Abstract
The Gibberellic Acid Stimulated Arabidopsis (GASA) proteins were investigated in the study to help understand their possible roles in fruit trees, particularly in Citrus. A total of 18 CcGASA proteins were identified and characterized in Citrus clementina via a genome-wide approach. It was shown that the CcGASA proteins structurally shared a conserved GASA domain but varied considerably in primary sequences and motif compositions. Thus, they could be classified into three major phylogenetic groups, G1~G3, and two groups, G1 and G3 could be further classified into subgroups. The cis- elements on all CcGASA promoters were identified and categorized, and the associated transcription factors were predicted. In addition, the possible interactions between the CcGASA proteins and other proteins were predicted. All the clues suggested that these genes should be involved in defense against biotic and abiotic stresses and in growth and development. The notion was further supported by gene expression analysis that showed these genes were more or less responsive to the treatments of plant hormones (GA3, SA, ABA and IAA), and infections of citrus canker pathogen Xanthomonas citri. It was noted that both the segmental and the tandem duplications had played a role in the expansion of the CcGASA gene family in Citrus. Our results showed that the members of the CcGASA gene family should have structurally and functionally diverged to different degrees, and hence, the representative group members should be individually investigated to dissect their specific roles.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-021-03326-6.
Keywords: GASA, Citrus, Phylogenetic tree, Promoter, Transcription factor, Protein interaction
Introduction
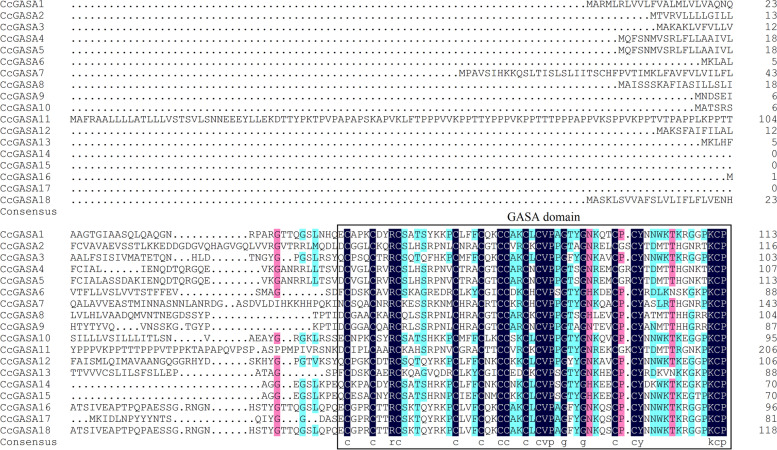
The Snakin/Gibberellic Acid Stimulated in Arabidopsis (GASA) is a unique multigene family. Since the isolation of GAST1 (Gibberellic Acid Stimulated Transcript 1) from tomato [43], many GASA protein family members have been characterized in different species, such as potato [42], common wheat [13], soybean [2], Arabidopsis [6, 38], petunia [8], rice [15], apple [14], grapevine [1] and maize [56]. A comprehensive genome sequence analysis of 33 plant species revealed approximately 445 Snakin/GASA protein encoding genes [45]. Further bioinformatics data mining showed that the Snakin/GASA genes were present in all well-characterized or sequenced plant species but were completely absent in moss and green algae [17]. It is known that the GASA family peptides share a conserved C-terminal domain, designated as GASA. The GASA domain contains 12 cysteine residues (Cys-motif) arranged in the pattern of “XnCX3CX3CX8(9)CX3CX2CCX2CX1(3)C11CPC11(14)KCP” (where: X represents any of the 20 non-cysteine amino acids; P and K represent proline and lysine, respectively) [48]. A typical GASA protein also possesses a putative signal peptide at the N-terminus and a variable region in the middle of the sequence.
GASA proteins are known to play diverse roles in plants. They are involved in the regulation of growth and development processes, including cell division [34], stem elongation [8], floral induction [36], seed germination [38, 39], lateral root formation [56] and fruit development [33]. The GASA proteins are also linked to stress responses, such as resistance to heat [26], drought, and paclobutrazol (PBZ) stresses [51], tolerance to salt and oxidative stresses [3], and modulation of reactive oxygen species (ROS) [47]. Moreover, GASA proteins have shown suppressive effects on a wide range of bacterial and fungal pathogens. Purified StSN1 peptide, for example, was found in an in vitro challenge experiment to be toxic to several fungal pathogens like Fusarium solani, Fusarium culmorum, Bipolaris maydis and Botrytis cinerea, and to bacterial pathogens such as Clavibacter michiganensis subsp. Sepedonicus [46]. The GASA family proteins even exhibit anti-viral and anti-nematode activities, as exemplified by GmSN1 that enhances soybean mosaic virus resistance in both Arabidopsis and soybean [19] and by CaSn that promotes nematode-resistance in pepper [31]. The citrus CcGASA4 was shown to be highly induced in citrus leaves following infection of Citrus tristeza virus [54].
GASA family genes are implicated in the responses of plants to hormones such as gibberellin (GA), abscisic acid (ABA), salicylic acid (SA), jasmonic acid (JA) and ethylene (ETH). Most GASA genes including Arabidopsis AtGASA4, 6, 7, 8 and 13 [39, 57], rice OsGASR1 and 2 [15], petunia GIP 1, 2, 4 and 5 [8], maize ZmGSL1, 2, 4, 6 and 9 [56], were induced by exogenous GA treatment. However, some other GASA genes, such as Arabidopsis AtGASA1, 5, 9 and 11 [57], and potato StSN2 [9] were repressed by GA. Interestingly, some GASA genes exhibited tissue-specific responses to GA applications. GsGASA1 expression was induced in leaves but repressed in roots by exogenous application of GA in Glycine soja [30]. AtGASA4 was up-regulated in most, if not all, meristematic regions, presumably in actively dividing cells, but was down-regulated in cotyledons and leaves following GA treatment [6]. ABA was shown to induce the expression of AtGASA2, 3, 5 and 14, and inhibit the expression of AtGASA7 and 9 in Arabidopsis [57]. The expression of StSN2 was induced [9] whereas Snakin-3 was down regulated by ABA treatment in potato [35]. In addition, some members of the GASA family, such as GAST1 [37, 43], StSN2 [9] and GASA5 [57], were regulated by GA and ABA antagonism. The GA mediated increase in GAST1 transcripts was partially inhibited by ABA in tomato [43]. AtGASA1 was up-regulated by GA and down-regulated by ABA in Arabidopsis [37]. StSN2 was up- and down- regulated by ABA and GA, respectively, in potato [9]. The expression of GASA5 was repressed by GA3 and enhanced by ABA [57]. GASA genes are also responsive to other hormones. Brassinoesteroid (BR) synthesis was activated by OsGSR1 by directly regulating a BR biosynthetic enzyme [52]. The transcription of HbGASA7-1, 14 and 16 was significantly increased after ETH, SA, or JA treatment in Hevea brasiliensis [4].
Less information about the GASA genes is available for trees, particularly for fruit trees. Nevertheless, 14 VvGASA genes in grapevine (Vitis vinifera L.) [1], 26 MdGASA genes in apple (Malus domestica) [14] and a CcGASA4 gene in citrus [54] were reported. Citrus is one of the most important fruit trees worldwide. A steady increase in global per capita consumption of citrus fruits has been witnessed in the past 30 years [32]. Citrus production is, however, being threatened by numerous biotic and abiotic stresses. Identification and functional analysis of citrus defense- and stress-related genes should deepen our understanding of the molecular mechanisms of stress-responses and lend helps in improving stress tolerance in plants. Considering that a citrus GASA gene was highly responsive to the Citrus tristiza virus, we performed a detailed bioinformatics analysis of the relevant gene family, aiming to provide first tier information for dissecting their exact roles in the defense of citrus against stresses.
Materials and methods
Identification of putative Citrus clementina GASA genes
The GASA protein sequences of Arabidopsis, apple [14] and grape [1] were downloaded from the EnsemblPlants online database (http://plants.ensembl.org/info/data/ftp/index.html) and analyzed using the HMMER (https://www.ebi.ac.uk/Tools/hmmer/) to build a model based on the GASA domain (Accession: pfam02704). The model was used to query the entire C. clementina genome to obtain all putative clementine mandarin GASA proteins (Supplementary Table S1). The integrity of the GASA domain of every CcGASA protein sequence was verified using the Simple Modular Architecture Research Tool (SMART: http://smart.embl.de/) [24]. All non-redundant putative protein sequences with conserved GASA domain were reserved and used for further analysis. GASA proteins from other three citrus species, pommelo (Citrus maxima), sweet orange (Citrus sinensis) and trifoliate orange (Poncirus trifoliata), were also identified following the same protocol.
All CcGASA protein coding sequences, genomic sequences and the associated information such as accession number and chromosomic position, were downloaded from the Phytozome database (https://phytozome.jgi.doe.gov/pz/portal.html#). In addition, the physical location of each CcGASA gene on the genome was mapped by the Mapchart software.
Analysis of physicochemical properties of the citrus GASA proteins
The physicochemical parameters of the CcGASA proteins were calculated by using the ProtParam (http://web.expasy.org/protparam) [16]. Protein putative subcellular locations and tertiary structures were predicted by using the WOLF PSORT II program (https://www.genscript.com/wolf-psort.html) [22] and the PHYRE2 engine (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index), respectively. The transmembrane helices were predicted using the TMHMM server v2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/).
Analysis for protein phylogenetic relationships and gene structures
A phylogenetic tree was generated based on 111 GASA protein sequences from different plant species including C. clementina (15), C. sinensis (14), C. maxima (13), P. trifoliata (14), Arabidopsis thaliana (15), M. domestica (26) and V. vinifera (14). The GASA sequences of C. sinensis, C. maxima and P. trifoliata were downloaded from the Orange (C. sinensis) Genome Annotation Project website (http://citrus.hzau.edu.cn/orange/). MEGA 7.0 software [28] was used to construct the phylogenetic tree by using the neighbor-joining (NJ) method. The parameters of the NJ method were as follows: 1000 bootstrap replications, “p-distance” model, “Uniform rates”, “partial deletion”, and 95% site coverage cutoff. The conserved regions within the CcGASA proteins were identified by using the MEME v5.2.0 (http://meme-suite.org/tools/meme) [7]. The GASA CDSs and their corresponding genomic sequences of the four Citrus species were compared to reveal exons and introns using the Gene Structure Display Server 2.0 (GSDS2.0, http://gsds.gao-lab.org/).
