Abstract
Cell biology is driven by complex networks of biomolecular interactions. Characterizing the kinetic and thermodynamic properties of these interactions is crucial to understanding their role in different physiological processes. Surface plasmon resonance (SPR)-based approaches have become a key tool in quantifying biomolecular interactions, however conventional approaches require isolating the interacting components from the cellular system. Cell-based SPR approaches have recently emerged, promising to enable precise measurements of biomolecular interactions within their normal biological context. Two major approaches have been developed, offering their own advantages and limitations. These approaches currently lack a systematic exploration of ‘best practices’ like those existing for traditional SPR experiments. Toward this end, we describe the two major approaches, and identify the experimental parameters that require exploration, and discuss the experimental considerations constraining the optimization of each. In particular, we discuss the requirements of future biomaterial development needed to advance the cell-based SPR technique.
Keywords: biomolecular interactions, surface plasmon resonance (SPR), cell-based SPR, systems biology
Introduction
Systems biology is a growing field that incorporates biological measurements with computational modeling to uncover new understandings of biological systems, measurements which require the use of advanced biomaterials to capture biologically-accurate conditions11,16. Different systems biology studies explore physiological systems under normal and pathological conditions. Computational systems biology approaches have been applied to describe endothelial cell apoptosis signaling pathways117, investigate vascular endothelial growth factor (VEGF) family activity36, explore and design better pro-angiogenic therapies37, and predict cell response from the protein-protein interactions occurring within a cell112. Thus, systems biology has advanced knowledge of the underpinning mechanisms behind cell processes.
Despite this progress, deterministic models based on mass action kinetics have been limited by a lack of quantitative data on biomolecular signaling and interactions. Mass action kinetics models are defined by both the amount of species (concentrations), and the probability of these species interacting (i.e. binding kinetics). Therefore, data needed to parameterize such models are both protein concentrations and protein-protein interaction kinetics. Although there is a plethora of qualitative data available on protein expression (e.g., Western blots) and protein-protein interactions (e.g., co-immunoprecipitation), there is a need to move from qualitative to quantitative characterizations of biomolecular interactions. To address the quantitative data limitation, systems biology researchers are developing new assays to measure protein concentrations9,18,53 and build databases72,76 that aggregate data and provide researchers with the information needed to build computational models. Indeed, we and others have led efforts to quantify protein concentrations on cell membranes17,18,51–54,112; thus supplying data to computationally model vascular signaling, which is critical to advance engineering goals of vascularizing tissues29,70,112–114.
However, current approaches for measuring binding kinetics for biomolecular interactions involving membrane-bound proteins are performed using recombinant versions or the full protein extracted from the membrane23,80. Such approaches, therefore, measure protein-protein binding outside of their biological environment, such as within a cell membrane, which can result in different protein confirmations. Since protein conformation differences can impact their binding and signaling abilities64,101, performing these measurements outside of their normal biological context could produce results that poorly reflect the actual dynamics in biological systems.
However, there are currently few experimental approaches to measure biomolecular kinetics in biologically native conditions. Recently, the surface plasmon resonance (SPR)-based biosensor approach has been expanded for use with whole cell samples instead of purified protein samples85,93. Cell-based SPR approaches offer the promise of high-throughput quantification of biomolecular interaction kinetics and affinities under biologically native conditions. While recent studies have measured membrane-bound protein-protein kinetics, there remain several critical questions unanswered and unexplored regarding assay optimization and best practices. We overview the different approaches developed to adapt SPR biosensor assays to measuring kinetics on whole cells, describe the key experimental conditions that ultimately require optimization, and layout a general guide towards establishing best practices for the major variants of cell-based SPR.
Measuring biomolecular kinetics via SPR
Kinetic and thermodynamic properties characterize biomolecular interactions
Biomolecular interaction dynamics are best characterized by: (1) binding kinetics and (2) binding affinities84. The binding kinetics represent the rate at which the proteins bind and dissociate. In a 1:1 protein interaction, the kinetics are characterized by two quantifiable properties: the association constant kon describes the rate that two proteins bind to form a complex; the dissociation constant koff, in turn, describes the rate this complex dissociates, back to the unbound molecues57. The binding affinity describes the ‘strength’ of the protein interaction82. Conventionally, binding affinities are expressed as the equilibrium dissociation constant KD; the higher the KD value, the lower the binding affinity. Conveniently, KD can be expressed in terms of the kinetic rate constants (Equation 1.1). The binding affinities and kinetics reflect intrinsic structural and chemical properties of the involved molecules, and are therefore altered by post-translational protein modifications72.
| (Equation 1.1) |
SPR to identify and measure biomolecular binding kinetics
The SPR-based assay is an ideal approach for identifying and measuring kinetic rate parameters for biomolecular interactions, like between growth factors and their receptors. SPR-based biosensors like the BIAcore83 detect protein-protein interactions utilizing an optical phenomenon that is sensitive to small changes in mass near the sensor surface3,100. By coupling a target protein on the sensor surface, binding kinetics and affinities can be measured by flowing the protein analyte through a flow channel over the sensor surface (Figure 1A) and recording the mass change over time while analyte binds and unbinds the target protein21,81,82. The binding kinetics and affinities are then determined by fitting these data to mathematical equations that represent specific chemical binding models, as described thoroughly in several excellent reviews48,79,81. Furthermore, SPR-based biosensors are capable of probing one analyte against multiple targets simultaneously, enabling faster measurements of different protein-protein pairs33. Therefore, SPR-based assays have proven an ideal approach for measuring binding kinetic parameters for biomolecular interactions.
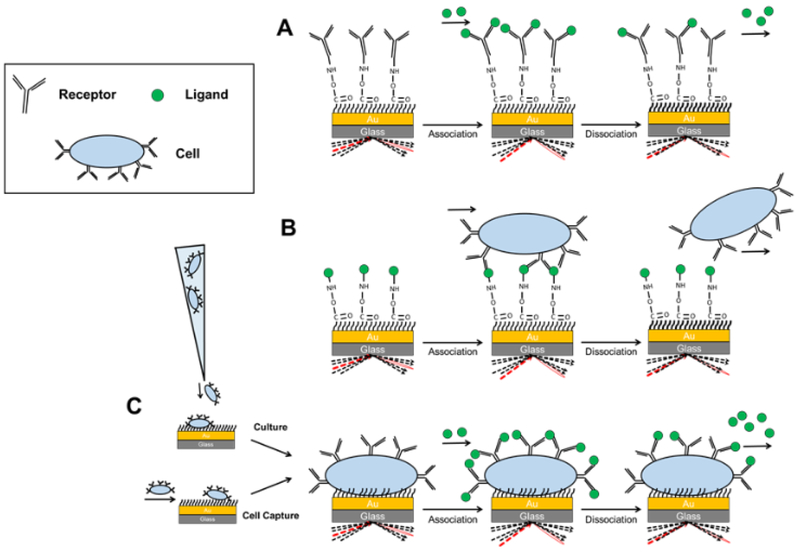
Figure 1.
SPR approaches for kinetics and affinity measurements. (a) Traditional SPR compared to the cell-based SPR approaches, (b) injected cell analysis (ICA) and (c) immobilized target cell (ITC) appraches.
