Abstract
Inexpensive, rapid, and reliable methods of detecting infection by and drug susceptibility of Mycobacterium tuberculosis (MTB) are crucial to the control of tuberculosis. The novel microscopic observation broth-drug susceptibility assay (MODS) detects early growth of MTB in liquid medium, allowing more timely diagnosis and drug susceptibility testing. Sputum samples from hospitalized patients in Peru were analyzed by using stains, culture, and PCR. Sensitivity of MODS (92%) compared favorably with the most sensitive of the other culture methods (93%). Sputum samples positive for tuberculosis were tested for susceptibility to isoniazid and rifampin with the microwell alamar blue assay (MABA) and MODS. In 89% of cases, there was concordance between MODS and MABA. Of the diagnostic and susceptibility testing methods used, MODS yielded results most rapidly (median, 9.0 and 9.5 days, respectively). MODS is a rapid, inexpensive, sensitive, and specific method for MTB detection and susceptibility testing; it is particularly appropriate for use in developing countries burdened by significant infection rates and increasing numbers of multiple-drug-resistant cases.
In the last decade, there has been a dramatic resurgence in the incidence of tuberculosis (TB) throughout the world. Contributing factors include population growth, the human immunodeficiency virus epidemic, and poorly implemented public health measures (19). In developing countries, efforts to curb TB are further hindered by overcrowding, poverty, poor infection control in hospitals, widespread AIDS, and severe lack of financial and technical resources for effective control programs. The number of TB patients infected with multiple-drug-resistant strains is also on the rise, particularly in developing countries; this is due in large part to the failure of control programs to provide adequate treatment and to the marked increase in high-risk groups, such as patients with AIDS.
To successfully control the spread of TB, cases must be detected and treated in a timely, effective manner. Well-funded clinical laboratories can detect TB cases within 7 to 14 days using sophisticated liquid culturing systems such as BACTEC or Mycobacteria Growth Indicator tubes (MGITs) (15, 24). TB laboratories in most developing countries lack the sophisticated and costly equipment required by these systems but generally have a centrifuge, a microscope, and an incubator. If cultures are attempted at all, most laboratories in developing countries use solid media such as Lowenstein-Jensen (LJ) and Ogawa because of their lower cost. With these traditional culture methods, Mycobacterium tuberculosis (MTB detection takes an average of 3 weeks under optimal conditions, and drug susceptibility testing takes an additional 3 to 4 weeks (9). Consequently, TB control programs in developing countries rely upon results of stained sputum smears for diagnosis, and treatment regimens are usually prescribed without assessing the drug susceptibility profiles of infecting strains (4, 5).
MTB grows more rapidly on liquid medium, where it grows as strings and tangles, than on solid medium (1). Based on this, we developed a new, efficient, reliable, and inexpensive method that permits simultaneous MTB detection and determination of drug susceptibility in less than 2 weeks. Sputum samples are first digested to reduce viscosity and kill other bacteria and then concentrated by centrifugation. They are diluted into 7H9 broth and distributed to wells of a 24-well plastic culture tray with and without antimicrobial agents. MTB growth is observed as the formation of characteristic strings and tangles by simple light microscopy. This microscopic observation broth drug susceptibility (MODS) assay, unlike other rapid detection systems, does not require either radioactive isotopes or fluorescent indicators. It is particularly well suited for use in developing countries, as it is rapid, reliable, and inexpensive.
MATERIALS AND METHODS
This study was carried out in a level P2 biosafety tuberculosis laboratory in the Department of Pathology at the Universidad Peruana Cayetano Heredia in Lima, Peru. In the United States and Western Europe, it is customary to handle MTB at a P3 level; in Peru, however, as in much of the developing world, the majority of MTB is handled in P1 containment facilities due to resource constraints.
Detection of mycobacteria. (i) Specimen processing.
Six to 8 ml of sputum was collected from 172 samples from patients who were admitted to six hospitals in Lima, Peru. Most patients were suspected of having tuberculosis based on clinical symptoms and/or a positive stain for acid-fast bacilli (AFB). Sputum samples collected first thing in the morning from 10 subjects with mild chronic bronchitis who were otherwise healthy (laboratory personnel) were assayed as negative controls. All specimens were digested and decontaminated of other bacteria by the standard N-acetyl-l-cysteine–NaOH–Na citrate method (4). After centrifugation for 15 min at 3,000 × g, the pellet was then resuspended into 2 ml of 7H9 broth.
(ii) Smear test.
A total of 100 μl of each decontaminated sample was placed on a microscope slide and stained with 0.1% auramine O by standard procedures (9). Slides were examined microscopically at ×1000 magnification. A stain was considered positive if it contained at least five bacilli per 300 fields.
(iii) MTB culture techniques.
