Abstract
A cohort of 77 renal transplant recipients was prospectively studied for comparison of two commercially available cytomegalovirus (CMV) load assays, i.e., the COBAS AMPLICOR CMV Monitor test (Amplicor), using plasma samples, and the Murex Hybrid Capture System (HCS), using whole blood. The manufacturer of the HCS assay changed the version of the test from 1.0 (HCS-1) to 2.0 (HCS-2) after the first 37 patients had been tested. Despite the differences in principle and type of specimen used, the Amplicor and HCS assays gave comparable results. The regression line correlating the HCS-1 assay to the Amplicor assay was similar to that correlating the HCS-2 assay to the Amplicor assay. The HCS results could be converted to Amplicor-equivalent units by using linear-regression equations [log10 HCS-1 result = 0.49 (log10 Amplicor result) + 2.58, and log10 HCS-2 result = 0.61 (log10 Amplicor result) + 2.18]. The HCS-2 assay appeared to have the lowest detection limit, followed by the Amplicor assay and then the HCS-1 assay. When a sliding scale of cutoff values in Amplicor-equivalent units (>1,000, >2,500, >6,000, >16,000, >40,000, and >100,000 copies/ml) was applied to diagnose CMV disease, similar patterns of sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were observed with the Amplicor and HCS assays. A cutoff value of >40,000 copies/ml has a low sensitivity (Amplicor, 29.4%; HCS, 41.2%) but is specific (Amplicor, 96.7%; HCS, 93.3%) and can be used for the differential diagnosis of CMV disease (PPV, 71.4% [Amplicor] or 63.6% [HCS]; NPV, 82.9% [Amplicor] or 84.8% [HCS]). A lower cutoff value of >1,000 copies/ml improves the sensitivity (Amplicor, 76.5%; HCS, 82.4%) and has a high NPV (Amplicor, 91.8%; HCS, 94.2%) but, due to the low PPV (Amplicor, 46.2%; HCS, 56%), is useful only for exclusion of CMV disease.
Cytomegalovirus (CMV) disease is a major infective cause of morbidity and mortality following organ transplantation (12). The pathogenesis of CMV disease has been carefully studied, and recent evidence suggests that the presence of viremia during active infection is a major risk factor for disease progression (11). As a result, there is great interest in the quantification of systemic CMV load for the prediction and monitoring of disease development and progression. A number of studies using DNA techniques have demonstrated that the amount of CMV DNA is significantly associated with disease development (1). However, most of these studies have relied on the use of in-house assays with differing cutoff levels (2–5, 9, 10, 13, 15). A recent multicenter study has revealed significant differences in the sensitivities of various in-house methods for qualitative CMV DNA detection (6). Due to the use of different methods, amplification targets, and calibration standards, the problem of variation is expected to be more serious with quantitative assays than with qualitative methods.
Leukocyte preparations are commonly used samples, but there is no consensus with regard to the amount of input DNA needed in each test, i.e., the denominator of the measurement. Gerdes et al. (4) suggested that the detection of 500 copies per 100 ng of extracted DNA could be used for the differential diagnosis of CMV disease versus other infections or rejection, whereas Toyoda et al. (15) suggested that the significant level for differential diagnosis was ≥500 copies/1 μg of total DNA, i.e., a possible 10-fold difference. Elsewhere, Kühn et al. (9) found that a CMV DNA level of ≥1,000 copies per 106 β-globin copies was indicative of CMV disease, and Imbert-Marcille et al. (7), using a capture assay, concluded that a CMV DNA level of >60 pg/ml (equivalent to 1.4 million genome copies per ml of whole blood) was predictive of severe or moderate disease. Roberts et al. (13), on the other hand, found that all renal transplant recipients who had a CMV DNA level above 1,000 copies per 100,000 leukocytes 4 weeks after transplantation developed CMV disease. The amount of input DNA was not always quantified; one study simply separated an unquantified number of leukocytes from 5 ml of blood and concluded that a cutoff level of 7,000 copies of CMV DNA in leukocytes improved the diagnosis of CMV disease in liver transplant recipients (10).
