Abstract
Objectives: The study aimed to characterize the clonal spread of resistant bacteria and dissemination of resistance plasmids among carbapenem-resistant Enterobacterales at a tertiary hospital in Catalonia, Spain.
Methods: Isolates were recovered from surveillance rectal swabs and diagnostic samples. Species identification was by matrix-assisted laser desorption ionization-time time of flight mass spectrometry (MALDI-TOF MS). Molecular typing was performed by pulsed-field gel electrophoresis (PFGE) and multi-locus sequence typing (MLST). Antimicrobial susceptibility was assessed by gradient-diffusion and carriage of bla genes was detected by PCR. Plasmid typing, conjugation assays, S1-PFGE studies and long-read sequencing were used to characterize resistance plasmids.
Results: From July 2018 to February 2019, 125 Klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacterales were recovered from 101 inpatients from surveillance (74.4%) or clinical samples (25.6%), in a tertiary hospital in Barcelona. Clonality studies identified a major clone of Klebsiella pneumoniae belonging to sequence type ST15 and additional isolates of K. pneumoniae, Escherichia coli and Enterobacter sp. from different STs. All isolates but one carried the blaKPC–2 allelic variant. The blaKPC–2 gene was located in an IncFIIk plasmid of circa 106 Kb in a non-classical Tn4401 element designated NTEKPC-pMC-2-1. Whole-genome sequencing revealed different rearrangements of the 106 Kb plasmid while the NTEKPC-pMC-2-1 module was highly conserved.
Conclusion: We report a hospital outbreak caused by the clonal dissemination of KPC-producing ST15 K. pneumoniae but also the intra- and inter-species transmission of the blaKPC–2 gene associated with plasmid conjugation and/or transposon dissemination. To our knowledge, this is the first report of an outbreak caused by KPC-producing Enterobacterales isolated from human patients in Catalonia and highlights the relevance of surveillance studies in the early detection and control of antibiotic resistant high-risk clones.
Keywords: Klebsiella, outbreak, antibiotic resistance, carbapenemase, epidemiology, plasmid, high-risk clone, KPC
Introduction
The rapid emergence of bacterial pathogens presenting resistance to multiple antimicrobial agents (MDR) together with the decreasing trend in the development of new antimicrobial compounds constitute an extremely serious threat to public health (Roca et al., 2015). Resistance rates are particularly alarming among Gram-negative bacteria (GNB) as available treatment options are severely impaired. In 2016 a report commissioned by the UK Prime minister, known as the “O’Neill report,” attributed 700.000 annual human deaths worldwide to microbial infections caused by antimicrobial resistant (AMR) pathogens (O’neill, 2016). The same report estimated that this figure would rise to 10 million annual deaths by 2050 unless there is a global effort to tackle AMR. Likewise, organizations such as the US Centers for Disease Control and Prevention (CDC), the European Centre for Disease Prevention and Control (ECDC) and the World Health Organization (WHO) are considering infections caused by MDR bacteria as an emergent global disease and a major public health problem (Roca et al., 2015). According to their mortality, health-care and community burden, prevalence of resistance and treatability, carbapenem-resistant Enterobacterales have been included into the 2017 WHO priority list of antibiotic-resistant bacterial pathogens and currently constitute one of the major public health threats worldwide (Tacconelli et al., 2017).
Among carbapenem-resistant Enterobacterales, Klebsiella pneumoniae stands out as a formidable nosocomial pathogen causing several infections associated with high mortality rates (Tumbarello et al., 2012). In Europe, carbapenem-resistant K. pneumoniae accounted for almost 16,000 infections and more than 2,000 deaths in 2015 (Cassini et al., 2018). Carbapenem-resistance is usually associated with the carriage of genes encoding carbapenem-hydrolyzing enzymes, most of which are commonly located within conjugative plasmids and, therefore, are easily disseminated among nosocomial pathogens (Bonomo et al., 2017). Predominant carbapenemases in Europe include OXA-48 oxacillinase, New Delhi metallo-β-lactamase (NDM) and Klebsiella pneumoniae carbapenemase (KPC), among others. In Spain, OXA-48 is currently the predominant mechanism associated with the dissemination of carbapenem-resistant Enterobacterales (Grundmann et al., 2017). NDM-producing bacteria are reported less frequently in Spain but their presence is increasing and associated with a few clonal lineages (Marí-Almirall et al., 2021) while KPC-producing Enterobacterales are also sporadic in Spain, and only a few outbreaks have been reported in Madrid and Andalusia (Oteo et al., 2016).
Klebsiella pneumoniae carbapenemase is likely one of the most common mechanisms of resistance to carbapenems in K. pneumoniae, and the worldwide spread of the blaKPC gene was initially associated with the dissemination of a few K. pneumoniae clonal lineages within the ST258 clonal group (CG258), a particular plasmid backbone (pKpQIL) and a highly conserved Tn4401 transposon (Chen et al., 2014). More recently blaKPC has been found on a variety of clonal lineages and plasmids from different incompatibility groups (Wyres et al., 2020), and although it is usually associated with a composite Tn4401 transposon, some structural variations have also been described (Shen et al., 2009; Naas et al., 2012; Chmelnitsky et al., 2014). In this retrospective study we have examined the rapid dissemination of KPC-producing Enterobacterales within a tertiary hospital in Barcelona and we have investigated the inter- and intra-species spread of carbapenem resistance.
Materials and Methods
This study included 125 carbapenem-resistant isolates of Enterobacterales collected from July 2018 to February 2019 at one tertiary hospital in Barcelona, Spain. Strains were recovered from surveillance and clinical samples (Supplementary Table 2). Identification at species level was performed by matrix-assisted laser desorption ionization-time time of flight mass spectrometry (MALDI-TOF MS) in a Microflex LT benchtop instrument (Bruker Daltonics) operated in linear positive mode.
Antimicrobial Susceptibility Testing and Detection of Resistance
Antimicrobial susceptibility was assessed by gradient diffusion (E-test, BioMérieux, Spain) on Muller-Hinton agar plates (Becton-Dickinson, Spain) for the following antimicrobials: imipenem, meropenem, ceftazidime, cefepime, cefotaxime, amikacin, tobramycin, gentamicin, kanamycin, tigecycline, ciprofloxacin, levofloxacin, ceftazidime-avibactam and fosfomycin. Susceptibility to colistin was assessed by gradient diffusion on Iso-Sensitest agar plates (ThermoFisher, United Kingdom). On note, colistin susceptibility was tested under research use only (RUO) acknowledgment since the recommended method for in vitro diagnostic (IVD) is broth microdilution (European Committee on Antimicrobial Susceptibility Testing (EUCAST), 2016). The MICs were interpreted according to EUCAST breakpoints for Enterobacterales (European Committee on Antimicrobial Susceptibility Testing (EUCAST), 2021). Escherichia coli ATCC 25922 was used for quality control.
Production of KPC, OXA-48-like, VIM, IMP or NDM carbapenemases was detected with the NG-Test®CARBA5 (NG-Biotech, France) and confirmed by PCR using previously described primers and conditions (Queenan and Bush, 2007; Solé et al., 2011; Bogaerts et al., 2013). Amplification products were purified from agarose gels and sent for Sanger sequencing (Genewiz, Germany) whenever necessary. The allelic identity of all genes was determined by sequence alignment with reference sequences retrieved from public repositories (PRJNA313047, last accessed June, 2021).
