Abstract
The coronavirus disease 2019 (COVID-19) pandemic unfolded due to the widespread severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) transmission reinforced the urgent need for affordable molecular diagnostic alternative methods for massive testing screening. We present the clinical validation of a pH-dependent colorimetric reverse transcription loop-mediated isothermal amplification (RT-LAMP) for SARS-CoV-2 detection. The method revealed a limit of detection of 19.3 ± 2.7 viral genomic copies/μL when using RNA extracted samples obtained from nasopharyngeal swabs collected in guanidine-containing viral transport medium. Typical RT-LAMP reactions were performed at 65°C for 30 min. When compared to reverse transcriptase–quantitative polymerase chain reaction (RT-qPCR), up to cycle-threshold (Ct) value 32, RT-LAMP presented 98% [95% confidence interval (CI) = 95.3–99.5%] sensitivity and 100% (95% CI = 94.5–100%) specificity for SARS-CoV-2 RNA detection targeting E and N genes. No cross-reactivity was detected when testing other non–SARS-CoV virus, confirming high specificity. The test is compatible with primary RNA extraction–free samples. We also demonstrated that colorimetric RT-LAMP can detect SARS-CoV-2 variants of concern and variants of interest, such as variants occurring in Brazil named gamma (P.1), zeta (P.2), delta (B.1.617.2), B.1.1.374, and B.1.1.371. The method meets point-of-care requirements and can be deployed in the field for high-throughput COVID-19 testing campaigns, especially in countries where COVID-19 testing efforts are far from ideal to tackle the pandemics. Although RT-qPCR is considered the gold standard for SARS-CoV-2 RNA detection, it requires expensive equipment, infrastructure, and highly trained personnel. In contrast, RT-LAMP emerges as an affordable, inexpensive, and simple alternative for SARS-CoV-2 molecular detection that can be applied to massive COVID-19 testing campaigns and save lives.
Keywords: COVID-19, RT-LAMP, SARS-CoV-2, molecular test, respiratory virus, diagnostic test
Introduction
Emerging viral infections continue to pose a major threat to global public health. In the past decades, different viral emergencies have been reported including the severe acute respiratory syndrome coronavirus (SARS-CoV), H1N1 influenza, Middle East respiratory syndrome coronavirus, Ebola vírus, Zika virus, and most recently, the new coronavirus has been described, which cause coronavirus disease 2019 (COVID-19; Wang et al., 2020; Zhu et al., 2020). COVID-19’s etiologic agent is SARS-CoV-2, which belongs to the Coronaviridae family, Betacoronavirus genus (Gorbalenya et al., 2020; Rambaut et al., 2020). People with COVID-19 have a wide range of symptoms reported such as fever, cough, anosmia, ageusia, headache, fatigue, muscle or body aches, sore throat, and shortness of breath or difficulty breathing. Some of these symptoms help spread the virus; however, human-to-human transmission from infected individuals with no or mild symptoms has been extensively reported (Bai et al., 2020; Rothe et al., 2020). This outbreak has spread rapidly; as of September 2021, there were more than 230 million confirmed COVID-19 cases with more than 4.7 million deaths recorded worldwide1. Isolation and quarantine of infected individuals are essential to viral spread and community dissemination of airborne pathogens and require an accurate, fast, affordable, readily available tests for massive population testing. In contrast to antibody detection, which may take weeks after the onset of the infection, detection of viral RNA is the best way to confirm the acute infection phase, the most important phase for viral shedding, so that rationally managed social distancing and lockdown can be implemented (Long et al., 2020; Wang et al., 2020).
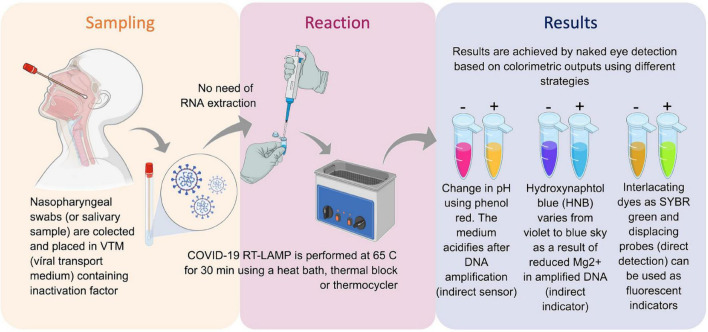
Reverse transcriptase–quantitative polymerase chain reaction (RT-qPCR) is considered the gold-standard method for SARS-CoV-2 RNA detection, mainly targeting combinations of viral genome regions that codes for nucleocapsid protein (N), envelope protein (E), RNA-dependent RNA polymerase (RdRp), and other targets on the open reading frame (ORF1ab; Esbin et al., 2020). Although RT-qPCR assays have played an important role in the SARS-CoV-2 diagnosis, the technique has limitations for massive population testing such as processing time; it requires sophisticated equipment, infrastructure, and highly trained staff, as well as costly reagents with high demand and shortages around the world. Thus, developing complementary, inexpensive point-of-care (PoC) methods that are rapid and simple and allowing the use of alternative reagents for COVID-19 diagnosis test are urgently needed. Methods gathering these features can make affordable massive testing campaigns, including contact tracing strategies in highly dense countries, saving lives (Baek et al., 2020; Dudley et al., 2020; Song et al., 2021; Godfrey et al., 2020; Park et al., 2020; Wang, 2020; Yan et al., 2020; Yu et al., 2020; Anahtar et al., 2021). In this regard, reverse transcription loop-mediated isothermal amplification (RT-LAMP) has been shown to be an affordable technique applied to detect different pathogens (Mori and Notomi, 2009; Li et al., 2017). RT-LAMP has been used during Ebola outbreak (Kurosaki et al., 2016a,b) and for tracking Zika virus (da Silva et al., 2019) or Wolbachia (Gonçalves et al., 2014) in Brazilian mosquitoes. The method relies on specific DNA amplification at constant temperature without the need for sophisticated thermal cyclers (Zhang et al., 2020a). The amplified products can be visually detected through magnesium pyrophosphate precipitation, fluorescence emission from DNA intercalating dyes, agarose gel electrophoresis, lateral flow immunochromatography, magnesium chelating color indicators (Bhadra et al., 2021), and pH-dependent colorimetric reaction that changes from fuchsia (pink) to yellow (positive result) due to proton release during nucleic acid amplification (Tanner et al., 2015; Figure 1). The possibility of accessing results by the naked eye made RT-LAMP an exciting alternative that facilitates the use of COVID-19 molecular testing. Simple, scalable, cost-effective RT-LAMP–based alternatives for SARS-CoV-2 detection have emerged during pandemics including protocols for viral inactivation, quick run, RNA extraction–free and LAMP-associated CRISPR/Cas strategies (Baek et al., 2020; Broughton et al., 2020; Chow et al., 2020; Dudley et al., 2020; Song et al., 2021; Godfrey et al., 2020; Joung et al., 2020; L’Helgouach et al., 2020; Park et al., 2020; Rabe and Cepko, 2020; Bektaş et al., 2021; Bokelmann et al., 2021). On April 14, 2020, the RT-LAMP received the emergency use authorization from the United States Food and Drug Administration (FDA) for SARS-CoV-2 detection in COVID-19 diagnostics.
FIGURE 1.
Reverse transcription loop-mediated isothermal amplification (RT-LAMP) for SARS-CoV-2 RNA detection and COVID-19 testing. Inactivated saliva samples or from nasopharyngeal swabs can processed for RNA extraction previously or be directly added to RT-LAMP reaction. Colorimetric output can be achieved by different sensors and can be read by naked eye. The whole procedure is rapid and simple and does not require complex infrastructures. Created with biorender.com.
In this study, we optimized and validated a colorimetric RT-LAMP assay to detect SARS-CoV-2 RNA in clinical samples collected in different parts of Brazil, including samples with known SARS-CoV-2 variants of interest (VOIs) and concern (VOCs). After testing different primer sets for SARS-CoV-2 RNA detection by RT-LAMP, best results were achieved when using N gene or N/E genes-based strategies. 367 nasopharyngeal swabs collected in a guanidine-containing viral transport medium (VTM; Faria et al., 2021) from suspect patients were tested. The clinical validation revealed a sensitivity of 98% [95% confidence interval (CI) = 95.3–99.5%] with samples of cycle-threshold (Ct) values ranging from 15 to 32 with 100% specificity. We also demonstrated that RT-LAMP is affordable for the detection of more transmissible SARS-CoV-2 variants encompassing a number of genomic nucleotide changes. Part of the results presented here is the research basis of OmniLAMP® SARS-CoV-2 kit, which was approved by the Brazilian Heath Regulatory Agency for COVID-19 molecular testing (Anvisa no: 10009010368) as an alternative for massive decentralized diagnostic in Brazil, which records the third-highest number of COVID-19 cases worldwide (see text footnote 1). Together with vaccination, RT-LAMP for COVID-19 diagnosis could help to improve better life quality during the pandemic, offering an alternative molecular testing for monitoring lockdown measures; traveling restrictions; the return of universities, schools, kindergartens; and sport league activities with worldwide impact.
Results
Reverse Transcription Loop-Mediated Isothermal Amplification Targeting SARS-CoV-2 N/E Genes Can Detect as Low as 19 Viral Copies/μL
In order to access absolute analytical sensitivity of the colorimetric RT-LAMP for SARS-CoV-2 detection, we calculated the limit of detection (LoD), which is the lowest detectable concentration of viral nucleic acid, here represented in viral copies per microliter (/μL), which was determined based on a calibration curve from a known copy number load standard E gene-harboring plasmid. Purified SARS-CoV-2, obtained from infected Vero E6 cells, revealed an LoD equivalent to 0.44 ± 0.2 copies/μL, whereas RNA obtained from clinical samples (nasopharyngeal swab in VTM) resulted in an LoD of 19.3 ± 2.7 copies/μL. Validation was performed using clinical samples, confirming the LoD by colorimetric RT-LAMP, as well as by the visualization of the amplified DNA in agarose gel (Figure 2).
FIGURE 2.
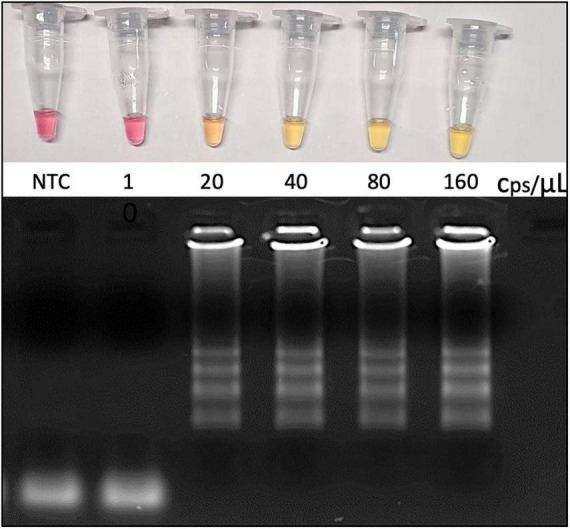
Analytical sensitivity as revealed by the limit of detection (LoD). RNA was extracted from VTM-nasopharyngeal swab, and the genome viral copies input was calculated based on SARS-CoV-2 E gene-harboring plasmid (Bioclin #K228-1) calibration curve. RT-LAMP reaction was performed at 65°C during 30 min using WarmStart® colorimetric master LAMP mix (NEB #M1800) in 20 μL final volume (upper panel). Amplicons were resolved in 2% agarose gel and stained with GelRed® (Biotium #41003) to confirm DNA amplification (bottom panel). cps/μL, viral genome copies per microliter; NTC, nontemplate control; VTM, viral transport medium (Bioclin #G092-1).
SARS-CoV-2 Detection by Reverse Transcription Loop-Mediated Isothermal Amplification on Clinical Samples Presents 100% Specificity, Whereas Sensitivity Varies From 100 to 84%, Depending on the Viral Load
The diagnostic accuracy for RT-LAMP was compared to the “gold-standard” technique RT-qPCR. The relative sensitivity was accessed in a panel of 367 clinical specimens from nasopharyngeal swab collected in VTM, including 254 positive and 113 negative samples according to the colorimetric RT-LAMP output that were previously characterized by RT-qPCR (Table 1). The colorimetric output was correlated with the visualization of amplified DNA after agarose gel electrophoresis (Figure 3).
TABLE 1.
