Abstract
Introduction:
This signal finding study was designed to evaluate the efficacy of talazoparib in advanced stage squamous cell lung cancer harboring Homologous Recombination Repair Deficiency (HRRD).
Patient and Method:
The full eligible population (FEP) had tumors with a deleterious mutation in any of the study-defined HRR genes and without prior exposure to a PARP inhibitor. The primary analysis population (PAP) is a subset of FEP with alteration in ATM, ATR, BRCA1, BRCA2, or PALB2]. Treatment consisted of talazoparib 1mg daily, continuously in 21-day cycles. A 2-stage design with exact 93% power and 1-sided 0.07 level type I error required enrollment of 40 patients in the PAP in order to rule out an ORR of 15% or less if the true ORR is ≥35%.
Result:
The study enrolled 47 patients in the FEP of whom 24 were in the PAP. The median age for the FEP was 66.7 years; male: 83% and 85% White. Overall response rate (ORR) in the PAP was 4% (95%CI: 0, 21) with disease control rate (DCR) of 54% (95%CI: 33, 74); median PFS and OS of 2.4 months (95%CI: 1.5-2.8) and 5.2 months (95%CI: 4.0-10), respectively. In the FEP, ORR was 11% (95%CI: 3.6, 23); DCR of 51% (95%CI: 36, 66) and median duration of response was 1.8 months (95% CI: 1.3, 4.2); median PFS and OS were 2.5 months and 5.7 months, respectively.
Conclusions:
S1400G failed to show sufficient level of efficacy for single agent talazoparib in a biomarker defined subset of squamous lung cancer with HRRD.
Keywords: PARP, talazoparib, squamous, non-small cell lung cancer, trial
Introduction
The Lung Master Protocol (Lung-MAP), developed as a cooperative effort by the National Cancer Institute (NCI) and National Clinical Trials Network (NCTN), is an umbrella protocol containing a screening component and multiple independently conducted and analyzed treatment sub-studies.1, 2 The overarching hypothesis is that the umbrella master protocol will establish genomic screening for a large population of previously treated squamous NSCLC (SqNSCLC) patients to evaluate targeted therapies (or combinations) in biomarker-matched subpopulations (biomarker-driven sub-studies) within a single infrastructure (“umbrella”). The ultimate goal of Lung-MAP is the efficient approval of efficacious regimens. We report the results of the Lung-MAP substudy S1400G, a phase 2 study that evaluated single agent talazoparib, a PARP inhibitor, in SqNSCLC harboring genomic alterations predicted to result in homologous recombination repair deficiency (HRRD). Cancer cells are genomically unstable and are therefore prone to deleterious changes in critical genes including those that regulate intracellular DNA damage response (DDR). DDR involves a set of well-orchestrated intracellular processes that occur in response to endogenous and exogenous insults to the cell genome.3 This network of protein-protein interaction that effects the repair of damaged DNA, in addition to maintaining cell viability, may also confer survival advantage. The specific nature of the DNA damage calls for different DDR mechanisms to effect the needed repair. The base excision repair or single-strand break repair (BER/SSBR), nucleotide excision repair (NER) and mismatch repair (MMR) pathways repair single-strand breaks while non-homologous end-joining (NHEJ) and homologous recombination repair (HRR) are responsible for the repair of double-strand breaks. Defects in DDR genes and or other DNA damage signaling proteins such as MMR, BRCA1, BRCA2, and FANCD2 occur in familial cancers but can also be encountered in sporadic cancers. Impaired DDR as a result of loss of an important element of a specific repair pathway can engender a compensatory dependence on another component of the repair machinery thereby leading to a therapeutic vulnerability. The classic example is the impairment of the HRR pathway in patients with BRCA1 or BRCA2 mutation leading to inability of the cell to repair double strand breaks. Such patients are very vulnerable to inhibitors of poly (ADP) ribose polymerase (PARP) enzyme, which by inactivating the BER pathway leads to persistence and progression of single strand breaks to the highly lethal double-strand breaks.4, 5 This therapeutic strategy referred to as synthetic lethality has been clinically validated leading to regulatory approval of PARP inhibitors in patients with ovarian, breast and pancreatic cancers harboring deleterious mutations in DNA repair genes.6–9 Besides BRCA1 and BRCA2 mutations, genomic alterations affecting other critical components of HRR pathway such as RAD51, RAD54, DSS1, RPA1, NBS1, ATR, ATM, CHK1, CHK2, PTEN, FANCD2, FANCA, or FANCC can also result in synthetic lethality with PARP inhibition in preclinical models.10, 11
PARP is highly expressed in lung cancer including the SqNSCLC subtype suggesting a potential dependency on this enzyme.12 The frequency of deleterious alterations (deletions and mutations and copy number variations) in the publicly available TCGA data showed that SqNSCLC is in the top 10 cancers with deleterious alterations in HRR pathway genes. Approximately 50% of cases of SqNSCLC harbored a genetic alteration in at least one of the Fanconi Anemia (FA) gene family that are implicated in HRR.13 Moreover, preclinical studies showed sensitivity to PARP inhibitor of NSCLC cell lines with inactivation of HRR genes such BRACA1, BRACA2, and FANCD.14–17
S1400G was designed as a signal finding study of the PARP inhibitor, talazoparib, when used as a single agent in previously treated SqNSCLC harboring deleterious mutations in HRR pathway genes.
