Abstract
Invariant natural killer T (iNKT) cells are lipid‐specific T lymphocytes endowed with cytotoxic activities and are thus considered important in antitumor immunity. While several studies have demonstrated iNKT cell cytotoxicity against different tumors, very little is known about their cell‐killing activities in human colorectal cancer (CRC). Our aim was to assess whether human iNKT cells are cytotoxic against colon cancer cells and the mechanisms underlying this activity. For this purpose, we generated stable iNKT cell lines from peripheral blood and colon specimens and used NK‐92 and peripheral blood natural killer cells as cell‐mediated cytotoxicity controls. In vitro cytotoxicity was assessed using a panel of well‐characterized human CRC cell lines, and the cellular requirements for iNKT cell cytotoxic functions were evaluated. We demonstrated that both intestinal and circulating iNKT cells were cytotoxic against the entire panel of CRC lines, as well as against freshly isolated patient‐derived colonic epithelial cancer cells. Perforin and/or granzyme inhibition impaired iNKT cell cytotoxicity, whereas T‐cell receptor (TCR) signaling was a less stringent requirement for efficient killing. This study is the first evidence of tissue‐derived iNKT cell cytotoxic activity in humans, as it shows that iNKT cells depend on the perforin–granzyme pathway and both adaptive and innate signal recognition for proper elimination of colon cancer cells.
Keywords: CD1d, colorectal cancer, cytotoxicity, iNKT, perforin
Invariant natural killer T (iNKT) cells are lipid‐specific T lymphocytes with important roles in antitumor immunity. Here, we demonstrated that intestinal and blood‐derived iNKT cells can eliminate human colon cancer (CRC) cells. We also showed that iNKT cells require the release of perforin and granzymes to kill CRC cells, using both innate and adaptive mechanisms to recognize them.
Abbreviations
- CD1d
cluster of differentiation 1d
- CRC
colorectal cancer
- iNKT
invariant natural killer T cells
- LPMC
lamina propria mononuclear cells
- NK
natural killer cells
- PBMC
peripheral blood mononuclear cells
- TCR
T‐cell receptor
1. Introduction
Colorectal cancer (CRC) is the third‐most common cancer worldwide and the fourth cancer type for mortality [1]. Surgery, chemotherapy, and radiotherapy are currently the most frequently used therapeutic approaches for metastatic and nonmetastatic diseases. Targeted therapies with monoclonal antibodies directed against growth and angiogenic factors have been effectively included in the therapeutic options for CRC patients [1]. More recently, immunotherapy has gained consideration for CRC treatment thanks to the advent of technologies such as immune checkpoint inhibition [2, 3, 4] and CAR‐T technology [5, 6, 7, 8, 9]. Alongside CD8+ cytotoxic T lymphocytes [10, 11, 12] and natural killer (NK) cells [13, 14], innate‐like T lymphocytes, including invariant natural killer T (iNKT) cells [15], mucosa‐associated invariant T cells (MAIT) [16, 17], and gamma delta T cells [18, 19, 20], have been taken into consideration for cancer immunotherapy. iNKT cells show unique features classically associated with innate immunity, such as tumor‐directed cytotoxicity via innate receptors (i.e., NKG2D), but they also express a T‐cell receptor which might specifically recognize tumor‐derived antigens [16, 21, 22].
Invariant natural killer T cells are a nonconventional T‐cell population coexpressing natural killer lineage receptors and a semi‐invariantly rearranged, CD1d‐restricted, lipid‐specific T‐cell receptor [23]. In fact, iNKT cells can be activated in an adaptive manner by microbial and endogenous lipids presented by the CD1d antigen‐presenting molecule [24, 25, 26] and in an innate manner via activating NK receptors or cytokines, such as IL‐12 and IL‐18 [27, 28]. Thanks to their cytotoxic activity, iNKT cells are involved in antitumor functions against many types of cancers, showing either a direct or indirect antitumor effect on cancer cells [29, 30, 31, 32, 33, 34]. Murine iNKT cells isolated from liver, thymus, and spleen can kill leukemia, melanoma, lung, breast, and colorectal cancer cells both in vitro and in vivo; they express Fas ligand (FasL), TNF‐related apoptosis‐inducing ligand (TRAIL), granzyme B (GZMB), and perforin (PRF) and can target tumor cells in a CD1d‐dependent or CD1d‐independent manner [29, 33, 35, 36, 37, 38, 39, 40, 41]. While hematopoietic tumors express CD1d, many solid tumors are CD1d negative but can nonetheless be targeted by other mechanisms, including NKG2D engagement [27, 37, 42], or by indirect activation of iNKT cells by other CD1d‐expressing, antigen‐presenting cells present within the tumor microenvironment [31]. In vitro studies have shown that the preferred killing mechanism adopted by iNKT cells is the granzyme/perforin pathway [27, 36, 42, 43, 44]. However, the death receptor pathways (involving FasL and TRAIL) are also used in vivo [35, 38, 41].
A role for iNKT cells in killing human cancers has been demonstrated [34, 36, 37, 42, 45, 46], and Va24+ NKT cell tumor infiltration is considered an independent positive prognostic factor in human CRC [47]. Nonetheless, the data generated so far in human cancers describe only the cytotoxic activity of blood‐derived iNKT cells [21, 33, 36, 39, 40, 45, 48], possibly as a consequence of the great difficulty to isolate and perform functional assays with human tissue‐derived iNKT cells. As iNKT cells are mostly tissue‐resident and not recirculating [49], and the presence of different iNKT cell subsets (analogous to the T helper classification) seems to differ in the mouse [50], it is of great interest to study how tissue‐derived and circulating iNKT cell activities might differ. Moreover, very little is known about iNKT cell‐mediated killing mechanisms in human colorectal cancer, and if differences exist in terms of cytotoxic activities between peripheral blood and gut‐derived iNKT cells. In murine models of colon cancer, iNKT cells seem to have contrasting roles, as they promote polyp formation in ApcMin−/+ mice by producing IL‐10 [51] but decrease tumor progression in AOM/DSS‐treated mice by inducing a reduction of IL‐13 [52]. Given the differences between mice and humans, it is important to address how iNKT cells behave in the context of human colon cancer.
Here, we demonstrate that not only circulating but also intestinal iNKT cells are cytotoxic against human colon cancer cell lines as well as patient‐derived CRC cells. We showed that upon coculture with colorectal cancer cells, peripheral and intestinal iNKT cells expressed TRAIL, Fas ligand, and released granzyme B and perforin. Mechanistically, iNKT cell cytotoxicity against human CRC is preferentially perforin and granzyme‐dependent, but not completely CD1d‐mediated. These results provide the first insights about the cytotoxic role of human intestinal iNKT cells against CRC and contribute to shed light on the implementation of these cells for immunotherapeutic approaches against colorectal cancer.
2. Materials and methods
2.1. Human CRC cell lines
Caco‐2, HT‐29, RKO, DiFi, Colo 205 (from American Type Culture Collection, ATCC), NCM460D (from INCELL), and NK‐92 (from DSMZ) were cultured with specific media according to Table S1.
2.2. Human PBMC and LPMC isolation
Healthy donors’ peripheral blood buffy coats were obtained from San Matteo Hospital, Pavia, Italy, whereas intestinal specimens were obtained from IRCCS Policlinico Ospedale Maggiore, Milan, Italy. Institutional Review Board (Milan, Area B) approved the study with permission number 566_2015. All methodologies were under full compliance with the Declaration of Helsinki, and informed consent was obtained from all subjects.
Peripheral blood mononuclear cells (PBMC) were isolated by density gradient centrifugation using Ficoll‐Paque as for standard protocol. Natural killer cells were isolated from PBMCs using the NK Isolation Kit (Miltenyi Biotec, Bergisch Gladbach, Germany). Human lamina propria mononuclear cells (LPMCs) were isolated as previously described [53]. Briefly, the dissected intestinal mucosa was depleted of mucus and epithelial cells in sequential washes with Hanks’ Balanced Salt Solution (HBSS, Euroclone, Pero, Italy) containing DTT (0.1 mmol·L−1, Sigma‐Aldrich, St Louis, MO, USA) and EDTA (1 mmol·L−1, Sigma‐Aldrich) and then digested with collagenase D (400 U·mL−1, Worthington Biochemical Corporation, Lakewood, NJ, USA) for 5 h at 37 °C. LPMCs were then separated with a Percoll gradient and cultured in RPMI‐1640 medium containing 5% human serum (Sigma‐Aldrich), penicillin/streptomycin, gentamicin, and 100 U·mL−1 IL‐2 (Proleukin).
2.3. Isolation of CRC cells from surgical specimens
Colorectal cancer specimens were obtained from IRCCS Policlinico Ospedale Maggiore, Milan, Italy, under the same permission number as in 2.2. Specimens’ mucosa underwent sequential washes with Hanks’ Balanced Salt Solution (HBSS, Euroclone) containing DTT (0.1 mmol·L−1, Sigma‐Aldrich) and EDTA (1 mmol·L−1, Sigma‐Aldrich) in the presence of penicillin/streptomycin and gentamicin. Epithelial cells were obtained after collecting the supernatants from washes with EDTA, centrifuging at 200× g for 5 min, and preserving the pellet. Purity was confirmed via flow cytometry.
2.4. Human iNKT cell isolation and stable line generation
All the iNKT cell lines generated for this study are listed in Table S2. iNKT cells were isolated from LPMC and PBMC cells by sorting CD45+ CD3+ CD1d : PBS‐57Tet+ cells as described in Fig S1 [54]. For stable line generation, sorted iNKT cells were stimulated with phytohemagglutinin (PHA, 1 µg·mL−1, Sigma‐Aldrich) and irradiated peripheral blood feeders in a 2 : 1 iNKT : feeder ratio. PBMCs used as feeders were irradiated at 12.5 Gy and not pulsed with α‐GalCer. We did not stimulate iNKT cells in the presence of α‐GalCer to prevent iNKT cells to skew toward a Th1 phenotype as previously reported [55]. Stimulated cells were then expanded for 15 days by subculturing them every 2–3 days and maintained in RPMI‐1640 medium with stable glutamine, 5% v/v human serum, and 100 IU·mL−1 IL‐2 (Proleukin).