Promoter analysis of CcGASA genes
Around 1.9-kb long promoter sequence upstream of the start codon (ATG) of each CcGASA gene was downloaded from the Phytozome12.0 database [18]. The cis-regulatory elements on the promoters were analyzed by using the Plant Cis-Acting Regulatory DNA Elements (PlantCARE) program (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [29].
Expression analysis of CcGASA genes
The 4-year-old C. clementina trees had been planted in plastic pots and used as experimental materials. For the biotic stress treatment, mature leaves of similar size and shape were picked and brought to lab in a humidified chamber, and immediately pierced at ten evenly spaced points at the back of each leaf with a syringe needle. The pierced leaves were placed face down in a try lined at the bottom with water-soaked filter papers. Ten micro litter of Xanthomonas citri subsp. Citri (Xcc) cell cultures, diluted to 5 × 108 cfu/mL (OD600 ≈ 0.3), was applied to cover the pierced holes. For controls, 10 μL of water was applied. The trays were then misted with water and covered with plastic films, and sampled at 0 h, 12 h, 24 h, 48 h, and 72 h. Samples were immediately frozen in liquid nitrogen, and then stored in a -80°C refrigerator until use. For hormone treatment, trees were sprayed with 100 μΜ of GA3, 2 mΜ of SA, 100 μM of indole-3-acetic acid (IAA) and 200 μΜ of ABA, and leaves were harvested after 3h, 6h, 12h and 24h, flash frozen in liquid nitrogen, and stored at −80°C. Three biological replicates were used in the study.
Total RNA was extracted from frozen leaf samples using the Polysaccharide Polyphenol plant total RNA rapid extraction kit (Bioteke, China) following the manufacturer’s instructions. RNA concentrations were determined using the NanoDrop2000C (ThermoFisher Scientific, USA), and RNA quality was evaluated by ratios of OD260 / OD280. cDNA was synthesized from total RNA using the PrimeScriptTM RT reagent kit with gDNA Eraser (TaKaRa, Japan). The quantitative real-time PCR (qRT-PCR) experiment was performed on a thermo-cycler, QuantStudio5 (ABI, USA). Each tube of qRT-PCR solution contained 10.0 μL iTaqTM Universal SYBR® Green Supermix (Bio-Rad, USA), 1.0 μL primer pair F/R, 2.0 μL cDNA, and 6.0 μL dH2O. The thermo-cycler was programed as follows: an initial incubation at 95°C for 10 min and followed by 40 cycles of 95°C for 5 s + 60°C for 20 s. For melting-curve analysis, the program was set to 95°C for 15 s, 60°C for 2 min and then the temperature was progressively increased to 95°C at a constant rate of 0.2°C/s. The primers used were shown in Supplementary Table S2. The Actin (GenBank accession: XM_006427792) was used as the reference gene. Since three proteins, CcGASA16, CcGASA17 and CcGASA18, were predicted to be derived from a single gene sequence (Ciclev10006243m.g) via alternative splicing, it was impossible to design primers to separate CcGASA18 from CcGASA16 and CcGASA17, and hence only CcGASA16 and CcGASA17 were eventually analyzed. For quality controls, three technical replicates were also used in addition to three biological replicates. The relative expression level, shown as the ratio of the analyzed gene to the reference gene, was determined by calculating the 2 −ΔΔCt (ΔCt = Ct CcGASA -Ct Actin). Data were statistically analyzed using ANOVA. Duncan’s LSD multiple range test (p ≤ 0.05) was performed to reveal significant changes. Figures were drawn by using the Origin 2019b.
The leaves, stems, young fruits and roots from healthy C. clementina trees were subjected to transcriptome profiling in a commercial biotech company, Oebiotech Company (China; https://www.oebiotech.com/) to reveal possible tissue-specific expression patterns. Briefly, total RNA was isolated from samples and used to prepare RNA-seq libraries. The libraries were then sequenced on Illumina Genome Analyzer platform. Clean reads were obtained by passing the row sequencing data through all quality control procedures, and were mapped to the C. clementina genome sequence using the HISAT2 software. The expression level of each gene in the RNA-seq libraries was calculated as the FPKM (Fragments Per Kilobase of transcript per Million fragments mapped) by using the Cuffquant and Cuffnorm software. The differentially expressed transcripts, defined by a fold change (|log2Fold Change|) of greater than 1 and a P value (false discovery rate, FDR) of less than 0.05, between samples were identified by using the DESeq2 [5] software.
Synteny analysis and calculation of Ka/Ks ratio for duplicated genes
CcGASA gene duplication events were identified according to the criteria proposed by Holub [21]. The Basic Circos function of TBtools software was used to show the interspersed segmental duplications using the data from the PLAZA (https://bioinformatics.psb.ugent.be/plaza/versions/plaza_v4_5_dicots/) [49]. The Ka (non-synonymous substitution rate) and Ks (synonymous substitution rate) between the duplicated genes were calculated using the online tool PAL2NAL (http://www.bork.embl.de/pal2nal/index.cgi?example=Yes#RunP2N). Mode of selection acting on the duplicated genes was evaluated through Ka/Ks ratio, and a positive, negative or neutral selection was considered when the ratio was > 1, < 1, or = 1, respectively. The gene loci of GASAs were extracted from the annotation gff3-file on the EnsemblPlants and the Orange (C. sinensis) Genome Annotation Project online database. Collinear pairs were extracted using TBtools [12] to identify syntenic blocks and duplications within the GASAs across the whole genomes of 7 species, C. clementina, C. sinensis, C. maxima, P. trifoliata, A. thaliana, M. domestica and V. vinifera. The collinearity map between these species was drawn with the help of MCScan X (TBtools software) program.
Analysis of transcription factor regulatory network and protein interaction network involving CcGASA proteins
Transcription factor (TFs) network prediction was performed online at the threshold parameter p-value ≤ 1e-5 on the Plant Transcriptional Regulatory Map (PTRM) website (http://plantregmap.gao-lab.org/regulation_prediction.php), using all the CcGASA sequences as an input. The Cytoscape 3.8 software was used to visualize the transcription factor regulatory network [27]. The predicted TFs were subjected to GO analyses on the Omicshare cloud platform (https://www.omicshare.com/tools/). The functional interacting network models of CcGASA proteins were predicted using the web program STRING 11.0 (http://string-db.org). The confidence parameter was set at a threshold of 0.40, and for other parameters the default values were used.
Results
Genome-wide distribution of C. clementina GASA genes and features of their deduced proteins
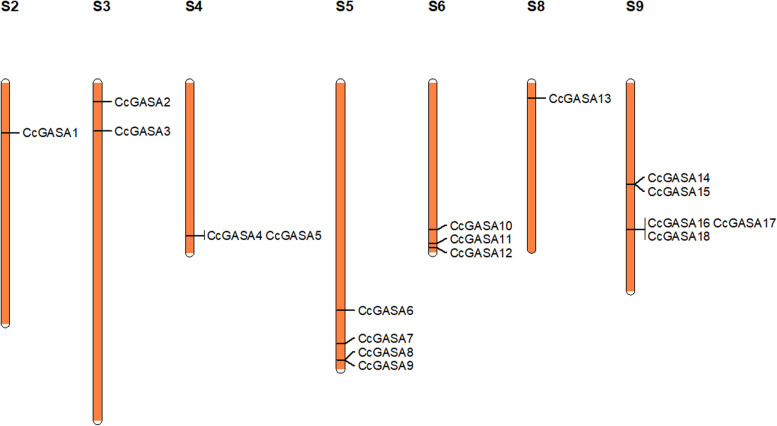
Eighteen putative CcGASA proteins were found by searching the C. clementine protein database against a model built from other known plant GASA proteins (Table 1), and correspondingly, 15 CcGASA genes were identified. The proteins were sequentially designated according to their chromosomal locations as CcGASA1–18 in this study (Fig. 1). Comparison between CcGASA4 and CcGASA5 showed that both are coded by the same gene, Ciclev10033115m.g. Similarly, CcGASA16, 17 and 18 are coded by Ciclev10006243m.g. It should be noted that Ciclev10013454m, previously designated as CcGASA4 [54], was re-designated as CcGASA12 in this study. As shown in Fig. 1, the 15 CcGASA genes were located on 7 scaffolds on the C. clementina genome. More specifically, 4 of them were on scaffold 5, 3 on scaffold 6, 3 on scaffold 9, 2 on Scaffold 3, 1 on scaffold 2 and 1 on scaffold 4.
Table 1.
Detailed information of citrus CcGASA genes
| Transcript ID | Protein Name | Scaffold | Start Sit | End Sit | Strand | No. of Exons | CDS (bp) | Protein (A.A) |
|---|---|---|---|---|---|---|---|---|
| Ciclev10017244m | CcGASA1 | S2 | 7477430 | 7479641 | forward | 4 | 342 | 113 |
| Ciclev10022925m | CcGASA2 | S3 | 2774465 | 2775592 | forward | 4 | 351 | 116 |
| Ciclev10023012m | CcGASA3 | S3 | 7235575 | 7236465 | forward | 3 | 312 | 103 |
| Ciclev10033135m | CcGASA4 | S4 | 23072592 | 23073840 | reverse | 3 | 342 | 107 |
| Ciclev10033115m | CcGASA5 | S4 | 23072592 | 23073840 | reverse | 3 | 342 | 113 |
| Ciclev10002979m | CcGASA6 | S5 | 34404147 | 34405161 | reverse | 2 | 267 | 88 |
| Ciclev10002796m | CcGASA7 | S5 | 39444681 | 39445917 | forward | 4 | 432 | 143 |
| Ciclev10002927m | CcGASA8 | S5 | 41913536 | 41914321 | reverse | 3 | 315 | 104 |
| Ciclev10002984m | CcGASA9 | S5 | 41920802 | 41921611 | reverse | 2 | 264 | 87 |
| Ciclev10013200m | CcGASA10 | S6 | 22210589 | 22211749 | forward | 3 | 288 | 95 |
| Ciclev10012786m | CcGASA11 | S6 | 24264177 | 24265636 | reverse | 4 | 621 | 206 |
| Ciclev10013454m | CcGASA12 | S6 | 24913375 | 24914177 | forward | 4 | 321 | 106 |
| Ciclev10029695m | CcGASA13 | S8 | 2308972 | 2309555 | forward | 2 | 267 | 88 |
| Ciclev10006931m | CcGASA14 | S9 | 15301341 | 15301778 | reverse | 2 | 213 | 70 |
| Ciclev10006668m | CcGASA15 | S9 | 15311772 | 15312184 | reverse | 2 | 213 | 70 |
| Ciclev10006310m | CcGASA16 | S9 | 22139872 | 22141728 | forward | 3 | 291 | 96 |
| Ciclev10006347m | CcGASA17 | S9 | 22140369 | 22141728 | forward | 4 | 246 | 81 |
| Ciclev10006243m | CcGASA18 | S9 | 22140369 | 22141728 | forward | 1 | 357 | 118 |
CDS coding sequence
Fig. 1.
