Abstract
Data-driven approaches have great potential to shape future practices in radiology. The most straightforward strategy to obtain clinically accurate models is to use large, well-curated and annotated datasets. However, patient privacy constraints, tedious annotation processes, and the limited availability of radiologists pose challenges to building such datasets. This review details model training strategies in scenarios with limited data, insufficiently labeled data, and/or limited expert resources. This review discusses strategies to enlarge the data sample, decrease the time burden of manual supervised labeling, adjust the neural network architecture to improve model performance, apply semisupervised approaches, and leverage efficiencies from pretrained models.
Keywords: Computer-aided Detection/Diagnosis, Transfer Learning, Limited Annotated Data, Augmentation, Synthetic Data, Semisupervised Learning, Federated Learning, Few-Shot Learning, Class Imbalance
Keywords: Computer-aided Detection/Diagnosis, Transfer Learning, Limited Annotated Data, Augmentation, Synthetic Data, Semisupervised Learning, Federated Learning, Few-Shot Learning, Class Imbalance
Summary
Model training strategies are described for use in scenarios in which there are limited datasets available.
Essentials
■ Models developed with data-driven approaches have great potential to shape the future practice of radiology.
■ A wide variety of strategies are available to enhance the performance of models in data-limited settings, including transfer learning, data augmentation, semisupervised training techniques, efficient annotation strategies, federated learning, few-shot learning, and different neural network architectures.
■ This review summarizes the model training strategies for radiologic image analysis in data-limited scenarios.
Introduction
Research in diagnostic radiology decision support has progressed rapidly because of the availability of large datasets, powerful machine learning (ML) techniques, and computers to efficiently run ML techniques (1–3). Advances in medical image analysis research include multiple systems that aim to help radiologists detect disease (4–7), identify disease progression (8), localize abnormalities (9), automate time-consuming tasks, and improve the radiology workflow.
The performance of deep learning–based algorithms depends on the availability of large-scale annotated data (3,10,11). A large dataset with diverse, high-quality images curated from multiple institutions and different geographic areas is preferable to ensure the generalizability of a model for clinical use (12). However, curating large datasets is challenging because of their volume, limited radiologist availability, and tedious annotation processes. It is particularly challenging to curate data for rare diseases. Additionally, many complexities are introduced in the data de-identification process to comply with patient privacy rules, institutional review board requirements, and local ethical committee protocols (13). If training data are limited, deep learning–based models may suffer from overfitting, which results in poor generalizability.
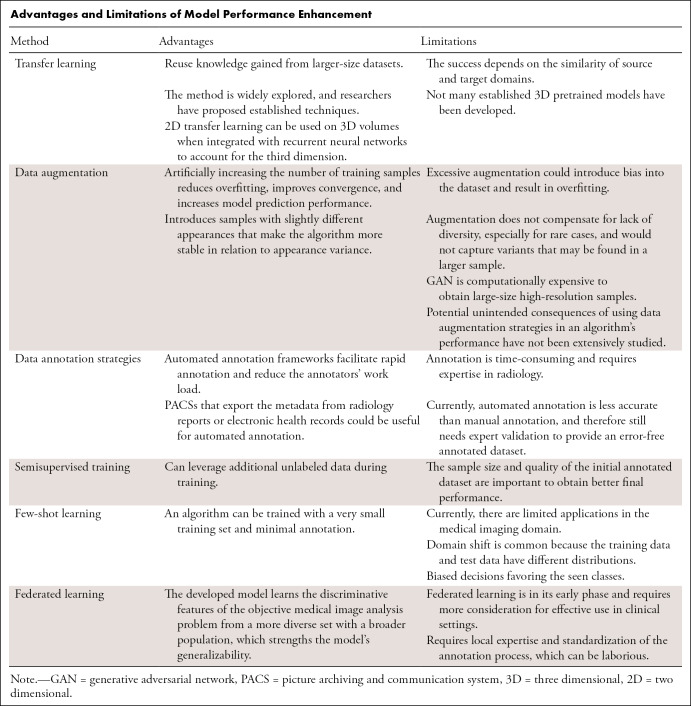
Several reviews have described deep learning–based frameworks for medical imaging (2,3,14). We focus on model training strategies that boost neural network (NN) performance in limited-data scenarios. We discuss transfer learning, data augmentation, semisupervised training techniques, efficient annotation strategies, federated learning, few-shot learning, and NN architectures to remedy data scarcity. We present use cases from the radiology research literature that use these strategies to improve model performance. The Table summarizes the advantages and limitations of the mentioned techniques.
Advantages and Limitations of Model Performance Enhancement
Transfer Learning
Two-dimensional Transfer Learning
Although the Radiological Society of North America, several research institutions, and hospitals worldwide have released multiple radiologic datasets for medical image analysis, the availability of large-scale medical images remains limited. Transfer learning is a common strategy to address limited data (10). In transfer learning, a NN is pretrained on a larger dataset (15), and learned features can be applied to the target domain with limited data (16). The underlying idea is that low-level features are common; therefore, they can be learned from the available large-scale dataset (17).
There are different types of transfer learning strategies, such as fixing the earlier layers, retraining only the higher layers (shallow tuning), or fine-tuning the whole architecture (deep tuning). Fine-tuning is a type of transfer learning technique in which all or part of the pretrained model is retrained with the domain data with a low learning rate. This approach helps adapt the learned features to the transfer domain. The level of fine-tuning is a hyperparameter that can be optimized during the training. One factor that affects fine-tuning performance is the domain similarity of the source and target datasets. If source and target domains are similar, fine-tuning of the final layers would be adequate to obtain accurate results. However, if the source and target domains are very different, fine-tuning of more layers provides better outcomes (10). The other factor is the amount of training data. If training data are limited, it would not be enough to fine-tune more layers. On the other hand, if there are adequate training data, even if the domains are similar, updating the weights of more layers to the new domain will help with convergence to a better solution (10). Off-the-shelf features (19), which use the pretrained model as a feature extractor, tend to perform well with limited data because the framework uses traditional ML methods as classifiers instead of fully connected layers, therefore reducing the number of trainable parameters. This approach has the potential to minimize overfitting while maintaining the discriminative power of deep features.
Transfer learning techniques are widely applicable to radiologic images. One of the earlier applications is a fine-tuned convolutional NN (CNN) that processes the different views of mammographic images (18). The model performance was compared with training from scratch on a mammographic dataset, with and without data augmentation. Researchers reported 5%–16% improvement in the volume under the receiver operating characteristic surface metric (an extension of the area under the curve metric for multiclass tasks) with the pretrained multiview model. Tajbakhsh et al (10) thoroughly investigated the performance of pretrained CNNs, incremental fine-tuning, training from scratch, and the hand-crafted approaches for pulmonary embolism detection. The fine-tuned CNN outperformed the CNNs trained from scratch and with the hand-crafted method. A recent study (19) compared three deep learning approaches to distinguish among breast cancer subtypes on MR images: (a) using learning from scratch when only tumor patches were used for training, (b) using transfer learning when a pretrained network was fine-tuned by using tumor patches, and (c) using off-the-shelf deep features when a pretrained network was used as a feature extractor and the extracted features were trained with a support vector machine. The off-the-shelf features approach achieved the highest area under the receiver operating characteristic curve performance for distinguishing breast cancer subtypes. The techniques mentioned in this section are illustrated in Figure 1. See the studies by Tajbakhsh et al (10) and Shin et al (20) for a comprehensive analysis of transfer learning in medical imaging.