Five hundred microliters of each decontaminated sample was inoculated into an MGIT (Becton Dickinson, Sparks, Md.) containing both 10% OADC (oleic acid, albumin, dextrose, and catalase; Becton Dickinson) and 100 μl of PANTA antimicrobial supplement (polymyxin B, amphotericin B, nalidixic acid, trimethoprim, and azlocillin; Becton Dickinson), as per manufacturer's instructions. Two hundred-fifty microliters of sample was used to inoculate one LJ slant (Difco, Detroit, Mich.), and another 250 μl was used to inoculate one plate of Middlebrook's 7H11 medium (Difco). MGITs were incubated at 37°C and examined for fluorescence every day for up to 40 days with a box containing two longwave UV light sources. LJ slants were incubated at 37°C and examined twice a week, from week 2 to 8, following inoculation. Microagar 7H11 plates were incubated at 37°C in an atmosphere of 5% CO2 in humidified air and then examined under an inverted light microscope. The plates were examined once in the first week and then 2 to 3 times per week, every week, for up to 6 weeks following inoculation (8). Plates and slants were interpreted according to criteria developed by the National Centers for Disease Control and Prevention (9). A culture was considered positive if it contained at least one colony. All positive MGITs were checked with an auramine O stain to confirm the presence of AFB. Any suspicious colonies observed as growing on LJ or 7H11 were also stained to check for acid fastness. The identities of all cultures were blinded to laboratory workers.
(iv) Growth detection in MODS.
Eight hundred microliters of each decontaminated sample was inoculated into 7.2 ml of Middlebrook 7H9 broth (Difco) containing 5.9 g of Middlebrook 7H9 broth base per liter, 0.31% glycerol, 1.25 g of Bacto Casitone (Difco) per liter, 10% OADC, and 160 μl of PANTA Antimicrobic Supplement Stock (20 μl/ml). For each sputum-medium mixture, aliquots of 1.2 ml (each) were distributed to 6 wells of a sterile 24-well plate (Falcon, Franklin Lakes, N.J.). A 24-well plate rather than a 96-well plate was used to provide increased growth medium and also to improve microscopic visualization of the bacteria. When three specimens had been run, the fourth set of 6 wells was filled with broth only to control for cross-contamination. Each run also included a susceptible control of MTB strain H37Rv in which a 1/50 dilution of 0.5 McFarland standard equivalent in broth was added to one well. This was examined daily for growth between 6 and 8 days. Plates were sealed with Scotch polyethylene tape (Fisher, Springfield, N.J.) and incubated at 37°C for up to 40 days. Aliquots were examined daily under an inverted light microscope at ×40 magnification. Each well was examined for approximately 30 to 45 s each. The identities of the cultures were blinded to laboratory workers.
Cultures of M. avium ATCC 35713 and 35718, Mycobacterium chelonae ATCC 35752, Mycobacterium smegmatis ATCC 19420, Mycobacterium kansasii ATCC 12478, Mycobacterium bovis ATCC 19210 (obtained from American Type Culture Collection, Washington, D.C.), and MTB strain H37Rv (ATCC 27294) were inoculated into coded wells containing 7H9 medium. The cultures were examined, and species morphology was described by laboratory personnel who did not know the identity of the cultures. The study was done to determine the ability of the microscopist to identify MTB accurately in comparison with the other strains.
(v) PCR.
PCR was performed using separate rooms and separate pipettes to avoid cross-contamination. Before DNA extraction, 500 μl of each decontaminated sample was incubated with 2 volumes of absolute ethanol for 2 h at room temperature. Samples were then centrifuged at 13,000 × g for 10 min; supernatants were discarded and pellets were submitted to a simple heat–and–Triton X-100 detergent treatment for DNA extraction (3). A heminested PCR was performed on each DNA sample with three primers specific for insertion sequence IS6110 from the MTB complex genome. Outer primers Pt 8 (5′-GTGCGGATGGTCGCAGAGAT-3′) (10) and Pt 9 (5′-GTGCGGATGGTCGCAGAGAT-3′) were used for the first amplification. The inner primer TB 290 (5′-GGCGGGACAACGCCGAATGCGAA-3′) (21) plus Pt 9 were used to amplify products of the first PCR (Pt 8-Pt 9). The first amplification was carried out according to the following program: hold at 94°C for 4 min; 20 cycles at 94°C for 20 s, 65°C for 35 s, and 72°C for 35 s. The program of the second amplification was similar to the first except that 35 cycles were used instead of 20. A sample was considered positive if this second PCR yielded a 337-bp product. DNA was extracted from the isolates, and the above procedure was used to confirm that these isolates were MTB.
(vi) Susceptibility testing.
In a separate study, 88 auramine O-positive sputum samples were selected randomly from specimens sent from various hospitals in Lima for laboratory analysis at Hospital Cayetano Heredia, Lima, Peru. Two methods were used to determine drug susceptibility to isoniazid (INH) and rifampin (RIF): the microwell alamar blue assay (MABA), an indirect susceptibility assay (2, 7, 28), and MODS, which tests susceptibility directly on 2 ml of sputum sample. With MABA, MTB isolates are grown and then inoculated into a series of wells of a 96-well microtiter plate containing twofold increments of the drug of interest; MTB growth is evaluated by adding alamar blue color indicator (TREK, Westlake, Ohio) (2, 7, 28). MABA provides a mean MIC. MODS uses only two antimicrobial concentrations, similar to the BACTEC format; INH concentrations were 0.4 μg/ml and 0.1 μg/ml and the RIF concentrations was 1.0 μg/ml; we added an additional concentration of RIF of 0.5 μg/ml (1, 7). H37Rv was used as a sensitive control each time susceptibilities were run.
