Abstract
The rapidly changing treatment paradigm for patients with metastatic oncogene-driven lung cancer continues to evolve, and consequently our understanding of the landscape of resistance must also advance. MET amplification is an established and frequent driver of resistance in EGFR-mutant non-small-cell lung cancer (NSCLC). Recently, the combination of MET proto-oncogene (MET) and epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) has shown promise in overcoming this molecularly defined resistance in clinical trials, and this combination strategy is being pursued in ongoing trials. Emerging data also demonstrate MET amplification as a resistance driver to TKI-treated ALK-, RET-, and ROS-1-fusion NSCLC, consistently at the range of 15%, while the resistance profiling data are maturing for other molecular targets. In this review, we discuss MET amplification as a driver of acquired resistance in well-defined molecular subsets of NSCLC, explore the biology behind this mechanism of resistance, and summarize the recently published clinical data, including the proposed combination strategies in the clinic achieving success in overcoming acquired MET amplification-dependent resistance.
Key words: EGFR, MET, amplification, resistance, NSCLC, targeted therapy
Highlights
-
•
Understanding mechanisms of resistance in oncogene-driven lung cancer is crucial.
-
•
MET amplification is a recurrent driver of resistance, across molecularly defined subsets of NSCLC.
-
•
Overcoming this resistance in clinical trials, using combination strategies, is currently being pursued.
-
•
We explore the biology behind this mechanism of resistance and summarize recent successes in the clinic.
Introduction
Genomic profiling and the implementation of prospective tumor molecular analysis have revolutionized the way that lung cancer is diagnosed and treated in the clinic. As of 2021, various clinical guidelines [National Comprehensive Cancer Network (NCCN)/College of American Pathologists (CAP)/International Association for the Study of Lung Cancer (IASLC)/Association for Molecular Pathology (AMP)/American Society of Clinical Oncology (ASCO)/European Society of Medical Oncology (ESMO)] recommend genetic profiling of non-small-cell lung cancer (NSCLC) for > 10 genetic alterations, including EGFR, BRAF, KRAS, ERBB2 (HER2) mutations; ALK, ROS1, NTRK, RET fusions; and METex14 skipping and amplification to capture actionable molecular targets.1, 2, 3, 4, 5, 6, 7 Seven of these oncogene alterations now have Food and Drug Administration (FDA)-approved targeted therapies. With targeted therapies, patients with the oncogene-driven NSCLC have remarkable response rates and documented improved progression-free survival (PFS); however, resistance to these targeted therapies inevitably develops despite initial responses.
Understanding and addressing the development of targeted therapy resistance that occurs with tyrosine kinase inhibitors (TKIs) represent a key challenge of the precision medicine era. MET amplification is a well-established mechanism of acquired resistance to epidermal growth factor receptor (EGFR) inhibitors,8,9 and now increasing evidence, reported recently, suggests MET amplification is a consistent mechanism of acquired resistance in a number of other oncogene-driven molecular subsets of NSCLC post-tyrosine kinase inhibition. Combination strategies may overcome the genomic heterogeneity of drug resistance; however, the risk of overlapping toxicities and resulting dose adjustments can often hamper effective drug combinations.10 In this review, we discuss MET amplification as a driver of acquired resistance in NSCLC, review the clinical evidence to date in well-defined molecular subsets of NSCLC, discuss the possible mechanisms driving MET amplification in this setting, and summarize the proposed TKI combination strategies to overcome MET amplification-dependent resistance.
MET structure and function
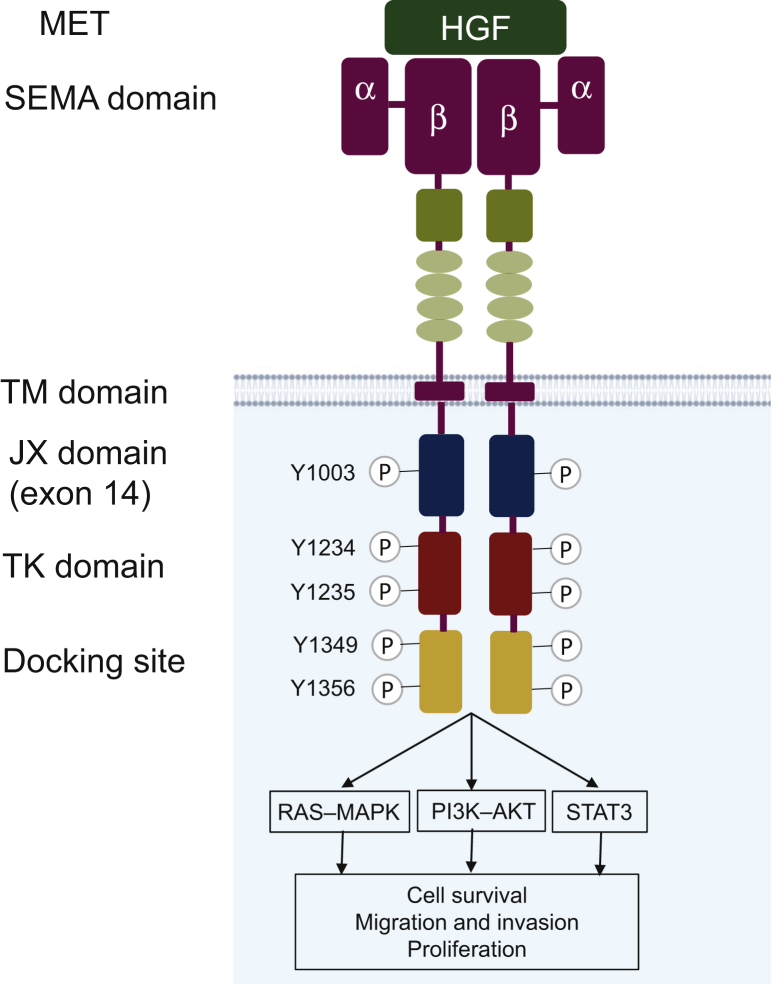
MET was first identified in a chemically transformed osteosarcoma-derived cell line.11 The proto-oncogene was later discovered to encode the receptor tyrosine kinase (RTK) MET, an RTK activated by an endogenous ligand, scatter factor, or hepatocyte growth factor (HGF).12, 13, 14, 15 In general, RTKs such as MET contain an N-terminal extracellular binding domain, a single transmembrane α helix, and a cytosolic C-terminal domain with tyrosine kinase activity.16 MET is a disulfide-linked heterodimeric RTK consisting of an extracellular α chain, a β chain that encompasses the remainder of extracellular domain, the juxtamembrane, and the kinase domains (Figure 1). The intracellular component contains a juxtamembrane region responsible for signal downregulation and receptor degradation, a catalytic region with the enzyme activity, and a C-terminal region acting as a docking site for adaptor proteins, which leads to downstream signaling via phosphoinositide 3-kinase (PI3K), signal transducer and activator of transcription proteins (STAT), and mitogen-activated protein kinase (MAPK).17, 18, 19 HGF, typically produced and secreted by mesenchymal cells, is the only natural ligand of MET (Figure 1); binding of HGF to MET leads to receptor dimerization and phosphorylation of tyrosine residues in the kinase domain and autophosphorylation of the carboxy-terminal bidentate substrate-binding sites.20, 21, 22 The resulting phosphotyrosines function as docking sites for other proteins involved in RTK-mediated signal transduction,20, 21, 22 and bind to activate distinct downstream signaling pathways,12,14,21 including PI3K/AKT (protein kinase B), MAPK, STAT, and nuclear factor-kB (Figure 1).23, 24, 25 MET signaling can become dysregulated through several mechanisms, including overexpression of MET protein or MET gene alterations, such as mutations, amplifications, or rearrangements.26,27 In this review, we focus on amplification of the MET gene in the setting of other oncogene-driven lung cancer.
Figure 1.
MET and its downstream signaling pathways.
