Abstract
Supplemental material is available for this article.
Keywords: Conventional Radiography, Thorax, Trauma, Ribs, Catheters, Segmentation, Diagnosis, Classification, Supervised Learning, Machine Learning
© RSNA, 2021
Keywords: Conventional Radiography, Thorax, Trauma, Ribs, Catheters, Segmentation, Diagnosis, Classification, Supervised Learning, Machine Learning
Summary
This large chest radiograph dataset has segmented annotations for pneumothoraces, acute rib fractures, and intercostal chest tubes. The dataset, which can be used to train and test machine learning algorithms in the identification of these features on chest radiographs, can be found by searching DOI: 10.17608/k6.auckland.14173982.
Key Points
■ The current study outlines the curation of a dataset of 19 237 chest radiographs with segmented annotations for pneumothoraces, acute rib fractures, and intercostal chest tubes. Ground truth labels were assigned for each image after being reviewed by multiple physicians and/or radiologists.
■ Corresponding anonymized, free-text radiology reports were included with the images. The temporal relationship between images of the same patient has been preserved.
■ The dataset includes a 1:5 positive-to-negative pneumothorax ratio.
Introduction
Pneumothoraces, acute rib fractures, and the placement of devices such as intercostal chest tubes are often confirmed by using a chest radiograph. However, increasing demands on imaging departments can result in considerable delays in reporting on chest radiographs (1,2), thereby increasing the risk of patient care being compromised. Artificial intelligence (AI) is an emerging technology with many potential applications for radiology, such as identifying life-threatening features on chest radiographs (3,4), improving reporting quality (5), and triaging concerning images for prompt reporting (6,7). A significant bottleneck in this field of research is in the development of large, well-annotated datasets of images to train and test AI algorithms, as this carries a significant time and labor cost (8). The current study outlines the process for curation of the first large pneumothorax segmentation dataset in a New Zealand population. This dataset could be used by the machine learning community to develop various computer vision deep learning models for use on adult chest radiographs.
Methods
This project has been approved by the University of Otago Human Ethics Committee, which waived individual patient informed consent because of the low-risk nature of the study and the use of anonymized data.
Image Collection and De-Identification
A total of 295 613 chest radiographs from the Dunedin Hospital picture archiving and communication system, in Digital Imaging and Communications in Medicine (DICOM) format, and their radiology reports were retrospectively acquired between January 2010 and April 2020 and were exported onto dedicated research workstations. Inclusion in the final dataset required the images to be frontal chest radiographs (including bedside images) from patients over 16 years of age. Exclusion criteria are listed in Figure 1. The image metadata and reports were de-identified by using algorithm-based tools (9,10), which removed any protected health information to satisfy New Zealand Ministry of Health standards (11). Further details of the anonymization process are provided in Appendix E1 (supplement).
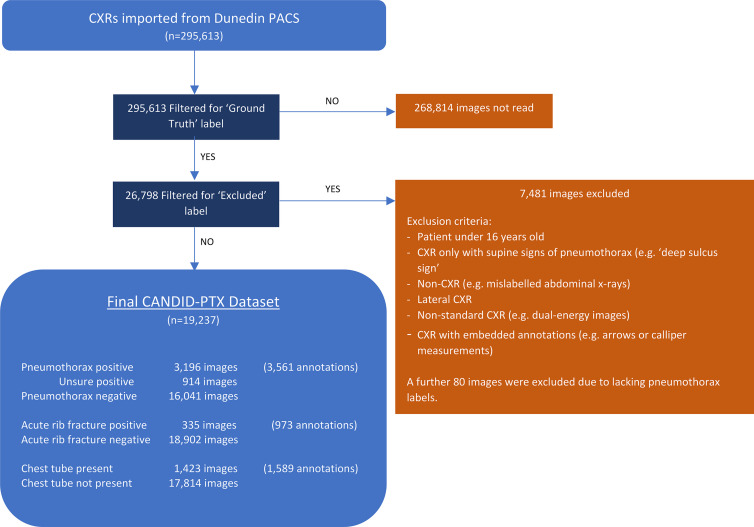
Figure 1:
Curation process and final characteristics of the Chest x-ray Anonymised New Zealand Dataset in Dunedin–Pneumothorax (CANDID-PTX) dataset. A total of 295 613 chest radiographs (CXRs) were imported from the Dunedin Hospital picture archiving and communication system (PACS). A total of 26 798 of these images were reviewed by a tier 3 annotator (either a consultant radiologist or registrar) as per the study method; 7481 of these were subsequently excluded as per the exclusion criteria listed, and a further 80 were excluded because they lacked the appropriate “pneumothorax” or “no pneumothorax” labels. The remaining 19 237 images comprise the final dataset, with the numbers of pneumothorax, acute rib fracture, and chest tube annotations being displayed in the figure. Some images contained multiple regions of visible pneumothorax, or multiple fractures or tubes, which were also expressed in the figure.
