This study expands the genotypic and phenotypic spectra of ichthyosis and delineates genotype-phenotype associations.
Key Points
Question
What is the genotypic and phenotypic spectrum of ichthyosis, and what are significant genotype-phenotype associations?
Findings
This cohort study identified disease-associated variants in 869 of 1000 unrelated kindreds in 59 genes, including 266 novel pathogenic variants in 32 genes, expanding the genotypic and phenotypic spectra and delineating clinical features that may serve as diagnostic clues.
Meaning
This study highlights the broad spectrum of phenotypes associated with individual disease-associated gene variants, phenotypic features that may serve as diagnostic clues, and the importance of detailed clinical and genetic characterization to define disease natural history in rare disorders.
Abstract
Importance
Ichthyoses are clinically and genetically heterogeneous disorders characterized by scaly skin. Despite decades of investigation identifying pathogenic variants in more than 50 genes, clear genotype-phenotype associations have been difficult to establish.
Objective
To expand the genotypic and phenotypic spectra of ichthyosis and delineate genotype-phenotype associations.
Design, Setting, and Participants
This cohort study recruited an international group of individuals with ichthyosis and describes characteristic and distinguishing features of common genotypes, including genotype-phenotype associations, during a 10-year period from June 2011 to July 2021. Participants of all ages, races, and ethnicities were included and were enrolled worldwide from referral centers and patient advocacy groups. A questionnaire to assess clinical manifestations was completed by those with a genetic diagnosis.
Main Outcomes and Measures
Genetic analysis of saliva or blood DNA, a phenotyping questionnaire, and standardized clinical photographs. Descriptive statistics, such as frequency counts, were used to describe the cases in the cohort. Fisher exact tests identified significant genotype-phenotype associations.
Results
Results were reported for 1000 unrelated individuals enrolled from around the world (mean [SD] age, 50.0 [34.0] years; 524 [52.4%] were female, 427 [42.7%] were male, and 49 [4.9%] were not classified); 75% were from the US, 12% from Latin America, 4% from Canada, 3% from Europe, 3% from Asia, 2% from Africa, 1% from the Middle East, and 1% from Australia and New Zealand. A total of 266 novel disease-associated variants in 32 genes were identified among 869 kindreds. Of these, 241 (91%) pathogenic variants were found through multiplex amplicon sequencing and 25 (9%) through exome sequencing. Among the 869 participants with a genetic diagnosis, 304 participants (35%) completed the phenotyping questionnaire. Analysis of clinical manifestations in these 304 individuals revealed that pruritus, hypohydrosis, skin pain, eye problems, skin odor, and skin infections were the most prevalent self-reported features. Genotype-phenotype association analysis revealed that the presence of a collodion membrane at birth (odds ratio [OR], 6.7; 95% CI, 3.0-16.7; P < .001), skin odor (OR, 2.8; 95% CI, 1.1-6.8; P = .02), hearing problems (OR, 2.9; 95% CI, 1.6-5.5; P < .001), eye problems (OR, 3.0; 95% CI, 1.5-6.0; P < .001), and alopecia (OR, 4.6; 95% CI, 2.4-9.0; P < .001) were significantly associated with TGM1 variants compared with other ichthyosis genotypes studied. Skin pain (OR, 6.8; 95% CI, 1.6-61.2; P = .002), odor (OR, 5.7; 95% CI, 2.0-19.7; P < .001), and infections (OR, 3.1; 95% CI, 1.4-7.7; P = .03) were significantly associated with KRT10 pathogenic variants compared with disease-associated variants in other genes that cause ichthyosis. Pathogenic variants were identified in 869 (86.9%) participants. Most of the remaining individuals had unique phenotypes, enabling further genetic discovery.
Conclusions and Relevance
This cohort study expands the genotypic and phenotypic spectrum of ichthyosis, establishing associations between clinical manifestations and genotypes. Collectively, the findings may help improve clinical assessment, assist with developing customized management plans, and improve clinical course prognostication.
Introduction
Disorders of cornification such as ichthyoses are diverse disorders featuring scale and erythema. Most are inherited and rare and manifest initially at, or shortly after, birth. Disease-associated variants in more than 50 genes result in barrier defects that increase transepidermal water loss and compensatory hyperproliferation.1
Despite advances in understanding of pathogenesis, diagnosis and classification remain challenging. Pathogenic variants have not been identified in 17% to 22% of participants in prior cohorts, and significant genetic heterogeneity is present.2,3 There is also a broad phenotypic spectrum, including palmoplantar keratoderma (PPK), hair and nail abnormalities, hypohydrosis, ectropion, recurrent infections, and dental and neurological abnormalities. Although genetic testing is the diagnostic standard, it is not uniformly performed.
To identify new genetic causes of ichthyosis and create a platform to advance basic, clinical, and translational research in these rare genodermatoses, we reestablished the National Registry for Ichthyosis and Related Skin Disorders, previously funded by the National Institutes of Health but closed to enrollment in 2012. With the Foundation for Ichthyosis and Related Skin Types (FIRST), we enrolled more than 1400 kindreds worldwide (eFigure 1 in the Supplement). Participants were screened for pathogenic variants using an ichthyosis gene panel before using exome sequencing to identify novel and rare genetic causes.
We report genotypic and phenotypic data of what is to our knowledge the largest cohort of ichthyosis and related disorders to date—1000 unrelated kindreds (eFigure 2 in the Supplement). We discovered novel disease-associated variants and systematically evaluated clinical characteristics with respect to genetic diagnosis. In a subset of individuals, we investigated genotype-phenotype associations, focusing on cardinal, characteristic, and distinguishing features. Our goal was to provide an overview of the phenotypic and genotypic landscape to set the stage for classification based on genotypes associated with clinical course.
Methods
Participants and Samples
This study was approved by the Yale Human Investigation Committee, consistent with Declaration of Helsinki guidelines. Participants of all ages were recruited over a 10-year period through family conferences, self-referrals, and referrals from physicians. Kindreds were identified by asking for the surname, names of both parents, and regional location on enrollment. Written informed consent was obtained from participants and/or their parents.
Individuals completed a questionnaire assessing clinical manifestations (eTable 1 in the Supplement) and provided a saliva or blood sample. Fully completed questionnaires were obtained from participants with a genetic diagnosis. When possible, standardized photographs were taken. Participants were characterized based on genotype. Those being treated with retinoids were only excluded from clinical analyses. Participants self-reported race and ethnicity per National Institutes of Health data collection guidelines.
We have enrolled more than 1400 kindreds to date, but only report kindreds for which we performed primary genotyping, as some received prior clinical genetic testing, which we do not include in this article.
