Y., Kita S., Yasuda K., Sasaki N., Komori J., Uemoto S. Am. J. Transplant. 2018;18:1351–1359. doi: 10.1111/ajt.14666. [DOI] [PubMed] [Google Scholar]
- 79.Kelly da Palma R., Naomi Nonaka P., Campillo N., Uriarte J.J., Urbano J.J., Navajas D., Farre R., Oliveira L.V.F. J. Biomech. 2016;49:1230–1232. doi: 10.1016/j.jbiomech.2016.02.043. [DOI] [PubMed] [Google Scholar]
- 80.Doryab A., Heydarian M., Amoabediny G., Sadroddiny E., Mahfouzi S. J. Med. Biol. Eng. 2017;37:53–62. [Google Scholar]
- 81.Nguyen D.T., O'Hara M., Graneli C., Hicks R., Miliotis T., Nystrom A.-C., Hansson S., Davidsson P., Gan L.-M., Magnone M.C., Althage M., Heydarkhan-Hagvall S. Sci. Rep. 2018;8:7458. doi: 10.1038/s41598-018-25883-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saghizadeh M., Winkler M.A., Kramerov A.A., Hemmati D.M., Ghiam C.A., Dimitrijevich S.D., Sareen D., Ornelas L., Ghiasi H., Brunken W.J., Maguen E., Rabinowitz Y.S., Svendsen C.N., Jirsova K., Ljubimov A.V. PloS One. 2013;8 doi: 10.1371/journal.pone.0079632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Forouzesh F., Rabbani M., Bonakdar S. J Med Signals Sens. 2019;9:227–233. doi: 10.4103/jmss.JMSS_64_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yusof F., Sha'Ban M., Azhim A. Int. J. Nanomed. 2019;14:5491–5502. doi: 10.2147/IJN.S207270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lin C.-H., Hsia K., Su C.-K., Chen C.-C., Yeh C.-C., Ma H., Lu J.-H. Polymers. 2021;13:1699. doi: 10.3390/polym13111699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang Y.-H., Tseng F.-W., Chang W.-H., Peng I.C., Hsieh D.-J., Wu S.-W., Yeh M.-L. Acta Biomater. 2017;58:238–243. doi: 10.1016/j.actbio.2017.05.060. [DOI] [PubMed] [Google Scholar]
- 87.Gil-Ramirez A., Spangenberg A., Spegel P., Rodriguez-Meizoso I. J. Co2 Util. 2019;34:700–708. [Google Scholar]
- 88.Gil-Ramirez A., Rosmark O., Spegel P., Sward K., Westergren-Thorsson G., Larsson-Callerfelt A.-K., Rodriguez-Meizoso I. Sci. Rep. 2020;10:4031. doi: 10.1038/s41598-020-60827-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Buse E.E., Hilbert S.L., Hopkins R.A., Converse G.L. Cardiovas. Eng. Technol. 2016;7:352–362. doi: 10.1007/s13239-016-0275-9. [DOI] [PubMed] [Google Scholar]
- 90.Park C.S., Kim Y.J., Lee J.R., Lim H.-G., Chang J.-E., Jeong S., Kwon N. Interact. Cardiovasc. Thorac. Surg. 2017;25:391–399. doi: 10.1093/icvts/ivx131. [DOI] [PubMed] [Google Scholar]
- 91.Taylor D.A., Sampaio L.C., Cabello R., Elgalad A., Parikh R., Wood R.P., Myer K.A., Yeh A.T., Lee P.-F. Jove-J. Vis. Exp. 2018;26 doi: 10.3791/58123. [DOI] [PubMed] [Google Scholar]
- 92.Reimer J.M., Syedain Z.H., Haynie B.H.T., Tranquillo R.T. Biomaterials. 2015;62:88–94. doi: 10.1016/j.biomaterials.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sierad L.N., Shaw E.L., Bina A., Brazile B., Rierson N., Patnaik S.S., Kennamer A., Odum R., Cotoi O., Terezia P., Branzaniuc K., Smallwood H., Deac R., Egyed I., Pavai Z., Szanto A., Harceaga L., Suciu H., Raicea V., Olah P., Simionescu A., Liao J., Movileanu I., Harpa M., Simionescu D.T. Tissue Eng. C Methods. 2015;21:1284–1296. doi: 10.1089/ten.tec.2015.0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Morimoto N., Mahara A., Jinno C., Ogawa M., Kakudo N., Suzuki S., Fujisato T., Kusumoto K., Yamaoka T. J. Biomed. Mater. Res. B Appl. Biomater. 2017;105:2653–2661. doi: 10.1002/jbm.b.33807. [DOI] [PubMed] [Google Scholar]
- 95.Shi W., Zhou Q., Gao H., Li S., Dong M., Wang T., Jia Y., Dong C., Wang X., Guo Z., Zhao L., Hu X., Xie L. Adv. Funct. Mater. 2019;29 [Google Scholar]
- 96.Golberg A., Bruinsma B.G., Jaramillo M., Yarmush M.L., Uygun B.E. PeerJ. 2016;4:e1571. doi: 10.7717/peerj.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Saini A., Breen I., Alzubaidi S., Pershad Y., Sheth R., Naidu S., Knuttinen M.G., Albadawi H., Oklu R. J. Clin. Med. 2019;8:22. doi: 10.3390/jcm8010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chang T.T., Zhou V.X., Rubinsky B. Biotechniques. 2017;62:229–231. doi: 10.2144/000114547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zager Y., Kain D., Landa N., Leor J., Maor E. PloS One. 2016;11 doi: 10.1371/journal.pone.0165475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McCrary M.W., Vaughn N.E., Hlavac N., Song Y.H., Wachs R.A., Schmidt C.E. Tissue Eng. C Methods. 2020;26:23–36. doi: 10.1089/ten.TEC.2019.0135. [DOI] [PubMed] [Google Scholar]
- 101.Dong M., Zhao L., Wang F., Hu X., Li H., Liu T., Zhou Q., Shi W. J. Tissue Eng. 2019;10 doi: 10.1177/2041731419875876. 2041731419875876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hassanpour A., Talaei-Khozani T., Kargar-Abarghouei E., Razban V., Vojdani Z. Stem Cell Res. Ther. 2018;9:252. doi: 10.1186/s13287-018-0971-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xing Q., Yates K., Tahtinen M., Shearier E., Qian Z., Zhao F. Tissue Eng. C Methods. 2015;21:77–87. doi: 10.1089/ten.tec.2013.0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li N., Li Y., Gong D., Xia C., Liu X., Xu Z. Interact. Cardiovasc. Thorac. Surg. 2018;26:768–776. doi: 10.1093/icvts/ivx416. [DOI] [PubMed] [Google Scholar]
- 105.Schneider C., Lehmann J., van Osch G.J.V.M., Hildner F., Teuschl A., Monforte X., Miosga D., Heimel P., Priglinger E., Redl H., Wolbank S., Nuernberger S. Tissue Eng. C Methods. 2016;22:1095–1107. doi: 10.1089/ten.TEC.2016.0380. [DOI] [PubMed] [Google Scholar]
- 106.Fernandez-Perez J., Ahearne M. Sci. Rep. 2019;9:14933. doi: 10.1038/s41598-019-49575-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li M., Zhang T., Jiang J., Mao Y., Zhang A., Zhao J. Mater. Sci. Eng. C-Mater. Biol. Appl. 2019;105:110039. doi: 10.1016/j.msec.2019.110039. [DOI] [PubMed] [Google Scholar]
- 108.Burk J., Erbe I., Berner D., Kacza J., Kasper C., Pfeiffer B., Winter K., Brehm W. Tissue Eng. C Methods. 2014;20:276–284. doi: 10.1089/ten.tec.2012.0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Poornejad N., Schaumann L.B., Buckmiller E.M., Momtahan N., Gassman J.R., Ma H.H., Roeder B.L., Reynolds P.R., Cook A.D. J. Biomater. Appl. 2016;31:521–533. doi: 10.1177/0885328216656099. [DOI] [PubMed] [Google Scholar]