(vii) Indirect testing on strain isolates by MABA.
Three hundred microliters of each concentrated sample was inoculated onto Middlebrook 7H11 agar (Becton Dickinson, Cockeysville, Md.), incubated at 37°C in an atmosphere of 5% CO2 in humidified air until colonies appeared, and then refrigerated. Bacterial suspensions were prepared and tested for susceptibility to INH and RIF by a standardized modification of the colorimetric method (7). Wells were tested for MTB growth by adding 50 μl of freshly prepared 1:1 mixture of 10× alamar blue reagent and 10% Tween 80. A change in color from blue to pink in medium with characteristic growth was interpreted as MTB growth. Medium that appeared turbid was considered to be contaminated (7). The MIC for each sample was defined as the lowest drug concentration that prevented a color change from blue to pink.
(viii) direct sputum testing by MODS.
Sputum samples obtained from patients at the seven hospitals were decontaminated using the procedure described above, except pellets were resuspended in 4.5 ml of Middlebrook 7H9 broth containing a final concentration of 20 μl of PANTA per ml of broth and 10% OADC. The four antimicrobial drug stock solutions were prepared at the following concentrations: INH, 4.0 and 1.0 μg/ml; RIF, 10.0 and 5.0 μg/ml. Five hundred-forty microliters of each sputum-medium solution was placed onto each of 8 microwells of a sterile 24-well plate; each plate was used to process three samples. A 24-well plate rather than a 96-well plate was used to minimize cross-contamination. For each sample, a 60-μl aliquot of each of the four drug stock solutions was added to a well containing sputum-medium solution. Sixty microliters of 7H9 media was added to each of the remaining four wells containing the sputum-media mixture to serve as antimicrobial agent-free controls. Plates were sealed with Scotch polyethylene tape, incubated at 37°C, and moved to the microscope without agitation. Wells were examined for the presence of mycobacteria every 1 or 2 days under an inverted light microscope at 40× magnification. A sample was considered susceptible if growth was observed in the drug-free control wells but not in wells containing antimicrobial agents. A sample was considered resistant if growth was observed in both control wells and wells containing antimicrobial agents. The final drug concentrations were 0.4 and 0.1 μg/ml for INH and 1.0 and 0.5 μg/ml for RIF. Concordance was determined by comparing MODS results at final high- and low-drug concentrations with MABA results at MICs comparable to the final MODS drug concentrations. MODS was repeated twice in 14 randomly selected specimens to determine reproducibility.
(ix) SSCP:
In order to determine whether results of rifampin antimicrobial susceptibility tests were accurate, we used a method that determines whether a mutation has occurred in the rpoB gene. Previous studies have reported encouraging results by using the single-stranded conformation polymorphism (SSCP) method to detect mutations on MTB cultures resistant to RIF (23). Ninety-five percent of RIF-resistant isolates have a mutation in a 69-bp region of the rpoB gene, making this a good target for molecular genotypic diagnostic methods (25, 26, 27) such as the SSCP or heteroduplex test. DNA from M. tuberculosis RIF-susceptible reference strain H37Rv and 14 M. tuberculosis cultured strains was extracted by proteinase K digestion, and phenol-chloroform purification was performed. The RIF-resistant region of the rpoB gene was amplified by primers TR9 (5′-TCGCCGCGATCAAGGAGT-3′) and TR8 (5′-TGCACGTCGCGGACCTCCA-3′) (23). The PCR (50 μl) contained 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 0.5 μM (each) primers, 1.25 U of Taq polymerase, and 5 ng of DNA. After a 4-min denaturation at 94°C, the reaction was subjected to 35 cycles of amplification (30 s at 94°C, 45 s at 55°C, and 45 s at 72°C), followed by a 4-min extension at 72°C. This reaction generates a 157-bp PCR product. After amplification, 20 μl of PCR product was mixed with 20 μl of SSCP dilution solution (95% formamide, 20 mM EDTA, 10 mM NaCl, 0.05% bromophenol blue), boiled for 10 min, cooled on ice, and loaded onto nondenaturing sequencing-format MDE gel (Hydrolink; AT Biochem, Inc., Malvern, Pa.) composed of 0.5× MDE, 0.6× Tris-borate-EDTA, 0.045% (vol/vol) N,N,N′,N′-tetramethylenediamine, and 0.40% (wt/vol) ammonium persulfate. Electrophoresis was performed at room temperature at the following settings: 1 W for 1 h, 3 W for 2 h, and 2.5 W for 3 h, using a 16- by 18- by 0.3-cm gel. Gels were stained by silver nitrate (20) and analyzed.
Statistical analysis.