Genetic Analysis
Standard protocols were followed for DNA extraction. Multiplex polymerase chain reaction amplification of exons plus approximately 50 flanking bases was performed for genes implicated in Mendelian disorders of cornification in prior publications, including AAGAB, ABCA12, ABHD5, ALDH3A2, ALOX12B, ALOXE3, AQP5, ATP2A2, ATP2C1, CARD14, CDSN, CERS3, CLDN1, CSTA, CYP4F22, DKC1, DSC2, DSG1, DSP, EBP, EDA, FLG, GJA1, GJB2, GJB3, GJB4, GJB6, KANK2, KRT1, KRT10, KRT16, KRT17, KRT2, KRT6C, KRT9, LOR, MBTPS2, NIPAL4, NSDHL, PNPLA1, POGLUT1, RHBDF2, RSPO1, SERPINB7, SLC27A4, SLURP1, SNAP29, SPINK5, SREBF1, STS, TGM1, and TRPV3 (GeneRead, Qiagen). Library preparation and pooled, paired-end Illumina sequencing generated approximately 10 million reads per 40 samples, mean coverage of greater than 800 times, and 96% bases covered more than 100 times, with variant calling by GATK (Broad Institute). For cases without compelling pathogenic variants identified in panel-based screening, exome sequencing was performed via capture with IDT xGen oligo pools and Illumina sequencing. Reads were aligned to the hg38 reference genome with BWA-MEM, and variants were called with GATK and annotated for impact and prevalence in control data sets with ANNOVAR. Data were examined for damaging variants (indels, missense, splice site, and nonsense) that were absent or extremely rare in control databases. Ortholog conservation was examined via PhyloP and prediction of damaging impact was assessed with SIFT and PolyPhen. Previously reported disease-associated variants were considered pathogenic, as were damaging novel mutations in known genes with a consistent clinical presentation and mode of inheritance.
Statistical Analysis
Fisher exact tests identified genotype-phenotype associations in 304 participants. Odds ratios (ORs) were calculated relative to others in the cohort for whom we have questionnaire data. Analyses were completed with R, version 1.2.1335 (R Foundation for Statistical Computing).4 P values are 2-tailed, and statistical significance was adjusted for multiple comparisons by the Benjamini-Hochberg procedure (false discovery rate of 0.1, resulting in a significance threshold P < .04).5
Results
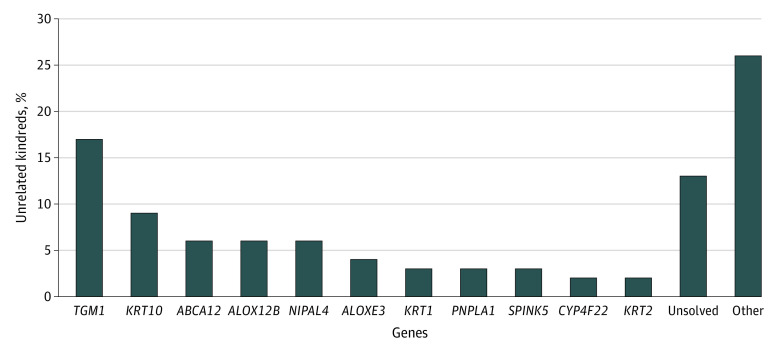
Within 1000 unrelated kindreds (mean [SD] age, 50.0 [34.0] years; 524 [52.4%] were female, 427 [42.7%] were male, and 49 [4.9%] were not classified), pathogenic variants were identified in 872 (87.2%). Demographics and genotypes are reported in Table 1 and Figure 1, respectively. Disease-associated variants were found in 869 kindreds. Recessive variants in 32 genes were found in 629 kindreds (eTable 2A in the Supplement). Dominant variants in 27 genes were found in 240 kindreds (eTable 2B in the Supplement). Genotype-phenotype association analyses were performed for genes with variants in more than 30 unrelated index cases from distinct kindreds (Table 2), comprising 73.9% (642 of 869) solved kindreds and 64.2% (642 of 1000) overall (Figure 1). Novel variants are unpublished and unannotated in the Single Nucleotide Polymorphism Database (eTable 3 in the Supplement). Herein, we describe characteristic and distinguishing features and significant genotype-phenotype associations for the most prevalent causes of ichthyosis and related disorders of cornification.
Table 1. Mendelian Disorders of Cornification Cohort Demographics (n = 1000).
| Category | Participants, No. (%) |
|---|---|
| Sex | |
| Female | 524 (52.4) |
| Male | 427 (42.7) |
| Unclassified | 49 (4.9) |
| Race and ethnicity | |
| Asian | 87 (8.9) |
| Black | 52 (5.3) |
| Hispanic/Latino | 161 (16.1) |
| Middle Eastern | 7 (0.7) |
| Mixed | 8 (0.8) |
| Native American | 3 (0.3) |
| White | 512 (51.2) |
| Unclassified | 170 (17.0) |
| Age, y | |
| <18 | 456 (45.5) |
| ≥18 | 502 (50.3) |
| Unclassified | 42 (4.2) |
| Ichthyosis subtype | |
| ARCI-LI and ARCI-CIE | 450 (44.6) |
| EI | 80 (8.5) |
| Ichthyosis vulgaris | 46 (4.6) |
| X-linked ichthyosis | 47 (4.7) |
| Harlequin ichthyosis | 12 (1.2) |
| Darier disease | 13 (1.2) |
| Hailey-Hailey disease | 18 (1.7) |
| Netherton syndrome | 29 (2.9) |
| Ichthyosis with confetti | 39 (3.9) |
| Other | 138 (13.9) |
| Unsolved | 128 (12.8) |
Abbreviations: ARCI, autosomal recessive congenital ichthyosis; CIE, congenital ichthyosiform erythroderma; EI, epidermolytic ichthyosis; LI, lamellar ichthyosis.
Figure 1. Genotypic Spectrum of 1000 Ichthyosis Unrelated Kindreds.
Table 2. Genotype-Phenotype Analysis for Participants Who Completed a Questionnaire.
| TGM1 (n = 71) | ABCA12 (n = 24) | ALOX12B (n = 32) | ALOXE3 (n = 19) | NIPAL4 (n = 31) | KRT10 (n = 37) | KRT1 (n = 14) | FLG (n = 38) | STS (n = 38) | Total (n = 304) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Collodion membrane | ||||||||||
| No. (%) | 60/69 (87) | 9/19 (47) | 27/29 (93) | 12/16 (75) | 11/29 (38) | 3/27 (11) | 4/13 (31) | 5/31 (16) | 4/27 (15) | 135/260 (52) |
| OR (95% CI) | 6.7 (3.0-16.7) | 0.5 (0.2-1.5) | 10.0 (2.4-89.6) | 1.9 (0.5-8.3) | 0.3 (0.1-0.7) | 0.1 (0.0-0.2) | 0.3 (0.1-0.9) | 0.2 (0.0-0.4) | 0.1 (0.0-0.4) | |
| P value | <.001 | .21 | <.001 | .42 | .006 | <.001 | .02 | <.001 | <.001 | |
| Skin pain | ||||||||||
| No. (%) | 39/66 (59) | 17/22 (77) | 17/27 (63) | 11/16 (69) | 22/29 (76) | 29/31 (94) | 10/11 (91) | 34/38 (89) | 16/38 (42) | 195/278 (70) |