- 110.Yamanaka H., Morimoto N., Yamaoka T. J. Artif. Organs : Off. J. Jpn. Soc. Artificial Org. 2020;23:156–162. doi: 10.1007/s10047-019-01152-0. [DOI] [PubMed] [Google Scholar]
- 111.Hsieh C.-M., Wang W., Chen Y.-H., Wei P.-S., Liu Y.-H., Sheu M.-T., Ho H.-O. Pharmaceutics. 2020;12:538. doi: 10.3390/pharmaceutics12060538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cornelison R.C., Wellman S.M., Park J.H., Porvasnik S.L., Song Y.H., Wachs R.A., Schmidt C.E. Acta Biomater. 2018;77:116–126. doi: 10.1016/j.actbio.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim J.K., Koh Y.-D., Kim J.O., Seo D.H. J. Plast. Reconstr. Aesthetic Surg. 2016;69:1690–1696. doi: 10.1016/j.bjps.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 114.Abedin E., Lari R., Shahri N.M., Fereidoni M. Tissue Cell. 2018;55:46–52. doi: 10.1016/j.tice.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 115.Sreypich S., Dugos N., Roces S., Mondragon J.M. MATEC Web Confer. 2019;268 [Google Scholar]
- 116.Roth S.P., Brehm W., Gross C., Scheibe P., Schubert S., Burk J. Int. J. Mol. Sci. 2019;20:5474. doi: 10.3390/ijms20215474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shin Y.H., Park S.Y., Kim J.K. J. Tissue Eng. Regen. Med. 2019;13:1241–1252. doi: 10.1002/term.2874. [DOI] [PubMed] [Google Scholar]
- 118.Rosmark O., Åhrman E., Müller C., Rendin L., Eriksson L., Malmström A., Hallgren O., Larsson-Callerfelt A.-K., Westergren-Thorsson G., Malmström J. Sci. Rep. 2018;8:5409. doi: 10.1038/s41598-018-23702-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rendin L., Lofdahl A., Muller C., Isaksson H., Bjermer L., Ahrman E., Malmstrom J., Westergren-Thorsson G. Transpl. Int. 2019;20:4013. [Google Scholar]
- 120.Simsa R., Padma A.M., Heher P., Hellstrom M., Teuschl A., Jenndahl L., Bergh N., Fogelstrand P. PloS One. 2018;13 doi: 10.1371/journal.pone.0209269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ghim J.H., Hussein K.H., Park K.-M., Woo H.M. Biomed. Eng. Lett. 2015;5:58–64. [Google Scholar]
- 122.Jones G., Herbert A., Berry H., Edwards J.H., Fisher J., Ingham E. Tissue Eng. 2017;23:124–134. doi: 10.1089/ten.tea.2016.0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mohiuddin O.A., Campbell B., Poche J.N., Thomas-Porch C., Hayes D.A., Bunnell B.A., Gimble J.M. Adv. Exp. Med. Biol. 2020;1212:57–70. doi: 10.1007/5584_2019_371. [DOI] [PubMed] [Google Scholar]
- 124.Zhang S., Lu Q., Cao T., Toh W.S. Plast. Reconstr. Surg. 2016;137:1171–1180. doi: 10.1097/PRS.0000000000002019. [DOI] [PubMed] [Google Scholar]
- 125.Paduano F., Marrelli M., White L.J., Shakesheff K.M., Tatullo M. PloS One. 2016;11 doi: 10.1371/journal.pone.0148225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Phan N.V., Wright T., Rahman M.M., Xu J., Coburn J.M. ACS Biomater. Sci. Eng. 2020;6:822–832. doi: 10.1021/acsbiomaterials.9b00870. [DOI] [PubMed] [Google Scholar]
- 127.Su M., Zhang Q., Zhu Y., Wang S., Lv J., Sun J., Qiu P., Fan S., Jin K., Chen L., Lin X. Adv. Healthcare Mater. 2019;8 doi: 10.1002/adhm.201900831. [DOI] [PubMed] [Google Scholar]
- 128.Ahmed E., Saleh T., Yu L., Kwak H.-H., Kim B.-M., Park K.-M., Lee Y.-S., Kang B.-J., Choi K.-Y., Kang K.-S., Woo H.M. J. Biosci. Bioeng. 2019;128:218–225. doi: 10.1016/j.jbiosc.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 129.Yam G.H.-F., Yusoff N.Z.B.M., Goh T.-W., Setiawan M., Lee X.-W., Liu Y.-C., Mehta J.S. Sci. Rep. 2016;6 doi: 10.1038/srep26339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li J., Cai Z., Cheng J., Wang C., Fang Z., Xiao Y., Feng Z.-G., Gu Y. J. Biomater. Sci. Polym. Ed. 2020;31:999–1023. doi: 10.1080/09205063.2020.1736741. [DOI] [PubMed] [Google Scholar]
- 131.Ramm R., Goecke T., Theodoridis K., Hoeffler K., Sarikouch S., Findeisen K., Ciubotaru A., Cebotari S., Tudorache I., Haverich A., Hilfiker A. Xenotransplantation. 2019;27 doi: 10.1111/xen.12571. [DOI] [PubMed] [Google Scholar]
- 132.Giraldo-Gomez D.M., Julieta Garcia-Lopez S., Tamay-de-Dios L., Sanchez-Sanchez R., Villalba-Caloca J., Sotres-Vega A., Luisa Del Prado-Audelo M., Gomez-Lizarraga K.K., Garciadiego-Cazares D., Cristina Pina-Barba M. Mater. Sci. Eng. C-Mater. Biol. Appl. 2019;105 [Google Scholar]
- 133.Lin C.-H., Kao Y.-C., Ma H., Tsay R.-Y. J. Mech. Behav. Biomed. Mater. 2018;86:199–207. doi: 10.1016/j.jmbbm.2018.06.029. [DOI] [PubMed] [Google Scholar]
- 134.Cornelison R.C., Wellman S.M., Park J.H., Porvasnikb S.L., Song Y.H., Wachs R.A., Schmidt C.E. Acta Biomater. 2018;77:116–126. doi: 10.1016/j.actbio.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gu Y., Wang F., Wang R., Li J., Wang C., Li L., Xu Z., Zhang J. Cell Tissue Bank. 2018;19:311–321. doi: 10.1007/s10561-017-9675-9. [DOI] [PubMed] [Google Scholar]
- 136.Lin C.-H., Kao Y.-C., Lin Y.-H., Ma H., Tsay R.-Y. Acta Biomater. 2016;46:101–111. doi: 10.1016/j.actbio.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 137.Van den Bogerd B., Ni Dhubhghaill S., Zakaria N. J. Tissue Eng. Regen. Med. 2018;12:E2020–E2028. doi: 10.1002/term.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Huh M.-I., Lee K.-P., Kim J., Yi S., Park B.-U., Kim H.K. J. Ophthalmol. 2018;2018 doi: 10.1155/2018/2590536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pesaraklou A., Mahdavi-Shahri N., Hassanzadeh H., Ghasemi M., Kazemi M., Mousavi N.S., Matin M.M. Biomed. Mater. 2019;14 doi: 10.1088/1748-605X/ab0679. [DOI] [PubMed] [Google Scholar]
- 140.Joszko K., Gzik-Zroska B., Kawlewska E., Klama-Baryla A., Suchon S., Burkacki M., Wolanski W. Acta Bioeng. Biomech. 2019;21:87–97. [PubMed] [Google Scholar]
- 141.Song M., Wang W., Ye Q., Bu S., Shen Z., Zhu Y. Mater. Sci. Eng. C-Mater. Biol. Appl. 2017;77:739–747. doi: 10.1016/j.msec.2017.03.232. [DOI] [PubMed] [Google Scholar]