Estimated values comparing clinimetric parameters between colorimetric RT-LAMP and RT-qPCR on the detection of SARS-CoV-2 for molecular diagnosis of COVID-19.
| RT-qPCR | Colorimetric RT-LAMP |
Metrics % (95% CI) |
|||||
| Ct value | Positive | Negative | Sensitivity | Specificity | Accuracy | PPV | NPV |
| 15–30 | 171 | 0 | 100 (98–100) | 100 (94.5–100) | 100 | 100 | 100 |
| 15–32 | 199 | 4 | 98 (95–99.5) | 100 (94.5–100) | 99.95 | 100 | 99.95 (99.8–100) |
| 15–34 | 221 | 13 | 94 (90.7–97) | 100 (94.5–100) | 99.90 | 100 | 99.9 (99.7–100) |
| 15–36 | 245 | 29 | 89 (85.1–93) | 100 (94.5–100) | 99.74 | 100 | 99.7 (99.6–99.8) |
| 15–40 | 254 | 48 | 84 (79.4–88) | 100 (94.5–100) | 99.60 | 100 | 99.6 (99.5–99.7) |
| Negative | 0 | 65 | |||||
Sensitivity: probability that the test result will be positive when the disease is present (true positive rate) = true positive/(true positives + false negatives); Specificity: probability that a test result will be negative when the disease is not present (true-negative rate) = true negatives/(true negatives + false positives); accuracy, PPV, and NPV depending on COVID-19 disease prevalence that was considered here as 2.5% according to the average value of two surveys during May and June 2020 (Hallal et al., 2020). PPV is the probability that the disease is present when the test is positive, whereas NPV is the probability that the disease is not present when the test is negative, and both are calculated as follows: PPV = sensitivity × prevalence/sensitivity × prevalence + (1 – specificity) × (1 – prevalence); NPV = specificity × (1 – prevalence)/(1 – sensitivity) × prevalence + specificity × (1 – prevalence); accuracy is the overall probability that a patient is correctly classified and is calculated as follows: =sensitivity × prevalence + specificity × (1 – prevalence). All calculations were performed using MedCalc (https://www.medcalc.org/) and VassarStats—Clinical Research Calculators (http://vassarstats.net/).
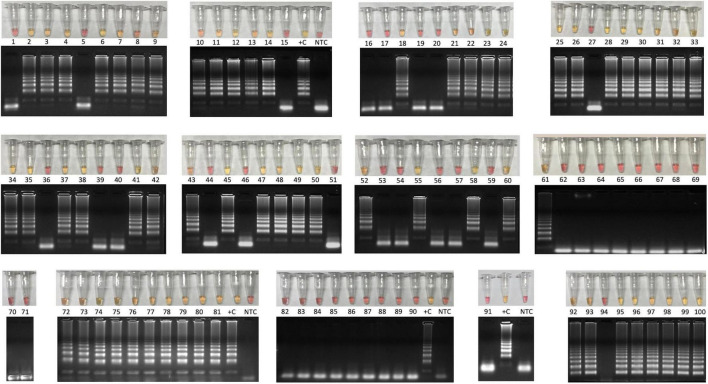
FIGURE 3.
Colorimetric RT-LAMP for COVID-19 diagnosis validation using 100 clinical samples. Clinical samples were collected from symptomatic and hospitalized patients by nasopharyngeal swabs in partnership with CT-Vacinas/UFMG, Belo Horizonte, Brazil. Samples were obtained from different parts including Brazilian Southeast and Northeast regions. The reaction was performed at 65°C during 30 min using WarmStart® colorimetric LAMP master mix (NEB #M1800) in 20 μL final volume. The RT-LAMP reaction targeted SARS-CoV-2 N gene. Yellow content indicates positive reaction, whereas the pink pattern reveals nonreagent samples. Amplicons were resolved in 2% agarose gel and stained with GelRed® (Biotium #41003) to confirm DNA amplification. Latter pattern confirmed specific SARS-CoV-2 amplification that matches with yellow output tubes, which is not observed in pink nonreagent tests. +C, positive control using RNA extracted from laboratory-Vero E6 cultured inactivated SARS-CoV-2; NTC, nontemplate control. Clinimetric parameters from these samples are presented in Supplementary Figure S1.
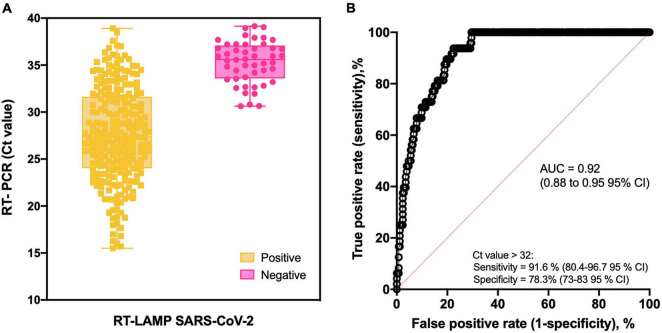
The overall accuracy of colorimetric RT-LAMP compared to RT-qPCR was 99%, considering Ct values ranging from 15 to 40, with relative sensitivity of 84% (95% CI = 79.4–88%) and 100% (95% CI = 94.5–100%) specificity (Table 1). However, considering samples with equivalent RT-qPCR Ct value ≤ 32, RT-LAMP sensitivity is 98% (95% CI = 95.3–99.5%) and reaches 100% (95% CI = 94.5–100%) in samples with Ct value ≤ 30, whereas specificity is always 100% (Table 1), which means there are no false-positive hits. It is noteworthy that Ct > 32 RT-LAMP starts to present false-negative outputs (Table 1 and Figure 4); however, 55 samples were detected as positive on RT-LAMP with RT-qPCR Ct values ranging from 32 to 39 (Figure 4A). Receiver operating characteristic curve confirmed high sensitivity at RT-PCR equivalent Ct value > 32 for RT-LAMP on COVID-19 diagnostics (Figure 4B). The aforementioned results were achieved when using a multiplexed set of primers targeting E and N genes combined. However, prior to this, we performed the evaluation of N gene alone in 100 clinical samples (60 positive and 40 negative results) derived from hospitalized patients (Supplementary Figure 1). In this set of samples, we were also able to validate high sensitivity/specificity, absence of cross-reactivity with non–SARS-CoV viruses, and the capacity of SARS-CoV-2 variant detection (Supplementary Figure 1).
FIGURE 4.
Colorimetric RT-LAMP for SARS-CoV-2 RNA detection. (A) Box-and-whisker representation of colorimetric RT-LAMP SARS-CoV-2–positive and –negative output (x axis) plotted in function of their respective RT-PCR Ct values (y axis). Forty-eight false negative samples were detected on RT-LAMP after Ct 32 despite other 55 being positive from Cts ranging from 32 to 39. (B) Receiver operating characteristic (ROC) curve constructed based on data presented in A. As summarized in Table 1, high-sensitivity values were obtained at the predicted cutoff.
Reverse Transcription Loop-Mediated Isothermal Amplification Targeting SARS-CoV-2 Does Not Cross-React With Other Viruses, Including Respiratory Ones
The analytical specificity was confirmed by performing RT-LAMP for SARS-CoV-2 on putative cross-reacting viruses such as pathogens that colonize the human upper respiratory tract or that are associated with seasonal outbreaks in Brazil. None of the tested viruses [human influenza A virus/H1N1, influenza B virus, human respiratory syncytial virus (hRSV), dengue, Zika, Chikungunya, and yellow fever viruses] presented cross-reactivity on RT-LAMP using E an N gene as SARS-CoV-2 target (Figure 5). Similar results were obtained when using N gene alone as target (Supplementary Figure 1E). It reinforces the high specificity observed on clinical validation with no false-positive results (Figure 3). Thermodynamic and alignment analyses were performed on SARS-CoV-2 N, E, and RdRp RT-LAMP primer sets, revealing that there is no cross-reactivity over more than 300 non–SARS coronaviruses–derived genomes (Supplementary Table 1).
FIGURE 5.
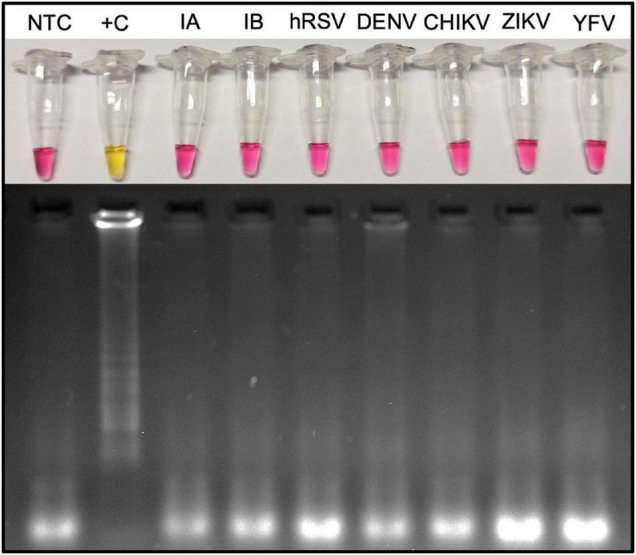
Microbial cross-reactivity assay to test SARS-CoV-2 RT-LAMP analytical sensitivity. The test was performed using potentially cross-reacting respiratory viruses or local occurring arboviruses. RT-LAMP reaction was performed at 65°C during 30 min, with additional 10 min, to confirm the absence of cross-reactivity when targeting SARS-CoV-2 E and N genes. The assay was performed using the WarmStart® colorimetric LAMP 2× master mix (NEB #M1800). Yellow (positive) reaction is observed only when the template is SARS-CoV-2 viral RNA. hRSV, human respiratory syncytial virus; NTC, nontemplate control; M, molecular size marker. RT-LAMP amplification products were resolved in 2% agarose gel and stained with GelRed® (Biotium #41003) to confirm DNA amplification. DENV3, dengue virus serotype 3; ZIKV, Zika virus; CHIKV, Chikungunya virus; YFV, yellow fever virus; Influenza A (H1N1/H3N2); and influenza B (Yamagata/Victoria).
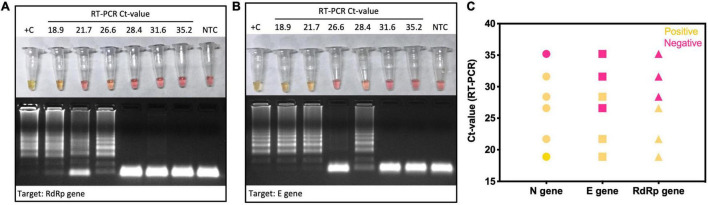
Six clinical samples previously confirmed as SARS-CoV-2 positive by RT-qPCR were subclassified as presenting low, medium, or high Ct values targeting E gene. All of them were tested by colorimetric RT-LAMP in independent reactions to test the performance of N, E, and RdRp genes as target to detect SARS-CoV-2. The samples with low Ct values (18.9 and 21.7) were positive for all tested primer sets, whereas E and RdRp genes started to present false-negative results from medium (26.6 and 28.4) Ct values (Figure 6). It indicates that the SARS-CoV-2 N gene is a better target for colorimetric RT-LAMP, detecting viral RNA in samples with equivalent RT-qPCR Ct values > 30 (Figure 6).
FIGURE 6.
Colorimetric RT-LAMP for SARS-CoV-2 detection using genes N, E, and RdRp as target. Selected SARS-CoV-2–positive clinical samples by RT-qPCR were classified as low (Ct 18.9 and 21.7), medium (Ct 26.6 and 28.4), and high (Ct 31.6 and 35.2) Ct values for E gene. They were included as input for colorimetric RT-LAMP reaction using primers targeting N, RdRp (A), and E genes (B). RT-LAMP SARS-CoV-2 false-negative samples were more frequent when using E and RdRp genes as target (C). RT-LAMP reaction was performed at 65°C during 30 min, using the WarmStart® colorimetric LAMP 2× master mix (NEB #M1800). RT-LAMP amplification products were resolved in 2% agarose gel and stained with GelRed® (Biotium #41003) to confirm DNA amplification. +C, positive control using SARS-CoV-2 RNA extracted from laboratory-cultured inactivated SARS-CoV-2; NTC, nontemplate control.
Colorimetric RT-LAMP sensitivity depends on the set of LAMP primers that can vary even within the same target. When RT-LAMP was performed on low viral load samples (Ct value for E gene ranging from 31.8 to 36.2), the N gene_Set1 was able to identify 4 of 12 (33.3%) true-positive samples. In contrast, N gene_Set2 or primer multiplex strategy (N gene Set1/Set2) allowed the detection of 11 of 12 (91.6%) true-positive samples (Supplemental Table 2).