Chromosomal locations and duplications of citrus CcGASA genes. The scaffolds(S) number is indicated above each bar. The scaffold size is indicated by its relative length using the information from Phytozome
The deduced protein sequences of the 18 CcGASA transcripts varied in length from 70 to 206 amino acids (Fig. 2). They were low molecular weight peptides, mostly less than 13kDa, although CcGASA7 (15.86 kDa) and CcGASA11 (21.78 kDa) were slightly larger (Table 2). All CcGASAs were relatively high in their isoelectric point (pI) values, for 7 of them had a pI value of higher than 8, and the others even higher than 9. They were mostly predicted to be unstable since 13 of them, except for CcGASA1, CcGASA2, CcGASA4, CcGASA5 and CcGASA6, had an instability index values of higher than 40. According to the Grand average of hydropathicity (GRAVY), the CcGASA proteins, excluding CcGASA2 and CcGASA8, were hydrophilic. Amino acid preference analysis showed that Cys, Lys, Ser, Leu and Pro were the preferable amino acids although Ala, Gly, Thr, Glu, Argand Tyr were also common. The aliphatic index values of the CcGASA proteins were different, varying from 25.14 to 84.74.
Fig. 2.
Amino acid sequence alignment of CcGASA proteins
Table 2.
Analysis of physicochemical properties of CcGASAs
| Protein name | MW | PI | Major amino acid% | Instability index | Aliphatic index | GRAVY | Localization predicted |
|---|---|---|---|---|---|---|---|
| CcGASA1 | 12.25 | 9.48 | A(11.5%), C(10.6%), L(8.8%) | 33.50 | 64.87 | -0.165 | chlo, extr, vacu |
| CcGASA2 | 12.44 | 8.75 | L(12.1%), C(11.2%), G(11.2%) | 28.19 | 84.74 | 0.011 | extr, vacu, ER |
| CcGASA3 | 11.42 | 9.28 | C(11.7%), K(8.7%), P(7.8%) | 42.68 | 57.77 | -0.095 | extr, chlo, nucl, mito |
| CcGASA4 | 11.76 | 9.02 | C(12.1%) , R (10.3%), L (8.4%),T(8.4%) | 26.38 | 71.03 | -0.079 | extr, vacu |
| CcGASA5 | 12.32 | 9.01 | C(11.5%), R(9.7%); L(8.0%); T(8.0%) | 26.47 | 69.03 | -0.123 | extr, vacu, chlo |
| CcGASA6 | 9.68 | 8.62 | C(13.6%), K(11.4%), S(9.1%) | 28.36 | 63.07 | -0.086 | extr, vacu |
| CcGASA7 | 15.86 | 9.59 | C(9.1%), A(8.4%), S(8.4%) | 67.18 | 81.19 | -0.122 | chlo, extr |
| CcGASA8 | 11.02 | 8.66 | A(10.6%), C(11.5%), S(9.6%) | 46.75 | 77.02 | 0.121 | extr |
| CcGASA9 | 9.38 | 8.79 | C(13.8%), T(10.3%), G(9.2%) | 47.05 | 38.16 | -0.574 | chlo, mito, cyto |
| CcGASA10 | 10.42 | 9.27 | C(12.6%), S(11.6%), L(10.5%), K(10.5%) | 49.34 | 69.79 | -0.093 | extr, vacu |
| CcGASA11 | 21.78 | 9.63 | P(25.7%), T(12.6%), K(8.7%) | 69.01 | 60.10 | -0.398 | extr, chlo, vacu |
| CcGASA12 | 11.74 | 9.29 | C(11.3%), K(11.3%), G(9.4%) | 40.13 | 51.60 | -0.263 | extr, vacu |
| CcGASA13 | 9.67 | 8.70 | C(14.8%), K(12.5%), G(6.8%), L(6.8%), P(6.8%), S(6.8%) | 40.22 | 59.77 | -0.242 | extr, golg |
| CcGASA14 | 7.78 | 8.75 | C(17.1%), K(15.7%), P(10.0%) | 51.43 | 25.14 | -0.863 | — |
| CcGASA15 | 7.71 | 8.04 | C(17.1%), E(10.0%), K(10.0%) | 46.37 | 25.14 | -0.791 | — |
| CcGASA16 | 10.43 | 9.20 | C(12.5%), P(10.4%), G(9.4%), T(9.4%) | 57.25 | 30.52 | -0.750 | chlo, nucl, mito, plas |
| CcGASA17 | 9.14 | 9.15 | C(14.8%), K(11.1%), G(8.6%), P(8.6%), Y(8.6%) | 50.33 | 34.94 | -0.668 | chlo, nucl, extr, mito, cyto |
| CcGASA18 | 12.86 | 9.19 | C(10.2%), P(8.5%), S(8.5%) | 47.72 | 56.19 | -0.307 | extr, vacu, golg |
MW molecular weight (kDa), pI isoelectric point, GRAVY grand average of hydropathicity. Hydrophilic is represented by negative value, hydrophobic is represented by positive value. A Ala, C Cys, E Glu, G Gly, K Lys, L Leu, P Pro, R Arg, S Ser, T Thr, Tyr Y, chlo chloroplast, mito mitochondria, cyto cytoplasm, extr extracellular, vacu vacuoles, nucl nucleus, golg golgiosome, plas plastid, ER endoplasmic reticulum
The WOLF PSORT II program predicted that the citrus CcGASA family proteins were mostly extracellular, or vacuole- and chloroplast-localized. A few of them might be located in endoplasmic reticulum, nucleus, mitochondria, cytoplasm, plastid and golgiosome (Table 2). The 3D model prediction, with a confidence of more than 99.9%, showed that all of them were relatively flexible for possessing random coils (Supplementary Fig. S1). As can be seen, a small α helix was located at the end of the N-terminal, which was connected next to a larger α helix, but the β-strand was only present on two of them, CcGASA7 and CcGASA17 (Supplementary Table S3). The transmembrane topology analysis showed that there was at least one transmembrane helix on CcGASA1, 3, 4, 5, 7, 8,10, and 12 (Supplementary Fig. S2).
Phylogenetic relationship of the GASA proteins
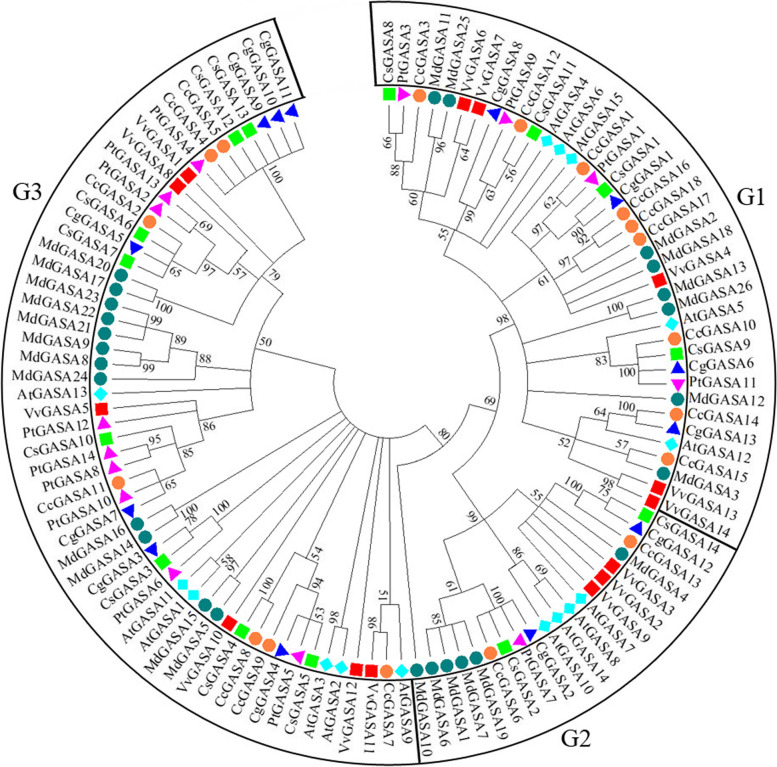
An unrooted NJ phylogenetic tree was established by aligning all GASA protein sequences from citrus (56), Arabidopsis (15), apple (26) and grape (14) (Fig. 3, Supplementary Table S4). Three branches, G1, G2, G3, were clearly shown on the tree. For citrus CcGASA proteins, CcGASA1, 3, 10, 12, 14, 15, 16, 17 and 18 were clustered with G1, CcGASA2, 4, 5, 7, 8, 9 and 11 were grouped with G3, whereas CcGASA6 and CcGASA13 were classified with G2. Clearly, all the six species analyzed contained homologs of the three branches.
Fig. 3.