- 142.Kuljanin M., Brown C.F.C., Raleigh M.J., Lajoie G.A., Flynn L.E. Biomaterials. 2017;144:130–143. doi: 10.1016/j.biomaterials.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 143.Isidan A., Liu S., Li P., Lashmet M., Smith L.J., Hara H., Cooper D.K.C., Ekser B. Xenotransplantation. 2019;26 doi: 10.1111/xen.12564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Walker S., Dittfeld C., Jakob A., Schonfelder J., Konig U., Tugtekin S.-M. Thorac. Cardiovasc. Surg. 2020 doi: 10.1055/s-0040-1705100. [DOI] [PubMed] [Google Scholar]
- 145.Nieto-Nicolau N., López-Chicón P., Fariñas O., Bolívar S., Udina E., Navarro X., Casaroli-Marano R., Vilarrodona A. Cell Tissue Res. 2021;384:167–177. doi: 10.1007/s00441-020-03317-3. [DOI] [PubMed] [Google Scholar]
- 146.Wu L.-C., Kuo Y.-J., Sun F.-W., Chen C.-H., Chiang C.-J., Weng P.-W., Tsuang Y.-H., Huang Y.-Y. Cell Tissue Bank. 2017;18:383–396. doi: 10.1007/s10561-017-9619-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Baert Y., Stukenborg J.B., Landreh M., De Kock J., Jornvall H., Soder O., Goossens E. Hum. Reprod. 2015;30:256–267. doi: 10.1093/humrep/deu330. [DOI] [PubMed] [Google Scholar]
- 148.Shi Q., Chen Y., Li M.Z., Zhang T., Ding S.L., Xu Y., Hu J.Z., Chen C., Lu H.B. Ann. Transl. Med. 2020;8 doi: 10.21037/atm-20-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang Z.H., Sun F., Lu Y., Zhang B.Y., Zhang G.Z., Shi H.C. ACS Omega. 2021;6:10637–10644. doi: 10.1021/acsomega.0c06247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rabbani M., Zakian N., Alimoradi N. J. Med. Sig. Sens. 2021;11:1–11. doi: 10.4103/jmss.JMSS_2_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Butler C.R., Hynds R.E., Crowley C., Gowers K.H.C., Partington L., Hamilton N.J., Carvalho C., Plate M., Samuel E.R., Burns A.J., Urbani L., Birchall M.A., Lowdell M.W., De Coppi P., Janes S.M. Biomaterials. 2017;124:95–105. doi: 10.1016/j.biomaterials.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Alizadeh M., Rezakhani L., Khodaei M., Soleimannejad M., Alizadeh A. J. Tissue Eng. Regen. Med. 2021;15:116–128. doi: 10.1002/term.3150. [DOI] [PubMed] [Google Scholar]
- 153.Alizadeh M., Rezakhani L., Soleimannejad M., Sharifi E., Anjomshoa M., Alizadeh A. Heliyon. 2019;5 doi: 10.1016/j.heliyon.2019.e02253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Song Y.H., Maynes M.A., Hlavac N., Visosevic D., Daramola K.O., Porvasnik S.L., Schmidt C.E. Biomater. Sci. 2021;9:3485–3498. doi: 10.1039/d1bm00032b. [DOI] [PubMed] [Google Scholar]
- 155.Novoseletskaya E., Grigorieva O., Nimiritsky P., Basalova N., Eremichev R., Milovskaya I., Kulebyakin K., Kulebyakina M., Rodionov S., Omelyanenko N., Efimenko A. Front. Cell Develop. Biol. 2020;8 doi: 10.3389/fcell.2020.555378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Goecke T., Theodoridis K., Tudorache I., Ciubotaru A., Cebotari S., Ramm R., Hoffler K., Sarikouch S., Rivera A.V., Haverich A., Wolkers W.F., Hilfiker A. Acta Biomater. 2018;68:41–52. doi: 10.1016/j.actbio.2017.11.041. [DOI] [PubMed] [Google Scholar]
- 157.Qiu S., Rao Z., He F., Wang T., Xu Y., Du Z., Yao Z., Lin T., Yan L., Quan D., Zhu Q., Liu X. J. Tissue Eng. Regen. Med. 2020;14:931–943. doi: 10.1002/term.3050. [DOI] [PubMed] [Google Scholar]
- 158.Mendes G.C.C., Brandão T.R.S., Silva C.L.M. Am. J. Infect. Contr. 2007;35:574–581. doi: 10.1016/j.ajic.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 159.Zhou C., Zhou L., Liu J., Xu L., Xu Z., Chen Z., Ge Y., Zhao F., Wu R., Wang X., Jiang N., Mao L., Jia R. Acta Biomater. 2020;115:250–263. doi: 10.1016/j.actbio.2020.07.056. [DOI] [PubMed] [Google Scholar]
- 160.Qing Q., Zhang Y.-J., Yang J.-L., Ning L.-J., Zhang Y.-J., Jiang Y.-L., Zhang Y., Luo J.-C., Qin T.-W. J. Biomed. Mater. Res. 2019;107:1476–1490. doi: 10.1002/jbm.a.36662. [DOI] [PubMed] [Google Scholar]
- 161.Wang W., Itoh S., Takakuda K. J. Biomed. Mater. Res. 2016;104:445–454. doi: 10.1002/jbm.a.35589. [DOI] [PubMed] [Google Scholar]
- 162.Bombelli S., Meregalli C., Scalia C., Bovo G., Torsello B., De Marco S., Cadamuro M., Vigano P., Strada G., Cattoretti G., Bianchi C., Perego R.A. Am. J. Pathol. 2018;188:184–195. doi: 10.1016/j.ajpath.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 163.Clapp P.A., Davies M.J., French M.S., Gilbert B.C. Free Radic. Res. 1994;21:147–167. doi: 10.3109/10715769409056566. [DOI] [PubMed] [Google Scholar]
- 164.Zvarova B., Uhl F.E., Uriarte J.J., Borg Z.D., Coffey A.L., Bonenfant N.R., Weiss D.J., Wagner D.E. Tissue Eng. C Methods. 2016;22:418–428. doi: 10.1089/ten.tec.2015.0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Moradi L., Jobania B.M., Jafarnezhad-Ansariha F., Ghorbani F., Esmaeil-Pour R., Zolbina M.M., Kajbafzadeh A.-M. Tissue Cell. 2020;66 doi: 10.1016/j.tice.2020.101396. [DOI] [PubMed] [Google Scholar]
- 166.Rowland C.R., Lennon D.P., Caplan A.I., Guilak F. Biomaterials. 2013;34:5802–5812. doi: 10.1016/j.biomaterials.2013.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gao S., Yuan Z., Guo W., Chen M., Liu S., Xi T., Guo Q. Mater. Sci. Eng. C-Mater. Biol. Appl. 2017;71:891–900. doi: 10.1016/j.msec.2016.10.074. [DOI] [PubMed] [Google Scholar]
- 168.Yu Y., Liu L., Zhang J., Wei Z., Mei J. Methods Mol. Biol. 2018;1577:111–119. doi: 10.1007/7651_2017_72. [DOI] [PubMed] [Google Scholar]
- 169.Browe D.C., Mahon O.R., Diaz-Payno P.J., Cassidy N., Dudurych I., Dunne A., Buckley C.T., Kelly D.J. J. Biomed. Mater. Res. 2019;107:2222–2234. doi: 10.1002/jbm.a.36731. [DOI] [PubMed] [Google Scholar]
- 170.Wu X., Wang Y., Wu Q., Li Y., Li L., Tang J., Shi Y., Bu H., Bao J., Xie M. Tissue Eng. Regen. Med. 2015;12:417–426. [Google Scholar]
- 171.Xing H., Yin H., Sun C., Ren X., Tian Y., Yu M., Jiang T. Mol. Med. Rep. 2019;20:1075–1084. doi: 10.3892/mmr.2019.10364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Eltom A., Zhong G., Muhammad A. Adv. Mater. Sci. Eng. 2019;2019:1–13. [Google Scholar]