2.5. Assessment of cell viability
Cell viability prior and after coincubation was addressed using the Zombie Green Fixable Viability Kit (BioLegend, San Diego, CA, USA) according to the manufacturer’s instructions. In brief, cells were stained with Zombie Green dye and incubated for 20 min at room temperature, and subsequently washed twice with 1% BSA in PBS.
2.6. Lactate dehydrogenase release assay
Cytotoxicity was assessed using the Cytotoxicity Lactate Dehydrogenase (LDH) Assay Kit‐WST (nonhomogeneous assay, Dojindo, EU) following the manufacturer’s instructions. All experimental conditions were performed in duplicate. Cancer cells (2.5 × 104 cells·well−1) were incubated at 37 °C for 4 h with effector cells at effector : target (E : T) ratios of 8 : 1, 4 : 1, 2 : 1, and 1 : 1. Supernatants were collected and plated in optically clear, 96‐well plates, and absorbance at 490 nm was measured using a GloMax Microplate Reader (Promega, Madison, WI, USA) after the colorimetric reaction for LDH detection was finished. The percentage of cytotoxicity was calculated as follows: (test well – spontaneous release control)/(maximal release control – spontaneous release control) × 100. For blocking experiments, iNKT cells were used at 8 : 1 E : T ratio. Target cells were pretreated for 30 min with anti‐CD1d antibody (10 µg·mL−1, clone CD1d42, R&D Systems); iNKT cells were pretreated with anti‐Fas Ligand antibody (30 min, 10 µg·mL−1, clone 100419, R&D Systems), anti‐TRAIL antibody (30 min, 10 µg·mL−1, clone 75411, R&D Systems), concanamycin A (2 h, 10 nm, Sigma‐Aldrich), or 3,4 dichloroisocoumarin (20 min, 40 µm, Enzo Life Sciences). Media from pretreated cells was then removed and replaced with fresh medium.
2.7. Calcein release assay
Target cell death was also addressed using the Calcein AM Assay Kit (Abcam, Cambridge, UK). All experimental conditions were performed in duplicate. In summary, 2.5 × 104 cells·well−1 were incubated with Calcein AM for 30 min at 37 °C and then washed with PBS. Then, iNKT cells were added to each well at 8 : 1 E : T ratio for 4h. Supernatants were collected and transferred to 96‐well, white opaque plates (Falcon), and fluorescence was measured at Ex: 475 nm, Em: 500–550 nm using a GloMax Discover Microplate reader (Promega). The percentage of cytotoxicity was calculated as follows: (test well – spontaneous release control)/(maximal release control – spontaneous release control) × 100.
2.8. Caspase‐3/7 activity assay
Caspase‐3 and caspase‐7 activity was measured using the ApoLive‐Glo Multiplex Assay (Promega) according to the manufacturer’s instructions. All experimental conditions were performed in duplicate. iNKT cells were used in an 8 : 1 E : T ratio and were incubated with cancer cells (12.5 × 104 cells·well−1) at 37 °C for 4 h in 96‐well, white opaque plates (Falcon). After coculture, luminescence was measured with a GloMax Microplate Reader (Promega). Ionomycin (Sigma‐Aldrich, 100 mm, 6 h) was used as necrosis control. Controls also included untreated cancer cells, effector cells alone, and wells with medium only. Luminescence upon coculture was calculated as follows: Test well – mean (effector cells alone wells).
2.9. Flow cytometry
Cells were stained with combinations of directly conjugated antibodies as specified in Table S3. The gating strategies to identify and analyze effector and target cells are described in Fig. S1. Effector cells were fixed and permeabilized with Cytofix/Cytoperm (BD) and treated with brefeldin A before the addition of antibodies, whereas colon cancer cells were fixed with 1% paraformaldehyde (PFA, Sigma‐Aldrich). All samples were acquired with a FACSCelesta Cytometer using the BD facsdiva software (BD Biosciences Milan, Italy). Analyses were performed using the flowjo software version 10.5.3 (BD Biosciences).
2.10. Cytotoxicity molecule release measurements
Granzyme B (GZMB, MabTech, Cambridge, UK) and perforin (PRF, MabTech) concentrations in coculture supernatants were measured by sandwich ELISA assays according to manufacturers’ protocols (for antibodies, Table S4). All experimental conditions were performed in duplicate. Absorbance was measured at 450 nm using a GloMax Microplate Reader (Promega).
2.11. Statistical analysis
Analysis of variance (ANOVA) and Tukey and Šidak’s tests for multiple comparisons or Kruskal–Wallis test with Dunn’s test for multiple comparisons and graphs were done with graphpad Prism version 8.0. Linear regressions and cytotoxicity percentages were calculated with Microsoft Excel version 16.25. P < 0.05 (*), 0.01 (**), 0.001 (***), and 0.0001 (****) were regarded as statistically significant.
3. Results
3.1. Human Intestinal and circulating iNKT cells are cytotoxic against colon cancer cell lines
Human peripheral blood iNKT cells exert cytotoxic functions against cancers of different origins [21, 33, 34, 39, 45, 46]. To test whether iNKT cells of either blood or intestinal origin are capable of cytotoxic against colon cancer cells, we isolated iNKT cells by cell sorting, expanded them in vitro, and generated stable lines to perform functional assays (Fig. S1). Several stable cell lines from healthy peripheral blood and intestinal specimens were obtained (n = 9, Table S2). Circulating (PB1, PB2, PB3, PB5, and PB6) and intestinal (CD1, CD2, CD3, and NUN) iNKT cells were coincubated with five colorectal cancer cell lines at different effector : target ratios for in vitro cytotoxicity evaluation by measuring lactate dehydrogenase release (Fig 1A). As this assay measures the release of the whole system, we also measured iNKT cell viability prior and after coincubation using Zombie staining (Fig. S2E). For these series of experiments, NK‐92 cells, a stable human natural killer cell line, and freshly isolated, circulating NK cells were used as cell‐mediated cytotoxicity controls (Figs. 1A, Fig. S2C & Fig. S3A). Both intestinal (Fig. 2A) and circulating (Fig. 2B) iNKT cells exerted efficient killing activity against all colon cancer cell lines tested. Effector : target ratio curves for iNKT cell killing were similar to that of blood natural killer cells, but killing was lower compared with NK‐92 cells (Fig. 1A). When iNKT cell lines were analyzed separately, we observed great donor variability in killing efficiency, being iNKT CD1‐3, PB1, and PB6 the most efficient (Fig. S2A–B).
Fig. 1.
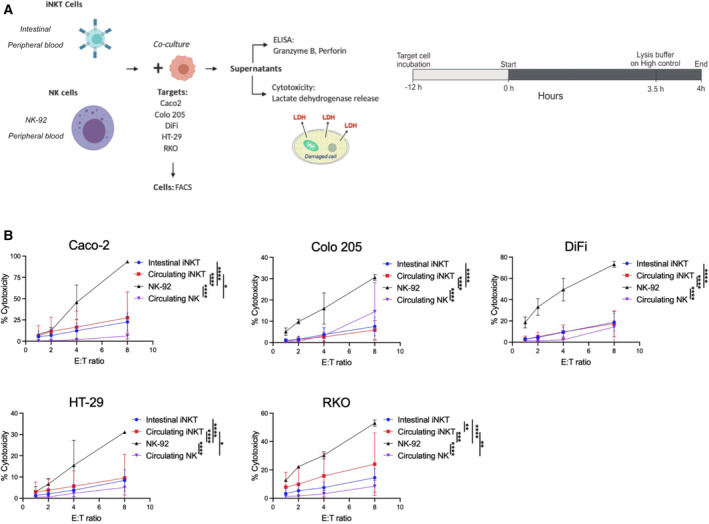
iNKT cell lines exert direct cytotoxic activities against colon cancer cell lines. (A) Experimental design. (B) Effector : target (E : T) ratio curve graphs comparing the killing efficiency of intestinal and circulating iNKT cells (pooled donors) with NK‐92 and peripheral blood natural killer (NK) cells. Two‐way ANOVA tests were used to assess statistical significance and Tukey tests for multiple comparisons. Data are means ± SD of at least 3 independent experiments for iNKT and NK‐92 cell experiments. ns. nonsignificant, P‐value < 0.05 (*), 0.01 (**), 0.001 (***), 0.0001 (****).
Fig. 2.
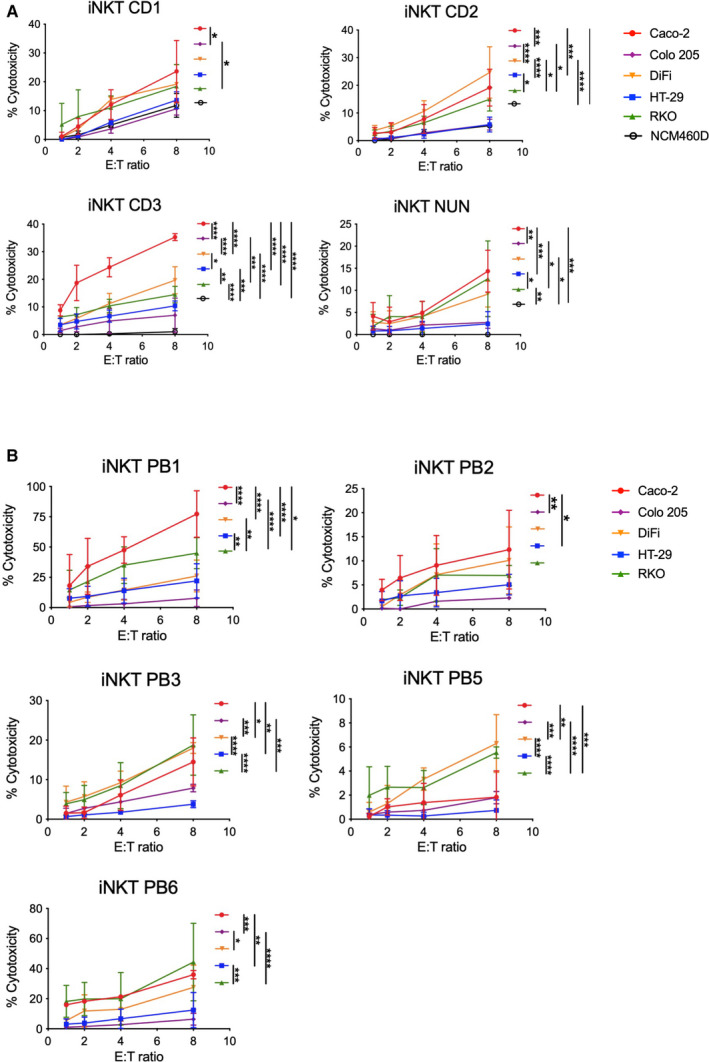
iNKT cells kill colon cancer cell lines with different efficiencies. (A–B) E : T ratio curves of each intestinal (A) and circulating (B) iNKT cell line comparing killing levels for each target cell line. Two‐way ANOVA tests were used to assess statistical significance and Tukey tests for multiple comparisons. Data are means ± SD of at least 3 independent experiments. ns, nonsignificant, P‐value < 0.05 (*), 0.01 (**), 0.001 (***), 0.0001(****).