Phylogenetic tree of citrus GASA protein and Arabidopsis AtGASA, apple MdGASA and grape VvGASA. Different groups (G1, G2, and G3) are located in different branches. Protein name: orange-colored dots represent Citrus clementina CcGASA proteins, green-colored squares represent Citrus sinensis CsGASA proteins, blue-colored squares represent Citrus maxima CgGASAs, purple-colored squares represent Poncirus trifoliata PtGASAs, blue-colored rhombuses represent Arabidopsis AtGASAs, dark green dots represent apple MdGASAs, and red-colored squares represent grape VvGASA proteins. The number near the branch indicates the bootstrap value
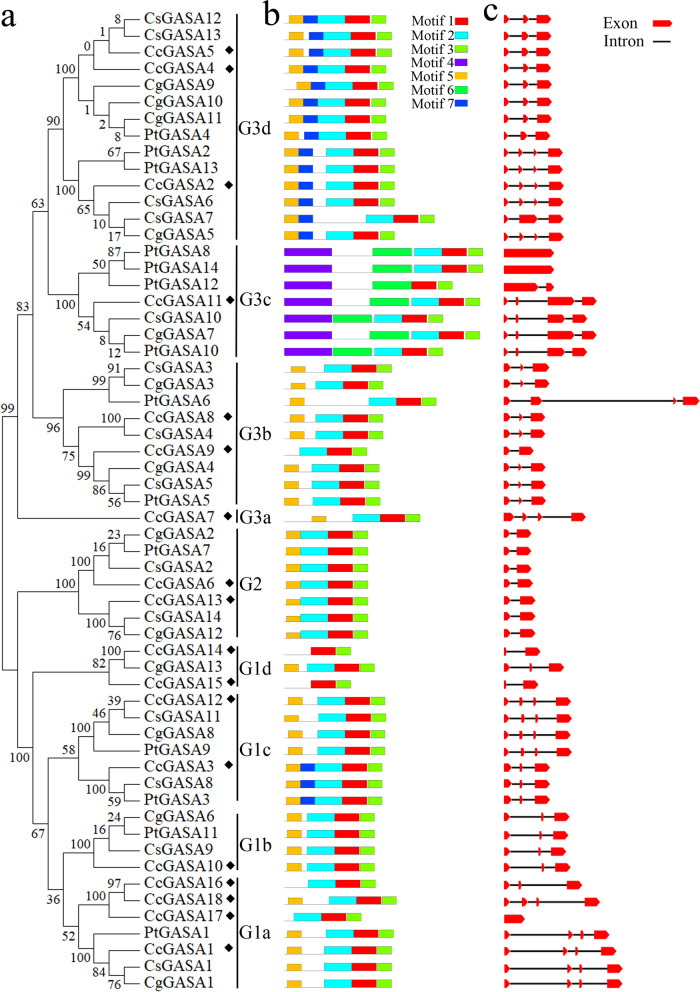
The phylogenetic tree for all citrus GASA proteins was shown in Fig. 4a. A total of 7 conserved protein motifs could be identified from the citrus GASA proteins analyzed (Fig. 4b). They were represented by CLRACGTCCARCLCVPPGTYGNKEVC (motif1), SGYTRGLLQSIDCGGLCAARCSLHSRPNP (motif2), CYTBMTTKGGKPKCP (motif3), MAFRAALLLLATLLLVSTSVLSNNEEEYLLEKDTTYPKTPVPAPAPPKAP (motif4), MASRVFLLLSJLLFC (motif5), PTVTPAPPLKPPTTYPPPVKPPTTTPPPVTPPKTAPAPQVP (motif6) and IAVIENQDTQRGZEV (motif7), respectively. It was found that the three motifs, motif1, motif2 and motif3, were universally presented on every member analyzed. Motif5 was missing only in G3c and while motif4 and 6 were only found in the same G3c. Motif7 was appeared only on G3d and on three members of G1. Apparently, the branching of the phylogenetic tree was related to the differences in the arrangement of these motifs. For example, the G2 group members contained 4 closely spaced motifs, motif1, motif2, motif3 and motif5. Similarly, most of the G1 members also contained the same 4 motifs but unlike G2, their motif5 and motif2 was interrupted by insertion of motif7 or a non-motif spacer. Clearly, the citrus G3 members were relatively more diverse than those of G1 and G2 as shown by their motif compositions and motif arrangements, and thus could be further classified into 4 subgroups G3a-d (Fig. 4a). The G1group could also be sub-classified into four sub-groups, G1a-d, Some citrus CcGASA proteins should be structurally or functionally impaired for missing one or two motifs as compared to their respective group members. For instance, motif2 and motif5 were deleted from CcGASA14 and 15, and motif5 was missing in CcGASA9, 16 and 17. The arrangements of exons and introns were relatively conserved in the same group (Fig. 4c). Only one intron was found in G2 while 2 to 3 introns were presented on most of the G1 and G3 group members. Uniquely, two trifoliate orange genes, PtGASA8 and 14, and one citrus gene, CcGASA17, did not contain any intron.
Fig. 4.
Phylogenetic relationships, exon-intron pattern and group designations in CcGASA proteins from citrus. a the neighbor-joining (NJ) tree based on the complete protein sequences of CcGASA. The tree shows the 6 phylogenetic groups (G1a-d, G2, G3a-d). b Conserved region analysis of CcGASA proteins. The different colors of boxes denote different motif numbers. The length of box indicates motif length. c the gene structure is presented by exon (red boxes) and intron (black line between the red boxes)
CcGASA promoters and their possible activators
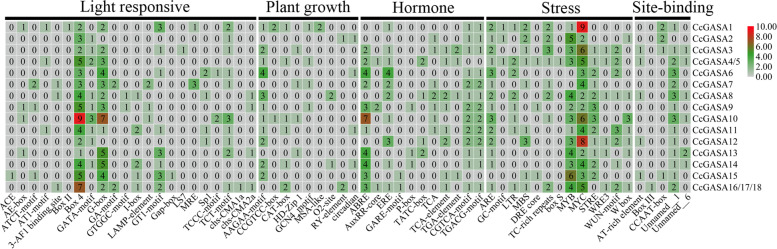
The cis-regulatory elements identified on the promoters of CcGASA genes were listed in Fig. 5 and Supplementary Table S5. It was shown that a large number of the elements were stress-related, such as ARE (antioxidant response element), GC-motif (enhancer-like element involved in anoxic specific inducibility), LTR (low-temperature responsiveness), MBS (drought-inducibility), DRE core (cold- and dehydration-responsiveness), TC-rich repeats (defense and stress responsiveness), box S (elicitation, wounding and pathogen responsievness), MYB(abiotic element), MYC(abiotic element), STRE (stress response element), WRE3 (wound-response element 3), WUN-motif (wound responsiveness) and W box (wounding and pathogen responsive). Notably, 2 to 9 MYCs were present on all CcGASA promoters. The ARE, essential for the anaerobic induction, was present on 17 CcGASA promoters but not on the CcGASA15 promoter. Hormone responsive elements were also abundant, including ABRE that is responsive to ABA, P-box, GARE-motif and TATC-box that are responsive to GA, AuxRR-core and TGA-element that are responsive to AUX, ERE that is responsive to ET, TCA and TCA that are responsive to SA, and TGACG-motif and CGTCA-motif that are responsive to MeJA. Light response cis-elements, including 3-AF1 binding site, ACE, AE-box, ATCT-motif, AT1-motif, Box II, Box 4, GATA-motif, G-box, GA-motif, GTGGC-motif, I-box, LAMP-element, GT1-motif, Gap-box, LS7, MRE, Sp1, TCCC-motif, TCT-motif, chs-CMA1a and chs-CMA2a, were also frequently found on CcGASA promoters. In addition, plant growth and development associated cis-elements, including meristem-specific expression elements, CCGTCC-box and CAT-box, plant seed and shoot development-related RY-element, circadian control element, MSA-like cell cycle control element, and the palisade mesophyll cells differentiation element HD-Zip 1, were also identified on CcGASA promoters.
Fig. 5.
Analysis of cis-elements in CcGASA genes. Grey indicates the absence of cis-elements upstream of the CcGASA gene. The different colour represents the number of cis-elements upstream of the CcGASA gene. The redder of the colour, the greater number of the cis-element
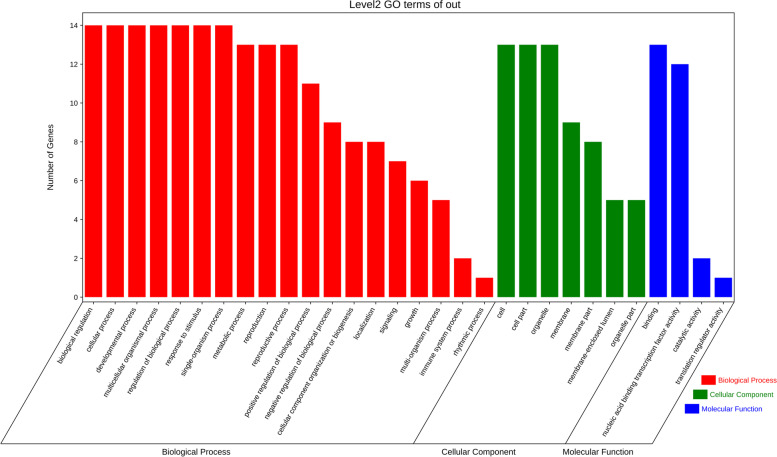
The potential transcriptional regulatory network of the CcGASA gene family was analyzed. It was shown that the TFs that might bind to the above-mentioned cis-elements were numerous (Supplementary Fig. S3 and Table S6). Most of the TFs were ERF, MYB and MIKC_MADS, for 23, 12 and 9 of them were respectively identified in the study. In addition, 5 Dofs, 4 ARFs, 4 C2H2s, and 4 HSF were also predicted to be the activators of the CcGASA genes. Functionally, these TFs were mostly associated with abiotic and biotic stresses such as pathogen attacks, heat shock and drought stresses. In addition, some TFs including ERF (Ciclev10009361m.g), ERF (Ciclev10025816m.g), ARF (Ciclev10000183m.g, Ciclev10011065m.g, Ciclev10014391m.g, Ciclev10030860m.g), GRAS (Ciclev10017466m.g), and C2H2 (Ciclev10002297m.g) that are responsive to plant hormones are known to be involved in plant growth and development regulations. Tissue specific TFs were also identified, including the lateral organ boundaries transcription factor LBD (Ciclev10024416m.g) and the root specific transcription factor NAC (Ciclev10010579m.g). Detailed GO function enrichment analysis showed that these putative transcription factors were mainly enriched in the cellular process (GO:0009987), multicellular organismal process (GO:0032501), developmental process (GO:0032502), single-organism process (GO:0044699), regulation of biological process (GO:0050789), biological regulation (GO:0065007), metabolic process (GO:0008152) and response to stimulus (GO:0050896) (Fig. 6).
Fig. 6.