SPR is a label-free, highly sensitive, and cost-effective approach to measure biomolecular interactions in real time.
SPR-based approaches have several fundamental advantages over other existing affinity and kinetics assays. Several of these assays have been reviewed extensively by others39,44,56, and include: fluorescence-based, radiolabeling, and enzyme immunoassays23,26,56,107. For measuring binding kinetics, SPR has four major advantages: (1) SPR is a label-free technique, unlike other approaches, which require coupling an additional reporting label, such as radioactive compounds or fluorescent tags, to one or both proteins. Such tags, therefore, can interfere with protein-protein binding90. (2) SPR biosensors detect binding in real-time: protein association and dissociation responses are detected as they occur, allowing straightforward binding kinetic measurements33,79. (3) SPR detects interactions with high sensitivity and can therefore measure binding kinetic and affinity constants to higher precision than other techniques. For example, binding affinities (KD) on the scale of picomolar (pM) can be measured using SPR, while fluorescence, absorption assays, and calorimetry assays measure binding affinity on the μM-mM scale23,56. (4) SPR requires relatively small sample quantities, using protein solution volumes of 10-20 μL per sample56,70,97, whereas calorimetry and absorption assays require mL quantities56,77. Altogether, SPR offers a reliable technique to accurately characterize binding kinetics and affinities of biomolecular interactions.
Conventional SPR limited to characterizing biomolecular interactions outside their native environments
Conventional SPR-based approaches have been primarily limited to measuring biomolecular interactions in isolation, outside of their biological context. For membrane-bound proteins, SPR experiments are typically performed using recombinant partial version of membrane receptors that often include only the extracellular domains, rather than including transmembrane domains78. Measurements with partial proteins can produce non-physiologically relevant results, because binding is often regulated by conformational changes in receptor subunits118. Additionally, the membrane-bound protein is typically covalently bound to the sensor surface via amine coupling, creating a physiologically inaccurate system, since the membrane protein environment should facilitate interactions with cholesterols, lipids, and other membrane-bound proteins89. An innovative workaround to this limitation is to perform these measurements on nanodiscs—self assembled lipid-bilayers—containing the target protein in an environment mimicking the cell membrane99,103. However, nanodiscs do not entirely mimic the cell membrane composition, as they lack cholesterol and other membrane proteins. These differences are critical, as studies have demonstrated that membrane protein binding properties can vary depending on membrane composition, such as the cholesterol concentrations41,62. Furthermore, purified or recombinant membrane proteins will lack the post-translational modifications, like N-linked glycosylation, which have been shown to alter binding properties104.
An additional improvement on these existing approaches would be to perform SPR measurements with actual cells. Cell-based SPR is an emerging technique that combines the experimental benefits of SPR-based bioassays with the ability to measure interactions on receptors within actual cell membranes. Optimizing these approaches, however, to obtain useful chemical kinetic and affinities remains unexplored and will require significant advancements in biomaterials to ensure existing SPR biosensors provide ideal conditions for use with whole cells.
Cell-based SPR Approaches
Two major approaches have been developed to adapt SPR approaches, using standard SPR instrumental setups, to measure interactions with live cells by substituting the cells for either: the analyte, by flowing the target cell through the system, referred here as the Injected Cell Analyte (ICA) approach (Figure 1B)—or the immobilized/target protein—i.e. the protein immobilized to the sensor surface, called here the Immobilized Target Cell (ITC) approach (Figure 1C).
Immobilized Target Cell Approach
The Immobilized Target Cell (ITC) approach monitors injected ligand binding to membrane or surface proteins on cells immobilized to the SPR sensor chip (Figure 1C). This approach provides the advantage of directly measuring the equilibrium dissociation constant KD, because known concentrations of analytes are injected before each experiment. Therefore, an ITC approach allows measuring kinetic rate constants directly. Moreover, the binding kinetic constants measured will reflect the effective binding between the ligand and the target receptor while incorporating the effects introduced by other modifications, such as differing membrane composition and non-specific ligand-membrane effects. Nevertheless, the ITC approach has disadvantages. First, due to inherent limitations of SPR100, the short penetration depth of the evanescent field cannot detect the whole cell and the physical binding activity. This leads to smaller apparent response levels as binding is only detected to the part of the cell that is in the evanescent field (about 300 – 400 nm) above the gold sensor chip surface. However, a novel SPR system that uses near-infrared incident light—instead of visible light, as used in conventional SPR systems—generate evanescent fields that extend 10 μm, vastly extending the detection range. These Fourier transform infrared spectroscopy (FTIR) SPR systems, therefore, would enable detecting activity across the entire cell121, and has already been used to monitor membrane composition changes in HeLa cells and detect endocytic processes in human melanoma cells119,121. Additionally, researchers recently demonstrated that the evanescent field depths could be extended to 2 mm using a graphene-based biosensor in place of the conventional gold sensors, and used the expanded signal depth to study drug-responses in whole cancer cells110. Additionally, attached cells can detach from the surface more readily than covalently bound receptors as found in traditional plasmon resonance-based approach. Both differences introduce challenges that require the selection of optimal flow rate conditions and biomaterial choices for the sensor surface.
A need for biomaterials: maximizing cell-sensor adhesion via sensor coating and functionalization
The adhesive strength—i.e. the attachment force between the cells and the surface in resistance to shear—of the chosen surface material is key to designing a cell-based SPR study using an ITC approach. A surface material with a weak adhesive strength will weakly immobilize cells and result in cell detachment when buffer or analytes are injected over the channel surface. Conversely, adhesive strength that is too strong may cause cells to spread abnormally74. With the ITC approach, there are typically two methods implemented to adhere cells to the surface: directly culturing cells on the sensor surface via overnight incubation15,118, or flowing cells onto the sensor surface122. In both cases, however, adhesion of cells can be greatly affected by surface coating. Typically, a short-chain surface such as a derivatized alkanethiol is used as the backbone of the surface to ensure that the captured cells are close to the sensor chip surface to optimize detection. Hydrogels such as dextran are not recommended because they usually extend 100 nm from the sensor chip surface, which would cause more of the cell to not be in the evanescent field. The chip with the short chain alkanethiol groups that also contain some carboxyl groups are typically derivatized with a biomaterial to provide an adhesion matrix for the cells. Therefore, cell adhesion to the chosen material must be tested. For example, cells adhered to poly-L-lysine (PLL) coated surfaces can flatten against the surface due to the interaction between positively-charged poly-L-lysine and anionic cell membrane74. Identifying the best material may be daunting when one couples the need for optimal adhesive strength with the many immobilization material choices. Amongst different approaches, some common ones include high-affinity biomolecules, like antibodies, engineered peptides, and aptamers5,6,43,61,109,120; extracellular matrix proteins, like fibronectin, collagen, and laminin; or cationic molecules, like lipids68, polymers66,74,96, and peptides49,69. When choosing adhesion molecules, one ought to consider the interactions between chosen molecules and membrane proteins, such as coating a sensor with an integral protein membrane like CD317. The approach and the adhesive molecules used to target cells should not compromise the need for optimal adhesive strength. There are several guides in literature for choosing optimal materials. For example, several molecules have been optimized for high cell binding specificity in the drug delivery field2,24,86,91. The biomaterial porosity should also be considered in context of the analyte molecular size, to prevent the injected analytes to leech into the surface, registering falsely as binding signal. Likewise, the chosen biomaterial should not incorporate chemical functional groups that resemble the analyte binding target sites. The cell patterning40, affinity microfluidics50 and biomaterials fields20 also offer immobilization material guidelines4. In these fields, extracellular microenvironment mimics have been engineered to enable optimal cell residence and honing1,71,92. Altogether, it is critical to identify the optimal surface material, which should facilitate cell immobilization with good adhesive strength while being specific to the cell and receptor biology.
