Abstract
Numerous studies in the last decades have provided evidence for the existence of a local renin-angiotensin system (RAS) in the central nervous system (CNS). Widespread distribution of the different RAS components in the brain demonstrates the pleiotropic role of this system in the structure and function of CNS. With the advent of new molecular techniques, a novel receptor has been identified within the beneficial arm of the RAS, the Mas-related G-protein coupled receptor D (MrgD), which can be stimulated by two heptapeptides, Ala1-(Ang-(1-7), also named alamandine, and Ang-(1-7). However, the biological and physiological relevance of this interaction remains obscure. Since several recent studies hinted at a role of MrgD in the CNS, we determined the distribution pattern of MrgD receptors in the adult mouse brain by using a genetic mouse model with tracers of MrgD expression. MrgD-positive cells could be identified in some forebrain areas, including cortex, hippocampus, amygdala, hypothalamus, habenular nuclei, striatum and pallidum, as well as in some mid-brain nuclei in a region-specific manner. The specific localization of MrgD in the reward- and limbic-related areas can hint at a role of MrgD in processes such as pain perception/modulation, synaptic plasticity, learning, memory and cognition.
Keywords: MrgD, Brain, Mouse
MrgD; Brain; Mouse.
1. Introduction
Aside from the systemic renin-angiotensin-system (RAS), a local independent RAS has been reported within the central nervous system (CNS) (von Bohlen und Halbach and Albrecht, 2006; Wright and Harding, 2013; Jackson et al., 2018). Widespread distribution of the members of the RAS (e.g. angiotensin (Ang) II and Ang IV), as well as their specific cognate receptors (e.g. Ang type-1 receptor [AT1R], AT2R, and AT4R) has been reported in the CNS (von Bohlen und Halbach and Albrecht, 2006). In addition, the expression of further RAS components, such as e.g. pro-renin and its receptor have been recently identified, and these members of the brain RAS seem to play a key role in adult hippocampal neurogenesis (Schäfer et al., 2015). Several enzymatic pathways have been shown to be involved in the generation of different members of the RAS (Wright and Harding, 2013; Jackson et al., 2018). Apart from the classical RAS cascade, in which the angiotensin-converting enzyme (ACE) cleaves Ang I to the active Ang II, there is another enzymatic pathway: This pathway, via ACE2, allows the formation of Ang-(1-7) (Hrenak et al., 2016). Ang-(1-7) signals through a specific receptor, the G-protein coupled receptor Mas (Santos et al., 2003).
The ACE2/Ang (1-7)/Mas receptor signalling pathway is highly expressed throughout the brain and is considered to play important roles in the context of neuroprotection (Wright and Harding, 2013; Jackson et al., 2018). Mas is expressed in various brain areas (Freund et al., 2012), and signalling via Mas seems to have an impact on neuronal activity, synaptic plasticity, learning and memory (von Bohlen und Halbach et al., 2000; Freund et al., 2014).
Multiple studies have identified more than 20 functional members of the Mas related G-protein coupled receptor family (Mrg) and based on their sequence homology they have been grouped into several subfamilies: MrgA1-22, MrgB1-13, MrgC1-14, and MrgD-G. So far, these subfamilies have only been identified in rodents, but they have human orthologs (Dong et al., 2001; Choi and Lahn, 2003; Zhang et al., 2005). Despite an increasing number of studies assessing the importance of the expression of Mrg receptors in nervous system, their physiological functions have been only partially elucidated so far. It is postulated that Mrg receptors are involved in pathways that contribute to the processing of pain and analgesia as well as in itch sensation (Dong et al., 2001; Choi and Lahn, 2003; He et al., 2014; Li et al., 2014; Liu and Dong, 2015; Li et al., 2017; Meixiong and Dong, 2017; Zhu et al., 2017; He et al., 2018; Schott et al., 2021; von Bohlen und Halbach, 2021).
Some years ago, a new member of RAS, Ala1-Ang-(1-7), also named alamandine, which is a decarboxylated form of Ang (1-7), has been identified (Villela et al., 2014; Hrenak et al., 2016; Shen et al., 2018; Schleifenbaum, 2019). Although Ala1-Ang-(1-7), has been reported to act mainly through binding to MrgD (Bader et al., 2014; Villela et al., 2014; Hrenak et al., 2016; Tetzner et al., 2016; Jackson et al., 2018; Schleifenbaum, 2019) it can also act through the Mas receptor (Tetzner et al., 2018) and thus shows clear similarities to Ang-(1-7) (Tetzner et al., 2016). Currently, the role of the Ala1-Ang-(1-7)/Ang-(1-7)/MrgD axis in the nervous system is rather unknown, and mapping of the expression of MrgD receptors on the protein level is hampered by the fact that specific antibodies directed against MrgD are missing. To overcome this problem, we mapped MrgD expression indirectly by taking advantage of a specific transgenic mouse line, in which MrgD expressing cells are marked by green fluorescence (Zylka et al., 2005).
2. Materials and methods
A mouse line has been generated by Zylka and coworkers (Zylka et al., 2005) in which the MrgD coding sequence was left intact and followed by an internal ribosomal entry site – farnesylated enhanced green fluorescent protein IRES-EGFPf cassette (MrgD IRES-EGFPf). Homozygous MrgD IRES-EGFPf (MrgDEGF) and corresponding control mice were bred, and adult female mice were used in this study. For analysis, mice were euthanized and transcardially perfused with phosphate buffered saline (PBS) and 4 % paraformaldehyde (PFA). The brains were explanted and stored in 4 % PFA at 4 °C. All experimental procedures were conducted in accordance with the protocols approved by the University College Cork Animal Experimentation Ethics Committee (AEEC; AE19130, 2018/018).
Immunohistochemistry: Thirty μm coronal sections were made using a vibration blade microtome (Type VT 1000 S, Leica, Germany). Sections were mounted on superfrost slides (R. Langenbrinck GmbH, Germany) and dried over night at 37 °C. Since the fluorescence signals from GFP were sometimes weak and hard to distinguish from background fluorescence in some brain areas, we performed immunohistochemical labelling using a specific antibody against GFP (1:200, rabbit; Catalog # A-11122; Thermo-Fisher, Germany). For visualization, sections were incubated with goat anti-rabbit secondary antibody labelled with Cy3 (1:2000; Dianova, Germany) for 2 h at room temperature. Finally, the sections were counterstained with 4,6-diamidino-2-phenylindole (DAPI, to visualize cell nuclei) and cover-slipped with Mowiol (Merck KGaA, Germany). Control mice did not show specific GFP signals, but sometimes a weak green background fluorescence, but no labelling with Cy3 was seen in these mice (data not shown).