All data were compiled and analyzed using SPSS (SPSS, Inc., Chicago, Ill.). P values for the difference in speeds of various methods were calculated using Wilcoxon's nonparametric test.
Cost.
All costs were calculated using recent purchase records or catalogue prices from biotechnology companies. Costs for decontamination of samples, shipping and handling, customs taxes, and labor were not included.
RESULTS
Detection of mycobacteria.
Ninety-eight (57%) of 172 sputum samples were positive for MTB by at least one standard culture method (MGIT, LJ, or microagar 7H11). The study was performed on 100 specimens from 84 patients from six Lima hospitals suspected clinically of having tuberculosis, of which 33 specimens from 27 patients were positive on culture. An enriched sample was obtained from a tuberculosis program which evaluated smear-positive patients from the community. This program provided 72 specimens from 64 patients, of which 65 specimens from 56 patients were positive. All positive specimens were confirmed to be MTB by PCR. In determining sensitivities of various methods (Table 1), we considered true positives to be samples which were positive by one or more standard culture methods. Of the 172 sputum samples collected, 4 (2%) of MODS, 7 (4%) of MGIT, 2 (1%) of LJ, and 2 (1%) of microagar 7H11 cultures were contaminated and therefore excluded from analysis. PCR was not possible in 13 (8%) cases due to insufficient patient sample. Auramine O smears were successfully performed on all samples. In eight cases, the speed of the MGIT test could not be determined due to a temporary failure of our UV light box.
TABLE 1.
MODS sensitivity and time to detection of MTB versus standard culture methods
| Method | No. of samples positive by method | No. of samples positive by at least one culturea | Sensitivity (%)b | Median detection time (range)c |
|---|---|---|---|---|
| Auramine O stain | 76 | 98 | 78 | |
| MODS | 89 | 97 | 92 | 9 (4–31) |
| MGIT | 88 | 95 | 93 | 10 (4–39) |
| LJ | 73 | 96 | 76 | 24 (6–59) |
| microagar 7H11 | 75 | 96 | 78 | 14.5 (4–28) |
| PCR | 81 | 90 | 90 |
Only samples which were positive by at least one culture (MODS, MGIT, LJ, or 7H11) and interpretable by the given method were considered in determining sensitivity. Of the 172 samples, 98 were positive by at least one culture. Denominators differ for different methods because not all results were interpretable due to contamination or (in the case of PCR) insufficient patient sample.
Sensitivity is based on the gold standard of detection by any culture.
Includes only interpretable results. Values shown are in days.
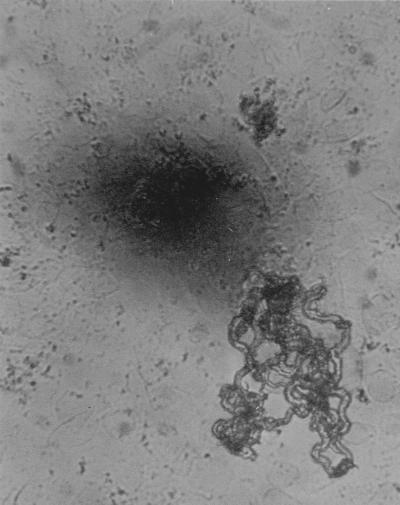
Figure 1 shows a microphotograph of MTB in the 7H9 liquid culture medium used for MODS. Small colonies have a characteristic string-and-tangles appearance that is easily distinguishable from the noncorded colonies of M. avium and M. kansasii. The MODS assay had a sensitivity of 92% (89 of 97), while MGIT had a sensitivity of 93% (88 of 95). Results for MODS and MGIT were concordant in all cases except for one, which was positive by MGIT but negative by MODS. Solid media were less sensitive with detection rates of 76% (73 of 96) and 78% (75 of 96) for LJ and microagar 7H11 methods, respectively. Using all culture methods collectively as the “gold standard,” PCR had a sensitivity of 90%. Eighteen samples were positive by PCR but negative by all culture methods. Specificity was high for both PCR and MODS, as none of the 10 control sputum samples were positive by either method. The auramine O staining method detected TB in 78% (76 of 98) of culture-positive samples. All samples that had positive auramine O stains were also positive by at least one culture method. A summary of the results obtained using various MTB detection methods is shown in Table 1.
FIG. 1.
Appearance of M. tuberculosis at 100× magnification after 25 days of culture in broth. Note the serpentine, ropy aspect of the microcolonies.
MODS detected MTB in sputum samples within a median time of 9.0 days after inoculation (range, 4 to 31 days). This was significantly faster than MGIT, which had a median of 10.0 days (P < 0.05) (range, 4 to 39 days). In contrast, 7H11 and LJ agar cultures took a median of 14 and 24 days, respectively, to detect MTB, as shown in Table 1.
Susceptibility testing.
Susceptibility results using MODS were available between 5 and 20 days (median, 9.5 days) after the inoculation of patient sputum samples directly into 7H9 broth containing antimicrobials. Susceptibility assays such as MABA require a pure MTB isolate (which typically takes 3 weeks to obtain); this is much slower than MODS.