Albeit all cancer cell lines were targeted and killed by iNKT cells of different origins, cytotoxicity was not equal against the target lines. In general, Caco‐2, DiFi, and RKO cells were the most efficiently eliminated, whereas Colo 205 and HT‐29 cells were more resistant to iNKT cell killing (Fig. 2). This was also confirmed by low caspase activity and calcein release upon coculture with iNKT cells (Fig. S2D & F). The same pattern was observed on NK‐92 cells, but not on blood‐derived NK cells, as each donor displayed different killing preferences (Fig. S3A). As intestinal iNKT cells were obtained from patients with inflammatory bowel diseases (Table S2), we wondered whether these cells were cytotoxic due to a skewed proinflammatory phenotype and not being able to distinguish between normal and tumor colon cells. To test this, we performed killing experiments using a normal colon cell line (NCM460D) as target cells. We observed that none of the intestinal iNKT cells could eliminate NCM460D cells with the exception of iNKT CD1, which showed a mild killing activity (Fig. 2A).
Altogether, these data indicate that the tissue of origin does not define the cytotoxic potential of iNKT cell lines against colon cancer cells. Furthermore, we showed that some target lines are more prone to iNKT cell killing, suggesting differences in how tumor cells can activate iNKT cell cytotoxic activity.
3.2. iNKT cells change the expression of TRAIL and Fas ligand upon interaction of CRC cells
Murine and human iNKT cells display on their surface the cytotoxicity‐related molecules Fas ligand and TRAIL, used for their cancer‐killing function in vitro and in vivo [35, 38, 41, 44]. Therefore, we analyzed the expression of these molecules on human iNKT cells before and after the exposure to CRC cells (Figs. 3 & 4). TRAIL levels at steady‐state conditions varied greatly among donors, ranging from 60% to less than 1% in average (Fig. S2I). Fas ligand expression, on the contrary, was considerably low in all the iNKT cells studied (Fig. S2J).
Fig. 3.
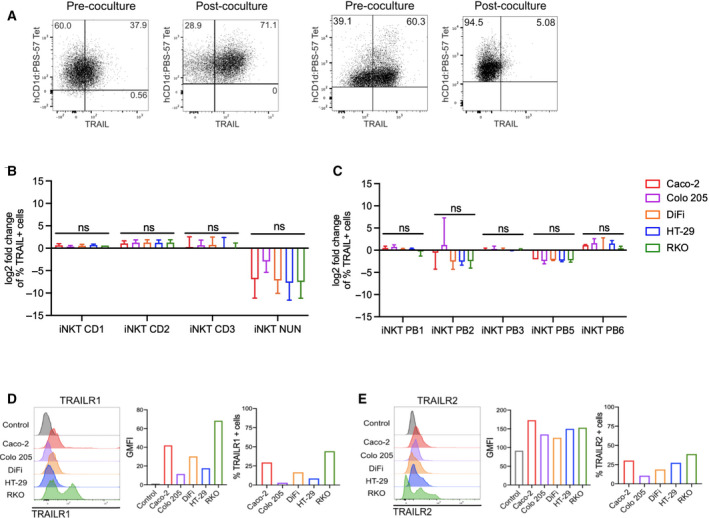
Surface TRAIL changes upon encounter with colon cancer cells. (A) Representative dot plots for iNKT cells prior and after coculture. (B–C) Fold change of TRAIL‐positive intestinal (B) and blood‐derived (C) iNKT cells respect to resting conditions. D‐E. TRAIL receptor 1 (TRAILR1, D) and TRAIL receptor 2 (TRAILR2, E) expression in colorectal cancer (CRC) cell lines. Kruskal–Wallis test was used to assess statistical significance, and Dunn’s test was used for multiple comparisons in B‐C. ns. nonsignificant, P‐value < 0.05 (*), 0.01 (**), 0.001 (***), 0.0001 (****). Data are means ± SD of at least 3 independent experiments in B and C.
Fig. 4.
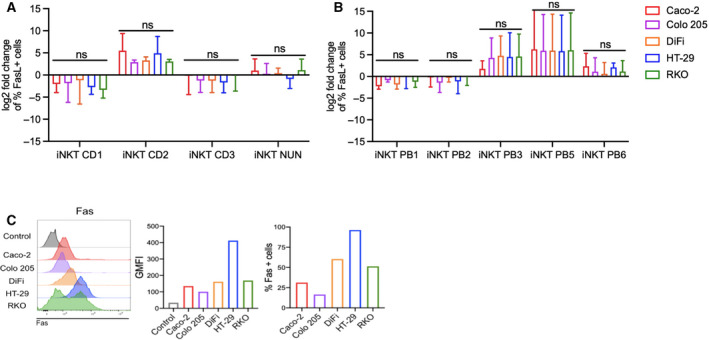
Fas ligand is modulated upon encounter with colon cancer cells. (A–B) Fold change of Fas ligand expression by colon (A) and circulating (B) iNKT cell lines at steady‐state conditions vs after 4‐h incubation with CRC cell lines. (C) Fas expression by colon cancer cells. Kruskal–Wallis test was used to assess statistical significance, and Dunn’s test was used for multiple comparisons. ns. nonsignificant, P‐value < 0.05 (*), 0.01(**), 0.001 (***), 0.0001 (****). Data are means ± SD of at least 3 independent experiments for A and B.
Upon encounter with CRC cells, TRAIL expression increased on some iNKT lines, namely CD1, CD2, and PB6, whereas it decreased on iNKT NUN, PB2, and PB5 cells (Fig. 3B & C). FasL expression was also modulated upon coculture, increasing on iNKT CD2, PB3, PB5, and modestly on iNKT PB6 cells, and decreasing on iNKT CD1, CD3, PB1, and PB2 cells (Fig. 4A & B). To note, also NK cells were characterized by a low expression of Fas ligand at steady‐state conditions, which was further reduced upon encounter with CRC cells (Fig. S3C). TRAIL expression varied among NK cells, being the highest on NK‐92 cells (Fig. S3B). Upon coculture, TRAIL was slightly upregulated on NK PB1 cells, while it was downregulated on NK PB2 cells (Fig. S3C).
The expression of Fas and TRAIL receptors 1 and 2 (TRAILR1 and TRAILR2) was variable on target cells (Figs. 3D–E & 4C). Differences among lines were observed in TRAILR1 (Caco‐2 29.5%, Colo 205 2.98%, DiFi 16.68%, HT‐29 8.61%, RKO 44.2%; Fig. 3D) and TRAILR2 (Caco‐2 30.3%, Colo 205 10.65%, DiFi 18.7%, HT‐29 27.37%, RKO 38.83%; Fig. 3E). Caco‐2 and Colo 205 cells showed relatively low Fas expression (Caco‐2 31.2%, Colo 205 16.58%), and DiFi and RKO cells had intermediate frequencies (DiFi 60.38%, RKO 51.39%) while most HT‐29 were Fas positive (96.25%; Fig. 4C). Taken together, these data confirm that human iNKT cells modulate TRAIL and Fas ligand membrane expression upon encounter with CRC cells.
3.3. Targeting of CRC cells induces secretion of perforin and granzyme B by iNKT cells
Next, we assessed the production of soluble cytotoxic molecules prior and after coincubation of iNKT cells with colon cancer cells. Granzyme B (GZMB) and perforin (PRF) were detected by flow cytometry analysis and in culture supernatants. As it occurred with TRAIL and Fas ligand, frequencies of GZMB+ cells changed after incubation with colon cancer cells. More specifically, frequencies increased in iNKT CD1‐3 cells, whereas there was a reduction in almost all circulating iNKT cells with the exception of PB6, on which changes were quite modest (Fig. 5B–C). Perforin‐positive cells did not vary greatly upon coculture, decreasing in almost all the iNKT cell lines we tested (Fig. 5A & B). More importantly, to evaluate whether these molecules were also efficiently secreted, we measured granzyme B and perforin concentrations in coculture supernatants by ELISA and compared them with levels secreted by iNKT cells alone during the time of the killing experiments (basal level). Even if some iNKT cell lines could release spontaneously perforin and granzyme B, their levels were lower when compared to the release in the presence of CRC cells, being particularly evident in intestinal iNKT cells (Figs. 5D–E & 6C–D).
Fig. 5.