Clonal Relatedness
Clonality was studied by pulsed-field gel electrophoresis (PFGE) using XbaI (New England BioLabs Inc., United States) genomic digestions and a CHEFF-DRIII system (Bio-Rad, Spain) (Marí-Almirall et al., 2021). Molecular patterns were analyzed with InfoQuestTMFP-v.5.4 (Bio-Rad, Spain) and the unweighted pair group method with arithmetic mean (UPGMA) to create dendrograms based on Dice’s similarity coefficient. Bandwidth tolerance and optimization values were set at 2% and isolates were considered within the same PFGE cluster (pulsotype) if their Dice similarity index was >85%.
Multi-locus sequence typing (MLST) was performed according to the Pasteur scheme for K. pneumoniae and the Achtman scheme for E. coli (Diancourt et al., 2005; Wirth et al., 2006).
Plasmid Analysis
Conjugation assays were carried out using E. coli MC1061 resistant to rifampicin and sodium azide as the recipient strain. Transconjugant strains were selected in LB agar plates supplemented with 1 mg/L of meropenem and 100 mg/L of sodium azide (Sigma-Aldrich, Spain). The location of resistant genes was determined by S1-nuclease digestion (New England BioLabs Inc., United States) followed by PFGE and Southern blot hybridization with digoxigenin-labeled PCR-probes against blaKPC (Bogaerts et al., 2013). Plasmid incompatibility groups were determined using the PBRT-2.0 kit (Diatheva, Italy) (Carattoli et al., 2005). The genetic environment of blaKPC was determined by PCR using primers listed in Supplementary Table 1 and confirmed by long-read sequencing.
Genomic DNA was extracted using the Wizard Genomic DNA purification kit (Promega, Spain) and sequenced on a MinION instrument (Oxford-Nanopore, United Kingdom) using the rapid barcoding kit (SQK-RBK004) for library preparation and a R9.4.1 flowcell following the manufacturer’s instructions. Basecalling was done with Guppy-v3.0.3 and demultiplexing with qcat-v1.1.01. FASTQ files were mapped using Minimap2-v2.17 against plasmids from Enterobacterales (Orlek et al., 2017). Mapping reads were assembled with Flye-v2.52. Annotation was done with Prokka-v.1.12 combined with BLASTP/BLASTN searches against the UniProtKB/Swiss-Prot and RefSeq databases (Seemann, 2014).
ResFinder3, PlasmidFinder4, and ISFinder5 were used to identify antimicrobial resistance genes, plasmid replicons and mobile elements, respectively. The presence of virulence genes in plasmid sequences was checked against the Pasteur MLST site6. Genetic diagrams were drawn using SnapGene®Viewer-v5.1.27 and CGViewAdvanced-v.0.0.1 (Stothard and Wishart, 2005).
FASTQ files of isolates MC-2-1, MC-2-207, MC-2-230, MC-2-240, MC-2-251, MC-2-304, MC-2-315, MC-2-316, MC-2-362, and MC-2-387, were deposited into the NCBI Sequence Read Archive under accession numbers SRX11379706, SRX11379707, SRX11379708, SRX11379709, SRX11379710, SRX11379711, SRX11379712, SRX11379713, SRX11379714, and SRX11379715, respectively; BioProject PRJNA744857.
Results
Bacterial Isolation and Clonal Relatedness
In mid-July 2018, five carbapenem-resistant K. pneumoniae isolates were recovered from surveillance samples (rectal swabs) upon the implementation of a “zero-resistance” surveillance program to screen all patients admitted to intensive care units within the hospital. All five isolates produced a KPC-type enzyme. During the following weeks, additional surveillance samples and an increasing number of clinical samples flagged positive for KPC-producing Enterobacterales. By the end of August, 87 KPC-producing isolates had been recovered and infection control measures were initiated, including exhaustive room cleaning twice a day, reinforcing surveillance measures, skin cleaning with 2% chlorhexidine wipes, and oral decolonization of patients (amikacin, colistin, nystatin) (Pellicé et al., 2021).
During the following months the number of KPC-producing isolates rapidly decreased and the outbreak was considered eradicated by November 2018, although some isolates were still recovered up until February 2019 (Figure 1). Overall, 125 isolates (112 K. pneumoniae, 12 E. coli, and 1 Enterobacter sp.) from 101 patients were collected and included for further studies. Thirty-two isolates (25.6%) were detected from diagnosis samples and 93 (74.4%) were isolated from surveillance samples (Supplementary Table 2). The outbreak involved the screening of 2031 patients. The Index case could not be identified.
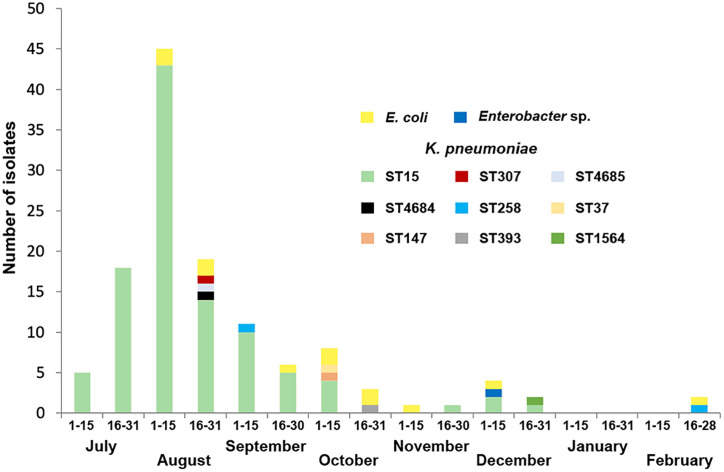
FIGURE 1.
Temporal distribution of KPC-producing K. pneumoniae isolates according to sequence types (STs) as well as KPC-producing E. coli and Enterobacter sp. isolates recovered in this study.
All K. pneumoniae strains and the single Enterobacter sp. isolate were classified into 11 PFGE clusters or pulsotypes (designated as PTk, to differentiate from PTc, derived from E. coli isolates, see below) with a major cluster, PTk1, that included 103 isolates while all other clusters just contained singletons. According to MLST, PTk-1 K. pneumoniae isolates were identified as ST15, PTk-2 as ST4684, PTk-3 as ST4685, PTk-5 as ST147, PTk-6 as ST1564, PTk-7 and PTk-8 as ST258, Ptk-9 as ST307, PTk-10 as ST393, PTk-11 as ST37, and PTk-4 corresponded to the single Enterobacter sp. isolate (Supplementary Figure 1).
Among E. coli isolates, PFGE showed higher clonal heterogeneity, since the 12 strains were grouped into 10 clusters (PTc 1-10), with only clusters 1 and 10 containing two strains each and all other pulsotypes corresponding to singletons (Supplementary Figure 2). According to MLST, the PTc-1 E. coli isolates were identified as ST101, PTc-2 and PTc-8 E. coli isolates were identified as ST410, PTc-3 as ST38, PTc-4 as ST8576, PTc-5 as ST1236, PTc-6 as ST1193, PTc-7 as ST131, PTc-9 as ST1431 and PTc-10 as ST1642.
One strain from each cluster of either K. pneumoniae or E. coli were selected for further phenotypic and genotypic characterization. A total of 24 isolates were eventually selected, 13 K. pneumoniae, 10 E. coli isolates, and 1 Enterobacter sp. (Table 1).
TABLE 1.