Image Analysis: The number of cells somata immunolabelled by MrgDΔEGFPf in the mouse brain was quantitatively estimated within a region of interest (ROI) selected from 4–6 sections. The cells were considered as specifically positive for MrgDEGFPf, when they fulfill two criteria: (1) due to the GFP-signal, the MrgD expressing cells must be detectable by their green fluorescence, and, in addition, (2) the cells positive for MrgDΔEGFPf must be identifiable by their red fluorescence due to the Cy3-labelling. Images were acquired (magnification, 40×) by an Olympus BX 63 fluorescence microscope (Olympus, Hamburg, Germany) equipped with DP80 camera (Olympus, Germany) with the same exposure time. The number of MrgDEGF reactive cells within the ROI was counted within one focal plane. The total number of the cells immunoreactive for MrgD per ROI was calculated by using ImageJ and expressed as mean number of cells per brain area (±SEM). In order to get further insight in the expression level of MrgD protein in each specific ROI, the intensity for each green and red fluorescence emission of the MrgD positive cells was measured using ImageJ software for windows version 1.51 (National Institutes of Health, rsb.info.nih.gov/ij). Thereafter, measurements were adjusted by the "corrected total cell fluorescence" (CTCF) method provided by Hammond (2014). The sum of the intensity of each pixel and the CTCF were calculated as follows:
| CTCF = Integrated Density – (ROI area × mean gray value fluorescence of background) |
This process was repeated for all cells and images selected for analysis. The CTCF value represents the overall fluorescence intensity in each ROI, calculated with the integrated density of fluorescence and corrected for the surface area examined and the background (Hammond, 2014; Ponnazhagan et al., 2016).
3. Results
We have examined the expression of MrgD- positive cells in the adult mouse brain. Scattered MrgD-expressing cells were detected in different brain areas, including cortical areas, limbic areas and e.g. basal ganglia (Figures 1, 2, and 3). The quantitative analysis revealed a relatively high fluorescence emission intensity of Cy3-labelled anti-GFP compared to EGFPf, whereas only a little difference in fluorescence emission values has been found between the GFP- and Cy3-conjugated secondary antibodies (Table 1), demonstrating a specific binding of primary anti-GFP antibody.
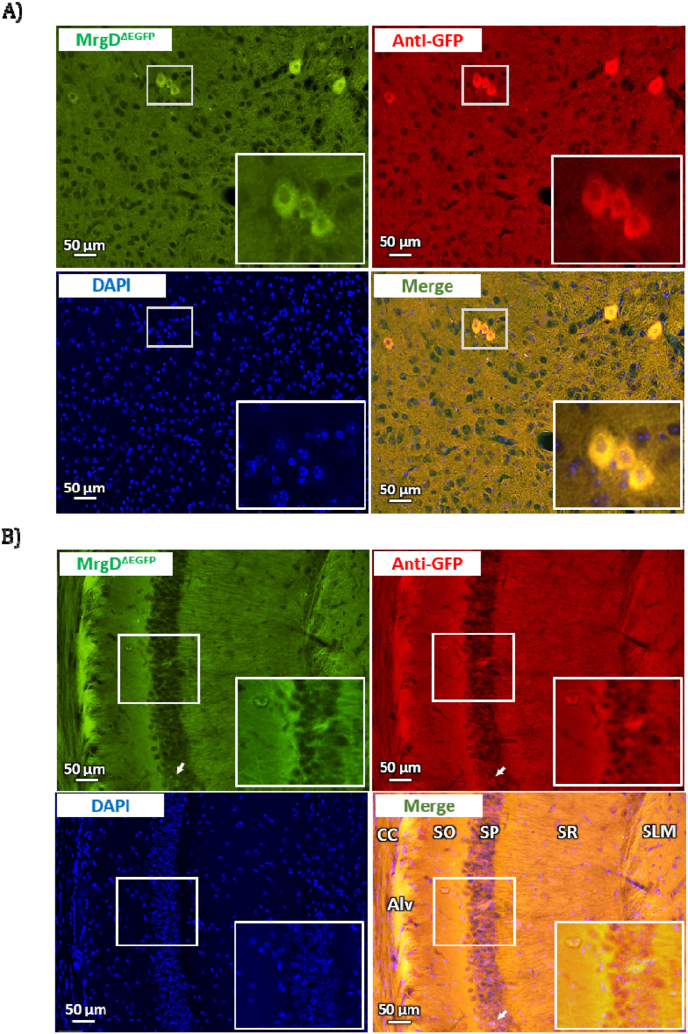
Figure 1.
Representative photomicrographs demonstrating the expression of MrgD immunoreactivity in the (A) midbrain trigeminal nucleus and (B) hippocampal area CA1 of the mouse brain. Vibratome sections (30 μm thick) were labeled with EGFP (Green), anti-GFP antibodies (visualized by Cy3, red) and DAPI (blue). Abbreviations: CC, corpus callosum; Alv, alveus; SO, stratum oriens; SP: stratum pyramidalis; SR: stratum radiatum; SLM: stratum lacunosum moleculare.
