Abstract
The paucity of microbiome studies at intestinal tissues has contributed to a yet limited understanding of potential viral and bacterial cofactors of colorectal cancer (CRC) carcinogenesis or progression. We analysed whole‐genome sequences of CRC primary tumours, their corresponding metastases and matched normal tissue for sequences of viral, phage and bacterial species. Bacteriome analysis showed Fusobacterium nucleatum, Streptococcus sanguinis, F. Hwasookii, Anaerococcus mediterraneensis and further species enriched in primary CRCs. The primary CRC of one patient was enriched for F. alocis, S. anginosus, Parvimonas micra and Gemella sp. 948. Enrichment of Escherichia coli strains IAI1, SE11, K‐12 and M8 was observed in metastases together with coliphages enterobacteria phage φ80 and Escherichia phage VT2φ_272. Virome analysis showed that phages were the most preponderant viral species (46%), the main families being Myoviridae, Siphoviridae and Podoviridae. Primary CRCs were enriched for bacteriophages, showing five phages (Enterobacteria, Bacillus, Proteus, Streptococcus phages) together with their pathogenic hosts in contrast to normal tissues. The most frequently detected, and Blast‐confirmed, viruses included human endogenous retrovirus K113, human herpesviruses 7 and 6B, Megavirus chilensis, cytomegalovirus (CMV) and Epstein–Barr virus (EBV), with one patient showing EBV enrichment in primary tumour and metastases. EBV was PCR‐validated in 80 pairs of CRC primary tumour and their corresponding normal tissues; in 21 of these pairs (26.3%), it was detectable in primary tumours only. The number of viral species was increased and bacterial species decreased in CRCs compared with normal tissues, and we could discriminate primary CRCs from metastases and normal tissues by applying the Hutcheson t‐test on the Shannon indices based on viral and bacterial species. Taken together, our results descriptively support hypotheses on microorganisms as potential (co)risk factors of CRC and extend putative suggestions on critical microbiome species in CRC metastasis.
Keywords: bacteriome, colorectal cancer, metastasis, phages, viruses
This work analysed the presence of sequences of bacteria and viruses in primary colorectal cancer (CRC) tissues, compared with corresponding normal colorectal tissues and metastases. We observed a higher viral species diversity and lower bacterial species diversity in primary CRCs than in matched normal tissues. Some microbial species were enriched in primary CRCs and some metastases, particularly phages.
Abbreviations
- AcMNPV
autographa californica nucleopolyhedrovirus
- CD
Crohn’s disease
- CMV
cytomegalovirus
- CRC
colorectal cancer
- EBV
Epstein–Barr virus
- EMCV
encephalomyocarditis virus
- HERV‐K113
human endogenous retrovirus K113
- HHV‐6B
human herpesvirus 6B
- HHV‐7
human herpesvirus 7
- HPV
human papillomavirus
- HPyV7
human polyomavirus 7
- IBD
inflammatory bowel disease
- JCV
JC virus
- SV40
simian virus 40
- UC
ulcerative colitis
- VHD
Virus‐Host Database
- WGS
whole‐genome sequencing
1. Introduction
Colorectal cancer (CRC) is the third most prevalent form of cancer worldwide, with a global incidence of over 1.4 million cases annually [1]. In Europe, it represents the second most frequent cause of death among people who have cancer and its incidence is increasing among young adults, who are not included yet in screening campaigns [2, 3]. About 3–5% of CRC cases show hereditary transmission [4, 5, 6], implying that the vast majority of patients develop this disease by accumulating molecular changes and further CRC‐promoting factors during their lifetime. Every person has a four per cent lifetime risk of developing sporadic CRC [7] but several studies have indicated the possibility of a microbial cofactor to increase such a risk [8, 9, 10]. In particular, it has been proposed that some bacterial species (microbial ‘drivers’), which carry genes encoding proteins that can induce chromosomal instability, might be able to initiate the oncogenic cascade in the intestinal cells [11, 12, 13]. Subsequently, other opportunistic bacteria (‘passengers’) could become more prevalent in this pretumour microenvironment, boosting inflammation and fostering oncogenesis or even progression. So far existing studies differ in drivers and passenger definitions, but the species Fusobacterium nucleatum, Bacteroides fragilis, Streptococcus bovis and Enterococcus faecalis are the most frequently cited [14].
Moreover, viruses could play a yet underestimated role in the carcinogenic process. In the past decades, several studies have pointed out an increased prevalence of viral infections in patients suffering from CRC or associated inflammatory conditions, such as inflammatory bowel disease (IBD), which include Crohn’s disease (CD) and ulcerative colitis (UC) [15]. IBD is a risk factor for CRC whose prevalence is currently increasing worldwide also [16, 17]. Among the viruses most frequently investigated for their possible association with CRC and IBD are Epstein–Barr virus (EBV, also known as human herpesvirus type 4, HHV‐4), cytomegalovirus (also known as human herpesvirus type 5, HHV‐5), human papillomavirus (HPV) and several members of the Polyomaviridae family [18, 19, 20, 21]. Nevertheless, a causative involvement of viruses in CRC oncogenesis or progression is still unclear [21].
Recent microbiome work has expanded beyond human‐infecting viruses and has highlighted, for example, the importance of bacteria‐infecting viruses (phages) in CRC and IBD. For instance, an increased prevalence of phage species belonging to the order of Caudovirales (which includes the families Siphoviridae, Myoviridae and Podoviridae) has been observed in patients affected by these diseases [22, 23, 24, 25]. Phages can indirectly contribute to CRC development by modulating the prevalence of driver and passenger bacteria [26, 27, 28, 29, 30, 31, 32]. Diet is also crucial in this context because it can alter the bacterial composition and, consequently, that of the phages [33]. This association can, at least in part, explain the epidemiological link observed between a western type of nutrition, which is usually poor in fibres but rich in fats and red meat, and a higher risk of CRC [10]. Furthermore, most microbiome studies have been based upon faecal samples, which can be considered a proxy to the actual microbial community present within the intestinal tissue. Since mucus covers the intestine walls, acting as a barrier to the microorganisms [34, 35], faecal and tissue specimens do not necessarily bear the same microbial species [36]. The few microbiome studies based on tumour tissues have shown a higher prevalence of phages in inflammatory lesions than in normal matched sections [37, 38].
The paucity of microbiome studies based on intestine tissues has contributed to the poor understanding of the conditions that foster CRC. Specifically, there is certainly a lack of research based on metastases. About one‐quarter of CRC cases progress towards metastasis [39, 40]; thus, understanding whether microorganisms modulate this progression could help to determine cofactors and consequently improve the diagnosis and treatment of metastases.
In this study, we (re‐)analysed whole‐genome sequences (WGS), which we generally described in previous work [41], to assess whether we could detect the enrichment of particular microbial sequences in primary colorectal carcinomas in comparison to matched control tissues, and whether metastases show additional microbial sequences that could potentially set them aside from colorectal primary tumours. This metagenomic analysis might help not only to further understand, or suggest, putative microbial cofactors in the oncogenic and metastatic process, but support in the diagnosis or even risk prediction for CRC carcinogenesis or metastasis.
2. Materials and methods
2.1. Tissues
All of the samples were completely anonymized and handled exclusively in strictly anonymized conditions. Tissues had been generally obtained from the biobank of the Medical Faculty Mannheim (Dept. of Surgery), University of Heidelberg, Germany, and the biobanking and tissue sampling approved by the Ethical Committee of this institution. The primary colorectal tumours, matched healthy colorectal tissues, and corresponding metastases re‐analysed here were described previously [41], the analysis having been undertaken with the understanding and written consent of each subject (or relatives if deceased), and the Declaration of Helsinki was followed. Samples were processed and anonymized data stored within the EGA database (accession number EGAS00001002717) as already described [41]. Genomic DNA had been isolated from frozen tissue sections with the QIAamp DNA mini kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. The extracted DNA had been quality controlled and sequenced as described previously [41] at the Genomics and Proteomics Core Facility of the German Cancer Research Center (DKFZ), Heidelberg, Germany, on the Illumina HiSeq2000 platform.
2.2. Virome analysis
Genomic sequences from the human assembly issue 38 built 12 (GChr38) were obtained from the Ensembl database. Viral reference sequences (n = 10 384) were retrieved from the National Center for Biotechnology Information (NCBI) database using Entrez utilities (https://www.ncbi.nlm.nih.gov/books/NBK25497/) and were concatenated into a single sequence (viral chromosome) using mingle v. 2.3 [42]. Human and viral chromosomes were combined into a single fusion reference genome indexed with bwa v. 0.7.17 [43].
Sequencing adapters and reads with quality below 33 phred‐score units were removed with trimmomatic v. 0.38 [44, 45, 46]. The remaining reads were aligned against the composite human/virus reference genome with bwa‐mem v. 0.7.17 [47]. De‐duplication was carried out with sambamba v. 0.6.7 [48], and only reads mapping to the virus chromosome, and with mapping quality of at least ten phred‐score units, were retained. The starting point of the remaining reads was used in combination with the index file provided by Mingle to determine the corresponding viral species. Reads mapping to phage φX174, which was used as quality control in the libraries’ preparation, were discarded.
The quality of the alignment and sample coverage was assessed with samtools v. 1.9‐18 and qualimap v. 2.2.1 [49, 50]. blast+ v. 2.9.0 [51, 52] was used to align the remaining reads against the human chromosome, the composite viral chromosome alone, or the individual virus genomes. Reads that aligned preferably to the human genome were removed. For each sample, overlapping reads belonging to the same viral species were merged into a single consensus sequence using clustal omega v. 1.2.4 [53]. To further remove possible false‐positive results, these consensuses were aligned against the NCBI protein database with blastx v. 2.9.0 [54]. Sequences identified as nonviral were discarded. For each specimen, remaining consensus sequences belonging to the same virus were concatenated into a single viral species. Structural variation (SV) was investigated with delly v. 0.8.3 [55].
In a series of 80 colorectal primary tumours and corresponding normal colorectal tissues, EBV DNA was detected as a validation by end‐point PCR targeting EBNA1 as previously described [56]. Briefly, total cellular RNA was isolated with RNeasy Mini Kit (Qiagen) according to the manufacturer’s instruction and the cDNA generated using SuperScript III (Thermo Fisher Scientific, Karlsruhe and Dreieich, Germany) as recommended by the manufacturer. Amplification of the cDNA was accomplished with the Taq polymerase kit provided by Thermo Fisher Scientific (Cat. No. EP0401) with the following conditions: 2.5 μL of reaction buffer 10×, 2 mm of each dNTP, 1 μm of forward primer (5´‐CCGCTCCTACCTGCAATATCA‐3´), 1 μm of reverse primer (5´‐CAATAACGGCAGCAAGCTTG −3´), 1 mm MgCl2, 1.25 units of enzyme, 100 ng template DNA and water to 25 μL. The amplification conditions were as follows: 5 min at 95 °C followed by 30 cycles of 1 min at 95 °C, 1 min at 58 °C, 1 min at 72 °C and final extension for 7 min at 72 °C.
To address the question of cell types primarily affected within these tissues, the presence of EBV was detected by in situ hybridization with an EBER‐specific peptide nucleic acid probe, in conjunction with a PNA detection kit (Dako) following the manufacturer’s protocol as described [57]. Images were taken with a camera attached to a light microscope (M2500; Leica).
2.3. Bacteriome analysis
Reads not mapping to either the human chromosomes or the composite virus chromosomes were assembled with spades v. 3.13.0 [58]. Sequence assemblies smaller than 1000 base pairs were discarded; assembled sequences identified as metazoan, artificial or environmental sequences were removed. The classification of the obtained sequences was kept at the species level and subspecies and strains were merged together. The bacteria‐phage infection network was assessed by inquiring the Virus‐Host Database (VHD) [59]. Classification of phages as lytic or virulent was based on the PhageAI database [60].
2.4. Statistical analysis
Data processing was performed with julia v. 1.3.1 (https://julialang.org/) and bash v. 5.0.3 languages. Plotting, statistical analysis and hierarchical clustering were performed with r v. 3.6.1 [61]. The Shannon index (or, more appropriately, Shannon‐Wiener index or entropy) [62] and its variance were computed with the r package Qsutils [63]. Species richness analysis [64, 65] was obtained with the r package iNEXT [66]. Statistical comparison between sample groups was carried out by paired t‐test. The Hutcheson t‐test [67] was used to compare Shannon indices, and it was implemented in r with the package ecoltest (https://github.com/hugosal/ecolTest).