Antimicrobial susceptibility and molecular characterization of representative KPC-producing K. pneumoniae, Enterobacter sp. and E. coli isolates.
| Strains | Bacterial Species | bla gene | ST | MIC (mg/L) | Tn Type | P Inc | P Size | ||||||||||||||
|
|
|||||||||||||||||||||
| IPM | MEM | CAZ | FEP | CTX | AMK | TOB | GEN | KAN | CST | TGC | CIP | LVX | CAZ/AVI | FOF | |||||||
| MC-2-1 | K. pneumoniae | KPC-2 | ST15 | 24 | >32 | 32 | 6 | >32 | 1 | 0,38 | 0,38 | 1 | 2 | 1,5 | 12 | 4 | 0,75 | 24 | pMC-2-1 | IncFIIk | 106 Kb |
| MC-2-16 | K. pneumoniae | KPC-2 | ST15 | >32 | >32 | 24 | 6 | >32 | 1,5 | 0,5 | 0,38 | 1,5 | 0,094 | 1 | >32 | 4 | 0,75 | 32 | pMC-2-1 | IncFIIk | 106 Kb |
| MC-2-20 | K. pneumoniae | KPC-2 | ST15 | >32 | >32 | 24 | 4 | >32 | 1 | 0,38 | 0,38 | 1,5 | 1 | 1,5 | 16 | 3 | 0,75 | 12 | pMC-2-1 | IncFIIk | 106 Kb |
| MC-2-146 | K. pneumoniae | KPC-2 | ST15 | >32 | >32 | 32 | 48 | >32 | 1 | 0,38 | 0,38 | 1 | 0,25 | 1,5 | 16 | 6 | 1 | 32 | pMC-2-1 | IncFIIk | 106 Kb |
| MC-2-177 | E. coli | KPC-2 | ST410 | 4 | 3 | 192 | 32 | >32 | 4 | 16 | 32 | 32 | 0,5 | 0,5 | >32 | >32 | 0,5 | 1 | pMC-2-1 | IncFIIk | 106 Kb |
| MC-2-196 | E. coli | KPC-2 | ST8576 | >32 | 16 | 48 | >256 | >32 | 1 | 0,38 | 0,5 | 1 | 0,125 | 0,75 | 0,008 | 0,023 | 0,125 | 1 | pMC-2-1 | IncFIIk | 106 Kb |
| MC-2-203 | K. pneumoniae | KPC-2 | ST4684 | 4 | >32 | 24 | >256 | >32 | 1 | 0,25 | 0,38 | 1 | 0,38 | 1 | 0,016 | 0,047 | 0,38 | 8 | pMC-2-1 | IncFIIk | 106 Kb |
| MC-2-207 | E. coli | KPC-2 | ST1236 | 32 | 16 | 192 | >256 | >32 | 2 | 0,5 | 0,5 | 2 | 0,325 | 0,25 | 0,016 | 0,032 | 0,38 | 0,5 | pMC-2-1 | IncFIIk | 106 Kb |
| MC-2-216 | K. pneumoniae | KPC-2 | ST4685 | 3 | 1 | 48 | 4 | >32 | 1,5 | 2 | 0,38 | 4 | 0,25 | 1 | 0,5 | 0,38 | 0,25 | 6 | pMC-2-1 | IncFIIk | 106 Kb |
| MC-2-230 | K. pneumoniae | KPC-2 | ST307 | 24 | >32 | 24 | 8 | >32 | 1 | 0,25 | 0,25 | 1 | 1 | 0,75 | 2 | 2 | 0,75 | 16 | pMC-2-1 | IncFIIk IncN | 281 Kb |
| MC-2–240 | E. coli | KPC-2 | ST38 | 1 | 1 | 8 | 6 | 32 | 1,5 | 0,75 | 0,5 | 1,5 | 0,38 | 0,19 | 0,008 | 0,16 | 0,125 | 0,75 | pMC-2-1 | IncFIIk | 190, 50 Kb |
| MC-2-251 | K. pneumoniae | KPC-3 | ST258 | >32 | >32 | >256 | >32 | >32 | 32 | 16 | 1 | >256 | 0,25 | 1 | >32 | >32 | 2 | 8 | Tn4401 | IncFIIk | 78 Kb |
| MC-2-285 | E. coli | KPC-2 | ST410 | 12 | 8 | 24 | 8 | >32 | 1 | 0,19 | 0,19 | >256 | 0,25 | 0,25 | >32 | >32 | 0,19 | >1024 | pMC-2-1 | IncFIIk | 106 Kb |
| MC-2-303 | K. pneumoniae | KPC-2 | ST37 | 4 | >32 | 24 | 12 | >32 | 1 | 0,19 | 0,19 | 0,75 | 0,38 | 2 | 0,125 | 0,38 | 1 | 32 | pMC-2-1 | IncFIIk | 106 Kb |
| MC-2-304 | E. coli | KPC-2 | ST1642 | 12 | 1,5 | 4 | 3 | >32 | 4 | 16 | 128 | >256 | 0,125 | >32 | >32 | >32 | 0,19 | 0,75 | pMC-2-1 | IncFIIk IncU | 170 Kb |
| MC-2-306 | K. pneumoniae | KPC-2 | ST147 | >32 | >32 | 16 | 8 | >32 | 2 | 8 | 16 | 16 | 3 | 3 | >32 | >32 | 1 | 32 | pMC-2-1 | IncFIIk | 106 Kb |
| MC-2-315 | E. coli | KPC-2 | ST101 | >32 | >32 | 192 | 128 | >32 | 1,5 | 0,5 | 0,25 | 1,5 | 0,125 | 0,75 | 0,125 | 0,5 | 0,75 | 1 | pMC-2-1 | IncFIIk | 101 Kb |
| MC-2-316 | K. pneumoniae | KPC-2 | ST393 | >32 | >32 | 16 | 16 | >32 | 1,5 | 0,25 | 0,38 | 1 | 0,25 | 0,75 | 0,016 | 0,047 | 0,75 | 16 | pMC-2-1 | IncFIIk | 109 Kb |
| MC-2-328 | E. coli | KPC-2 | ST1431 | 4 | 4 | 12 | 192 | >32 | 1 | 0,5 | 0,25 | 2 | 0,25 | 0,5 | >32 | >32 | 0,125 | 0,75 | pMC-2-1 | IncFIIk | 106 Kb |
| MC-2-350 | E. coli | KPC-2 | ST1193 | 3 | 24 | 16 | 24 | >32 | 2 | 16 | >256 | 16 | 0,125 | 0,38 | >32 | >32 | 0,25 | 0,75 | pMC-2-1 | IncFIIk | 106 Kb |
| MC-2-362 | Enterobacter sp. | KPC-2 | ND | >32 | >32 | 48 | 48 | >32 | 2 | 12 | 32 | 16 | 1 | 4 | >32 | >32 | 1,5 | 12 | pMC-2-1 | IncFIIk IncN IncU | 408 Kb |
| MC-2-382 | E. coli | KPC-2 | ST131 | 24 | 24 | 96 | 256 | >32 | 6 | 16 | 16 | 16 | 1 | 0,25 | >32 | >32 | 0,25 | 1 | pMC-2-1 | IncFIIk | 106 Kb |
| MC-2-387 | K. pneumoniae | KPC-2,-3 | ST258 | >32 | >32 | >256 | >32 | >32 | 32 | >256 | 16 | >256 | 0,38 | 1,5 | >32 | >32 | 1 | 32 | pMC-2-1 Tn4401 | IncFIIk | 120; 28 Kb |
| MC-22-164 | K. pneumoniae | KPC-2 | ST1564 | >32 | >32 | 24 | 64 | >32 | 1 | 0,19 | 0,38 | 1 | 1 | 2 | 1,5 | 3 | 1 | >1024 | pMC-2-1 | IncFIIk | 106 Kb |
ST, sequence type; IPM, imipenem; MEM, meropenem; CAZ, ceftazidime; FEP, cefepime; CTX, cefotaxime; AMK: amikacin; TOB: tobramycin; GEN, gentamicin; KAN, kanamycin; CST, colistin; TGC, tigecycline; CIP, ciprofloxacin; LVX, levofloxacin; CAZ/AVI, ceftazidime/avibactam; FOF, fosfomycin; Tn Type, type of transposon containing the blaKPC gene; P Inc, Incompatibility group of plasmids containing the blaKPC gene. Inc groups from sequenced plasmids are shown in bold face; P Size, Size of plasmids containing the blaKPC gene; ND, not determined.