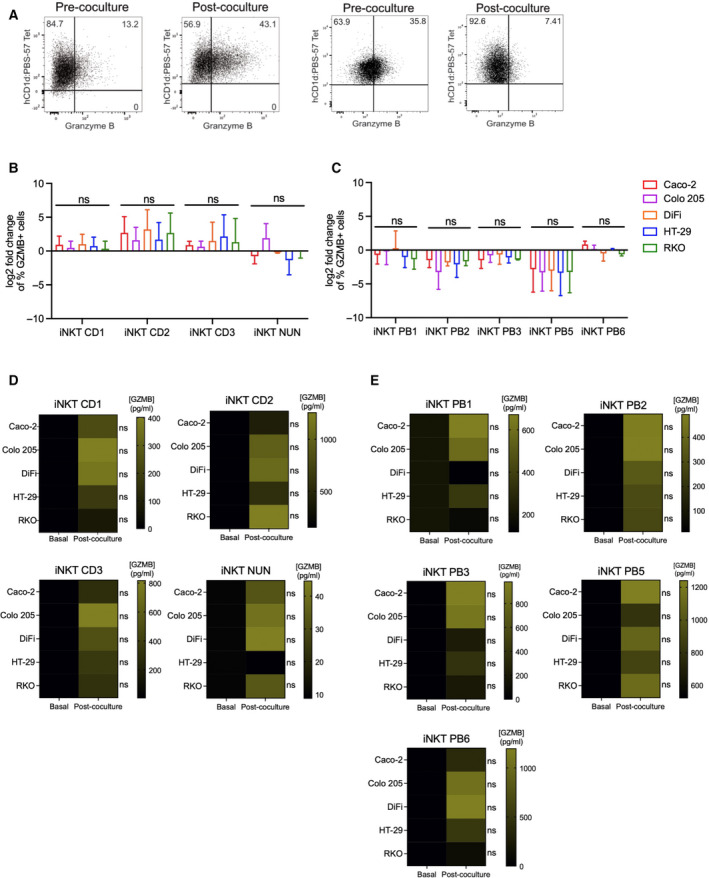
iNKT cell lines release granzyme B upon with CRC cell recognition. (A) Representative dot plots for iNKT cells prior and after coculture. (B–C) Fold change of frequencies of granzyme B (GZMB)‐positive intestinal (B) and circulating (C) iNKT cells respect to steady‐state conditions. D‐E. Granzyme B concentration in iNKT cell supernatants at 8 : 1 E : T ratio alone vs after incubation with colon cancer cells for intestinal (D) and peripheral blood (E) iNKT cells. Kruskal–Wallis test was used to assess statistical significance, and Dunn’s test was used for multiple comparisons. ns. nonsignificant, P‐value < 0.05 (*), 0.01 (**), 0.001 (***), 0.0001 (****). Data are means ± SD for (B–C) and means for (D–E) of 3 independent experiments.
Fig. 6.
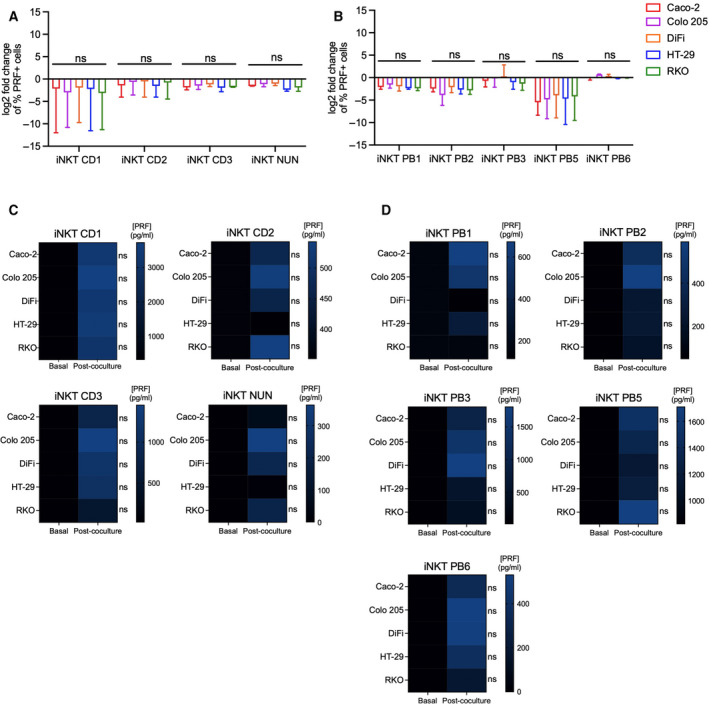
iNKT cell lines release perforin upon CRC cell encounter. (A–B) Frequencies of perforin (PRF)‐positive intestinal (A) and circulating (B) iNKT cells before and after 4‐h coincubation. (C–D) Perforin concentration in iNKT cell supernatants at 8 : 1 E : T ratio alone vs after incubation with colon cancer cells for intestinal (C) and peripheral blood (D) iNKT cells. Kruskal–Wallis test was used to assess statistical significance, and Dunn’s test was used for multiple comparisons. ns. nonsignificant, P‐value < 0.05 (*), 0.01 (**), 0.001 (***), 0.0001 (****). Data are means ± SD for B–C and means for D–E of 3 independent experiments.
On the other hand, natural killer cells showed high frequencies of GRZB and PRF+ positive cells in steady state in some cases (Fig. S3D & F). Upon coculture, granzyme B‐positive cells mildly increased on NK‐92 and NK PB2 cells (Fig. S3D), whereas PRF‐positive cell frequencies increased in NK‐92 cells, but there was a reduction in NK PB2 cells (Fig. S3F). Interestingly, fresh NK cells secreted similar amounts of granzyme B as compared to iNKT cells, while perforin levels were higher (Fig. S3E & G). These results indicate that intestinal and circulating iNKT cells increase their secretion of granzyme B and perforin when they interact with CRC cells.
3.4. iNKT cells recognize CRC cells in CD1d‐dependent and CD1d‐independent manners, requiring perforin and granzymes for proper elimination
Murine and human iNKT cells can target and kill solid tumors by either antigen‐dependent or antigen‐independent mechanisms [21, 27, 33, 37, 38, 39, 42, 48]. We thus wondered which could be the case for human iNKT cells in the context of colorectal cancer. To test this, we used five of the most efficient iNKT cell lines and the three most sensitive CRC cell lines as target cells at the highest E : T ratio (8 : 1).
We first evaluated whether colon cancer cells expressed the CD1d molecule on their surfaces. All cell lines were CD1d positive, albeit differences in frequencies were found, with DiFi and RKO cells being the lines with most CD1d‐positive cells (DiFi 41.4%, RKO 40.3%) and the metastatic line Colo 205 having the lowest number of positive cells (3.75%; Fig. 7B). CD1d expression levels on these cells were lower than on professional antigen‐presenting cells such as monocyte‐derived dendritic cells and THP‐1 cells, a monocytic cell line (Fig. S1E–F). Interestingly, iNKT cell cytotoxicity was not profoundly affected by CD1d blockade that only induced a partial decrease in cytotoxic activities (Fig. 7C). These results suggest that iNKT cells can recognize CRC cells by both CD1d‐dependent and CD1d‐independent mechanisms.
Fig. 7.
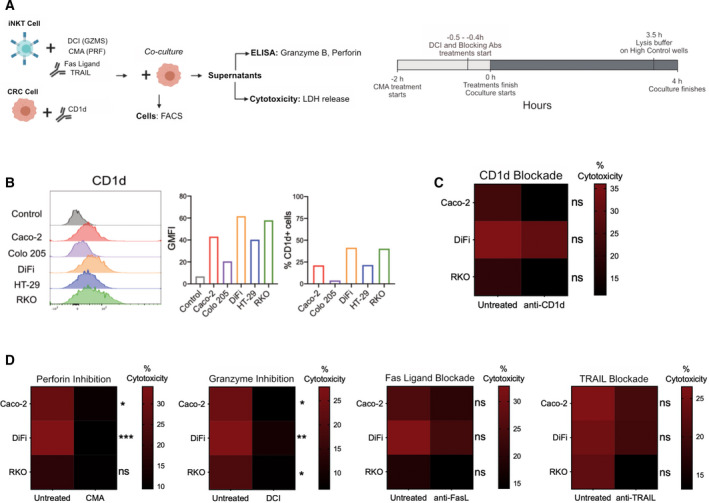
iNKT cells recognize CRC cells in CD1d‐dependent and CD1d‐independent manners, requiring perforin and granzymes for proper elimination. (A) Experimental design. All experiments were performed at an E : T ratio of 8 : 1. (B) CD1d expression in CRC cell lines. (C) Effect of CD1d blockade on iNKT cell elimination of CRC cells. (D) Effect of perforin, granzymes, Fas ligand, and TRAIL inhibition iNKT cell cytotoxicity. Two‐way ANOVA test was used to assess statistical significance, and Šidak’s test for multiple comparisons. ns. nonsignificant, P‐value < 0.05(*), 0.01 (**), 0.001 (*** 0.0001 (****). Data are means of five pooled iNKT cell lines with 3 independent experiments.
Since antigen‐dependent mechanisms seemed to be only partially involved, to unravel the mechanism of iNKT cell cytotoxicity, we used inhibitors and blocking antibodies against different cytotoxicity‐related molecules prior to the coculture with cancer cells. Concanamycin A (CMA) was added to inhibit perforin, 3,4 dichloroisocoumarin (DCI) to inhibit granzymes, and neutralizing antibodies were used to block TRAIL (anti‐TRAIL) and Fas ligand (anti‐FasL) (Fig. 7D), for which we also assessed potential toxicity on iNKT cells by Zombie staining. At the concentrations used here, none of the inhibitors was toxic against iNKT cells (Fig. S4E). Perforin and granzyme inhibition drastically affected the cytotoxic functions of all the iNKT cell lines tested. Conversely, neutralizing antibodies induced a mild reduction in killing activity, being of particular interest the decrease induced by anti‐TRAIL antibody against RKO cells (Fig. 7D). To note, Fas Ligand, TRAIL, and CD1d blockade partially affected the release of granzyme B and perforin (Fig. S4A–B), while DCI treatment increased the frequencies of Fas ligand‐positive cells (Fig. S4C). Taken together, these results indicate that the perforin/granzyme pathway is required for iNKT cell cytotoxic activity against colon cancer cells.
3.5. iNKT cells kill patient‐derived colon cancer cells with production of perforin and granzyme B
As the previous experiments were performed with well‐established CRC cell lines, we wondered whether intestinal and peripheral blood iNKT cells were also efficient at eliminating cancer cells freshly isolated from CRC patients. To address this point, we isolated cancer cells from surgical specimens of four patients (Fig. S5A) and cocultured them with three intestinal and two circulating iNKT cell lines at 8 : 1 E : T ratio for 4 h. As shown in Fig. 8A, iNKT NUN and PB6 efficiently eliminated patient CRC cells, at comparable levels shown for CRC cell lines. More interestingly, iNKT CD1, CD3, and PB5 were more efficient at killing freshly isolated CRC cells than any of the cell lines previously tested. Strikingly, iNKT CD1 and CD3 reached 100% of killing efficiency for some patients (Figs. 8A & 2A–B). To note, all iNKT cell lines were characterized by the expression and release of granzyme B and perforin (Fig. 8D & F), whereas some of them upregulated death ligands (Fig. 8B). These data confirm that human intestinal and circulating iNKT cells have the ability to eliminate freshly isolated, patient‐derived colon cancer cells by releasing granzyme B and perforin.