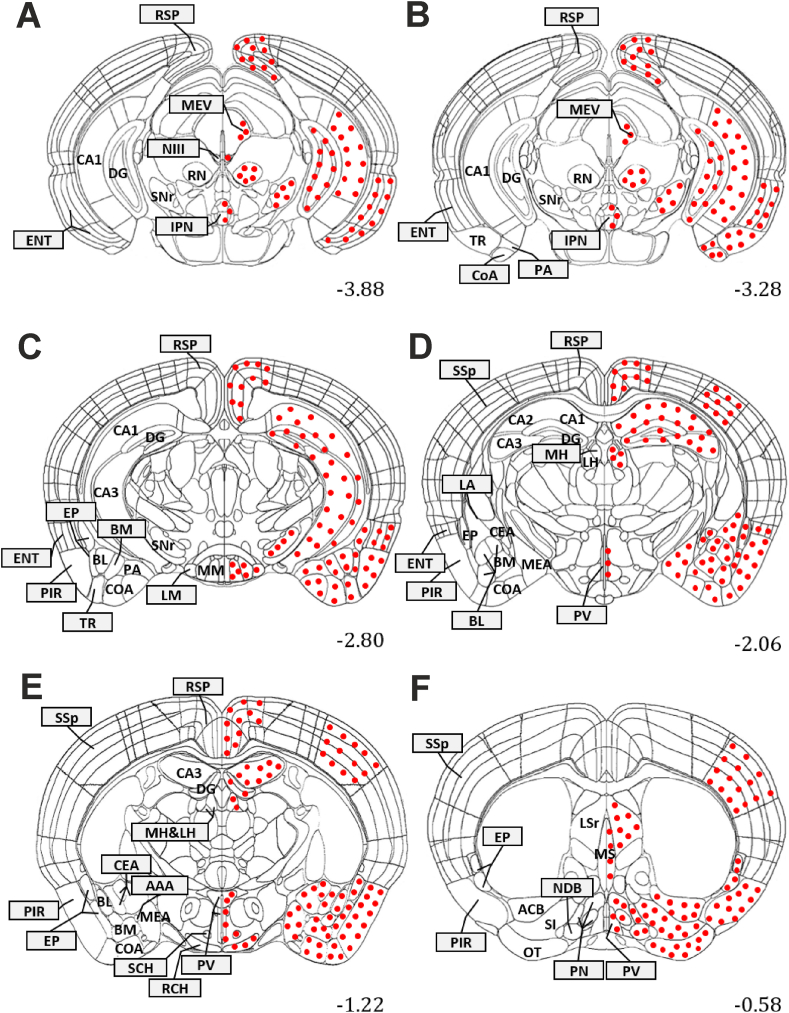
Figure 2.
Diagrams of coronal sections from Allen Mouse Brain Atlas at different levels (A–F: number in the lower right corner of the section) showing the localization of MrgD expressing cell bodies in the mouse brain. Filled circles represent the presence of MrgDΔEGFP and Anti-GFP-immunoreactive somata. The densities of the filled circles do not indicate the relative cells density in the brain regions. Abbreviations: AAA, Ant. amygdalar Ncl.; ACB, Ncl. accumbens; BL, basolateral amygdalar Ncl.; BM, basomedial amygdalar Ncl.; CA1, hippocampal CA1 field; CA2, hippocampal CA2 field; CA3, hippocampal CA3 field; CEA, central amygdalar Ncl.; CoA, cortical amygdalar area; DG, dentate gyrus; ENT, entorhinal cortex; EP, endopiriform Ncl.; IPN, Interpeduncular Ncl.; LH, lateral habenula.; LM, lateral mammilary Ncl.; LPN, Lateral preoptic Ncl.; LSr, Lateral septal Ncl, rostral part; MEA, medial amygdalar Ncl.; MH, medial habenula.; MM, medial mammilary Ncl.; MPN, medial preoptic Ncl.; MS, Medial septal Ncl.; N III, oculomotor Ncl.; NDB, diagonal band Ncl.; OT, olfactory tubercle; PA, posterior amygdalar Ncl.; PIR, piriform cortex; PV, periventricular hypothalamic Ncl.; RCH, retrochiasmatic Ncl.; RN, Red Ncl.; RSP, retrosplenial cortex; SCH, suprachiasmatic Ncl.; SI, substantia innominata; SNr, substantia nigra, pars reticulata; SSp, primary somatosensory cortex.
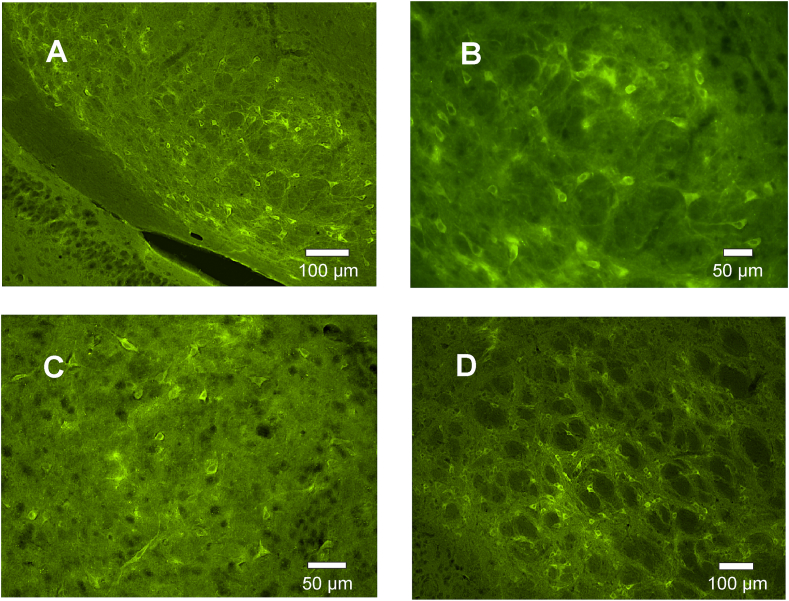
Figure 3.
Green fluorescent cells, due to MrgDΔEGFP, could be detected in different brain areas, as e.g. in the substantia nigra (overview in a lower magnification in A and in a higher magnification in B), the nucleus accumbens (C) or the lateral globus pallidus (D). Images were made using an Olympus BX3, fitted for fluorescence. Z-stacks were generated and the image-stacks were subjected to extended focus imaging (EFI) projection.
Table 1.
Fluorescence intensity and the number of MrgD expressing cells in the mouse brain.