| OR (95% CI) | 0.4 (0.2-0.8) | 1.4 (0.5-5.0) | 0.6 (0.2-1.6) | 0.9 (0.3-3.3) | 1.3 (0.5-3.8) | 6.8 (1.6-61.2) | 4.1 (0.6-183.0) | 2.6 (0.9-10.5) | 0.3 (0.1-0.5) | |
| P value | .007 | .63 | .36 | .78 | .66 | .002 | .19 | .09 | <.001 | |
| Pruritus | ||||||||||
| No. (%) | 62/65 (95) | 21/22 (95) | 25/29 (86) | 13/16 (81) | 27/31 (87) | 28/31 (90) | 7/10 (70) | 36/37 (97) | 27/30 (90) | 246/271 (91) |
| OR (95% CI) | 3.1 (0.9-16.8) | 2.6 (0.4-112.5) | 0.7 (0.2-3.0) | 0.5 (0.1-2.8) | 0.7 (0.2-3.2) | 1.1 (0.3-6.1) | 0.2 (0.1-1.6) | 4.1 (0.6-173.7) | 0.9 (0.3-5.0) | |
| P value | .08 | .71 | .51 | .22 | .53 | >.99 | .07 | .22 | .75 | |
| Skin odor | ||||||||||
| No. (%) | 29/38 (76) | 13/24 (54) | 7/31 (23) | 5/16 (31) | 15/31 (48) | 31/36 (86) | 11/14 (79) | 2/37 (5) | 4/32 (13) | 117/259 (45) |
| OR (95% CI) | 2.8 (1.1-6.8) | 0.8 (0.3-2.2) | 0.2 (0.1-0.4) | 0.3 (0.1-1.0) | 0.6 (0.3-1.4) | 5.7 (2.0-19.7) | 2.8 (0.7-16.0) | 0.1 (0.0-0.2) | 0.2 (0.0-0.4) | |
| P value | .02 | .66 | <.001 | .03 | .24 | <.001 | .16 | <.001 | <.001 | |
| Skin infections | ||||||||||
| No. (%) | 31/70 (44) | 7/24 (29) | 14/32 (44) | 8/17 (47) | 18/29 (62) | 27/37 (73) | 8/14 (57) | 13/37 (35) | 5/32 (16) | 131/272 (48) |
| OR (95% CI) | 0.7 (0.3-1.3) | 0.4 (0.1-1.0) | 0.7 (0.3-1.6) | 0.9 (0.3-2.6) | 1.7 (0.7-4.2) | 3.1 (1.4-7.7) | 1.3 (0.4-4.8) | 0.6 (0.3-1.4) | 0.2 (0.1-0.5) | |
| P value | .25 | .03 | .45 | .09 | .23 | .03 | .78 | .22 | <.001 | |
| Hypohydrosis | ||||||||||
| No. (%) | 58/65 (89) | 15/20 (75) | 23/28 (82) | 11/13 (85) | 26/30 (87) | 21/32 (66) | 6/11 (55) | 25/37 (68) | 13/28 (46) | 198/264 (75) |
| OR (95% CI) | 2.6 (1.0-7.4) | 0.4 (0.1-1.1) | 1.1 (0.4-4.1) | 1.4 (0.3-13.2) | 1.6 (0.5-6.7) | 0.4 (0.2-1.0) | 0.3 (0.1-1.2) | 0.7 (0.3-1.5) | 0.2 (0.1-0.6) | |
| P value | .04 | .55 | >.99 | >.99 | .61 | .03 | .04 | .31 | <.001 | |
| Hearing problems | ||||||||||
| No. (%) | 45/71 (63) | 13/24 (54) | 11/29 (38) | 5/17 (29) | 13/31 (42) | 11/36 (31) | 3/14 (21) | 11/38 (29) | 6/38 (16) | 118/298 (40) |
| OR (95% CI) | 2.9 (1.6-5.5) | 1.5 (0.6-3.8) | 0.7 (0.3-1.7) | 0.5 (0.1-1.5) | 0.9 (0.4-2.0) | 0.5 (0.2-1.1) | 0.4 (0.1-1.5) | 0.6 (0.3-1.3) | 0.3 (0.1-0.6) | |
| P value | <.001 | .39 | .48 | .21 | .70 | .07 | .17 | .16 | .001 | |
| Eye problems | ||||||||||
| No. (%) | 48/68 (71) | 15/24 (63) | 15/30 (50) | 8/17 (47) | 16/31 (52) | 13/37 (35) | 3/14 (21) | 20/38 (53) | 12/38 (32) | 150/297 (51) |
| OR (95% CI) | 3.0 (1.5-6.0) | 1.5 (0.6-4.1) | 0.9 (0.4-2.0) | 0.8 (0.2-2.3) | 0.8 (0.4-1.9) | 0.4 (0.2-0.9) | 0.2 (0.0-0.9) | 1.1 (0.5-2.3) | 0.4 (0.2-0.9) | |
| P value | <.001 | .39 | .70 | .62 | .70 | .02 | .02 | .86 | .01 | |
| Alopecia | ||||||||||
| No. (%) | 41/68 (60) | 12/24 (50) | 6/31 (19) | 5/14 (36) | 4/30 (13) | 9/37 (24) | 1/14 (7) | 13/38 (34) | 7/29 (24) | 98/285 (34) |
| OR (95% CI) | 4.6 (2.4-9.0) | 1.9 (0.7-5.0) | 0.4 (0.1-1.0) | 1.0 (0.3-3.5) | 0.2 (0.1-0.7) | 0.5 (0.2-1.2) | 0.1 (0.0-0.9) | 1.0 (0.4-2.1) | 0.6 (0.2-1.5) | |
| P value | <.001 | .17 | .04 | >.99 | .007 | .13 | .02 | >.99 | .30 |
Abbreviation: OR, odds ratio.
Clinical manifestations were available for 304 individuals (30%) with complete survey data. Analysis revealed that pruritus (246 of 247 [91%]), hypohydrosis (198 of 264 [75%]), skin pain (195 of 278 [70%]), eye problems (150 of 297 [51%]), skin odor (117 of 259 [45%]), and skin infections (131 of 272 [45%]) were the most prevalent self-reported features.
Recessive Ichthyoses
Ichthyoses include nonsyndromic and syndromic forms. In the former, phenotypes are limited to the skin; in the latter, additional organ systems are involved.1 Autosomal recessive congenital ichthyosis encompasses several rare nonsyndromic ichthyosis phenotypes—harlequin ichthyosis (HI), lamellar ichthyosis (LI), and congenital ichthyosiform erythroderma (CIE). These rare disorders have an estimated incidence of 1 in 200 000 births.6
Other phenotypes include Sjögren-Larsson syndrome, Netherton syndrome, neutral lipid storage disease, peeling skin syndrome 1, exfoliative ichthyosis, CEDNIK (cerebral dysgenesis, neuropathy, ichthyosis, keratoderma) syndrome, neonatal ichthyosis-sclerosing cholangitis syndrome, and ichthyosis prematurity syndrome. Overall, 470 of 629 (74.7%) recessive cases in the cohort were associated with variants in 8 genes: TGM1, ABCA12, ALOX12B, ALOXE3, NIPAL4, PNPLA1, SPINK5, and CYP4F22.
TGM1
The TGM1 gene encodes transglutaminase-1, an enzyme that cross-links proteins to form the cornified cell envelope.7 Variants in TGM1 accounted for 26.9% of recessive cases (169 kindreds, 54 novel variants).2,8
Phenotypes are in eFigure 3 in the Supplement. Most participants (60 of 69 [87.0%]) were born with a collodion, a tight, shiny, parchment-like membrane encasing the neonate, resulting in ectropion and eclabion. Variants in TGM1 were found in both LI phenotype cases with flat, dark, plate-like scale and fine, white, superficial scales with mild to moderate erythema and ectropion in CIE cases. Individuals with TGM1 variants were more likely to have collodion membrane (OR, 6.7; 95% CI, 3.0-16.7) and to experience alopecia (OR, 4.6; 95% CI, 2.4-9.0) than those with other variants. They were nearly 3 times more likely to experience hearing problems, eye problems, and skin odor. Skin pain was less likely to occur (Table 2).
ABCA12
The ABCA12 gene encodes an ATP-binding cassette (ABC) transporter that transports lipids into lamellar bodies.9,10 Sixty-six kindreds had ABCA12 variants (41 novel).
