Abstract
Gastric cancer (GC) is one of the most prevalent malignant tumor types and the third leading cause of cancer-related death worldwide. Its morbidity and mortality are very high due to a lack of understanding about its pathogenesis and the slow development of novel therapeutic strategies. Long non-coding RNAs (lncRNAs) are a class of non-coding RNAs with a length of more than 200 nt. They play crucial roles in a wide spectrum of physiological and pathological processes by regulating the expression of genes involved in proliferation, differentiation, apoptosis, cell cycle, invasion, metastasis, DNA damage, and carcinogenesis. The aberrant expression of lncRNAs has been found in various cancer types. A growing amount of evidence demonstrates that lncRNAs are involved in many aspects of GC pathogenesis, including its occurrence, metastasis, and recurrence, indicating their potential role as novel biomarkers in the diagnosis, prognosis, and therapeutic targets of GC. This review systematically summarizes the biogenesis, biological properties, and functions of lncRNAs and highlights their critical role and clinical significance in GC. This information may contribute to the development of better diagnostics and treatments for GC.
Keywords: long non-coding RNA, biogenesis, biomarker, therapeutic target, gastric cancer
Graphical Abstract
In this article, Liu et al. summarize the biological features, biogenesis, and functions of lncRNAs and focus on their mechanisms in gastric cancer (GC) progression and their potential as diagnostic and prognostic biomarkers and therapeutic targets for GC.
Introduction
Gastric cancer (GC) is one of the most common malignant tumors of the digestive system and the third leading cause of cancer-related death in the world.1 Its clinical outcomes are poor, which seriously affects patients’ quality of life. Although the incidence and mortality of GC have decreased slightly in recent years, it is still a major global health problem.2 Most patients with GC are usually diagnosed at an advanced stage due to the difficulty of determining early symptoms, limitations in screening techniques, and a lack of efficient biomarkers.2, 3, 4 In addition, the local recurrence rate of GC is also high, ranging from 2.8% to 12.5%.5 Therefore, continued in-depth research into the molecular mechanisms of GC progression and the identification of specific and efficient biomarkers and therapeutic targets are urgently required to provide novel therapeutic strategies for GC patients.
Non-coding RNAs (ncRNAs) are recognized as an emerging class of transcripts without protein-coding potential that are transcribed by more than 80% of the entire human genome.6 They play crucial roles in both normal development and diseases, including GC.7,8 Based on their size, ncRNAs are divided into two classes, namely long ncRNAs (>200 nt) and small ncRNAs (≤200 nt).9 Many types of small ncRNAs, such as microRNAs (miRNAs),10 circular RNAs (circRNAs),11 and piwi-interacting RNAs,12 have been identified and many aspects of their physiological and pathological functions have been elucidated. However, little is known about the biological function of lncRNAs.
lncRNAs were previously considered to be transcriptional “junk” or “noise” but, in recent years, increasing studies revealed that they play crucial roles in various physiological and pathological processes, such as gene expression regulation, embryonic development, and carcinogenesis.5,13 The dysregulation of lncRNAs has been observed in many types of cancer, including glioblastoma, colon adenocarcinoma, breast cancer, prostate cancer, and GC.14, 15, 16, 17, 18, 19 lncRNAs participate in GC progression by regulating the gene expression networks involved in cell cycle, proliferation, apoptosis, invasion, metastasis, and the epithelial-to-mesenchymal transition (EMT) of GC cells at the epigenetic, transcriptional, and post-transcriptional levels.4,20 The aberrant expressions of lncRNAs have been shown to have clinical significance in GC, indicating their potential role as biomarkers for GC patients.21, 22, 23 In addition, lncRNAs have also attracted increasing attention due to their benefits in GC therapies. For instance, targeting specific GC-related lncRNAs can effectively reverse drug resistance and enhance the sensitivity of GC cells to chemotherapeutic drugs.24 N6-methyladenosine (m6A)-related lncRNAs have been shown to remodel tumor microenvironment and alter immune checkpoint blockers sensitivity, indicating the great potential of m6A-related lncRNA as an indicator for the response to immunotherapy in GC.25 Although great progress has been made in investigating the role of lncRNAs in GC, more studies are still required to further elucidate their detailed mechanisms in GC progression, thereby providing novel insights into the application of lncRNAs in GC clinical treatment.
In this review, we focus on recent findings regarding the biogenesis and molecular mechanisms of lncRNAs and highlight their clinical significance in the diagnosis and treatment of GC to provide in-depth insights into the functions of lncRNAs being potential biomarkers and therapeutic targets for GC.
Overview of lncRNAs
Biological features of lncRNAs
With the rapid development of RNA sequencing and bioinformatics, a large number of lncRNAs have been identified. These lncRNAs share a few biological features, including the following. (1) lncRNAs have been shown to be highly abundantly expressed in all organisms, from prokaryotes to mammals.26 (2) lncRNAs are widely expressed in prokaryotic and eukaryotic cells and have a wide diversity, with Bermúdez et al. reporting that there are about 5,400 to more than 10,000 lncRNA transcripts in humans.27 Some classes of them are generated from distinct DNA elements in genomes, including promoters, enhancers, intergenic regions, and the opposite strand of protein-coding genes, while others are produced from long primary transcripts with noncanonical RNA processing pathways.28 (3) The expression of lncRNAs includes stronger cell type, and tissue and spatial-temporal specificity compared with protein-coding genes.14,29 (4) lncRNA genes have lower expression frequency in human beings than protein-coding genes.30 (5) Most lncRNA genes are relatively conserved during the evolutionary progress, and they evolve faster than protein-coding genes. lncRNA is less conserved in primary sequences, and its sequence similarity is mainly preserved in secondary structures.6
Biogenesis of lncRNAs
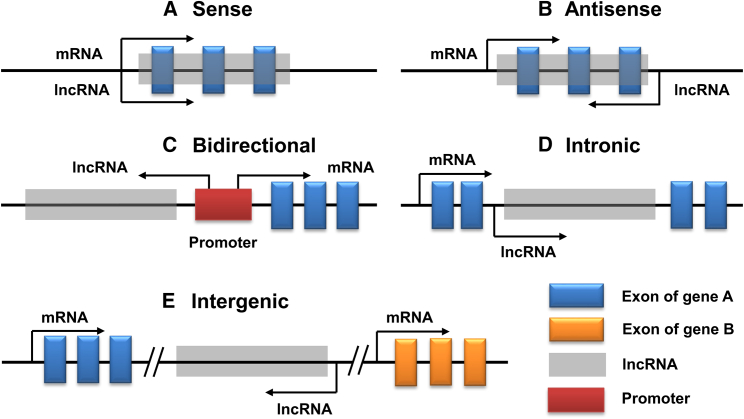
lncRNAs are typical RNA-type molecules transcribed by RNA polymerase II (Pol II) and harbor a 5′ methyl-cytosine cap and 3′ poly(A) tail.31 According to their different characteristics, lncRNAs are classified into many different types. For instance, lncRNAs can be divided into five classes based on their distinct genomic origins, including sense, antisense, bidirectional, intronic, and intergenic (Figure 1). According to their function, lncRNAs can be classified into three types, namely rRNA, tRNA, and cRNA. Moreover, according to their subcellular localization, lncRNAs can also be divided into nuclear lncRNAs, cytoplasmic lncRNAs, and mitochondrial lncRNAs.32
Figure 1.
Classification of lncRNAs on their genomic origins
(A) Sense or (B) antisense lncRNA locates within or overlaps with the exons of the associated protein-coding gene on the same, or opposite strand. Antisense lncRNA transcribes in the opposite direction of protein-coding gene. (C) Bidirectional lncRNA locates nearby the promoter (within 1 kb) of the associated protein-coding gene and transcribes in the opposite direction. (D) Intronic lncRNA arises from long introns and transcribes from inside of an intron of the associated protein-coding gene. (E) Intergenic lncRNA originates from intergenic segment of two protein-coding genes.
The biogenesis of lncRNAs is comparable with that of mRNA with some differences in the mechanisms. Most of lncRNAs have been shown to be capped, polyadenylated, and spliced by the canonical mode.33 They also can be processed by several noncanonical mechanisms, including cleavage by ribonuclease P (RNase P) to produce mature 3′ ends, capping by snoRNA-protein (snoRNP) complexes at their ends, and the generation of circular structures.28 For instance, some lncRNAs (e.g., MALAT1 and Menβ) are processed at their 3′ ends through the recognition and cleavage of tRNA-like structures by RNase P, leading to the formation of their mature 3′ ends.28,34 snoRNA is one of the members of the conserved nuclear RNA family, which plays crucial roles during ribosome subunit maturation as guide RNAs. It has been reported that snoRNAs can be capped by snoRNPs at both ends, leading to their enhanced stability. In addition to liner lncRNAs, two types of circRNA have been shown to be processed from Pol II-transcribed RNA precursors by some specific mechanisms. Their non-polyadenylated circular structures protect them from degradation.35
The biogenesis of lncRNAs is regulated by different epigenetic modifications and several kinds of regulators. For instance, the transcription of antisense lncRNA is promoted by H3K56 acetylation and the chromatin remodeler SWI/SNF, while, this process is repressed by chromatin assembly factor complex CAF-1.36 The methylation of paternal allele significantly inhibits the transcription of lncRNA Air within the imprinted gene cluster, thereby activating the expression of flanking protein-coding genes, including Igf2r, Slc22a2, and Slc22a3.37 In addition, the degradation of lncRNA is mediated by exosomes with Nrd1-Nab3-Sen1 and TRAMP complexes in the nucleus or by Xrn1 in the cytoplasm. This process is suppressed by the nonsense-mediated decay protein UPF1.36,38 These findings suggest that epigenetic modifications play crucial roles in the regulation of lncRNA biogenesis. However, the detailed mechanisms are still not fully understood. More in-depth research is required to elucidate the exact mechanisms of epigenetic modification in the regulation of lncRNA biogenesis in future studies.
Functions of lncRNAs
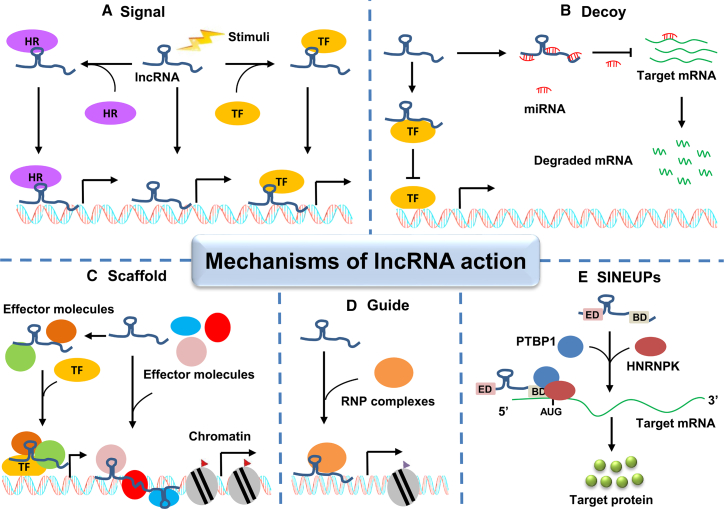
lncRNAs play crucial roles in a series of pathological and physiological processes through regulating the timing and degree of gene expression. This is realized by different molecular mechanisms that can be summarized as the five manners: signal, decoy, scaffold, guide, and SINEUPs (Figure 2). All these mechanisms are based on lncRNAs and their interactions with DNA, RNA, and proteins.
Figure 2.
Molecular functions of lncRNAs
(A) Signal lncRNAs can act as molecular signals in response to diverse stimuli. lncRNAs in this archetype regulate gene expression in space and time by directly binding to DNA or combining with hormone receptor (HR) or transcription factor (TF). (B) Decoy. lncRNAs in this archetype serve as protein decoys for miRNAs or TF to prevent these molecules from binding to their targets. (C) Scaffold. lncRNAs can act as a scaffold as to form ribonucleoprotein complexes with different protein partners, which regulates the transcription of their target genes. (D) Guide. lncRNAs in this archetype can modulate gene expression by guiding the ribonucleoprotein complex to their target genes. (E) SINEUPs. lncRNAs in this archetype can increase translation of their target mRNAs by recruiting PTBP1 and HNRNPK to form translational initiation complexes with no effects on target mRNA levels.
Signal lncRNAs are recognized as signal molecules that correlated with specific signaling pathways and their expression means an active signaling event, regardless of their functions in the signaling process.39 For instance, the high expression of lncRNA Xist has been shown to be a signal of X chromosome inactivation in females. Mechanistically, the expansion of Xist on the X chromosome induces DNA methylation and histone modification, leading to X chromosome inactivation.40 Decoy lncRNAs act as a sponge or molecular sink for microRNAs, transcription factors (TFs), or RNA binding proteins to promote their target genes’ activation or silencing. These lncRNAs are also called competing endogenous RNAs (ceRNAs). NKX2-1-AS1 is a typical decoy lncRNA. Teng et al. showed that NKX2-1-AS1 upregulates the expression of serpin family E member 1 (SERPINE1) by sponging miR-145-5p, thus activating the VEGFR-2 signaling pathway to facilitate the angiogenesis and progression of GC.41
An archetype of lncRNAs can serve as scaffolds for the assembly of scaffolding complexes with TFs or effector molecules to regulate chromosome rearrangement, RNA Pol II activity, or histone modifications. These lncRNAs often act as regulators involved in the epigenetic and transcriptional control of gene expression. For instance, Sun et al. found that lncRNA HOXA11-AS downregulates the expression of PRSS8 and KLF2 by scaffolding the chromatin modification factors polycomb repressive complex 2, lysine-specific demethylase 1, and DNA methyltransferase-1, leading to promotion of GC cell proliferation and invasion.42 Guide lncRNAs can bind to the target proteins to form a ribonucleoprotein complex and then guide the newly formed complex to a specific target gene promoter or genomic loci, consequently regulating the gene expression at the transcription level. HOTTIP is a classic example of guide lncRNA that transcribed from the 5′ tip of the HOXA locus. Wang et al. showed that HOTTIP directly interacts with the adaptor protein WDR5 and guides the WDR5/MLL complexes to the 5′ HOXA locus, leading to the trimethylation of histone H3 lysine 4 and target gene transcription.43
SINEUPs is a new functional class of natural and synthetic antisense lncRNAs, which can facilitate translation of target mRNAs with no effects on their mRNA levels.44 This type of lncRNA contains an embedded inverted SINE element (effector domain) conferring biological activity and a binding domain conferring target specificity.45 AS-Uchl1 is the representative member of natural SINEUPs. Carrieri et al. found that AS-Uchl1 was able to promote the translation of its sense Uchl1 mRNA in mouse dopaminergic neuronal cells through enhancing Uchl1 mRNA association to heavy polysomes. This process depends on the inverted SINEB2 element of AS-Uchl1, which has no effect on Uchl1 mRNA quantities.46 Their other study revealed that PTBP1 and HNRNPK acted as RNA binding proteins to interact with SINEUP, thereby contributing to SINEUP RNA subcellular distribution and to assembly of translational initiation complexes.47 Synthetic SINEUPs have been shown to be the first scalable tool to increase protein production of a gene of interest. For instance, in-vitro-transcribed SINEUP-cox7B is a synthetic SINEUP designed against endogenous cox7B mRNA, which can effectively and specifically increase COX7B protein synthesis, thereby rescuing eye and brain size in cox7B morphants.48 These findings suggest that SINEUPs possesses great therapeutic potential to be applied in various disorders caused by insufficient protein production.