| Area | Region | Intensity of Fluorescence (Mean ± SD) |
Reactive cells per ROI (Mean ± SD) |
||
|---|---|---|---|---|---|
| MrgDΔEGFP | Anti-GFP | ||||
| Cortex | Retrosplenial cortex (RSP) | Layer I | 39.98 ± 14.06 | 48.47 ± 5.96 | 0.62 ± 0.22 |
| Layer II/III | 92.47 ± 19.33 | 107.35 ± 27.89 | 7.03 ± 0.38 | ||
| Layer IV–VI | 82.58 ± 22.61 | 94.17 ± 33.71 | 4.70 ± 0.53 | ||
| Primary somatosensory cortex (SSp) | Layer I | – | – | – | |
| Layer II/III | 102.74 ± 21.48 | 119.27 ± 30.98 | 1.08 ± 0.48 | ||
| Layer IV–VI | 96.11 ± 25.49 | 104.64 ± 37.46 | 3.85 ± 1.08 | ||
| Cortical amygdalar area (CoA) | Layer I | 40.85 ± 7.43 | 76.47 ± 13.38 | 5.00 ± 3.30 | |
| Layer II | 87.91 ± 14.85 | 112.68 ± 17.87 | 16.34 ± 5.27 | ||
| Layer III | 54.90 ± 11.97 | 105.16 ± 15.25 | 7.72 ± 2.99 | ||
| Piriform cortex (PIR) | Layer I | 70.84 ± 10.53 | 82.75 ± 4.20 | 1.06 ± 0.65 | |
| Layer II | 58.40 ± 8.03 | 71.64 ± 6.41 | 4.80 ± 1.21 | ||
| Layer III | 52.70 ± 6.93 | 80.02 ± 1.80 | 8.19 ± 3.13 | ||
| Entorhinal cortex (ENT) | Layer I | 73.18 ± 11.46 | 84.15 ± 4.27 | 5.11 ± 0.86 | |
| Layer II | 51.32 ± 5.96 | 71.49 ± 6.48 | 2.83 ± 0.69 | ||
| Layer III | 58.02 ± 7.92 | 79.10 ± 3.92 | 3.47 ± 1.25 | ||
| Olfactory tubercle (OT) | Layer I | 56.19 ± 7.07 | 98.06 ± 8.49 | 0.78 ± 0.32 | |
| Layer II | 77.97 ± 11.14 | 110.69 ± 11.36 | 8.23 ± 2.02 | ||
| Layer III | 62.67 ± 8.55 | 101.11 ± 10.76 | 3.11 ± 1.32 | ||
| Hippocampus | CA1 | SO | 126.37 ± 18.60 | 130.23 ± 11.28 | 1.02 ± 0.31 |
| SP | 92.90 ± 13.70 | 122.31 ± 18.68 | 3.12 ± 0.55 | ||
| SR | 93.38 ± 12.35 | 121.18 ± 11.90 | 0.78 ± 0.31 | ||
| SLM | 105.72 ± 9.53 | 123.96 ± 18.93 | 0.87 ± 0.44 | ||
| CA2 | SO | 85.33 ± 22.33 | 91.59 ± 9.82 | 0.82 ± 0.28 | |
| SP | 50.77 ± 20.08 | 83.79 ± 11.48 | 4.84 ± 0.68 | ||
| SR | 73.97 ± 9.16 | 92.85 ± 8.14 | 0.71 ± 0.27 | ||
| SLM | 74.20 ± 19.42 | 93.32 ± 6.08 | 0.83 ± 0.34 | ||
| CA3 | SO | 69.81 ± 6.80 | 64.47 ± 7.70 | 1.27 ± 0.45 | |
| SP | 65.24 ± 10.60 | 61.86 ± 18.42 | 3.98 ± 0.54 | ||
| SR | 66.17 ± 9.81 | 71.68 ± 5.95 | 1.49 ± 0.81 | ||
| SLM | 57.87 ± 6.15 | 67.52 ± 10.01 | 1.19 ± 0.65 | ||
| DG | SM | 116.51 ± 9.08 | 140.22 ± 8.97 | 0.50 ± 0.18 | |
| SG | 88.29 ± 5.88 | 113.50 ± 11.94 | 2.40 ± 0.27 | ||
| SPo | 127.88 ± 19.61 | 143.30 ± 10.39 | 1.06 ± 0.33 | ||
| Amygdala | Basolateral amygdalar nucleus (BL) | 91.10 ± 17.70 | 142.93 ± 26.98 | 16.17 ± 2.74 | |
| Basomedial amygdalar nucleus (BM) | 78.70 ± 15.29 | 138.21 ± 18.12 | 18.12 ± 2.41 | ||
| Medial amygdalar nucleus (MEA) | 86.17 ± 9.60 | 127.34 ± 12.53 | 21.62 ± 3.12 | ||
| Central amygdalar nucleus (CEA) | 106.19 ± 17.91 | 155.48 ± 11.47 | 23.27 ± 2.67 | ||
| Posterior amygdalar nucleus (PA) | 84.50 ± 14.35 | 138.98 ± 7.92 | 19.39 ± 2.23 | ||
| Anterior amygdalar nucleus (AAA) | 82.42 ± 14.33 | 135.15 ± 15.16 | 12.38 ± 2.29 | ||
| Endopiriform nucleus (EP) | 98.26 ± 5.45 | 104.41 ± 9.28 | 14.87 ± 3.53 | ||
| Nucleus accumbens (ACB) | 61.81 ± 9.59 | 94.31 ± 7.47 | 10.86 ± 1.69 | ||
| Substantia innominata (SI) | 86.94 ± 9.79 | 111.66 ± 6.74 | 12.78 ± 2.75 | ||
| Diagonal band nucleus (NDB) | 62.40 ± 13.27 | 91.36 ± 20.58 | 9.48 ± 2.34 | ||
| Medial septal nucleus (MS) | 77.00 ± 17.48 | 91.21 ± 20.86 | 10.02 ± 2.44 | ||
| Lateral septal nucleus, rostral (LSr) | 70.91 ± 13.32 | 92.49 ± 12.65 | 7.40 ± 1.45 | ||
| Thalamus | Medial habenular nucleus (MH) | 83.15 ± 17.19 | 91.46 ± 11.68 | 17.68 ± 1.89 | |
| Lateral habenular nucleus (LH) | 90.22 ± 14.51 | 96.45 ± 12.89 | 16.66 ± 2.32 | ||
| Hypothalamus | Medial mammilary nucleus (MM) | 47.64 ± 3.20 | 69.00 ± 8.06 | 35.98 ± 1.80 | |
| Lateral mammilary nucleus (LM) | 48.19 ± 3.32 | 71.74 ± 3.63 | 33.11 ± 2.95 | ||
| Periventricular nucleus (PV) | 91.83 ± 14.26 | 116.21 ± 17.77 | 7.08 ± 1.21 | ||
| Retrochiasmatic nucleus (RCH) | 70.64 ± 10.97 | 118.54 ± 18.22 | 16.37 ± 3.91 | ||
| Suprachiasmatic nucleus (SCH) | 74.17 ± 11.51 | 132.04 ± 10.45 | 21.29 ± 5.08 | ||
| Medial Preoptic nucleus (MPN) | 66.90 ± 12.57 | 87.25 ± 11.94 | 11.84 ± 2.32 | ||
| Lateral Preoptic nucleus (LPN) | 70.24 ± 13.20 | 94.31 ± 7.47 | 19.15 ± 4.05 | ||
| Mid-brain | Midbrain trigeminal nucleus (MEV) | 120.98 ± 8.72 | 117.0 ± 10.80 | 2.08 ± 1.05 | |
| Oculomotor nucleus (NIII) | 74.58 ± 7.97 | 97.90 ± 13.78 | 6.63 ± 1.29 | ||
| Red nucleus (RN) | 98.41 ± 4.73 | 104.95 ± 9.60 | 7.65 ± 0.94 | ||
| Substantia nigra, pars reticulata (SNr) | 77.85 ± 5.31 | 102.51 ± 10.37 | 9.35 ± 1.76 | ||
| Interpeduncular nucleus (IPN) | 70.64 ± 10.97 | 95.22 ± 7.34 | 13.52 ± 2.77 | ||
Abbreviations: SG: stratum granulare; SLM: stratum lacunosum moleculare; SM: stratum moleculare; SO: stratum oriens; SP: stratum pyramidalis; SPo: stratum Polymorph; SR: stratum radiatum.
