Abstract
The presence of lactose-fermenting Salmonella strains in clinical case materials presented to microbiology laboratories presents problems in detection and identification. Failure to detect these strains also presents a public health problem. The laboratory methods used in detecting lactose-fermenting Salmonella enterica serotype Typhimurium from six outbreaks of salmonellosis in veal calves are described. Each outbreak was caused by a multiply-resistant and lactose-fermenting strain of S. enterica serotype Typhimurium. The use of Levine eosin-methylene blue agar in combination with screening of suspect colonies for C8 esterase enzyme and inoculation of colonies into sulfide-indole-motility medium for hydrogen sulfide production was particularly effective for their detection. A hypothesis for the creation of lactose-fermenting salmonellae in the environment is presented. It is proposed that the environment and husbandry practices of veal-raising barns provide a unique niche in which lactose-fermenting salmonellae may arise.
To differentiate Salmonella from other Enterobacteriaceae, bacteriologists use lactose fermentation as a key biochemical test. As early as 1887, it was known that Escherichia coli was a lactose fermenter and that Salmonella was not a lactose fermenter. Therefore, most differential plating media commonly developed and used today for the isolation of Salmonella contain lactose (16, 17, 31).
It has been reported that less than 1% of all salmonellae ferment lactose (17). Since 1907, there have been various reports of the occurrence of lactose-fermenting (Lac+) Salmonella in humans, such as Lac+ Salmonella enterica serotype Virchow, S. enterica serotype Tennessee, S. enterica serotype Indiana, S. enterica serotype Agona, S. enterica serotype Typhimurium, S. enterica serotype Oranienburg, S. enterica serotype Tuebingen, S. enterica serotype Newport, S. enterica serotype Typhi, S. enterica serotype Java, and S. enterica serotype Toulon (1, 7, 12, 14, 19–22, 26, 27, 35, 38, 48, 55, 58–60). Lac+ S. enterica serotype Typhimurium and S. enterica serotype Anatum have been reported from dried milk products and milk-drying equipment (5, 57). There have been, however, only a few accounts of animal isolations; e.g., from 1969 to 1971 in Arizona, Lac+ S. enterica serotype Typhimurium was isolated from avian, bovine, canine, and porcine hosts, with most of the Lac+ isolates coming from calves that had both high morbidity and mortality rates (52). In 1976, an epidemic in 4-day-old calves of salmonellosis due to Lac+ S. enterica serotype Typhimurium was reported in Australia (33). In 1978, S. enterica serotype Indiana was isolated in turkeys in Great Britain (30, 55, 57). Also, in 1978, we reported the first veal calf outbreak due to Lac+ S. enterica serotype Typhimurium in the northeastern United States (43, 57). In 1981 and 1983, respectively, Corbion in France (9) and Rodriguez et al. in Cuba (51) described Lac+ Salmonella in cattle.
It is apparent that there are only a few reports that convey details of animal outbreaks caused by Lac+ S. enterica serotype Typhimurium. The purpose of our work is to present the clinical microbiology characteristics of six outbreaks of Lac+ S. enterica serotype Typhimurium occurring in intensively raised, milk-fed veal calves in the late 1970s and early 1980s in the northeastern United States. Also, we present a testing protocol for use by clinical laboratories concerned with the detection of biochemically aberrant isolates. The public health significance of these outbreaks is highlighted. The results of antimicrobial resistance found in the isolates is discussed in the context of the genetic events that likely occurred to produce the Lac+ Salmonella isolates. A hypothesis is proposed as to the creation of Lac+ Salmonella in the environment and is related to the epidemiology of salmonellosis in veal calves.
MATERIALS AND METHODS
Case material.
The isolates of S. enterica serotype Typhimurium used in this study originated from New York and Pennsylvania and were from diarrheal disease samples submitted to the New York State Diagnostic Laboratory as part of its mission to conduct laboratory investigations of spontaneously occurring disease in food-fiber animals, other animals, and birds. Fecal samples obtained from live animals and submitted in various transport media and internal tissues obtained at postmortem examination of calves were the usual specimens to be cultured. Field trip visits to three of the six outbreak sites were made to gather additional animal and environmental specimens for culturing. A questionnaire was developed for use in the field to gather demographic and calf information, including the use of antimicrobial agents. Further information was gathered by telephone interviews.
Viral fluorescent-antibody testing.
For detecting bovine rotaviruses and coronaviruses, sections of calf intestine were frozen at −70°C for 2 h. A cryostat microtome was used to cut tissue sections, which were then mounted on glass slides (two sections per slide), air dried, and fixed in acetone at −20°C for 15 min. Next, the acetone-fixed slides were dried in a biological safety cabinet and stored at −70°C before staining. An fluorescein isothiocyanate-conjugated calf anti-bovine rotavirus reagent and a fluorescein isothiocyanate-conjugated calf anti-bovine coronavirus reagent were used in a direct fluorescent-antibody assay to detect the presence of bovine rotaviruses and coronaviruses (conjugates came from the Reagents Division, National Veterinary Services Laboratories, Animal and Plant Health Inspection Service, U.S. Department of Agriculture, Ames, Iowa, and were produced in gnotobiotic calves). Slides were read on a fluorescent microscope at 450 nm.
Bacterial culturing.
All fecal and tissue specimen manipulations were performed with a biological safety cabinet (type 2A) by trained microbiology staff following aseptic techniques. Standard microbiological procedures were used throughout the study (15, 16, 23, 25, 28, 32). The following bacteriological media were used: Levine eosin-methylene blue agar (BBL, Cockeysville, Md.), brilliant green agar (BBL), MacConkey agar (BBL) for direct plating of specimens, Selenite-F broth (BBL), and gram-negative broth (GN broth; Hajnu; Difco Laboratories, Inc., Detroit, Mich.) for enrichment culturing.