Materials and Methods
The primary objective of this phase II trial was to evaluate the overall response rate (ORR) with single agent talazoparib in HRRD SqNSCLC patients. The study was approved by the Institutional Review Board at all participating sites and all participants provided written informed consent. The study was conducted in accordance with the ethical principles enshrined in the declaration of Helsinki and the US common rules. This study was registered at www.clinicaltrials.gov with study reference number NCT02154490.
Eligible patients identified through the parent S1400 Lung-MAP screening platform were required to have tumors harboring a somatic deleterious mutation in any of the study-defined HRR genes [ATM, ATR, BARD1, BRCA1, BRCA2, BRIP1, CHEK1, CHEK2, FANCA, FANCC, FANCD2, FANCF, FANCM, NBN (NBS1), PALB2, RAD51, RAD51B (RAD51L1), RAD54L, RPA1] defined as the full eligible population (FEP). The primary analysis population (PAP) was defined by a subset of genes [ATM, ATR, BRCA1, BRCA2, and PALB2]. Other eligibility requirements included prior benefit from a platinum-based chemotherapy and progression of disease on the most recent line of systemic therapy, a Zubrod performance status of 0-1, adequate organ function, and no previous exposure to a PARP inhibitor.
Study design and treatment:
The study was designed as a single-arm, 2-stage phase II clinical trial to study the efficacy of talazoparib. Talazoparib was administered orally at a dose of 1 mg once daily in a 21-day treatment cycle. Treatment was continued until evidence of disease progression, intolerable toxicity, or withdrawal of consent by the patient.
Efficacy:
Treatment effect was assessed using measurable target lesions according to RECIST 1.1 criteria 18 based on cross-sectional anatomic imaging scans obtained at baseline and repeated at the end of every two cycles of treatment until progression or withdrawal from study. Efficacy was classified as complete response (CR), partial response (PR), stable disease (SD), or progressive disease (PD). Progression free survival (PFS) was calculated from beginning of treatment to the time of disease progression or death while overall survival (OS) was calculated from treatment initiation until date of death or censoring.
Mutational analysis was performed on archival formalin-fixed paraffin-embedded (FFPE) tumor specimens in a Clinical Laboratory Improvement Amendments (CLIA)-certified and College of American Pathologists (CAP)-accredited laboratory (Foundation Medicine, Cambridge, MA). Genomic DNA (⩾50 ng) was extracted from FFPE specimens and sonicated to ~200bp fragments. Material underwent whole-genome shotgun library construction and hybridization-capture of at least 236 genes and selected introns of 19 genes involved in rearrangements. Using the Illumina® HiSeq 2000, 2500, and 4000 platforms, hybrid-capture-selected libraries were sequenced using 49 × 49bp paired-end reads to high uniform depth. Sequence data were processed using a customized analysis pipeline designed to accurately detect base substitutions, small insertions and deletions, focal copy number amplifications, homozygous gene deletions, and genomic rearrangements. Patients assigned to the S1400G substudy must meet biomarker eligibility of Homologous Recombination Repair Deficiency (HRRD) defined as truncating mutation, frameshift deletions, indels, missense and nonsense mutations predicted to have functional consequence in any of the specified HRR genes using the Foundation Medicine genomic sequencing platform.
Statistical Considerations
The primary objective was to evaluate the RECIST 1.1 overall response rate (ORR; confirmed and unconfirmed, complete and partial) in patients with SqNSCLC in the PAP treated with talazoparib. The accrual goal was 40 response-evaluable PAP patients. It was estimated that 67% of patients would satisfy the criteria to be in the PAP for an estimated accrual goal of 60 patients in the full eligible population (FEP). The sample size in the PAP was based on a design with 91% power to rule out a RR of 15% at the 1-sided 5% level, if the true rate were 35%. The observation of 10/40 (25% ORR) in the PAP would be considered evidence to rule out the null RR and evidence to pursue an independent randomized phase III trial. The design included an interim analysis at 20 PAP patients evaluable for response, with no hold in accrual. If 2 or fewer responses were observed, accrual to the study would be terminated with the conclusion of insufficient evidence to continue. A key secondary objective was an investigator assessment of median PFS (mPFS) in the PAP. If the study continued to full accrual and the RR rate in the PAP was less than 25% but the mPFS was at least 4.5 months, this would be considered sufficient evidence to continue to the follow-on Phase III. With 40 PAP patients, this design had 90% power to rule out a median PFS of 3 months or less, if the true mPFS were 6 months, at the 0.05 1-sided level. The observation of an mPFS of at least 4.5 months would be considered evidence to rule out a mPFS of 3 months or less. Binary proportions and associated 95% confidence intervals were estimated. Survival distributions were estimated using the method of Kaplan-Meier and the Brookmeyer-Crowley method was used to estimate confidence intervals.