3. Results
3.1. Virome analysis
Twelve cases previously described [41] with matched primary colorectal tumours, corresponding liver (or lung in one case) metastasis and normal colorectal tissues were available for the present analysis. After sequencing, the subset of reads mapping to viral genomes had a mean coverage of 28.33 ± 16.21 (Table S1). We initially identified 662 viral species of which 272 (41.1%) were phagial with the families Myoviridae (n = 154), Siphoviridae (n = 79) and Podoviridae (n = 29) as the most represented (Fig. 1, Table 1). The most prevalent species were human endogenous retrovirus K113 (HERV‐K113), Synechococcus phage S‐SM2, Enterobacteria phage λ and autographa californica nucleopolyhedrovirus (AcMNPV). Among the most represented DNA viruses, human herpesvirus 7 (HHV‐7) was observed in eight, and HHV‐6B in six specimens, besides Ebstein‐Barr virus (EBV, HHV‐4) and Cytomegalovirus (CMV or HHV‐5). Several genotypes of torque teno (TT) virus were identified: 5, 9, 16, 24, 7, 11 and 5. One sample showed sequences of simian virus 40 (SV40) and another bore human polyomavirus 7 (HPyV7). We also identified several giant viruses such as Pandoravirus salinus and dulcis, Megavirus chiliensis, Cafeteria roenbergensis and Acanthamoeba polyphaga. Among the phages, we observed the uncultured crAssphage in six tissues. There were also several RNA species (n = 189), including the aforementioned HERV‐K113 (an endogenous retrovirus) and encephalomyocarditis virus (EMCV). There were 73, 404 and 24 viral species present either in normal tissues, primary colorectal tumours, or metastases only, respectively, and an additional three (27.3%) were common to both primary colorectal tumours and metastases: EBV, Qinghai Himalayan marmot astrovirus and Tipula oleracea nudivirus. EBV was present in the lung metastasis together with AcMNPV, Synechococcus phage S‐SM2 and HERV‐K113.
Fig. 1.
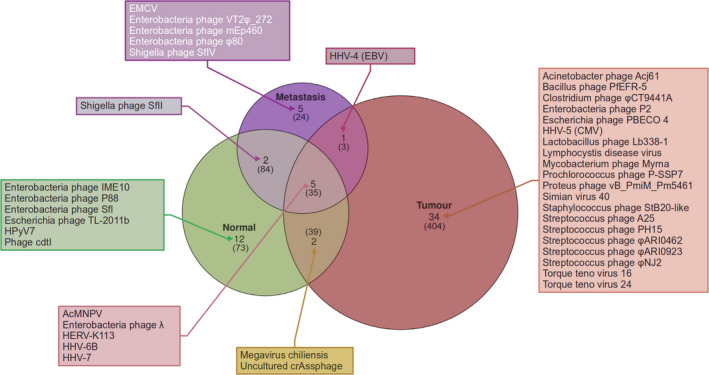
Stratification of viral species sequences observed in the present sample set. Venn diagram showing the number of virus species present within each tissue type (normal tissue, primary tumour and metastasis) based on the Blast‐filtered data. The number of species observed prior to this filtering step is given in parentheses. For each group, the most representative species are reported.
Table 1.
Selection of the most represented viral species based on raw data (reads aligned only by BWA‐MEM) and assignment to their respective families.
| Group | Virus | Family | Type |
|---|---|---|---|
| Normal, tumour and metastasis | HERV‐K113 | Retroviridae | Endogenous retrovirus |
| AcMNPV | Baculoviridae | DNA virus | |
| Enterobacteria phage λ | Siphoviridae | Bacteriophage | |
| Synechococcus phage S‐SM2 | Myoviridae | Bacteriophage | |
| Escherichia phage TL‐2011b | Podoviridae | Bacteriophage | |
| Pandoravirus neocaledonia | Pandoraviridae | Giant virus | |
| HHV‐7 | Herpesviridae | DNA virus | |
| Cafeteria roenbergensis virus | Mimiviridae | Giant virus | |
| Pandoravirus salinus | Pandoraviridae | Giant virus | |
| Phage cdtI DNA | Siphoviridae | Bacteriophage | |
| HHV‐6B | Herpesviridae | DNA virus | |
| Pandoravirus dulcis | Pandoraviridae | Giant virus | |
| Normal and metastasis | Torque teno midi virus 5 | Anelloviridae | DNA virus |
| Torque teno midi virus 9 | Anelloviridae | DNA virus | |
| Encephalomyocarditis virus | Picornaviridae | RNA virus | |
| Hepatitis C virus genotype 1 | Flaviviridae | RNA virus | |
| Enterobacteria phage VT2φ_272 | Podoviridae | Bacteriophage | |
| Shigella phage SfII | Myoviridae | Bacteriophage | |
| Escherichia phage pro483 | Myoviridae | Bacteriophage | |
| Shigella phage SfIV | Myoviridae | Bacteriophage | |
| Enterobacteria phage mEp460 | Siphoviridae | Bacteriophage | |
| Normal and tumour | Megavirus chiliensis | Mimiviridae | Giant virus |
| Uncultured crAssphage | Unassigned | Bacteriophage | |
| Aeromonas phage PX29 | Myoviridae | Bacteriophage | |
| Enterobacteria phage P88 | Myoviridae | Bacteriophage | |
| Enterobacteria phage P2 | Myoviridae | Bacteriophage | |
| Acanthamoeba polyphaga mouvirus | Mimiviridae | Giant virus | |
| Tumour only | CMV | Herpesviridae | DNA virus |
| Streptococcus phage φARI0462 | Adenoviridae | Bacteriophage | |
| Bacillus phage PfEFR‐5 | Siphoviridae | Bacteriophage | |
| Proteus phage vB_PmiM_Pm5461 | Myoviridae | Bacteriophage | |
| Streptococcus phage φARI0923 | Siphoviridae | Bacteriophage | |
| Simian virus 40 | Polyomaviridae | DNA virus | |
| Acinetobacter phage Acj61 | Myoviridae | Bacteriophage | |
| Clostridium phage phiCT9441A | Myoviridae | Bacteriophage | |
| Escherichia phage PBECO 4 | Myoviridae | Bacteriophage | |
| Lactobacillus phage Lb338‐1 | Herelleviridae | Bacteriophage | |
| Lymphocystis disease virus | Iridoviridae | DNA virus | |
| Mycobacterium phage Myrna | Myoviridae | Bacteriophage | |
| Prochlorococcus phage P‐SSP7 | Autographiviridae | Bacteriophage | |
| Staphylococcus phage StB20‐like | Siphoviridae | Bacteriophage | |
| Streptococcus phage A25 | Siphoviridae | Bacteriophage | |
| Streptococcus phage PH15 | Siphoviridae | Bacteriophage | |
| Streptococcus phage phiNJ2 | Siphoviridae | Bacteriophage | |
| Torque teno virus 16 | Anelloviridae | DNA virus | |
| Torque teno virus 24 | Anelloviridae | DNA virus | |
| Metastasis only | Enterobacteria phage HK629 | Siphoviridae | Bacteriophage |
| Enterobacteria phage HK97 | Siphoviridae | Bacteriophage | |
| Enterobacteria phage M13 | Inoviridae | Bacteriophage | |
| Enterobacteria phage P1 | Myoviridae | Bacteriophage | |
| Enterobacteria phage φ80 | Siphoviridae | Bacteriophage | |
| Tumour and metastasis | EBV | Herpesviridae | DNA virus |
| Tipula oleracea nudivirus | Nudiviridae | DNA virus | |
| Qinghai Himalayan marmot astrovirus | Astroviridae | RNA virus | |
| Normal only | Enterobacteria phage IME10 | Podoviridae | Bacteriophage |
| Enterobacteria phage SfI | Myoviridae | Bacteriophage | |
| Human polyomavirus 7 | Polyomaviridae | DNA virus |
To avoid the possibility of false‐positive detection, we further filtered the data with the highly sensitive Blast alignment (see Materials and methods, [51, 52]). After Blast filtering, we found 61 viral species across all of the tissue entities that passed the threshold, corresponding to 9.2% of the initial species identified, of which 28 (45.9%) were phages, with the families Siphoviridae (n = 11), Myoviridae (n = 10) and Podoviridae (n = 7) as the most represented (Fig. 1). Of the 28 phagial species, 10 (35.7%) had E. coli as a host (coliphages) and 15 (53.6%) were observed in primary colon tumours. Stratification of the phagial species by tissue type showed that one species (Enterobacteria phage λ) was common to all tissues types, one (Shigella phage SfII) was common to normal colorectal tissues and liver metastases. Enterobacteria phage VT2φ_272 and enterobacteria phage φ80, Shigella phage SfIV and enterobacteria phage mEp460 were confirmed specifically in one metastasis each. An additional 16 species were present only in primary carcinomas, and six were observed only in normal colorectal tissues. Overall, 87.5%, 70.6% and 100.0% of phages in in normal tissues, primary colorectal tumours and liver metastases were temperate, respectively. Filtering confirmed HERV‐K113, AcMNPV, phage λ (but not Synechococcus phage S‐SM2) as the most preponderant viral species across all tissue entities. HHV‐7 was present collectively in five patients, and specifically in the normal colorectal tissue and primary colorectal carcinomas of two patients as well as in the primary colorectal carcinoma and liver metastasis of another patient. HHV‐6B was confirmed collectively in four patients, and specifically in the normal colon tissue and primary colorectal carcinoma of one patient as well as the normal colon tissue and liver metastasis of another patient. CMV was present specifically in the primary colorectal carcinoma tissues of two patients. Torque teno (TT) viruses 16 and 24 were also confirmed in primary colorectal carcinomas. We also confirmed SV40 and HPyV7 in one case each (in a primary colorectal tumour and one normal colon tissue, respectively). EMCV was specifically present in liver metastasis tissue. Of the aforementioned giant viruses, only megavirus chiliensis was confirmed in two patients. The phage crAssphage was confirmed in the normal colon tissues and the primary colorectal tumours of three patients. EBV was confirmed after BLAST filtering in the primary colorectal tumour and the lung metastasis of the same patient. Himalayan marmot astrovirus and Tipula oleracea nudivirus were not confirmed in the lung metastasis after Blast filtering.
Given the recognized oncogenic potential of EBV, we further investigated, and sought to validate, the frequency of presence of EBV by PCR on a group of 80 independent primary colorectal tumour and matched corresponding normal colorectal tissues (Table 2). Of 41 tumour samples positive in PCR, in 21 cases (51.2%) EBV was detectable specifically in the primary colorectal carcinomas but not in corresponding normal tissues, with a significant paired t‐test (P = 0.048). To gain additional information on putative cell types infected with EBV within colorectal carcinoma tissues, the presence of EBV was further investigated in a subset of 5 samples using EBER staining (ISH) (representative examples shown in Fig. 2). EBER staining of individually positive EBV‐cases (in PCR) showed positivity for the ISH probe in isolated cells with a small nucleus, which were tightly associated with neoplastic CRC glands. These images suggest infection of cancer‐associated lymphocytes, without definite evidence for epithelial cell infection, although, in a few instances, staining appeared projected onto single epithelial cells.
Table 2.
Prevalence of EBV by PCR, n = 80 validation cases.
| Tissue entity | Presence of EBV | |
|---|---|---|
| Yes | No | |
| Primary tumour | 41 | 39 |
| Normal tissue | 30 | 50 |
Fig. 2.
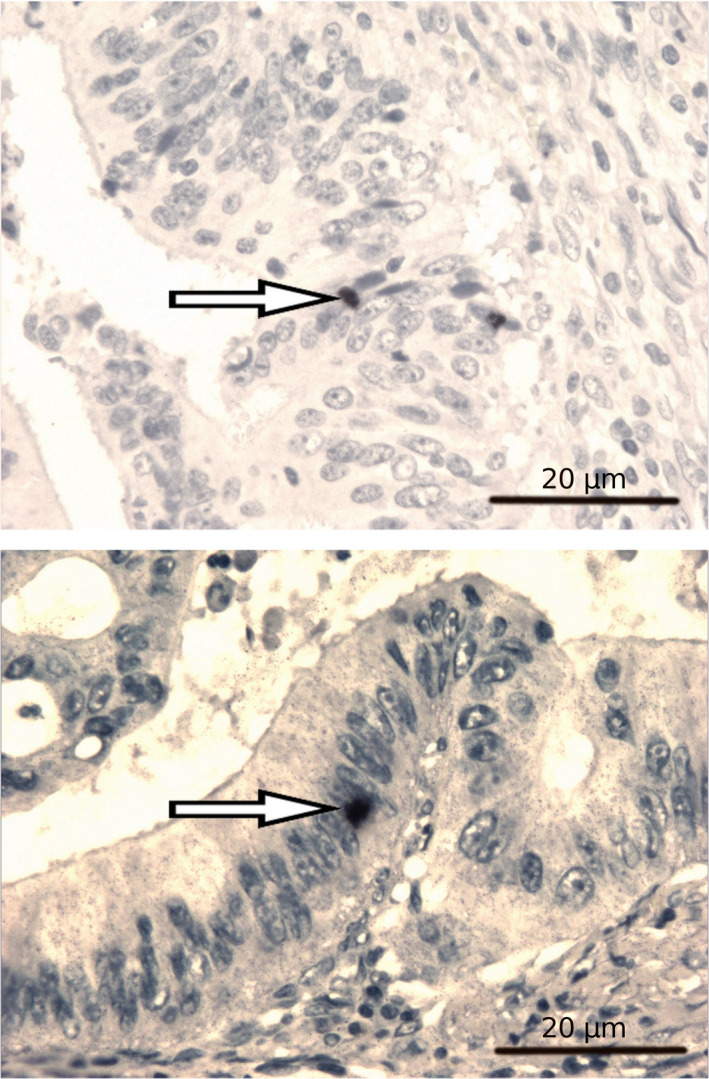
Visualization of EBV in primary colorectal cancer tissues with EBER staining. The pictures show an in situ hybridization with an Epstein–Barr expression region (EBER)‐specific probe performed on histological sections. EBV‐infected cells are depicted in black (examples shown with arrows). Magnification 400×, scale bar 20 μm.
