Abstract
Objective
A vast amount of medical data is still stored in unstructured text documents. We present an automated method of information extraction from German unstructured clinical routine data from the cardiology domain enabling their usage in state-of-the-art data-driven deep learning projects.
Methods
We evaluated pre-trained language models to extract a set of 12 cardiovascular concepts in German discharge letters. We compared three bidirectional encoder representations from transformers pre-trained on different corpora and fine-tuned them on the task of cardiovascular concept extraction using 204 discharge letters manually annotated by cardiologists at the University Hospital Heidelberg. We compared our results with traditional machine learning methods based on a long short-term memory network and a conditional random field.
Results
Our best performing model, based on publicly available German pre-trained bidirectional encoder representations from the transformer model, achieved a token-wise micro-average F1-score of 86% and outperformed the baseline by at least 6%. Moreover, this approach achieved the best trade-off between precision (positive predictive value) and recall (sensitivity).
Conclusion
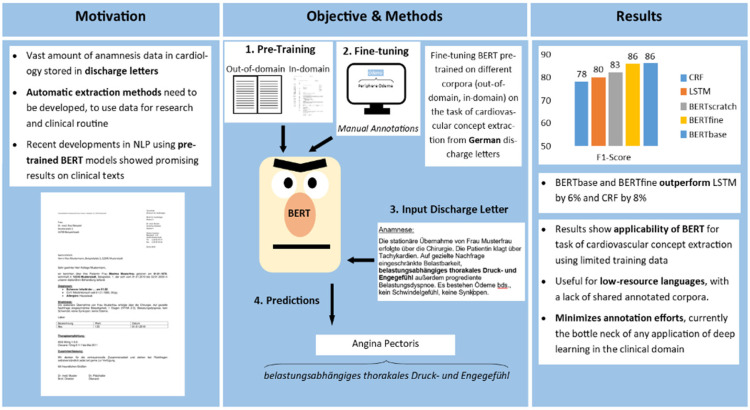
Our results show the applicability of state-of-the-art deep learning methods using pre-trained language models for the task of cardiovascular concept extraction using limited training data. This minimizes annotation efforts, which are currently the bottleneck of any application of data-driven deep learning projects in the clinical domain for German and many other European languages.
Keywords: Deep learning, pre-trained language models, bidirectional encoder representations from transformer, fine-tuning, medical information extraction, natural language processing
Introduction
While structured reporting is an emerging field in the clinical domain, a vast amount of clinical data is still stored in unstructured text. In particular, information about patient anamnesis, cardiovascular risk factors or patient therapy is often stored as free text in discharge letters. To make these data available for research and clinical routine, we need to automatically extract relevant clinical information and store it in structured formats using methods of natural language processing (NLP).
Extracting clinical information from unstructured texts was mostly done via different pattern matching-based methods1–3 and statistical machine learning.4–6 Later, deep learning methods primarily based on recurrent neural networks (RNNs) became more and more popular,7,8 particularly in the field of clinical concept extraction.9–12 Regarding medical texts from the cardiovascular domain there had been a number of publications on English data (e.g., Small et al., 13 Nath et al., 14 Patterson et al., 15 and Khalifa and Meystre 16 ) just a few studies focused on German texts.17,18 Tasks performed in the cardiovascular domain range from text classification tasks 13 to concept and concept-value pair extraction tasks covering up to 10 concepts19,20 (e.g. risk factors), 16 to a broad range of cardiovascular concepts (CCs) covering up to 80 different clinical values.15,17,18,21 All of the cardiovascular-related publications used either rule- and pattern-based approaches or commercial text mining tools.
Annotated clinical text corpora are rare and mostly of limited size (for details, see Supplemental Report, section-related work). This made the use of state-of-the-art supervised deep learning methods a challenge, in particular for non-English clinical texts. To overcome this obstacle transfer-learning-based methods using pre-trained language models gained recently more and more popularity in clinical information extraction (e.g. Li et al., 22 Beltagy et al., 23 Lee et al., 24 Si et al., 25 Bressem et al., 26 Scheible et al. 27 and Sänger et al. 28 ).
Several publications pre-trained such models, primarily based on the architecture of bidirectional encoder representations from transformers (BERTs), on English biomedical and clinical texts23,24,29,30 and fine-tuned them on various clinical downstream tasks (e.g. Li et al. 22 and Si et al. 25 ).
Besides general German pre-trained BERT models (https://deepset.ai/german-bert, https://github.com/dbmdz/berts, https://huggingface.co/uklfr/gottbert-base, and https://huggingface.co/deepset/gbert-large) and a model fine-tuned on medical texts collected from German internet forums and evaluated on a data set containing animal experiment reports (Non-technical Summaries of Animal Experiments) 31 (https://huggingface.co/smanjil/German-MedBERT) there are no publicly available German pre-trained language models available.27,32,33 Sänger et al. 28 used a multilingual BERT model and fine-tuned it for text classification of German animal experiment summaries. Bressem et al. 26 pre-trained a local BERT model on chest radiographic reports and evaluated it with promising results on a text classification downstream task. To the best of our knowledge, there is no study existing, evaluating pre-training and fine-tuning language models on a concept extraction task on German discharge letters from the cardiovascular domain.
This raised the need for an in-depth evaluation of pre-training and fine-tuning BERT on a German clinical routine corpus from the cardiovascular domain to investigate the applicability of deep-learning-based NLP methods for clinical information extraction tasks.
Objective of the study
The objective of this study was to evaluate fine-tuning of transformer-based language models based on the BERT architecture pre-trained on three different corpus types for the task of CC extraction (CCE) on limited training data. We performed our concept extraction task as a token classification task, by assigning each token to a CC or to a negative class ‘O’.
