Significance Statement
Autosomal dominant polycystic kidney disease (ADPKD) is caused by mutations in PKD1 and PKD2 (PKD1/2) in renal tubular epithelium. PKD1/2 somatic mutations were previously implicated in cyst formation, but studies of this second-hit model in ADPKD had significant technical limitations. Comprehensive analysis of renal cyst epithelium by whole-genome sequencing identified pathogenic inactivating somatic mutations of PKD1/2 in all 24 patients and in 93% of their 90 cysts. Short variant mutations occurred in 77% of cysts, and another 18% acquired chromosomal loss of heterozygosity encompassing PKD1/2, frequently at chromosomal fragile sites or in regions comprising chromosome microdeletion diseases/syndromes. These findings support a cellular recessive mechanism for renal cystogenesis in ADPKD caused by inactivating germline and somatic variants of PKD1/2.
Keywords: genetic renal disease, ADPKD, cystic kidney, end-stage renal disease, polycystic kidney disease, polycystic kidney, autosomal dominant, whole genome sequencing, epithelial cells, cysts
Visual Abstract

Abstract
Background
Autosomal dominant polycystic kidney disease (ADPKD) is a genetic disorder characterized by the development of multiple cysts in the kidneys. It is often caused by pathogenic mutations in PKD1 and PKD2 genes that encode polycystin proteins. Although the molecular mechanisms for cystogenesis are not established, concurrent inactivating germline and somatic mutations in PKD1 and PKD2 have been previously observed in renal tubular epithelium (RTE).
Methods
To further investigate the cellular recessive mechanism of cystogenesis in RTE, we conducted whole-genome DNA sequencing analysis to identify germline variants and somatic alterations in RTE of 90 unique kidney cysts obtained during nephrectomy from 24 unrelated participants.
Results
Kidney cysts were overall genomically stable, with low burdens of somatic short mutations or large-scale structural alterations. Pathogenic somatic “second hit” alterations disrupting PKD1 or PKD2 were identified in 93% of the cysts. Of these, 77% of cysts acquired short mutations in PKD1 or PKD2; specifically, 60% resulted in protein truncations (nonsense, frameshift, or splice site) and 17% caused non-truncating mutations (missense, in-frame insertions, or deletions). Another 18% of cysts acquired somatic chromosomal loss of heterozygosity (LOH) events encompassing PKD1 or PKD2 ranging from 2.6 to 81.3 Mb. 14% of these cysts harbored copy number neutral LOH events, while the other 3% had hemizygous chromosomal deletions. LOH events frequently occurred at chromosomal fragile sites, or in regions comprising chromosome microdeletion diseases/syndromes. Almost all somatic “second hit” alterations occurred at the same germline mutated PKD1/2 gene.
Conclusions
These findings further support a cellular recessive mechanism for cystogenesis in ADPKD primarily caused by inactivating germline and somatic variants of PKD1 or PKD2 genes in kidney cyst epithelium.
Autosomal dominant polycystic kidney disease (ADPKD) is the most common inherited kidney disease, with a prevalence of approximately 0.1%–0.25% of the general population, affecting approximately 12 million people worldwide.1–3 ADPKD is primarily caused by pathogenic variants of PKD1/2 genes. Germline pathogenic variants of PKD1/2 account for approximately 78% and approximately 13% of pedigrees, respectively.4–6
PKD1 and PKD2 encode polycystin 1 (PC1, also known as TRPP1) and polycystin 2 (PC2, also known as TRPP2) proteins, respectively, which localize to the cilia of cells of the renal tubular epithelium (RTE).7 Inactivating somatic alterations of PKD genes, which lead to loss-of-function of the corresponding polycystins, cause clonal expansion of RTE and cyst formation in ADPKD.8 However, previous studies reported a relatively low prevalence (20%–70%) of pathogenic somatic mutations of PKD1/2 due to methodologic limitations, including the inability to detect large structure alterations on chromosomes, and short mutations in PKD1 segmental duplicated regions (Exons 1–33).6,9–11
We previously identified inactivating somatic alterations of PKD1/2 in 90% of kidney cysts analyzed by a combination of whole-exome sequencing (WES) and long-range PCR–based next-generation sequencing (LR-PCR-NGS).12 This higher detection rate of somatic pathogenic mutations, compared with earlier reports, reflects the extensive and sensitive screening methods utilized.12–15 However, WES alone detected PKD1 somatic mutations in only 43% of cysts harboring PKD1 constitutional mutations, significantly below the PKD1 mutation detection rate (91%) by LR-PCR-NGS.12 All PKD1 WES-negative mutations were located in the duplicated region of PKD1, reflecting the low coverage by WES in this region, which was also reported by others.16 Whole-genome sequencing (WGS) is more sensitive than WES for detecting genomic alterations, especially within the duplicated region of PKD1, allowing the detection of both short variants (i.e., single nucleotide variants [SNVs], small insertions/deletions) and large-scale structural variants.16–18
Our previous finding that PKD1/2 somatic alterations were highly prevalent in RTE in ADPKD supported a primary role for a cellular recessive mechanism for renal cystogenesis primarily caused by inactivating germline and somatic variants of these genes.12 However, given the limited diagnostic sensitivity of WES, particularly in the duplicated region of PKD1, we employed WGS to further analyze the prevalence and characteristics of somatic alterations in 90 unique cysts obtained from 24 unrelated patients with ADPKD.
Methods
Study Subjects
This was a single-center study of somatic variants of RTE from individuals bearing PKD1 or PKD2 germline variants. Twenty-four unrelated patients with ADPKD enrolled in the ADPKD Data Repository of the Rogosin Institute (NIH clinicaltrials.gov website; http://clinicaltrials.gov identifier: NCT00792155) undergoing simultaneous native nephrectomy and living donor transplantation at the New York-Presbyterian Hospital/Weill Cornell Medicine12 participated in this study. Kidney volumes were obtained by manually contouring each kidney on the preoperative CT scan using itk-SNAP version 3.8.0 (www.itksnap.org).19 All subjects provided written, informed consent. The study was approved by the Institutional Review Board Committee at Weill Cornell Medicine (New York, NY).
Sample Processing
Cyst epithelial cells were isolated after nephrectomy and gross examination by pathologists as previously described.8,12 Cysts were selected on the basis of size (≥10 mm in diameter) and appearance. Hemorrhagic cysts were excluded. Typically, 5–20 cysts per kidney were processed. The cyst lumen was washed repeatedly with cold PBS and then incubated with PBS/EDTA to separate RTE from the basement membrane. RTE cells were collected by centrifugation and stored at −80°C. DNA was isolated from the cyst epithelial cells and peripheral blood leukocytes (PBLs) using the Gentra Puregene DNA extraction kit (Qiagen, Germantown, MD). Aliquots of cyst genomic DNAs (at least 300 ng per sample on the basis of Picogreen quantification) were sent to the Broad Institute Genomic Services (Cambridge, MA) for library preparation, sequencing, and read preprocessing.
Cyst and PBL DNAs from the same subjects were processed during the same batch of library preparation. The libraries were then sequenced on Illumina HiSeq instruments to produce 151-bp paired-end reads. Reads were processed for variant discovery following the Genome Analysis Toolkit (GATK, version 3.5) Best Practices Workflow to generate BAM files, which includes mapping to the human reference genome (GRCh38) using the Burrows–Wheeler Aligner (BWA) and subsequent processing using the Picard pipeline.20,21
WGS Constitutional Variant Detection
To detect constitutional pathogenic variants in PKD1 (NM_001009944.3) and PKD2 (NM_000297.4), we performed germline short variant discovery (SNVs and small insertions and deletions [indels]) using the joint calling process in GATK and germline structural variant discovery using Manta (version 1.6).22(preprint),23 Predicted functional consequences of the detected variants were annotated using the Ensembl Variant Effect Predictor (VEP, version 93).24 To filter for high-quality germline variants, those of low quality (FILTER other than PASS) or resided within previously reported low-complexity genomic regions were excluded.25 To confirm the absence of any hidden relatedness among the 24 donors, we ran the Peddy program on high-quality variants across all sequenced samples.26 To identify likely pathogenic variants in PKD1/2, those with >1% allele frequency in gnomAD (a database of >141,000 whole genomes or exomes, version 2) were further removed.27 Besides previously reported pathogenic variants in the Mayo Clinic PKD Mutation Database,28 novel protein-truncating variants, novel missense variants with Combined Annotation–Dependent Depletion (CADD) score ≥15, and novel in-frame insertion and deletion with PROVEAN score <−2.5 were deemed pathogenic.29,30
WGS Somatic Variant Detection
To detect somatic short variants (SNVs and small indels) present in cysts but not matched PBL DNAs, we followed the GATK Best Practices Workflow using Mutect2.31(preprint) Short alterations and affected PC1/2 domains were visualized using ProteinPaint.32 To discover large-scale somatic structural alterations, we used two complementary approaches—a split read–based method as implemented in Manta23 and a read coverage ratio and variant allele frequency (VAF)–based method as implemented in GATK and DNAcopy package in R (version 3.6.1).33 After similar variant annotation and filtration steps described above, high-quality somatic chromosomal amplifications, deletions, copy number neutral loss of heterozygosity (LOH) events, and translocations were identified. Subsequent data analysis and visualization were performed using R program and associated software packages.