Figure 1:
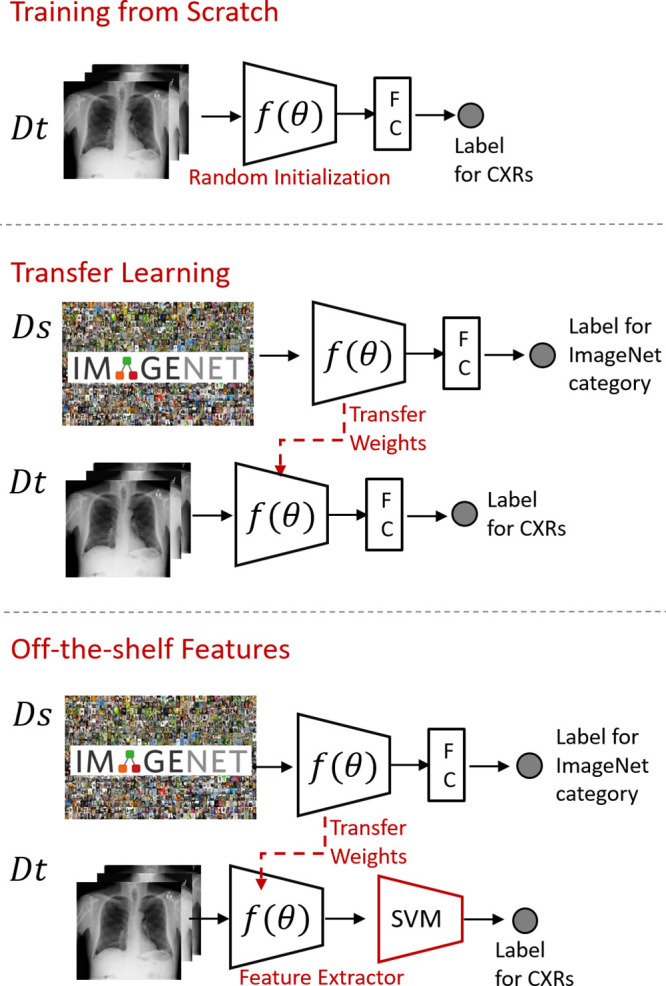
Training from scratch: only the data in the target domain is used for training. Transfer learning: the weights of a pretrained network at the source domain (Ds) are fine-tuned with the target domain data (Dt). Off-the-shelf deep features: the pretrained network is used as a feature extractor on the target domain data, and a traditional machine learning algorithm such as support vector machine (SVM) uses the extracted features to classify the imaging findings. CXRs = chest radiographs, FC = fully connected layer.
Three-dimensional Transfer Learning
Transfer learning is easily applicable to two-dimensional (2D) images because of the abundance of pretrained models but is not widely used for three-dimensional (3D) modalities because there are not many established 3D pretrained models. It is also more challenging to curate a large-scale 3D medical dataset because of the laborious annotation process when including the third dimension. In addition, NNs that are developed for 3D analysis have a larger number of parameters (2); therefore, NNs that process volumes require larger-scale datasets to optimize the weights.
Several approaches have been proposed to process 3D radiologic volumes. Chen et al (21) built a large-scale 3D medical dataset containing various modalities, target organs, and pathologic conditions. The collected data were used to train the Med3D network that learned 3D features from large-scale radiologic data. Researchers transferred pretrained models to lung segmentation and pulmonary nodule classification. To our knowledge, there is not any other published research that curates large-scale 3D medical data and trains a deep network with it to serve as a base model. However, researchers have followed alternative strategies to process 3D volumes. A few studies converted volumes to frames to use established 2D pretrained models. For instance, for automated plaque detection, the vessel volumes from CT sequences were projected onto frames, which contained vessel views from different angles (22). In another study (23), 3D data were sectioned into axial, coronal, and sagittal planes, and 2D pretrained models were applied to the planes. Although transfer learning has been successfully used from 2D to 3D space, projected views are sparse representations of volumes, therefore containing less information than 3D. An additional disadvantage of 2D CNNs is their inability to leverage context from adjacent sections. As in recurrent CNN (24), the approach needs additional mechanisms to preserve the sequential knowledge.
Transfer Learning with a Same-Modality Dataset
Pretrained models typically are trained with ImageNet (15), which consists of general nonmedical images. However, there is a substantial difference between general and medical images. The image contents and, therefore, histogram distributions of nonmedical and radiologic images are different. This difference may cause redundant pretrained features and an excessive number of parameters for the medical imaging domain (25). Additionally, the intensity values, scale, and location of the region of interest (eg, lesion) have more meaning in the medical imaging domain. For instance, a CT pixel value denotes a quantifiable physical characteristic of a tissue or structure. Bright or dark spots on a dog would not affect its semantic classification, but how bright or dark a head CT lesion appears has a substantial impact on diagnosis. The size and the location of the detected lesion might be additional clues regarding the disease. Radiologic images typically are presented in a fixed, predetermined orientation, in contrast to objects on nonmedical images; orientation of the image can be crucial for some diagnoses, such as situs inversus on a chest radiograph.
The success of transfer learning depends on the similarity of the source and target domains (10,17). Several researchers employed transfer learning in which the source and target domains had similar feature spaces. For instance, a 3D CNN trained to distinguish between MR images of patients with Alzheimer disease and MR images of healthy controls was later fine-tuned to differentiate between patients with mild cognitive impairment and healthy controls, a more challenging scenario (7). In another study, the kidney regions were delineated by using a 3D CNN pretrained with segmented hippocampus images (26). With the availability of large-scale chest radiographic datasets, a NN pretrained with a large-scale radiographic dataset was fine-tuned for cardiomegaly detection (27) and pneumonia prediction (4).
Efficient Usage of Data in Model Development
Data Augmentation
Data augmentation is the process of artificially increasing the number of training samples by altering the existing images or synthetically creating new images.