Next, we sought to determine whether we could differentiate primary colorectal tumour tissues, corresponding normal colorectal tissues, and metastases based on the viral content. We used rarefaction analysis to calculate the estimated number of species normalized for the sample size, and the Shannon index to have a comparative value. Overall, there were an estimated 124 (95% CI: 60–296), 232 (95% CI: 99–674) and 57 (95% CI: 26–157) normalized viral species, in the normal, primary colorectal tumour and metastatic tissues, respectively (Fig. 3). The respective species richness corresponded to Shannon indices of 2.56, 3.31 and 1.99, respectively. The Hutcheson t‐test allowed to discriminate the normal colorectal tissues from the primary colorectal tumours (P‐value < 0.001) and either liver or lung metastases (P‐value = 0.006) as well as primary colorectal tumours from either liver or lung metastases (P‐value < 0.001).
Fig. 3.
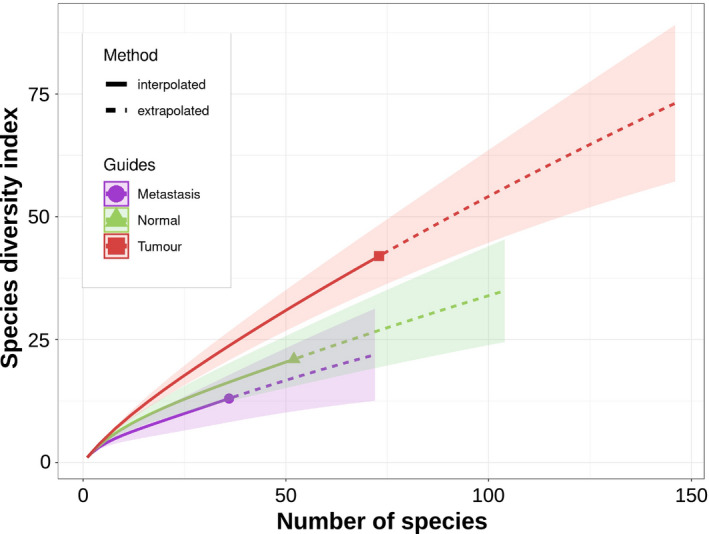
Viral richness. Rarefaction analysis of the viral species normalized for the sample size. Rarefaction is a bootstrap method that allows the direct comparison of samples by giving a count of species normalized for the sample size. The curves represent the mean measures of the number of species identified during the sampling process; the shaded areas depict the 95 confidence intervals of the measurements. Subspecies and strains were merged within the same species.
3.2. Bacteriome analysis
We identified 518 bacterial species, corresponding to 143 bacterial families in our sample set. Overall, there were 391, 217 and 10 bacterial species in normal colorectal tissues, primary colorectal tumours and liver metastases, respectively, corresponding to a ratio of phages over bacteria of 0.02, 0.08 and 0.60. There were 122 species present only in primary colon tumour tissues, four strains of Escherichia coli specifically in liver metastasis (IAI1, K‐12, M8 and SE11) and Klebsiella pneumoniae concomitantly in primary colon tumour and liver metastasis. There were no bacterial species observed in the lung metastatic tissue. We found Fusobacterium nucleatum enriched only in the primary colorectal tumour sections of three patients in contrast to corresponding normal colorectal tissues, whereas Fusobacterium hwasookii was present in the primary colorectal tumour sections of two patients. Porphyromonas gingivalis was present in the normal colorectal tissue and primary colon carcinoma sections of one patient. Anaerococcus mediterraneensis and Prevotella denticola were observed in the primary colorectal tumours of two patients, as opposed to corresponding normal tissues. Among the 122 species observed only in primary colorectal tumours, the families Bacillaceae and Clostridiaceae represented the most represented, with 20 species each (16.4%), followed by the Streptococcaceae with 17 species (13.9%). Bacteroides fragilis was prevalent in normal colorectal tissues and also present in primary colorectal tumour sections. Streptococcus anginosus was observed in the normal colorectal tissue of one patient, and in the primary colorectal tumours as opposed to their matched normal tissues of two patients. Furthermore, among the species observed only in primary colorectal tumours, S. sanguinis, Filifactor alocis, Gemella sp. oral taxon 928 and Parvimonas micra were present in high frequency in one patient and were represented by a number of sequence assemblies in the range of 468–147, compared to a mean of 5.7, indicating their highly specific representation.
The normalized species count, obtained by rarefaction analysis, was 2370 (95% CI: 1701–3383), 1011 (95% CI: 693–1543) and 40 (95% CI: 14–209) for the normal, primary colorectal carcinoma and metastatic tissues (Fig. 4), corresponding to Shannon indices of 5.87, 5.27 and 1.95, respectively. The Hutcheson t‐test allowed discriminating normal tissues from both primary colorectal tumours (P‐value < 0.001) and metastases (P‐value < 0.001); similarly, primary colorectal tumours could be discriminated from metastases (P‐value < 0.001).
Fig. 4.
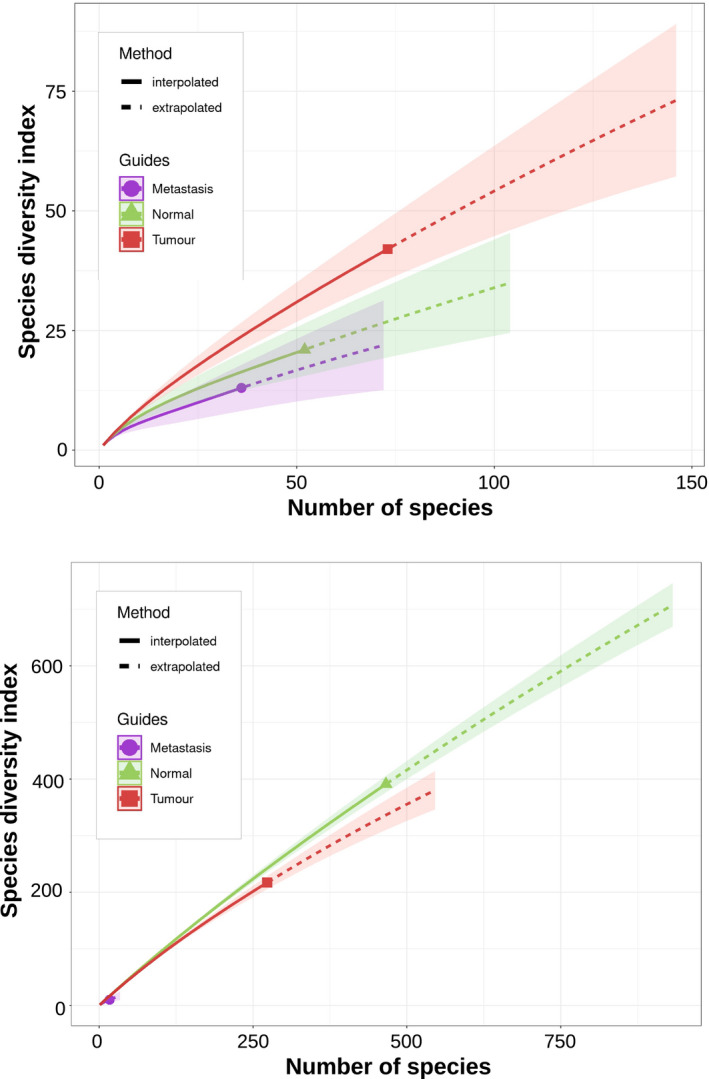
Bacterial richness. Rarefaction analysis of the bacterial species normalized for the sample size. Rarefaction is a bootstrap method that allows the direct comparison of samples by giving a count of species normalized for the sample size. The number of bacterial species in the metastases is much less than in normal colorectal and colorectal carcinoma tissues, reducing the curve to close to the coordinates 0, 0. The curves represent the mean measures of the number of species identified during the sampling process; the shaded areas depict the 95 confidence intervals of the measurements. Subspecies and strains were merged within the same species.
We further investigated the relationship between phages and bacteria by enumerating them in the tissues. The phagial hosts of the observed phages were derived from the Virus‐Host Database (VHD). Our results indicated a certain level of parallelism between the presence of the host and that of the associated phage (Table 3). Half of the eight phages observed most frequently had E. coli as a host. Enterobacteria phage λ was observed in all tissue types with a peak in the primary colorectal tumour sections. Enterobacteria phage P2 and φ80 were observed concomitantly with their host E. coli in one primary colorectal carcinoma and one liver metastasis, respectively. The primary tumour tissues also contained Bacillus phage PfEFR‐5 together with its host Bacillus cereus, as well as Streptococcus phage φARI0462 and φARI0923 together with their host Streptococcus pneumoniae.
Table 3.
Parallelism between phages and their hosts. The simultaneous presence of sequences belonging to phages and their host within the same patient is reported.
| Phage | Host | ||
|---|---|---|---|
| Enterobacteria phage λ | (N, T, M) | Escherichia coli | (N, T, M) |
| Enterobacteria phage P88 | (N) | Escherichia coli | (N) |
| Enterobacteria phage P2 | (T) | Escherichia coli | (T) |
| Enterobacteria phage φ80 | (M) | Escherichia coli | (M) |
| Bacillus phage PfEFR‐5 | (T) | Bacillus cereus | (T) |
| Proteus phage vB_PmiM_Pm5461 | (T) | Proteus mirabilis | (T) |
| Streptococcus phage phiARI0462 | (T) | Streptococcus pneumoniae | (T) |
| Streptococcus phage phiARI0923 | (T) | Streptococcus pneumoniae | (T) |
M, liver metastasis; N, normal colon tissue; T, primary colon tumour.
4. Discussion
The present work is one of the very few, if not the first so far, that carried out microbiome analysis on primary colorectal carcinoma and corresponding metastases in comparison to matched normal tissues. Although, certainly, our analysis from whole genomes so far is largely bioinformatic, our study suggests that some microorganisms, at least their sequences, are more prevalent in some primary colorectal tumours or metastases, this pertaining to viruses and bacteria, and—potentially most interesting since these were not yet systematically investigated in such a setting—(bacterio‐)phages.
In both primary colorectal cancers and metastases, we observed several species of phages, for example phage λ or crAssphage, crAssphage having been reported as the most common virus in the colon [68, 69, 70]. The high prevalence of phages in general and, specifically, their enrichment in primary colorectal tumours matches previous studies indicating an increased prevalence of phages in CRC and IBD [22, 23, 24, 25, 37, 38]. The majority (over 45%) of the phages we observed had E. coli as a host, for example enterobacteria phage φ80 and Escherichia phage VT2φ 272 which, after Blast filtering, we even found specifically in metastatic tissues. We observed over half of the most frequent phages especially in primary tumours, two of which (Bacillus prophage PfEFR‐5 and Streptococcus phage φARI0462) showing their specific host (Bacillus cereus and Streptococcus pneumoniae, respectively) in primary tumour tissues only, not in corresponding normal tissues, which suggests that this might not have been due to contamination. This is hypothesized since the primary colorectal cancers and corresponding normal colorectal tissues were both acquired simultaneously, being exposed to the same microbiologic milieu of the same intestinal lumen within the same patient under the same settings, and therefore, they would have been exposed to the same contaminants. Interestingly, some of the hosts of the phages specifically observed in primary colorectal tumour tissues are known pathogens. For instance, Bacillus prophage PfEFR‐5 infects Bacillus cereus, a pathogen associated with food poisoning [71]. The observation of B. cereus and S. pneumoniae in tumour samples only, in contrast to the corresponding normal colorectal tissue of the very same patient, suggested a sort of ‘linkage disequilibrium’ between these tissue types.
The bacterial species we observed enriched, or exclusively, in primary colorectal carcinomas have been reported to show oncogenic or pro‐inflammatory potential. Our tumour‐specific findings of F. nucleatum, S. anginosus and F. alocis, S. sanguinis, P. micra and Gemella oral taxon 928 confirm the results of other research groups [72, 73, 74, 75, 76, 77, 78, 79, 80]. In particular, a recent microbiome analysis of CRC tissues found the species F. nucleatum, S. anginosus and S. sanguinis at high prevalence in cancer tissues (false discovery rate lower than 105) [81]. Fusobacterium spp. are included in the passenger/driver model of CRC and have been reported to promote the formation of metastases [11, 82]. F. nucleatum and P. micra are commonly enriched in CRC sections [83, 84, 85], and a recent microbiome study reported the enrichment of Fusobacterium hwasookii and Porphyromonas gingivalis in colorectal cancer tissues, together with high prevalence of Fusobacteria and Bacteroidetes [86]. F. alocis and P. micra are prevalent at sites of periodontitis (a chronic inflammation of the gum that results in tooth loss) and in oral squamous cell carcinomas [79, 87, 88]. S. sanguinis has been reported in cases of occult colon carcinomas [89, 90, 91]. Prevotella denticola was enriched in colorectal cancer tissues [92]. Klebsiella pneumoniae, although commonly associated with pneumonia, has been reported as a risk factor for colorectal cancer [93, 94]. S. anginosus is an opportunistic bacterium commonly observed in the oral and intestinal flora, but it is also enriched in esophageal and gastric cancers [95, 96], and has been reported in a case of rectal adenocarcinoma [97]. Gemella is a genus of commensal bacteria of the mouth and gut whose main members (G. morbillorum and G. haemolysans) are usually isolated in abscesses and endocarditis [98, 99, 100]. Anaerococcus mediterraneensis has been recently isolated in a case of vaginosis [101], suggesting a potential pathogenic role or an association with dysbiosis.