Results
Patient population and duration of study:
The study was activated February 2017 and closed to accrual in July 2018. There were 72 patients (4.2% of patients screened on S1400 as of July 23, 2018) assigned to the S1400G substudy and 51 patients were enrolled. Of the 21 biomarker positive patients who did not register after assignment to the study, eight were not eligible, five had symptomatic deterioration, three succumbed to death, two were investigator decision, one was not eligible for the screening study, and one was enrolled to another Lung-MAP substudy. Three of the 51 patients registered to S1400G were ineligible due to progressive disease as best response on first line platinum-based chemotherapy in two patients and one patient who never received prior platinum based chemotherapy. Additionally, one patient passed away before receiving any protocol treatment. Thus, these four were not included in the safety and efficacy analyses leaving a total of 47 patients who were eligible for the study. There were 27 patients assigned to the primary analysis population based on protocol defined somatic alterations in BRCA1, BRCA2, ATM, ATR and PALB2. The details of patient demographics and tumor characteristics for the full eligibility population and the primary analysis population are summarized in Table 1.
Table 1:
Demographics and tumor characteristics in FEP and PAP
| Full Eligible Patients (FEP) (N=47) |
Primary Analysis Population (PAP) (N=24*) |
|
|---|---|---|
|
Age Median (Range) |
66.7 (37.5-82.1) | 68 (41.8-82) |
|
Male Gender
|
39 (83%) | 21 (88%) |
|
Performance Status
|
||
| 0 |
10 (21%) | 6 (25%) |
| 1 |
37 (79%) | 18 (75%) |
|
Race/Ethnicity
|
||
| White |
40 (85%) | 22 (92%) |
| Black |
7 (15%) | 2 (8%) |
|
Hispanic ethnicity
|
||
| Yes |
2 (4%) | 2 (8%) |
| No |
44 (94%) | 21 (88%) |
| Unknown |
1 (2%) | 1 (4%) |
|
Number of Prior Lines of Therapy For Stage IV Disease
|
||
| 0 |
11 (23%) | 5 (21%) |
| 1 |
13 (28%) | 7 (29%) |
| 2 or more |
23 (49%) | 12 (50%) |
|
Smoking Status
|
||
| Current Smoker |
7 (15%) | 5 (21%) |
| Recent Smoker |
8 (17%) | 3 (13%) |
| Former Smoker |
31 (66%) | 15 (63%) |
| Never smoker |
1 (2%) | 1 (4%) |
: three of 27 patients assigned to the study did not start treatment
Mutation analysis and patient assignment to S1400G substudy
Alterations in the Fanconi Anemia (FA) pathway gene family were the most frequently observed HRRD mutation in the 47 eligible patients enrolled on this substudy (FANCM: 15%; FANCD2: 2%; FANCF: 4% and FANCA: 9%). Overall, short variant alterations in TP53 gene was the most frequently reported genetic alteration in 43 (91.5%) patients. Other common alteration occurring in >10% of patients included MLL2 (21.2%), BRCA2, CDKNA2A (17%), BRCA1 and LRP1B (12.8%) and PTEN in 5 (10.6%) patients. Copy number alterations were reported in the CDKN2A and SOX2 genes (27.7%); CDKN2B (25.5%); PIK3CA (23.4%) and FGF12 (21.3%) patients. Full details of the HRRD mutation summary and the specific types of genetic mutations are presented in Supplemental Table 1.
Protocol Treatment
Eligible patients started treatment with single agent talazoparib within 21 days of registration to the study and continued until evidence of disease progression or intolerable toxicity. The median number of cycles of treatment received was (3 cycles). Overall, treatment was well tolerated without any new safety signal for talazoparib and most adverse events were grade 1 or 2 in severity. The most frequent treatment emergent adverse events of all grades regardless of attribution and occurring in > 10% of patients were: anemia (55%), thrombocytopenia (43%), fatigue (38%), nausea (34%), lymphopenia (23%), leukopenia (17%), anorexia (15%), vomiting (15%) and weight loss (13%). The most frequent grades 3/4 adverse events were anemia (7 patients), thrombocytopenia (6 patients) and lymphopenia (4 patients). There was a single case of grade 5 adverse event of respiratory failure considered related to disease progression. Details of the treatment emergent adverse events are summarized in Table 2.