Alteration of Images
The standard augmentation technique is to alter the existing images in a dataset. Example alteration strategies are random rotation, brightness variation, noise injection, contrast changes, blurring, cropping, and mirroring. Obtaining new samples increases the amount of data and introduces samples with slightly different appearances that make the algorithm more stable in relation to appearance variance. The augmentation strategy should be carefully designed. Excessive augmentation of training data could introduce bias into the dataset and result in overfitting. Additionally, the distinctive features of a disease should not be distorted during the augmentation. For instance, the organ ratios at chest radiography are important features in cardiomegaly prediction. In Candemir et al (27), limited augmentation was applied to chest radiographs to preserve the ratios. In Esmaeilzadeh et al (7), the researchers only applied flipping to the MRI sequences to not distort the shapes of brain structures.
Synthetic Data Augmentation
Another strategy to increase sample size is to generate synthetic samples by using a generative adversarial network (GAN) (28). A GAN consists of two networks: a generator and a discriminator. The generator produces synthetic samples with a similar distribution of the existing images; the discriminator predicts whether the instance is original or synthesized. The discriminator forces the generator to create realistic samples until the discriminator can no longer differentiate between actual and synthetic samples.
In radiology, GANs have been used to synthesize medical images such as chest radiographs (6), liver lesion samples (29), CT scans with lung nodules (30), and images from brain MRI sequences (31). These studies reported improved NN performance with additional synthetic data. For instance, for liver lesions on CT images (29), with the alteration of the images, a NN achieved 78.6% sensitivity and 88.4% specificity. Incrementally adding synthetic samples in the threefold cross-validation process improved model performance as the number of training examples increased, with saturation occurring at 5000 samples per fold. The system’s classification performance increased to 85.7% sensitivity and 92.4% specificity with additional synthetic samples. Synthetic data augmentation is an active research area. A comprehensive review of data augmentation can be found in Shorten et al (32). GAN usage for data augmentation is reviewed in Yi et al (33) and Salimans et al (34).
Limitations of Data Augmentation
Artificially increasing the number of training samples reduces overfitting, improves convergence, and increases model prediction performance. However, augmentation adds slight variability by altering the existing data or producing new samples derived from the available data distribution. Some instances in the data might be poorly represented or unrepresented. Augmentation does not compensate for a lack of diversity in biologic variability, especially for rare cases, and would not capture variants that may be found in a larger sample.
GANs have limitations. They require a substantial amount of annotated data to synthesize realistic samples (34). It is also computationally expensive to obtain large-sized, high-resolution samples. One recent study (35) reported similar false-positive and sensitivity results when a GAN was trained only with synthetic MRI data (2000 synthetic volumes), compared with the results obtained by using original data. More use cases are needed to assess whether synthesized images provide model training performance that is similar to that provided by authentic images, especially when abnormalities are present (36).
The extent to which data augmentation can adversely affect algorithm performance is difficult to quantify. The potential, unintended algorithm-performance consequences of using data augmentation strategies have not been extensively studied.
Efficient Usage of Data in Model Evaluation: Cross-Validation
Splitting the size-limited dataset into training, validation, and test sets further reduces the amount of data that is available for model training and reliable performance validation. Evaluation of a model with a small, nonrepresentative test set causes unreliable generalization results (37). Cross-validation is a resampling evaluation approach to provide a more accurate estimation of generalization error (38). In one such technique, the k-fold strategy, the training and validation are repeated k times. The average validation errors obtained from k iterations provide more reliable results because all data have been used for prediction. The leave-one-out technique is recommended for small or imbalanced datasets (39): for each case in the dataset, train the model with the other n − 1 cases and test on that single case.
Data Annotation Strategies
Image annotation is the process of labeling images with identifiers (eg, class labels as a benign or malignant tumor; the delineated boundary of a tumor region) (12,13). It is a time-consuming process and requires expertise in radiology. For general applications, such as labeling photographs of natural scenes, crowd-sourcing over the Web can be used to collect annotations from nonexperts. However, reference standards for medical images need to be decided by domain experts. Additionally, it is important to determine the intra- and interrater variability when dealing with multiple annotators, as poorer performance has been observed for models trained on datasets with lower interrater agreement (40).
Recent efforts have focused on automatically assigning annotations from available information. For instance, metadata from radiology reports or electronic health records can be useful for automated annotation (41). Structured information and disease labels can be extracted by using natural language processing (42). The diseased regions on the images can be highlighted by extracting the medical subject headings from the radiology reports (43). A recent study integrated eye-tracking and speech recognition algorithms to annotate MRI brain lesions automatically (44) by matching the time of spoken keywords with the time of gaze points. The study reports 92% accuracy for extracting the lesion locations. Although automated annotation frameworks facilitate rapid annotation and reduce the annotators’ workload, they are currently less accurate than manual annotation (42). Therefore, automated annotation methods still need expert validation to provide an error-free annotated dataset. The iterative labeling strategies such as active learning or self-training can be employed for automated labeling (45). A radiologist can validate the automated labels and provide annotations only for the wrong annotation predictions. The model training can be repeated with corrected annotations.
Radiologists have been more frequently connecting reports to stored radiologic images with their annotations, particularly in clinical trials that require measurements of target lesions. This “interactive content” reporting has been used clinically since 2015 (46); in this type of reporting, radiologists automatically create hyperlinked texts imported from the annotated images in x, y image space and with z table space acting as 3D expert labeling. Hyperlinking annotations within reports (47) have the potential to increase the amount of labeled data for model training, which would help improve federated learning strategies (13). Hyperlinked annotations have been made publicly available (48) with this technology, in which the measurements are easily converted to bounding boxes. Last, some researchers have leveraged simulated artificial intelligence workflows (41), with radiologic preprocessors identifying and measuring lesions following cross-sectional anatomy training (49) and the use of automated tools having already been incorporated in some picture archiving and communication systems in advance of radiologists opening the examinations. Integration of artificial intelligence tools within the picture archiving and communication system will eventually help radiologists to identify and measure lesions, identify incidental findings, and facilitate earlier notification and will save radiologists’ time for time-consuming tasks.
Semisupervised Training
If curated unlabeled data are abundant, but the labeled portion is insufficient, the semisupervised technique can be employed. This strategy combines labeled and unlabeled data to train a classifier. One popular approach is predicting pseudolabels (Fig 2) (50). An initial model (teacher) is trained with the available labeled images. The teacher model then predicts the labels for unlabeled data. The confident predictions are considered pseudolabels (51). The training process continues using labeled and predicted pseudolabeled datasets and iteratively repeats this process until a predetermined termination condition.
