Abstract
Circulating tumor DNA (ctDNA) is a critical biomarker not only important for the early detection of tumors but also invaluable for personalized treatments. Currently ctDNA detection relies on sequencing. Here, a platform termed three-dimensional-coded interlocked DNA rings (3D-coded ID rings) was created for multiplexed ctDNA identification. The ID rings provide a ctDNA recognition ring that is physically interlocked with a reporter ring. The specific binding of ctDNA to the recognition ring initiates target-responsive cutting via a restriction endonuclease; the cutting then triggers rolling circle amplification on the reporter ring. The signals are further integrated with internal 3D codes for multiplexed readouts. ctDNAs from non-invasive clinical specimens including plasma, feces, and urine were detected and validated at a sensitivity much higher than those obtained through sequencing. This 3D-coded ID ring platform can detect any multiple DNA fragments simultaneously without sequencing. We envision that our platform will facilitate the implementation of future personalized/precision medicine.
Keywords: 3D-coded ID rings, Multiplexed detection, Circulating tumor DNA, Colorectal cancer
Graphical abstract
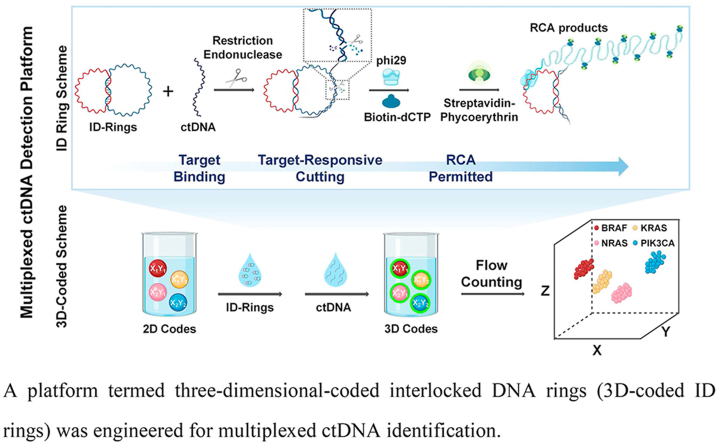
A platform termed three-dimensional-coded interlocked DNA rings (3D-coded ID rings) was engineered for multiplexed ctDNA identification.
Highlights
-
•
A platform termed 3D-coded ID rings was created for multiplexed ctDNA detection.
-
•
This platform was integrated with two schemes: the ID ring scheme and the 3D-coded scheme.
-
•
The platform could achieve multiplexed detection with detection limit of 500 copies per million in non-invasive specimens.
1. Introduction
Circulating tumor DNA (ctDNA) with tumor-specific fragmented variations is a fraction of circulating cell-free DNA released from the cancer cells [1]. Different tumor cells release ctDNA with unique mutated sequences, making ctDNA one of the earliest biomarkers for the occurrence of specific tumors [2]. In addition to the early diagnosis of tumors, specific ctDNAs have also become hallmarks for implementing individual treatments and also for the prediction of tumor prognosis [3]. Indeed, according to the US National Comprehensive Cancer Network (NCCN) guidelines [4], sequence variants of ctDNA from V-Ki-ras2 Kirsten ratsarcoma viral oncogene homolog (KRAS), neuroblastoma RAS viral oncogene homolog (NRAS), V-raf murine sarcoma viral oncogene homolog B1 (BRAF) are sufficient clinical indicators for specific therapeutic regimes. In addition, ctDNA aberration of phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) serves as a prognostic indicator for colorectal cancer (CRC) patients [[5], [6], [7]]. Since different patients with different tumors have specific ctDNA mutations, detecting these mutations have become critically important to implement personalized medicine and also precision medicine [8]. Currently ctDNA mutations are detected by DNA sequencing or droplet digital polymerase chain reaction (ddPCR), which are time-consuming, complicated and cannot implement multiplexed detection in one pot [9,10]. Also, non-invasive clinical samples from blood, feces, and urine contain too few ctDNAs to be detected by the sequencing approach. In addition, in hospital practices, ctDNA sequencing is routinely carried out in a batch mode from pooled patients samples mainly because it is too time-consuming to sequence individual patients' ctDNAs [11,12]. And ddPCR needs three complicated operating steps including droplets making, PCR in droplets and interpretation of results [13]. Thus, a new detection system beyond DNA sequencing and ddPCR is urgently needed for ctDNA-based diagnoses.
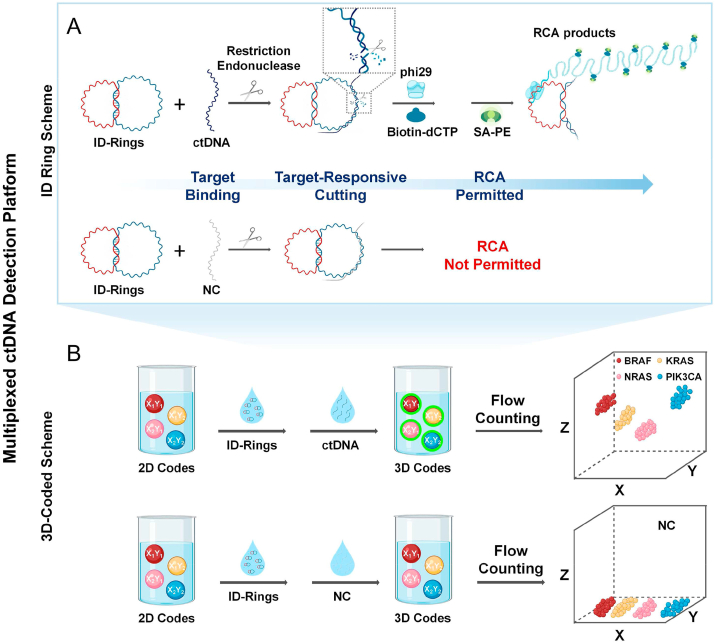
Using DNA nanostructure to detect DNA is an achievable approach that can potentially transform ctDNA diagnoses. In the last several decades, DNA molecules themselves have been demonstrated to be both precise, arbitrary nanostructures [14,15] and bulk-scale, functional materials [16,17], based mostly on the Watson-Crick base pairing principle and DNA enzymes. For example, DNA strands have been used for the fabrication of nanomechanics and nanorobotics [18,19], including DNA “tweezers” [20], “walkers” [21], “gears” [22], “interlocked nanoarchitectures” [23,24], etc. Branched DNA has been used to create DNA nanobarcodes for molecular sensing [25]. Compared with DNA “tweezers” and “tetrahedron”, the DNA interlocked structures have simpler structures and also easier to prepare due to having only simple rings [26]. In particular, interlocked nanoarchitectures have been used in supramolecular chemistry, nanotechnology, and nanodevices due to its dynamic properties [[27], [28], [29], [30]]. Interlocked structures function as reversible logic circuits [31], rotary motors [32], and nano-joints [33] to sense different stimuli [34], such as ions and pH alternations, via strand displacement [35,36]. Inspired by the impressive achievements of DNA nanostructures and DNA nanomaterials, and in order to identify ctDNA without sequencing, we designed an Interlocked DNA ring (ID ring) scheme which contained double interlocked DNA functional rings, one for recognition and one for reporting (Fig. 1A and Fig. 2A). The existence of the mechanically interlocked structure poses a powerful topological constraint on functions of reporter ring and recognition ring [37,38]. The recognition ring had a specific region for ctDNA binding, after which the bound duplexes were automatically cleaved by a chosen, specific restriction endonuclease (RE) [[39], [40], [41]], resulting in opening the recognition ring and releasing the reporter ring. And the topological constraint between reporter ring and recognition ring becomes weak so that the function of the released reporter ring can be regained [42]. ctDNA-specific amplifications were subsequently realized via the rolling circle amplification (RCA), an isothermal nucleic acid amplification technique with high fidelity and extreme processivity [43,44]. Please note that a sequence specific RE is extremely specific down to recognition sites. Hence, a sequence specific recognition ring and RE digestion can guarantee the specificity of ID ring scheme. Besides high specificity and high sensitivity, clinically it is routinely needed that multiple ctDNA samples from individual patients can be detected immediately and simultaneously [45]. Towards that end, we designed a unique three-dimensional-coded (3D-coded) scheme in suspension arrays that has a potential to detect more than hundreds of ctDNAs in a short time (less than 3 h) and simultaneously (Fig. 1B) [46,47]. In this 3D scheme, we first coded microspheres with two fluorescent dyes as X and Y coordinates to create hundreds of microspheres with different codes whose colors can be easily detected by fluorescence-based, existing equipments in most hospitals [48]. More specifically, we decorated single microspheres with two distinct fluorescent dyes such that the ratio between the two dyes was tunable and the ratio was fixed once tuned [49,50]. Integrated with the ID ring scheme, hundreds of ctDNAs could bind with different code specifically, resulting in each coding (X, Y) for each target ctDNA. We then utilized the specifically amplified ctDNA signal (i.e., the fluorescence intensity) as the third dimension (Z), making the entire coded scheme three dimensional. In other words, the X–Y colors are 2D codes determining which ctDNAs, and the Z color is the third dimension indicating the existence of the specific ctDNA and if needed, its quantity.
