Abstract
Multilocus Sequence Typing (MLST) is a precise microbial typing approach at the intra-species level for epidemiologic and evolutionary purposes. It operates by assigning a sequence type (ST) identifier to each specimen, based on a combination of alleles of multiple housekeeping genes included in a defined scheme. The use of MLST has multiplied due to the availability of large numbers of genomic sequences and epidemiologic data in public repositories. However, data processing speed has become problematic due to the massive size of modern datasets. Here, we present FastMLST, a tool that is designed to perform PubMLST searches using BLASTn and a divide-and-conquer approach that processes each genome assembly in parallel. The output offered by FastMLST includes a table with the ST, allelic profile, and clonal complex or clade (when available), detected for a query, as well as a multi-FASTA file or a series of FASTA files with the concatenated or single allele sequences detected, respectively. FastMLST was validated with 91 different species, with a wide range of guanine-cytosine content (%GC), genome sizes, and fragmentation levels, and a speed test was performed on 3 datasets with varying genome sizes. Compared with other tools such as mlst, CGE/MLST, MLSTar, and PubMLST, FastMLST takes advantage of multiple processors to simultaneously type up to 28 000 genomes in less than 10 minutes, reducing processing times by at least 3-fold with 100% concordance to PubMLST, if contaminated genomes are excluded from the analysis. The source code, installation instructions, and documentation of FastMLST are available at https://github.com/EnzoAndree/FastMLST
Keywords: MLST, microbial typing, genome analysis, parallel computing, divide-and-conquer approach
Introduction
Multilocus Sequence Typing (MLST) was a revolutionary attempt initially proposed for molecular typing of Neisseria meningitidis 1 and subsequently replicated for multiple pathogens with global relevance.2-5 This technique assigns a sequence type (ST) to bacterial isolates based on a combination of alleles from an optimal set of housekeeping genes defined for each species. Even though there are methods with higher discrimination power (cgMLST/wgMLST), MLST offers several advantages, such as (1) a common language for typing, (2) simple implementation, and (3) a vast amount of information collected in dedicated databases throughout history. 6
Currently available programs for ST determination from assembled genomes, that is, mlst, 7 CGE/MLST, 8 MLSTar, 9 and the online tool PubMLST, 10 deliver results in 1 to 9 minutes when dealing with hundreds of genomes. However, as most of them do not (or inefficiently) implement parallel computing, processing of tens of thousands of genomes can be extremely time-consuming. In addition, most current programs do not generate a file with concatenated allele sequences, despite its common requirement in downstream processes, such as evaluation of genetic diversity and phylogenetic analyses. 11
In response to these limitations, we developed FastMLST, a tool focused on parallel computing via a divide-and-conquer approach 12 that performs BLASTn searches 13 against the sequences of MLST schemes deposited in the PubMLST database 10 using genome assemblies of isolates in (multi-) FASTA format as input. FastMLST delivers 2 main outputs: (1) a table with the allelic profile detected for each genome query and (2) a (multi)-FASTA file with the (concatenated) sequences of the housekeeping genes included in each MLST scheme, suitable for downstream multilocus sequence analyses (MLSAs).
Methods
Implementation
FastMLST is an open-source, stand-alone software implemented in Python 3, that only requires BLASTn, Biopython, 14 tqdm, 15 and pandas 16 as an external program. It is easy to install via the Anaconda Package Manager (https://anaconda.org/bioconda/fastmlst) and is in constant development. A divide-and-conquer app tasks but in High-Performance Computing (HPC) clusters. 17 Instead, our specific implementation for MLST typing makes it available in HPC clusters, workstations, or personal computers.
Key analysis steps
PubMLST database setup
FastMLST can retrieve and update the PubMLST database 10 with its “–update-mlst” option. It automatically processes the 153 currently available schemes (September 2021) and prepares the system for subsequent deployment.
Allele searches
Allele searches are locally performed with BLASTn using the public, regularly updated, and centralized database maintained at PubMLST. 10 FastMLST is based on a divide-and-conquer approach, whereby each genome is processed in parallel and BLASTn runs in only one central processing unit (CPU). This is a major difference from some of the other programs that use BLASTn for MLST typing. In other programs, the genome processing is serial and uses multiple processes to do the BLASTn search in multiple CPUs.