Results
Clinical Cohort
Twenty-four unrelated patients with ADPKD (ten women and 14 men) had a median age of 30.5 years at ADPKD diagnosis and 52 years for nephrectomy (Supplemental Table 1). Patient sequencing data from our previous WES analysis of PKD1/2 somatic mutations were not included in this study.12 Kidney volumes were not normally distributed, as expected for ADPKD (Supplemental Table 1, Supplemental Figure 1). Each kidney contained innumerable cysts. A total of 459 intact cysts were collected, ranging from 1- to 20-cm-diameter with an average of ten cysts per kidney (Supplemental Table 1). Ninety cysts from 24 patients were selected for WGS, where sufficient DNA was available and qualified, regardless of the size of the cysts, including one previously analyzed cyst using WES that was negative for any somatic “second hit” alteration in PKD1/212 (Table 1, Supplemental Table 2). For quality control, six cysts with known PKD1/2 somatic alterations previously characterized by WES were tested by WGS12 (Supplemental Table 3). Moreover, 16 of the cysts evaluated by WGS from nine patients were repeatedly tested to further confirm their somatic alterations (Supplemental Table 4).
Table 1.
Summary of PKD1 and PKD2 germline and somatic variants identified by WGS in renal cyst epithelia (n=90)
| Patient/Gene | Cyst ID# | Germline Varianta | Exon/Intron | Somatic Alteration (VAF)b | Exon/Intron | Variant Type | Variant Designation | Cyst Semimajor Diameter, mm |
|---|---|---|---|---|---|---|---|---|
| FH9002294/PKD1 | CECDNA0125 | c.2806_2807insTC, p.Arg936Leufs*16 | Exon 11 | c.9097del p.Leu3033Trpfs*41 (0.46) | Exon 25 | Frameshift | Pathogenic | 10.0 |
| CECDNA0121 | c.11501T>C p.Leu3834Pro (CADD=25.9) (0.31) | Exon 41 | Missense | Likely pathogenic | 10.0 | |||
| CECDNA0120 | c.348_351del p.Asn116Lysfs*4 (0.46) | Exon 3 | Frameshift | Pathogenic | 10.0 | |||
| CECDNA0118 | c.10284_10300del p.Ile3429Alafs*36 (0.12) | Exon 33 | Frameshift | Pathogenic | 30.0 | |||
| CECDNA0117 | c.10237_10280dup p.Ser3428Profs*60 (0.43) | Exon 33 | Frameshift | Pathogenic | 50.0 | |||
| CECDNA0116 | c.4545del p.His1515Glnfs*19 (0.36) | Exon 15 | Frameshift | Pathogenic | 20.0 | |||
| CECDNA0115 | c.7619C>G p.Pro2540Arg (CADD=23.4) (0.33) | Exon 19 | Missense | Likely pathogenic | 20.0 | |||
| CECDNA0114 | None | NA | None | None | 30.0 | |||
| CECDNA0112 | c.1433_1436dup p.Pro480Glyfs*40 (0.45) | Exon 7 | Frameshift | Pathogenic | 15.0 | |||
| CECDNA0108 | c.12109_12121del p.Gly4037Profs*157 (0.43) | Exon 44 | Frameshift | Pathogenic | 40.0 | |||
| BJA001578/PKD1 | CECDNA0039 | c.3929_3930del, p.Asp1310Glyfs*120 | Exon 15 | c.1111G>T p.Glu371* (0.38) | Exon 5 | Nonsense | Pathogenic | 25.0 |
| CECDNA0010c | c.6364del p.Val2122Cysfs*3 (0.38) | Exon 5 | Frameshift | Pathogenic | 40.0 | |||
| c.9504C>G, p.Phe3168Leu (CADD=25.5) (0.17) | Exon 27 | Missense | Likely pathogenic | |||||
| CECDNA0007 | c.9296A>T p.Asp3099Val (CADD=25.1) (0.36) | Exon 26 | Missense | Likely pathogenic | 30.0 | |||
| CECDNA0006 | c.1578_1587del p.Ser527Alafs*28 (0.1) | Exon 7 | Frameshift | Pathogenic | 30.0 | |||
| MA9002292/PKD1 | CECDNA0093 | chr16:2132291–214257 1del, 10.28 kb | Promoter & Exon 1 | c.11758_11779dup p.Arg3927Profs*41 (0.55) | Exon 43 | Frameshift | Pathogenic | 30.0 |
| CECDNA0092 | c.11898_11914del p.Phe3966Leufs*5 (0.4) | Exon 43 | Frameshift | Pathogenic | 20.0 | |||
| CECDNA0091 | c.5722C>T p.Gln1908* (0.25) | Exon 15 | Nonsense | Pathogenic | 20.0 | |||
| CECDNA0089 | c.398T>C p.Leu133Pro (CADD=21.6) (0.25) | Exon 4 | Missense | Likely pathogenic | 50.0 | |||
| CECDNA0088 | c.935_936dup p.Glu313Profs*22 (0.18) | Exon 5 | Frameshift | Pathogenic | 50.0 | |||
| CECDNA0087 | chr16:35250–15206250 neutral LOH (0.99) | Promoter & exon 1 | LOH | Pathogenic | 30.0 | |||
| CECDNA0083 | c.5016_5036del p.Arg1672_Gly1678del (PROVEAN=−9.12) (0.49) | Exon 15 | In-frame deletion | Likely pathogenicd | 20.0 | |||
| PJ9002394/PKD1 | CECDNA0077 | c.11766G>A, p.Trp3922* | Exon 43 | chr16:34250–21861736 neutral LOH (1.00) | Exon 43 | LOH | Pathogenic | 150.0 |
| CECDNA0073 | chr16:34250–26370736 neutral LOH (0.92) | Exon 43 | LOH | Pathogenic | 20.0 | |||
| CECDNA0072 | c.11842_11875del p.Thr3948Leufs*25 (0.58) | Exon 43 | Frameshift | Pathogenic | 20.0 | |||
| CECDNA0068 | c.7288C>T p.Arg2430* (0.44) | Exon 18 | Nonsense | Pathogenic | 20.0 | |||
| CECDNA0066 | chr16:34250–22576736 neutral LOH (0.92) | Exon 43 | LOH | Pathogenic | 20.0 | |||
| PM9002387/PKD1 | CECDNA0263 | c.11293_11378dup, p.Thr3794Profs*61 | Exon 40 | c.81del p.Arg28Alafs*45 (0.45) | Exon 1 | Frameshift | Pathogenic | 20.0 |
| CECDNA0249 | c.906_912dup p.Trp305Hisfs*68 (0.54) | Exon 5 | Frameshift | Pathogenic | 60.0 | |||
| CECDNA0246 | chr16:51250–36715408 neutral LOH (0.84) | Exon 40 | LOH | Pathogenic | 40.0 | |||
| DA9002295/PKD1 | CECDNA0311 | c.6736C>T, p.Gln2246* | Exon 15 | c.755dup p.Pro253Alafs*8 (0.48) | Exon 5 | Frameshift | Pathogenic | 30.0 |
| CECDNA0302 | chr16:11250–36793408 neutral LOH (0.82) | Exon 15 | LOH | Pathogenic | 50.0 | |||
| CECDNA0298 | c.6979_6980del p.Arg2327Glyfs*92 (0.47) | Exon 16 | Frameshift | Pathogenic | 40.0 | |||
| LES002455/PKD1 | CECDNA0147 | c.7749_7770del, p.Leu2584Serfs*29 | Exon 20 | c.11467 C>T p.Arg3753Gln (CADD=28.1) (0.27) | Exon 41 | Missense | Likely pathogenic | 20.0 |
| CECDNA0146 | c.1529_1541del,p Arg510Profs*44 (0.43) | Exon 7 | Frameshift | Pathogenic | 40.0 | |||
| CECDNA0145 | c.3219del p.Glu1073Aspfs*31 (0.45) | Exon 14 | Frameshift | Pathogenic | 15.0 | |||
| VM9002450/PKD1 | CECDNA0197 | c.9325_9326insGCGCC, p.Ile3109Serfs*209 | Exon 26 | c.4177C>T p.Gln1393* (0.48) | Exon 15 | Nonsense | Pathogenic | 30.0 |
| CECDNA0188 | chr16:10250–15265250 neutral LOH (0.87) | Exon 26 | LOH | Pathogenic | 35.0 | |||
| CECDNA0187 | c.3948C>A p.Tyr1316* (0.41) | Exon 15 | Nonsense | Pathogenic | 35.0 | |||