Thirty-three of the 88 patients analyzed for susceptibility were taking anti-TB therapy, 22 for less than 1 week and 11 for over 1 week. By MABA, there were 64 patient isolates sensitive to both INH and RIF, 8 resistant to INH alone, 2 resistant to RIF alone, and 14 resistant to both. The results obtained with MODS were concordant in 78 of these 88 cases (89%) and discordant in 10 cases (11%). Discordant results occurred in both the treated and untreated groups. MODS classified all resistant strains correctly but misclassified 10 isolates as resistant that MABA classified as susceptible. Seventy-two RIF-sensitive strains were detected by MABA. Of these, MODS classified nine (13%) strains resistant to RIF; seven samples were scored resistant to RIF at 0.5 μg/ml but sensitive at 1.0 μg/ml, and two samples were resistant to RIF at both low and high concentrations by MODS. Only 1 of the 66 (2%) strains sensitive to INH by MABA was misclassified as INH resistant by MODS at both high and low concentrations (Table 2). MODS yielded the same results in all 14 specimens that were repeated.
TABLE 2.
Comparison of MODS and MABA in evaluating MTB sensitivity to INH and RIF
| Method | INH
|
RIF
|
Total (%) specimens | ||
|---|---|---|---|---|---|
| No. sensitive (%) | No. resistant (%) | No. sensitive (%) | No. resistant (%) | ||
| MODS | 65 (74) | 23 (26)a | 63 (72) | 25 (28)b | 88 (100) |
| MABA | 66 (75) | 22 (25) | 72 (82) | 16 (18) | 88 (100) |
Resistant at both low (0.1 μg/ml) and high (0.4 μg/ml) concentrations of INH.
Seven specimens were resistant at low (0.5 μg/ml) but susceptible at high (1.00 μg/ml) concentrations of RIF; two specimens were resistant at both low and high concentrations.
SSCP.
Fourteen of the MTB strains for which rifampin susceptibility was previously tested by MABA and MODS of MTB were also evaluated by SSCP.
As seen on Table 3, over 90% of the strains that were resistant to RIF by MABA had a mutant found in the rpoB gene by SSCP. Three sensitive isolates were also studied by SSCP. MODS did not misclassify any SSCP-resistant strains but classified three SSCP-sensitive strains as resistant when tested with a 0.5 μg/ml cutoff. Two of these strains were correctly classified when the cutoff was increased to 1.00 μg/ml. It appears that MODS at a 0.5 μg/ml cutoff will falsely classify sensitive strains as RIF resistant.
TABLE 3.
Comparison of MODS, MABA, and SSCP in evaluating MTB sensitivity to RIF
| Response | No. (%) of specimens showing RIF susceptibility by
|
||
|---|---|---|---|
| MABA | MODS | SSCP | |
| Resistance | 11 (79) | 14 (100)a | 10 (71) |
| Sensitivity | 3 (21) | 0 (0) | 4 (29) |
| Total | 14 (100) | 14 (100) | 14 (100) |
The cutoff level used to determine RIF resistance was 0.5 μg/ml. When a concentration of 1.00 μg/ml was used, two of the three strains initially classified by MODS as resistant were reclassified as sensitive.
Cost.
The current estimated cost per sample with MODS is $0.77 for detection and $1.72 for direct susceptibility testing with two drugs at two different concentrations (Table 4). This is substantially cheaper than MGIT, which costs $7.00 for detection testing and $35.02 for susceptibility testing with two drugs. It is also substantially less expensive than the BACTEC, which costs $2.55 per sample for detection and $12.75 per sample for susceptibility testing with two drugs (not including the costly investment of a BACTEC machine). Table 4 shows the estimated costs per sample for TB detection and susceptibility testing. Susceptibility costs were calculated for two drugs (INH and RIF) at two concentrations (high and low) each, and four drugs (INH, RIF, ethambutol, and streptomycin) at two concentrations each. Items included in cost analysis are noted below the table.
TABLE 4.
Costs per sample for TB detection and susceptibility testing
| Method | Detection cost ($) | Susceptibilitya cost ($)
|
|
|---|---|---|---|
| Two drugs | Four drugs | ||
| MODSb | 0.77 | 1.72 | 1.80 |
| MGITc | 7.00 | 35.02 | 63.03 |
| BACTECd | 2.55 | 12.75 | 23.00 |
| LJe | 0.14 | 1.60 | 1.57 |
| Microagar 7H11f | 0.29 | 1.60 | 2.92 |
| MABAg | 1.23–2.43 | 5.62 | 6.87 |
| PCRh | 2.90 | ||
| Acid-fast stain | 0.10 | ||
| Smeari | |||
Calculated for two different concentrations of each drug. Two drugs, RIF and INH; four drugs, RIF, INH, ethambutol, and streptomycin.
Includes costs of Middlebrook 7H9 medium, glycerol, Bacto Casitone, OADC, PANTA, 24-well plate, Scotch polyethylene tape, pipette tips, and antimicrobial drugs.
Includes costs of MGIT (prepared tubes), OADC, PANTA, pipette tips, and antimicrobial drugs.