Clearly, standardized assays are necessary so that results can be compared. Here, a cohort of 77 renal transplant recipients was prospectively studied for comparison of two commercially available quantitative assays, i.e., the COBAS AMPLICOR CMV Monitor test (Amplicor; Roche, Basel, Switzerland) and the Hybrid Capture System (HCS; Murex, Dartford, United Kingdom).
MATERIALS AND METHODS
Patients.
Seventy-seven consecutive renal (n = 75) or pancreatic-renal (n = 2) transplant recipients who had given informed consent to participate in the study were prospectively enrolled at the time of transplantation and followed up for 12 weeks, with weekly blood samples being obtained from them. CMV-seronegative recipients who received seronegative kidneys were excluded from the study. Of the 77 patients, 21 were CMV-seronegative (R−) recipients of a CMV-seropositive (D+) kidney and 56 were CMV seropositive (R+) at the time of transplantation (43 D+ R+, 13 D− R+). No prophylaxis or preemptive therapy against CMV was given. The test results from this study were not available to the clinical team, and all quantitative tests were done retrospectively after each patient had completed the 12-week follow-up period. One of us (A.B.) who had no knowledge of the study results reviewed the case note of each patient at 6 months after transplantation. The qualitative CMV test results for the first 37 patients have been previously reported (14).
Sample preparation.
Between 15 and 20 ml of EDTA-treated whole blood was obtained by venepuncture from each patient at each visit, and the samples were processed immediately on receipt in the laboratory. An aliquot of 3.5 ml was removed for the HCS assay. The remainder of each sample was separated into plasma and peripheral blood leukocytes (PBL) by a density gradient method (14).
Qualitative CMV surveillance tests.
Each weekly blood sample was tested by four different PCR formats: an in-house PCR on PBL and plasma, and a commercial PCR (Amplicor) on PBL and plasma. The in-house PCR method, which amplified the CMV major capsid protein gene of 10,000 PBL or 10 μl of plasma, was described previously (14). The commercial test amplified the DNA polymerase gene (UL54) of CMV, and 500,000 PBL or 5 μl of plasma was used in each reaction in accordance with the protocol recommended by the manufacturer. The sensitivity and specificity of these two assays for the prediction of CMV disease have been reported previously (14).
HCS.
The HCS is a signal-amplified solution hybridization antibody capture assay for the quantification of CMV DNA in whole blood. All of the samples obtained from each patient were tested by this method. The HCS version 1.0 assay (HCS-1) was used on the samples from the first 37 patients, after which the manufacturer changed the assay (version 2.0 [HCS-2]). Both assays were performed in accordance with the manufacturer's instructions. Briefly, 3.5 ml of whole blood was lysed with the manufacturer's lysis solution, subjected to denaturation conditions, and hybridized with an RNA probe specific for CMV DNA. The RNA-DNA hybrid that was formed was captured with anti-RNA-DNA antibodies bound to the surface of capture tubes, and an alkaline phosphatase-conjugated secondary antibody specific for RNA-DNA hybrids was used for detection with a chemiluminescent substrate. The number of relative light units (RLU) for each sample was determined with a luminometer. A negative control and three positive standards were used in each assay in duplicate. The CMV probe supplied was complementary to approximately 40,000 bp, or 17%, of the total (230,000-bp) CMV genome. Thus, the concentration of the target detected, as shown by the RLU output, could be used to determine the number of viral genomes in the volume of sample tested, employing the known concentrations of the three standards as calibrators. The positive cutoff value was calculated as twice the mean number of RLU for the negative control, in accordance with the manufacturer's recommendation. The cutoff value differed from run to run, depending on the RLU of the negative control. Readings below the cutoff value in each run were considered to be below the detection limit. The format and methodology of the HCS-2 assay were similar to that of the HCS-1 assay. According to the manufacturer, the probe diluent of the HCS-2 assay was reformulated to be less viscous and the detection reagent was changed to a more stable formulation. The two assays used different calibration standards, i.e., 4 × 105, 4 × 106, and 4 × 107 genomes per ml for the HCS-1 assay and 5 × 104, 1 × 106, and 2 × 107 genomes per ml for the HCS-2 assay. The results obtained with the two versions were initially analyzed separately. They were analyzed in combination after it was established that the regression equations relating HCS-1 and HCS-2 data to that obtained by the Amplicor assay were similar and that the levels from both HCS assays could be similarly converted to Amplicor-equivalent units.