| FTA002290/PKD1 | CECDNA0130 | c.6493C>T, p.Gln2165* | Exon 15 | chr16:17250–2605250 deletion het (1.00) | Exon 15 | LOH | Pathogenic | 40.0 |
| CECDNA0129 | chr16:17250–2605250 neutral LOH (0.99) | Exon 15 | LOH | Pathogenic | 35.0 | |||
| MJ9000363/PKD1 | CECDNA0333 | c.12263_12264del, p.Val4088Glyfs*68 | Exon 45 | None | NA | None | None | 30.0 |
| CECDNA0328 | None | NA | None | None | 30.0 | |||
| FC9002445/PKD1 | CECDNA0270 | c.7861G>T, p.Glu2621* | Exon 20 | c.403_404del p.Trp135Alafs*43 (0.4) | Exon 4 | Frameshift | Pathogenic | 50.0 |
| JS9001595/PKD1 | CECDNA0127 | c.6743dup, p.Asn2248Lysfs*14 | Exon 15 | chr16:17250–16419250 neutral LOH (0.99) | Exon 15 | LOH | Pathogenic | 70.0 |
| RP9001591/PKD1 | CECDNA0019 | c.10552G>T, p.Glu3518* | Exon 35 | chr16:17250–32002736 deletion het (0.81) | Exon 35 | LOH | Pathogenic | 50.0 |
| CMJ001593/PKD1 | CECDNA0142 | c.10405dup, p.Asp3469Glyfs*2 | Exon 34 | c.1889del p.Pro630Argfs*155 (0.38) | Exon 10 | Frameshift | Pathogenic | 30.0 |
| CECDNA0136 | c.10264del p.Asp3422Thrfs*51 (0.37) | Exon 33 | Frameshift | Pathogenic | 80.0 | |||
| CECDNA0135 | c.4802dup p.Arg1602Profs*13 (0.42) | Exon 15 | Frameshift | Pathogenic | 40.0 | |||
| CECDNA0133 | chr16:17250–36715408 neutral LOH (0.74) | Exon 34 | LOH | Pathogenic | 100.0 | |||
| LA9001480/PKD1 | CECDNA0018 | chr16:2105263–2122734 del, 17.471 kb | Exon 2–21del | c.11225_11245del p.Pro3742_Arg3748del (PROVEAN=−20.96) (0.46) | Exon 39 | In-frame deletion | Likely pathogenicd | 35.0 |
| LA9002451/PKD1 | CECDNA0168 | c.5878C>T, p.Gln1960* | Exon 15 | c.402_406dup p.Leu136Argfs*156 (0.45) | Exon 4 | Frameshift | Pathogenic | 80.0 |
| GCA002396/PKD1 | CECDNA0065 | c.8792–2A>T | Intron 23 | c.417G>A p.Trp139* (0.38) | Exon 4 | Nonsense | Pathogenic | Not measured |
| CECDNA0064 | c.11818_11819dup p.Leu3941Cysfs*5 (0.41) | Exon 43 | Frameshift | Pathogenic | Not measured | |||
| CECDNA0062 | c.407T>C p.Leu136Pro (CADD=22.4) (0.52) | Exon 4 | Missense | Likely pathogenic | Not measured | |||
| CECDNA0060 | c.862del p.Gln288Serfs*2 (0.42) | Exon 5 | Frameshift | Pathogenic | 20.0 | |||
| CECDNA0058 | c.11830_11838dup p.Leu3944_Ala3946dup (PROVEAN=−9.12) (0.13) | Exon 43 | In-frame duplication | Likely pathogenicd | 25.0 | |||
| CECDNA0056 | c.417G>A p.Trp139* (0.3) | Exon 4 | Nonsense | Pathogenic | Not measured | |||
| CECDNA0055 | c.4796dup p.Tyr1599* (0.48) | Exon 15 | Nonsense | Pathogenic | 12.0 | |||
| CECDNA0052 | c.7915C>T p.Arg2639* (0.38) | Exon 21 | Nonsense | Pathogenic | 20.0 | |||
| CECDNA0051 | chr16:2133655–2137225del (3.57 kb del) (0.25) | Promoter & Exon 1 | Structural variations | Pathogenic | 30.0 | |||
| CECDNA0049 | c.2889_2908dup p.Asn970Argfs*13 (0.61) | Exon 12 | Frameshift | Pathogenic | 90.0 | |||
| DR9001565/PKD1 | CECDNA0047 | c.9504C>G, p.Phe3168Leu (CADD=25.5) | Exon 27 | c.1523G>A p.Cys508Tyr (CADD=24.6) (0.41) | Exon 7 | Missense | Likely pathogenic | 20.0 |
| CECDNA0045 | c.1048del p.Val350Cysfs*115 (0.37) | Exon 5 | Frameshift | Pathogenic | 20.0 | |||
| CECDNA0044 | c.11538–2A>C (0.31) | Intron 41 | RNA splice | Pathogenic | 20.0 | |||
| CECDNA0043 | chr16:21250–15209250 neutral LOH (0.85) | Exon 27 | LOH | Likely pathogenic | 20.0 | |||
| CECDNA0016 | c.9241T>C p.Cys3081Arg (CADD=23.7) (0.68) | Exon 26 | Missense | Likely pathogenic | 30.0 | |||
| CECDNA0015 | None | NA | None | None | 80.0 | |||
| CECDNA0013 | None | NA | None | None | 35.0 | |||
| CECDNA0012c |
PKD1: c.4916dup, p.Gly1640Argfs*18 (0.47); PKD2: c.203dup, p.Ala69Glyfs*23 (0.15) |
Exon 15 Exon 1 |
PKD1 frameshift PKD2 frameshift |
Pathogenic Pathogenic |
20.0 | |||
| CECDNA0011 | c.11853dup p.Arg3952Thrfs*9 (0.37) | Exon 43 | Frameshift | Pathogenic | 25.0 | |||
| SL9001502/PKD1 | CECDNA0032 | c.8927A>G, p.Tyr2976Cys (CADD=27.7) | Exon 24 | chr16:10250–16623250 neutral LOH (0.98) | Exon 24 | LOH | Likely pathogenic | 10.0 |
| CECDNA0031 | None | NA | None | None | 20.0 | |||
| CECDNA0030 | c.348_352del p.Asn116Lysfs*2 (0.54) | Exon 4 | Frameshift | Pathogenic | 15.0 | |||
| CECDNA0028 | c.5017_5033del p.Gly1673Glnfs*92 (0.49) | Exon 15 | Frameshift | Pathogenic | 20.0 | |||
| CECDNA0026 | c.5383G>T p.Glu1795* (0.48) | Exon 15 | Nonsense | Pathogenic | Not measured | |||
| CECDNA0025 | c.10405 + 2T>A (0.4) | Intron 33 | RNA splice | Pathogenic | Not measured | |||
| CECDNA0024 | c.348_352del p.Asn116Lysfs*2 (0.03) | Exon 3 | Frameshift | Pathogenic | Not measured | |||
| VKZ002452/PKD1 | CECDNA0164 | c.11015G>A, p.Arg3672Gln (CADD=17.76) | Exon 37 | c.10952G>A p.Gly3651Asp (CADD=25.6) (0.43) | Exon 37 | Missense | Likely pathogenic | 55.0 |
| CECDNA0157 | c.4336_4343del p.Thr1446Glnfs*8 (0.49) | Exon 15 | Frameshift | Pathogenic | 60.0 | |||
| CECDNA0155 | chr16:21250–9107250 neutral LOH (0.06) | Exon 37 | LOH | VUSe | 20.0 | |||
| KJ9001580/PKD1 | CECDNA0232 | c.8318C>T, p.Pro2773Leu (CADD=26.7); c.8509G>T, p.Gly2837Cys (CADD=28.6) |
Exon 23 | c.9046C>T p.Gln3016* (0.08) | Exon 25 | Nonsense | Pathogenic | 30.0 |
| SG9000393/PKD2 | CECDNA0200 | c.357_368delCCCG GGCAGCCGGins. AGGACGTAGAG TGGAGATGGAC p.Pro120 Glyfs*96 | Exon 1 | chr4:840 79631–92 710631 deletion het (0.71)) | Exon 1 | LOH | Pathogenic | >100 |
| BH9002280/PKD2 | CECDNA0106 | c.923del p.Phe308 Serfs*9 | Exon 4 | c.2684del p.Gly895Valfs*14 (0.18) | Exon 15 | Frameshift | Pathogenic | 50.0 |
| CECDNA0105 | chr4:50386350–131727631 neutral LOH (0.68) | Exon 4 | LOH | Pathogenic | 50.0 | |||
| CECDNA0102 | c.165_166del p.Glu55Aspfs*36 (0.48) | Exon 1 | Frameshift | Pathogenic | 60.0 | |||
| CECDNA0094* | c.1484del p.Arg496Alafs*18 (0.18) | Exon 6 | Frameshift | Pathogenic | 65.0 | |||
| CECDNA0001 | c.1843G>A p.Ala615Thr (CADD=26.7) (0.3) | Exon 8 | Missense | Likely pathogenic | 40.0 | |||
| GE9002273/PKD2 | CECDNA0288 | c.1716 + 2T>A | Intron 7 | c.1135_1136dup p.Trp380Thrfs*73 (0.27) | Exon 5 | Frameshift | Pathogenic | 70.0 |
| CECDNA0284 | c.964C>T p.Arg322Trp (CADD=28.1) (0.47) | Exon 4 | Missense | Likely pathogenic | 45.0 | |||