Figure 2:
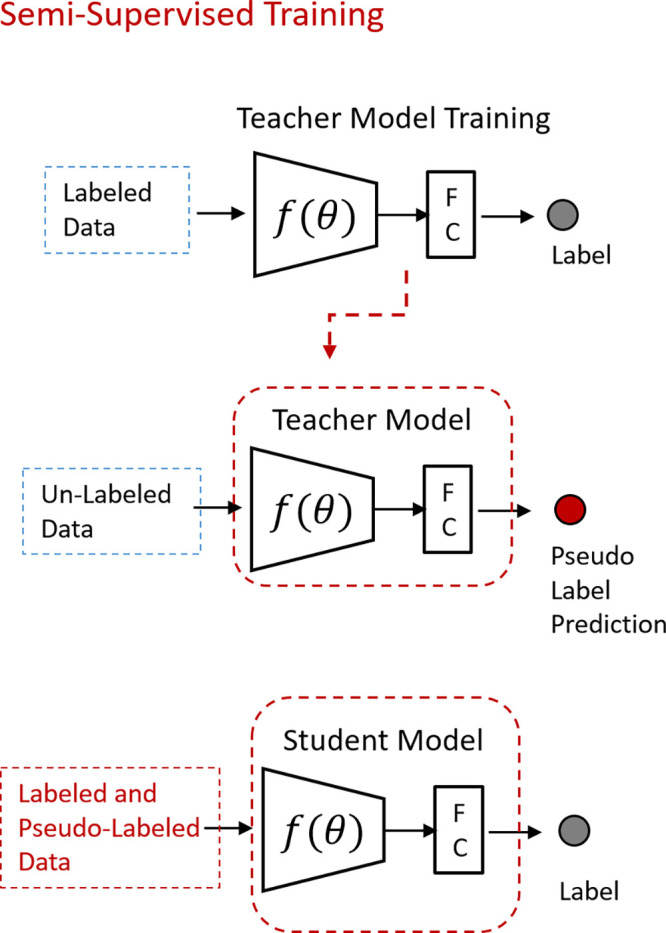
Illustration of semisupervised learning. An initial model (teacher) trained with labeled data predicts pseudolabels for the unlabeled dataset. The training process continues using the labeled and predicted pseudolabeled dataset until a predetermined termination condition. FC = fully connected layer.
Semisupervised learning embraces the heterogeneity of a patient population. Some diseases progress in a continuous manner or in small increments without a clear method for diagnosing the disease’s incremental stages. Therefore, semisupervised learning could add finer granularity to a labeled dataset by detecting homogeneous subpopulations within a heterogeneous dataset. A semisupervised method based on the clustering technique has been applied to brain MRI (52). The authors employed a dataset from the Baltimore Longitudinal Study of Aging, which had labels of normal or mild cognitive impairment. However, they used clustering for fine-tuned detection of more subtle cognitive differences within each broad classification to detect brain regions involved in predicting cognitive stability or decline within these populations. A recent study used the teacher–student method for COVID-19 severity prediction (53). A teacher model was trained initially with limited COVID-19 cases with corresponding severity labels and was then used to predict pseudolabels for unlabeled data. Incorporating unlabeled data into the training process tends to improve model performance. Another study on chest radiographic disease classification described a semisupervised approach in which radiologists annotated the location of disease by using bounding boxes on a small subset of the data, resulting in improvement in both classification and localization performance (54).
The semisupervised strategy helps to improve performance when annotation is limited; however, large numbers of unannotated images are still needed (55,56). The sample size and quality of the initial annotated dataset are also important to obtain better final performance. See the detailed review of semisupervised approaches in medical image analysis in Cheplygina et al (57).
Few-Shot Learning
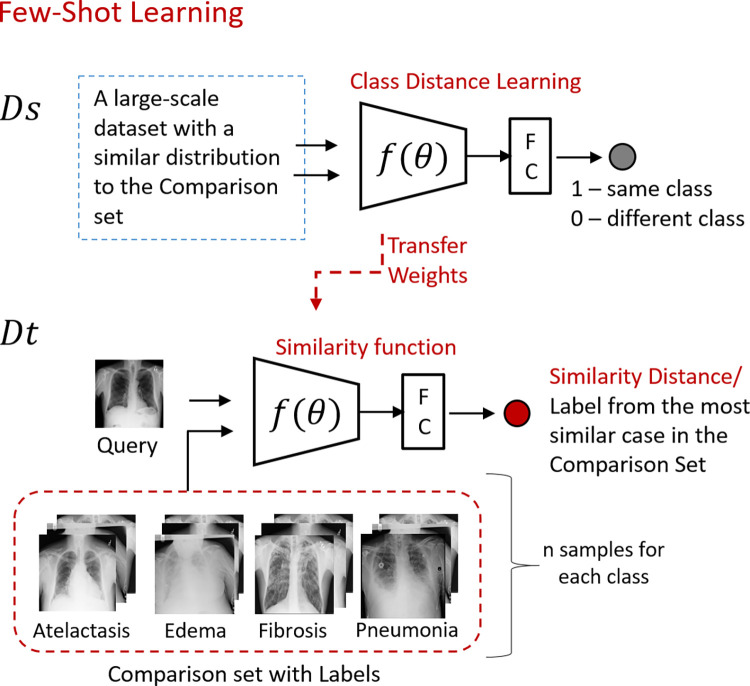
In few-shot learning, the classification task is posed as an image-matching problem (58,59). Therefore, the training process turns into building a model that learns the similarity between images instead of learning the specific features of objects. Few-shot learning uses a large-scale dataset as a source domain and trains the model for class similarities (Fig 3). The trained model is then applied to a query image whose class is unknown and may not necessarily be one of the trained classes. As the model learns the similarities between images, running the model on a query requires a comparison set, considered prior knowledge for the objective problem. The limited data in the target domain are used as an annotated comparison set. The model then predicts the object class for the query image by comparing it with the images in the comparison set. The highest similarity class label is associated with the query. If the comparison set contains n instances for each class, it is called the n shot. In a one-shot learning setup (59,60), the comparison set contains a single image representing the classes. The literature has limited applications of few-shot learning in the medical imaging domain. For instance, healthy and tumoral tissues of the colon, breast, and lung were classified by using a few-shot learning strategy (61) with the Siamese NN (59). The researchers trained the architecture with histopathologic images to learn class distances. Few-shot learning was then applied in the target domain with healthy and tumoral samples of tissues.
Figure 3:
Illustration of the few-shot learning strategy. The model starts with a large-scale dataset in the source domain (Ds) and learns to differentiate between two given inputs. The trained model is then applied to query images. The limited labeled dataset in the target domain (Dt) is used as a comparison set. The model predicts the class label by comparing the query image with the comparison set. The highest similarity class label is associated with the query. FC = fully connected layer.
The other limited-shot learning paradigm is zero-shot learning, which aims to recognize new categories whose instances may not have been seen during training. The method compensates for the lack of labeled data for the new categories with auxiliary information, which is composed of the semantic descriptions of the images. The semantic descriptions are one-shot–encoded, distinguishing the descriptive attributes of classes. The training process involves learning a function from the image space and applying it to the semantic space by using the seen data with the corresponding semantic descriptions. Zero-shot learning has recently been applied to radiology for chest radiographic classification (62). The study constructed three semantic spaces containing disease-specific auxiliary information. One challenge of the method is extracting accurate auxiliary semantic descriptions from the unstructured text found in most radiology reports. Chen et al (63) summarized the free-text analysis problem in radiology reports, provided a CNN-based method for free-text analysis, and compared it with conventional natural language processing approaches. The other important limitation of zero-shot learning is the domain shift, as the training and test data have different distributions. Biased decisions favoring the seen classes is another limitation due to the same problem. The transductive zero shot, which incorporates the unseen images into the training process, may mitigate the domain shift and bias.