Several MrgD-positive (MrgD+) cells have been identified through the cortical layers in both neocortical and allocortical parts of mouse cerebral cortex (Figure 2). Among all of the cortical areas, our analysis has revealed that the highest fluorescence intensity and the highest number of MrgD+ cells could be found in the primary somatosensory cortex and in the cortical amygdala area, respectively (Table 1).
Within the neocortex, some MrgD expressing cells were seen in the retrosplenial (RSP) and primary somatosensory (SSp) cortices (Figure 2; Table 1). The MrgD+ cells were differentially distributed in these cortices. The highest number of MrgD+ cells has been observed in middle cortical layers of RSP, whereas in the SSp cortex, the deeper layers displayed higher densities of reactive cells (Table 1). Additionally, scattered MrgD reactive cells have also been discovered in the three-layered allocortical regions, such as piriform (PIR) and entorhinal (ENT) cortex as well as in the cortical amygdala (CoA) and the olfactory tubercle (OT; Figure 2). The highest MrgD expression level was seen in layers II and III of the CoA and OT (Table 1).
Within the limbic system, some MrgD+ cells have been detected in the hippocampus as well as in different nuclei of the amygdala (Figure 2). Thus, MrgD-positive signals were found in all layers of the different hippocampal subregions (CA1, CA2, CA3, and dentate gyrus (DG); Figure 1). Concerning the amygdala, MrgD expression was mapped to the basolateral (BL), basomedial (BM), medial (MEA), central (CEA), posterior (PA) and anterior (AAA) amygdalar nuclei (Figure 2). Quantitative analysis has demonstrated the higher density of MrgD expressing cells in CEA and MEA nuclei, and the lowest number in AAA area (Table 1). In addition, we found higher fluorescence intensities in the CEA and BL nuclei, whereas lower intensities have been detected in BM and AAA (Table 1). Furthermore, the MrgD+ signals have also been found in the endopiriform nuclei (EP; Figure 2).
Some MrgD-expressing cells were detected in the nucleus accumbens (Figure 3), as well as in the substantia innominata (SI), diagonal band nucleus (NDB), medial (MS) and rostral part of lateral (LSr) septal nuclei (Figure 2).
Among the thalamic nuclei analyzed, we only observed low MrgD expression in the medial and lateral habenula (MH and LH; Figure 2). MrgD+ signals were also identified in some hypothalamic nuclei, including the lateral and medial mamillary (LM and MM), periventricular (PV), retrochiasmatic (RCH), suprachiasmatic (SCH), and medial and lateral preoptic (MPN and LPN) nuclei (Figure 2; Table 1).
Some scattered MrgD positive cells were seen in the midbrain, as e.g. in the lateral and anterior regions near the periaqueduct grey matter (PAG; Figure 1), corresponding to the location of the mid-brain trigeminal nucleus (MEV) and oculomotor nucleus (NIII; Table 1).
4. Discussion
In the present study, we provided evidence that the MrgD receptor is expressed in the murine forebrain in a region-specific manner, although on a very low level. MrgD expression was found in cortical and subcortical structures, including the limbic system. However, MrgD is not restricted to areas involved in the nociception, but can also be seen in brain areas involved in reward or motor functions. Nevertheless, the overall expression of MrgD in the brain is rather low and mainly only some scattered cells were found per brain region. The most prominent staining with the most intense stained cells could be seen in the trigeminal nucleus (Figure 1). Aside from the CNS, MrgD expression was detected in vascular endothelium, arterial smooth muscle cells, and cardiomyocytes (Habiyakare et al., 2014; Oliveira et al., 2019), demonstrating a possible role of MrgD in blood pressure regulation and cardiovascular function, as also supported by functional studies (Tetzner et al., 2016). Furthermore, the identification of MrgD protein in the skin epidermis and in a distinct subset of nociceptive neurons in the DRGs, trigeminal ganglia, and dorsal horn of spinal cord has suggested a modulatory role of this receptor in neuropathic pain and itch sensation, as well as, in the mechanical and thermal nociception (Dong et al., 2001; Zylka et al., 2003; Rau et al., 2009; Wang and Zylka, 2009; Liu and Dong, 2015; Tiwari et al., 2016; Meixiong and Dong, 2017). Along this line, mice lacking MrgD gene (MrgD−/−) exhibit reduced excitability to thermal and mechanical stimuli compared to their wildtype littermates (Rau et al., 2009).