Although E. coli was commonly observed in our samples, we also observed an enrichment of some E. coli strains specifically in metastatic tissue. E. coli is both a commensal of the human gut but also an opportunistic species [102]. Thus, lytic phages might remove commensal strains of E. coli, unbalancing the gut microbiome. On the other hand, phages can piggyback pathogenic species invading inner tissues. Therefore, our detection of coliphages might reflect these scenarios, and further data are needed to discriminate between these possibilities. Interestingly, it has been shown that this bacterium can use α‐haemolysin to invade the colon epithelium in a process known as ‘focal leak’ [103]. Specific strains of E. coli (C25 and HBTEC‐1) have been reported to transcytose colonic cells [104]. The portal vein directly connects the human gut to the liver, establishing a special connection between these two organs [105]. It has been shown that dysbiosis and inflammation enhance bacterial translocation, that is, the movement of bacteria and their products from the intestinal lumen to the mesenteric lymph nodes, the bloodstream and, consequently, the liver [106]. The intestinal barrier is also negatively affected by excessive food intake [107, 108], which also can trigger dysbiosis [109]. In turn, increased bacterial translocation can affect the liver, for instance by causing cirrhosis [110]. An impaired intestinal barrier can facilitate, alongside bacteria, the translocation of phages as well, an event that is believed to occur naturally [111].
The gut and the liver share a special relationship due to their connection via the portal vein, the biliary tract and systemic circulation. Hence, the presence of pathogenic bacteria in the gut might be reflected in the liver especially if there are leakages in the mucosal barrier in the intestine. Such a hypothesis is backed by extensive literature, demonstrating a linkage between intestinal dysbiosis and liver disease due to an increased permeability of the intestinal barrier. If certain bacteria or phages (viruses), or their disbalance, can be pathogenic for the colorectal tissue, they might affect the liver, too, in a similar or also slightly different way as compared to the intestinal tissue. Specifically, if particular pathogens, or their interaction, are able to induce inflammation in the gut, they might also induce inflammatory processes in the liver. However, with the exception of some specific systemically infecting viruses such as HCV, EBV, and others which are able to induce pro‐oncogenic pathways in cells including hepatocytes [112, 113, 114], still very little data are available showing how microbiobal pathogens translocated into the liver might foster cancer or cancer metastasis.
Nevertheless, the implications for the presence of bacteria and phages in the liver are still poorly understood. It is feasible to assume that these microorganisms might induce a chronic inflammation that can foster liver disease, albeit more experimental data are required to determine whether microorganisms might also induce metastasis. The fact that the ratio of phages to bacteria was much higher in metastases than in colorectal tissues might reflect the fact that phages are more likely to cross the gut epithelial barrier. Recent metagenomic analysis suggested that the induction of temperate phages might be responsible for propagating intestinal dysbiosis, resulting in an increased amount of phages in relation to bacteria [27, 115, 116]. The higher phage‐to‐bacteria ratio observed in primary colorectal tumours and liver metastases, including 100% temperate phages in the latter, might be explained in this context. Further experiments might help determine the phage‐to‐bacteria ratio with greater accuracy than the present work.
Furthermore, recent metagenomic studies have reported the increased prevalence of members of the Enterobacteriaceae family (thus, including E. coli) in the inflamed gut [117]. Our observations, therefore, can be explained in such a context, but more experimental work is needed to prove that such presence is not coincidental or to analyse whether the transmission of suchlike species, including possibly their phages, through leaky gut situations into other organs is contributing to the ‘seed’ or ‘soil’ component within any metastatic process. Along these lines, strains of E. coli, in addition to B. fragilis, E. faecalis and F. nucleatum, all of which we found represented in our tissues analysed, have been suggested to be able to act as pathogens, questioning their in part uncritical use as probiotics [78, 118, 119, 120, 121]. Several members of the Enterobacteriaceae family, including strains of E. coli and K. pneumoniae, have been shown to release a genotoxin (colibactin) able to induce genetic damage, fostering the insurgence of cancer [122, 123].
The oncogenic potential of these bacteria, together with, at least in part, an enrichment in primary colorectal carcinomas we observed herein, suggest a putatively complex scenario of several infectious components which, in their interaction, might lead to a netto support of carcinogenesis and/or CRC progression. Towards this end, phages such as the ones we identified here could further support the growth of pathobionts, which can cause damage to intestinal cells, local inflammation and more. Thus, phages are increasingly acknowledged as a putative risk factor for CRC, whose importance has been underestimated in the past [31]. As to further mechanisms, phages might change homeostasis of the bacterial microbiome, targeting bacterial species or commensals, that are not pathogenic or oncogenic per se, favouring the expansion of bacteria that promote inflammation or even carcinogenesis [30]. Again, several studies indicate that phages are capable of inducing a ‘leaky gut’ [124], areas of increased intestinal permeability that enable the infiltration of pathogenic bacteria, further promoting chronic inflammation and possibly contributing to a spread of some species to immediate metastatic target organs that generally are considered to be sterile (see below) [124]. Finally, it has been shown that phage‐induced bacteriolysis causes the release of cellular debris into the microenvironment, inducing inflammation, whereby bacterial DNA and lipopolysaccharides are able to act as a pathogen‐associated molecular pattern (PAMP) that triggers immune response [30]. Phages, therefore, could represent an immunogenic stimulus in their own right and be associated with the release of PAMPs that further stimulate the immune system. Specifically, PAMPs will reach the liver first, inducing inflammation [125, 126]. The phage‐mediated bacteriolysis might induce a wave of endotoxins able to ignite the immune response, albeit its extent is not completely understood. Nevertheless, it has been hypothesized that such stimulation might be involved in establishing chronic inflammation [127, 128, 129, 130]. Tetz and Tetz [124] are the principal advocates of the possible pathogenic consequences of phage activity, and given the preliminary state of research on phage interaction with human physiology, it is not surprising that there is little literature besides the one cited of these two authors. Moreover, most of the descriptions of phage immunity have been rather inferred than clinically demonstrated. Still, it is logical to assume that phage‐mediated bacteriolysis will release endotoxins (including bacterial DNA, LPS, and peptidoglycan) in the intestinal lumen that are capable of activating immune cells of, for example of the intestinal wall. In contrast, intact whole bacteria, being adapted to their environment within their host, might minimize their immunogenicity to cause a low as possible activation of the immune system. Thus, we believe it to be more likely that the consumption of whole bacteria due to phage‐mediated bacteriolysis will rather increase than reduce the number of immunogens, and inflammation, in the intestinal milieu, although it is certainly acknowledged that other theories might be true as well, this maybe also depending on the type of phage and corresponding type of bacterial host.
Taken together, there are several mechanisms by which phages, and phage‐host interactions, impact the microbiome and pro‐inflammatory, or even pro‐carcinogenic/‐metastatic, conditions in the intestine. Thus, our findings can encourage mechanistic studies on particular bacteriophages as to their putative function in CRC carcinogenesis or progression in the future, especially since, despite the intense research ongoing to describe the microbial communities in the human intestinal tract of CRC and IBD [22, 24, 36, 38, 131], the characterization of the microbial ecosystem itself is still poorly understood.
Similar to the bacteria, many of the viral species we observed enriched in primary colorectal tumours and liver metastases, besides phages, have been reported to possess oncogenic and/or pro‐inflammatory potential. This is certainly true for EBV which we found in whole‐genome sequencing, including even in one metastasis after Blast filtering, and also by PCR in an independent series in which about a quarter of the analysed primary tumour/normal colorectal tissue sample pairs showed a specific positivity in the carcinomas. Regarding putative pro‐carcinogenic mechanisms, EBV has been reported to act several fold [132, 133]: it produces oncogenic proteins and micro‐interfering RNAs (miRNAs) which could cause pro‐oncogenic pathway switches within a human cell. Moreover, EBV can induce ‘hit‐and‐run mutagenesis’, thereby increasing frequencies of genetic mutations in the infected cell, hyper‐methylate tumour suppressor genes and disrupt cellular miRNA expression [132, 133]. In particular, EBV activates the Wnt/β‐catenin signalling pathway which is fundamental in CRC carcinogenesis and progression [134, 135]. Certainly, our own data are descriptive only and do not give experimental or functional evidence for EBV causing or promoting CRC, and in general, it is known that EBV infects up to 90% of the population [136]. Still, it is estimated that about 10% of all gastric carcinomas are causally associated with EBV infection [137]. In CRC primary tumour tissues, EBV has been recovered by others with a prevalence of between 5% and 60%, depending on the sensitivity of the method used [138, 139, 140], thus in ranges we have found in our sets. This is not surprising as the gut has been found to be an important reservoir of infected resting B cells [141]. These cells usually do not proliferate, but can be periodically reactivated in that they start producing virus, or initiate a short burst of cell growth during which viral products with oncogenic properties are generated [142]. Based on our data, we still consider it unlikely that, in most instances, EBV directly infects CRC cells and contributes to transformation through endogenous expression of viral oncogenes or of viral noncoding RNAs. However, EBV‐infected cells are known to secrete microvesicles that contain many viral products including LMP1, the main EBV oncogene, as well as noncoding RNAs such as the EBERs [143, 144]. Thus, it is theoretically possible that microvesicles laden with EBV‐derived molecules are captured by the neighbouring colonic epithelial cells and contribute to the acquisition of the malignant phenotype. Such a scenario remains entirely speculative but is worth investigating experimentally. Regarding further (microenvironmental) interactions of EBV, interestingly, it has been reported that EBV increases the infectivity of torque teno (TT) virus, a feature that might contribute to multiple sclerosis [145]. In our analysis, we retrieved torque teno virus sequences in a few CRC liver metastases and, even confirmed after stringent Blast filtering, primary CRCs, and although biases such as the known high blood volume in the liver could have impacted on these observations, a potential long‐term cooperation between these two viruses in the CRC context might be an interesting speculation, which needs to be investigated in future functional studies, especially since both torque teno virus and EBV can be present in the blood without causing obvious clinical symptoms of infection [146, 147, 148, 149, 150]. Indeed, TT virus is commonly encountered in both blood and faecal samples, and it is reported to be more prevalent in CRC and chronic inflammation than in normal tissues [151, 152].
We observed other viruses with oncogenic potential. CMV infects preferentially neoplastic epithelium, where it can reach a prevalence of over 40% compared to < 6% in the surrounding normal tissues [153], and tumour tissues have a much higher risk of being infected with CMV than normal tissues (OR = 6.6) [154]. CMV infection activates Wnt signalling pathways, promoting cell proliferation and migration [155]. HHV‐6B infects over 90% of the human population, can cause gastroenteritis, and it is detected in about 6% of colon carcinomas and 4% of rectal adenocarcinomas [156]. HHV‐6B infects mainly T lymphocytes (but also macrophages, dendritic cells, fibroblasts and epithelia) and has been associated with lymphoproliferative diseases as well as oral and cervical carcinomas [157]. HHV‐6 has a prevalence of about 90% [158, 159], but its association with IBD is controversial. For example, no significant difference has been reported for the prevalence of this virus between IBD patients and healthy controls [160]. Further studies also indicated a nonsignificant difference in HHV‐6 prevalence between IBD and controls, but, interestingly, co‐infection with EBV was significantly higher in IBD cases [161]. Other studies reported a prevalence of 4–44% in IBD [162, 163] and in five out of eight (62.5%) patients with colonic adenomas [164] but in none of healthy matched controls. HPyV7 has been isolated about one decade ago from the sera of healthy volunteers [165] and not extensively associated with human diseases yet; still, one study reported its presence in 54–62% of 37 thymic tumours, with 17 samples associated with a high expression of the large T antigen, as compared to no evidence for this virus in 20 fetal thymic tissues [166]. Although this comparison, certainly, might be biased by the use of nonmatched controls, it provides a first hint for a possible oncogenic role of HPyV7, potentially mediated by the large T antigen. Our observation of this virus in CRC in our present work certainly needs to be extended, and validated, by epidemiological and functional studies in the future [167]. Polyomavirus large T (which increases the stability of β‐catenin and, consequently, the activation of the Wnt signal pathway) and small t antigens (which can affect the expression of several modulators of the cell cycle including cyclin D, c‐myc, and survivin) [168], mediate the oncogenic ability of these viruses. Another polyomavirus, SV40, has been associated directly with colorectal cancer. A survey of 94 colon cancers identified the presence of SV40 in 6% of them while it was absent in the colon tissues of healthy controls, but infection was not statistically significant as a risk factor (OR = 3.91, P‐value = 0.115) [169].