Phenotypic and Molecular Characterization of Resistance
The MICs of selected isolates are shown in Table 1. All isolates were non-susceptible to cephalosporins and carbapenems, but were susceptible to ceftazidime-avibactam, fosfomycin and colistin, except for MC-2-285 and MC-22-164, that were highly resistant to fosfomycin.
Sanger sequencing identified the blaKPC–2 variant in all isolates except for the MC-2-251 and MC-2-387 isolates, that carried blaKPC–3 (note that MC-2-387 also carried blaKPC–2). The genes for CTX-M-group 1 enzymes were also detected in 7 of the 24 strains, while CTX-M-group 2, 9, and 25 were only found in a few strains (Supplementary Table 2). CTX-M-group 8 was not detected. All isolates were negative for the presence of blaOXA–48, blaNDM, blaVIM, or blaIMP.
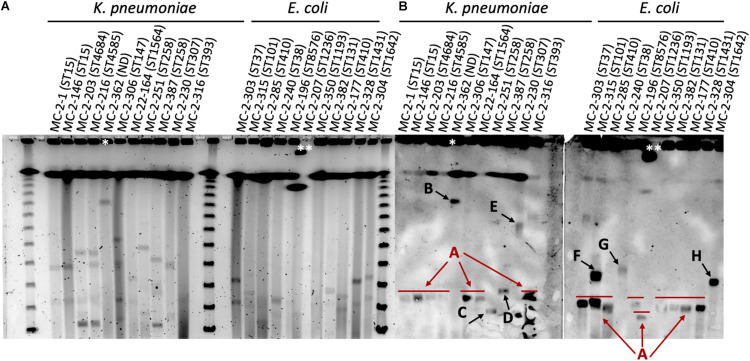
S1-nuclease-PFGE profiles of selected strains revealed the presence of different plasmids (according to size) that contained the blaKPC gene. Eight of the selected K. pneumoniae strains harbored blaKPC–2 within a plasmid of circa 110 kb in size (plasmid type A, Figure 2), including the ST15 strains. The two ST258 strains MC-2-251 and MC-2-387 carrying blaKPC–3, however, showed hybridization signals with plasmids of circa 80 Kb and 120 Kb, respectively (plasmid types C and D, Figure 2), and the single ST307 strain (MC-2-230) carried blaKPC–2 within a plasmid of circa 300 kb (plasmid type E, Figure 2). Likewise, the Enterobacter sp. isolate harbored blaKPC–2 in a plasmid of approximately 400 Kb (plasmid type B, Figure 2).
FIGURE 2.
(A) S1-nuclease-PFGE profiles and (B) corresponding southern hybridization membranes with blaKPC probes of selected K. pneumoniae and E. coli isolates in this study. The ST of both K. pneumoniae and E. coli isolates is shown in brackets. Arrows indicate the different arbitrarily designated plasmid types. Lambda ladder PFGE Marker (New England Biolabs, United States). *Enterobacter sp. isolate. **The PFGE plug moved from the loading site, so the band is shifted. ND, not determined.
Among selected E. coli isolates, eight isolates also carried blaKPC–2 in plasmids of similar size as that of type A plasmids in ST15 K. pneumoniae (110 Kb), although the MC-2-315 isolate presented a second hybridization signal at circa 180 Kb (plasmid type F, Figure 2). Two additional strains, MC-2-240 and MC-2-304, carried blaKPC–2 in plasmids of different sizes. MC-2-240 showed a hybridization band at approximately 190 Kb, and MC-2-304 at 170 Kb (plasmid types G and H, respectively, Figure 2).
Notably, all E. coli isolates but one and the single Enterobacter sp. isolate recovered during the outbreak, originated from patients that previously carried a ST15 KPC-2-producing K. pneumoniae. On the other hand, only three out of the nine non-ST15 K. pneumoniae isolates were recovered from patients also co-carrying a ST15 isolate (Supplementary Figure 4 and Supplementary Table 2). Plasmid replicon typing identified the presence of multiple plasmids in some of the strains (Table 1), but all strains were positive for a replicon belonging to the IncFIIk incompatibility group. Conjugation assays using the ST15 isolate MC-2-1 as donor and the E. coli strain MC1061 as recipient, showed that the acquisition of a blaKPC–2 gene was associated with the transfer of an IncFIIk plasmid (not shown).
Plasmid Sequencing
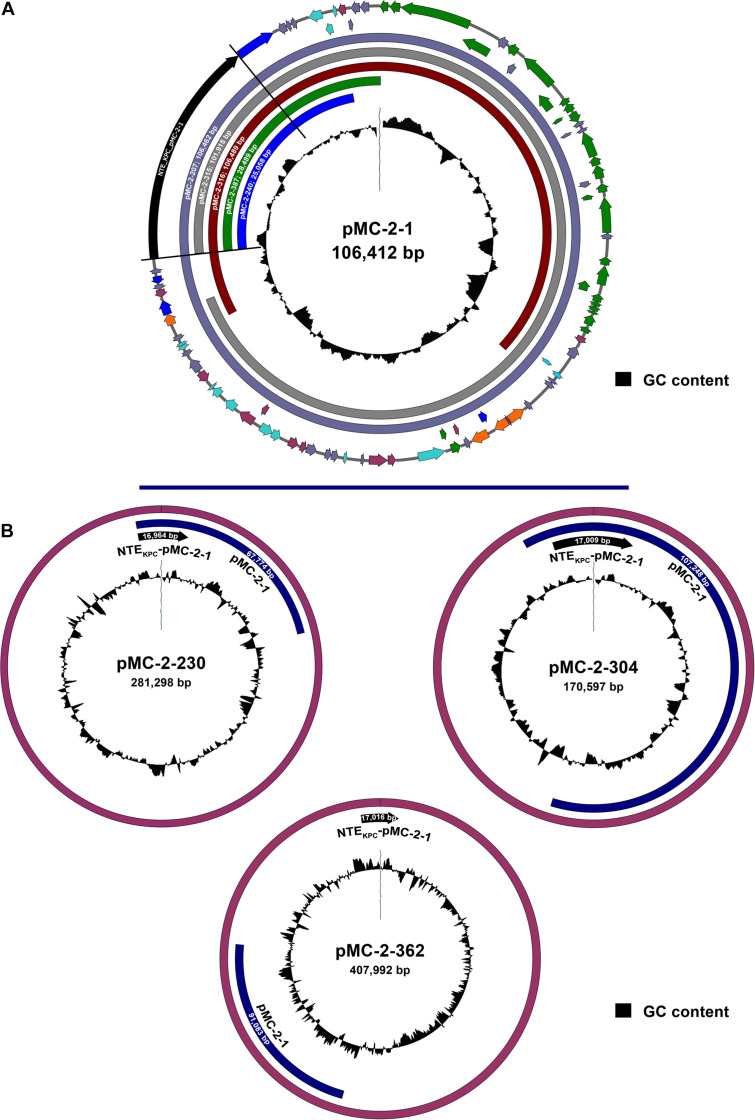
Ten isolates, 5 K. pneumoniae, 4 E. coli, and 1 Enterobacter sp., representative of strains carrying all the different plasmid types identified with hybridization probes, were further selected for long-read plasmid sequencing. Plasmid sequencing identified an IncFIIk plasmid of 106,412 bp carrying blaKPC–2 in the ST15 K. pneumoniae MC-2-1 isolate, designated as pMC-2-1 (Supplementary Figure 3). The blaKPC–2 gene was the only antibiotic resistance gene present in the pMC-2-1 plasmid that also harbored several genes involved in conjugative transfer. The presence of plasmid-associated virulence genes was not detected in plasmid pMC-2-1 (Supplementary Figure 3). An almost identical plasmid of 106,462 bp was found in the E. coli strain MC-2-207, and highly similar IncFIIk plasmids of 109,489 and 101,915 bp were located in the K. pneumoniae and E. coli isolates MC-2-316 and MC-2-315, respectively, both recovered from the same patient (Figure 3A), in good agreement with the carriage of blaKPC–2 in a plasmid of circa 110 Kb (plasmid type A), as identified by S1-digestion. Unfortunately, plasmid type F of circa 180 Kb, also present in strain MC-2-315 according to S1-digestion, could not be identified by long-read sequencing analysis.