MET is a disulfide-linked heterodimeric RTK consisting of an extracellular α chain, a β chain that encompasses the remainder of extracellular domain, the juxtamembrane, and the kinase domains. HGF is an MET ligand. The intracellular component contains a juxtamembrane region, a catalytic region with the enzyme activity, and a C-terminal region that acts as a docking site for adaptor proteins. MET activates RAS–MAPK, PI3K–AKT, and other oncogenic signaling pathways. AKT, protein kinase B; HGF, hepatocyte growth factor; MAPK, mitogen-activated protein kinase; MET, MET proto-oncogene; NSCLC, non-small-cell lung cancer; PI3K, phosphoinositide 3-kinase; RAS, rat sarcoma virus; RTK, receptor tyrosine kinase.
Detection of MET amplification
MET amplification can occur as a focal amplification or as a result of chromosome 7 polysomy. Polysomy occurs when there are multiple copies of chromosome 7 in tumor cells, secondary to factors such as chromosomal duplication, whereas true amplification occurs in the setting of focal or regional gene duplication, via processes such as breakage–fusion–bridge mechanisms.28 Clinically, focal high-copy amplification of MET represents an oncogenic driver event for cancer, while polysomy is typically not.29 Traditionally, MET amplification has been detected using a FISH method, with the challenges of the technical complexity and interpretation of the test.30 Using FISH, the mean MET per cell and chromosome 7 centromere ratio (MET/CEP7) ratio, the ratio of MET relative to chromosome 7 centromere, is used to distinguish between polysomy and true amplification. In polysomy, each copy of MET is associated with a corresponding centromere, preserving the MET/CEP7 ratio as copy number increases, while in true MET amplification, copy number increases without an increase in CEP7, and the MET/CEP7 ratio increases.31 In general, a cut-off of MET/CEP ≥2.0 is now used as the FISH criteria for MET amplification; however, historically various cut-offs have been used in studies for MET amplification and MET copy number gain: for instance, a MET/CEP7 threshold of 5 was set as minimum for high MET (FISH ≥5 MET signals/cell) in a study by Cappuzzo et al.,32 MET/CEP7 ratio of ≥2 was used by Tanaka et al.,33 and MET gene copy number (GCN) ≥ 10 was ultimately used in the GEOMETRY study.34
Broad, hybrid-capture next-generation sequencing (NGS) assays are able to detect amplification events and are now increasingly used in clinical practice for MET amplification detection. In contrast to FISH for single-gene testing, NGS may provide additional information on other, potentially clinically relevant, concurrent genomic alterations.35 However, some NGS-based assays do not control for CEP7, and therefore may detect increase in copy number as in polysomy rather than true MET amplification. As with FISH, copy number gains detected via NGS are reported as continuous variables, and cut-offs can vary significantly between assays.
An ongoing difficulty with MET copy number studies has been to define a threshold for any given methodology for which MET-directed therapy will likely be active. In theory, increases in MET copy number are postulated to cause excessive amounts of MET protein, and subsequent auto-aggregation, ligand-independent MET signaling, and subsequent oncogenic addiction to the MET pathway.27 The challenge is that changes in MET copy number represent a continuous variable, and this variable has been assessed in different ways, defined as either the ratio of MET relative to another region of chromosome 7, such as CEP7, or the absolute number of MET copies.27 A persistent challenge with any defined cut-off criteria, however, is that a more flexible criteria could include more patients but then dilute the clinical benefit, and thus conversely, a more stringent criteria while identifying fewer patients, may include patients who could potentially derive the greatest clinical benefit. The risk is always the potential of excluding patients who may still derive some benefit.
Very high level of MET amplification (MET/CEP7 ≥5 by FISH) or GCN ≥ 10 by NGS often has an absence of other oncogenic drivers, and is therefore viewed as a de novo primary oncogene driver in NSCLC.36, 37, 38 In the setting of acquired resistance, the definition of MET amplification can be different; ongoing and future studies will inform the selection criteria for this important biomarker.
MET amplification as a resistance mechanism
MET-dependent resistance is triggered by the activation of common downstream pathways of oncogene receptors, directly by the homodimer formation or indirectly by trans-activating other tyrosine kinase receptors. In the setting of EGFR-mutant NSCLC, MET amplification leads to resistance by persistent activation of signaling pathways downstream of EGFR, such as those mediated by MAPK, STAT, and PI3K/AKT, independent of EGFR activation and signaling.39 Signaling occurs through two adaptors: human epidermal growth factor receptor 3 (HER3 or ERBB3), when MET is triggered by genomic amplification, or Grb2-associated binder 1 (GAB1), when MET is activated by HGF.39 Higher levels of HGF expression have also been detected in tumor samples from patients resistant to the EGFR TKI gefitinib or erlotinib than in treatment-naïve tumor specimens.39 Resistance has been found even in the absence of MET amplification via HGF-induced activation of the AKT pathway.40,41 Thus, EGFR signaling becomes redundant and, as preclinical and recent clinical studies suggest, targeting both receptors by adding an anti-MET agent to EGFR TKIs is required to obtain an effective antitumor activity.42 In other oncogene-driven lung cancers, similar downstream signaling pathways were activated, and MET amplification functions similarly in a redundant manner to render resistance to TKI treatment to the original driver oncogene.
MET amplification in EGFR-mutant lung cancer
Genomic alterations in the EGFR gene account for up to 50% of NSCLC in Asian patients and 10% in Western patients.43 While the resistance mechanism spectrum evolves with the introduction of third-generation EGFR TKIs, amplification of the MET gene as an acquired resistance mechanism to EGFR TKI treatment has been reported in all generations of TKIs.8,9,44, 45, 46 MET amplification as an acquired mechanism resistance occurs in approximately 10%-15% of patients with NSCLC who have received erlotinib, gefitinib, or afatinib.26,39,45 Osimertinib, a third-generation EGFR TKI, is now used in the first-line setting for patients with advanced EGFR-mutant NSCLC.47, 48, 49 Despite this more potent inhibition to EGFR signaling, MET amplification remains a major resistance mechanism, occurring still in about 15% of patients with treatment failure of first-line osimertinib,50,51 and 10%-22% of patients following second-line osimertinib.8,52,53 In the AURA3 study, MET amplification was the most common (19%) resistance mechanism,52 where it co-occurred with EGFR C797S in 7% of cases, and was also likely to be associated with CDK6 and BRAF amplifications.54 Because of the relative proximity of CDK6, MET, and BRAF on chromosome 7q (7q21.2, 7q31.2, and 7q34, respectively), a single genomic event could be hypothesized to be responsible for gene amplifications.54 In the FLAURA study, MET amplification was also the most common acquired resistance mechanism (15%).55 MET amplification can also occur as a mechanism of resistance to third-generation EGFR TKIs, with or without loss of the T790M mutation (Table 1).54,56,57 It is important to note that using diverse patient materials, such as tissue or plasma for analysis of MET amplification, may result in different levels of MET amplification being detected, and so a caveat should be that different levels of MET amplification detected using different samples and techniques in various studies may simply reflect these differences.
Table 1.
Summary of key clinical studies identifying MET amplification as a mechanism of resistance in oncogene-driven NSCLC
| Molecular subset of NSCLC | Number of lung cancer samples + Type | Prior targeted therapy | Incidence of MET amplification | Method of MET amplification testing | Reference |
|---|---|---|---|---|---|
| EGFR | Following second-line osimertinib: range 10%-22% | ||||
| 83 Plasma |
19% (14/83) | NGS | 52 | ||
| 32 Tumor tissue |
22% (7/32) | FISH | 8 | ||
| 42 Tumor tissue |
14% (6/42) | FISH and/or NGS | 54 | ||
| 41 Tumor tissue |
10% (4/41) | NGS and FISH | 53 | ||
| EGFR | Following first-line osimertinib: range 7%-15% | ||||
| 91 Plasma |
15% (14/91) | NGS | 55 | ||
| 27 Tumor tissue |
7.4% (2/27) | NGS | 51 | ||
| ALK | Post-treatment tissue (n = 101) or | Crizotinib, or next-generation ALK inhibitors (e.g. lorlatinib) | 11 (13%) | FISH and/or NGS | 77 |
| Plasma (n = 106) | |||||
| RET | 23 | Selpercatinib or pralsetinib | 15% | FISH or NGS | 82 |
| ROS1 | 17 | Lorlatinib | 6% | NGS and FISH | 85 |
| KRAS | 10 Tumor tissue and/or plasma |
Adagrasib | 20% | NGS | 95 |
NGS, next-generation sequencing; NSCLC, non-small-cell lung cancer.