Concept of Learning the Normal
In a curated dataset, positive cases are generally scarce. One alternative method that uses data with limited positive but abundant negative samples is the concept of learning the normal. In this approach, the classifier is trained only with normal images to learn the representative normal features to subsequently distinguish abnormal from normal findings on the basis of deviations of features from the learned representation of the normal class (57). In Wong et al (9), researchers trained a CNN by using CT scans with normal heart anatomy. The feature maps of the trained model provide information on the learned relationship between imaging data and semantic labels to detect deviations from normal anatomy. This information can be transferred to pericardial effusion and cardiac septal defect classifiers to improve the detection performance on limited positive samples. In another study, the researchers trained an autoencoder only with negative patches extracted from mammographic images to detect microcalcifications (5). The test images were fed into the trained autoencoder as patches. The reconstruction error, which indicates the deviation from the learned representation of what is normal, can then be set to a threshold to detect suspicious areas containing anomalies.
Federated Learning
One potential solution to train a model with a larger dataset is federated learning. In this collaborative training strategy, the model, weights, and parameters are sent to different institutions. Each institution trains the model with its local data and submits the updated model weights to a shared server. The aggregated model weights are sent back to the institutions for another round of training (64).
Various training strategies have been proposed for federated learning (13). In parallel training, the models are trained in parallel, and model weights are transferred to the central server. In nonparallel training, the model is sequentially updated at each institution. One of the main benefits of federated learning is the opportunity to train models with larger and more diverse datasets through multisite collaboration while preserving patient privacy and strengthening the model’s generalizability. Collaborative training also provides the opportunity for research and development in rare diseases that require multi-institutional efforts for data curation.
Federated learning is in its early phase and requires more consideration for effective use in clinical settings. One challenge is the variations in data types, data formats, acquisition protocols, annotation formats, and terminology across institutions (13). These impediments can be overcome with agreements in standardization among the involved institutions. The other barriers to the effective use of federated learning are technical limitations. The computational infrastructures needed to train the models efficiently may be limited at some institutions.
Despite the challenges, preliminary efforts show the applicability of cooperative training in medical image analysis (64). It has also been described as one of the fundamental techniques in the future of digital health (65). A comprehensive analysis of federated learning can be found in Yang et al (66).
Regularization in Network Architecture
Researchers have also developed techniques to construct more data-efficient NN architectures. Regularization techniques can mitigate the overfitting seen in data-limited scenarios and increase the model’s generalizability. Dropout (67) is one of the most employed regularization techniques. In this strategy, random neurons are dropped out by assigning zeros to the activation values at each training iteration to force learning of more discriminative features. The dropout rate is a hyperparameter, which, according to the thorough review by Srivastava et al (67), should be between 0.5 and 0.8. The optimal value can be chosen by using a validation set or can be set at 0.5, which has been empirically found to be near optimal (67). The study also found no improvement of dropout in extremely small datasets (eg, 100–500), but as the data size increases, the gain from dropout increases up to a point and then declines. The researchers concluded that for any given architecture, the dropout necessity and the dropout rate can be determined by using a validation set. Batch normalization is another explicit regularization technique that subtracts the batch mean from each activation and divides by the batch standard deviation (68). The regularization can be applied implicitly by tuning the loss function with weight decays (eg, lasso regression and ridge regression) that constrain the model’s capacity (69). See the comprehensive review by Kukačka et al (69) for regularization methods in deep learning.
Addressing Class Imbalance
As we previously mentioned when discussing the concept of learning the normal, positive cases are sometimes scarce in a curated dataset, which creates an imbalanced training set. The class imbalance causes a biased model that makes decisions in favor of the majority class. The class-balancing techniques are well studied in ML (70,71).
One of the well-known class-balancing techniques is based on data sampling. The minority class can be oversampled with augmentation by altering the existing images or synthetically creating new images. The samples undergoing augmentation can be selected randomly or strategically (72). Care must be taken when augmenting the minority class synthetically because of the challenges of learning the minority class distribution (73), as unsuccessful augmentation might distort the class boundary. To efficiently learn the distribution of the minority class, an autoencoder framework that estimates class distributions in the latent space (74) can be used by a GAN to generate minority class samples on the basis of the predicted class distribution. Oversampling techniques increase the training data, which can therefore increase model accuracy, but excessive oversampling might lead to overfitting (70).
Another class-balancing technique is increasing the importance of the minority class. The training process can be altered by penalizing the majority samples during loss computation or by increasing the weights of minority samples with a weight multiplication (75). Lin et al (76) proposed use of focal loss that reduces the weights of the well-classified examples, assuming that the dominant class becomes well classified because of larger representative samples.
One alternative training strategy to work with imbalanced datasets is learning the majority class (ie, learning normal anatomy). In this approach, the classifier is trained only with the majority class and classifies the minority class on the basis of the deviation from the majority class (57). Comprehensive studies that address the class-imbalance strategies in ML can be found in He and Garcia (71).
Conclusion
Models developed with deep NNs have great potential to shape the future practice of radiology, but their success will depend on their ability to generalize to different settings (1). A wide variety of approaches are available to enhance the generalization capabilities of deep learning–based models in data-limited settings. This review discussed strategies to enlarge the data sample, decrease the time burden of manual labeling, adjust the NN architecture to improve model accuracy, apply semisupervision approaches, and leverage efficiencies from pretrained models. Most of these described strategies have been routinely implemented in medical imaging artificial intelligence research and can be readily incorporated into an ML pipeline to enhance model performance.
Supported by the Department of Radiology of The Ohio State University College of Medicine and in part by the Intramural Research Program of the National Institutes of Health Clinical Center.
Disclosures of Conflicts of Interest: S.C. No relevant relationships. X.V.N. Equity ownership in and dividends from multiple publicly traded companies that may be considered “broadly relevant to artificial intelligence” (NVIDIA, Amazon, Microsoft, AMD, Apple). L.R.F. Patents issued, no royalties: Radiographic marker that displays upright angle on portable radiographs (Patent no. 9,541,822 B2) and multiscale universal CT window (Patent no. 8,406,493 B2); royalties from Springer (combat radiology, unrelated); research agreement with Philips. L.M.P. associate editor of Radiology: Artificial Intelligence.