HERV‐K113 is an endogenous retrovirus with a prevalence of up to 30% [170], and, unlike many other viruses of this class, it is still capable of reactivation and production of infectious virions [171]. Increased prevalence of HERV‐K113 has been reported in breast cancer tissues (16.7%) in comparison to matched normal tissues (12.7%) [172] and in multiple sclerosis (11.9%) compared to 4.6% in healthy controls, suggesting a role in cancer as well as in autoimmunity [173]. Reactivation of a related virus (HERV‐K‐T47D), measured by the expression of its reverse transcriptase, has been suggested as a biomarker for breast cancer [174]. Given the potential reactivation of this virus and its connection to cancer, it would have been interesting to measure the expression of HERV‐K113 at the protein level but, unfortunately, we had no tissue material left to perform this additional analysis. It is also possible to foresee HERV‐K113 and further infection‐associated markers being included in examining circulating tumour cells (CTC). These cells are gaining popularity as CRC diagnostic indicators [175]. Potentially, a study on the methylation status of HERV‐K113 genes or HERV‐K113 mRNAs within CTCs might help with cancer precursor diagnosis. Likewise, identifying genetic material belonging to oncoviruses either in the bloodstream or inside CTCs might help to stratify patients with CRC. Nevertheless, extensive clinical research is needed to sustain such possibilities.
We also observed EMCV, a zoonotic pathogen that has a seroprevalence ranging from two per cent to over 30% [176, 177]. EMCV is not known to cause cancer but it has been shown that it can induce autoimmune reactions against T cells, fostering the development of type 1 diabetes [178]. It can also interfere with the cellular E3 ubiquitin ligase E6‐associated protein (E6AP) [179], which regulates cell proliferation via the PI3K‐AKT signalling pathway [180]. In the present study, the detection of EMCV sequences in liver metastasis again raises the question whether this virus might be contributing to aspects of inflammation, carcinogenesis or microenvironmental interactions in CRC.
Taking advantage of the observed presence of microorganisms within our samples, we tried to apply statistical methods to distinguish between normal colorectal, primary colorectal carcinoma and metastasis tissue types. Previous studies have applied the Shannon index for diagnostic purposes, for instance applying the genetic variability of c‐MYC to identify breast cancer patients at a higher risk of mortality [181, 182, 183]. In our attempt, both viral and bacterial species proved to be useful in differentiating primary colorectal carcinoma from normal paired colorectal tissues, which supports the notion that our microbial findings in CRC primary tumours were not due to mere contamination by contents of the intestinal lumen or normal tissue fractions, but that microbial or metagenomic findings might aid in the differential diagnosis of malignant versus normal colorectal tissues.
The much lower Shannon indices observed in metastatic sections was unsurprising since these tissues are expected to be rather sterile. Thus, the presence of some microorganisms in liver metastases still appears counter‐intuitive. Only HERV‐K113, an endogenous retrovirus, displayed a profile characterized by high sequence coverage associated with integration in the host’s chromosomes (Fig. S1), thus, the other microorganisms must have gained access to the liver cells by means other than vertical transmission. The species we observed in metastases were not reported as environmental contaminants [184, 185]. Thus, the most likely explanation, as already indicated above, is systemic access through conditions like a ‘leaky gut’ or ‘focal leak’ (see above) and/or general access via the blood stream (see above). Indeed, an increasing number of studies is reporting the presence of, for example, phages in tissues previously considered sterile [30, 186, 187, 188]. We could not compare our results in metastases with previously existing metagenome/microbiome studies in these tissues since, to the best of our knowledge, our study has pioneering character in this regard so far. Therefore, future studies at also larger metastasis sample sizes are mandatory.
Certainly, our study has limitations, including methodological ones. First, it was initially designed for whole‐genome screening. Consequently, no viral enrichment, or general enrichment for microbial genomes, had been carried out, nor included, in the processing of the samples. Thus, the low microbial biomass could have introduced a bias in the results, generating lower than expected findings in sequence copies, or in individual sequence coverage, of microbial sequences as compared to sequences from human chromosomes [185]. Lower sensitivity might also have led to a lower than expected detection of some species, for example JCV or HPV, or a lower detection of positivity as compared to other methods such as PCR, although, still, our results on EBV correspond to the range of frequencies reported by others [138, 139, 189, 190, 191]. Second, except for EBV we had no further tissue available to perform further wet laboratory validations. Nonetheless, the fact that the species we observed have not been reported as contaminants, and that some of the species have been observed by others with similar frequencies, still strengthen our findings. Also, the parallelism between the presence of some bacterial hosts and phages in the same specimens support the validity of our results, since it is highly likely that host and guest species are present simultaneously in the same tissue. Such parallelism would not be expected if the sequences were mapped randomly or derived from environmental contaminants.
5. Conclusion
In conclusion, this is the first work carrying out metagenomic analysis on CRC metastases as compared to corresponding primary CRC tumours and normal colorectal tissues and that attempted to apply microbial richness calculations to differentiate these tissues. We showed that the microbial landscape might be used to differentiate primary colorectal tumours from nonmalignant tissue and metastases, in particular by using the Shannon index. Moreover, we highlighted particular species, especially including the previously not extensively considered species of (bacterio‐)phages, for future functional studies as to how they could contribute to colorectal carcinogenesis or even progression and metastasis, for example by creating permissive or nonpermissive microbial environments. Extending this analysis to broader sets of available samples, for example in multicentre approaches, could foster a better understanding of infectious agents as a potentially complex interplay of cofactors for CRC carcinogenesis and progression, and assess whether the use of microbial analysis could support precision medicine to pinpoint patients at increased risk for CRC or CRC metastasis.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
LM mainly performed and coordinated the analyses within this paper. LM, TR, and JJML performed all aspects of the bioinformatics analysis, MLA acquired the samples and consents, coordinated the aspects associated with their collection, and led wet laboratory sample analysis and PCR. SD and HJD performed and interpreted EBER stainings. HA initiated the idea of virome, phage and bacteriome analysis in association with CRC carcinogenesis and metastasis, and coordinated the research and funding. All authors participated in the discussion and interpretation of data, the writing and/or revision, and approval of the manuscript.
Supporting information
Fig. S1. Visualization of viral integration of human endogenous retrovirus K113 (HERV‐K113).
Table S1. Read counts stratified by tissue type.
Acknowledgements
HA was supported by the Alfried Krupp von Bohlen und Halbach Foundation, Essen; the Deutsche Krebshilfe, Bonn (grant number 70112168, also to MLA); the Deutsche Forschungsgemeinschaft (DFG, grant number AL 465/9‐1); the HEiKA Initiative (Karlsruhe Institute of Technology/University of Heidelberg collaborative effort); Dr Hella‐Buehler‐Foundation, Heidelberg; the DKFZ‐MOST Cooperation, Heidelberg (grant number CA149); the the NCT Molecular Precision Oncology Program, Heidelberg (technical support and funding through HIPO032; HIPO/DKFZ/NCT and its employees not bearing responsibility for the further analysis or interpretation of the data). We are grateful to the EMBL Genomics Core Facility, Heidelberg, Germany, for technical support and advice on the analysis. A special thanks goes to Prof. Scott A. Handley, Department of Pathology and Immunology, Washington University School of Medicine, St. Louis, Missouri, USA, for his valuable comments on the manuscript. We would like also to thank Dr. Svetlana Hetjens, Department of Statistics of the Medical Faculty in Mannheim of the Ruprecht‐Karls University of Heidelberg, for her precious help on statistical analysis. A special thanks goes to Prof. Dr. Ingo Plag, Institute for English Language and Linguistics of the University of Düsseldorf, for his advice on the adjective ‘phagial’, which is still not officially recognized. Open Access funding enabled and organized by Projekt DEAL.
Data accessibility
Anonymized sequence data are stored within the EGA database (accession number EGAS00001002717) as already described previously [41].
References
- 1. Favoriti P, Carbone G, Greco M, Pirozzi F, Pirozzi REM & Corcione F (2016) Worldwide burden of colorectal cancer: a review. Updates Surg 68, 7–11. [DOI] [PubMed] [Google Scholar]
- 2. Ferlay J, Shin HR, Bray F, Forman D, Mathers C & Parkin DM (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 127, 2893–2917. [DOI] [PubMed] [Google Scholar]
- 3. Song M, Chan AT & Sun J (2020) Influence of the gut microbiome, diet, and environment on risk of colorectal cancer. Gastroenterology 158, 322–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brennan CA & Garrett WS (2016) Gut microbiota, inflammation, and colorectal cancer. Annu Rev Microbiol 70, 395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Butt J, Romero‐Hernandez B, Perez‐Gomez B, Willhauck‐Fleckenstein M, Holzinger D, Martin V, Moreno V, Linares C, Dierssen‐Sotos T, Barricarte A et al. (2016) Association of Streptococcus gallolyticus subspecies gallolyticus with colorectal cancer: serological evidence. Int J Cancer 138, 1670–1679. [DOI] [PubMed] [Google Scholar]
- 6. Leslie A, Carey FA, Pratt NR & Steele RJC (2002) The colorectal adenoma‐carcinoma sequence. Br J Surg 89, 845–860. [DOI] [PubMed] [Google Scholar]
- 7. Ma H, Brosens LAA, Offerhaus GJA, Giardiello FM, de Leng WWJ & Montgomery EA (2018) Pathology and genetics of hereditary colorectal cancer. Pathology 50, 49–59. [DOI] [PubMed] [Google Scholar]
- 8. Collins D, Hogan AM & Winter DC (2011) Microbial and viral pathogens in colorectal cancer. Lancet Oncol 12, 504–512. [DOI] [PubMed] [Google Scholar]
- 9. De Paoli P & Carbone A (2013) Carcinogenic viruses and solid cancers without sufficient evidence of causal association. Int J Cancer 133, 1517–1529. [DOI] [PubMed] [Google Scholar]
- 10. zur Hausen H, Bund T & de Villiers EM (2017) Infectious agents in bovine red meat and milk and their potential role in cancer and other chronic diseases. Curr Top Microbiol Immunol 407, 83–116. [DOI] [PubMed] [Google Scholar]
- 11. Candela M, Turroni S, Biagi E, Carbonero F, Rampelli S, Fiorentini C & Brigidi P (2014) Inflammation and colorectal cancer, when microbiota‐host mutualism breaks. World J Gastroenterol 20, 908–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li S, Konstantinov SR, Smits R & Peppelenbosch MP (2017) Bacterial biofilms in colorectal cancer initiation and progression. Trends Mol Med 23, 18–30. [DOI] [PubMed] [Google Scholar]
- 13. Tjalsma H, Boleij A, Marchesi JR & Dutilh BE (2012) A bacterial driver‐passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol 10, 575–582. [DOI] [PubMed] [Google Scholar]
- 14. Yu YN & Fang JY (2015) Gut microbiota and colorectal cancer. Gastrointest Tumors 2, 26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kappelman MD, Moore KR, Allen JK & Cook SF (2013) Recent trends in the prevalence of Crohn's disease and ulcerative colitis in a commercially insured US population. Dig Dis Sci 58, 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bounthavong M, Li M & Watanabe JH (2017) An evaluation of health care expenditures in Crohn's disease using the United States Medical Expenditure Panel Survey from 2003 to 2013. Res Soc Adm Pharm 13, 530–538. [DOI] [PubMed] [Google Scholar]
- 17. von Roon A, Reese G, Teare J, Constantinides V, Darzi A & Tekkis P (2007) The risk of cancer in patients with Crohn's disease. Dis Colon Rectum 50, 839–855. [DOI] [PubMed] [Google Scholar]