Ichthyosis caused by ABCA12 pathogenic variants included significant, often severe erythema; fine, white, or thick lamellar scale, commonly localized to extremities or face; ectropion; and PPK (eFigure 4 in the Supplement). Distinguishing features in almost all participants with ABCA12 variants, rarely found in others, included tapered digits, hyperconvex nails, and auricle malformation (eFigure 4D-G in the Supplement).
At least 50% of participants with ABCA12 variants reported pruritus (21 of 22), skin pain (17 of 22), hypohydrosis (15 of 20), eye problems (15 of 24), hearing problems (13 of 24), and alopecia (12 of 24), though these symptoms did not occur at a significantly higher rate in those with other disease-associated variants. Skin infections were less likely to occur in participants with ABCA12 variants (Table 2).
Consistent with previous reports, phenotypic variation may be partially explained by variant type.11 Nonsense variants in ABCA12 have been reported to result in HI, characterized by neonatal encasement in a thick, armor-like scale overlying intense erythroderma, eclabium, ectropion, and flattened ears. Missense variants were consistently found in those with milder phenotypes similar to those seen in CIE, while those with a nonsense variant had a spectrum of phenotypes, including CIE and HI.12 Twelve participants had truncating variants on both alleles, resulting in HI.
ABCA12 Pathogenic Variants in Erythrokeratodermia Variabilis et Progressiva
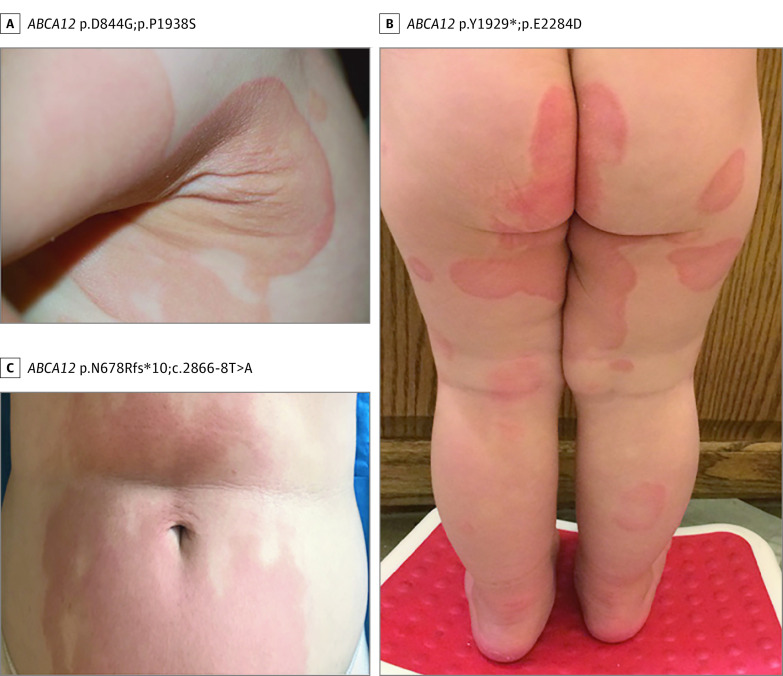
Unexpectedly, 2 individuals with pathogenic ABCA12 variants developed annular, migratory red patches shortly after birth, which developed into symmetric hyperkeratotic plaques on the extremities, trunk, and buttocks in childhood (Figure 2A and B). A third individual developed sharply demarcated, fixed, red-brown hyperkeratotic plaques distributed symmetrically (Figure 2C). All lacked corneal abnormalities and hearing loss. Clinical presentations suggested erythrokeratodermia variabilis et progressiva (EKVP), but all harbored compound heterozygous variants in ABCA12, a gene not previously associated with EKVP. The first individual was heterozygous for frameshift and splice-site variants (p.N678Rfs*10;c.2866-8T>A). The second was heterozygous for nonsense and missense variants (p.Y1929*;p.E2284D). The third had compound heterozygous missense variants (p.D844G;p.P1938S). No other pathogenic variants were found.
Figure 2. Phenotypes of Erythrokeratodermia Variabilis et Progressiva Associated With ABCA12 Pathogenic Variants.
These findings establish that ABCA12 pathogenic variants were associated with recessive EKVP, an otherwise dominant disorder. As has been previously reported, the ABCA12 protein has an indirect but critical role in linoleic ester production from glucosyl ceramide precursors to maintain the epidermal permeability barrier.13 In ABCA12-deficient mice, absent lamellar organelles in epidermis and defective ceramide distribution led to severe skin barrier defects.13,14 Variants in KDSR cause progressive symmetrical erythrokeratodermia and defects in primary ceramide biosynthesis, providing a potential mechanism for EKVP phenotypes associated with ABCA12 variants.15,16
ALOX12B and ALOXE3
The ALOX12B and ALOXE3 genes encode 12R-lipoxygenase (12R-LOX) and epidermis-type lipoxygenase 3 (eLOX-3), respectively. These enzymes sequentially oxygenate ceramides necessary for the corneocyte lipid envelope.17 Sixty-three kindreds had ALOX12B variants (33 novel), and 40 kindreds had ALOXE3 variants (10 novel).
Ichthyosis associated with ALOX12B or ALOXE3 pathogenic variants demonstrated mild to moderate fine, white or light-brown scale; absent to mild PPK; and mild erythema (eFigures 5 and 6 in the Supplement). Neonatal collodion was more likely in those with ALOX12B variants (OR, 10.0; 95% CI, 2.4-89.6). Participants with ALOX12B and ALOXE3 variants also reported pruritus, hypohydrosis, and skin pain, while skin odor was less likely (Table 2).
NIPAL4
The NIPAL4 gene encodes NIPA-like domain containing 4 protein. This membrane transporter likely functions as a Mg2+ transporter with roles in lipid processing and lamellar body formation.18,19 Variants in NIPAL4 were identified in 56 kindreds (4 novel). As in prior reports,20 the p.A176D variant was most prevalent, representing 87.0% (13 of 15) of all NIPAL4 variants found.
The characteristic appearance of NIPAL4 ichthyosis included fine, white scale; yellow keratoderma; and varying degrees of erythema in the CIE spectrum (eFigure 7 in the Supplement). Larger brown scales, similar to those of LI, were also found. There was significant clinical variability without apparent variant correlation. While some participants with homozygous p.A176D disease-associated variants exhibited barely perceptible pink erythema and areas of slight smoothening (eFigure 7F in the Supplement), others had pink erythema with confluent scale (eFigure 7H in the Supplement).
Additional features reported by participants with NIPAL4 pathogenic variants included pruritus, hypohydrosis, skin pain, and skin infections. Collodion membrane at birth and alopecia were significantly less common among those with NIPAL4 disease-associated variants (Table 2).
PNPLA1
The PNPLA1 gene encodes patatin-like phospholipase domain–containing 1, which is central to acylceramide biosynthesis.21,22 Twenty-nine kindreds had PNPLA1 pathogenic variants (13 novel).
The characteristic appearance of PNPLA1 ichthyosis included fine or plate-like scale and variable erythema (eFigure 8 in the Supplement). Clear genotype-phenotype associations for PNPLA1 are lacking.23 Disease-associated variants in PNPLA1 manifested as mild or moderate erythema with fine, white scale and mild erythema with more severe scale (eFigure 8F-H in the Supplement). Palmoplantar keratoderma and ectropion were either absent or mild.