Strategies to overcome resistance – MET TKI + EGFR TKI
Dual inhibition of EGFR and MET is a rationale treatment strategy for overcoming acquired EGFR TKI resistance due to MET amplification.26 Several preclinical studies have demonstrated that concomitant use of MET inhibitors with osimertinib overcame resistance in osimertinib-resistant EGFR-mutant NSCLC cell lines with MET amplification.54,58 More recently, preclinical models using the potent and selective MET TKI tepotinib with EGFR TKI (erlotinib, gefitinib, afatinib, or rociletinib) overcame resistance to EGFR TKIs in EGFR-mutant NSCLC with MET amplification and high MET expression in in vivo and in vitro models,59 and led to tumor shrinkage, and even complete regression of established tumors.59
The INSIGHT study confirms these data clinically60: Wu and colleagues investigated the combination of tepotinib and gefitinib in patients with resistant EGFR-mutant NSCLC and with MET overexpression [immunohistochemistry (IHC)2+ or IHC3+] or MET amplification (or both). Nearly 67% of patients with MET amplification, treated with tepotinib plus gefitinib, achieved an objective response in both phase Ib and phase II of the study. The combination was also found to be safe and tolerable.60 Median PFS was 16.6 months with tepotinib plus gefitinib versus 4.2 months with chemotherapy (hazard ratio 0.13 90% confidence interval 0.04-0.43) and median overall survival was 37.3 months with tepotinib plus gefitinib versus 13.1 months with chemotherapy (hazard ratio 0.08, 90% confidence interval 0.01-0.51).60 This study highlights the need for tailored therapy, as the combination of tepotinib and gefitinib was less effective in patients with low MET expression (IHC2+ or less) but greatly beneficial for high MET amplification.
In the TATTON study, a number of novel combinations were investigated in previously treated EGFR-mutant NSCLC, including the combination of MET TKI savolitinib with osimertinib.61 In this trial, patients with EGFR-mutant NSCLC had received prior EGFR TKI and had evidence of MET amplification, defined using FISH as ≥5 copies of MET averaged over 50 cells scored. The objective response rate was 44% (22%-69%) and 30% of patients progressing on a third-generation EGFR TKI showed an objective response. An expansion cohort of this trial included patients with EGFR-mutant T790M-negative NSCLC who had not previously received a third-generation EGFR TKI, and objective partial responses of 64% were observed in 23 patients.62
There are additional early phase trial data to support the promising combination of MET TKI with EGFR TKI in patients with MET amplification-dependent resistance: in a phase Ib/II single-arm trial, capmatinib plus gefitinib in patients with EGFR-mutant NSCLC who progressed on an EGFR TKI showed activity in patients with MET-dysregulated NSCLC, particularly in MET-amplified tumors [47% of patients with MET-amplified tumors (GCN ≥6) and 32% patients with MET overexpression (IHC3+) achieved an objective response].63 In another phase Ib study investigating MET TKI savolitinib plus gefitinib, up to 52% of patients with EGFR-mutant NSCLC with MET-amplified tumors (defined as MET to CEP7 ratio ≥2 or GCN ≥5), having relapsed on previous EGFR TKI treatment, achieved an objective response.64 Some studies have investigated MET-targeted antibodies onartuzumab and emibetuzumab in combination with erlotinib; however, these trials relied on MET overexpression as a stratification marker, and as such did not meet their primary endpoints.65,66 These data support the importance of selecting the appropriate drug–drug combination for the appropriate patient (here high-level MET amplification).
There are a number of ongoing studies aiming to bring the EGFR plus MET TKI combination to this EGFR-mutant MET-amplified NSCLC patient population, including INSIGHT 2 (NCT03940703; tepotinib and osimertinib) and SAVANNAH (NCT03778229; savolitinib and osimertinib).67
MET amplification in ALK-fusion-positive lung cancer
ALK-fusion-positive NSCLC is another established molecular subtype of lung cancer occurring in 3%-5% of lung adenocarcinomas.68 Similar to EGFR-mutant NSCLC, the clinical benefit of targeting anaplastic lymphoma kinase (ALK) using TKIs is limited by the emergence of drug resistance. Historically, the standard first-line therapy for advanced ALK-fusion NSCLC has been the multikinase ALK/ROS1/MET TKI crizotinib,49 which has recently been surpassed by more potent and selective second- and third- generation ALK TKIs, such as ceritinib, alectinib, brigatinib, and lorlatinib.69, 70, 71, 72 Despite initial sensitivity to ALK TKIs, ALK-fusion-positive tumors invariably develop resistance, and a number of diverse mechanisms of resistance to ALK TKIs have now been discovered.73 In ∼50% of cases, resistance to second-generation ALK TKIs is due to ALK-independent resistance mechanisms, most often due to activation of bypass signaling pathways, including activation of MET, EGFR, SRC, and IGF-1R.74,75 The availability of potent MET TKIs makes MET a particularly attractive target.74, 75, 76
MET amplification has emerged as a prominent ALK-independent mediator of resistance, and recently MET amplification was detected in 15% of tumor biopsies from patients relapsing on selective ALK inhibitors, including 12% and 22% of biopsies from patients progressing on second-generation inhibitors and lorlatinib, respectively.77 Several case reports have also confirmed this, and suggest that the ALK/ROS1/MET TKI crizotinib may be able to overcome MET-dependent resistance.78,79
In a comprehensive analysis of MET alterations in ALK-fusion-positive NSCLC,77 FISH and/or NGS were performed on 207 post-treatment tissue (n = 101) or plasma (n = 106) specimens from patients with ALK-fusion-positive lung cancer to detect MET genetic alterations. The analysis also evaluated ALK inhibitor sensitivity in cell lines with MET alterations and assessed antitumor activity of the ALK/MET dual blockade in ALK-fusion cell lines and two subsequent patients with MET-dependent resistance. Eleven (13%) biopsies harbored MET amplification, including four with low-level MET amplification (MET/CEP7 2.4–3.9) and six with high-level MET amplification based on FISH (MET/CEP7 5.2 to >25) or NGS (16–19 MET copies). One sample had focal MET amplification by NGS, and FISH was too variable to estimate copy number. Of note, no coalterations in other genes potentially associated with resistance or bypass signaling were identified in the 11 biopsies, and MET amplification was mutually exclusive with ALK-resistance mutations, with the exception of one case.77 Patients treated with a second-generation ALK inhibitor in the first-line setting were more likely to develop MET amplification than those who had received next-generation ALK inhibitors after crizotinib (P = 0.019).77 This study demonstrated in preclinical models that treatment with MET-specific TKIs capmatinib or savolitinib, none of which have anti-ALK activity, partially suppressed proliferation.77 However, combination therapy using lorlatinib with capmatinib/savolitinib, or crizotinib potently suppressed cell proliferation and only dual inhibition of ALK and MET by crizotinib or by utilizing the combination of lorlatinib plus a MET TKI effectively suppressed both ALK and downstream signaling pathways.77 Based on these preclinical findings, two TKI-resistant ALK-fusion-positive lung cancer patients with acquired MET alterations were treated with a combination of lorlatinib and crizotinib and achieved rapid responses to ALK/MET combination therapy, validating that MET was the resistance driver and the combination's therapeutic potential.77 Although this study confirmed the rationale for exploring ALK/MET combinations in the clinic, in the setting of MET-dependent resistance to ALK TKIs, the optimal combination needs to be evaluated in prospective clinical trials, especially taking into consideration newer generation MET inhibitors' potency and activities for brain metastasis.