Abbreviations:
- CNN
- convolutional NN
- GAN
- generative adversarial network
- ML
- machine learning
- NN
- neural network
- 3D
- three dimensional
- 2D
- two dimensional
References
- 1. Chartrand G , Cheng PM , Vorontsov E , et al . Deep learning: a primer for radiologists . RadioGraphics 2017. ; 37 ( 7 ): 2113 – 2131 . [DOI] [PubMed] [Google Scholar]
- 2. Litjens G , Kooi T , Bejnordi BE , et al . A survey on deep learning in medical image analysis . Med Image Anal 2017. ; 42 ( 60 ): 88 . [DOI] [PubMed] [Google Scholar]
- 3.Zhou SK, Greenspan H, Davatzikos C, et al. A review of deep learning in medical imaging: image traits, technology trends, case studies with progress highlights, and future promises. In: Proceeding of the IEEE. Vol 109, issue 5; 2021:820–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rajpurkar P , Irvin J , Ball RL , et al . Deep learning for chest radiograph diagnosis: a retrospective comparison of the CheXNeXt algorithm to practicing radiologists . PLoS Med 2018. ; 15 ( 11 ): e1002686 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei Q, Ren Y, Hou R, Shi B, Lo JY, Carin L. Anomaly detection for medical images based on a one-class classification. In: Petrick N, Mori K, eds.Proceedings of SPIE: medical imaging 2018—computer-aided diagnosis. Vol 10575.Bellingham, Wash:International Society for Optics and Photonics,2018;105751M. [Google Scholar]
- 6.Madani A, Moradi M, Karargyris A, Syeda-Mahmood T. Chest x-ray generation and data augmentation for cardiovascular abnormality classification. In: Angelini ED J, Landman BA, eds.Proceedings of SPIE: medical imaging 2018—image processing. Vol 10574.Bellingham, Wash:International Society for Optics and Photonics,2018;105741M. [Google Scholar]
- 7.Esmaeilzadeh S, Belivanis DI, Pohl KM, Adeli E. End-to-end Alzheimer’s disease diagnosis and biomarker identification. In: Shi Y, Suk HI, Liu M, eds. Machine learning in medical imaging. MLMI 2018. Vol 11046,Lecture notes in computer science.Cham, Switzerland:Springer,2018;337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Candemir S , Nguyen XV , Prevedello LM , et al . Predicting rate of cognitive decline at baseline using a deep neural network with multidata analysis . J Med Imaging (Bellingham) 2020. ; 7 ( 4 ): 044501 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong KC, Karargyris A, Syeda-Mahmood T, Moradi M. Building disease detection algorithms with very small numbers of positive samples. In: Descoteaux M, Maier-Hein L, Franz A, Jannin P, Collins D, Duchesne S, eds.Medical image computing and computer assisted intervention—MICCAI 2017. MICCAI 2017. Vol 10435,Lecture notes in computer science.Cham, Switzerland:Springer,2017;471–479. [Google Scholar]
- 10. Tajbakhsh N , Shin JY , Gurudu SR , et al . Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imaging 2016. ; 35 ( 5 ): 1299 – 1312 . [DOI] [PubMed] [Google Scholar]
- 11. Esteva A , Robicquet A , Ramsundar B , et al . A guide to deep learning in healthcare . Nat Med 2019. ; 25 ( 1 ): 24 – 29 . [DOI] [PubMed] [Google Scholar]
- 12. Park SH , Han K . Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction . Radiology 2018. ; 286 ( 3 ): 800 – 809 . [DOI] [PubMed] [Google Scholar]
- 13. Willemink MJ , Koszek WA , Hardell C , et al . Preparing medical imaging data for machine learning . Radiology 2020. ; 295 ( 1 ): 4 – 15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shen D , Wu G , Suk HI . Deep learning in medical image analysis . Annu Rev Biomed Eng 2017. ; 19 ( 1 ): 221 – 248 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. ImageNet . http://www.image-net.org/. ImageNet Web site. Updated 2020. Accessed July 7, 2020. .
- 16. Russakovsky O , Deng J , Su H , et al . ImageNet large scale visual recognition challenge . Int J Comput Vis 2015. ; 115 ( 3 ): 211 – 252 . [Google Scholar]
- 17.Yosinski J, Clune J, Bengio Y, Lipson H. How transferable are features in deep neural networks? In: Ghahramani Z, Welling M, Cortes C, Lawrence N, Weinberger KQ, eds.Advances in neural information processing systems 27 (NIPS 2014).Red Hook, NY:Curran Associates,2014;3320–3328. [Google Scholar]
- 18. Carneiro G , Nascimento J , Bradley AP . Unregistered multi-view mammogram analysis with pre-trained deep learning models . In: Navab N , Hornegger J , Wells W , Frangi A , eds. Medical image computing and computer-assisted intervention—MICCAI 2015 . MICCAI 2015. . Vol 9351 , Lecture notes in computer science . Cham, Switzerland: : Springer; , 2015. ; 652 – 660 . [Google Scholar]
- 19. Zhu Z , Albadawy E , Saha A , Zhang J , Harowicz MR , Mazurowski MA . Deep learning for identifying radiogenomic associations in breast cancer . Comput Biol Med 2019. ; 109 ( 85 ): 90 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shin HC , Roth HR , Gao M , et al . Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning . IEEE Trans Med Imaging 2016. ; 35 ( 5 ): 1285 – 1298 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen S , Ma K , Zheng Y . Med3D: transfer learning for 3D medical image analysis . ArXiv 1904.00625[preprint] https://arxiv.org/abs/1904.00625. Posted April 1, 2019. Accessed October 2021 . [Google Scholar]
- 22. Gupta V , Demirer M , Bigelow M , et al . Performance of a deep neural network algorithm based on a small medical image dataset: incremental impact of 3D-to-2D reformation combined with novel data augmentation, photometric conversion, or transfer learning . J Digit Imaging 2020. ; 33 ( 2 ): 431 – 438 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Han X . Automatic liver lesion segmentation using a deep convolutional neural network method . ArXiv 1704.07239 [preprint] https://arxiv.org/abs/1704.07239. Posted April 24, 2017. Accessed October 2021 . [Google Scholar]
- 24. Liang M , Hu X . Recurrent convolutional neural network for object recognition . In: 2015 IEEE conference on computer vision and pattern recognition (CVPR) . Piscataway, NJ: : Institute of Electrical and Electronics Engineers; , 2015. ; 3367 – 3375 . [Google Scholar]
- 25. Raghu M , Zhang C , Kleinberg J , Bengio S . Transfusion: understanding transfer learning for medical imaging . ArXiv 1902.07208 [preprint] https://arxiv.org/abs/1902.07208. Posted February 14, 2019. Accessed October 2021 . [Google Scholar]