GO enrichment analysis results of predicted TFs. Red bar indicates GO term related to biological process; green bar indicate GO term related to cellular component; Blue bar indicate GO term related to molecular function
Expression of CcGASAs under treatments of Xcc and plant hormones
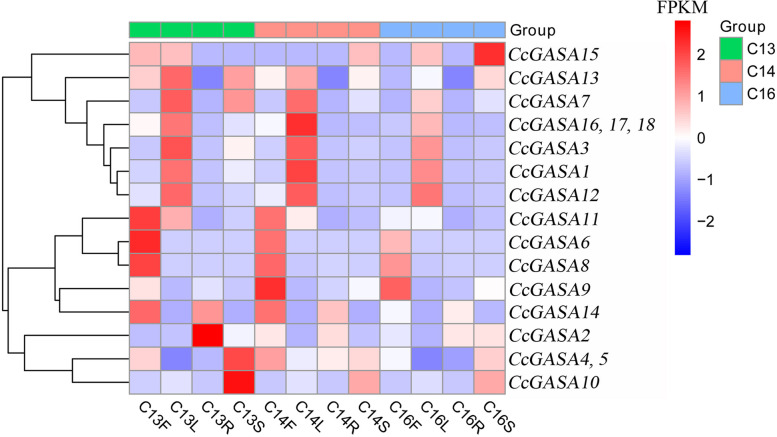
The basal expression of all CcGASA genes was analyzed in different organs of C. clementina (Fig. 7). The results showed that CcGASA1, CcGASA3, CcGASA7, CcGASA12, CcGASA16, CcGASA17 and CcGASA18 were preferably expressed in leaves. CcGASA6, CcGASA8, CcGASA9 and CcGASA11 were highly expressed in fruits. CcGASA2 was mainly expressed in roots. CcGASA4, CcGASA5 and CcGASA10 were most abundantly expressed in stems. CcGASA14 was predominantly expressed in fruits and, to a lesser extent, in roots. Similarly, CcGASA13 was most highly expressed in leaves and next highly expressed in stems.
Fig. 7.
Transcript analysis of CcGASAs in fruits (F), leaves (L), roots(R) and stems (S) of citrus. The C13, C14 and C15 represent three independent C. clementina plants. The expression abundance of genes is showed by the FPKM value, and from blue to red, the higher the expression level of genes
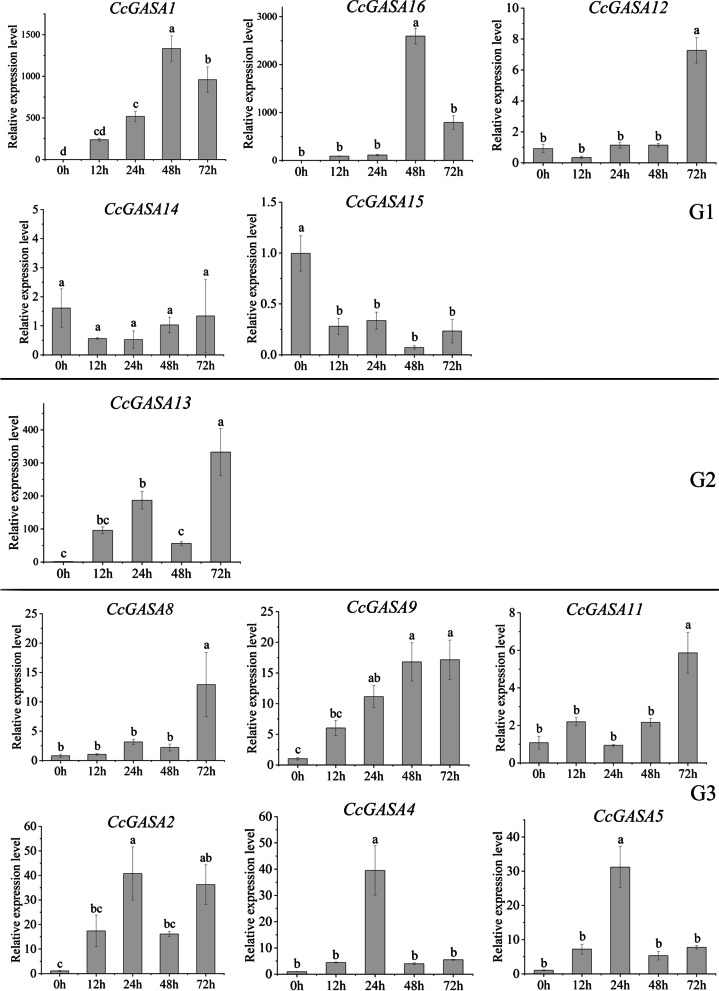
The inducibility of the CcGASA genes by Xcc was investigated. As shown in Supplementary Fig. S4, the detached C. clementina leaf explants developed water-soaked symptoms and white spots around the pinholes within 7d following inoculation with Xcc, the citrus canker pathogen. As expected, no such symptom was observed on control leaves. The qRT-PCR analysis showed that the CcGASA genes were mostly induced by Xcc (Fig. 8). Ten of the genes, CcGASA1, 2, 4, 5, 8, 9, 11, 12, 13, 16 were highly induced by Xcc, as exemplified by peak induction of more than 1000-fold for CcGASA1 and CcGASA16, and 300-fold for CcGASA13. Three clear patterns, gradual induction, later-stage induction and middle-stage induction, were observed. Gradual induction occurred to CcGASA1, 2, 9, 13. Later-stage induction occurred to 4 genes, CcGASA8, 11, 12, 16, for their expression was mostly peaked at 72 h. The expression of two genes, CcGASA4 and CcGASA5, was peaked at 24h, i.e., in the middle of the treatment. Two genes, CcGASA14 and CcGASA15, was shown to be moderately repressed or not induced by Xcc infection (Fig. 8).
Fig. 8.
Relative expression level of CcGASAs in citrus leaves inoculation with Xcc. Mean±standard error of three replicates is shown. Different lowercases letters (a-e) on the bars indicate statistically significant differences (P<0.05) based on Duncan’s LSD multiple range test
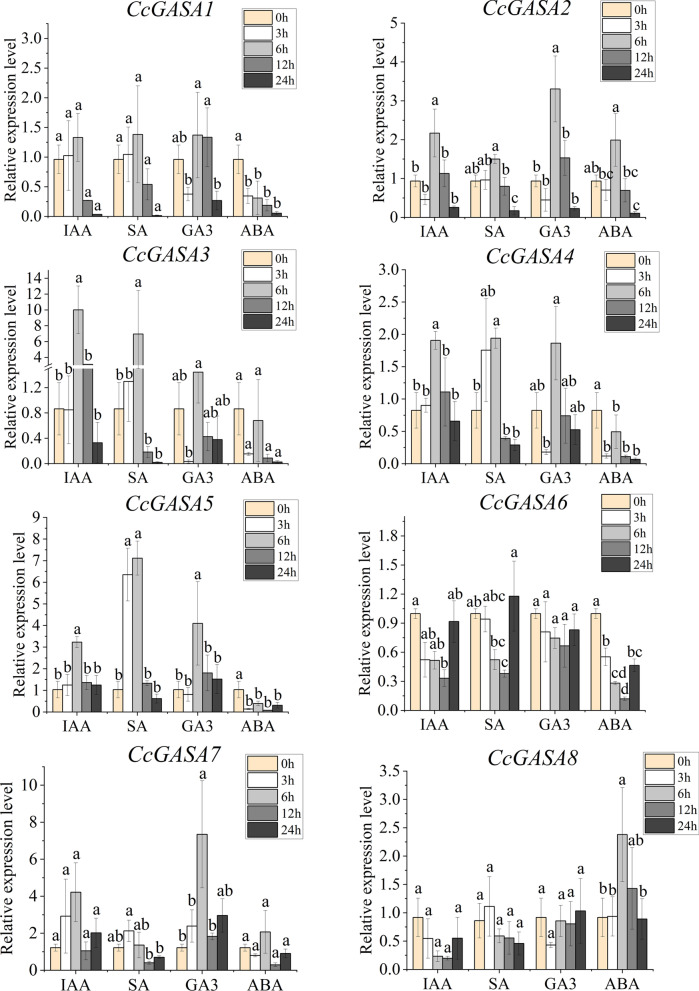
The expression patterns of CcGASA genes were found to be modulated by hormone treatments (Fig. 9, Supplementary Figs. S5 and S6). It was shown that CcGASA3, 4, 5, 10, 11, 12 and CcGASA14 were significantly up-regulated by IAA treatment. The highest up-regulation, about 12-fold, was shown by CcGASA3. The second highest induction, about 10-folds, was shown by CcGASA12. CcGASA11 was up-regulated by about 9-folds. In contrast, CcGASA13, 15, 16 and CcGASA17 were significantly down-regulated by IAA. Under SA treatment, the expression of CcGASA2, 3, 4, 5, 10, 11, 12 and CcGASA13 followed an inverted V-shape pattern whereas CcGASA16 and CcGASA17 were significantly down-regulated. Under GA3 treatment, the expression of CcGASA1, 2, 11 and CcGASA12 was increased at the beginning, peaked in the middle and subdued at the end of the treatment while the expression of CcGASA3, 4 and CcGASA5 were decreased at 3h, but reversed to increase at 6h and 12h, and then decreased again at the end of the treatment. Notably, 3 genes, CcGASA7, 10 and CcGASA15 were always up-regulated whereas 3 other genes, CcGAS13, 16 and CcGASA17 were always down-regulated under GA3 treatment. Under ABA treatment, CcGASA8, 10 and 14 were up-regulated, CcGASA2, 7 and 9 were initially increased and then decreased, whereas the other remaining genes were always down-regulated.
Fig. 9.
The expression abundance of CcGASA1-8 during IAA, SA, GA3 and ABA treatment. Data are mean ± SE of 3 qRT-PCR experiments and 3 biological replicates. Different lowercases letters (a-c) on the bars indicate statistically significant differences (P<0.05) based on Duncan’s LSD multiple range test
Evolution of citrus CcGASA genes
The possible tandem duplication events were inferred from analyzing the sequences of all CcGASA genes according to Holub et al. [21]. It was shown that a recent whole gene duplication event might be responsible for the cluster of two genes, CcGASA8 and CcGASA9, on scaffold 5. A more ancient whole gene duplication event should have generated another cluster on scaffold 9, CcGASA14 and CcGASA15. Interspersed segmental duplications were also detected across different scaffolds as shown in Fig. S7. They should be responsible for the generation of 4 pairs, CcGASA2-CcGASA5, CcGASA3-CcGASA12, CcGASA5-CcGASA11 and CcGASA6-CcGASA13 (Table 3). Ka/Ks ratios were calculated to measure the selection pressures between each of the 6 gene pairs (Table 3). The results showed that the Ka/Ks ratios were all less than 1, suggesting that the duplicates had experienced purifying selection.