FIGURE 3.
(A) Schematic representation of the 106,412 bp IncFIIk plasmid from the ST15 K. pneumoniae strain MC-2-1 (pMC-2-1) carrying blaKPC–2 within a NTEKPC-pMC-2-1 genetic structure and sequence alignment with similar IncFIIk plasmid backbones from strains MC-2-207, MC-2-315, MC-2-316, MC-2-387, and MC-2-240 (only the aligned region is shown). Arrows are proportional to the lengths of the genes and oriented in the direction of transcription. Red arrows represent resistance genes, orange arrows represent full-length transposon-related genes and ISs, dark blue arrows represent partial or truncated transposon-related genes and ISs, green arrows indicate genes involved in plasmid conjugation and light blue arrows shown genes related to plasmid replication/maintenance. Blue-gray arrows show putative or hypothetical genes and plum arrows show genes involved in other functions. The inner circle shows the GC content. The fully annotated sequence of plasmid pMC-2-1 is shown in Supplementary Figure 3. (B) Schematic representation of the larger IncFIIk plasmids carrying blaKPC–2 recovered from strains MC-2-230, MC-2-304, and MC-2-363. The location of the genetic structure NTEKPC-pMC-2-1 harboring blaKPC–2 is shown as well as the region matching the pMC-2-1 sequence. The inner circle shows the GC content.
The K. pneumoniae isolate MC-2-230, the Enterobacter sp. isolate MC-2-362 and the E. coli isolate MC-2-304, carried blaKPC–2 within IncFIIk plasmids of 281,298, 407,992, and 170,594 bp, respectively, much larger than that of ST15 strains and in good agreement with results from S1-nuclease digestion. Additional replicon types from IncN and/or IncU incompatibility groups were also identified in these plasmids. Interestingly, the entire 106 Kb sequence that made up for the pMC-2-1 plasmid was also found inserted within the plasmids harboring KPC in strains MC-2-304 and MC-2-362, and the 281 Kb plasmid from strain MC-2-230 also carried a 67 kb fragment from pMC-2-1 (Figure 3B). On the other hand, the two ST258 K. pneumoniae isolates (MC-2-251 and MC-2-387) harbored blaKPC–3 within IncFIIk plasmids of 78,515 and 120,395 bp, and there was no similarity at all between these two plasmids and those of ST15 strains as there was no resemblance between them either.
In the E. coli strain MC-2-240 we only detected blaKPC–2 within a 25,058 bp plasmid showing 100% similarity with pMC-2-1, and a similar plasmid of 28,498 bp was also detected in strain MC-2-387, which also carried blaKPC–3 in the 120,395 bp plasmid (Figure 3A).
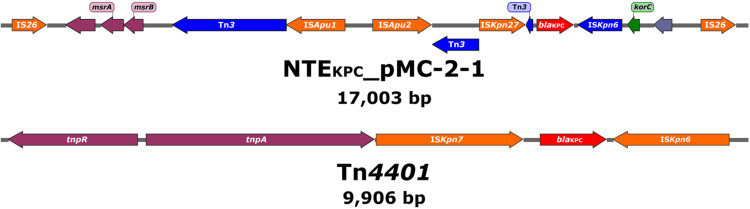
Notably, in all plasmids blaKPC–2 was not located within the canonical Tn4401 element but inside an IS26-based composite transposon of roughly 17 Kb containing an ISKpn27-blaKPC–2-ΔISKpn6-korC core structure as well as Tn3-associated sequences upstream from ISKpn27 (Figure 4). This rearrangement was tentatively designated as NTEKPC-pMC-2-1. The genetic structures surrounding the blaKPC–3 gene in both ST258 strains, however, did match that of a classical Tn4401 element (Figure 4). Specific primers to amplify the structures associated with NTEKPC-pMC-2-1 were designed (Supplementary Table 1 and Supplementary Figure 5) and used to verify the carriage of either the canonical Tn4401 transposon or the NTEKPC-pMC-2-1 variant in all 125 isolates of the outbreak. Interestingly, NTEKPC-pMC-2-1 was identified in all isolates but in the ST258 K. pneumoniae isolate MC-2-251.
FIGURE 4.
Schematic drawing showing the genetic elements surrounding the blaKPC genes in the NTEKPC-pMC-2-1 variant and Tn4401, respectively. Arrows are oriented in the direction of transcription. Red arrows show the blaKPC gene, orange arrows represent full-length insertion sequences (IS), dark blue arrows represent partial or truncated transposon-related regions and ISs, and green arrows indicate genes involved in gene regulation. Blue-gray arrows show putative or hypothetical genes and plum arrows show genes involved in other functions.
In addition, all plasmid sequences were checked for the presence of plasmid-mediated virulence genes producing salmochelin, aerobactin, the hypermucoviscosity factor and/or other virulence genes (Russo et al., 2018), but none of the plasmid sequences carrying blaKPC harbored any known virulence determinant.
Discussion
The emergence of KPC-producing Enterobacterales has been increasing worldwide during the last decades (Lee et al., 2019). Since its first description in 1996 in the United States, KPC has spread through the globe, becoming endemic in areas such as the United States, Israel, Greece, and Italy, and sporadic spread being reported in additional European countries as well as in the Asia-Pacific region (Lee et al., 2019). Many of the descriptions from European countries have been related to patients traveling to endemic areas.
In Spain, the first KPC report dates back to 2009 and it was associated with the rapid spread of a ST384 K. pneumoniae strain carrying blaKPC–3 within a Tn4401a structure (Curiao et al., 2010). KPC-2 and KPC-3-producing Enterobacterales were later reported in the central region of Spain and Andalusia mainly associated with K. pneumoniae isolates belonging to the CG258 or ST101 (López-Cerero et al., 2014; Porres-Osante et al., 2014; Oteo et al., 2016). In Catalonia, a single KPC-3-producing K. pneumoniae isolate belonging to ST258 was recovered from a human patient in 2015 (Piedra-Carrasco et al., 2018) but, to our knowledge, our study constitutes the first hospital outbreak caused by KPC-producing Enterobacterales in this region.