SPINK5
The SPINK5 gene encodes a serine protease inhibitor, and pathogenic variants result in Netherton syndrome.24 Thirty kindreds had SPINK5 disease-associated variants (17 novel).
Netherton syndrome features generalized or localized scaly skin, hair shaft abnormalities, and atopy.24 Netherton syndrome is frequently misdiagnosed as atopic dermatitis, especially if distinguishing features such as ichthyosis linearis circumflexa, which presents as polycyclic erythematous plaques with a double-edge configuration (eFigure 9A-F in the Supplement), are absent.25 Persons with SPINK5 pathogenic variants can also have LI or CIE phenotypes, as in 1 individual in this cohort with congenital fine white scale, erythema, and moderate PPK associated with p.K420E, p.E825Rfs*117 pathogenic variants (eFigure 9G-L in the Supplement). Trichorrhexis invaginata, or “bamboo hair,” resulting from intussusception of the distal hair shaft into the proximal hair shaft, is another pathognomonic feature. The hairs are dry, brittle, and lusterless. Atopic features include elevated serum immunoglobulin E, atopic dermatitis, urticaria, and angioedema.25
CYP4F22
The CYP4F22 gene encodes a fatty acid ω-hydroxylase necessary for the formation of acylceramides.26,27 Seventeen kindreds had CYP4F22 disease-associated variants (9 novel).
The characteristic appearance of ichthyosis associated with pathogenic variants in CYP4F22 included fine, white or plate-like scale, mild to severe erythema, and absence of PPK (eFigure 10 in the Supplement). Participants with CYP4F22 disease-associated variants exhibited a wide phenotypic spectrum, which ranged from imperceptible pink erythema and plate-like scale to moderately severe erythema with fine white scale (eFigure 10F and 10G in the Supplement).
Dominant Ichthyoses
Keratinopathic ichthyosis accounted for 143 of 240 (59.6%) dominant cases. Keratins 10 and 1 are intermediate filaments that comprise the cytoskeleton.28,29 Defects in this network were associated with compromised suprabasal epidermis structural integrity and induced cell stress, resulting in skin fragility, blistering, and hyperkeratosis. Pathogenic variants in KRT10 and KRT1 accounted for 94 of 240 (39.1%) and 31 of 240 (12.9%) dominant cases, respectively.
Heterozygous KRT10 and KRT1 disease-associated variants have been previously reported to cause epidermolytic ichthyosis (EI), characterized by diffuse erythroderma and skin fragility with blisters during the neonatal period, evolving into hyperkeratosis and blistering at trauma sites in adulthood.30,31,32,33 Heterozygous KRT10 and KRT1 pathogenic variants have been previously reported to cause ichthyosis with confetti (IWC), distinguished by confetti-like spots of healthy skin on a background of ichthyosiform erythroderma.34,35 In this study, IWC-causative variants in KRT10 and KRT1 were associated with similar but distinct phenotypes, denoted as IWC-I and IWC-II, respectively.
KRT10
Ninety-four kindreds had KRT10 pathogenic variants; 28 were novel. The characteristic appearance of EI caused by KRT10 disease-associated variants included piled scale (eFigure 11 in the Supplement). Corrugation, particularly in flexural areas, and skin fragility, especially at trauma sites, were additional characteristics. Erythema ranged from imperceptible to severe, even among individuals with identical pathogenic variants (eFigure 11G and 11H in the Supplement).
Of the 94 KRT10 variants, 29 manifested as IWC-I. Among 39 patients with IWC in the study, 29 (74.4%) were heterozygous for a frameshift or splice-site KRT10 pathogenic variant affecting the tail domain. Most exhibited CIE with evolving phylloid-shaped white spots of normal skin, but there was some phenotypic variability. For example, PPK was absent in some but mild to moderate in others; similarly, ectropion, hypertrichosis, and contractures with gait abnormality were present only in some. Other characteristic features of IWC-I included ear abnormalities, hypertrichosis, and hypoplasia of mammillae. Age at onset of white spots ranged from early to late childhood.
Overall, individuals with a KRT10 disease-associated variant were significantly more likely to experience skin pain, odor, and infections and significantly less likely to report eye problems and collodion membrane (Table 2). Skin pain (OR, 6.8; 95% CI, 1.6-61.2; P = .002), odor (OR, 5.7; 95% CI, 2.0-19.7; P < .001), and infections (OR, 3.1; 95% CI, 1.4-7.7; P = .03) were significantly associated with KRT10 pathogenic variants compared with disease-associated variants in other genes associated with ichthyosis.
KRT1
Thirty-one kindreds had a KRT1 disease-associated variant; 9 were novel. As with KRT10 pathogenic variants, the characteristic appearance of KRT1 EI included skin fragility, especially at sites of trauma, and columns of scale (eFigure 12 in the Supplement). Scale was typically accentuated over extensor surfaces of joints, particularly knees and elbows (eFigure 12A and 12B in the Supplement). Erythema ranged from mild to severe (eFigure 12G and 12H in the Supplement).
A distinguishing feature of all participants with KRT1 EI, found only rarely in others, was thick, often functionally limiting PPK. The range of PPK included moderate and confluent scale with yellow thickening, moderate and focal piled scale with yellow thickening, and thick, confluent yellow piled scale (eFigure 12J-L in the Supplement).
Of 31 KRT1 pathogenic variants, 10 manifested as IWC-II. Among 39 participants with IWC in the study, 10 (26.0%) were heterozygous for a frameshift or splice-site KRT1 pathogenic variant affecting the tail domain. In contrast to IWC-I, every participant with IWC-II had moderate or severe PPK. Other IWC-I features, such as malformed ears, hypoplastic nipples, ectropion, and hypertrichosis, were absent among participants with IWC-II. Onset of white spots typically occurred at later ages than IWC-I, frequently at or after late teenage years.
Additional features reported by at least 50% of participants with KRT1 pathogenic variants include skin pain (10 of 11), skin odor (11 of 14), pruritus (7 of 10), skin infections (8 of 14), and hypohydrosis (6 of 11). Participants with KRT1 variants were significantly less likely to have collodion membrane, eye problems, or alopecia (Table 2).
PPK and Skin Findings Associated With KRT1 and KRT10 Disease-Associated Variants
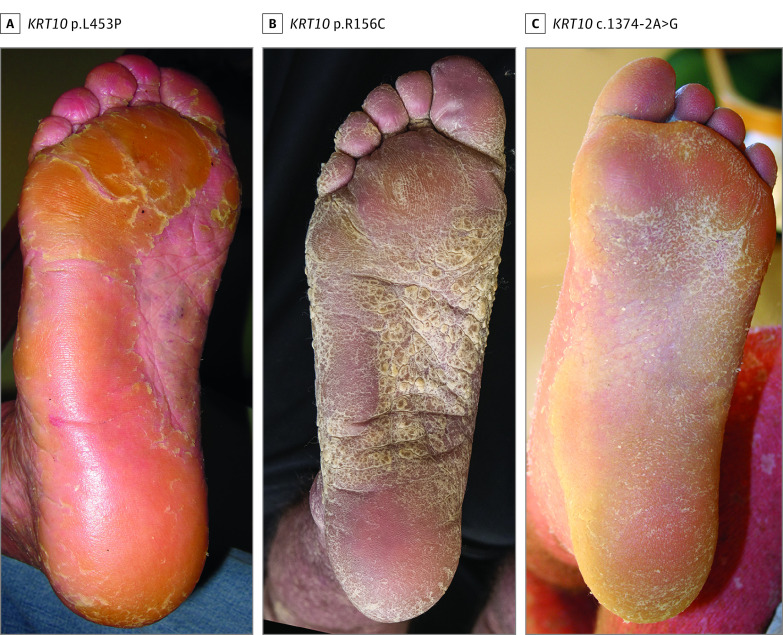
Palmoplantar keratoderma, when present in those with a KRT10 pathogenic variant, was typically mild and smooth with occasional focal accentuation, because KRT10 has limited expression in the palmoplantar suprabasal epidermis.36 Prior studies36 have suggested that KRT1 pathogenic variants were associated with a limited cutaneous phenotype with predominant palmoplantar presentation, while KRT10 variants were associated with a more widespread cutaneous involvement. In this study, while PPK tended to be more severe in EI associated with KRT1 than KRT10 pathogenic variants, there was significant overlap. For instance, severe PPK with thick, confluent, yellow-piled scale was not exclusive to KRT1 pathogenic variants and can also manifest in individuals with KRT10 pathogenic variants (Figure 3). Those with a KRT1 disease-associated variant can in turn present with significant cutaneous findings aside from PPK (eFigure 12H and 12I in the Supplement).