Nevertheless, the detailed mechanisms of MrgD action are not fully understood yet, but it has recently been shown that MrgD promotes the activation of canonical NF-κB signalling pathway and enhances the development of inflammatory pain in mouse DRGs and RAW 264.7 cells (Lan et al., 2020). In consistence with the action of MrgD in the perception and modulation of pain (Guan et al., 2010; Tiwari et al., 2016; Lan et al., 2020), populations of MrgD+ cells have observed through the neocortical layers of RSP and SSp cortices, which are commonly activated by noxious stimuli, as revealed in electrophysiological and imaging studies (Apkarian et al., 2005; Gregory et al., 2013). Accordingly, the multiple pain-related brain pathways are important for different aspects of the pain experience. Thus, the somatosensory cortices encode information about sensory features, such as the location and duration of pain (Kenshalo and Isensee, 1983). Alternatively, the RSP (a limbic part of the cortex), is more important for encoding the emotional and motivational aspects of pain (Berthier et al., 1988). Moreover, MrgD immunoreactive cells have also been detected in different nuclei of the amygdala and in the nucleus accumbens, with extensive nociceptive inputs through spinoparabrachial–amygdala projections (Becerra et al., 2001; Baliki et al., 2010). Furthermore, MrgD-positive cells were also found cortical and subcortical elements of the reward circuity in the brain such as e.g. nucleus accumbens, hippocampus and amygdala. The reward system is a collection of brain structures that not only involves GABA and glutamate neurotransmission, but is also modulated by dopamine (Mitsi and Zachariou, 2016; DosSantos et al., 2017). The reward neural circuits might play a significant role not only in decision making, pain perception and/or modulation, but also in the transition from acute to chronic pain (Borsook et al., 2016; Mitsi and Zachariou, 2016; DosSantos et al., 2017). Interestingly, we have only discovered a small population of MrgD reactive cells in the lateral and anterolateral portions of PAG, which is described as a primary anatomical pathway mediating pain and opioid-based analgesia in a wide range of vertebrate species (Behbehani, 1995; Loyd and Murphy, 2009). This part of midbrain receives nociceptive input through the spinoreticular pathways (Basbaum and Fields, 1984; Dunckley et al., 2005). Beyond that, the lateral and ventrolateral PAG heavily project to the rostral ventromedial medulla (RVM) and dorsal horn of the spinal cord in rats (Loyd and Murphy, 2006). Above all, the PAG is also involved in much more than just analgesia. For example, the PAG appears to play roles in the regulation of heart rate and blood pressure, and processing of emotional responses (i.e. fear, anxiety and defensive reactions) by interacting with the amygdala (Walker and Carrive, 2003; Green et al., 2007). In addition, there is growing evidence that the hippocampal formation is involved in pain processing (Liu and Chen, 2009; Mutso et al., 2012). Accordingly, chronic pain could lead to deficits in learning/memory and emotional decision-making. A study by Mutso and colleagues (2012) has reported that mice with nerve injury (a model of neuropathic pain) show defects in hippocampal-mediated behaviours, synaptic plasticity, and neurogenesis (Mutso et al., 2012).
The localization of MrgD in the reward- and limbic-related areas can hint for a role of MrgD in processes such as pain perception/modulation, synaptic plasticity, learning, memory and cognition. However, MrgD expression in the adult murine brain is not as strong as the expression of Mas. The Ang-(1-7)/Mas system is, among others, involved in mechanisms related to learning and memory. Aside from these well-known actions of Ang-(1-7)/Mas system, the roles of the Ang-(1-7)/MrgD system in the brain is far from being understood. It would be of special interest whether both, Mas and MrgD are activated mainly by Ang-(1-7) and whether Mas and MrgD would cooperate in intracellular signalling or whether they induce antagonistic functions.
Declarations
Author contribution statement
Hami, Javad, von Bohlen und Halbach, Viola: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Tetzner, Anja: Contributed reagents, materials, analysis tools or data.
Walther, Thomas: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
von Bohlen und Halbach, Oliver: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data will be made available on request.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We wish to thank Sabine Hanisch for excellent technical assistance.
References
- Apkarian A.V., Bushnell M.C., Treede R.D., Zubieta J.K. Human brain mechanisms of pain perception and regulation in health and disease. Eur. J. Pain. 2005;9:463–484. doi: 10.1016/j.ejpain.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Bader M., Alenina N., Andrade-Navarro M.A., Santos R.A. Mas and its related G protein-coupled receptors, Mrgprs. Pharmacol. Rev. 2014;66:1080–1105. doi: 10.1124/pr.113.008136. [DOI] [PubMed] [Google Scholar]
- Baliki M.N., Geha P.Y., Fields H.L., Apkarian A.V. Predicting value of pain and analgesia: nucleus accumbens response to noxious stimuli changes in the presence of chronic pain. Neuron. 2010;66:149–160. doi: 10.1016/j.neuron.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basbaum A.I., Fields H.L. Endogenous pain control systems: brainstem spinal pathways and endorphin circuitry. Annu. Rev. Neurosci. 1984;7:309–338. doi: 10.1146/annurev.ne.07.030184.001521. [DOI] [PubMed] [Google Scholar]
- Becerra L., Breiter H.C., Wise R., Gonzalez R.G., Borsook D. Reward circuitry activation by noxious thermal stimuli. Neuron. 2001;32:927–946. doi: 10.1016/s0896-6273(01)00533-5. [DOI] [PubMed] [Google Scholar]
- Behbehani M.M. Functional characteristics of the midbrain periaqueductal gray. Prog. Neurobiol. 1995;46:575–605. doi: 10.1016/0301-0082(95)00009-k. [DOI] [PubMed] [Google Scholar]
- Berthier M., Starkstein S., Leiguarda R. Asymbolia for pain: a sensory-limbic disconnection syndrome. Ann. Neurol. 1988;24:41–49. doi: 10.1002/ana.410240109. [DOI] [PubMed] [Google Scholar]
- Borsook D., Linnman C., Faria V., Strassman A.M., Becerra L., Elman I. Reward deficiency and anti-reward in pain chronification. Neurosci. Biobehav. Rev. 2016;68:282–297. doi: 10.1016/j.neubiorev.2016.05.033. [DOI] [PubMed] [Google Scholar]