Scheme identification
The MLST scheme to use for each query genome is inferred in FastMLST by an automatic scoring system, similar to previous software. 7 This system assigns known, full-length alleles a maximum score of 100 points, 70 points to alleles of the expected length but with Single Nucleotide Polymorphisms (SNPs) (95% identity by default), or 20 points to alleles with unexpected lengths (less than 99% of coverage by default). At the end, the MLST scheme with the highest overall score is selected for further analyses. Alternatively, it is possible to manually choose the scheme to use with the argument “–scheme.” When multiple perfect hits to a single scheme are found, FastMLST reports all hits to alert the user of potential contaminations.
Outputs
FastMLST delivers 2 output types. The first one is a table with the ST, allele classification, and—when available—the clonal complex or clade to which the ST belongs. The second type of output is a FASTA file of the MLST alleles concatenated in alphabetical order, or a series of independent files containing one gene of the scheme each.
Concordance and speed analysis
Concordance analysis
We assessed the capability of FastMLST to correctly assign STs for 33 464 isolates of 91 bacterial species that were selected and downloaded using ncbi-genome-download v0.3.0. 18 Typing by PubMLST was also done and considered a gold standard. Cohen’s kappa was calculated with the cohen_kappa_score method implemented in scikit-learn V0.24.2. 19 The code to reproduce this analysis is available in a Jupyter Notebook in the FastMLST-Concordance repository (https://github.com/EnzoAndree/FastMLST-Concordance), along with the intermediate results of FastMLST and PubMLST typing and the genome assembly stats. The quality of the assemblies used was assessed using assembly-stats v1.0.1. 20
Speed analysis
The Unix time program was used to compare the processing speed of FastMLST with that of the other 4 widely used software: mlst v2.11, CGE/MLST v2.0.4, MLSTar v1.0, and the online PubMLST tool. Only the MLSTar run time was measured with the R function proc.time(). The species on which the speed test was done were carefully chosen to evaluate the impact of genome size on speed performance: Burkholderia cepacia, median genome length (MGL) = 8 544 148 bp; n = 186; scheme = bcepacia_complex; Clostridium botulinum (MGL = 3 943 908 bp; n = 250; scheme = cbotulinum); and Mycoplasma pneumoniae (MGL = 816 465 bp; n = 175; scheme = mpneumoniae). The code to reproduce the speed test is available in a Jupyter Notebook in the FastMLST-Concordance repository (https://github.com/EnzoAndree/FastMLST-Concordance).
Results
Compared with PubMLST, FastMLST reached an overall concordance of 99.06% (n = 27 101 of 27 359) (Table 1). The 258 inconsistencies detected were due to incorrect ST assignment by PubMLST. These 258 genomes were flagged as suspected contaminated genomes and were not typed by FastMLST because they had more than one perfect hit for an allele (Supplementary Tables S1-S6). This situation remained unnoticed by PubMLST and could be misleading for users. After removal of these possibly contaminated genomes, the overall concordance of FastMLST increased to 100% (Cohen’s kappa = 1.000).
Table 1.
Concordance analysis between FastMLST and PubMLST using 91 different species.