Roles of lncRNAs in GC
With the rapid development of RNA high-throughput sequencing technology, a large number of lncRNAs have been identified in GC. A deeper understanding of lncRNA mechanisms in GC progression will be extremely beneficial to the diagnosis and treatment of GC patients. In this review, we summarize the dysregulated lncRNAs that have been identified to be related to GC.
lncRNAs and oncogenic signaling pathways in GC
A growing number of studies have demonstrated that lncRNAs play crucial roles in GC progression by targeting signaling pathways, such as PI3K/AKT, mitogen-activated protein kinase (MAPK), Wnt/β-catenin, and STAT3 signaling pathways (Table 1). Understanding the mechanisms of lncRNAs in signaling pathway regulation may provide us with new insights into GC progression.
Table 1.
lncRNAs and their target signaling pathways in GC
| Signaling pathways | lncRNAs | Expression | Key message(s) | References |
|---|---|---|---|---|
| PI3K/AKT | OGFRP1 | up | the knockdown of OGFRP1 downregulates the expression of p-AKT, leading to the inhibition of cell-cycle progression and induction of apoptosis of GC cells | Zhang et al.49 |
| PCAT18 | down | PCAT18 inhibits the PTEN/PI3K/AKT signaling pathway by sponging miR-107, leading to the suppression of GC progression | Chen et al.50 | |
| ADAMTS9-AS2 | down | the suppression of ADAMTS9-AS2 increases the expression of p-PI3K and p-AKT. The administration of the PI3K inhibitor LY294002 reverses the negative effect of ADAMTS9-AS2 on GC progression | Cao et al.51 | |
| LOC101928316 | down | LOC101928316 overexpression decreases the expression levels of PI3K, p-AKT, mTOR, and p-mTOR. However, the knockdown of LOC101928316 upregulates AKT3, mTOR, and p-mTOR expression and suppresses PTEN expression | Li et al.52 | |
| LINC01419 | up | the silencing LINC01419 in GC cells decreases the expression of p-AKT1 and p-mTOR but does not affect their total levels | Wang et al.53 | |
| XLOC_006753 | up | XLOC_006753 knockdown decreases the expression of PI3K, p-AKT (Thr308), p-AKT (Ser473), and p-mTOR (Ser2448), leading to the promotion of MDR in GC cells | Zeng et al.54 | |
| PICART1 | down | the overexpression of PICART1 decreases p-AKT expression, whereas PICART1 knockdown increases p-AKT expression | Li et al.55 | |
| TMPO-AS1 | up | the knockdown of TMPO-AS1 inhibits the PI3K/AKT/mTOR signaling pathway by downregulating the expression of BRCC3 via releasing miR-126-5p in GC cells | Hu et al.56 | |
| LINC01559 | up | silencing LINC01559 downregulates the expression of PGK1, p-PI3K, p-AKT, and p-mTOR. IGF-1 treatment (PI3K activator) significantly reverses the LINC01559 knockdown-induced phosphorylation of PI3K, AKT, and mTOR | Wang et al.57 | |
| FOXD1-AS1 | up | FOXD1-AS1 activates the PI3K/AKT/mTOR pathway via the upregulation of PIK3CA, leading to an aggravation of GC progression and chemoresistance | Wu et al.23 | |
| CRNDE | up | silencing CRNDE significantly downregulates p-PI3K and p-Akt expression in GC | Du et al.58 | |
| MAPK | AOC4P | up | the knockdown of AOC4P decreases the expression of ERK1, JNK, and p38 in GC cells | Qu et al.59 |
| linc00483 | up | Linc00483 knockdown downregulates the expression of c-Jun without affecting the p-Jnk, p53, and p-p38 expression in GC cells | Li et al.60 | |
| PICART1 | down | the overexpression of PICART1 decreases the p-ERK expression, whereas PICART1 knockdown reverses the expression of p-ERK | Li et al.55 | |
| LINC00152 | up | silencing LINC00152 significantly decreases the expression of p-ERK-1/2, p-MEK1/2, and c-fos without affecting the total ERK-1/2 and MEK1/2 expressions. SA (ERK/MAPK signaling pathway activator) treatment reverses the role of LINC00152 in GC cells | Shi and Sun61 | |
| CASC2 | down | the overexpression of CASC2 downregulates the expression of p-ERK1/2 and p-JNK without affecting p-p38 in GC cells. The administration of U0126 and SP600125 (MAPK inhibitors) inhibits the proliferation of CASC2-overexpressing GC cells | Li et al.62 | |
| Wnt/β-catenin | MIR4435-2HG | up | the knockdown of MIR4435-2HG decreases β-catenin expression in GC xenografts and inhibits the transactivating activity of β-catenin in GC cells | Wang et al.63 |
| ZEB2-AS1 | up | ZEB2-AS1 activates the Wnt/β-catenin signaling pathway by upregulating ZEB2 expression in GC cells | Wang et al.64 | |
| LINC01133 | down | LINC01133 suppresses the nuclear accumulation of β-catenin in GC cells by sponging miR-106a-3p and promoting APC expression | Yang et al.65 | |
| HCG11 | up | the knockdown of HCG11 inhibits the proliferation of GC cells by suppressing the activity of the Wnt signaling pathway. The administration of LiCl (Wnt signaling activator) reverses the effect of HCG11 knockdown on the proliferation of GC cells | Zhang et al.66 | |
| GASL1 | down | the overexpression of GASL1 decreases β-catenin expression in GC cells, while GASL1 knockdown increases β-catenin expression. The administration of a Wnt agonist reduces the negative role of GASL1 on GC cells | Peng et al.67 | |
| LINC00665 | up | LINC00665 knockdown decreases the expression of β-catenin and cyclin D1 in GC cells, whereas it increases GSK-3β expression | Yang et al.68 | |
| GATA6-AS1 | down | the overexpression of GATA6-AS1 downregulates β-catenin levels and decreases intranuclear β-catenin expression. In GATA6-AS1-silenced GC cells treated with LiCl, β-catenin expression is upregulated | Li et al.69 | |
| TOB1-AS1 | down | the knockdown of TOB1-AS1 increases the expression of β-catenin, c-Myc, cyclin D1, and N-cadherin in GC cells | Jiang et al.70 | |
| LINC01314 | down | the overexpression of LINC01314 downregulates the expression of Wnt-1, β-catenin, cyclin D1, and N-cadherin, while it upregulates E-cadherin expression in GC cells | Tang et al.71 | |
| LINC01225 | up | LINC01225 knockdown decreases the expression of Wnt1 and β-catenin in GC cells, whereas it does not affect the expression or Ser9 phosphorylation of GSK-3β | Xu et al.72 | |
| LINC01503 | up | silencing LINC01503 in GC cells reduces the expression of β-catenin, cyclin D1, and c-Myc, whereas LINC01503 overexpression reverses their expression | Ding et al.73 | |
| lincRNA-p21 | down | the overexpression of lincRNA-p21 decreases the expression of β-catenin and c-Myc in GC cells | Chen et al.74 | |
| FAM83H-AS1 | up | silencing FAM83H-AS1 decreases the expression of β-catenin in GC cells | Wang et al.75 | |
| STAT3 | PVT1 | up | PVT1 overexpression in GC cells promotes the accumulation of p-STAT3 in the nucleus through blocking its ubiquitin-dependent degradation and increases the transcriptional activity of STAT3 | Zhao et al.76 |
| HOTAIR | up | silencing HOTAIR decreases the expression of STAT3 and cyclin D1 in GC cells | Jiang et al.77 | |
| SNHG16 | up | the knockdown of SNHG16 in GC cells reduces the expression of JAK2 and p-STAT3 by sponging miR-135a | Wang et al.78 | |
| NEAT1 | up | silencing NEAT1 decreases STAT3 expression by sponging miR-506 in GC | Tan and Wang79 | |
| HOXD-AS1 | up | silencing HOXD-AS1 downregulates the expression of p-JAK2 and p-STAT3 in GC cells | Zheng et al.80 | |
| NF-kB | ANRIL | N/A | the knockdown of ANRIL in GC cells decreases the protein levels of p65 in the nucleus and the mRNA levels of NF-kB target genes | Deng et al.81 |
| LINC01410 | N/A | the overexpression of LINC01410 increases the nuclear signals of NF-κB p65 and the expression of p-IKK-β, p-IκBα, and c-FLIP in GC cells, whereas LINC01410 reverses their expression | Zhang et al.82 | |
| KRT19P3 | down | KRT19P3 inactivates the NF-κB signaling pathway by promoting the degradation of IκBα induced by COPS7A knockdown | Zheng et al.83 | |
| BANCR | up | BANCR silence decreases the expression of NF-κB1 (P50/105) and inhibits the activity of NF-κB1 3′ UTR | Zhang et al.84 | |
| ASB16-AS1 | up | ASB16-AS1 activates the NF-kB signaling pathway through upregulating TRIM37 expression in GC cells | Fu et al.85 | |
| Hedgehog | EGOT | up | the knockdown EGOT decreases the expression of Shh, SUFU, and Gli1 at both the transcription and protein levels in GC cells | Peng et al.86 |
| NOTCH | NALT1 | up | NALT1 knockdown decreases the expression of NOTCH1, NICD, and the downstream target genes of the notch signaling pathway, including HES1 and HES5 | Piao et al.87 |
| MIR22HG | down | silencing MIR22HG increases the expression of HEY1 and nucleus NOTCH2 in GC cells | Li and Wang88 | |
| RhoA | HOTAIR | up | the overexpression of HOTAIR increases the expression of CXCR4, RhoGEF, PI3K, ROCK, PAK, and PKN in GC cells | Xiao et al.89 |
| CTC-497E21.4 | up | the knockdown of CTC-497E21.4 regulates the expression of total and active RhoA, CDC42, and Rac1 in GC cells | Zong et al.90 | |
| NORAD | up | silencing NORAD in GC cells decreases the expression of RhoA and ROCK1 | Yu et al.91 |
The PI3K/AKT signaling pathway
The PI3K/AKT signaling pathway has been shown to regulate various cellular functions of GC cells, such as proliferation, metastasis, and drug resistance. The dysregulation of the PI3K/AKT signaling pathway is closely correlated to GC progression.92 It has also been reported that lncRNAs are key regulators of the PI3K/AKT signaling pathway. For instance, the overexpression of lncRNA LOC101928316 significantly inhibits the expression of PI3K, p-AKT, mTOR, and p-mTOR in human GC cell line SGC-7901, indicating that LOC101928316 is involved in GC progression by suppressing the PI3K/AKT signaling pathway.52 Another study showed that lncRNA XLOC_006753 is significantly upregulated in GC samples and the multidrug resistance (MDR) GC cell lines. The knockdown of XLOC_006753 decreases the expression of PI3K, p-AKT, and p-AKT in MDR GC cells.54 In addition, some other lncRNAs, such as OGFRP1, TMPO-AS1, and FOXD1-AS1, have also been reported to be involved in the regulation of the PI3K/AKT signaling pathway in GC.23,49,56,93,94 Collectively, these studies suggest that lncRNAs may contribute to GC progression by positively or negatively regulating the PI3K/AKT signaling pathway.
The MAPK signaling pathway
It is well known that the MAPK signaling pathway is closely associated with fundamental cellular functions, such as cell proliferation, apoptosis, migration, and senescence. The dysregulation of the MAPK signaling pathway has been observed in a number of cancers, including GC.95,96 In GC cells, silencing lncRNA LINC00152-1 significantly decreases the expression of p-ERK-1/2 and p-MEK1/2, while no effect is observed on the total ERK-1/2 and MEK1/2 expression, indicating the promotion role of LINC00152-1 in the MAPK signaling pathway. The administration of staurosporine aglycone (ERK/MAPK signaling pathway activator) reverses the effect of LINC00152-1 on the cellular functions of GC cells.61 Moreover, Li et al. showed that the overexpression of lncRNA CASC2 in the GC BGC-823 cell line significantly inhibits the expression of p-ERK1/2 and p-JNK, whereas no differences are observed in p-p38 expression.62 In another study, lncRNA AK025387 overexpression significantly upregulates the expression of Raf-1, MEK2, and ERK in MKN45 and SGC7901 GC cells, whereas the downregulation of AK025387 reverses the expression of these proteins in GC cells.97 In addition, some lncRNAs, such as AOC4P, LINC00483, and BCAR4, have also been reported to play a role in GC progression via direct or indirect regulation on the MAPK signaling pathway.59,60
The Wnt/β-catenin signaling pathway
The Wnt/β-catenin signaling pathway plays an important role in regulating the biological processes of normal gastric mucosa, such as proliferation, stem cell maintenance, and homeostasis. Recent studies showed that the Wnt/β-catenin signaling pathway is aberrantly activated in more than 30% of GC patients, which contributes to many aspects of GC progression.98 Some lncRNAs, such as GASL1, HCG11, LINC00665, LINC01503, lincRNA-p21, LINC01314, LINC01225, and FAM83H-AS1, have been shown to affect the carcinogenesis of GC by regulating the expression of key components in the Wnt/β-catenin signaling pathway.66, 67, 68,71, 72, 73, 74, 75 Some other lncRNAs, such as CASC15, SNHG11, and NCK1-AS1, have been found to indirectly modulate the Wnt/β-catenin signaling pathway through sponging miRNAs as ceRNAs.4,99 In addition, lncRNA MIR4435-2HG has been reported to activate the Wnt/β-catenin signaling pathway by targeting desmoplakin (DSP).63 Zhou et al. demonstrated that lncRNA HOXC-AS1 upregulates the expression of β-catenin by inhibiting eIF4AIII.100 LncRNA GATA6-AS1 has been shown to inactivate the Wnt/β-catenin signaling pathway by inhibiting Frizzled 4 expression.69 Taken together, these studies indicate that the Wnt/β-catenin signaling pathway is a key target of lncRNAs in GC.
The STAT3 signaling pathway
The STAT3 signaling pathway has been shown to promote the proliferation, angiogenesis, and invasion of cancer cells and contribute to the development of chemotherapy resistance, making STAT3 a potential therapeutic target in GC.101 A growing amount of evidence shows that lncRNAs are crucial upstream regulators of the STAT3 signaling pathway in GC. For instance, lncRNA PVT1 has been shown to directly interact with the activated p-STAT3 protein to enhance its stability, contributing to the accumulation of p-STAT3 in the nucleus of GC cells and leading to the activation of the STAT3 signaling pathway.76 lncRNA AC093818.1 promotes the transcriptional activation of the STAT3 target gene PDK1 by guiding STAT3 to its promoter.102 Moreover, Zhou et al. demonstrated that lncRNA OLC8 promotes the STAT3 signaling pathway by enhancing the stability of IL-11 mRNA.103 Some lncRNAs (e.g., HOTAIR, SNHG16, NEAT1, and GACAT3) have been found to indirectly regulate the STAT3 signaling pathway by sponging miRNAs as ceRNAs.77, 78, 79 In addition, some lncRNAs, such as HOXD-AS1, TRPM2-AS, and LINC00691, have been reported to regulate the expression of key components in the STAT3 signaling pathway.80,104,105
Other signaling pathways
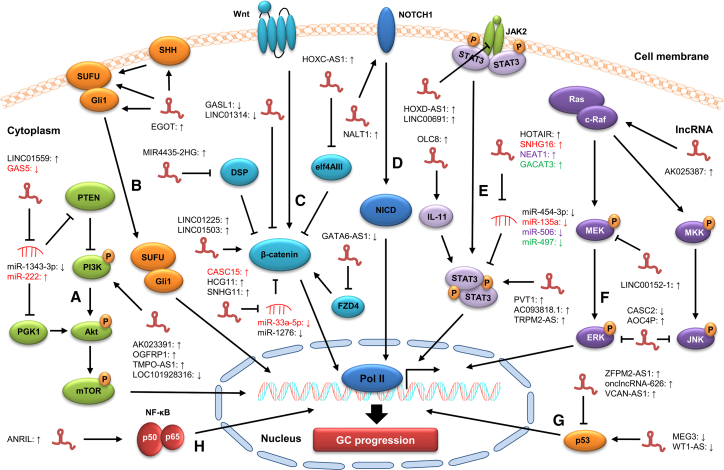
Recent studies have shown that lncRNAs can also regulate GC progression by targeting some other signaling pathways. For instance, lncRNA antisense non-coding RNA at the INK4 locus promotes the entrance of p65 from the cytoplasm to the nucleus, leading to the enhancement of the NF-kB signaling pathways in GC cells.81 Peng et al. showed that silencing lncRNA EGOT downregulates the expression of Shh, SUFU, and Gli1 at both the transcription and protein levels, indicating the negative regulation of EGOT on the Hedgehog signaling pathway.86 Some lncRNAs, such as ZFPM2-AS1, onclncRNA-626, and VCAN-AS1, facilitate GC progression by inhibiting the p53 signaling pathway, whereas other lncRNAs (e.g., MEG3 and WT1-AS) have been found to suppress GC progression by inactivating the p53 signaling pathway.106, 107, 108, 109, 110, 111 In addition, the knockdown of lncRNA NALT1 has been shown to decrease the expression of NICD and HES1, and downstream target gene HES5 in the Notch signaling pathway.87 These findings suggest that lncRNAs are crucial regulators of oncogenic signaling pathways in GC (Figure 3).