Fig. 1.
Schematic illustration of the multiplexed ctDNA detection platform integrating A) the ID ring scheme and B) the 3D-coded scheme.
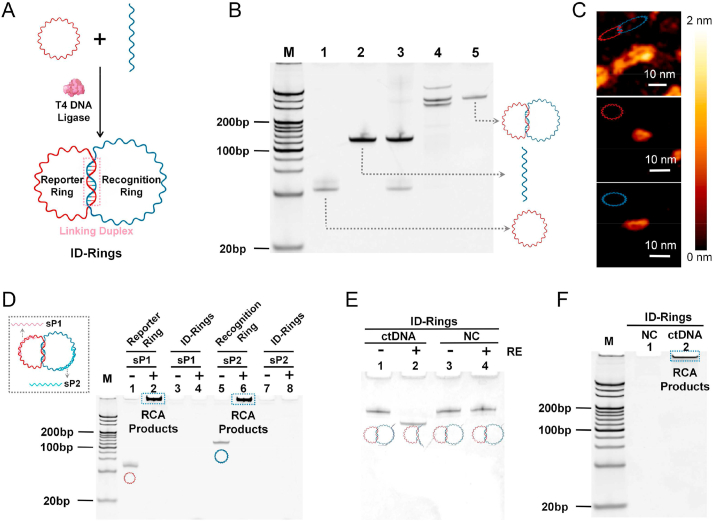
Fig. 2.
Preparation and characterization of the ID-rings. A. Schematic illustration of the ID-ring synthesis with the linking duplex via T4 DNA ligase. B. PAGE analysis of the ID-ring assembly processes. Lane M: 20 bp DNA ladder, lane 1: the reporter ring, lane 2: the linear recognition sequence, lane 3: the reporter ring hybridized with the linear recognition sequence without T4 DNA ligase, lane 4: the reporter ring hybridized with the linear recognition sequence with T4 DNA ligase, lane 5: the ID-rings. C. AFM images of the ID-rings, the reporter ring and the recognition ring. D. PAGE analysis of RCA reactions of the reporter ring, the recognition ring and the ID-rings as the RCA template, whether or not the sP1 and sP2 as the RCA primer. E. PAGE analysis of the ID-rings cleavage by RE in the presence of ctDNA. F. PAGE analysis of ctDNA-triggered reporter ring RCA.
By integrating the aforementioned two schemes: the ID ring scheme and the 3D-coded scheme, we created a multiplexed ctDNA detection platform (Fig. 1). We used non-invasive clinical specimens including plasma, feces and urine to achieve multiplexed ctDNA detection with high sensitivity of only 500 copies per million (1.2 pg/mL) and specificity to only one mutation. We believe that our platform is among the first that achieved multiplexed ctDNA detection without sequencing with high sensitivity and specificity. In addition, non-invasively obtained clinical samples can be used directly in our ctDNA detection platform. The platform is adaptable to other DNA-based detection/diagnosis (e.g., single nucleotide polymorphism). We envision that this 3D-coded ID ring platform will provide an early diagnostic tool coming to the forefront of treatment and prognosis monitoring of tumor in the areas of personalized medicine and precise medicine.
2. Materials and methods
2.1. Materials
All oligonucleotides (Table S1), purified using high-performance liquid chromatography (HPLC), and deoxy-ribonucleoside triphosphate (dNTP, 10 mM each) were purchased from Takara Biomedical Technology Co., Ltd. (Beijing, China). T4 DNA ligase (400,000 units/mL), exonuclease I (20,000 units/mL) and exonuclease III (100,000 units/mL), HpyCH4IV (10,000 units/mL), BccI (10,000 units/mL), NcoI (8000 units/mL), MluCI (10,000 units/mL), EcoRV (20,000 units/mL), phi29 DNA Polymerase (10,000 units/mL), and Bst DNA Polymerase (8000 units/mL) were purchased from New England Biolabs, Inc. (MA, USA). Super EvaGreen was obtained from US Everbright Inc. (Suzhou, China). Dimethyl sulfoxide (DMSO), KCl, 3,3′,5,5′-tetramethylbenzidine (TMB), streptavidin-phycoerythrin (SA-PE, 1 g/L) and 3-aminopropyl triethoxysilane (APTES) were purchased from Sigma-Aldrich Co. (MO, USA). EZ-10 Spin Column DNA PAGE Gel Extraction Kit, 1 × TE buffer (10 mmol/L Tris-HCl, 1 mmol/L EDTA, pH 7.8–8.2), and hemin were purchased from Sangon Biotech Co., Ltd. (Shanghai, China). Carboxylated 2D codes (polystyrene microspheres; diameter: 5.6 μm) and fluorescent dyes of these 2D codes were all provided by the Luminex Corporation (TX, USA). The cross-linker 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC, 10 g/L) was purchased from Pierce (IL, USA). OmniPur reagents including 2-[N-Morpholino]ethanesulfonic acid (MES, 0.1 M, pH 4.5), sodium dodecyl sulfate (SDS, 1 g/L), Tween 20 (0.2 mL/L), and Tris-EDTA buffer (10 mM Tris, 1 mM EDTA, pH 8.0) were purchased from EM Science (NJ, USA). Biotin-14-dCTP (0.4 mM) was acquired from Invitrogen (CA, USA). Circulating Nucleic Acid Kit and Fast DNA Stool Mini Kit were obtained from Qiagen Bioinformatics (NRW, Germany). The plasma, fecal, urine specimens of patients were collected from the Southwest Hospital of Army Medical University (Chongqing, China), and this study was approved by the ethics committee of the hospital (KY2020146).
2.2. Instruments
Polyacrylamide gel electrophoresis (PAGE) was performed on an electrophoresis system and imaged on the ChemiDoc XRS+ (Bio-Rad, CA, USA). Fluid mode atomic force microscopy (AFM) was carried on NanoWizard® BioAFM (Bruker, MA, USA). The colorimetric assays and the fluorescence spectra were recorded using a Thermo Scientific Varioskan Flash (MA, USA). A hemocytometer (Bright Line) was sourced from VWR (Radnor, PA, USA). Surface morphology images of 3D-codes were analyzed using scanning electron microscopy (SEM) (S–3400 N, Tokyo, Japan) and flow counted using luminex 200 (TX, USA). Sequencing was carried out in sequenator (3730XL, ABI, CA, USA).
2.3. Preparation and characterization of the ID-rings
The ID-rings consisted of two single-stranded oligonucleotides; a reporter ring (10 μM; 4 μL) and a recognition sequence (10 μM; 16 μL) with a fixed molar ratio of 1:4 mixed in the buffer (10 mM Tris-HCl, 1 mM EDTA, and 20 mM MgCl2). After heating at 95 °C for 5 min and slowly cooling down to 25 °C, T4 DNA ligase (1 μL) was added and incubated at 16 °C overnight (40 μL), and inactivated at 65 °C for 10 min. Subsequently, exonuclease I (1 μL) and exonuclease III (2 μL) were incubated at 37 °C for 30 min (100 μL) and inactivated at 80 °C for 20 min. Then ID-rings were extracted and purified using a DNA PAGE Gel Extraction Kit following the manufacturer's protocol, and the products were stored at 4 °C for further use. The ID-rings (10 μL), the reporter ring (10 μM; 1 μL), and the recognition ring (10 μM; 1 μL) were incubated with either a specific primer of the reporter ring (sP1; 10 μM; 1 μL) or a specific primer of the recognition ring (sP2; 10 μM; 1 μL), supplemented with 2 μL of dNTPs and 1 μL of phi29 DNA polymerase to initiate RCA reactions (50 μL) at 30 °C for 60 min. And RCA was heat inactivated at 65 °C for 10 min. Then, the successful constructions and confirmations of the ID-rings with interlocked duplex were verified utilizing 12% PAGE. Moreover, the ID-rings (20 μL) were dropped on the preconditioned dry mica substrate, the surface of which had been previously deposited with 80 μL of 1% (V/V) APTES, and after 5 min of static state, 20 μL of TE replaced residual fluids. Finally, samples were scanned using AFM.