- 18. Baandrup L, Thomsen LT, Olesen TB, Andersen KK, Norrild B & Kjaer SK (2014) The prevalence of human papillomavirus in colorectal adenomas and adenocarcinomas: a systematic review and meta‐analysis. Eur J Cancer 50, 1446–1461. [DOI] [PubMed] [Google Scholar]
- 19. Burnett‐Hartman AN, Newcomb PA & Potter JD (2008) Infectious agents and colorectal cancer: a review of Helicobacter pylori, Streptococcus bovis, JC virus, and human papillomavirus. Cancer Epidemiol Biomarkers Prev 17, 2970–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen H, Chen X‐Z, Waterboer T, Castro FA & Brenner H (2015) Viral infections and colorectal cancer: a systematic review of epidemiological studies. Int J Cancer 137, 12–24. [DOI] [PubMed] [Google Scholar]
- 21. Tolentino YF, Fogaca HS, Zaltman C, Ximenes LL & Coelho HS (2008) Hepatitis B virus prevalence and transmission risk factors in inflammatory bowel disease patients at Clementino Fraga Filho university hospital. World J Gastroenterol 14, 3201–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hannigan GD, Duhaime MB, Ruffin MT, Koumpouras CC & Schloss PD (2018) Diagnostic potential and interactive dynamics of the colorectal cancer virome. MBio 9, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakatsu G, Zhou H, Wu WKK, Wong SH, Coker OO, Dai Z, Li X, Szeto CH, Sugimura N, Lam TYT et al. (2018) Alterations in enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology 155, 529–541.e525. [DOI] [PubMed] [Google Scholar]
- 24. Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, Kambal A, Monaco CL, Zhao G, Fleshner P et al. (2015) Disease‐specific alterations in the enteric virome in inflammatory bowel disease. Cell 160, 447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zuo T, Lu XJ, Zhang Y, Cheung CP, Lam S, Zhang F, Tang W, Ching JYL, Zhao R, Chan PKS et al. (2019) Gut mucosal virome alterations in ulcerative colitis. Gut 68, 1169–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Caldwell JA (1928) Bacteriologic and bacteriophagic study of infected urines. J Infect Dis 43, 353–362. [Google Scholar]
- 27. Dieterich W, Schink M & Zopf Y (2018) Microbiota in the gastrointestinal tract. Med Sci 6, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fortier LC (2018) Bacteriophages contribute to shaping clostridioides (Clostridium) difficile species. Front Microbiol 9, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Handley SA & Devkota S (2019) Going viral: a novel role for bacteriophage in colorectal cancer. MBio 10, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hsu BB, Gibson TE, Yeliseyev V, Liu Q, Lyon L, Bry L, Silver PA & Gerber GK (2019) Dynamic modulation of the gut microbiota and metabolome by bacteriophages in a mouse model. Cell Host Microbe 25, 803–814.e805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Manrique P, Dills M & Young MJ (2017) The human gut phage community and its implications for health and disease. Viruses 9, 9–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tetz G & Tetz V (2018) Bacteriophages as new human viral pathogens. Microorganisms 6, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Keku TO, Dulal S, Deveaux A, Jovov B & Han X (2015) The gastrointestinal microbiota and colorectal cancer. Am J Physiol Gastrointest Liver Physiol 308, G351–G363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Capaldo CT, Powell DN & Kalman D (2017) Layered defense: how mucus and tight junctions seal the intestinal barrier. J Mol Med 95, 927–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sicard J‐F, Le Bihan G, Vogeleer P, Jacques M & Harel J (2017) Interactions of intestinal bacteria with components of the intestinal mucus. Front Cell Infect Microbiol 7, 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Perez‐Brocal V, Garcia‐Lopez R, Nos P, Beltran B, Moret I & Moya A (2015) Metagenomic analysis of Crohn's disease patients identifies changes in the virome and microbiome related to disease status and therapy, and detects potential interactions and biomarkers. Inflamm Bowel Dis 21, 2515–2532. [DOI] [PubMed] [Google Scholar]
- 37. Ungaro F, Massimino L, Furfaro F, Rimoldi V, Peyrin‐Biroulet L, D'Alessio S & Danese S (2019) Metagenomic analysis of intestinal mucosa revealed a specific eukaryotic gut virome signature in early‐diagnosed inflammatory bowel disease. Gut Microbes 10, 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wagner J, Maksimovic J, Farries G, Sim WH, Bishop RF, Cameron DJ, Catto‐Smith AG & Kirkwood CD (2013) Bacteriophages in gut samples from pediatric Crohn's disease patients. Inflamm Bowel Dis 19, 1598–1608. [DOI] [PubMed] [Google Scholar]
- 39. Engstrand J, Nilsson H, Strmberg C, Jonas E & Freedman J (2018) Colorectal cancer liver metastases – a population‐based study on incidence, management and survival. BMC Cancer 18, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Luo Q, O'Connell DL, Kahn C & Yu XQ (2017) Colorectal cancer metastatic disease progression in Australia: a population‐based analysis. Cancer Epidemiol 49, 92–100. [DOI] [PubMed] [Google Scholar]
- 41. Ishaque N, Abba ML, Hauser C, Patil N, Paramasivam N, Huebschmann D, Leupold JH, Reszec J, Toprak UH, Kozlowski M et al. (2018) Whole genome sequencing puts forward hypotheses on metastasis evolution and therapy in colorectal cancer. Nat Commun 9, 4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Marongiu L & Allgayer H (2019) Mingle: a command line utility for merging multi‐fasta files. J Comput Biol 26, 396–404. [DOI] [PubMed] [Google Scholar]
- 43. Li H & Durbin R (2009) Fast and accurate short read alignment with Burrows‐Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bolger AM, Lohse M & Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liao P, Satten GA & Hu YJ (2017) PhredEM: a phred‐score‐informed genotype‐calling approach for next‐generation sequencing studies. Genet Epidemiol 41, 375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. MacManes MD (2014) On the optimal trimming of high‐throughput mRNA sequence data. Front Genet 5, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA‐MEM. arXiv:13033997v1, 10.1186/s13756-018-0352-y [DOI]
- 48. Tarasov A, Vilella AJ, Cuppen E, Nijman IJ & Prins P (2015) Sambamba: fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G & Durbin R (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Okonechnikov K, Conesa A & Garca‐Alcalde F (2015) Qualimap 2: advanced multi‐sample quality control for high‐throughput sequencing data. Bioinformatics 32, 292–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Altschul SF, Gish W, Miller W, Myers EW & Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 52. Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K & Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Sding J et al. (2011) Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. States DJ & Gish W (1994) Combined use of sequence similarity and codon bias for coding region identification. J Comput Biol 1, 39–50. [DOI] [PubMed] [Google Scholar]
- 55. Rausch T, Zichner T, Schlattl A, Sttz AM, Benes V & Korbel JO (2012) DELLY: structural variant discovery by integrated paired‐end and split‐read analysis. Bioinformatics 28, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Birdwell CE, Prasai K, Dykes S, Jia Y, Munroe TGC, Bienkowska‐Haba M & Scott RS (2018) Epstein‐Barr virus stably confers an invasive phenotype to epithelial cells through reprogramming of the WNT pathway. Oncotarget 9, 10417–10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Antsiferova O, Müller A, Rämer PC, Chijioke O, Chatterjee B, Raykova A, Planas R, Sospedra M, Shumilov A, Tsai MH et al. (2014) Adoptive transfer of EBV specific CD8+ T cell clones can transiently control EBV infection in humanized mice. PLoS Pathog 10, e1004333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD et al. (2012) SPAdes: a new genome assembly algorithm and its applications to single‐cell sequencing. J Comput Biol 19, 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mihara T, Nishimura Y, Shimizu Y, Nishiyama H, Yoshikawa G, Uehara H, Hingamp P, Goto S & Ogata H (2016) Linking virus genomes with host taxonomy. Viruses 8, 10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tynecki P, Guziński A, Kazimierczak J, Jadczuk M, Dastych J & Onisko A (2020) PhageAI – bacteriophage life cycle recognition with machine learning and natural language processing. BioRxiv, 10.1101/2020.07.11.198606 [DOI] [Google Scholar]
- 61. R Core Team (2020) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R‐project.org/. [Google Scholar]
- 62. Shannon CE (1948) A mathematical theory of communication. Bell Syst Tech J 27, 379–423. [Google Scholar]
- 63. Gregori J, Salicrú M, Domingo E, Sanchez A, Esteban JI, Rodríguez‐Frías F & Quer J (2014) Inference with viral quasispecies diversity indices: Clonal and NGS approaches. Bioinformatics 30, 1104–1111. [DOI] [PubMed] [Google Scholar]
- 64. Gotelli J & Colwell K (2001) Quantifying biodversity: procedures and pitfalls in the measurement and comparison of species richness. Ecol Lett 4, 379–391. [Google Scholar]
- 65. Lande R, DeVries PJ & Walla TR (2000) When species accumulation curves intersect: implications for ranking diversity using small samples. Oikos 89, 601–605. [Google Scholar]
- 66. Hsieh TC, Ma KH & Chao A (2016) iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol Evol 7, 1451–1456. [Google Scholar]
- 67. Hutcheson K (1970) A test for comparing diversities based on the Shannon formula. J Theor Biol 29, 151–154. [DOI] [PubMed] [Google Scholar]
- 68. Dutilh BE, Cassman N, McNair K, Sanchez SE, Silva GGZ, Boling L, Barr JJ, Speth DR, Seguritan V, Aziz RK et al. (2014) A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat Commun 5, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Guerin E, Shkoporov A, Stockdale SR, Clooney AG, Ryan FJ, Sutton TDS, Draper LA, Gonzalez‐Tortuero E, Ross RP & Hill C (2018) Biology and taxonomy of crAss‐like bacteriophages, the most abundant virus in the human gut. Cell Host Microbe 24, 653–664.e656. [DOI] [PubMed] [Google Scholar]
- 70. Shkoporov AN, Khokhlova EV, Fitzgerald CB, Stockdale SR, Draper LA, Ross RP & Hill C (2018) PhiCrAss001 represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis . Nat Commun 9, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Enosi Tuipulotu D, Mathur A, Ngo C & Man SM (2020) Bacillus cereus: epidemiology, virulence factors, and host‐pathogen interactions. Trends Microbiol 29, 458–471. [DOI] [PubMed] [Google Scholar]
- 72. Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss S, Barnes R, Watson S, Allen‐Vercoe E, Moore R et al. (2012) Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 22, 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J et al. (2012) Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 22, 292–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Masood U, Sharma A, Lowe D, Khan R & Manocha D (2016) Single case colorectal cancer associated with Streptococcus anginosus bacteremia and liver abscesses. Case Rep Gastroenterol 10, 769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. McCoy AN, Arajo‐Prez F, Azcrate‐Peril A, Yeh JJ, Sandler RS & Keku TO (2013) Fusobacterium is associated with colorectal adenomas. PLoS One 8, e53653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mulita A & Ajayi T (2014) Streptococcus viridians bacteraemia and colonic adenocarcinoma. BMJ Case Rep. 10.1136/bcr-2014-203695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sasaki H, Ishizuka T, Muto M, Nezu M, Nakanishi Y, Inagaki Y, Watanabe H, Watanabe H & Terada M (1998) Presence of Streptococcus anginosus DNA in esophageal cancer, dysplasia of esophagus, and gastric cancer. Cancer Res 1, 2991–2995. [PubMed] [Google Scholar]
- 78. Shang F‐M & Liu H‐L (2018) Fusobacterium nucleatum and colorectal cancer: a review. World J Gastrointest Oncol 10, 71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Shoaib K, Ishaq M, Hinson M, Potugari B & Rehman A (2019) Case report Parvimonas micra bacteremia in a patient with colonic carcinoma. Caspian J Intern Med 10, 472–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yang CY, Yeh YM, Yu HY, Chin CY, Hsu CW, Liu H, Huang PJ, Hu SN, Liao CT, Chang KP et al. (2018) Oral microbiota community dynamics associated with oral squamous cell carcinoma staging. Front Microbiol 9, 1–15. 10.3389/fmicb.2018.00862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, Fleck JS, Voigt AY, Palleja A, Ponnudurai R et al. (2019) Meta‐analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med 25, 679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yang Y & Jobin C (2020) Far reach of Fusobacterium nucleatum in cancer metastasis. Gut 70, 1507–1519. 10.1136/gutjnl-2020-323496 [DOI] [PubMed] [Google Scholar]