Reducing non-specific binding and preserving cell surface receptors contributes to the selection of cell immobilization approaches
One important parameter to control in SPR experiments is non-specific binding: the interactions between analytes and non-targeted molecules and/or the sensor surface47. Traditional SPR-based kinetics approaches are prone to signal associated with non-specific interaction42 which requires reference correction. This consideration carries over to ITC approach-based cell-based SPR. The incorporation of a reference channel—i.e. a separate sensor channel where the ligands have no specific interaction target—is the standard approach to obtaining a background reference signal, which is subsequently subtracted as correction42,70,98. Selecting a background reference target, however, is challenging (as described previously42), and deciding on an appropriate reference in cell-based SPR is dependent on the ITC sub-approach taken. When immobilizing cells on the chip via direct culturing on the chip46,49, it is difficult to separate the experimental side of the chip from the reference side of the chip since culturing different groups of cells on the same sensor chip can be problematic. When cells are immobilized onto the chip by injecting the cells over the sensor surface, a reference can be easily achieved. A reference channel is ideally created by immobilizing non-active cells that are not expressed with analyte receptor at a surface density similar to that achieved for the active cells immobilized onto the sample channel. Alternatively, a reference channel could be left as the surface matrix backbone itself. For adherent cells or cells from tissue, a single cell suspension can be obtained via enzymatic dissociation from flasks or tissue, respectively. This must be tested, because enzymatic agents may be disrupt the membrane proteins to be studied via SPR18. Before cell-based SPR-based approaches can be utilized more commonly, therefore, the question of an ideal background reference signal source must be answered.
Minimizing rebinding effects through an optimized cell density
An ideal cell density for studying kinetics should result in a measurable increase in SPR signal compared to the background signal while minimizing rebinding effects. If the sensor surface cell density is too low, then injected analyte may result in a low binding signal, whereby differentiating the true binding signal from the background, non-specific signal becomes increasingly difficult73. Conversely, injecting over a high-density surface can result in target-rebinding effects and promote significant non-specific cell attachment85. In each case, the unwanted effects will interfere with measuring the true binding kinetics. Another consideration is the receptor density on the surface of the cells and the molecular weight of the analyte that binds to the cells. If the cells are enriched with receptor, then a lower cell density can potentially be used. In addition, for large analytes (> 100 kDa), a lower density can also be used in comparison to a smaller analyte, as the SPR signal is sensitive to the total mass that binds to the surface. While appropriate immobilized protein ranges have been determined for traditional SPR experiments25,38,45, no comparable systematic study has been performed for cell-based SPR approaches. Researchers have investigated this indirectly, by varying the cell concentration range they inject to coat the sensor surface, but these covered a narrow window (600 cells/mL49 to 1600 cells/ml118), and do not provide researchers with guidelines for surface densities. Future work, therefore, is required to systematically test cell injection concentrations to determine ranges that optimize the detected signal while minimizing the negative effect of non-specific cell adhesion and re-binding effects.
Optimizing analyte flow rates to minimize cell shear stress and avoid mass transport limiting conditions
The analyte flow rate—the rate at which analyte is injected through the microfluidic system—is already an important optimization parameter in traditional SPR experiments33,38,75, and takes an additional importance for experiments injecting across captured cells. Flow rate serves as a critical element in fluid dynamics, and many biological processes take place in solution13. Analyte flow rates have previously been optimized to be fast enough to avoid mass transport limitation (MTL) effects59,98,102. But because shear stress is proportional to the flow rate87, setting the flow rate arbitrarily high could result in cells detaching from the sensor chip67. Therefore, flow rates must be optimized to be sufficiently high as to avoid MTL effects–which distort analyte :receptor binding kinetic measurements83–while minimizing the shear stress thus minimizing cell detachment rate. Analyte injection flow rates have been explored across a narrow flow rate range–50 μl/min to 20 μl/min–and chosen apparently arbitrarily46,118. A systemic study is required to establish criteria to optimize flow rate to minimize MTL effects while reducing cell shear stress.
Optimizing sensor regeneration conditions to minimize cell loss
In a traditional SPR analysis, five (5) concentrations of analyte are injected over the immobilized target that span a concentration range centered around the interaction affinity. If the rate of dissociation is slow (i.e., signal does not decay back to the starting baseline in 10 min), a regeneration solution is injected that disrupts the interaction between the analyte and target and returns the baseline back to the original starting value. If the target is covalently immobilized, the surface is regenerated back to free target and another analyte concentration can be injected. However, if the target is captured via a non-covalent means, the target is removed from the surface along with the analyte and would need to be reloaded for each analyte concentration. In the case of the ITC approach, the surface would be regenerated with a solution that would remove the cells from the surface along with the analyte, but the cells would then need to be recaptured for each analyte concentration. This approach would consume a large quantity of cells and it can be very difficult to remove all the bound cells from the chip surface. Alternatively, there is a different tactic that can be implemented instead of regenerating between each analyte concentration. This approach is called a kinetic titration whereby analyte is injected sequentially from low to high concentration without regenerating between injections22. This option is very attractive because it eliminates the need for regeneration, which would save on sample consumption and time, which is an important consideration for cell-based SPR.
Injected Cell Analyte Approach
The second general approach currently utilized in cell-based SPR is the Injected Cell Analyte (ICA) approach. This is opposite to the ITC approach in that the target cell is injected in place of the analyte protein over the immobilized target receptor. In the ICA approach: (1) the interactant to the cells —e.g. growth factors like VEGFA—are immobilized instead of the cells to the sensor surface. (2) Cells are injected across the immobilized ligand. (3) The surface is ‘regenerated’ to remove the bound cells from the surface before re-injecting at a different cell concentration (Figure 1B)32. While the protocols related to ligand immobilization and regeneration are well-established by traditional SPR analysis75, the use of cells as the analyte has its advantages and limitations. Both ligand immobilization and regeneration steps for cell-as-analyte approach can be adapted from traditional SPR techniques. The main drawback of this tactic is that since a molar concentration of cells cannot be determined, an association rate constant cannot be calculated as it is a function of molarity and time. However, qualitative information can still be learned from this approach. In addition, the number of regeneration cycles can be limited due to the potential loss of cell binding capacity93. Cell debris may affect SPR signal if the regeneration approach is not thorough. Like the ITC approach, several experimental conditions require optimization to ensure useful binding parameters are obtained.