For pre-training our BERT models, we used a corpus containing German discharge letters from the cardiology department at the Heidelberg HiGHmed (consortium of the Medical Informatics Initiative Germany) partner site. For fine-tuning BERT on our CC extraction task, we used a subset of this corpus manually annotated with 12 CCs (graphical abstract, Figure 1). This study applies existing publicly available transfer-learning methods on a novel data set in a German clinical site. This task is based on an application-driven approach. Before setting up our annotation project, we had intensive discussions with physicians to define relevant concepts for their specific domain. It was of priority to embed this task as close as possible to the clinical routine needs. In this context, this study is not aiming at benchmarking-related work results, as the selection of the concepts and the data set are use-case specific. Rather, this study is an initial investigation of the applicability of transfer-learning-based NLP methods on a local clinical use case in the cardiovascular domain.
Figure 1.
Graphical abstract: automatic extraction of 12 cardiovascular concepts (CCs) from German discharge letters using pre-trained language models.
Methods
Data
Our main corpus contained ∼200,000 German discharge letters in binary MS doc format covering the time period 2004 to 2020 (see Supplemental Figure S1 for an example). We applied the following pre-processing steps to the corpus:
Converting every discharge letter into a utf-8 encoded raw text file using the LibreOffice command-line tool (version 6.2.8). This step preserves new lines and blank lines.
As the task did not require the personal data of patients, we automatically de-identified each discharge letter using a deep learning approach trained on in-house data (Richter-Pechanski et al. 34 ).
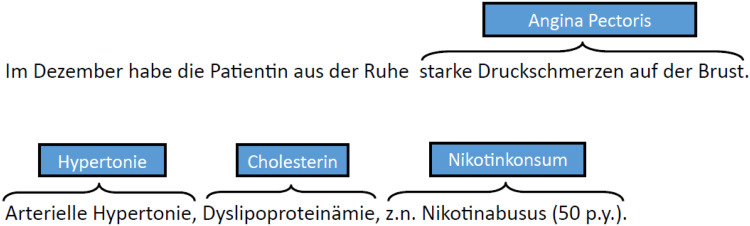
For pre-training BERTscratch and fine-tuning BERTfine with a language model objective, we concatenated all discharge letters into a single text file. Splitting the corpus into a single token using whitespace separation, the final corpus covered ∼2 GB of text, 218,084,192 tokens in total and 667,903 unique tokens. For manual annotation and fine-tuning the models on the task of CC extraction, we selected a corpus of 204 German discharge letters (called CardioAnno) from the main corpus, covering the time period 2004 to 2016, using stratified sampling. For details on our corpus and our sampling method, see Supplemental Section 1. We tokenized the corpus using whitespace separation. We did not perform sentence splitting, due to error-prone results. Instead, we split the text by newline, thus performing a paragraph splitting. The final corpus contained 381,628 tokens in 36,355 paragraphs. Due to time limitations, we restricted the annotations to the anamnesis and the cardiovascular risk factor sections. Cardiologists involved in clinical routine carefully selected and annotated the documents with a set of 12 CCs: Angina Pectoris (AP), Dyspnoe (dyspnea), Nykturie (nycturia), Ödeme (edema), Palpitationen (palpitation), Schwindel (vertigo), Synkope (syncope), Arterielle Hypertonie (Hypertonie, arterial hypertension), Hypercholesterinämie (Cholesterin, hypercholesterolemia), Diabetes mellitus (DM), Positive Familienanamnese für kardiovaskuläre Erkrankungen ( familial anamnesis (FA)) and Nikotinkonsum (nicotine consumption) (Table 1). There are no nested or overlapping concepts in the data set. Figure 2 shows an annotated text snippet of a discharge letter.
Table 1.
CCs – data analysis.
| CC | ICD-10 | Description | Instances | Uniqueness (%) |
|---|---|---|---|---|
| AP | I20 | Describes a chest pain or pressure. | 211 | 54 |
| Dyspnoe | R06.0 | Dyspnoe describes a feeling of not being able to breathe sufficiently. | 215 | 22 |
| Nykturie | R35 | Nocturia describes the need of a patient to wake up in the night to urinate. | 72 | 4 |
| Ödeme | R60 | Edema is the swelling of body tissue due to fluid retention. | 127 | 28 |
| Palpitationen | R00.2 | Palpitation describes the conscious awareness of your own heartbeat. | 136 | 17 |
| Schwindel | H81-82 | Vertigo describes the feeling of turning or swaying. | 149 | 10 |
| Synkope | R55 | Syncope describes the sudden loss of consciousness. | 168 | 8 |
| Arterielle Hypertonie | I10.* | Hypertension describes the disease when the blood pressure in the arteries is persistently elevated. | 175 | 5 |
| Hypercholesterinämie | E78.* | This describes all appearances of cholesterols or lipids, mostly expressed as cardiovascular risk factors. | 128 | 9 |
| DM | E10-14 | DM is a metabolic disorder characterized by high blood sugar levels. | 65 | 8 |
| FA | – | FA is a kind of anamnesis, which gives information about specific disease of family members. | 74 | 11 |
| Nikotinkonsum | F17.* | Describes a state of dependence on nicotine. | 111 | 11 |
Description: Distribution of CCs in CardioAnno corpus (first column) including ICD-10 code (second column), short description (third column), number of instances (fourth column) and proportion of unique instances (fifth column).
CC: cardiovascular concept; ICD-10: International Classification of Diseases, Tenth Edition; AP: Angina Pectoris; DM: diabetes mellitus; FA: familial anamnesis.
Figure 2.
Discharge letter snippet annotated with CCs: text snippet of a discharge letter annotated with CCs. For example, the sequence ‘starke Druckschmerzen auf der Brust’ is annotated with the concept AP.