Screening protocol used to identify Lac+ colonies.
Up to 5 suspicious Lac+ colonies from each of the three types of selective media were tested for each tissue or environmental specimen that was cultured; i.e., up to 15 total colonies were checked for each processed specimen.
(i) C8 esterase test.
A quick presumptive test to detect the C8 esterase enzyme of Salmonella (32) was performed directly on suspect Lac+ colonies that grew on any of the various differential and/or selective agars. This test uses 4-methylumbelliferyl caprilate to detect C8 esterase. A drop of reagent is placed on a suspect colony, the plate is allowed to sit for 3 min at room temperature, and then the plate is observed with a long-wavelength UV light source (365 nm) in a darkened room (Bactidrop Mucap test; REMEL, Lenexa, Kans.). Salmonella but not Citrobacter fluoresces.
(ii) Hydrogen sulfide detection.
If the C8 esterase enzyme test was positive for a colony, the colony was then screened for H2S production and concurrently for lysine decarboxylase activity (see below). Screening for H2S production (black precipitate in the medium) was done with sulfide-indole-motility (SIM; BBL) agar medium, triple sugar iron agar (BBL), and Kligler agar (BBL). In early experiments, peptone iron agar and lysine iron agar were used (BBL).
(iii) Lysine decarboxylase.
Colonies that produced H2S in SIM agar (and that were indole negative) were concurrently tested in tubes of lysine decarboxylase broth (strains should show a positive reaction in this overnight test) (43).
(iv) Polyvalent salmonella antisera and bacteriophage lysis.
Other quick screening or presumptive tests that were used were direct testing of colonies by slide agglutination with polyvalent salmonella O antisera; colonies are slide positive. Another colony-screening technique which worked well was the polyvalent salmonella bacteriophage O-1 group E assay, which produced same-day presumptive results (24, 29).
Identification of Salmonella.
Colonies thought to be Salmonella were tested biochemically with a standard conventional biochemical protocol (17, 18, 32). Finally, biochemically confirmed salmonellae were serogrouped with commercially available rabbit antisera (BBL) (18).
Salmonella serotyping.
Salmonellae were sent to the Veterinary Services, National Veterinary Services Laboratories, Animal and Plant Health Inspection Service, U.S. Department of Agriculture, for complete serotyping by standard techniques (18).
Bacteriophage typing of Lac+ isolates.
Isolates of S. enterica serotype Typhimurium were phage typed with an updated serotype Typhimurium scheme (2, 6, 34, 45). Phage typing studies were performed at the Laboratory Center for Disease Control, Bureau of Microbiology, National Laboratory for Enteric Pathogens, Ottawa, Canada.
Biotyping of Lac+ isolates.
Biotyping of isolates was performed by the methods of Duguid et al. (13) and McDonough et al. (45) for S. enterica serotype Typhimurium.
Plasmid profile.
Plasmid DNA was isolated by an adaptation of the Birnboim-Doly extraction procedure (4) and analyzed by vertical gel electrophoresis by the technique of Crosa and Falkow (11).
Antimicrobial susceptibility testing.
The disk diffusion method of Bauer et al. (3) was used for susceptibility testing. Fourteen drugs were routinely used to test gram-negative enteric bacteria: ampicillin (10 μg), chloramphenicol (30 μg), colistin (10 μg), sulfisoxazole (Gantrisin) (300 μg), gentamicin (10 μg), kanamycin (30 μg), neomycin (30 μg), nitrofurantoin (300 μg), polymyxin B (300 μg), streptomycin (10 μg), tetracycline (30 μg), triple sulfa (300 μg), trimethoprim (1.25 μg)-sulfadiazine (23.75 μg), and cephalothin (30 μg). The Mueller-Hinton agar and drug disks were BBL products, except for sulfisoxazole (Difco). Standard bacterial strains were used as quality controls in these assays (45), and National Committee for Clinical Laboratory Standards guidelines were used for interpretation of results (46). A mnemonic was used to reduce the resistances detected to a number score by the method of McDonough et al. (45).
Gross pathology and histopathology.
Gross and microscopic examinations of any calves and their tissues submitted to the Necropsy Service, Department of Pathology, College of Veterinary Medicine, Cornell University, were made.
RESULTS
Clinical case and treatment histories.
A series of six outbreaks of salmonellosis in veal calf farms occurred from October 1977 to February 1982 in northern Pennsylvania and the southern tier of New York. In all cases, both morbidity and mortality rates were high; calves, 7 to 10 days old on average, presented with clinical signs of depression, anorexia, fever, bloody diarrhea, and severe dehydration. Most affected calves died within 24 to 36 h of the onset of illness. The antimicrobial treatment histories, including medicated milk replacer diets, are shown in Table 1.
TABLE 1.
Antimicrobial drugs used in veal calves and in their food
| Outbreak | Drugs used in feed and for treatment of sick calves
|
|
|---|---|---|
| Milk replacer | Ill calves (oral and injectable drugs) | |
| 1 | Nitrofurantoin, oxytetracycline | Oxytetracycline, chloramphenicol |
| 2 | Chlortetracycline, sulfamethazine, penicillin | Gentamicin, lincomycin, spectinomycin |
| 3 | Neomycin, oxytetracycline | Nitrofurantoin, penicillin, penicillin-streptomycin, gentamicin, neomycin-tetracycline, nitrofurazone, lincomycin-spectinomycin |
| 4 | Neomycin, oxytetracycline | Trimethoprim-sulfadiazine |
| 5 | Neomycin, oxytetracycline, furazolidone | Sulfamethazine, lincomycin-spectinomycin |
| 6 | None (nonmedicated) | Neomycin, polymyxin B sulfate, tetracycline |
Clinical bacteriology and clinical virology.