| CECDNA0282 | c.1135_1136dup p.Trp380Thrfs*73 (0.39) | Exon 5 | Frameshift | Pathogenic | 30.0 |
The germline variants were confirmed in all 90 cysts, whereas pathogenic somatic alterations were found in 84 cysts. The remaining six cysts, in which no pathogenic variants were detected, were recorded with “none”. Of the 90 cysts, one was enrolled in the previous WES study where no somatic second hit was found12 and marked with “*”. NA, not applicable.
Missense variants were predicted to be likely pathogenic if CADD value is >15 (http://cadd.gs.washington.edu/).
For LOH events, VAF represents the VAF of the somatic pathogenic variant.
CECDNA0010 from patient BJA001578 carried two somatic mutations in PKD1, and cyst CECDNA0012 from patient DR9001565 acquired somatic mutations in both PKD1 and PKD2.
The effect of amino acid introduction or removal caused by DNA in-frame deletion/insertion is evaluated by PROVEAN (http://provean.jcvi.org/index.php). The in-frame deletion/insertion was predicted to be “deleterious” (likely pathogenic) when the score is <−2.5.
VUS (variant of unknown significance) is defined as a variation in a genetic sequence for which the association with disease risk is unclear (National Cancer Institute).
Cyst samples were sequenced to an average of 83±2 SEM fold coverage (median of 79, IQR 18), whereas matched blood DNAs were sequenced to an average of 45±1 SEM (median of 45, IQR 9). For each sample, a minimum of 99.7% of the reads successfully mapped to the human reference genome. We also observed overall uniform sequencing coverage of PKD1, including segmental duplicated regions, and PKD2. All PKD1 and PKD2 exons reached a minimum coverage of 41- and 67-fold in cyst samples, respectively (Figure 1).
Figure 1.
WGS shows a deep and consistent coverage of PKD1 and PKD2 exons. The horizontal axis represents genomic coordinates. The vertical axis indicates median WGS read coverage across 90 cysts for the PKD1 (top panel) and PKD2 (bottom panel) regions. Exonic regions are colored in purple whereas intronic regions are in green. Segmental duplicated regions of PKD1 (exons 1–33) and previously known low-complexity genomic regions are marked. WGS reached consistent coverage of all exons, including segmentally duplicated regions of PKD1.
Confirmation of Germline Pathogenic PKD1 and PKD2 Variants Using WGS
Using WGS, pathogenic germline heterozygous PKD1/2 variants were detected in all 24 subjects: 87.5% (21 of 24) in PKD1 and 12.5% (three of 24) in PKD2 (Table 1). Twenty patients (83.3%) carried germline protein-truncating variants in PKD1/2; the other four patients (16.7%) had nontruncating, missense variants (Table 1). All constitutional short variants (21 of 24) were consistent with previous genotyping results of the same patients by LR-PCR-NGS/WES.12 Notably, large exonic deletions or duplications were detected in three patients (MA9002292, LA9001480, and PM9002387), in which the germline pathogenic variants were previously not detected by LR-PCR-NGS. In patient MA9002292, a 10.28-kb deletion extended from the promoter to exon 1 of PKD1, leading to null expression of that allele. Patient LA9001480 had a 17.47-kb deletion of PKD1 exons 2 through 21, which encode the extracellular domain (leucine-rich repeats to REJ-module domains) of PC1. Patient PM9002387 had an 86-bp duplication of exon 40 of PKD1 (c.11293_11378dup), causing a frameshift in the Polycystin Domain (PCD) (Table 1). Additionally, in patient VKZ002452, we detected a pathogenic missense variant in PKD1 (c.11015G>A; p.Arg3672Gln; CADD score=17.8), located 2 bp upstream of the canonical splice donor site of exon 37 that potentially interferes with RNA splicing.34 Patient KJ9001580 had two heterozygous, pathogenic PKD1 missense variants, p.Pro2773Leu and p.Gly2837Cys; both variants were predicted to be pathogenic (CADD scores of 26.7 and 28.6, respectively) and neither was documented in the Mayo Clinic PKD and gnomAD databases.27,28 Although patients with the rare germline biallelic inheritance of hypomorphic PKD1 alleles tend to develop severe disease appearing in utero or during infancy,5,35–38 we could not computationally determine from the WGS data whether these two missense variants were on the same allele (Table 1).
Detection of Somatic Short Mutations in PKD1 and PKD2
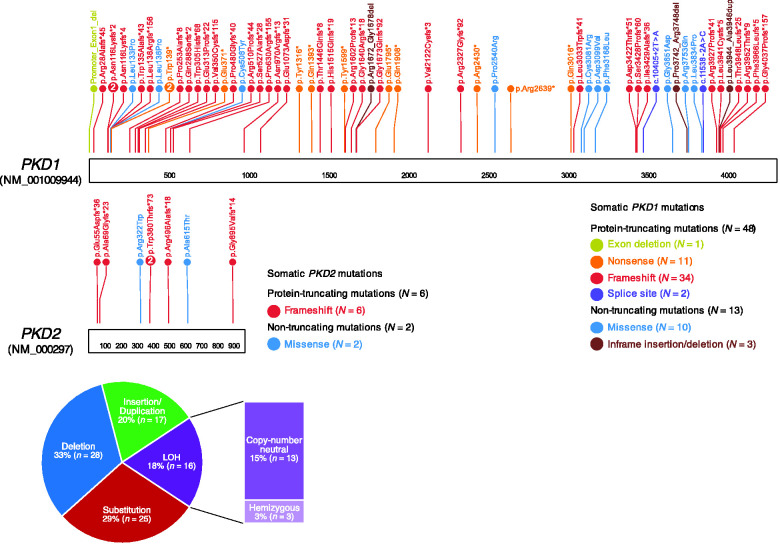
Pathogenic somatic alterations in PKD1/2, either short mutations or large structural alterations (e.g., chromosomal copy number alterations), were newly detected in 93% of cysts (84 out of 90 cysts), leading to biallelic inactivation (Table 1). Across the genome, cysts acquired an average of 1033±118 SEM (median of 738 and IQR of 696) somatic short mutations (Supplemental Table 4). When looking specifically at somatic short mutations that result in protein sequence changes, ADPKD cysts harbored an average of 10±3 SEM (median of 5 and IQR of 5) mutations (Supplemental Table 4). PKD1 and PKD2 had predicted pathogenic short mutations observed in 68% and 9% of cysts (61 and 8 out of 90), respectively. The somatic mutations were distributed throughout the entire length of the PKD genes, without a hotspot region (Figure 2). Of these mutations, 28 were deletion mutations ranging from a single nucleotide to 3.57 kb; 17 were duplication/insertion mutations; and 25 were SNVs (Table 1, Figure 2). Seventy-eight percent (54 out of 69) of these mutations resulted in protein truncation—exon deletion (1.4%), nonsense (15.9%), frameshift (58%), and splice site (2.9%) mutations. The remaining 22% (15 out of 69) were missense (17%) or in-frame insertions or deletions (4%), of which the protein function is partially affected (Table 2). We did not observe significant correlation between the VAFs of the somatic “second hit” mutations and cyst size (Pearson’s correlation P=0.15) (Supplemental Table 2).