Table 2.:
Treatment Emergent Adverse Events Regardless of Attribution
| ADVERSE EVENTS (N=47) | Grade | ||||||
|---|---|---|---|---|---|---|---|
| 1 (N) | 2 (N) | 3 (N) | 4 (N) | 5 (N) | Total N (%) | ||
| Anemia | 11 | 8 | 6 | 1 | 0 | 26 | 55% |
| Platelet count decreased | 11 | 3 | 4 | 2 | 0 | 20 | 43% |
| Fatigue | 8 | 8 | 2 | 0 | 0 | 18 | 38% |
| Nausea | 12 | 1 | 3 | 0 | 0 | 16 | 34% |
| Lymphocyte count decreased | 4 | 3 | 4 | 0 | 0 | 11 | 23% |
| White blood cell decreased | 5 | 2 | 1 | 0 | 0 | 8 | 17% |
| Vomiting | 4 | 2 | 1 | 0 | 0 | 7 | 15% |
| Hypoalbuminemia | 1 | 2 | 1 | 0 | 0 | 4 | 9% |
| Hyponatremia | 2 | 0 | 1 | 1 | 0 | 4 | 9% |
| Hypertension | 0 | 2 | 1 | 0 | 0 | 3 | 6% |
| Neutrophil count decreased | 1 | 1 | 1 | 0 | 0 | 3 | 6% |
| ALT increased | 1 | 0 | 1 | 0 | 0 | 2 | 4% |
| AST increased | 0 | 1 | 1 | 0 | 0 | 2 | 4% |
| Dehydration | 0 | 1 | 1 | 0 | 0 | 2 | 4% |
| Generalized muscle weakness | 0 | 1 | 1 | 0 | 0 | 2 | 4% |
| Dyspnea | 0 | 0 | 1 | 0 | 0 | 1 | 2% |
| Hypotension | 0 | 0 | 1 | 0 | 0 | 1 | 2% |
| Hypoxia | 0 | 0 | 0 | 1 | 0 | 1 | 2% |
| Lung infection | 0 | 0 | 0 | 1 | 0 | 1 | 2% |
| Respiratory failure | 0 | 0 | 0 | 0 | 1 | 1 | 2% |
| Sepsis | 0 | 0 | 0 | 1 | 0 | 1 | 2% |
| Wound infection | 0 | 0 | 1 | 0 | 0 | 1 | 2% |
| Maximum Grade Any Adverse Event | 14 | 11 | 14 | 4 | 1 | ||
Efficacy
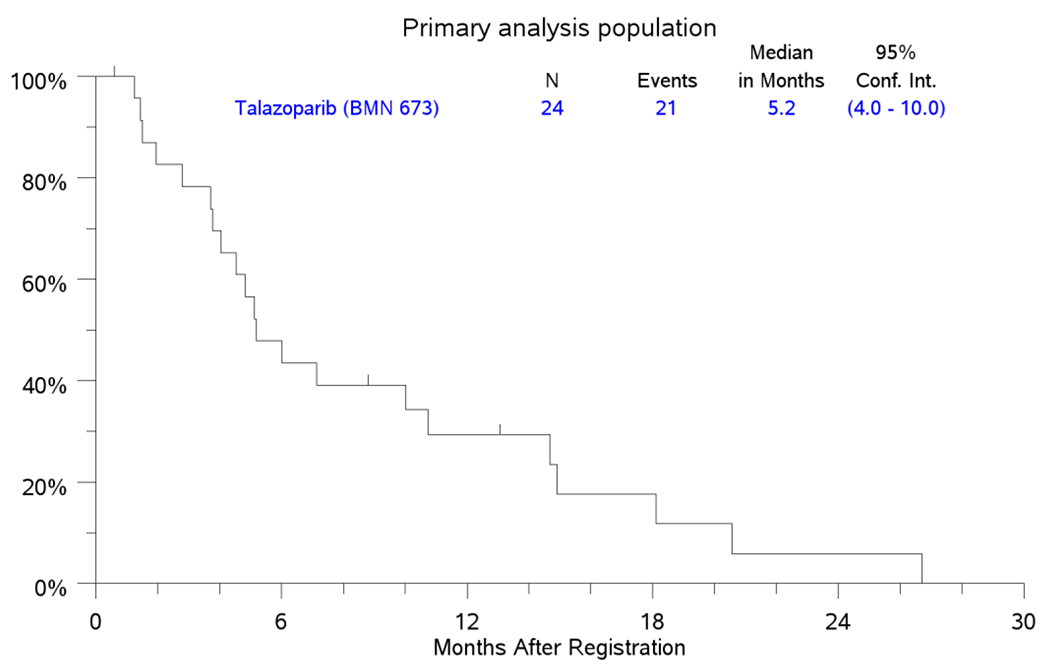
Assessment for efficacy of treatment in the 24 eligible patients in the PAP showed a CR rate of 0%, PR (4%), SD 50% and PD in 25% of patient; three patients did not meet the minimum assessment required (Figure 1). The median PFS in the PAP was 2.4 months (95% CI: 1.5-2.8) while the median OS was 5.2 months (95%CI: 4-10); Figure 2A and 2B. The study was therefore closed to further enrolment at the end of stage I accrual due to failure to demonstrate the minimum efficacy signal in terms of ORR.