CC: cardiovascular concept; AP: Angina Pectoris.
The documents were manually annotated using a well-established iterative approach including redundant annotation, an inter-annotator agreement and guideline adaptation.35–37 Two annotators (assistant physicians from cardiology) achieved a token-wise inter-annotator agreement using an F1-score of 89.8%. They annotated a total of 1631 concepts. An in-depth description of the annotated corpus and the annotation process including the annotation guidelines, see Supplemental Report.
Baseline
To compare our three pre-trained BERT models, we used two baseline classifiers, well-established methods for token classification: (i) a statistical machine learning method based on a conditional random field (CRF) 38 and (ii) an RNN approach based on long short-term memory networks (LSTM) 39 (we did not perform hyperparameter optimization steps for the baseline models, for more details, Supplemental Section 2).
Pre-trained language models
Our project uses BERT, a deep learning architecture for language representation based on transformers. In contrast to traditional RNNs, transformers process input sequences in parallel. They use self-attention and positional embeddings to extract the relation between words in an input sequence and to capture its order. To apply BERT, our concept extraction task is conducted in two separate steps: (i) a pre-training step and (ii) a fine-tuning step. A randomly initialized BERT model is pre-trained on large amounts of unannotated text with two language model objectives: masked language modeling and next sentence prediction modeling. The masked language modeling objective targets predicting randomly masked (removed) tokens in an input sequence. The sentence prediction objective targets predicting, if input sentence A is followed by input sentence B.
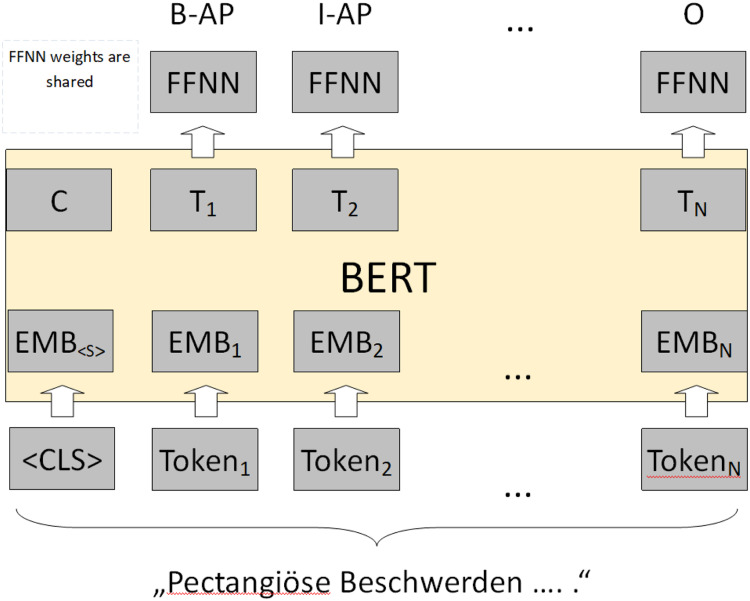
After pre-training, the BERT model is fine-tuned as a supervised downstream task on annotated training data. In our task, we seek and classify phrases containing CCs. Each output vector of the BERT model is used as input to a feed-forward neural network with shared weights and a softmax layer as a final layer to classify each input token into our set of 12 concepts (Figure 3).
Figure 3.
Fine-tuning BERT for cardiovascular concept extraction: input sequence ‘pectangiöse Beschwerden….’ is tokenized and embedded into a numerical representation. Each output representation T is used as input to an FFNN with a final softmax layer. For example, the token pectangiöse is labelled as a B-AP, the token Beschwerden is labelled as an I-AP sequence. 40
BERT: bidirectional encoder representations from transformer; FFNN: feed-forward neural network; B-AP: beginning token of Angina Pectoris; I-AP: inner token of an Angina Pectoris.
We evaluated three different pre-trained BERT models:
BERTbase: based on a publicly available German BERT model, trained on a German Wikipedia dump, an OpenLegalData dump and various news texts (https://deepset.ai/german-bert). In total, the training data size covered ∼12 GB of free text (the authors did not publish further information regarding token count in the corpus or pre-preprocessing steps).
BERTfine: based on BERTbase but fine-tuned with the BERT language model objectives using the complete corpus of German discharge letters at the cardiology department covering ∼2 GB of free text. We did not adapt the vocabulary of the BERTbase model.
BERTscratch: based on randomly initialized BERT architecture using our 2 GB corpus of discharge letters to pre-train a language model from scratch.
To perform training of BERTfine, we used the language modeling script of the HuggingFace transformer library (https://github.com/huggingface/transformers/blob/v4.0.1-release/examples/language-modeling/run_mlm.py). To perform training BERTscratch, we used the following script as a template (https://huggingface.co/blog/how-to-train). Hyperparameters: BERTfine and BERTscratch: vocab_size: 30,000; max_seq_length: 512; num_train_epochs: 3; BERTfine: per_gpu_train_batch_size: 12; BERTscratch: per_gpu_train_batch_size: 80. We used 4 × RTX6000 graphics processing units (GPUs) with each 24 GB video random access memory (VRAM). Training time BERTfine: ∼20 h, BERTscratch: ∼65 h. We did not perform hyperparameter optimization for pre-training and fine-tuning the BERT models under a language model objective.
Results
To evaluate our CC classifiers, including the baseline classifiers, we used identical 4-fold cross-validation splits on the CardioAnno corpus. We calculated token-wise F1-score (the harmonic mean between precision and recall) per concept and a micro-average F1-score per classifier. We used the HuggingFace command-line script for token classification to fine-tune the BERT models on our concept extraction task (https://github.com/huggingface/transformers/blob/v4.0.1-release/examples/token-classification/run_ner_old.py). Training time was ∼1 h per fold using 2 × RTX6000 GPUs with each 24 GB VRAM. The training was performed for 30 epochs with a batch size of 16. We did not perform hyperparameter optimization steps during our experiments.