In the first outbreak, eight of eight dead calves had Lac+ S. enterica serotype Typhimurium isolated from intestines and mesenteric lymph nodes; in six of eight, isolates were obtained from gallbladder swabs; in five of eight, isolates were obtained from livers and spleens; two of eight calves had Pasteurella multocida and Arcanobacterium pyogenes isolated from lungs; the environment (room drains and milk mixer apparatus) were culture positive for Lac+ S. enterica serotype Typhimurium; and no salmonellae were found in the milk replacer. Fluorescent-antibody tests were negative for rotavirus and coronavirus for eight of eight tested calf intestines.
In the second outbreak, the intestines of 2 calves were tested and were found culture positive for Lac+ S. enterica serotype Typhimurium; no bacteria were isolated from joints; feces from 4 of 12 live calves were culture positive for Lac+ Salmonella; and the milk replacer was culture negative for Salmonella. No virology testing was conducted on these calves.
In the third outbreak, the ileum tested from a single calf contained Lac+ S. enterica serotype Typhimurium; environmental specimens (room drains, milk mixer vat, environmental surfaces, and milk pails) contained no salmonellae; and four of nine fecal specimens from live calves contained Lac+ S. enterica serotype Typhimurium. Fluorescent-antibody tests were negative for rotavirus and coronavirus for the single tested dead calf intestines.
In the fourth outbreak, only two fecal specimens were received from live calves; Lac+ S. enterica serotype Typhimurium was cultured from these. Virology testing was not attempted for these calves.
In the fifth outbreak, Lac+ S. enterica serotype Typhimurium was recovered from three dead calf hock joints, ilea, spleens, and mesenteric lymph nodes, as well as from fecal samples from six live calves. The intestine from one of the three autopsied calves was tested by a fluorescent-antibody test for rotavirus and coronavirus, with negative results.
In the sixth outbreak, cultures from two calves grew Lac+ S. enterica serotype Typhimurium from the intestines, mesenteric lymph nodes, spleens, and lungs. All intestinal tissue sections were found negative for rotavirus and coronavirus by fluorescent-antibody testing.
Appearance of colonies of Lac+ S. enterica serotype Typhimurium on agar in vitro.
The overall appearance of the Lac+ Salmonella colonies on differential media was similar to that of E. coli; i.e., they appeared as rough, flat, lactose-fermenting bacterial colonies. All isolates recovered from plating on brilliant green agar appeared as rough, yellowish green colonies; MacConkey agar yielded rough, red colonies; and on Levine eosin-methylene blue agar, the colonies looked like small, rough, 2- to 4-mm green sequins with a metallic sheen. Levine EMB agar was the best medium for distinguishing Lac+ enteric bacteria, including Lac+ Salmonella, from other bacteria due to their very typical appearance on this medium.
Screening tests for the detection of Lac+ Salmonella.
On average, a total of 15 colonies were screened (five from each of three selective agars) from each specimen (feces, internal organ, or environmental swab). In total, 45 calves were tested and over 10 environmental swabs were screened.
(i) C8 esterase enzyme test.
All of the colonies that ultimately were identified as Lac+ Salmonella were positive in the C8 esterase enzyme test; i.e., the colonies fluoresced.
(ii) H2S production.
Early testing demonstrated that screening of suspect colonies for H2S production was not possible with triple sugar iron agar and Kligler agar slants (the reaction was acid slant/acid butt/gas), because the black iron sulfide precipitate that would normally be seen from non-lactose-fermenting salmonellae in these media was dissolved due to acid production in the media. However, in SIM medium, peptone iron agar, and lysine iron agar, H2S production in the form of a black precipitate was visible from all colonies that subsequently were identified as Salmonella.
(iii) Lysine decarboxylase broth.
All colonies that subsequently were identified as Salmonella were positive in the lysine decarboxylase broth test.
(iv) Agglutination with polyvalent salmonella O antisera.
All colonies that subsequently were identified as Salmonella agglutinated in the polyvalent salmonella O antiserum assay.
(v) Polyvalent salmonella bacteriophage O1-OE assay.
All colonies that subsequently were identified as Salmonella were lysed by bacteriophage O1-OE.
Bacteriophage typing of Lac+ S. enterica serotype Typhimurium.
Only a single isolate was tested from each outbreak, except for outbreak 6, in which the isolate was found not to be viable at the testing laboratory. Of the five isolates, only one was typeable to a definitive phage type, and the other four were either untypeable or produced atypical phage reactions (Table 2). Reference 45 provides a thorough analysis of the phage types of the population of S. enterica serotype Typhimurium strains found in New York, of which these Lac+ strains were a part.
TABLE 2.
Phage type, biotype, antibiogram, and plasmid profile of Lac+ S. enterica serotype Typhimurium strains
| Outbreak | Phage typea/biotypeb | Antibiogram mnemonicc | Plasmid size (MDa) |
|---|---|---|---|
| 1 | U276/26ei | 64272 | 100, 62, 5.3, 3.7, 2.2, 1.7 |
| 2 | Atypical/26ei | 65672 | 100, 62, 5.3, 3.7, 2.2, 1.7 |
| 3 | Atypical/26ei | 65672 | 62, 5.3, 3.7, 2.2, 1.7 |
| 4 | Untypeable/26ei | 65672 | 100, 62, 54, 5.3, 3.7, 2.2, 1.7 |
| 5 | Atypical/26ei | 65672 | 100, 62, 5.3, 3.7, 2.2, 1.7 |
| 6 | —/— | 65672 | 100, 62, 54, 5.3, 3.7, 2.2, 1.7 |
U, provisional phage type classification; atypical, the isolates reacted with the typing phages but did not conform to a recognizable type; untypeable, the isolate was resistant to all of the typing phages (see also reference 45); —, not tested.