Figure 1:
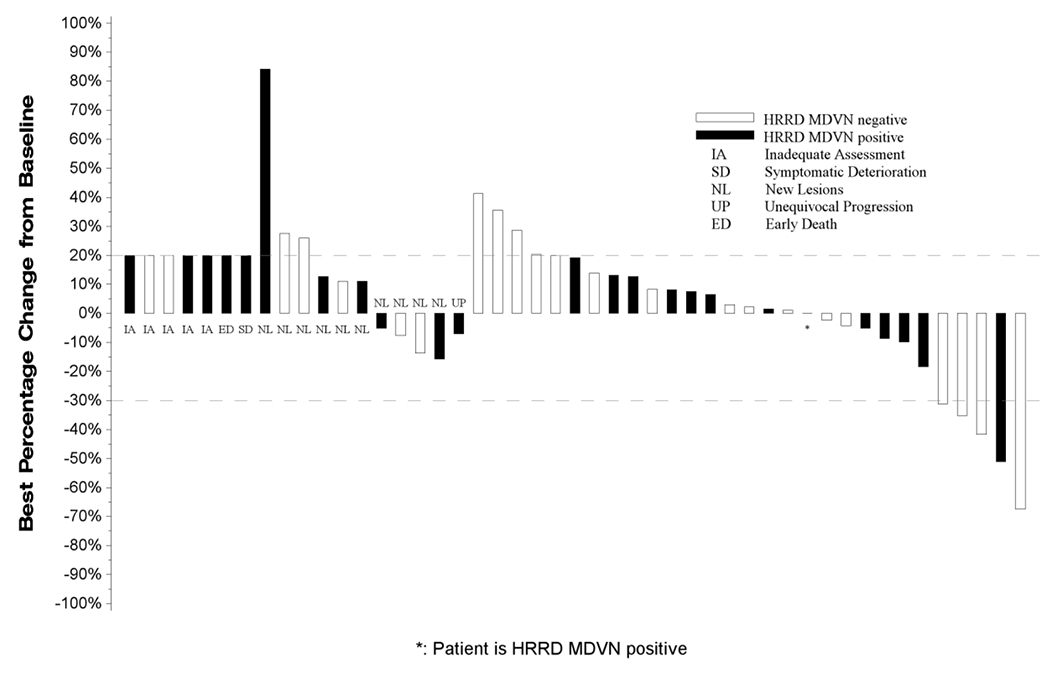
Waterfall Plot with Annotations for Individual Alterations
All eligible patients who received at least one dose of Talazoparib (BMN673) are represented in this plot. From left to right, best change in tumor measurements included in the plot are displayed as;
• IA: Measurements for patients who did not have any follow-up tumor disease assessments are labeled with ‘IA’ and displayed as a 20% increase, the threshold for progressive disease.
• SD: Measurements for patients who had symptomatic deterioration at first disease assessment are labeled as “SD” and displayed as a 20% increase, the threshold for progressive disease.
• ED: Measurements for patients who expired prior to disease assessment, but the death was not due to disease are labeled as “ED” and displayed as a 20% increase, the threshold for progressive disease.
• NL: Measurements for patients who had progressive disease at their first assessment based on new lesions are labeled by “NL”. The percentage change from baseline to first disease assessment is displayed.
• UP: Measurements for patients who had unequivocal progression in non-target lesions at their first follow-up assessment are labeled as “UP”. The percentage change from baseline to first disease assessment is displayed.
• Unlabeled bars: To the right of the labeled bars are bars representing the best change in measurements for patients not coded as IA, SD, ED, NL, or UP with at least one follow-up disease assessment.
Bars extending above the dashed line at +20% or labeled as IA, SD, ED, NL, or UP are coded as progressive disease. Unlabeled bars between the dashed lines (+20% to −30%) are coded as best response of stable disease and unlabeled bars extending below the −30% line are coded as best response of partial or complete response.
Figure 2.
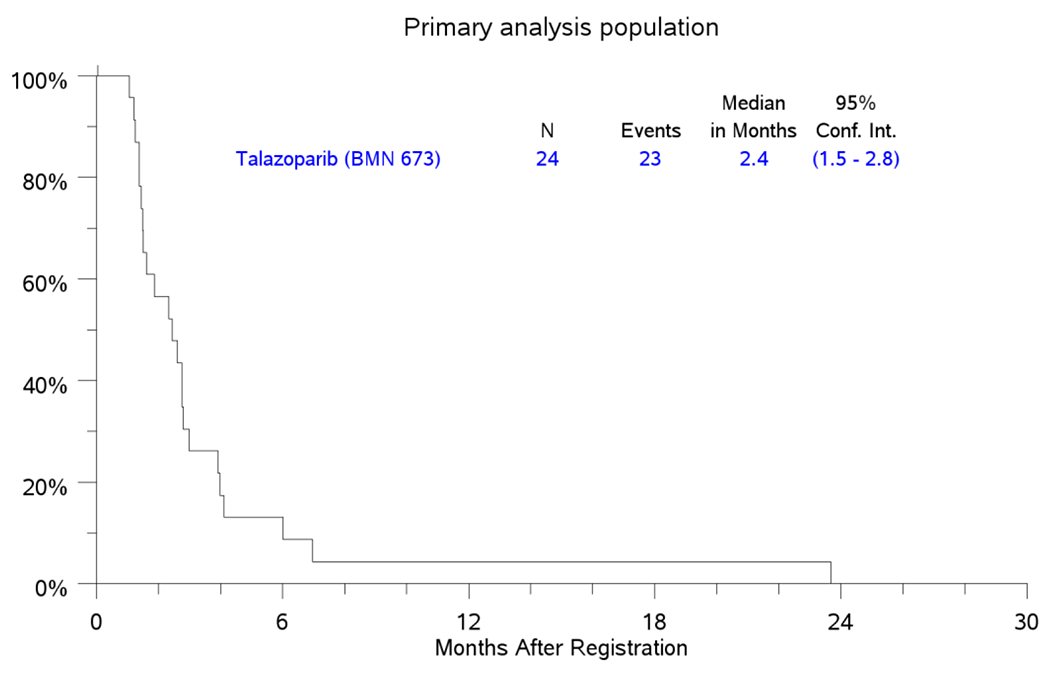
PFS and OS for Primary Analysis Population Patients
2a. PFS
2B. OS
Kaplan-Meier Curves for PFS and OS in the primary analysis population (PAP) counting from the date of registration
Secondary analysis for efficacy in the FEP employed data from 47 eligible patients and showed a CR rate of 0%; PR in 5 patients (11%); SD in 19 (40%) patients and best response of PD in 16 (34%) patients (Figure 1). Five patients (11%) in the FEP were not eligible for efficacy assessment. The median PFS in the FEP was 2.5 months (95%CI: 1.6-3.0) while the median OS was 5.7 months (95%CI: 4.5-8.7); Figure 3A and 3B. Five patients in the FEP, including 1 patient who was included in the PAP achieved objective response by RECIST criteria (2 confirmed and 3 unconfirmed responses). These patients harbored mutations in BRCA2 (1), FANCM (3), FANCC (1) and CHEK1 (1); one patient had more than one HRRD mutation (FANCM and FANCC).