Table 2 shows the results of the baseline classifiers CRF and LSTM and the three BERT classifiers. Overall, the BERTbase and BERTfine models achieved both a similar micro-average F1-score of ∼86% and outperformed the baseline classifiers and the BERTscratch model. Per concept highest F1-scores are achieved by BERTbase (7 of 13 concepts) and BERTfine (4 of 13 concepts) and BERTscratch (3 of 13 concepts), (for more details, see Supplemental Table S1).
Table 2.
CCE – F1-score.
| CC | CRF | LSTM | BERTbase | BERTfine | BERTscratch |
|---|---|---|---|---|---|
| AP | 69 | 73 | 83 | 82 | 78 |
| Dyspnoe | 70 | 72 | 74 | 73 | 70 |
| Nykturie | 96 | 92 | 97 | 91 | 97 |
| Ödeme | 57 | 79 | 91 | 94 | 84 |
| Palpitation | 79 | 74 | 80 | 79 | 77 |
| Schwindel | 87 | 87 | 95 | 98 | 92 |
| Synkope | 87 | 85 | 88 | 89 | 88 |
| Hypertonie | 89 | 90 | 93 | 87 | 92 |
| Cholesterin | 86 | 89 | 92 | 90 | 89 |
| DM | 86 | 90 | 90 | 91 | 91 |
| FA | 81 | 77 | 82 | 74 | 80 |
| Nikotin | 86 | 87 | 92 | 90 | 94 |
| Micro average/standard deviation | 78/0.83 | 80/1.87 | 86/1.43 | 86/1.32 | 83/1.98 |
Note: Highest values are highlighted in bold type.
Description: Mean average F1-score per concept and micro average F1-score including standard deviation of the baseline classifiers (CRF and LSTM) and the three pre-trained language models (BERTbase, BERTfine and BERTscratch) in percent. F1-score is calculated by summing up F1-scores per fold and dividing it by four.
CC: cardiovascular concept; CCE: CC extraction; CRF: conditional random field; LSTM: long short-term memory; AP: Angina Pectoris; DM: Diabetes mellitus; FA: familial anamnesis.
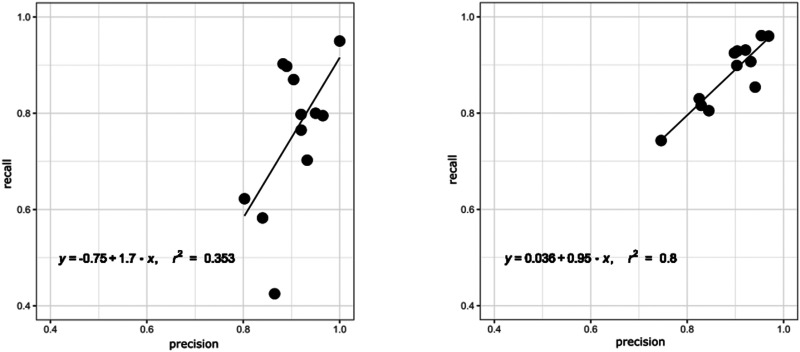
In addition to F1-scores, we investigated the balance between precision and recall of all models per concept as visualized in Figure 4 for the worst (CRF) and the best (BERTfine) performing models. The BERTfine model achieved a slope coefficient of 0.95, with r2 of 0.8 and a bias of 0.036, while the CRF performs worse in all parameters with a bias of −0.75, a slope of 1.7 and r2 of only 0.353. In general, all BERT models achieved a better precision/recall balance than the baseline models reflecting their ability to increase recall, while keeping precision high (for detailed information see Supplemental Figure S2).
Figure 4.
Precision/recall balance of CRF and BERTfine: balance between precision and recall per cardiovascular concept of the CRF and BERTfine model. Each data point in the scatter plots represents a CC. Defining the regression line with y = b + ax, an optimal result would be r2 = 1, a slope coefficient of a = 1 and a bias b = 0.
CRF: conditional random field; CC: cardiovascular concept.
Discussion
All three BERT models outperform the baseline models regarding micro average F1-score. However, BERTscratch achieved the lowest performance improvement to the baseline classifiers. While BERTbase and BERTfine show similar performance over most concepts, both models significantly outperform the baseline classifiers by 8% (CRF) and 6% (LSTM) regarding token-wise micro-average F1-score. We applied significant tests comparing the F1-scores of each model combination. We used approximate randomization using a threshold p-value <0.5 (script by Dmitry Ustalov; https://gistgithub.com/dustalov/e6c3b9d3b5b83c81ecd92976e0281d6c). 41 The highest F1-score improvements of these models we observed for the Ödeme (>10%), AP (∼10%) and Schwindel (>8%) concepts. Significant improvements can be observed as well for the concepts: Palpitation, Nikotin, Dyspnoe and Synkope (for more details regarding significance tests on our results, see Supplemental Table S2). These concepts contain the highest number of unique instances in the corpus: AP (54%), Ödeme (28%), Dyspnoe (22%), Palpitation (17%), Nikotin (11%) and Schwindel (10%) (Table 1).