Here we report an outbreak caused by KPC-producing K. pneumoniae and E. coli in a tertiary hospital in Barcelona that initiated during the summer of 2018. It involved 125 Enterobacterales isolates recovered from surveillance (74.4%) or diagnosis (25.6%) samples. Outbreak identification was possible upon the implementation of an active surveillance program in ICU patients, but it is likely that KPC-producing K. pneumoniae isolates had already been circulating in asymptomatic fecal carriers for some time. The outbreak was caused by the rapid spread of a carbapenem-resistant ST15 K. pneumoniae strain carrying blaKPC–2 in an IncFIIk plasmid of approximately 106 Kb and located within a non-Tn4401 genetic element (NTEKPC-pMC-2-1).
Nevertheless, upon the initial stage of clonal spread, we speculate that the IncFIIk plasmid was successfully transferred first to E. coli isolates in patients either infected or colonized with the original ST15 strain, but later to other strains of K. pneumoniae or even Enterobacter spp. (Figure 1 and Supplementary Figure 4). This is supported by the fact that in at least 14 instances the same patient co-carried a blaKPC–2-ST15 K. pneumoniae strain together with either a blaKPC–2-K. pneumoniae belonging to a different sequence type or a blaKPC–2-E. coli or Enterobacter sp. strain, but also because all plasmids carrying blaKPC–2 in K. pneumoniae or E. coli strains shared high similarity with the IncFIIk plasmids from the ST15 K. pneumoniae strains, including the NTEKPC-pMC-2-1 structure surrounding blaKPC–2. Some genetic rearrangements, however, seem to have occurred, mainly in non-K. pneumoniae strains where the 106 Kb IncFIIk plasmid either co-integrated with other plasmids or suffered extensive genetic reduction. Therefore, intra- and inter-species dissemination of resistance also contributed to the spread of the outbreak. KPC transfer to other K. pneumoniae strains was not as heavily associated with co-carriage within the same patient, as opposite to inter-species dissemination, but such finding may also reflect a selection bias in the microbiology laboratory. Interestingly though, the clonal dissemination of non-ST15 strains was not detected. Recently, San Millán and co-workers proposed a similar hospital transmission dynamic for pOXA-48, where patient-to-patient transmission was tightly associated with the dissemination of a particular high-risk clone, while intra- and inter-species transmission of the plasmid was linked to concurrent gut colonization (León-Sampedro et al., 2021). As in the case of OXA-48, this genetic exchange represents an opportunity for the resistance gene to rearrange and shuffle into new plasmids and/or hosts, some of which may become more successful (Millan, 2018).
The genetic structures associated with the core structure of NTEKPC-pMC-2-1 identified in this work (ISKpn27-blaKPC–2-ΔISKpn6-korC) had already been reported by other studies and seem to constitute a common rearrangement associated with blaKPC–2 mostly in isolates from China (Wang et al., 2015; Zhang et al., 2017). In Spain though, this structure has also been identified in IncP-6 plasmids from Citrobacter freundii, Enterobacter cloacae, and Klebsiella oxytoca but, to our knowledge, have never been associated with K. pneumoniae isolates of human origin, most likely suggesting a recent acquisition (Yao et al., 2017; Pérez-Vazquez et al., 2019).
Two sporadic K. pneumoniae strains belonging to the clonal group CG258 and carrying the blaKPC–3 gene within a canonical Tn4401 in two different non-related plasmids were also detected in the study and, most likely, constituted an independent event unrelated to the spread of the outbreak. Nevertheless, one of such strains also managed to acquire a partial plasmid sequence containing the entire NTEKPC-pMC-2-1 element.
The identification of a carbapenem resistant ST307 K. pneumoniae isolate in this study is also worth mentioning, as strains from this epidemic clonal group have recently been responsible for a hospital outbreak in Germany associated with multidrug resistance but also with the hypervirulent Klebsiella pneumoniae phenotype (hvKp). The hvKp phenotype was identified among ST307 isolates upon the acquisition of several plasmid-mediated virulence genes that merged with a resistance plasmid, hence creating a mosaic plasmid carrying both resistance and virulence genes (Heiden et al., 2020). In our study the ST307 strain (MC-2-230) carried blaKPC–2 in a plasmid of 280 Kb also likely resulting from the merging of two different plasmids and, hence, the potential carriage of plasmid-mediated virulence genes was investigated. Fortunately, none of the plasmid-mediated virulence factors associated with the hvKp phenotype were present in plasmid pMC-2-230 nor in any other plasmid sequenced in this study. Likewise, none of the K. pneumoniae strains showed a hypermucoid phenotype, which is characteristic (although not exclusive) of hvKp (Lan et al., 2021).
We acknowledge several limitations in our study. First, WGS was performed under a single long-read sequencing approach, and we acknowledge that the use of a hybrid approach would have allowed for additional and more accurate comparisons. In addition, only a selected group of isolates were sequenced and sequence similarity was, therefore, assumed for the remaining isolates on the basis of PFGE, MLST and conventional PCR data. Unfortunately, further WGS analyses were beyond our possibilities but we expect that results from this study will contribute to a better implementation of WGS pipelines in our institution.
Conclusion
We report a hospital outbreak caused by the clonal dissemination of KPC-producing ST15 K. pneumoniae mainly among colonized carriers but also by the intra- and inter-species transmission of the blaKPC–2 gene associated with plasmid conjugation and/or transposon dissemination. The ST15 clonal lineage is considered a high-risk clone and has been associated with KPC-2-producing isolates in Bulgaria and Vietnam (Markovska et al., 2015; Berglund et al., 2019), and it has also been reported in Portugal and Italy (Rodrigues et al., 2016; Fasciana et al., 2019). In Spain, ST15 has been reported but only associated with the production of OXA-48 (Madueño et al., 2017). Nevertheless, ST15 has been attributed with a high potential for horizontal gene acquisition and dissemination and it is critical that active surveillance strategies are prolonged over time to allow for the rapid detection and eradication of these highly resistant and virulent clones (Berglund et al., 2019).
MERCyCAT Study Group
Pepa Pérez Jove, Emma Padilla, and Mónica Ballestero-Téllez (Catlab, Centre Analítiques Terrassa AIE), Yuliya Zboromyrska, Miguel Ángel Benítez, Raquel Clivillé, Sabina González, and Iolanda Calvet (Consorci del Laboratori Intercomarcal de l’Alt Pendès, l’Anoia i el Garraf), Carmen Gallés (Corporació de Salut del Maresme i la Selva), Goretti Sauca (Hospital de Mataró), Carmina Martí-Sala and Ma Angeles Pulido (Hospital General de Granollers), Anna Vilamala (Hospital General de Vic), Araceli González-Cuevas (Hospital General del Parc Sanitari Sant Joan de Déu), Amadeu Gené (Hospital Sant Joan de Déu de Barcelona), Gloria Trujillo and Joan Lopez Madueño (Hospital Sant Joan de Déu de Manresa), Xavier Raga (Hospital Sant PauI Santa Tecla), Frederic Gómez, Ester Picó, and Carolina Sarvisé (Hospital Universitari Joan XXIII de Tarragona), Isabel Pujol and Xesca Font (Hospital Universitari Sant Joan de Reus).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
MM-A, NF, and MF contributed to the conception, design, and implementation of the study, acquisition of laboratory and clinical data, analysis of the results, drafting the manuscript, and approval of the final version of the manuscript. CC, JViñ, ER, AC, and LM contributed to the acquisition of laboratory data, analysis of the results, and review and approval of the final version of the manuscript. MP, AV, IC, LR-S, GS, and AD contributed to the design of the study, acquisition of clinical data, analysis of the results, and review and approval of the final version of the manuscript. OF, PC, FB, FM, JM, and ÁS contributed to the design of the study, analysis of the results, and review and approval of the final version of the manuscript. CP, JVil, and IR contributed to the conception, design, and implementation of the study, analysis of the results, drafting the manuscript, and approval of the final version of the manuscript. All authors critically revised the manuscript for intellectual content and read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the team of curators of the Institute Pasteur MLST and whole genome MLST databases for curating the data and making them publicly available at http://bigsdb.pasteur.fr/. We also thank the clinical and laboratory staff of the participating hospitals for obtaining clinical samples and the initial identification of microorganisms.