| Species | Scheme | %GC | Median genome length (bp) | Total number of isolates | ST assigned by PubMLST a | ST assigned by FastMLST b | New ST detected by FastMLST | Genomes with new alleles detected by FastMLST | Concordance (%) c | Cohen’s kappa | Possibly contaminated genomes d | Concordance when possibly contaminated genomes are removed (%) | Cohen’s kappa when possibly contaminated genomes are removed |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clostridioides difficile | cdifficile | 29 | 4 147 183 | 2044 | 1991 | 1991 | – | 1 | 100.00 | 1.000 | – | 100.00 | 1.000 |
| Enterococcus faecium | efaecium | 38 | 2 923 827 | 2339 | 1953 | 1953 | 120 | 113 | 100.00 | 1.000 | – | 100.00 | 1.000 |
| Enterococcus faecalis | efaecalis | 37 | 2 964 918 | 1828 | 1645 | 1645 | 62 | 106 | 100.00 | 1.000 | – | 100.00 | 1.000 |
| Burkholderia pseudomallei | bpseudomallei | 68 | 7 124 009 | 1697 | 1567 | 1567 | 96 | 17 | 100.00 | 1.000 | – | 100.00 | 1.000 |
| Streptococcus suis | ssuis | 41 | 2 106 098 | 1539 | 1359 | 1359 | 68 | 84 | 100.00 | 1.000 | – | 100.00 | 1.000 |
| Vibrio parahaemolyticus | vparahaemolyticus | 45 | 5 112 626 | 1574 | 1254 | 1254 | 103 | 141 | 100.00 | 1.000 | – | 100.00 | 1.000 |
| Vibrio cholerae | vcholerae | 47 | 4 025 109 | 1476 | 1239 | 1239 | 65 | 142 | 100.00 | 1.000 | – | 100.00 | 1.000 |
| Other 78 species | – | 25-67 | 761 051-8 544 149 | 13 548 | 9416 | 9416 | 610 | 2765 | 100.00 | 1.000 | – | 100.00 | 1.000 |
| Listeria monocytogenes | lmonocytogenes | 38 | 2 969 897 | 3849 | 3813 | 3812 | 6 | 8 | 99.97 | 1.000 | 1 | 100.00 | 1.000 |
| Campylobacter jejuni | cjejuni | 31 | 1 676 498 | 1763 | 1704 | 1703 | 6 | 15 | 99.94 | 0.999 | 1 | 100.00 | 1.000 |
| Enterobacter cloacae | ecloacae | 55 | 4 991 140 | 341 | 275 | 272 | 6 | 25 | 98.91 | 0.989 | 3 | 100.00 | 1.000 |
| Helicobacter cinaedi | hcinaedi | 39 | 2 131 745 | 41 | 34 | 33 | – | 2 | 97.06 | 0.961 | 1 | 100.00 | 1.000 |
| Porphyromonas gingivalis | pgingivalis | 48 | 2 334 097 | 67 | 14 | 13 | 19 | 24 | 92.86 | 0.915 | 1 | 100.00 | 1.000 |
| Streptococcus agalactiae | sagalactiae | 36 | 2 093 532 | 1358 | 1 095 | 844 | 2 | 10 | 77.08 | 0.758 | 251 | 100.00 | 1.000 |
| Total | 33 464 | 27 359 | 27 101 | 1163 | 3453 | 99.06 | 0.990 | 258 | 100.00 | 1.000 | |||
Abbreviations: MLST, Multilocus Sequence Typing; ST, sequence type.
A genome was included in this category if an ST was assigned using PubMLST.
A genome was included in this category if an ST was assigned using FastMLST and PubMLST.
An ST assignment was considered concordant if the STs assigned by PubMLST and FastMLST were identical.
FastMLST found evidence of contamination due to multiple alleles for a single gene.
FastMLST reported 1163 novel STs and 3453 isolates with novel alleles that PubMLST overlooked. The number of downloaded and typed genomes are different and is explained by the fact that some genomes are not typable because of missing alleles due to genome fragmentation. Therefore, ST assignment in these isolates was not possible by either PubMLST or FastMLST. An extended version of Table 1 is shown in Supplementary Table S7.
Our results with real-world data regarding isolates of different %GC, genome lengths, fragmentation levels (Supplementary Table S8), and possibly contamination levels confirm that MLST typing by FastMLST and PubMLST is of comparable quality. Nonetheless, FastMLST provides additional information about possible contaminations and identifies new alleles and STs.