We quantitatively analysed the most common misclassifications on token level per model. We focused on false-negative classifications, as the recall was lower than precision in the majority of our evaluations. In addition, we filtered the misclassifications for the lowest-performing concepts AP, Ödeme and Dyspnea. The analysis has shown that the BERT models, especially BERTbase and BERTfine are more resistant regarding spelling mistakes. The CRF and the LSTM frequently misclassify spelling mistakes such as Belastungsdysnpoe, Dypnoe, Blstungsdyspnoe, etc. The same is true regarding inflected words such as pectanginöse/m/n or restrosternale/m. Explicit and frequent concept-related tokens in the data set such as Angina, Dyspnoe, Beschwerden or Ödeme are more frequently misclassified by the LSTM and CRF models. We observed frequent false-negative errors in all model predictions in the context of function words (in, der, bei, und, etc.) and punctuations (for a detailed quantitative analysis, see Supplemental Figure S3).
While the micro average recall score of the CRF (70.3) is the lowest in comparison with the LSTM (72.8) and the BERT models (81.5–85.3), the precision score of the CRF (88.5) is higher than the score of all BERT models (85.2–86.8). A similar result can be observed for the LSTM model (precision: 89.5; recall: 72.8). We observed that standard deviation of the micro average scores precision, recall and F1-score of the CRF model achieved the lowest values (0.5/0.83/0.83) suggesting the highest stability of this model, on the opposite BERTscratch had the highest standard deviation and so the lowest stability in all scores (5.2/2.5/1.98) (for details on precision and recall mean average and per fold per model, see Supplemental Table S1).
BERTfine did not achieve a significant performance gain in comparison with the general domain BERTbase model. Comparing our results with different pre-training studies, we see that the BioBERT model, fine-tuned on medical domain data sets (e.g. PubMed, PubMed + PMC) could improve performance on a biomedical entity recognition task in comparison with a general domain BERT model by increasing the data set size up to 1 billion tokens. 24 Li et al. 22 EhrBERT was fine-tuned on a BioBERT model on 500,000 discharge letters containing a comparable amount of token, as our data set (295 million tokens). In contrast to our observations, this model improved the results significantly in an entity normalization task on four different corpora in comparison with BioBERT. An important finding was the effect of closely related domains on the performance of BERT models. In contrast to our approach, their initial BioBERT model was already pre-trained on a closely related biomedical corpus. For future work, this should be investigated on German clinical text data: pre-training a general domain German BERT model on large amounts of publicly available German medical texts, as proposed by Borchert et al. 42 and fine-tune this model on a local clinical corpus.
BERTscratch performed worse in comparison with BERTfine and BERTbase. Bressem et al. 26 showed similar results on German clinical data containing radiographic reports. Performing pre-training on a data set containing ∼415 million words and 3.36 G of text their BERT model trained from scratch showed the lowest performance on a text classification task. Their best performing BERT model was initialized from the general domain German BERT model and fine-tuned on their radiographic report corpus. In contrast to BioBERT and EhrBERT, they adapted the vocabulary of their fine-tuned model to the domain-specific data set.
The effect of vocabulary adaptations needs to be investigated in future work studies. Taking the two most frequent sequences containing CCs into account (for an overview of unique token sequences per CC occurring at least two times in the corpus, see Supplemental Figure S4), we observed that BERTbase and BERTfine tokenizers split all tokens into sub-tokens, as they have not been part of the vocabulary (e.g. the sequence ‘Angina Pectoris' is split into sub-tokens Ang, ##ina, Pe, ##ctor, and ##is). Regarding the 840B GloVe embeddings used for the LSTM model, six out of the 27 most frequent tokens in CC tokens are in the vocabulary. As GloVe embeddings do not use subword tokenization, out of vocabulary issues might have a more severe effect on the performance of the LSTM model. To address the out-of-vocabulary issue using traditional deep learning architectures, a valuable future work will be to compare or combine different embedding approaches, for example, FastText 43 (using subword-level information) and character embeddings.
In the context of balance between precision and recall, the BERT models outperform the baseline classifiers in general and the CRF classifier in particular. While the CRF results per concept are skewed to a higher precision score, the BERTfine model improved recall while keeping precision high.
While our BERT approaches showed promising results, we propose a few improvements for the experimental setup. Significance tests and standard deviations of precision, recall and F1-score showed instabilities between different pieces of training/test splits for evaluation (Supplemental Table S2). Therefore, to improve the representability of the results, we need to increase the amount of training and test data. This applies particularly to the concepts with <100 instances: DM (65), Nykturie (72) and FA (74). In addition, we assume that instances such as AP, Dyspnoe and Ödeme will benefit from larger training sets, due to their high number of unique instances. To increase annotation speed, we currently apply weak supervision and active learning to support manual annotation workflows. 44
In the annotation guidelines, we defined the constraint to restrict the physicians to exclusively annotate anamnesis and risk factor sections in discharge letters, to minimize annotation time. After discussions with the annotators followed by the manual data assessment, we could confirm that the majority of the CCs are located in these sections. Still, our manual review of the predictions showed that this constraint led to several false-positive classifications in other sections leading to ∼18% for the BERT models. To overcome this pitfall, we currently train an automatic section segmentation model for our corpus, as already done on a different corpus by Lohr et al. 45
Conclusion
In this study, we performed an in-depth evaluation of transfer-learning approaches using language models based on the BERT architecture pre-trained on three different corpora. We fine-tune them on German discharge letters from the cardiology domain, which are manually annotated with 12 CCs. We show that pre-trained language models outperform conventional strategies for automatic CC extraction in use-case scenarios where only limited-size training data are available. In the clinical domain annotation projects face various challenges. They rely on the knowledge of clinical experts with limited time resources. In addition data protection regulations in the European Union often prevent sharing clinical corpora with external domain experts. We are certain that local models can support the collaboration between different clinical sites by sharing deep learning architectures and foster optimization of tedious manual extraction processes in clinical daily routine, by training powerful deep learning models per clinical site, as demonstrated by our BERT CCE models. By just sharing model architectures, data protection issues can be avoided. However, sharing of a trained model may imply sharing of model vocabularies and weights, which may contain sensitive patient data.