COBAS AMPLICOR CMV Monitor test.
The Amplicor assay is a PCR-based method for the quantification of CMV DNA in plasma and is similar to the qualitative assay produced by the same manufacturer. Nucleic acid was extracted from the equivalent of 25 μl of plasma per PCR by using the reagents and methodology provided by the manufacturer. An automated gene amplification and detection system (COBAS; Roche, Basel, Switzerland) was used for DNA amplification, and the amount of PCR product present was compared to that of an internal standard coamplified in the same reaction with the same primer set. The protocol recommended by the manufacturer was followed throughout. Results were calculated with the computer software provided with the COBAS machine, using the signal detected from each PCR product, and were expressed as DNA copies per milliliter of plasma. The lower detection limit was 400 copies/ml. Only samples which were positive by one or more of the four qualitative PCR methods in this study were used for quantitative Amplicor analysis. Samples that were negative by all four qualitative methods were considered negative and below the detection limit of the Amplicor assay.
Statistical analysis.
The CMV load values determined by the Amplicor assay were correlated with those of the HCS assays by the use of a scatter diagram incorporating data from samples with measurable CMV loads by both assays. Regression equations relating the Amplicor results to the HCS-1 and HCS-2 results were constructed by the least-squares method, and the HCS measurements were converted to Amplicor-equivalent values. These were then used to calculate the sensitivity and specificity at different cutoff levels. Differences between assays and variables were evaluated by the χ2 test and the resultant 95% confidence intervals (95% CI). Geometric mean CMV load values were compared by using the Student t test. Statistically significant differences were set at the 5% level.
Definitions.
In this study, CMV disease was defined as a viral syndrome in which, concurrent with detection of CMV DNA in a patient's blood by at least two methods, at least two of the following were demonstrated: unexplained pyrexia for 2 or more days, malaise, back pain, abdominal pain, leucopenia (<3 × 109/liter), or thrombocytopenia (<75% of baseline). The sensitivity of a quantitative assay was defined as the proportion of patients with CMV disease who had CMV loads above the given cutoff level; the specificity was defined as the proportion of patients without CMV disease who had CMV loads below the same cutoff value in all samples during the follow-up period. The positive predictive value (PPV) of a test was defined as the proportion of patients with CMV loads above the given cutoff value who had CMV disease, and the negative predictive value (NPV) was defined as the proportion of patients with CMV loads below the cutoff value at all times and who had no CMV disease.
RESULTS
A total of 854 samples from 77 patients were available for study: 425 from the first 37 patients when the HCS-1 assay was used, and 429 from the last 40 patients when the HCS-2 assay was used. Seventeen patients developed CMV disease as defined in this study. Table 1 shows the number of samples with CMV loads measurable by the Amplicor assay and the HCS assays.
TABLE 1.
Numbers of samples with measurable CMV loads by the Amplicor, HCS-1, and HSC-2 assays during the two parts of the study
| Total no. of samples | No. (%) of patients with measurable CMV load determined by assay with:
|
|||
|---|---|---|---|---|
| Both Amplicor and HCS | Amplicor only | HCS-1 only | HCS-2 only | |
| 425a | 51 (12%) | 17 (4%) | 3 (0.7%) | NTc |
| 429b | 62 (14.5%) | 9 (2.1%) | NT | 28 (6.5%) |
The HCS-1 assay was used for the first 425 samples.
The HCS-2 assay was used for the last 429 samples.
NT, not tested.