This study demonstrated a comparable prevalence of MET amplification in patients relapsing on ALK TKIs to that seen in EGFR-mutant NSCLC, and interestingly, presented similar findings of increased frequency of MET amplification in tumors resistant to the third-generation, broad-spectrum ALK TKI lorlatinib.77
MET amplification in RET-fusion lung cancer
The RET proto-oncogene encodes an RTK which is activated by gene fusion in 1%-2% of NSCLC, and has been recently defined as a druggable molecular subset of NSCLC. Selpercatinib80 and pralsetinib81 are highly selective RET kinase inhibitors that have recently been FDA approved for advanced RET-fusion NSCLC based on impressive efficacy data from the LIBRETTO-001 and ARROW studies, respectively. Molecular mechanisms of acquired resistance to RET inhibitors are not yet fully understood; however, recent data confirm MET amplification as a recurrent mechanism of resistance to targeted therapy in NSCLC patients treated with selpercatinib. Lin et al.82 recently performed a multi-institutional analysis of repeat tumor or plasma biopsies from a cohort of patients with RET-fusion NSCLC following treatment with selpercatinib and pralsetinib, to systematically characterize acquired resistance mechanisms to these inhibitors (patients received pralsetinib or selpercatinib in clinical trials).
A strikingly similar prevalence was noted, comparable to EGFR-/ALK-resistance cases, as three resistant cases (15%) harbored acquired MET amplification without concurrent RET-resistance mutations82; acquired RET mutations were identified in two cases (10%) and KRAS amplification in one case. A recently published work confirmed these data, and provided further preclinical evidence that that MET amplification is sufficient to cause selpercatinib resistance in in vitro models.80 Rosen et al.80 identified that patients treated with selpercatinib have MET amplification associated with resistance to selpercatinib, and proposed a dual targeting strategy to overcome MET-dependent resistance to RET-directed therapy, piloting a combination strategy using selpercatinib and crizotinib to rescue the phenotype.
Rosen and investigators80 demonstrated that this combination strategy with selpercatinib and crizotinib overcame MET-dependent resistance to selective RET inhibition in patients with RET-fusion lung cancer (one case of clinical activity and tolerability, with response lasting 10 months), and that this strategy was tolerable and feasible. The numbers in this report are admittedly small (n = 4) and further prospective work is needed, as our collective clinical experience with RET-fusion NSCLC is increased. Interestingly, while the level of MET gene amplification was shown to clearly increase during selpercatinib monotherapy, in three of four cases, some degree of MET gain was already present before exposure to selpercatinib: this is reminiscent of EGFR-mutant NSCLC, in which rare clones with high-level MET amplification may be detected at baseline, before EGFR inhibitor therapy.32,66
ROS1-fusion-positive lung cancer
Genetic rearrangements of the ROS1 gene account for 1%-2% of NSCLC. ROS1 can be targeted by TKIs such as crizotinib, entrectinib, and lorlatinib, which results in dramatic responses6,83,84; however, ROS1-independent resistance mechanisms remain poorly characterized. Recent evidence suggests a role for MET amplification.85
Lin et al. recently analyzed repeat tumor biopsies derived from advanced ROS1-fusion-positive NSCLC patients progressing on lorlatinib using NGS (n = 17) or whole-exome sequencing (n = 1) to detect potential drivers of resistance.85 While on-target ROS1 kinase domain mutations featured prominently as a mechanism of resistance, 38.9% harbored a ROS1-resistance mutation. NGS analyses also identified MET copy number gain in a lorlatinib-resistant case, validated by FISH as high-level focal MET amplification without a concomitant ROS1-resistance mutation. There are no published data investigating combination strategies in MET-amplified ROS1-positive lung cancer. Thus there is a clear need to further elucidate ROS1-independent resistance mechanisms and develop strategies to tackle resistance.
NTRK-fusion-positive lung cancer
Similar to ALK- and ROS1-fusion-positive lung cancers, neurotrophic tropomyosin-related kinase (NTRK)-fusion-positive cancers can develop on- and off-target resistance to TKI therapy. Recently, published data suggest that resistance to TRK inhibition is mediated by genomic alterations that converge to activate the mitogen-activated protein kinase (MAPK) pathway, and MET amplification has been identified as a mediator of resistance,86 together with BRAF V600E mutation or hotspot mutations involving KRAS. To date, there are a paucity of published reports regarding NTRK-fusion-positive lung cancer resistance after TKI therapy; a recent combination of a TRK and MET inhibitor achieved a confirmed response to therapy in a patient with a NTRK-fusion-positive cholangiocarcinoma with MET amplification-dependent resistance to a first-generation TRK inhibitor, accompanied by the disappearance of detectable NTRK fusion and MET amplification in circulating free DNA.86
BRAF-mutant-positive lung cancer
BRAF gene is mutated in up to 5% of lung adenocarcinomas87 and represents another actionable target in lung cancer. Scant evidence has been available concerning the mechanisms of resistance to BRAF/MEK inhibitors in BRAF V600E NSCLC. Recently, circulating tumor DNA genomics have demonstrated that mutations in effectors of the MAPK and PI3K pathways may play a role in resistance.88 Oncogenic mutations in KRAS, which encode immediate upstream regulators of the RAF kinases have also been suggested.89 MET amplification has not been identified as a driver of resistance; however, complete molecular analyses were available for only seven patients.90
KRAS G12C-mutant NSCLC
The KRAS G12C mutation occurs in ∼13%-34% of NSCLC91,92 and is emerging as the newest actionable target in lung cancer.93 While previously KRAS has proven to be an elusive target, Hong and colleagues7 recently demonstrated that a potent and selective small-molecule KRAS G12C inhibitor, sotorasib (AMG 510), can induce impressive, durable responses in KRAS G12C-mutant NSCLCs. The phase II CodeBreak 100 trial has recently further validated these impressive data: a response rate of 37.1%, a disease control rate of 80.6%, and a median PFS of 6.8 months.94 Therefore understanding the resistance spectrum will become critically important.
Serial biopsy data and serial circulating tumor DNA data have yet to be reported in the CodeBreak study; however, recently published data from Awad et al.95 have confirmed MET amplification as a mechanism for resistance in patients with KRAS G12C-mutant NSCLC previously treated with the KRAS G12C inhibitor adagrasib. Preclinical models have highlighted the importance of RTK and SHP2 activation in acquired resistance to KRAS inhibitors,96 and there are published data which show that METex14 and KRAS G12 mutations can co-occur, at a rate of co-occurrence significantly higher than in other major driver-defined lung cancer subsets.97 In the study by Awad et al.,95 10 patients with KRAS G12C-mutant NSCLC had samples available for assessment on progression of therapy; furthermore, acquired MET amplification was the only potential genomic mechanism of adagrasib resistance, which was identified in two of these patients (20%). This is again a frequency rate similar to other oncogene-driven NSCLCs, and suggests that MET amplification may also be a driver of resistance to selective small-molecule KRAS G12C inhibitors.
Conclusions
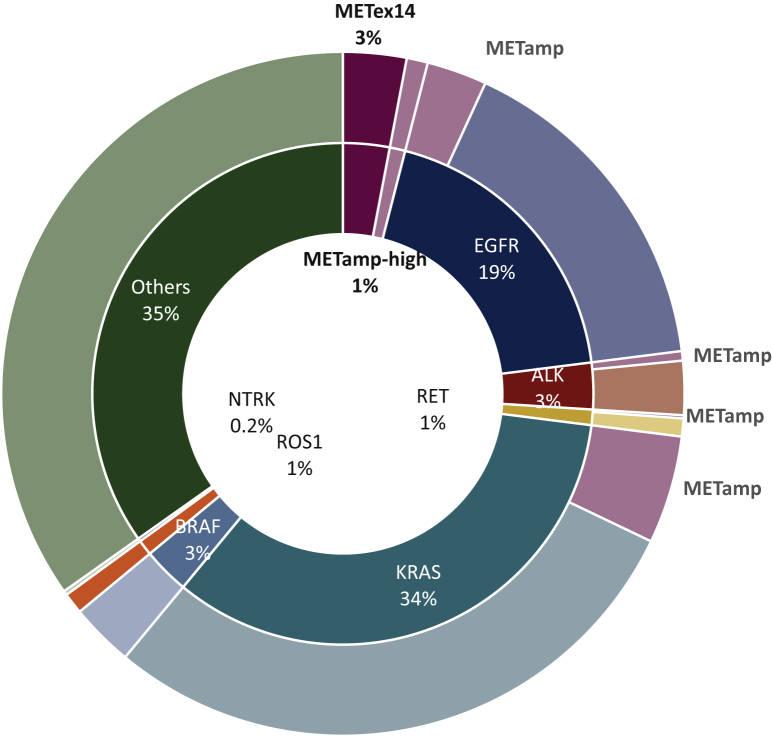
MET amplification is a well-defined mechanism of resistance in EGFR-mutant NSCLCs, and in this review, we highlight the increasing number of reports which have underscored that MET amplification plays a role in acquired resistance in a number of other oncogene-driven NSCLCs, including ALK-, RET-, ROS-fusion-positive and more recently, KRAS G12C-mutant lung cancers. The presence and strikingly similar prevalence of MET amplification as a driver of resistance in ∼15% of the TKI-treated population across various oncogene-driven NSCLCs is remarkable (Figure 2). In the more recently defined rare molecular subsets of NSCLCs, such as TRK fusion, BRAF mutant, the relative frequency of off-target resistance, such as MET amplification, has yet to be elucidated. Recent data have proved that in KRAS G12C-mutant NSCLCs following adagrasib therapy, MET amplification is a consistent mechanism of resistance.95 We eagerly await the serial biopsy data and serial circulating tumor DNA data from the seminal sotorasib KRAS G12C study,7 which will likely confirm adagrasib findings.