Includes costs of BACTEC vials and antimicrobial drugs; does not include the cost of syringes and reconstituting fluid.
Includes costs of LJ medium, glycerol, eggs, Scotch polyethylene tape, pipette tips, and antimicrobial drugs.
Includes costs of Middlebrook 7H11 medium, glycerol, OADC, cyclohexamide, Scotch polyethylene tape, pipette tips, and antimicrobial drugs.
Includes costs of Middlebrook 7H9 medium, glycerol, Bacto Casitone, OADC, PANTA, Tween-80, alamar blue reagent, 24- or 96-well plates, Scotch polyethylene tape, pipette tips, Combitips, and antimicrobial drugs.
Includes costs of one DNA extraction (Tris, EDTA, ethanol, 1.5-ml microcentrifuge tubes, and pipette tips), and two regular PCR reactions (Taq polymerase, 10× PCR buffer, deoxynucleoside triphosphates, MgCl, distilled water, 0.5-ml microcentrifuge tubes, pipette tips, primers, and agarose) and one nested PCR reaction (equal in cost to two regular PCR reactions).
Includes costs of microscope slides, auramine O or Ziehl-Neelsen stain, phenol, HCl, and ethanol.
Identification.
Three strains, M. chelonae, M. smegmatis, and M. bovis, had a rope-type growth pattern similar to that of MTB. M. chelonae and M. smegmatis, both rapid growers, were observed on the second day of growth; this contrasts with MTB, which was never detected earlier than 4 days after inoculation. M. bovis was not distinguishable from MTB, but all other strains were correctly identified and easily separated from MTB and from each other by their colony morphology.
DISCUSSION
Each year there are an estimated 8 million new cases of clinical tuberculosis, and 3 million deaths caused by TB (17, 19). Most cases occur in developing, resource-poor countries, where the conventional MGIT and BACTEC rapid detection and susceptibility testing techniques are simply not feasible because of their high costs and equipment requirements. The present study demonstrated that MODS is a sensitive and specific new method for detecting and determining drug susceptibility of MTB. MODS uses microscopy to detect early growth of MTB as strings and tangles of bacterial cells in Middlebrook 7H9 broth medium with or without antimicrobial agents of interest. Given its simplicity, low cost, speed (under 2 weeks in most cases), and accuracy, MODS would be an excellent method for routine tuberculosis testing in developing nations such as Peru, where tuberculosis is highly prevalent (12, 22).
At present, the only affordable method for the definitive diagnosis of MTB in most resource-poor nations is in vitro culture on solid medium. However, MTB grows better in liquid than on agar; diagnosis using solid medium culture is both less sensitive and slower than liquid medium culture (1). MODS was developed as a simple adaptation of the microagar 7H11 test, which uses microscopic observation to detect MTB on agar. In MODS, liquid medium is used to replace agar. Our results confirm the advantage of using liquid medium: both liquid culture methods used, MODS and MGIT, were at least 14% more sensitive and 4 to 5 days faster than either the microagar 7H11 or LJ culture.
To our knowledge, microscopic observation of MTB growing in broth culture has not been evaluated as a diagnostic method, although it has served as a research tool (6). The appearance of MTB in liquid medium is distinct (16). Our blinded controls indicate that contaminating bacteria or fungi are not mistaken for Mycobacteria species in our test. We readily distinguished the characteristic nonropy growth of M. avium complex and M. kansasii from MTB with the MODS assay. Rapid growers such as M. chelonae and M. smegmatis also grew as stringy tangles, similar to MTB, but were distinguished by speed of appearance (<3 versus ≥4 days for MTB). The minimum major equipment required for performing a simple test such as MODS (as should be required for any TB work in a developed country) include a P2 biosafety cabinet, a 37°C incubator, and a light microscope (preferably inverted). Although the techniques we have used, such as PCR and SSCP, are not applicable to many developing country laboratories, they were useful to demonstrate the validity of our results.
The auramine O and Ziehl-Neelsen acid-fast staining techniques are cheap and rapid but not very sensitive and not suited for drug susceptibility testing. Various studies have reported that a single sputum smear test has a sensitivity ranging from 22 to 78% (11). In this study, the auramine O-stained smear test had a sensitivity of 78%, compared to 93% of MGIT, the most sensitive method. PCR is a rapid, sensitive, and relatively inexpensive method that could be used for the detection of MTB in midlevel developing countries. However, in practice, positive PCR results are often discordant with results obtained from in vitro cultures, both in our laboratory and in laboratories with extensive MTB and PCR experience (18).
MODS offers a superb alternative for susceptibility testing in developing countries. In these countries, drug resistance was relatively low, and susceptibility tests were mainly performed for surveillance than for patient management. Now, with the rise in multiple-drug-resistant tuberculosis, timely drug susceptibility test results are a must for proper patient management. Yet presently, susceptibility tests are not performed in many areas where TB is endemic, often because rapid testing systems are expensive, and the time needed to obtain results via the less-costly solid culture tests often renders the results clinically irrelevant. Susceptibility results with solid medium testing are generally unavailable until 4 to 5 weeks after inoculation, even when done directly from sputum concentrates (13). With MODS, the patient sample is used directly for susceptibility testing, and results are generally obtained within 2 weeks.