Figure 3.
Regulation of lncRNAs on signaling pathways in GC
lncRNAs are involved in GC progression by modulating the expression of key components involved in oncogenic signaling pathways, including the PI3K/AKT (A), Hedgehog (B), Wnt/β-catenin (C), Notch (D), STAT3 (E), MAPK (F), p53 (G), and NF-kB (H) signaling pathways.
lncRNAs and GC progression
The most distinctive features of cancer cells are their sustained proliferation and evasion from apoptosis.12,112 Complex signaling pathways and various factors are involved in these cellular processes of GC cells. However, the detailed mechanisms are still inconclusive. A large number of oncogenic lncRNAs, such as SNHG7, CCHE1, and lncMIF-AS1, have been shown to promote proliferation and inhibit apoptosis in GC cells.23,113, 114, 115, 116 Consistent with this, the expression of these lncRNAs is upregulated in GC samples and cell lines. Conversely, some lncRNAs (e.g., MT1JP and BG981369) have been reported to be downregulated in GC progression. The overexpression of these lncRNAs inhibits proliferation and promotes apoptosis in GC cells,117,118 indicating their anti-tumor functions. Guo et al. found that the knockdown of lncRNA AFAP1-AS1 inhibits the viability of GC cell line SGC7901 and increases the number of apoptotic SGC7901 cells. Further investigation revealed that AFAP1-AS1 knockdown decreases the expression of p-AKT and increases the expression of PTEN, indicating that AFAP1-AS1 regulates the proliferation and apoptosis of GC cells by targeting the PTEN/p-AKT signaling pathway.119 Linc00483 and AOC4P have been found to affect the proliferation and apoptosis of GC cells by regulating the MAPK signaling pathway.59,60 Moreover, some lncRNAs, such as ZEB2-AS1 and TOB1-AS1, have been shown to regulate proliferation and apoptosis in GC progression in a Wnt/βcatenin signaling pathway-dependent manner.64,70
Invasion and metastasis are the pathological processes closely associated with the mortality of GC patients. Invasion is recognized as the first step toward metastasis. It has been reported that lncRNAs are crucial regulators of invasion and metastasis in GC.120 For instance, LINC00163 acts as a ceRNA to inhibit the invasion and metastasis of GC cells by targeting the miR-183/AKAP12 axis.121 LncRNA PCGEM1 promotes the invasion and metastasis of GC cells by sponging miR-129-5p to increase the P4HA2 expression.122 In our previous study, we demonstrated that lncRNA GCRL1 is upregulated in GC tissues and cell lines. The overexpression of GCRL1 increases the number of invasive GC cells, whereas GCRL1 knockdown significantly decreases the number of migrative and invasive cells. The study further showed that the knockdown of GCRL1 suppresses the metastasis of GC cells in a mice lung metastasis model. It also found that GCRL1 promotes invasion and metastasis in GC cells through sponging miR-885-3p.123 The aberrant activation of EMT has been shown to endow cancer cells with migratory and invasive properties. Growing evidence has revealed that lncRNAs modulate invasion and metastasis in GC cells by inducing EMT progress.124 For instance, lncRNA PCGEM1 promotes the invasion and metastasis of GC cells by upregulating the expression of SNAI1 (a key TF of EMT).125 The knockdown of lncRNA MALAT-1 inhibits GC cell migration and invasion. Other studies have shown that MALAT-1 knockdown decreases the expression of EMT-associated marker vimentin and increases the expression of E-cadherin.126
In addition, the characteristics of lncRNAs in GC progression may be partially due to their involvement in the regulation of the cell-cycle process. Cyclin-dependent kinase 4 (CDK4) is a crucial intracellular modulator involved in the regulation of the cell cycle. Our previous work showed that the high expression of GCRL1 facilitates the proliferation and metastasis of GC cells by upregulating CDK4 expression through directly targeting miR-885-3p.123 Moreover, the knockdown of lncRNA HOXA11-AS has been reported to induce GC cell G0/G1 phase arrest and inhibit the migration, invasion, and metastasis of GC cells. Mechanistically, HOXA11-AS regulates GC progression by affecting β-catenin transcription and the expression of P21, KLF2, cyclin D1, and CDK2.127 In fact, lncRNAs have been shown to modulate the expression of many crucial genes involved in cell-cycle progression, such as CDK1, CDK6, and cyclin E1.128, 129, 130, 131 These findings indicate that lncRNAs exert their oncogenic or anti-tumoral functions in GC by regulating the expression of distinct genes involved in different signaling pathways. Further studies are required to identify key regulators by analyzing differentially expressed lncRNAs in GC tissues or cell lines, which may provide new insights into lncRNA-based therapeutics strategies in GC.
lncRNAs and GC cells immune escape
Cancer immune escape is a central factor of clinical outcomes due to its influence on tumor dormancy versus progression, invasion and metastasis, and therapeutic response.132 It has been recognized as a major stumbling block in designing effective anticancer therapeutic strategies. Recent studies suggested that lncRNAs are involved in the regulation of the immune escape of GC cells. Some oncogenic lncRNAs, such as UCA1, H19, and SNHG15, have been shown to promote immune escape in GC. Wang et al. found that UCA1 contributes to the immune escape of GC cells by upregulating programmed cell death ligand 1 (PDL1) expression via sponging miR-193a and miR-214.133 Sun et al. revealed that overexpression of H19 promotes immune escape of GC cells by modulating the activity of immune cells (γδT cells, Jurkat cells, and TAMs) via miR-519d-3p/LDHA axis.134 Moreover, upregulation of SNHG15 is reported to increase the expression of PDL1 by targeting miR-141, thus facilitating the immune escape of GC cells.135 In contrast to these oncogenic lncRNAs, some anti-tumoral lncRNAs show the opposite effect on the immune escape of GC cells. For instance, linc00936 inhibits immune escape of GC cells by sponging microRNA-425-3 to upregulate ZC3H12A expression.136 These findings indicate that lncRNAs play crucial roles in regulating the immune escape of GC cells. However, the exact functions are still unclear. Further studies are required to address the detailed mechanisms of lncRNA in GC immune escape.
lncRNAs and cancer angiogenesis
It is well known that aberrant angiogenesis contributes to cancer progression and is closely associated with tumor growth and metastatic spread.137 LncRNAs have been reported to play crucial roles in the regulation of angiogenesis in GC (Table 2). For instance, Zhao et al. found that lncRNA PVT1 is closely correlated with a high microvessel density in GC. PVT1 induces the angiogenesis of GC by upregulating the expression of VEGFA. This process is dependent on the activation of the STAT3 signaling pathway induced by PVT1.76 LncRNA LINC01410 has been shown to promote GC angiogenesis and metastasis by directly targeting miR-532-5p, which resulted in the upregulation of NCF2 and continuous NF-κB signaling pathway activation.82 Li et al. revealed that the silencing of lncRNA MALAT1 significantly decreases the vasculogenic mimicry of GC cells, blocks angiogenesis, and enhances vascular permeability by modulating the expression of classical markers of vasculogenic mimicry and angiogenesis as well as key components of related signaling pathways, including VE-cadherin, β-catenin, MMPs 2 and 9, MT1-MMP, p-ERK, p-FAK, and p-paxillin.138 Moreover, lncRNA LINC01314 is identified as a tumor suppressor in GC. LINC01314 overexpression reduces the microvessel density of transplanted tumors and inhibits angiogenesis by downregulating the expression of VEGF-C and VEGFR-3.71 Currently, the exact functions of lncRNAs in GC angiogenesis are still unclear, and further research is required to elucidate their regulation mechanisms, which will provide novel strategies for the development of lncRNAs-based anti-angiogenesis therapeutics.
Table 2.
Role of lncRNAs in angiogenesis of GC
| lncRNAs | Expression | Mechanisms of actions | References |
|---|---|---|---|
| PVT1 | up | PVT1 induces angiogenesis in GC and is significantly correlated with a high microvessel density. Mechanistically, PVT1 upregulates VEGFA expression by activating the STAT3 signaling pathway, leading to the angiogenesis of GC | Zhao et al.76 |
| LINC01410 | up | LINC01410 overexpression accelerates the angiogenesis of GC through inhibiting miR-532-5p, which leads to the upregulation of NCF2 and the continuous activation of the NF-κB signaling pathway | Zhang et al.82 |
| MALAT1 | up | MALAT1 promotes angiogenesis in GC by targeting the VE-cadherin/β-catenin complex and ERK/MMP and the FAK/paxillin signaling pathways | Li et al.138 |
| CASC2 | down | the overexpression of CASC2 suppresses the angiogenesis of GC cells | Zhou et al.139 |
| LINC01314 | down | the upregulation of LINC01314 inhibits angiogenesis in GC by decreasing the expression of VEGF-C and VEGFR-3 | Tang et al.71 |
lncRNAs and stemness of GC cells
Cancer stem cells (CSCs) are tumor cells with “stem-like” properties, including self-renewal, metastasis, and tumor initiation. Recent studies have suggested that CSCs are one of the main causes of therapeutic resistance, metastasis, and cancer recurrence.140 Therefore, the investigation of CSCs’ regulation mechanism will provide new insights into the development of GC treatment strategies. Accumulating evidence has shown that lncRNAs play crucial roles in regulating the stemness of gastric CSCs (Table 3). For instance, Xiao et al. reported that the overexpression of lncRNA MALAT1 increases the stemness of GC cells, whereas MALAT1 knockdown decreases their stemness. Mechanistically, MALAT1 upregulated the expression of SOX2 by enhancing the stability of SOX2 mRNA, leading to the promotion of gastric CSC stemness.141 Another study showed that lncRNA THOR promotes the stemness of GC cells by enhancing SOX9 mRNA stability. THOR knockdown significantly decreases the expression of stemness markers ALDH1, Nanog, Oct1/2/4, SOX2, SOX9, and CD44 in GC cells.142 Hui et al. found that knockdown of FEZF1-AS1 reduces the expression of stem factors and markers in GC cells, including ALDH1, CD133, Nanog, SOX2, and Oct4. Moreover, FEZF1-AS1 knockdown inhibits the proliferation, viability, invasion, and migration of gastric CSCs by targeting the miR-363-3/HMGA2 axis.143 In addition, a number of lncRNAs, such as regulator of reprogramming (ROR), HCP5, and ADAMTS9-AS2, have been shown to modulate the expression of stemness markers in GC cells, indicating their regulation of gastric CSCs stemness.144, 145, 146
Table 3.
Regulation of lncRNAs on stemness of GC cells
| lncRNAs | Expression | Mechanisms of actions | References |
|---|---|---|---|
| MALAT1 | up | the knockdown of MALAT1 reduces the stemness of non-adherent GC cells, whereas MALAT1 overexpression enhances the stemness of adherent GC cells. Mechanistically, MALAT1 upregulates the expression of the master stemness factor sox2 by enhancing its mRNA stability | Xiao et al.141 |
| MACC1-AS1 | down | the overexpression of MACC1-AS1 promotes the stemness of GC cells by upregulating the expression levels of stemness genes (e.g., OCT4 and sox2) in a FAO pathway-dependent manner | He et al.147 |
| PTCSC3 | down | PTCSC3 overexpression reduces the stemness of GC cells through cooperating with lncRNA Linc-pint | Hong et al.148 |
| THOR | up | silencing THOR decreases the stemness of GC cells with the downregulation of stemness marker expression and the formation of spheroid cells. Mechanistically, the knockdown of THOR downregulates SOX9 expression by decreasing its mRNA stability | Song et al.142 |
| LOXL1-AS1 | up | LOXL1-AS1 increases the expression of USF1 by targeting miR-708-5p, leading to an enhancement of the stemness marker SOX2 transcription | Sun et al.149 |
| HCP5 | the overexpression of HCP5 in GC cells upregulates the expression of stemness genes and increases the rate of CD44+ and CD133+ GC cells. Mechanistically, HCP5 promotes the stemness of GC cells by sponging miR-3619-5p | Wu et al.145 | |
| ROR | up | ROR overexpression upregulates the expression of core stemness transcriptional factors in gastric CSCs, including OCT4, SOX2, NANOG, and CD133, whereas the knockdown of ROR reverses their expression | Wang et al.144 |
| SNHG11 | up | SNHG11 promotes cell stemness in GC by upregulating the expression of CTNNB1 and ATG12 via sponging miR-483-3p/miR-1276 | Wu et al.4 |
| ASB16-AS1 | up | ASB16-AS1 enhances the stem cell-like characteristics of GC cells. Mechanistically, ASB16-AS1 promotes the expression of TRIM37 by sponging miR-3918 and miR4676-3p and the phosphorylation of TRIM37 by activating the NF-κB pathway | Fu et al.85 |
| LINC01559 | down | LINC01559 promotes the stemness of GC cells. Mechanistically, LINC01559 upregulates PGK1 expression by sponging miR-1343-3p and downregulates PTEN expression by promoting the methylation of the PTEN promoter, leading to the activation of the PI3K/AKT signaling pathway | Wang et al.57 |
| SNHG3 | up | the overexpression of SNHG3 increases the stemness of GC cells, whereas SNHG3 knockdown has the opposite effect. Mechanistically, SNHG3 promotes ARL2 expression by directly targeting miR-3619-5p | Sun et al.150 |
lncRNAs and chemotherapy sensitivity in GC
Chemotherapy is one of the main clinical therapeutic methods against cancers, including GC. Chemotherapy drugs are considered as cytotoxic drugs that kill cancer cells by damaging the DNA double-strand structure or induce the apoptosis of cancer cells. However, chemotherapy resistance and the development of MDR always lead to chemotherapy failure in GC patients during the treatment process.151 It has been reported that the aberrant expression of lncRNAs is one of the main reasons for the resistance of GC cells to chemotherapeutic agents.19,23,152 For instance, lncRNA ARHGAP5-AS1 is upregulated in chemoresistant GC cells. The data from MTT and flow cytometry assay showed that ARHGAP5-AS1 silencing significantly inhibits cell viability (p < 0.05), enhances drug-induced apoptosis (p < 0.05), and increases the intracellular drug concentration in MDR GC cell lines. Consistent with this, ARHGAP5-AS1 overexpression in sensitive GC cells significantly attenuates drug-induced viability suppression (p < 0.05), decreases drug-activated apoptosis (p < 0.05), and reduces intracellular drug concentration. These results demonstrate that ARHGAP5-AS1 facilitates chemoresistance in GC. Mechanistically, ARHGAP5-AS1 promotes ARHGAP5 transcription in the nucleus by directly binding to the ARHGAP5 promoter, and it also stabilizes ARHGAP5 mRNA in the cytoplasm by recruiting METTL3, leading to the chemoresistance of GC cells.153 lncRNA SNHG5 is reported to be upregulated in cisplatin-resistant GC patients compared with cisplatin-sensitive GC patients. The data from flow cytometry assay revealed that SNHG5 knockdown significantly increases the apoptotic rate of cisplatin-resistant GC cells, whereas overexpression of SNHG5 significantly decreases cisplatin-induced GC cell apoptosis. Moreover, overexpression of SNHG5 downregulates Bax expression and upregulates Bcl-2, MDR1, and MRP1 expression in GC cells. These results show that lncRNA SNHG5 plays a critical role in promoting chemoresistance of GC cells by modulating the expression of drug resistance-related genes and apoptosis-related genes.154 In our previous study, we found that lncRNA D63785 is highly expressed in both GC samples and cell lines, and its expression is negatively correlated with miR-422a expression. Silencing of D63785 significantly increases DOX-induced apoptosis of BGC823 cells (p < 0.05). Moreover, D63785 knockdown reduces the size and volume of the tumor nodules in DOX-treated mice (p < 0.05). The tumor weight in DOX-treated mice was 1.03 ± 0.22 g, whereas it was 0.5 ± 0.23 g in sh-D63785 plus DOX-treated mice. These data demonstrate that D63785 contributes to the enhancement of DOX-induced chemoresistance in GC. Further mechanism studies revealed that D63785 promotes the DOX resistance of GC cells by blocking the miR-422a-dependent inhibition of myocyte enhancer factor-2D.155 In addition, lncRNA XLOC_006753 is found to upregulate in MDR GC tissues and cell lines. The knockdown of XLOC_006753 in MDR GC cells promotes apoptosis and inhibits cell proliferation, viability, the cell-cycle G1/S transition, and migration by targeting the PI3K/AKT/mTOR signaling pathway.54 Therefore, elucidating lncRNA-related regulation mechanisms in chemotherapy resistance is crucial for effectively developing new drugs to treat chemoresistant tumors and exploring new compound combination therapies to enhance the therapeutic effects of chemotherapy drugs.