- Choi S.S., Lahn B.T. Adaptive evolution of MRG, a neuron-specific gene family implicated in nociception. Genome Res. 2003;13:2252–2259. doi: 10.1101/gr.1431603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Han S., Zylka M.J., Simon M.I., Anderson D.J. A diverse family of GPCRs expressed in specific subsets of nociceptive sensory neurons. Cell. 2001;106:619–632. doi: 10.1016/s0092-8674(01)00483-4. [DOI] [PubMed] [Google Scholar]
- DosSantos M.F., Moura B.S., DaSilva A.F. Reward circuitry plasticity in pain perception and modulation. Front. Pharmacol. 2017;8:790. doi: 10.3389/fphar.2017.00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley P., Wise R.G., Fairhurst M., Hobden P., Aziz Q., Chang L., Tracey I. A comparison of visceral and somatic pain processing in the human brainstem using functional magnetic resonance imaging. J. Neurosci. : Off. J. Soc. Neurosci. 2005;25:7333–7341. doi: 10.1523/JNEUROSCI.1100-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund M., Walther T., von Bohlen und Halbach O. Immunohistochemical localization of the angiotensin-(1-7) receptor Mas in the murine forebrain. Cell Tissue Res. 2012;348:29–35. doi: 10.1007/s00441-012-1354-3. [DOI] [PubMed] [Google Scholar]
- Freund M., Walther T., von Bohlen und Halbach O. Effects of the angiotensin-(1-7) receptor Mas on cell proliferation and on the population of doublecortin positive cells within the dentate gyrus and the piriform cortex. Eur. Neuropsychopharmacol. 2014;24:302–308. doi: 10.1016/j.euroneuro.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Green A.L., Wang S., Owen S.L., Aziz T.Z. The periaqueductal grey area and the cardiovascular system. Acta Neurochir. Suppl. 2007;97:521–528. doi: 10.1007/978-3-211-33081-4_60. [DOI] [PubMed] [Google Scholar]
- Gregory N.S., Harris A.L., Robinson C.R., Dougherty P.M., Fuchs P.N., Sluka K.A. An overview of animal models of pain: disease models and outcome measures. J. Pain. 2013;14:1255–1269. doi: 10.1016/j.jpain.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Liu Q., Tang Z., Raja S.N., Anderson D.J., Dong X. Mas-related G-protein-coupled receptors inhibit pathological pain in mice. Proc. Natl. Acad. Sci. U. S. A. 2010;107:15933–15938. doi: 10.1073/pnas.1011221107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habiyakare B., Alsaadon H., Mathai M.L., Hayes A., Zulli A. Reduction of angiotensin A and alamandine vasoactivity in the rabbit model of atherogenesis: differential effects of alamandine and Ang(1-7) Int. J. Exp. Pathol. 2014;95:290–295. doi: 10.1111/iep.12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond L. The University of Queensland; Australia: 2014. Measuring Cell Fluorescence Using ImageJ. [Google Scholar]
- He S.Q., Han L., Li Z., Xu Q., Tiwari V., Yang F., Guan X., Wang Y., Raja S.N., Dong X., Guan Y. Temporal changes in MrgC expression after spinal nerve injury. Neuroscience. 2014;261:43–51. doi: 10.1016/j.neuroscience.2013.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S.Q., Xu Q., Tiwari V., Yang F., Anderson M., Chen Z., Grenald S.A., Raja S.N., Dong X., Guan Y. Oligomerization of MrgC11 and mu-opioid receptors in sensory neurons enhances morphine analgesia. Sci. Signal. 2018;11 doi: 10.1126/scisignal.aao3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrenak J., Paulis L., Simko F. Angiotensin A/Alamandine/MrgD Axis: another clue to understanding cardiovascular pathophysiology. Int. J. Mol. Sci. 2016;17 doi: 10.3390/ijms17071098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L., Eldahshan W., Fagan S.C., Ergul A. Within the brain: the renin angiotensin system. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19030876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenshalo D.R., Jr., Isensee O. Responses of primate SI cortical neurons to noxious stimuli. J. Neurophysiol. 1983;50:1479–1496. doi: 10.1152/jn.1983.50.6.1479. [DOI] [PubMed] [Google Scholar]
- Lan L., Xu M., Li J., Liu L., Xu M., Zhou C., Shen L., Tang Z., Wan F. Mas-related G protein-coupled receptor D participates in inflammatory pain by promoting NF-kappaB activation through interaction with TAK1 and IKK complex. Cell. Signal. 2020;76:109813. doi: 10.1016/j.cellsig.2020.109813. [DOI] [PubMed] [Google Scholar]
- Li Z., He S.Q., Xu Q., Yang F., Tiwari V., Liu Q., Tang Z., Han L., Chu Y.X., Wang Y., Hin N., Tsukamoto T., Slusher B., Guan X., Wei F., Raja S.N., Dong X., Guan Y. Activation of MrgC receptor inhibits N-type calcium channels in small-diameter primary sensory neurons in mice. Pain. 2014;155:1613–1621. doi: 10.1016/j.pain.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Tseng P.Y., Tiwari V., Xu Q., He S.Q., Wang Y., Zheng Q., Han L., Wu Z., Blobaum A.L., Cui Y., Tiwari V., Sun S., Cheng Y., Huang-Lionnet J.H., Geng Y., Xiao B., Peng J., Hopkins C., Raja S.N., Guan Y., Dong X. Targeting human Mas-related G protein-coupled receptor X1 to inhibit persistent pain. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E1996–E2005. doi: 10.1073/pnas.1615255114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.G., Chen J. Roles of the hippocampal formation in pain information processing. Neurosci. Bull. 2009;25:237–266. doi: 10.1007/s12264-009-0905-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Dong X. The role of the Mrgpr receptor family in itch. Handb. Exp. Pharmacol. 2015;226:71–88. doi: 10.1007/978-3-662-44605-8_5. [DOI] [PubMed] [Google Scholar]
- Loyd D.R., Murphy A.Z. Sex differences in the anatomical and functional organization of the periaqueductal gray-rostral ventromedial medullary pathway in the rat: a potential circuit mediating the sexually dimorphic actions of morphine. J. Comp. Neurol. 2006;496:723–738. doi: 10.1002/cne.20962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyd D.R., Murphy A.Z. The role of the periaqueductal gray in the modulation of pain in males and females: are the anatomy and physiology really that different? Neural Plast. 2009;2009:462879. doi: 10.1155/2009/462879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meixiong J., Dong X. Mas-related G protein-coupled receptors and the biology of itch sensation. Annu. Rev. Genet. 2017;51:103–121. doi: 10.1146/annurev-genet-120116-024723. [DOI] [PubMed] [Google Scholar]