Figure 3. Significant Palmoplantar Keratoderma Associated With KRT10 Pathogenic Variants.
Discussion
We gathered data on what is to our knowledge the largest cohort of kindreds with ichthyosis and related disorders to date. Prior studies of the phenotypic and genotypic spectra of these Mendelian disorders of cornification were limited to specific ethnicities and subtypes.8,37,38,39 Our cohort was racially and ethnically heterogeneous with representation of all major forms of ichthyosis, and we report 266 novel variants (eTable 3 in the Supplement). During the past decade we have identified and described 8 new genetic causes of ichthyosis. These include KRT1 and KRT10 C-terminal frameshift pathogenic variants in IWC; GJA1 pathogenic variants in EKVP; DSP pathogenic variants in erythrokeratodermia cardiomyopathy; KDSR pathogenic variants in recessive progressive symmetrical erythrokeratodermia; PERP pathogenic variants in dominant and recessive keratoderma; AP1B1 pathogenic variants in ichthyosis, deafness, and photophobia; and ASPRV1 pathogenic variants in dominantly inherited ichthyosis.16,34,35,40,41,42,43,44 Seven additional genetic causes of ichthyosis have been described by others over this same time period.21,45,46,47,48,49,50
Genotype-phenotype associations in ichthyoses are difficult to establish. Significant clinical variations exist among individuals with disease-associated variants in the same gene. Even identical pathogenic variants, such as NIPAL4 p.A176D, can have diverse phenotypic consequences, with varying degrees of erythema and scale. Despite this, we aimed to capture general trends, analyze characteristic and distinguishing features, and detail the range of disease manifestations for each major gene.
We identified several clinical characteristics associated with certain genotypes (Table 2). Collodion membrane was more likely in individuals with TGM1 or ALOX12B disease-associated variants. Hearing loss, eye problems, alopecia, and skin odor were more prevalent among those with TGM1 disease-associated variants. These findings corroborate other studies, although the mechanism is unknown.39,51,52 Individuals with KRT10 disease-associated variants had a higher risk of skin pain, odor, and infections compared with those with other genotypes.
Despite clinical associations with specific causative genes, our analyses underscored the complexities of the ichthyoses. For instance, although collodion membrane is a common feature in autosomal recessive congenital ichthyosis, 3 participants with keratinopathic ichthyosis were born as collodion, a rarely reported, unusual presentation among individuals with a KRT10 or KRT1 variant.37 Further research is needed to elucidate the outcome of genetic and environmental modifiers.
Limitations
One limitation is the study’s retrospective nature, which prevented follow-up to delineate phenotypes (eg, isolate unique eye problems). Another is that our cohort included many individuals who remained pathogenic variant–unknown in clinical testing, enriching our cohort for less common genotypes. Finally, symptoms were self-reported in a subset of study participants. We did not formally assess phenotypic associations with specific pathogenic variants because in most cases, sample sizes would be inadequate. Nonetheless, we examined overall frequency of the most common clinical manifestations of ichthyosis, including pruritus (91%), hypohydrosis (75%), skin pain (70%), eye problems (51%), skin odor (45%), and skin infections (45%). Awareness of these symptoms is important not only for prognostication but also because they can negatively influence mental health and quality of life.53
We have identified distinguishing features for several genotypes, including tapered digits, hyperconvex nails, and auricle malformation in participants with ABCA12 disease-associated variants. These can serve as diagnostic clues. However, clinical criteria alone may be insufficient to discriminate among subtypes, such as EKVP phenotypes linked with recessive ABCA12 pathogenic variants, or severe PPK linked with KRT10 pathogenic variants. Genetic analysis is central to arriving at a precise diagnosis and crucial to understanding disease pathogenesis.
Conclusions
In this cohort study of 1000 kindreds, the genotypic and phenotypic landscape of the ichthyoses was expanded, but nearly 13% of the cohort remain unsolved, indicating significant potential for discovery of new genetic causes. Systematic characterization of this rare group of disorders is the first step toward developing customized management plans, generating targeted therapeutics, and improving prognostication.