- 26. Lundervold AS , Rorvik J , Lundervold A . Fast semi-supervised segmentation of the kidneys in DCE-MRI using convolutional neural networks and transfer learning . Presented at the 2nd meeting of the International Scientific Symposium: functional renal imaging: where physiology, nephrology, radiology and physics meet , Berlin, Germany , October 11–13, 2017 . [Google Scholar]
- 27. Candemir S , Rajaraman S , Thoma G , Antani S . Deep learning for grading cardiomegaly severity in chest x-rays: an investigation . In: 2018 IEEE life sciences conference (LSC) . Piscataway, NJ : Institute of Electrical and Electronics Engineers; , 2018. ; 109 – 113 . [Google Scholar]
- 28. Goodfellow I , Pouget-Abadie J , Mirza M , et al . Generative adversarial nets . In: Advances in neural information processing systems 27 (NIPS 2014) . Red Hook, NY: : Curran Associates; , 2014. ; 2672 – 2680 . [Google Scholar]
- 29. Frid-Adar M , Diamant I , Klang E , Amitai M , Goldberger J , Greenspan H . GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification . Neurocomputing 2018. ; 321 ( 321 ): 331 . [Google Scholar]
- 30. Chuquicusma MJ , Hussein S , Burt J , Bagci U . How to fool radiologists with generative adversarial networks? a visual Turing test for lung cancer diagnosis . In: 2018 IEEE 15th international symposium on biomedical imaging (ISBI 2018) . Piscataway, NJ: : Institute of Electrical and Electronics Engineers; , 2018. ; 240 – 244 . [Google Scholar]
- 31.Calimeri F, Marzullo A, Stamile C, Terracina G. Biomedical data augmentation using generative adversarial neural networks. In: Lintas A, Rovetta S, Verschure P, Villa A, eds.Artificial neural networks and machine learning—ICANN 2017. ICANN 2017. Vol 10614,Lecture notes in computer science.Cham, Switzerland:Springer,2017;626–634. [Google Scholar]
- 32. Shorten C , Khoshgoftaar TM . A survey on image data augmentation for deep learning . J Big Data 2019. ; 6 ( 1 ): 60 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yi X , Walia E , Babyn P . Generative adversarial network in medical imaging: a review . Med Image Anal 2019. ; 58 101552 . [DOI] [PubMed] [Google Scholar]
- 34.Salimans T, Goodfellow I, Zaremba W, Cheung V, Radford A, Chen X. Improved techniques for training GANs. In: Lee D, Sugiyama M, Luxburg U, Guyon I, Garnett R, eds.Advances in neural information processing systems 29 (NIPS 2016).Red Hook, NY:Curran Associates,2016;2234–2242. [Google Scholar]
- 35. Dikici E , Bigelow M , White RD , Erdal BS , Prevedello LM . Constrained generative adversarial network ensembles for sharable synthetic medical images . J Med Imaging (Bellingham) 2021. ; 8 ( 2 ): 024004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Salehinejad H , Colak E , Dowdell T , Barfett J , Valaee S . Synthesizing chest x-ray pathology for training deep convolutional neural networks . IEEE Trans Med Imaging 2019. ; 38 ( 5 ): 1197 – 1206 . [DOI] [PubMed] [Google Scholar]
- 37. Beam AL , Manrai AK , Ghassemi M . Challenges to the reproducibility of machine learning models in health care . JAMA 2020. ; 323 ( 4 ): 305 – 306 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maleki F , Muthukrishnan N , Ovens K , Reinhold C , Forghani R . Machine learning algorithm validation: from essentials to advanced applications and implications for regulatory certification and deployment . Neuroimaging Clin N Am 2020. ; 30 ( 4 ): 433 – 445 . [DOI] [PubMed] [Google Scholar]
- 39. Wong TT . Performance evaluation of classification algorithms by k-fold and leave-one-out cross validation . Pattern Recognit 2015. ; 48 ( 9 ): 2839 – 2846 . [Google Scholar]
- 40. Lampert TA , Stumpf A , Gancarski P . An empirical study into annotator agreement, ground truth estimation, and algorithm evaluation . IEEE Trans Image Process 2016. ; 25 ( 6 ): 2557 – 2572 . [DOI] [PubMed] [Google Scholar]
- 41. Do HM , Spear LG , Nikpanah M , et al . Augmented radiologist workflow improves report value and saves time: a potential model for implementation of artificial intelligence . Acad Radiol 2020. ; 27 ( 1 ): 96 – 105 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang X , Peng Y , Lu L , Lu Z , Bagheri M , Summers RM . ChestX-Ray8: hospital-scale chest x-ray database and benchmarks on weakly supervised classification and localization of common thorax diseases . In: 2017 IEEE conference on computer vision and pattern recognition (CVPR) . Piscataway, NJ : Institute of Electrical and Electronics Engineers; , 2017. ; 2097 – 2106 . [Google Scholar]
- 43.Moradi M, Madani A, Gur Y, Guo Y, Syeda-Mahmood T. Bimodal network architectures for automatic generation of image annotation from text. In: Frangi A, Schnabel J, Davatzikos C, Alberola-López C, Fichtinger G, eds.Medical image computing and computer assisted intervention—MICCAI 2018. MICCAI 2018. Vol 11070,Lecture notes in computer science.Cham, Switzerland:Springer,2018;449–456. [Google Scholar]
- 44. Stember JN , Celik H , Gutman D , et al . Integrating eye tracking and speech recognition accurately annotates MR brain images for deep learning: proof of principle . Radiol Artif Intell 2020. ; 3 ( 1 ): e200047 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gur Y , Moradi M , Bulu H , Guo Y , Compas C , Syeda-Mahmood T . Towards an efficient way of building annotated medical image collections for big data studies . In: Cardoso MJ , Arbel T , Lee SL , et al. , eds. Intravascular imaging and computer assisted stenting, and large-scale annotation of biomedical data and expert label synthesis . Cham, Switzerland: : Springer; , 2017. ; 87 – 95 . [Google Scholar]
- 46. Folio LR , Nelson CJ , Benjamin M , Ran A , Engelhard G , Bluemke DA . Quantitative radiology reporting in oncology: survey of oncologists and radiologists . AJR Am J Roentgenol 2015. ; 205 ( 3 ): W233 – W243 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Beesley SD , Patrie JT , Gaskin CM . Radiologist adoption of interactive multimedia reporting technology . J Am Coll Radiol 2019. ; 16 ( 4 Pt A ): 465 – 471 . [DOI] [PubMed] [Google Scholar]
- 48. Yan K , Wang X , Lu L , Summers RM . DeepLesion: automated mining of large-scale lesion annotations and universal lesion detection with deep learning . J Med Imaging (Bellingham) 2018. ; 5 ( 3 ): 036501 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Radiology preprocessing curriculum. Radiology Preprocessing Course Web site . https://folio47.wixsite.com/rp-course/. Accessed August 24, 2020 . [Google Scholar]
- 50. Xie Q , Luong MT , Hovy E , Le QV . Self-training with noisy student improves ImageNet classification . In: 2020 IEEE/CVF conference on computer vision and pattern recognition (CVPR) . Piscataway, NJ: : Institute of Electrical and Electronics Engineers; , 2020. ; 10684 – 10695 . [Google Scholar]