2.4. ctDNA-triggered reporter ring RCA
After ctDNA pretreatment by heating to 95 °C, the double-stranded DNA (dsDNA) were denatured to form single-stranded DNA (ssDNA). And during the annealing process, the ID-rings (10 μL) were added and hybridized with ssDNA of ctDNA (10 μM; 1 μL) at 37 °C for 30 min. After adding 1 μL of HpyCH4IV (for KRAS) or BccI (for NRAS) or NcoI (for PIK3CA), or MluCI (for BRAF), cleavage up to 20 μL was carried out at 37 °C for 30 min. And the cleavage process was terminated at 65 °C for 20 min. Following cleavage, RCA reactions (50 μL) were initiated by sP1 (10 μM; 1 μL), 2 μL of dNTPs, and 1 μL of phi29 DNA polymerase at 30 °C for 60 min, and inactivated at 65 °C for 10 min. Then, RCA products (5 μL) were confirmed to bind with 5 μL of the primer (CS, complementary to the repetitive sequence of RCA products; 10 μM) at 37 °C for 30 min and cleaved by 1 μL of EcoRV (20 μL) at 37 °C for 15 min, and inactivated at 65 °C for 20 min. The above processes were all certified utilizing 12% PAGE.
2.5. Colorimetric detections
Different ctDNA ratios of samples (total 1011 copies/mL) were prepared at 0%, 0.05%, 0.1%, 0.2%, 0.3%, 0.4%, 0.5%, 1%, 10%, 100% according to the following mutation type/wild type ratios: 0:100, 0.05:99.95, 0.1:99.9, 0.2:99.8, 0.3:99.7, 0.4:99.6, 0.5:99.5, 1:99, 10:90, and 100:0 [51]. Following the cleavage and RCA reactions described above, RCA products (25 μL) were incubated at 37 °C for 30 min with 10 μL of KCl (500 mM) and 15 μL of TE, and then added to 2 μL of hemin (2 μM, dissolved in DMSO) at 37 °C for 30 min. Then, 50 μL of TMB was added to perform colorimetric reactions at 37 °C for 30 min, results of which were observed with the naked eye, and optical densities were recorded at 350–700 nm.
2.6. Hyperbranched RCA and a fluorescent color
The ctDNA-responsive cleavage later, hyperbranched RCA reactions (50 μL) were then triggered by addition of the forward primer and the reverse primer (10 μM; 1 μL), 2 μL of dNTPs, and 1 μL of Bst DNA Polymerase at 65 °C for 30 min. In addition, hyperbranched RCA reactions were inactivated at 80 °C for 20 min. These amplified products were carried out on 12% PAGE, and then added to 5 μL of Super EvaGreen and 45 μL of water to proceed the measurements of fluorescence spectra from 515 nm to 650 nm at an excitation wavelength of 500 nm.
2.7. Construction of the 3D-coded ID ring platform
For each population, 5 × 106 2D codes (X1Y1, X1Y2, X2Y1, X2Y2) were combined with 1 nmol of primer-BRAF, KRAS, NRAS, and PIK3CA respectively in a total volume of 50 μL of MES. Moreover, 2.5 μL of EDC was added to the above mixture and incubated for 30 min twice to conjugate primer-BRAF, KRAS, NRAS, and PIK3CA to carboxylated microspheres to obtain 2D codes, washed in 1 mL Tween 20 and 1 mL SDS, and each primer-coupled 2D code population was then resuspended in 100 μL of Tris-EDTA buffer, enumerated using a hemacytometer and stored in the dark at 4 °C. The ID-rings were bound with the corresponding primers on the surface of 2D codes at an equal molar ratio at 37 °C for 30 min, and then ctDNA-responsive cleavage was conducted based on the above-mentioned methods. Next, 1 μL of phi29 DNA polymerase, 2 μL of dATP, dGTP, dTTP, and biotin-14-dCTP were added and incubated at 30 °C for 60 min to launch RCA reactions (50 μL). RCA reactions were inactivated at 65 °C for 10 min. Further, 50 μL of SA-PE was added to the RCA products and incubated at 48 °C for 15 min to produce 3D codes. 3D codes were observed using luminex 200 at double lasers, the red laser (635 nm), and green laser (525 nm) without any purification. The surface morphology images of 2D codes and 3D codes were surveyed via SEM coating with a 200 Å of gold-palladium layer with a step voltage of 10 kV.
2.8. Clinical sample measurements
We collected 187 plasma, 20 fecal, and 20 urine specimens histologically confirmed as colorectal cancer from inpatients at the Southwest Hospital of Army Medical University (Chongqing, China) between April 2020 and July 2020 following ethical approval. None of the patients had received any therapies before sampling. The blood specimens (5 mL per person) were centrifuged at 1600×g for 10 min at 4 °C and the faint yellow supernatant was collected. The ctDNAs were concentrated and purified in accordance with the manufacturer's instructions with use of Circulating Nucleic Acid Kit. The ctDNAs of fecal (200 mg per person) and urine (15 mL per person) specimens were purified according to the Fast DNA Stool Mini Kit and Circulating Nucleic Acid Kit instructions.
2.9. Sequencing of clinical samples
The extracted ctDNAs were amplified by nested polymerase chain reaction (PCR) before sequencing. The first-round amplification was performed using the outer primers F-out and R-out. Conditions for the first round of PCR were 95 °C for 5 min, followed by 38 cycles of 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 1 min, and a final extension at 72 °C for 10 min. Utilizing the first-round products as the template, the second round of PCR was proceeded with the inner primers F-in and R-in as follows: 96 °C for 1 min, followed by 25 cycles at 96 °C for 10 s, 50 °C for 5 s, and 60 °C for 4 min and thermal insulation at 4 °C. Then, the PCR products were purified and then sequenced by sequenator.
2.10. Statistical analysis
Statistical analyses were performed using SPSS (version 20.0). A P value of less than 0.05 was considered statistically significant differences. For parametric tests, comparison of groups were performed using Analysis of Variance (ANOVA) with post-hoc analysis (Least Significant Difference, LSD). For non-parametric tests, rank sum test (Kruskal-Wallis H test) was used for multiple comparison. Chi-square (χ2) test and McNemar’ s test were employed to compare ctDNA detection rates between 3D-coded ID ring platform and sequencing. The concordance analyses between the two methods were calculated using the Kappa test.
3. Results and discussion
3.1. Preparation and characterization of the ID-rings
The ID rings comprised two DNA rings, functioning as a recognition and a reporter ring (Fig. 2A). A small portion of the sequences of the recognition ring was hybridized to the corresponding small portion of the reporter ring, forming an interlocking duplex region. Circularization was then achieved through a ligation reaction. Intermediate and final products were analyzed using a 12% PAGE (Fig. 2B). In more detail, we designed and circularized a ssDNA ring as the reporter ring (Fig. 2B, lane 1, red circle). A linear ssDNA was also designed (Fig. 2B, lane 2) such that a portion of it was complementarily hybridized to the corresponding region of the reporter ring. Then upon adding T4 ligase, the linear portion of the recognition ring closed, forming ID-rings (Fig. 2B, lane 4; as a control, the ring was not closed without T4 ligase, see lane 3). Non-circularized and intermediate products were subject to the digestion by exonucleases I and III, and the final ID-rings were purified (Fig. 2B, lane 5). The successful formation of the ID-rings was further confirmed in solution by using the tapping mode of AFM (Fig. 2C).
3.2. Optimizing the interlocking duplex sequences of the ID-rings
In order to detect different ctDNA targets (i.e., KRAS, NRAS, PIK3CA, and BRAF), we designed specific recognition sequences corresponding to each individual ctDNA target. Three lengths (from 24 to 28 to 32 bp) of the interlocking duplex sequences between the recognition and reporter rings for each ctDNA were prepared in Fig. S1. For KRAS, the bands of ID-rings with the length of 24 bp in lane 5 were relatively faint indicating that efficacy was not high. So, this option (KRAS-24 bp) could be removed. Furthermore, RCA reactions of several ID-rings with linking duplexes from 24 to 28 to 32 bp to bind with corresponding ctDNAs or normal controls (NCs) were compared in Fig. S2. The optimal ID-rings should possess strong interlocked linking duplexes to be capable of RCA when detecting ctDNA but incapable of RCA when detecting NC. Based on the above principle, the ID-rings with optimum linking duplexes of 28 bp, 24 bp, 28 bp and 32 bp for KRAS, NRAS, PIK3CA and BRAF respectively were selected.
The selected ID-rings had three bands indicating three isomers (lane 5 of Fig. S1) [52]. Afterwards, the three bands of isomers were extracted and purified to detect ctDNA or NC DNA (Fig. S3). The lower band of the isomer could initiate RCA triggered both by ctDNA and NC due to its loose interlocking structure. And the upper band of the isomer could't initiate RCA triggered by neither ctDNA nor NC. But the middle band of the isomer could meet the demand. Moreover, the yield of the middle band was the highest. Therefore, the middle band was chosen for the final ID-rings in the subsequent steps.
3.3. Reporter ring amplification by RCA
After the ID-rings form, the DNA amplification is accomplished through RCA of the reporter ring. Thus, it is ultimately important that the RCA reaction only happens when a specific ctDNA target presents, and no RCA should occur even in the presence of other, non-target ctDNAs. In addition, it is equally important that no RCA should occur without any target ctDNA. Towards that end, we tested the specific amplification of RCA by using two primers: a specific primer of the reporter ring (sP1) and a specific primer of the recognition ring (sP2). Indeed, our results revealed that RCA products of specific reporter ring or recognition ring were evident in the presence of sP1 or sP2 (Fig. 2D, lanes 2 and 6) and that no RCA products were observed in the presence of no primer (reporter ring and recognition ring, Fig. 2D, lanes 1 and 5). Notably, RCA did not happen with the ID-rings themselves regardless of whether or not the sP1 and sP2 were present, suggesting that the ID-rings were quite stable when interlocked (Fig. 2D, lanes 3 and 4, lanes 7 and 8).