Figure 2.
Frequency of MET dependency in lung cancer.
The inner ring represents known primary oncogenic driver alterations in metastatic lung cancers, such as EGFR, ALK, RET, KRAS, and BRAF. The outer ring illustrates known resistance mechanisms in these oncogenic-driven NSCLC subsets: frequency of MET amplification (red) is ∼15% in EGFR, KRAS, and ALK- and RET-fusion-positive NSCLC. Taken together, these data highlight that ∼7%-10% of NSCLC tumors are MET dependent, including de novo METex14 and high MET amplification. EGFR, epidermal growth factor receptor; NSCLC, non-small-cell lung cancer.
As our collective understanding of the molecular mechanisms underpinning TKI resistance in oncogene-driven NSCLCs continues to mature, the development of rationale combination strategies will be crucial in overcoming acquired resistance. For combination therapy, the EGFR story provides the highest level of evidence and proof of concept, where combining potent and selective MET inhibitor with EGFR inhibitor in patients with EGFR-mutant MET-amplified resistant tumors has produced clinically meaningful responses and tolerable safety profiles.61,98 MET amplification-dependent TKI resistance therefore demands attention from the field because of the need to prospectively identify and treat these patients with efficacious and safe combinations.
Acknowledgments
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Disclosure
XL is supported by Paul Calabresi Award at MDACC (K12/NIH), Rexanna's Foundation, Khalifa Scholarship, and Conquer Cancer Foundation; received research funds from Eli Lilly, Boehringer Ingelheim, and Spectrum Pharmaceuticals (all outside of the submitted work); received consultant and advisory fee from Eli Lilly, AstraZeneca, and EMD Serono (all outside of the submitted work). DH reports receiving consulting fees and research/grant funding from AbbVie, Adaptimmune, Adlai Nortye, Amgen, Astra-Zeneca, Bayer, Bristol Myers Squibb, Daiichi-Sankyo, Eisai, Eli Lilly, EMD Serono, Erasca, Fate Therapeutics, Genentech, Genmab, GlaxoSmithKline, Ignyta, Infinity, Kite, Kyowa, LOXO, Merck, MedImmune, Millennium, Mirati, miRNA, Molecular Templates, Mologen, Navier, NCI-CTEP, Novartis, Numab, Pfizer, Seattle Genetics, Takeda, Turning Point Therapeutics, Verstatem, VM Oncology; travel, accommodations, and expenses covered by AACR, Amgen, ASCO, Astra Zeneca, Bayer, Celgene, Eli Lilly, Genentech, Genmab, GlaxoSmithKline, Janssen, LOXO, miRNA, Pfizer, Philips, SITC, Takeda; consulting, speaker, or advisory role with Alpha Insights, Acuta, Amgen, Axiom, Adaptimmune, Baxter, Bayer, Boxer Capital, COG, Ecor1, Genentech, GLG, Group H, Guidepoint, HCW Precision, Infinity, Janssen, Merrimack, Medscape, Numab, Pfizer, Prime Oncology, Seattle Genetics, ST Cube, Takeda, Tavistock, Trieza Therapeutics, and WebMD; other ownership interests in Molecular Match (Advisor), OncoResponse (Founder), and Presagia Inc (Advisor). JH reports advisory/consulting fees from Bristol-Myers Squibb, GlaxoSmithKline, Kairos Venture Investments, BrightPath Therapeutics, Hengrui Therapeutics, Eli Lilly, EMD Serono, and Foundation One Medicine, Spectrum, AstraZeneca; and research funding from NIH/NCI, American Cancer Society, Checkmate Pharmaceuticals, AstraZeneca, and Spectrum; and royalties and patents from Spectrum. JZ is supported by MD Anderson Physician Scientist Award (NIH R01), AACR Johnson and Johnson Innovative Cancer Research Award, Khalifa Scholarship, and Conquer Cancer Foundation; receives research funding from Merck and Johnson and Johnson; personal fees from AstraZeneca, Bristol-Myers Squibb, Geneplus-Beijing Institute, and InnoVent outside the submitted work. NC and LH have no disclosures.
References
- 1.Rosell R., Carcereny E., Gervais R., et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13(3):239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 2.Yang J.C.H., Wu Y.L., Schuler M., et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol. 2015;16(2):141–151. doi: 10.1016/S1470-2045(14)71173-8. [DOI] [PubMed] [Google Scholar]
- 3.Shaw A.T., Ou S.-H.I., Bang Y.-J., et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2014;371(21):1963–1971. doi: 10.1056/NEJMoa1406766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solomon B.J., Mok T., Kim D.-W., et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. 2014;371(23):2167–2177. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 5.Drilon A., Siena S., Ou S.H.I., et al. Safety and antitumor activity of the multitargeted pan-TRK, ROS1, and ALK inhibitor entrectinib: combined results from two phase I trials (ALKA-372-001 and STARTRK-1) Cancer Discov. 2017;7:400–409. doi: 10.1158/2159-8290.CD-16-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaw A.T., Felip E., Bauer T.M., et al. Lorlatinib in non-small-cell lung cancer with ALK or ROS1 rearrangement: an international, multicentre, open-label, single-arm first-in-man phase 1 trial. Lancet Oncol. 2017;18:1590–1599. doi: 10.1016/S1470-2045(17)30680-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong D.S., Fakih M.G., Strickler J.H., et al. KRAS G12C inhibition with sotorasib in advanced solid tumors. N Engl J Med. 2020;383:1207–1217. doi: 10.1056/NEJMoa1917239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piotrowska Z., Isozaki H., Lennerz J.K., et al. Landscape of acquired resistance to osimertinib in EGFR-mutant NSCLC and clinical validation of combined EGFR and RET inhibition with osimertinib and BLU-667 for acquired RET fusion. Cancer Discov. 2018;8:1529–1539. doi: 10.1158/2159-8290.CD-18-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu H.A., Arcila M.E., Rekhtman N., et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19:2240–2247. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yap T.A., Omlin A., de Bono J.S. Development of therapeutic combinations targeting major cancer signaling pathways. J Clin Oncol. 2013;31:1592–1605. doi: 10.1200/JCO.2011.37.6418. [DOI] [PubMed] [Google Scholar]
- 11.Cooper C.S., Park M., Blair D.G., et al. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature. 1984;311(5981):29–33. doi: 10.1038/311029a0. [DOI] [PubMed] [Google Scholar]
- 12.Ponzetto C., Bardelli A., Zhen Z., et al. A multifunctional docking site mediates signaling and transformation by the hepatocyte growth factor/scatter factor receptor family. Cell. 1994;77(2):261–271. doi: 10.1016/0092-8674(94)90318-2. [DOI] [PubMed] [Google Scholar]
- 13.Naldini L., Vigna E., Narsimhan R.P., et al. Hepatocyte growth factor (HGF) stimulates the tyrosine kinase activity of the receptor encoded by the proto-oncogene c-MET. Oncogene. 1991;6:501–504. [PubMed] [Google Scholar]
- 14.Fixman E.D., Fournier T.M., Kamikura D.M., Naujokas M.A., Park M. Pathways downstream of Shc and Grb2 are required for cell transformation by the Tpr-Met oncoprotein. J Biol Chem. 1996;271:13116–13122. doi: 10.1074/jbc.271.22.13116. [DOI] [PubMed] [Google Scholar]