Optimizing cell injection flow rates and concentrations to minimize MTL effects and maximize response signal
As in the ITC approach, the quality of the obtained data for the injected cell analyte approach is dependent on optimizing the cell injection flow rate in order to minimize mass transport limit effects. At high injection flow rates, the bulk flow concentration is higher than the cell concentrations at the binding surface. Analyte depletion during association phase can be induced at the surface. If the bulk concentration is lower than the cell concentrations at the binding surface due to a low cell injection flow rate, a retention zone can be formed during dissociation phase102. With both conditions, the SPR signal will be altered, exhibiting slower binding and unbinding curve102. There is a need, therefore, for a systematic study of the optimal injection flow rate. However, no such systematic study has established an optimal cell injection flow rate range. Previous studies using this approach having used a wide range of rates, from 3 μl/min106 to 70 μl/min73, but no research has determined a protocol that optimizes these rates for specific cell types. Future studies, therefore, are needed to identify the ideal conditions
Another major challenge in cell-based SPR is identifying the cell concentrations injected through the system, since both cell density and size ultimately impact viscosity and flow resistance. These effects have been observed in therapeutic fields, where the size and concentration of red blood cells alter blood viscosity58,67,87. High RBC concentrations, for example, increase blood viscosity and impair drug delivery34. These factors, therefore, will influence whether injected cells will effectively reach the sensor surface to bind immobilized target proteins and produce a signal. In cell-based SPR, an ideal cell concentration is the cell concentration that can produce a reliable signal. The reliability is determined by how easily we can differentiate specific binding signals from non-specific binding signals73. A higher cell concentration can produce a higher difference between ligand-receptor binding induced signal and background signal, yet a high cell concentration can lead to higher viscosity, causing a clog in the SPR system. Some early work has begun investigating the importance of injected cell concentrations in such studies: the relationship between binding rate and cell concentration were described as an exponential curve in a red blood cell binding study93. Further work is needed to determine the optimal cell concentrations for different cell sizes.
The future of cell-based SPR
Cell-based SPR has been used to characterize the interactions between ligands and membrane protein receptors, and these membrane proteins are important for biological processes and are linked with certain diseases89. Understanding these interactions is critical for drug development. For instance, cell-based SPR can be performed to obtain the binding affinity and study dosage-dependent responses.(e.g. anti-TNF agents85). In addition, cell-based SPR offers the opportunity to obtain biological signals triggered by agonists and antagonists. Cell-based SPR allows for the evaluation of pharmacodynamic parameters and for the prediction of the potency of new drugs63. Cell-based SPR can advance computational models of complex biological systems by enabling high-precision measurements of ligand:receptor kinetics that better reflect biological reality. Computational models serve as powerful tools to study complex biological systems117, because physiologically-relevant phenomena—such as tumor metastasis, wound-healing, or immune reactions—emerge from many cell-level interactions10,12,16,28. Modeling cell signaling pathways—whereby ligands bind membrane-bound receptors to trigger interwoven signaling networks to modulate cell activity—has provided insight into several growth factor-receptor families known to mediate physiologic and pathological processes19,28–31, including epidermal growth factors (EGFs)115, fibroblast growth factors (FGFs)27, platelet-derived growth factors (PDGFs)88, and vascular endothelial growth factors (VEGFs)35,37,55,94,95,108,113.
Such models are often constructed using the law of mass action, where an interaction rate is proportional to the interacting species’ concentration and their underlying kinetics3,28,60,70,111,116; their predictive power is therefore limited by how accurately the experimental measurements of binding kinetics reflect biological reality. Traditional SPR assays rely on measuring protein-protein interactions removed from biological systems; e.g. VEGF-A:VEGFR and PDGF:PDGFR ligand:receptor interaction kinetics are measured by observing the ligand binding a recombinant receptor protein representing the extracellular portion only65,70,105. These experimental models, therefore, are limited because they cannot reflect factors that modulate ligand binding, such as membrane composition62 and post-translational modifications (e.g. receptor protein n-glycosylation14). By measuring these interactions under more biologically comparable conditions, we can construct more accurate, useful models. Cell-based SPR is well-suited for these measurements by enabling highly sensitive kinetic measurements of the interactions between proteins and native cell membranes in a label-free environment.
Cell-based SPR achieves the measurement of protein-protein interactions within a biologically native environment. Both approaches—the immobilized target cell and injected cell analyte approaches—offer advantages towards obtaining biologically-representative parameters for computational modeling. The ITC approach allows measure binding affinities and kinetic parameters using the mathematical fitting approaches used in conventional SPR but requires careful optimization to ensure stable cell adhesion across experiments. The ICA approach allows conventional chemical coupling techniques to immobilize target proteins to cell sensors, but injected cells face significant mass transport limitations due to cell size that require careful flow rate and cell concentration optimization to reduce. The ICA approach, therefore, may be better applied for small cell types, like with bacteria, while the ITC approach may be a better choice for cell sizes too large to effective flow as analyte. To enhance the outcomes of cell-based SPR and establish a standard procedure, each critical parameter should be optimized and a standard for assigning values to these parameters should be established.
Conclusions
The next steps should be to establish the optimal experimental conditions and standards of the cell-based SPR procedures. Although several different studies investigating living cell reactions in response to stimuli have been carried out using cell-based SPR approaches, there are no “best practices” for cell-based SPR throughout the literature. Experiments need to be performed to optimize critical parameters, such as cell density, ligand flow rate, and cell capture surface in the Immobilized Target Cell approach, as well as both cell concentration and cell flow rate in the Injected Cell Analyte approach. Developing a framework to optimize the key experimental parameters in cell-based SPR, can help researchers perform experiments in a more effective and meaningful manner. By establishing these optimal conditions, we can also better understand the effects of these parameters on the binding kinetics.