- 173.Li M., Khadim R.R., Nagayama M., Shinohara M., Inamura K., Danoy M., Nishikawa M., Furukawa K., Sakai Y., Niino T. Appl. Sci. 2021;11:2473. [Google Scholar]
- 174.Li Y., Xu Y., Liu Y., Wang Z., Chen W., Duan L., Gu D. Mater. Sci. Eng. C. 2019;105 doi: 10.1016/j.msec.2019.110139. [DOI] [PubMed] [Google Scholar]
- 175.Goldberg-Bockhorn E., Schwarz S., Subedi R., Elsässer A., Riepl R., Walther P., Körber L., Breiter R., Stock K., Rotter N. Laser Med. Sci. 2018;33:375–384. doi: 10.1007/s10103-017-2402-8. [DOI] [PubMed] [Google Scholar]
- 176.Oh H.J., Kim S.H., Cho J.-H., Park S.-H., Min B.-H. Tissue Eng. Regen. Med. 2018;15:287–299. doi: 10.1007/s13770-018-0114-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Muller W.E.G., Wang S.F., Ackermann M., Gerich T., Neufurth M., Wiens M., Schroder H.C., Wang X.H. Adv. Funct. Mater. 2019;29 [Google Scholar]
- 178.Chen G.B., Lv Y.G. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-01938-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Aubin H., Mas-Moruno C., Iijima M., Schutterle N., Steinbrink M., Assmann A., Gil F.J., Lichtenberg A., Pegueroles M., Akhyari P. Tissue Eng. C Methods. 2016;22:496–508. doi: 10.1089/ten.tec.2015.0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yamanaka H., Yamaoka T., Mahara A., Morimoto N., Suzuki S. Biomaterials. 2018;179:156–163. doi: 10.1016/j.biomaterials.2018.06.022. [DOI] [PubMed] [Google Scholar]
- 181.Jia J., Jeon E.J., Li M., Richards D.J., Lee S., Jung Y., Barrs R.W., Coyle R., Li X., Chou J.C., Yost M.J., Gerecht S., Cho S.-W., Mei Y. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aaz5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wang C.Y., Liu Y., Fan Y.B., Li X.M. Regen. Biomater. 2017;4:191–206. doi: 10.1093/rb/rbx011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bras L.E.D., Frangogiannis N.G. Matrix Biol. 2020;91–92:176–187. doi: 10.1016/j.matbio.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yue B. J. Glaucoma. 2014;23:S20–S23. doi: 10.1097/IJG.0000000000000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hillebrandt K.H., Everwien H., Haep N., Keshi E., Pratschke J., Sauer I.M. Transpl. Int. 2019 doi: 10.1111/tri.13462. [DOI] [PubMed] [Google Scholar]
- 186.Kenry W., C Lee K., Loh P., Lim C.T. Biomaterials. 2018;155:236–250. doi: 10.1016/j.biomaterials.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 187.Neves J., Sousa-Victor P., Jasper H. Cell Stem Cell. 2017;20:161–175. doi: 10.1016/j.stem.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sart S., Yan Y., Li Y., Lochner E., Zeng C., Ma T., Li Y. Acta Biomater. 2016;30:222–232. doi: 10.1016/j.actbio.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 189.Hong X., Yuan Y., Sun X., Zhou M., Guo G., Zhang Q., Hescheler J., Xi J. Cell. Physiol. Biochem. 2018;45:319–331. doi: 10.1159/000486813. [DOI] [PubMed] [Google Scholar]
- 190.Sambi M., Chow T., Whiteley J., Li M.R., Chua S., Raileanu V., Rogers I.M. Stem Cell Reviews and Reports. 2017;13:513–531. doi: 10.1007/s12015-016-9712-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Schwan J., Kwaczala A.T., Ryan T.J., Bartulos O., Ren Y.M., Sewanan L.R., Morris A.H., Jacoby D.L., Qyang Y.B., Campbell S.G. Sci. Rep. 2016;6 doi: 10.1038/srep32068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yang J., Park J.W., Zheng D.J., Xu R.H. Transl. Vis. Sci. Technol. 2018;7:23. doi: 10.1167/tvst.7.5.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hong X., Yuan Y., Sun X.X., Zhou M.L., Guo G.Y., Zhang Q., Hescheler J., Xi J.Y. Cell. Physiol. Biochem. 2018;45:319–331. doi: 10.1159/000486813. [DOI] [PubMed] [Google Scholar]
- 194.Brouwer M., Zhou H.Q., Kasri N.N. Stem Cell Reviews and Reports. 2016;12:54–72. doi: 10.1007/s12015-015-9622-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Shi Y., Inoue H., Wu J.C., Yamanaka S. Nat. Rev. Drug Discov. 2017;16:115–130. doi: 10.1038/nrd.2016.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wang B., Jakus A.E., Baptista P.M., Soker S., Soto-Gutierrez A., Abecassis M.M., Shah R.N., Wertheim J.A. Stem Cells Transl. Med. 2016;5:1257–1267. doi: 10.5966/sctm.2015-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Schick R., Mekies L.N., Shemer Y., Eisen B., Hallas T., Ben Jehuda R., Ben-Ari M., Szantai A., Willi L., Shulman R., Gramlich M., Pane L.S., My I., Freimark D., Murgia M., Santamaria G., Gherghiceanu M., Arad M., Moretti A., Binah O. PloS One. 2018;13 doi: 10.1371/journal.pone.0205719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Montserrat N., Garreta E., Belmonte J.C.I. FEBS J. 2016;283:3303–3324. doi: 10.1111/febs.13704. [DOI] [PubMed] [Google Scholar]
- 199.Ghaedi M., Le A.V., Hatachi G., Beloiartsev A., Rocco K., Sivarapatna A., Mendez J.J., Baevova P., Dyal R.N., Leiby K.L., White E.S., Niklason L.E. J. Tissue Eng. Regen. Med. 2018;12:E1623–E1635. doi: 10.1002/term.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wan J., Huang Y., Zhou P.C., Guo Y.B., Wu C., Zhu S.J., Wang Y., Wang L., Lu Y.H., Wang Z.W. BioMed Res. Int. 2017;2017 doi: 10.1155/2017/4276928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Guyette J.P., Charest J.M., Mills R.W., Jank B.J., Moser P.T., Gilpin S.E., Gershlak J.R., Okamoto T., Gonzalez G., Milan D.J., Gaudette G.R., Ott H.C. Circ. Res. 2016;118:56–72. doi: 10.1161/CIRCRESAHA.115.306874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bhumiratana S., Bernhard J.C., Alfi D.M., Yeager K., Eton R.E., Bova J., Shah F., Gimble J.M., Lopez M.J., Eisig S.B., Vunjak-Novakovic G. Sci. Transl. Med. 2016;8 doi: 10.1126/scitranslmed.aad5904. 343ra383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Paduano F., Marrelli M., Alom N., Amer M., White L.J., Shakesheff K.M., Tatullo M. J. Biomater. Sci. Polym. Ed. 2017;28:730–748. doi: 10.1080/09205063.2017.1301770. [DOI] [PubMed] [Google Scholar]