The biotype designations conform to standard nomenclature in the system of Duguid et al. (13); —, not tested.
Antibiogram mnemonic code: 64272, resistance to ampicillin, chloramphenicol, sulfisoxazole, nitrofurantoin, streptomycin, tetracycline, triple sulfa, and cephalothin and sensitivity to colistin, gentamicin, kanamycin, neomycin, polymyxin B, and trimethoprim-sulfadiazine; 65672, resistance to ampicillin, chloramphenicol, sulfisoxazole, kanamycin, neomycin, nitrofurantoin, streptomycin, tetracycline, triple sulfa, and cephalothin and sensitivity to colistin, gentamicin, polymyxin B, and trimethoprim-sulfadiazine.
Biotyping of Lac+ S. enterica serotype Typhimurium.
Five of five strains tested were biotype 26ei, which is a commonly found biotype in New York (Table 2) (45).
Plasmid profile.
Five of the six strains of Lac+ S. enterica serotype Typhimurium (one from each outbreak) contained a very large plasmid of about 100 MDa. The Birnboim-Doly extraction method (4) does not optimally recover very large plasmids, so it is possible that the other salmonella strain also contained the same plasmid, which went undetected. All six strains contained the 62-MDa serotype-specific or vir plasmid of S. enterica serotype Typhimurium. In addition, all six strains tested had small plasmids of unknown significance (Table 2).
Antimicrobial susceptibility test results.
Antibiograms for the Lac+ S. enterica serotype Typhimurium strains are shown in Table 2. All strains tested were multiply resistant to at least eight drugs.
DISCUSSION
From the points of view of human clinical and veterinary microbiologists, it is important to recognize that Lac+ strains exist and then to screen case materials for these biochemically aberrant bacteria. From our varied case load in a large veterinary diagnostic laboratory screening thousands of specimens each month, we have detected Lac+ salmonellae only from veal calves and their barn environments. We have not statistically shown that we have found a higher frequency of Lac+ salmonellae than the literature reports (less than 1%). However, we have demonstrated that in the northeastern United States, there exists a somewhat unique niche for Lac+ S. enterica serotype Typhimurium in milk-fed veal populations. We are well aware of the pitfalls of using diagnostic submissions and the data derived from them because of potential submission bias; however, this report is at the same time a useful indicator of what is occurring in the field, although perhaps not useful for statistical statements of prevalence. As long as Salmonella isolates are found in cattle and bull calves are sold from dairy farms to be raised as veal, the potential exists for Lac+ S. enterica serotype Typhimurium to occur in veal farms.
While a microbiologist may screen for salmonellae, whether or not they are biochemically aberrant, by using the PCR test on broth enrichment cultures of clinical specimens, in order to carry out serotyping and susceptibility testing of Salmonella, the Lac+ Salmonella would still have to be successfully isolated on agar and correctly identified. Hence, it is important to recognize that these strains are found in animal populations.
Salmonella is a zoonotic bacterial agent, and S. enterica serotype Typhimurium is the most common serotype found in animals and in humans (44). During outbreak 3, a child helping to manage calves became ill with an acute gastrointestinal disease manifested by vomiting, fever, diarrhea, and cramps. Stool specimens were cultured from this child, but Salmonella was not isolated in vitro. Her presumptive diagnosis, however, was salmonellosis, based in part on the estimated incubation period of her disease, on knowledge of the salmonellosis situation on her family farm, and on the fact that laboratory cultures had ruled out other enteric bacterial pathogens. It is plausible that the hospital clinical laboratory performing the enteric culturing did not detect or did not recognize Lac+ Salmonella in her stool cultures; the laboratory staff was not routinely using bismuth sulfite agar, which does not use lactose as the sugar in the medium. It is noteworthy that calves recovering from clinical disease but whose salmonella shedding status was unknown were sent to market after these outbreaks occurred; exposure of humans in contact with these calves during marketing also may have occurred. Perhaps there are relatively more reports in the literature of human cases of Lac+ salmonellae than animal cases due to the more widespread use of bismuth sulfite agar in laboratories testing specimens from humans.