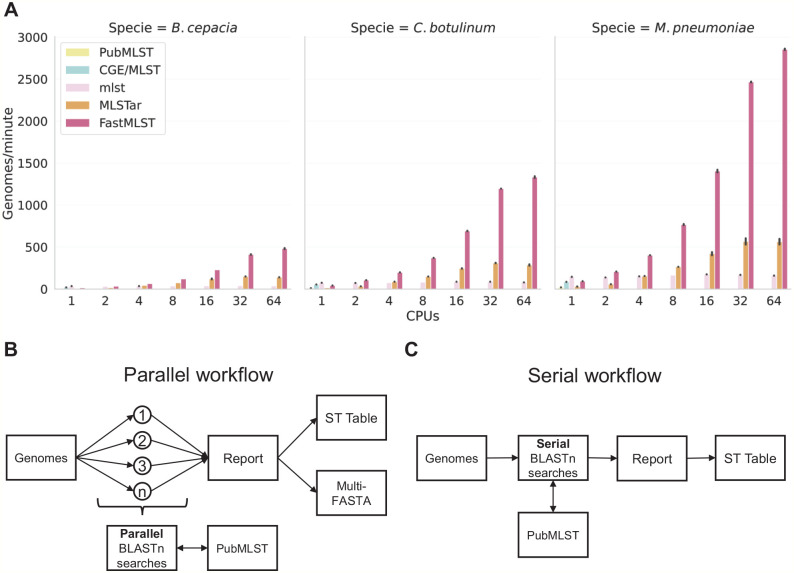
The speed of FastMLST, MLSTar, PubMLST, and mlst (expressed in Genomes/minute) was determined with a workstation equipped with 2 Intel(R) Xeon(R) CPU E5-2683 v4 processors @ 2.10 GHz using 1, 2, 4, 8, 16, 32, or 64 cores. Each program was run in triplicate for each test datasets. The PubMLST’s results were obtained remotely because it does not offer a stand-alone version. In all evaluated programs, processing speed and genome size were inversely related (Figure 1A). When a single core was used, mlst v2.11 achieved the best performance for all 3 datasets, with a max speed of approximately 145 genomes/min (a total run time of approximately 1.2 minutes) (Figure 1A). However, mlst v2.11 did not speed up when its multiple processing option was enabled (Figure 1A), as it inefficiently implements parallel computing (Figure 1C). MLSTar can also use multiple processors, but it was at least 3-fold slower than FastMLST when 64 CPUs was used (Figure 1A and B). FastMLST’s processing speed grew exponentially as more cores were deployed (Figure 1A and B), reaching a max speed of 2855 genomes/min using 64 cores (a total run time of approximately 3.7 seconds). This allows the processing of 28 000 genomes in less than 10 minutes.
Figure 1.
Processing speed and simplified comparative-workflow. (A) Processing speed (genomes/minute) of FastMLST and standard software for MLST determination (mlst, PubMLST, CGE/MLST, and MLSTar) was compared by triplicate using 3 Burkholderia cepacia (n = 186), Clostridium botulinum (n = 250), and Mycoplasma pneumoniae (n = 175) datasets and 1, 2, 4, 8, 16, 32, and 64 CPUs. Error bars represent the standard error of 3 replicates. Only mlst, MLSTar, and FastMLST natively support parallel processing. (B) FastMLST and MLSTar workflow. (C) mlst, PubMLST, and CGE/MLST workflow. CPU indicates central processing unit; MLST, Multilocus Sequence Typing.
Although speed will depend on the length of the genome queries, the speed of FastMLST in the worst-case scenario will be at least 14 times higher than the speed offered by mlst (the fastest single-core program). Raw data of run time for each speed test are available in Supplementary Table S8.
Along with the improvement in processing speed, FastMLST allows the user to generate a file with concatenated MLST gene fragments that can be used without further modification in subsequent phylogenetic analyses.
Conclusions
Compared with PubMLST, FastMLST assigns STs to thousands of genomes in minutes with 100% concordance in genomes without suspected contamination in a wide variety of species with different genome lengths, %GC, and assembly fragmentation levels. In addition, FastMLST can generate a multi-FASTA file with concatenated allele sequences suitable for downstream phylogenetic analysis. This addition allows the user to focus on downstream analyses and the biological aspect of the data much sooner. These unique features have the potential to boost future research.