- 204.Brouki Milan P., Lotfibakhshaiesh N., Joghataie M.T., Ai J., Pazouki A., Kaplan D.L., Kargozar S., Amini N., Hamblin M.R., Mozafari M., Samadikuchaksaraei A. Acta Biomater. 2016;45:234–246. doi: 10.1016/j.actbio.2016.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Gholipourmalekabadi M., Sameni M., Radenkovic D., Mozafari M., Mossahebi-Mohammadi M., Seifalian A. Cell Prolif. 2016;49:115–121. doi: 10.1111/cpr.12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lovati A.B., Bottagisio M., Moretti M. Stem Cell. Int. 2016;2016:7276150. doi: 10.1155/2016/7276150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kitahara H., Yagi H., Tajima K., Okamoto K., Yoshitake A., Aeba R., Kudo M., Kashima I., Kawaguchi S., Hirano A., Kasai M., Akamatsu Y., Oka H., Kitagawa Y., Shimizu H. Interact. Cardiovasc. Thorac. Surg. 2016;22:571–579. doi: 10.1093/icvts/ivw022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Yuan X., Wei Y., Villasante A., Ng J.J.D., Arkonac D.E., Chao P.-h. G., Vunjak-Novakovic G. Biomaterials. 2017;132:59–71. doi: 10.1016/j.biomaterials.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Song H.X., Yin Z., Wu T., Li Y.Z., Luo X., Xu M.Z., Duan L.H., Li J.H. Cell Transplant. 2018;27:1634–1643. doi: 10.1177/0963689718805383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Guo W., Xu W., Wang Z., Chen M., Hao C., Zheng X., Huang J., Xiang S., Yuan Z., Zhang Y. Stem Cell. Int. 2018;2018:1–10. [Google Scholar]
- 211.Wang X., Wang G., Sarah Z., Zhao B. vol. 24. 2018. (Tissue Engineering Part B Reviews). ten.TEB.2018.0012. [Google Scholar]
- 212.Yuan Z., Liu S., Hao C., Guo W., Gao S., Wang M., Chen M., Sun Z., Xu Y., Wang Y. Biomaterials. 2016;111:13–26. doi: 10.1016/j.biomaterials.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 213.Clark R., Ghosh K., Tonnesen M. J. Invest. Dermatol. 2007;127:1018–1029. doi: 10.1038/sj.jid.5700715. [DOI] [PubMed] [Google Scholar]
- 214.Zhang Q., Johnson J.A., Dunne L.W., Chen Y., Iyyanki T., Wu Y., Chang E.I., Branch-Brooks C.D., Robb G.L., Butler C.E. Acta Biomater. 2016;35:166–184. doi: 10.1016/j.actbio.2016.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Choi J.S., Kim J.D., Yoon H.S., Cho Y.W. Tissue Eng. 2013;19:329–339. doi: 10.1089/ten.tea.2011.0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Brouki Milan P., Pazouki A., Joghataei M.T., Mozafari M., Amini N., Kargozar S., Amoupour M., Latifi N., Samadikuchaksaraei A. Methods. 2020;171:62–67. doi: 10.1016/j.ymeth.2019.07.005. [DOI] [PubMed] [Google Scholar]
- 217.Takami Y., Yamaguchi R., Ono S., Hyakusoku H. J. Nippon Med. Sch. 2014;81:356–363. doi: 10.1272/jnms.81.356. [DOI] [PubMed] [Google Scholar]
- 218.Sotnichenko A.S., Gilevich I.V., Melkonian K.I., Yutskevich Y.A., Karakulev A.V., Bogdanov S.B., Bykov I.M., Redko A.N., Porhanov V.A., Alekseenko S.N. Vestnik Transplantologii I Iskusstvennyh Organov. 2019;21:81–87. [Google Scholar]
- 219.Zhang X., Yang J., Li Y., Liu S., Long K., Zhao Q., Zhang Y., Deng Z., Jin Y. Tissue Eng. C Methods. 2011;17:423–433. doi: 10.1089/ten.TEC.2010.0466. [DOI] [PubMed] [Google Scholar]
- 220.Nyame T.T., Chiang H.A., Leavitt T., Ozambela M., Orgill D.P. Plast. Reconstr. Surg. 2015;136:1379–1388. doi: 10.1097/PRS.0000000000001748. [DOI] [PubMed] [Google Scholar]
- 221.Groeber F., Engelhardt L., Lange J., Kurdyn S., Schmid F.F., Ruecker C., Mielke S., Walles H., Hansmann J. Altex-Alt. Anim. Exp. 2016;33:415–422. doi: 10.14573/altex.1604041. [DOI] [PubMed] [Google Scholar]
- 222.Olekson M.A.P., Faulknor R., Bandekar A., Sempkowski M., Hsia H.C., Berthiaume F. Wound Repair Regen. 2015;23:711–723. doi: 10.1111/wrr.12334. [DOI] [PubMed] [Google Scholar]
- 223.Gholipourmalekabadi M., Seifalian A.M., Urbanska A.M., Omrani M.D., Hardy J.G., Madjd Z., Hashemi S.M., Ghanbarian H., Milan P.B., Mozafari M., Reis R.L., Kundu S.C., Samadikuchaksaraei A. Biomacromolecules. 2018;19:2409–2422. doi: 10.1021/acs.biomac.7b01807. [DOI] [PubMed] [Google Scholar]
- 224.Climov M., Bayer L.R., Moscoso A.V., Matsumine H., Orgill D.P. Plast. Reconstr. Surg. 2016;138:148S–157S. doi: 10.1097/PRS.0000000000002652. [DOI] [PubMed] [Google Scholar]
- 225.Tondreau M.Y., Laterreur V., Gauvin R., Vallieres K., Bourget J.-M., Lacroix D., Tremblay C., Germain L., Ruel J., Auger F.A. Acta Biomater. 2015;18:176–185. doi: 10.1016/j.actbio.2015.02.026. [DOI] [PubMed] [Google Scholar]
- 226.Hoganson D.M., O'Doherty E.M., Owens G.E., Harilal D.O., Goldman S.M., Bowley C.M., Neville C.M., Kronengold R.T., Vacanti J.P. Biomaterials. 2010;31:6730–6737. doi: 10.1016/j.biomaterials.2010.05.019. [DOI] [PubMed] [Google Scholar]
- 227.Shepherd B.R., Enis D.R., Wang F., Suarez Y., Pober J.S., Schechner J.S. Faseb. J. 2006;20:1739–1741. doi: 10.1096/fj.05-5682fje. [DOI] [PubMed] [Google Scholar]
- 228.Parmaksiz M., Dogan A., Odabas S., Elçin A.E., Elçin Y.M. Biomed. Mater. 2016;11 doi: 10.1088/1748-6041/11/2/022003. [DOI] [PubMed] [Google Scholar]
- 229.Su Z., Ma H., Wu Z., Zeng H., Li Z., Wang Y., Liu G., Xu B., Lin Y., Zhang P., Wei X. Mater. Sci. Eng. C-Mater. Biol. Appl. 2014;44:440–448. doi: 10.1016/j.msec.2014.07.039. [DOI] [PubMed] [Google Scholar]
- 230.Bernhard J., Ferguson J., Rieder B., Heimel P., Nau T., Tangl S., Redl H., Vunjak-Novakovic G. Biomaterials. 2017;139:202–212. doi: 10.1016/j.biomaterials.2017.05.045. [DOI] [PubMed] [Google Scholar]
- 231.Mansour A., Mezour M.A., Badran Z., Tamimi F. Tissue Eng. 2017;23:1436–1451. doi: 10.1089/ten.TEA.2017.0026. [DOI] [PubMed] [Google Scholar]
- 232.Caetano G., Violante R., Sant'Ana A.B., Murashima A.B., Domingos M., Gibson A., Bartolo P., Frade M.A. Mater. Lett. 2016;182:318–322. [Google Scholar]
- 233.He J., Li Z., Yu T., Wang W., Tao M., Ma Y., Wang S., Fan J., Tian X., Wang X., Lin Y., Ao Q. J. Biomed. Mater. Res. 2020;108:19–29. doi: 10.1002/jbm.a.36787. [DOI] [PubMed] [Google Scholar]
- 234.Wei W., Li J., Chen S., Chen M., Xie Q., Sun H., Ruan J., Zhou H., Bi X., Zhuang A., You Z., Gu P., Fan X. J. Mater. Chem. B. 2017;5:2468–2482. doi: 10.1039/c6tb03150a. [DOI] [PubMed] [Google Scholar]