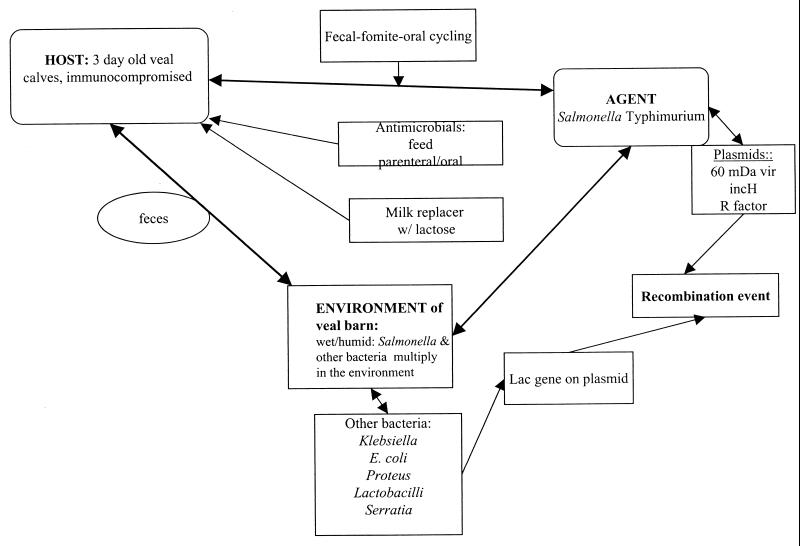
Figure 1 provides an overview of the hypothesis for the creation of Lac+ S. enterica serotype Typhimurium. The epidemiological triad of disease consists of the host, the agent, and the environment, with all of its stress factors. In the New York and Pennsylvania outbreaks, the host is the 3-day-old immunocompromised veal calf (hypogammaglobulinemic due to lack of colostrum or insufficient colostrum) that is trucked at least twice in its journey from the dairy farm of origin to the sale yard to the final destination at the veal farm. This calf potentially is exposed to the agent, i.e., Salmonella, in the environments of all the places it has been moved. Calves tend to suckle and lick each other and environmental surfaces during trucking. Depending on the design of the veal barn, there also may be extensive licking between adjacent calves. During conditions of intensified husbandry (crowding), rapid buildup of Salmonella in feces occurs in the wet environments of the veal barns, so ample opportunity exists for a neonatal population of calves to be exposed to Salmonella by the fecal-fomit-oral route of infection (42, 43, 49, 63, 64). Calves with acute salmonellosis shed Salmonella in their salivary glands, so in addition to the cross-licking problem, if individual calf-feeding buckets are not provided, one can easily see how additional cross-infection may occur between calves (36, 47, 50, 62). If farms do not effectively clean and disinfect the veal barn between the exit of the 14- to 16-week-old calves going to market and the incoming 3- to 5-day-old calves, the neonatal calves literally walk into a plethora of bacterial pathogens in the veal barn environment. The most frequent causes of this management problem are (i) failure to practice an “all in-all out” style of management (i.e., practice of having only one age group at a time in the veal barn) and/or (ii) failure to allot sufficient time to clean, disinfect, and dry the veal barn before restocking with a new group of calves.
FIG. 1.
Hypothesis for the development of Lac+ S. enterica serotype Typhimurium in the environment of veal-raising barns. The hypothesis is based on the interaction of the veal calf host, the salmonella agent, and the environment.
The feces accumulating in the environment of the veal barn contain not only salmonellae but also other enteric bacteria, such as Klebsiella pneumoniae (39, 40, 61), Lactobacillus (8), Proteus (14, 61), and Serratia (61), which may carry the lactose (lac) operon on a plasmid or, in the case of some Klebsiella strains, on the chromosome (40). One theory for the origin of lac in these plasmids involves the unlikely transfer of the operon from the chromosome of another enteric bacterium (E. coli) via a transposon (10). The β-galactosidase enzyme produced by some strains of Lac+ Salmonella differs from E. coli β-galactosidase, offering further evidence that the operon did not originate from E. coli but could have originated from an enterobacterial ancestor common to E. coli (10, 14, 40). Some strains of Klebsiella carry the lac gene on the chromosome; there is evidence for these strains that the lac gene of Klebsiella differs from that of E. coli and that they both diverged from a common ancestor (40). The lac plasmids also may contain determinants of antimicrobial resistance, and there have been reports of outbreaks of Lac+ and antimicrobial agent-resistant Salmonella in the literature (19, 55, 57).
In addition, the source of food for the veal calves, i.e., milk replacer, contains abundant lactose sugar and is often supplemented with viable lactobacillus-containing products.
From our previous work with the agent S. enterica serotype Typhimurium, we know that many strains of S. enterica serotype Typhimurium isolated from calves in New York and Ontario, Canada, carry IncH1 plasmids (57). IncH1 plasmids are of high molecular weight, have a low copy number in the bacterial cell, and are thermosensitive for transfer (optimally between 22 and 30°C) (41, 54). We documented the presence of IncH1 plasmids in S. enterica serotype Typhimurium and E. coli strains from outbreak 1 (57). The ultimate source of the IncH1 plasmids is unknown, but they have been reported to occur naturally in Citrobacter, Klebsiella, Enterobacter, E. coli, Salmonella, Serratia, and Shigella. The presence of these plasmids that transfer at a low temperature in the veal barn environment but not in the intestine of the calf at body temperature suggests that they may be important vectors for disseminating antibiotic resistance genes in soil and in aquatic environments (41, 53). The presence of high-molecular-weight plasmids (100 MDa) (Table 2) in Lac+ S. enterica serotype Typhimurium strains from outbreaks 2, 4, 5, and 6 (in addition to outbreak 1, previously reported in reference 57) provides preliminary evidence that these also may be IncH1 plasmids.
We have proposed that a recombination event occurred between the IncH plasmid of Salmonella and a lac plasmid in the feces in the environment of the veal barn and that this recombination event was facilitated by the selective pressure of antimicrobial agent usage in the veal calves. Because of the efficient fecal-(fomite)-oral cycling of Salmonella in the veal barn environment, the calves would be expected to become infected easily with a salmonella strain that carried a recombinant lac-IncH1 plasmid.
In each of these outbreaks, the strain of Lac+ S. enterica serotype Typhimurium appeared to be more virulent, i.e., killed calves within hours after clinical signs were noted. In E. coli, IncH plasmids confer the ability to persist longer in the intestine of the calf (37, 41, 56). One may consider that the same effect could occur in a salmonella-infected calf whose Salmonella strain carried the same type of plasmid. Also, the newly acquired ability of Salmonella to use lactose may confer on it a selective advantage in an environment where lactose is plentiful, i.e., milk replacer in the environment and intestine of the calf; thus, the calves may have died more quickly because of exposure to overwhelming numbers of Salmonella in the veal barn environment.