3.4. ctDNA-triggered reporter ring RCA
Thus far, we have demonstrated that our ID-rings were pure and stable, and that the reporter ring can be amplified upon specific primers. We hypothesized that a highly specific ctDNA identification scheme can be established without sequencing if we can release the reporter ring only in the presence of a specific ctDNA sequence. Towards that end, we utilized REs which precisely cut the duplexed DNA after recognizing specific sequences. We designed the recognition ring in our ID-rings so that it can base pair with a specific ctDNA. Once recognized and bound, the ctDNA-recognition ring duplex became the substrate of a unique RE for precise cutting by that RE, resulting in the releasing of the reporter ring from the ID-rings. The released reporter ring then served as the template for RCA reactions. This specific release of reporter rings for amplification only happens in the presence of mutation-type ctDNA. Wild-type DNAs from non-tumor patients, designated as a NC will not release any rings and thus will not result in any amplification (Fig. 2E).
In the presence of ctDNAs and the RE, the recognition ring was cleaved thus becoming linearized (Fig. 2E, lane 2). When the ctDNA was absent or when RE was absent, the recognition ring was not linearized, suggesting that indeed, RE cleavage could be initiated only in the presence of ctDNAs. Once cleaved, the reporter ring was released, and it served as a template for RCA (Fig. 2F, lane 2). These RCA products were not detected when only NC was present, indicating a ctDNA-specific amplification. To further confirm the specific amplified RCA products were indeed from the reporter ring, we subjected the products to EcoRV digestion, which by design would yield monomeric fragments that were tandemly replicated during the RCA reaction. As expected, we detected these monomeric repeating fragments through gel electrophoresis (Fig. S4).
3.5. ctDNA detection using the ID ring scheme with a DNAzyme and a visible color
Afterwards, we tested the performance of our ID rings, especially for determining the detection sensitivity. To simplify the experimental procedures and to visualize the results with the naked eye, we employed a hemin-guanine (G)-quadruplex DNAzyme that produced colorimetric signals for readouts (Fig. 3A). Briefly, we designed the reporter ring with cytosine (C)-rich sequences which became G-rich sequences after RCA. These G-rich, amplified products folded automatically into G-quadruplexes in the presence of potassium ions (K+). Upon further adding hemin, the G-quadruplexes became a DNAzyme that catalyzed the colorless substrate, TMB, into a blue-colored product (oxidized TMB, or oxTMB) [53,54]. Note that in addition to the aforementioned signal amplification via RCA, this approach added one more amplification step through a DNAzyme-catalyzed substrate conversion.
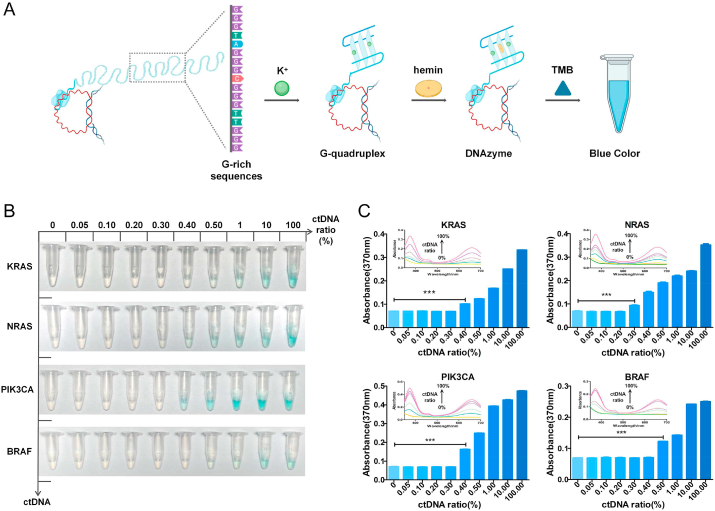
Fig. 3.
ctDNA detection using the ID ring scheme with a DNAzyme and a visible color. A. Schematic illustration of the ctDNA detection using the ID ring scheme with a DNAzyme and a visible color. B. Determination of ctDNA ratios of KRAS, NRAS, PIK3CA and BRAF via the visible color change. C. Histograms with an error bar at absorbance of 370 nm of different ctDNA ratios of KRAS, NRAS, PIK3CA and BRAF. Each column bar represents the average value of three independent assays with error bars indicated. ***P < 0.001. The inset is the image of the corresponding absorption spectra from 350 to 700 nm.
To evaluate the sensitivity of ctDNA detection using our ID rings, we mixed mutation ctDNAs (KRAS, NRAS, PIK3CA, and BRAF) with NC DNAs at various ratios. With the increase of the mutated/NC DNA ratio, the blue color from oxTMB intensified as expected (Fig. 3B). Quantitative analyses on the value at the 370 nm, the characteristic peak of oxTMB, indicated that the detection limits (actually minimum detectable ctDNA ratio) of specific ctDNAs reached to 0.4%, 0.3%, 0.4%, and 0.5% for KRAS, NRAS, PIK3CA, and BRAF, respectively. And corresponding sensitivities of these ctDNAs reached to 9.2 pg/mL, 6.9 pg/mL, 9.2 pg/mL and 11.5 pg/mL for KRAS, NRAS, PIK3CA, and BRAF, respectively. The conversion of detection limit from ctDNA ratio to concentration is demonstrated in Supplementary Materials. These detection limits are on par with those obtained through sequencing [55].
3.6. ctDNA detection using the ID ring scheme with hyperbranched RCA and a fluorescent color
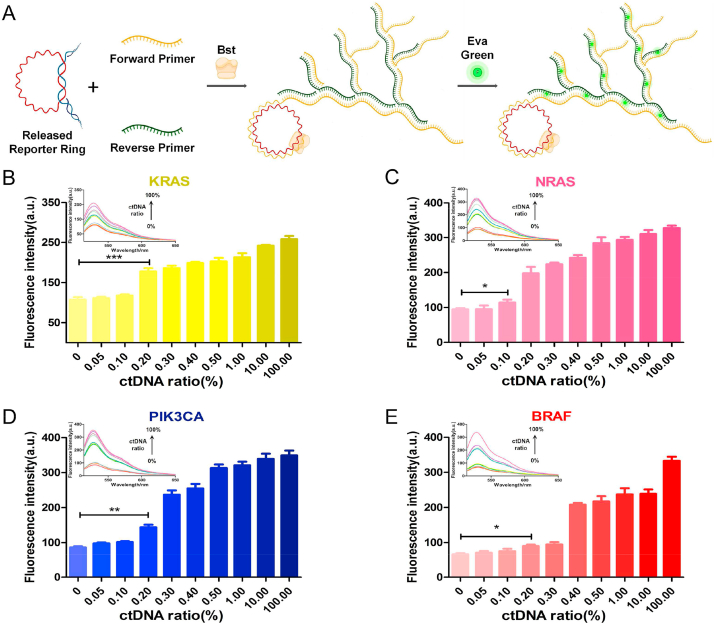
In addition to using DNAzyme and a visible color to detect ctDNA with the naked eye, our ID ring scheme can be adapted to other detection modalities including fluorescent-based detections. In this case, we utilized a totally different way of DNA amplification, hyperbranched amplification [56], along with a fluorescent dye to achieve a lower ctDNA detection limit. Briefly, instead of a previously used single primer, we designed two primers, a forward primer and a reverse primer, for the hyperbranched amplification. After the ctDNA initiated release of the reporter ring DNA (i.e., the template DNA) and after one round of amplification of the template DNA using the forward primer, the amplified products served as new templates for a second round of amplification using the reverse primer. This product-template-product cycle continued until a fluorescent dye was added for the final detection (Fig. 4A).
Fig. 4.
ctDNA detection using the ID ring scheme with hyperbranched RCA and a fluorescent color. A. Schematic illustration of the ctDNA detection using the ID ring scheme with hyperbranched RCA and a fluorescent color. B, C, D, E. Histograms with an error bar of fluorescence intensity at 530 nm of different ctDNA ratios of KRAS, NRAS, PIK3CA and BRAF, respectively. Each column bar represents the average value of three independent assays with error bars indicated. *P < 0.05, **P < 0.01, ***P < 0.001. The inset is the image of the corresponding fluorescence spectra from 515 to 650 nm.