Footnotes
Contributor Information
MERCyCAT Study Group:
Pepa Pérez Jove, Emma Padilla, Mónica Ballestero-Téllez, Yuliya Zboromyrska, Miguel Ángel Benítez, Raquel Clivillé, Sabina González, Iolanda Calvet, Carmen Gallés, Goretti Sauca, Carmina Martí-Sala, Mᵃ Angeles Pulido, Anna Vilamala, Araceli González-Cuevas, Amadeu Gené, Gloria Trujillo, Joan Lopez Madueño, Xavier Raga, Frederic Gómez Ester Picó, Carolina Sarvisé, Isabel Pujol, and Xesca Font
Funding
This study was supported by Plan Nacional de I+D+i 2013–2016, Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI RD16/0016/0010); the 2017 Call for Strategic Action on Health (PI17/01932), co-financed by European Development Regional Fund “A way to achieve Europe” and the operative program Intelligent Growth 2014–2020; and grant 2017 SGR 0809 from the Departament d’Universitats, Recerca i Societat de la Informació, of the Generalitat de Catalunya. MM-A and CC were supported by grants FPU 14/06357 and FPU 13/02564, respectively, from the Spanish Ministry of Education, Culture and Sports. IR was supported by the Department of Health, Generalitat de Catalunya, grant SLT002/16/00349. We also acknowledge support from the Spanish Ministry of Science, Innovation and Universities through the “Centro de Excelencia Severo Ochoa 2019–2023” Program (CEX2018-000806-S), and support from the Generalitat de Catalunya through the CERCA Program.” The funders had no role in the study design, data collection, analysis and interpretation of data, decision to publish, or preparation of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.781127/full#supplementary-material
References
- Berglund B., Hoang N. T. B., Tärnberg M., Le N. K., Nilsson M., Khu D. T. K., et al. (2019). Molecular and phenotypic characterization of clinical isolates belonging to a KPC-2-producing strain of ST15 Klebsiella pneumoniae from a Vietnamese pediatric hospital. Antimicrob. Resist. Infect. Control 8:156. 10.1186/s13756-019-0613-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogaerts P., de Castro R. R., de Mendonca R., Huang T. D., Denis O., Glupczynski Y. (2013). Validation of carbapenemase and extended-spectrum β-lactamase multiplex endpoint PCR assays according to ISO 15189. J. Antimicrob. Chemother. 68 1576–1582. 10.1093/jac/dkt065 [DOI] [PubMed] [Google Scholar]
- Bonomo R. A., Burd E. M., Conly J., Limbago B. M., Poirel L., Segre J. A., et al. (2017). Carbapenemase-producing organisms: a global scourge. Clin. Infect. Dis. 20:440. 10.1371/journal.pmed.1001021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Bertini A., Villa L., Falbo V., Hopkins K. L., Threlfall E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63 219–228. 10.1016/j.mimet.2005.03.018 [DOI] [PubMed] [Google Scholar]
- Cassini A., Högberg L. D., Plachouras D., Quattrocchi A., Hoxha A., Simonsen G. S., et al. (2018). Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect. Dis. 19 56–66. 10.1016/s1473-3099(18)30605-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Chavda K. D., Melano R. G., Jacobs M. R., Koll B., Hong T., et al. (2014). Comparative genomic analysis of KPC-encoding pKpQIL-like plasmids and their distribution in New Jersey and New York hospitals. Antimicrob. Agents Chemother. 58 2871–2877. 10.1128/aac.00120-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmelnitsky I., Shklyar M., Leavitt A., Sadovsky E., Navon-Venezia S., Dalak M. B., et al. (2014). Mix and match of KPC-2 encoding plasmids in Enterobacteriaceae-comparative genomics. Diagn. Microbiol. Infect. Dis. 79 255–260. 10.1016/j.diagmicrobio.2014.03.008 [DOI] [PubMed] [Google Scholar]
- Curiao T., Morosini M. I., Ruiz-Garbajosa P., Robustillo A., Baquero F., Coque T. M., et al. (2010). Emergence of blaKPC-3-Tn4401a associated with a pKPN3/4-like plasmid within ST384 and ST388 Klebsiella pneumoniae clones in Spain. J. Antimicrob. Chemother. 65 1608–1614. 10.1371/journal.pone.0008601 [DOI] [PubMed] [Google Scholar]
- Diancourt L., Passet V., Verhoef J., Grimont P. A. D., Brisse S. (2005). Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43 4178–4182. 10.1128/jcm.43.8.4178-4182.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST) (2016). Recommendations for MIC Determination of Colistin (Polymyxin E) as Recommended by the Joint CLSI-EUCAST Polymyxin Breakpoints Working Group. Växjö: EUCAST. [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST) (2021). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 11.0. Växjö: EUCAST. [Google Scholar]
- Fasciana T., Gentile B., Aquilina M., Ciammaruconi A., Mascarella C., Anselmo A., et al. (2019). Co-existence of virulence factors and antibiotic resistance in new Klebsiella pneumoniae clones emerging in south of Italy. BMC Infect. Dis. 19:928. 10.1186/s12879-019-4565-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundmann H., Glasner C., Albiger B., Aanensen D. M., Tomlinson C. T., Andrasevic A. T., et al. (2017). Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect. Dis. 17 153–163. 10.1016/s1473-3099(16)30257-2 [DOI] [PubMed] [Google Scholar]
- Heiden S. E., Hübner N.-O., Bohnert J. A., Heidecke C.-D., Kramer A., Balau V., et al. (2020). A Klebsiella pneumoniae ST307 outbreak clone from Germany demonstrates features of extensive drug resistance, hypermucoviscosity, and enhanced iron acquisition. Genome Med. 12:113. 10.1186/s13073-020-00814-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan P., Jiang Y., Zhou J., Yu Y. (2021). A global perspective on the convergence of hypervirulence and carbapenem-resistance in Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. 25 26–34. 10.1016/j.jgar.2021.02.020 [DOI] [PubMed] [Google Scholar]
- Lee Y.-L., Lu M.-C., Shao P.-L., Lu P.-L., Chen Y.-H., Cheng S.-H., et al. (2019). Nationwide surveillance of antimicrobial resistance among clinically important Gram-negative bacteria, with an emphasis on carbapenems and colistin: results from the Surveillance of Multicenter Antimicrobial Resistance in Taiwan (SMART) in 2018. Int. J. Antimicrob. Agents 54 318–328. 10.1016/j.ijantimicag.2019.06.009 [DOI] [PubMed] [Google Scholar]
- León-Sampedro R., DelaFuente J., Díaz-Agero C., Crellen T., Musicha P., Rodríguez-Beltrán J., et al. (2021). Pervasive transmission of a carbapenem resistance plasmid in the gut microbiota of hospitalized patients. Nat. Microbiol. 6 606–616. 10.1038/s41564-021-00879-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Cerero L., Egea P., Gracia-Ahufinger I., Gonzalez-Padilla M., Rodríguez-López F., Rodriguez-Bano J., et al. (2014). Characterisation of the first ongoing outbreak due to KPC-3-producing Klebsiella pneumoniae (ST512) in Spain. Int. J. of Antimicrob. Agents 44 538–540. 10.1016/j.ijantimicag.2014.08.006 [DOI] [PubMed] [Google Scholar]