Fig. 8.
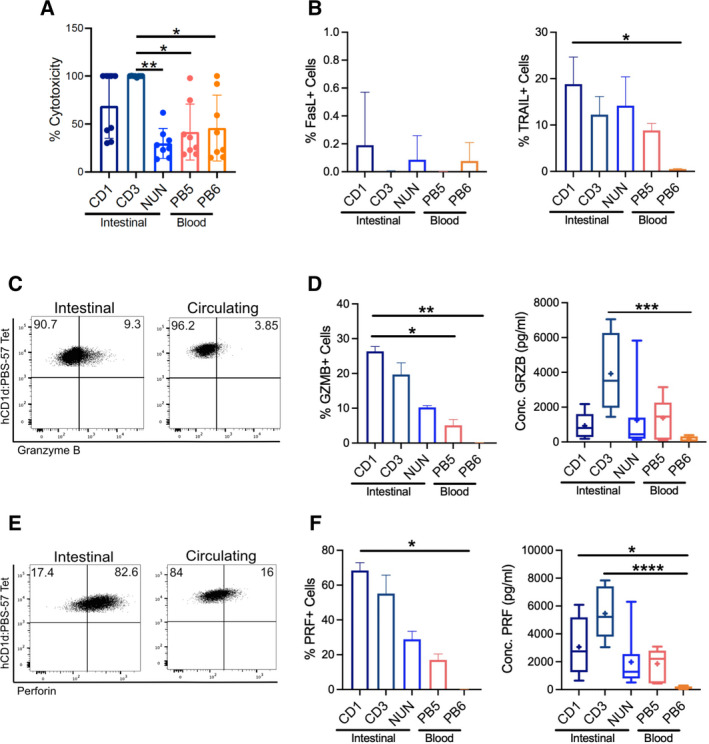
iNKT cells efficiently kill patient‐derived colon cancer cells. (A) Cytotoxicity at 8 : 1 E : T ratio. (B) Frequencies of Fas ligand (left) and TRAIL (right)‐positive iNKT cells after incubation with CRC cells (C) Representative dot plots showing granzyme B expression. (D) Frequencies of granzyme B‐positive cells (left) and granzyme B levels (right) after CRC cell encounter. (E) Representative dot plots showing perforin expression. (F) Frequencies of PRF‐positive cells (left) and perforin levels (right) after CRC cell encounter. Kruskal–Wallis test was used to assess statistical significance, and Dunn’s test was used for multiple comparisons. ns. nonsignificant, P‐value < 0.05 (*), 0.01 (**), 0.001 (***), 0.0001 (****). Data are means ± SD of 4 pooled patients.
4. Discussion
The antitumor functions of iNKT cells have been of particular interest since the emergence of cancer immunotherapies. They are peculiar in several ways, since they are capable of being activated by lipid antigens, innate signals, and cytokines, and not being affected by histocompatibility issues, as CD1d is a nonpolymorphic molecule [21, 37, 56, 57]. Nonetheless, the mechanisms of human iNKT cell cytotoxicity are still largely unknown. Here, we showed that human intestinal and peripheral blood‐derived iNKT cells exerted killing activities against colon cancer cells through different mechanisms, including granzyme B and perforin release and death receptor pathways.
Previous studies showed that human peripheral blood iNKT cells could have killing activities against cell lines from different origins, including colorectal cancer [34, 39, 42, 44, 45, 58]. Here, we observed that both circulating and intestinal iNKT cells could kill all the colon cancer cell lines they were exposed to. This study is the first that demonstrates the cytotoxic activity of human intestinal iNKT cells. Furthermore, we showed that the perforin/granzyme pathway is required for iNKT cell cytotoxic functions against colorectal cancer cells.
iNKT cells are usually divided into subsets depending on their transcriptional programs and cytokines released [49]. The NKT1 subset is of particular importance in cancer research as it is the only iNKT cell subset endowed of antitumor activities, both through cytokine release and cytotoxic activity [49]. Studies in mice have shown that these cells are mainly located in the liver, intestine, thymus, lung, and spleen [49, 59],suggesting that iNKT cells from many organs are potentially capable of killing transformed cells [38, 40, 41, 44, 46]. In addition, most of the human iNKT cell cytotoxicity studies have been performed with cells derived from peripheral blood [21, 34, 36, 39, 45, 46, 48], indicating that iNKT cells from different tissues preserve their cytotoxic activity, being capable of eliminating tumor cells even if these do not belong to the same tissue/organ of origin. In line with these findings, we observed that peripheral blood and intestinal iNKT cell lines were similarly efficient at killing colorectal cancer cells. It is noteworthy that intestinal iNKT cell lines were obtained from tissue samples from inflammatory bowel disease (IBD) patients, known to be characterized by the intestinal infiltration of pathogenic Th1/17 lymphocytes [60], including iNKT cells [54]. However, even if our intestinal iNKT lines have the potential of being proinflammatory and tissue‐damaging, we observed that they were not cytotoxic against normal colon cells, discarding the possibility that the observed cytotoxicity was due to their proinflammatory origin.
From a mechanistic point of view, iNKT cells, similarly to other cytotoxic populations, are known for inducing tumor cell death both by granzyme B/perforin production and by expression of Fas ligand (FasL) and TRAIL on their plasma membranes [38, 42, 44, 61, 62, 63]. Here, all the iNKT cell lines, regardless their origin, produced granzyme B and perforin to eliminate CRC cell lines. In particular, we confirmed that the prevailing killing mechanism of iNKT cells was based on the presence of functional perforin and granzymes, as treatment with concanamycin A and DCI significantly impaired iNKT cell killing against the colon cancer lines tested. While the perforin/granzyme pathway does not require the presence of specific receptors on target cells, the death ligand pathways require death receptor expression and are slower than the granzyme/perforin mechanism, requiring more degranulation events [61, 64]. This might explain why the PRF/GZMB mechanism was prevalent in this model. Even if the presence of functional perforin and granzyme B is fundamental for iNKT cell killing of CRC cells, some iNKT lines expressed TRAIL on their membranes, and some of them upregulated Fas ligand upon encounter with colon cancer cells. Furthermore, Fas and TRAIL receptors were present in CRC cell lines. Thus, it is possible to envisage that upon cancer cell encounter, cytotoxic cells might choose the best killing mechanism according to their activation state and/or by switches from PRF/GZMB to death receptor pathways over time, as it occurs with natural killer cells [63].
Although iNKT cell cytotoxicity was not profoundly affected by CD1d blockade, killing was still reduced. These results indicate that epithelial cancer cells not only express CD1d on their membranes, but they might also present antigens involved in the adaptive activation of iNKT cell cytotoxicity, even if not as strongly as other innate signals. In this regard, iNKT cells are known to be activated by a range of self‐antigens [56, 65, 66, 67, 68]. Altered pathways observed in cancer might affect lipid presentation and therefore iNKT cell activation. For example, it has been observed that endoplasmic reticulum stress, process commonly found in cancer cells, induces the enhancement of immunogenic self‐lipid antigens and activation of iNKT cells [56, 68]. While CD1d presence is essential for iNKT cell killing in some cancer types [38, 40, 42, 44, 45] other studies, including this one, have demonstrated that in some contexts, antigen presentation by cancer cells might not be as important as triggering by innate mechanisms [27, 36, 37, 46, 69].
Here, we also observed that CRC lines were not eliminated with the same efficiency, being some lines more sensitive than others to iNKT cell‐mediated killing. These patterns of sensitivity were considerably preserved among iNKT cell lines and NK‐92 cells, suggesting similar mechanisms of detection of tumor cells by iNKT cell lines from different tissue origins. Moreover, iNKT cell cytotoxicity was not substantially affected by the inhibition of CD1d. Both phenomena can be explained by the innate nature of iNKT cells, and the characteristics displayed by different CRC cells. One of the main features of iNKT cells is the expression of some NK receptors. On natural killer cells, the balance given by the interactions of activating and inhibitory receptors with their respective ligands establishes their activation state [70]. iNKT cells might be also regulated by the same mechanisms, as they express NKG2D, NKG2A, NKp30, NKp46, and some killer immunoglobulin (KIR) receptors [27, 49, 71, 72, 73].
Furthermore, CRC lines express different arrays of NK receptor ligands. Regarding the most resistant colon cancer lines, some evidence has demonstrated that Colo 205 cells are generally resistant to killing by natural killer cells due to high expression of HLA‐A, HLA‐B, and HLA‐C molecules (ligands of KIR receptors, [74]), CD155 (ligand for CD96 and TIGIT, inhibitory receptors; [75]), and low expression of MICA (ligand for NKG2D, one of the main activating NK receptors; [76]). HT‐29 cells express MICA on their membranes, but they are also characterized by a high expression of HLA‐ABC molecules, as well as HLA‐E, ligand of the inhibitory receptor NKG2A [77, 78, 79]. On the other hand, many studies have shown that RKO cells are sensitive to NK killing due to the expression of the NKG2D ligands MICB, ULBP1, ULBP2, and ULBP3, as well as the lack of HLA‐E expression [80, 81, 82, 83]. Caco‐2 line shows low expression of the MHC‐I complex and HLA‐E [79, 83], while no data are available on the expression of NK ligands on DiFi cells.
Another explanation for the differential sensitivity to iNKT cell killing might be given by the aneuploidy status of colon cancer cells. It has been observed that chromosome mis‐segregation causes the insurgence of complex karyotypes, and cells with these karyotypes are more efficiently killed by natural killer cells [84]. In line with these findings, we observed that two out of three of the most sensitive CRC lines (DiFi and Caco‐2) have the highest ploidy of the whole panel (Table S1). In the case of the RKO line, it must be considered that these cells are microsatellite instable, and it has been widely recognized that microsatellite‐instable tumors are usually more immunocompetent than the microsatellite‐stable counterparts [85, 86, 87]. Overall, this heterogeneity might also reflect the heterogeneity commonly found among patients, some responding better than others to immunotherapies [88].