In more detail, we chose the Bst DNA polymerase for the hyperbranched DNA amplification [57]. The amplification products were analyzed on PAGE (Fig. S5). The results indicated that the reporter ring DNA was indeed hyper-amplified according to the evident and expected appearance of DNA ladder bands starting at the length of the reporter ring (Fig. S5). In order to determine the detection sensitivity using this fluorescence-based approach, we used Super EvaGreen, a fluorescent dye that specifically binds to and stains dsDNA. The detection was quantified using a fluorometer. Using a similar procedure to that of DNAzyme and a visible dye, we first mixed mutated ctDNAs (KRAS, NRAS, PIK3CA, and BRAF) with NC DNA at various ratios. With the increase of the mutated/NC DNA ratio, the fluorescence intensity, as expected, increased (Fig. 4B–E). Quantitative analyses of the value at 530 nm, the characteristic peak of Super EvaGreen, indicated that actual detection limits of the specific ctDNA reached to 0.2%, 0.1%, 0.2% and 0.2% for KRAS, NRAS, PIK3CA, and BRAF, respectively. And corresponding sensitivities of these ctDNAs reached to 4.6 pg/mL, 2.3 pg/mL, 4.6 pg/mL and 4.6 pg/mL for KRAS, NRAS, PIK3CA, and BRAF, respectively. These detection limits are lower than those obtained through visible dyes.
3.7. A multiplexed ctDNA detection platform using 3D-coded ID-rings
In order to detect multiple ctDNAs simultaneously, we created a 3D-coded scheme to integrate with ID rings. Our 3D codes were in solution (a one-pot reaction) and were decoded using conventional flow cytometery. The 3D coded scheme started with 2D coded microspheres, termed X and Y (Fig. 5A). Briefly, to construct 2D coded X–Y microspheres, we simply labelled microspheres with two fluorescent dyes (XiYj) and tuned the concentrations of these two dyes according to our design. The two fluorescent dyes were chosen such that they had the same excitation wavelength (at 635 nm using a red laser beam) but two distinctly different emission wavelengths (at 680 nm for X and at 705 nm for Y). Since the tuning of 2D codes was based on different concentrations of dyes, the possibility of different combinations would be enormous. To demonstrate the principle and the feasibility of our platform in a succinct manner, we designed a total of four 2D codes, termed X1Y1, X1Y2, X2Y1 and X2Y2. Unique to our platform, we attached our ID-rings onto the surface of these microspheres through the reporter ring. Thus, each unique ctDNA recognition ring was interlocked with its reporter ring which in turn was attached to the microspheres
Fig. 5.
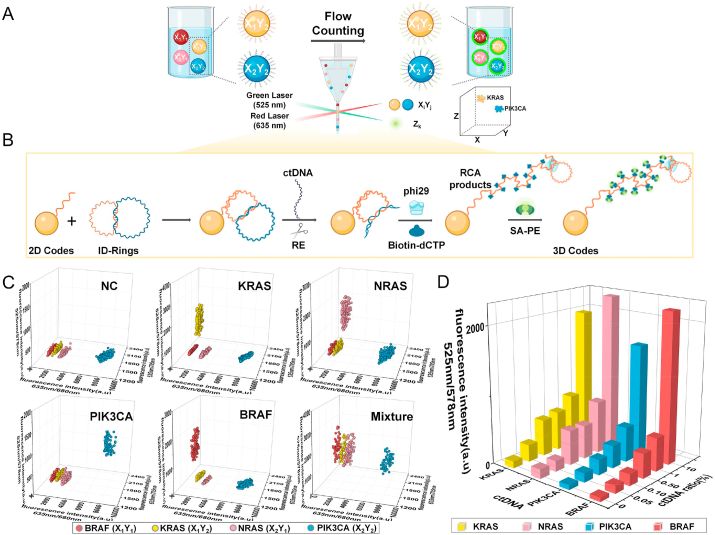
The multiplexed ctDNA detection platform using 3D-coded ID-rings. A, B. Schematic illustration of the principle of the 3D-coded ID ring platform. C. 3D scatter plots of mixed codes in the presence of KRAS, NRAS, PIK3CA, BRAF, and a mixture of the four ctDNAs as indicated. D. 3D bar plot of the mean fluorescence intensities under indicated conditions. Each column bar represents the average value of three independent assays.
With a unique 2D codes (XiYj). The addition of a target ctDNA triggered the cutting by the specific RE which subsequently initiated a RCA reaction on the attached reporter ring (Fig. 5B). The signals of amplified DNA from RCA were further intensified by another fluorescence dye Z, specially chosen for its emission only under a green (not red) laser beam (SA-PE, excitation wavelength at 525 nm and emission wavelength at 578 nm). The fluorescence intensity of RCA amplified signal served as a third dimension, termed Zk. Thus, different 3D codes were established (XiYjZk) that were unique to different ctDNAs. A two-laser beam (red and green) flow cytometer was used to both detect and decode, in one pot, our 3D-coded ID ring system.
The implementation of the 3D-coded ID ring platform was relatively simple, starting with the construction of four different ID-rings whose recognition rings and reporter rings were specific to target ctDNAs (KRAS, NRAS, PIK3CA, and BRAF). Four different types of microspheres were also fabricated with four XY codes, each code corresponding to one microsphere and only one ctDNA. In one pot, ID-rings were bound to X–Y microspheres specifically, forming the 3D-coded ID ring platform. In the presence of any target ctDNA, alone or combined, RE spontaneously cut the bound ctDNA-recognition ring complex which initiated the subsequent RCA reaction on the remaining reporter rings. Biotinylated building blocks, biotin-dCTP, were deliberately included in the system so that the amplified DNA products would bind a streptavidin-conjugated fluorescent dye (Z signals), further enhancing the amplified signals (Fig. 5B). The post-amplified microspheres were visualized and qualitatively analyzed through SEM (Fig. S6).
A total of six samples, including five controls and one testing sample, were assembled for the multiplexed detection. The five control samples consisted of one negative control (with only NC DNA) plus four positive controls (with only one specific ctDNA). The testing sample included all four ctDNA premixed together in one pot. Please note that all six samples contained the same amount of DNA. Please also note that four types of ID-rings along with four types of 2D-coded microspheres (X1Y1, X1Y2, X2Y1 and X2Y2 corresponding to microspheres that were specific for BRAF, KRAS, NRAS, and PIK3CA, respectively) were always present in the pot.
After running each sample through the two-colored flow cytometer, fluorescence intensities were recorded and 3D scatter plots were automatically generated with X–Y coordinates representing microspheres' 2D codes and Z coordinate representing the RCA results of the reporter ring (Fig. 5C and Figs. S7A–D). Thus, through X and Y, we successfully identified the type(s) of specific ctDNAs present (or absent) and through Z, we quantitatively measured the amounts of that specific ctDNAs. We would like to emphasize that different ctDNAs formed individual clusters across the X–Y coordinate system with each ctDNA landing on a specific cluster.
The third dimension in our 3D-coded ID ring platform was the Z signal, which conveniently and effectively represented the minimum detectable amount, serving as an accurate parameter for the determination of the platform's sensitivity (Fig. 5D). With the decreasing ratios of ctDNA in the sample, the Z signals decreased as well until reaching to the level of background noise as determined from statistical tests. Our results indicated that the actual ctDNA detection limits for KRAS, NRAS, PIK3CA and BRAF were 0.05%, 0.1%, 0.1%, and 0.05%, respectively. Especially, an ultrahigh sensitivity of detecting as low as 1.2 pg/mL of KRAS point mutation and BRAF could be achieved. And 2.4 pg/mL of NRAS and PIK3CA could be actually detectable. These detection limits were better than those using the sequencing method and were very much clinically relevant and valuable. Also, linear correlations of the fluorescence intensity and the ctDNA ratio were established (Figs. S8A–D).
3.8. Clinical applications of the 3D-coded ID ring platform
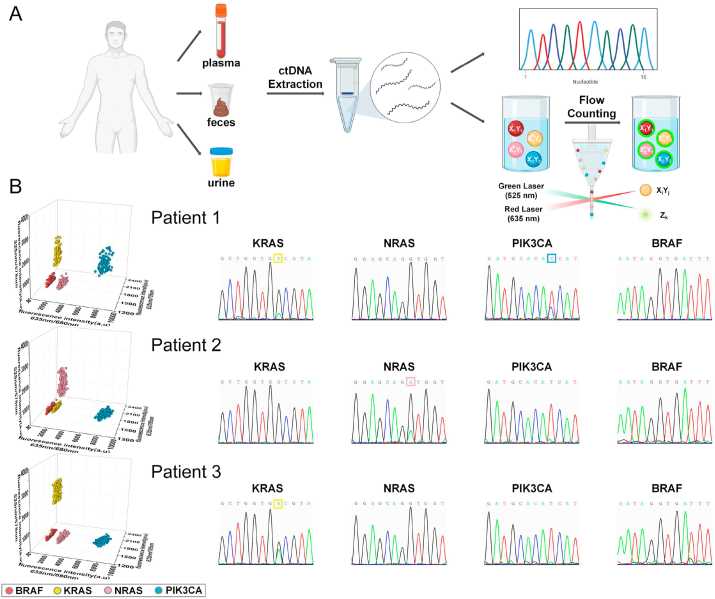
To further demonstrate the feasibility and advantages of our platform in real-world applications, we tested real patient samples instead of mimetic samples (Fig. 6). We particularly chose those samples that were obtained non-invasively from patients. Altogether, we obtained ctDNAs from 187 plasma, 20 fecal, and 20 urine specimens from colorectal cancer patients. Targeted ctDNAs were KRAS, NRAS, PIK3CA and BRAF. Using our multiplexed 3D-coded ID ring platform, we tested all 227 patient samples. Meanwhile, all 227 samples were subjected to the conventional sequencing method for the detection of ctDNAs; the sequencing results served as reliable benchmarks for validating our platform. It can be clearly concluded that there were no evident distinctions between the two methods for the detection of ctDNAs (Table 1). The concordance between the two methods were also analyzed and strongly established (Table 2, Table S2-3 and Figs. S9–10). And compared with sequencing and ddPCR, our platform not only can implement multiplexed detection, but also show shorter time and better sensitivity (Table S4).