Cell-based SPR has proven to be a powerful tool to study both ligand-receptor binding and its subsequent signaling pathways in each study. An optimized method to perform cell-based SPR is necessary to ensure the meaningfulness of the outcome and expedite the applications of cell-based SPR. Regardless of the challenges that it may face, cell-based SPR has the capability of monitoring the dynamic changes at the binding site and cellular changes in a real-time and label-free setting. The advantageous capabilities of cell-based SPR can result in scientific breakthroughs for brain therapy and enhancements in novel therapeutics.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- 1.Aguado BA, Caffe JR, Nanavati D, Rao SS, Bushnell GG, Azarin SM, and Shea LD. Extracellular matrix mediators of metastatic cell colonization characterized using scaffold mimics of the pre-metastatic niche. Acta Biomater. 33:13–24, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen TM, and Cullis PR. Drug delivery systems: entering the mainstream. Science 303:1818–22, 2004. [DOI] [PubMed] [Google Scholar]
- 3.Anderson J, and Papachristodoulou A. On validation and invalidation of biological models. BMC Bioinformatics 10:132, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ansari A, and Imoukhuede PI. Plenty more room on the glass bottom: Surface functionalization and nanobiotechnology for cell isolation. , 2018. [Google Scholar]
- 5.Ansari A, Lee-Montiel FT, Amos J, and Imoukhuede PI. Secondary anchor targeted cell release. Biotechnol. Bioeng 112:2214–2227, 2015. [DOI] [PubMed] [Google Scholar]
- 6.Ansari A, Patel R, Schultheis K, Naumovski V, and Imoukhuede PI. A method of targeted cell isolation via glass surface functionalization. J. Vis. Exp 115:, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asahara T, Murohara T, Sullivan A, Silver M, van der Zee R, Li T, Witzenbichler B, Schatteman G, and Isner JM. Isolation of Putative Progenitor Endothelial Cells for Angiogenesis. Science (80-, ). 275:964–966, 1997. [DOI] [PubMed] [Google Scholar]
- 8.Beseničar M, Maček P, Lakey JH, and Anderluh G. Surface plasmon resonance in protein-membrane interactions. Chem. Phys. Lipids 141:169–178, 2006. [DOI] [PubMed] [Google Scholar]
- 9.Bose AK, and Janes KA. A high-throughput assay for phosphoprotein-specific phosphatase activity in cellular extracts. Mol. Cell. Proteomics 12:797–806, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bray D Advances in Systems Biology. 736:193–198, 2012. [DOI] [PubMed] [Google Scholar]
- 11.Breitling R What is systems biology? Front. Physiol. 1:9, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burrage K, Hood L, and Ragan M. a.. Advanced computing for systems biology. Brief. Bioinform. 7:390–398, 2006. [DOI] [PubMed] [Google Scholar]
- 13.Cartwright JHE, Piro O, and Tuval I. Fluid dynamics in developmental biology: moving fluids that shape ontogeny. HFSP J. 3:77–93, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandler KB, Leon DR, Meyer RD, Rahimi N, and Costello CE. Site-Specific N-Glycosylation of Endothelial Cell Receptor Tyrosine Kinase VEGFR-2. J. Proteome Res. 16:677–688, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen K, Obinata H, and Izumi T. Detection of G protein-coupled receptor-mediated cellular response involved in cytoskeletal rearrangement using surface plasmon resonance. Biosens. Bioelectron. 25:1675–1680, 2010. [DOI] [PubMed] [Google Scholar]
- 16.Chen S, Ansari A, Sterrett W, Hurley K, Kemball J, Weddell JC, Imoukhuede PI, Kemball K, Weddell JC, and Imoukhuede PI. Current state-of-the-art and future directions in systems biology. Prog. Commun. Sci 1:12–26, 2014. [Google Scholar]
- 17.Chen S,Guo X, Imarenezor O, and Imoukhuede PI. Quantification of VEGFRs, NRP1, and PDGFRs on Endothelial Cells and Fibroblasts Reveals Serum, Intra-Family Ligand, and Cross-Family Ligand Regulation. Cell. Mol. Bioeng 8:383–403, 2015. [Google Scholar]
- 18.Chen S, Weddell J, Gupta P, Conard G, Parkin J, and Imoukhuede PI. qFlow Cytometry-Based Receptoromic Screening: A High-Throughput Quantification Approach Informing Biomarker Selection and Nanosensor Development. In: Biomedical Nanotechnology: Methods and Protocols, edited by Petrosko SH, and Day ES. New York, NY: Springer New York, 2017, pp. 117–138 .doi: 10.1007/978-1-4939-6840-4_8 [DOI] [PubMed] [Google Scholar]
- 19.Chu L-H, Ganta VC, Choi MH, Chen G, Finley SD, Annex BH, and Popel AS. A multiscale computational model predicts distribution of anti-angiogenic isoform VEGF165b in peripheral arterial disease in human and mouse. Sci. Rep 6:37030, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D’Souza SF Immobilization and stabilization of biomaterials for biosensor applications. Appl. Biochem. Biotechnol 96:225–38, 2001. [DOI] [PubMed] [Google Scholar]
- 21.Drake AW, Myszka DG, and Klakamp SL. Characterizing high-affinity antigen/antibody complexes by kinetic- and equilibrium-based methods. Anal. Biochem 328:35–43, 2004. [DOI] [PubMed] [Google Scholar]
- 22.Drake AW, Tang ML, Papalia G. a., Landes G, Haak-Frendscho M, and Klakamp SL. Biacore surface matrix effects on the binding kinetics and affinity of an antigen/antibody complex. Anal. Biochem 429:58–69, 2012. [DOI] [PubMed] [Google Scholar]
- 23.Du X, Li Y, Xia Y-L, Ai S-M, Liang J, Sang P, Ji X-L, and Liu S-Q. Insights into Protein-Ligand Interactions: Mechanisms, Models, and Methods. Int. J. Mol. Sci 17:, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eniola AO, and Hammer DA. Characterization of biodegradable drug delivery vehicles with the adhesive properties of leukocytes II: effect of degradation on targeting activity. Biomaterials 26:661–670, 2005. [DOI] [PubMed] [Google Scholar]
- 25.Evaluation D PROTOCOL FOR MEASURING SMALL MOLECULE INTERACTIONS USING BIACORE A Practical Guide. Symp. A Q. J. Mod. Foreign Lit. 1–16, 2002. [Google Scholar]
- 26.Favicchio R, Dragan AI, Kneale GG, and Read CM. Fluorescence Spectroscopy and Anisotropy in the Analysis of DNA-Protein Interactions. In: Methods in Molecular Biology. Humana Press, 2009, pp. 589–611. [DOI] [PubMed] [Google Scholar]
- 27.Filion RJ, and Popel AS. A reaction-diffusion model of basic fibroblast growth factor interactions with cell surface receptors. Ann. Biomed. Eng. 32:645–663, 2004. [DOI] [PubMed] [Google Scholar]