- 235.Kara A., Tamburaci S., Tihminlioglu F., Havitcioglu H. Int. J. Biol. Macromol. 2019;130:266–279. doi: 10.1016/j.ijbiomac.2019.02.067. [DOI] [PubMed] [Google Scholar]
- 236.Engler A.J., Sen S., Sweeney H.L., Discher D.E. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 237.Bracey D.N., Seyler T.M., Jinnah A.H., Lively M.O., Willey J.S., Smith T.L., van Dyke M.E., Whitlock P.W. J. Funct. Biomater. 2018;9:45. doi: 10.3390/jfb9030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Xie X., Wang W., Cheng J., Liang H., Lin Z., Zhang T., Lu Y., Li Q. Colloids Surf. B Biointerfaces. 2020;190 doi: 10.1016/j.colsurfb.2020.110903. [DOI] [PubMed] [Google Scholar]
- 239.Lee M.S., Lee D.H., Jeon J., Tae G., Shin Y.M., Yang H.S. J. Ind. Eng. Chem. 2020;83:323–332. [Google Scholar]
- 240.Spiller K.L., Nassiri S., Witherel C.E., Anfang R.R., Ng J., Nakazawa K.R., Yu T., Vunjak-Novakovic G. Biomaterials. 2015;37:194–207. doi: 10.1016/j.biomaterials.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Qiu P., Li M., Chen K., Fang B., Chen P., Tang Z., Lin X., Fan S. Biomaterials. 2020;227:119552. doi: 10.1016/j.biomaterials.2019.119552. [DOI] [PubMed] [Google Scholar]
- 242.Wu R.-X., He X.-T., Zhu J.-H., Yin Y., Li X., Liu X., Chen F.-M. Mater. Sci. Eng. C-Mater. Biol. Appl. 2019;101:330–340. doi: 10.1016/j.msec.2019.03.107. [DOI] [PubMed] [Google Scholar]
- 243.Grue B.H., Veres S.P. J. Biomed. Mater. Res. B Appl. Biomater. 2020;108:845–856. doi: 10.1002/jbm.b.34438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Sladkova M., Cheng J., Palmer M., Chen S., Lin C., Xia W., Yu Y.E., Zhou B., Engqvist H., de Peppo G.M. Tissue Eng. 2019;25:288–301. doi: 10.1089/ten.TEA.2018.0149. [DOI] [PubMed] [Google Scholar]
- 245.Hesse E., Kluge G., Atfi A., Correa D., Haasper C., Berding G., Shin H.O., Viering J., Langer F., Vogt P.M., Krettek C., Jagodzinski M. Bone. 2010;46:1457–1463. doi: 10.1016/j.bone.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 246.Scotti C., Tonnarelli B., Papadimitropoulos A., Scherberich A., Schaeren S., Schauerte A., Lopez-Rios J., Zeller R., Barbero A., Martin I. Proc. Natl. Acad. Sci. U.S.A. 2010;107:7251–7256. doi: 10.1073/pnas.1000302107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Goldring M., Tsuchimochi K., Ijiri K. J. Cell. Biochem. 2006;97:33–44. doi: 10.1002/jcb.20652. [DOI] [PubMed] [Google Scholar]
- 248.Einhorn T. Clin. Orthop. Relat. Res. 1998;355:S7. doi: 10.1097/00003086-199810001-00003. [DOI] [PubMed] [Google Scholar]
- 249.Liu M., Lv Y. Nanomaterials. 2018;8:999. doi: 10.3390/nano8120999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Gupta S.K., Kumar R., Mishra N.C. Mater. Sci. Eng. C-Mater. Biol. Appl. 2017;71:919–928. doi: 10.1016/j.msec.2016.10.085. [DOI] [PubMed] [Google Scholar]
- 251.Tian T., Yu Z.H., Zhang N.L., Chang Y.W., Zhang Y.Q., Zhang L.P., Zhou S., Zhang C.L., Feng G.Y., Huang F. Biomater. Sci. 2017;5:2480–2492. doi: 10.1039/c7bm00485k. [DOI] [PubMed] [Google Scholar]
- 252.Harris G.M., Madigan N.N., Lancaster K.Z., Enquist L.W., Windebank A.J., Schwartz J., Schwarzbauer J.E. Matrix Biol. 2017;60–61:176–189. doi: 10.1016/j.matbio.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Zhao B., Sun X.L., Li X.L., Yang Q., Li Y.J., Zhang Y., Li B., Ma X.L. Neurosci. Lett. 2013;556:52–57. doi: 10.1016/j.neulet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 254.Hudson T.W., Liu S.Y., Schmidt C.E. Tissue Eng. 2004;10:1346–1358. doi: 10.1089/ten.2004.10.1641. [DOI] [PubMed] [Google Scholar]
- 255.Guo S.Z., Ren X.J., Wu B., Jiang T. Spinal Cord. 2010;48:576–581. doi: 10.1038/sc.2009.170. [DOI] [PubMed] [Google Scholar]
- 256.Heng B.C., Gong T., Wang S., Lim L.W., Wu W.T., Zhang C.F. J. Endod. 2017;43:409–416. doi: 10.1016/j.joen.2016.10.033. [DOI] [PubMed] [Google Scholar]
- 257.Baiguera S., Del Gaudio C., Lucatelli E., Kuevda E., Boieri M., Mazzanti B., Bianco A., Macchiarini P. Biomaterials. 2014;35:1205–1214. doi: 10.1016/j.biomaterials.2013.10.060. [DOI] [PubMed] [Google Scholar]
- 258.Sage E.H. Trends Cell Biol. 1997;7:182–186. doi: 10.1016/S0962-8924(97)01037-4. [DOI] [PubMed] [Google Scholar]
- 259.Homandberg G.A., Kramer-Bjerke J., Grant D., Christianson G., Eisenstein R. Biochim. Biophys. Acta. 1986;874:61–71. doi: 10.1016/0167-4838(86)90102-0. [DOI] [PubMed] [Google Scholar]
- 260.Agrawal V., Tottey S., Johnson S.A., Freund J.M., Siu B.F., Badylak S.F. Tissue Eng. 2011;17:2435–2443. doi: 10.1089/ten.tea.2011.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Adair-Kirk T.L., Senior R.M. Int. J. Biochem. Cell Biol. 2008;40:1101–1110. doi: 10.1016/j.biocel.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Ribatti D., Conconi M.T., Nico B., Baiguera S., Corsi P., Parnigotto P.P., Nussdorfer G.G. Brain Res. 2003;989:9–15. doi: 10.1016/s0006-8993(03)03225-6. [DOI] [PubMed] [Google Scholar]
- 263.Crapo P.M., Medberry C.J., Reing J.E., Tottey S., van der Merwe Y., Jones K.E., Badylak S.F. Biomaterials. 2012;33:3539–3547. doi: 10.1016/j.biomaterials.2012.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Koci Z., Vyborny K., Dubisova J., Vackova I., Jager A., Lunov O., Jirakova K., Kubinova S. Tissue Eng. C Methods. 2017;23:333–345. doi: 10.1089/ten.TEC.2017.0089. [DOI] [PubMed] [Google Scholar]
- 265.Cornelison R.C., Gonzalez-Rothi E.J., Porvasnik S.L., Wellman S.M., Park J.H., Fuller D.D., Schmidt C.E. Biomed. Mater. 2018;13 doi: 10.1088/1748-605X/aaab82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Arslan Y.E., Efe B., Arslan T.S. Biotechnol. Prog. 2019;35 doi: 10.1002/btpr.2814. [DOI] [PubMed] [Google Scholar]
- 267.DeQuach J.A., Yuan S.H., Goldstein L.S.B., Christman K.L. Tissue Eng. 2011;17:2583–2592. doi: 10.1089/ten.tea.2010.0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Wang Q.B., Zhang C.L., Zhang L.P., Guo W., Feng G.Y., Zhou S., Zhang Y.Q., Tian T., Li Z.F., Huang F. J. Biomed. Mater. Res. 2014;102:4301–4308. doi: 10.1002/jbm.a.35103. [DOI] [PubMed] [Google Scholar]