Supplemental Material
Supplemental material, sj-pdf-1-bbi-10.1177_11779322211059238 for FastMLST: A Multi-core Tool for Multilocus Sequence Typing of Draft Genome Assemblies by Enzo Guerrero-Araya, Marina Muñoz, César Rodríguez and Daniel Paredes-Sabja in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-1-bbi-10.1177_11779322211059238 for FastMLST: A Multi-core Tool for Multilocus Sequence Typing of Draft Genome Assemblies by Enzo Guerrero-Araya, Marina Muñoz, César Rodríguez and Daniel Paredes-Sabja in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-2-bbi-10.1177_11779322211059238 for FastMLST: A Multi-core Tool for Multilocus Sequence Typing of Draft Genome Assemblies by Enzo Guerrero-Araya, Marina Muñoz, César Rodríguez and Daniel Paredes-Sabja in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-3-bbi-10.1177_11779322211059238 for FastMLST: A Multi-core Tool for Multilocus Sequence Typing of Draft Genome Assemblies by Enzo Guerrero-Araya, Marina Muñoz, César Rodríguez and Daniel Paredes-Sabja in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-4-bbi-10.1177_11779322211059238 for FastMLST: A Multi-core Tool for Multilocus Sequence Typing of Draft Genome Assemblies by Enzo Guerrero-Araya, Marina Muñoz, César Rodríguez and Daniel Paredes-Sabja in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-5-bbi-10.1177_11779322211059238 for FastMLST: A Multi-core Tool for Multilocus Sequence Typing of Draft Genome Assemblies by Enzo Guerrero-Araya, Marina Muñoz, César Rodríguez and Daniel Paredes-Sabja in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-6-bbi-10.1177_11779322211059238 for FastMLST: A Multi-core Tool for Multilocus Sequence Typing of Draft Genome Assemblies by Enzo Guerrero-Araya, Marina Muñoz, César Rodríguez and Daniel Paredes-Sabja in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-7-bbi-10.1177_11779322211059238 for FastMLST: A Multi-core Tool for Multilocus Sequence Typing of Draft Genome Assemblies by Enzo Guerrero-Araya, Marina Muñoz, César Rodríguez and Daniel Paredes-Sabja in Bioinformatics and Biology Insights
Footnotes
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by a Doctoral fellowship 21181536 from ANID to EGA, the EU-Lac project “Genomic Epidemiology of Clostridium difficile in Latin America (T020076),” ANID—Millennium Science Initiative Program—NCN17_093, and Start-up funds of Texas A&M University to D.P.-S.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: EG-A performed the design, software development, testing, implementation of FastMLST, and drafted the manuscript. MM carried out the concordance analysis and drafted the manuscript. CR participated in design of the concordance and speed analysis, and draft, and corrected the manuscript. DP-S participated in draft and corrected the manuscript.
ORCID iDs: Enzo Guerrero-Araya  https://orcid.org/0000-0003-2282-7722
https://orcid.org/0000-0003-2282-7722
César Rodríguez  https://orcid.org/0000-0001-5599-0652
https://orcid.org/0000-0001-5599-0652
Daniel Paredes-Sabja  https://orcid.org/0000-0002-6176-9943
https://orcid.org/0000-0002-6176-9943
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Maiden MC, Bygraves JA, Feil E, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dingle KE, Colles FM, Wareing DR, et al. Multilocus sequence typing system for Campylobacter jejuni. J Clin Microbiol. 2001;39:14–23. doi: 10.1128/JCM.39.1.14-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Meats E, Feil EJ, Stringer S, et al. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J Clin Microbiol. 2003;41:1623–1636. doi: 10.1128/jcm.41.4.1623-1636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Griffiths D, Fawley W, Kachrimanidou M, et al. Multilocus sequence typing of Clostridium difficile. J Clin Microbiol. 2010;48:770–778. doi: 10.1128/JCM.01796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Martin-Rodriguez AJ, Suarez-Mesa A, Artiles-Campelo F, Romling U, Hernandez M. Multilocus sequence typing of Shewanella algae isolates identifies disease-causing Shewanella chilikensis strain 6I4. FEMS Microbiol Ecol. 2019;95:fiy210. doi: 10.1093/femsec/fiy210. [DOI] [PubMed] [Google Scholar]
- 6. Kimura B. Will the emergence of core genome MLST end the role of in silico MLST. Food Microbiol. 2018;75:28–36. doi: 10.1016/j.fm.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 7. mlst. https://github.com/tseemann/mlst. Updated 2015.