Figure 3.
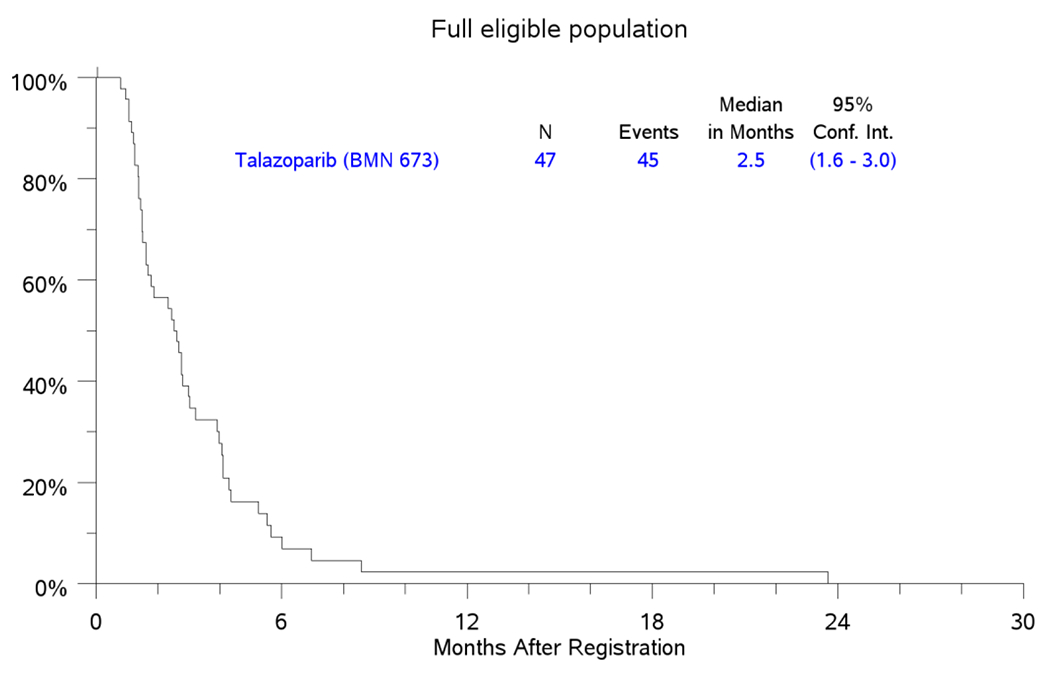
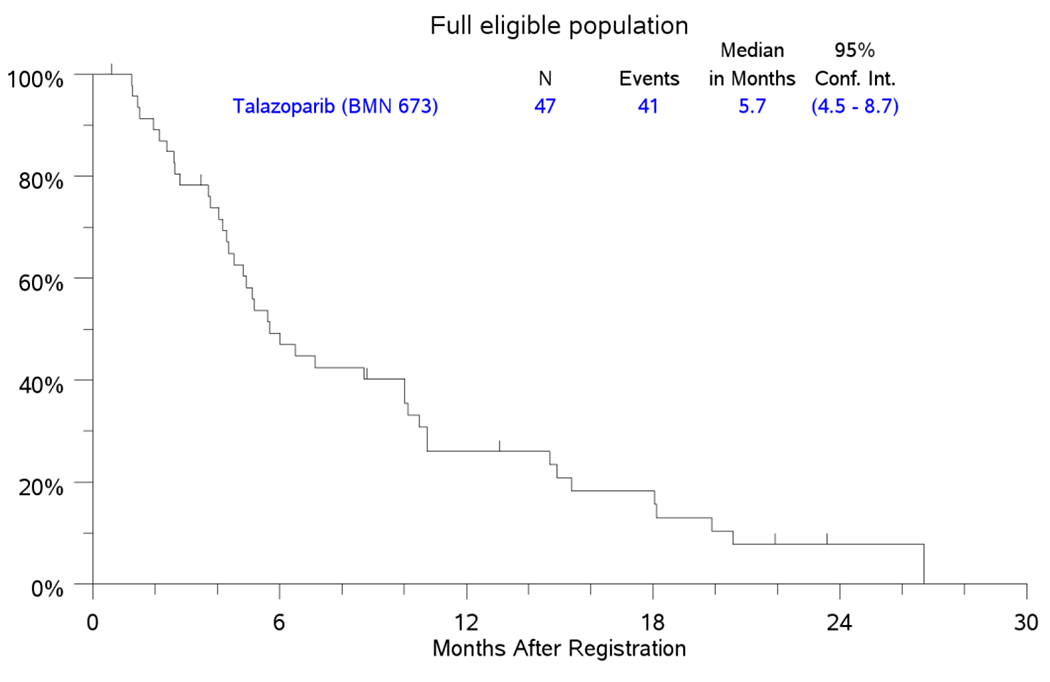
PFS and OS in FEP
3A. KM Plot for PFS.