CMV DNA was detectable by qualitative PCR in 25 of the first 37 patients (67.6%); 15 of those patients had measurable CMV loads in one or more samples by both assays, and 12 developed CMV disease. Of the 425 samples, the results of the Amplicor assay and the HCS-1 assay were discrepant in 20 (Table 1); 3 (15%) had CMV loads measurable by the HCS-1 assay only (HCS-1 loads of 9,096, 9,695, and 18,323 genomes per ml of whole blood), and 17 (85%) had loads measurable by the Amplicor assay only (median Amplicor loads, 820 copies per ml of plasma [range, 410 to 65,400]).
CMV DNA was detectable by qualitative PCR in 25 of the last 40 patients (62.5%); 21 patients had measurable CMV loads, and 5 developed CMV disease. Of the 21 patients, 19 had at least one concordant sample with CMV load measurable by both assays. Discordant samples among these patients generally were obtained either at the beginning or toward the end of the infection. Two patients had measurable CMV loads only by the HCS-2 assay. The peak HCS-2 loads in these two patients were not high (3,817 and 2,722 genomes per ml of whole blood). Of the 429 samples, results of the Amplicor assay and the HCS-2 assay were discrepant for 37 (Table 1); 28 had measurable CMV loads by the HCS-2 assay only (median HCS-2 load, 3,973 genomes per ml of whole blood [range, 1,330 to 34,732]) and 9 by the Amplicor assay only (median Amplicor load, 1,330 copies per ml of plasma [range, 612 to 5,750]).
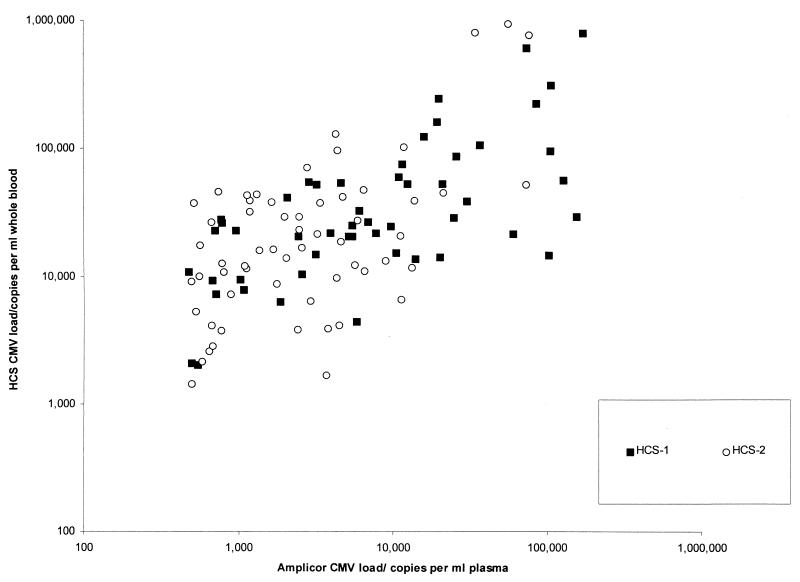
Figure 1 shows the scatter diagram of CMV loads measured by the Amplicor assay plotted against those measured by the HCS-1 and HCS-2 assays. The regression equation relating the Amplicor measurements to the HCS-1 and HCS-2 measurements were, respectively, log10 HCS-1 reading = 0.49 (log10 Amplicor reading) + 2.58 (95% CI of slope, 0.34 to 0.66; y intercept, 1.98 to 3.22) and log10 HCS-2 reading = 0.61 (log10 Amplicor reading) + 2.18 (95% CI of slope, 0.38 to 0.84; y intercept, 1.39 to 2.97).
FIG. 1.
HCS-1 and HCS-2 CMV load measurements plotted against those obtained by the Amplicor assay.
Analysis of the residuals for the fitted HCS-1 and HCS-2 measurements in the regression lines showed a random distribution (r2 = 0.01), indicating that the fitted-observed value differences were not related to the size of the viral load. There was no statistically significant difference between the Amplicor measurements and the HCS-1 or HCS-2 measurements due to the overlapping 95% CI of their slopes and y intercepts. Using these equations, at the lower detection limit of the Amplicor assay (400 copies/ml of plasma), the corresponding HCS-1 and HCS-2 measurements were 1.2 to 1.3 logs higher in numerical value than that of the Amplicor value (7,244 genomes/ml of whole blood with the HCS-1 assay and 5,888 genomes/ml of whole blood with the HCS-2 assay). At an Amplicor reading of 100,000 copies/ml of plasma, the corresponding measurements by the HCS-1 and HCS-2 assays were 107,152 and 169,824 genomes/ml of whole blood, respectively.