In addition, we demonstrated that iNKT cells can eliminate patient‐derived CRC cells. This finding raises the question of how their cytotoxic activity is impaired during colon cancer progression. We speculate that this occurs as a consequence of changes in tissue architecture that might impede iNKT cells to form the immunological synapse with target cells, or the involvement of other components of the tumor microenvironment (tumor‐associated macrophages, neutrophils, gut microbiota, metabolites, etc.) in the modulation of iNKT cell activity toward an exhausted or protumorigenic phenotype. Nonetheless, the actual contribution of these factors will need further studies.
Altogether, these results might become of interest in the light of potential iNKT cell‐based immunotherapies against colon cancer. It has been observed that, even if iNKT cells are found in considerably low numbers [50], there is a higher infiltration in human CRC tumors [47]. Therapeutic strategies could be targeted to potentiate the killing activities of iNKT cells already infiltrating the tumor, or selecting iNKT cells with high cytotoxic potential for cell transfer interventions. Regarding the latter, as circulating and colon‐derived iNKT cells show similar killing efficiency, it would be more advantageous to obtain cells from peripheral blood, as numbers would be higher for further expansion, and obtaining surgical specimens might logistically become more difficult.
In conclusion, this study sheds light on the effector roles of human circulating and intestinal iNKT cells against colorectal cancer. Furthermore, it provides new knowledge about the activation and mechanistic cues of iNKT cell cytotoxicity, elucidating the importance of studying iNKT cell behavior to propose them as a new immunotherapeutic approach against colon cancer.
5. Conclusions
In this work, we demonstrated that both intestinal and circulating iNKT cells kill colon cancer cells in a similar manner as circulating NK cells. Furthermore, we showed that iNKT cells increase the release of soluble cytotoxic molecules upon encounter with target cells, being functional perforin and granzymes essential for iNKT cell cytotoxicity. In addition, we observed that antigen presentation by CRC cells is not required for their recognition by iNKT cell lines. To sum up, we explored the cellular requirements of human iNKT cell cytotoxicity in the context of colorectal cancer and concluded that iNKT cells might have a role in immunosurveillance in the colon via the recognition and killing of tumor cells.
Conflict of interest
The authors declare no conflict of interest.
Peer Review
The peer review history for this article is available at https://publons.com/publon/10.1002/1878‐0261.13104.
Author contributions
ADB curated the data, analyzed, investigated, and wrote–original draft; CB investigated and conceptualized the study; GL curated the data and investigated the study; LB, FB, and AC provided resources; FC provided resources and wrote–review and editing; FF conceptualized and supervised the study, involved in funding acquisition and project administration, and wrote–original draft, review, and editing.
Supporting information
Table S1. Cell lines used in the study.
Table S2. List of iNKT cell lines generated for functional studies.
Table S3. List of antibodies used for flow cytometry analysis.
Table S4. Antibodies used for ELISA assays.
Fig. S1. Flow cytometry analyses conducted in this study.
Fig. S2. iNKT cell lines are cytotoxic against colon cancer cell lines.
Fig. S3. Cytotoxicity effectors on NK cells.
Fig. S4. Effect of cytotoxicity mechanism and CD1d inhibition on cytotoxic mediators.
Fig. S5. iNKT cell killing of patient‐derived CRC cells.
Acknowledgments
This work was possible through AIRC (Associazione Italiana per la Ricerca sul Cancro, IG‐22923) grant to F. Facciotti. We are grateful to Abbass Darwich and Francesco Strati for the scientific support, the San Matteo Hospital for the buffy‐coat samples, Erika Mileti for technical support, and the NIH Tetramer Facility for providing hCD1d:PBS‐57 Tetramers. Authors acknowledge support from the University of Milan (Università degli Studi di Milano) through the APC initiative.
Data accessibility
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- 1. Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, van de Velde CJH & Watanabe T (2015) Colorectal cancer. Nat Rev Dis Primers 1, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chung K, Gore I, Fong L, Venook A, Beck S, Dorazio P, Criscitiello PJ, Healey DI et al. (2010) Phase II study of the anti‐cytotoxic T‐lymphocyte‐associated antigen 4 monoclonal antibody, tremelimumab, in patients with refractory metastatic colorectal cancer. J Clin Oncol 28, 3485–3490. [DOI] [PubMed] [Google Scholar]
- 3. Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM, Blosser RL, Fan H, Wang H et al. (2015) The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter‐inhibitory checkpoints. Cancer Discov 5, 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Overman MJ, Lonardi S, Wong KYM, Lenz H, Gelsomino F, Aglietta M, Morse MA, Van Cutsem E, McDermott R et al. (2018) Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair–deficient/microsatellite instability‐high metastatic colorectal cancer. J Clin Oncol 36, 773–779. [DOI] [PubMed] [Google Scholar]
- 5. Teng R, Zhao J, Zhao Y, Gao J, Li H, Zhou S, Wang Y, Sun Q, Lin Z et al. (2019) Chimeric antigen receptor–modified T cells repressed solid tumors and their relapse in an established patient‐derived colon carcinoma xenograft model. J Immunother 42, 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Darcy PK, Haynes NM, Snook MB, Trapani JA, Cerruti L, Jane SM & Smyth MJ (2000) Redirected perforin‐dependent lysis of colon carcinoma by ex vivo genetically engineered CTL. J Immunol 164, 3705–3712. [DOI] [PubMed] [Google Scholar]
- 7. Magee MS, Abraham TS, Baybutt T, Flickinger JC, Ridge NA, Marszalowicz GP, Prajapati P, Hersperger A, Waldman SA et al. (2018) Human GUCY2C‐targeted chimeric antigen receptor (CAR)‐expressing T cells eliminate colorectal cancer metastases. Cancer Immunol Res 6, 509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Magee MS, Kraft CL, Abraham TS, Baybutt TR, Marszalowicz GP, Li P, Waldman SA & Snook AE (2016) GUCY2C‐directed CAR‐T cells oppose colorectal cancer metastases without autoimmunity. OncoImmunology 5, e1227897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hege KM, Bergsland EK, Fisher GA, Nemunaitis JJ, Warren RS, McArthur JG, Lin AA, Schlom J, June CH et al. (2017) Safety, tumor trafficking and immunogenicity of chimeric antigen receptor (CAR)‐T cells specific for TAG‐72 in colorectal cancer. J Immunother Cancer 5, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lu YC, Yao X, Crystal JS, Li Y, El‐Gamil M, Gross C, Davis L, Dudley ME, Yang JC et al. (2014) Efficient identification of mutated cancer antigens recognized by T Cells associated with durable tumor regressions. Clin Cancer Res 20, 3401–3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rosenberg S, Yang JC, Sherry RM, Kammula US, Hughes S, Phan GQ, Citrin DE, Restifo NP, Robbins PF et al. (2011) Durable complete responses in heavily pretreated patients with metastatic melanoma using T‐cell transfer immunotherapy. Clin Cancer Res 17, 4550–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dudley ME, Gross CA, Somerville RPT, Hong Y, Schaub NP, Rosati SF, White DE, Nathan D, Restifo NP et al. (2013) Randomized selection design trial evaluating CD8+‐enriched versus unselected tumor‐infiltrating lymphocytes for adoptive cell therapy for patients with melanoma. J Clin Oncol 31, 2152–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miller JS, Soignier Y, Panoskaltsis‐Mortari A, McNearney SA, Yun GH, Fautsch SK, McKenna D, Le C, Defor TE et al. (2005) Successful adoptive transfer and in vivo expansion of human haploidentical NK cells in patients with cancer. Blood 105, 3051–3057. [DOI] [PubMed] [Google Scholar]
- 14. Korde N, Carlsten M, Lee MJ, Minter A, Tan E, Kwok M, Manasanch E, Bhutani M, Tageja N et al. (2014) A phase II trial of pan‐KIR2D blockade with IPH2101 in smoldering multiple myeloma. Haematologica 99, e81–e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nieda M, Okai M, Tazbirkova A, Lin H, Yamaura A, Ide K, Abraham R, Juji T, Macfarlane D et al. (2004) Therapeutic activation of Vα24+Vβ11+ NKT cells in human subjects results in highly coordinated secondary activation of acquired and innate immunity. Blood 103 (2), 383–389. [DOI] [PubMed] [Google Scholar]
- 16. Lepore M, Kalinichenko A, Calogero S, Kumar P, Paleja B, Schmaler M, Narang V, Zolezzi F, Poidinger M et al. (2017) Functionally diverse human T cells recognize non‐microbial antigens presented by MR1. eLife 6, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Won EJ, Ju JK, Cho YN, Jin HM, Park KJ, Kim TJ, Kwon YS, Kee HJ, Kim JC et al. (2016) Clinical relevance of circulating mucosal‐associated invariant T cell levels and their anti‐cancer activity in patients with mucosal‐associated cancer. Oncotarget 7, 76274–76290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wilhelm M, Kunzmann V, Eckstein S, Reimer P, Weissinger F, Ruediger T & Tony HP (2003) γδ T cells for immune therapy of patients with lymphoid malignancies. Blood 102, 200–206. [DOI] [PubMed] [Google Scholar]