3B. KM Plot for OS.
Kaplan-Meier Curves for PFS and OS in the full eligible population (FEP) counting from the date of registration.
Discussion
Treatment options for patients with SqNSCLC are much more limited than for patients with non-squamous NSCLC, especially patients whose tumor has a driver alterations. To date no driver alteration informs the use of targeted therapy specifically in SqNSCLC. The Lung-MAP trial is an umbrella trial designed to bridge this gap in our knowledge of genomics of SqNSCLC and to use such knowledge to efficiently develop newer and effective treatment options for this disease.1, 2 The S1400G substudy was designed to test the efficacy of talazoparib as a single agent in patients whose tumors have deficient HRR machinery. While 4.2% of patients with SqNSCLC screened on the Lung-MAP protocol were assigned to S1400G, the total prevalence of HRRD was approximately 19% indicating a sizeable subset of lung cancer patients in this category.
The study pre-specified a PAP as those harboring somatic alterations in BRCA1, BRCA2, ATM, ATR and PALB2 genes. However, four of the five patients with objective response had alterations in FANCM, FANCC or CHEK1 genes, which were not included in the genomic definition of our PAP. The study did not meet the efficacy threshold in the PAP required for a larger definitive trial of single agent talazoparib but efficacyin tumors with genomic alterations affecting the Fanconi Anemia complementation (FANC) gene family was intriguing. FANC mutations were the most prevalent HRRD mutation in the FEP occurring in approximately 39% of cases; and in more than 60% of patients when we factor in PALB2 and BRCA1, which have recently been included in the FA gene family (FANCN or PALB2 and FANCS or BRCA1).13, 19 This rate is comparable to that described in an analysis of the TCGA database where approximately 50% of SqNSCLC harbored somatic mutations in the FANC pathway genes.13 Interestingly, approximately 35% of lung adenocarcinoma samples in the TCGA database also harbored alterations in the same FA pathway genes.13 Further support for the biological relevance of the signal observed in this clinical trial is the finding from a recent analysis of publicly available sequencing data showing an enrichment for germline mutation in FANC in SqNSCLC.20
The FA gene family consists of 22 known members that encode 22 FA proteins.13, 19 These proteins are directly involved in an integrated DNA repair pathway, the FA/BRCA pathway that is critical for the repair of DNA interstrand crosslinks.21, 22 Following DNA damage, FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, and FANCM form a nuclear core complex, which interacts with FANCL and leads to the recruitment and ubiquitination of FANCD2 at sites of DNA damage where it associates with BRCA1 and BRCA2.21–23 Germline mutations in FA pathway genes result in a heterogeneous disease phenotype including increased risk of cancer.13, 21 The central role of FA pathway genes in DNA repair and the prevalence of genetic alterations in the FA pathway genes is a vulnerability that could be synthetically lethal with PARP inhibitors in SqNSCLC patients.
Talazoparib is the most potent PARP inhibitor due to its superior PARP trapping ability.24, 25 It was effective against FANCD2-deficient lung cancer cell lines.17 The clinical efficacy of talazoparib has also been established leading to its regulatory approval for treatment of locally advanced/metastatic breast cancer patients with a deleterious/suspected deleterious germline BRCA1/2 mutation.6 Failure to show efficacy of talazoparib in the current study is therefore unlikely to be due to a lack of efficacy of talazoparib but more likely a result of an imperfect biomarker definition. While all the eligible genes for study are critical components of the HRR pathway, it is conceivable that the degree of redundancy in the repair pathways makes certain mutations insufficient by themselves to induce synthetic lethality when combined with a PARP inhibitor. Future iteration of this strategy may benefit from additional clinical validation of the HRRD biomarker definition as well as combination strategies with DNA damaging cytotoxic chemotherapy,immunce checkpoint blockade, ionizing radiation or agents targeting other DNA repair proteins such as DNA-PK and ATR inhibitors.
Supplementary Material
Funding:
The Lung-MAP trial was supported in part by the National Institutes of Health NCI [grant numbers CA180888, CA180819, CA180820, CA180821, CA180868, CA189971, CA189858, CA180846, CA180835, CA189809, CA189812, CA180828, CA189997, CA189854, CA189821, CA189972, CA180798,CA233230, CA189808, CA180826]; Pfizer through the Foundation for the National Institutes of Health, in partnership with Friends of Cancer Research.