The HCS-2 assay appeared to have a lower detection limit than the HCS-1 assay. Of the 20 discrepant samples in the first part of the study, 3 (15%) had measurable CMV loads by the HCS-1 assay only and 17 (85%) had measurable CMV loads by the Amplicor assay only (Table 1). In contrast, in the later part of the study, 28 of 37 (75.7%) discrepant samples had measurable CMV loads by the HCS-2 assay only and 9 (24.3%) had measurable CMV loads by the Amplicor assay only (discrepancy rate between the Amplicor and HCS-1 assays versus the Amplicor and HCS-2 assays; P < 0.001). The CMV loads of the discrepant cases were generally low. During the first part of the study, the median Amplicor CMV load of the 17 discrepant samples that were below the detection limit of the HCS-1 assay was 820 copies/ml. In the later part of the study, the median Amplicor-equivalent CMV load of the 28 discrepant samples that were measurable by the HCS-2 assay but below the detection limit of the Amplicor assay was 212 copies/ml. Taking the Amplicor assay detection limit of 400 copies/ml as a reference point, the median detection limit of the Amplicor assay was 2.1 times (820/400) lower than that of the HCS-1 assay. Likewise, the median detection limit of the HCS-2 assay was approximately 3.9 times (820/212) lower than that of the HCS-1 assay and 1.9 (400/212) times lower than that of the Amplicor assay.
The peak CMV loads of the D+ R− and R+ recipients were compared by using the two assays. The geometric mean CMV loads of the two groups were not significantly different (t test; P > 0.05). Of the 15 D+ R− patients who had CMV infections, only 2 did not develop CMV disease. Both of these two patients had CMV loads that were comparable to those of other D+ R− patients with CMV disease (P > 0.05). However, among the R+ recipients, patients with CMV disease had significantly higher CMV loads by both the Amplicor and the HCS assays than those who did not develop CMV disease (Amplicor; P < 0.01; HCS, P < 0.03).
The performances of the two assays in the diagnosis of CMV disease in individual patients were compared by using a sliding scale of cutoff levels. In order to compare them directly, the HCS measurements were converted to Amplicor-equivalent units. Due to the differences in the lower limits of detection of the three assays, the sliding scale started at 1,000 copies/ml, with a 0.4 log difference between each cutoff, i.e., >1,000, >2,500, >6,000, >16,000, >40,000, and >100,000. Since the HCS-1 and HCS-2 assays were similar at cutoff levels above their detection limits, these data were pooled in the analysis. Since HCS-2 is the currently marketed assay, its sensitivity, specificity, PPV, and NPV were compared with those of the Amplicor assay, using their respective detection limits as cutoff points (Table 2). The difference between the Amplicor and the HCS assays was not statistically significant over all the selected cutoff points since their 95% CIs overlapped at all levels.
TABLE 2.