- 51. Lee DH . Pseudo-label: the simple and efficient semi-supervised learning method for deep neural networks . Presented at the ICML 2013 workshop: challenges in representation learning (WREPL) , Atlanta, Ga , June 16–21, 2013 . [Google Scholar]
- 52. Filipovych R , Resnick SM , Davatzikos C . Semi-supervised cluster analysis of imaging data . Neuroimage 2011. ; 54 ( 3 ): 2185 – 2197 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Signoroni A , Savardi M , Benini S , et al . BS-Net: learning COVID-19 pneumonia severity on a large chest x-ray dataset . Med Image Anal 2021. ; 71 102046 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li Z , Wang C , Han M , et al . Thoracic disease identification and localization with limited supervision . In: 2018 IEEE/CVF conference on computer vision and pattern recognition . Piscataway, NJ: : Institute of Electrical and Electronics Engineers; , 2018. ; 8290 – 8299 . [Google Scholar]
- 55. Kingma DP , Mohamed S , Rezende DJ , Welling M . Semi-supervised learning with deep generative models . In: Advances in neural information processing systems 27 (NIPS 2014) . Red Hook, NY: : Curran Associates; , 2014. ; 3581 – 3589 . [Google Scholar]
- 56. Schlegl T , Seebӧck P , Waldstein SM , Schmidt-Erfurth U , Langs G . Unsupervised anomaly detection with generative adversarial networks to guide marker discovery . In: Niethammer M , Styner M , Aylward S , et al. , eds. Information processing in medical imaging. IPMI 2017 . Vol 10265 , Lecture notes in computer science . Cham, Switzerland: : Springer; , 2017. ; 146 – 157 . [Google Scholar]
- 57. Cheplygina V , de Bruijne M , Pluim JPW . Not-so-supervised: a survey of semi-supervised, multi-instance, and transfer learning in medical image analysis . Med Image Anal 2019. ; 54 ( 280 ): 296 . [DOI] [PubMed] [Google Scholar]
- 58. Li FF , Fergus R , Perona P . One-shot learning of object categories . IEEE Trans Pattern Anal Mach Intell 2006. ; 28 ( 4 ): 594 – 611 . [DOI] [PubMed] [Google Scholar]
- 59.Koch G, Zemel R, Salakhutdinov R. Siamese neural networks for one shot image recognition. Presented at the ICML 2015 deep learning workshop,Lille, France,July 6–11, 2015. [Google Scholar]
- 60. Santoro A , Bartunov S , Botvinick M , Wierstra D , Lillicrap T . One-shot learning with memory-augmented neural networks . ArXiv 1605.06065 [preprint] https://arxiv.org/abs/1605.06065. Posted May 19, 2016. Accessed October 2021. [Google Scholar]
- 61. Medela A , Picon A , Saratxaga CL , et al . Few shot learning in histopathological images: reducing the need of labeled data on biological datasets . In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019) . Piscataway, NJ: : Institute of Electrical and Electronics Engineers; , 2019. ; 1860 – 1864 . [Google Scholar]
- 62. Paul A , Shen TC , Lee S , et al . Generalized zero-shot chest x-ray diagnosis through trait-guided multi-view semantic embedding with self-training . IEEE Trans Med Imaging 2021. ; 40 ( 10 ): 2642 – 2655 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chen MC , Ball RL , Yang L , et al . Deep learning to classify radiology free-text reports . Radiology 2018. ; 286 ( 3 ): 845 – 852 . [DOI] [PubMed] [Google Scholar]
- 64.Sheller MJ, Reina GA, Edwards B, Martin J, Bakas S. Multi-institutional deep learning modeling without sharing patient data: a feasibility study on brain tumor segmentation. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T, eds. Brainlesion: glioma, multiple sclerosis, stroke and traumatic brain injuries. Vol 11383, Lecture notes in computer science. Cham, Switzerland:Springer,2018;92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rieke N , Hancox J , Li W , et al . The future of digital health with federated learning . NPJ Digit Med 2020. ; 3 ( 119 ). 10.1038/s41746-020-00323-1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yang Q , Liu Y , Chen T , Tong Y . Federated machine learning: concept and applications . ACM Trans Intell Syst Technol 2019. ; 10 ( 2 ): 1 – 19 . [Google Scholar]
- 67. Srivastava N , Hinton G , Krizhevsky A , Sutskever I , Salakhutdinov R . Dropout: a simple way to prevent neural networks from overfitting . J Mach Learn Res 2014. ; 15 ( 56 ): 1929 – 1958 . [Google Scholar]
- 68. Ioffe S , Szegedy C . Batch normalization: accelerating deep network training by reducing internal covariate shift . Presented at the ICML 2015 deep learning workshop , Lille, France , July 6–11, 2015 . [Google Scholar]
- 69. Kukačka J , Golkov V , Cremers D . Regularization for deep learning: a taxonomy . ArXiv 1710.10686 [preprint] https://arxiv.org/abs/1710.10686. Posted October 29, 2017. Accessed October 2021. [Google Scholar]
- 70.Chawla NV. Data mining for imbalanced datasets: an overview. In: Maimon O J, Rokach L, eds.Data mining and knowledge discovery handbook. Boston, Mass:Springer,2009;875–886. [Google Scholar]
- 71. He H , Garcia EA . Learning from imbalanced data . IEEE Trans Knowl Data Eng 2009. ; 21 ( 9 ): 1263 – 1284 . [Google Scholar]
- 72. Pouyanfar S , Tao Y , Mohan A , et al . Dynamic sampling in convolutional neural networks for imbalanced data classification . In: 2018 IEEE conference on multimedia information processing and retrieval (MIPR) . Piscataway, NJ : Institute of Electrical and Electronics Engineers; , 2018. ; 112 – 117 . [Google Scholar]
- 73. Mullick SS , Datta S , Das S . Generative adversarial minority oversampling . In: 2019 IEEE/CVF international conference on computer vision (ICCV) . Piscataway, NJ : Institute of Electrical and Electronics Engineers; , 2019. ; 1695 – 1704 . [Google Scholar]
- 74. Cai Z , Wang X , Zhou M , Xu J , Jing L . Supervised class distribution learning for GANs-based imbalanced classification . In: 2019 IEEE international conference on data mining (ICDM) . Piscataway, NJ : Institute of Electrical and Electronics Engineers; , 2019. ; 41 – 50 . [Google Scholar]
- 75. Krawczyk B . Learning from imbalanced data: open challenges and future directions . Prog Artif Intell 2016. ; 5 ( 4 ): 221 – 232 . [Google Scholar]
- 76. Lin TY , Goyal P , Girshick R , He K , Dollar P . Focal loss for dense object detection . In: 2017 IEEE international conference on computer vision (ICCV) . Piscataway, NJ : Institute of Electrical and Electronics Engineers; , 2017. ; 2980 – 2988 . [Google Scholar]