- 15.Bottaro D.P., Rubin J.S., Faletto D.L., et al. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science. 1991;251:802–804. doi: 10.1126/science.1846706. [DOI] [PubMed] [Google Scholar]
- 16.Lemmon M.A., Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaeper U., Gehring N.H., Fuchs K.P., Sachs M., Kempkes B., Birchmeier W. Coupling of Gab1 to c-Met, Grb2, and Shp2 mediates biological responses. J Cell Biol. 2000;149:1419–1432. doi: 10.1083/jcb.149.7.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paliouras G.N., Naujokas M.A., Park M. Pak4, a Novel Gab1 binding partner, modulates cell migration and invasion by the met receptor. Mol Cell Biol. 2009;29:3018–3032. doi: 10.1128/MCB.01286-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maroun C.R., Naujokas M.A., Holgado-Madruga M., Wong A.J., Park M. The tyrosine phosphatase SHP-2 is required for sustained activation of extracellular signal-regulated kinase and epithelial morphogenesis downstream from the met receptor tyrosine kinase. Mol Cell Biol. 2000;20:8513–8525. doi: 10.1128/mcb.20.22.8513-8525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai A.Z., Abella J.V., Park M. Crosstalk in met receptor oncogenesis. Trends Cell Biol. 2009;19:542–551. doi: 10.1016/j.tcb.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 21.Weidner K.M., Di Cesare S., Sachs M., Brinkmann V., Behrens J., Birchmeier W. Interaction between Gab1 and the c-Met receptor tyrosine kinase is responsible for epithelial morphogenesis. Nature. 1996;384:173–176. doi: 10.1038/384173a0. [DOI] [PubMed] [Google Scholar]
- 22.Birchmeier C., Birchmeier W., Gherardi E., Vande Woude G.F. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 23.Sipeki S., Bander E., Buday L., et al. Phosphatidylinositol 3-kinase contributes to Erk1/Erk2 MAP kinase activation associated with hepatocyte growth factor-induced cell scattering. Cell Signal. 1999;11(12):885–890. doi: 10.1016/s0898-6568(99)00060-1. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y.W., Wang L.M., Jove R., Vande Woude G.F. Requirement of Stat3 signaling for HGF/SF-Met mediated tumorigenesis. Oncogene. 2002;21:217–226. doi: 10.1038/sj.onc.1205004. [DOI] [PubMed] [Google Scholar]
- 25.van der Steen N., Pauwels P., Gil-Bazo I., et al. cMET in NSCLC: can we cut off the head of the hydra? From the pathway to the resistance. Cancers (Basel) 2015;7(2):556–573. doi: 10.3390/cancers7020556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Y.L., Soo R.A., Locatelli G., Stammberger U., Scagliotti G., Park K. Does c-Met remain a rational target for therapy in patients with EGFR TKI-resistant non-small cell lung cancer? Cancer Treat Rev. 2017;61:70–81. doi: 10.1016/j.ctrv.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Drilon A., Cappuzzo F., Ou S.H.I., Camidge D.R. Targeting MET in lung cancer: will expectations finally be MET? J Thorac Oncol. 2017;12:15–26. doi: 10.1016/j.jtho.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hellman A., Zlotorynski E., Scherer S.W., et al. A role for common fragile site induction in amplification of human oncogenes. Cancer Cell. 2002;1:89–97. doi: 10.1016/s1535-6108(02)00017-x. [DOI] [PubMed] [Google Scholar]
- 29.Drilon A., Clark J.W., Weiss J., et al. Antitumor activity of crizotinib in lung cancers harboring a MET exon 14 alteration. Nat Med. 2020;26(1):47–51. doi: 10.1038/s41591-019-0716-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cappuzzo F., Marchetti A., Skokan M., et al. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J Clin Oncol. 2009;27:1667–1674. doi: 10.1200/JCO.2008.19.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawakami H., Okamoto I., Okamoto W., Tanizaki J., Nakagawa K., Nishio K. Targeting MET amplification as a new oncogenic driver. Cancers (Basel) 2014;6:1540–1552. doi: 10.3390/cancers6031540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cappuzzo F., Jänne P.A., Skokan M., et al. MET increased gene copy number and primary resistance to gefitinib therapy in non-small-cell lung cancer patients. Ann Oncol. 2009;20:298–304. doi: 10.1093/annonc/mdn635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaka A., Sueoka-Aragane N., Nakamura T., et al. Co-existence of positive MET FISH status with EGFR mutations signifies poor prognosis in lung adenocarcinoma patients. Lung Cancer. 2012;75(1):89–94. doi: 10.1016/j.lungcan.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 34.Wolf J., Seto T., Han J.Y., et al. Capmatinib in MET Exon 14-mutated or MET-amplified non-small-cell lung cancer. N Engl J Med. 2020;383(10):944–957. doi: 10.1056/NEJMoa2002787. [DOI] [PubMed] [Google Scholar]
- 35.Zheng Z., Liebers M., Zhelyazkova B., et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20:1479–1484. doi: 10.1038/nm.3729. [DOI] [PubMed] [Google Scholar]
- 36.Tong J.H., Yeung S.F., Chan A.W.H., et al. MET amplification and exon 14 splice site mutation define unique molecular subgroups of non-small cell lung carcinoma with poor prognosis. Clin Cancer Res. 2016;22:3048–3056. doi: 10.1158/1078-0432.CCR-15-2061. [DOI] [PubMed] [Google Scholar]
- 37.Le X. Heterogeneity in MET-aberrant NSCLC. J Thorac Oncol. 2021;16:504–506. doi: 10.1016/j.jtho.2021.01.1609. [DOI] [PubMed] [Google Scholar]
- 38.Kron A., Scheffler M., Heydt C., et al. Genetic heterogeneity of MET-aberrant NSCLC and its impact on the outcome of immunotherapy. J Thorac Oncol. 2021;16:572–582. doi: 10.1016/j.jtho.2020.11.017. [DOI] [PubMed] [Google Scholar]
- 39.Turke A.B., Zejnullahu K., Wu Y.L., et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamada T., Matsumoto K., Wang W., et al. Hepatocyte growth factor reduces susceptibility to an irreversible epidermal growth factor receptor inhibitor in EGFR-T790M mutant lung cancer. Clin Cancer Res. 2010;16:174–183. doi: 10.1158/1078-0432.CCR-09-1204. [DOI] [PubMed] [Google Scholar]
- 41.Yano S., Wang W., Li Q., et al. Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor-activating mutations. Cancer Res. 2008;68:9479–9487. doi: 10.1158/0008-5472.CAN-08-1643. [DOI] [PubMed] [Google Scholar]
- 42.Wang W., Li Q., Takeuchi S., et al. Met kinase inhibitor E7050 reverses three different mechanisms of hepatocyte growth factor-induced tyrosine kinase inhibitor resistance in EGFR mutant lung cancer. Clin Cancer Res. 2012;18:1663–1671. doi: 10.1158/1078-0432.CCR-11-1171. [DOI] [PubMed] [Google Scholar]
- 43.Castellanos E., Feld E., Horn L. Driven by mutations: the predictive value of mutation subtype in EGFR-mutated non–small cell lung cancer. J Thorac Oncol. 2017;12:612–623. doi: 10.1016/j.jtho.2016.12.014. [DOI] [PubMed] [Google Scholar]