- 8. Larsen MV, Cosentino S, Rasmussen S, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50:1355–1361. doi: 10.1128/jcm.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ferrés I, Iraola G. MLSTar: automatic multilocus sequence typing of bacterial genomes in R. PeerJ. 2018;6:e5098. doi: 10.7717/peerj.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Glaeser SP, Kampfer P. Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst Appl Microbiol. 2015;38:237–245. doi: 10.1016/j.syapm.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 12. Smith DR. The design of divide and conquer algorithms. Sci Comp Prog. 1985;5:37–58. doi: 10.1016/0167-6423(85)90003. [DOI] [Google Scholar]
- 13. Camacho C, Coulouris G, Avagyan V, et al. BLAST+: architecture and applications. BMC Bioinf. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cock PJ, Antao T, Chang JT, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Casper da, Costa-Luis SKL, Altendorf K, Mary H, et al. tqdm: a fast, extensible progress bar for Python and CLI. https://zenodo.org/record/5202772#.YYVCfGBBzIU. Updated 2021.
- 16. pandas-dev/pandas: Pandas. Version latest. Zenodo. https://zenodo.org/record/5574486#.YYVCy2BBzIU. Updated 2020.
- 17. Yim WC, Cushman JC. Divide and Conquer (DC) BLAST: fast and easy BLAST execution within HPC environments. PeerJ. 2017;5:e3486. doi: 10.7717/peerj.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. NCBI genome downloading scripts. Version 0.3.0. https://github.com/kblin/ncbi-genome-download. Updated 2020.
- 19. Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 20. assembly-stats. https://github.com/sanger-pathogens/assembly-stats. Updated 2014.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-bbi-10.1177_11779322211059238 for FastMLST: A Multi-core Tool for Multilocus Sequence Typing of Draft Genome Assemblies by Enzo Guerrero-Araya, Marina Muñoz, César Rodríguez and Daniel Paredes-Sabja in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-1-bbi-10.1177_11779322211059238 for FastMLST: A Multi-core Tool for Multilocus Sequence Typing of Draft Genome Assemblies by Enzo Guerrero-Araya, Marina Muñoz, César Rodríguez and Daniel Paredes-Sabja in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-2-bbi-10.1177_11779322211059238 for FastMLST: A Multi-core Tool for Multilocus Sequence Typing of Draft Genome Assemblies by Enzo Guerrero-Araya, Marina Muñoz, César Rodríguez and Daniel Paredes-Sabja in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-3-bbi-10.1177_11779322211059238 for FastMLST: A Multi-core Tool for Multilocus Sequence Typing of Draft Genome Assemblies by Enzo Guerrero-Araya, Marina Muñoz, César Rodríguez and Daniel Paredes-Sabja in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-4-bbi-10.1177_11779322211059238 for FastMLST: A Multi-core Tool for Multilocus Sequence Typing of Draft Genome Assemblies by Enzo Guerrero-Araya, Marina Muñoz, César Rodríguez and Daniel Paredes-Sabja in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-5-bbi-10.1177_11779322211059238 for FastMLST: A Multi-core Tool for Multilocus Sequence Typing of Draft Genome Assemblies by Enzo Guerrero-Araya, Marina Muñoz, César Rodríguez and Daniel Paredes-Sabja in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-6-bbi-10.1177_11779322211059238 for FastMLST: A Multi-core Tool for Multilocus Sequence Typing of Draft Genome Assemblies by Enzo Guerrero-Araya, Marina Muñoz, César Rodríguez and Daniel Paredes-Sabja in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-7-bbi-10.1177_11779322211059238 for FastMLST: A Multi-core Tool for Multilocus Sequence Typing of Draft Genome Assemblies by Enzo Guerrero-Araya, Marina Muñoz, César Rodríguez and Daniel Paredes-Sabja in Bioinformatics and Biology Insights