Figure 2.
Somatic alterations of PKD1 and PKD2 were detected in 90 cysts by WGS. The somatic short mutations were distributed throughout the entire length of the PKD genes, without a hotspot region (top panel). Taking somatic structural alterations into account, 33% were deletion mutations, 20% were insertions/duplications, 29% were substitutions (SNVs), and the remaining 18% were LOH—either copy number neutral (15%) or hemizygous deletions (3%) (bottom panel).
Table 2.
Summary of somatic alterations in PKD1 and PKD2 in 24 patients with ADPKD by WGS
| Patient ID | Total WGS’ed Cysts, N | Short Somatic Alterations, N | Structural Alteration LOH, N | Total Somatic Second Hit | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Exon Deletion | Nonsense | Frameshift | Splice Site | Missense | In-Frame Deletion/Insertion | N | % | |||
| FH9002294 | 10 | 0 | 0 | 7 | 0 | 2 | 0 | 0 | 9 | 90 |
| BJA001578a | 4 | 0 | 1 | 2 | 0 | 2 | 0 | 0 | 4 | 100 |
| MA9002292 | 7 | 0 | 1 | 3 | 0 | 1 | 1 | 1 | 7 | 100 |
| PJ9002394 | 5 | 0 | 1 | 1 | 0 | 0 | 0 | 3 | 5 | 100 |
| PM9002387 | 3 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 3 | 100 |
| DA9002295 | 3 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 3 | 100 |
| LES002455 | 3 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 3 | 100 |
| VM9002450 | 3 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 3 | 100 |
| FTA002290 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 100 |
| MJ9000363 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| FC9002445 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 100 |
| JS9001595a | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 100 |
| RP9001591a | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 100 |
| CMJ001593a | 4 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 4 | 100 |
| LA9001480 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 100 |
| LA9002451 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 100 |
| GCA002396 | 10 | 1 | 4 | 3 | 0 | 1 | 1 | 0 | 10 | 100 |
| DR9001565a | 9 | 0 | 0 | 4 | 1 | 2 | 0 | 1 | 8 | 80 |
| SL9001502 | 7 | 0 | 1 | 3 | 1 | 0 | 0 | 1 | 6 | 86 |
| SG9000393 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 100 |
| VKZ002452 | 3 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 67 |
| KJ9001580 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 100 |
| BH9002280a | 5 | 0 | 0 | 3 | 0 | 1 | 0 | 1 | 5 | 100 |
| GE9002273 | 3 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 3 | 100 |
| Total | 90 | 1 | 11 | 40 | 2 | 12 | 3 | 16 | 84 | 93 |
Somatic alterations in PKD1 and PKD2 were analyzed by WGS in 90 cysts from 24 ADPKD participants and pathogenic somatic variants were detected. The number of newly identified variants in the cysts was listed by the participant without confirmatory data.
Notably, among the participants, six were enrolled in the previous WES study as indicated (BJA001578, JS9001595, RP9001591; CMJ001593, DR9001565, BH9002280)12 but their cysts listed here were not analyzed before.
To assess the reproducibility of WGS, replicate DNA aliquots for 16 cysts from nine patients were sequenced in duplicate. We found identical somatic second hit alterations in these replicate samples as in the original DNA aliquots, with similar VAFs (Supplemental Table 3). To further confirm the accuracy of WGS, six cysts in the previously published WES/LR-PCR-NGS study were also analyzed by WGS and revealed consistent somatic second hit alterations (Supplemental Table 5).12
Detection of Somatic Structural Alterations Disrupting PKD1 and PKD2
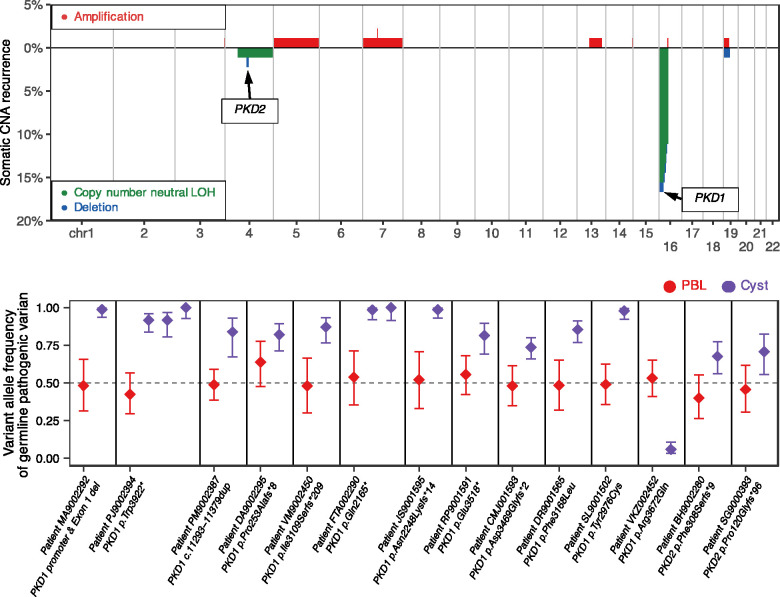
In addition to short mutations, we also detected large somatic structural alterations such as chromosomal copy number alterations in cysts, exhibiting homozygosity of the mutant PKD1 or PKD2 alleles.39 On average, 0.39%±0.14% SEM of the cysts’ genomes, or 12.1±4.3 Mb, were affected by somatic structural alterations. Interestingly, we found recurrent somatic LOH events disrupting chromosomes 16p (PKD1 locus) and 4q (PKD2 locus) in 15.5% (14 of 90 cysts) and 2.2% (two of 90) of cysts, respectively (Figure 3, Supplemental Table 6). All LOH events were observed in cysts without somatic short mutations in PKD1 or PKD2 (Fisher’s exact test P<2.2×10−16).
Figure 3.
Large-scale somatic structural alterations disrupt PKD1 and PKD2 in 90 cysts. Top panel: genome-wide patterns of large-scale somatic alterations in 90 cysts. The horizontal axis represents the genome. The vertical axis indicates the frequency of somatic copy number neutral LOH events (green) or deletions (blue), and chromosomal amplifications (red). Recurrently affected somatic regions where PKD1 and PKD2 genes reside are indicated. Bottom panel: shift in VAFs of germline pathogenic PKD1 and PKD2 variants in somatic LOH regions of cysts. In 17 cysts (purple) with somatic LOH events at PKD1 and PKD2 loci, the VAFs of the germline pathogenic variants shifted away from heterozygosity observed in matched PBLs (red). In all except one cyst, the LOH events led to increased VAFs of the pathogenic allele, suggesting biallelic inactivation of PKD1 or PKD2.
Specifically, we found two types of somatic LOH events at PKD1 and PKD2 loci: 13 events (14.4% of cysts) were copy-number neutral LOHs, where the deletion of one allele was followed by compensatory duplication of the remaining allele (Supplemental Figures 2 and 3, Supplemental Table 6), and the other three events (3.3% of cysts) were hemizygous deletions without compensatory duplication39 (Supplemental Figure 4). By assessing the VAFs of the constitutional pathogenic PKD1 or PKD2 variants in these 16 cysts, we found that all LOH events, except one (15 of 16, binomial P<2.7×10−4), disrupted the germline wild-type allele, resulting in biallelic gene inactivation. In the remaining cyst (CECDNA0155), the presumed germline pathogenic allele (PKD1 p.R3672Q) was replaced with the wild-type allele. (Table 1, Figure 3).