The images at their original size and their corresponding radiology reports were uploaded by using a command line client-library interface onto MD.ai (12), a Health Insurance Portability and Accountability Act–compliant, commercial, cloud-based image annotation platform.
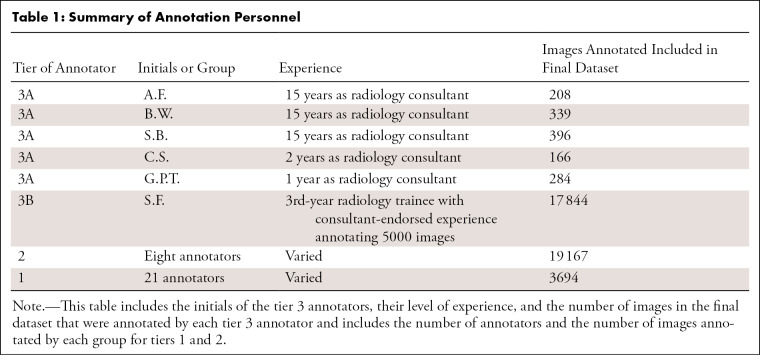
Annotation team structure.— A team of 35 doctors were separated into three tiers to coordinate the varying levels of pneumothorax chest radiograph reading competencies more efficiently. Tier 1 included junior doctors (1–5 years of postgraduate medical experience). Tier 2 included experienced junior doctors who had annotated at least 1000 images in the initial 4 weeks and had endorsement from a tier 3 member. Tier 3 annotators included Royal Australian and New Zealand College of Radiologists radiology consultants (tier 3A; A.F., B.W., and S.B., all with 15 years of experience, C.S., with 2 years of experience, and G.P.T., with 1 year of experience) and a 3rd-year radiology trainee (tier 3B; S.F.) after 5000 images had been annotated with consultant endorsement. All tier 2 and tier 3 annotators obtained a score of 100% on a 73-image practice test set, with multiple attempts being allowed prior to participation (Table 1). Tier 3A annotators’ consensus opinions were used as the correct answers for this set.
Table 1:
Summary of Annotation Personnel
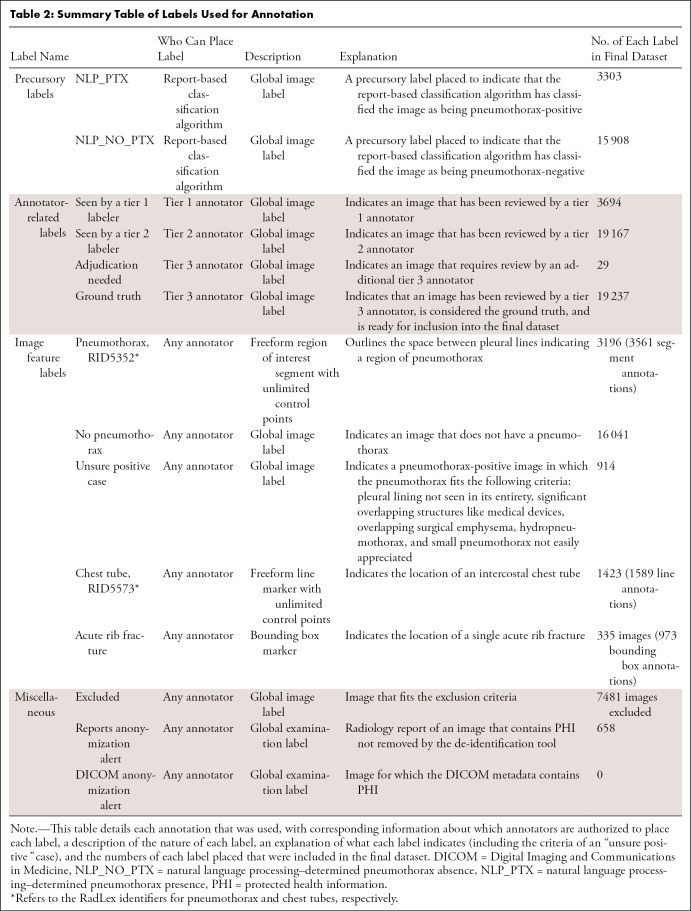
Annotation process.— Fourteen labels were created on the MD.ai platform to facilitate image categorization and annotation; a detailed description of each label is provided in Table 2, and an example of an annotated image is provided in Figure 2. A text analysis–based algorithm (13) detected and identified the context for a range of synonyms and abbreviations for “pneumothorax” in each report to give the corresponding image a preliminary classification as pneumothorax-positive or pneumothorax-negative. All preliminary positive cases were reviewed by team members, along with approximately five times the number of preliminary negative cases.