Based on our results, we suggest that isolates scored by MODS as resistant to RIF alone be retested by a more standard technique, such as MABA. Moreover, we suggest that a concentration of 1.0 μg/ml be the lowest concentration of RIF used when testing drug susceptibility with MODS, to improve concordance with MABA. Also, the agreement between SSCP and MABA RIF susceptibility results were considerably higher than that obtained between the SSCP and MODS. This is evidence that MODS is less reliable for determinating RIF resistance. There was no discordance between MODS and MABA for the 14 multiply resistant strains. The high agreement between the MABA and SSCP also supports the accuracy of the MABA test. We now perform all susceptibility testing in our laboratory using MABA because of its simplicity and speed. Subjects for this study were considered to be at high risk for MTB, and many had tested positive by AFB stain before being referred to us. Further testing of MODS should be performed in an unselected patient population; such a study will assist in determining the true sensitivity of MODS.
It is well recognized that the resurgence of TB calls for second-generation laboratory methods for more rapid detection, identification, and drug susceptibility testing (14). Unfortunately, all such second-generation methods developed to date are for use in wealthy developed countries and are inappropriate for settings with limited resources. Our MODS method provides the first rapid, sensitive, and specific method for TB detection and drug susceptibility testing that is clinically relevant and appropriate for use in developing countries.
ACKNOWLEDGMENTS
We are grateful to R. Black, J. Friedland, and C. Evans for their comments and to J. B. Phu and D. Sara for their assistance.
REFERENCES
- 1.Cheng A F, Li M S, Chan C Y, Lyon D, Wise R, Lee J C. Evaluation of three culture media and their combinations for the isolation of Mycobacterium tuberculosis from pleural aspirates of patients with tuberculous pleurisy. J Trop Med Hyg. 1994;97:249–253. [PubMed] [Google Scholar]
- 2.Collins L, Franzblau S G. Microplate alamar blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob Agents Chemother. 1997;41:1004–1009. doi: 10.1128/aac.41.5.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalovisio J R, Montenegro-James S, Kemmerly S A, Genre C F, Chambers R, Greer D, Pankey G A, Failla D M, Haydel K G, Hutchinson L, Lindley M F, Nunez B M, Praba A, Eisenach K D, Cooper E S. Comparison of the amplified Mycobacterium tuberculosis (MTB) direct test, Amplicor MTB PCR, and IS6110-PCR for detection of MTB in respiratory specimens. Clin Infect Dis. 1996;23:1099–1106. doi: 10.1093/clinids/23.5.1099. , 1107–1108. [DOI] [PubMed] [Google Scholar]
- 4.De Cock K M, Wilkinson D. Tuberculosis control in resource-poor countries: alternative approaches in the era of HIV. Lancet. 1995;346:675–677. doi: 10.1016/s0140-6736(95)92284-9. [DOI] [PubMed] [Google Scholar]
- 5.Enarson D A, Rieder H L, Arnadottir T. Tuberculosis guide for low income countries. 3rd ed. Paris, France: International Union Against Tuberculosis and Lung Disease; 1994. [Google Scholar]
- 6.Fazal N, Bartlett R, Lammas D A, Kumararatne D S. A comparison of the different methods available for determining BCG-macrophage interactions in vitro, including a new method of colony counting in broth. FEMS Microbiol Immunol. 1992;5:355–362. doi: 10.1111/j.1574-6968.1992.tb05921.x. [DOI] [PubMed] [Google Scholar]
- 7.Franzblau S G, Witzig R S, McLaughlin J C, Torres P, Madico G, Hernandez A, Degnan M T, Cook M B, Quenzer V K, Ferguson R M, Gilman R H. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis by using the Microplate Alamar Blue Assay. J Clin Microbiol. 1998;36:362–366. doi: 10.1128/jcm.36.2.362-366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Idigoras P, Perez-Trallero E, Alcorta M, Gutierrez C, Munoz-Baroja I. Rapid detection of tuberculosis and non-tuberculosis mycobacteria by microscopic observation of growth on Middlebrook 7H11 agar. Eur J Clin Microbiol Infect Dis. 1995;14:6–10. doi: 10.1007/BF02112611. [DOI] [PubMed] [Google Scholar]
- 9.Kent B D, Kubica G P. Public health mycobacteriology. A guide for the level III laboratory. 1985. pp. 36–39. , 47–69, and 185–187. U.S. Department of Health and Human Services, Centers for Disease Control, Atlanta, Ga. [Google Scholar]
- 10.Kolk A H, Kox L F, Kuijper S, Richter C. Detection of Mycobacterium tuberculosis in peripheral blood. Lancet. 1994;344:694. doi: 10.1016/s0140-6736(94)92135-0. [DOI] [PubMed] [Google Scholar]