- Madueño A., García J. G., Fernandez-Romero S., Oteo J., Lecuona M. (2017). Dissemination and clinical implications of multidrug-resistant Klebsiella pneumoniae isolates producing OXA-48 in a Spanish hospital. J. Hosp. Infect. 96 112–122. 10.1016/j.jhin.2017.02.024 [DOI] [PubMed] [Google Scholar]
- Marí-Almirall M., Cosgaya C., Pitart C., Viñes J., Muñoz L., Campo I., et al. (2021). Dissemination of NDM-producing Klebsiella pneumoniae and Escherichia coli high-risk clones in Catalan healthcare institutions. J. Antimicrob. Chemother. 76 345–354. 10.1093/jac/dkaa459 [DOI] [PubMed] [Google Scholar]
- Markovska R., Stoeva T., Schneider I., Boyanova L., Popova V., Dacheva D., et al. (2015). Clonal dissemination of multilocus sequence type ST15 KPC-2-producing Klebsiella pneumoniae in Bulgaria. Apmis 123 887–894. 10.1111/apm.12433 [DOI] [PubMed] [Google Scholar]
- Millan A. S. (2018). Evolution of plasmid-mediated antibiotic resistance in the clinical context. Trends Microbiol. 26 978–985. 10.1016/j.tim.2018.06.007 [DOI] [PubMed] [Google Scholar]
- Naas T., Cuzon G., Truong H.-V., Nordmann P. (2012). Role of ISKpn7 and deletions in blaKPC gene expression. Antimicrob. Agents Chemother. 56 4753–4759. 10.1128/aac.00334-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’neill J. (2016). Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Available online at: http://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf (accessed June, 2021). [Google Scholar]
- Orlek A., Phan H., Sheppard A. E., Doumith M., Ellington M., Peto T., et al. (2017). A curated dataset of complete Enterobacteriaceae plasmids compiled from the NCBI nucleotide database. Data Brief 12 423–426. 10.1016/j.dib.2017.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oteo J., Pérez-Vázquez M., Bautista V., Ortega A., Zamarrón P., Saez D., et al. (2016). The spread of KPC-producing Enterobacteriaceae in Spain: WGS analysis of the emerging high-risk clones of Klebsiella pneumoniae ST11/KPC-2, ST101/KPC-2 and ST512/KPC-3. J. Antimicrob. Chemother. 71 3392–3399. 10.1093/jac/dkw321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicé M., Rodríguez-Núñez O., Rico V., Agüero D., Morata L., Cardozo C., et al. (2021). Factors associated with short-term eradication of rectal colonization by KPC-2 producing Klebsiella pneumoniae in an outbreak setting. Front. Microbiol. 12:630826. 10.3389/fmicb.2021.630826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Vazquez M., Oteo-Iglesias J., Sola-Campoy P. J., Carrizo-Manzoni H., Bautista V., Lara N., et al. (2019). Characterization of carbapenemase-producing Klebsiella oxytoca in Spain, 2016–2017. Antimicrob. Agents Chemother. 63:e02529-18. 10.1128/aac.02529-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piedra-Carrasco N., Miguel L., Fàbrega A., Viñado B., Campany D., Mir A., et al. (2018). Effectiveness of a double-carbapenem regimen in a KPC-producing Klebsiella pneumoniae infection in an immunocompromised patient. Microb. Drug Resist. 24 199–202. 10.1089/mdr.2017.0129 [DOI] [PubMed] [Google Scholar]
- Porres-Osante N., Azcona-Gutiérrez J. M., Rojo-Bezares B., Undabeitia E., Torres C., Sáenz Y. (2014). Emergence of a multiresistant KPC-3 and VIM-1 carbapenemase-producing Escherichia coli strain in Spain. J. Antimicrob. Chemother. 65 1608–1614. 10.1093/jac/dku055 [DOI] [PubMed] [Google Scholar]
- Queenan A. M., Bush K. (2007). Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20 440–458. 10.1128/cmr.00001-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca I., Akóva M., Baquero F., Carlet J., Cavaleri M., Coenen S., et al. (2015). The global threat of antimicrobial resistance: science for intervention. New Microbes New Infect. 6 22–29. 10.1016/j.nmni.2015.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues C., Bavlovič J., Machado E., Amorim J., Peixe L., Novais A. (2016). KPC-3-Producing Klebsiella pneumoniae in Portugal linked to previously circulating Non-CG258 lineages and uncommon genetic platforms (Tn4401d-IncFIA and Tn4401d-IncN). Front. Microbiol. 7:1000. 10.3389/fmicb.2016.01000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo T. A., Olson R., Fang C.-T., Stoesser N., Miller M., MacDonald U., et al. (2018). Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J. Clin. Microbiol. 56:e00776-18. 10.1128/jcm.00776-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Shen P., Wei Z., Jiang Y., Du X., Ji S., Yu Y., et al. (2009). Novel genetic environment of the carbapenem-hydrolyzing β-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob. Agents Chemother. 53 4333–4338. 10.1128/aac.00260-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solé M., Pitart C., Roca I., Fàbrega A., Salvador P., Muñoz L., et al. (2011). First description of an Escherichia coli strain producing NDM-1 carbapenemase in Spain. Antimicrob. Agents Chemother. 55 4402–4404. 10.1128/aac.00642-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard P., Wishart D. S. (2005). Circular genome visualization and exploration using CGView. Bioinformatics 21 537–539. 10.1093/bioinformatics/bti054 [DOI] [PubMed] [Google Scholar]
- Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D. L., et al. (2017). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18 318–327. 10.1016/s1473-3099(17)30753-3 [DOI] [PubMed] [Google Scholar]
- Tumbarello M., Viale P., Viscoli C., Trecarichi E. M., Tumietto F., Marchese A., et al. (2012). Predictors of mortality in bloodstream infections caused by Klebsiella pneumoniae carbapenemase–producing K. pneumoniae: importance of combination therapy. Clin. Infect. Dis. 55 943–950. 10.1093/cid/cis588 [DOI] [PubMed] [Google Scholar]
- Wang L., Fang H., Feng J., Yin Z., Xie X., Zhu X., et al. (2015). Complete sequences of KPC-2-encoding plasmid p628-KPC and CTX-M-55-encoding p628-CTXM coexisted in Klebsiella pneumoniae. Front. Microbiol. 6:838. 10.3389/fmicb.2015.00838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T., Falush D., Lan R., Colles F., Mensa P., Wieler L. H., et al. (2006). Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60 1136–1151. 10.1111/j.1365-2958.2006.05172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyres K. L., Lam M. M. C., Holt K. E. (2020). Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 18 344–359. 10.1038/s41579-019-0315-1 [DOI] [PubMed] [Google Scholar]
- Yao Y., Lazaro-Perona F., Falgenhauer L., Valverde A., Imirzalioglu C., Dominguez L., et al. (2017). Insights into a novel blaKPC-2-encoding IncP-6 plasmid reveal carbapenem-resistance circulation in several Enterobacteriaceae species from wastewater and a hospital source in Spain. Front. Microbiol. 8:3579. 10.1016/j.diagmicrobio.2012.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Liu L., Zhou H., Chan E. W., Li J., Fang Y., et al. (2017). Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. Ebiomedicine 19 98–106. 10.1016/j.ebiom.2017.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.