Table 2:
Summary Table of Labels Used for Annotation
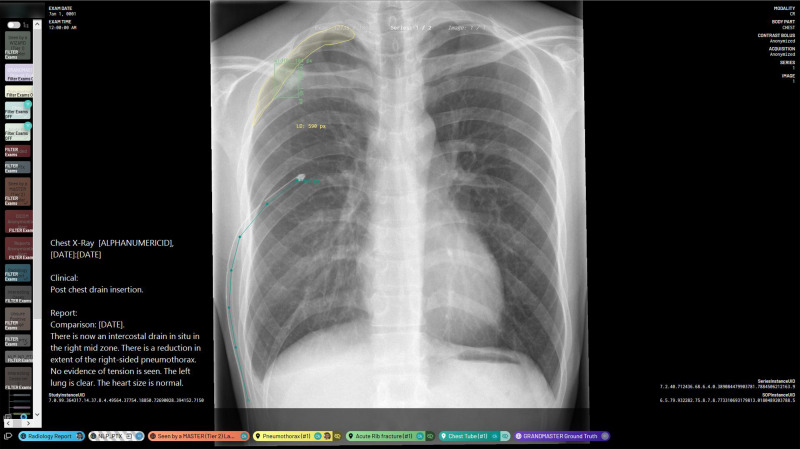
Figure 2:
Annotation platform interface and example annotations. This image shows a chest radiograph that has segmented annotations of a right apical pneumothorax, a single acute rib fracture, and one intercostal chest tube. It also features the image’s fully anonymized Digital Imaging and Communications in Medicine metadata and radiology report. The “NLP_PTX” label indicates that the report-based classification algorithm gives this image a preliminary “pneumothorax-positive” classification. There are also global image-level annotations indicating that this image has been reviewed by both a tier 2 annotator and a tier 3 annotator. NLP_PTX = natural language processing–determined pneumothorax presence.
Tier 1 and/or tier 2 members performed the first read and applied the appropriate labels (Table 2) regarding the presence and location of pneumothoraces, acute rib fractures, and chest tubes. Pneumothorax as diagnosed by using chest radiographs was defined as the presence of a visceral pleural lining visibly separate from the chest wall, as per the RadLex® (14) RID5352 definition. This included cases of tension pneumothorax but did not include cases that only demonstrated supine signs of pneumothorax such as the “deep sulcus sign.” Chest tubes were defined as any intercostal thoracostomy tube inserted into the pleural space to drain air or fluid as per the RadLex (14) RID5573 definition, and an acute rib fracture was defined as any rib with cortical disruption visible on a chest radiograph without evidence of healing such as callus formation.
Additionally, images fitting the exclusion criteria were labeled “excluded,” and those with remaining protected health information were labeled “reports anonymization alert” or “DICOM anonymization alert” for subsequent manual review and de-identification. Cases with a clear diagnosis of a pneumothorax or not were labeled as “pneumothorax” or “no pneumothorax,” respectively. These required review by a tier 3B annotator, whose opinion and labeling were considered final. Cases in which the pneumothorax had been difficult to appreciate were given the labels of “pneumothorax” and “unsure positive” (defined as in Table 2). They required review by a tier 3A member, whose opinion was considered final. Cases that were still difficult for a tier 3A annotator were passed on to another tier 3A member. If opinions differed, the image was labeled as “adjudication needed.” A thoracic radiologist (B.W., 15 years of consultant radiology experience) provided the final opinion, and the image was given a “ground truth” label (Fig 3).