Colorimetric Reverse Transcription Loop-Mediated Isothermal Amplification Can Be Performed on Clinical Samples Without RNA Extraction
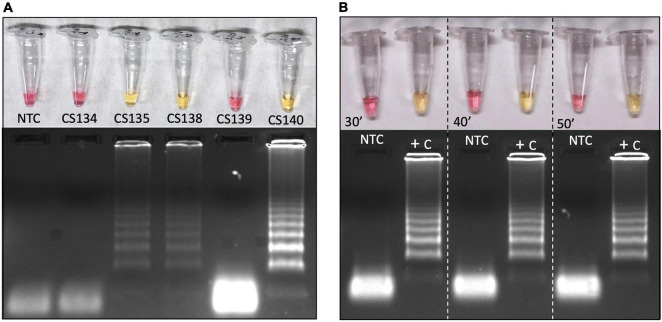
Reverse transcription loop-mediated isothermal amplification performed in clinical samples, without any chemical or physical pretreatment or RNA extraction, returned positive output color in three of five samples (Figure 7A). In this assay, we used laboratory-cultured and inactivated SARS-CoV-2 and clinical samples without previous RNA extraction, showing that it is possible to use direct patients’ samples without preprocessing (Figure 7A). However, this should be taken with caution, as crude clinical samples may contain interferents that can block RT-LAMP reaction. Previous heat inactivation can be used to reduce this possibility. Here, only 1 μL of 1:10 solution of hydrochloride guanidine-containing VTM from nasopharyngeal swabs was added as a template to the SARS-CoV-2 LAMP reaction. Further analyses are being performed to establish the method sensitivity and feasibility for massive patient screening. All five samples had previous RNA extraction, for RT-PCR analysis, supporting that extraction process can increase detection sensitivity. We also tested the incubation time at 65°C reaction temperature. All SARS-CoV-2 control samples turned reaction color from fuchsia to yellow as indicative of DNA amplification, confirming positive reaction from the earliest time point tested (Figure 7B). In all tested intervals nontemplate controls were pink/fuchsia (negative) as expected, without any spurious late amplification, as confirmed by agarose gel electrophoresis showing no amplification bands on it (Figure 7B).
FIGURE 7.
Colorimetric RT-LAMP to detect SAR-CoV-2 in RNA extraction–free clinical samples (A) or laboratory-cultured virus (B). Clinical samples were derived from nasopharyngeal swabs placed on guanidine-containing viral transport medium, diluted 1:10. The RT-PCR Ct values for SARS-CoV-2 based on E gene are as follows: CS134 = 31.8, CS135 = 15.3, CS138 = 18.4, CS139 = 21.7, and CS140 = 24.6. RT-LAMP reaction was performed in 20 μL final volume, incubated at 65°C during 30, 40, or 50 min (inactivated virus) using WarmStart® colorimetric LAMP master mix (NEB #M1800). Both clinical samples and viruses are RNA extraction–free samples. The amplification products (amplicons) were migrated in agarose gel at 2% to confirm amplification, as indicated by the characteristic ladder highlighted by GelRed® staining. NTC, nontemplate control; CS, clinical sample; and +C, positive control.
Colorimetric Reverse Transcription Loop-Mediated Isothermal Amplification Allows the Detection of New SARS-CoV-2 Variants of Interests and Variants of Concern
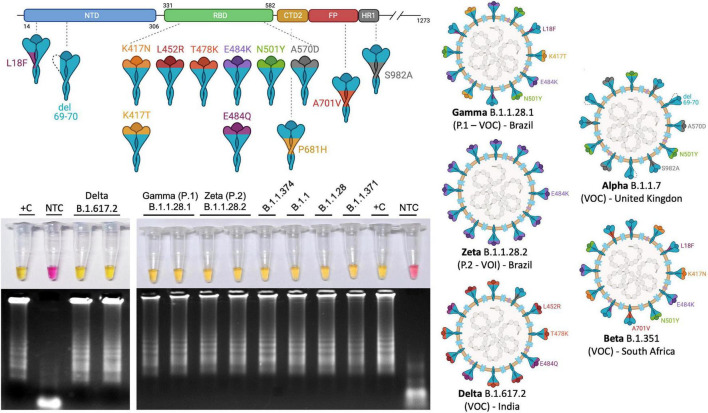
As a worldwide concern, SARS-CoV-2 VOI and VOC molecular detection could fail when applying S region–based RT-qPCR diagnostic methods due to mutations that would prevent primer annealing. In order to provide experimental evidences that RT-LAMP is a powerful molecular tool for detecting SARS-CoV-2 RNA, including VOCs and VOIs, we performed the tests on clinical samples that were previously identified as VOCs/VOIs by complete genome sequencing. All tested variants, including gamma (P.1 or B.1.1.28.1) and zeta (P.2 or B.1.1.28.2), originally reported in Brazil, and delta (B.1.167.2), first detected in India, were detected in colorimetric SARS-CoV-2 RT-LAMP (Figure 8), either by N gene alone as target (Supplementary Figure 1D) or by multiplex strategy using N2/E1 primer set, indicating that none of the mutant polymorphisms prevent specific primer annealing on RT-LAMP COVID-19 diagnosis (Figure 8).
FIGURE 8.
Colorimetric RT-LAMP allows the detection of SARS-CoV-2 VOCs and VOIs. RT-LAMP reaction was performed at 65°C for 30 min, using the WarmStart® colorimetric LAMP 2× master mix (NEB #M1804), using multiplex N2/E1 primer sets. The amplicons were migrated in agarose gel at 2% to confirm amplification, as indicated by the characteristic ladder highlighted by GelRed® staining. NTC, nontemplate control; CS, clinical sample; and +C, positive control. The top panel shows a schematic representation of SARS-CoV-2 spike protein (upper) and where the main mutations are highlighted and represented in SARS-CoV-2 virions (right hand side) present in VOC gamma (B.1), delta (B.1.167.2), and VOI zeta (P.2). The VOCs alpha (B.1.1.7) and beta (B.1.3.51), first reported in the United Kingdom and South Africa, respectively, are also represented. K417N: lysine-to-asparagine substitution at position 417 of spike protein at the receptor biding domain (RBD); V445A: valine-to-alanine substitution at position 445 and so on. L, leucine; Q, glutamine; E, glutamic acid; Y, tyrosine; T, threonine; P, proline; H, histidine; D, aspartic acid; S, serine; F, phenylalanine. del, deletion. Segments of SARS-CoV-2 protein NTD, N-terminal domain; CTD2, C-terminal domain 2 or C terminus of S1 fragment after furin cleavage; FP, fusion peptide; HR1, heptad repeat region 1. SARS-CoV-2 variants were previously sequenced. Variants of interest B.1.1.371 and B.1.1.374 were first reported in Saudi Arabia and Finland, respectively, (https://cov-lineages.org/). Created with biorender.com.
Discussion
The COVID-19 pandemics demanded a rapid global response in massive diagnostic solution to face the worldwide crisis. In this context, the RT-qPCR—considered the gold-standard technique for SARS-CoV-2 RNA detection—requires high-cost equipment, trained staff, and specialized laboratory infrastructure. In addition, during the COVID-19 pandemic, several health care centers and private laboratories competed for RT-qPCR kits and related products to meet the high diagnostic demand. In order to overcome the lacking of molecular testing and provide affordable alternatives, RT-LAMP had become one of the main hopes. Because of its simplicity, accuracy comparable with RT-qPCR to detect SARS-CoV-2 RNA, the fact that it does not require PCR machine, and for offering a naked eye readable colorimetric output, RT-LAMP is the focus of massive testing campaigns (Chow et al., 2020; Dudley et al., 2020; Song et al., 2021). This screening strategy is compatible with home, primary care clinics, point of entry (borders), schools, universities, sport leagues, and companies and can help to achieve a safe back-to-work and quarantine monitoring (Chow et al., 2020; Dudley et al., 2020; Song et al., 2021; Godfrey et al., 2020; Bokelmann et al., 2021). Since April 14, 2020, the United States FDA issued the emergency use authorization of Color SARS-CoV-2 RT-LAMP Diagnostic Assay from Color Health, Inc. (EUA no. EUA200539).
In order to provide an affordable SARS-CoV-2 detection tool, we validate a colorimetric RT-LAMP for the COVID-19 diagnosis using clinical samples collected from different parts of Brazil. The country has a flawed screening performance, testing fewer than 220 individuals per 1,000 people (May 2021)2 where the majority of tests rely on antibodies-based rapid tests, which are not the most reliable and recommended for mass screening and decision making to control local outbreaks. The test sensitivity of RT-LAMP is comparable to the gold standard RT-qPCR and clearly relies on the target choice, incubation time, viral load (asymptomatic patients, days of symptoms, and correct sampling), output reading, sample integrity, and quality (viral transport media, sample storage condition, preanalytical treatments, extraction procedure, and crude RNA extraction–free samples), and sample type (nasal, nasopharyngeal, saliva, sputum, and gargle lavage; Table 2).
TABLE 2.
Comparison of SARS-CoV-2 RT-LAMP solutions, including key parameters on clinical validation.
| Commercial name or acronym | Sample source | Transport medium | Target | Internal control | RNA extraction | Kit/output | Program | Sensitivity/specificity | LoD | Clinical sample tested | Local | References |
| OmniLAMP | Nasopharyngeal | VTM | N, E and RdRp genes | NA | Yes | NEB #M1800; #M1804 Color | 65°C/30 min | 100%/100% Up to RT-qPCR Ct value 30 | 20 copies/μL using clinical samples | 467 | CT-Vacinas-Fiocruz/UFMG, Belo Horizonte, Brazil | This study |
| Nasopharyngeal | Saline | N gene and ORF1a | Human actin B gene | No | NEB #M1800 Color | 65°C/30 min | 87.5%/100% | 25 copies/μL | 62 | Massachusetts General Hospital, Boston, MA, United States | Anahtar et al., 2021 | |
| Yes | 90%/100% | 40 | ||||||||||
| NI | NI | N gene; ORF1ab | NI | Yes | NEB #M1800 Color | 65°C/30 min | 100%/100% | 240 copies/Rx | 62 | Paraná Central Laboratory, Curitiba, Brazil | Aoki et al., 2021 | |
| Saliva | NA | N gene | NI | No, heated | NI | 63°C/30 min | 78.9%/100% | NI | 244 | Sírio-Libanês Hospital, São Paulo, Brazil | Asprino et al., 2020 | |
| Nasal | NI | N gene | NI | Yes | NEB #M1800 Color | 65°C/30 min | NI | 10–7 (equivalent to Ct 34 in RT-PCR) | 14 | National Medical Center, Republic of Korea | Baek et al., 2020 | |
| ALERT | Nasal, saliva | PBS | N gene | BPIFA1 gene | Yes or without extraction (lysate samples) | NEB Bst 3.0 and RTx; #M1800 Fluorescence | 63°C/45 min | 95%/97%–100% | 2 copies/μL | 47 | Hôpital Saint Louis, Paris, France; Pontifica Universidad Católica, Santiago, Chile | Bektaş et al., 2021 |
| Throat and nasopharyngeal | UTM | N gene; | NI | No (lysate samples) | NEB #M1800 | 65°C/30–40 min | 71.15%/96.77% (30 min) 76,9%/96/77% (35 or 40 min) | NI | 180 | Rambam Health Care Campus, Haifa, Israel | Ben-Assa et al., 2020 | |
| LAMP-OSD | Nasopharyngeal and oropharyngeal and SARS-CoV-2 spiked saliva | N gene; ORF1ab (NSP3 and RdRp genes) | NI | Yes | NEB Bst 2.0 polymerase; WarmStart RTx + betaine and additional MgCl2 + FAM Fluorescence | 65°C/90 min | NI | 10 copies/Rx | NA | NA | Bhadra et al., 2021 | |
| Cap-iLAMP | Gargle lavage | NA | N gene; ORF1ab | No, heated samples | NEB #M1800 Color + SYTO9 Fluorescence | 65°C/25–30 min | 97,1%/ | 500 copies/Rx | 192 | NI | Bokelmann et al., 2021 | |
| Nasopharyngeal and oropharyngeal | BD UVTM | N gene and E gene | Human actin B gene | Yes | NEB #M1800 Color + QuantiFluor (Fluorescence) | 65°C/30–40 min | 95.6%/99.2% | 8 copies/μL | 857 | New York Presbyterian Hospital Weill Cornell Medical Center, NY, United States | Butler et al., 2020 | |