In summary, we have described the clinical microbiology characteristics of a potentially zoonotic bacterial pathogen that presents the veterinary and medical communities with a difficult diagnostic and treatment problem. Our hypothesis as to the evolution of Lac+ Salmonella suggests that the veal barn environment, like the human hospital environment, is one with many selective pressures brought on by the juxtaposition of antimicrobial agent use, abundant plasmids, and an immunocompromised host population. The fact that these hosts are also human food presents a potential food-borne disease problem. Our future work will deal with understanding the genetics of the lac gene and the resistance determinants found in Lac+ strains of Salmonella. The veal industry of the 1990s has in place residue avoidance and quality assurance programs that should help to address many of the drug usage and management problems highlighted in our discussion of these six outbreaks of salmonellosis in the late 1970s and early 1980s.
ACKNOWLEDGMENTS
This work was supported by the New York State Department of Agriculture and Markets, Albany.
We thank Rasik Khakhria, Federal Laboratories, Health Canada, and the Canadian Food Inspection Agency, Health Protection Branch, Laboratory Center for Disease Control, Bureau of Microbiology, National Laboratory for Enteric Pathogens, Winnipeg, Manitoba, Canada, for bacteriophage typing studies.
REFERENCES
- 1.Anand C M, Finlayson M C, Garson J Z, Larson M L. An institutional outbreak of salmonellosis due to lactose-fermenting Salmonella newport. Am J Clin Pathol. 1980;74:657–660. doi: 10.1093/ajcp/74.5.657. [DOI] [PubMed] [Google Scholar]
- 2.Anderson E S, Ward L R, deSaxe M J, deSa J D S. Bacteriophage-typing designations of Salmonella typhimurium. J Hyg. 1977;78:297–300. doi: 10.1017/s0022172400056187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer A W, Kirby W M M, Sherris J C, Turck M. Antibiotic susceptibility testing by a standardized single disc method. Am J Clin Pathol. 1966;45:493–496. [PubMed] [Google Scholar]
- 4.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackburn B O, Ellis E M. Lactose-fermenting Salmonella from dried milk and milk-drying plants. Appl Microbiol. 1973;26:672–674. doi: 10.1128/am.26.5.672-674.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Callow B R. A new phage-typing scheme for Salmonella typhimurium. J Hyg. 1959;57:346–359. doi: 10.1017/s0022172400020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camara F P, Cardoso M A, de Almeida D F. Genetic analysis of lactose-fermenting Salmonella typhimurium isolated in Rio de Janeiro. Rev Soc Bras Med Trop. 1989;22:81–83. doi: 10.1590/s0037-86821989000200003. [DOI] [PubMed] [Google Scholar]
- 8.Chassy B M, Gibson E M, Giuffrida A. Evidence for plasmid-associated lactose metabolism in Lactobacillus casei subsp. casei. Curr Microbiol. 1978;1:141–144. doi: 10.1007/BF02601666. [DOI] [PubMed] [Google Scholar]
- 9.Corbion B. Differential diagnosis of lactose fermenting Salmonella and certain enterobacteria. Bull Inf Lab Serv Vet. 1981;81:165–169. [Google Scholar]
- 10.Cornelis G. Sequence relationships between plasmids carrying genes for lactose utilization. J Gen Microbiol. 1981;124:91–97. doi: 10.1099/00221287-124-1-91. [DOI] [PubMed] [Google Scholar]
- 11.Crosa J H, Falkow S. Plasmids. In: Gerhardt P, Murray R G E, Costilow R N, Nester E W, Wood W A, Krieg N R, Phillips G B, editors. Manual of methods for general bacteriology. Washington, D.C.: American Society for Microbiology; 1981. pp. 266–282. [Google Scholar]
- 12.Dube S D. Outbreak of food poisoning caused by lactose-fermenting Salmonella tuebingen. J Clin Microbiol. 1983;17:698–699. doi: 10.1128/jcm.17.4.698-699.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duguid J P, Anderson E S, Alfredsson G A, Barker R, Old D C. A new biotyping scheme for Salmonella typhimurium and its phylogenetic significance. J Med Microbiol. 1975;8:149–166. doi: 10.1099/00222615-8-1-149. [DOI] [PubMed] [Google Scholar]
- 14.Easterling S B, Johnson E M, Wohlhilter J A, Baron L S. Nature of lactose-fermenting Salmonella strains obtained from clinical sources. J Bacteriol. 1969;100:35–41. doi: 10.1128/jb.100.1.35-41.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ewing W H. Isolation and identification of Salmonella and Shigella. Atlanta, Ga: U.S. Department of Health, Education, and Welfare Center for Disease Control; 1972. . publication no. (CDC) 77-8098. [Google Scholar]
- 16.Ewing W H. Edwards and Ewing's identification of the Enterobacteriaceae. 4th ed. New York, N.Y: Elsevier Science Publishing Co., Inc.; 1986. Isolation and preliminary identification; pp. 27–45. [Google Scholar]
- 17.Ewing W H. Edwards and Ewing's identification of the Enterobacteriaceae. 4th ed. New York, N.Y: Elsevier Science Publishing Co., Inc.; 1986. Differentiation of Enterobacteriaceae by biochemical reactions; pp. 47–72. [Google Scholar]
- 18.Ewing W H. Edwards and Ewing's identification of the Enterobacteriaceae. 4th ed. New York, N.Y: Elsevier Science Publishing Co., Inc.; 1986. The genus Salmonella; pp. 181–245. [Google Scholar]