Sensitivities, specificities, PPVs, and NPVs of the Amplicor and HCS assays
| Assay | Cutoff | Sensitivity
|
Specificity
|
PPV
|
NPV
|
||||
|---|---|---|---|---|---|---|---|---|---|
| % | 95% CI | % | 95% CI | % | 95% CI | % | 95% CI | ||
| Amplicor | Any positive (>400)a | 88.2 | 66.3–97.9 | 76.7 | 64.8–86.1 | 51.7 | 33.8–69.3 | 95.8 | 70.8–91.9 |
| >1,000 | 82.4 | 59.1–95.3 | 81.7 | 70.4–90 | 56.0 | 36.4–74.3 | 94.2 | 85.1–98.5 | |
| >2,500 | 76.5 | 52.5–92 | 83.3 | 72.3–91.2 | 56.5 | 36.1–75.4 | 92.6 | 83.1–97.6 | |
| >6,000 | 58.8 | 35–79.9 | 86.7 | 76.3–93.6 | 55.6 | 32.7–76.8 | 88.1 | 77.9–94.7 | |
| >16,000 | 47.0 | 24.8–70.3 | 91.7 | 82.5–96.9 | 65.5 | 34.1–84.3 | 85.9 | 75.8–92.9 | |
| >40,000 | 29.4 | 11.7–53.7 | 96.7 | 89.4–99.4 | 71.4 | 33–94.9 | 82.9 | 72.7–90.4 | |
| >100,000 | 11.8 | 2–33.7 | 100 | 95.1–100 | 100 | 22.4–100 | 80 | 69.8–87.9 | |
| HCSb | Any positive (HCS-2 only; ≈ >200)c | 80.0 | 33.4–98.9 | 51.4 | 35.1–67.5 | 19.0 | 6.4–39.8 | 94.7 | 76.7–99.7 |
| >1,000 | 76.5 | 52.5–92 | 75.0 | 62.9–84.7 | 46.2 | 28.8–64.8 | 91.8 | 81.5–97.4 | |
| >2,500 | 76.5 | 52.5–92 | 80.0 | 68.5–88.7 | 52.0 | 32.8–70.8 | 92.3 | 82.5–97.5 | |
| >6,000 | 70.6 | 46.3–88.3 | 86.7 | 76.3–93.6 | 60.0 | 37.9–79.4 | 91.2 | 81.6–96.7 | |
| >16,000 | 47.0 | 24.8–70.3 | 88.3 | 78.3–94.8 | 53.3 | 28.7–76.8 | 85.5 | 75–92.7 | |
| >40,000 | 41.2 | 20.1–65.0 | 93.3 | 84.7–97.8 | 63.6 | 33.6–87.2 | 84.8 | 74.7–92.0 | |
| >100,000 | 29.4 | 11.7–53.7 | 96.7 | 89.4–99.4 | 71.4 | 33.0–94.9 | 82.9 | 72.7–90.4 | |
The performance of the Amplicor assay when used at its lower detection limit.
HSC measurements were converted to Amplicor-equivalent units by using regression equations (see text).
The performance of the HCS-2 assay when used at its lower detection limit.
DISCUSSION
There have been many attempts to define a significant CMV load either to diagnose or to predict CMV disease after renal transplantation. Most of these studies were based on in-house assays with nonstandardized calibrators and against variable denominators for quantification (2–5, 9, 10, 13, 15). In this study, we compared two commercial assays and tested their sensitivities, specificities, PPVs, and NPVs over a range of cutoff levels for the diagnosis of symptomatic CMV disease in a cohort of 77 renal transplant recipients.
The Amplicor and the HCS assays differ in principle as well as in the type of specimens used. The Amplicor assay is a quantitative PCR, whereas the HCS assay is a hybridization method using signal amplification. The Amplicor assay detects CMV DNA in plasma, whereas the HCS assay uses whole blood, with the main source of CMV DNA expected to be leukocytes. Most studies of CMV loads have used either leukocytes or urine specimens (2–5, 7, 9, 10, 13, 15). One study showed that PCR of leukocytes was a better indicator of CMV infection than PCR of plasma for some patients (5). It was not possible to study the HCS assays using separate plasma and leukocyte specimens since the sample preparation protocol required the use of whole blood. Likewise, the manufacturer of the Amplicor assay recommended the use of plasma at the time of the study. Theoretically, it is possible to use leukocytes for the Amplicor assay. Another recent study (8) compared the measurement of CMV loads in plasma and leukocytes by using the Amplicor assay and found a high correlation between measurements for the two sources. Although leukocytes generally had more detectable viral loads, the plasma results had better agreement with those of cultures (8). Moreover, the use of plasma has advantages in that it is a simpler specimen to process and is independent of a patient's leukocyte count. In this study, the HCS and the Amplicor assays gave comparable results. Although the measured values differed substantially, a constant relationship denoted by a linear-regression equation was shown to exist. The HCS-2 assay, which replaced the HCS-1 assay halfway through the study, had the lowest detection limit, and this was followed by the Amplicor assay and then the HCS-1 assay. The regression line relating the HCS-2 assay to the Amplicor assay was similar to that relating the HCS-1 assay to the Amplicor assay. Thus, the two HCS assays are expected to yield similar results at CMV loads that are above the detection limit of the less-sensitive HCS-1 assay.