| COVID-19-LAMP | Nasopharyngeal, sputum, and throat | ORF3a, E gene | NI | Yes | NEB colorimetric WarmStart | 63°C/60–90 min | 98.2%/100% | 42 copies/Rx | 223 | University of Hong Kong Hospital, China | Chow et al., 2020 | |
| Nasal and oral | PBS | ORF1a | human 18S RNA | Yes | NEB colorimetric WarmStart | 63°C/30 min | 93.8%/90.4% | 100 copies/Rx up to Ct 35 | 466 | Erasto Gaertner Hospital, Curitiba, PR, Brazil | de Oliveira Coelho et al., 2021 | |
| Nasopharyngeal | SPS | N gene and ORF1ab | NI | Yes | NEB Bst 2.0, 3.0, RTx WarmStart +EvaGreen Color and Fluorescence | 8.3%–100%/100% | 200 copies/Rx | University Hospital of Salamanca, Spain | García-Bernalt Diego et al., 2021 | |||
| Nasopharyngeal | UTM, VTM or PBS | ORF1a | NI | No | NEB #E1700 Fluorescence | 63°C/40 min | 81%/100% | 62.5 copies/μL | 137 | University of Wisconsin – Madison Hospital and Clinics, United States | Dudley et al., 2020 | |
| Penn-RAMP RPA+ LAMP | Nasal (spiked samples) | NA | ORF1ab | NI | NA | OptiGene Isothermal Mastermix (ISO-001) + EvaGreen dye Loopamp 2019-SARS-CoV-2 Detection Reagent Kit (Eiken Chemical, Tokyo, Japan) + Leuco Crystal Violet | 63°C/50 min | 100%/NI | 7 copies/Rx | NA | NA | Song et al., 2021 |
| Saliva; throat and nasal | VTM | N gene | NI | No, chelating agent treatment | NEB Color | 65°C/30 min | 90%100% | 105 copies/mL | 62 | Rambam Health Care Campus in Haifa, Israel | Flynn et al., 2020 | |
| OptiGene COVID-19 RT-LAMP | Nasopharyngeal | VTM | ORF1a | NI | Yes, also tested without RNA extraction | OptiGene GspSSD 2.0 Opti-RT Fluorescence | 65°C/20 min | 97%/99% | 100–200 copies/Rx | 196 | Hampshire Hospitals NHS Foundation Trust, United Kingdom | Fowler et al., 2021 |
| Nasopharyngeal | VTM | ORF1a, ORF8, S and N genes | No | NEB Bst 2.0 +EvaGreen Fluorescence | 65°C/60 min | 100%/100% | 50 copies/μL | 20 | OSF Healthcare. Peoria, IL, United States | Ganguli et al., 2020 | ||
| Nasopharyngeal | NI | N gene | NI | Yes | NEB #M1800 Color +EvaGreen Fluorescence | 65°C/50 min | NI | 625 copies/Rx | 14 | Hospital Alfa Medical Center, Guadalupe, México | González-González et al., 2021 | |
| Throat | VTM | ORF1ab, S gene and N gene | Human actin B gene | Yes | NEB #M1800 Color | 65°C/30 min | NI | 2 copies/25 μL | 16 | Shenzhen Luohu People’s Hospital in China. | Huang et al., 2020 | |
| Nasopharyngeal | BD UVTM | NI | NI | Yes | SARS-CoV-2 detection kit (Eiken Chemical Co.) Turbidimetry Fluorescence | 62.5°C/35 min | 56.6%/98.4% | 6.7 copies/Rx | 124 | University Hospital, Japan | Inaba et al., 2021 | |
| Nasopharyngeal | NI | NI | NI | Yes | Loopamp 2019-SARS-CoV-2 Detection Reagent Kit (Eiken Chemical, Tokyo, Japan) Turbidity | 62.5°C/35 min | 100%/97.6% | 101 copies/μL | 76 | National Institute of Infectious Diseases, Japan | Kitagawa et al., 2020 | |
| EasyCOV | Saliva | VTM | NI | NI | No | NEB E1700 + 1 M betaine/fluorescence | 65°C/30 min | 72.7%/95.7% | Equivalent to Ct 35 in RT-PCR | 123 | Montpellier University Hospital, France | L’Helgouach et al., 2020 |
| Saliva | Saline | N-A gene | NI | No, heat + Prot. K lysis | NEB #M1800 Color | 62.5°C/30–60 min | NI | < 10 copies/μL (200 copies/Rx) | 5 | Washington University School of Medicine; Barnes-Jewish Hospital; the Institute of Clinical and Translational Sciences; Tissue Procurement Core, United States | Lalli et al., 2021 | |
| Throat | NI | N gene | NI | Yes | NEB Bst 3.0; WarmStart RTx; Q5 HF DNA polymerase Color or fluorescence | 62.5°C/30–40 min | Sensitivity was 100% for 393 copies/Rx; 80% for 79 copies/Rx and 60% for 16 copies/Rx | 118.6 copies/25 μL or 4.7 copies/μL | 56 | Nantong Third Hospital, China | Lu et al., 2020 | |
| Nasopharyngeal and throat | NI | RdRp | NI | Yes | NEB #M1800 Color | 65°C/60 min | 95.74%/99.95% | 25 copies/Rx | 2,120 | Ramathibodi Hospital, Mahidol University, Bangkok, Thailand | Nawattanapaiboon et al., 2021 | |
| Nasopharyngeal | VTM | ORF1ab | NI | Yes, magnetic bead extraction | MicrosensDx RapiPrep | 65°C/25 min | 80%/100% | Not determined | 21 | National Health Service Care Home, United Kingdom | Österdahl et al., 2020 | |
| SARS-CoV-2 isolated from MRC-5 infected cells | NA | NSP3 gene (ORF1ab) S gene; N gene | NI | Yes | NEB Bst 3.0; + SuperScript IV RT Invitrogen + or leuko–crystal violet Color NEB #M1800 + SYTO-9 Fluorescence | 69°C or 65°C/30 or 60 min | NI | 100 copies/Rx or 10–6 RNA dilution | NA | NA | Park et al., 2020 | |
| Saliva, nasal and nasopharyngeal | Saline | ORF1ab (As1/1e); ORF1a-C and N gene | NI | No | NEB #M1800 and #E1700 Color and fluorescence | 65°C/30–60 min | NI | 1 copy/μL | NA | NA | Rabe and Cepko, 2020 | |
| LAMP-BEAC | Nasopharyngeal and saliva | NI | E, N genes; Orf1ab (As1/1e); | Human statherin mRNA | No, TCEP/EDTA and heat treated | NEB #E1700 and labmade Bst FL Fluorescence | 60°C–65°C/45 min | NI | More than 100 copies/μL | 82 | NA | Sherrill-Mix et al., 2021a |
| Nasal and nasopharyngeal | Amies medium | ORF1a and N gene | NI | Yes | NEB #M1800 Color | 65°C/30 min | 100%/99.7% up to Ct 25 | 100 copies/Rx or 4 copies/μL | 792 | University Hospital Heidelberg and municipal COVID-19 testing station, Germany | Thi et al., 2020 | |
| Dry swab | No, heat treated or directly included in LAMP reaction | 65°C/30 min | 90.5%/99.5% up to Ct 25 in RT-PCR (hot swab) | 343 | ||||||||
| 93.8%/94.1% up to Ct 25 in RT-LAMP (direct swab) | 235 | |||||||||||
| One-pot RT-LAMP | NA | NA | N gene | NA | pUC57-N gene (synthetic) | NEB Bst 3.0 DNA/RNA polymerase + EvaGreen +Rox Fluorescence | 59°C/50 min | NI | 6 copies/μL | NA | NA | Wang, 2020 |
| Nasopharyngeal | VTM | ORF1ab | NI | No | NEB #M1800 Color | 63°C/30 min | 75%/100% | 2.5 copies/uL Spiked samples on VTM | 20 | Columbia University Irving Medical Center | Wei et al., 2021 | |
| Respiratory swabs and bronchoalveolar lavage fluid | ORF1ab and S gene | NI | Yes | Loopamp? 2019-SARS-CoV-2 Detection Reagent Kit (Eiken Chemical, Tokyo, Japan) Turbidity or fluorescence (+ calcein) | 63°C/18–60 min | 100%/100% | 20–110 copies/Rx | 130 | PLA General Hospital, Beijing, China | Yan et al., 2020 | ||
| iLACO | Respiratory (not detailed) | NI | ORF1ab | NI | Yes | NEB #M1800 Color | 65°C/ ≥20 min | 89.9%/NI | 10 copies/μL (detection threshold of 60 copies/μL); equivalent to 35–37 Ct in RT-PCR | 248 | Shenyang province, China | Yu et al., 2020 |
| Respiratory swabs (not detailed) | VTM | N gene and ORF1a | NI | Yes | NEB #M1800 + SYTO-9 Color and fluorescence | 65°C/30 min | NI | 4.8 copies/μL | 6 | Wuhan Institute of Virology, China | Zhang et al., 2020a | |
| Synthetic SARS-CoV-2 RNA | NA | N gene; E gene and As1e gene (ORF1a) | Human actin B gene | Yes | NEB #M1800 + SYTO-9 Color and fluorescence | 65°C/20 min | 87.5% | 2 copies/μL | NA | NA | Zhang et al., 2020b |
NEB #M1800, WarmStart Colorimetric RT-LAMP 2 × Master Mix; NEB #E1700, WarmStart LAMP kit (DNA and RNA); RdRp, RNA-dependent RNA polymerase (harbored by ORF1ab SARS-COV-2 genome region); NI, noninformed; NA, not applied; UTM, Universal Transport medium; VTM, viral transport medium, commonly containing, Hank’s balanced salt solution at pH 7.4 containing BSA (1%), amphotericin (15 μg/mL), penicillin G (100 units/mL), and streptomycin (50 μg/mL); Prot K, proteinase K; BD UVTM, Becton–Dickinson Universal Viral Transport Media system; SPS, sample preservation solution; QuantiFluor, Promega system for dsDNA quantification using a DNA intercalating dye; Prot. K, proteinase K; Cps, copies; Rx, reaction; RdRp, RNA-dependent RNA polymerase; LoD, limit of detection; and BPIFA1, bactericidal/permeability-increasing fold-containing family A1.
Upon RNA extraction from nasopharyngeal swab–derived clinical samples, we found an LoD of 20 viral genomic copies/μL, confirming previous studies based on N SARS-CoV-2 target (Anahtar et al., 2021; Bokelmann et al., 2021; González-González et al., 2021). It is worth noting that when using nonclinical SARS-CoV-2 extracted RNA or synthetic target, the LoD reaches less than 0.5 copies/μL. This can be explained by the presence of interferents such as VTM, host cells, and enzymes that could reduce the yield (Dudley et al., 2020; Nawattanapaiboon et al., 2021). In this regard, we have to be careful when interpreting LoD calculated using nonclinical samples. Nevertheless, extracted samples are rich enough in viral genomic copies to meet SARS-CoV-2 clinically relevant levels.
Clinical validation of RT-LAMP for COVID-19 diagnosis relies on calculating parameters, such as sensitivity, specificity, positive predictive value, negative predictive value, and accuracy compared to the gold standard RT-qPCR. We have to be careful when associating the RT-LAMP sensitivity, and indirect assumption on RT-qPCR viral load is not straightforward because of some technical concerns. It is well accepted that Ct values can be representative of viral load. However, this parameter could lead to misinterpretation when comparing different kits, targets, and nonstandardized samples. A survey conducted by the College of American Pathologists on more than 700 laboratories, reported a variation as much as 14 cycles among different methods on the same batch material. Single laboratories using different platforms and targets in SARS-CoV-2 molecular testing can represent a potential variability on Ct values (Rhoads et al., 2020). Considering previous convergent reports and presuming different targets and platforms, the data from the literature show that with an RT-qPCR Ct 30 cutoff, RT-LAMP sensitivity for SARS-CoV-2 detection is close to 100% (Schermer et al., 2020; Thi et al., 2020; de Oliveira Coelho et al., 2021; García-Bernalt Diego et al., 2021) and eventually with a higher threshold Ct 35 as well (L’Helgouach et al., 2020; de Oliveira Coelho et al., 2021). Indeed, we confirm that up to Ct 30 RT-LAMP returned 100% sensitivity for SARS-CoV-2 detection, reaching 98 and 94% when considering Ct values up to 32 and 34, respectively. Curiously, Kim et al. (2021) found that samples from hospitalized patients presenting Ct value of 28.4 or less were infective to human cell culture (Kim et al., 2021), an evidence based on in vitro extrapolation that RT-LAMP sensitivity is compatible with the threshold of infectivity. This reinforces that simple, robust, and reliable RT-LAMP meets clinical requirements, presenting similar COVID-19 diagnostic accuracy as RT-qPCR (Österdahl et al., 2020; Inaba et al., 2021; Jang et al., 2021).
