Abstract
Spoligotyping, a method based on the variability of distribution of the 43 inter-direct repeat (DR) spacers of Mycobacterium tuberculosis and Mycobacterium bovis BCG, is useful to study the molecular epidemiology of bovine and human tuberculosis. Recently, a major family of M. tuberculosis clinical isolates named the Haarlem family, which did not contain spacers 31 and 33 to 36, was reported in a multicenter study. Independently, a data bank containing all the published spoligotypes showed that the two most prevalent spoligotypes in the world differed only by the presence or absence of spacer 31. A careful analysis of the DR locus sequence led us to hypothesize that spacer 31 may not have been amplified in some isolates with the primer sets DRa and DRb currently used for spoligotyping. Consequently, a modified spoligotyping method based on different combinations of the 36-bp DR and IS6110 primers was devised that was able to discriminate between the left and the right parts of the DR locus and demonstrated the presence of the previously unamplified spacer 31 for some of the clinical isolates. By analogy, we suggest that a single-spacer difference in some epidemiologically linked cases of tuberculosis may simply arise due to the insertion of an extra copy of IS6110 within the DR locus, leading to its asymmetrical disruption and subsequent lack of the DRa or DRb targets. The influence of the IS6110 preferential insertion sites within the DR locus on spoligotyping results should be further investigated.
Originally described in Mycobacterium bovis (12), the direct repeat (DR) locus of the Mycobacterium tuberculosis complex genome consists of a variable number of copies of a 36-bp DR interspersed with specific spacer sequences, which has been used to develop a rapid molecular fingerprinting technique called spoligotyping (14), often associated with the IS6110 restriction fragment-length polymorphism (RFLP) for epidemiological purposes (9, 10). A recent multicenter study involving comparison of various molecular markers for the study of epidemiology of M. tuberculosis concluded that, along with other techniques, spoligotyping was a method of choice for reproducible PCR-based genotyping (15). In parallel, this study also underlined that several separate genotype families within the M. tuberculosis complex could be recognized on the basis of multiple genetic markers, namely, the Beijing, Africa, and Haarlem families (15). A careful examination of these families showed that the Haarlem family was always characterized by the lack of spacer 31. This observation, along with the fact that epidemiologically related M. tuberculosis clinical isolates may sometimes differ by a single spacer (14), and our independent observations that the two most prevalent spoligotypes around the world (named types 53 and 50 in Fig. 2) differ by the presence or absence of spacer 31 (18), prompted us to have a closer look at this spacer by using a modified spoligotyping method.
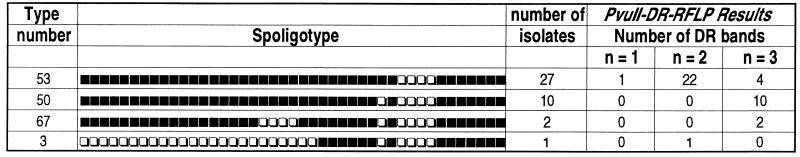
FIG. 2.
Correlation between spoligotypes and PvuII-DR RFLP results showing a link between the absence of spacer 31, the presence of a third DRr band, and the consequent presence of a second copy of IS6110 inserted within the DR locus.
Bacterial isolates.
All of the experimentally studied M. tuberculosis isolates were obtained from the collection of the Pasteur Institute of Guadeloupe and were isolated and identified locally by standard mycobacteriological procedures.
Spoligotyping.
Standard spoligotyping was performed with the DRa and DRb primers as described previously (14). For the left-right (L-R) spoligotyping, primers IS6 and IS3 (4, 5) were used in combination with biotinylated DRb or biotinylated DRa to amplify either the left or the right part of the IS6110 boundaries or the central part of the DR locus for strains harboring two IS6110 copies within the DR locus. The following primers were used: DRa, GGTTTTGGGTCTGACGAC (14); DRb, CCGAGAGGGGACGGAAAC (14); IS3, GCTGCCTACTACGCTCAAC (4, 5); and IS6, CAAGTAGACGGGCGACCTC (4, 5). The primer combinations are shown in Table 1. The PCR protocol and the conditions for membrane preparation, hybridization, and detection were the same as those reported earlier (14), except for the elongation time, which was set to 3 min.
TABLE 1.