- 11.Lipsky B A, Gates J, Tenover F C, Plorde J J. Factors affecting the clinical value of microscopy for acid-fast bacilli. Rev Infect Dis. 1984;6:214–222. doi: 10.1093/clinids/6.2.214. [DOI] [PubMed] [Google Scholar]
- 12.Madico G, Gilman R, Checkley W, Cabrera L, Kohlstadt I, Kacena K, Diaz J F, Black R. Community infection ratio as an indicator for tuberculosis control. Lancet. 1995;345:416–419. doi: 10.1016/s0140-6736(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 13.Matthew S, Paramasivan C N, Rehman F, Balambal R, Rajaram K, Prabhakar R. A direct rifampicin sensitivity test for tubercle bacilli. Indian J Med Res. 1995;102:99–103. [PubMed] [Google Scholar]
- 14.McGowan J E, Jr, Mechock B, Nolte F S. Laboratory diagnosis of tuberculosis: past, present and future. J Med Assoc Ga. 1995;84:215–220. [PubMed] [Google Scholar]
- 15.Middlebrook G, Reggiardo Z, Tigertt W D. Automatable radiometric detection of growth of Mycobacterium tuberculosis in selective media. Am Rev Respir Dis. 1977;115:1066–1069. doi: 10.1164/arrd.1977.115.6.1066. [DOI] [PubMed] [Google Scholar]
- 16.Morris A J, Reller L B. Reliability of cord formation in BACTEC media for presumptive identification of mycobacteria. J Clin Microbiol. 1993;31:2533–2534. doi: 10.1128/jcm.31.9.2533-2534.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murray C J L, Lopez A D. Mortality by cause for eight regions of the world: global burden of disease study. Lancet. 1997;349:1269–1276. doi: 10.1016/S0140-6736(96)07493-4. [DOI] [PubMed] [Google Scholar]
- 18.Noordhoek G T, van Embden J D, Kolk A H J. Reliability of nucleic acid amplification for detection of Mycobacterium tuberculosis: an international collaborative quality control study among 30 laboratories. J Clin Microbiol. 1996;34:2522–2525. doi: 10.1128/jcm.34.10.2522-2525.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raviglione M C, Snider D E, Kochi A. Global epidemiology of tuberculosis: morbidity and mortality of a worldwide epidemic. JAMA. 1995;273:220–226. [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 21.Sechi L A, Pinna M P, Sanna A, Pirina P, Ginesu F, Saba F, Aceti A, Turrini F, Zanetti S, Fadda G. Detection of Mycobacterium tuberculosis by PCR analysis of urine and other clinical samples from AIDS and non-HIV-infected patients. Mol Cell Probes. 1997;11:281–285. doi: 10.1006/mcpr.1997.0119. [DOI] [PubMed] [Google Scholar]
- 22.Shangavi D, Gilman R, Lescano-Guevara G, Checkley W, Cabrera L, Cardenas V. Hyperendemic pulmonary tuberculosis in a Peruvian shantytown. Am J Epidemiol. 1998;148:384–389. doi: 10.1093/oxfordjournals.aje.a009657. [DOI] [PubMed] [Google Scholar]
- 23.Telenti A, Imboden P, Marchesi F, Schmidheini T, Bodmer T. Direct, automated detection of rifampin-resistant Mycobacterium tuberculosis by polymerase chain reaction and single-strand conformation polymorphism analysis. Antimicrob Agents Chemother. 1993;37:2054–2058. doi: 10.1128/aac.37.10.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walters S B, Hanna B A. Testing of susceptibility of Mycobacterium tuberculosis to isoniazid and rifampin by mycobacterium growth indicator tube method. J Clin Microbiol. 1996;34:1565–1567. doi: 10.1128/jcm.34.6.1565-1567.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watterson S, Wilson S, Yates M, Drobniewski F. Comparison of three molecular assay for rapid detection of rifampin resistance in Mycobacterium tuberculosis. J Clin Microbiol. 1998;36:1969–1973. doi: 10.1128/jcm.36.7.1969-1973.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams D L, Waguespack C, Eisenach K, Crawford J T, Portaels F, Salfinger M, Nolan C M, Abe C, Sticht-Groh V, Gillis T P. Characterization of rifampin resistance in pathogenic Mycobacteria. Antimicrob Agents Chemother. 1994;38:2380–2386. doi: 10.1128/aac.38.10.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams D L, Spring L, Gillis T P, Salfinger M, Persing D H. Evaluation of a polymerase chain reaction-based universal heteroduplex generator assay for direct detection of rifampin susceptibility of Mycobacterium tuberculosis from sputum specimens. Clin Infect Dis. 1998;26:446–450. doi: 10.1086/516313. [DOI] [PubMed] [Google Scholar]
- 28.Yajko D M, Madej J J, Lancaster M V, Sanders C A, Cawthon V L, Gee B, Babst A, Hadley W K. Colorimetric method for determining MICs of antimicrobial agents for Mycobacterium tuberculosis. J Clin Microbiol. 1995;33:2324–2327. doi: 10.1128/jcm.33.9.2324-2327.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]