- 269.Jensen G., Morrill C., Huang Y. Acta Pharm. Sin. B. 2018;8:756–766. doi: 10.1016/j.apsb.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.De Waele J., Reekmans K., Daans J., Goossens H., Berneman Z., Ponsaerts P. Biomaterials. 2015;41:122–131. doi: 10.1016/j.biomaterials.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 271.Modo M., Badylak S.F. Brain Res. Bull. 2019;150:136–149. doi: 10.1016/j.brainresbull.2019.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Ceyssens F., Deprez M., Turner N., Kil D., van Kuyck K., Welkenhuysen M., Nuttin B., Badylak S., Puers R. J. Neural. Eng. 2017;14 doi: 10.1088/1741-2552/14/1/014001. [DOI] [PubMed] [Google Scholar]
- 273.Wu Y., Wang J., Shi Y., Pu H., Leak R.K., Liou A.K.F., Badylak S.F., Liu Z., Zhang J., Chen J., Chen L. Cell Transplant. 2017;26:1224–1234. doi: 10.1177/0963689717714090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Sood D., Cairns D.M., Dabbi J.M., Ramakrishnan C., Deisseroth K., Black L.D., III, Santaniello S., Kaplan D.L. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-54248-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Zhu T.M., Tang Q.S., Shen Y.W., Tang H.L., Chen L.P., Zhu J.H. Brain Res. Bull. 2015;113:48–57. doi: 10.1016/j.brainresbull.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 276.Sart S., Yan Y.W., Li Y., Lochner E., Zeng C.C., Ma T., Li Y. Acta Biomater. 2016;30:222–232. doi: 10.1016/j.actbio.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 277.Volpato F.Z., Fuehrmann T., Migliaresi C., Hutmacher D.W., Dalton P.D. Biomaterials. 2013;34:4945–4955. doi: 10.1016/j.biomaterials.2013.03.057. [DOI] [PubMed] [Google Scholar]
- 278.Meng F., Modo M., Badylak S.F. Regen. Med. 2014;9:367–383. doi: 10.2217/rme.14.9. [DOI] [PubMed] [Google Scholar]
- 279.Laflamme M.A., Murry C.E. Nature. 2011;473:326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Wang Z., Long D.W., Huang Y., Chen W.C.W., Kim K., Wang Y. Acta Biomater. 2019;87:140–151. doi: 10.1016/j.actbio.2019.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Boroumand S., Asadpour S., Akbarzadeh A., Faridi-Majidi R., Ghanbari H. Regen. Med. 2018;13:41–54. doi: 10.2217/rme-2017-0061. [DOI] [PubMed] [Google Scholar]
- 282.Baecker H., Polgar L., Soos P., Lajko E., Lang O., Merkely B., Szabo G., Dohmen P.M., Weymann A., Kohidai L. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. 2017;23:2232–2240. doi: 10.12659/MSM.901527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Bai R., Tian L., Li Y., Zhang J., Wei Y., Jin Z., Liu Z., Liu H. Stem Cell. Int. 2019;2019:6708435. doi: 10.1155/2019/6708435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Rajabi S., Pahlavan S., Ashtiani M.K., Ansari H., Abbasalizadeh S., Sayahpour F.A., Varzideh F., Kostin S., Aghdami N., Braun T., Baharvand H. Biomaterials. 2018;154:99–112. doi: 10.1016/j.biomaterials.2017.10.054. [DOI] [PubMed] [Google Scholar]
- 285.Pawan K.C., Shah M., Liao J., Zhang G. ACS Appl. Mater. Interfaces. 2017;9:2196–2204. doi: 10.1021/acsami.6b15291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Qian Z., Sharma D., Jia W., Radke D., Kamp T., Zhao F. Theranostics. 2019;9:2143–2157. doi: 10.7150/thno.29552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Taylor D.A., Frazier O.H., Elgalad A., Hochman-Mendez C., Sampaio L.C. Artif. Organs. 2018;42:970–982. doi: 10.1111/aor.13336. [DOI] [PubMed] [Google Scholar]
- 288.Roderjan J.G., de Noronha L., Stimamiglio M.A., Correa A., Leitolis A., Loures Bueno R.R., Affonso da Costa F.D. Xenotransplantation. 2019;26 doi: 10.1111/xen.12503. [DOI] [PubMed] [Google Scholar]
- 289.Perea-Gil I., Galvez-Monton C., Prat-Vidal C., Jorba I., Segu-Verges C., Roura S., Soler-Botija C., Iborra-Egea O., Revuelta-Lopez E., Fernandez M.A., Farre R., Navajas D., Bayes-Genis A. Sci. Rep. 2018;8:6708. doi: 10.1038/s41598-018-25115-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Blazeski A., Lowenthal J., Zhu R., Ewoldt J., Boheler K.R., Tung L. Stem Cell Reports. 2019;12:982–995. doi: 10.1016/j.stemcr.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Blazeski A., Lowenthal J., Wang Y., Teuben R., Zhu R., Gerecht S., Tomaselli G., Tung L. Tissue Eng. 2019;25:725–735. doi: 10.1089/ten.tea.2018.0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Leor J., Amsalem Y., Cohen S. Pharmacol. Ther. 2005;105:151–163. doi: 10.1016/j.pharmthera.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 293.Silva A.C., Rodrigues S.C., Caldeira J., Nunes A.M., Sampaio-Pinto V., Resende T.P., Oliveira M.J., Barbosa M.A., Thorsteinsdottir S., Nascimento D.S., Pinto-do-O P. Biomaterials. 2016;104:52–64. doi: 10.1016/j.biomaterials.2016.06.062. [DOI] [PubMed] [Google Scholar]
- 294.Bejleri D., Davis M.E. Adv. Healthcare Mater. 2019;8 doi: 10.1002/adhm.201801217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Wang B., Borazjani A., Tahai M., Curry A.L.d.J., Simionescu D.T., Guan J., To F., Elder S.H., Liao J. J. Biomed. Mater. Res. 2010;94A:1100–1110. doi: 10.1002/jbm.a.32781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Reis L.A., Chiu L.L.Y., Feric N., Fu L., Radisic M. J. Tissue Eng. Regen. Med. 2016;10:11–28. doi: 10.1002/term.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Arslan Y.E., Galata Y.F., Arslan T.S., Derkus B. J. Mater. Sci. Mater. Med. 2018;29:127. doi: 10.1007/s10856-018-6135-4. [DOI] [PubMed] [Google Scholar]
- 298.Zheng C.X., Sui B.D., Hu C.H., Qiu X.Y., Zhao P., Jin Y. J. Tissue Eng. Regen. Med. 2018;12:1432–1447. doi: 10.1002/term.2676. [DOI] [PubMed] [Google Scholar]
- 299.De Santis M.M., Bolukbas D.A., Lindstedt S., Wagner D.E. Eur. Respir. J. 2018;52 doi: 10.1183/13993003.01355-2016. [DOI] [PubMed] [Google Scholar]