Table 3.
Duplicate information in the Citrus Clementina GASA family
| Seq 1 | Seq 2 | Ka | Ks | Ka/Ks | Selection pressure | Gene duplications |
|---|---|---|---|---|---|---|
| Ciclev10002927m.g-CcGASA8 | Ciclev10002984m.g-CcGASA9 | 0.1947 | 0.4914 | 0.3962 | purifying | tandem |
| Ciclev10006931m.g-CcGASA14 | Ciclev10006668m.g-CcGASA15 | 0.0910 | 1.2137 | 0.0750 | purifying | tandem |
| Ciclev10033115m.g-CcGASA5 | Ciclev10022925m.g-CcGASA2 | 0.2781 | 1.7066 | 0.1630 | purifying | interspersed |
| Ciclev10013454m.g-CcGASA12 | Ciclev10023012m.g-CcGASA3 | 0.2924 | 2.7325 | 0.1070 | purifying | interspersed |
| Ciclev10012786m.g-CcGASA11 | Ciclev10033115m.g-CcGASA5 | 0.4009 | 2.5934 | 0.1546 | purifying | interspersed |
| Ciclev10029695m.g-CcGASA13 | Ciclev10002979m.g-CcGASA6 | 0.2638 | 1.1030 | 0.2392 | purifying | interspersed |
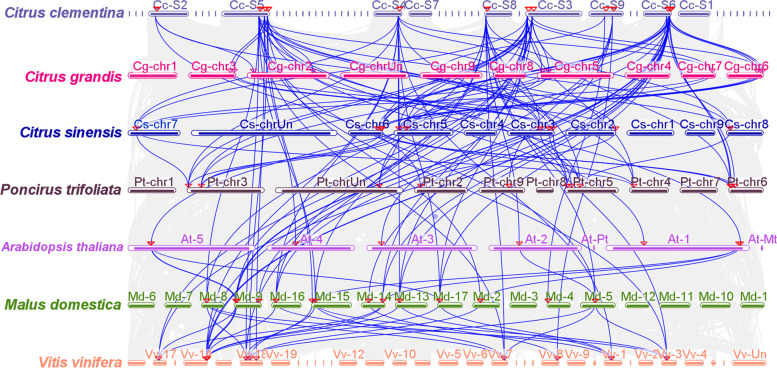
A synergy analysis of the orthologous GASA genes from C. clementina, C. sinensis, C. maxima, P. trifoliata, A. thaliana, M. domestica and V. vinifera genomes identified 96 collinearity events between C. clementina and the other six species. It was found that 6, 12, 11, 11, 11 and 10 CcGASA genes had synonyms in A. thaliana, C. maxima, C. sinensis, P. trifoliata, apple, and grape, respectively (Fig. 10 and Supplementary Table S7). In addition, we found that CcGASA8, 13, 12 were collinearity with the GASA genes of the other 6 species, indicating that these 3 genes play a very important role in the expansion of the GASA family.
Fig. 10.
The synteny analysis of GASA genes among citrus, Arabidopsis, apple and grape. According to the chromosomes of citrus, Arabidopsis, apple and grape, the relative positive positions are depicted. Gray lines in the background indicate the collinear blocks within citrus, Arabidopsis, apple and grape, while the blue lines highlight the syntenic GASA gene pairs
Protein interaction network of CcGASA proteins
The possible interactions between the 18 CcGASAs and other proteins were analyzed. As shown in Supplementary Fig. S8, only 8 CcGASAs were predicted to have 10 functional partners which were Cellulose synthase (XP_006439113.1), ATPase ASNA1 homolog (XP_006445186.1) and uncharacterized proteins, XP_006440347.1, XP_006440529.1, XP_006427852.1, XP_006443889.1, XP_006437427.1, XP_006437429.1, XP_006430408.1 and XP_006433793.1. The remaining 10 CcGASA proteins (CcGASA1, 2, 4, 5, 9, 14, 15, 16, 17, 18) were not successfully predicted to have a functional partner. CcGASA11 might have a potential relationship with CcGASA3 and CcGASA12 respectively. Both CcGASA12 and CcGASA3 were predicted to interact with XP_006445186.1 and XP_006445186.1 that participate in the peroxisome pathway and the proteasome pathway. The CcGASA6, CcGASA8 and CcGASA13 could respectively interact with XP_006443889.1. CcGASA10 was predicted to interact with 5 proteins, XP_006430408.1, XP_006440529.1, XP_006437427.1, XP_006437429.1 and XP_006439113.1. CcGASA7 was predicted to interact with XP_006427852.1.
Discussion
The low molecular-weight GASA proteins have been identified in different plants, and a large number of functional studies have shown their crucial roles in regulating plant growth, development and defense against pathogens [46]. However, the detailed regulation mechanisms through which GASAs operate have not yet been established. We previously found that a Citrus GASA gene was induced by Citrus tristiza virus infection [54]. To go a step deeper in understanding the role of the gene and, in a broad sense, other GASA genes in citrus, we set out to conduct a genome-wide bioinformatics study on the C. clementina GASA gene family.
In this study, 15 CcGASA genes were identified from the whole-genome sequence of C. clementina, including the gene previous reported by us [54]. Their genomic DNA and deduced protein sequences were compared with each other and with their homologous genes from other plant species, allowing the establishment of the inter-genomic and the intragenic phylogenetic trees. Detailed analysis on gene structures, cis-elements, chromosomal locations was performed. Primary sequences, physiochemical properties, subcellular localizations, 3D structures, transmembrane domains, motifs of the proteins were also analyzed. In addition, evolution of the CcGASA genes was investigated and possible duplication events were thus identified. Collinear relationships were revealed between the CcGASA genes and those from other citrus species, arabidopsis, apple and grape. The transcription factors possibly binding to CcGASA promoters, and the proteins possibly interacting with CcGASAs were also predicted and their associated functions were analyzed.
Our results indicated that CcGASA might be involved in plant defense processes. Firstly, these small hydrophilic and unstable proteins were mostly predicted to be in the extracellular space (Table 2), strongly indicating their association with plant cell walls that constitute the first line of defense in plants [44]. Secondly, their involvement in stress regulation was also suggested by the presence of many stress responsive cis-acting elements on their promoters (Fig. 5), such as ABRE involved in ABA-regulated osmotic stress [25], CGTCA-motif and TGACG-motif involved in the regulation in seed germination, senescence, and stress responses [10], ERE required for the expression of most ethylene-induced genes [23]. In addition, anaerobic (ARE and/or GC-motif), abiotic (MYB, MYC and/or STRE), low temperature (LTR), drought (MBS/DRE core), pathogens (TC-rich repeats/box S/W box) and wounding (WUN-motif/W box) induced/responsive elements were also very abundant. Furthermore, analysis for TFs interacting with these elements also indicated that many of the TFs were associated with stress responses (Supplementary Fig. S3). Moreover, the possible involvement of some CcGASA proteins in defense was also shown by their predicted interactions with defense-related proteins (Supplementary Fig. S8). Thirdly, the expression of the CcGASAs was induced by treatments of stress-related hormones and citrus canker bacterium Xcc (Figs. 8 and 9 and Supplementary Figs. S5 and S6).
The GASA genes have also been known to play important roles in plant growth and development [34]. In this regard, the nature of some cis-elements found on their promoters (Supplementary Table S5), such as tissue specific expression elements, AAGAA-motif (driving endosperm-specific negative expression), CAT-box (specifying meristem expression) and many light responsive elements, among others, implied that the CcGASA genes should indeed play important roles in the growth and development of citrus. Moreover, GA (GARE-motif/P-box/TATC-box), SA (TCA/TCA-element) and auxin (AuxRR-core/TGA-element) responsive elements were found in the promoter regions of most GASA genes. GO analysis also showed that some TFs identified by bioinformatics method in the study are associated with plant development (GO: 0032502), biological regulation (GO: 0050789 and GO: 0065007), responses to stimuli (GO: 0050896) (Fig. 6). That several flowering-related TFs were identified indicated that the citrus CcGASA genes or at least some of them should play a role in reproduction processes in Citrus, which is similar to Arabidopsis AtGASA4, a flowering promotion gene [38], and AtGASA5, a flowering delaying gene [58]. It should be noted that both Arabidopsis genes are actually involved in GA-mediated flowering.
The above notions that CcGASA proteins should play multiple roles in citrus were further supported by phylogenetic classification results. As shown in Fig. 4, the proteins were classified into 3 large groups and the classification was apparently related to their motif compositions (Fig. 4b) and primary sequences (Fig 2). It has long been known that structure and function of a protein is interrelated [20]. Thus, variations occurred to GASA protein structures should have allowed them to evolve different functions although they still share the highly conserved GASA domain. This could be exemplified by two G1d genes, CcGASA14 and 15 that have lost two motifs in their primary sequences (Fig. 4b). The possible consequence of such a large structural variation might be far-reaching, i.e., the pathogen inducibility they shared with other paralogs in the same G1 group was lost forever (Fig. 8). Structural differentiations among GASA proteins should also have resulted in functional differentiations. In this respect, the Xcc induction of G1a members, represented by CcGASA1 and CcGASA16, was 2 and 3 orders of magnitude higher than that of G2 and G3 members (Fig. 8), respectively, which strongly suggested that the G1a members have been specialized to cope with biotic stresses. However, we need to do more investigations to demonstrate this speculation.