- 28.Finley SD, Chu L-H, and Popel AS. Computational systems biology approaches to anti-angiogenic cancer therapeutics. Drug Discov Today 00:, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finley SD, Engel-Stefanini MO, Imoukhuede PI, Popel AS, Dokun AO, Annex BH, Popel AS, Finley SD, Engel-Stefanini MO, Imoukhuede PI, and Popel AS. Pharmacokinetics and pharmacodynamics of VEGF-neutralizing antibodies. Am. J. Physiol. Heart Circ. Physiol 5:193, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finley SD, and Popel AS. Predicting the effects of anti-angiogenic agents targeting specific VEGF isoforms. AAPS J. 14:500–9, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finley SD, Popel AS, S F, AS P, Finley SD, and AS P Effect of tumor microenvironment on tumor VEGF during anti-VEGF treatment: systems biology predictions. J. Natl. Cancer Inst. 105:802–811,2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischer MJE. Surface Plasmon Resonance. 2010, 55–73 pp. [Google Scholar]
- 33.Fivash M, Towler EM, and Fisher RJ. BIAcore for macromolecular interaction. Curr. Opin. Biotechnol 9:97–101, 1998. [DOI] [PubMed] [Google Scholar]
- 34.Fullstone G, Wood J, Holcombe M, and Battaglia G. Modelling the Transport of Nanoparticles under Blood Flow using an Agent-based Approach. Sci. Rep 5:10649, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mac Gabhann F, Ji JW, Popel AS, Mac PAS Gabhann F Ji JW, J. J. W. Mac Gabhann F Popel AS, Mac Gabhann F, J. W. Ji, and Popel AS. VEGF gradients, receptor activation, and sprout guidance in resting and exercising skeletal muscle. J Appl Physiol 102:722–734, 2007. [DOI] [PubMed] [Google Scholar]
- 36.Gabhann, Mac F, Popel AS, Mac Gabhann F, and Popel AS. Systems Biology of Vascular Endothelial Growth Factors. Microcirculation 15:715–738, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mac Gabhann F, Qutub A. a., Annex BH, and Popel AS. Systems biology of pro-angiogenic therapies targeting the VEGF system. Wiley Interdiscip. Rev. Syst. Biol. Med 2:694–707, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.GE Healthcare. Biacore Sensor Surface Handbook. 8–10, 2008.at <http://www.gelifesciences.com/gehcls_images/GELS/RelatedContent/Files/1363789281999/litdoc14100571_20130430000159.pdf>
- 39.Goh WL, Yen Lee M, Joseph TL, Tng Quah S, Brown CJ, Verma C, Brenner S, Ghadessy FJ, and Nah Teo Y. Molecular Rotors As Conditionally Fluorescent Labels for Rapid Detection of Biomolecular Interactionsdoi: 10.1021/ja413031h [DOI] [PubMed] [Google Scholar]
- 40.Goubko CA, and Cao X. Patterning multiple cell types in co-cultures: A review. Mater. Sci. Eng. C 29:1855–1868, 2009. [Google Scholar]
- 41.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EYT, Velasquez J, Kuhn P, and Stevens RC. A Specific Cholesterol Binding Site Is Established by the 2.8 Å Structure of the Human α2-Adrenergic Receptor. Structure 16:897–905, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haseley SR, Talaga P, Kamerling JP, and Vliegenthart JFG. Characterization of the Carbohydrate Binding Specificity and Kinetic Parameters of Lectins by Using Surface Plasmon Resonance. Anal. Biochem 274:203–210, 1999. [DOI] [PubMed] [Google Scholar]
- 43.Hassan U, Ghonge T, Reddy B, Patel M, Rappleye M, Taneja I, Tanna A, Healey R, Manusry N, Price Z, Jensen T, Berger J, Hasnain A, Flaugher E, Liu S, Davis B, Kumar J, White K, and Bashir R. A point-of-care microfluidic biochip for quantification of CD64 expression from whole blood for sepsis stratification. Nat. Commun 8:15949, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He D, He X, Wang K, Yang X, Yang X, Li X, and Zou Z. Nanometer-sized manganese oxide-quenched fluorescent oligonucleotides: an effective sensing platform for probing biomolecular interactions. Chem. Commun. Chem. Commun 50:11049–11052, 2014. [DOI] [PubMed] [Google Scholar]
- 45.Healthcare GE, and Sciences L. Biacore 3000. 29–30. [Google Scholar]
- 46.Hide M, Tsutsui T, Sato H, Nishimura T, Morimoto K, Yamamoto S, and Yoshizato K. Real-Time Analysis of Ligand-Induced Cell Surface and Intracellular Reactions of Living Mast Cells Using a Surface Plasmon Resonance-Based Biosensor. Anal. Biochem 302:28–37, 2002. [DOI] [PubMed] [Google Scholar]
- 47.Homola J Present and future of surface plasmon resonance biosensors. Anal. Bloanal. Chem 377:528–539, 2003. [DOI] [PubMed] [Google Scholar]
- 48.Horn F, and Jackson R. General mass action kinetics. Arch. Ration. Mech. Anal 47:81–116, 1972. [Google Scholar]
- 49.Hun Lee S, Jin Ko H, and Hyun Park T. Real-time monitoring of odorant-induced cellular reactions using surface plasmon resonance. Biosens. Bioelectron 25:55–60, 2009. [DOI] [PubMed] [Google Scholar]
- 50.Hyun K-AA, and Jung H-I II. Microfluidic devices for the isolation of circulating rare cells: A focus on affinity-based, dielectrophoresis, and hydrophoresis. 2013. [DOI] [PubMed] [Google Scholar]
- 51.Imoukhuede PI, Dokun AO, Annex BH, and Popel AS. Endothelial cell-by-cell profiling reveals the temporal dynamics of VEGFR1 and VEGFR2 membrane localization after murine hindlimb ischemia. Am J Physiol Hear. Circ Physiol 304:H1085–H1093, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Imoukhuede PIPI, and Popel ASAASAS. Expression of VEGF receptors on endothelial cells in mouse skeletal muscle. PLoS One 7:e44791,2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Imoukhuede PI, and Popel AS. Quantitative fluorescent profiling of VEGFRs reveals tumor cell and endothelial cell heterogeneity in breast cancer xenografts. Cancer Med. 3:225–244, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Imoukhuede PI, and Popel ASAS. Quantification and cell-to-cell variation of vascular endothelial growth factor receptors. Exp. Cell Res. 317:955–965,2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ji JW, Mac Gabhann F, and Popel AS. Skeletal muscle VEGF gradients in peripheral arterial disease: simulations of rest and exercise. Am. J. Physiol. Heart Circ. Physiol. 293:H3740–H3749, 2007. [DOI] [PubMed] [Google Scholar]
- 56.Jing M, and Bowser MT. Methods for measuring aptamer-protein equilibria: a review. Anal. Chim. Acta 686:9–18, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Joss L, a Morton T, Doyle ML, and Myszka DG. Interpreting kinetic rate constants from optical biosensor data recorded on a decaying surface. Anal. Biochem 261:203–210, 1998. [DOI] [PubMed] [Google Scholar]
- 58.Karimi A, Yazdi S, and Ardekani AM. Hydrodynamic mechanisms of cell and particle trapping in microfluidics. Biomicrofluidics 7:, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karlsson R Affinity analysis of non-steady-state data obtained under mass transport limited conditions using BIAcore technology. J. Mol. Recognit 12:285–292, 1999. [DOI] [PubMed] [Google Scholar]
- 60.Kenakin T Quantifying biological activity in chemical terms: A pharmacology primer to describe drug effect. ACS Chem. Biol 4:249–260, 2009. [DOI] [PubMed] [Google Scholar]
- 61.Khademhosseini A, Suh KY, Yang JM, Eng G, Yeh J, Levenberg S, and Langer R. Layer-by-layer deposition of hyaluronic acid and poly-L-lysine for patterned cell co-cultures. Biomaterials 25:3583–3592, 2004. [DOI] [PubMed] [Google Scholar]