Figure 3:
Workflow diagram of the annotation process for producing the ground truth. For pneumothorax annotations, images were first given a preliminary positive or negative classification by an algorithm that employed text analysis of images’ radiology reports. All preliminary positive images and five times this number of preliminary negative images randomly selected from each year were then presented to the annotators. These images were first read by tier 1 and/or tier 2 annotators and were then passed on to the appropriate tier 3 annotator, depending on whether the image was clearly pneumothorax-positive or pneumothorax-negative (the image was then reviewed by a tier 3B annotator [radiology registrar]) or on whether the image fit the criteria of “unsure positive” (the image was then reviewed by a tier 3A annotator [radiology consultant]). Images that met the exclusion criteria were tagged as such. When a tier 3 annotator was not confident, the image was then referred for adjudication by a thoracic radiologist (who also served as a tier 3A annotator). Once the appropriate tier 3 annotator had given their ground truth opinion, each image went through a quality validation process, whereby it was manually reviewed to ensure the quality of annotation and anonymization. The result of this validation process was considered the ground truth as expressed in the final public dataset. Annotators, when reading images at all stages of this process, also marked images as being positive or negative for rib fractures and chest tubes, with higher-tier annotators being able to overrule lower-tier annotators when they deemed appropriate.
Quality Assurance
Following initial annotation, a validation process was undertaken by a smaller team of six members from each tier. They reviewed every image with the “ground truth” label. Any errors underwent the same process outlined in the annotation process. Attention was paid to any images or reports with the “reports anonymization alert” or “DICOM anonymization alert” labels, ensuring the quality of anonymization by removing any remaining protected health information.
Data Postprocessing
After validation, all annotations were exported to the dedicated research workstation and filtered to include those with a “ground truth” label and to remove those with an “excluded” label. These DICOM images, along with their segmented annotations, were resized to a pixel dimension of 1024 × 1024.
File names of the images in the dataset containing images’ date of acquisition were anonymized consistently and irreversibly by translating them into future dates, with the temporal relationship between the images being preserved.
Results
Image Annotation
A total of 295 613 images from the Dunedin Hospital picture archiving and communication system were uploaded onto the MD.ai platform. “Ground truth” labels were placed on 26 798 images by six tier 3 members. A total of 7481 images were labeled as “excluded,” as per the study exclusion criteria, and a further 80 were excluded, as they lacked clear “pneumothorax” or “no pneumothorax” labels, which was most likely due to human error. After these exclusions, there were 19 237 images in the final Chest x-ray Anonymised New Zealand Dataset in Dunedin–Pneumothorax (CANDID-PTX) dataset (Fig 1). A total of 658 of these were labeled with “reports anonymization alert” and had their remaining protected health information manually removed from their corresponding reports.
The CANDID-PTX dataset contains 3561 pneumothorax segmentation annotations placed on 3196 pneumothorax-positive images (of which 914 were also labeled “unsure positive”), and 16 041 pneumothorax-negative images. A total of 973 acute rib fractures were labeled on 335 images with bounding boxes. A total of 1589 chest tubes were labeled on 1423 images with freeform line markings.
Dataset Characteristics
The 19 237-image CANDID-PTX dataset contains 10 278 images obtained from male patients and 8929 images obtained from female patients. The remaining 30 images had no information on sex when extraction from the original DICOM metadata was attempted. The mean age of the patients was 60.1 years (standard deviation, 20.1 years; range, 16–101 years). A total of 13 550, 5669, and 15 of the images were acquired on x-ray machines manufactured by Philips, GE Healthcare, and Kodak, respectively, and three images did not have x-ray machine information. Further results are provided in Appendix E1 (supplement).
Discussion
This study outlined the curation of a dataset of 19 237 images. It is publicly available, anonymized, adequately prepared, and of appropriate size (15) for use by machine learning algorithms featuring pneumothorax segmentation annotations, acute rib fracture bounding box annotations, and intercostal chest tube line annotations.