- 19. Bennouna J, Levy V, Sicard H, Senellart H, Audrain M, Hiret S, Rolland F, Bruzzoni‐Giovanelli H, Rimbert M et al. (2010) Phase I study of bromohydrin pyrophosphate (BrHPP, IPH 1101), a Vγ9Vδ2 T lymphocyte agonist in patients with solid tumors. Cancer Immunol Immunother 59, 1521–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nicol AJ, Tokuyama H, Mattarollo SR, Hagi T, Suzuki K, Yokokawa K & Nieda M (2011) Clinical evaluation of autologous gamma delta T cell‐based immunotherapy for metastatic solid tumours. Br J Cancer 105, 778–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Metelitsa LS, Naidenko OV, Kant A, Wu HW, Loza MJ, Perussia B, Kronenberg M & Seeger RC (2001) Human NKT cells mediate antitumor cytotoxicity directly by recognizing target cell CD1d with bound ligand or indirectly by producing IL‐2 to activate NK cells. J Immunol 167, 3114–3122. [DOI] [PubMed] [Google Scholar]
- 22. Crowther MD, Dolton G, Legut M, Caillaud ME, Lloyd A, Attaf M, Galloway SAE, Rius C, Farrell CP et al. (2020) Genome‐wide CRISPR–Cas9 screening reveals ubiquitous T cell cancer targeting via the monomorphic MHC class I‐related protein MR1. Nat Immunol 21, 178–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bendelac A, Savage PB & Teyton L (2007) The Biology of NKT Cells. Annu Rev Microbiol 25, 297–336. [DOI] [PubMed] [Google Scholar]
- 24. Exley M, Garcia J, Wilson SB, Spada F, Gerdes D, Tahir SMA, Patton KT, Blumberg RS, Porcelli RS et al. (2000) CD1d structure and regulation on human thymocytes, peripheral blood T cells, B cells and monocytes. Immunology 100, 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bannai M, Kawamura T, Naito T, Kameyama H, Abe T, Kawamura H, Tsukada C, Watanabe H, Hatakeyama K et al. (2001) Abundance of unconventional CD8+ natural killer T cells in the large intestine. Eur J Immunol 31, 3361–3369. [DOI] [PubMed] [Google Scholar]
- 26. Panja BA, Blumberg RS, Balk SP & Mayer L (1993) CD1d is involved in T cell‐intestinal epithelial cell interactions. J Exp Med 178, 1115–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kuylenstierna C, Björkström NK, Andersson SK, Sahlström P, Bosnjak L, Paquin‐Proulx D, Malmberg J, Ljunggren HG, Moll M et al. (2011) NKG2D performs two functions in invariant NKT cells: Direct TCR‐independent activation of NK‐like cytolysis and co‐stimulation of activation by CD1d. Eur J Immunol 41, 1913–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baxevanis CN, Gritzapis AD & Papamichail M (2003) In Vivo Antitumor Activity of NKT Cells Activated by the Combination of IL‐12 and IL‐18. J Immunol 171, 2953–2959. [DOI] [PubMed] [Google Scholar]
- 29. Dou R, Hong Z, Tan X, Hu F, Ding Y, Wang W, Liang Z, Zhong R, Wu X et al. (2018) Fas/FasL interaction mediates imbalanced cytokine/cytotoxicity responses of iNKT cells against Jurkat cells. Mol Immunol 99, 145–153. [DOI] [PubMed] [Google Scholar]
- 30. Song L, Asgharzadeh S, Salo J, Engell K, Wu HW, Sposto R, Ara T, Silverman AM, DeClerck YA et al. (2009) Vα24‐invariant NKT cells mediate antitumor activity via killing of tumor‐associated macrophages. Journal of Clinical Investigation 119, 1524–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cortesi F, Delfanti G, Grilli A, Calcinotto A, Gorini F, Pucci F, Lucianò R, Grioni M, Recchia A et al. (2018) Bimodal CD40/Fas‐dependent crosstalk between iNKT cells and tumor‐associated macrophages impairs prostate cancer progression. Cell Rep 22, 3006–3020. [DOI] [PubMed] [Google Scholar]
- 32. Bellone M, Ceccon M, Grioni M, Jachetti E, Calcinotto A, Napolitano A, Freschi M, Casorati G & Dellabona P (2010) iNKT cells control mouse spontaneous carcinoma independently of tumor‐specific cytotoxic T cells. PLoS One 5, e8646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mattarollo SR, Kenna T, Nieda M & Nicol AJ (2006) Chemotherapy pretreatment sensitizes solid tumor‐derived cell lines to Vα24+ NKT cell‐mediated cytotoxicity. Int J Cancer 119, 1630–1637. [DOI] [PubMed] [Google Scholar]
- 34. Kawano T, Nakayama T, Kamada N, Kaneko Y, Harada M, Ogura N, Akutsu Y, Motohashi S, Iizasa T et al. (1999) Antitumor cytotoxicity mediated by ligand‐activated human Va24 NKT cells. Cancer Res 59, 5102–5105. [PubMed] [Google Scholar]
- 35. Nieda M, Nicol A, Koezuka Y, Kikuchi A, Lapteva N, Tanaka Y, Tokunaga K, Suzuki K, Kayagaki N et al. (2001) TRAIL expression by activated human CD4+Vɑ24 NKT cells induces in vitro and in vivo apoptosis of human acute myeloid leukemia cells. Blood 97, 2067–2074. [DOI] [PubMed] [Google Scholar]
- 36. Konishi J, Yamazaki K, Yokouchi H, Shinagawa N, Iwabuchi K & Nishimura M (2004) The characteristics of human NKT cells in lung cancer—CD1d independent cytotoxicity against lung cancer cells by NKT cells and decreased human NKT cell response in lung cancer patients. Hum Immunol 65, 1377–1388. [DOI] [PubMed] [Google Scholar]
- 37. Aoki T, Takami M, Takatani T, Motoyoshi K, Ishii A, Hara A, Toyoda T, Okada R, Hino M et al. (2020) Activated invariant natural killer T cells directly recognize leukemia cells in a CD1d‐independent manner. Cancer Sci 111, 2223–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wingender G, Krebs P, Beutler B & Kronenberg M (2010) Antigen‐specific cytotoxicity by invariant NKT cells in vivo is CD95/CD178 dependent and is correlated with antigenic potency. J Immunol 185, 2721–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ni C, Wu P, Wu X, Zhang T, Liu Y, Wang Z, Zhang S, Qiu F & Huang J (2015) Thymosin alpha1 enhanced cytotoxicity of iNKT cells against colon cancer via upregulating CD1d expression. Cancer Lett 356, 579–588. [DOI] [PubMed] [Google Scholar]
- 40. Hix LM, Shi YH, Brutkiewicz RR, Stein PL, Wang CR & Zhang M (2011) CD1d‐expressing breast cancer cells modulate NKT cell‐mediated antitumor immunity in a murine model of breast cancer metastasis. PLoS One 6, e20702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lisbonne M, Hachem P, Tonanny MB, Fourneau JM, Sidobre S, Kronenberg M, Van Endert P, Dy M, Schneider E et al. (2004) In vivo activation of invariant Vα14 natural killer T cells by α‐galactosylceramide sequentially induces Fas‐dependent and ‐independent cytotoxicity. Eur J Immunol 34, 1381–1388. [DOI] [PubMed] [Google Scholar]
- 42. Metelitsa LS, Weinberg KI, Emanuel PD & Seeger RC (2003) Expression of CD1d by myelomonocytic leukemias provides a target for cytotoxic NKT cells. Leukemia 17, 1068–1077. [DOI] [PubMed] [Google Scholar]
- 43. Nicol A, Nieda M, Koezuka Y, Porcelli S, Suzuki K, Tadokoro K, Durrant S & Juji T (2000) Human invariant Vα24+ natural killer T cells activated by α‐galactosylceramide (KRN7000) have cytotoxic anti‐tumour activity through mechanisms distinct from T cells and natural killer cells. Immunology 99, 229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bassiri H, Das R, Guan P, Barrett DM, Brennan PJ, Banerjee PP, Wiener SJ, Orange JS & Brenner MB (2014) iNKT cell cytotoxic responses control T‐lymphoma growth in vitro and in vivo . Cancer Immunol Res 2, 59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dhodapkar KM, Cirignano B, Chamian F, Zagzag D, Miller DC, Finlay JL & Steinman RM (2004) Invariant natural killer T cells are preserved in patients with glioma and exhibit antitumor lytic activity following dendritic cell‐mediated expansion. Int J Cancer 109, 893–899. [DOI] [PubMed] [Google Scholar]
- 46. Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Sato H, Kondo E, Harada M, Koseki H et al. (1998) Natural killer‐like nonspecific tumor cell lysis mediated by specific ligand‐activated V14 NKT cells. Proc Natl Acad Sci USA 95, 5690–5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tachibana T, Onodera H, Tsuruyama T, Mori A, Nagayama S, Hiai H & Imamura M (2005) Increased intratumor Vα24‐positive natural killer T cells: a prognostic factor for primary colorectal carcinomas. Clin Cancer Res 11, 7322–7327. [DOI] [PubMed] [Google Scholar]
- 48. Dockry É, O'Leary S, Gleeson LE, Lyons J, Keane J, Gray SG & Doherty DG (2018) Epigenetic induction of CD1d expression primes lung cancer cells for killing by invariant natural killer T cells. OncoImmunology 7, e1428156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Engel I, Seumois G, Chavez L, Samaniego‐Castruita D, White B, Chawla A, Mock D, Vijayanand P & Kronenberg M (2016) Innate‐like functions of natural killer T cell subsets result from highly divergent gene programs. Nat Immunol 17, 728–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Crosby CM & Kronenberg M (2018) specific functions of invariant natural killer T cells. Nat Rev Immunol 18, 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang Y, Sedimbi S, Löfbom L, Singh AK, Porcelli SA & Cardell SL (2018) Unique invariant natural killer T cells promote intestinal polyps by suppressing TH1 immunity and promoting regulatory T cells. Mucosal Immunol 11, 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yoshioka K, Ueno Y, Tanaka S, Nagai K, Onitake T, Hanaoka R, Watanabe H & Chayama K (2012) Role of natural killer T cells in the mouse colitis‐associated colon cancer model. Scand J Immunol 75, 16–26. [DOI] [PubMed] [Google Scholar]
- 53. Caprioli F, Pallone F & Monteleone G (2008) Th17 immune response in IBD: A new pathogenic mechanism. J Crohn’s Colitis 2, 291–295. [DOI] [PubMed] [Google Scholar]
- 54. Burrello C, Pellegrino G, Giuffrè MR, Lovati G, Magagna I, Bertocchi A, Cribiù FM, Boggio F, Botti F et al. (2019) Mucosa‐associated microbiota drives pathogenic functions in IBD‐derived intestinal iNKT cells. Life Sci Alliance 2, e201800229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Singh AK, Gaur P & Das SN (2014) Natural killer T cell anergy, co‐stimulatory molecules and immunotherapeutic interventions. Hum Immunol 75, 250–260. [DOI] [PubMed] [Google Scholar]