- 300.Peloso A., Dhal A., Zambon J.P., Li P., Orlando G., Atala A., Soker S. Stem Cell Res. Ther. 2015;6:107. doi: 10.1186/s13287-015-0089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Balestrini J.L., Gard A.L., Gerhold K.A., Wilcox E.C., Liu A., Schwan J., Le A.V., Baevova P., Dimitrievska S., Zhao L.P., Sundaram S., Sun H.X., Rittie L., Dyal R., Broekelmann T.J., Mecham R.P., Schwartz M.A., Niklason L.E., White E.S. Biomaterials. 2016;102:220–230. doi: 10.1016/j.biomaterials.2016.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Bellezzia M.A., Cruz F.F., Martins V., de Castro L.L., Lopes-Pacheco M., Vilanova E.P., Mourao P.A., Rocco P.R.M., Silva P.L. Regen. Med. 2018;13:519–530. doi: 10.2217/rme-2018-0008. [DOI] [PubMed] [Google Scholar]
- 303.Guenthart B.A., O'Neill J.D., Kim J., Fung K., Vunjak-Novakovic G., Bacchetta M. J. Heart Lung Transplant. 2019;38:215–224. doi: 10.1016/j.healun.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Uriarte J.J., Uhl F.E., Enes S.E.R., Pouliot R.A., Weiss D.J. Curr. Opin. Organ Transplant. 2018;23:673–678. doi: 10.1097/MOT.0000000000000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Skolasinski S.D., Panoskaltsis-Mortari A. Expet Rev. Respir. Med. 2019;13:665–678. doi: 10.1080/17476348.2019.1624163. [DOI] [PubMed] [Google Scholar]
- 306.Morais A.D., Vieira S., Zhao X.L., Mao Z.W., Gao C.Y., Oliveira J.M., Reis R.L. Adv. Healthcare Mater. 2020;9 doi: 10.1002/adhm.201901435. [DOI] [PubMed] [Google Scholar]
- 307.Croce S., Peloso A., Zoro T., Avanzini M.A., Cobianchi L. Biomolecules. 2019;9:813. doi: 10.3390/biom9120813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Struecker B., Raschzok N., Sauer I.M. Nat. Rev. Gastroenterol. Hepatol. 2014;11:166–176. doi: 10.1038/nrgastro.2013.204. [DOI] [PubMed] [Google Scholar]
- 309.Ansari T., Southgate A., Obiri-Yeboa I., Jones L.G., Greco K., Olayanju A., Mbundi L., Somasundaram M., Davidson B., Sibbons P.D. Stem Cell. Dev. 2020;29:314–326. doi: 10.1089/scd.2019.0069. [DOI] [PubMed] [Google Scholar]
- 310.Mattei G., Magliaro C., Pirone A., Ahluwalia A. Artif. Organs. 2017;41:E347–E355. doi: 10.1111/aor.12925. [DOI] [PubMed] [Google Scholar]
- 311.Mazza G., Al-Akkad W., Telese A., Longato L., Urbani L., Robinson B., Hall A., Kong K., Frenguelli L., Marrone G., Willacy O., Shaeri M., Burns A., Malago M., Gilbertson J., Rendell N., Moore K., Hughes D., Notingher I., Jell G., Hernandez A.D., De Coppi P., Rombouts K., Pinzani M. Sci. Rep. 2017;7:5534. doi: 10.1038/s41598-017-05134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Zhou P.C., Huang Y., Guo Y.B., Wang L., Ling C.C., Guo Q.S., Wang Y., Zhu S.J., Fan X.J., Zhu M.Y., Huang H., Lu Y.H., Wang Z.W. Artif. Organs. 2016;40:E25–E38. doi: 10.1111/aor.12645. [DOI] [PubMed] [Google Scholar]
- 313.Petrosyan A., Zanusso I., Lavarreda-Pearce M., Leslie S., Sedrakyan S., De Filippo R.E., Orlando G., Da Sacco S., Perin L. Tissue Eng. B Rev. 2016;22:183–192. doi: 10.1089/ten.TEB.2015.0368. [DOI] [PubMed] [Google Scholar]
- 314.Hussein K.H., Saleh T., Ahmed E., Kwak H.H., Park K.M., Yang S.R., Kang B.J., Choi K.Y., Kang K.S., Woo H.M. J. Biomed. Mater. Res. 2018;106:2034–2047. doi: 10.1002/jbm.a.36407. [DOI] [PubMed] [Google Scholar]
- 315.He M., Callanan A., Lagaras K., Steele J.A.M., Stevens M.M. J. Biomed. Mater. Res. B Appl. Biomater. 2017;105:1352–1360. doi: 10.1002/jbm.b.33668. [DOI] [PubMed] [Google Scholar]
- 316.Caralt M., Uzarski J.S., Iacob S., Obergfell K.P., Berg N., Bijonowski B.M., Kiefer K.M., Ward H.H., Wandinger-Ness A., Miller W.M., Zhang Z.J., Abecassis M.M., Wertheim J.A. Am. J. Transplant. 2015;15:64–75. doi: 10.1111/ajt.12999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Leuning D.G., Witjas F.M.R., Maanaoui M., de Graaf A.M.A., Lievers E., Geuens T., Avramut C.M., Wiersma L.E., van den Berg C.W., Sol W.M.P.J., de Boer H., Wang G., LaPointe V.L.S., van der Vlag J., van Kooten C., van den Berg B.M., Little M.H., Engelse M.A., Rabelink T.J. Am. J. Transplant. 2019;19:1328–1343. doi: 10.1111/ajt.15200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Zambon J.P., Ko I.K., Abolbashari M., Huling J., Clouse C., Kim T.H., Smith C., Atala A., Yoo J.J. Acta Biomater. 2018;75:226–234. doi: 10.1016/j.actbio.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 319.Hu D., Zhang D., Liu B., Zhou Y., Yu Y., Shen L., Long C., Liu X., Lin T., He D., Wei G. Sheng wu gong cheng xue bao = Chin. J. Biotechnol. 2019;35:307–318. doi: 10.13345/j.cjb.180272. [DOI] [PubMed] [Google Scholar]
- 320.Lin C.H., Hsia K., Ma H., Lee H., Lu J.H. Int. J. Mol. Sci. 2018;19:2101. doi: 10.3390/ijms19072101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Chaudhari A.A., Vig K., Baganizi D.R., Sahu R., Dixit S., Dennis V., Singh S.R., Pillai S.R. Int. J. Mol. Sci. 2016;17:1974. doi: 10.3390/ijms17121974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Juhasz I., Kiss B., Lukacs L., Erdei I., Peter Z., Remenyik E. vol. 2010. 2010. (Dermatology Research and Practice). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Mostow E.N., Haraway G.D., Dalsing M., Hodde J.P., King D. J. Vasc. Surg. 2005;41:837–843. doi: 10.1016/j.jvs.2005.01.042. [DOI] [PubMed] [Google Scholar]
- 324.Vashi A.V., White J.F., Mclean K.M., Neethling W., Rhodes D.I., Ramshaw J., Werkmeister J.A. J. Biomed. Mater. Res. 2015;103 doi: 10.1002/jbm.a.35335. [DOI] [PubMed] [Google Scholar]
- 325.Simon P., Kasimir M.T., Seebacher G., Weigel G., Ullrich R., Salzer-Muhar U., Rieder E., Wolner E. Eur. J. Cardio. Thorac. Surg. 2003;23:1002–1006. doi: 10.1016/s1010-7940(03)00094-0. [DOI] [PubMed] [Google Scholar]
- 326.Yang D., Xiao J., Wang B., Li L., Kong X., Liao J. Mater. Sci. Eng. C-Mater. Biol. Appl. 2019;104 doi: 10.1016/j.msec.2019.109927. [DOI] [PubMed] [Google Scholar]
- 327.Tan J., Zhang Q.-Y., Huang L.-P., Huang K., Xie H.-Q. Biomater. Sci. 2021;9:4803–4820. doi: 10.1039/d1bm00470k. [DOI] [PubMed] [Google Scholar]
- 328.Rowley A.T., Nagalla R.R., Wang S.W., Liu W.F. Adv. Healthcare Mater. 2019;8 doi: 10.1002/adhm.201801578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Dziki J.L., Huleihel L., Scarritt M.E., Badylak S.F. Tissue Eng. 2017;23:1152–1159. doi: 10.1089/ten.tea.2016.0538. [DOI] [PMC free article] [PubMed] [Google Scholar]