- 62.Klein U, Gimpl G, and Fahrenholz F. Alteration of the Myometrial Plasma Membrane Cholesterol Content with β-Cyclodextrin Modulates the Binding Affinity of the Oxytocin Receptor. Biochemistry 34:13784–13793, 1995. [DOI] [PubMed] [Google Scholar]
- 63.Kosaihira A, and Ona T. Rapid and quantitative method for evaluating the personal therapeutic potential of cancer drugs. Anal. Bioanal. Chem 391:1889–1897, 2008. [DOI] [PubMed] [Google Scholar]
- 64.Li E, and Hristova K. Receptor tyrosine kinase transmembrane domains: Function, dimer structure and dimerization energetics. Cell Adhes. Migr. 4:249–254, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin X, Takahashi K, Liu Y, Derrien A, and Zamora PO. A synthetic, bioactive PDGF mimetic with binding to both alpha-PDGF and beta-PDGF receptors. Growth Factors 25:87–93, 2007. [DOI] [PubMed] [Google Scholar]
- 66.Lungwitz U, Breunig M, Blunk T, and Göpferich A. Polyethylenimine-based non-viral gene delivery systems. Eur. J. Pharm. Biopharm 60:247–266, 2005. [DOI] [PubMed] [Google Scholar]
- 67.LUO ZY, XU F, LU TJ, and BAI BF. DIRECT NUMERICAL SIMULATION OF DETACHMENT OF SINGLE CAPTURED LEUKOCYTE UNDER DIFFERENT FLOW CONDITIONS. J. Mech. Med. Biol 11:273–284, 2011. [Google Scholar]
- 68.Mahato RI, Rolland A, and Tomlinson E. Cationic Lipid-Based Gene Delivery Systems: Pharmaceutical Perspectives. Pharm. Res 14:853–859, 1997. [DOI] [PubMed] [Google Scholar]
- 69.Maltais J-S, Denault J-B, Gendron L, and Grandbois M. Label-free monitoring of apoptosis by surface plasmon resonance detection of morphological changes. Apoptosis 17:916–925, 2012. [DOI] [PubMed] [Google Scholar]
- 70.Mamer SB, Chen S, Weddell JC, Palasz A, Wittenkeller A, Kumar M, and Imoukhuede PI. Discovery of High-Affinity PDGF-VEGFR Interactions: Redefining RTK Dynamics. Sci. Rep 7:16439, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.March S, Hui EE, Underhill GH, Khetani S, and Bhatia SN. Microenvironmental regulation of the sinusoidal endothelial cell phenotype in vitro. Hepatology 50:920–928, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Matlock MK, Holehouse AS, and Naegle KM. ProteomeScout: a repository and analysis resource for post-translational modifications and proteins. Nucleic Acids Res. 43:, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mauriz E, Carbajo-Pescador S, Ordoñez R, Garcia-Fernández MC, Mauriz JL, Lechuga LM, and Gonzalez-Gallego J. On-line surface plasmon resonance biosensing of vascular endothelial growth factor signaling in intact-human hepatoma cell lines. Analyst 139:1426, 2014. [DOI] [PubMed] [Google Scholar]
- 74.Mazia D, Schatten G, and Sale W. Adhesion of cells to surfaces coated with polylysine. Applications to electron microscopy. J. Cell Biol. 66:, 1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Merwe, Van Der PA, De Mol NJ, Fischer MJE, and Van Der Merwe PA. Surface Plasmon Resonance. Methods Mol. Biol. 627:1–14, 2010. [DOI] [PubMed] [Google Scholar]
- 76.Mooradian AD, Held JM, and Naegle KM. Using ProteomeScout: A Resource of Post-Translational Modifications, Their Experiments, and the Proteins That They Annotate. In: Current Protocols in Bioinformatics. Hoboken, NJ, USA: John Wiley & Sons, Inc., 2017, pp. 13.32.1–13.32.27.doi: 10.1002/cpbi.31 [DOI] [PubMed] [Google Scholar]
- 77.Müller M, Weigand JE, Weichenrieder O, and Suess B. Thermodynamic characterization of an engineered tetracycline-binding riboswitch. Nucleic Acids Res. 34:2607–17, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Murphy M Using SPR to Analyze Cell-Binding Interactions. Genet. Eng. Biotechnol. News 37:18–19, 2017. [Google Scholar]
- 79.Murphy M, Jason-Moller L, and Bruno J. Using Biacore to measure the binding kinetics of an antibody-antigen interaction. Curr. Protoc. protein Sci. Editor. board John E Coligan al Chapter 19:Unit 19.14--Unit 19.14, 2006. [DOI] [PubMed] [Google Scholar]
- 80.Myszka DG Kinetic analysis of macromolecular interactions using surface plasmon resonance biosensors. Curr. Opin. Biotechnol 8:50–57, 1997. [DOI] [PubMed] [Google Scholar]
- 81.Myszka DG Improving biosensor analysis. J. Mol. Recognit 12:279–284, 1999. [DOI] [PubMed] [Google Scholar]
- 82.Myszka DG Kinetic, equilibrium, and thermodynamic analysis of macromolecular interactions with BIACORE. , 2000. [DOI] [PubMed] [Google Scholar]
- 83.Nguyen B, Tanious F. a., and Wilson WD. Biosensor-surface plasmon resonance: Quantitative analysis of small molecule-nucleic acid interactions. Methods 42:150–161,2007. [DOI] [PubMed] [Google Scholar]
- 84.Nucleic L Biochemical applications kinetics. Reactions 2:366–372, 1975. [Google Scholar]
- 85.Ogura T, Tanaka Y, and Toyoda H. Whole cell-based surface plasmon resonance measurement to assess binding of anti-TNF agents to transmembrane target. Anal. Biochem 508:73–77, 2016. [DOI] [PubMed] [Google Scholar]
- 86.Onyskiw PJ, and Eniola-Adefeso O. Effect of PEGylation on Ligand-Based Targeting of Drug Carriers to the Vascular Wall in Blood Flow. Langmuir 29:11127–11134, 2013. [DOI] [PubMed] [Google Scholar]
- 87.Papaioannou TG, and Stefanadis C. Vascular wall shear stress: basic principles and methods. Hell. J Cardiol 46:9–15, 2005. [PubMed] [Google Scholar]
- 88.Park CS, Schneider IC, and Haugh JM. Kinetic analysis of platelet-derived growth factor receptor/phosphoinositide 3-kinase/Akt signaling in fibroblasts. J. Biol. Chem 278:37064–37072, 2003. [DOI] [PubMed] [Google Scholar]
- 89.Patching SG Surface plasmon resonance spectroscopy for characterisation of membrane protein–ligand interactions and its potential for drug discovery. Biochim. Biophys. Acta - Biomembr. 1838:, 2014. [DOI] [PubMed] [Google Scholar]
- 90.Patel VJ, Thalassinos K, Slade SE, Connolly JB, Crombie A, Murrell JC, and Scrivens JH. A comparison of labeling and label-free mass spectrometry-based proteomics approaches. J. Proteome Res. 8:3752–3759, 2009. [DOI] [PubMed] [Google Scholar]
- 91.Patil SD, Rhodes DG, and Burgess DJ. DNA-based therapeutics and DNA delivery systems: A comprehensive review. AAPS J. 7:E61–E77, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pedron S, and Harley BAC. Impact of the biophysical features of a 3D gelatin microenvironment on glioblastoma malignancy. J. Biomed. Mater. Res. Part A 101:3404–3415, 2013. [DOI] [PubMed] [Google Scholar]
- 93.Quinn JG, O’Neill S, Doyle A, McAtamney C, Diamond D, MacCraith BD, and O’Kennedy R. Development and application of surface plasmon resonance-based biosensors for the detection of cell-ligand interactions. Anal. Biochem 281:135–143, 2000. [DOI] [PubMed] [Google Scholar]