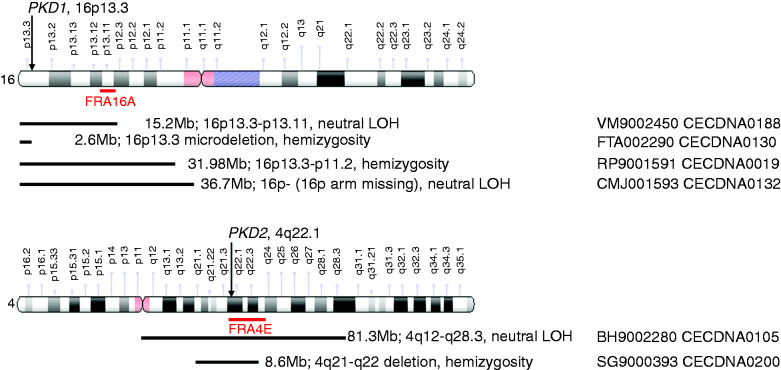
The size of the LOH events ranged from 2.6 to 36.7 Mb in PKD1 and 8.6–81.3 Mb in PKD2 (Supplemental Table 6). In seven of the events, the genetic rearrangement occurred at chromosomal fragile sites (CFSs)—FRA16A (five of seven), FRA16E (one of seven), and FRA4E (one of seven), whereas in five LOH events, the rearrangement occurred in the regions involved in genetic chromosome microdeletion diseases or syndromes, such as microdeletions of chromosomes 16p13.3, 16p12.2, and 16p11.2.40 The remaining five LOH events were large fragment rearrangement, disrupting the entirety (three cysts) or most of (one cyst, 16p13.3–11.2) chromosome 16p or nearly half of chromosome 4q (one cyst, 4q12–28.3) (Figure 4, Supplemental Table 6). We did not observe a significant relationship between the size and type (copy-number neutral versus hemizygous deletion) of LOH events (t test P=0.48).
Figure 4.
Large chromosome fragment rearrangement involved in LOH. The size and location of the chromosomal fragments involved in the rearrangement are marked on the chromosomes encompassing PKD1 and PKD2. The black lines indicate the positions where the chromosome rearrangement occurred. The CFSs are highlighted with red lines. The original schematics for chromosomes 4 and 16 were obtained and modified from https://ghr.nlm.nih.gov/chromosome/4#idiogram_chromosome_4 and https://ghr.nlm.nih.gov/chromosome/4#idiogram_chromosome_16.
Trans-Heterozygous and Concurrent Somatic Mutations
Rare cases of somatic trans-heterozygous mutations in cysts, where a patient with a germline PKD1 mutation acquired a somatic PKD2 mutation, or vice versa, have been reported.15,41 In this study, almost all PKD1 and PKD2 somatic mutations were limited to germline PKD1 and PKD2 carriers, respectively (Fisher’s exact test P<2.2×10−16), except for cyst CECDNA0012 (from patient DR9001565) (Table 1). In this germline PKD1 mutant (p.Phe3168Leu) cyst, the somatic mutations were acquired in both PKD1 (p.Gly1640Argfs*18, VAF=0.47, suggesting mutation present in approximately 94% of cyst cells) and PKD2 (p.Ala69Glyfs*23, VAF=0.15, mutation present in approximately 30% of cyst cells) (Table 1). Because the somatic PKD2 mutation had significantly lower VAF than the PKD1 mutation (P=2.5×10−5), the former was likely present in a subclone of the latter.
Additionally, concurrent somatic mutations in PKD1 were detected in cyst CECDNA0010 from patient BJA001578. In the cyst harboring germline variant PKD1 p.Asp1310Glyfs*120, somatic PKD1 mutations were detected at PKD1.p.Val2122Cysfs*3 (VAF=0.38) and PKD1.p.Phe3168Leu (VAF=0.17) (Table 1). However, from the current sequencing and computational method, we cannot determine the allelic location and the clonal origin of these concurrent mutations.42–44
Discussion
To circumvent the limitations of WES and to further assess the role of somatic second hit alterations in renal cystogenesis, we evaluated by WGS 90 kidney cysts from 24 unrelated patients with ADPKD who had kidney transplantation. We observed uniform WGS coverage across the entire PKD1/2 genes, including segmental duplicated regions of PKD1, with a minimum coverage of 41- and 67-fold in PKD1 and PKD2 exons, respectively. We then identified somatic second hit alterations in PKD1/2, either short mutations or large structural alterations, in every patient, affecting 93% (84 out of 90) of the cysts (Table 2). WGS reproducibility studies of duplicate samples and our previously published WES/LR-PCR-NGS samples12 have confirmed second hit alterations in all DNA aliquots tested, with similar VAFs. These results further support the accuracy and high sensitivity of WGS for detecting somatic alterations in renal cyst epithelia. Altogether with our previous study,12 we have sequenced a total of 152 unique kidney cysts from 27 patients with ADPKD (Supplemental Table 7, Supplemental Figure 5). To our knowledge, this is the largest cyst somatic sequencing cohort to date, with an overall detection rate of 92% (140 out of 152), providing an accurate measure of the somatic second hit frequency.
Using WGS, we found that ADPKD kidney cysts acquired an average of approximately 1000 somatic short mutations across the genome. This low somatic mutation burden in cysts is comparable to that previously observed in benign neoplasms, such as benign bone and myeloproliferative neoplasia, and much lower than that of malignant neoplasms, such as renal cell carcinoma (approximately 7000).45 In terms of protein-sequence–changing mutations, ADPKD cysts harbored an average of only 10 (median of 5) such somatic mutations. We evaluated all genes and found PKD1 and PKD2 were the only two that were recurrently mutated between cysts and patients—characteristic of somatic driver genes. No other genes, including those associated with cancer, ciliopathies, and N-glycosylation, met these criteria. We further performed pathway enrichment analysis on all mutated genes and did not find any significantly enriched pathways.
PKD1 and PKD2 are somatic drivers specifically for ADPKD cysts; they are not cancer-driving genes on the basis of the COSMIC Cancer Gene Census46 or previously published pan-cancer analyses of whole genomes.45 By assessing 30,828 tumor samples across 46 different cancer types in the cBioPortal,47 we found the overall frequency of somatic, nonsynonymous PKD1 and PKD2 mutations to be only 2.2% and 0.7%, respectively—much lower than that of 69% and 9% in our ADPKD kidney cysts. Specifically, somatic PKD1/2 mutations were found in <20% of any given cancer type, among which the top-ranked cancer types (skin, endometrial, and colorectal) are known to have the highest overall somatic mutation burden. These findings suggested that PKD1/2 are likely passenger mutations rather than cancer drivers (Supplemental Figure 6). Importantly, PKD1/2 mutations were found in <5% of any of the kidney cancer types examined. Moreover, although 83% of the PKD1/2 somatic mutations detected in ADPKD kidney cysts result in protein truncation, only 13% of PKD gene mutations in tumor samples are protein-truncating—further suggesting PKD1/2 mutations have less functional effect and are therefore passenger mutations in cancer.