Primer combinations used in this study and expected results according to the number of IS6110 copies inserted within the DR locus of M. tuberculosis
| Primer set | Primera
|
Result with given no. of IS6110 copies
|
||
|---|---|---|---|---|
| Forward | Reverse | 1 copy | 2 copiesb | |
| Ac | DRb | Biot-DRa | All spacers present | All spacers present |
| B | Biot-DRb | IS3 | No amplification | No amplification |
| C | IS3 | Biot-DRa | Right DR spacers | Right and central DR spacers |
| D | Biot-DRb | IS6 | Left DR spacers | Left and central DR spacers |
| E | IS6 | Biot-DRa | No amplification | No amplification |
The directions of the forward and reverse primers are deduced from previous DR sequence data (11). Biot, biotinylated.
If the two copies of IS6110 are inserted in the same direction.
Positive control.
IS6110 and DRr hybridization.
IS6110 and DRr RFLPs were performed according to published procedures (12, 21) by either direct or indirect labeling. Detection by the enhanced chemoluminescence (ECL) systems was performed according to the supplier's protocols (Amersham, Buckinghamshire, United Kingdom).
Modified LR spoligotyping: basis for development and results.
The spoligotyping results for M. tuberculosis isolates from our laboratory were pooled with those published previously to construct a database containing 993 individual patterns. The nomenclature of specific spoligotypes numbered 1 to 69 was recently described (18), to which new types 70 to 72 were added during the course of this investigation. The description of these spoligotypes is available on request. Statistical analysis of spacer distribution on these 993 isolates underlined a region (spacers 21 to 32) that is characterized by successive decreases for some of the spacers, particularly spacer 31 (results not shown). Based on the available sequence of the DR locus (11), a physical map was redrawn for a strain containing two copies of IS6110 (Fig. 1), with one copy being inserted after the spacer 31. As shown in Fig. 1, a second copy inserted after spacer 31 may lead to an unequal split of the DR, leading to the consequent loss of the DRa primer target, which in turn will not permit the amplification of spacer 31 with primers DRa and DRb, since there was no target for primer DRa at the right side of this spacer. Inversely, the bordering spacers 24 and 25 would be detected (Fig. 1). To experimentally verify this hypothesis, we searched in our collection for isolates lacking spacer 31 after routine spoligotyping (types 3, 50, and 67 [Fig. 2]). Type 50, which represents the second most frequent spoligotype in our database and is overrepresented in our region, differs only by the lack of spacer 31 as compared to type 53, another very common spoligotype (18). The PvuII-DR RFLPs of these types of isolates lacking spacer 31 (spoligotype patterns 3, 50, and 67) mostly showed the presence of three DRr-hybridizing bands (Fig. 2), suggesting the presence of a second IS6110 copy within the DR locus, very likely inserted asymmetrically after spacer 31 (Fig. 1). This assumption is strengthened by the observation that nearly 82% of the strains of type 53 (universal type; presence of all spacers except 33 to 36) (18) harbored only two DRr-hybridizing bands (Fig. 2). Assessment of the percentage of strains of spoligotype 50 harboring a second copy of IS6110 inserted asymmetrically and located between spacers 31 and 25 of the DR locus is currently in progress.
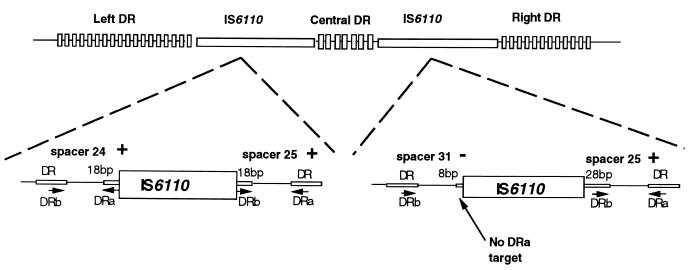
FIG. 1.
Schematic representation of the DR locus of M. tuberculosis with an enlargement of the two IS6110 insertion sites. The top of the figure shows the general organization of the DR locus, whereas the bottom of the figure is an enlargement showing the flanking regions of IS6110 insertions. In the bottom part, the left-hand copy of IS6110 is inserted exactly in the middle of the DRr sequence between spacers 24 and 25, splitting the DRr into two equal 18-bp fragments which serve as PCR targets for primers DRa and DRb and give a positive hybridization signal for spacers 24 and 25. On the other hand, an asymmetrical disruption of the DRr sequence due to the insertion of a second copy of IS6110 in the right-hand side of the DR locus between spacers 31 and 25 results in two unequal parts in such a way that only an 8-bp fragment of the DRa target is conserved toward spacer 31, not enough for successful amplification and subsequent detection of this spacer.