- 94.Qutub AA, Popel AS, Qutub A PA, Qutub AA, and Popel AS. Elongation, proliferation & migration differentiate endothelial cell phenotypes and determine capillary sprouting. BMC Syst. Biol. 3:13, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Qutub A, Gabhann F, Karagiannis E, Vempati P, and Popel A. Multiscale models of angiogenesis. , 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rainaldi G, Calcabrini A, and Santini MT. Positively charged polymer polylysine-induced cell adhesion molecule redistribution in K562 cells. J. Mater. Sci. Mater. Med 9:755–760, 1998. [DOI] [PubMed] [Google Scholar]
- 97.Renaud JP, Chung CW, Danielson UH, Egner U, Hennig M, Hubbard RE, and Nar H. Biophysics in drug discovery: Impact, challenges and opportunities. Nat. Rev. Drug Discov. 15:679–698, 2016. [DOI] [PubMed] [Google Scholar]
- 98.Roden LD, and Myszka DG. Global analysis of a macromolecular interaction measured on BIAcore. Biochem. Biophys. Res. Commun 225:1073–1077, 1996. [DOI] [PubMed] [Google Scholar]
- 99.Rouck JE, Krapf JE, Roy J, Huff HC, and Das A. Recent advances in nanodisc technology for membrane protein studies (2012–2017). FEBS Lett. 591:2057–2088, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Salamon Z, and Tollin G. Surface Plasmon Resonance, Theory. Elsevier; 3:2311–2319, 1999. [Google Scholar]
- 101.Sarabipour S, Ballmer-Hofer K, and Hristova K. VEGFR-2 conformational switch in response to ligand binding. Elife 5:1–23, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schuck P, and Zhao H. The role of mass transport limitation and surface heterogeneity in the biophysical characterization of macromolecular binding processes by SPR biosensing. , 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schuler MA, Denisov IG, and Sligar SG. Nanodiscs as a new tool to examine lipid-protein interactions. In: Methods in Molecular Biology, edited by Kleinschmidt JH. Totowa, NJ: Humana Press, 2013, pp. 415–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shintani Y, Takashima S, Asano Y, Kato H, Liao Y, Yamazaki S, Tsukamoto O, Seguchi O, Yamamoto H, Fukushima T, Sugahara K, Kitakaze M, and Hori M. Glycosaminoglycan modification of neuropilin-1 modulates VEGFR2 signaling. EMBO J. 25:3045–3055, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.von Tiedemann B, and Bilitewski U. Characterization of the vascular endothelial growth factor-receptor interaction and determination of the recombinant protein by an optical receptor sensor. Biosens. Bioelectron 17:983–991,2002. [DOI] [PubMed] [Google Scholar]
- 106.Uchida H, Fujitani K, Kawai Y, Kitazawa H, Horii A, Shiiba K, Saito K, and Saito T. A new assay using surface plasmon resonance (SPR) to determine binding of the Lactobacillus acidophilus group to human colonic mucin. Biosci. Biotechnol. Biochem 68:1004–10, 2004. [DOI] [PubMed] [Google Scholar]
- 107.Velazquez-Campoy A, and Freire E. Isothermal titration calorimetry to determine association constants for high-affinity ligands. Nat. Protoc 1:186–191,2006. [DOI] [PubMed] [Google Scholar]
- 108.Vempati P Popel AS, M. G. F., P. A. S Vempati P Mac Gabhann F, Vempati P, Mac Gabhann F, and Popel AS. Quantifying the Proteolytic Release of Extracellular Matrix-Sequestered VEGF with a Computational Model. PLoS One 5:e11860, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wan Y, Kim YT, Li N, Cho SK, Bachoo R, Ellington AD, and Iqbal SM. Surface-immobilized aptamers for cancer cell isolation and microscopic cytology. Cancer Res. 70:9371–9380, 2010. [DOI] [PubMed] [Google Scholar]
- 110.Wang Y, Zhang S, Xu T, Zhang T, Mo Y, Liu J, Yan L, and Xing F. Ultra-sensitive and ultra-fast detection of whole unlabeled living cancer cell responses to paclitaxel with a graphene-based biosensor. Sensors Actuators B Chem. 263:417–425, 2018. [Google Scholar]
- 111.Weddell JC Predicting angiogenic receptor trafficking and signaling via computational systems biology , 2016.at <http://hdl.handle.net/2142/95356%0A>
- 112.Weddell JC, Chen S, and Imoukhuede PI. VEGFR1 promotes cell migration and proliferation through PLCγ and PI3K pathways, npj Syst. Biol. Appl 4:1, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Weddell JC, and Imoukhuede PI. Quantitative Characterization of Cellular Membrane-Receptor Heterogeneity through Statistical and Computational Modeling. PLoS One 9:e97271, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Weddell JC, and Imoukhuede PI. Integrative meta-modeling identifies endocytic vesicles, late endosome and the nucleus as the cellular compartments primarily directing RTK signaling. Integr. Biol , 2017.doi: 10.1039/C7IB00011A [DOI] [PubMed] [Google Scholar]
- 115.Wiley HS, Shvartsman SY, Lauffenburger D. a., Steven Wiley H, Shvartsman SY, and Lauffenburger D. a.. Computational modeling of the EGF-receptor system: a paradigm for systems biology. Trends Cell Biol. 13:43–50, 2003. [DOI] [PubMed] [Google Scholar]
- 116.Witelski T, and Bowen M. Methods of Mathematical Modelling: Continuous Systems and Differential Equations. Switzerland: Springer International Publishing, 2015, xviii—305 pp.doi: 10.1007/978-3-319-23042-9 [DOI] [Google Scholar]
- 117.Wu FTH, Stefanini MO, Gabhann F. Mac, Popel AS, Gabhann FM, Popel AS, and A. S. P Wu FTH Gabhann FM Stefanini MO, F. M. G Wu FTH Stefanini MO, and AS. P Modeling of Growth Factor-Receptor Systems:: From Molecular-Level Protein Interaction Networks to Whole-Body Compartment Models. Methods Enzym. 467:461–497, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yanase Y, Suzuki H, Tsutsui T, Hiragun T, Kameyoshi Y, and Hide M. The SPR signal in living cells reflects changes other than the area of adhesion and the formation of cell constructions. Biosens. Bioelectron 22:1081–1086, 2007. [DOI] [PubMed] [Google Scholar]
- 119.Yashunsky V, Shimron S, Lirtsman V, Weiss AM, Melamed-Book N, Golosovsky M, Davidov D, and Aroeti B. Real-time monitoring of transferrin-induced endocytic vesicle formation by mid-infrared surface plasmon resonance. Biophys. J 97:1003–1012, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhu J, Nguyen T, Pei R, Stojanovic M, and Lin Q. Specific capture and temperature-mediated release of cells in an aptamer-based microfluidic device. Lab Chip 12:3504–3513, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ziblat R, Lirtsman V, Davidov D, and Aroeti B. Infrared surface plasmon resonance: a novel tool for real time sensing of variations in living cells. Biophys. J 90:2592–2599, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Reichert SPR Endothelial Cell Attachment to Matrix Proteins and Hypersmolar Response Quantified using Surface Plasmon Resonance (SPR) - Reichert Technologies | Life Sciences, Surface Plasmon Resonance, Single Channel, Dual Channel and Modular System Platf.