- 83. Drewes JL, White JR, Dejea CM, Fathi P, Iyadorai T, Vadivelu J, Roslani AC, Wick EC, Mongodin EF, Loke MF et al. (2017) High‐resolution bacterial 16S rRNA gene profile meta‐analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes 3, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Löwenmark T, Löfgren‐Burström A, Zingmark C, Eklöf V, Dahlberg M, Wai SN, Larsson P, Ljuslinder I, Edin S, Palmqvist R et al. (2020) Parvimonas micra as a putative non‐invasive faecal biomarker for colorectal cancer. Sci Rep 10, 15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, Tang L, Zhao H, Stenvang J, Li Y et al. (2017) Metagenomic analysis of faecal microbiome as a tool towards targeted non‐invasive biomarkers for colorectal cancer. Gut 66, 70–78. [DOI] [PubMed] [Google Scholar]
- 86. Purcell RV, Visnovska M, Biggs PJ, Schmeier S & Frizelle FA (2017) Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci Rep 7, 11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fine DH, Markowitz K, Fairlie K, Tischio‐Bereski D, Ferrendiz J, Furgang D, Paster BJ & Dewhirst FE (2013) A consortium of Aggregatibacter actinomycetemcomitans, Streptococcus parasanguinis, and Filifactor alocis is present in sites prior to bone loss in a longitudinal study of localized aggressive periodontitis. J Clin Microbiol 51, 2850–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lundmark A, Hu YOO, Huss M, Johannsen G, Andersson AF & Yucel‐Lindberg T (2019) Identification of salivary microbiota and its association with host inflammatory mediators in periodontitis. Front Cell Infect Microbiol 9, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kampe CE, Vovan T, Alim A & Berenson J (1995) Streptococcus sanguis bacteremia and colorectal cancer: a case report. Med Pediatr Oncol 24, 67–68. [DOI] [PubMed] [Google Scholar]
- 90. Macaluso A, Simmang C & Anthony T (1998) Streptococcus sanguis bacteremia and colorectal cancer. South Med J 91, 206–207. [DOI] [PubMed] [Google Scholar]
- 91. Thomas R, Gupta V & Kwan B (2018) Second look at Streptococcus sanguinis and the colon. BMJ Case Rep 2018. 10.1136/bcr-2018-224799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yang Y, Cai Q, Shu XO, Steinwandel MD, Blot WJ, Zheng W & Long J (2019) Prospective study of oral microbiome and colorectal cancer risk in low‐income and African American populations. Int J Cancer 144, 2381–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Huang WK, Chang JW, See LC, Tu HT, Chen JS, Liaw CC, Lin YC & Yang TS (2012) Higher rate of colorectal cancer among patients with pyogenic liver abscess with Klebsiella pneumoniae than those without: an 11‐year follow‐up study. Colorectal Dis 14, e794–e801. [DOI] [PubMed] [Google Scholar]
- 94. Strakova N, Korena K & Karpiskova R (2021) Klebsiella pneumoniae producing bacterial toxin colibactin as a risk of colorectal cancer development – a systematic review. Toxicon 197, 126–135. [DOI] [PubMed] [Google Scholar]
- 95. Morita E, Narikiyo M, Yano A, Nishimura E, Igaki H, Sasaki H, Terada M, Hanada N & Kawabe R (2003) Different frequencies of Streptococcus anginosus infection in oral cancer and esophageal cancer. Cancer Sci 94, 492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sasaki M, Yamaura C, Ohara‐Nemoto Y, Tajika S, Kodama Y, Ohya T, Harada R & Kimura S (2005) Streptococcus anginosus infection in oral cancer and its infection route. Oral Dis 11, 151–156. [DOI] [PubMed] [Google Scholar]
- 97. Masood U, Sharma A, Lowe D, Khan R, Manocha D, McQuinn M & Horswell BB (2016) Colorectal cancer associated with Streptococcus anginosus bacteremia and liver abscesses. Case Rep Gastroenterol 10, 769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Brouqui P & Raoult D (2001) Endocarditis due to rare and fastidious bacteria. Clin Microbiol Rev 14, 177–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Chotai S, Moon HJ, Kim JH, Kim JH, Chung HS, Park YK & Kwon TH (2012) Brain abscess caused by Gemella morbillorum: case report and review of the literature. Turk Neurosurg 22, 374–377. [DOI] [PubMed] [Google Scholar]
- 100. McQuinn M & Horswell BB (2019) First case of cutaneous orbital abscess caused by gemella: a case report and review of the literature. J Oral Maxillofac Surg 77, 1414–1417. [DOI] [PubMed] [Google Scholar]
- 101. Diop K, Bretelle F, Fournier PE & Fenollar F (2017) 'Anaerococcus mediterraneensis' sp. nov., a new species isolated from human female genital tract. New Microbes New Infect 17, 75–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Denamur E, Clermont O, Bonacorsi S & Gordon D (2021) The population genetics of pathogenic Escherichia coli . Nat Rev Microbiol 19, 37–54. [DOI] [PubMed] [Google Scholar]
- 103. Bucker R, Schulz E, Gnzel D, Bojarski C, Lee IFM, John LJ, Wiegand S, Janen T, Wieler LH, Dobrindt U et al. (2014) Alpha‐haemolysin of Escherichia coli in IBD: a potentiator of inflammatory activity in the colon. Gut 63, 1893–1901. [DOI] [PubMed] [Google Scholar]
- 104. Macutkiewicz C, Carlson G, Clark E, Dobrindt U, Roberts I & Warhurst G (2008) Characterisation of Escherichia coli strains involved in transcytosis across gut epithelial cells exposed to metabolic and inflammatory stress. Microbes Infect 10, 424–431. [DOI] [PubMed] [Google Scholar]
- 105. Delzenne N, Knudsen C, Beaumont M, Rodriguez J, Neyrinck A & Bindels L (2019) Contribution of the gut microbiota to the regulation of host metabolism and energy balance: a focus on the gut‐liver axis. Proc Nutr Soc 78, 319–328. [DOI] [PubMed] [Google Scholar]
- 106. Plaza‐Díaz J, Solís‐Urra P, Rodríguez‐Rodríguez F, Olivares‐Arancibia J, Navarro‐Oliveros M, Abadía‐Molina F & Álvarez‐Mercado A (2020) The gut barrier, intestinal microbiota, and liver disease: molecular mechanisms and strategies to manage. Int J Mol Sci 21, 8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Thuy S, Ladurner R, Volynets V, Wagner S, Strahl S, Königsrainer A, Maier K, Bischoff S & Bergheim I (2008) Nonalcoholic fatty liver disease in humans is associated with increased plasma endotoxin and plasminogen activator inhibitor 1 concentrations and with fructose intake. J Nutr 138, 1452–1455. [DOI] [PubMed] [Google Scholar]
- 108. Álvarez‐Mercado A, Navarro‐Oliveros M, Robles‐Sánchez C, Plaza‐Díaz J, Sáez‐Lara M, Muñoz‐Quezada S, Fontana L & Abadía‐Molina F (2019) Microbial population changes and their relationship with human health and disease. Microorganisms 7, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Panasevich M, Peppler W, Oerther D, Wright D & Rector R (2017) Microbiome and NAFLD: potential influence of aerobic fitness and lifestyle modification. Physiol Genomics 49, 385–399. [DOI] [PubMed] [Google Scholar]
- 110. Wiest R, Lawson M & Geuking M (2014) Pathological bacterial translocation in liver cirrhosis. J Hepatol 60, 197–209. [DOI] [PubMed] [Google Scholar]
- 111. Łusiak‐Szelachowska M, Weber‐Dąbrowska B, Jończyk‐Matysiak E, Wojciechowska R & Górski A (2017) Bacteriophages in the gastrointestinal tract and their implications. Gut Pathog 9, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Akram N, Imran M, Noreen M, Ahmed F, Atif M, Fatima Z & Bilal Waqar A (2017) Oncogenic role of tumor viruses in humans. Viral Immunol 30, 20–27. [DOI] [PubMed] [Google Scholar]
- 113. Gaglia M & Munger K (2018) More than just oncogenes: mechanisms of tumorigenesis by human viruses. Curr Opin Virol 32, 48–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Stern J, Miller G, Li X & Saxena D (2019) Virome and bacteriome: two sides of the same coin. Curr Opin Virol 37, 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Mills S, Shanahan F, Stanton C, Hill C, Coffey A & Ross R (2013) Movers and shakers: influence of bacteriophages in shaping the mammalian gut microbiota. Gut Microbes 4, 4–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Clooney AG, Sutton TDS, Shkoporov AN, Holohan RK, Daly KM, O’Regan O, Ryan FJ, Draper LA, Plevy SE, Ross RP et al. (2019) Whole‐virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host Microbe 26, 764–778.e5. [DOI] [PubMed] [Google Scholar]
- 117. Darfeuille‐Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, Gambiez L, Joly B, Cortot A & Colombel JF (1998) Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology 115, 1405–1413. [DOI] [PubMed] [Google Scholar]
- 118. de Almeida CV, Taddei A & Amedei A (2018) The controversial role of Enterococcus faecalis in colorectal cancer. Ther Adv Gastroenterol 11, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Franz CM, Huch M, Abriouel H, Holzapfel W & Gálvez A (2011) Enterococci as probiotics and their implications in food safety. Int J Food Microbiol 151, 125–140. [DOI] [PubMed] [Google Scholar]
- 120. Li R, Helbig L, Fu J, Bian X, Herrmann J, Baumann M, Stewart AF, Müller R, Li A, Zips D et al. (2019) Expressing cytotoxic compounds in Escherichia coli Nissle 1917 for tumor‐targeting therapy. Res Microbiol 170, 74–79. [DOI] [PubMed] [Google Scholar]
- 121. Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, Huso DL, Brancati FL, Wick E, McAllister F et al. (2009) A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med 15, 1016–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Cougnoux A, Dalmasso G, Martinez R, Buc E, Delmas J, Gibold L, Sauvanet P, Darcha C, Dchelotte P, Bonnet M et al. (2014) Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence‐associated secretory phenotype. Gut 63, 1932–1942. [DOI] [PubMed] [Google Scholar]
- 123. Faïs T, Delmas J, Barnich N, Bonnet R & Dalmasso G (2018) Colibactin: more than a new bacterial toxin. Toxins (Basel) 10, 151–167. 10.3390/toxins10040151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Tetz G & Tetz V (2016) Bacteriophage infections of microbiota can lead to leaky gut in an experimental rodent model. Gut Pathog 8, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Mencin A, Kluwe J & Schwabe R (2009) Toll‐like receptors as targets in chronic liver diseases. Gut 58, 704–720. 10.1136/gut.2008.156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Wang L, Llorente C, Hartmann P, Yang A, Chen P & Schnabl B (2015) Methods to determine intestinal permeability and bacterial translocation during liver disease. J Immunol Methods 421, 44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Mueller C (2012) Danger‐associated molecular patterns and inflammatory bowel disease: is there a connection? Dig Dis 30(Suppl 3), 40–46. [DOI] [PubMed] [Google Scholar]
- 128. Walsh D, McCarthy J, O'Driscoll C & Melgar S (2013) Pattern recognition receptors–molecular orchestrators of inflammation in inflammatory bowel disease. Cytokine Growth Factor Rev 24, 91–104. [DOI] [PubMed] [Google Scholar]
- 129. Aguilera M, Darby T & Melgar S (2014) The complex role of inflammasomes in the pathogenesis of Inflammatory Bowel Diseases ‐ lessons learned from experimental models. Cytokine Growth Factor Rev 25, 715–730. [DOI] [PubMed] [Google Scholar]
- 130. Maronek M, Link R, Ambro L & Gardlik R (2020) Phages and their role in gastrointestinal disease: focus on inflammatory bowel disease. Cells 9, 1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Perez‐Brocal V, Garcia‐Lopez R, Vazquez‐Castellanos JF, Nos P, Beltran B, Latorre A & Moya A (2013) Study of the viral and microbial communities associated with Crohn's disease: a metagenomic approach. Clin Transl Gastroenterol 4, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Morales‐Sanchez A & Fuentes‐Panana EM (2017) Epstein‐Barr virus‐associated gastric cancer and potential mechanisms of oncogenesis. Curr Cancer Drug Targets 17, 534–554. 10.2174/1568009616666160926124923 [DOI] [PubMed] [Google Scholar]
- 133. Shinozaki‐ushiku A, Kunita A & Fukayama M (2015) Update on Epstein‐Barr virus and gastric cancer (Review). Int J Oncol 46, 1421–1434. [DOI] [PubMed] [Google Scholar]
- 134. Jang KL, Shackelford J, Seo SY & Pagano JS (2005) Up‐regulation of beta‐catenin by a viral oncogene correlates with inhibition of the seven in absentia homolog 1 in B lymphoma cells. Proc Natl Acad Sci USA 102, 18431–18436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. QingLing Z, LiNa Y, Li L, Shuang W, YuFang Y, Yi D, Divakaran J, Xin L & YanQing D (2011) LMP1 antagonizes WNT/β‐catenin signalling through inhibition of WTX and promotes nasopharyngeal dysplasia but not tumourigenesis in LMP1(B95–8) transgenic mice. J Pathol 223, 574–583. [DOI] [PubMed] [Google Scholar]
- 136. Lieberman PM (2014) Virology. Epstein‐Barr virus turns 50. Science 343, 1323–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Takada K (2000) Epstein‐Barr virus and gastric carcinoma. Mol Pathol 53, 255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Cho YJ, Chang MS, Park SH, Kim HS & Kim WH (2001) In situ hybridization of Epstein‐Barr virus in tumor cells and tumor‐infiltrating lymphocytes of the gastrointestinal tract. Hum Pathol 32, 297–301. [DOI] [PubMed] [Google Scholar]