Supplemental Material
Supplemental material, sj-docx-1-dhj-10.1177_20552076211057662 for Automatic extraction of 12 cardiovascular concepts from German discharge letters using pre-trained language models by Phillip Richter-Pechanski, Nicolas A Geis, Christina Kiriakou, Dominic M Schwab and Christoph Dieterich in Digital Health
Supplemental material, sj-svg-2-dhj-10.1177_20552076211057662 for Automatic extraction of 12 cardiovascular concepts from German discharge letters using pre-trained language models by Phillip Richter-Pechanski, Nicolas A Geis, Christina Kiriakou, Dominic M Schwab and Christoph Dieterich in Digital Health
Supplemental material, sj-tif-3-dhj-10.1177_20552076211057662 for Automatic extraction of 12 cardiovascular concepts from German discharge letters using pre-trained language models by Phillip Richter-Pechanski, Nicolas A Geis, Christina Kiriakou, Dominic M Schwab and Christoph Dieterich in Digital Health
Supplemental material, sj-docx-4-dhj-10.1177_20552076211057662 for Automatic extraction of 12 cardiovascular concepts from German discharge letters using pre-trained language models by Phillip Richter-Pechanski, Nicolas A Geis, Christina Kiriakou, Dominic M Schwab and Christoph Dieterich in Digital Health
Supplemental material, sj-docx-5-dhj-10.1177_20552076211057662 for Automatic extraction of 12 cardiovascular concepts from German discharge letters using pre-trained language models by Phillip Richter-Pechanski, Nicolas A Geis, Christina Kiriakou, Dominic M Schwab and Christoph Dieterich in Digital Health
Supplemental material, sj-docx-6-dhj-10.1177_20552076211057662 for Automatic extraction of 12 cardiovascular concepts from German discharge letters using pre-trained language models by Phillip Richter-Pechanski, Nicolas A Geis, Christina Kiriakou, Dominic M Schwab and Christoph Dieterich in Digital Health
Supplemental material, sj-xlsx-7-dhj-10.1177_20552076211057662 for Automatic extraction of 12 cardiovascular concepts from German discharge letters using pre-trained language models by Phillip Richter-Pechanski, Nicolas A Geis, Christina Kiriakou, Dominic M Schwab and Christoph Dieterich in Digital Health
Supplemental material, sj-xlsx-8-dhj-10.1177_20552076211057662 for Automatic extraction of 12 cardiovascular concepts from German discharge letters using pre-trained language models by Phillip Richter-Pechanski, Nicolas A Geis, Christina Kiriakou, Dominic M Schwab and Christoph Dieterich in Digital Health
Supplemental material, sj-xlsx-9-dhj-10.1177_20552076211057662 for Automatic extraction of 12 cardiovascular concepts from German discharge letters using pre-trained language models by Phillip Richter-Pechanski, Nicolas A Geis, Christina Kiriakou, Dominic M Schwab and Christoph Dieterich in Digital Health
Acknowledgements
We would like to thank all members of the Dieterich Lab for their great input and insightful discussions.
Footnotes
Conflict of interest: The authors declared no potential conflicts of interest with respect to the research, authorship and/or publication of this article.
Contributorship: PR-P was lead in conceptualization, data curation, formal analysis, investigation, methodology and initial manuscript draft writing. Together with NG and CD, he equally contributed to project administration. All authors equally contributed to software, data and results validation, data visualization and reviewing and editing the manuscript. NG, DS and ChK equally contributed to data curation. DS and ChK supported the investigation. NG was lead in resources and clinical methodology. Together with CD, he equally contributed to conceptualization together with PR-P to supervision. NG was supporting in investigation and funding acquisition. CD was lead in funding acquisition and supervision.
Ethical approval: The authors state that this study complies with the Declaration of Helsinki. Our task has been performed with respect to Section 46 Abs.2 Nr.2a (LKHG) and Section 13 Abs.1 Landesdatenschutzgesetz BW. In this context, we had the possibility to use the data for the purpose of optimizing internal clinical procedures.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The work of PR-P, NG and CD was kindly supported by Informatics for Life funded by the Klaus Tschira Foundation and for PR-P and CD by the BMBF-funded HiGHmed consortium (www.highmed.org, consortium of the Medical Informatics Initiative Germany).
Guarantor: CD.
ORCID iD: Phillip Richter-Pechanski https://orcid.org/0000-0003-0121-373X
Supplemental material: Supplemental material for this article is available online.