- Mitsi V., Zachariou V. Modulation of pain, nociception, and analgesia by the brain reward center. Neuroscience. 2016;338:81–92. doi: 10.1016/j.neuroscience.2016.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutso A.A., Radzicki D., Baliki M.N., Huang L., Banisadr G., Centeno M.V., Radulovic J., Martina M., Miller R.J., Apkarian A.V. Abnormalities in hippocampal functioning with persistent pain. J. Neurosci. : Off. J. Soc. Neurosci. 2012;32:5747–5756. doi: 10.1523/JNEUROSCI.0587-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira A.C., Melo M.B., Motta-Santos D., Peluso A.A., Souza-Neto F., da Silva R.F., Almeida J.F.Q., Canta G., Reis A.M., Goncalves G., Cerri G., Coutinho D., Guedes de Jesus I.C., Guatimosim S., Linhares N.D., Alenina N., Bader M., Campagnole-Santos M.J., Santos R.A.S. Genetic deletion of the alamandine receptor MRGD leads to dilated cardiomyopathy in mice. Am. J. Physiol. Heart Circ. Physiol. 2019;316:H123–H133. doi: 10.1152/ajpheart.00075.2018. [DOI] [PubMed] [Google Scholar]
- Ponnazhagan R., Harms A.S., Thome A.D., Jurkuvenaite A., Gogliotti R., Niswender C.M., Conn P.J., Standaert D.G. The metabotropic glutamate receptor 4 positive allosteric modulator ADX88178 inhibits inflammatory responses in primary microglia. J. Neuroimmune Pharmacol. 2016;11:231–237. doi: 10.1007/s11481-016-9655-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau K.K., McIlwrath S.L., Wang H., Lawson J.J., Jankowski M.P., Zylka M.J., Anderson D.J., Koerber H.R. Mrgprd enhances excitability in specific populations of cutaneous murine polymodal nociceptors. J. Neurosci. : Off. J. Soc. Neurosci. 2009;29:8612–8619. doi: 10.1523/JNEUROSCI.1057-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos R.A., Simoes e Silva A.C., Maric C., Silva D.M., Machado R.P., de Buhr I., Heringer-Walther S., Pinheiro S.V., Lopes M.T., Bader M., Mendes E.P., Lemos V.S., Campagnole-Santos M.J., Schultheiss H.P., Speth R., Walther T. Angiotensin-(1-7) is an endogenous ligand for the G protein-coupled receptor Mas. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8258–8263. doi: 10.1073/pnas.1432869100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer S.T., Han J., Pena M., von Bohlen und Halbach O., Peters J., Gage F.H. The Wnt adaptor protein ATP6AP2 regulates multiple stages of adult hippocampal neurogenesis. J. Neurosci. 2015;35:4983–4998. doi: 10.1523/JNEUROSCI.4130-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleifenbaum J. Alamandine and its receptor MrgD pair up to join the protective arm of the renin-angiotensin system. Front. Med. 2019;6:107. doi: 10.3389/fmed.2019.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott B.H., Kronenberg G., Schmidt U., Dusedau H.P., Ehrentraut S., Geisel O., von Bohlen und Halbach O., Gass P., Dunay I.R., Hellweg R. Robustly high hippocampal BDNF levels under acute stress in mice lacking the full-length p75 neurotrophin receptor. Pharmacopsychiatry. 2021;54:205–213. doi: 10.1055/a-1363-1680. [DOI] [PubMed] [Google Scholar]
- Shen Y.H., Chen X.R., Yang C.X., Liu B.X., Li P. Alamandine injected into the paraventricular nucleus increases blood pressure and sympathetic activation in spontaneously hypertensive rats. Peptides. 2018;103:98–102. doi: 10.1016/j.peptides.2018.03.014. [DOI] [PubMed] [Google Scholar]
- Tetzner A., Gebolys K., Meinert C., Klein S., Uhlich A., Trebicka J., Villacanas O., Walther T. G-Protein-Coupled receptor MrgD is a receptor for angiotensin-(1-7) involving adenylyl cyclase, cAMP, and phosphokinase A. Hypertension. 2016;68:185–194. doi: 10.1161/HYPERTENSIONAHA.116.07572. [DOI] [PubMed] [Google Scholar]
- Tetzner A., Naughton M., Gebolys K., Eichhorst J., Sala E., Villacanas O., Walther T. Decarboxylation of Ang-(1-7) to Ala(1)-Ang-(1-7) leads to significant changes in pharmacodynamics. Eur. J. Pharmacol. 2018;833:116–123. doi: 10.1016/j.ejphar.2018.05.031. [DOI] [PubMed] [Google Scholar]
- Tiwari V., Tiwari V., He S., Zhang T., Raja S.N., Dong X., Guan Y. Mas-related G protein-coupled receptors offer potential new targets for pain therapy. Adv. Exp. Med. Biol. 2016;904:87–103. doi: 10.1007/978-94-017-7537-3_7. [DOI] [PubMed] [Google Scholar]
- Villela D.C., Passos-Silva D.G., Santos R.A. Alamandine: a new member of the angiotensin family. Curr. Opin. Nephrol. Hypertens. 2014;23:130–134. doi: 10.1097/01.mnh.0000441052.44406.92. [DOI] [PubMed] [Google Scholar]
- von Bohlen und Halbach O. The angiotensin converting enzyme 2 (ACE2) system in the brain: possible involvement in Neuro-Covid. Histol. Histopathol. 2021:18356. doi: 10.14670/HH-18-356. [DOI] [PubMed] [Google Scholar]
- von Bohlen und Halbach O., Albrecht D. The CNS renin-angiotensin system. Cell Tissue Res. 2006;326:599–616. doi: 10.1007/s00441-006-0190-8. [DOI] [PubMed] [Google Scholar]
- von Bohlen und Halbach O., Walther T., Bader M., Albrecht D. Interaction between Mas and the angiotensin AT1 receptor in the amygdala. J. Neurophysiol. 2000;83:2012–2021. doi: 10.1152/jn.2000.83.4.2012. [DOI] [PubMed] [Google Scholar]
- Walker P., Carrive P. Role of ventrolateral periaqueductal gray neurons in the behavioral and cardiovascular responses to contextual conditioned fear and poststress recovery. Neuroscience. 2003;116:897–912. doi: 10.1016/s0306-4522(02)00744-3. [DOI] [PubMed] [Google Scholar]
- Wang H., Zylka M.J. Mrgprd-expressing polymodal nociceptive neurons innervate most known classes of substantia gelatinosa neurons. J. Neurosci. : Off. J. Soc. Neurosci. 2009;29:13202–13209. doi: 10.1523/JNEUROSCI.3248-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J.W., Harding J.W. The brain renin-angiotensin system: a diversity of functions and implications for CNS diseases. Pflügers Archiv. 2013;465:133–151. doi: 10.1007/s00424-012-1102-2. [DOI] [PubMed] [Google Scholar]
- Zhang L., Taylor N., Xie Y., Ford R., Johnson J., Paulsen J.E., Bates B. Cloning and expression of MRG receptors in macaque, mouse, and human. Brain Res. Mol. Brain Res. 2005;133:187–197. doi: 10.1016/j.molbrainres.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Hanson C.E., Liu Q., Han L. Mrgprs activation is required for chronic itch conditions in mice. Itch (Phila) 2017;2 doi: 10.1097/itx.0000000000000009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylka M.J., Dong X., Southwell A.L., Anderson D.J. Atypical expansion in mice of the sensory neuron-specific Mrg G protein-coupled receptor family. Proc. Natl. Acad. Sci. U. S. A. 2003;100:10043–10048. doi: 10.1073/pnas.1732949100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylka M.J., Rice F.L., Anderson D.J. Topographically distinct epidermal nociceptive circuits revealed by axonal tracers targeted to Mrgprd. Neuron. 2005;45:17–25. doi: 10.1016/j.neuron.2004.12.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.