- 56. Bedard M, Shrestha D, Priestman DA, Wang Y, Schneider F, Matute JD, Iyer SS, Gileadi U, Prota G et al. (2019) Sterile activation of invariant natural killer T cells by ER‐stressed antigen‐presenting cells. Proc Natl Acad Sci USA 116, 23671–23681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Smyth MJ, Wallace ME, Nutt SL, Yagita H, Godfrey DI & Hayakawa Y (2005) Sequential activation of NKT cells and NK cells provides effective innate immunotherapy of cancer. J Exp Med 201, 1973–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ghnewa YG, O'Reilly VP, Vandenberghe E, Browne PV, McElligott AM & Doherty DG (2017) Retinoic acid induction of CD1d expression primes chronic lymphocytic leukemia B cells for killing by CD8 + invariant natural killer T cells. Clin Immunol 183, 91–98. [DOI] [PubMed] [Google Scholar]
- 59. Georgiev H, Ravens I, Benarafa C, Förster R & Bernhardt G (2016) Distinct gene expression patterns correlate with developmental and functional traits of iNKT subsets. Nat Commun 7, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nizzoli G, Burrello C, Cribiù FM, Lovati G, Ercoli G, Botti F, Trombetta E, Porretti L, Todoerti K et al. (2018) Pathogenicity of in vivo generated intestinal Th17 lymphocytes is IFNγ dependent. J Crohns Colitis 12, 981–992. [DOI] [PubMed] [Google Scholar]
- 61. Hassin D, Garber OG, Meiraz A, Schiffenbauer YS & Berke G (2011) Cytotoxic T lymphocyte perforin and Fas ligand working in concert even when Fas ligand lytic action is still not detectable. Immunology 133, 190–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zamai L, Ahmad M, Bennett IM, Azzoni L, Alnemri ES & Perussia B (1998) Natural Killer (NK) cell–mediated cytotoxicity: differential use of TRAIL and Fas ligand by immature and mature primary human NK cells. J Exp Med 188, 2375–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Prager I, Liesche C, van Ooijen H, Urlaub D, Verron Q, Sandström N, Fasbender F, Claus M, Eils R et al. (2019) NK cells switch from granzyme B to death receptor–mediated cytotoxicity during serial killing. J Exp Med 216, 2113–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Prager I & Watzl C (2019) Mechanisms of natural killer cell‐mediated cellular cytotoxicity. J Leukoc Biol 105, 1319–1329. [DOI] [PubMed] [Google Scholar]
- 65. Facciotti F, Ramanjaneyulu GS, Lepore M, Sansano S, Cavallari M, Kistowska M, Forss‐Petter S, Ni G, Colone A et al. (2012) Peroxisome‐derived lipids are self antigens that stimulate invariant natural killer T cells in the thymus. Nat Immunol 13, 474–480. [DOI] [PubMed] [Google Scholar]
- 66. Fox LM, Cox DG, Lockridge JL, Wang X, Chen X, Scharf L, Trott DL, Ndonye RM, Veerapen N et al. (2009) Recognition of lyso‐phospholipids by human natural killer T lymphocytes. PLoS Biol 7, e1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Darmoise A, Teneberg S, Bouzonville L, Brady RO, Beck M, Kaufmann SHE & Winau F (2010) Lysosomal α‐galactosidase controls the generation of self lipid antigens for natural killer T cells. Immunity 33, 216–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Govindarajan S, Verheugen E, Venken K, Gaublomme D, Maelegheer M, Cloots E, Gysens F, De Geest BG, Cheng TY et al. (2020) ER stress in antigen‐presenting cells promotes NKT cell activation through endogenous neutral lipids. EMBO Rep 21, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Leite‐de‐Moraes MC, Hameg A, Machavoine F, Koezuka Y, Schneider E, Herbelin A & Dy M (1999) A distinct IL‐18‐induced pathway to fully activate NK T lymphocytes independently from TCR engagement. J Immunol 163, 5871–5876. [PubMed] [Google Scholar]
- 70. Huntington ND, Cursons J & Rautela J (2020) The cancer–natural killer cell immunity cycle. Nat Rev Cancer 20, 437–454. [DOI] [PubMed] [Google Scholar]
- 71. Patterson S, Chaidos A, Neville DCA, Poggi A, Butters TD, Roberts IAG & Karadimitris A (2008) Human invariant NKT cells display alloreactivity instructed by invariant TCR‐CD1d interaction and killer Ig receptors. J Immunol 181, 3268–3276. [DOI] [PubMed] [Google Scholar]
- 72. Kawamura T, Takeda K, Kaneda H, Matsumoto H, Hayakawa Y, Raulet DH, Ikarashi Y, Kronenberg M, Yagita H et al. (2009) NKG2A inhibits invariant NKT Cell activation in hepatic injury. J Immunol 182, 250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nguyen KD, Vanichsarn C & Nadeau KC (2008) Increased cytotoxicity of CD4+ invariant NKT cells against CD4+CD25hiCD127lo/‐ regulatory T cells in allergic asthma. Eur J Immunol 38, 2034–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hibi T, Ohara M, Watanabe M, Kanai T, Takaishi H, Hayashi A, Hosoda Y, Ogata H, Iwao Y et al. (1993) Interleukin 2 and interferon‐gamma augment anticolon antibody dependent cellular cytotoxicity in ulcerative colitis. Gut 34, 788–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Okumura G, Iguchi‐Manaka A, Murata R, Yamashita‐Kanemaru Y, Shibuya A & Shibuya K (2020) Tumor‐derived soluble CD155 inhibits DNAM‐1–mediated antitumor activity of natural killer cells. J Exp Med 217, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ostberg JR, Dayanc BE, Yuan M, Oflazoglu E & Repasky EA (2007) Enhancement of natural killer (NK) cell cytotoxicity by fever‐range thermal stress is dependent on NKG2D function and is associated with plasma membrane NKG2D clustering and increased expression of MICA on target cells. J Leukoc Biol 82, 1322–1331. [DOI] [PubMed] [Google Scholar]
- 77. Bae JH, Kim SJ, Kim MJ, Oh SO, Chung JS, Kim SH & Kang CD (2012) Susceptibility to natural killer cell‐mediated lysis of colon cancer cells is enhanced by treatment with epidermal growth factor receptor inhibitors through UL16‐binding protein‐1 induction. Cancer Sci 103, 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bhat R & Rommelaere J (2013) NK‐cell‐dependent killing of colon carcinoma cells is mediated by natural cytotoxicity receptors (NCRs) and stimulated by parvovirus infection of target cells. BMC Cancer 13, 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Veluchamy JP, Spanholtz J, Tordoir M, Thijssen VL, Heideman DAM, Verheul HMW, de Gruijl TD & van der Vliet HJ (2016) Combination of NK cells and cetuximab to enhance anti‐tumor responses in RAS mutant metastatic colorectal cancer. PLoS One 11, e0157830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bauman Y, Nachmani D, Vitenshtein A, Tsukerman P, Drayman N, Stern‐Ginossar N, Lankry D, Gruda R & Mandelboim O (2011) An identical miRNA of the human JC and BK polyoma viruses targets the stress‐induced ligand ULBP3 to escape immune elimination. Cell Host Microbe 9, 93–102. [DOI] [PubMed] [Google Scholar]
- 81. Nachmani D, Zimmermann A, Oiknine Djian E, Weisblum Y, Livneh Y, Le Khanh VT, Galun E, Horejsi V, Isakov O et al. (2014) MicroRNA editing facilitates immune elimination of HCMV infected cells. PLoS Pathog 10, e1003963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Seidel E, Le VTK, Bar‐On Y, Tsukerman P, Enk J, Yamin R, Stein N, Schmiedel D, Oiknine Djian E et al. (2015) Dynamic co‐evolution of host and pathogen: hcmv downregulates the prevalent allele MICA∗008 to escape elimination by NK cells. Cell Rep 10, 968–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lanuza PM, Vigueras A, Olivan S, Prats AC, Costas S, Llamazares G, Sanchez‐Martinez D, Ayuso JM, Fernandez L et al. (2018) Activated human primary NK cells efficiently kill colorectal cancer cells in 3D spheroid cultures irrespectively of the level of PD‐L1 expression. OncoImmunology 7, e1395123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Santaguida S, Richardson A, Iyer DR, M'Saad O, Zasadil L, Knouse KA, Wong YL, Rhind N, Desai A et al. (2017) Chromosome mis‐segregation generates cell‐cycle‐arrested cells with complex karyotypes that are eliminated by the immune system. Dev Cell 41, 638–651.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lee LH, Cavalcanti MS, Segal NH, Hechtman JF, Weiser MR, Smith JJ, Garcia‐Aguilar J, Sadot E, Ntiamoah P et al. (2016) Patterns and prognostic relevance of PD‐1 and PD‐L1 expression in colorectal carcinoma. Mod Pathol 29, 1433–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Galon J, Costes A, Sanchez‐Cabo F, Kirilovsky A, Mlecnik B, Lagorce‐Pages C, Tosolini M, Camus M, Berger A et al. (2006) Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313, 1960–1964. [DOI] [PubMed] [Google Scholar]
- 87. Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L et al. (2013) Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39, 782–795. [DOI] [PubMed] [Google Scholar]
- 88. O’Donnell JS, Teng MWL & Smyth MJ (2019) Cancer immunoediting and resistance to T cell‐based immunotherapy. Nat Rev Clin Oncol 16, 151–167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Cell lines used in the study.
Table S2. List of iNKT cell lines generated for functional studies.
Table S3. List of antibodies used for flow cytometry analysis.
Table S4. Antibodies used for ELISA assays.
Fig. S1. Flow cytometry analyses conducted in this study.
Fig. S2. iNKT cell lines are cytotoxic against colon cancer cell lines.
Fig. S3. Cytotoxicity effectors on NK cells.
Fig. S4. Effect of cytotoxicity mechanism and CD1d inhibition on cytotoxic mediators.
Fig. S5. iNKT cell killing of patient‐derived CRC cells.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