- 139. Grinstein S, Preciado MV, Gattuso P, Chabay PA, Warren WH, De Matteo E & Gould VE (2002) Demonstration of Epstein‐Barr virus in carcinomas of various sites. Cancer Res 62, 4876–4878. [PubMed] [Google Scholar]
- 140. Mehrabani‐Khasraghi S, Ameli M & Khalily F (2016) Demonstration of Herpes Simplex Virus, Cytomegalovirus, and Epstein‐Barr Virus in colorectal cancer. Iran Biomed J 20, 302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Delecluse S, Tsai MH, Shumilov A, Bencun M, Arrow S, Beshirova A, Cottignies‐Calamarte A, Lasitschka F, Bulut OC, Münz C et al. (2019) Epstein‐Barr Virus induces expression of the LPAM‐1 integrin in B cells in vitro and in vivo. J Virol 93, e01618‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Thorley‐Lawson DA (2015) EBV persistence‐introducing the virus. Curr Top Microbiol Immunol 390, 151–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Dukers DF, Meij P, Vervoort MB, Vos W, Scheper RJ, Meijer CJ, Bloemena E & Middeldorp JM (2000) Direct immunosuppressive effects of EBV‐encoded latent membrane protein 1. J Immunol 165, 663–670. [DOI] [PubMed] [Google Scholar]
- 144. Ahmed W, Philip P, Tariq S & Khan G (2014) Epstein‐Barr virus‐encoded small RNAs (EBERs) are present in fractions related to exosomes released by EBV‐transformed cells. PLoS One 9, e99163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Borkosky SS, Whitley C, Kopp‐Schneider A, zur Hausen H & deVilliers E‐M (2012) Epstein‐Barr virus stimulates torque teno virus replication: a possible relationship to multiple sclerosis. PLoS One 7, e32160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Decker LL, Klaman LD & Thorley‐Lawson DA (1996) Detection of the latent form of Epstein‐Barr virus DNA in the peripheral blood of healthy individuals. J Virol 70, 3286–3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Takeuchi H, Kobayashi R, Hasegawa M & Hirai K (1996) Detection of latent infection by Epstein‐Barr virus in peripheral blood cells of healthy individuals and in non‐neoplastic tonsillar tissue from patients by reverse transcription‐polymerase chain reaction. J Virol Methods 58, 81–89. [DOI] [PubMed] [Google Scholar]
- 148. Bekker V, Scherpbier H, Beld M, Piriou E, van Breda A, Lange J, van Leth F, Jurriaans S, Alders S, Wertheim‐van Dillen P et al. (2006) Epstein‐Barr virus infects B and non‐B lymphocytes in HIV‐1‐infected children and adolescents. J Infect Dis 194, 1323–1330. [DOI] [PubMed] [Google Scholar]
- 149. AbuOdeh R, Al‐Mawlawi N, Al‐Qahtani AA, Bohol MF, Al‐Ahdal MN, Hasan HA, AbuOdeh L & Nasrallah GK (2015) Detection and genotyping of torque teno virus (TTV) in healthy blood donors and patients infected with HBV or HCV in Qatar. J Med Virol 87, 1184–1191. [DOI] [PubMed] [Google Scholar]
- 150. Fatholahi M & Bouzari M (2015) Torque Teno Midi Virus/small anellovirus in sera of healthy, HIV/HCV and HIV infected individuals in lorestan province, Iran. Jundishapur J Microbiol 8, e25368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Hino S & Miyata H (2007) Torque teno virus (TTV): current status. Rev Med Virol 17, 45–57. [DOI] [PubMed] [Google Scholar]
- 152. Pinho‐Nascimento CA, Leite JP, Niel C & Diniz‐Mendes L (2011) Torque teno virus in fecal samples of patients with gastroenteritis: prevalence, genogroups distribution, and viral load. J Med Virol 83, 1107–1111. [DOI] [PubMed] [Google Scholar]
- 153. Chen HP, Jiang JK, Chen CY, Chou TY, Chen YC, Chang YT, Lin SF, Chan CH, Yang CY, Lin CH et al. (2012) Human cytomegalovirus preferentially infects the neoplastic epithelium of colorectal cancer: a quantitative and histological analysis. J Clin Virol 54, 240–244. [DOI] [PubMed] [Google Scholar]
- 154. Bai B, Wang X, Chen E & Zhu H (2016) Human cytomegalovirus infection and colorectal cancer risk: a meta‐analysis. Oncotarget 7, 76735–76742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Teo WH, Chen H‐P, Huang JC & Chan Y‐J (2017) Human cytomegalovirus infection enhances cell proliferation, migration and upregulation of EMT markers in colorectal cancer‐derived stem cell‐like cells. Int J Oncol 51, 1415–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Eliassen E, Lum E, Pritchett J, Ongradi J, Krueger G, Crawford JR, Phan TL, Ablashi D & Hudnall SD (2018) Human herpesvirus 6 and malignancy: a review. Front Oncol 8, 512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Dockrell DH (2003) Human herpesvirus 6: molecular biology and clinical features. J Med Microbiol 52, 5–18. [DOI] [PubMed] [Google Scholar]
- 158. Clark DA (2002) Human herpesvirus 6 and human herpesvirus 7: emerging pathogens in transplant patients. Int J Hematol 76(S2), 246–252. [DOI] [PubMed] [Google Scholar]
- 159. Sura R, Gavrilov B, Flamand L, Ablashi D, Cartun R, Colombel J‐F & Van kruiningen HJ (2010) Human herpesvirus‐6 in patients with Crohn's disease. Apmis 118, 394–400. [DOI] [PubMed] [Google Scholar]
- 160. Collin V & Flamand L (2017) HHV‐6A/B integration and the pathogenesis associated with the reactivation of chromosomally integrated HHV‐6A/B. Viruses 9, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Wakefield A, Fox J, Sawyerr A, Taylor J, Sweenie C, Smith M, Emery V, Hudson M, Tedder R & Pounder R (1992) Detection of herpesvirus DNA in the large intestine of patients with ulcerative colitis and Crohn’s disease using the nested polymerase chain reaction. J Med Virol 38, 183–190. [DOI] [PubMed] [Google Scholar]
- 162. Knösel T, Schewe C, Petersen N, Dietel M & Petersen I (2009) Prevalence of infectious pathogens in Crohn's disease. Pathol Res and Practice 205, 223–230. [DOI] [PubMed] [Google Scholar]
- 163. Sipponen T, Turunen U, Lautenschlager I, Nieminen U, Arola J & Halme L (2011) Human herpesvirus 6 and cytomegalovirus in ileocolonic mucosa in inflammatory bowel disease. Scand J Gastroenterol 46, 1324–1333. [DOI] [PubMed] [Google Scholar]
- 164. Halme L, Loginov R, Arola J, Turunen U & Lautenschlager I (2013) HHV‐6 antigen and HHV‐6 DNA expression in sporadic adenomatous polyps of the colon. Scand J Gastroenterol 48, 1423–1427. [DOI] [PubMed] [Google Scholar]
- 165. Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL & Buck CB (2010) Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe 7, 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Nicol J, Robinot R, Carpentier A, Carandina G, Mazzoni E, Tognon M, Touzé A & Coursaget P (2013) Age‐specific seroprevalences of merkel cell polyomavirus, human polyomaviruses 6, 7, and 9, and trichodysplasia spinulosa‐associated polyomavirus. Clin Vaccine Immunol 20, 363–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Rennspiess D, Pujari S, Keijzers M, Abdul‐Hamid MA, Hochstenbag M, Dingemans AM, Kurz AK, Speel EJ, Haugg A, Pastrana DV et al. (2015) Detection of human polyomavirus 7 in human thymic epithelial tumors. J Thorac Oncol 10, 360–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Wu JH, Simonette RA, Hsiao T, Doan HQ, Rady PL & Tyring SK (2015) Cutaneous human polyomavirus small T antigens and 4E‐BP1 targeting. Intervirology 58, 382–385. [DOI] [PubMed] [Google Scholar]
- 169. Campello C, Comar M, Zanotta N, Minicozzi A, Rodella L & Poli A (2010) Detection of SV40 in colon cancer: a molecular case‐control study from northeast Italy. J Med Virol 82, 1197–1200. [DOI] [PubMed] [Google Scholar]
- 170. Beimforde N, Hanke K, Ammar I, Kurth R & Bannert N (2008) Molecular cloning and functional characterization of the human endogenous retrovirus K113. Virology 371, 216–225. [DOI] [PubMed] [Google Scholar]
- 171. Boller K, Schnfeld K, Lischer S, Fischer N, Hoffmann A, Kurth R & Tnjes RR (2008) Human endogenous retrovirus HERV‐K113 is capable of producing intact viral particles. J Gen Virol 89, 567–572. [DOI] [PubMed] [Google Scholar]
- 172. Burmeister T, Ebert AD, Pritze W, Loddenkemper C, Schwartz S & Thiel E (2004) Insertional polymorphisms of endogenous HERV‐K113 and HERV‐K115 retroviruses in breast cancer patients and age‐matched controls. AIDS Res Hum Retroviruses 20, 1223–1229. [DOI] [PubMed] [Google Scholar]
- 173. Moyes DL, Martin A, Sawcer S, Temperton N, Worthington J, Griffiths DJ & Venables PJ (2005) The distribution of the endogenous retroviruses HERV‐K113 and HERV‐K115 in health and disease. Genomics 86, 337–341. [DOI] [PubMed] [Google Scholar]
- 174. Golan M, Hizi A, Resau JH, Yaal‐Hahoshen N, Reichman H, Keydar I & Tsarfaty I (2008) Human endogenous retrovirus (HERV‐K) reverse transcriptase as a breast cancer prognostic marker. Neoplasia 10, 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Marcuello M, Vymetalkova V, Neves RPL, Duran‐Sanchon S, Vedeld HM, Tham E, van Dalum G, Flügen G, Garcia‐Barberan V, Fijneman RJA et al. (2019) Circulating biomarkers for early detection and clinical management of colorectal cancer. Mol Aspects Med 69, 107–122. [DOI] [PubMed] [Google Scholar]
- 176. Kirkland PD, Gleeson AB, Hawkes RA, Naim HM & Boughton CR (1989) Human infection with encephalomyocarditis virus in New South Wales. Med J Aust 151, 178. [DOI] [PubMed] [Google Scholar]
- 177. Feng R, Wei J, Zhang H, Fan J, Li X, Wang D, Xie J, Qiao Z, Li M, Bai J et al. (2015) National serosurvey of encephalomyocarditis virus in healthy people and pigs in China. Arch Virol 160, 2957–2964. [DOI] [PubMed] [Google Scholar]
- 178. Yoon JW & Jun HS (2004) Viruses in type 1 diabetes: brief review. ILAR J 45, 343–348. [DOI] [PubMed] [Google Scholar]
- 179. Bandilovska I, Keam SP, Gamell C, Machicado C, Haupt S & Haupt Y (2019) E6AP goes viral: the role of E6AP in viral‐ and non‐viral‐related cancers. Carcinogenesis 40, 707–714. [DOI] [PubMed] [Google Scholar]
- 180. Srinivasan S & Nawaz Z (2011) E3 ubiquitin protein ligase, E6‐associated protein (E6‐AP) regulates PI3K‐Akt signaling and prostate cell growth. Biochim Biophys Acta 1809, 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Haegeman B, Hamelin J, Moriarty J, Neal P, Dushoff J & Weitz JS (2013) Robust estimation of microbial diversity in theory and in practice. ISME J 7, 1092–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Chung YR, Kim HJ, Kim YA, Chang MS, Hwang KT & Park SY (2017) Diversity index as a novel prognostic factor in breast cancer. Oncotarget 8, 97114–97126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Kim BR, Shin J, Guevarra RB, Lee JHH, Kim DW, Seol KH, Lee JHH, Kim HB & Isaacson RE (2017) Deciphering diversity indices for a better understanding of microbial communities. J Microbiol Biotechnol 27, 2089–2093. [DOI] [PubMed] [Google Scholar]
- 184. Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ & Walker AW (2014) Reagent and laboratory contamination can critically impact sequence‐based microbiome analyses. BMC Biol 12, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Eisenhofer R, Minich JJ, Marotz C, Cooper A, Knight R & Weyrich LS (2019) Contamination in low microbial biomass microbiome studies: issues and recommendations. Trends Microbiol 27, 105–117. [DOI] [PubMed] [Google Scholar]
- 186. Barr JJ (2017) A bacteriophages journey through the human body. Immunol Rev 279, 106–122. [DOI] [PubMed] [Google Scholar]
- 187. Lehti TA, Pajunen MI, Skog MS & Finne J (2017) Internalization of a polysialic acid‐binding Escherichia coli bacteriophage into eukaryotic neuroblastoma cells. Nat Commun 8, 1915–1928. 10.1038/s41467-017-02057-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Nguyen S, Baker K, Padman BS, Patwa R, Dunstan RA, Weston TA, Schlosser K, Bailey B, Lithgow T, Lazarou M et al. (2017) Bacteriophage transcytosis provides a mechanism to cross epithelial cell layers. MBio 8, e01874‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Yuen ST, Chung LP, Leung SY, Luk IS, Chan SY & Ho J (1994) In situ detection of Epstein‐Barr virus in gastric and colorectal adenocarcinomas. Am J Surg Pathol 18, 1158–1163. [DOI] [PubMed] [Google Scholar]
- 190. Wong NA, Herbst H, Herrmann K, Kirchner T, Krajewski AS, Moorghen M, Niedobitek F, Rooney N, Shepherd NA & Niedobitek G (2003) Epstein‐Barr virus infection in colorectal neoplasms associated with inflammatory bowel disease: detection of the virus in lymphomas but not in adenocarcinomas. J Pathol 201, 312–318. [DOI] [PubMed] [Google Scholar]
- 191. Vanoli A, Di Sabatino A, Martino M, Dallera E, Furlan D, Mescoli C, Macciomei MC, Biancone L, Neri B, Grillo F et al. (2017) Epstein‐Barr virus‐positive ileal carcinomas associated with Crohn's disease. Virchows Arch 471, 549–552. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Visualization of viral integration of human endogenous retrovirus K113 (HERV‐K113).
Table S1. Read counts stratified by tissue type.
Data Availability Statement
Anonymized sequence data are stored within the EGA database (accession number EGAS00001002717) as already described previously [41].