The choice of SARS-CoV-2 genomic target plays an important role when selecting the RT-LAMP method for COVID-19 diagnosis. Several research groups have tested different regions on SARS-CoV-2 genome with the potential to generate RT-LAMP primers. Once the majority of primers were designed using the open source software Primer Explorer, it is expected that at some point, the default algorithm returned the same result or overlapping regions, independently identified in a context where molecular biology scientists everywhere in the world are working to tackle COVID-19 (Figure 9). According to our data compilation, N gene and ORF1ab regions (overlapping NSP3, As1e, and RdRp-coding sequences) were the most frequent targets chosen for SARS-CoV-2 RT-LAMP (Table 2 and Figure 9). Ganguli et al. (2020) and Zhang et al. (2020b) arrived at the same conclusion when selecting the SARS-CoV-2 N gene-targeting primer set after confirming better performances for RNA viral detection when compared to other targets (Ganguli et al., 2020; Zhang et al., 2020b). When testing N, E, and RdRp genes in true-positive, previously RT-qPCR–characterized clinical samples, we observed more false-negative outputs from assays using E and RdRp genes, corroborating what was previously reported. We also highlight that primer subsets within the same N target gene can contribute differentially to RT-LAMP test sensitivity (Supplementary Table 2). Furthermore, multiplexing different primer sets is encouraged in order to increase sensitivity (Figure 9; Kim et al., 2019; Mautner et al., 2020; Zhang et al., 2020b).
FIGURE 9.
Schematic representation of SARS-CoV-2 genome indicating the amplicons for the COVID-19 molecular diagnostics by RT-LAMP. Structural representation of SARS-CoV-2 virion shows the main particle parts. LAMP primer regions are indicated as previously reported (Baek et al., 2020; Ben-Assa et al., 2020; Butler et al., 2020; Chow et al., 2020; Dudley et al., 2020; Song et al., 2021; Ganguli et al., 2020; Huang et al., 2020; Lamb et al., 2020; Lu et al., 2020; Mohon et al., 2020; Park et al., 2020; Rabe and Cepko, 2020; Thi et al., 2020; Yan et al., 2020; Yu et al., 2020; Zhang et al., 2020a,b; Anahtar et al., 2021; Bhadra et al., 2021; Bokelmann et al., 2021; González-González et al., 2021). ORF, open reading frame; RdRp, RNA-dependent RNA polymerase; NSP, nonstructural protein. Schematic representation created using Snap Gene Viewer software version 5.0.7; N1, N2, and N3_CDC correspond to the amplicons for SARS-CoV-2 detection by RT-PCR. Created with biorender.com.
Another important (almost neglected) point is the fact that, although inspired by RT-qPCR target selection, few SARS-CoV-2 RT-LAMP approaches reported an internal control target to confirm the presence of human RNA and monitor sampling or extraction process (de Oliveira Coelho et al., 2021). Wilson-Davies et al. (2021) pointed out that the lack of amplification can happen for different reasons concerning the whole reaction, a specific well, or due to inhibitory substances, highlighting the importance of including internal control even before nucleic acid extraction, in order to be considered a reliable SARS-CoV-2 LAMP assay (Wilson-Davies et al., 2021). In this study, all clinical samples were previously characterized by RT-qPCR, including human RNAse P as housekeeping gene (internal control). In the current OmniLAMP® assay, we included human b-actin RNA (rACTB) as internal control. Other constitutive targets for SARS-CoV-2 RT-LAMP include BPIFA1 (Bektaş et al., 2021), human 18S RNA (de Oliveira Coelho et al., 2021), and Statherin RNA (Sherrill-Mix et al., 2021a; Table 2).
Similar to its high sensitivity, obtained in this work and by other studies, the SARS-CoV-2 RT-LAMP specificity is undoubtedly high and is frequently reported as 100% without any cross-reactivity with other respiratory or SARS-CoV–unrelated viruses (Chow et al., 2020; Mohon et al., 2020; Park et al., 2020; Aoki et al., 2021; Nawattanapaiboon et al., 2021). We also confirm that the SARS-CoV-2 RT-LAMP solution presented here is highly specific and does not cross-react with Brazilian occurring seasonal influenza A and B, hRSV, or arboviruses.
Despite the advantages presented by purified and nucleic acid–enriched samples for SARS-CoV-2 RT-LAMP, RNA extraction–free protocols have attracted attention as they can be noninvasive (saliva-based), do not require additional steps and equipment, and fulfill point-of-sampling requirements. Indeed, the preanalytical phase on RT-LAMP is the bottleneck for PoC applications. For this reason, several studies highlighted the feasibility of primary RNA extraction–free approaches for SARS-CoV-2 RNA detection (Asprino et al., 2020; Ben-Assa et al., 2020; Dudley et al., 2020; Esbin et al., 2020; L’Helgouach et al., 2020; Schermer et al., 2020; Srivatsan et al., 2020; Anahtar et al., 2021; Lalli et al., 2021; Wei et al., 2021). Pretreatment of saliva samples includes heat sample inactivation, and the use of lysis/stabilizing buffers that can contain proteinase K, TCEP, EDTA, and DTT could help the viral RNA assessment maintaining its integrity (Ben-Assa et al., 2020; Lamb et al., 2020; L’Helgouach et al., 2020; Smyrlaki et al., 2020; Lalli et al., 2021; Newman et al., 2021; Yang et al., 2021). Caution must be taken when running colorimetric RT-LAMP as pretreatment could interfere on result outputs. One of the main limitations for direct sample test by colorimetric RT-LAMP based on pH-sensing is the false-positive result upon input sample addition (previous to amplification) because of naturally acidic samples (Thi et al., 2020; Bokelmann et al., 2021). To prevent spurious amplification due to the presence of DNA from oral microbiome, food, or host cells on primary samples, Bokelmann et al. (2021) treated samples with λ exonuclease that acts by preferentially digesting 5′-phosphorylated DNA, leaving nonphosphorylated primers or LAMP products intact (Bokelmann et al., 2021). Here we showed the preliminary results on RNA extraction–free (also pretreatment free) diluted 10× in hydrochloride guanidine-containing VTM nasopharyngeal samples directly accessed to compare colorimetric results. Three of five RT-qPCR true-positive, directly accessed samples returned positive yellow output on colorimetric RT-LAMP for SARS-CoV-2 detection. This provides clues on the use of unextracted samples for massive COVID-19 testing campaigns with a trade-off on cost-benefits for LoD and test sensitivity. A recent study on 559 swabs and 86,760 saliva samples performed a sample preparation method for RNA extraction–free and found diagnostic sensitivity of 70.35 and 84.62%, respectively, for swab and saliva samples (Kidd et al., 2021). Most of the high and medium viral load samples will be detected on unextracted protocols. However, to meet RT-qPCR detection sensitivity levels, this requires some type of purification step and RNA concentration (Broughton et al., 2020; Park et al., 2020; Zhang et al., 2020a).
We are currently observing rapid converging evolution of SARS-CoV-2 during the COVID-19 pandemic worldwide. Several reports alert for the emergence of VOIs and VOCs such as the alpha (B.1.1.7), first detected in England (ECDC threat assessment brief on December 20, 2020; European Centre for Disease Prevention and Control, 2020); Beta (B.1.351), initially reported from South Africa (Tegally et al., 2021); gamma (P.1 or B.1.1.28.1), which was identified in Japan but obtained from a traveler from Brazil (Faria et al., 2021); and more recently, the VOI kappa (B.1.617.1) and VOC delta (B.1.617.2) detected in India, responsible for the majority of new COVID-19 cases in many countries in different parts of the world. The regional selection of SARS-CoV-2 VOC is associated with higher transmissibility, mortality and reduced neutralizing antibody response (Shah et al., 2020; Davies et al., 2021a,b; Li et al., 2021). In Brazil, we observed the emergence of different SARS-CoV-2 VOCs and VOIs, including gamma (P.1), zeta (P.2; Resende et al., 2021a; Voloch et al., 2021), B.1.1.33.9 (N.9; Resende et al., 2021c), and B.1.1.33.10 (N.10; Resende et al., 2020, 2021b). A plethora of mutations is observed in these variants, including N501Y, E484K/Q, K417N/T, A570D, and the Δ69–70 at the SARS-CoV-2 S protein sequence, which was associated with detection failures by S-target RT-qPCR methods (Brown et al., 2021). For SARS-CoV-2 RT-LAMP detection, few studies selected S-coding protein region as a target (Figure 9). In addition, isothermal amplification for SARS-CoV-2 RNA detection strategies is commonly addressed as multiplex targeted, making RT-LAMP a good choice even for SARS-CoV-2 variant detection. Indeed, here we reported that singleplex N gene-based or multiplex N2/E1-based RT-LAMP was able to perfectly detect VOCs and VOIs circulating in Brazil such as gamma (P.1), zeta (P.2), B.1.1.374, and B.1.1.371 (Figure 8 and Supplementary Figure 1D), the two latter first detected in Finland and Saudi Arabia3. Recent efforts made by Sherrill-Mix et al. (2021a,b) showed a beacon-based RT-LAMP strategy designed to precisely identify alpha (B.1.1.7) SARS-CoV-2 variant (Sherrill-Mix et al., 2021a,b), a promising tool not only for massive screening but also to monitor VOC/VOI SARS-CoV-2 spreading.
The colorimetric RT-LAMP is a reliable molecular tool for detecting SARS-CoV-2, providing rapid and easy-to-read results, compatible with high-throughput screenings and PoC requirements. This test is especially important for nations with poor diagnostic conditions, such as Brazil, where RT-qPCR COVID-19 diagnostic is far from ideal to control disease spreading. The RT-LAMP sensitivity can be equivalent to those reported from the gold standard RT-qPCR method and also present 100% specificity. Results are commonly obtained after 30-min reaction and if needed, additional 20 min was not associated with spurious unspecific amplification. Sample collection in guanidine-containing VTM has been described as a useful strategy to avoid contamination of health care workers during sample manipulation. RT-LAMP primer selection can directly interfere on sensitivity, being N genes the best target for SARS-CoV-2 RNA detection with fewer false-negative results, especially in low viral load samples, which is improved upon multiplexing E/N targets. Colorimetric RT-LAMP is also compatible with detecting SARS-CoV-2 VOIs and VOCs, being robust to cope with the monitoring of emerging new SARS-CoV-2 variants and that can be easily adapted. We thus reinforce and recommend the use of RT-LAMP for massive testing as a decentralized PoC alternative to avoid SARS-CoV-2 spread and to tackle COVID-19.
Materials and Methods
Clinical Samples, Reverse Transcriptase–Quantitative Polymerase Chain Reaction, and Ethics Statement
In total, 467 clinical samples were included in this study. Initially, 100 nasopharyngeal clinical samples were obtained from hospitalized patients in different parts of Brazil from April to July 2020. The samples derived from this first batch were tested by RT-LAMP using N gene alone as target, and the group presented a median age of 60 years, and 60% of patients were male. An additional 367 samples were included in the study. They were obtained from symptomatic patients, considered COVID-19 suspected cases, during September until November 2020 in Belo Horizonte, Minas Gerais, Brazil. The samples from the latter group were validated by RT-LAMP targeting E and N genes combined and were characterized with a median age of 46 years old, in whom 75% of patients were female.
Nasopharyngeal swabs were collected and maintained in 2 mL VTM (Bioclin, Belo Horizonte, Brazil #G092-1) at room temperature until RNA extraction or direct dilution for LAMP reaction. The VTM contains guanidine chloride as inactivation agent and to preserve viral RNA. All procedures were performed inside a biosafety level 2 cabinet. RNA extraction was performed using the QIAamp® Viral RNA Mini Kit (Qiagen #52906), following manufacturer instructions. The molecular diagnostic routine was performed by RT-qPCR using the SARS-CoV-2 commercial kits produced at Fundação Oswaldo Cruz [Kit Molecular SARS-CoV-2 E/RP, from Bio-Manguinhos/Fiocruz, based on Charité/Berlin protocol, and Kit Biomol OneStep/COVID-19 from IBMP/Fiocruz, based on China/Centers for Disease Control and Prevention (CDC) protocol with recommended targets polyprotein ORF1ab and N gene]. RT-qPCR was carried out using the 7500, ViiA 7 real-time PCR systems (Applied Biosystems, Foster City, CA, United States) or the dual-channel Open qPCR machine (Chai, Santa Clara, CA, United States), following the temperature program profile of 95°C for 3 min, followed by 40 cycles of amplification (95°C/15 s and 60°C/1 min). Influenza and hRSV samples were kindly provided by IOM/FUNED, and the arbovirus samples are part of the collection from the Laboratório de Imunologia de Doenças Virais at Oswaldo Cruz Foundation. All procedures involving human participants and collection and use of clinical samples and data were in accordance with ethical standards and approved by the local Research Ethics Committee involving human beings at Instituto René Rachou, Fundação Oswaldo Cruz, under license protocol no. 4084902 and CAAE (certificate of presentation for ethical appreciation): 31984720300005091. The ethics approval was issued on June 12, 2020. SARS-CoV-2 VOCs and VOIs included in this study were isolated from symptomatic patients (Ct value < 25, using E gene as target on RT-qPCR—Kit Molecular SARS-CoV-2 E/RP Bio-Manguinhos Fiocruz), in the State Pernambuco, Northeast Brazil (Bezerra et al., 2021; Supplementary Table 3). The study was approved by the local Human Research Ethics Committee (CAAE: 32333120.4.0000.5190). The genomes of SARS-CoV-2 VOI and VOCs generated are deposited on GISAID according to the following accession codes: EPI_ISL_2221860, EPI_ISL_2221850, EPI_ISL_2221873, EPI_ISL_2221890, EPI_ISL_2221902, EPI_ISL_2221885, EPI_ISL_2221844, and EPI_ISL_2221866.