lncRNAs and Helicobacter pylori infection in GC
Helicobacter pylori (HP) infection has been identified as the most frequent risk factor for GC, which infects approximately over 70% of all cases.156 HP contributes to carcinogenesis and progression of GC by inducing cell autophagy, epigenetic modifications, and disruption of the balance between cell proliferation and apoptosis as well as cancer cell invasion and metastasis.157 However, the detailed mechanism of HP infection in GC progression is still unclear. Increasing evidence suggests that lncRNAs involve in the pathogenesis of HP-associated GC. Aberrant expression of lncRNAs has been observed in HP-infected gastric epithelial cells and GC tissues. Yang et al. found that 23 lncRNAs are upregulated and 21 are downregulated in HP-infected gastric epithelial cells, indicating the potential role of these differentially expressed lncRNAs in the immune system response against HP.158 Zhong et al. showed that lncRNA NR_026827 is downregulated in all stages of GC associated with HP infection and exhibits the potential as a diagnostic biomarker for GC.159 Furthermore, overexpression of lncRNA HOXA-AS2 has been demonstrated to be closely correlated with HP infection in GC patients.160 In addition, Zhang et al. revealed that the expression of lncRNA H19 is significantly upregulated in HP-infected GC tissues and cell lines. Overexpression of H19 facilitates HP-induced proliferation, migration, and invasion of GC cells by enhancing NF-κB-mediated inflammation.161 In another study, Jia et al. found that HP infection promotes the proliferation and migration of GC cells by increasing the expression of lncRNA THAP9-AS1.162 Collectively, these findings indicate that lncRNAs are key factors in HP-induced GC progression. Understanding the functions of lncRNAs in pathogenesis of HP-associated GC may provide new insights in the development of GC therapeutic strategies.
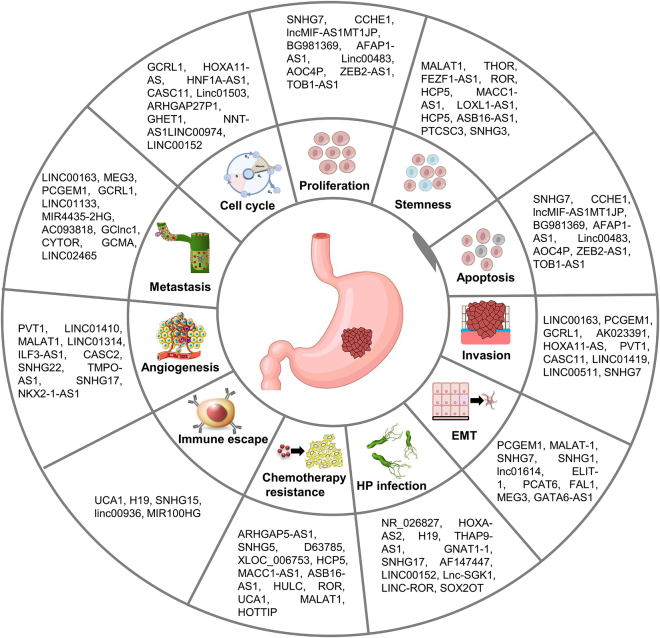
In summary, lncRNAs act as oncogenes or tumor suppressors to regulate GC biological behaviors, including cell proliferation, apoptosis, invasion, metastasis, EMT, cell cycle, stemness, immune escape, angiogenesis, chemotherapy resistance, and HP infection (Figure 4). The dual effects of lncRNAs on GC progression might be dependent on their diversity of function.
Figure 4.
Effect of lncRNAs on biological behaviors of GC
lncRNAs function as oncogenes or tumor suppressors to modulate GC biological behaviors, including cell proliferation, apoptosis, invasion, metastasis, EMT, cell cycle, stemness, immune escape, angiogenesis, chemotherapy resistance, and HP infection.
Clinical implications of lncRNAs in GC
Clinically, it is difficult to detect the early symptoms of GC patients due to the complicated pathophysiological features of the disease, most of which are diagnosed at an advanced stage of cancer with poor prognosis. Some biomarkers, such as CEA, CA 19-9, and CA72-4, have been used in clinical diagnosis and prognostic evaluation of GC.163 However, the unideal specificity and sensitivity of these biomarkers limit their further application. Therefore, the identification of new biomarkers with high specificity and sensitivity is crucial for the early screening and prognostic judgment of GC patients. A growing number of studies indicate that lncRNAs possess great potential to be biomarkers for the diagnosis, prognosis and treatment of GC due to their high stability, specificity, and detectability.164
lncRNAs and GC diagnosis
lncRNAs freely circulate in the plasma, and their levels are almost the same as those in primary tumor tissues, which can exactly reflect the features of the corresponding tumors.165 These characteristics make lncRNAs in the plasma ideal diagnostic biomarkers in clinical applications. A large number of lncRNAs in the plasma have been identified as diagnostic biomarkers of GC. For instance, Elsayed et al. showed that the expression of lncRNA HOTAIR was significantly upregulated in the plasma of GC patients compared with that of healthy controls. Receiver operating characteristic curve (ROC) analysis revealed that plasma HOTAIR can diagnose GC with 88% sensitivity and 84% specificity,166 Zhou et al. found that the plasma level of lncRNA H19 in GC patients is significantly higher than that in healthy controls. ROC analysis showed that the area under the curve (AUC) value for H19 is 0.838 with 82.9% sensitivity and 72.9% specificity. In addition, H19 expression distinguishes GC patients from healthy controls with an AUC of 0.877, 85.5% sensitivity, and 80.1% specificity.167 These data suggest plasma H19 as a promising biomarker for GC diagnosis. In addition, Feng et al. revealed that lncRNA B3GALT5-AS1 in plasma can also act as a diagnostic biomarker to distinguish GC patients from normal controls with an AUC of 0.816, which indicates better sensitivity and specificity than CEA and CA19-9.168
Exosomal lncRNAs have been recognized as promising biomarkers for GC diagnosis due to their intracellular origin and high quantities in the plasma.169 For instance, Lin et al. demonstrated that almost all lncUEGC1 in the plasma are packaged into exosomes. Further analysis revealed that exosomal lncUEGC1 has an AUC of 0.8760 and 0.8406 in discriminating early GC patients from normal controls and those with premalignant chronic atrophic gastritis, respectively, indicating its better diagnostic accuracy than CEA.170 Li et al. showed that the expression of exosomal lnc-GNAQ-6:1 in GC patients is significantly downregulated and the AUC values for lnc-GNAQ-6:1 was 0.732, which is higher than the diagnostic accuracy of CEA, CA 19-9, and CA72-4.171 Moreover, the expression of exosomal lncRNA PCSK2-2:1 has also been found to be downregulated in GC patients and its expression is closely associated with tumor size, tumor stage, and venous invasion. The AUC values for PCSK2-2:1 was 0.896, with 86.5% sensitivity and 84% specificity.172
In addition, multiple lncRNA combinations show better diagnostic biomarker values for GC. For instance, Ke et al. found that the plasma levels of lncRNAs AK001058, INHBA-AS1, MIR4435-2HG, and CEBPA-AS1 are significantly upregulated in GC patients compared with healthy controls. The combination of these plasma lncRNAs increases the AUC value to 0.921, indicating that their combination might be used as better diagnostic biomarkers of GC than their individuals.173 Yang et al. showed that the expressions of lncRNAs PANDAR, FOXD2-AS1, and SMARCC2 plasma are significantly increased in GC patients than in normal controls. The AUC value for PANDAR is 0.767, for FOXD2-AS1 0.700, and for SMARCC2 0.748. Interestingly, the AUC value of the three lncRNAs combined is up to 0.839.174 Liu et al. demonstrated that the plasma levels of lncRNAs FEZF1-AS1 and AFAP1-AS1 are upregulated in GC patients compared with healthy controls. The combination of FEZF1-AS1 and AFAP1-AS1 increases the AUC value to 0.866, whereas the combination of FEZF1-AS1, AFAP1-AS1, CEA, and CA19-9 increases the diagnostic sensitivity to 95.5%.175
lncRNAs and GC prognosis
Prognostic judgment is important for cancer patients in terms of evaluating their treatment status and adjust their therapeutic plan.176,177 It has been reported that lncRNAs play crucial roles in regulating prognosis-related factors, including tumor size, stage, depth of invasion, lymph node metastasis, distant metastasis, and pathological type,165 indicating their great potential as prognostic biomarkers in GC. Zhang et al. found that high expression of DQ786243 is closely associated with invasion depth, TNM stage, and lymphatic metastasis of GC. GC patients with high DQ786243 levels have higher survival rates than those with low levels. Moreover, multivariate analysis showed that DQ786243 is a significant and independent prognostic biomarker in GC patients.178 Shi et al. revealed that lncRNA CADM1-AS1 expression is significantly downregulated in GC samples, and its expression is closely associated with tumor differentiation, N stage, M stage, and TNM stage. The overall survival and disease-free survival of GC patients with high CADM1-AS1 expression are significantly better than for patients with low CADM1-AS1 expression. Both univariate and multivariate analysis showed that CADM1-AS1 expression is an independent prognostic biomarker of GC patients.179
In addition, multiple lncRNA combinations show better prognostic biomarker value for GC in some cases.180,181 This may due to their joint action. As the key regulators of ceRNA network, lncRNAs are involved in almost all aspects of GC progression. Thus, the combination of lncRNAs could efficiently improve the prediction accuracy of GC. For instance, Song et al. identified a set of three lncRNAs (LINC01140, TGFB2-OT1, and RP11-347C12.10) form the Gene Expression Omnibus datasets consisted of 492 GC patients, which exhibits a great potential as prognostic biomarker for GC patients. High expression of LINC01140 and TGFB2-OT1 was significantly associated with a shorter survival, whereas high expression of RP11-347C12.10 was significantly associated with a longer survival. ROC analysis revealed that the AUC value for LINC01140 was 0.620, for TGFB2-OT1 0.677, and for RP11-347C12.10 0.610. Interestingly, the combination of these three lncRNAs increased the AUC value to 0.688. These data indicate that the combination of LINC01140, TGFB2-OT1, and RP11-347C12.10 has better prediction accuracy than that of each single lncRNA for the prognosis of GC.182
Therapeutic potential of lncRNAs in GC
lncRNAs exhibit great potential as efficient therapeutic targets due to their crucial roles in GC progression. Targeting specific lncRNAs using nucleic acid-based knockdown techniques may destroy multiple cancer-related pathways, leading to the reduction of chemotherapy resistance.17 RNAi targeting aberrant GC-specific lncRNAs is an effective strategy in this regard. For instance, the expression of the lncRNA ROR has been found to be positively correlated with increased MDR and poor prognosis of GC patients. The knockdown of ROR by shRNA has been shown to promote the apoptosis of drug-resistant GC cells through downregulating multidrug resistance-associated protein 1 (MRP1).183 Similar to RNAi, antisense oligonucleotides (ASOs) can form a DNA-RNA structure with the target lncRNA, leading to its degradation in an RNase-H-dependent manner. Li et al. showed that the expression of lncRNA PVT1 is positively associated with larger tumor size, lymph node metastases, and short survival duration in GC. Silencing of PVT1 by ASOs significantly inhibits the growth and invasion of GC cells.184 Moreover, the CRISPR-Cas9 genome editing technique has been recognized as a potential therapeutic tool for cancer treatment by reactivating or silencing lncRNAs. For instance, GC metastasis associated long non-coding RNA (GMAN) can be disrupted by CRISPR-Cas9 through deleting the MFR region of GMAN, leading to downregulation of GC cell invasive activity.185 In addition, the screening of natural products or the synthesis of chemical molecules specifically targeting lncRNAs may provide a valuable strategy for GC treatment. Although lncRNAs have shown great potential as possible therapeutic targets for GC, there are still some challenges that limit their further application, such as off-target effects, side effects, and modes of targeted delivery. Therefore, more in-depth studies should be conducted to resolve these issues prior to their clinical application in GC treatment.
Methods for lncRNA identification and functional characterization
Analysis tools for lncRNA functions
With the continuous development of high-throughput sequencing technologies, a large number of lncRNA expression data are accumulating from GC samples and cell lines. It is crucial to evaluate the potential functions of differentially expressed lncRNAs by bioinformatic analysis for the development of lncRNA-based GC therapy. Currently, some research tools have been developed to explore the underlying biological processes and signaling pathways regulated by lncRNAs. For instance, LncRNAs2Pathways is a novel computational method based on a global network propagation algorithm, which can help researchers to identify the signaling pathways regulated by the combinatorial effects of a set of lncRNAs.186 LncRNA2Function allows researchers to search the lncRNAs correlated with a specific functional term or the functions of a specific lncRNA, or to annotate functionally a set of human lncRNA genes.187 In addition, the enrichment analysis has also been widely used to assess the potential functions of lncRNAs. Lv et al. performed the gene ontology (GO) enrichment analysis to predict the function of differentially m6A-methylated and expressed lncRNAs in GC (dme-lncRNAs). Four dme-lncRNAs, including RASAL2-AS1, LINC00910, SNHG7, and LINC01105, were found to play a potential role in the cellular processes and biological behaviors involved in mitosis and cell cycle.188 In another study, Xiao et al. identified 520 differentially expressed GC-associated lncRNAs from a TCGA dataset. GO and Kyoto Encyclopedia of Genes and Genomes pathway analysis showed that these lncRNAs in were closely associated with functions, such as cell signaling, cell cycle, immune response, metabolic processes, angiogenesis, and regulation of retinoic acid receptors.189 Taken together, the application of these bioinformatic tools greatly accelerates the research progress of lncRNA functions in GC progression.