The modified L-R spoligotyping method, which is theoretically able to selectively amplify the right or the left border of the IS6110 sequence(s) of the DR locus, was initially tested on reference strains of M. tuberculosis (H37Rv) and M. bovis BCG (BCG-Pasteur), known to contain only a single IS6110 copy linked to the DR. As expected, each of the reference strains was split into two distinct left and right spoligotyping patterns by L-R spoligotyping (Fig. 3 [data for BCG not shown]), confirming that the IS6110 copy was indeed inserted immediately after spacer 24. Accordingly, the primer combinations C and D resulted in a strongly positive spoligotyping result in all cases, each distinctly covering half of the positive control as amplified with primer set A. Primer set B always gave negative results (Fig. 3), whereas primer set E produced no signals or weak and nonreproducible signals (results not shown). These results conclusively showed that primer sets C and D were able to discriminate the right and the left parts of the DR region in the reference M. tuberculosis and M. bovis BCG strains. However, due to the lesser sensitivity of the L-R spoligotyping technique (explained by the smaller amount of PCR targets [Fig. 1]), spacers located on the extremities of the DR locus were sometimes undetected (results not shown).
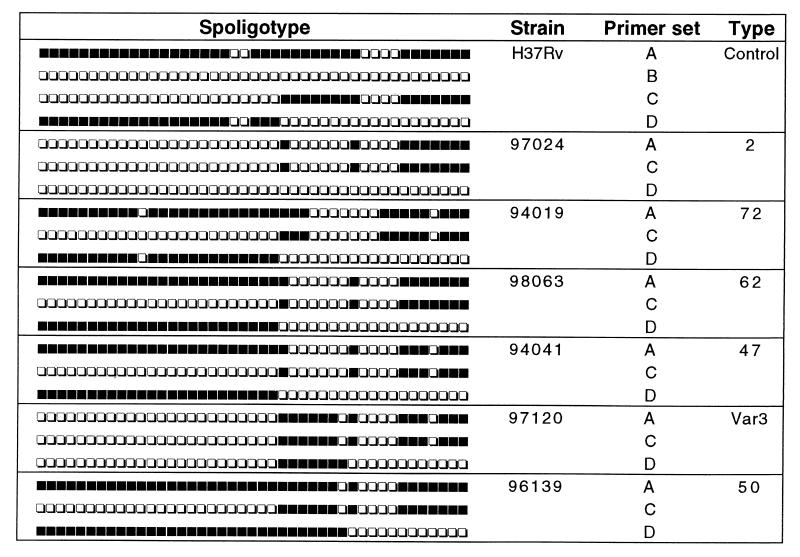
FIG. 3.
Schematic representation of results obtained by L-R spoligotyping of M. tuberculosis H37Rv and by the routine spoligotyping method of representative clinical isolates that lacked spacer 31. The primer sets A, B, C, and D have been detailed in the text and in Table 1.
This experimental approach was later extended to investigate the DR loci of various clinical isolates lacking spacer 31 that were typed in parallel by PvuII-DR RFLP; a total of 12 isolates selected comprised either strains found to harbor a single copy of IS6110 within the DR locus (types 2, 72, 62, and 47) or those found to possess two copies within the DR locus (types 50, 3, and var3; the latter differed from type 3 by absence of a single spacer, spacer 40). Representative experimental results for some of the isolates are shown in Fig. 3; for types 2, 72, 62, and 47, the primer sets C and D showed a split in position 24 resulting in distinct right and left amplifications of the DR locus starting from the point of insertion, whereas for types 50 and var3, an additional overlapping of spacers 25 to 31 by the primer set D corresponded to the insertion of the second IS6110 copy (Table 1 and Fig. 3). The fact that the left part of the DR pattern was absent in the type 2 isolate when primer set D was used suggests that this portion was effectively missing and that a single copy of IS6110 was inserted immediately after spacer 31 (Fig. 1). Finally, the observation that previously undetected spacer 31 was revealed with primer set D in types 50 and var3 was in agreement with an asymmetric insertion of the second copy of IS6110 within the DR immediately after spacer 31 (Fig. 1 and 3).