The expansion of the GASA gene family was shown to be mainly through DNA duplication, either interspersed segmental duplication or tandem duplication [40]. In this study a total of 15 CcGASA genes were identified in C. clementina. Comparatively, there are about 15, 14, 26, and 37 GASA genes in Arabidopsis, grape, apple and soybean, respectively [1, 2, 14]. As can be seen, there are approximately two times more GASA genes in apple and soybean than in citrus, grape and Arabidopsis. Such a large discrepancy in GASA gene numbers between different species could be better explained by that only apple and soybean respectively experienced a recent whole genome duplication (WGD) while the others did not [41, 50]. Similarly, it was found that the EIN3/EIL genes were doubled from 4~5 in Arabidopsis, tobacco, tomato, rice, peach, mei and strawberry that did not have a recent WGD, to 10 in pear that shared a recent WGD with apple [11, 53, 55]. The Ka/Ks ratios between the 6 paralogous GASA gene pairs were all less than 1 (Table 3), indicating that they have been undergoing a purifying selection rather than a positive or neutral selection. Comparison of the two tandem duplication originated pairs, CcGASA14 / 15 and CcGASA8 / 9, with other CcGASA genes revealed that both CcGASA14 and 15 lost two motifs while CcGASA9 lost one motif (Fig. 4b), indicating that the three genes might either undergo degeneration or evolve new functions.
Conclusions
Eighteen CcGASA proteins from the C. clementina genome were identified and analyzed in this study, with emphasis on their possible roles in defense in citrus. Results from bioinformatics analysis showed that the members of the gene family have structurally and functionally diverged to different degrees and thus may play different roles in the growth and development of Citrus. Experimental evidence showed that the expression of the G1a subgroup members was highly sensitive to bacterial infection, strongly suggesting that they may play an important role in the responses of citrus to biotic stresses.
Supplementary Information
Additional file 1: Table S1. List of CcGASA proteins along-with their protein, CDS and genomic sequence. Table S2. Primers used for qRT-PCR analysis. Table S3. Secondary structures of CcGASA protein. Table S4. Protein, CDS and genomic sequences of Citrus sinensis, Citrus maxima, Poncirus trifoliata, Arabidopsis, apple and grape. Table S5. Analysis of cis-elements in the CcGASA genes. Table S6. Sequence information of predicted transcription factors (TF) that interacts with CcGASA genes. Table S7. The collinearity genelink of GASA family genes in citrus, Arabidopsis, apple and grape genomes.
Additional file 2: Figure S1. Predicted three- dimensional (3D) structures of CcGASA proteins. Ribbon representation of the structural model obtained by Phyre2, illustrating the mainly helical structure, characteristic of the GASA protein fold.
Additional file 3: Figure S2. Transmembrane topology analysis of CcGASA proteins. The Y axis represents probability, and the X axis represents the number of amino acid residues. The red peaks indicate the predicted transmembrane helices.
Additional file 4: Figure S3. The putative transcription factor regulatory network of the CcGASA genes. The transcriptional regulatory network was constructed with the PTRM tool and Cytoscape 3.8 software. The same color represents transcription factors of the same family. For example, pink represents ERF, green represents MYB, and blue represents MIKC_MADS.
Additional file 5: Figure S4. Citrus clementina leaves inoculated with Xcc.
Additional file 6: Figure S5. The expression abundance of CcGASA9-16 genes during IAA, SA, GA3 and ABA treatment. Data are mean ± SE of 3 qRT-PCR experiments and 3 biological replicates. Different lowercases letters (a-c) on the bars indicate statistically significant differences (P<0.05) based on Duncan’s LSD multiple range test.
Additional file 7: Figure S6. The expression abundance of CcGASA17 during IAA, SA, GA3 and ABA treatment. Data are mean ± SE of 3 qRT-PCR experiments and 3 biological replicates. Different lowercases letters (a-c) on the bars indicate statistically significant differences (P<0.05) based on Duncan’s LSD multiple range test.
Additional file 8: Figure S7. Chromosomal distribution and synteny analysis of citrus CcGASA gene family members. Syntenic regions and chromosomal regions are depicted in different colors.
Additional file 9: Figure S8. Putative protein-protein interaction network of CcGASA proteins in C.clementina. Colored nodes: query proteins and first shell of interactors. white nodes: second shell of interactors. Empty nodes: proteins of unknown 3D structure. Filled nodes: some 3D structure is known or predicted.
Acknowledgements
The authors are thankful to Dr. Yanyan Ma for her valuable help and advice in writing this article.
Abbreviations
- GASA
Gibberellic Acid Stimulated in Arabidopsis
- MCScanX
Multiple Collinearity Scan toolkit
- SDs
Segmental duplications
- Ka
Non-synonymous
- Ks
Synonymous
- WGD
Whole genome duplication
Authors’ contributions
TLW was responsible for most of the experiments and analyzed, wrote, and revised the original manuscript. YZ coordinated the team in writing the manuscript. MC participated in the qRT-PCR experiment for CcGASA gene expression analysis. BW, TW and BJ participated in the writing of the manuscript. GYZ participated in revising the manuscript and obtained funding. All authors have read and agreed to the published version of the manuscript.
Funding
This research was financially supported by the Guangdong Provincial Science and Technology Program [2018B020202009,2019B030316005], the National Natural Sciences Foundation of China (31601722, 32002016), and the Guangdong Provincial Special Fund for Modern Agriculture Industry Technology Innovation Special Teams[2021KJ108].
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Tianli Wu and Yun Zhong contributed equally to this work
References
- 1.Ahmad B, Yao J, Zhang S, Li X, Zhang X, Yadav V, et al. Genome-wide characterization and expression profiling of GASA genes during different stages of seed development in grapevine (Vitis vinifera L.) predict their involvement in seed development. Int J Mol Sci. 2020;21(3):1088. doi: 10.3390/ijms21031088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad MZ, Sana A, Jamil A, Nasir JA, Ahmed S, Hameed MU, et al. A genome-wide approach to the comprehensive analysis of GASA gene family in Glycine max. Plant Mol Biol. 2019;100(6):607–620. doi: 10.1007/s11103-019-00883-1. [DOI] [PubMed] [Google Scholar]
- 3.Alonso-Ramírez A, Rodríguez D, Reyes D, Jiménez JA, Nicolás G, López-Climent M, et al. Evidence for a role of gibberellins in salicylic acid-modulated early plant responses to abiotic stress in Arabidopsis seeds. Plant Physiol. 2009;150:1335–1344. doi: 10.1104/pp.109.139352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.An B, Wang Q, Zhang X, Zhang B, Luo H, He C. Comprehensive transcriptional and functional analyses of HbGASA genes reveal their roles in fungal pathogen resistance in Hevea brasiliensis. Tree Genet Genomes. 2018;14:1–13. [Google Scholar]
- 5.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aubert D, Chevillard M, Dorne AM, Arlaud G, Herzog M. Expression patterns of GASA genes in Arabidopsis thaliana: the GASA4 gene is up-regulated by gibberellins in meristematic regions. Plant Mol Biol. 1998;36:871–883. doi: 10.1023/a:1005938624418. [DOI] [PubMed] [Google Scholar]
- 7.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben-Nissan G, Lee JY, Borohov A, Weiss D. GIP, a Petunia hybrida GA-induced cysteine-rich protein: a possible role in shoot elongation and transition to flowering. Plant J. 2004;37:229–238. doi: 10.1046/j.1365-313x.2003.01950.x. [DOI] [PubMed] [Google Scholar]
- 9.Berrocal-lobo M, Segura A, Moreno M, López G, García-Olmedo F, Molina A. Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant Physiol. 2002;128(3):951–961. doi: 10.1104/pp.010685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao J, Li M, Chen J, Liu P, Li Z. Effects of MeJA on Arabidopsis metabolome under endogenous JA deficiency. Sci Rep. 2016;6:37674. doi: 10.1038/srep37674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao Y, Han Y, Meng D, Li D, Jin Q, Lin Y, Cai Y. EIN3/EILGenome-wide analysis suggests high level of microsynteny and purifying selection affect the evolution of family in Rosaceae. PeerJ. 2017;5:e3400. doi: 10.7717/peerj.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C, Chen H, Zhang Y, Thomas Hannah R, Frank Margaret H, He Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Cheng X, Wang S, Xu D, Liu X, Li X, Xiao W, et al. Identification and analysis of the GASR gene family in common wheat (Triticum aestivum L.) and characterization of TaGASR34, a gene associated with seed dormancy and germination. Front Genet. 2019;10:1–20. doi: 10.3389/fgene.2019.00980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan S, Zhang D, Zhang L, Gao C, Xin M, Tahir MM, et al. Comprehensive analysis of GASA family members in the Malus domestica genome: identification, characterization, and their expressions in response to apple flower induction. BMC Gen. 2017;18(1):1–19. doi: 10.1186/s12864-017-4213-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furukawa T, Sakaguchi N, Shimada H. Two OsGASR genes, rice GAST homologue genes that are abundant in proliferating tissues, show different expression patterns in developing panicles. Genes Genet Syst. 2006;81:171–180. doi: 10.1266/ggs.81.171. [DOI] [PubMed] [Google Scholar]
- 16.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.García AN, Ayub ND, Fox AR, Gómez MC, Diéguez MJ, Pagano EM, et al. Alfalfa snakin-1 prevents fungal colonization and probably coevolved with rhizobia. BMC Plant Biol. 2014;14:1–13. doi: 10.1186/s12870-014-0248-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40(D1):D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He H, Yang X, Xun H, Lou X, Li S, Zhang Z, et al. Over-expression of GmSN1 enhances virus resistance in Arabidopsis and soybean. Plant Cell Rep. 2017;36:1441–1455. doi: 10.1007/s00299-017-2167-3. [DOI] [PubMed] [Google Scholar]
- 20.Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC, et al. Mol Biol Evol. 2003;20(5):735–747. doi: 10.1093/molbev/msg088. [DOI] [PubMed] [Google Scholar]
- 21.Holub EB. The arms race is ancient history in Arabidopsis, the wild flower. Nat Rev Genet. 2001;2(7):516–527. doi: 10.1038/35080508. [DOI] [PubMed] [Google Scholar]