This dataset adds to the existing repository (16) on which AI algorithms can be trained and evaluated, allowing researchers to develop algorithms with better classification and segmentation performance and improving algorithm generalizability across datasets. In the future, this may allow AI to play a role in clinical practice by aiding in areas of diagnosis, reporting quality assurance, and image triaging (3–7). The inclusion of each image’s anonymized radiology report also allows researchers to perform text-based machine learning to correlate findings stated in the reports with findings annotated in the images. The inclusion of rib fractures and chest tubes allows researchers to perform interpretability studies on pneumothorax classification and segmentation with reference to these confounding factors.
This project’s three-tiered annotation team structure with subsequent validation required each image to be read by at least three members with differing experience levels. This was an efficient way to annotate a substantial number of images and establish a reliable ground truth.
One limitation of the CANDID-PTX dataset is that all images were obtained from a single institution. This reduces the heterogeneity of the patient demographics and technical image characteristics included in the dataset, which may reduce the generalizability of AI algorithms trained and tested on this dataset alone. Another limitation is the lack of CT confirmation of pneumothoraces when determining the ground truth. Adding this standard would have severely limited the size of the dataset. In addition, a CT examination is not required in routine clinical practice, as most pneumothoraces are diagnosed and managed according to radiographic findings alone.
Conclusion
The development of robust AI algorithms requires large, accurately annotated datasets. This study outlines the curation of a publicly available CANDID-PTX dataset with segmented annotations for pneumothoraces, chest tubes, and rib fractures that is ready for use by AI algorithms.
To access the dataset, an online ethics course and a data use agreement are both required to be completed and emailed to the corresponding author before a link to the dataset will be provided. Detailed instructions on this can be found by searching DOI: 10.17608/k6.auckland.14173982 in a digital object identifier search engine such as https://www.doi.org/.
Acknowledgments
Acknowledgments
We thank Anouk Stein and George Shih for providing access to MD.ai annotation platform; Jacob Han and Chris Anderson for data preprocessing; Arvenia Anis Boyke Berahmana, Sophie Lee, Amy Clucas, Andy Kim, and Aamenah Al Ani for dataset annotation; Richard Kelly for quality assurance; Qixiu Liu for interannotator variability analysis; and Colleen Bergin for manuscript editing. Author roles/job titles at the time of the study were as follows: radiology registrar (S.F., J.S.K.), resident medical officer (D.A., C.K.J., S.P.G., J.Y., E.K., M.H.), trainee intern (A.L.), medical student (A.P.), research staff member (J.W.), lecturer (M.U.), and consultant radiologist (A.F., C.S., G.P.T., S.B., B.W.).
Supported by a Royal Australia and New Zealand College of Radiologists (RANZCR) research grant in 2020.
Disclosures of Conflicts of Interest: S.F. RANZCR research grant (payment made to research team account held by Health Research South at Southern District Health Board). D.A. No relevant relationships. J.S.K. No relevant relationships. C.K.J. No relevant relationships. S.P.G. No relevant relationships. J.Y. No relevant relationships. E.K. No relevant relationships. M.H. No relevant relationships. A.L. No relevant relationships. A.P. No relevant relationships. J.W. No relevant relationships. M.U. No relevant relationships. A.F. No relevant relationships. C.S. No relevant relationships. G.P.T. No relevant relationships. S.B. No relevant relationships. B.W. No relevant relationships.