DISCUSSION
In M. tuberculosis, various repetitive loci have been described, among which are numerous insertion sequences, such as IS6110 (20), as well as the variable number tandem repeats (8, 13), the DR (12), and the polymorphic GC-rich sequence (16). All of these markers have recently been used in a multicenter study of M. tuberculosis epidemiology (15). Spoligotyping based on the variability of inter-DR sequences has been proposed as a method of choice for reproducible PCR-based genotyping (15). In this context, the existence of preferential IS6110 insertion sites in the DR locus of M. tuberculosis (4, 7) may indirectly contribute to the variability of spoligotyping results, which may have important epidemiological consequences. Moreover, clinical isolates of M. tuberculosis differing by a single DR spacer or a single IS6110 copy that were either epidemiologically related (14) or were serial isolates from the same patient (17) have been reported. Consequently, we decided to investigate the fine structure of the DR locus in relation to the insertion of IS6110 and its consequences in terms of spoligotyping results.
In this investigation, a modified spoligotyping method based on different combinations of DRr and IS6110 primers was able to discriminate between the left and the right parts of the DR locus and demonstrated the presence of previously undetected spacer 31 for some of the clinical isolates studied. Although limited to spacer 31 in this study, this interpretation of a missing spacer provides us with a new interpretation for a single negative hybridization spot in spoligotyping. However, this interpretation of the absence of spacer 31 does not mean that all single-spacer absences are caused by an asymmetrical IS6110 insertion: e.g., the lack of spacers 3, 9, and 16 for M. bovis BCG is not linked to any IS6110 insertion (11).
The study of preferential IS6110 transposition sites may have important applications in the study of the evolutionary genetics of M. tuberculosis, particularly because the DR locus is likely to evolve more slowly than IS6110, whose evolution has recently been shown to be too fast for the evolution of multi-drug-resistant strains of M. tuberculosis to be monitored efficiently (1). A recent study has evaluated the rate of change of IS6110 RFLP patterns in serial M. tuberculosis patient isolates as being 3.2 years (3). Seen from an evolutionary point of view, various mechanisms, such as the transposition of IS6110 and other mobile genetic elements (2, 5, 19), homologous recombination (11), and slipped-strand mispairing (15), are believed to drive genomic diversification in M. tuberculosis. Our study underlines the role of IS6110 transpositional events in the evolution of the DR locus. Alternatively, a one-spacer difference may also be due to mutations in the spacer sequence itself; however, if the DR locus is neutral (no environmental and/or selective pressure), point mutations in this locus should be extremely rare, as shown by the restricted allelic diversity shown in M. tuberculosis (19). More likely, IS6110-mediated deletions and insertions and homologous recombination may have driven the evolution of the DR locus (6, 11). Our experimental approach may increase the discriminatory power of spoligotyping to study isolates differing by a single missing spacer and thus result in a better evaluation of strain relatedness in epidemiological studies of tuberculosis.
REFERENCES
- 1.Alito A, Morcillo N, Scipioni S, Dolmann A, Romano M I, Cataldi A, van Soolingen D. The IS6110 restriction fragment length polymorphism in particular multidrug-resistant Mycobacterium tuberculosis strains may evolve too fast for reliable use in outbreak investigation. J Clin Microbiol. 1999;37:788–791. doi: 10.1128/jcm.37.3.788-791.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bifani P J, Plikaytis B B, Kapur V, Stockbauer K, Pan X, Lutfey M L, Moghazeh S L, Eisner W, Daniel T M, Kaplan M H, Musser J M, Kreiswirth B N. Origin and interstate spread of a New York City multidrug-resistant Mycobacterium tuberculosis clone family. JAMA. 1996;275:452–457. [PubMed] [Google Scholar]
- 3.De Boer A S, Borgdoff M W, de Haas P E, Nagelkerke N J, van Embden J D A, van Soolingen D. Analysis of rate of change of IS6110 RFLP patterns of Mycobacterium tuberculosis based on serial patient isolates. J Infect Dis. 1999;180:1238–1244. doi: 10.1086/314979. [DOI] [PubMed] [Google Scholar]