Since HCS-1 is no longer marketed, it seemed sensible only to analyze the later (HCS-2) version. However, only 5 patients in the later (HCS-2) part of this series developed CMV disease, and this resulted in very wide confidence intervals. By demonstrating that the HCS-1 and -2 assays were similar except in their detection limits, it was possible to pool the HCS-1 and -2 results. We have also compared the performance of the HCS-2 assay directly with that of the Amplicor assay at their respective detection limits. When the HCS-2 and Amplicor assays were used at their lowest detection limits, the sensitivities increased but the PPVs were very low (Amplicor, 51.7%; HCS-2, 19%). At a cutoff of 1,000 to 2,500 copies/ml, the sensitivity and specificity of both assays were about 70 to 80% (Table 2). However, the PPVs were low (46.2 to 52%). For the differential diagnosis of CMV disease and other causes of infection or rejection, a higher specificity and PPV are desired. This can be achieved by using a higher cutoff. At >40,000 copies per ml, the specificity and PPV of the Amplicor assay were 96.7 and 71.4%, respectively, and those of the HCS assay were 93.3 and 63.3%, respectively. At an even higher cutoff of >100,000 copies/ml, the specificity and PPV of the Amplicor assay were both 100% and those of the HCS assay were 96.7 and 71.4%, respectively. However, this increased specificity and PPV is at the expense of a poor sensitivity. The sensitivity of the Amplicor assay at a cutoff of >100,000 copies/ml was only 11.8%, and that of the HCS assay at the same cutoff was only 29.4%. In this cohort of renal transplant recipients, the PPVs of both assays were low throughout the range of cutoffs except at the higher cutoff point of above 40,000 copies/ml. However, the NPVs were high throughout the entire range of cutoffs. This effect of changing sensitivity, specificity, and PPV over different cutoffs should be taken into account when viral load results are being used for patient management.
To conclude, we have compared two commercial assays for the measurement of CMV load in blood. Despite the apparent differences in principle and type of specimen used, the results were comparable and could be converted from one to another by using a simple equation. One cannot extrapolate the results of this study to other patient groups, such as liver transplant patients or individuals with AIDS, since the CMV DNA copy level may not have the same distribution pattern in other situations. In this group of renal transplant recipients, a high CMV DNA cutoff (>40,000 copies/ml) is specific for CMV disease and is predictive of the diagnosis, but with a low sensitivity. A lower cutoff (>1,000 copies/ml) improves the sensitivity but is only useful for the exclusion of CMV disease.
ACKNOWLEDGMENTS
This study was supported by the Mersey Association for Kidney Research.
We thank R. Sells and M. Brown for their help in this study and G. Colucci from Roche Diagnostic for supplying the Amplicor kits.