Abbreviations:
- AI
- artificial intelligence
- CANDID-PTX
- Chest x-ray Anonymised New Zealand Dataset in Dunedin–Pneumothorax
- DICOM
- Digital Imaging and Communications in Medicine
References
- 1. Rimmer A . Radiologist shortage leaves patient care at risk, warns royal college . BMJ 2017. ; 359 : j4683 . [DOI] [PubMed] [Google Scholar]
- 2. Bastawrous S , Carney B . Improving patient safety: avoiding unread imaging exams in the national VA enterprise electronic health record . J Digit Imaging 2017. ; 30 ( 3 ): 309 – 313 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rajpurkar P , Irvin J , Ball R2 , et al . Deep learning for chest radiograph diagnosis: a retrospective comparison of the CheXNeXt algorithm to practicing radiologists . PLoS Med 2018. ; 15 ( 11 ): e1002686 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang R , Tie X , Qi Z , et al . Diagnosis of coronavirus disease 2019 pneumonia by using chest radiography: value of artificial intelligence . Radiology 2021. ; 298 ( 2 ): E88 – E97 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schaffter T , Buist DSM , Lee CI , et al . Evaluation of combined artificial intelligence and radiologist assessment to interpret screening mammograms . JAMA Netw Open 2020. ; 3 ( 3 ): e20026 . [Published correction appears in JAMA Netw Open 2020;3(3):e204429.]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Annarumma M , Withey SJ , Bakewell RJ , Pesce E , Goh V , Montana G . Automated triaging of adult chest radiographs with deep artificial neural networks . Radiology 2019. ; 291 ( 1 ): 196 – 20 . [Published correction appears in Radiology 2019;291(1):272.]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yates EJ , Yates LC , Harvey H . Machine learning “red dot”: open-source, cloud, deep convolutional neural networks in chest radiograph binary normality classification . Clin Radiol 2018. ; 73 ( 9 ): 827 – 831 . [DOI] [PubMed] [Google Scholar]
- 8. Chartrand G , Cheng PM , Vorontsov E , et al . Deep learning: a primer for radiologists . RadioGraphics 2017. ; 37 ( 7 ): 2113 – 2131 . [DOI] [PubMed] [Google Scholar]
- 9. Lam L , Drucker D . Dicomanonymizer v1.0.4 . GitHub . https://github.com/KitwareMedical/dicom-anonymizer. Published February 25, 2020. Updated February 24, 2020. Accessed April 23, 2020 . [Google Scholar]
- 10. Kayaalp M , Way M , Jones J , et al. NLM-Scrubber . U.S. National library of Medicine . https://scrubber.nlm.nih.gov. Published May 3, 2019. Accessed May 3, 2020 . [Google Scholar]
- 11. National Ethics Advisory Committee. National ethical standards for health and disability research and quality improvement . New Zealand Ministry of Health . https://neac.health.govt.nz/publications-and-resources/neac-publications/national-ethical-standards-for-health-and-disability-research-and-quality-improvement/. Published December 19, 2019. Accessed February 23, 2020 .
- 12. MD.ai security and privacy policy . MD.ai . https://www.md.ai/privacy/ Updated August 10, 2018. Accessed April 23, 2020 .
- 13. Wu JT , Syed AB , Ahmad H , et al . AI accelerated human-in-the-loop structuring of radiology reports . Presented at the AMIA 2020 virtual annual symposium, virtual , November 14–18, 2020 . [PMC free article] [PubMed] [Google Scholar]
- 14. RadLex version 4.1. Radiological Society of North America . http://radlex.org/. Accessed August 1, 2021 .
- 15. Dunnmon JA , Yi D , Langlotz CP , Ré C , Rubin DL , Lungren MP . Assessment of convolutional neural networks for automated classification of chest radiographs . Radiology 2019. ; 290 ( 2 ): 537 – 544 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Filice RW , Stein A , Wu CC , et al . Crowdsourcing pneumothorax annotations using machine learning annotations on the NIH chest x-ray dataset . J Digit Imaging 2020. ; 33 ( 2 ): 490 – 496 . [DOI] [PMC free article] [PubMed] [Google Scholar]



![Workflow diagram of the annotation process for producing the ground truth. For pneumothorax annotations, images were first given a preliminary positive or negative classification by an algorithm that employed text analysis of images’ radiology reports. All preliminary positive images and five times this number of preliminary negative images randomly selected from each year were then presented to the annotators. These images were first read by tier 1 and/or tier 2 annotators and were then passed on to the appropriate tier 3 annotator, depending on whether the image was clearly pneumothorax-positive or pneumothorax-negative (the image was then reviewed by a tier 3B annotator [radiology registrar]) or on whether the image fit the criteria of “unsure positive” (the image was then reviewed by a tier 3A annotator [radiology consultant]). Images that met the exclusion criteria were tagged as such. When a tier 3 annotator was not confident, the image was then referred for adjudication by a thoracic radiologist (who also served as a tier 3A annotator). Once the appropriate tier 3 annotator had given their ground truth opinion, each image went through a quality validation process, whereby it was manually reviewed to ensure the quality of annotation and anonymization. The result of this validation process was considered the ground truth as expressed in the final public dataset. Annotators, when reading images at all stages of this process, also marked images as being positive or negative for rib fractures and chest tubes, with higher-tier annotators being able to overrule lower-tier annotators when they deemed appropriate.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/17e5/8637219/512e77181deb/ryai.2021210136fig3.jpg)