Besides somatic short mutations, we found a higher frequency (18%) and larger fragment size of somatic LOH events in the current WGS study compared with previous WES/LR-PCR-NGS and microsatellite studies.12,48 Somatic LOH as the hallmark of tumorigenesis is frequently observed in various types of cancer.39,49 Although its detailed genetic mechanism is still unclear, it has been associated with mitotic missegregation and defective DNA repair responding to DNA replicative stress, resulting in chromosome interstitial deletions or translocations.39,49–52 In this study, besides five LOH events encompassing the entirety or most of chromosome 16p arm, or nearly half of chromosome 4q arm, all other LOH events occurred in CFSs or in the regions involved in genetic chromosome microdeletion diseases/syndromes. This suggests that LOH and genetic chromosome microdeletion may share a similar mechanism during DNA repair, chromosome recombination, and/or rearrangement.39,40,49
Except for one cyst, all LOH events detected in this study resulted in the loss of the germline wild-type allele, often followed by compensatory duplication of the remaining germline mutant, which leads to biallelic inactivation of PKD1 or PKD2 (Figure 3). However, in cyst CECDNA0155, the presumed germline pathogenic allele (PKD1 p.Arg3672Gln) was replaced by the germline wild-type allele instead.53 It is possible that this patient has an even more deleterious germline mutation on the wild-type allele that was undetected. However, we do not have additional data to support this hypothesis. Accordingly, we have labeled this LOH event as a variant of unknown significance (VUS).53
Although it is clear, on the basis of the VAFs of the germline pathogenic variants, that 94% of the LOH events resulted in biallelic inactivation of PKD1 or PKD2, indirectly supporting the in trans loss-of-function model of PKD genes, we could not computationally determine the phase of the somatic short mutations to their germline variants (i.e., in trans or cis) because the physical distances between the two were larger than the NGS read length of 151 bp. In previous studies of ADPKD cysts, direct phase assessment was performed using cloning techniques and found all somatic PKD1/2 mutations tested (ten out of ten) to be trans to their germline variants.8,13–15 Conceptually, somatic inactivation of PKD1/2 in ADPKD cysts is also analogous to tumor suppressor genes in cancers; the shared assumption is that the somatic alterations need to be trans to their germline variants in order to provide an additional selective advantage to cells to drive clonal expansion.8,13,14
Taken together, strengths of this study include: (1) patients with ADPKD were recruited according to standard selection criteria in a single health system that draws from a wide geographic area around New York City; (2) a relatively large number (n=90) of cysts from 24 patients were evaluated, expanding observations from our previous study; (3) use of WGS yielded more uniform read coverage than WES, enabling more sensitive and accurate detection of both somatic short mutations and large-scale structural alterations; and (4) confirmation of WGS findings by duplicate testing. To our knowledge, this is the largest study performed thus far to comprehensively analyze somatic alterations in renal cyst epithelia.
A limitation of this study was the selection of relatively large cysts (>10 mm) that were accessible by dissection and yielded a sufficient amount of DNA materials for WGS analysis. This study was not designed to determine when during cystogenesis the somatic second hits occurred, or whether PKD1/2 somatic mutations are sufficient to initiate cystogenesis or are a secondary driver for cyst expansion. Additional studies of smaller size cysts are warranted to gain additional understanding of the pathogenic mechanisms that initiate renal cyst development.
In summary, WGS provides a sensitive sequencing technique for identifying multiple genetic alterations including short mutations and large-scale structural alterations. The sensitivity of WGS was significantly higher than WES for identifying novel mutations in the segmentally duplicated region of PKD1. This study confirmed and extended our earlier observation that >90% of patients with ADPKD had somatic mutations of PKD1 or PKD2 in renal cyst epithelium.12 Altogether, these findings support a cellular recessive mechanism for cystogenesis in ADPKD that is caused by inactivating germline and somatic mutations of PKD1 or PKD2 in kidney cyst epithelium.
Disclosures
H. Bai is employed by and has ownership interest in Vertex Pharmaceuticals, Inc. I. Barash is employed by IQVIA; has consultancy agreements with Caraway, Otsuka, and Sanofi; and has received honoraria from Caraway, Otsuka, and Sanofi. J. Blumenfeld reports research funding from Vertex Pharmaceuticals, Inc.; and other interests/relationships as Secretary on the Board of Directors of the American Journal of Hypertension. S. Giannakopoulos is employed by Silence Therapeutics; and has ownership interest in Biomarin Pharmaceuticals and Vertex Pharmaceuticals. S. Hughes is employed by Pathios Therapeutics; has consultancy agreements with 30FiveBio and Grey Wolf Therapeutics; and has ownership interest in 30FiveBio, Grey Wolf Therapeutics, and Pathios Therapeutics. S. Kapur reports research funding from Astellas Pharma, Bristol Myers Squibb, Medeor Therapeutics, Quark Pharmaceuticals, Shire Therapeutics, and Talaris Therapeutics. K. Larbi is employed by Vertex Pharmaceuticals Ltd. M. Prince reports patents and inventions via GE Healthcare paying royalties for Magnetic Resonance Patents; and other interests/relationships as Adjunct Professor of Radiology at Columbia University. H. Rennert reports research funding from Vertex. Z. Zhang reports research funding from Vertex Pharmaceuticals.
Funding
Reseach reported in this publication was supported by Vertex Pharmaceuticals and the National Center for Advancing Translational Science of the National Institute of Health under award number UL1TR002384.
Supplementary Material
Acknowledgments
We are grateful to all patients and their families for their invaluable participation.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2021050690/-/DCSupplemental.
Supplemental Table 1. ADPKD patient cohort clinical and pathologic characteristics.
Supplemental Table 2. List of cysts previously analyzed by WES and resequenced by WGS in this study.
Supplemental Table 3. Replicate cyst samples sequenced by WGS.
Supplemental Table 4. Summary of genome-wide somatic alterations in 90 cysts detected by WGS.
Supplemental Table 5. Correlation analysis between cyst diameter and somatic mutation frequency.
Supplemental Table 6. Summary of somatic loss of heterozygosity (LOH) alterations in PKD1 or PKD2 in renal cyst epithelia.
Supplemental Table 7. Summary of somatic PKD1 and PKD2 alterations in 152 cysts analyzed in the current and previous studies.
Supplemental Figure 1. Statistical analysis of kidney volume measured by imaging.
Supplemental Figure 2. Example somatic copy number neutral LOH event of the PKD1 locus.
Supplemental Figure 3. Example somatic copy number neutral LOH event of the PKD2 locus.
Supplemental Figure 4. Example somatic chromosomal deletion of the PKD2 locus.
Supplemental Figure 5. Summary and distribution of somatic short mutations in PKD1 and PKD2 in 152 cysts analyzed by LR-PCR/WES or WGS.
Supplemental Figure 6. Frequency of somatic, nonsynonymous PKD1/2 mutations across 46 cancer types.