Application of single-cell RNA sequencing for lncRNA identification in GC
GC is a highly heterogeneous malignant disease and its heterogeneity origins remain poorly understood, which brings a great challenge for GC research and treatment.190 Interestingly, the expression of lncRNAs has shown stronger cell type, tissue, and spatial-temporal specificity than protein-coding genes,29 indicating that lncRNAs may play crucial roles in the origins of tumor heterogeneity. Thus, the precise detection of lncRNA expression profiles in GC samples may help researchers and clinicians better understand GC heterogeneity. However, it is difficult to decipher complete lncRNA expression pattern across each single cell, particularly for unannotated lncRNAs, due to the large number of lncRNAs in a genome.191 Single-cell RNA sequencing (scRNA-seq) technology is recognized as a robust and unbiased tool to evaluate tumor heterogeneity, which can efficiently detect the specific expression patterns of lncRNA at the single-cell resolution.192,193 Currently, scRNA-seq has been used to investigate the transcriptional heterogeneity in primary GC, cellular reprogramming of GC microenvironment, and identification of GC lymph node metastasis markers and GC evolution-driving genes;192,194,195 however, its application in detecting the lncRNA expression profile of GC samples has not been reported. Therefore, we believe that scRNA-seq possesses great potential to investigate lncRNA expression profiles at the single-cell level in GC. The lncRNA expression profile obtained by scRNA-seq will help us to further understand molecular characteristics of GC heterogeneity, which may provide new insights into the development of GC therapeutic strategies.
Conclusions
GC is a life-threatening malignant disease with poor prognosis. Genetic mutations, epigenetic changes, and environmental factors are recognized as the main factors driving GC, but a detailed pathogenesis of this disease is still unclear.155 An elucidation of the GC regulation mechanisms will be very helpful in the screening of diagnostic and prognostic biomarkers and the identification of therapeutic targets, both of which bring great benefits to the precise treatment of GC patients. In recent years, increasing numbers of lncRNAs have been found to be aberrantly expressed in GC tissues. Most of them have been shown to promote GC progression, with only a few lncRNAs exhibiting anti-tumor functions, such as PWRN1 and TUBA4B.17 lncRNAs are involved in the regulation of GC development and progression by modulating the expression of genes associated with cell proliferation, apoptosis, invasion, metastasis, angiogenesis, and chemotherapy resistance. Moreover, a complex crosstalk network has been observed between lncRNAs and multiple key signaling pathways in GC progression. Due to the crucial role of lncRNAs in GC development and progression, they have been recognized as valuable therapeutic targets for GC. Therefore, targeting specific lncRNAs shows great potential in GC treatment. Currently, lncRNA-based methods for GC treatment, such as RNAi, ASOs, and CRISPR-Cas9, are being explored, and some interesting progress has been made. In addition, the tumor specificity and plasma stability of lncRNAs make them potential noninvasive biomarkers for the diagnosis and prognosis of GC patients.
However, there are still some unsolved challenges that block the clinical application of lncRNAs in GC treatment. For instance, lncRNAs have been shown to be key regulators of gene expression, functional proteins, and RNA molecules. Targeting specific lncRNAs may trigger a series of unknown physiological and pathological reactions. Therefore, it is important to elucidate the pathophysiological mechanism of lncRNAs before they enter formal clinical application. Furthermore, every lncRNA possesses multiple target genes. Off-target effects and lower delivery efficiencies may delay lncRNA-based GC therapies. The development of more efficient delivery systems may improve this situation. Moreover, the conservation of lncRNAs is poor across different species. This may lead to a serious consequence in that the information and promising therapeutic strategies developed based on in vitro and animal models may not be easily extended to human clinical applications. Therefore, more detailed clinical studies are needed to solve this problem.
In conclusion, recent findings provide in-depth insights on the diagnosis and treatment of GC. However, our current understanding on lncRNAs in GC progression is still insufficient, and the translational process from basic research to clinical application is usually very long. With the continuous development of high-throughput sequencing technologies, novel lncRNAs in GC are being identified and their important functions will be revealed. Overall, we believe that lncRNAs will be widely used in the early diagnosis, prognosis, and clinical treatment of GC in the future.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81802822), the Natural Science Foundation of Shandong Province (grant no. ZR2018BH017 and ZR202102220061), the China Postdoctoral Science Foundation (grant no. 2018M642607), and the Qingdao Applied Basic Research Project (19-6-2-50-cg).
Author contributions
W.D., W.Y., and Y.Z. collected the related paper. Y.L. drafted and wrote the manuscript. Y.L. and X.A. revised the manuscript. Y.L., X.A., and J.W. participated in the design of the review and helped to draft and revise the manuscript. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Xiang Ao, Email: xiangao2016@163.com.
Jianxun Wang, Email: wangjx@qdu.edu.cn.
References
- 1.Shan C., Zhang Y.F., Hao X.D., Gao J.N., Chen X.Z., Wang K. Biogenesis, functions and clinical significance of circRNAs in gastric cancer. Mol. Cancer. 2019;18:136. doi: 10.1186/s12943-019-1069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu X., Meltzer S.J. Gastric cancer in the era of precision medicine. Cell. Mol. Gastroenterol. Hepatol. 2017;3:348–358. doi: 10.1016/j.jcmgh.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun W., Yang Y., Xu C., Xie Y., Guo J. Roles of long noncoding RNAs in gastric cancer and their clinical applications. J. Cancer Res. Clin. Oncol. 2016;142:2231–2237. doi: 10.1007/s00432-016-2183-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Q., Ma J., Wei J., Meng W., Wang Y., Shi M. lncRNA SNHG11 promotes gastric cancer progression by activating the Wnt/beta-catenin pathway and oncogenic autophagy. Mol. Ther. 2021;29:1258–1278. doi: 10.1016/j.ymthe.2020.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin M.T., Song H.J., Ding X.Y. Long non-coding RNAs involved in metastasis of gastric cancer. World J. Gastroenterol. 2018;24:3724–3737. doi: 10.3748/wjg.v24.i33.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martens-Uzunova E.S., Böttcher R., Croce C.M., Jenster G., Visakorpi T., Calin G.A. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur. Urol. 2014;65:1140–1151. doi: 10.1016/j.eururo.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Zhao M., Zhu N., Hao F., Song Y., Wang Z., Ni Y., Ding L. The regulatory role of non-coding RNAs on programmed cell death four in inflammation and cancer. Front. Oncol. 2019;9:919. doi: 10.3389/fonc.2019.00919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liyanage K.I.P., Ganegoda G.U. Therapeutic approaches and role of ncRNAs in cardiovascular disorders and insulin resistance. Biomed. Res. Int. 2017;2017:4078346. doi: 10.1155/2017/4078346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Svoboda P. Long and small noncoding RNAs during oocyte-to-embryo transition in mammals. Biochem. Soc. Trans. 2017;45:1117–1124. doi: 10.1042/BST20170033. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y., Ao X., Ding W., Ponnusamy M., Wu W., Hao X., Yu W., Wang Y., Li P., Wang J. Critical role of FOXO3a in carcinogenesis. Mol. Cancer. 2018;17:104. doi: 10.1186/s12943-018-0856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y., Jia D.D., Zhang Y.F., Cheng M.D., Zhu W.X., Li P.F., Zhang Y.F. The emerging function and clinical significance of circRNAs in thyroid cancer and autoimmune thyroid diseases. Int. J. Biol. Sci. 2021;17:1731–1741. doi: 10.7150/ijbs.55381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y., Ding W., Ge H., Ponnusamy M., Wang Q., Hao X., Wu W., Zhang Y., Yu W., Ao X., et al. FOXK transcription factors: regulation and critical role in cancer. Cancer Lett. 2019;458:1–12. doi: 10.1016/j.canlet.2019.05.030. [DOI] [PubMed] [Google Scholar]
- 13.Sun W., Yang Y., Xu C., Guo J. Regulatory mechanisms of long noncoding RNAs on gene expression in cancers. Cancer Genet. 2017;216-217:105–110. doi: 10.1016/j.cancergen.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Begolli R., Sideris N., Giakountis A. LncRNAs as chromatin regulators in cancer: from molecular function to clinical potential. Cancers. 2019;11:1524. doi: 10.3390/cancers11101524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y., Liu X., Cui X., Tan Y., Wang Q., Wang Y., Xu C., Fang C., Kang C. LncRNA PRADX-mediated recruitment of PRC2/DDX5 complex suppresses UBXN1 expression and activates NF-kappaB activity, promoting tumorigenesis. Theranostics. 2021;11:4516–4530. doi: 10.7150/thno.54549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu P., He F., Hou Y., Tu G., Li Q., Jin T., Zeng H., Qin Y., Wan X., Qiao Y., et al. A novel hypoxic long noncoding RNA KB-1980E6.3 maintains breast cancer stem cell stemness via interacting with IGF2BP1 to facilitate c-Myc mRNA stability. Oncogene. 2021;40:1609–1627. doi: 10.1038/s41388-020-01638-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan H., Zhang S., Zhang J., Zhu L., Chen Y., Yang H., Chen Y., An Y., Liu B. Long non-coding RNAs in gastric cancer: new emerging biological functions and therapeutic implications. Theranostics. 2020;10:8880–8902. doi: 10.7150/thno.47548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao Y., Wang J.W., Ren J.Y., Guo M., Guo C.W., Ning S.W., Yu S. Long noncoding RNAs in gastric cancer: from molecular dissection to clinical application. World J. Gastroenterol. 2020;26:3401–3412. doi: 10.3748/wjg.v26.i24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang F., Wang H., Yu J., Yao X., Yang S., Li W., Xu L., Zhao L. LncRNA CRNDE attenuates chemoresistance in gastric cancer via SRSF6-regulated alternative splicing of PICALM. Mol. Cancer. 2021;20:6. doi: 10.1186/s12943-020-01299-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan H., Ding Y., Jiang Y., Wang X., Rao J., Zhang X., Yu H., Hou Q., Li T. LncRNA LIFR-AS1 promotes proliferation and invasion of gastric cancer cell via miR-29a-3p/COL1A2 axis. Cancer Cell Int. 2021;21:7. doi: 10.1186/s12935-020-01644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin S., Yang L., Kong S., Xu Y., Liang B., Ju S. LncRNA HCP5: a potential biomarker for diagnosing gastric cancer. Front. Oncol. 2021;11:684531. doi: 10.3389/fonc.2021.684531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma S., Kong S., Gu X., Xu Y., Tao M., Shen L., Shen X., Ju S. As a biomarker for gastric cancer, circPTPN22 regulates the progression of gastric cancer through the EMT pathway. Cancer Cell Int. 2021;21:44. doi: 10.1186/s12935-020-01701-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Q., Ma J., Wei J., Meng W., Wang Y., Shi M. FOXD1-AS1 regulates FOXD1 translation and promotes gastric cancer progression and chemoresistance by activating the PI3K/AKT/mTOR pathway. Mol. Oncol. 2021;15:299–316. doi: 10.1002/1878-0261.12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y., Lu L., Wu X., Li Q., Zhao Y., Du F., Chen Y., Shen J., Xiao Z., Wu Z., et al. The multifaceted role of long non-coding RNA in gastric cancer: current status and future perspectives. Int. J. Biol. Sci. 2021;17:2737–2755. doi: 10.7150/ijbs.61410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han T., Xu D., Zhu J., Li J., Liu L., Deng Y. Identification of a robust signature for clinical outcomes and immunotherapy response in gastric cancer: based on N6-methyladenosine related long noncoding RNAs. Cancer Cell Int. 2021;21:432. doi: 10.1186/s12935-021-02146-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu H., Wan J., Chu J. Long non-coding RNAs and endometrial cancer. Biomed. Pharmacother. 2019;119:109396. doi: 10.1016/j.biopha.2019.109396. [DOI] [PubMed] [Google Scholar]
- 27.Bermúdez M., Aguilar-Medina M., Lizárraga-Verdugo E., Avendaño-Félix M., Silva-Benítez E., López-Camarillo C., Ramos-Payán R. LncRNAs as regulators of autophagy and drug resistance in colorectal cancer. Front. Oncol. 2019;9:1008. doi: 10.3389/fonc.2019.01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu H., Yang L., Chen L.L. The diversity of long noncoding RNAs and their generation. Trends Genet. 2017;33:540–552. doi: 10.1016/j.tig.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 29.Jiang M.C., Ni J.J., Cui W.Y., Wang B.Y., Zhuo W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am. J. Cancer Res. 2019;9:1354–1366. [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Q., Song Z., Zhu C., Tao C., Kang L., Liu W., He F., Yan J., Sang T. Systematic comparison of lncRNAs with protein coding mRNAs in population expression and their response to environmental change. BMC Plant Biol. 2017;17:42. doi: 10.1186/s12870-017-0984-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X., Hong R., Chen W., Xu M., Wang L. The role of long noncoding RNA in major human disease. Bioorg. Chem. 2019;92:103214. doi: 10.1016/j.bioorg.2019.103214. [DOI] [PubMed] [Google Scholar]
- 32.Alessio E., Bonadio R.S., Buson L., Chemello F., Cagnin S. A single cell but many different transcripts: a journey into the world of long non-coding RNAs. Int. J. Mol. Sci. 2020;21:302. doi: 10.3390/ijms21010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gourvest M., Brousset P., Bousquet M. Long noncoding RNAs in acute myeloid leukemia: functional characterization and clinical relevance. Cancers. 2019;11:1638. doi: 10.3390/cancers11111638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilusz J.E., Freier S.M., Spector D.L. 3' end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 36.Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 37.Sleutels F., Zwart R., Barlow D.P. The non-coding air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 38.Tam C., Wong J.H., Tsui S.K.W., Zuo T., Chan T.F., Ng T.B. LncRNAs with miRNAs in regulation of gastric, liver, and colorectal cancers: updates in recent years. Appl. Microbiol. Biotechnol. 2019;103:4649–4677. doi: 10.1007/s00253-019-09837-5. [DOI] [PubMed] [Google Scholar]