References
- 1.Long W. Extracting diagnoses from discharge summaries. AMIA Annu Symp Proc 2005; 2005: 470–474. [PMC free article] [PubMed] [Google Scholar]
- 2.Turchin A, Kolatkar NS, Grant RW, et al. Using regular expressions to abstract blood pressure and treatment intensification information from the text of physician notes. J Am Med Inform Assoc 2006; 13: 691–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roller R, Rethmeier N, Thomas P, et al. Detecting named entities and relations in German clinical reports. Cham: Springer, 2017, pp.146–154. [Google Scholar]
- 4.Bashyam V, Taira RK. Indexing anatomical phrases in neuro-radiology reports to the UMLS 2005AA. AMIA Annu Symp Proc 2005; 2005: 26–30. [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Wang L, Rastegar-Mojarad M, et al. Clinical information extraction applications: A literature review. J Biomed Inform 2018; 77: 34–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng S, Lu JJ, Ghasemzadeh N, et al. Effective information extraction framework for heterogeneous clinical reports using online machine learning and controlled vocabularies. JMIR Med Inform 2017; 5: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shickel B, Tighe P, Bihorac A, et al. Deep EHR: A survey of recent advances in deep learning techniques for electronic health record (EHR) analysis. IEEE J Biomed Heal Inform 2018; 22: 1589–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hahn U, Oleynik M. Medical information extraction in the age of deep learning. Yearb Med Inform 2020; 29: 208–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jagannatha A, Yu H. Structured prediction models for RNN based sequence labeling in clinical text. In: EMNLP 2016 – conference on empirical methods in natural language processing proceedings 2016, vol. 2016, pp. 856–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jagannatha A, Yu H. Bidirectional RNN for medical event detection in electronic health records. In: Proceedings of the 2016 conference of the north American chapter of the association for computational linguistics: Human language technologies 2016: vol. 2016, 473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Y, Jiang M, Lei J, et al. Named entity recognition in Chinese clinical text using deep neural network. In: Studies in health technology and informatics. Amsterdam: IOS Press, 2015, pp.624–628. [PMC free article] [PubMed] [Google Scholar]
- 12.Kittner M, Lamping M, Rieke DT, et al. Annotation and initial evaluation of a large annotated German oncological corpus. JAMIA Open 2021; 4: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Small AM, Kiss DH, Zlatsin Y, et al. Text mining applied to electronic cardiovascular procedure reports to identify patients with trileaflet aortic stenosis and coronary artery disease. J Biomed Inform 2017; 72: 77–84. [DOI] [PubMed] [Google Scholar]
- 14.Nath C, Albaghdadi MS, Jonnalagadda SR. A natural language processing tool for large-scale data extraction from echocardiography reports. PLoS One 2016; 11: e0153749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patterson O V, Freiberg MS, Skanderson M, et al. Unlocking echocardiogram measurements for heart disease research through natural language processing. BMC Cardiovasc Disord 2017; 17: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khalifa A, Meystre S. Adapting existing natural language processing resources for cardiovascular risk factors identification in clinical notes. J Biomed Inform 2015; 58: S128–S132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaspar M, Morbach C, Fette G, et al. Information extraction from echocardiography reports for a clinical follow-up study—comparison of extracted variables intended for general use in a data warehouse with those intended specifically for the study. Methods Inform Med 2019; 58: 140–150. Epub ahead of print 30 January 2020. DOI: 10.1055/s-0039-3402069. [DOI] [PubMed] [Google Scholar]
- 18.Toepfer M, Corovic H, Fette G, et al. Fine-grained information extraction from German transthoracic echocardiography reports. BMC Med Inform Decis Mak 2015; 15. Epub ahead of print 2015. DOI: 10.1186/s12911-015-0215-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garvin JH, DuVall SL, South BR, et al. Automated extraction of ejection fraction for quality measurement using regular expressions in unstructured information management architecture (UIMA) for heart failure. J Am Med Inform Assoc 2012; 19: 859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung J, Murphy S. Concept-Value pair extraction from semi-structured clinical narrative: a case study using echocardiogram reports. AMIA Annu Symp Proc 2005; 2005: 131. [PMC free article] [PubMed] [Google Scholar]
- 21.Mykowiecka A, Marciniak M, Kupść K. Rule-based information extraction from patients’ clinical data. J Biomed Inform 2009; 42: 923–936. [DOI] [PubMed] [Google Scholar]
- 22.Li F, Jin Y, Liu W, et al. Fine-tuning bidirectional encoder representations from transformers (BERT)–based models on large-scale electronic health record notes: an empirical study. JMIR Med Inform 2019; 7: e14830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beltagy I, Lo K, Cohan A. SciBERT: A pretrained language model for scientific text. EMNLP-IJCNLP 2019–2019 conference on empirical methods in natural language processing–9th international joint conference on natural language processing proceedings, 2019, vol. 2019, pp. 3615–3620. [Google Scholar]
- 24.Lee J, Yoon W, Kim S, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2019; 36: 1234–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Si Y, Wang J, Xu H, et al. Enhancing clinical concept extraction with contextual embeddings. J Am Med Inform Assoc 2019; 26: 1297-1304. DOI: 10.1093/jamia/ocz096. [DOI] [PMC free article] [PubMed]
- 26.Bressem KK, Adams LC, Gaudin RA, et al. Highly accurate classification of chest radiographic reports using a deep learning natural language model pre-trained on 3.8 million text reports. Bioinformatics 2021; 36: 5255–5261. [DOI] [PubMed] [Google Scholar]
- 27.Scheible R, Thomczyk F, Tippmann P, et al. GottBERT: A pure German language model. arXiv, http://arxiv.org/abs/2012.02110 (2020, accessed 29 October 2021).
- 28.Sänger M, Weber L, Kittner M, et al. Classifying German animal experiment summaries with multi-lingual BERT at CLEF eHealth 2019 Task 1. 2019.
- 29.Alsentzer E, Murphy JR, Boag W, et al. Publicly available clinical BERT embeddings. arXiv, http://arxiv.org/abs/1904.03323 (2019, accessed 24 March 2021).
- 30.Gururangan S, Marasović A, Swayamdipta S, et al. Don’t stop pretraining: Adapt language models to domains and tasks. arXiv, http://arxiv.org/abs/2004.10964 (2020, accessed 24 March 2021).