Conflicts of Interest
List for each author declaring potential conflicts. Follow order of author list. TKO (grant funding to institution from Pfizer Inc.). VAP worked at MD Anderson Cancer Center at the time of study conduct but is currently an employee of Pfizer Inc.
Footnotes
No Conflicts of Interest
The following authors declare no potential conflicts of interest: MWR, LAB, FRH, PCM, LHS, JDB, TES, NBL, TAB, PL, JM, KK, SSR, RSH, and DRG
References
- 1.Herbst RS, Gandara DR, Hirsch FR, et al. Lung Master Protocol (Lung-MAP)-A Biomarker-Driven Protocol for Accelerating Development of Therapies for Squamous Cell Lung Cancer: SWOG S1400. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:1514–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lam VK, Papadimitrakopoulou V. Master protocols in lung cancer: experience from Lung Master Protocol. Current opinion in oncology. 2018;30:92–97. [DOI] [PubMed] [Google Scholar]
- 3.Curtin NJ. Inhibiting the DNA damage response as a therapeutic manoeuvre in cancer. British journal of pharmacology. 2013;169:1745–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. [DOI] [PubMed] [Google Scholar]
- 5.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. [DOI] [PubMed] [Google Scholar]
- 6.Litton JK, Rugo HS, Ettl J, et al. Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. The New England journal of medicine. 2018;379:753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robson M, Im SA, Senkus E, et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. The New England journal of medicine. 2017;377:523–533. [DOI] [PubMed] [Google Scholar]
- 8.Golan T, Hammel P, Reni M, et al. Maintenance Olaparib for Germline BRCA-Mutated Metastatic Pancreatic Cancer. The New England journal of medicine. 2019;381:317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pujade-Lauraine E, Ledermann JA, Selle F, et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): a double-blind, randomised, placebo-controlled, phase 3 trial. The Lancet. Oncology. 2017;18:1274–1284. [DOI] [PubMed] [Google Scholar]
- 10.Bolderson E, Richard DJ, Zhou BB, Khanna KK. Recent advances in cancer therapy targeting proteins involved in DNA double-strand break repair. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:6314–6320. [DOI] [PubMed] [Google Scholar]
- 11.McCabe N, Turner NC, Lord CJ, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–8115. [DOI] [PubMed] [Google Scholar]
- 12.Byers LA, Wang J, Nilsson MB, et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer discovery. 2012;2:798–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nalepa G, Clapp DW. Fanconi anaemia and cancer: an intricate relationship. Nature reviews. Cancer. 2018;18:168–185. [DOI] [PubMed] [Google Scholar]
- 14.Ji W, Weng X, Xu D, Cai S, Lou H, Ding L. Non-small cell lung cancer cells with deficiencies in homologous recombination genes are sensitive to PARP inhibitors. Biochemical and biophysical research communications. 2019. [DOI] [PubMed] [Google Scholar]
- 15.Paul I, Savage KI, Blayney JK, et al. PARP inhibition induces BAX/BAK-independent synthetic lethality of BRCA1-deficient non-small cell lung cancer. The Journal of pathology. 2011;224:564–574. [DOI] [PubMed] [Google Scholar]
- 16.Morra F, Luise C, Visconti R, et al. New therapeutic perspectives in CCDC6 deficient lung cancer cells. International journal of cancer. Journal international du cancer. 2015;136:2146–2157. [DOI] [PubMed] [Google Scholar]
- 17.Duan W, Gao L, Aguila B, Kalvala A, Otterson GA, Villalona-Calero MA. Fanconi anemia repair pathway dysfunction, a potential therapeutic target in lung cancer. Frontiers in oncology. 2014;4:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). European journal of cancer. 2009;45:228–247. [DOI] [PubMed] [Google Scholar]
- 19.Niraj J, Farkkila A, D’Andrea AD. The Fanconi Anemia Pathway in Cancer. Annu Rev Cancer Biol. 2019;3:457–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esai Selvan M, Klein RJ, Gumus ZH. Rare, Pathogenic Germline Variants in Fanconi Anemia Genes Increase Risk for Squamous Lung Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2019;25:1517–1525. [DOI] [PubMed] [Google Scholar]
- 21.D’Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nature reviews. Cancer. 2003;3:23–34. [DOI] [PubMed] [Google Scholar]
- 22.Crossan GP, Patel KJ. The Fanconi anaemia pathway orchestrates incisions at sites of crosslinked DNA. The Journal of pathology. 2012;226:326–337. [DOI] [PubMed] [Google Scholar]
- 23.Shakeel S, Rajendra E, Alcon P, et al. Structure of the Fanconi anaemia monoubiquitin ligase complex. Nature. 2019;575:234–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hopkins TA, Shi Y, Rodriguez LE, et al. Mechanistic Dissection of PARP1 Trapping and the Impact on In Vivo Tolerability and Efficacy of PARP Inhibitors. Molecular cancer research : MCR. 2015;13:1465–1477. [DOI] [PubMed] [Google Scholar]
- 25.Murai J, Huang SY, Renaud A, et al. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Molecular cancer therapeutics. 2014;13:433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