- 44.Benedettini E., Sholl L.M., Peyton M., et al. Met activation in non-small cell lung cancer is associated with de novo resistance to EGFR inhibitors and the development of brain metastasis. Am J Pathol. 2010;177:415–423. doi: 10.2353/ajpath.2010.090863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Remon J., Morán T., Majem M., et al. Acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in EGFR-mutant non-small cell lung cancer: a new era begins. Cancer Treat Rev. 2014;40:93–101. doi: 10.1016/j.ctrv.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 46.Ou S.H.I., Agarwal N., Ali S.M. High MET amplification level as a resistance mechanism to osimertinib (AZD9291) in a patient that symptomatically responded to crizotinib treatment post-osimertinib progression. Lung Cancer. 2016;98:59–61. doi: 10.1016/j.lungcan.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 47.Soria J.-C., Ohe Y., Vansteenkiste J., et al. Osimertinib in untreated EGFR-mutated advanced non–small-cell lung cancer. N Engl J Med. 2018;378:113–125. doi: 10.1056/NEJMoa1713137. [DOI] [PubMed] [Google Scholar]
- 48.Mok T.S., Wu Y.-L., Ahn M.-J., et al. Osimertinib or platinum–pemetrexed in EGFR T790M–positive lung cancer. N Engl J Med. 2017;376:629–640. doi: 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Planchard D., Popat S., Kerr K., et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29:iv192–iv237. doi: 10.1093/annonc/mdy275. [DOI] [PubMed] [Google Scholar]
- 50.Cho B.C., Cheng Y., Zhou C., et al. Mechanisms of acquired resistance to first-line osimertinib: preliminary data from the phase III FLAURA study. Ann Oncol. 2018;29:VIII740. [Google Scholar]
- 51.Schoenfeld A.J., Chan J.M., Kubota D., et al. Tumor analyses reveal squamous transformation and off-target alterations as early resistance mechanisms to first-line osimertinib in EGFR-mutant lung cancer. Clin Cancer Res. 2020;26:2654–2663. doi: 10.1158/1078-0432.CCR-19-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Papadimitrakopoulou V.A., Wu Y.-L., Han J.-Y., et al. Analysis of resistance mechanisms to osimertinib in patients with EGFR T790M advanced NSCLC from the AURA3 study. Ann Oncol. 2018;29:VIII741. [Google Scholar]
- 53.Oxnard G.R., Hu Y., Mileham K.F., et al. Assessment of resistance mechanisms and clinical implications in patients with EGFR T790M-positive lung cancer and acquired resistance to osimertinib. JAMA Oncol. 2018;4:1527–1534. doi: 10.1001/jamaoncol.2018.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Le X., Puri S., Negrao M.V., et al. Landscape of EGFR-dependent and -independent resistance mechanisms to osimertinib and continuation therapy beyond progression in EGFR-Mutant NSCLC. Clin Cancer Res. 2018;24(24):6195–6203. doi: 10.1158/1078-0432.CCR-18-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramalingam S.S., Cheng Y., Zhou C., et al. Mechanisms of acquired resistance to first-line osimertinib: preliminary data from the phase III FLAURA study. Ann Oncol. 2018;29:VIII740. [Google Scholar]
- 56.O'Kane G.M., Barnes T.A., Leighl N.B. Resistance to epidermal growth factor receptor tyrosine kinase inhibitors, T790M, and clinical trials. Curr Oncol. 2018;25(suppl 1):S28–S37. doi: 10.3747/co.25.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ortiz-Cuaran S., Scheffler M., Plenker D., et al. Heterogeneous mechanisms of primary and acquired resistance to third-generation EGFR inhibitors. Clin Cancer Res. 2016;22(19):4837–4847. doi: 10.1158/1078-0432.CCR-15-1915. [DOI] [PubMed] [Google Scholar]
- 58.Shi P., Oh Y.T., Zhang G., et al. Met gene amplification and protein hyperactivation is a mechanism of resistance to both first and third generation EGFR inhibitors in lung cancer treatment. Cancer Lett. 2016;380:494–504. doi: 10.1016/j.canlet.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 59.Friese-Hamim M., Bladt F., Locatelli G., Stammberger U., Blaukat A. The selective c-Met inhibitor tepotinib can overcome epidermal growth factor receptor inhibitor resistance mediated by aberrant c-Met activation in NSCLC models. Am J Cancer Res. 2017;7(4):962–972. [PMC free article] [PubMed] [Google Scholar]
- 60.Wu Y.L., Cheng Y., Zhou J., et al. Tepotinib plus gefitinib in patients with EGFR-mutant non-small-cell lung cancer with MET overexpression or MET amplification and acquired resistance to previous EGFR inhibitor (INSIGHT study): an open-label, phase 1b/2, multicentre, randomised trial. Lancet Respir Med. 2020;8:1132–1143. doi: 10.1016/S2213-2600(20)30154-5. [DOI] [PubMed] [Google Scholar]
- 61.Oxnard G.R., Yang J.C.H., Yu H., et al. TATTON: a multi-arm, phase Ib trial of osimertinib combined with selumetinib, savolitinib, or durvalumab in EGFR-mutant lung cancer. Ann Oncol. 2020;31:507–516. doi: 10.1016/j.annonc.2020.01.013. [DOI] [PubMed] [Google Scholar]
- 62.Sequist L.V., Han J.Y., Ahn M.J., et al. Osimertinib plus savolitinib in patients with EGFR mutation-positive, MET-amplified, non-small-cell lung cancer after progression on EGFR tyrosine kinase inhibitors: interim results from a multicentre, open-label, phase 1b study. Lancet Oncol. 2020;21:373–386. doi: 10.1016/S1470-2045(19)30785-5. [DOI] [PubMed] [Google Scholar]
- 63.Wu Y.L., Zhang L., Kim D.W., et al. Phase Ib/II study of capmatinib (INC280) plus gefitinib after failure of epidermal growth factor receptor (EGFR) inhibitor therapy in patients with EGFR-mutated, MET factor–dysregulated non–small-cell lung cancer. J Clin Oncol. 2018;36(31):3101–3109. doi: 10.1200/JCO.2018.77.7326. [DOI] [PubMed] [Google Scholar]
- 64.Yang J.J., Fang J., Shu Y.Q., et al. A phase Ib trial of savolitinib plus gefitinib for Chinese patients with EGFR-mutant MET-amplified advanced NSCLC. J Thorac Oncol. 2017;12(11 Suppl 2):S1769. [Google Scholar]
- 65.Spigel D.R., Edelman M.J., O'Byrne K., et al. Results from the phase III randomized trial of onartuzumab plus erlotinib versus erlotinib in previously treated stage IIIB or IV non-small-cell lung cancer: METLung. J Clin Oncol. 2017;35(4):412–420. doi: 10.1200/JCO.2016.69.2160. [DOI] [PubMed] [Google Scholar]
- 66.Scagliotti G., Moro-Sibilot D., Kollmeier J., et al. A randomized-controlled phase 2 study of the MET antibody emibetuzumab in combination with erlotinib as first-line treatment for EGFR mutation–positive NSCLC patients. J Thorac Oncol. 2020;15:80–90. doi: 10.1016/j.jtho.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 67.Oxnard G.R., Cantarini M., Frewer P., et al. SAVANNAH: A Phase II trial of osimertinib plus savolitinib for patients (pts) with EGFR-mutant, MET-driven (MET+), locally advanced or metastatic non-small cell lung cancer (NSCLC), following disease progression on osimertinib. J Clin Oncol. 2019;37(15_suppl) doi: 10.1200/jco.2019.37.15_suppl.tps9119. [DOI] [Google Scholar]
- 68.Shaw A.T., Yeap B.Y., Mino-Kenudson M., et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27(26):4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shaw A.T., Bauer T.M., de Marinis F., et al. First-line lorlatinib or crizotinib in advanced ALK-positive lung cancer. N Engl J Med. 2020;383:2018–2029. doi: 10.1056/NEJMoa2027187. [DOI] [PubMed] [Google Scholar]
- 70.Peters S., Camidge D.R., Shaw A.T., et al. Alectinib versus crizotinib in untreated ALK-positive non–small-cell lung cancer. N Engl J Med. 2017;377:829–838. doi: 10.1056/NEJMoa1704795. [DOI] [PubMed] [Google Scholar]
- 71.Camidge D.R., Kim H.R., Ahn M.J., et al. Brigatinib versus crizotinib in ALK-positive non-small-cell lung cancer. N Engl J Med. 2018;379:2027–2039. doi: 10.1056/NEJMoa1810171. [DOI] [PubMed] [Google Scholar]