Fig. 6.
Clinical sample measurements. A. Schematic illustration of ctDNA extraction from plasma, fecal, and urine specimens of CRC patients and measurements with sequencing and the 3D-coded ID ring platform. B. 3D scatter plots and sequencing results of Patient 1–3.
Table 1.
Comparison of positive detection rates between the 3D-coded ID ring platform and sequencing.
| ctDNA | Method | Number of samplese |
Total number | Positive rate (%) | |
|---|---|---|---|---|---|
| ctDNA | NC | ||||
| KRASa | 3D-coded ID ring platform | 24 (23) | 163 (164) | 187 | 12.8 |
| Sequencing | 22 (23) | 165 (164) | 11.8 | ||
| NRASb | 3D-coded ID ring platform | 3 (3) | 184 (184) | 187 | 1.6 |
| Sequencing | 3 (3) | 184 (184) | 1.6 | ||
| PIK3CAc | 3D-coded ID ring platform | 5 (4.5) | 182 (182.5) | 187 | 2.7 |
| Sequencing | 4 (4.5) | 183 (182.5) | 2.1 | ||
| BRAFd | 3D-coded ID ring platform | 1 (1) | 186 (186) | 187 | 0.5 |
| Sequencing | 1 (1) | 186 (186) | 0.5 | ||
χ2 = 0.099, P>0.05.
χ2 = 0.000, P>0.05.
χ2 = 0.000, P>0.05.
P>0.05.
The values in parentheses represent the expected count of samples according to χ2 test.
Table 2.
Concordance between the 3D-coded ID ring platform and sequencing.
| ctDNA | Determination by 3D-coded ID ring platform | Number of samples |
Total number | |
|---|---|---|---|---|
| ctDNA | NC | |||
| KRASa | ctDNA | 22 | 2 | 24 |
| NC | 1 | 162 | 163 | |
| Total | 23 | 164 | 187 | |
| NRASb | ctDNA | 3 | 0 | 3 |
| NC | 0 | 184 | 184 | |
| Total | 3 | 184 | 187 | |
| PIK3CAc | ctDNA | 4 | 1 | 5 |
| NC | 0 | 182 | 182 | |
| Total | 4 | 183 | 187 | |
| BRAFd | ctDNA | 1 | 0 | 1 |
| NC | 0 | 186 | 186 | |
| Total | 1 | 186 | 187 | |
McNemar’ s test for KRAS, P>0.05; Kappa value = 0.927 (P = 0.000).
McNemar’ s test for NRAS, P>0.05; Kappa value = 1.000 (P = 0.000).
McNemar’ s test for PIK3CA, P>0.05; Kappa value = 0.886 (P = 0.000).
McNemar’ test for BRAF, P>0.05; Kappa value = 1.000 (P = 0.000).
4. Conclusion
We have designed an innovative ctDNA ID ring scheme that can be employed to detect any ctDNA without sequencing. Any ctDNA could be detected by choosing appropriate RE and redesigning the sequences of the recognition ring according to its own sequence of the target ctDNA. We further developed our scheme such that it can be exploited with different modalities of signal readouts, from the naked eye detection of colors to fluorometer-based fluorescence. We further integrated the ID ring scheme with a 3D-coded scheme, creating for the first time a 3D-coded ID ring platform that successfully detected multiple ctDNAs simultaneously in one pot. In addition to the mimetic samples, we successfully detected multiple ctDNAs from 227 real patient samples in a hospital setting, benchmarking better than the conventional sequencing method. Our platform is unique and advantageous due to the following designs that have minimized RCA non-specific reactions: Specificity-wise, our platform employs three independent mechanisms to ensure specificity down to one base. These three steps include a sequence specific recognition ring, a sequence specific binding probe on microspheres, and a sequence specific RE. Sensitivity-wise, our platform successfully executes three consecutive amplifications at three stages. These three stages include the initial microspheres, the subsequent RCA reactions, and the final color conversion.
We believe that our 3D-coded ID ring platform reported here is not only the first successful attempt to identify ctDNA in a multiplexed manner without sequencing, but also an intriguing demonstration of detecting other DNA beyond ctDNA without sequencing, including but not limited to single nucleotide polymorphism, diseases caused by gene mutations, and drug-resistant genes of pathogens. We envision that the 3D-coded ID ring platform points to an alternative pathway forward for almost any DNA detection without sequencing and that this platform will become invaluable in clinical settings from multiplexed diagnoses, to individualized therapies, and to precision medicine.
Credit Author Statement
M. C. designed the overall approach with K. C. carried out experiments. YX. W.. carried out experiments S. Y. carried out experiments and XY. Z. carried out experiments and S. Y. wrote the manuscript with D. L. Funding acquisition, Formal analysis, contributed to the acquisition, analysis, or interpretation of data. and MX. G.. Funding acquisition, Formal analysis, contributed to the acquisition, analysis, or interpretation of data. XQ. T., Funding acquisition, Formal analysis, contributed to the acquisition, analysis, or interpretation of data. S. Z. Funding acquisition, Formal analysis, contributed to the acquisition, analysis, or interpretation of data. LY. Y. Funding acquisition, Formal analysis, contributed to the acquisition, analysis, or interpretation of data. All the authors discussed the results and commented on the manuscript.
Declaration of competing interest
The authors declare that they have no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 81972027, 82030066, 82122042, 81430053).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2021.09.007.
Contributor Information
Yunxia Wang, Email: wyxtmmu@foxmail.com.
Kai Chang, Email: changkai0203@163.com.
Ming Chen, Email: chming1971@126.com.