REFERENCES
- 1.Boeckh M, Boivin G. Quantitation of cytomegalovirus: methodologic aspects and clinical applications. Clin Microbiol Rev. 1998;11:533–554. doi: 10.1128/cmr.11.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cope A V, Sweny P, Sabin C, Rees L, Griffiths P D, Emery V C. Quantity of cytomegalovirus viruria is a major risk factor for cytomegalovirus disease after renal transplantation. J Med Virol. 1997;52:200–205. [PubMed] [Google Scholar]
- 3.Fox J C, Kidd I M, Griffiths P D, Sweny P, Emery V C. Longitudinal analysis of cytomegalovirus load in renal transplant recipients using a quantitative polymerase chain reaction: correlation with disease. J Gen Virol. 1995;76:309–319. doi: 10.1099/0022-1317-76-2-309. [DOI] [PubMed] [Google Scholar]
- 4.Gerdes J C, Spees E K, Fitting K, Hiraki J, Sheehan M, Jarvi D, Roehl C, Robertson A D. Prospective study utilizing a quantitative polymerase chain reaction for detection of cytomegalovirus DNA in the blood of renal transplant patients. Transpl Proc. 1993;25:1411–1413. [PubMed] [Google Scholar]
- 5.Gerna G, Furione M, Baldanti F, Sarasini A. Comparative quantitation of human cytomegalovirus DNA in blood leukocytes and plasma of transplant and AIDS patients. J Clin Microbiol. 1994;32:2709–2717. doi: 10.1128/jcm.32.11.2709-2717.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grundy J E, Ehrnst A, Einsele H, Emery V C, Hebart H, Prentice H G, Ljungman P. A three-center European external quality control study of PCR for detection of cytomegalovirus DNA in blood. J Clin Microbiol. 1996;34:1166–1170. doi: 10.1128/jcm.34.5.1166-1170.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imbert-Marcille B-M, Cantarovich D, Ferre-Aubineau V, Richet B, Soulillou J-P, Billaudel S. Usefulness of DNA viral load quantification for cytomegalovirus disease monitoring in renal and pancreas/renal transplant recipients. Transplantation. 1997;63:1467–1481. doi: 10.1097/00007890-199705270-00018. [DOI] [PubMed] [Google Scholar]
- 8.Jabs D A, Forman M, Enger C, Jackson J B for The Cytomegalovirus Retinitis and Viral Resistance Study Group. Comparison of cytomegalovirus loads in plasma and leukocytes of patients with cytomegalovirus retinitis. J Clin Microbiol. 1999;37:1431–1435. doi: 10.1128/jcm.37.5.1431-1435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kühn J E, Wendland T, Schäfer P, Möhring K, Wieland U, Elgas M, Eggers H J. Monitoring of renal allograft recipients by quantitation of human cytomegalovirus genomes in peripheral blood leukocytes. J Med Virol. 1994;44:398–405. doi: 10.1002/jmv.1890440416. [DOI] [PubMed] [Google Scholar]
- 10.Mendez J, Espy M, Smith T F, Wilson J, Wiesner R, Paya C V. Clinical significance of viral load in the diagnosis of cytomegalovirus disease after liver transplantation. Transplantation. 1998;65:1477–1481. doi: 10.1097/00007890-199806150-00012. [DOI] [PubMed] [Google Scholar]
- 11.Meyers J D, Ljungman P, Fisher L D. Cytomegalovirus excretion as a predictor of cytomegalovirus disease after marrow transplantation: importance of cytomegalovirus viremia. J Infect Dis. 1990;162:373–380. doi: 10.1093/infdis/162.2.373. [DOI] [PubMed] [Google Scholar]
- 12.Patel R, Paya C V. Infections in solid-organ transplant recipients. Clin Microbiol Rev. 1997;10:86–124. doi: 10.1128/cmr.10.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts T C, Brennan D C, Buller R S, Gaudreault-Keener M, Schnitzler M A, Sternhell K E, Garlock K A, Singer G G, Storch G A. Quantitative polymerase chain reaction to predict occurrence of symptomatic cytomegalovirus infection and assess response to ganciclovir therapy in renal transplant recipients. J Infect Dis. 1998;178:626–635. doi: 10.1086/515383. [DOI] [PubMed] [Google Scholar]
- 14.Tong C Y W, Cuevas L, Williams H, Bakran A. Use of laboratory assays to predict cytomegalovirus disease in renal transplant recipients. J Clin Microbiol. 1998;36:2681–2685. doi: 10.1128/jcm.36.9.2681-2685.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toyoda M, Carlos J B, Galera O A, Galfayan K, Zhang X, Sun Z, Czer L S C, Jordan S C. Correlation of cytomegalovirus DNA levels with response to antiviral therapy in cardiac and renal allograft recipients. Transplantation. 1997;63:957–963. doi: 10.1097/00007890-199704150-00009. [DOI] [PubMed] [Google Scholar]