Reverse Transcription Loop-Mediated Isothermal Amplification Primer Design
Reverse transcription loop-mediated isothermal amplification primers were designed based on SARS-CoV-2 reference genome (GenBank accession NC_045512.2) using the open source software Primer Explorer V54 or the New England Biolabs (NEB) LAMP primer design tool5. The free energy (ΔG) of selected primers was less than –4 kcal/mol, as a parameter chosen based on oligo stability (Parida et al., 2008). The set of primers used in this study is listed in Table 3 and additional information can be found in Figure 9 and Supplementary Figures 4–6. We designed and validated different LAMP primer sets, such as N gene Set1 and Set2 that appeared in other independent researches (Figure 9 and Table 3). N2 and E1 primer sets were previously designed by Zhang et al. (2020b). The oligos were purchased from Integrated DNA technologies (IDT, Coralville, IA, United States) and from Exxtend (Paulínia, SP, Brazil). All oligos were synthesized at 25 nanomole scale and purified by standard desalting. Thermodynamic evaluation of primers targeting SARS-CoV-2 N, E, and RdRp genes was performed as previously described (Miranda and Weber, 2021). Briefly, hybridization temperature of F3, FIP (F1c+F2), BIP (B1c+B2), LF, and LB primer sets were calculated upon aligning to SARS-CoV or other coronavirus (non-SARS) genomes, considering potential mismatches. The SARS-CoV-2 coverage for each primer was also obtained (Supplementary Table 1).
TABLE 3.
Sets of LAMP oligonucleotides used in this study.
| LAMP primer | Sequence (5′–3′) | References |
| N_Set1_F3 | TGGCTACTACCGAAGAGCT | Ben-Assa et al., 2020; Bhadra et al., 2021; Song et al., 2021; Rabe and Cepko, 2020; Thi et al., 2020; Zhang et al., 2020a; Anahtar et al., 2021; this study |
| N_Set1_B3 | TGCAGCATTGTTAGCAGGAT | |
| N_Set1_FIP | TCTGGCCCAGTTCCTAGGTA GTGACGAATTCGTGGTGGTGA | |
| N_Set1_BIP | AGACGGCATCATATGGGTTGC ACGGGTGCCAATGTGATCT | |
| N_Set1_LF | TGGACTGAGATCTTTCATTTTACCG | |
| N_Set1_LB | ACTGAGGGAGCCTTGAATACA | |
| N_Set2_F3 | TGGACCCCAAAATCAGCG | Huang et al., 2020; González-González et al., 2021; this study |
| N_Set2_B3 | GCCTTGTCCTCGAGGGAAT | |
| N_Set2_FIP | CCACTGCGTTCTCCATTCTGGTAA ATGCACCCCGCATTACG | |
| N_Set2_BIP | CGCGATCAAAACAACGTCGGCCC TTGCCATGTTGAGTGAGA | |
| N_Set2_LF | TTGAATCTGAGGGTCCACCAAA | |
| N_Set2_LB | GGTTTACCCAATAATACTGCGTCTT | |
| E_Set1_F3 | TGATGAGCCTGAAGAACATG | This study |
| E_Set1_B3 | CGCTATTAACTATTAACGTACCT | |
| E_Set1_FIP | TCGGTTCATCATAAATTGGTTCCAT CAAATTCACACAATCGACGG | |
| E_Set1_BIP | ACGACTACTAGCGTGCCTTTGTCT CTTCCGAAACGAATG | |
| E_Set1_LF | ACTGGATTAACAACTCCGGATGA | |
| E_Set1_LB | GTAAGCACAAGCTGATGAGTACGAA | |
| RdRp_F3 | CTGTCAAATTACAGAATAATGAGC | This study |
| RdRp_B3 | TCCATCACTCTTAGGGAATC | |
| RdRp_FIP | TGTCATCAGTGCAAGCAGTTTGCTG TTGCACTACGACAGA | |
| RdRp_BIP | ATGCGTTAGCTTACTACAACACACC CATTTCAAATCCTGTAAATCG | |
| RdRp_LF | ACCGGCAGCACAAGACA | |
| RdRp_LB | ACAAAGGGAGGTAGGTTTGTACT | |
| N2_F3 | ACCAGGAACTAATCAGACAAG | Butler et al., 2020; Zhang et al., 2020b |
| N2_B3 | GACTTGATCTTTGAAATTTGGATCT | |
| N2_FIP | TTCCGAAGAACGCTGAAGCGGAAC TGATTACAAACATTGGCC | |
| N2_BIP | CGCATTGGCATGGAAGTCACAATTT GATGGCACCTGTGTA | |
| N2_LF | GGGGGCAAATTGTGCAATTTG | |
| N2_LB | CTTCGGGAACGTGGTTGACC | |
| E1_F3 | TGAGTACGAACTTATGTACTCAT | Butler et al., 2020; Zhang et al., 2020b |
| E1_B3 | TTCAGATTTTTAACACGAGAGT | |
| E1_FIP | ACCACGAAAGCAAGAAAAAGAAG TTCGTTTCGGAAGAGACAG | |
| E1_BIP | TTGCTAGTTACACTAGCCATCCTTA GGTTTTACAAGACTCACGT | |
| E1_LF | CGCTATTAACTATTAACG | |
| E1_LB | GCGCTTCGATTGTGTGCGT |
Ref, references where the DNA oligos where originally published or share the same set of primers; F3/B3, outer forward (F) and backward primers; FIP/BIP, inner primers; LF/LB, loop primers. For detailed information on targeted SARS-CoV-2 sequence used, refer to Supplementary Figures 4–6 and Figure 9.
Reverse Transcription Loop-Mediated Isothermal Amplification Assays
All mix preparations for RT-LAMP reaction were performed on ice inside a biosafety level 2 cabinet. RT-LAMP reactions were performed according to NEB recommendations, containing the following components: 10 μL of WarmStart® Colorimetric LAMP 2× Master Mix [NEB #M1800 or #M1804, the latter contains dUTP UDG (uracil-DNA-glycosylase) to avoid carryover contamination; composition of both are NEB’s proprietary]—ready-to-use mixture of WarmStart® Bst 2.0 DNA polymerase and WarmStart® RTx (reverse transcriptase for one-step transcription/amplification reaction) in presence of a pH sensor that turns from fuchsia (pink) to yellow in presence of increased proton (acid pH) during DNA polymerization on isothermal amplification, 1.6 μmol/L forward inner/backward inner primers (FIP/BIP); 0.2 μmol/L forward and backward outer primers (F3/B3), and 0.4 μmol/L loop forward and loop backward primers (LF/LB); Ultra-pureTM DNAse/RNase-free distilled water (InvitrogenTM #10977015) was added in quantity enough to complete the final volume reaction of 20 μL; isothermal amplification was performed on VeritiTM thermal cycler (Applied Biosystems, Foster City, CA, United States) at 65°C for 30 min. From clinical samples in the first batch, we used as input, 1 μL of RNA extracted from nasopharyngeal swab placed on guanidine-containing VTM, whereas upon optimization, 5 μL source template was considered from the samples in the second group.
When using raw RNA extraction–free samples, we initially prepared a 1:10 ultrapure water diluted clinical sample (1 μL of VTM sample in 9 μL water) and used 1 μL as RT-LAMP reaction input. A similar strategy was applied to SARS-CoV-2 VOC/VOI samples. Positive controls were performed either by RNA extraction from Vero E6-derived inactivated SARS-CoV-2, using synthetic SARS-CoV-2 N gene-harboring plasmid (ECRA Biotech, Campinas, SP, Brazil #EB14-20) or inactivated laboratory-cultured SARS-CoV-2, when aiming the RNA extraction–free tests. For optimization purposes, incubation time tested varied from 30 to 50 min. The first 100 RT-LAMP reaction products were migrated in 2% agarose gel to confirm specific amplification in positive reactions and amplicon-free nontemplate controls. Gel images were taken using the ImageQuantTM LAS 4000 with GelRedTM (Biotium #41003) as intercalating dye. Non–SARS-CoV-2 RNA extracted samples of influenza A, influenza B, hRSV, dengue, Zika, Chikungunya, and yellow fever viruses were also added as 1-μL input.
Analytical Sensitivity
Absolute quantification was performed based on a calibration curve prepared using the standard SARS-CoV-2 E gene–harboring plasmid (2 × 105 copies/μL; Biogene COVID-19 PCR, Bioclin/Quibasa #K228-1; Lot: 0007), SARS-CoV-2 (2019-nCoV) Charité/Berlin primer probe panel (IDT, #10006804), and the GoTaq® Probe 1-step RT-qPCR System (Promega #A6120), according to manufacturer instructions, as indicated by the US CDC. Real-time RT-PCR program was performed as follows: first stage (×1) 15 min at 45°C, second stage (×1) 2 min at 95°C, and third stage (×40) 3 s at 95°C followed by 30 s at 55°C. Linear regression was performed using Prism software, version 9 (GraphPad Software, San Diego, CA, United States) leading to the equation: Y = –3.6383X + 38.771 and coefficient of correlation R2 = 0.9938 (Supplementary Figure 3). Viral RNA either from Vero E6-derived SARS-CoV-2 (SARS-CoV-2 isolate HIAE-02: SARS-CoV2/SP02/human/2020/BRA GenBank accession no. MT126808.1) or obtained from clinical nasopharyngeal swabs was quantitated based on the Ct value for E gene.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.gisaid.org/, The genomes of SARS-CoV-2 VOI and VOCs generated are deposited on GISAID according to the following accession codes: EPI_ISL_2221860, EPI_ISL_2221850, EPI_ISL_2221873, EPI_ISL_2221890, EPI_ISL_2221902, EPI_ISL_2221885, EPI_ISL_2221844, and EPI_ISL_2221866.
Ethics Statement
The studies involving human participants were reviewed and approved by Research Ethics Committee involving human beings at Instituto René Rachou, Fundação Oswaldo Cruz, under license protocol number: 4084902 and CAAE (certificate of presentation for ethical appreciation): 31984720300005091. The ethics approval was issued on June 12, 2020. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
PA, IB, and RM-N: conceptualization and experimental design. EO, AF-L, LA, AG, IB, and FR: investigation and performed experiments. AF-L, LA, AG, EO, PA, and RM-N: analyzed the data. MB, PM, FC, HM, GW, ST, and GLW: contributed to reagents, materials, and analysis and tools. PA and RM-N: supervision. RM-N, EO, AF-L, and RR: writing—original draft. PA, ST, GLW, GW, MB, and RM-N: writing, review, and editing. All authors discussed the results and contributed to the final manuscript.
Conflict of Interest
HM is part of Visuri company. Results presented here are the basis of a COVID-19 RT-LAMP diagnostic test offered by Visuri named OmniLAMP® SARS-CoV-2 kit. PA and RM-N are co-founders and scientific advisors at CEPHA Biotech. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Professor Édison L. Durigon, Laboratório de Virologia Clínica e Molecular, ICB/USP, São Paulo, Brazil, who kindly provided inactivated samples of SARS-CoV-2 isolate HIAE-02: SARS-CoV2/SP02/human/2020/BRA (GenBank accession number MT126808.1). We thank Luciano Moreira, Instituto René Rachou – Fiocruz Minas, for providing the first colorimetric LAMP buffers during the testing and optimization phase. We also thank Marluce Aparecida Assunção Oliveira – Former Director of Instituto Octávio Magalhães, Fundação Ezequiel Dias, Laboratório Central de Saúde Pública de Minas Gerais – LACEN-MG; and Marcos Vinicius Ferreira da Silva, head of virology service at LACEN-MG, for providing samples of Influenza A and B viruses and human respiratory syncytial virus (hRSV). We are grateful to Cristiane P. Gomes and Patrícia P. N. Miranda for resources management and excellent technical assistance. PA, GLW, and RM-N are part of Fiocruz COVID-19 Genomic Surveillance Network (http://www.genomahcov.fiocruz.br).