- 19.Ezaki T, Liu S L, Yabuuchi E, Sasakawa C, Yoshikawa M. Molecular characterization of a conjugative R-lac plasmid in Salmonella typhi isolated from a patient with typhoid fever. Ann Inst Pasteur Microbiol. 1987;138:303–311. doi: 10.1016/0769-2609(87)90118-9. [DOI] [PubMed] [Google Scholar]
- 20.Falcao D P, Trabulsi L R, Hickman F W, Farmer J J. Unusual Enterobacteriaceae: lactose-positive Salmonella typhimurium which is endemic in São Paulo, Brazil. J Clin Microbiol. 1975;2:349–353. doi: 10.1128/jcm.2.4.349-353.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falkow S, Baron L S. Episomic element in a strain of Salmonella typhosa. J Bacteriol. 1962;84:581–589. doi: 10.1128/jb.84.3.581-589.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farley J D, Ghesquiere W, MacLean D R, Moulton G S. Endemic institutional salmonellosis due to lactose-fermenting Salmonella newport in Nova Scotia. Can Med Assoc J. 1988;138:434–436. [PMC free article] [PubMed] [Google Scholar]
- 23.Farmer J J., III . Enterobacteriaceae: introduction and identification. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: American Society for Microbiology; 1995. pp. 438–449. [Google Scholar]
- 24.Fey H, Burgi E, Margadant A, Boller E. An economic and rapid diagnostic procedure for the detection of Salmonella/Shigella using the polyvalent Salmonella phage O-1. Zentbl Bakteriol Parasitenkd Infektionskr Hyg Abt 1 Orig Reihe A. 1978;240:7–15. [PubMed] [Google Scholar]
- 25.Forbes B A, Granato P A. Processing specimens for bacteria. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: American Society for Microbiology; 1995. pp. 265–281. [Google Scholar]
- 26.Glosnicka R, Pienkowska D, Dera-Tomaszewska B, Ilnicka M J. Characteristics of lactose-fermenting Salmonella strains from Poland. Bull Inst Mar Trop Med Gdynia. 1987;38:69–75. [PubMed] [Google Scholar]
- 27.Gonzalez A B. Lactose-fermenting Salmonella. J Bacteriol. 1966;91:1661. doi: 10.1128/jb.91.4.1661-1662.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gray L D. Escherichia, Salmonella, Shigella, and Yersinia. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: American Society for Microbiology; 1995. pp. 450–464. [Google Scholar]
- 29.Gudel K, Fey H. Improvement of the polyvalent Salmonella phage's O-1 diagnostic value by addition of a phage specific for the O groups E1–E4. Zentbl Bakteriol Parasitenkd Infektionskr Hyg Abt 1 Orig Reihe A. 1981;249:220–224. [PubMed] [Google Scholar]
- 30.Hall M L M, Threlfall E J, Rowe B, Pinegar J A, Gibson G L. Lactose-fermenting Salmonella indiana from turkeys in Britain. Lancet. 1978;ii:1197–1198. doi: 10.1016/s0140-6736(78)92175-x. [DOI] [PubMed] [Google Scholar]
- 31.Janda J M, Abbot S L. The enterobacteria. Philadelphia, Pa: Lippincott-Raven Publishers; 1998. Historical perspectives on the family Enterobacteriaceae; pp. 1–12. [Google Scholar]
- 32.Janda J M, Abbot S L. The enterobacteria. Philadelphia, Pa: Lippincott-Raven Publishers; 1998. The salmonellae; pp. 80–109. [Google Scholar]
- 33.Johnston K G, Jones R T. Salmonellosis in calves due to lactose fermenting Salmonella typhimurium. Vet Rec. 1976;98:276–278. doi: 10.1136/vr.98.14.276. [DOI] [PubMed] [Google Scholar]
- 34.Khakhria R, Lior H. Distribution of phagovars of Salmonella typhimurium in Canada (1969–1976) Zentbl Bakteriol Parasitenkd Infektionskr Hyg Abt 1 Orig Reihe A. 1980;248:50–63. [PubMed] [Google Scholar]
- 35.Kukulska D, Zimmer A. Selected properties of lactose-fermenting and non-fermenting Salmonella agona strains isolated from specimens from hospitalized infants. Med Dosw Mikrobiol. 1989;41:115–120. [PubMed] [Google Scholar]
- 36.Linton A H, Howe K, Pethiyagoda S, Osborne A D. Epidemiology of Salmonella infection in calves. 1. Its relation to their husbandry and management. Vet Rec. 1974;94:581–585. doi: 10.1136/vr.94.25.581. [DOI] [PubMed] [Google Scholar]
- 37.Linton A H, Timoney J F, Hinton M. The ecology of chloramphenicol-resistance in Salmonella typhimurium and Escherichia coli in calves with endemic Salmonella infection. J Appl Bacteriol. 1981;50:115–129. doi: 10.1111/j.1365-2672.1981.tb00876.x. [DOI] [PubMed] [Google Scholar]
- 38.Louie K K, Paccagnella A M, Osei W D, Lior H, Francis B J, Osterholm M T. Salmonella serotype Tennessee in powdered milk products and infant formula—Canada and United States, 1993. Morbid Mortal Weekly Rep. 1993;42:516–517. [PubMed] [Google Scholar]
- 39.Lucain C, Piffaretti J-C. Characterization of a high molecular mass resistance plasmid isolated from Klebsiella pneumoniae and coding for lactose degradation. FEMS Microbiol Lett. 1983;20:131–134. [Google Scholar]
- 40.MacDonald C, Riley M. Cloning chromosomal lac genes of Klebsiella pneumoniae. Gene. 1983;24:341–345. doi: 10.1016/0378-1119(83)90096-3. [DOI] [PubMed] [Google Scholar]
- 41.Maher D, Taylor D E. Host range and transfer efficacy of incompatibility group HI plasmids. Can J Microbiol. 1993;39:581–587. doi: 10.1139/m93-084. [DOI] [PubMed] [Google Scholar]