eTable 1. Questionnaire questions
eTable 2a. Kindreds with recessive inheritance (n=629)
eTable 2b. Kindreds with dominant inheritance (n=240)
eTable 3. Novel mutations (n=265)
eFigure 1. Geographic distribution of 1000 kindreds
eFigure 2. Distribution of trios, dyads and probands among 1000 kindreds
eFigure 3. Phenotypes of ARCI due to TGM1 mutations
eFigure 4. Phenotypes of ARCI due to ABCA12 mutations
eFigure 5. Phenotypes of ARCI due to ALOX12B mutations
eFigure 6. Phenotypes of ARCI due to ALOXE3 mutations
eFigure 7. Phenotypes of ACRI due to NIPAL4 mutations
eFigure 8. Phenotypes of ARCI due to PNPLA1 mutations
eFigure 9. Phenotypes of Netherton syndrome due to SPINK5 mutations
eFigure 10. Phenotypes of ARCI due to CYP4F22 mutations
eFigure 11. Phenotypes of EI due to KRT10 mutations
eFigure 12. Phenotypes of EI due to KRT1 mutations
References
- 1.Oji V, Tadini G, Akiyama M, et al. Revised nomenclature and classification of inherited ichthyoses: results of the First Ichthyosis Consensus Conference in Sorèze 2009. J Am Acad Dermatol. 2010;63(4):607-641. doi: 10.1016/j.jaad.2009.11.020 [DOI] [PubMed] [Google Scholar]
- 2.Fischer J. Autosomal recessive congenital ichthyosis. J Invest Dermatol. 2009;129(6):1319-1321. doi: 10.1038/jid.2009.57 [DOI] [PubMed] [Google Scholar]
- 3.Simpson JK, Martinez-Queipo M, Onoufriadis A, et al. Genotype-phenotype correlation in a large English cohort of patients with autosomal recessive ichthyosis. Br J Dermatol. 2020;182(3):729-737. doi: 10.1111/bjd.18211 [DOI] [PubMed] [Google Scholar]
- 4.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2018. [Google Scholar]
- 5.McDonald JH. Handbook of Biological Statistics. Vol 3. Sparky House Publishing; 2014. [Google Scholar]
- 6.Richard G, Bale SJ. Autosomal recessive congenital ichthyosis. In: Pagon RA, Adam MP, Ardinger HH, et al. , eds. GeneReviews. University of Washington; 1993. [Google Scholar]
- 7.Thacher SM, Rice RH. Keratinocyte-specific transglutaminase of cultured human epidermal cells: relation to cross-linked envelope formation and terminal differentiation. Cell. 1985;40(3):685-695. doi: 10.1016/0092-8674(85)90217-X [DOI] [PubMed] [Google Scholar]
- 8.Pigg MH, Bygum A, Gånemo A, et al. Spectrum of autosomal recessive congenital ichthyosis in Scandinavia: clinical characteristics and novel and recurrent mutations in 132 patients. Acta Derm Venereol. 2016;96(7):932-937. doi: 10.2340/00015555-2418 [DOI] [PubMed] [Google Scholar]
- 9.Lefévre C, Audebert S, Jobard F, et al. Mutations in the transporter ABCA12 are associated with lamellar ichthyosis type 2. Hum Mol Genet. 2003;12(18):2369-2378. doi: 10.1093/hmg/ddg235 [DOI] [PubMed] [Google Scholar]
- 10.Akiyama M. The roles of ABCA12 in epidermal lipid barrier formation and keratinocyte differentiation. Biochim Biophys Acta. 2014;1841(3):435-440. doi: 10.1016/j.bbalip.2013.08.009 [DOI] [PubMed] [Google Scholar]
- 11.Takeichi T, Akiyama M. Inherited ichthyosis: non-syndromic forms. J Dermatol. 2016;43(3):242-251. doi: 10.1111/1346-8138.13243 [DOI] [PubMed] [Google Scholar]
- 12.Loo BKG, Batilando MJ, Tan EC, Koh MJA. Compound heterozygous mutations with novel missense ABCA12 mutation in harlequin ichthyosis. BMJ Case Rep. 2018;2018:bcr2017222025. doi: 10.1136/bcr-2017-222025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuo Y, Zhuang DZ, Han R, et al. ABCA12 maintains the epidermal lipid permeability barrier by facilitating formation of ceramide linoleic esters. J Biol Chem. 2008;283(52):36624-36635. doi: 10.1074/jbc.M807377200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yanagi T, Akiyama M, Nishihara H, et al. Harlequin ichthyosis model mouse reveals alveolar collapse and severe fetal skin barrier defects. Hum Mol Genet. 2008;17(19):3075-3083. doi: 10.1093/hmg/ddn204 [DOI] [PubMed] [Google Scholar]
- 15.Takeichi T, Torrelo A, Lee JYW, et al. Biallelic mutations in KDSR disrupt ceramide synthesis and result in a spectrum of keratinization disorders associated with thrombocytopenia. J Invest Dermatol. 2017;137(11):2344-2353. doi: 10.1016/j.jid.2017.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyden LM, Vincent NG, Zhou J, et al. Mutations in KDSR cause recessive progressive symmetric erythrokeratoderma. Am J Hum Genet. 2017;100(6):978-984. doi: 10.1016/j.ajhg.2017.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jobard F, Lefèvre C, Karaduman A, et al. Lipoxygenase-3 (ALOXE3) and 12(R)-lipoxygenase (ALOX12B) are mutated in non-bullous congenital ichthyosiform erythroderma (NCIE) linked to chromosome 17p13.1. Hum Mol Genet. 2002;11(1):107-113. doi: 10.1093/hmg/11.1.107 [DOI] [PubMed] [Google Scholar]
- 18.Wajid M, Kurban M, Shimomura Y, Christiano AM. NIPAL4/ichthyin is expressed in the granular layer of human epidermis and mutated in two Pakistani families with autosomal recessive ichthyosis. Dermatology. 2010;220(1):8-14. doi: 10.1159/000265757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mauldin EA, Crumrine D, Casal ML, et al. Cellular and metabolic basis for the ichthyotic phenotype in NIPAL4 (ichthyin)–deficient canines. Am J Pathol. 2018;188(6):1419-1429. doi: 10.1016/j.ajpath.2018.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dahlqvist J, Klar J, Hausser I, et al. Congenital ichthyosis: mutations in ichthyin are associated with specific structural abnormalities in the granular layer of epidermis. J Med Genet. 2007;44(10):615-620. doi: 10.1136/jmg.2007.050542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grall A, Guaguère E, Planchais S, et al. PNPLA1 mutations cause autosomal recessive congenital ichthyosis in golden retriever dogs and humans. Nat Genet. 2012;44(2):140-147. doi: 10.1038/ng.1056 [DOI] [PubMed] [Google Scholar]
- 22.Hirabayashi T, Anjo T, Kaneko A, et al. PNPLA1 has a crucial role in skin barrier function by directing acylceramide biosynthesis. Nat Commun. 2017;8:14609. doi: 10.1038/ncomms14609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyden LM, Craiglow BG, Hu RH, et al. Phenotypic spectrum of autosomal recessive congenital ichthyosis due to PNPLA1 mutation. Br J Dermatol. 2017;177(1):319-322. doi: 10.1111/bjd.15570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chavanas S, Bodemer C, Rochat A, et al. Mutations in SPINK5, encoding a serine protease inhibitor, cause Netherton syndrome. Nat Genet. 2000;25(2):141-142. doi: 10.1038/75977 [DOI] [PubMed] [Google Scholar]
- 25.Saleem HMK, Shahid MF, Shahbaz A, Sohail A, Shahid MA, Sachmechi I. Netherton syndrome: a case report and review of literature. Cureus. 2018;10(7):e3070-e3070. doi: 10.7759/cureus.3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lefèvre C, Bouadjar B, Ferrand V, et al. Mutations in a new cytochrome P450 gene in lamellar ichthyosis type 3. Hum Mol Genet. 2006;15(5):767-776. doi: 10.1093/hmg/ddi491 [DOI] [PubMed] [Google Scholar]
- 27.Ohno Y, Nakamichi S, Ohkuni A, et al. Essential role of the cytochrome P450 CYP4F22 in the production of acylceramide, the key lipid for skin permeability barrier formation. Proc Natl Acad Sci U S A. 2015;112(25):7707-7712. doi: 10.1073/pnas.1503491112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huber M, Scaletta C, Benathan M, et al. Abnormal keratin 1 and 10 cytoskeleton in cultured keratinocytes from epidermolytic hyperkeratosis caused by keratin 10 mutations. J Invest Dermatol. 1994;102(5):691-694. doi: 10.1111/1523-1747.ep12374270 [DOI] [PubMed] [Google Scholar]