- 22.Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:585–587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itzhaki H, Maxson JM, Woodson WR. An ethylene-responsive enhancer element is involved in the senescence-related expression of the carnation glutathione-S-transferase (GST1) gene. Proc Natl Acad Sci S A. 1994;91(19):8925–8929. doi: 10.1073/pnas.91.19.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Letunic I, Khedkar S, Bork P. SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 2021;49(D1):D458–D460. doi: 10.1093/nar/gkaa937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JS, Mizoi J, Yoshida T, Fujita Y, Nakajima J, Ohori T, et al. An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A Gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol. 2011;52(12):2136–2146. doi: 10.1093/pcp/pcr143. [DOI] [PubMed] [Google Scholar]
- 26.Ko CB, Woo YM, Lee DJ, Lee MC, Kim CS. Enhanced tolerance to heatstress in transgenic plants expressing the GASA4 gene. Plant Physiol Biochem. 2007;45:722–728. doi: 10.1016/j.plaphy.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 27.Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol. 2011;696:291–303. doi: 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- 28.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(1):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li K, Bai X, Li Y, Cai H, Ji W, Tang LL, et al. GsGASA1 mediated root growth inhibition in response to chronic cold stress is marked by the accumulation of DELLAs. J Plant Physiol. 2011;168:2153–2160. doi: 10.1016/j.jplph.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 31.Mao Z, Zheng J, Wang Y, Chen G, Yang Y, Feng D, et al. The new CaSn gene belonging to the snaking family induces resistance against root-knot nematode infection in pepper. Phytoparasitica. 2011;39:151–164. [Google Scholar]
- 32.Matheyambath AC, Padmanabhan P, Paliyath G. Citrus fruits, encyclopedia of food and health. 2016. pp. 136–140. [Google Scholar]
- 33.Moyano-Canete E, Bellido ML, Garcia-Caparros N, Medina-Puche L, Amil-Ruiz F, Gonzalez-Reyes JA, et al. FaGAST2, a strawberry ripening-related gene, acts together with FaGAST1 to determine cell size of the fruit receptacle. Plant Cell Physiol. 2013;54:218–236. doi: 10.1093/pcp/pcs167. [DOI] [PubMed] [Google Scholar]
- 34.Nahirñak V, Almasia NI, Fernandez PV, Hopp HE, Estevez M, Carrari F, et al. Potato snakin-1 gene silencing affects cell division, primary metabolism, and cell wall composition. Plant Physiol. 2012;158:252–263. doi: 10.1104/pp.111.186544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nahirñak V, Rivarola M, Urreta MG, De Paniego N, Hopp HE, Almasia NI, et al. Genome-wide analysis of the Snakin/GASA gene family in Solanum Tuberosum cv. Kennebec. Am J Potato Res. 2016;93:172–188. [Google Scholar]
- 36.Qu J, Kang SG, Hah C, Jang J. Molecular and cellular characterization of GA-Stimulated TranscriptsGASA4 and GASA6 in Arabidopsis thaliana. Plant Sci. 2016;246:1–10. doi: 10.1016/j.plantsci.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 37.Raventos D, Meier C, Mattsson O, Jensen AB, Mundy J. Fusion genetic analysis of gibberellin signaling mutants. Plant J. 2000;22:427–438. doi: 10.1046/j.1365-313x.2000.00759.x. [DOI] [PubMed] [Google Scholar]
- 38.Roxrud I, Lid SE, Fletcher JC, Schmidt EDL, Opsahl-Sorteberg HG. GASA4, one of the 14-member Arabidopsis GASA family of small polypeptides, regulates flowering and seed development. Plant Cell Physiol. 2007;48(3):471–483. doi: 10.1093/pcp/pcm016. [DOI] [PubMed] [Google Scholar]
- 39.Rubinovich L, Weiss D. The Arabidopsis cysteine-rich protein GASA4 promotes GA responses and exhibits redox activity in bacteria and in planta. Plant J. 2010;64:1018–1027. doi: 10.1111/j.1365-313X.2010.04390.x. [DOI] [PubMed] [Google Scholar]
- 40.Samonte RV, Eichler EE. Segmental duplications and the evolution of the primate genome. Nat Rev Genet. 2002;3(1):65–72. doi: 10.1038/nrg705. [DOI] [PubMed] [Google Scholar]
- 41.Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463(7278):178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 42.Segura A, Moreno M, Madueño F, Molina A, García-olmedo F. Snakin-1, a peptide from potato that is active against plant pathogens. Mol Plant Microbe In. 1999;12:16–23. doi: 10.1094/MPMI.1999.12.1.16. [DOI] [PubMed] [Google Scholar]
- 43.Shi L, Gast RT, Gopalraj M, Olszewski NE. Characterization of a shoot-specific, GA3- and ABA-regulated gene from tomato. Plant J. 1992;2:153–159. [PubMed] [Google Scholar]
- 44.Showalter AM. Structure and function of plant cell wall proteins. Plant Cell. 1993;5(1):9–23. doi: 10.1105/tpc.5.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silverstein KAT, Moskal WA, Wu HC, Underwood BA, Graham MA, Town CD, et al. Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 2007;51:262–280. doi: 10.1111/j.1365-313X.2007.03136.x. [DOI] [PubMed] [Google Scholar]
- 46.Su T, Han M, Cao D, Xu M. Molecular and biological properties of snakins: the foremost cysteine-rich plant host defense peptides. J Fungi. 2020;6(4):220. doi: 10.3390/jof6040220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun S, Wang H, Yu H, Zhong C, Zhang X, Peng J, et al. GASA14 regulates leaf expansion and abiotic stress resistance by modulating reactive oxygen species accumulation. J Exp Bot. 2013;64:1637–1647. doi: 10.1093/jxb/ert021. [DOI] [PubMed] [Google Scholar]
- 48.Tam JP, Wang S, Wong KH, Tan WL. Antimicrobial peptides from plants. Pharmaceuticals. 2015;8:711–757. doi: 10.3390/ph8040711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Bel M, Diels T, Vancaester E, Kreft L, Botzki A, Van de Peer Y, et al. PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res. 2018;46(D1):D1190–D1196. doi: 10.1093/nar/gkx1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, et al. The genome of the domesticated apple (Malus × domestica Borkh.) Nat Genet. 2010;42(10):833–839. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- 51.Wang H, Wei T, Wang X, Zhang L, Yang M, Chen L, et al. Transcriptome analyses from mutant salvia miltiorrhiza reveals important roles for SmGASA4 during plant development. Int J Mol Sci. 2018;19:2088. doi: 10.3390/ijms19072088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang L, Wang Z, Xu Y, Joo S, Kim S, Xue Z, et al. OsGSR1 is involved in crosstalk between gibberellins and brassinosteroids in rice. Plant J. 2009;57:498–510. doi: 10.1111/j.1365-313X.2008.03707.x. [DOI] [PubMed] [Google Scholar]
- 53.Wu J, Wang Z, Shi Z, Zhang S, Ming R, Zhu S, Khan MA, Tao S, Korban SS, Wang H. The genome of the pear (Pyrus bretschneideri Rehd.) Genome Res. 2013;23:396–408. doi: 10.1101/gr.144311.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu T, Cheng C, Zhong Y, Lv Y, Ma Y, Zhong G. Molecular characterization of the gibberellin-stimulated transcript of GASA4 in Citrus. Plant Growth Regul. 2020;91:89–99. [Google Scholar]
- 55.Xu Q, Chen LL, Ruan X, Chen D, Zhu A, Chen C, et al. The draft genome of sweet orange (Citrus sinensis) Nat Genet. 2013;45(1):59–66. doi: 10.1038/ng.2472. [DOI] [PubMed] [Google Scholar]
- 56.Zimmermann R, Sakai H, Hochholdinger F. The gibberellic acid stimulated-like gene family in maizeand its role in lateral root development. Plant Physiol. 2010;152:356–365. doi: 10.1104/pp.109.149054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang S, Wang X. Expression pattern of GASA, downstream genes of DELLA, in Arabidopsis. Chin Sci Bull. 2008;53:3839–3846. [Google Scholar]
- 58.Zhang S, Yang C, Peng J, Sun S, Wang X. GASA5, a regulator of flowering time and stem growth in Arabidopsis thaliana. Plant Mol. Biol. 2009;69:745–759. doi: 10.1007/s11103-009-9452-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. List of CcGASA proteins along-with their protein, CDS and genomic sequence. Table S2. Primers used for qRT-PCR analysis. Table S3. Secondary structures of CcGASA protein. Table S4. Protein, CDS and genomic sequences of Citrus sinensis, Citrus maxima, Poncirus trifoliata, Arabidopsis, apple and grape. Table S5. Analysis of cis-elements in the CcGASA genes. Table S6. Sequence information of predicted transcription factors (TF) that interacts with CcGASA genes. Table S7. The collinearity genelink of GASA family genes in citrus, Arabidopsis, apple and grape genomes.
Additional file 2: Figure S1. Predicted three- dimensional (3D) structures of CcGASA proteins. Ribbon representation of the structural model obtained by Phyre2, illustrating the mainly helical structure, characteristic of the GASA protein fold.
Additional file 3: Figure S2. Transmembrane topology analysis of CcGASA proteins. The Y axis represents probability, and the X axis represents the number of amino acid residues. The red peaks indicate the predicted transmembrane helices.
Additional file 4: Figure S3. The putative transcription factor regulatory network of the CcGASA genes. The transcriptional regulatory network was constructed with the PTRM tool and Cytoscape 3.8 software. The same color represents transcription factors of the same family. For example, pink represents ERF, green represents MYB, and blue represents MIKC_MADS.
Additional file 5: Figure S4. Citrus clementina leaves inoculated with Xcc.
Additional file 6: Figure S5. The expression abundance of CcGASA9-16 genes during IAA, SA, GA3 and ABA treatment. Data are mean ± SE of 3 qRT-PCR experiments and 3 biological replicates. Different lowercases letters (a-c) on the bars indicate statistically significant differences (P<0.05) based on Duncan’s LSD multiple range test.
Additional file 7: Figure S6. The expression abundance of CcGASA17 during IAA, SA, GA3 and ABA treatment. Data are mean ± SE of 3 qRT-PCR experiments and 3 biological replicates. Different lowercases letters (a-c) on the bars indicate statistically significant differences (P<0.05) based on Duncan’s LSD multiple range test.
Additional file 8: Figure S7. Chromosomal distribution and synteny analysis of citrus CcGASA gene family members. Syntenic regions and chromosomal regions are depicted in different colors.
Additional file 9: Figure S8. Putative protein-protein interaction network of CcGASA proteins in C.clementina. Colored nodes: query proteins and first shell of interactors. white nodes: second shell of interactors. Empty nodes: proteins of unknown 3D structure. Filled nodes: some 3D structure is known or predicted.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.