- 39.Bhan A., Soleimani M., Mandal S.S. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y.Y., Chen M., Li B. Dosage compensation mechanism of X chromosome. Yi Chuan. 2012;34:977–984. doi: 10.3724/sp.j.1005.2012.00977. [DOI] [PubMed] [Google Scholar]
- 41.Teng F., Zhang J.X., Chen Y., Shen X.D., Su C., Guo Y.J., Wang P.H., Shi C.C., Lei M., Cao Y.O., et al. LncRNA NKX2-1-AS1 promotes tumor progression and angiogenesis via upregulation of SERPINE1 expression and activation of the VEGFR-2 signaling pathway in gastric cancer. Mol. Oncol. 2021;15:1234–1255. doi: 10.1002/1878-0261.12911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun M., Nie F., Wang Y., Zhang Z., Hou J., He D., Xie M., Xu L., De W., Wang Z., et al. LncRNA HOXA11-AS promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299–6310. doi: 10.1158/0008-5472.CAN-16-0356. [DOI] [PubMed] [Google Scholar]
- 43.Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B.R., Protacio A., Flynn R.A., Gupta R.A., et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toki N., Takahashi H., Zucchelli S., Gustincich S., Carninci P. Synthetic in vitro transcribed lncRNAs (SINEUPs) with chemical modifications enhance target mRNA translation. FEBS Lett. 2020;594:4357–4369. doi: 10.1002/1873-3468.13928. [DOI] [PubMed] [Google Scholar]
- 45.Zucchelli S., Cotella D., Takahashi H., Carrieri C., Cimatti L., Fasolo F., Jones M.H., Sblattero D., Sanges R., Santoro C., et al. SINEUPs: a new class of natural and synthetic antisense long non-coding RNAs that activate translation. RNA Biol. 2015;12:771–779. doi: 10.1080/15476286.2015.1060395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carrieri C., Cimatti L., Biagioli M., Beugnet A., Zucchelli S., Fedele S., Pesce E., Ferrer I., Collavin L., Santoro C., et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 47.Toki N., Takahashi H., Sharma H., Valentine M.N.Z., Rahman F.M., Zucchelli S., Gustincich S., Carninci P. SINEUP long non-coding RNA acts via PTBP1 and HNRNPK to promote translational initiation assemblies. Nucleic Acids Res. 2020;48:11626–11644. doi: 10.1093/nar/gkaa814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Indrieri A., Grimaldi C., Zucchelli S., Tammaro R., Gustincich S., Franco B. Synthetic long non-coding RNAs [SINEUPs] rescue defective gene expression in vivo. Sci. Rep. 2016;6:27315. doi: 10.1038/srep27315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J., Xu X., Yin J., Tang J., Hu N., Hong Y., Song Z., Bian B., Wu F. lncRNA OGFRP1 promotes tumor progression by activating the AKT/mTOR pathway in human gastric cancer. Aging. 2021;13:9766–9779. doi: 10.18632/aging.202731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen P., Zhao X., Wang H., Zheng M., Wang Q., Chang W. The down-regulation of lncRNA PCAT18 promotes the progression of gastric cancer via MiR-107/PTEN/PI3K/AKT signaling pathway. Onco Targets Ther. 2019;12:11017–11031. doi: 10.2147/OTT.S225235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cao B., Liu C., Yang G. Down-regulation of lncRNA ADAMTS9-AS2 contributes to gastric cancer development via activation of PI3K/Akt pathway. Biomed. Pharmacother. 2018;107:185–193. doi: 10.1016/j.biopha.2018.06.146. [DOI] [PubMed] [Google Scholar]
- 52.Li C., Liang G., Yang S., Sui J., Wu W., Xu S., Ye Y., Shen B., Zhang X., Zhang Y. LncRNA-LOC101928316 contributes to gastric cancer progression through regulating PI3K-Akt-mTOR signaling pathway. Cancer Med. 2019;8:4428–4440. doi: 10.1002/cam4.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang L.L., Zhang L., Cui X.F. Downregulation of long noncoding RNA LINC01419 inhibits cell migration, invasion, and tumor growth and promotes autophagy via inactivation of the PI3K/Akt1/mTOR pathway in gastric cancer. Ther. Adv. Med. Oncol. 2019;11 doi: 10.1177/1758835919874651. 1758835919874651. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Zeng L., Liao Q., Zou Z., Wen Y., Wang J., Liu C., He Q., Weng N., Zeng J., Tang H., et al. Long non-coding RNA XLOC_006753 promotes the development of multidrug resistance in gastric cancer cells through the PI3K/AKT/mTOR signaling pathway. Cell. Physiol. Biochem. 2018;51:1221–1236. doi: 10.1159/000495499. [DOI] [PubMed] [Google Scholar]
- 55.Li J.F., Li W.H., Xue L.L., Zhang Y. Long non-coding RNA PICART1 inhibits cell proliferation by regulating the PI3K/AKT and MAPK/ERK signaling pathways in gastric cancer. Eur. Rev. Med. Pharmacol. Sci. 2019;23:588–597. doi: 10.26355/eurrev_201901_16871. [DOI] [PubMed] [Google Scholar]
- 56.Hu Y., Zhang Y., Ding M., Xu R. Long noncoding RNA TMPO-AS1/miR-126-5p/BRCC3 axis accelerates gastric cancer progression and angiogenesis via activating PI3K/Akt/mTOR pathway. J. Gastroenterol. Hepatol. 2021;36:1877–1888. doi: 10.1111/jgh.15362. [DOI] [PubMed] [Google Scholar]
- 57.Wang L., Bo X., Yi X., Xiao X., Zheng Q., Ma L., Li B. Exosome-transferred LINC01559 promotes the progression of gastric cancer via PI3K/AKT signaling pathway. Cell Death Dis. 2020;11:723. doi: 10.1038/s41419-020-02810-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Du D.X., Lian D.B., Amin B.H., Yan W. Long non-coding RNA CRNDE is a novel tumor promoter by modulating PI3K/AKT signal pathways in human gastric cancer. Eur. Rev. Med. Pharmacol. Sci. 2017;21:5392–5398. doi: 10.26355/eurrev_201712_13925. [DOI] [PubMed] [Google Scholar]
- 59.Qu C.X., Shi X.C., Bi H., Zhai L.Q., Yang Q. LncRNA AOC4P affects biological behavior of gastric cancer cells through MAPK signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019;23:8852–8860. doi: 10.26355/eurrev_201910_19280. [DOI] [PubMed] [Google Scholar]
- 60.Li D., Yang M., Liao A., Zeng B., Liu D., Yao Y., Hu G., Chen X., Feng Z., Du Y., et al. Linc00483 as ceRNA regulates proliferation and apoptosis through activating MAPKs in gastric cancer. 2018;22:3875–3886. doi: 10.1111/jcmm.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi Y., Sun H. Down-regulation of lncRNA LINC00152 suppresses gastric cancer cell migration and invasion through inhibition of the ERK/MAPK signaling pathway. Onco Targets Ther. 2020;13:2115–2124. doi: 10.2147/OTT.S217452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li P., Xue W.J., Feng Y., Mao Q.S. Long non-coding RNA CASC2 suppresses the proliferation of gastric cancer cells by regulating the MAPK signaling pathway. Am. J. Transl Res. 2016;8:3522–3529. [PMC free article] [PubMed] [Google Scholar]
- 63.Wang H., Wu M., Lu Y., He K., Cai X., Yu X., Lu J., Teng L. LncRNA MIR4435-2HG targets desmoplakin and promotes growth and metastasis of gastric cancer by activating Wnt/β-catenin signaling. Aging. 2019;11:6657–6673. doi: 10.18632/aging.102164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang F., Zhu W., Yang R., Xie W., Wang D. LncRNA ZEB2-AS1 contributes to the tumorigenesis of gastric cancer via activating the Wnt/β-catenin pathway. Mol. Cell. Biochem. 2019;456:73–83. doi: 10.1007/s11010-018-03491-7. [DOI] [PubMed] [Google Scholar]
- 65.Yang X.Z., Cheng T.T., He Q.J., Lei Z.Y., Chi J., Tang Z., Liao Q.X., Zhang H., Zeng L.S., Cui S.Z. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/β-catenin pathway. Mol. Cancer. 2018;17:126. doi: 10.1186/s12943-018-0874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang H., Huang H., Xu X., Wang H., Wang J., Yao Z., Xu X., Wu Q., Xu F. LncRNA HCG11 promotes proliferation and migration in gastric cancer via targeting miR-1276/CTNNB1 and activating Wnt signaling pathway. Cancer Cell Int. 2019;19:350. doi: 10.1186/s12935-019-1046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peng C., Li X., Yu Y., Chen J. LncRNA GASL1 inhibits tumor growth in gastric carcinoma by inactivating the Wnt/β-catenin signaling pathway. Exp. Ther. Med. 2019;17:4039–4045. doi: 10.3892/etm.2019.7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang B., Bai Q., Chen H., Su K., Gao C. LINC00665 induces gastric cancer progression through activating Wnt signaling pathway. J. Cell. Biochem. 2020;121:2268–2276. doi: 10.1002/jcb.29449. [DOI] [PubMed] [Google Scholar]
- 69.Li Z.T., Zhang X., Wang D.W., Xu J., Kou K.J., Wang Z.W., Yong G., Liang D.S., Sun X.Y. Overexpressed lncRNA GATA6-AS1 inhibits LNM and EMT via FZD4 through the Wnt/β-catenin signaling pathway in GC. Mol. Ther. Nucleic Acids. 2020;19:827–840. doi: 10.1016/j.omtn.2019.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 70.Jiang K., Zhi X.H., Ma Y.Y., Zhou L.Q. Long non-coding RNA TOB1-AS1 modulates cell proliferation, apoptosis, migration and invasion through miR-23a/NEU1 axis via Wnt/b-catenin pathway in gastric cancer. Eur. Rev. Med. Pharmacol. Sci. 2019;23:9890–9899. doi: 10.26355/eurrev_201911_19554. [DOI] [PubMed] [Google Scholar]
- 71.Tang L., Wen J.B., Wen P., Li X., Gong M., Li Q. Long non-coding RNA LINC01314 represses cell migration, invasion, and angiogenesis in gastric cancer via the Wnt/beta-catenin signaling pathway by down-regulating KLK4. Cancer Cell Int. 2019;19:94. doi: 10.1186/s12935-019-0799-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu Y., Zhang G., Zou C., Qi W., Gong Z., Zhang G., Ma G., Zhang W. Long non-coding RNA LINC01225 promotes proliferation, invasion and migration of gastric cancer via Wnt/β-catenin signalling pathway. J. Cell. Mol. Med. 2019;23:7581–7591. doi: 10.1111/jcmm.14627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ding J., Shi F., Xie G., Zhu Y. Long non-coding RNA LINC01503 promotes gastric cancer cell proliferation and invasion by regulating Wnt signaling. Dig. Dis. Sci. 2020;66:452–459. doi: 10.1007/s10620-020-06215-4. [DOI] [PubMed] [Google Scholar]
- 74.Chen L., Yuan D., Yang Y., Ren M. LincRNA-p21 enhances the sensitivity of radiotherapy for gastric cancer by targeting the β-catenin signaling pathway. J. Cell. Biochem. 2019;120:6178–6187. doi: 10.1002/jcb.27905. [DOI] [PubMed] [Google Scholar]
- 75.Wang B., Guan G., Zhao D. Silence of FAM83H-AS1 promotes chemosensitivity of gastric cancer through Wnt/β-catenin signaling pathway. Biomed. Pharmacother. 2020;125:109961. doi: 10.1016/j.biopha.2020.109961. [DOI] [PubMed] [Google Scholar]
- 76.Zhao J., Du P., Cui P., Qin Y., Hu C., Wu J., Zhou Z., Zhang W., Qin L., Huang G. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene. 2018;37:4094–4109. doi: 10.1038/s41388-018-0250-z. [DOI] [PubMed] [Google Scholar]
- 77.Jiang D., Li H., Xiang H., Gao M., Yin C., Wang H., Sun Y., Xiong M. Long chain non-coding RNA (lncRNA) HOTAIR knockdown increases miR-454-3p to suppress gastric cancer growth by targeting STAT3/cyclin D1. Med. Sci. Monit. 2019;25:1537–1548. doi: 10.12659/MSM.913087. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 78.Wang X., Kan J., Han J., Zhang W., Bai L., Wu H. LncRNA SNHG16 functions as an oncogene by sponging MiR-135a and promotes JAK2/STAT3 signal pathway in gastric cancer. J. Cancer. 2019;10:1013–1022. doi: 10.7150/jca.29527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tan H.Y., Wang C. Long noncoding RNA NEAT1-modulated miR-506 regulates gastric cancer development through targeting STAT3. 2019;120:4827–4836. doi: 10.1002/jcb.26691. [DOI] [PubMed] [Google Scholar]
- 80.Zheng L., Chen J., Zhou Z., He Z. Knockdown of long non-coding RNA HOXD-AS1 inhibits gastric cancer cell growth via inactivating the JAK2/STAT3 pathway. Tumour Biol. 2017;39 doi: 10.1177/1010428317705335. 1010428317705335. [DOI] [PubMed] [Google Scholar]
- 81.Deng W., Zhang Y., Cai J., Zhang J., Liu X. LncRNA-ANRIL promotes gastric cancer progression by enhancing NF-kB signaling. Exp. Bio Med. 2019;244:953–959. doi: 10.1177/1535370219860207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang J.X., Chen Z.H., Chen D.L., Tian X.P., Wang C.Y., Zhou Z.W., Gao Y., Xu Y., Chen C., Zheng Z.S., et al. LINC01410-miR-532-NCF2-NF-kB feedback loop promotes gastric cancer angiogenesis and metastasis. Oncogene. 2018;37:2660–2675. doi: 10.1038/s41388-018-0162-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 83.Zheng J., Zhang H., Ma R., Liu H., Gao P. Long non-coding RNA KRT19P3 suppresses proliferation and metastasis through COPS7A-mediated NF-κB pathway in gastric cancer. Oncogene. 2019;38:7073–7088. doi: 10.1038/s41388-019-0934-z. [DOI] [PubMed] [Google Scholar]
- 84.Zhang Z.X., Liu Z.Q., Jiang B., Lu X.Y., Ning X.F., Yuan C.T., Wang A.L. BRAF activated non-coding RNA (BANCR) promoting gastric cancer cells proliferation via regulation of NF-κB1. Biochem. Biophys. Res. Commun. 2015;465:225–231. doi: 10.1016/j.bbrc.2015.07.158. [DOI] [PubMed] [Google Scholar]
- 85.Fu T., Ji K., Jin L., Zhang J., Wu X., Ji X., Fan B., Jia Z., Wang A., Liu J., et al. ASB16-AS1 up-regulated and phosphorylated TRIM37 to activate NF-κB pathway and promote proliferation, stemness, and cisplatin resistance of gastric cancer. Gastric Cancer. 2020;24:45–59. doi: 10.1007/s10120-020-01096-y. [DOI] [PubMed] [Google Scholar]
- 86.Peng W., Wu J., Fan H., Lu J., Feng J. LncRNA EGOT promotes tumorigenesis via Hedgehog pathway in gastric cancer. Pathol. Oncol. Res. 2019;25:883–887. doi: 10.1007/s12253-017-0367-3. [DOI] [PubMed] [Google Scholar]
- 87.Piao H.Y., Guo S., Wang Y., Zhang J. Long noncoding RNA NALT1-induced gastric cancer invasion and metastasis via NOTCH signaling pathway. World J. Gastroenterol. 2019;25:6508–6526. doi: 10.3748/wjg.v25.i44.6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li H., Wang Y. Long noncoding RNA (lncRNA) MIR22HG suppresses gastric cancer progression through attenuating NOTCH2 signaling. Med. Sci. Monit. 2019;25:656–665. doi: 10.12659/MSM.912813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xiao J., Lai H., Wei S.H., Ye Z.S., Gong F.S., Chen L.C. lncRNA HOTAIR promotes gastric cancer proliferation and metastasis via targeting miR-126 to active CXCR4 and RhoA signaling pathway. Cancer Med. 2019;8:6768–6779. doi: 10.1002/cam4.1302. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 90.Zong W., Feng W., Jiang Y., Cao Y., Ke Y., Shi X., Ju S., Cong H., Wang X., Cui M. LncRNA CTC-497E21.4 promotes the progression of gastric cancer via modulating miR-22/NET1 axis through RhoA signaling pathway. Gastric Cancer. 2020;23:228–240. doi: 10.1007/s10120-019-00998-w. [DOI] [PubMed] [Google Scholar]