Abbreviations
- ctDNA
circulating tumor DNA
- 3D-coded ID rings
three-dimensional-coded interlocked DNA rings
- NCCN
National Comprehensive Cancer Network
- KRAS
V-Ki-ras2 Kirsten ratsarcoma viral oncogene homolog
- NRAS
neuroblastoma RAS viral oncogene homolog
- BRAF
V-raf murine sarcoma viral oncogene homolog B1
- PIK3CA
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha
- CRC
colorectal cancer
- dPCR
digital polymerase chain reaction
- ID ring
Interlocked DNA ring
- RE
restriction endonuclease
- RCA
rolling circle amplification
- 3D-coded
three-dimensional-coded
- HPLC
high-performance liquid chromatography
- dNTP
deoxy-ribonucleoside triphosphate
- DMSO
dimethyl sulfoxide;
- TMB
3,3′,5,5′-tetramethylbenzidine;
- SA-PE
streptavidin-phycoerythrin
- APTES
3-Aminopropyl triethoxysilane
- EDC
1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride;
- MES
2-[N-Morpholino]ethanesulfonic acid
- SDS
sodium dodecyl sulfate
- PAGE
polyacrylamide gel electrophoresis
- AFM
atomic force microscopy
- SEM
scanning electron microscopy
- dsDNA
double-stranded DNA
- ssDNA
single-stranded DNA
- NC
normal control
- PCR
polymerase chain reaction
- bp
base pair
- G
guanine;
- C
cytosine.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Siravegna G., Mussolin B., Venesio T., Marsoni S., Seoane J., Dive C., Papadopoulos N., Kopetz S., Corcoran R.B., Siu L.L., Bardelli A. How liquid biopsies can change clinical practice in oncology. Ann. Oncol. 2019;30:1580–1590. doi: 10.1093/annonc/mdz227. [DOI] [PubMed] [Google Scholar]
- 2.Kinde I., Wu J., Papadopoulos N., Kinzler K.W., Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. U. S. A. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pittella-Silva F., Chin Y.M., Chan H.T., Nagayama S., Miyauchi E., Low S.K., Nakamura Y. Plasma or serum: which is preferable for mutation detection in liquid biopsy? Clin. Chem. 2020;66:946–957. doi: 10.1093/clinchem/hvaa103. [DOI] [PubMed] [Google Scholar]
- 4.Benson A.R., Venook A.P., Cederquist L., Chan E., Chen Y.J., Cooper H.S., Deming D., Engstrom P.F., Enzinger P.C., Fichera A., Grem J.L., Grothey A., Hochster H.S., Hoffe S., Hunt S., Kamel A., Kirilcuk N., Krishnamurthi S., Messersmith W.A., Mulcahy M.F., Murphy J.D., Nurkin S., Saltz L., Sharma S., Shibata D., Skibber J.M., Sofocleous C.T., Stoffel E.M., Stotsky-Himelfarb E., Willett C.G., Wu C.S., Gregory K.M., Freedman-Cass D. Colon cancer, version 1.2017, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Canc. Netw. 2017;15:370–398. doi: 10.6004/jnccn.2017.0036. [DOI] [PubMed] [Google Scholar]
- 5.Osumi H., Shinozaki E., Yamaguchi K. Circulating tumor DNA as a novel biomarker optimizing chemotherapy for colorectal cancer. Cancers. 2020;12:1566. doi: 10.3390/cancers12061566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santos C., Azuara D., Vieitez J.M., Paez D., Falco E., Elez E., Lopez-Lopez C., Valladares M., Robles-Diaz L., Garcia-Alfonso P., Buges C., Duran G., Salud A., Navarro V., Capella G., Aranda E., Salazar R. Phase II study of high-sensitivity genotyping of KRAS, NRAS, BRAF and PIK3CA to ultra-select metastatic colorectal cancer patients for panitumumab plus FOLFIRI: the ULTRA trial. Ann. Oncol. 2019;30:796–803. doi: 10.1093/annonc/mdz082. [DOI] [PubMed] [Google Scholar]
- 7.Normanno N., Rachiglio A.M., Lambiase M., Martinelli E., Fenizia F., Esposito C., Roma C., Troiani T., Rizzi D., Tatangelo F., Botti G., Maiello E., Colucci G., Ciardiello F. Heterogeneity of KRAS, NRAS, BRAF and PIK3CA mutations in metastatic colorectal cancer and potential effects on therapy in the CAPRI GOIM trial. Ann. Oncol. 2015;26:1710–1714. doi: 10.1093/annonc/mdv176. [DOI] [PubMed] [Google Scholar]
- 8.Alves I., Condinho M., Custodio S., Pereira B.F., Fernandes R., Goncalves V., Da C.P., Lacerda R., Marques A.R., Martins-Dias P., Nogueira G.R., Neves A.R., Pinho P., Rodrigues R., Rolo E., Silva J., Travessa A., Leite R.P., Sousa A., Romao L. Genetics of personalized medicine: cancer and rare diseases. Cell. Oncol. 2018;41:335–341. doi: 10.1007/s13402-018-0379-3. [DOI] [PubMed] [Google Scholar]
- 9.Koessler T., Addeo A., Nouspikel T. Implementing circulating tumor DNA analysis in a clinical laboratory: a user manual. Adv. Clin. Chem. 2019;89:131–188. doi: 10.1016/bs.acc.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 10.Feng W., Jia N., Jiao H., Chen J., Chen Y., Zhang Y., Zhu M., Zhu C., Shen L., Long W. Circulating tumor DNA as a prognostic marker in high-risk endometrial cancer. J. Transl. Med. 2021;19:51. doi: 10.1186/s12967-021-02722-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tarazona N., Gimeno-Valiente F., Gambardella V., Zuniga S., Rentero-Garrido P., Huerta M., Rosello S., Martinez-Ciarpaglini C., Carbonell-Asins J.A., Carrasco F., Ferrer-Martinez A., Bruixola G., Fleitas T., Martin J., Tebar-Martinez R., Moro D., Castillo J., Espi A., Roda D., Cervantes A. Targeted next-generation sequencing of circulating-tumor DNA for tracking minimal residual disease in localized colon cancer. Ann. Oncol. 2019;30:1804–1812. doi: 10.1093/annonc/mdz390. [DOI] [PubMed] [Google Scholar]
- 12.Amelio I., Bertolo R., Bove P., Buonomo O.C., Candi E., Chiocchi M., Cipriani C., Di Daniele N., Ganini C., Juhl H., Mauriello A., Marani C., Marshall J., Montanaro M., Palmieri G., Piacentini M., Sica G., Tesauro M., Rovella V., Tisone G., Shi Y., Wang Y., Melino G. Liquid biopsies and cancer omics. Cell Death Dis. 2020;6:131. doi: 10.1038/s41420-020-00373-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tzanikou E., Markou A., Politaki E., Koutsopoulos A., Psyrri A., Mavroudis D., Georgoulias V., Lianidou E. PIK3CA hotspot mutations in circulating tumor cells and paired circulating tumor DNA in breast cancer: a direct comparison study. MOL ONCOL. 2019;13:2515–2530. doi: 10.1002/1878-0261.12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seeman N.C. From genes to machines: DNA nanomechanical devices. Trends Biochem. Sci. 2005;30:119–125. doi: 10.1016/j.tibs.2005.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan S.J., Campolongo M.J., Luo D., Cheng W. Building plasmonic nanostructures with DNA. Nat. Nanotechnol. 2011;6:268–276. doi: 10.1038/nnano.2011.49. [DOI] [PubMed] [Google Scholar]
- 16.Roh Y.H., Ruiz R.C., Peng S., Lee J.B., Luo D. Engineering DNA-based functional materials. Chem. Soc. Rev. 2011;40:5730–5744. doi: 10.1039/c1cs15162b. [DOI] [PubMed] [Google Scholar]
- 17.Yuan Y., Gu Z., Yao C., Luo D., Yang D. Nucleic acid-based functional nanomaterials as advanced cancer therapeutics. Small. 2019;15 doi: 10.1002/smll.201900172. [DOI] [PubMed] [Google Scholar]
- 18.Pinheiro A.V., Han D., Shih W.M., Yan H. Challenges and opportunities for structural DNA nanotechnology. Nat. Nanotechnol. 2011;6:763–772. doi: 10.1038/nnano.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong J., Zhou C., Wang Q. Towards active self-assembly through DNA nanotechnology. Top. Curr. Chem. 2020;378:33. doi: 10.1007/s41061-020-0297-5. [DOI] [PubMed] [Google Scholar]
- 20.Zhou L., Gao M., Fu W., Wang Y., Luo D., Chang K., Chen M. Three-dimensional DNA tweezers serve as modular DNA intelligent machines for detection and regulation of intracellular microRNA. SCI ADV. 2020;6:b695. doi: 10.1126/sciadv.abb0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W., Wang L., Jiang W. A catalytic assembled enzyme-free three-dimensional DNA walker and its sensing application. Chem. Commun. 2017;53:5527–5530. doi: 10.1039/c7cc02306e. [DOI] [PubMed] [Google Scholar]
- 22.Liu X., Zhang J., Fadeev M., Li Z., Wulf V., Tian H., Willner I. Chemical and photochemical DNA "gears" reversibly control stiffness, shape-memory, self-healing and controlled release properties of polyacrylamide hydrogels. Chem. Sci. 2019;10:1008–1016. doi: 10.1039/c8sc04292f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phan A.T., Do N.Q. Engineering of interlocked DNA G-quadruplexes as a robust scaffold. Nucleic Acids Res. 2013;41:2683–2688. doi: 10.1093/nar/gks1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Z.S., Shen Z., Tram K., Li Y. Engineering interlocking DNA rings with weak physical interactions. Nat. Commun. 2014;5:4279. doi: 10.1038/ncomms5279. [DOI] [PubMed] [Google Scholar]
- 25.Li Y., Cu Y.T., Luo D. Multiplexed detection of pathogen DNA with DNA-based fluorescence nanobarcodes. Nat. Biotechnol. 2005;23:885–889. doi: 10.1038/nbt1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu C.H., Cecconello A., Willner I. Recent advances in the synthesis and functions of reconfigurable interlocked DNA nanostructures. J. Am. Chem. Soc. 2016;138:5172–5185. doi: 10.1021/jacs.6b00694. [DOI] [PubMed] [Google Scholar]