Footnotes
Funding
This work was funded by Fundação de Amparo à Pesquisa do Estado de Minas Gerais – Fapemig, grant number #APQ-00485-20, to RM-N; grant number #APQ-00262-20 to FC and Fundação Oswaldo Cruz – Inova Fiocruz Program – Innovative Products (grant number VPPIS-004-FIO-18-51) to RM-N; Innovative Products to face COVID-19 pandemics (grant number VPPIS-005-FIO-20-2-45) to PA and from the MCTI – Brazilian Ministry of Science, Technology and Innovation, through the “Rede Virus” initiative to PA (grant number – FINEP 01.20.0005.00). EO received a Master’s fellowship from the Vice presidency of Education, Formation and communication VPEIC – Fundação Oswaldo Cruz. IB and AF-L received post-doctoral research fellowships from Inova Fiocruz Program – Fundação Oswaldo Cruz. AG holds a science, technology and innovation development scholarship from Fapemig (BDCTI-I). LA is a post-doctoral fellow from MCTI (CNPq – DTI-A). RM-N, ST, GW, and GLW are CNPq Research Fellows (#310640/2017-2; #302961/2017-8; #303902/2019-1; and #307538/2019-2). PM holds a scholarship from Coordenação de Aperfeiçoamento de Nível Superior (CAPES/Ação Emergencial). The funders had no role in the study design, data collection and analysis, decision to publish, or manuscript preparation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.713713/full#supplementary-material
References
- Anahtar M. N., McGrath G. E. G., Rabe B. A., Tanner N. A., White B. A., Lennerz J. K. M., et al. (2021). Clinical assessment and validation of a rapid and sensitive SARS-CoV-2 test using reverse transcription loop-mediated isothermal amplification without the need for RNA extraction. Open Forum Infect. Dis. 8:ofaa631. 10.1093/ofid/ofaa631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki M. N., de Oliveira Coelho B., Góes L. G. B., Minoprio P., Durigon E. L., Morello L. G., et al. (2021). Colorimetric RT-LAMP SARS-CoV-2 diagnostic sensitivity relies on color interpretation and viral load. Sci. Rep. 11:9026. 10.1038/s41598-021-88506-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asprino P., Bettoni F., Camargo A., Coelho D., Coppini G., Correa I., et al. (2020). A scalable saliva-based, extraction-free rt-lamp protocol for sars-cov-2 diagnosis. medRxiv [Preprint]. 10.1101/2020.10.27.20220541 [DOI] [Google Scholar]
- Baek Y. H., Um J., Antigua K. J. C., Park J.-H., Kim Y., Oh S., et al. (2020). Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2. Emerg. Microbes Infect. 9 998–1007. 10.1080/22221751.2020.1756698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Yao L., Wei T., Tian F., Jin D.-Y., Chen L., et al. (2020). Presumed asymptomatic carrier transmission of COVID-19. JAMA 323 1406–1407. 10.1001/jama.2020.2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bektaş A., Covington M. F., Aidelberg G., Arce A., Matute T., Núñez I., et al. (2021). Accessible LAMP-enabled rapid test (ALERT) for detecting SARS-CoV-2. Viruses 13:742. 10.3390/v13050742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Assa N., Naddaf R., Gefen T., Capucha T., Hajjo H., Mandelbaum N., et al. (2020). Direct on-the-spot detection of SARS-CoV-2 in patients. Exp. Biol. Med. 245 1187–1193. 10.1177/1535370220941819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezerra M. F., Machado L. C., do Carmo Vasconcelos de Carvalho V., Docena C., Brandão-Filho S. P., Ayres C. F. J., et al. (2021). A sanger-based approach for scaling up screening of SARS-CoV-2 variants of interest and concern. Infect. Genet. Evol. 92:104910. 10.1016/j.meegid.2021.104910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadra S., Riedel T. E., Lakhotia S., Tran N. D., Ellington A. D. (2021). High-surety isothermal amplification and detection of SARS-CoV-2. mSphere 6 e911–e920. 10.1128/mSphere.00911-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokelmann L., Nickel O., Maricic T., Pääbo S., Meyer M., Borte S., et al. (2021). Point-of-care bulk testing for SARS-CoV-2 by combining hybridization capture with improved colorimetric LAMP. Nat. Commun. 12:1467. 10.1038/s41467-021-21627-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J. P., Deng X., Yu G., Fasching C. L., Servellita V., Singh J., et al. (2020). CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 38 870–874. 10.1038/s41587-020-0513-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K. A., Gubbay J., Hopkins J., Patel S., Buchan S. A., Daneman N., et al. (2021). S-gene target failure as a marker of variant B.1.1.7 among SARS-CoV-2 isolates in the greater Toronto Area, December 2020 to March 2021. JAMA. 325 2115–2116. 10.1001/jama.2021.5607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler D. J., Mozsary C., Meydan C., Danko D., Foox J., Rosiene J., et al. (2020). Shotgun transcriptome and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions. medRxiv [Preprint]. 10.1101/2020.04.20.048066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow F. W.-N., Chan T. T.-Y., Tam A. R., Zhao S., Yao W., Fung J., et al. (2020). A rapid, simple, inexpensive, and mobile colorimetric assay COVID-19-LAMP for mass on-site screening of COVID-19. Int. J. Mol. Sci. 21:5380. 10.3390/ijms21155380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N. G., Abbott S., Barnard R. C., Jarvis C. I., Kucharski A. J., Munday J. D., et al. (2021a). Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 372:eabg3055. 10.1126/science.abg3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N. G., Jarvis C. I., Edmunds W. J., Jewell N. P., Diaz-Ordaz K., Keogh R. H. (2021b). Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature 593 270–274. 10.1038/s41586-021-03426-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Coelho B., Sanchuki H. B. S., Zanette D. L., Nardin J. M., Morales H. M. P., Fornazari B., et al. (2021). Essential properties and pitfalls of colorimetric Reverse transcription loop-mediated isothermal amplification as a point-of-care test for SARS-CoV-2 diagnosis. Mol. Med. 27:30. 10.1186/s10020-021-00289-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley D. M., Newman C. M., Weiler A. M., Ramuta M. D., Shortreed C. G., Heffron A. S., et al. (2020). Optimizing direct RT-LAMP to detect transmissible SARS-CoV-2 from primary nasopharyngeal swab samples. PLoS One 15:e0244882. 10.1371/journal.pone.0244882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control (2020). Rapid Increase of a SARS-CoV-2 Variant with Multiple Spike Protein Mutations Observed in the United Kingdom. Stockholm: ECDC. [Google Scholar]
- Esbin M. N., Whitney O. N., Chong S., Maurer A., Darzacq X., Tjian R. (2020). Overcoming the bottleneck to widespread testing: a rapid review of nucleic acid testing approaches for COVID-19 detection. RNA 26 771–783. 10.1261/rna.076232.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria N. R., Claro I. M., Candido D., Franco L. A. M., Andrade P. S., Coletti T. M., et al. (2021). Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings – SARS-CoV-2 coronavirus / nCoV-2019 Genomic Epidemiology. Virological. Available online at: https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586 (Accessed May 14, 2021). [Google Scholar]
- Flynn M. J., Snitser O., Flynn J., Green S., Yelin I., Szwarcwort-Cohen M., et al. (2020). A simple direct RT-LAMP SARS-CoV-2 saliva diagnostic. medRxiv [Preprint]. 10.1101/2020.11.19.20234948 [DOI] [Google Scholar]
- Fowler V. L., Armson B., Gonzales J. L., Wise E. L., Howson E. L. A., Vincent-Mistiaen Z., et al. (2021). A highly effective reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay for the rapid detection of SARS-CoV-2 infection. J. Infect. 82, 117–125. 10.1016/j.jinf.2020.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguli A., Mostafa A., Berger J., Aydin M. Y., Sun F., de Ramirez S. A. S., et al. (2020). Rapid isothermal amplification and portable detection system for SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 117 22727–22735. 10.1073/pnas.2014739117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Bernalt Diego J., Fernández-Soto P., Domínguez-Gil M., Belhassen-García M., Bellido J. L. M., Muro A. (2021). A simple, affordable, rapid, stabilized, colorimetric, versatile RT-LAMP assay to detect SARS-CoV-2. Diagnostics 11:438. 10.3390/diagnostics11030438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey K., Bagust L., Baird J., Barker M., Batchelor J., Bryant S., et al. (2020). Evaluation of the Expanded Southampton Pilot Study (Phase 2) for Use of Saliva-Based Lamp Testing in Asymptomatic Populations: Final Report, 16th November 2020. Southampton: University of Southampton. 10.5258/SOTON/P0045 [DOI] [Google Scholar]
- Gonçalves D. S., Cassimiro A. P. A., de Oliveira C. D., Rodrigues N. B., Moreira L. A. (2014). Wolbachia detection in insects through LAMP: loop mediated isothermal amplification. Parasit Vectors 7:228. 10.1186/1756-3305-7-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-González E., Lara-Mayorga I. M., Rodríguez-Sánchez I. P., Zhang Y. S., Martínez-Chapa S. O., Santiago G. T., et al. (2021). Colorimetric loop-mediated isothermal amplification (LAMP) for cost-effective and quantitative detection of SARS-CoV-2: the change in color in LAMP-based assays quantitatively correlates with viral copy number. Anal. Methods 13 169–178. 10.1039/d0ay01658f [DOI] [PubMed] [Google Scholar]
- Gorbalenya A. E., Baker S. C., Baric R. S., de Groot R. J., Drosten C., Gulyaeva A. A., et al. (2020). The species severe acute respiratory syndrome-related coronavirus?: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 5 536–544. 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallal P. C., Hartwig F. P., Horta B. L., Silveira M. F., Struchiner C. J., Vidaletti L. P., et al. (2020). SARS-CoV-2 antibody prevalence in Brazil: results from two successive nationwide serological household surveys. Lancet Glob. Health 8:e01390-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W. E., Lim B., Hsu C.-C., Xiong D., Wu W., Yu Y., et al. (2020). RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 13 950–961. 10.1111/1751-7915.13586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba M., Higashimoto Y., Toyama Y., Horiguchi T., Hibino M., Iwata M., et al. (2021). Diagnostic accuracy of LAMP versus PCR over the course of SARS-CoV-2 infection. Int. J. Infect. Dis. 107 195–200. 10.1016/j.ijid.2021.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang W. S., Lim D. H., Yoon J., Kim A., Lim M., Nam J., et al. (2021). Development of a multiplex loop-mediated isothermal amplification (LAMP) assay for on-site diagnosis of SARS CoV-2. PLoS One 16:e0248042. 10.1371/journal.pone.0248042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J., Ladha A., Saito M., Segel M., Bruneau R., Huang M. W., et al. (2020). Point-of-care testing for COVID-19 using SHERLOCK diagnostics. Infectious diseases (except HIV/AIDS). medRxiv [Preprint]. 10.1101/2020.05.04.20091231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd S. P., Burns D., Armson B., Beggs A. D., Howson E. L. A., Williams A., et al. (2021). RT-LAMP has high accuracy for detecting SARS-CoV-2 in saliva and naso/oropharyngeal swabs from asymptomatic and symptomatic individuals. medRxiv [Preprint]. 10.1101/2021.06.28.21259398 [DOI] [Google Scholar]
- Kim J. H., Kang M., Park E., Chung D. R., Kim J., Hwang E. S. (2019). A simple and multiplex loop-mediated isothermal amplification (LAMP) assay for rapid detection of SARS-CoV. Biochip J. 13 341–351. 10.1007/s13206-019-3404-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.-C., Cui C., Shin K.-R., Bae J.-Y., Kweon O.-J., Lee M.-K., et al. (2021). Duration of culturable SARS-CoV-2 in hospitalized patients with Covid-19. N. Engl. J. Med. 384 671–673. 10.1056/NEJMc2027040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa Y., Orihara Y., Kawamura R., Imai K., Sakai J., Tarumoto N., et al. (2020). Evaluation of rapid diagnosis of novel coronavirus disease (COVID-19) using loop-mediated isothermal amplification. J. Clin. Virol. 129:104446. 10.1016/j.jcv.2020.104446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki Y., Magassouba N., Bah H. A., Soropogui B., Doré A., Kourouma F., et al. (2016a). Deployment of a reverse transcription loop-mediated isothermal amplification test for ebola virus surveillance in remote areas in Guinea. J. Infect. Dis. 214 S229–S233. 10.1093/infdis/jiw255 [DOI] [PubMed] [Google Scholar]