- 72.Soria J.C., Tan D.S.W., Chiari R., et al. First-line ceritinib versus platinum-based chemotherapy in advanced ALK-rearranged non-small-cell lung cancer (ASCEND-4): a randomised, open-label, phase 3 study. Lancet. 2017;389:917–929. doi: 10.1016/S0140-6736(17)30123-X. [DOI] [PubMed] [Google Scholar]
- 73.Recondo G., Mezquita L., Facchinetti F., et al. Diverse resistance mechanisms to the third-generation ALK inhibitor lorlatinib in ALK-rearranged lung cancer. Clin Cancer Res. 2020;26:242–255. doi: 10.1158/1078-0432.CCR-19-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yoda S., Lin J.J., Lawrence M.S., et al. Sequential ALK inhibitors can select for lorlatinib-resistant compound ALK mutations in ALK-positive lung cancer. Cancer Discov. 2018;8(6):714–729. doi: 10.1158/2159-8290.CD-17-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gainor J.F., Dardaei L., Yoda S., et al. Molecular mechanisms of resistance to first- and second-generation ALK inhibitors in ALK -rearranged lung cancer. Cancer Discov. 2016;6(10):1118–1133. doi: 10.1158/2159-8290.CD-16-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Isozaki H., Ichihara E., Takigawa N., et al. Non-small cell lung cancer cells acquire resistance to the ALK inhibitor alectinib by activating alternative receptor tyrosine kinases. Cancer Res. 2016;76:1506–1516. doi: 10.1158/0008-5472.CAN-15-1010. [DOI] [PubMed] [Google Scholar]
- 77.Dagogo-Jack I., Yoda S., Lennerz J.K., et al. MET alterations are a recurring and actionable resistance mechanism in ALK-positive lung cancer. Clin Cancer Res. 2020;26:2535–2545. doi: 10.1158/1078-0432.CCR-19-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gouji T., Takashi S., Mitsuhiro T., Yukito I. Crizotinib can overcome acquired resistance to CH5424802: is amplification of the MET gene a key factor? J Thorac Oncol. 2014;9:e27–e28. doi: 10.1097/JTO.0000000000000113. [DOI] [PubMed] [Google Scholar]
- 79.Sakakibara-Konishi J., Kitai H., Ikezawa Y., et al. Response to crizotinib re-administration after progression on lorlatinib in a patient with ALK-rearranged non–small-cell lung cancer. Clin Lung Cancer. 2019;20:e555–e559. doi: 10.1016/j.cllc.2019.06.021. [DOI] [PubMed] [Google Scholar]
- 80.Rosen E.Y., Johnson M.L., Clifford S.E., et al. Overcoming MET-dependent resistance to selective RET inhibition in patients with RET fusion–positive lung cancer by combining selpercatinib with crizotinib. Clin Cancer Res. 2021;27:34–42. doi: 10.1158/1078-0432.CCR-20-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Subbiah V., Hu M.I.-N., Gainor J.F., et al. Clinical activity of the RET inhibitor pralsetinib (BLU-667) in patients with RET fusion+ solid tumors. J Clin Oncol. 2020;38:109. [Google Scholar]
- 82.Lin J.J., Liu S.V., McCoach C.E., et al. Mechanisms of resistance to selective RET tyrosine kinase inhibitors in RET fusion-positive non-small-cell lung cancer. Ann Oncol. 2020;31:1725–1733. doi: 10.1016/j.annonc.2020.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ou S.I., Camidge D.R., Engelman J., et al. Non-small cell lung cancer, locally advanced. Ann Oncol. 2012;23:IX389. [Google Scholar]
- 84.Drilon A., Siena S., Dziadziuszko R., et al. Entrectinib in ROS1 fusion-positive non-small-cell lung cancer: integrated analysis of three phase 1–2 trials. Lancet Oncol. 2020;21:261–270. doi: 10.1016/S1470-2045(19)30690-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lin J.J., Johnson T., Lennerz J.K., et al. Resistance to lorlatinib in ROS1 fusion-positive non-small cell lung cancer. J Clin Oncol. 2020;38(suppl 15):9611. [Google Scholar]
- 86.Cocco E., Schram A.M., Kulick A., et al. Resistance to TRK inhibition mediated by convergent MAPK pathway activation. Nat Med. 2019;25:1422–1427. doi: 10.1038/s41591-019-0542-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Leonetti A., Facchinetti F., Rossi G., et al. BRAF in non-small cell lung cancer (NSCLC): pickaxing another brick in the wall. Cancer Treat Rev. 2018;66:82–94. doi: 10.1016/j.ctrv.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 88.Ortiz-Cuaran S., Mezquita L., Swalduz A., et al. Circulating tumor DNA genomics reveal potential mechanisms of resistance to BRAF-targeted therapies in patients with BRAF -mutant metastatic non–small cell lung cancer. Clin Cancer Res. 2020;26:6242–6253. doi: 10.1158/1078-0432.CCR-20-1037. [DOI] [PubMed] [Google Scholar]
- 89.Rudin C.M., Hong K., Streit M. Molecular characterization of acquired resistance to the BRAF inhibitor dabrafenib in a patient with BRAF-mutant non-small-cell lung cancer. J Thorac Oncol. 2013;8:e41–e42. doi: 10.1097/JTO.0b013e31828bb1b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Facchinetti F., Lacroix L., Mezquita L., et al. Molecular mechanisms of resistance to BRAF and MEK inhibitors in BRAFV600E non–small cell lung cancer. Eur J Cancer. 2020;132:211–223. doi: 10.1016/j.ejca.2020.03.025. [DOI] [PubMed] [Google Scholar]
- 91.El Osta B., Behera M., Kim S., et al. Characteristics and outcomes of patients with metastatic KRAS-mutant lung adenocarcinomas: the lung cancer mutation consortium experience. J Thorac Oncol. 2019;14:876–889. doi: 10.1016/j.jtho.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Aredo J.V., Padda S.K., Kunder C.A., et al. Impact of KRAS mutation subtype and concurrent pathogenic mutations on non-small cell lung cancer outcomes. Lung Cancer. 2019;133:144–150. doi: 10.1016/j.lungcan.2019.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Amgen. Amgen's sotorasib granted breakthrough therapy designation for advanced or metastatic non-small cell lung cancer patients with KRAS G12C mutation. News release. Available at https://www.amgen.com/newsroom/press-releases/2020/12/amgens-sotorasib-granted-breakthrough-therapy-designation-for-advanced-or-metastatic-nonsmall-cell-lung-cancer-patients-with-kras-g12c-mutation.
- 94.Skoulidis F., Li B.T., Dy G.K., et al. Sotorasib for lung cancers with KRAS p.G12C mutation. N Engl J Med. 2021;384(25):2371–2381. doi: 10.1056/NEJMoa2103695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Awad M.M., Liu S., Rybkin, et al. Acquired resistance to KRAS G12C inhibition in cancer. N Engl J Med. 2021;384(25):2382–2393. doi: 10.1056/NEJMoa2105281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dunnett-Kane V., Nicola P., Blackhall F., Lindsay C. Mechanisms of resistance to KRASG12C inhibitors. Cancers (Basel) 2021;13:151. doi: 10.3390/cancers13010151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Suzawa K., Offin M., Lu D., et al. Activation of KRAS mediates resistance to targeted therapy in MET exon 14–mutant non–small cell lung cancer. Clin Cancer Res. 2019;25:1248–1260. doi: 10.1158/1078-0432.CCR-18-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang J.C.H., Ellers-Lenz B., Straub J., Johne A., Wu Y.-L. INSIGHT 2: tepotinib plus osimertinib in patients with EGFR-mutant NSCLC having acquired resistance to EGFR TKIs due to MET-amplification: a phase II trial in progress study. Ann Oncol. 2019;30:IX181. [Google Scholar]