- 27.Liao H., Huang T., Hu L., Wang M. Fluorescent aptasensors for parallel analysis of biomolecules based on interlocked DNA catenane nanomachines. Anal. Chim. Acta. 2020;1114:1–6. doi: 10.1016/j.aca.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Lu C.H., Qi X.J., Cecconello A., Jester S.S., Famulok M., Willner I. Switchable reconfiguration of an interlocked DNA olympiadane nanostructure. Angew Chem. Int. Ed. Engl. 2014;53:7499–7503. doi: 10.1002/anie.201403202. [DOI] [PubMed] [Google Scholar]
- 29.Li Q., Wu G., Yang Y., An R., Li J., Liang X., Komiyama M. Topology- and linking number-controlled synthesis of a closed 3 link chain of single-stranded DNA. Chem. Commun. 2018;54:10156–10159. doi: 10.1039/c8cc04965c. [DOI] [PubMed] [Google Scholar]
- 30.Liu M., Zhang Q., Li Z., Gu J., Brennan J.D., Li Y. Programming a topologically constrained DNA nanostructure into a sensor. Nat. Commun. 2016;7:12074. doi: 10.1038/ncomms12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li T., Lohmann F., Famulok M. Interlocked DNA nanostructures controlled by a reversible logic circuit. Nat. Commun. 2014;5:4940. doi: 10.1038/ncomms5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu C.H., Cecconello A., Elbaz J., Credi A., Willner I. A three-station DNA catenane rotary motor with controlled directionality. Nano Lett. 2013;13:2303–2308. doi: 10.1021/nl401010e. [DOI] [PubMed] [Google Scholar]
- 33.Ma Y., Centola M., Keppner D., Famulok M. Interlocked DNA nanojoints for reversible thermal sensing. Angew Chem. Int. Ed. Engl. 2020;59:12455–12459. doi: 10.1002/anie.202003991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou W., Li D., Yuan R., Xiang Y. Programmable DNA ring/hairpin-constrained structure enables ligation-free rolling circle amplification for imaging mRNAs in single cells. Anal. Chem. 2019;91:3628–3635. doi: 10.1021/acs.analchem.8b05613. [DOI] [PubMed] [Google Scholar]
- 35.Liu X., Lu C.H., Willner I. Switchable reconfiguration of nucleic acid nanostructures by stimuli-responsive DNA machines. Acc. Chem. Res. 2014;47:1673–1680. doi: 10.1021/ar400316h. [DOI] [PubMed] [Google Scholar]
- 36.Zhang D.Y., Seelig G. Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. 2011;3:103–113. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- 37.Elbaz J., Wang Z.G., Wang F., Willner I. Programmed dynamic topologies in DNA catenanes. Angew Chem. Int. Ed. Engl. 2012;51:2349–2353. doi: 10.1002/anie.201107591. [DOI] [PubMed] [Google Scholar]
- 38.Spruell J.M., Coskun A., Friedman D.C., Forgan R.S., Sarjeant A.A., Trabolsi A., Fahrenbach A.C., Barin G., Paxton W.F., Dey S.K., Olson M.A., Benitez D., Tkatchouk E., Colvin M.T., Carmielli R., Caldwell S.T., Rosair G.M., Hewage S.G., Duclairoir F., Seymour J.L., Slawin A.M., Goddard W.R., Wasielewski M.R., Cooke G., Stoddart J.F. Highly stable tetrathiafulvalene radical dimers in [3]catenanes. Nat. Chem. 2010;2:870–879. doi: 10.1038/nchem.749. [DOI] [PubMed] [Google Scholar]
- 39.Smith M., Smith K., Olstein A., Oleinikov A., Ghindilis A. Restriction endonuclease-based assays for DNA detection and isothermal exponential signal amplification. Sensors. 2020;20:3873. doi: 10.3390/s20143873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma L., Zhu Z., Li T., Wang Z. Assaying multiple restriction endonucleases functionalities and inhibitions on DNA microarray with multifunctional gold nanoparticle probes. Biosens. Bioelectron. 2014;52:118–123. doi: 10.1016/j.bios.2013.08.045. [DOI] [PubMed] [Google Scholar]
- 41.Xu H., Zhang Y., Zhang S., Sun M., Li W., Jiang Y., Wu Z.S. Ultrasensitive assay based on a combined cascade amplification by nicking-mediated rolling circle amplification and symmetric strand-displacement amplification. Anal. Chim. Acta. 2019;1047:172–178. doi: 10.1016/j.aca.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 42.Kay E.R., Leigh D.A., Zerbetto F. Synthetic molecular motors and mechanical machines. Angew Chem. Int. Ed. Engl. 2007;46:72–191. doi: 10.1002/anie.200504313. [DOI] [PubMed] [Google Scholar]
- 43.Bjorkesten J., Patil S., Fredolini C., Lonn P., Landegren U. A multiplex platform for digital measurement of circular DNA reaction products. Nucleic Acids Res. 2020;48:e73. doi: 10.1093/nar/gkaa419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soares R., Varela J.C., Neogi U., Ciftci S., Ashokkumar M., Pinto I.F., Nilsson M., Madaboosi N., Russom A. Sub-attomole detection of HIV-1 using padlock probes and rolling circle amplification combined with microfluidic affinity chromatography. Biosens. Bioelectron. 2020;166:112442. doi: 10.1016/j.bios.2020.112442. [DOI] [PubMed] [Google Scholar]
- 45.Zhang X., Zhao W., Wei W., You Z., Ou X., Sun M., Yin Y., Tang X., Zhao Z., Hu C., Liu F., Deng J., Mao L., Zhou D., Ren Y., Li X., Zhang S., Liu C., Geng J., Yao G., Song B., Liu Y., Li D., Jiang Y., Chen Y., Zhao Y., Yu S., Pang D. Parallel analyses of somatic mutations in plasma circulating tumor DNA (ctDNA) and matched tumor tissues in early-stage breast cancer. CLIN CANCER RES. 2019;25:6546–6553. doi: 10.1158/1078-0432.CCR-18-4055. [DOI] [PubMed] [Google Scholar]
- 46.Leng Y., Sun K., Chen X., Li W. Suspension arrays based on nanoparticle-encoded microspheres for high-throughput multiplexed detection. Chem. Soc. Rev. 2015;44:5552–5595. doi: 10.1039/c4cs00382a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu W., Wang X., Shen M., Li L., Yin Y., Shen L., Wang W., Cui D., Ni J., Chen X., Li W. AIEgens barcodes combined with AIEgens nanobeads for high-sensitivity multiplexed detection. THERANOSTICS. 2019;9:7210–7221. doi: 10.7150/thno.36525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma C.S., Tangye S.G. Flow cytometric-based analysis of defects in lymphocyte differentiation and function due to inborn errors of immunity. Front. Immunol. 2019;10:2108. doi: 10.3389/fimmu.2019.02108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo Q., Wang Y., Chen C., Wei D., Fu J., Xu H., Gu H. Multiplexed luminescence oxygen channeling immunoassay based on dual-functional barcodes with a host-guest structure: a facile and robust suspension array platform. Small. 2020;16 doi: 10.1002/smll.201907521. [DOI] [PubMed] [Google Scholar]
- 50.Smith L.D., Liu Y., Zahid M.U., Canady T.D., Wang L., Kohli M., Cunningham B.T., Smith A.M. High-fidelity single molecule quantification in a flow cytometer using multiparametric optical analysis. ACS Nano. 2020;14:2324–2335. doi: 10.1021/acsnano.9b09498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu P., Zhang S., Wu T., Ni D., Fan W., Zhu Y., Qian R., Shi J. Fe-Au nanoparticle-coupling for ultrasensitive detections of circulating tumor DNA. Adv. Mater. 2018;30:1801690. doi: 10.1002/adma.201801690. [DOI] [PubMed] [Google Scholar]
- 52.Li Q., Wu G., Yang Y., An R., Li J., Liang X., Komiyama M. Topology- and linking number-controlled synthesis of a closed 3 link chain of single-stranded DNA. Chem. Commun. 2018;54:10156–10159. doi: 10.1039/c8cc04965c. [DOI] [PubMed] [Google Scholar]
- 53.Yang X., Li T., Li B., Wang E. Potassium-sensitive G-quadruplex DNA for sensitive visible potassium detection. Analyst. 2010;135:71–75. doi: 10.1039/b913036e. [DOI] [PubMed] [Google Scholar]
- 54.Huang R., He L., Xia Y., Xu H., Liu C., Xie H., Wang S., Peng L., Liu Y., Liu Y., He N., Li Z. A sensitive aptasensor based on a hemin/G-quadruplex-assisted signal amplification strategy for electrochemical detection of gastric cancer exosomes. Small. 2019;15 doi: 10.1002/smll.201900735. [DOI] [PubMed] [Google Scholar]
- 55.Paweletz C.P., Sacher A.G., Raymond C.K., Alden R.S., O'Connell A., Mach S.L., Kuang Y., Gandhi L., Kirschmeier P., English J.M., Lim L.P., Janne P.A., Oxnard G.R. Bias-corrected targeted next-generation sequencing for rapid, multiplexed detection of actionable alterations in cell-free DNA from advanced lung cancer patients. CLIN CANCER RES. 2016;22:915–922. doi: 10.1158/1078-0432.CCR-15-1627-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang D.Y., Brandwein M., Hsuih T.C., Li H. Amplification of target-specific, ligation-dependent circular probe. Gene. 1998;211:277–285. doi: 10.1016/s0378-1119(98)00113-9. [DOI] [PubMed] [Google Scholar]
- 57.Yi J., Zhang W., Zhang D.Y. Molecular Zipper: a fluorescent probe for real-time isothermal DNA amplification. Nucleic Acids Res. 2006;34:e81. doi: 10.1093/nar/gkl261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.