- 42.McDonough P L. Salmonellosis: diagnostic approach to disease control and epidemiology in the bovine animal. Bovine Proc. 1995;27:61–68. [Google Scholar]
- 43.McDonough P L, Shin S J, Fairbrother J M, Kradel D C, Timoney J F. 21st Annual Proceedings of the American Association of Veterinary Laboratory Diagnosticians. 1978. Salmonellosis due to lactose-fermenting Salmonella typhimurium; pp. 253–264. Buffalo, N.Y. [Google Scholar]
- 44.McDonough P L, Shin S J, Timoney J F. Salmonella serotypes from animals in New York State, 1978–1983. Cornell Vet. 1986;76:30–37. [PubMed] [Google Scholar]
- 45.McDonough P L, Timoney J F, Jacobson R H, Khakhria R. Clonal groups of Salmonella typhimurium in New York State. J Clin Microbiol. 1989;27:622–627. doi: 10.1128/jcm.27.4.622-627.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.National Committee for Clinical Laboratory Standards. Approved standard M2-A6. Performance standards for antimicrobial disk diffusion susceptibility tests. 6th ed. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 47.Osborne A D, Linton A H, Pethiyagoda S. Epidemiology of Salmonella infection in calves. 2. Detailed study in a large beef rearing unit. Vet Rec. 1974;94:604–610. doi: 10.1136/vr.94.26.604. [DOI] [PubMed] [Google Scholar]
- 48.Proschen R K, Hale D, Goodman Z. Misdiagnosed Salmonella septicemia and endarteritis due to a lactose-fermenting strain: bacteriologic and epidemiologic considerations. Am J Clin Pathol. 1977;68:416–419. doi: 10.1093/ajcp/68.3.416. [DOI] [PubMed] [Google Scholar]
- 49.Rebhun W C •••, editors. Diseases of dairy cattle. Baltimore, Md: The William & Wilkins Co.; 1995. Infections of the gastrointestinal tract; pp. 155–223. [Google Scholar]
- 50.Richardson A. Salmonellosis in cattle. Vet Rec. 1975;96:329–331. doi: 10.1136/vr.96.15.329. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez D, Troncoso M, Ancizar J A. Lactose-fermenting salmonellae from calves with diarrhoea. Rev Cubana Cien Vet. 1983;14:55–59. [Google Scholar]
- 52.Rokey N W, Mecca M D. Lactose fermenting salmonellae—a dilemma for diagnostic laboratories? Proc Annu Meet US Anim Health Assoc. 1972;75:509–514. [Google Scholar]
- 53.Smith H W. Thermosensitive transfer factors in chloramphenicol-resistant strains of Salmonella typhi. Lancet. 1974;ii:281–282. doi: 10.1016/s0140-6736(74)91435-4. [DOI] [PubMed] [Google Scholar]
- 54.Taylor D E, Shermer M, Grant R B. Incidence of the H2 group of plasmids in chloramphenicol-sensitive Salmonella isolated in 1974 from clinical sources in Ontario. Can J Microbiol. 1978;24:600–607. doi: 10.1139/m78-098. [DOI] [PubMed] [Google Scholar]
- 55.Threlfall E J, Hall M L M, Rowe B. Lactose-fermenting salmonellae in Britain. FEMS Microbiol Lett. 1983;17:127–130. [Google Scholar]
- 56.Timoney J F, Linton A H. Experimental ecological studies on H2 plasmids in the intestine and feces of the calf. J Appl Bacteriol. 1982;52:417–424. doi: 10.1111/j.1365-2672.1982.tb05072.x. [DOI] [PubMed] [Google Scholar]
- 57.Timoney J F, Taylor D E, Shin S, McDonough P. pJT2: unusual H1 plasmid in a highly virulent lactose-positive and chloramphenicol-resistant Salmonella typhimurium strain from calves. Antimicrob Agents Chemother. 1980;18:480–482. doi: 10.1128/aac.18.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Twort F W. The fermentation of glucosides by bacteria of the typhoid coli group and the acquisition of new fermenting powers by Bacillus dysenteriae and other micro-organisms. Proc R Soc London Ser B. 1907;79:329–336. [Google Scholar]
- 59.Usera M A, Echeita A, Aladuena A, Blanco M C, Reymundo R, Prieto M I, Tello O, Cano R, Herrera D, Martinez-Navarro F. Interregional foodborne salmonellosis outbreak due to powdered infant formula contaminated with lactose-fermenting Salmonella virchow. Eur J Epidemiol. 1996;12:377–381. doi: 10.1007/BF00145301. [DOI] [PubMed] [Google Scholar]
- 60.Usera M A, Rodriguez A, Echeita A, Cano R. Multiple analysis of a foodborne outbreak caused by infant formula contaminated by an atypical Salmonella virchow strain. Eur J Clin Microbiol Infect Dis. 1998;17:551–555. doi: 10.1007/BF01708617. [DOI] [PubMed] [Google Scholar]
- 61.Walia S K, Madhavan T, Chugh T D, Sharma K B. Characterization of self-transmissible plasmids determining lactose fermentation and multiple antibiotic resistance in clinical strains of Klebsiella pneumoniae. Plasmid. 1987;17:3–12. doi: 10.1016/0147-619x(87)90003-5. [DOI] [PubMed] [Google Scholar]
- 62.Williams B M. Environmental considerations in salmonellosis. Vet Rec. 1975;96:318–321. doi: 10.1136/vr.96.14.318. [DOI] [PubMed] [Google Scholar]
- 63.Wray C, Todd N, McLaren I M, Beedell Y E. The epidemiology of Salmonella in calves: the role of markets and vehicles. Epidemiol Infect. 1991;107:521–525. doi: 10.1017/s0950268800049219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wray C, Todd N, McLaren I, Beedell Y, Rowe B. The epidemiology of Salmonella infection of calves: the role of dealers. Epidemiol Infect. 1990;105:295–305. doi: 10.1017/s0950268800047890. [DOI] [PMC free article] [PubMed] [Google Scholar]