- 4.Fang Z, Forbes K J. A Mycobacterium tuberculosis IS6110 preferential locus (ipl) for insertion into the genome. J Clin Microbiol. 1997;35:479–481. doi: 10.1128/jcm.35.2.479-481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang Z, Morrison N, Watt B, Doig C, Forbes K J. IS6110 transposition and evolutionary scenario of the direct repeat locus in a group of closely related Mycobacterium tuberculosis strains. J Bacteriol. 1998;180:2102–2109. doi: 10.1128/jb.180.8.2102-2109.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang Z, Doig C, Kenna D T, Smittipat N, Palittapongarnpim P, Watt B, Forbes K J. IS6110-mediated deletions of wild-type chromosomes of Mycobacterium tuberculosis. J Bacteriol. 1999;181:1014–1020. doi: 10.1128/jb.181.3.1014-1020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fomukong N, Beggs M, Hajj H E, Templeton G, Eisenach K, Cave M D. Differences in the prevalence of IS6110 insertion sites in Mycobacterium tuberculosis strains: low and high copy number of IS6110. Tuber Lung Dis. 1998;78:109–116. doi: 10.1016/s0962-8479(98)80003-8. [DOI] [PubMed] [Google Scholar]
- 8.Frothingham R, Meeker-O'Connell W A. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology. 1998;144:1189–1196. doi: 10.1099/00221287-144-5-1189. [DOI] [PubMed] [Google Scholar]
- 9.Goguet de la Salmonière Y-O, Li H M, Torrea G, Bunschoten A, van Embden J, Gicquel B. Evaluation of spoligotyping in a study of the transmission of Mycobacterium tuberculosis. J Clin Microbiol. 1997;35:2210–2214. doi: 10.1128/jcm.35.9.2210-2214.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goyal M, Saunders N A, van Embden J D A, Young D B, Shaw R J. Differentiation of Mycobacterium tuberculosis isolates by spoligotyping and IS6110 restriction fragment length polymorphism. J Clin Microbiol. 1997;35:647–651. doi: 10.1128/jcm.35.3.647-651.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groenen P M A, Bunschoten A E, van Soolingen D, van Embden J D A. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis, application for strain differentiation by a novel typing method. Mol Microbiol. 1993;10:1057–1065. doi: 10.1111/j.1365-2958.1993.tb00976.x. [DOI] [PubMed] [Google Scholar]
- 12.Hermans P W M, van Soolingen D, Bik E M, de Haas P E W, Dale J W, van Embden J D A. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect Immun. 1991;59:2695–2705. doi: 10.1128/iai.59.8.2695-2705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hermans P W M, van Soolingen D, van Embden J D A. Characterization of a major polymorphic tandem repeat in Mycobacterium tuberculosis and its potential use in the epidemiology of Mycobacterium kansasii and Mycobacterium gordonae. J Bacteriol. 1992;174:4157–4165. doi: 10.1128/jb.174.12.4157-4165.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kremer K, van Soolingen D, Frothingham R, Haas W H, Hermans P W M, Martín C, Palittapongarnpim P, Plikaytis B B, Riley L W, Yakrus M A, Musser J M, van Embden J D A. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J Clin Microbiol. 1999;37:2607–2618. doi: 10.1128/jcm.37.8.2607-2618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poulet S, Cole S T. Characterization of the highly abundant polymorphic GC-rich repetitive sequence (PGRS) present in Mycobacterium tuberculosis. Arch Microbiol. 1995;163:87–95. doi: 10.1007/BF00381781. [DOI] [PubMed] [Google Scholar]
- 17.Rastogi N, Ross B C, Dwyer B, Goh K S, Clavel-Sérès S, Jeantils V, Cruaud P. Emergence during unsuccessful chemotherapy of multiple drug resistance in a strain of Mycobacterium tuberculosis. Eur J Clin Microbiol Infect Dis. 1992;11:901–907. doi: 10.1007/BF01962370. [DOI] [PubMed] [Google Scholar]
- 18.Sola C, Devallois A, Horgen L, Maïsetti J, Filliol I, Legrand E, Rastogi N. Tuberculosis in the Carribean: using spacer oligonucleotide typing to understand strain origin and transmission. Emerg Infect Dis. 1999;5:404–414. doi: 10.3201/eid0503.990311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sreevatsan S, Pan X, Stockbauer K, Connell N, Kreiswirth B, Whittam T, Musser J. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci USA. 1997;97:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thierry D, Cave M D, Eisenach K D, Crawford J T, Bates J H, Gicquel B, Guesdon J L. IS6110, an IS-like element of Mycobacterium tuberculosis complex. Nucleic Acids Res. 1990;18:188. doi: 10.1093/nar/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]