- 29.Pulkkinen L, Christiano AM, Knowlton RG, Uitto J. Epidermolytic hyperkeratosis (bullous congenital ichthyosiform erythroderma). genetic linkage to chromosome 12q in the region of the type II keratin gene cluster. J Clin Invest. 1993;91(1):357-361. doi: 10.1172/JCI116193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith FJD, Kreuser-Genis IM, Jury CS, Wilson NJ, Terron-Kwiatowski A, Zamiri M. Novel and recurrent mutations in keratin 1 cause epidermolytic ichthyosis and palmoplantar keratoderma. Clin Exp Dermatol. 2019;44(5):528-534. doi: 10.1111/ced.13800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothnagel JA, Dominey AM, Dempsey LD, et al. Mutations in the rod domains of keratins 1 and 10 in epidermolytic hyperkeratosis. Science. 1992;257(5073):1128-1130. doi: 10.1126/science.257.5073.1128 [DOI] [PubMed] [Google Scholar]
- 32.Cheng J, Syder AJ, Yu QC, Letai A, Paller AS, Fuchs E. The genetic basis of epidermolytic hyperkeratosis: a disorder of differentiation-specific epidermal keratin genes. Cell. 1992;70(5):811-819. doi: 10.1016/0092-8674(92)90314-3 [DOI] [PubMed] [Google Scholar]
- 33.Fuchs E, Esteves RA, Coulombe PA. Transgenic mice expressing a mutant keratin 10 gene reveal the likely genetic basis for epidermolytic hyperkeratosis. Proc Natl Acad Sci U S A. 1992;89(15):6906-6910. doi: 10.1073/pnas.89.15.6906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choate KA, Lu Y, Zhou J, et al. Frequent somatic reversion of KRT1 mutations in ichthyosis with confetti. J Clin Invest. 2015;125(4):1703-1707. doi: 10.1172/JCI64415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choate KA, Lu Y, Zhou J, et al. Mitotic recombination in patients with ichthyosis causes reversion of dominant mutations in KRT10. Science. 2010;330(6000):94-97. doi: 10.1126/science.1192280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DiGiovanna JJ, Bale SJ. Clinical heterogeneity in epidermolytic hyperkeratosis. Arch Dermatol. 1994;130(8):1026-1035. doi: 10.1001/archderm.1994.01690080092014 [DOI] [PubMed] [Google Scholar]
- 37.Hotz A, Oji V, Bourrat E, et al. Expanding the clinical and genetic spectrum of KRT1, KRT2 and KRT10 mutations in keratinopathic ichthyosis. Acta Derm Venereol. 2016;96(4):473-478. doi: 10.2340/00015555-2299 [DOI] [PubMed] [Google Scholar]
- 38.Israeli S, Goldberg I, Fuchs-Telem D, et al. Non-syndromic autosomal recessive congenital ichthyosis in the Israeli population. Clin Exp Dermatol. 2013;38(8):911-916. doi: 10.1111/ced.12148 [DOI] [PubMed] [Google Scholar]
- 39.Farasat S, Wei MH, Herman M, et al. Novel transglutaminase-1 mutations and genotype-phenotype investigations of 104 patients with autosomal recessive congenital ichthyosis in the USA. J Med Genet. 2009;46(2):103-111. doi: 10.1136/jmg.2008.060905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boyden LM, Zhou J, Hu R, et al. Mutations in ASPRV1 cause dominantly inherited ichthyosis. Am J Hum Genet. 2020;107(1):158-163. doi: 10.1016/j.ajhg.2020.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyden LM, Craiglow BG, Zhou J, et al. Dominant de novo mutations in GJA1 cause erythrokeratodermia variabilis et progressiva, without features of oculodentodigital dysplasia. J Invest Dermatol. 2015;135(6):1540-1547. doi: 10.1038/jid.2014.485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyden LM, Atzmony L, Hamilton C, et al. Recessive mutations in AP1B1 cause ichthyosis, deafness, and photophobia. Am J Hum Genet. 2019;105(5):1023-1029. doi: 10.1016/j.ajhg.2019.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boyden LM, Kam CY, Hernández-Martín A, et al. Dominant de novo DSP mutations cause erythrokeratodermia-cardiomyopathy syndrome. Hum Mol Genet. 2016;25(2):348-357. doi: 10.1093/hmg/ddv481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duchatelet S, Boyden LM, Ishida-Yamamoto A, et al. Mutations in PERP cause dominant and recessive keratoderma. J Invest Dermatol. 2019;139(2):380-390. doi: 10.1016/j.jid.2018.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shigehara Y, Okuda S, Nemer G, et al. Mutations in SDR9C7 gene encoding an enzyme for vitamin A metabolism underlie autosomal recessive congenital ichthyosis. Hum Mol Genet. 2016;25(20):4484-4493. doi: 10.1093/hmg/ddw277 [DOI] [PubMed] [Google Scholar]
- 46.Fuchs-Telem D, Sarig O, van Steensel MAM, et al. Familial pityriasis rubra pilaris is caused by mutations in CARD14. Am J Hum Genet. 2012;91(1):163-170. doi: 10.1016/j.ajhg.2012.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kutkowska-Kaźmierczak A, Rydzanicz M, Chlebowski A, et al. Dominant ELOVL1 mutation causes neurological disorder with ichthyotic keratoderma, spasticity, hypomyelination and dysmorphic features. J Med Genet. 2018;55(6):408-414. doi: 10.1136/jmedgenet-2017-105172 [DOI] [PubMed] [Google Scholar]
- 48.Li M, Cheng R, Liang J, et al. Mutations in POFUT1, encoding protein O-fucosyltransferase 1, cause generalized Dowling-Degos disease. Am J Hum Genet. 2013;92(6):895-903. doi: 10.1016/j.ajhg.2013.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Basmanav FB, Oprisoreanu AM, Pasternack SM, et al. Mutations in POGLUT1, encoding protein O-glucosyltransferase 1, cause autosomal-dominant Dowling-Degos disease. Am J Hum Genet. 2014;94(1):135-143. doi: 10.1016/j.ajhg.2013.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H, Humbatova A, Liu Y, et al. Mutations in SREBF1, encoding sterol regulatory element binding transcription factor 1, cause autosomal-dominant IFAP syndrome. Am J Hum Genet. 2020;107(1):34-45. doi: 10.1016/j.ajhg.2020.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harting M, Brunetti-Pierri N, Chan CS, et al. Self-healing collodion membrane and mild nonbullous congenital ichthyosiform erythroderma due to 2 novel mutations in the ALOX12B gene. Arch Dermatol. 2008;144(3):351-356. doi: 10.1001/archderm.144.3.351 [DOI] [PubMed] [Google Scholar]
- 52.Putterman E, Zaki T, Milstone L, Choate K, Castelo-Soccio L. Association of the severity of alopecia with the severity of ichthyosis. JAMA Dermatol. 2019;155(9):1077-1078. doi: 10.1001/jamadermatol.2019.1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun Q, Ren I, Zaki T, Maciejewski K, Choate K. Ichthyosis affects mental health in adults and children: a cross-sectional study. J Am Acad Dermatol. 2020;83(3):951-954. doi: 10.1016/j.jaad.2020.01.052 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable 1. Questionnaire questions
eTable 2a. Kindreds with recessive inheritance (n=629)
eTable 2b. Kindreds with dominant inheritance (n=240)
eTable 3. Novel mutations (n=265)
eFigure 1. Geographic distribution of 1000 kindreds
eFigure 2. Distribution of trios, dyads and probands among 1000 kindreds
eFigure 3. Phenotypes of ARCI due to TGM1 mutations
eFigure 4. Phenotypes of ARCI due to ABCA12 mutations
eFigure 5. Phenotypes of ARCI due to ALOX12B mutations
eFigure 6. Phenotypes of ARCI due to ALOXE3 mutations
eFigure 7. Phenotypes of ACRI due to NIPAL4 mutations
eFigure 8. Phenotypes of ARCI due to PNPLA1 mutations
eFigure 9. Phenotypes of Netherton syndrome due to SPINK5 mutations
eFigure 10. Phenotypes of ARCI due to CYP4F22 mutations
eFigure 11. Phenotypes of EI due to KRT10 mutations
eFigure 12. Phenotypes of EI due to KRT1 mutations