References
- 1.Paul BM, Vanden Heuvel GB: Kidney: Polycystic kidney disease. Wiley Interdiscip Rev Dev Biol 3: 465–487, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kidney Disease Improving Global Outcomes (KDIGO) . Autosomal-dominant polycystic kidney disease (ADPKD): executive summary from a Kidney Disease: Improving Global Outcomes (KDIGO) Controversies Conference. Available at https://www.kidney-international.org/action/showPdf?pii=S2157-1716%2815%2932146-8. Accessed November 5, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heyer CM, Sundsbak JL, Abebe KZ, Chapman AB, Torres VE, Grantham JJ, et al. ; HALT PKD and CRISP Investigators : Predicted mutation strength of nontruncating PKD1 mutations aids genotype-phenotype correlations in autosomal dominant polycystic kidney disease. J Am Soc Nephrol 27: 2872–2884, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris PC, Torres VE: Genetic mechanisms and signaling pathways in autosomal dominant polycystic kidney disease. J Clin Invest 124: 2315–2324, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornec-Le Gall E, Audrézet MP, Le Meur Y, Chen JM, Férec C: Genetics and pathogenesis of autosomal dominant polycystic kidney disease: 20 years on. Hum Mutat 35: 1393–1406, 2014 [DOI] [PubMed] [Google Scholar]
- 6.Audrézet MP, Cornec-Le Gall E, Chen JM, Redon S, Quéré I, Creff J, et al. : Autosomal dominant polycystic kidney disease: comprehensive mutation analysis of PKD1 and PKD2 in 700 unrelated patients. Hum Mutat 33: 1239–1250, 2012 [DOI] [PubMed] [Google Scholar]
- 7.Hoffmeister H, Babinger K, Gürster S, Cedzich A, Meese C, Schadendorf K, et al. : Polycystin-2 takes different routes to the somatic and ciliary plasma membrane. J Cell Biol 192: 631–645, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian F, Watnick TJ, Onuchic LF, Germino GG: The molecular basis of focal cyst formation in human autosomal dominant polycystic kidney disease type I. Cell 87: 979–987, 1996 [DOI] [PubMed] [Google Scholar]
- 9.Rossetti S, Consugar MB, Chapman AB, Torres VE, Guay-Woodford LM, Grantham JJ, et al. ; CRISP Consortium : Comprehensive molecular diagnostics in autosomal dominant polycystic kidney disease. J Am Soc Nephrol 18: 2143–2160, 2007 [DOI] [PubMed] [Google Scholar]
- 10.Tan YC, Michaeel A, Blumenfeld J, Donahue S, Parker T, Levine D, et al. : A novel long-range PCR sequencing method for genetic analysis of the entire PKD1 gene. J Mol Diagn 14: 305–313, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan AY, Michaeel A, Liu G, Elemento O, Blumenfeld J, Donahue S, et al. : Molecular diagnosis of autosomal dominant polycystic kidney disease using next-generation sequencing. J Mol Diagn 16: 216–228, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan AY, Zhang T, Michaeel A, Blumenfeld J, Liu G, Zhang W, et al. : Somatic mutations in renal cyst epithelium in autosomal dominant polycystic kidney disease. J Am Soc Nephrol 29: 2139–2156, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watnick TJ, Torres VE, Gandolph MA, Qian F, Onuchic LF, Klinger KW, et al. : Somatic mutation in individual liver cysts supports a two-hit model of cystogenesis in autosomal dominant polycystic kidney disease. Mol Cell 2: 247–251, 1998 [DOI] [PubMed] [Google Scholar]
- 14.Pei Y, Watnick T, He N, Wang K, Liang Y, Parfrey P, et al. : Somatic PKD2 mutations in individual kidney and liver cysts support a “two-hit” model of cystogenesis in type 2 autosomal dominant polycystic kidney disease. J Am Soc Nephrol 10: 1524–1529, 1999 [DOI] [PubMed] [Google Scholar]
- 15.Watnick T, He N, Wang K, Liang Y, Parfrey P, Hefferton D, et al. : Mutations of PKD1 in ADPKD2 cysts suggest a pathogenic effect of trans-heterozygous mutations. Nat Genet 25: 143–144, 2000 [DOI] [PubMed] [Google Scholar]
- 16.Ali H, Al-Mulla F, Hussain N, Naim M, Asbeutah AM, AlSahow A, et al. : PKD1 duplicated regions limit clinical utility of whole exome sequencing for genetic diagnosis of autosomal dominant polycystic kidney disease. Sci Rep 9: 4141, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belkadi A, Bolze A, Itan Y, Cobat A, Vincent QB, Antipenko A, et al. : Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc Natl Acad Sci U S A 112: 5473–5478, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mallawaarachchi AC, Hort Y, Cowley MJ, McCabe MJ, Minoche A, Dinger ME, et al. : Whole-genome sequencing overcomes pseudogene homology to diagnose autosomal dominant polycystic kidney disease. Eur J Hum Genet 24: 1584–1590, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, et al. : User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 31: 1116–1128, 2006 [DOI] [PubMed] [Google Scholar]
- 20.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. : The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Durbin R: Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poplin R, Ruano-Rubio V, DePristo MA, Fennell TJ, Carneiro MO, Van der Auwera GA, et al. : Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv: 201178, 2018
- 23.Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, et al. : Manta: Rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32: 1220–1222, 2016 [DOI] [PubMed] [Google Scholar]
- 24.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. : The ensembl variant effect predictor. Genome Biol 17: 122, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H: Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics 30: 2843–2851, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pedersen BS, Quinlan AR: Who’s who? Detecting and resolving sample anomalies in human DNA sequencing studies with Peddy. Am J Hum Genet 100: 406–413, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. ; Genome Aggregation Database Consortium : The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581: 434–443, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gout AM, Martin NC, Brown AF, Ravine D: PKDB: Polycystic Kidney Disease Mutation Database–A gene variant database for autosomal dominant polycystic kidney disease. Hum Mutat 28: 654–659, 2007 [DOI] [PubMed] [Google Scholar]
- 29.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J: A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46: 310–315, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP: Predicting the functional effect of amino acid substitutions and indels. PLoS One 7: e46688, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benjamin D, Sato T, Cibulskis K, Getz G, Stewart C, Lichtenstein L: Calling somatic SNVs and indels with Mutect2. bioRxiv: 861054, 2019
- 32.Zhou X, Edmonson MN, Wilkinson MR, Patel A, Wu G, Liu Y, et al. : Exploring genomic alteration in pediatric cancer using ProteinPaint. Nat Genet 48: 4–6, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. : A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anna A, Monika G: Splicing mutations in human genetic disorders: Examples, detection, and confirmation. J Appl Genet 59: 253–268, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vujic M, Heyer CM, Ars E, Hopp K, Markoff A, Orndal C, et al. : Incompletely penetrant PKD1 alleles mimic the renal manifestations of ARPKD. J Am Soc Nephrol 21: 1097–1102, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossetti S, Kubly VJ, Consugar MB, Hopp K, Roy S, Horsley SW, et al. : Incompletely penetrant PKD1 alleles suggest a role for gene dosage in cyst initiation in polycystic kidney disease [published correction appears in Kidney Int 75: 1359, 10.1038/ki.2009.151]. Kidney Int 75: 848–855, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lanktree MB, Haghighi A, Di Bari I, Song X, Pei Y: Insights into autosomal dominant polycystic kidney disease from genetic studies. Clin J Am Soc Nephrol 16: 790–799, 2021. 32690722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Durkie M, Chong J, Valluru MK, Harris PC, Ong ACM: Biallelic inheritance of hypomorphic PKD1 variants is highly prevalent in very early onset polycystic kidney disease. Genet Med 23: 689–697, 2021. 33168999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Keefe C, McDevitt MA, Maciejewski JP: Copy neutral loss of heterozygosity: A novel chromosomal lesion in myeloid malignancies. Blood 115: 2731–2739, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watson CT, Marques-Bonet T, Sharp AJ, Mefford HC: The genetics of microdeletion and microduplication syndromes: An update. Annu Rev Genomics Hum Genet 15: 215–244, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koptides M, Mean R, Demetriou K, Pierides A, Deltas CC: Genetic evidence for a trans-heterozygous model for cystogenesis in autosomal dominant polycystic kidney disease. Hum Mol Genet 9: 447–452, 2000 [DOI] [PubMed] [Google Scholar]
- 42.Paguirigan AL, Smith J, Meshinchi S, Carroll M, Maley C, Radich JP: Single-cell genotyping demonstrates complex clonal diversity in acute myeloid leukemia. Sci Transl Med 7: 281re2, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morita K, Wang F, Jahn K, Hu T, Tanaka T, Sasaki Y, et al. : Clonal evolution of acute myeloid leukemia revealed by high-throughput single-cell genomics. Nat Commun 11: 5327, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miles LA, Bowman RL, Merlinsky TR, Csete IS, Ooi AT, Durruthy-Durruthy R, et al. : Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature 587: 477–482, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium : Pan-cancer analysis of whole genomes. Nature 578: 82–93, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sondka Z, Bamford S, Cole CG, Ward SA, Dunham I, Forbes SA: The COSMIC Cancer Gene Census: Describing genetic dysfunction across all human cancers. Nat Rev Cancer 18: 696–705, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. : Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6: pl1, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Badenas C, Torra R, Pérez-Oller L, Mallolas J, Talbot-Wright R, Torregrosa V, et al. : Loss of heterozygosity in renal and hepatic epithelial cystic cells from ADPKD1 patients. Eur J Hum Genet 8: 487–492, 2000 [DOI] [PubMed] [Google Scholar]
- 49.Couto SS: The pathologist’s slide reveals more than meets the eye: Loss of heterozygosity and cancer biology. Vet Pathol 48: 236–244, 2011 [DOI] [PubMed] [Google Scholar]
- 50.Symington LS, Rothstein R, Lisby M: Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics 198: 795–835, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minocherhomji S, Ying S, Bjerregaard VA, Bursomanno S, Aleliunaite A, Wu W, et al. : Replication stress activates DNA repair synthesis in mitosis. Nature 528: 286–290, 2015 [DOI] [PubMed] [Google Scholar]
- 52.Hazan I, Hofmann TG, Aqeilan RI: Tumor suppressor genes within common fragile sites are active players in the DNA damage response. PLoS Genet 12: e1006436, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Naeim F, Nagesh Rao P, Song SX, Phan RT: Cancer Cytogenetics. In: Atlas of Hematopathology, edited by Naeim F, Nagesh Rao P, Song SX, Phan RT, 2nd Ed., Amsterdam, Academic Press, 2018, pp 57–68 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.