- Kurosaki Y., Magassouba N., Oloniniyi O. K., Cherif M. S., Sakabe S., Takada A., et al. (2016b). Development and evaluation of reverse transcription-loop-mediated isothermal amplification (RT-LAMP) assay coupled with a portable device for rapid diagnosis of ebola virus disease in Guinea. PLoS Negl. Trop. Dis. 10:e0004472. 10.1371/journal.pntd.0004472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalli M. A., Langmade J. S., Chen X., Fronick C. C., Sawyer C. S., Burcea L. C., et al. (2021). Rapid and extraction-free detection of SARS-CoV-2 from saliva by colorimetric reverse-transcription loop-mediated isothermal amplification. Clin. Chem. 67 415–424. 10.1093/clinchem/hvaa267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb L. E., Bartolone S. N., Ward E., Chancellor M. B. (2020). Rapid detection of novel coronavirus/Severe Acute Respiratory Syndrome coronavirus 2 (SARS-CoV-2) by reverse transcription-loop-mediated isothermal amplification. PLoS One 15:e0234682. 10.1371/journal.pone.0234682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Helgouach N., Champigneux P., Santos-Schneider F., Molina L., Espeut J., Alali M., et al. (2020). EasyCOV?: LAMP based rapid detection of SARS-CoV-2 in saliva. medRxiv [Preprint]. 10.1101/2020.05.30.20117291 [DOI] [Google Scholar]
- Li C., Tian X., Jia X., Wan J., Lu L., Jiang S., et al. (2021). The impact of receptor-binding domain natural mutations on antibody recognition of SARS-CoV-2. Signal Trans. Target. Ther. 6:132. 10.1038/s41392-021-00536-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Fan P., Zhou S., Zhang L. (2017). Loop-mediated isothermal amplification (LAMP): a novel rapid detection platform for pathogens. Microb. Pathog. 107 54–61. 10.1016/j.micpath.2017.03.016 [DOI] [PubMed] [Google Scholar]
- Long Q.-X., Tang X.-J., Shi Q.-L., Li Q., Deng H.-J., Yuan J., et al. (2020). Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 26 1200–1204. 10.1038/s41591-020-0965-6 [DOI] [PubMed] [Google Scholar]
- Lu R., Wu X., Wan Z., Li Y., Jin X., Zhang C. (2020). A Novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Int. J. Mol. Sci. 21:2826. 10.3390/ijms21082826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mautner L., Baillie C.-K., Herold H. M., Volkwein W., Guertler P., Eberle U., et al. (2020). Rapid point-of-care detection of SARS-CoV-2 using reverse transcription loop-mediated isothermal amplification (RT-LAMP). Virol. J. 17:160. 10.1186/s12985-020-01435-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda P., Weber G. (2021). Thermodynamic evaluation of the impact of DNA mismatches in PCR-type SARS-CoV-2 primers and probes. Mol. Cell. Probes 56:101707. 10.1016/j.mcp.2021.101707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohon A. N., Oberding L., Hundt J., van Marle G., Pabbaraju K., Berenger B. M., et al. (2020). Optimization and clinical validation of dual-target RT-LAMP for SARS-CoV-2. J. Virol. Methods 286:113972. 10.1016/j.jviromet.2020.113972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y., Notomi T. (2009). Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Infect. Chemother. 15 62–69. 10.1007/s10156-009-0669-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawattanapaiboon K., Pasomsub E., Prombun P., Wongbunmak A., Jenjitwanich A., Mahasupachai P., et al. (2021). Colorimetric reverse transcription loop-mediated isothermal amplification (RT-LAMP) as a visual diagnostic platform for the detection of the emerging coronavirus SARS-CoV-2. Analyst 146 471–477. 10.1039/d0an01775b [DOI] [PubMed] [Google Scholar]
- Newman C. M., Ramuta M. D., McLaughlin M. T., Wiseman R. W., Karl J. A., Dudley D. M., et al. (2021). Initial evaluation of a mobile SARS-CoV-2 RT-LAMP testing strategy. medRxiv [Preprint]. 10.1101/2020.07.28.20164038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Österdahl M. F., Lee K. A., Lochlainn M. N., Wilson S., Douthwaite S., Horsfall R., et al. (2020). Detecting SARS-CoV-2 at point of care: preliminary data comparing loop-mediated isothermal amplification (LAMP) to polymerase chain reaction (PCR). BMC Infect. Dis. 20:783. 10.1186/s12879-020-05484-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida M., Sannarangaiah S., Dash P. K., Rao P. V. L., Morita K. (2008). Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev. Med. Virol. 18 407–421. 10.1002/rmv.593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G.-S., Ku K., Baek S.-H., Kim S.-J., Kim S. I., Kim B.-T., et al. (2020). Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). J. Mol. Diagn. 22 729–735. 10.1016/j.jmoldx.2020.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabe B. A., Cepko C. (2020). SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc. Natl. Acad Sci. U.S.A. 117 24450–24458. 10.1073/pnas.2011221117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E. C., O’Toole Á, Hill V., McCrone J. T., Ruis C., et al. (2020). A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 5 1403–1407. 10.1038/s41564-020-0770-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende P. C., Bezerra J. F., Teixeira Vasconcelos R. H., Arantes I., Appolinario L., Mendonça A. C., et al. (2021a). Severe acute respiratory syndrome coronavirus 2 P.2 lineage associated with reinfection case, Brazil, June-October 2020. Emerg. Infect. Dis. 27 1789–1794. 10.3201/eid2707.210401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende P. C., Gräf T., Paixão A. C. D., Appolinario L., Lopes R. S., Mendonça A. C., et al. (2021c). A potential SARS-CoV-2 Variant of Interest (VOI) harboring mutation E484K in the spike protein was identified within lineage B.1.1.33 circulating in Brazil. Viruses 13:724. 10.3390/v13050724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende P. C., Delatorre E., Gräf T., Mir D., Motta F. C., Appolinario L. R., et al. (2020). Evolutionary dynamics and dissemination pattern of the SARS-CoV-2 lineage B.1.1.33 during the early pandemic phase in Brazil. Front. Microbiol. 11:615280. 10.3389/fmicb.2020.615280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende P. C., Gräf T., Lima Neto L. G. L., da Silva F. V., Paixão A. C. D., Appolinario L., et al. (2021b). Identification of a new B.1.1.33 SARS-CoV-2 Variant of Interest (VOI) circulating in Brazil with mutation E484K and multiple deletions in the amino (N)-terminal domain of the Spike protein – SARS-CoV-2 coronavirus / nCoV-2019 genomic epidemiology. Virological. Available online at: https://virological.org/t/identification-of-a-new-b-1-1-33-sars-cov-2-variant-of-interest-voi-circulating-in-brazil-with-mutation-e484k-and-multiple-deletions-in-the-amino-n-terminal-domain-of-the-spike-protein/675 (accessed May 14, 2021). [Google Scholar]
- Rhoads D., Peaper D. R., She R. C., Nolte F. S., Wojewoda C. M., Anderson N. W., et al. (2020). College of American Pathologists (CAP) Microbiology Committee Perspective: Caution must be used in interpreting the Cycle Threshold (Ct) value. Clin. Infect. Dis. 72 e685–e686. 10.1093/cid/ciaa1199 [DOI] [PubMed] [Google Scholar]
- Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., et al. (2020). Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 382 970–971. 10.1056/NEJMc2001468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermer B., Fabretti F., Damagnez M., Cristanziano V. D., Heger E., Arjune S., et al. (2020). Rapid SARS-CoV-2 testing in primary material based on a novel multiplex RT-LAMP assay. PLoS One 15:e0238612. 10.1371/journal.pone.0238612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M., Ahmad B., Choi S., Woo H. G. (2020). Mutations in the SARS-CoV-2 spike RBD are responsible for stronger ACE2 binding and poor anti-SARS-CoV mAbs cross-neutralization. Comput. Struct. Biotechnol. J. 18 3402–3414. 10.1016/j.csbj.2020.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrill-Mix S., Duyne G. D. V., Bushman F. D. (2021a). Molecular beacons allow specific RT-LAMP detection of B.1.1.7 variant SARS-CoV-2. medRxiv [Preprint] 10.1101/2021.03.25.21254356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrill-Mix S., Hwang Y., Roche A. M., Glascock A., Weiss S. R., Li Y., et al. (2021b). Detection of SARS-CoV-2 RNA using RT-LAMP and molecular beacons. Genome Biol. 22:169. 10.1186/s13059-021-02387-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva S. J. R., Paiva M. H. S., Guedes D. R. D., Krokovsky L., de Melo F. L., da Silva M. A. L., et al. (2019). Development and validation of reverse transcription loop-mediated isothermal amplification (RT-LAMP) for rapid detection of ZIKV in mosquito samples from Brazil. Sci. Rep. 9:4494. 10.1038/s41598-019-40960-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyrlaki I., Ekman M., Lentini A., Rufino de Sousa N., Papanicolaou N., Vondracek M., et al. (2020). Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-PCR. Nat. Commun. 11:4812. 10.1038/s41467-020-18611-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., El-Tholoth M., Li Y., Graham-Wooten J., Liang Y., Li J., et al. (2021). Single- and two-stage, closed-tube, point-of-care, molecular detection of SARS-CoV-2. Anal. Chem. 93, 13063–13071. 10.1021/acs.analchem.1c03016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivatsan S., Han P. D., van Raay K., Wolf C. R., McCulloch D. J., Kim A. E., et al. (2020). Preliminary support for a “dry swab, extraction free” protocol for SARS-CoV-2 testing via RT-qPCR. medRxiv [Preprint] 10.1101/2020.04.22.056283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner N. A., Zhang Y., Evans T. C. (2015). Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. Biotechniques 58 59–68. 10.2144/000114253 [DOI] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., et al. (2021). Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 592 438–443. 10.1038/s41586-021-03402-9 [DOI] [PubMed] [Google Scholar]
- Thi V. L. D., Herbst K., Boerner K., Meurer M., Kremer L. P., Kirrmaier D., et al. (2020). A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 12:eabc7075. 10.1126/scitranslmed.abc7075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voloch C. M., da Silva Francisco R., Almeida L. G. P., de, Cardoso C. C., Brustolini O. J., et al. (2021). Genomic characterization of a novel SARS-CoV-2 Lineage from Rio de Janeiro, Brazil. J. Virol. 95 e119–e121. 10.1128/JVI.00119-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P. W., Hayden F. G., Gao G. F. (2020). A novel coronavirus outbreak of global health concern. Lancet 395 470–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D. (2020). One-pot detection of COVID-19 with real-time reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay and visual RT-LAMP assay. medRxiv [Preprint] 10.1101/2020.04.21.052530 [DOI] [Google Scholar]
- Wei S., Kohl E., Djandji A., Morgan S., Whittier S., Mansukhani M., et al. (2021). Direct diagnostic testing of SARS-CoV-2 without the need for prior RNA extraction. Sci. Rep. 11:2402. 10.1038/s41598-021-81487-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Davies E. S. W., Mahanama A. I. K., Samaraweera B., Ahmed N., Friar S., Pelosi E. (2021). Concerning the OptiGene Direct LAMP assay, and it’s use in at-risk groups and hospital staff. J. Infect 82 282–327. 10.1016/j.jinf.2021.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Cui J., Huang L., Du B., Chen L., Xue G., et al. (2020). Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 26 773–779. 10.1016/j.cmi.2020.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Meyerson N. R., Clark S. K., Paige C. L., Fattor W. T., Gilchrist A. R., et al. (2021). Saliva TwoStep for rapid detection of asymptomatic SARS-CoV-2 carriers. medRxiv [Preprint] 10.1101/2020.07.16.20150250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Wu S., Hao X., Dong X., Mao L., Pelechano V., et al. (2020). Rapid detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin. Chem. 66 975–977. 10.1093/clinchem/hvaa102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Odiwuor N., Xiong J., Sun L., Nyaruaba R., Wei H., et al. (2020a). Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRxiv [Preprint] 10.1101/2020.02.26.20028373 [DOI] [Google Scholar]
- Zhang Y., Ren G., Buss J., Barry A. J., Patton G. C., Tanner N. A. (2020b). Enhancing colorimetric loop-mediated isothermal amplification speed and sensitivity with guanidine chloride. BioTechniques 69 178–185. 10.2144/btn-2020-0078 [DOI] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. (2020). A novel coronavirus from patients with Pneumonia in China, 2019. N. Engl. J. Med. 382 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.gisaid.org/, The genomes of SARS-CoV-2 VOI and VOCs generated are deposited on GISAID according to the following accession codes: EPI_ISL_2221860, EPI_ISL_2221850, EPI_ISL_2221873, EPI_ISL_2221890, EPI_ISL_2221902, EPI_ISL_2221885, EPI_ISL_2221844, and EPI_ISL_2221866.