- 91.Yu S.Y., Peng H., Zhu Q., Wu Y.X., Wu F., Han C.R., Yan B., Li Q., Xiang H.G. Silencing the long noncoding RNA NORAD inhibits gastric cancer cell proliferation and invasion by the RhoA/ROCK1 pathway. Eur. Rev. Med. Pharmacol. Sci. 2019;23:3760–3770. doi: 10.26355/eurrev_201905_17802. [DOI] [PubMed] [Google Scholar]
- 92.Dai Q., Zhang T., Li C. LncRNA MALAT1 regulates the cell proliferation and cisplatin resistance in gastric cancer via PI3K/AKT pathway. Cancer Manag. Res. 2020;12:1929–1939. doi: 10.2147/CMAR.S243796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ma F., An K., Li Y. Silencing of long non-coding RNA-HCG18 inhibits the tumorigenesis of gastric cancer through blocking PI3K/Akt pathway. Onco Targets Ther. 2020;13:2225–2234. doi: 10.2147/OTT.S240965. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 94.Huang Y., Zhang J., Hou L., Wang G., Liu H., Zhang R., Chen X., Zhu J. LncRNA AK023391 promotes tumorigenesis and invasion of gastric cancer through activation of the PI3K/Akt signaling pathway. J. Exp. Clin. Cancer Res. 2017;36:194. doi: 10.1186/s13046-017-0666-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Guo Y.J., Pan W.W., Liu S.B., Shen Z.F., Xu Y., Hu L.L. ERK/MAPK signalling pathway and tumorigenesis. Exp. Ther. Med. 2020;19:1997–2007. doi: 10.3892/etm.2020.8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang M., Huang C.Z. Mitogen-activated protein kinase signaling pathway and invasion and metastasis of gastric cancer. World J. Gastroenterol. 2015;21:11673–11679. doi: 10.3748/wjg.v21.i41.11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sun Y.Y., Zhang H., Ma R.R., Zhang G.H., Tian Y.R., Liu L., Liu L., Gao P. Long non-coding RNA AK025387 promotes cell migration and invasion of gastric cancer. Front. Oncol. 2020;10:633. doi: 10.3389/fonc.2020.00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chiurillo M.A. Role of the Wnt/beta-catenin pathway in gastric cancer: an in-depth literature review. World J. Exp. Med. 2015;5:84–102. doi: 10.5493/wjem.v5.i2.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Guan B., Ma J., Yang Z., Yu F., Yao J. LncRNA NCK1-AS1 exerts oncogenic property in gastric cancer by targeting the miR-22-3p/BCL9 axis to activate the Wnt/beta-catenin signaling. Environ. Toxicol. 2021;36:1640–1653. doi: 10.1002/tox.23160. [DOI] [PubMed] [Google Scholar]
- 100.Zhou C., An N., Cao C., Wang G. lncRNA HOXC-AS1 promotes gastric cancer via binding eIF4AIII by activating Wnt/β-catenin signaling. J. Gene Med. 2020;22:e3202. doi: 10.1002/jgm.3202. [DOI] [PubMed] [Google Scholar]
- 101.Ashrafizadeh M., Zarrabi A. STAT3 pathway in gastric cancer: signaling. Ther. Target. Future Prospects. 2020;9:126. doi: 10.3390/biology9060126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ba M.C., Ba Z., Long H., Cui S.Z., Gong Y.F., Yan Z.F., Lin K.P., Wu Y.B., Tu Y.N. LncRNA AC093818.1 accelerates gastric cancer metastasis by epigenetically promoting PDK1 expression. Cell Death Dis. 2020;11:64. doi: 10.1038/s41419-020-2245-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhou R., Wu Z., Deng X., Chen H. The long non-coding RNA OLC8 enhances gastric cancer by interaction with IL-11. J. Clin. Lab. Anal. 2019;33:e22962. doi: 10.1002/jcla.22962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cui N., Sun Q., Liu H., Li L., Guo X., Shi Y., Jing C., Qin C., Zhao Y. Long non-coding RNA LINC00511 regulates the expression of microRNA-625-5p and activates signal transducers and activators of transcription 3 (STAT3) to accelerate the progression of gastric cancer. Bioengineered. 2021;12:2915–2927. doi: 10.1080/21655979.2021.1940611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang X., Jiang Y., Xie Y., Leng X., He M., Song F. Inhibition of gastric cancer cell apoptosis by long noncoding RNA TRPM2-AS via mitogen-activated protein kinase and activators of transduction-3. J. Gastroenterol. Hepatol. 2021;36:186–195. doi: 10.1111/jgh.15108. [DOI] [PubMed] [Google Scholar]
- 106.Kong F., Deng X., Kong X., Du Y., Li L., Zhu H., Wang Y., Xie D., Guha S., Li Z., et al. ZFPM2-AS1, a novel lncRNA, attenuates the p53 pathway and promotes gastric carcinogenesis by stabilizing MIF. Oncogene. 2018;37:5982–5996. doi: 10.1038/s41388-018-0387-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu Z.H., Liu C.C., Zhou Y.Q., Hu L.N., Guo W.J. OnclncRNA-626 promotes malignancy of gastric cancer via inactivated the p53 pathway through interacting with SRSF1. Am. J. Cancer Res. 2019;9:2249–2263. [PMC free article] [PubMed] [Google Scholar]
- 108.Feng L., Li J., Li F., Li H., Bei S., Zhang X., Yang Z. Long noncoding RNA VCAN-AS1 contributes to the progression of gastric cancer via regulating p53 expression. J. Cell. Physiol. 2020;235:4388–4398. doi: 10.1002/jcp.29315. [DOI] [PubMed] [Google Scholar]
- 109.Wei G.H., Wang X. lncRNA MEG3 inhibit proliferation and metastasis of gastric cancer via p53 signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2017;21:3850–3856. [PubMed] [Google Scholar]
- 110.Zhang Y., Na R., Wang X. LncRNA WT1-AS up-regulates p53 to inhibit the proliferation of cervical squamous carcinoma cells. BMC Cancer. 2019;19:1052. doi: 10.1186/s12885-019-6264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lu H., Shi C., Liu X., Liang C., Yang C., Wan X., Li L., Liu Y. Identification of ZG16B as a prognostic biomarker in breast cancer. Open Med. (Wars) 2021;16:1–13. doi: 10.1515/med-2021-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xue M., Liang H., Ji X., Zhou Z., Liu Y., Sun T., Zhang L. Effects of fucoidan on gut flora and tumor prevention in 1,2-dimethylhydrazine-induced colorectal carcinogenesis. J. Nutr. Biochem. 2020;82:108396. doi: 10.1016/j.jnutbio.2020.108396. [DOI] [PubMed] [Google Scholar]
- 113.Wang M.W., Liu J., Liu Q., Xu Q.H., Li T.F., Jin S., Xia T.S. LncRNA SNHG7 promotes the proliferation and inhibits apoptosis of gastric cancer cells by repressing the P15 and P16 expression. Eur. Rev. Med. Pharmacol. Sci. 2017;21:4613–4622. [PubMed] [Google Scholar]
- 114.Xu G., Zhang Y., Li N., Zhang J.B., Xu R. LncRNA CCHE1 in the proliferation and apoptosis of gastric cancer cells. Eur. Rev. Med. Pharmacol. Sci. 2018;22:2631–2637. doi: 10.26355/eurrev_201805_14957. [DOI] [PubMed] [Google Scholar]
- 115.Li L., Li Y., Huang Y., Ouyang Y., Zhu Y., Wang Y., Guo X., Yuan Y., Gong K. Long non-coding RNA MIF-AS1 promotes gastric cancer cell proliferation and reduces apoptosis to upregulate NDUFA4. Cancer Sci. 2018;109:3714–3725. doi: 10.1111/cas.13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu Y., Ao X., Wang Q., Wang J., Ge H. PiViewer: an open-source tool for automated detection and display of pi-pi interactions. Chem. Biol. Drug Des. 2018;92:1809–1814. doi: 10.1111/cbdd.13340. [DOI] [PubMed] [Google Scholar]
- 117.Zhang G., Li S., Lu J., Ge Y., Wang Q., Ma G., Zhao Q., Wu D., Gong W., Du M., et al. LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol. Cancer. 2018;17:87. doi: 10.1186/s12943-018-0829-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dong X., Chen R., Lin H., Lin T., Pan S. lncRNA BG981369 inhibits cell proliferation, migration, and invasion, and promotes cell apoptosis by SRY-related high-mobility group box 4 (SOX4) signaling pathway in human gastric cancer. Med. Sci. Monit. 2018;24:718–726. doi: 10.12659/MSM.905965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Guo J.Q., Li S.J., Guo G.X. Long noncoding RNA AFAP1-AS1 promotes cell proliferation and apoptosis of gastric cancer cells via PTEN/p-AKT pathway. Dig. Dis. Sci. 2017;62:2004–2010. doi: 10.1007/s10620-017-4584-0. [DOI] [PubMed] [Google Scholar]
- 120.Sun W., Jiang C., Ji Y., Xiao C., Song H. Long noncoding RNAs: new regulators of resistance to systemic therapies for gastric cancer. Biomed. Res. Int. 2021;2021:8853269. doi: 10.1155/2021/8853269. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 121.Zhang J., Piao H.Y. LINC00163 inhibits the invasion and metastasis of gastric cancer cells as a ceRNA by sponging miR-183 to regulate the expression of AKAP12. 2020;25:570–583. doi: 10.1007/s10147-019-01604-w. [DOI] [PubMed] [Google Scholar]
- 122.Zhang T., Piao H.Y., Guo S., Zhao Y., Wang Y., Zheng Z.C., Zhang J. LncRNA PCGEM1 enhances metastasis and gastric cancer invasion through targeting of miR-129-5p to regulate P4HA2 expression. Exp. Mol. Pathol. 2020;116:104487. doi: 10.1016/j.yexmp.2020.104487. [DOI] [PubMed] [Google Scholar]
- 123.Lin Z., Zhou Z., Guo H., He Y., Pang X., Zhang X., Liu Y., Ao X., Li P., Wang J. Long noncoding RNA gastric cancer-related lncRNA1 mediates gastric malignancy through miRNA-885-3p and cyclin-dependent kinase 4. Cell Death Dis. 2018;9:607. doi: 10.1038/s41419-018-0643-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bure I.V., Nemtsova M.V., Zaletaev D.V. Roles of E-cadherin and noncoding RNAs in the epithelial-mesenchymal transition and progression in gastric cancer. Int. J. Mol. Sci. 2019;20:2870. doi: 10.3390/ijms20122870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang J., Jin H.Y. Hypoxia-induced LncRNA PCGEM1 promotes invasion and metastasis of gastric cancer through regulating SNAI1. Clin. Transl.Oncol. 2019;21:1142–1151. doi: 10.1007/s12094-019-02035-9. [DOI] [PubMed] [Google Scholar]
- 126.Chen D., Liu L., Wang K., Yu H., Wang Y., Liu J., Guo Y., Zhang H. The role of MALAT-1 in the invasion and metastasis of gastric cancer. Scand. J. Gastroenterol. 2017;52:790–796. doi: 10.1080/00365521.2017.1280531. [DOI] [PubMed] [Google Scholar]
- 127.Liu Z., Chen Z., Fan R., Jiang B., Chen X., Chen Q., Nie F., Lu K., Sun M. Over-expressed long noncoding RNA HOXA11-AS promotes cell cycle progression and metastasis in gastric cancer. Mol. Cancer. 2017;16:82. doi: 10.1186/s12943-017-0651-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang L., Kang W., Lu X., Ma S., Dong L., Zou B. LncRNA CASC11 promoted gastric cancer cell proliferation, migration and invasion in vitro by regulating cell cycle pathway. Cell Cycle. 2018;17:1886–1900. doi: 10.1080/15384101.2018.1502574. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 129.Gao H., Yin Y., Qian A., Guo R., Qi J. LncRNA LINC00974 upregulates CDK6 to promote cell cycle progression in gastric carcinoma. Cancer Biother. Radiopharm. 2019;34:666–670. doi: 10.1089/cbr.2019.2904. [DOI] [PubMed] [Google Scholar]
- 130.Chen B., Zhao Q., Guan L., Lv H., Bie L., Huang J., Chen X.B. Long non-coding RNA NNT-AS1 sponges miR-424/E2F1 to promote the tumorigenesis and cell cycle progression of gastric cancer. J. Cell. Mol. Med. 2018;22:4751–4759. doi: 10.1111/jcmm.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ao X., Ding W., Ge H., Zhang Y., Ding D., Liu Y. PBX1 is a valuable prognostic biomarker for patients with breast cancer. Exp. Ther. Med. 2020;20:385–394. doi: 10.3892/etm.2020.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Prendergast G.C. Immune escape as a fundamental trait of cancer: focus on IDO. Oncogene. 2008;27:3889–3900. doi: 10.1038/onc.2008.35. [DOI] [PubMed] [Google Scholar]
- 133.Wang C.J., Zhu C.C., Xu J., Wang M., Zhao W.Y., Liu Q., Zhao G., Zhang Z.Z. The lncRNA UCA1 promotes proliferation, migration, immune escape and inhibits apoptosis in gastric cancer by sponging anti-tumor miRNAs. Mol. Cancer. 2019;18:115. doi: 10.1186/s12943-019-1032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sun L., Li J., Yan W., Yao Z., Wang R., Zhou X., Wu H., Zhang G., Shi T., Chen W. H19 promotes aerobic glycolysis, proliferation, and immune escape of gastric cancer cells through the microRNA-519d-3p/lactate dehydrogenase A axis. Cancer Sci. 2021;112:2245–2259. doi: 10.1111/cas.14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dang S., Malik A., Chen J., Qu J., Yin K., Cui L., Gu M. LncRNA SNHG15 contributes to immuno-escape of gastric cancer through targeting miR141/PD-L1. Onco Targets Ther. 2020;13:8547–8556. doi: 10.2147/OTT.S251625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Li H., Zhao C., Zhao H., Liu G., Mao H., Liu Y. Elevated linc00936 or silenced microRNA-425-3p inhibits immune escape of gastric cancer cells via elevation of ZC3H12A. Int. Immunopharmacol. 2021;95:107559. doi: 10.1016/j.intimp.2021.107559. [DOI] [PubMed] [Google Scholar]
- 137.Ao X., Ding W., Zhang Y., Ding D., Liu Y. TCF21: a critical transcription factor in health and cancer. J. Mol. Med. 2020;98:1055–1068. doi: 10.1007/s00109-020-01934-7. [DOI] [PubMed] [Google Scholar]
- 138.Li Y., Wu Z., Yuan J., Sun L., Lin L., Huang N., Bin J., Liao Y., Liao W. Long non-coding RNA MALAT1 promotes gastric cancer tumorigenicity and metastasis by regulating vasculogenic mimicry and angiogenesis. Cancer Lett. 2017;395:31–44. doi: 10.1016/j.canlet.2017.02.035. [DOI] [PubMed] [Google Scholar]
- 139.Zhou J., Huang H., Tong S., Huo R. Overexpression of long non-coding RNA cancer susceptibility 2 inhibits cell invasion and angiogenesis in gastric cancer. Mol. Med. Rep. 2017;16:5235–5240. doi: 10.3892/mmr.2017.7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liu Y. Targeting the non-canonical AKT-FOXO3a axis: a potential therapeutic strategy for oral squamous cell carcinoma. EBioMedicine. 2019;49:6–8. doi: 10.1016/j.ebiom.2019.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Xiao Y., Pan J., Geng Q., Wang G. LncRNA MALAT1 increases the stemness of gastric cancer cells via enhancing SOX2 mRNA stability. FEBS Open Bio. 2019;9:1212–1222. doi: 10.1002/2211-5463.12649. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 142.Song H., Xu Y., Shi L., Xu T., Fan R., Cao M., Xu W., Song J. LncRNA THOR increases the stemness of gastric cancer cells via enhancing SOX9 mRNA stability. Biomed. Pharmacother. 2018;108:338–346. doi: 10.1016/j.biopha.2018.09.057. [DOI] [PubMed] [Google Scholar]
- 143.Hui Y., Yang Y., Li D. LncRNA FEZF1-AS1 modulates cancer stem cell properties of human gastric cancer through miR-363-3p/HMGA2. Cell Transpl. 2020;29 doi: 10.1177/0963689720925059. 963689720925059. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 144.Wang S., Liu F., Deng J., Cai X., Han J., Liu Q. Long noncoding RNA ROR regulates proliferation, invasion, and stemness of gastric cancer stem cell. Cell. Reprogram. 2016;18:319–326. doi: 10.1089/cell.2016.0001. [DOI] [PubMed] [Google Scholar]