- 31.Neves M, Butzke D, Dörendahl A, et al. Non-technical summaries (NTS) of animal experiments indexed with ICD-10 codes (Version 1.0). In: Overview of the CLEF eHealth 2019 Multilingual information extraction, CLEF, Lugano Sept. 2019, pp. 322-339. Epub ahead of print 18 January 2019. DOI: 10.17590/20190118-134645-0. [DOI]
- 32.Becker R, Gilz L, Shrestha M. Development of a Language model for the medical domain . MSc Thesis Report, Hochschule Rhein-Waal, https://opus4.kobv.de/opus4-rhein-waal/frontdoor/index/index/docId/740 (2021, accessed 5 July 2021). [Google Scholar]
- 33.Chan B, Schweter S, Möller T. German's next language model. In: Proceedings of the 28th international conference on computational linguistics. Barcelona publicly available medical datasets (e.g. PubMed): International Committee on Computational Linguistics, Barcelona, Dec. 2020, pp.6788–6796. [Google Scholar]
- 34.Richter-Pechanski P, Amr A, Katus HA, et al. Deep learning approaches outperform conventional strategies in de-identification of German medical reports. Stud Health Technol Inform 2019; 267: 101–109. [DOI] [PubMed] [Google Scholar]
- 35.Wilbur WJ, Rzhetsky A, Shatkay H. New directions in biomedical text annotation: Definitions, guidelines and corpus construction. BMC Bioinform 2006; 7: 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts A, Gaizauskas R, Hepple M, et al. Building a semantically annotated corpus of clinical texts. J Biomed Inform 2009; 42: 950–966. [DOI] [PubMed] [Google Scholar]
- 37.Lohr C, Modersohn L, Hellrich J, et al. An evolutionary approach to the annotation of discharge summaries. In: Studies in health technology and informatics. Amsterdam: IOS Press, 2020, pp.28–32. [DOI] [PubMed] [Google Scholar]
- 38.Lafferty J, Mccallum A, Pereira FCN, et al. Conditional random fields: Probabilistic models for segmenting and labeling sequence , http://repository.upenn.edu/cis_papers (2001, accessed 14 March 2021).
- 39.Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput 1997; 9: 1735–1780. [DOI] [PubMed] [Google Scholar]
- 40.Devlin J, Chang M-W, Lee K, et al. BERT: Pre-training of deep bidirectional transformers for language understanding, http://arxiv.org/abs/1810.04805 (2018, accessed 7 March 2019).
- 41.Yeh A. More accurate tests for the statistical significance of result differences* , https://www.aclweb.org/anthology/C00-2137 (2000, accessed 10 June 2021).
- 42.Borchert F, Lohr C, Modersohn L, et al. GGPONC: A corpus of German medical text with rich metadata based on clinical practice guidelines. arXiv Prepr arXiv200706400.
- 43.Bojanowski P, Grave E, Joulin A, et al. Enriching word vectors with subword information. Trans Assoc Comput Linguist 2016; 5: 135–146. [Google Scholar]
- 44.Ratner A, Bach SH, Ehrenberg H, et al. Snorkel: Rapid training data creation with weak supervision. Proceedings of Very Large Data Base Endowment, 2017, vol. 11, pp. 269–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lohr C, Luther S, Matthies F, et al. CDA-compliant section annotation of German-language discharge summaries: Guideline development, annotation campaign, section classification. AMIA Annu Symp Proc 2018; 2018: 770–779. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-dhj-10.1177_20552076211057662 for Automatic extraction of 12 cardiovascular concepts from German discharge letters using pre-trained language models by Phillip Richter-Pechanski, Nicolas A Geis, Christina Kiriakou, Dominic M Schwab and Christoph Dieterich in Digital Health
Supplemental material, sj-svg-2-dhj-10.1177_20552076211057662 for Automatic extraction of 12 cardiovascular concepts from German discharge letters using pre-trained language models by Phillip Richter-Pechanski, Nicolas A Geis, Christina Kiriakou, Dominic M Schwab and Christoph Dieterich in Digital Health
Supplemental material, sj-tif-3-dhj-10.1177_20552076211057662 for Automatic extraction of 12 cardiovascular concepts from German discharge letters using pre-trained language models by Phillip Richter-Pechanski, Nicolas A Geis, Christina Kiriakou, Dominic M Schwab and Christoph Dieterich in Digital Health
Supplemental material, sj-docx-4-dhj-10.1177_20552076211057662 for Automatic extraction of 12 cardiovascular concepts from German discharge letters using pre-trained language models by Phillip Richter-Pechanski, Nicolas A Geis, Christina Kiriakou, Dominic M Schwab and Christoph Dieterich in Digital Health
Supplemental material, sj-docx-5-dhj-10.1177_20552076211057662 for Automatic extraction of 12 cardiovascular concepts from German discharge letters using pre-trained language models by Phillip Richter-Pechanski, Nicolas A Geis, Christina Kiriakou, Dominic M Schwab and Christoph Dieterich in Digital Health
Supplemental material, sj-docx-6-dhj-10.1177_20552076211057662 for Automatic extraction of 12 cardiovascular concepts from German discharge letters using pre-trained language models by Phillip Richter-Pechanski, Nicolas A Geis, Christina Kiriakou, Dominic M Schwab and Christoph Dieterich in Digital Health
Supplemental material, sj-xlsx-7-dhj-10.1177_20552076211057662 for Automatic extraction of 12 cardiovascular concepts from German discharge letters using pre-trained language models by Phillip Richter-Pechanski, Nicolas A Geis, Christina Kiriakou, Dominic M Schwab and Christoph Dieterich in Digital Health
Supplemental material, sj-xlsx-8-dhj-10.1177_20552076211057662 for Automatic extraction of 12 cardiovascular concepts from German discharge letters using pre-trained language models by Phillip Richter-Pechanski, Nicolas A Geis, Christina Kiriakou, Dominic M Schwab and Christoph Dieterich in Digital Health
Supplemental material, sj-xlsx-9-dhj-10.1177_20552076211057662 for Automatic extraction of 12 cardiovascular concepts from German discharge letters using pre-trained language models by Phillip Richter-Pechanski, Nicolas A Geis, Christina Kiriakou, Dominic M Schwab and Christoph Dieterich in Digital Health