- 145.Wu H., Liu B., Chen Z., Li G., Zhang Z. MSC-induced lncRNA HCP5 drove fatty acid oxidation through miR-3619-5p/AMPK/PGC1alpha/CEBPB axis to promote stemness and chemo-resistance of gastric cancer. Cell Death Dis. 2020;11:233. doi: 10.1038/s41419-020-2426-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang F., Tang C., Xu D., Tang Y., Jiang Y., Gao X., Xu J. LncRNA ADAMTS9-AS2 suppresses the proliferation of gastric cancer cells and the tumorigenicity of cancer stem cells through regulating SPOP. J. Cell. Mol. Med. 2020;24:4830–4838. doi: 10.1111/jcmm.15161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.He W., Liang B., Wang C., Li S., Zhao Y., Huang Q., Liu Z. MSC-regulated lncRNA MACC1-AS1 promotes stemness and chemoresistance through fatty acid oxidation in gastric cancer. Oncogene. 2019;38:4637–4654. doi: 10.1038/s41388-019-0747-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hong L., Wang H., Wang J., Wei S., Zhang F., Han J., Liu Y., Ma M., Liu C., Xu Y., et al. LncRNA PTCSC3 inhibits tumor growth and cancer cell stemness in gastric cancer by interacting with lncRNA linc-pint. Cancer Manag. Res. 2019;11:10393–10399. doi: 10.2147/CMAR.S231369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sun Q., Li J., Li F., Li H., Bei S., Zhang X., Feng L. LncRNA LOXL1-AS1 facilitates the tumorigenesis and stemness of gastric carcinoma via regulation of miR-708-5p/USF1 pathway. Cell Prolif. 2019;52:e12687. doi: 10.1111/cpr.12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sun B., Han Y., Cai H., Huang H., Xuan Y. Long non-coding RNA SNHG3, induced by IL-6/STAT3 transactivation, promotes stem cell-like properties of gastric cancer cells by regulating the miR-3619-5p/ARL2 axis. Cell. Oncol. 2020;44:179–192. doi: 10.1007/s13402-020-00560-2. [DOI] [PubMed] [Google Scholar]
- 151.Fattahi S., Kosari-Monfared M., Golpour M., Emami Z., Ghasemiyan M., Nouri M., Akhavan-Niaki H. LncRNAs as potential diagnostic and prognostic biomarkers in gastric cancer: a novel approach to personalized medicine. J. Cell. Physiol. 2020;235:3189–3206. doi: 10.1002/jcp.29260. [DOI] [PubMed] [Google Scholar]
- 152.Miao X., Liu Y., Fan Y., Wang G., Zhu H. LncRNA BANCR attenuates the killing capacity of cisplatin on gastric cancer cell through the ERK1/2 pathway. Cancer Manag. Res. 2021;13:287–296. doi: 10.2147/CMAR.S269679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhu L., Zhu Y., Han S., Chen M., Song P., Dai D., Xu W., Jiang T., Feng L., Shin V.Y., et al. Impaired autophagic degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in gastric cancer. Cell Death Dis. 2019;10:383. doi: 10.1038/s41419-019-1585-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Li M., Zhang Y.Y., Shang J., Xu Y.D. LncRNA SNHG5 promotes cisplatin resistance in gastric cancer via inhibiting cell apoptosis. Eur. Rev. Med. Pharmacol. Sci. 2019;23:4185–4191. doi: 10.26355/eurrev_201905_17921. [DOI] [PubMed] [Google Scholar]
- 155.Zhou Z., Lin Z., He Y., Pang X., Wang Y., Ponnusamy M., Ao X., Shan P., Tariq M.A., Li P., et al. The long noncoding RNA D63785 regulates chemotherapy sensitivity in human gastric cancer by targeting miR-422a. Mol. Ther. Nucleic Acids. 2018;12:405–419. doi: 10.1016/j.omtn.2018.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dastmalchi N., Khojasteh S.M.B., Nargesi M.M., Safaralizadeh R. The correlation between lncRNAs and Helicobacter pylori in gastric cancer. Pathog. Dis. 2019;77:ftaa004. doi: 10.1093/femspd/ftaa004. [DOI] [PubMed] [Google Scholar]
- 157.Meng W., Bai B., Sheng L., Li Y., Yue P., Li X., Qiao L. Role of Helicobacter pylori in gastric cancer: advances and controversies. Discov. Med. 2015;20:285–293. [PubMed] [Google Scholar]
- 158.Yang L., Long Y., Li C., Cao L., Gan H., Huang K., Jia Y. Genome-wide analysis of long noncoding RNA profile in human gastric epithelial cell response to Helicobacter pylori. Jpn. J. Infect. Dis. 2015;68:63–66. doi: 10.7883/yoken.JJID.2014.149. [DOI] [PubMed] [Google Scholar]
- 159.Zhong F., Zhu M., Gao K., Xu P., Yang H., Hu D., Cui D., Wang M., Xie X., Wei Y., et al. Low expression of the long non-coding RNA NR_026827 in gastric cancer. Am. J. Transl. Res. 2018;10:2706–2711. [PMC free article] [PubMed] [Google Scholar]
- 160.Rajabi A., Riahi A., Shirabadi-Arani H., Moaddab Y., Haghi M., Safaralizadeh R. Overexpression of HOXA-AS2 LncRNA in patients with gastric cancer and its association with Helicobacter pylori infection. J. Gastrointest. Cancer. 2020 doi: 10.1007/s12029-020-00549-y. [DOI] [PubMed] [Google Scholar]
- 161.Zhang Y., Yan J., Li C., Wang X., Dong Y., Shen X., Zhang X. LncRNA H19 induced by Helicobacter pylori infection promotes gastric cancer cell growth via enhancing NF-kappaB-induced inflammation. J. Inflamm. 2019;16:23. doi: 10.1186/s12950-019-0226-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jia W., Zhang J., Ma F., Hao S., Li X., Guo R., Gao Q., Sun Y., Jia J., Li W. Long noncoding RNA THAP9-AS1 is induced by Helicobacter pylori and promotes cell growth and migration of gastric cancer. Onco Targets Ther. 2019;12:6653–6663. doi: 10.2147/OTT.S201832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Shimada H., Noie T., Ohashi M., Oba K., Takahashi Y. Clinical significance of serum tumor markers for gastric cancer: a systematic review of literature by the Task Force of the Japanese Gastric Cancer Association. Gastric Cancer. 2014;17:26–33. doi: 10.1007/s10120-013-0259-5. [DOI] [PubMed] [Google Scholar]
- 164.Ghafouri-Fard S., Taheri M. Long non-coding RNA signature in gastric cancer. Exp. Mol. Pathol. 2020;113:104365. doi: 10.1016/j.yexmp.2019.104365. [DOI] [PubMed] [Google Scholar]
- 165.Yuan L., Xu Z.Y., Ruan S.M., Mo S., Qin J.J., Cheng X.D. Long non-coding RNAs towards precision medicine in gastric cancer: early diagnosis, treatment, and drug resistance. Mol. Cancer. 2020;19:96. doi: 10.1186/s12943-020-01219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Elsayed E.T., Salem P.E., Darwish A.M., Fayed H.M. Plasma long non-coding RNA HOTAIR as a potential biomarker for gastric cancer. Int. J. Biol. Markers. 2018 doi: 10.1177/1724600818760244. [DOI] [PubMed] [Google Scholar]
- 167.Zhou X., Yin C., Dang Y., Ye F., Zhang G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci. Rep. 2015;5:11516. doi: 10.1038/srep11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Feng W., Zong W., Li Y., Shen X., Cui X., Ju S. Abnormally expressed long noncoding RNA B3GALT5-AS1 may serve as a biomarker for the diagnostic and prognostic of gastric cancer. 2020;121:557–565. doi: 10.1002/jcb.29296. [DOI] [PubMed] [Google Scholar]
- 169.Wang J., Liu Y., Sun W., Zhang Q., Gu T., Li G. Plasma exosomes as novel biomarker for the early diagnosis of gastric cancer. Cancer Biomark. 2018;21:805–812. doi: 10.3233/CBM-170738. [DOI] [PubMed] [Google Scholar]
- 170.Lin L.Y., Yang L., Zeng Q., Wang L., Chen M.L., Zhao Z.H., Ye G.D., Luo Q.C., Lv P.Y., Guo Q.W., et al. Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol. Cancer. 2018;17:84. doi: 10.1186/s12943-018-0834-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Li S., Zhang M., Zhang H., Hu K., Cai C., Wang J., Shi L., Ma P., Xu Y., Zheng P. Exosomal long noncoding RNA lnc-GNAQ-6:1 may serve as a diagnostic marker for gastric cancer. Clin. Chim. Acta. 2020;501:252–257. doi: 10.1016/j.cca.2019.10.047. [DOI] [PubMed] [Google Scholar]
- 172.Cai C., Zhang H., Zhu Y., Zheng P., Xu Y., Sun J., Zhang M., Lan T., Gu B., Li S., et al. Serum exosomal long noncoding RNA pcsk2-2:1 as a potential novel diagnostic biomarker for gastric cancer. Onco Targets Ther. 2019;12:10035–10041. doi: 10.2147/OTT.S229033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ke D., Li H., Zhang Y., An Y., Fu H., Fang X., Zheng X. The combination of circulating long noncoding RNAs AK001058, INHBA-AS1, MIR4435-2HG, and CEBPA-AS1 fragments in plasma serve as diagnostic markers for gastric cancer. Oncotarget. 2017;8:21516–21525. doi: 10.18632/oncotarget.15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yang Z., Sun Y., Liu R., Shi Y., Ding S. Plasma long noncoding RNAs PANDAR, FOXD2-AS1, and SMARCC2 as potential novel diagnostic biomarkers for gastric cancer. Cancer Manag. Res. 2019;11:6175–6184. doi: 10.2147/CMAR.S201935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Liu W., Li Y., Zhang Y., Shen X., Su Z., Chen L., Cai W., Wang F., Ju S. Circulating long non-coding RNA FEZF1-AS1 and AFAP1-AS1 serve as potential diagnostic biomarkers for gastric cancer. Pathol. Res. Pract. 2020;216:152757. doi: 10.1016/j.prp.2019.152757. [DOI] [PubMed] [Google Scholar]
- 176.Liu X., Wang Q., Song S., Feng M., Wang X., Li L., Liu Y., Shi C. Epithelial splicing regulatory protein 1 is overexpressed in breast cancer and predicts poor prognosis for breast cancer patients. Med. Sci. Monit. 2021;27:e931102. doi: 10.12659/MSM.931102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Liu X., Liu Y., Wang Q., Song S., Feng L., Shi C. The alterations and potential roles of MCMs in breast cancer. J. Oncol. 2021;2021:7928937. doi: 10.1155/2021/7928937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhang M.H., Yang Y., Zhao Y., Wei H.B., Ma Y.Q., Yang C.J., Zhang X.J., Sun Y.L. LncRNA DQ786243 expression as a biomarker for assessing prognosis in patients with gastric cancer. Eur. Rev. Med. Pharmacol. Sci. 2018;22:2304–2309. doi: 10.26355/eurrev_201804_14819. [DOI] [PubMed] [Google Scholar]
- 179.Shi X.Y., Sun Y.Z., Li M., Li H.Y. LncRNA CADM1-AS1 serves as a new prognostic biomarker for gastric cancer. Eur. Rev. Med. Pharmacol. Sci. 2019;23:232–238. doi: 10.26355/eurrev_201908_18652. [DOI] [PubMed] [Google Scholar]
- 180.Zhu X., Tian X., Yu C., Shen C., Yan T., Hong J., Wang Z., Fang J.Y., Chen H. A long non-coding RNA signature to improve prognosis prediction of gastric cancer. Mol. Cancer. 2016;15:60. doi: 10.1186/s12943-016-0544-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Cheng C., Wang Q., Zhu M., Liu K., Zhang Z. Integrated analysis reveals potential long non-coding RNA biomarkers and their potential biological functions for disease free survival in gastric cancer patients. Cancer Cell Int. 2019;19:123. doi: 10.1186/s12935-019-0846-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Song P., Jiang B., Liu Z., Ding J., Liu S., Guan W. A three-lncRNA expression signature associated with the prognosis of gastric cancer patients. Cancer Med. 2017;6:1154–1164. doi: 10.1002/cam4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wang S., Chen W., Yu H., Song Z., Li Q., Shen X., Wu Y., Zhu L., Ma Q., Xing D. lncRNA ROR promotes gastric cancer drug resistance. Cancer Control. 2020;27 doi: 10.1177/1073274820904694. 1073274820904694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Li Y., Song S., Pizzi M.P., Han G., Scott A.W., Jin J., Xu Y. LncRNA PVT1 is a poor prognosticator and can be targeted by PVT1 antisense oligos in gastric adenocarcinoma. 2020;12:2995. doi: 10.3390/cancers12102995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhuo W., Liu Y., Li S., Guo D., Sun Q., Jin J., Rao X., Li M., Sun M., Jiang M., et al. Long noncoding RNA GMAN, up-regulated in gastric cancer tissues, is associated with metastasis in patients and promotes translation of ephrin A1 by competitively binding GMAN-AS. Gastroenterology. 2019;156:676–691.e11. doi: 10.1053/j.gastro.2018.10.054. [DOI] [PubMed] [Google Scholar]
- 186.Han J., Liu S., Sun Z., Zhang Y., Zhang F., Zhang C., Shang D., Yang H., Su F., Xu Y., et al. LncRNAs2Pathways: identifying the pathways influenced by a set of lncRNAs of interest based on a global network propagation method. Sci. Rep. 2017;7:46566. doi: 10.1038/srep46566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Jiang Q., Ma R., Wang J., Wu X., Jin S., Peng J., Tan R., Zhang T., Li Y., Wang Y. LncRNA2Function: a comprehensive resource for functional investigation of human lncRNAs based on RNA-seq data. BMC Genomics. 2015;16:S2. doi: 10.1186/1471-2164-16-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lv Z., Sun L., Xu Q., Xing C., Yuan Y. Joint analysis of lncRNA m(6)A methylome and lncRNA/mRNA expression profiles in gastric cancer. Cancer Cell Int. 2020;20:464. doi: 10.1186/s12935-020-01554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Xiao N., Hu Y., Juan L. Comprehensive analysis of differentially expressed lncRNAs in gastric cancer. Front. Cell Dev. Biol. 2020;8:557. doi: 10.3389/fcell.2020.00557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ho S.W.T., Tan P. Dissection of gastric cancer heterogeneity for precision oncology. Cancer Sci. 2019;110:3405–3414. doi: 10.1111/cas.14191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Xu H., Yang X., Huang W., Ma Y., Ke H., Zou L., et al. Single-cell profiling of long noncoding RNAs and their cell lineage commitment roles via RNA-DNA-DNA triplex formation in mammary epithelium. Stem Cells. 2020;38:1594–1611. doi: 10.1002/stem.3274. [DOI] [PubMed] [Google Scholar]
- 192.Zhang M., Hu S., Min M., Ni Y., Lu Z., Sun X., Wu J., Liu B., Ying X., Liu Y. Dissecting transcriptional heterogeneity in primary gastric adenocarcinoma by single cell RNA sequencing. Gut. 2021;70:464–475. doi: 10.1136/gutjnl-2019-320368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zheng L.L., Xiong J.H., Zheng W.J., Wang J.H., Huang Z.L., Chen Z.R., Sun X.Y., Zheng Y.M., Zhou K.R., Li B., et al. ColorCells: a database of expression, classification and functions of lncRNAs in single cells. Brief. Bioinform. 2021;22:bbaa325. doi: 10.1093/bib/bbaa325. [DOI] [PubMed] [Google Scholar]
- 194.Sathe A., Grimes S.M., Lau B.T., Chen J., Suarez C., Huang R.J., Poultsides G., Ji H.P. Single-cell genomic characterization reveals the cellular reprogramming of the gastric tumor microenvironment. Clin. Cancer Res. 2020;26:2640–2653. doi: 10.1158/1078-0432.CCR-19-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wang B., Zhang Y., Qing T., Xing K., Li J., Zhen T., Zhu S., Zhan X. Comprehensive analysis of metastatic gastric cancer tumour cells using single-cell RNA-seq. Sci. Rep. 2021;11:1141. doi: 10.1038/s41598-020-80881-2. [DOI] [PMC free article] [PubMed] [Google Scholar]